- 1The State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources, South China Agricultural University, Guangzhou, China
- 2The Key Laboratory of Plant Molecular Breeding of Guangdong Province, College of Agriculture, South China Agricultural University, Guangzhou, China
- 3The State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources, College of Life Science and Technology, Guangxi University, Nanning, China
Aluminum (Al)-resistant plant cultivars can recruit beneficial microbes to alleviate the stresses. However, the mechanism of how rhizobacterial communities strengthen Al tolerance of wild soybean has not been addressed. The aim of this study was to investigate the bacterial community structure in the rhizosphere of Al-tolerant (BW69) and Al-sensitive (W270) wild soybean germplasm subjected to three Al concentrations. We analyzed the rhizobacterial communities of the two genotypes by high-throughput sequencing of 16S rRNA genes. The results showed that high Al stress recruited different rhizobacterial communities between two genotypes. In total, 49 OTUs, such as OTU15 (Gammaproteobacteria_KF-JG30-C25_norank), OTU23 (Mizugakiibacter), and OTU93 (Alkanibacter), were enriched in the rhizosphere of BW69 at the low and high Al concentrations. Moreover, bacterial community in the rhizosphere of BW69 had a more complex co-occurrence network than did W270 at the high Al concentration. Overall, our findings highlighted that high Al concentration magnified the difference in rhizobacterial community structure between two genotypes. However, the lower modularity of the co-occurrence network in rhizosphere of BW69 than W270 under Al stress may cause the rhizobacterial community to be less resistant and more influenced by disturbance. This study emphasizes the possibility of using rhizobacteria as an improved crop breeding or gene to produce crops that are more resistant to the toxicity of heavy metal.
Introduction
Acidic soils with pH less than 5.5 account for about 30% of the world’s arable soils (von Uexküll and Mutert, 1995). Aluminum (Al) toxicity is one of the crucial factors limiting the growth of crops in acidic soils (Kochian et al., 2005). The main toxicity of Al to plants inhibits root growth and the absorption of water and nutrients (Liang et al., 2001). For instance, Al3+ could bind with phosphorus (P) in the soil and lead to the P deficiency (Verma et al., 2005; Zelinová et al., 2009). Al also reduces the conversion efficiency of NO3– to NH4+ and thus further increases the toxicity of Al because NH4+-N could mitigate the toxicity of Al (Matsumoto and Yamaya, 1986; Rengel and Elliott, 1992; Ma et al., 2002; Zhao and Shen, 2018).
The secretion of organic acids from roots, such as citric acid, oxalic acid, and malic acid, is one of the important mechanisms for plant to alleviate Al toxicity (Ma et al., 2001). These organic acids released from plant roots can chelate Al ions, reducing the toxicity of Al to plants (Ma et al., 2001; Kochian et al., 2005). The type and quantity of organic acids vary among different genotypes (Miyasaka et al., 1991). In wheat, maize, and soybean, Al-tolerant (Al-T) varieties secrete much more organic acids than Al-sensitive (Al-S) varieties (Pellet et al., 1995; Ryan et al., 1995; Yang et al., 2001). Furthermore, the remarkable differences in organic acids between Al-T and Al-S varieties directly affect the microbial community structure, abundance, and diversity in the rhizosphere (Germida and Siciliano, 2001; Yang et al., 2012; Peiffer et al., 2013). The root exudates secreted from stress-resistant genotypes can recruit beneficial microbes to alleviate the stresses, via regulating microbial metabolic activities and changing nutrient availability in soil (Fierer and Jackson, 2006; Parniske, 2008; West et al., 2010; Shi et al., 2011; Bulgarelli et al., 2013; Li et al., 2014; Kwak et al., 2018). For instance, some secondary metabolites from root exudates likely attract some arbuscular mycorrhizal fungi and rhizobia, which may produce compounds and in turn increase the secretion of organic acids to alleviate Al toxicity (Pellet et al., 1995; Phillips et al., 2004; Besserer et al., 2006; Berendsen et al., 2012; Yang et al., 2012). Moreover, some Al-tolerant bacteria (e.g., Klebsiella and Serratia) in soil could form Al3+–siderophore complexes and promote P uptake to alleviate Al phytotoxicity (Mora et al., 2017). In addition, exudates such as organic acids, protons, and acid phosphatases produced by the microbes can also reduce the impact of Al toxicity on plants or increase the available phosphorus in the rhizosphere, indirectly help plants reduce aluminum toxicity and adapt to acidic soils (Cabral et al., 2015).
Wild soybean (Glycine soja L.) is a close ancestor of cultivated soybean (Glycine max L.) (Hymowitz, 1970). Compared with cultivated soybean, wild soybean has stronger resistance to biotic and abiotic stresses, such as drought, cold, Al, and attack by viruses and insects (Dong et al., 2001; Chen et al., 2006; Singh and Hymowitz, 2011). In addition to the abundant resistance genes in wild soybeans, microorganisms in the rhizosphere also play an important role in resisting Al toxicity (Luo et al., 2013; Ma et al., 2018; Lian et al., 2019). Although many plant growth–promoting bacteria (PGPR) exert beneficial effects on various crops, their performance can be strongly cultivar specific (Montalban et al., 2017; Rodriguez et al., 2019). Previous research found that the composition and diversity of microorganisms in the rhizosphere of wild rice were significantly different from those in cultivated rice, and the enriched taxa affiliated to Anaerolineae and Nitrospirae in the rhizosphere of wild rice might benefit the disease resistance of the plants (Shenton et al., 2016). Moreover, our previous study reported that the impact of high Al stress on bacterial community structure differed in the rhizosphere between Al-T and Al-S cultivated soybeans, and Al-T soybean could recruit specific microorganisms to resist Al stress (Lian et al., 2019; Shi et al., 2020). However, the effect of wild soybean genotypes on the bacterial community in the rhizosphere has not been clarified, and the bacterial composition in the rhizosphere in response to different levels of Al stress was undefined either. Such studies may provide an important strategy for the identification of beneficial bacteria that may improve plants tolerance to Al stress. Therefore, this study could create a new window for research under acidic soil condition for explore the microbial resources of wild soybeans and crop production.
To investigate the mechanism of how rhizobacterial communities strengthen the Al tolerance of wild soybean, we analyzed the bacterial community structures in the rhizosphere of the Al-T (BW69) and Al-S (W270) genotypes of wide soybean, under different Al concentrations. The chemical properties of the rhizosphere were measured and their correlations with bacterial communities were examined. Given that BW69 is better adapted to Al stress than W270, we hypothesized that (1) there would be significant difference in bacterial structure between BW69 and W270, and (2) BW69 would recruit specific microorganisms in the rhizosphere that may play important roles in the resistance to Al stress.
Materials and Methods
Soil and Plant Materials
The soil used in the experiment was collected at Yingde County (113°40’N, 24°18’E), Guangdong Province, China. The soil was classified as Ali-Udic Argosol according to USDA soil taxonomy. The soil had a pH of 5.19, 10.1 g kg–1 total carbon, 0.39 g kg–1 total nitrogen, 19.40 g kg–1 total potassium, 1.27 g kg–1 total phosphorus, 60.4 mg kg–1 available phosphorus, 40.1 mg kg–1 ammonium nitrogen (NH4+-N), 12.6 mg kg–1 nitrate nitrogen (NO3–-N), 0.01 cmol kg–1 exchangeable H+, and 0.15 cmol kg–1 exchangeable Al3+. In this study, soybeans used in the experiment include Al-T (BW69) and Al-S (W270), two wild genotypes (Ma et al., 2018).
Experimental Design and Soil Sampling
A pot experiment was carried out in a greenhouse at South China Agricultural University in Guangzhou, China. In this study, there were three concentrations of Al in the form of pelletized Al2(SO4)3⋅18H2O, i.e., (1) 0 Al3+ g kg–1, (2) 0.2 Al3+ g kg–1, and (3) 0.4 Al3+ g kg–1, representing zero, low, and high Al concentrations, respectively. A no-plant control (CK) was set at each Al concentration. There were five replicates for each treatment. Each pot was filled with 1.5 kg of soil (sieved with 4 mm mesh) and four seeds with uniform size were sown into each pot on June 15, 2018. The pots were placed in a greenhouse with a temperature range of 16–20°C at night and 28–32°C during the daytime. The soil moisture content was controlled at 80% of the field water capacity by weighing and watering. The loose attached soil was removed by shaking the roots at the flowering stage (60 days after sowing), and then the roots were washed with phosphate-buffered saline (Shi et al., 2015). A total of 45 rhizosphere soil samples were collected. From each sample, 2 g was placed into a micro-centrifuge tube and kept at -80°C until DNA extraction. The remaining rhizosphere soil was kept at 4°C before the determination of soil characteristics. Meanwhile, the wild soybean plants were harvested and dried 70°C for 3 days (Lian et al., 2017).
Determination of Soil Properties
Soil pH was determined in a 5:1 water-to-soil suspension using a pH meter (FE20-FiveEasy pH, Germany) (Wang et al., 2013). Total carbon and total nitrogen were determined with an elemental analyzer (VarioEL III, Germany) (Jones and Willett, 2006). Soil total potassium was measured by inductively coupled plasma-atomic emission spectrometry (I-7500; Shimadzu, Japan) (Jiao et al., 2019). The soil-exchangeable Al3+ and H+ were measured by a titrimetric method (Abreu et al., 2003). Soil NO3–-N and NH4+-N levels were determined with a continuous flow chemistry analyzer (skalar SAN++, Netherlands) (Miranda et al., 2001). Available phosphorus and total potassium were determined using an atomic absorption spectrometer (GFA-6800, Japan) (Sun et al., 2015).
Extraction of Soil DNA and Real-Time Quantitative PCR
A Fast DNA SPIN Kit for Soil (MP Biomedicals, Santa Ana, CA) was used to extract DNA following the manufacturer’s specifications. The bacterial community abundance was determined by quantitative real-time PCR (q-PCR) (ABI 7900) using the primers 515f (5′-GTGCCAGCMGCCGCGGTAA-3′) and 907r (5′-CCGTCAATTCMTTTRAGTTT-3′) (Osburn et al., 2011). The 20 μl PCR mixture included 1.0 μl of extracted DNA, 1.0 μl forward and reverse primers (10 nM), 7.0 μl of sterile water, and 10 μl SYBR Premix Ex Taq (Takara, Dalian, China). The q-PCR amplification program was as follows: initial denaturation for 1 min at 98°C, followed by 30 cycles of denaturation for 10 s at 98°C, annealing for 30 s at 50°C, elongation for 60 s at 72°C, and the final cooling for 5 min at 72°C to perform a cycle. The copy number of 16S rRNA gene was calculated based on standard curves (Yao et al., 2014).
High-Throughput Sequencing and Data Analysis
Based on Illumina MiSeq sequencing, the 515F/907R primer sequence was used to amplify the V4 hypervariable region of the 16S rRNA gene (Osburn et al., 2011). Equimolar amounts of amplicons from the same sample were pooled, then the Illumina paired-end (PE 250) sequencing platform was used for double-end sequencing analysis. All raw sequences are available in the NCBI short-read archive under accession number PRJNA531335.
After sequencing, the FASTQ original sequence files of bacteria were processed by USEARCH 10 (Edgar, 2013). The unoise3 algorithm was conducted for denoising (error-correction) amplicon reads. Briefly, reads with sequencing errors were detected and corrected, and chimeras were identified and removed using the Uchime algorithm (Edgar et al., 2011). To obtain the corresponding species classification information for each operational taxonomic unit (OTU), a classification analysis of OTU representative sequences with 97% similarity was conducted using the RDP classifier with a Bayesian algorithm (Cole et al., 2009). Chao1 richness and Shannon index were calculated using Mothur V1.30.1 (Kwak et al., 2018).
