
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 20 August 2020
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.01967
 Shun-Chung Hsueh1
Shun-Chung Hsueh1 Chih-Cheng Lai2
Chih-Cheng Lai2 Yu-Tsung Huang3,4
Yu-Tsung Huang3,4 Chun-Hsing Liao3,5
Chun-Hsing Liao3,5 Ming-Tang Chiou6,7
Ming-Tang Chiou6,7 Chao-Nan Lin6,7*
Chao-Nan Lin6,7* Po-Ren Hsueh4,8*
Po-Ren Hsueh4,8*From January 2013 to December 2018, 90 Escherichia coli isolates were collected from 90 sick pigs on 58 farms in seven cities in Taiwan. The minimum inhibitory concentrations (MICs) of the isolates to 14 antimicrobial agents were determined on the VITEK 2 system (bioMérieux, Marcy-l’Etoile, France), and the resistance to colistin was assessed via the reference broth microdilution (BMD) method. The mobilized colistin resistance genes (mcr) were determined for the colistin-resistant isolates, which displayed BMD MICs ≥ 4 mg/L. Genotypes of the mcr-positive E. coli isolates were determined by multilocus sequence typing, arbitrarily primed polymerase chain reaction (PCR), and pulsed-field gel electrophoresis. All of the isolates were tested for susceptibility to carbapenems. Fifty isolates (55.6%) were resistant to colistin, 39 of which (78%) were positive for the mcr-1 gene. E. coli isolates harboring mcr-1 were most frequent in 2017 (15/18, 83.3%), followed by 2018 (13/23, 56.5%), 2015 (7/21, 33.3%), and 2016 (3/24, 12.5%). A total of 18 sequence types (STs) were identified among the 39 porcine mcr-1-carrying E. coli isolates; 13 were ST2521 (33.3%) isolated in 2017 and 2018. Five genotypes (clones) were identified, and the same genotypes were in sick pigs on the same farm and different farms. This suggests intra- and inter-farm spread of porcine mcr-1-carrying E. coli. The results presented here indicate high colistin resistance and wide mcr-1 E. coli prevalence among the sick pigs sampled in 2015–2018 in different regions of Taiwan.
The plasmid-borne mobilized colistin resistance-1 gene (mcr-1) confers resistance to polymyxin E (colistin) in Escherichia coli (Liu et al., 2016) and has been reported on nearly every continent (Nordmann et al., 2016; Meinersmann et al., 2017; Lai et al., 2018; Rebelo et al., 2018). Enterobacteriaceae isolates positive for mcr-1 have been recovered from humans and animals, including chickens, turkeys, swine, and pets (Brennan et al., 2016; Kempf et al., 2016; Kuo et al., 2016; Khan et al., 2017; Pulss et al., 2017; Lai et al., 2018). The gene is readily transferable, which limits the treatment options, especially if it resides in multidrug-resistant organisms (Lai et al., 2017; Pulss et al., 2017; Lin et al., 2018; Palmeira et al., 2018). Previous studies have shown that food animals are common sources of human infections due to colistin-resistance in these organisms (Kuo et al., 2016; Chiou et al., 2017). Moreover, the horizontal transmission of a colistin-resistant E. coli from a domesticated pig to a boy may have occurred (the boy fed the pig without protective equipment) (Olaitan et al., 2015).
The use of colistin in animal husbandry in Taiwan has been restricted since July 2007, but mcr-1-harboring E. coli isolates are still a concern. Liu et al. (2018) documented high rates of mcr-1-harboring E. coli (mainly enterohemolytic E. coli) in sick pigs from southern Taiwan. A high rate (46.2%) of colistin resistance among zoonotic Salmonella spp. has also been reported (Chiou et al., 2017). The mcr-1 gene has been identified in humans and retail meats (Kuo et al., 2016; Lai et al., 2018). Kuo et al. (2016) described 18 colistin-resistant E. coli isolates positive for mcr-1 in ground beef, chicken, and pork. The prevalence of mcr-1 in meat-associated E. coli isolates has increased in Taiwan, with rates of 1.1, 6.6, and 8.7% in 2012, 2013, and 2015, respectively (Kuo et al., 2016).