Genstat V12 was used to perform a one-way ANOVA to identify significant differences in soil chemical properties among all treatments (Lian et al., 2016). Non-metric multi-dimensional scaling (NMDS), canonical correspondence analysis (CCA), Adonis test, and Mantel test were conducted using R v.3.5.1 with the “vegan” package (R Development Core Team, 2006). To identify the OTUs that were significantly different between two genotypes, a generalized linear model with a negative binomial distribution of the OTU relative abundance was created to normalize the values of each OTU (Edwards et al., 2015). A Venn analysis was conducted to show the co-enriched OTUs according to the results of the generalized linear model (Edwards et al., 2015). Based on the 95% confidence interval, significantly different genera between BW69 and W270 were analyzed by Stamp v.2.1.3 (Parks et al., 2014).
Co-occurrence Network Analysis of Bacterial Community
We constructed six networks to analyze pairwise correlations of bacterial OTUs (average abundance > 0.1%). Spearman’s rank correlation and p-values were calculated in the “psych” R package, using the Gephi for visualization (Jiang et al., 2017a). A correlation was considered as significance with Spearman’s correlation coefficient > 0.8 and p < 0.05 (Jiang et al., 2017b). The nodes in the co-occurrence network represent bacteria OTUs, and edges represent robust and significant correlations between OTUs. A set of network topological properties (e.g., average degree, average path length, and graph density) and node features (e.g., degree, closeness centrality, and betweenness centrality) were calculated in Gephi. Nodes with high degree, high closeness centrality, and high betweenness centrality values are considered as keystone species (Zhou et al., 2010; Berry and Widder, 2014; Agler et al., 2016).
Results
Soybean Biomass, Bacterial Abundance, and Soil Chemical Properties
Overall, Al stress decreased the biomass of both soybean genotypes, and the biomass of BW69 was significantly higher than W270 at the low and high Al concentration (Figure 1A). The bacterial abundance ranged from 4.3 × 109 to 10.9 × 109 gene copies g–1 dry soil across all samples. A significant difference in bacterial abundance between BW69 and W270 was observed at the high Al concentration only (p < 0.01) (Figure 1B).
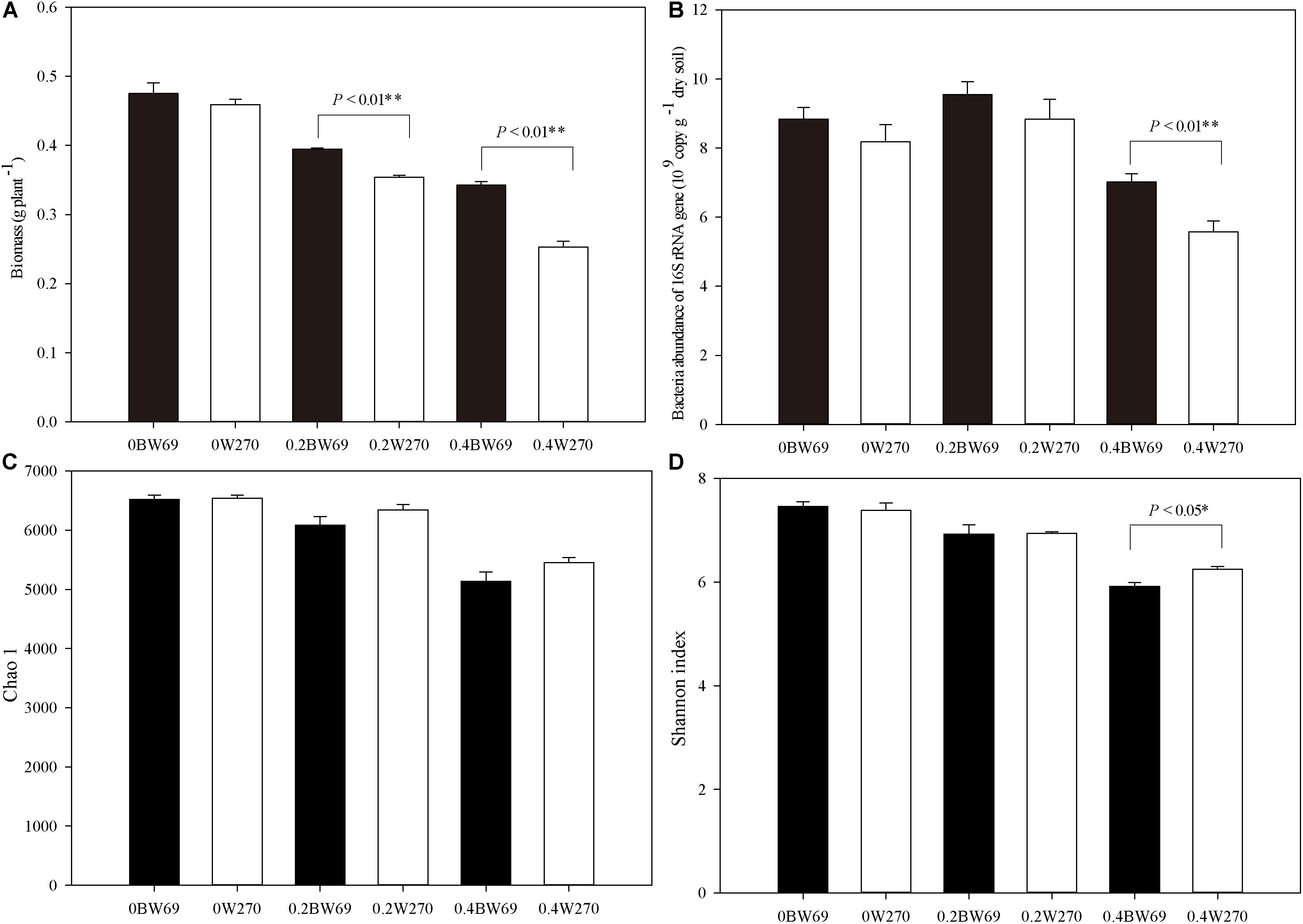
Figure 1. Effects of Al addition (0, 0.2, and 0.4 g kg–1) on the biomass (A), abundance of bacterial 16S rRNA gene copies (B), Chao1 index (C), and Shannon index in the rhizosphere of wild soybean (D). One-way ANOVA with Student’s t-test showed significant differences between BW69 and W270 (p < 0.05). Error bars on data points represent the SEM (n = 5).
The effect of Al stress on soil chemical properties was summarized in Table 1. Generally, soil available phosphorus, NH4+-N and NO3–-N were significantly higher in the rhizosphere of BW69 than W270 at the low and high Al concentrations. However, the concentrations of soil total potassium, exchangeable Al3+, and pH were significantly different between two genotypes only at the high Al concentration, with higher values for W270. Moreover, the soil-exchangeable H+ and Al3+ concentrations increased with the increase of Al concentration supplied, whereas pH showed the opposite trend (Table 1).
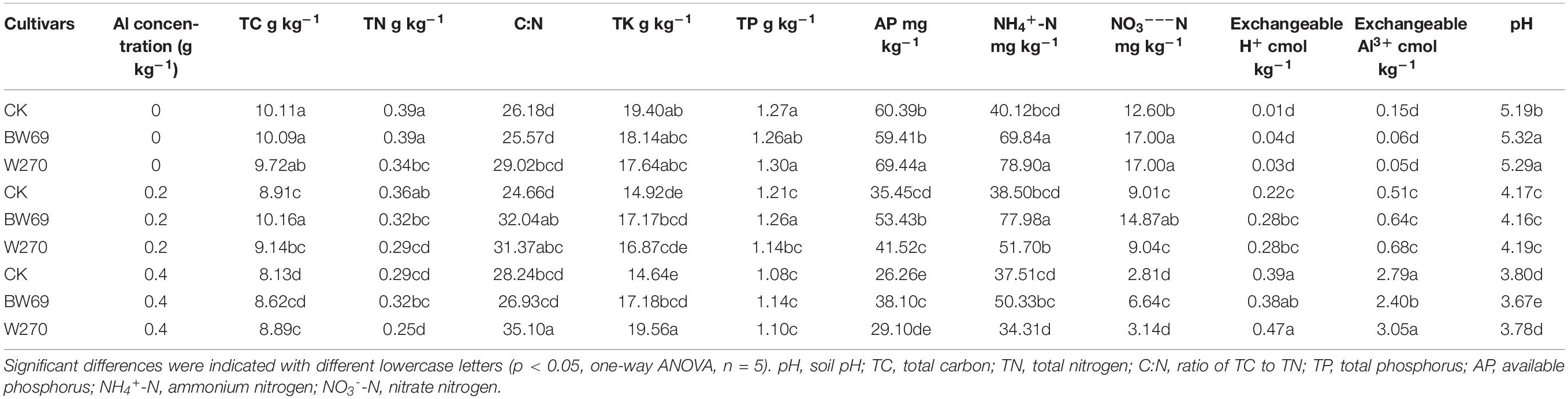
Table 1. Effects of different wild soybean cultivars and Al concentrations on soil chemical properties in an acidic soil.
Taxonomic Classification and Soil Bacterial Diversity
A total of 2,071,056 high-quality sequences were obtained in this study, and the average reading count for each sample was 28,309. When clustered with 97% sequence identity, there were 8108 OTUs for all samples, with a mean of 4222 OTU for each sample. The Chao1 and Shannon index of the bacterial community in the rhizosphere of two genotypes decreased with the increase of Al concentration. However, Shannon index only showed a significant difference between two genotypes at the high Al concentration, with the higher value for W270 (Figures 1C,D).
Structure of the Soybean Rhizobacterial Community
Based on the NMDS, the bacterial communities at three Al concentrations were significantly separated (Adonis test, p < 0.05) (Figure 2A). The rhizobacterial community for no-plant control samples significantly separated from BW69 and W270 at each Al concentration (Adonis test, p < 0.05) (Figure 2A). Significant differences between BW69 and W270 were only observed under the high Al concentration (Adonis test, p < 0.01) (Figures 2B–D). The bacterial community structure in the rhizosphere varied with Al concentration and genotype, and high Al concentration magnified the differences of bacterial community structure between two genotypes.
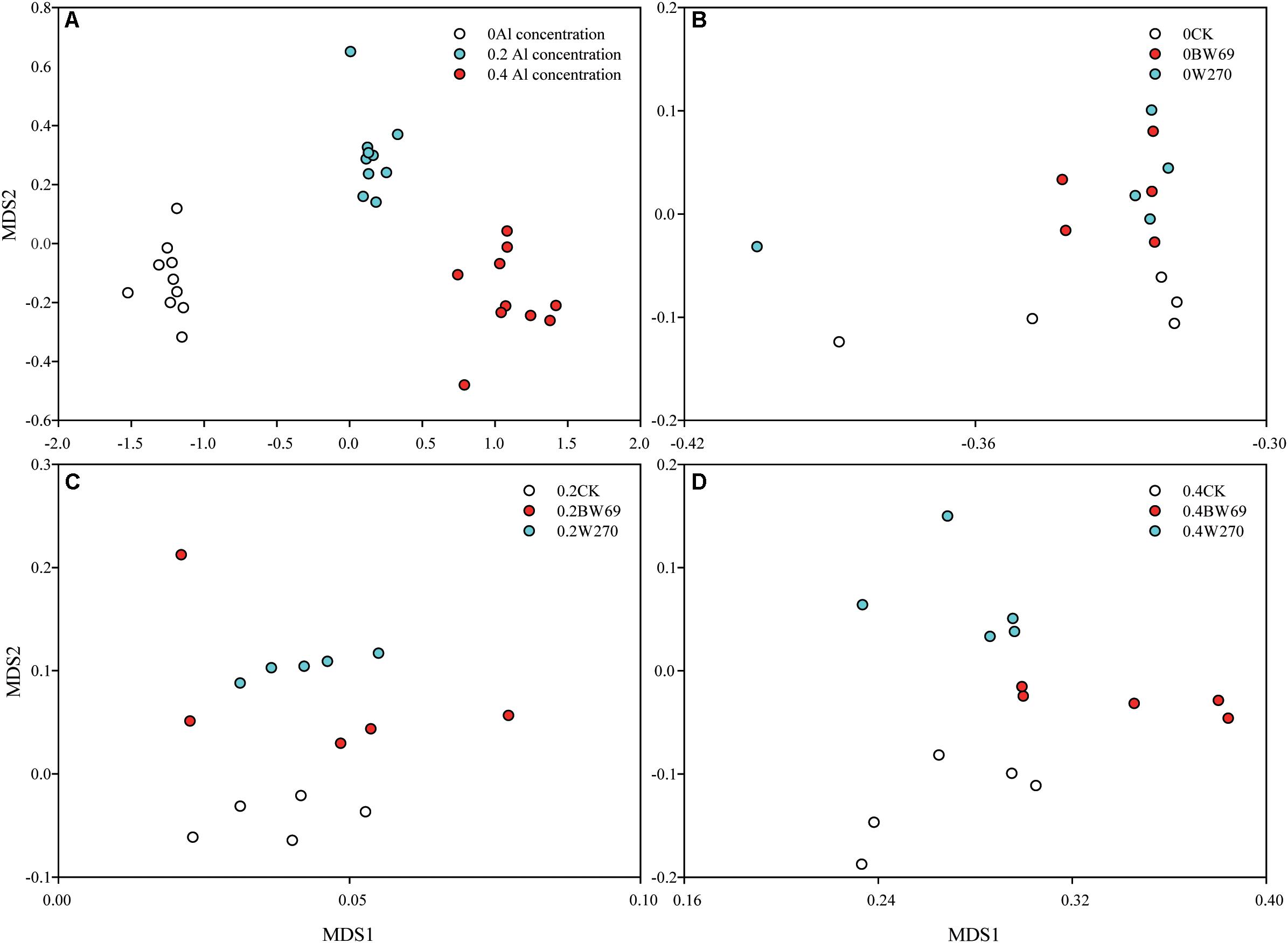
Figure 2. Non-metric multidimensional scale (NMDS) analysis based on Bray–Curtis dissimilarity showing that the rhizobacterial communities under 0, 0.2, and 0.4 g kg–1 of Al concentrations separated from each other (Adonis test, p < 0.05) (A). NMDS analysis based on Bray–Curtis dissimilarity that rhizobacterial communities of BW69 mixed those of W270 under the 0 g kg–1 Al concentration (Adonis test, p > 0.05) (B). NMDS analysis based on Bray–Curtis dissimilarity that rhizobacterial communities of BW69 mixed with those of W270 under the 0.2 g kg–1 Al concentration (Adonis test, p > 0.05) (C). NMDS analysis based on Bray–Curtis dissimilarity that rhizobacterial communities of BW69 separated from those of W270 under the 0.4 g kg–1 of Al concentration (Adonis test, p < 0.01) (D) (n = 5).
The CCA was used to determine the relationship between soil chemical properties and the bacterial communities in the rhizosphere. The CCA and Mantel test showed that the bacterial community structures were significantly related to the soil properties in rhizospheres of the two genotypes, which were total carbon, total nitrogen, total potassium, available phosphorus, NH4+-N, NO3–-N, exchangeable H+, exchangeable Al3+, and pH (Figures 3A,B and Supplementary Table S1). Moreover, the total nitrogen, C:N ratio, NH4+-N, available phosphorus, total potassium, and pH were significantly related to the bacterial community structure of the two genotypes at the high Al concentration (Figure 3C and Supplementary Table S1).
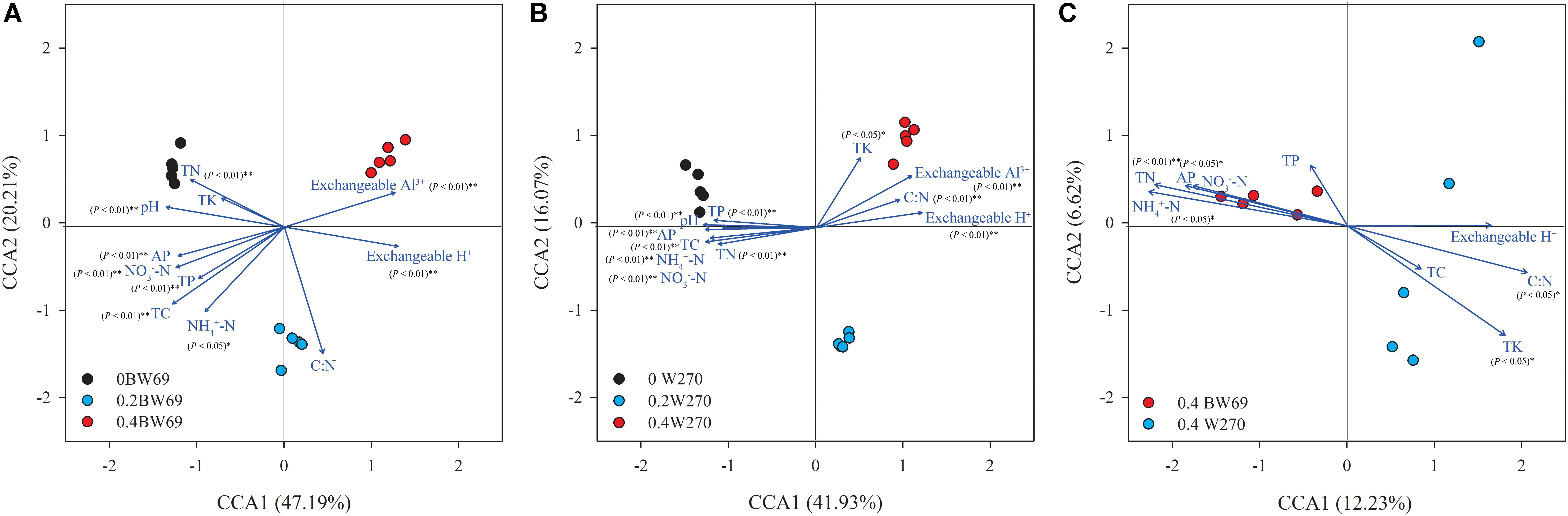
Figure 3. A canonical correspondence analysis (CCA) based on OTU level reveals the influence of soil properties on rhizobacterial communities under among BW69 samples (A), W270 samples (B), and high Al concentration samples (C). Relationships between rhizosphere bacterial communities and soil properties. Arrows indicate the direction and magnitude of environmental parameters associated with bacterial community structure. pH, soil pH; TC, total carbon; TN, total nitrogen; C:N, ratio of TC to TN; TP, total phosphorus; AP, available phosphorus; NH4+-N, ammonium nitrogen; NO3–-N, nitrate nitrogen.
The Relative Abundance of Soybean Rhizosphere Bacteria
Proteobacteria, Actinobacteria, Chloroflexi, Firmicutes, and Acidobacteria were the dominant phyla in all soil samples, with total relative abundance over 75%; the relative abundance of the five phyla ranged from 21.4 to 47.7%, 11.1 to 35.6%, 47.7 to 11.1%, 4.8 to 31.8%, and 16.3 to 31.8%, respectively (Figure 4A and Supplementary Figure S1). Moreover, the phyla of Planctomycetes, Bacteroidetes, Cyanobacteria, Gemmatimonadetes, and Nitrospirae exhibited low abundances in all soil samples (relative abundance > 0.1%, but < 5%) (Figure 4A and Supplementary Figure S1). There was no significant difference between BW69 and W270 for the five dominant phyla. However, at the class level, the relative abundances of Alphaproteobacteria, Deltaproteobacteria, and Acidobacteria in the rhizosphere of W270 was higher than BW69 under the high Al concentration, whereas the Gammaproteobacteria were more abundant in BW69 (p < 0.05, p < 0.01) (Figure 4B).
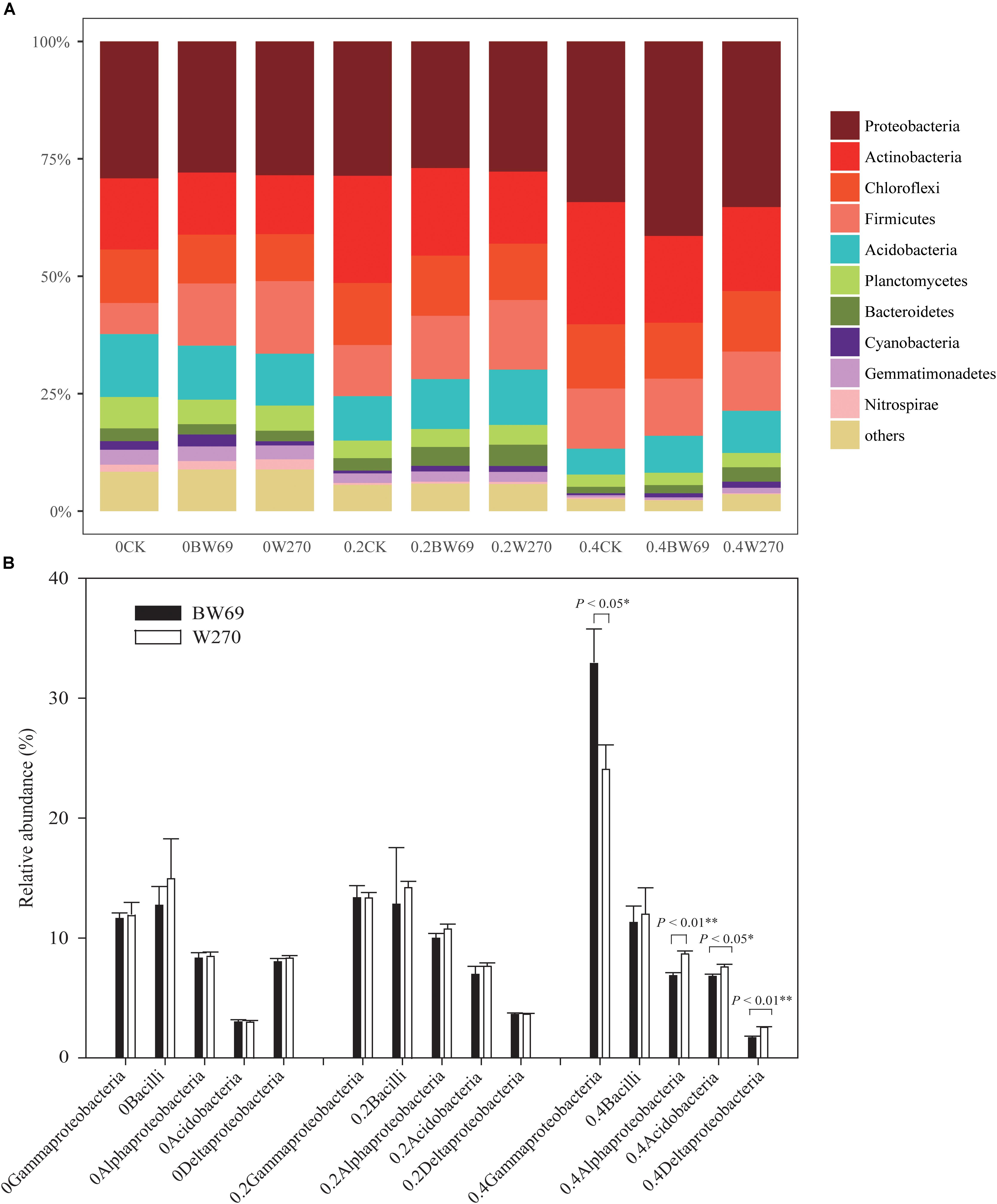
Figure 4. Relatively abundance of bacteria at the phylum level (A) and class level (top five dominant classes) (B) of the BW69 and W270 in 0, 0.2, and 0.4 g kg–1 Al concentration addition. One-way ANOVA with Student’s t-test showed significant differences between BW69 and W270 (p < 0.05). Error bars on data points represent the SEM (n = 5).