To our knowledge, no longitudinal studies have examined the prevalence of colistin-resistant E. coli in sick pigs. This study investigated the annual prevalence of colistin resistance and mcr-harboring E. coli in sick pigs from different farms in Taiwan.
A total of 90 E. coli isolates were obtained from 90 sick pigs from 58 farms in different geographical regions of Taiwan between January 2013 and December 2018. A total of 88 isolates were collected from seven counties in southern (n = 4), northern (n = 1), middle (n = 1), and eastern Taiwan (two isolates were from unknown sources; Table 1). Most of the isolates were collected between 2015 and 2018 (n = 84, 93.3%), from southern Taiwan (n = 82, 91.1%). The age of the sick pigs was known for 79 isolates, 78 (98.7%) were <10 weeks old. Isolates from rectal swabs (n = 32) and mesenteric lymph nodes (n = 32) comprised 71.2% of the samples. The isolates were sent to the Animal Disease Diagnostic Center, College of Veterinary Medicine, National Pingtung University of Science and Technology, Pingtung, Taiwan, for isolation and identification of the enterohemolytic E. coli.

Table 1. Characteristics of the 90 E. coli isolates from 90 sick pigs sampled in 2013 to 2018 in Taiwan.
Escherichia coli isolates were identified as enterohemolytic strains by the presence of β-hemolysis on Trypticase soy agar supplemented with 5% sheep blood and by the presence of at least one virulent gene (STa, STb, LT, F18, or aidA) (Chapman et al., 2006; Lai et al., 2017, 2018).
The minimum inhibitory concentrations (MICs) of 14 antimicrobial agents, including colistin, were determined on the VITEK 2 system (bioMérieux, Marcy-l’Etoile, France) (Table 2) and were interpreted according to guidelines of the Clinical and Laboratory Standards Institute (CLSI). The MICs of colistin were determined via the reference broth microdilution (BMD) method (Clinical and Laboratory Standards Institute [CLSI], 2020). Susceptibility to colistin was defined as intermediate (MICs of ≤2 mg/L) or resistant (MICs ≥ 4 mg/L) as in the MIC interpretive criteria from CLSI.
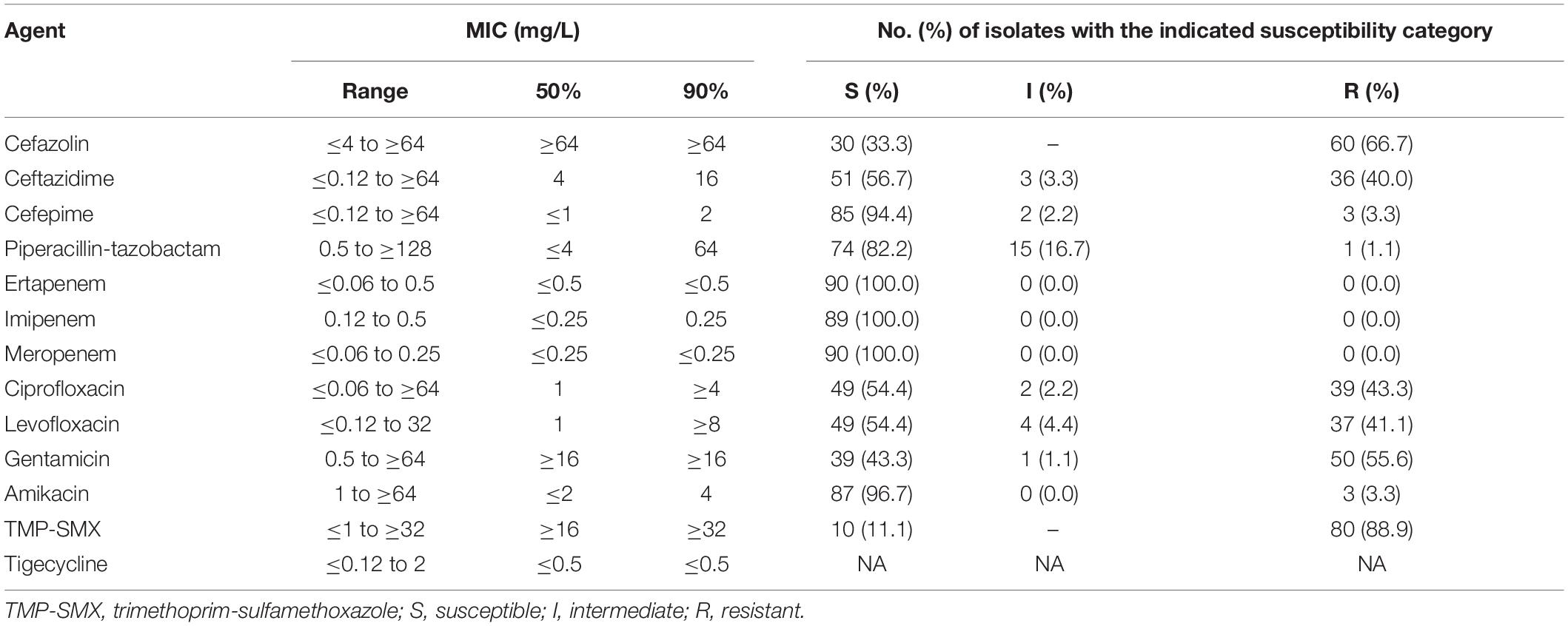
Table 2. Susceptibilities of the 90 E. coli isolates as determined by the VITEK 2 susceptibility method.