Using the 95% confidence interval, the bacterial genera between BW69 and W270 at three Al concentrations were compared. The relative abundance of the top-100 genera were selected for further statistical analyses. The number of genera with significant differences between BW69 and W270 were 4, 11, and 35 at the zero, low, and high Al concentrations, respectively. Among these genera, two and five genera had higher relative abundance in the rhizosphere of BW69 compared with W270 at the zero and low Al concentrations, respectively (Figures 5A,B). Eleven genera, such as Gammaproteobacteria_KF-JG30-C25_norank, Ktedonobacteraceae_uncultured, and Chujaibacter, were significantly higher in BW69 compared with W270 at the high Al concentrations (Figure 5C).
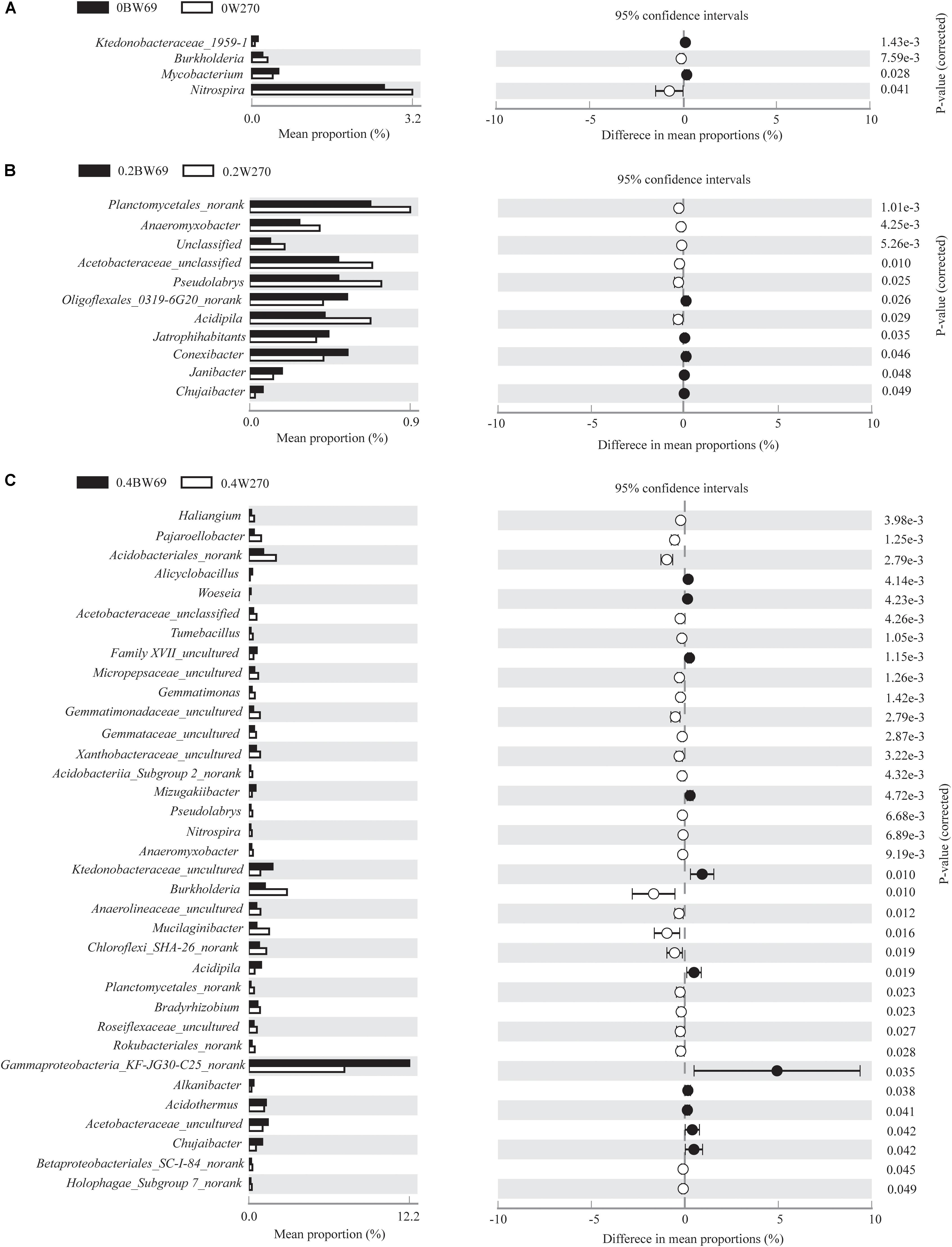
Figure 5. Stamp analysis of the relative abundance of genera between BW69 and W270 at the 95% confidence interval level under Al concentrations of 0 g kg–1 (A), 0.2 g kg–1 (B), and 0.4 g kg–1 (C). P-values were calculated using Welch’s t-test.
To evaluate the difference of OTU between BW69 and W270 at different Al concentrations, we created a generalized linear model of the negative binomial distribution. OTU counts in the rhizospheres of BW69 and W270 under the zero Al concentration were used as the controls to compare the enriched or depleted OTUs under the low and high Al concentrations, respectively. There were 504 and 541 OTUs enriched for BW69 and W270 at the low Al concentration, and 294 and 372 for BW69 and W270 at the high Al concentration, respectively (Figures 6A–D). There were 49 OTUs that overlapped in the enriched OTUs in the rhizosphere of BW69 at both the low and high Al concentrations (Figure 6E and Supplementary Table S2). The effect of different Al concentrations on relative abundance of OTUs (>0.1%) is shown in Figure 7. The relative abundances of OTU15 (Gammaproteobacteria_KF-JG30-C25_norank), OTU23 (Mizugakiibacter), OTU55 (Ktedonobacteraceae_uncultured), OTU65 (Dyella), OTU93 (Alkanibacter), and OTU195 (Ktedonobacterales_JG30-KF-AS9_norank) increased with the increase of Al concentration, with higher values in the rhizosphere of W69compared with W270 under the high Al concentration (Figure 7).
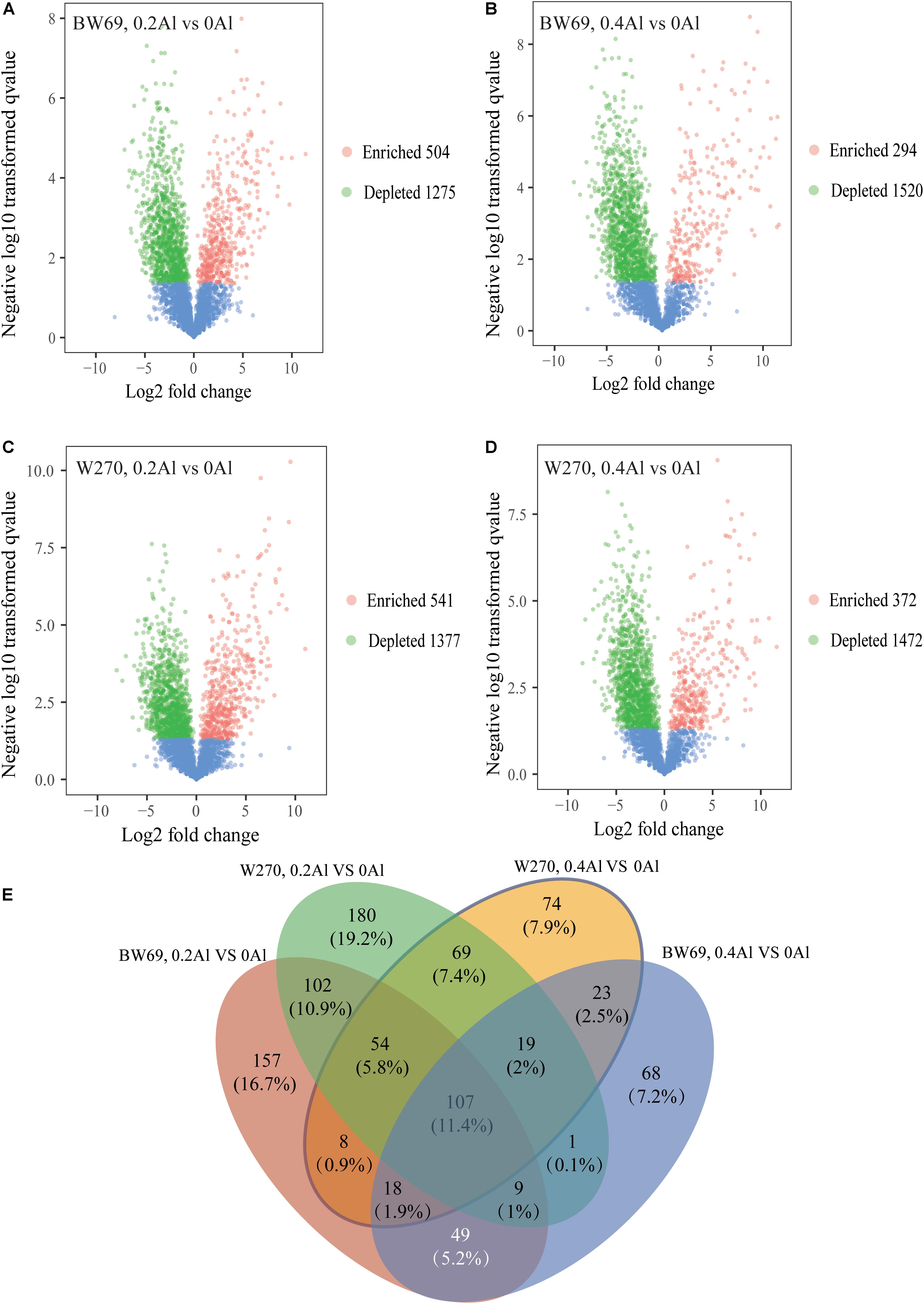
Figure 6. Enrichment and depletion of OTUs in 0.2 g kg–1 vs. 0 g kg–1 Al concentration of BW69 (A), 0.4 g kg–1 vs. 0 g kg–1 Al concentration of BW69 (B), 0.2 g kg–1 vs. 0 g kg–1 Al concentration of W270 (C) and 0.4 g kg–1 vs. 0 g kg–1 Al concentration of W270 (D). Each point represents an individual OTU, and the position along the y-axis represents the abundance fold change compared with BW69 and W270, respectively, under the 0 g kg–1 Al concentration. The Venn diagram shows the number of unique enriched and co-enriched OTUs in the rhizospheres of BW69 and W270 under different Al concentrations (E).
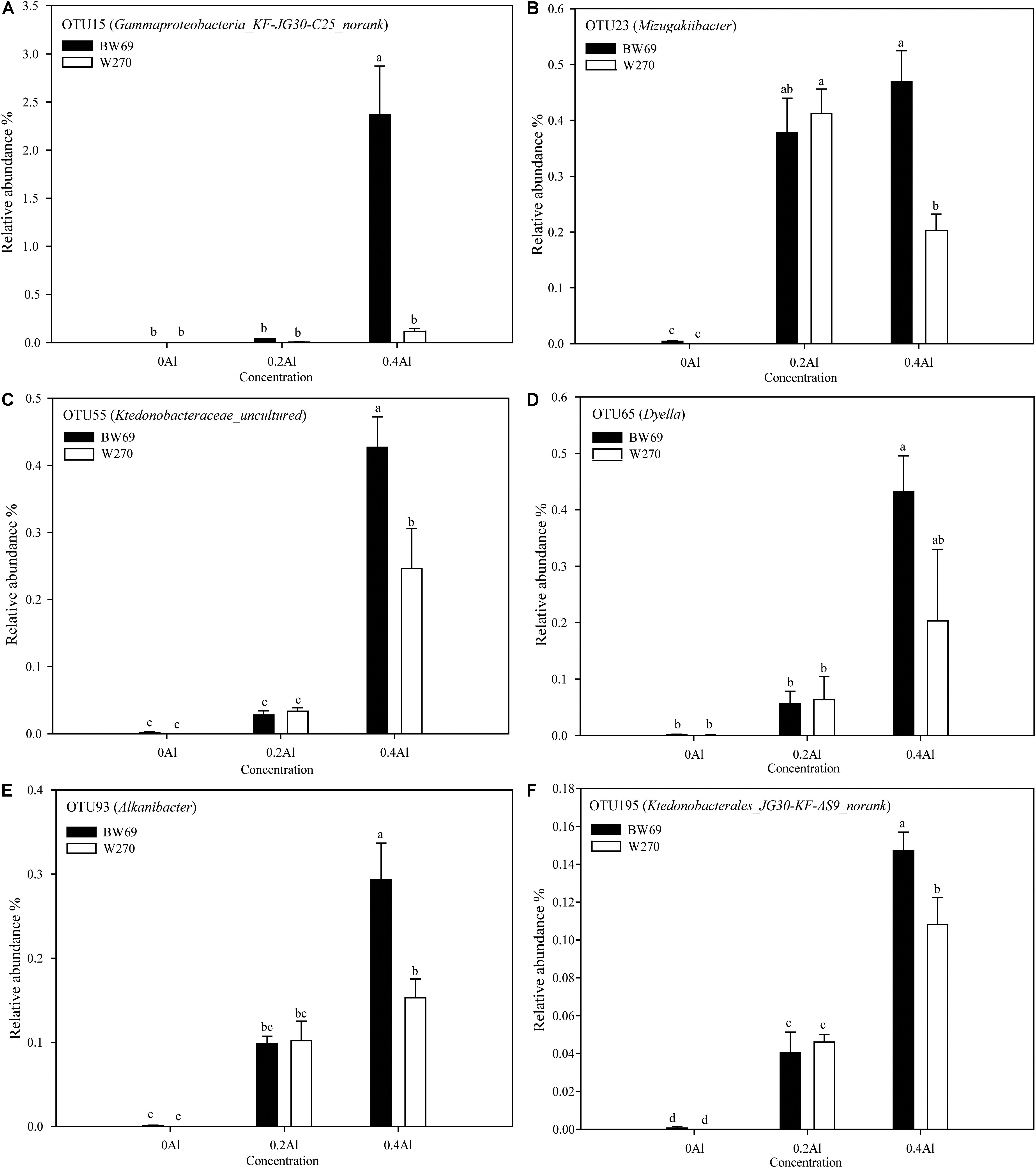
Figure 7. The relative abundance of dominant OTU15 (A), OTU23 (B), OTU55 (C), OTU65 (D), OTU93 (E), and OTU195 (F). Significant differences are indicated with different lowercase letters (p < 0.05, one-way ANOVA with Student’s t-test). Error bars on data points represent the standard error of the mean (n = 5).
Co-occurrence Network Analysis of Wild Soybean Rhizosphere Bacteria
To determine the symbiosis and competitive mode of bacterial consortia in the rhizosphere of BW69 and W270, the OTUs with relative abundance >0.1% were screened as nodes (Supplementary Table S3). Six networks were constructed, and the network structure of two genotypes at different Al concentrations was analyzed (Figure 8 and Table 2). The network topology properties, such as the number of edges, positively and negatively correlated edges, and graph density, were similar between BW69 and W270 at the zero Al concentration (Table 2). The number of edges, positively correlated edges, graph density, and average degree in the rhizosphere of W270 were greater than BW69 at the low Al concentration, whereas an opposite tendency was observed at the high Al concentration (Table 2). Keystone bacterial species of the two genotypes at different Al concentrations are presented in Table 3. The OTU496 (Occallatibacter) and OTU118 (Singulisphaera) were identified as keystone species in the BW69 network at the high Al concentration, whereas OTU16 (Bradyrhizobium) and OTU28 (Pandoraea) were identified in the rhizosphere of W270 (Table 3).
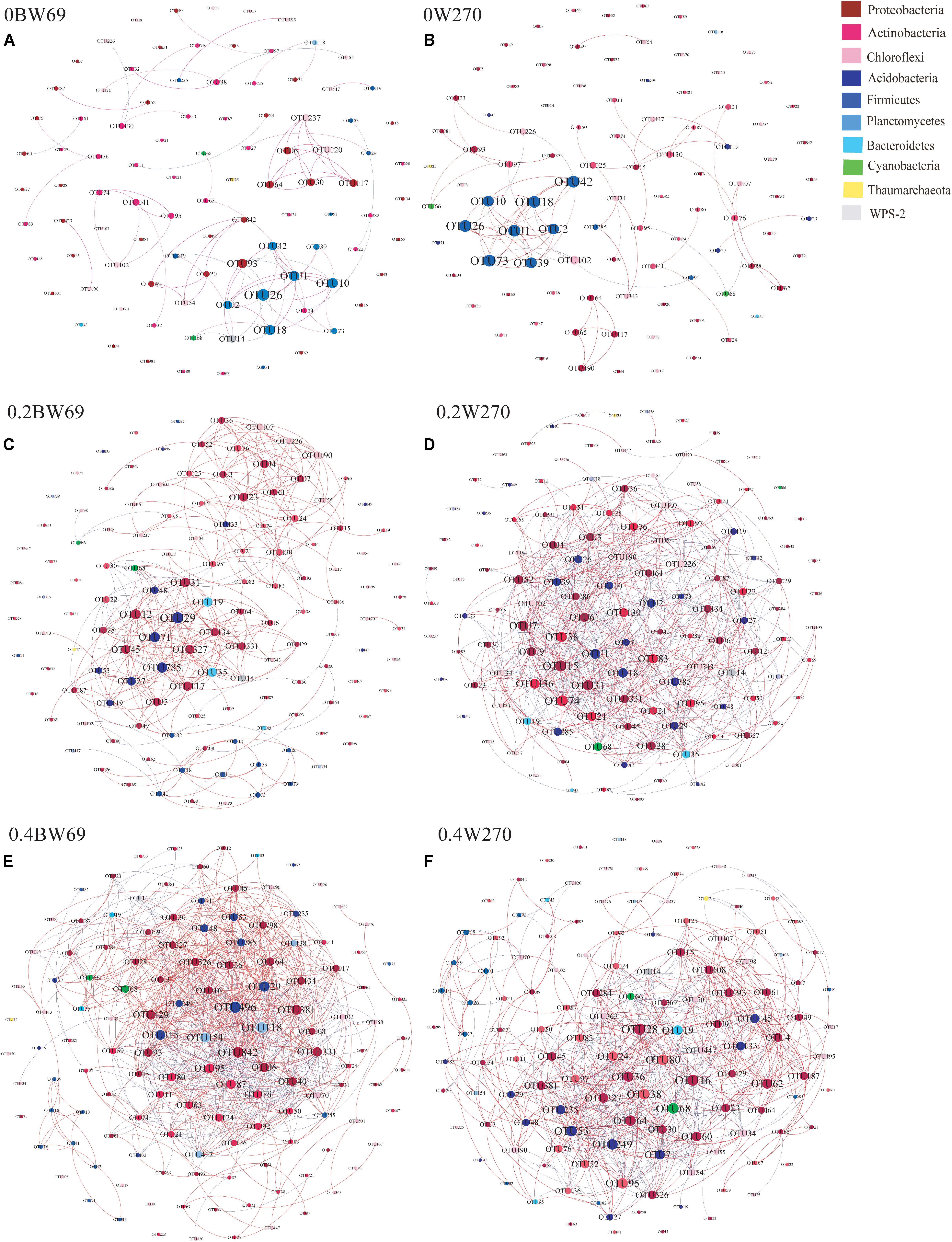
Figure 8. Co-occurrence network of bacterial community in the rhizosphere of genotypes under three Al concentrations, i.e., 0BW69 (A), 0W270 (B), 0.2BW69 (C), 0.2W270 (D), 0.4BW69 (E), and 0.4W270 (F). A connection stands for a strong (Spearman’s r > 0.8) and significant (p < 0.05) correlation. The co-occurring networks are colored by phyla. For each graph, the size of each node is proportional to the number of connections (degree). A red edge indicates a positive interaction between two individual nodes, whereas a green edge indicates a negative interaction.
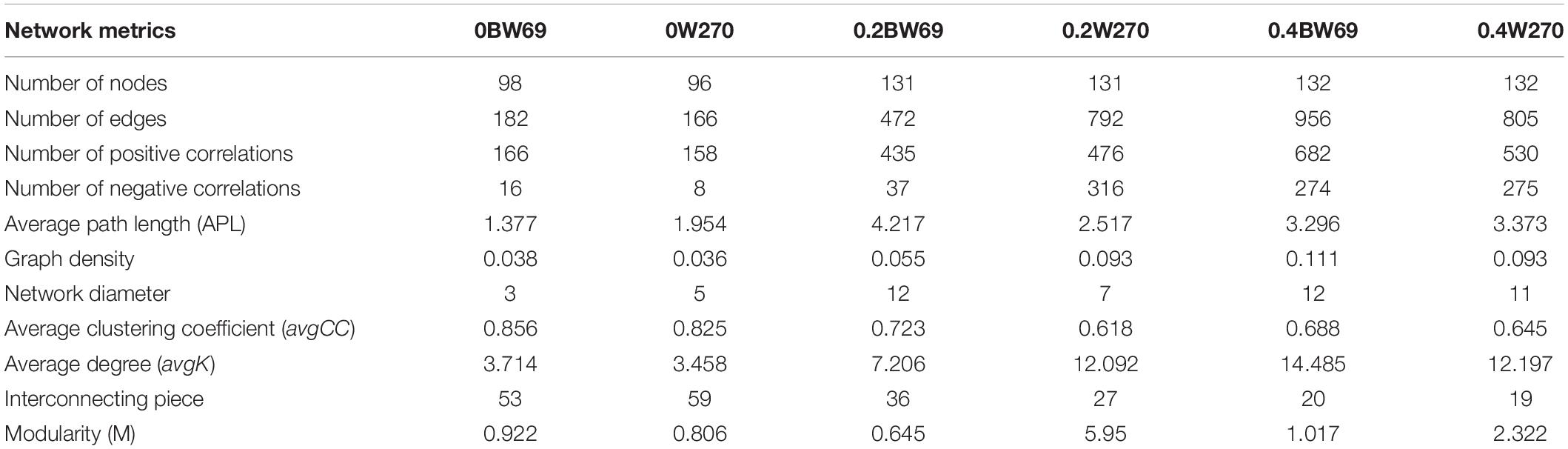
Table 2. Topological properties of co-occurring bacterial networks in the rhizosphere of two soybean genotypes under three Al concentrations (0BW69, 0W270, 0.2BW69, 0.2W270, 0.4BW69, and 0.4W270).