To examine the inter-test agreement between the methods for determining colistin susceptibility, the essential and categorical agreements and very major error (VME) were evaluated. The essential agreement between the BMD method and VITEK 2 was measured as the difference between MICs ≤ ± 1 log2 dilution, using the BMD method as the reference standard. Categorical agreement between the two methods was measured as the percentage of isolates with concordant test results (i.e., intermediate vs. resistant). A VME was defined as inconsistent results, e.g., a colistin-resistant isolate from the BMD method that was considered colistin-intermediate by VITEK 2 (Chen et al., 2014).
Polymerase chain reaction (PCR) amplification of the whole-cell DNA from colistin-resistant isolates (via the BMD method) was performed using previously described primers for mcr-1, mcr-2, mcr-3, mcr-4, and mcr-5 (Rebelo et al., 2018). The PCR products were sequenced.
The sequence types (STs) of the mcr-positive E. coli isolates were determined by multilocus sequencing typing (MLST) (Wirth et al., 2006). The genetic relationships of the mcr-positive E. coli isolates exhibiting the main STs (≥3 isolates) were further evaluated by arbitrarily primed PCR (AP-PCR) and pulsed-field gel electrophoresis (PFGE), as described previously (Hsueh et al., 2002). Randomly amplified polymorphic DNA (RAPD) patterns of the mcr-positive E. coli isolates were generated by AP-PCR using four primers: M13 (5’-TTATGTAAAACGACGGCCAGT-3’), ERIC1 (5’-GTGAATCCCCAGGAGCTTACAT-3’), ERIC2 (5’-AAGTAAGTGACTGGGGTGAGCG-3’), and 1254 (5’-CCGCAGCCAA-3’) (Operon Technologies, Inc., Alameda, CA, United States). Isolates with RAPD patterns containing one or more discrete bands were considered different.
The DNA extraction and purification for PFGE followed previous descriptions (Tenover et al., 1995; Hsueh et al., 2002). DNA was digested using the SmaI restriction enzyme, the restriction fragments were separated using a CHEF-DR III unit (Bio-Rad Laboratories, Hercules, CA, United States), and the pulsotypes were analyzed on the Bio-Rad CHEF-Mapper apparatus (Bio-Rad Laboratories). Cluster analysis was performed in BioNumerics version 5.0 (Applied Maths, Sint-Martens-Latem, Belgium) using the unweighted pair-group method with arithmetic averages. The Dice correlation coefficient was used to analyze similarities between the banding patterns (tolerance = 1%). Isolates with identical PFGE patterns were considered the same strain (same pulsotype) and isolates with PFGE patterns >80% similar were considered closely related. Isolates exhibiting identical ST, RAPD patterns, and pulsotypes, or closely related strains by PFGE, were considered identical strains (clones).