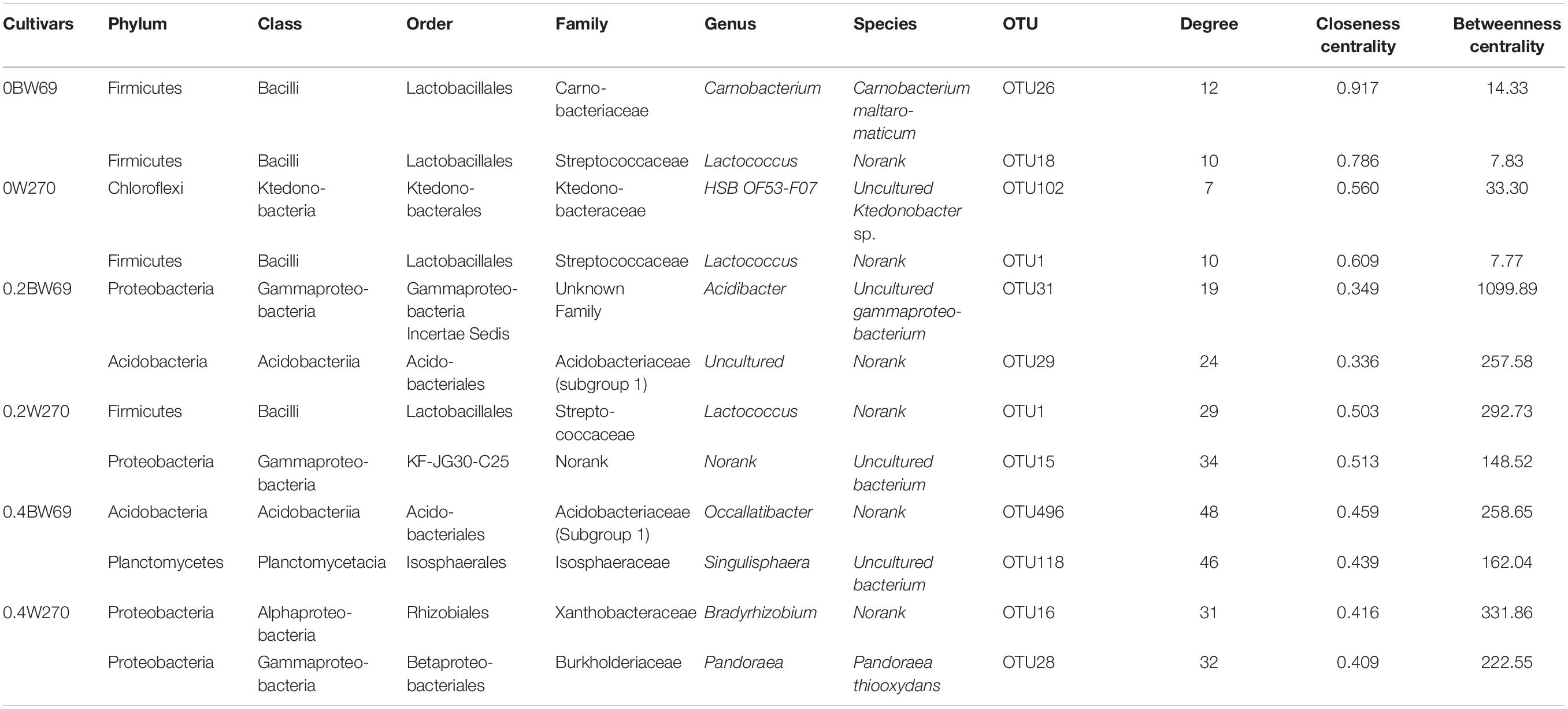
Table 3. Topological properties of keystone species in the rhizosphere of two soybean genotypes under three Al concentrations (0BW69, 0W270, 0.2BW69, 0.2W270, 0.4BW69, and 0.4W270).
Discussion
The purpose of this study was to reveal how different levels of Al stress affect bacterial community characteristics in the rhizosphere of wild soybean genotypes differing in the Al tolerance. The results were consistent with our first hypothesis, of which the bacterial community structures in the rhizosphere of BW69 were different from that of W270 at the high Al concentration. There were greater differences in bacterial abundance, Shannon index, and abundances of rhizobacteria at the class, genus, and OTU levels between BW69 and W270 under the high Al concentration than the low Al concentration. However, the second hypothesis was not verified according to our results because no bacteria species that related to Al resistance were recruited in the rhizosphere of BW69.
Response of Rhizosphere Bacterial Abundance and Diversity to Al Stress
Soil nutrients were positively correlated with bacterial abundance (Sun et al., 2015). In our study, the higher bacterial abundance in the rhizosphere of BW69 compared with W270 might be attributed to more abundant nutrients in the rhizosphere, such as more available phosphorus, NH4+-N, and NO3–-N (Table 1). For bacterial diversity, the Shannon index was significantly lower in the rhizosphere of BW69 compared with W270 at the high Al concentration. This finding was inconsistent with our previous research in which the Al-T cultivated soybean genotype had greater bacterial diversity than that of the Al-S soybean genotype (Lian et al., 2019). These inconsistent results may be caused by two reasons. One would be the lower soil pH in the rhizosphere of BW69 compared with W270, posing a negative impact on bacterial diversity that might conceal the positive contribution to more root exudates secreted by the Al-T genotype (Fierer and Jackson, 2006; Rousk et al., 2010; Bartram et al., 2014). Another reason could be associated with the different soybean varieties used in the two studies because the bacterial diversity was significantly different between wild and cultivated crops (Shenton et al., 2016).
Response of Rhizosphere Bacterial Community Structure to Al Stress
The NMDS showed significant differences in bacterial community structure in the rhizosphere between BW69 and W270 at the high Al stress. This finding was consistent with our previous study which illustrated that high Al stress magnified the differences of bacterial community structure between Al-T and Al-S soybean genotypes (Lian et al., 2019). This may be attributed to the fact that various genotypes have different levels of Al-related gene expression when exposed to Al stress, and these expression levels increased with the increase of Al concentration, especially in the Al-T genotype (Maron et al., 2010; Tovkach et al., 2012). The greater expression of these genes in the Al-T genotypes lead to more secretion of organic acids that could chelate more Al3+ to reduce Al toxicity (Yang et al., 2012; Li et al., 2017). For example, the GsMATE gene cloned from BW69 was related to the synthesis and secretion of citric acids, which enhanced the tolerance of Arabidopsis to Al stress (Ma et al., 2018). However, more secreted organic acids in the Al-T genotype may result in decreased pH, providing a complex environment, and consequently affecting a wide variety of bacteria in the rhizosphere (Table 1) (Clarkson and Marschner, 1995; Jones, 1998; Teplitski et al., 2000; Wang et al., 2013).
We conducted a differential analysis on OTU relative abundance and observed that 49 OTUs were co-enriched in BW69 soybean under the low and high Al concentrations. These OTUs mainly belong to Proteobacteria, Actinobacteria, and Chloroflexi. Some OTUs in BW69 had greater relative abundance than in W270 at the high Al concentration, such as OTU15 (Gammaproteobacteria_KF-JG30-C25_norank), OTU23 (Mizugakiibacter), OTU55 (Ktedonobacteraceae_uncultured), OTU65 (Dyella), OTU93 (Alkanibacter), and OTU195 (Ktedonobacterales_JG30-KF-AS9_norank) (Figure 7). Various functions of these bacteria have been reported. For instance, Mizugakiibacter has potential use for the remediation of soil with heavy metal pollution (Guo et al., 2017) and Alkanibacter can degrade hexane and other short-chain alkanes (Davis-Belmar et al., 2013). Ktedonobacteria, which predominate in acidic soil, may act as a microbial resource with the potential to produce secondary metabolites (Yabe et al., 2017). These bacteria were different from those identified in our previous study in which the co-enriched OTUs of cultivated Al-T soybeans mainly belonged to Tumebacillus and Burkholderia (Lian et al., 2019). These results indicated that wild and cultivated soybeans recruited different species of bacteria to resist Al stress. However, the relationship between these bacteria and soil Al toxicity needs further study. Whether exogenous addition of these bacteria improves soybean resistance to Al toxicity warrants further test.
The CCA showed that the bacterial community structures of two genotypes significantly correlated with soil properties, except for total potassium. This pattern could be explained by the fact that Al stress had a greater impact on the environmental factors than genotypes, which reflected that some environmental factors such as pH and exchangeable H+ and Al3+ were significantly different among Al concentrations, without a corresponding difference between two genotypes (Table 1). Moreover, the CCA for the high Al concentration showed that the differences in the bacterial community structure between BW69 and W270 were mainly linked with total nitrogen, NH4+-N and C:N ratio (Figure 3C). This finding was consistent with our previous study showing that NH4+-N mainly drove differences in bacterial community structure in the rhizosphere of Al-T and Al-S genotype under high Al concentration (Lian et al., 2019). These results provided more evidence on NH4+-N mitigating the toxicity of Al to plants (Zhao and Shen, 2018). An increase in NH4+-N decreases pH, thereby increasing the content of Al3+ and H+ (Zhao and Shen, 2018). However, Al-T plants may be able to take up more H+ and suppress Al3+ to enter the plant roots, consequently reducing Al phytotoxicity (Zhao et al., 2009, 2014).
Response of Bacterial Co-occurrence Networks to Al Stress
A network analysis was constructed to explore the interrelationship among bacteria and compare the structure of the networks in the rhizosphere of BW69 and W270. In this study, the network topology properties, such as the number of edges, positive correlation edges, graph density, and average degree in the rhizosphere of W270, were greater than those of BW69 at the low Al concentration, whereas an opposite trend was observed at the high Al concentration (Figure 8 and Table 2). This may be attributed to the absence of difference in soil pH between two genotypes at the low Al concentration, which might lead to the similar secretion of organic acids between BW69 and W270 (Chen and Liao, 2016; Liu et al., 2016). Combined with the results of network in this study, W270 may even be better adapted to low Al stress. Furthermore, a significant lower value of pH in BW69 than W270 may be caused by the more organic acid expression of BW69 (Liang et al., 2013; Liu et al., 2016; Zhou et al., 2017). More organic acids not only chelate more Al3+ but also provide a better living environment (Hu et al., 2018; Lian et al., 2019) for more complicated network structure of BW69 than W270. In addition, the mechanism for recruiting Al-tolerant microorganisms of Al-T and Al-S genotypes were different (Yang et al., 2012; Lian et al., 2019). The Al-tolerant microorganisms recruited by BW69 could cope with different degrees of Al stress and therefore form a more complicated network structure than W270. W270 may stimulate most of the Al-tolerant microorganisms to be involved in the physiological processes associated with the low Al stress. However, many microorganisms in the W270 rhizosphere cannot adapt to high Al stress, and only a small number of Al-tolerant bacteria were recruited in the rhizosphere, which simplified the network structure.