Among the 90 isolates, 45 (50.0%) were enterohemolytic E. coli. The rate of enterohemolytic E. coli among all of the isolates ranged from 0% in 2013–2014 to 66.7% (n = 12) in 2017 (Table 1).
The MICs of 13 antimicrobial agents against the 90 porcine E. coli isolates are shown in Table 2. The susceptibility to ceftazidime and cefepime was 56.7 and 94.4%, respectively, and the susceptibility to ciprofloxacin and levofloxacin were both 54.4%. Only 11.1% of the isolates were susceptible to trimethoprim-sulfamethoxazole. All of the isolates were susceptible to ertapenem, imipenem, and meropenem, and all were inhibited by tigecycline at 2 mg/L. The colistin-resistant isolates and those harboring mcr-1 displayed similar susceptibilities to ciprofloxacin, levofloxacin, and amikacin (Table 3). The colistin-intermediate isolates had higher susceptibility to gentamicin than the colistin-resistant or mcr-1 positive isolates (both p < 0.0001) (Table 3). The strains did not differ in their resistance to the other antibiotics.
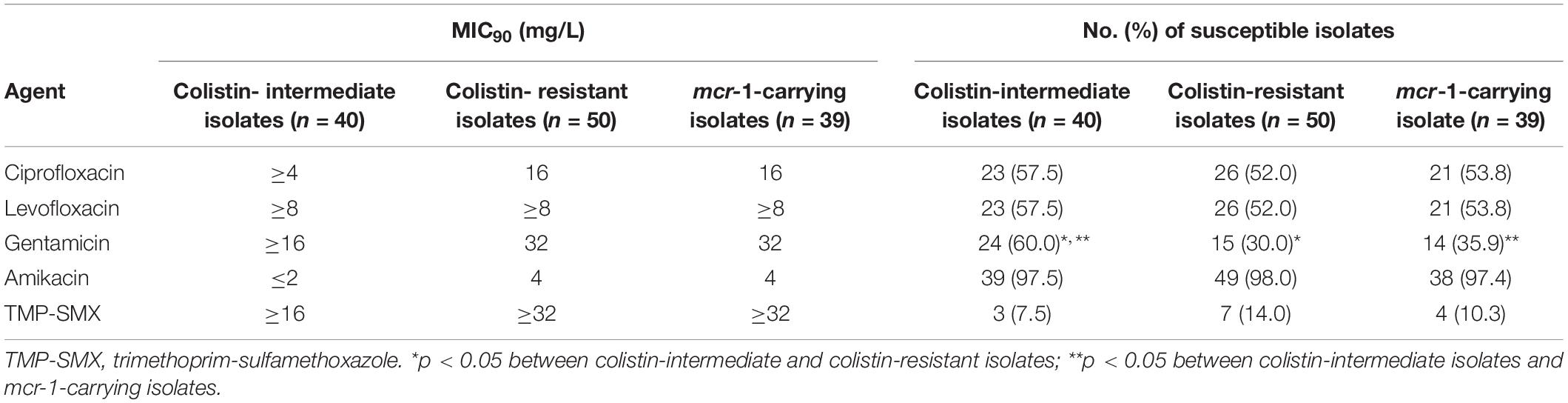
Table 3. Susceptibility of selected antimicrobial agents against the colistin-intermediate, -resistant, and mcr-1-carrying E. coli isolates from sick pigs.
The distribution of colistin MICs determined by the VITEK 2 susceptibility system and the reference BMD method are depicted in Figure 1. The respective rates of colistin-intermediate isolates were 54.1% (n = 48) and 44.4% (n = 40). The essential agreement of MICs was 97.8%, and the rate of agreement was 91.1% (Table 4). A VME occurred in 16.7% (8/48) of the VITEK 2 susceptibility determinations.