BW69 had lower modularity compared with that of Al-S under Al stress, which could be interpreted as increased inter-species competition, and less resistant to Al stress in the rhizosphere of BW69 than W270 (Saavedra et al., 2011; Fan et al., 2018). Moreover, the keystone species being associated with BW69 and W270, and the difference in keystone species, regardless of their abundance, would be a critical determinant of the other community compositions in the rhizosphere (Zhou et al., 2010; Ma et al., 2016; Jiang et al., 2017a). The difference in network structure between BW69 and W270 under different Al concentrations were inconsistent to our previous study which showed that the Al-S cultivated soybean had a simpler network at the low Al concentration and a more complex network at the high Al concentration compared with Al-T cultivated soybean (Lian et al., 2019). This might be attributed to that wild soybean has stronger resistance to biotic and abiotic stress than cultivated soybean (Dong et al., 2001; Chen et al., 2006; Singh and Hymowitz, 2011). The difference in the root architecture and microhabitat between wild and cultivated soybean could influence the root exudates and the plant immune system, and consequently causing the different bacterial network structure (Shenton et al., 2016). However, more specific reasons of the opposite trend of the network structure between wild and cultivated soybeans should be further studied.
In conclusion, our findings highlighted that high Al concentration magnified the differences of bacterial community structure between two genotypes. Furthermore, 49 OTUs were identified to be co-enriched in the rhizosphere of BW69 at the low and high Al concentrations. Moreover, total nitrogen and NH4+-N were the major factors affecting the bacterial community structure in the rhizospheres of the two genotypes under the high Al concentration. However, the lower modularity in BW69 under Al stress may cause the rhizobacterial community to be less resistant and more influenced by disturbance. Collectively, this study emphasizes the possibility of using rhizobacteria as an improved crop breeding or gene to produce crops that are more resistant to the toxicity of heavy metal. However, the relationship between these “enriched” bacterial species and soil aluminum toxicity has not been clarified. Further tests should be carried out to evaluate the universality of the results that whether these bacteria can improve the resistance of soybean to aluminum toxicity in different soil types.
Data Availability Statement
The datasets generated for this study can be found in the NCBI short-read archive under accession number PRJNA531335.
Author Contributions
TL, HN, YC, and QM designed the research. TL, QS, YL, and ZC performed the research. QS and TL analyzed the data and wrote the manuscript. JJ, RW, and YZ improved the English. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key Research and Development Program of China (Grant No. 2018YFD1000903).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01996/full#supplementary-material
References
Abreu, C. H. Jr., Muraoka, T., and Lavorante, A. F. (2003). Exchangeable aluminum evaluation in acid soils. Sci. Agric. 60, 543–548. doi: 10.1590/s0103-90162003000300020
Agler, M. T., Ruhe, J., Kroll, S., Morhenn, C., Kim, S. T., Weigel, D., et al. (2016). Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol. 14:e1002352. doi: 10.1371/journal.pbio.1002352
Bartram, A. K., Jiang, X. P., Lynch, M. D. J., Masella, A. P., Nicol, G. W., Jonathan, D., et al. (2014). Exploring links between pH and bacterial community composition in soils from the craibstone experimental farm. FEMS Microbiol. Ecol. 87, 403–415. doi: 10.1111/1574-6941.12231
Berendsen, R. L., Pieterse, C. M. J., and Bakker, P. A. H. M. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Berry, D., and Widder, S. (2014). Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front. Microbiol. 5:219. doi: 10.3389/fmicb.2014.00219
Besserer, A., Puech-Pages, V., Kiefer, P., Gomez, R. M. V., Jauneau, A., Roy, S., et al. (2006). Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol. 4:e226. doi: 10.1371/journal.pbio.0040226
Bulgarelli, D., Schlaeppi, K., Spaepen, S., Ver, L. T. E., and Schulze-Lefert, P. (2013). Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 64, 807–838. doi: 10.1146/annurev-arplant-050312-120106
Cabral, L., Soares, C. R. F. S., Giachini, A. J., and Siqueira, J. O. (2015). Arbuscular mycorrhizal fungi in phytoremediation of contaminated areas by trace elements: mechanisms and major benefits of their applications. World J. Microb. Biot. 31, 1655–1664. doi: 10.1007/s11274-015-1918-y
Chen, Y., Chen, P., and Reyes, B. G. D. L. (2006). Differential responses of the cultivated and wild species of soybean to dehydration stress. Crop Sci. 46, 2041–2046. doi: 10.2135/cropsci2005.12.0466
Chen, Z. C., and Liao, H. (2016). Organic acid anions: an effective defensive weapon for plants against aluminum toxicity and phosphorus deficiency in acidic soils. J. Genet. Genomics 43, 631–638. doi: 10.1016/j.jgg.2016.11.003
Clarkson, D. T., and Marschner, H. (1995). Mineral nutrition of higher plants. second edition. 889pp. London: academic Press, ε29.95 (paperback). Ann. Bot. 78, 527–528. doi: 10.1006/anbo.1996.0155
Cole, J., Wang, Q., Cardenas, E., Fish, J., Chai, B., Farris, R. J., et al. (2009). The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37, D141–D145.
Davis-Belmar, C., Pinto-Figueroa, E., Demergasso, C., and Rautenbach, G. (2013). Proteo and actinobacteria diversity at a sulfide, salt and acid-rich lake in the north of chile. Adv. Mater. 825, 37–41. doi: 10.4028/www.scientific.net/amr.825.37
Dong, Y. S., Zhuang, B. C., Zhao, L. M., Sun, H., and He, M. Y. (2001). The genetic diversity of annual wild soybeans grown in China. Theor. Appl. Genet. 103, 98–103. doi: 10.1007/s001220000522
Edgar, R. C. (2013). Uparse: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Edwards, J., Johnson, C., Santos-Medellín, C., Lurie, E., Podishetty, N. K., Bhatnagar, S., et al. (2015). Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. U.S.A. 112:E911.
Fan, K., Weisenhorn, P., Gilbert, J. A., Shi, Y., Bai, Y., and Chu, H. (2018). Soil pH correlates with the co-occurrence and assemblage process of diazotrophic communities in rhizosphere and bulk soils of wheat fields. Soil Biol. Biochem. 121, 185–192. doi: 10.1016/j.soilbio.2018.03.017
Fierer, N., and Jackson, B. R. (2006). The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. U.S.A. 103, 626–631. doi: 10.1073/pnas.0507535103
Germida, J. J., and Siciliano, S. (2001). Taxonomic diversity of bacteria associated with the roots of modern, recent and ancient wheat cultivars. Biol. Fert. Soils 33, 410–415. doi: 10.1007/s003740100343
Guo, H., Nasir, M., Lv, J., Dai, Y., and Gao, J. (2017). Understanding the variation of microbial community in heavy metals contaminated soil using high throughput sequencing. Ecotox. Environ. Saf. 144, 300–306. doi: 10.1016/j.ecoenv.2017.06.048
Hu, L., Robert, C. A. M., Cadot, S., Zhang, X., Ye, M., Li, B., et al. (2018). Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the Rhizosphere microbiota. Nat. Commun. 9:2738.
Hymowitz, T. (1970). On the domestication of the soybean. Econ. Bot. 24, 408–421. doi: 10.1007/bf02860745
Jiang, Y., Li, S., Li, R., Zhang, J., Liu, Y., Lv, L., et al. (2017a). Plant cultivars imprint the rhizosphere bacterial community composition and association networks. Soil Biol. Biochem. 109, 145–155. doi: 10.1016/j.soilbio.2017.02.010
Jiang, Y., Liu, M., Zhang, J., Chen, Y., Chen, X., Chen, L., et al. (2017b). Nematode grazing promotes bacterial community dynamics in soil at the aggregate level. ISME J. 11:2705. doi: 10.1038/ismej.2017.120
Jiao, S., Yang, Y., Xu, Y., Zhang, J., and Lu, Y. (2019). Balance between community assembly processes mediates species coexistence in agricultural soil microbiomes across eastern China. ISME J. 14, 202–216. doi: 10.1038/s41396-019-0522-9
Jones, D. L., and Willett, V. B. (2006). Experimental evaluation of methods to quantify dissolved organic nitrogen (DON) and dissolved organic carbon (DOC) in soil. Soil Biol. Biochem. 38, 991–999. doi: 10.1016/j.soilbio.2005.08.012
Kochian, L. V., Piñeros, M. A., and Hoekenga, O. A. (2005). The physiology, genetics and molecular biology of plant aluminium resistance and toxicity. Plant Soil 274, 175–195. doi: 10.1007/1-4020-4099-7_9
Kwak, M. J., Kong, H. G., Choi, K., Kwon, S. K., Song, J. Y., Lee, J., et al. (2018). Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat. Biotechnol. 36, 1117–1117. doi: 10.1038/nbt1118-1117
Li, N., Meng, H., Xing, H., Liang, L., Zhao, X., Luo, K., et al. (2017). Genome-wide analysis of MATE transporters and molecular characterization of aluminium resistance in populus. J. Exp. Bot. 68, 5669–5683. doi: 10.1093/jxb/erx370
Li, X., Rui, J., Xiong, J., Li, J., He, Z., Zhou, J., et al. (2014). Functional potential of soil microbial communities in the maize rhizosphere. PLoS One 9:e112609. doi: 10.1371/journal.pbio.112609
Lian, T., Jin, J., Wang, G., Tang, C., Yu, Z., Li, Y., et al. (2017). The fate of soybean residue-carbon links to changes of bacterial community composition in mollisols differing in soil organic carbon. Soil Biol. Biochem. 109, 50–58. doi: 10.1016/j.soilbio.2017.01.026
Lian, T., Ma, Q. B., Shi, Q. H., Cai, Z. D., Zhang, Y. F., Cheng, Y. B., et al. (2019). High aluminium stress drives different rhizosphere soil enzyme activities and bacterial community structure between aluminium-tolerant and aluminium-sensitive soybean genotypes. Plant Soil 440, 409–425. doi: 10.1007/s11104-019-04089-8
Lian, T., Wang, G., Yu, Z., Li, Y., Liu, X., and Jin, J. (2016). Carbon input from 13C-labelled soybean residues in particulate organic carbon fractions in a mollisol. Biol. Fert. Soils 52, 331–339. doi: 10.1007/s00374-015-1080-6
Liang, C., Pineros, M. A., Tian, J., Yao, Z., Sun, L., Liu, J., et al. (2013). Low pH, aluminum, and phosphorus coordinately regulate malate exudation through gmalmt1 to improve soybean adaptation to acid soils. Plant Physiol. 161, 1347–1361. doi: 10.1104/pp.112.208934
Liang, Y., Yang, C., and Shi, H. (2001). Effects of silicon on growth and mineral composition of barley grown under toxic levels of aluminium. J. Plant Nutr. 24, 229–243. doi: 10.1081/pln-100001384