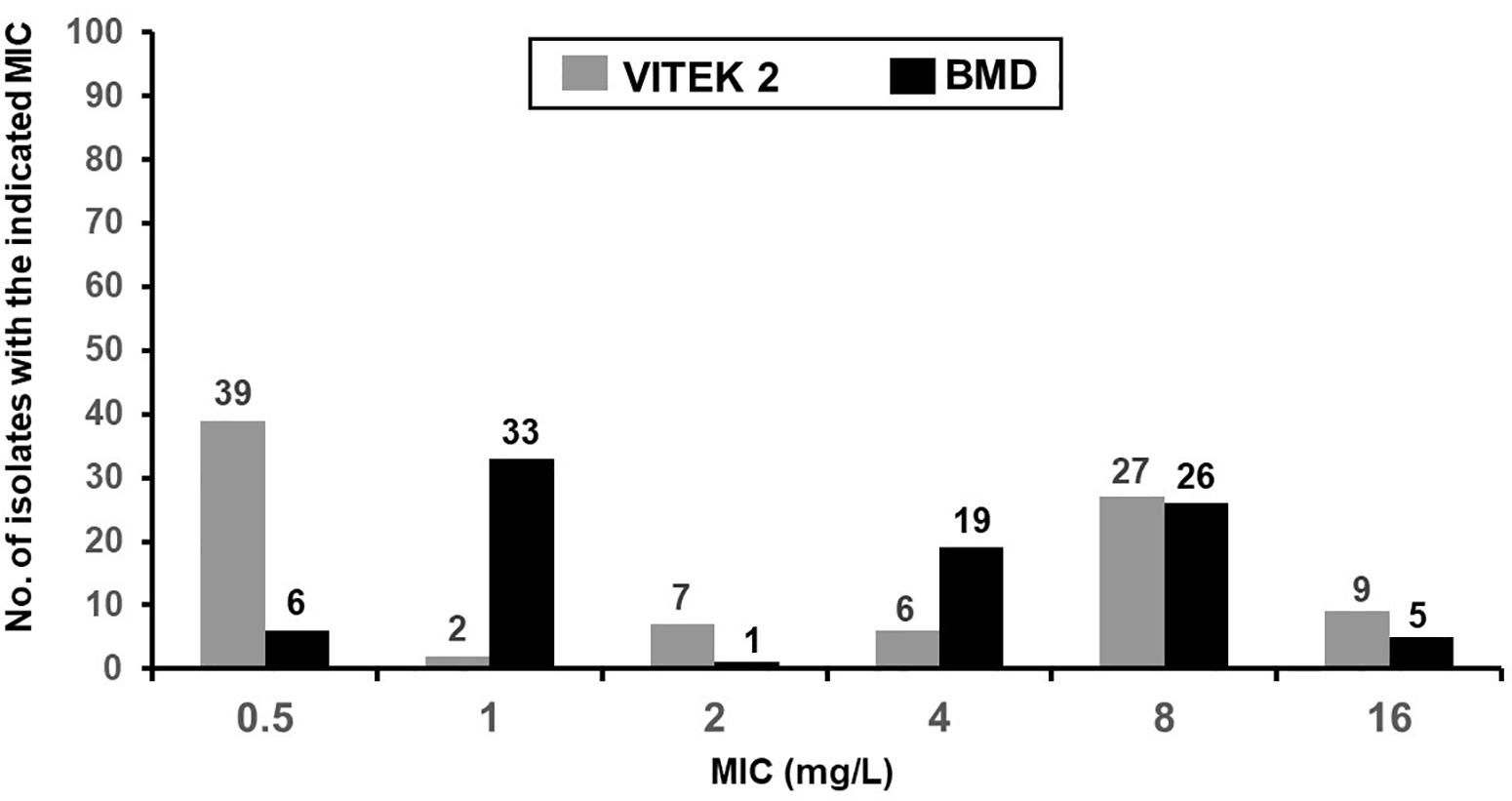
Figure 1. MIC distribution of colistin from the VITEK 2 and reference broth microdilution methods for 90 isolates of E. coli from sick pigs.