Liu, J., Yang, L., Wei, W., Gai, J., and Yan, L. (2016). Genome-wide analysis of MATE transporters and expression patterns of a subgroup of MATE genes in response to aluminum toxicity in soybean. BMC Genomics 17:223. doi: 10.1186/s12864-016-2559-8
Luo, X., Bai, X., Sun, X., Zhu, D., Liu, B., Ji, W., et al. (2013). Expression of wild soybean WRKY20 in Arabidopsis enhances drought tolerance and regulates ABA signalling. J. Exp. Bot. 64, 2155–2169. doi: 10.1093/jxb/ert073
Ma, B., Wang, H., Dsouza, M., Lou, J., He, Y., Dai, Z., et al. (2016). Geographic patterns of co-occurrence network topological features for soil microbiota at continental scale in eastern China. ISME J. 10:1891. doi: 10.1038/ismej.2015.261
Ma, J. F., Ryan, P. R., and Delhaize, E. (2001). Aluminium tolerance in plants and the complexing role of organic acids. Trends Plant Sci. 6, 273–278. doi: 10.1016/s1360-1385(01)01961-6
Ma, Q., Yi, R., Li, L., Liang, Z., Zeng, T., Zhang, Y., et al. (2018). GsMATE encoding a multidrug and toxic compound extrusion transporter enhances aluminium tolerance in Arabidopsis thaliana. BMC Plant Biol. 18:212. doi: 10.1186/s12870-018-1397-z
Ma, Q. F., Rengel, Z., and Kuo, J. (2002). Aluminium toxicity in rye (Secale cereale): root growth and dynamics of cytoplasmic Ca2+ in intact root tips. Ann. Bot. 89, 241–244. doi: 10.1093/aob/mcf017
Maron, L. G., Piñeros, M. A., Guimarães, C. T., Magalhaes, J. V., Pleiman, J. K., Mao, C., et al. (2010). Two functionally distinct members of the MATE (multi-drug and toxic compound extrusion) family of transporters potentially underlie two major aluminium tolerance QTLs in maize. Plant J. 61, 728–740. doi: 10.1111/j.1365-313x.2009.04103.x
Matsumoto, H., and Yamaya, T. (1986). Inhibition of potassium uptake and regulation of membrane-associated Mg2+-ATPase activity of pea roots by aluminium. Soil Sci. Plant Nutr. 32, 179–188. doi: 10.1080/00380768.1986.10557495
Miranda, K. M., Espey, M. G., and Wink, D. A. (2001). A rapid, simple spectrophotometric method for simultaneous detection of nitrate and nitrite. Nitric Oxide Biol. 5, 62–71. doi: 10.1006/niox.2000.0319
Miyasaka, S., Buta, J. G., Howell, R. K., and Foy, C. D. (1991). Mechanism of aluminium tolerance in snapbeans: root exudation of citric acid. Plant Physiol. 96:737. doi: 10.1104/pp.96.3.737
Montalban, B., Thijs, S., Lobo, M. C., Weyens, N., Ameloot, M., Vangronsveld, J., et al. (2017). Cultivar and metalspecific effects of endophytic bacteria in Helianthus tuberosus exposed to Cd and Zn. Int. J. Mol. Sci. 18, 2026. doi: 10.3390/ijms18102026
Mora, M. D. L. L., Demanet, R., Acuña, J. J., Viscardi, S., Jorquera, M., Rengel, Z., et al. (2017). Aluminium-tolerant bacteria improve the plant growth and phosphorus content in ryegrass grown in a volcanic soil amended with cattle dung manure. Appl. Soil Ecol. 115, 19–26. doi: 10.1016/j.apsoil.2017.03.013
Osburn, M., Sessions, A., Pepe-Ranney, C., and Spear, J. (2011). Hydrogen-isotopic variability in fatty acids from Yellowstone National park hot spring microbial communities. Geochim. Cosmochim. Acc. 75, 4830–4845. doi: 10.1016/j.gca.2011.05.038
Parks, D. H., Tyson, G. W., Philip, H., and Beiko, R. G. (2014). Stamp: statistical analysis of taxonomic and functional profiles. Bioinformatics 30, 3123–3124. doi: 10.1093/bioinformatics/btu494
Parniske, M. (2008). Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat. Rev. Microbiol. 6, 763–775. doi: 10.1038/nrmicro1987
Peiffer, J., Spor, A., Koren, O., Jin, Z., Tringe, S., Dangl, J. L., et al. (2013). Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. U.S.A. 110, 6548–6553. doi: 10.1073/pnas.1302837110
Pellet, D. M., Grunes, D. L., and Kochian, L. (1995). Organic acid exudation as an aluminium-tolerance mechanism in maize (Zea mays L.). Planta 196, 788–795. doi: 10.1007/bf01106775
Phillips, D., Fox, T. C., King, M., Bhuvaneswari, T. V., and Teuber, L. R. (2004). Microbial products trigger amino acid exudation from plant roots. Plant Physiol. 136, 2887–2894. doi: 10.1104/pp.104.044222
R Development Core Team (2006). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for statistical computing.
Rengel, Z., and Elliott, D. C. (1992). Mechanism of aluminium inhibition of net ca uptake by amaranthus protoplasts. Plant Physiol. 98, 632–638. doi: 10.1104/pp.98.2.632
Rodriguez, P. A., Rothballer, M., Chowdhury, S. P., Nussbaumer, T., Gutjahr, C., and Falter-Braun, P. (2019). Systems biology of plant-microbiome interactions. Mol. Plant 012, 804–821. doi: 10.1016/j.molp.2019.05.006
Rousk, J., Bååth, E., Brookes, P., Lauber, C. L., Lozupone, C., Caporaso, J. G., et al. (2010). Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 4, 1340–1351. doi: 10.1038/ismej.2010.58
Ryan, P., Delhaize, E., and Randall, P. (1995). Malate efflux from root apices and tolerance to aluminium are highly correlated in wheat. Aust. J. Plant Physiol. 22, 531–536.
Saavedra, S., Stouffer, D. B., Uzzi, B., and Bascompte, J. (2011). Strong contributors to network persistence are the most vulnerable to extinction. Nature 478, 233–235. doi: 10.1038/nature10433
Shenton, M., Iwamoto, C., Kurata, N., and Ikeo, K. (2016). Effect of wild and cultivated rice genotypes on rhizosphere bacterial community composition. Rice 9:42.
Shi, J., Yuan, X. F., Lin, H. R., Yang, Y. Q., and Li, Z. Y. (2011). Differences in soil properties and bacterial communities between the rhizosphere and bulk soil and among different production areas of the medicinal plant Fritillaria thunbergii. Int. J. Mol. Sci. 12, 3770–3785. doi: 10.3390/ijms12063770
Shi, Q., Liu, Y., Shi, A., Cai, Z., Nian, H., and Lian, T. (2020). Rhizosphere soil fungal communities of aluminum-tolerant and -sensitive soybean genotypes respond differently to aluminum stress in an acid soil. Front. Microbiol. 11:1177. doi: 10.3389/fmicb.2014.01177
Shi, S., Nuccio, E., Herman, D. J., Rijkers, R., Estera, K., Li, J., et al. (2015). Successional trajectories of rhizosphere bacterial communities over consecutive seasons. mBio 6:e00746-15. doi: 10.1128/mBio.00746-15
Singh, R., and Hymowitz, T. (2011). Soybean genetic resources and crop improvement. Genome 42, 605–616.
Sun, R., Zhang, X. X., Guo, X., Wang, D., and Chu, H. (2015). Bacterial diversity in soils subjected to long-term chemical fertilization can be more stably maintained with the addition of livestock manure than wheat straw. Soil Biol. Biochem. 88, 9–18. doi: 10.1016/j.soilbio.2015.05.007
Teplitski, M., Robinson, J. B., and Bauer, W. D. (2000). Plants secrete substances that mimic bacterial n-acyl homoserine lactone signal activities and affect population density-dependent behaviors in associated bacteria. Mol. Plant Microb. Int. 13, 637–648. doi: 10.1094/mpmi.2000.13.6.637
Tovkach, A., Ryan, P., Richardson, A., Lewis, D. C., Rathjen, T., Ramesh, S., et al. (2012). Transposon-mediated alteration of TaMATE1B expression in wheat confers constitutive citrate efflux from root Apices. Plant Physiol. 161, 880–892. doi: 10.1104/pp.112.207142
Verma, S., Subehia, S., and Sharma, S. P. (2005). Phosphorus fractions in an acid soil continuously fertilized with mineral and organic fertilizers. Biol. Fert. Soils 41, 295–300. doi: 10.1007/s00374-004-0810-y
von Uexküll, H. R., and Mutert, E. (1995). Global extent, development and economic impact of acid soils. Plant Soil 171, 1–15. doi: 10.1007/bf00009558
Wang, C., Zhao, X. Q., Chen, R. F., Chu, H. Y., and Shen, R. F. (2013). Aluminium tolerance of wheat does not induce changes in dominant bacterial community composition or abundance in an acidic soil. Plant Soil 367, 275–284. doi: 10.1007/s11104-012-1473-3
West, E. R., Cother, E. J., Steel, C. C., and Ash, G. J. (2010). The characterization and diversity of bacterial endophytes of grapevine. Revue Can. Microbiol. 56, 209–216. doi: 10.1139/w10-004
Yabe, S., Sakai, Y., Abe, K., and Yokota, A. (2017). Diversity of Ktedonobacteria with actinomycetes-like morphology in terrestrial environments. Microb. Environ. 32, 61–70. doi: 10.1264/jsme2.me16144
Yang, T., Liu, G., Li, Y., Zhu, S., Zou, A., Qi, J., et al. (2012). Rhizosphere microbial communities and organic acids secreted by aluminium-tolerant and aluminium-sensitive soybean in acid soil. Biol. Fert. Soils 48, 97–108. doi: 10.1007/s00374-011-0608-7
Yang, Z. M., Sivaguru, M., Horst, W., and Matsumoto, H. (2001). Aluminium tolerance is achieved by exudation of citric acid from roots of soybean (Glycine max). Physiol. Plant. 110, 72–77. doi: 10.1034/j.1399-3054.2000.110110.x
Yao, M., Rui, J., Li, J., Dai, Y., Bai, Y., Hedìnec, P., et al. (2014). Rate-specific responses of prokaryotic diversity and structure to nitrogen deposition in the Leymus chinensis steppe. Soil Biol. Biochem. 79, 81–90. doi: 10.1016/j.soilbio.2014.09.009
Zelinová, V., Huttová, J., Mistrík, I., Palove-Balang, P., and Tamás, L. (2009). Impact of aluminium on phosphate uptake and acid phosphatase activity in root tips of Lotus japonicus. J. Plant Nutr. 32, 1633–1641. doi: 10.1080/01904160903150891
Zhao, X. Q., Chen, R. F., and Shen, R. F. (2014). Coadaptation of plants to multiple stresses in acidic soils. Soil Sci. 179, 503–513. doi: 10.1097/ss.0000000000000086
Zhao, X. Q., and Shen, R. F. (2018). Aluminium-nitrogen interactions in the soil-plant system. Front. Plant Sci. 9:807. doi: 10.3389/fmicb.2014.00807
Zhao, X. Q., Shen, R. F., and Sun, Q. B. (2009). Ammonium under solution culture alleviates aluminium toxicity in rice and reduces aluminium accumulation in roots compared with nitrate. Plant Soil 315, 107–121. doi: 10.1007/s11104-008-9736-8
Zhou, J., Deng, Y., Luo, F., He, Z., Tu, Q., and Zhi, X. (2010). Functional molecular ecological networks. mBio 1, 1592–1601.
Keywords: aluminum toxicity, wild soybean, 16S rRNA high-throughput sequencing, bacterial communities, co-occurrence network
Citation: Shi Q, Jin J, Liu Y, Zhang Y, Cai Z, Ma Q, Cheng Y, Wen R, Nian H and Lian T (2020) High Aluminum Drives Different Rhizobacterial Communities Between Aluminum-Tolerant and Aluminum-Sensitive Wild Soybean. Front. Microbiol. 11:1996. doi: 10.3389/fmicb.2020.01996
Received: 21 October 2019; Accepted: 28 July 2020;
Published: 19 August 2020.
Edited by:
Michel Chalot, Université Bourgogne Franche-Comté, FranceCopyright © 2020 Shi, Jin, Liu, Zhang, Cai, Ma, Cheng, Wen, Nian and Lian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hai Nian, aG5pYW5Ac2NhdS5lZHUuY24=; Tengxiang Lian, bGlhbnR4QHNjYXUuZWR1LmNu
 Qihan Shi1,2
Qihan Shi1,2 Qibin Ma
Qibin Ma Yanbo Cheng
Yanbo Cheng Ronghui Wen
Ronghui Wen Hai Nian
Hai Nian Tengxiang Lian
Tengxiang Lian