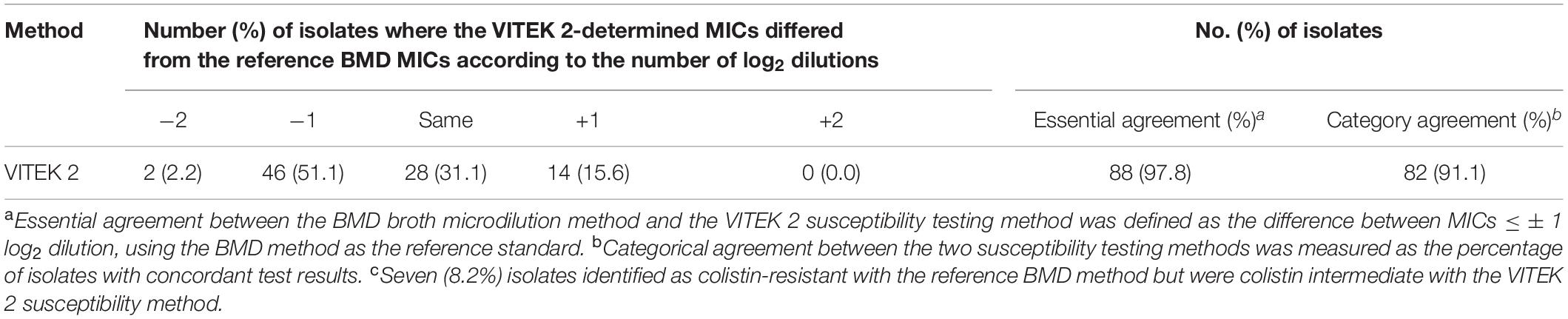
Table 4. Essential and categorical agreements of the colistin MICs from the VITEK 2 and reference broth microdilution (BMD) methods.
Among the 50 colistin-resistant E. coli isolates determined by the reference BMD method, 39 (78.0%) were positive for the mcr-1 gene. The mcr-2 to mcr-5 genes were not detected in any of the isolates. E. coli isolates harboring mcr-1 comprised 53.3% (24/45) of the enterohemolytic isolates and 33.3% (15/45) of the non-enterohemolytic isolates (p = 0.09). The rates of mcr-1 positive E. coli isolates rose each year and peaked in 2017 (83.3%) (Figure 2). The respective rates of colistin-resistant and mcr-1-positive colistin-resistant E. coli isolates were 52.8% (28/53) and 75% (21/28) in Pingtung county and 60% (9/15) and 88.9% (8/9) in Yulin county (Figure 3).
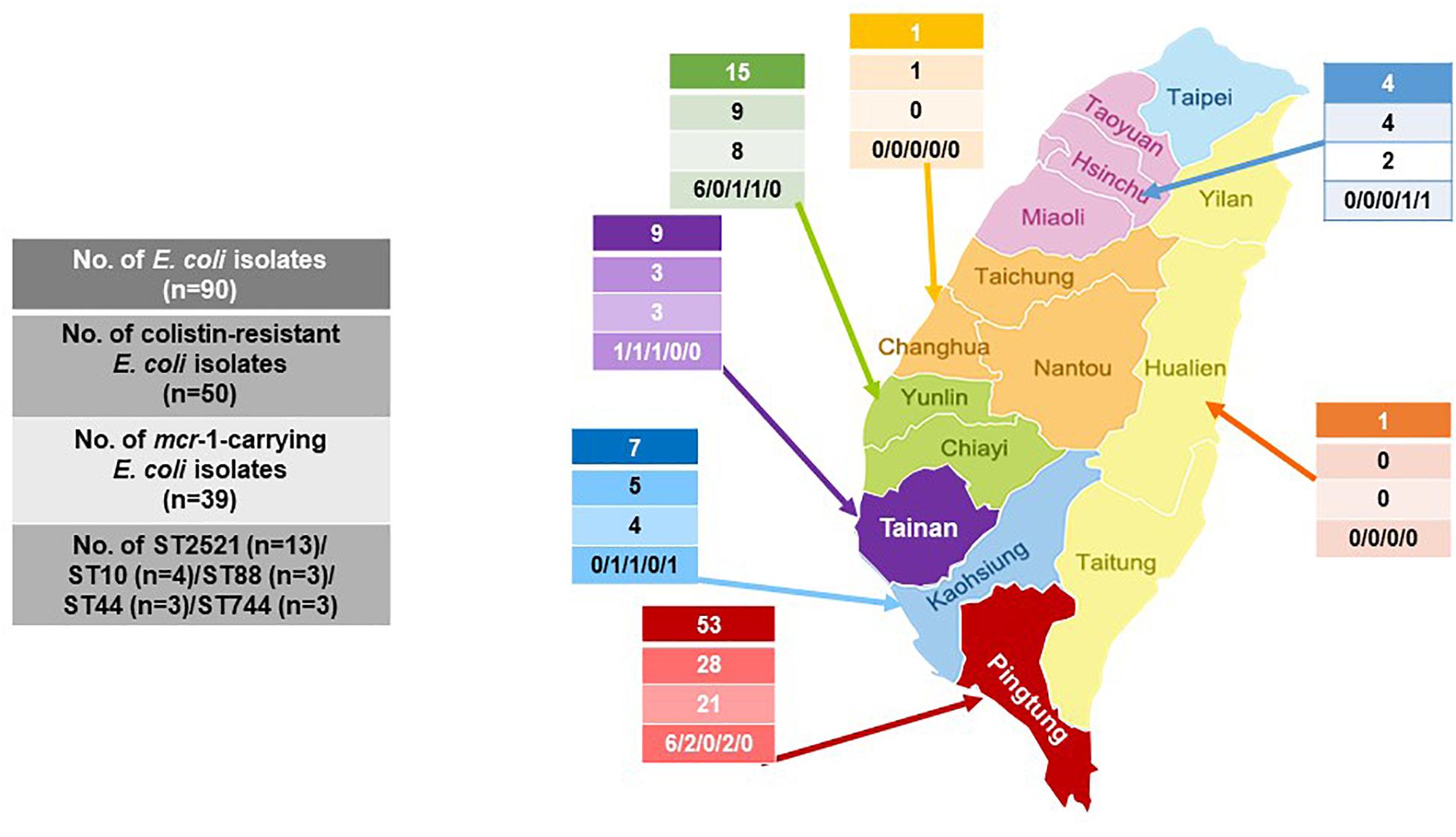
Figure 3. Geographical distribution of the colistin-resistant and mcr-1-carrying E. coli and the five sequence types (ST) of mcr-1 E. coli from 2013 to 2018.
Among the 18 STs identified from the 39 porcine E. coli isolates harboring mcr-1, 13 (33.3%) belonged to ST2521 (all from 2017 and 2018), 4 (10.3%) were ST10, and 3 (7.7%) each were ST88 (all from 2017), ST44, and ST744 (Table 5). AP-PCR and PFGE were performed for all 26 isolates; ST2521 (A1–A13), ST10 (B1–B4), ST88 (C1–C3), ST44 (D1–D3), and ST744 (E1–E3) (Table 6). The isolates were obtained from sick pigs on 18 different farms (identified as farms a to r) (Table 6). A total of 15 RAPD patterns were identified, including ST2521 (RA-1–RA-4), ST10 (RB-1–RB-4), ST88 (RC-1–RC-2), ST44 (RD-1 and RD-2), and ST744 (RE-1–RE-3). PFGE revealed 10 pulsotypes (P-I–P-X) with closely related isolates (Table 6). Five clones (C-1–C-5) were identified from the MLST results, RAPD patterns, and pulsotypes (Figure 4). Three isolates of clone C-2 were from farm g, three isolates of clone C-3 were from farm h, and two isolates of clone C-4 were from farm a. Clones C-1 and C-5 were from different farms.
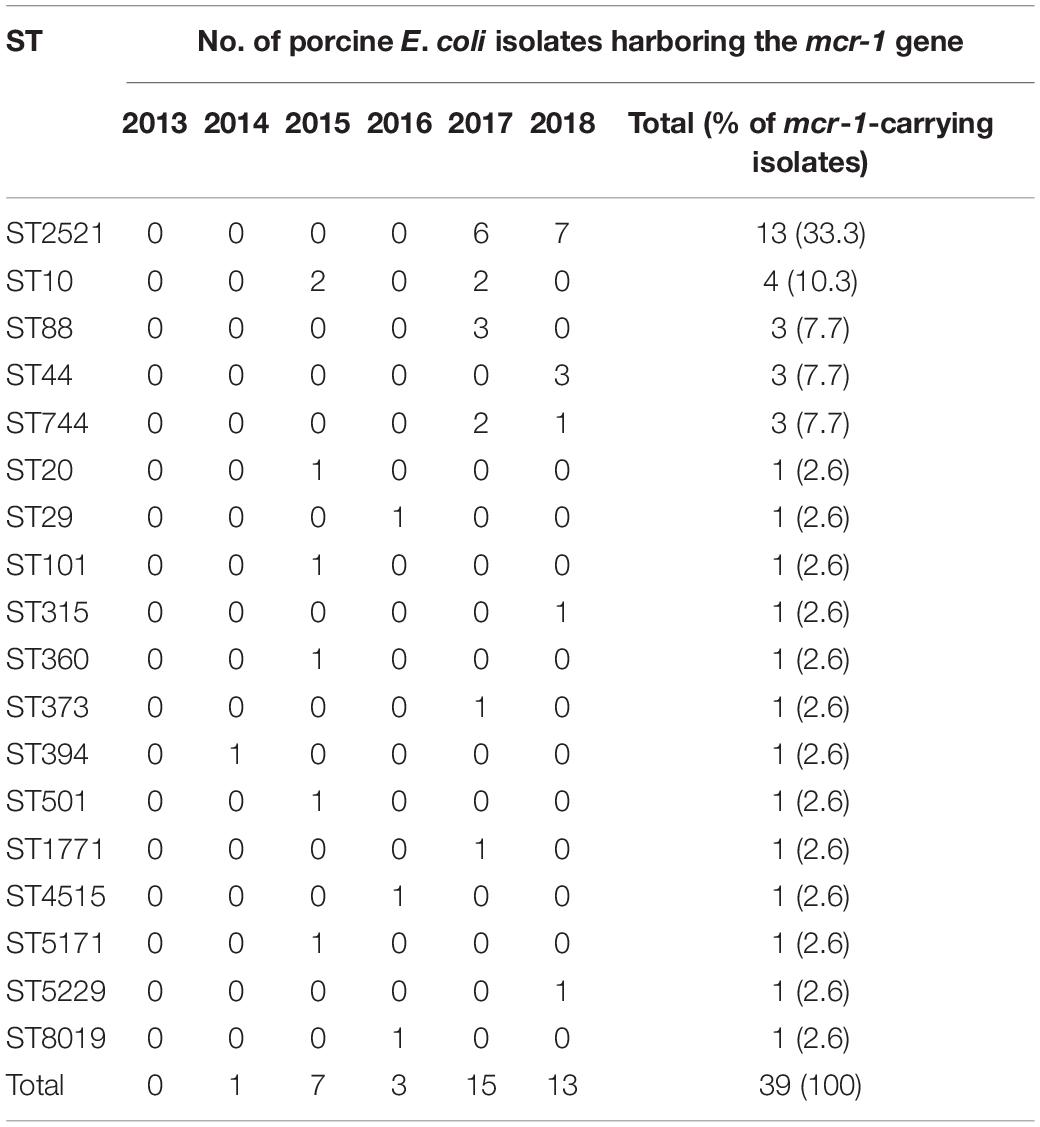
Table 5. Distribution of the sequence types (STs) from multilocus sequence typing of the 39 porcine E. coli isolates carrying the mcr-1 gene.
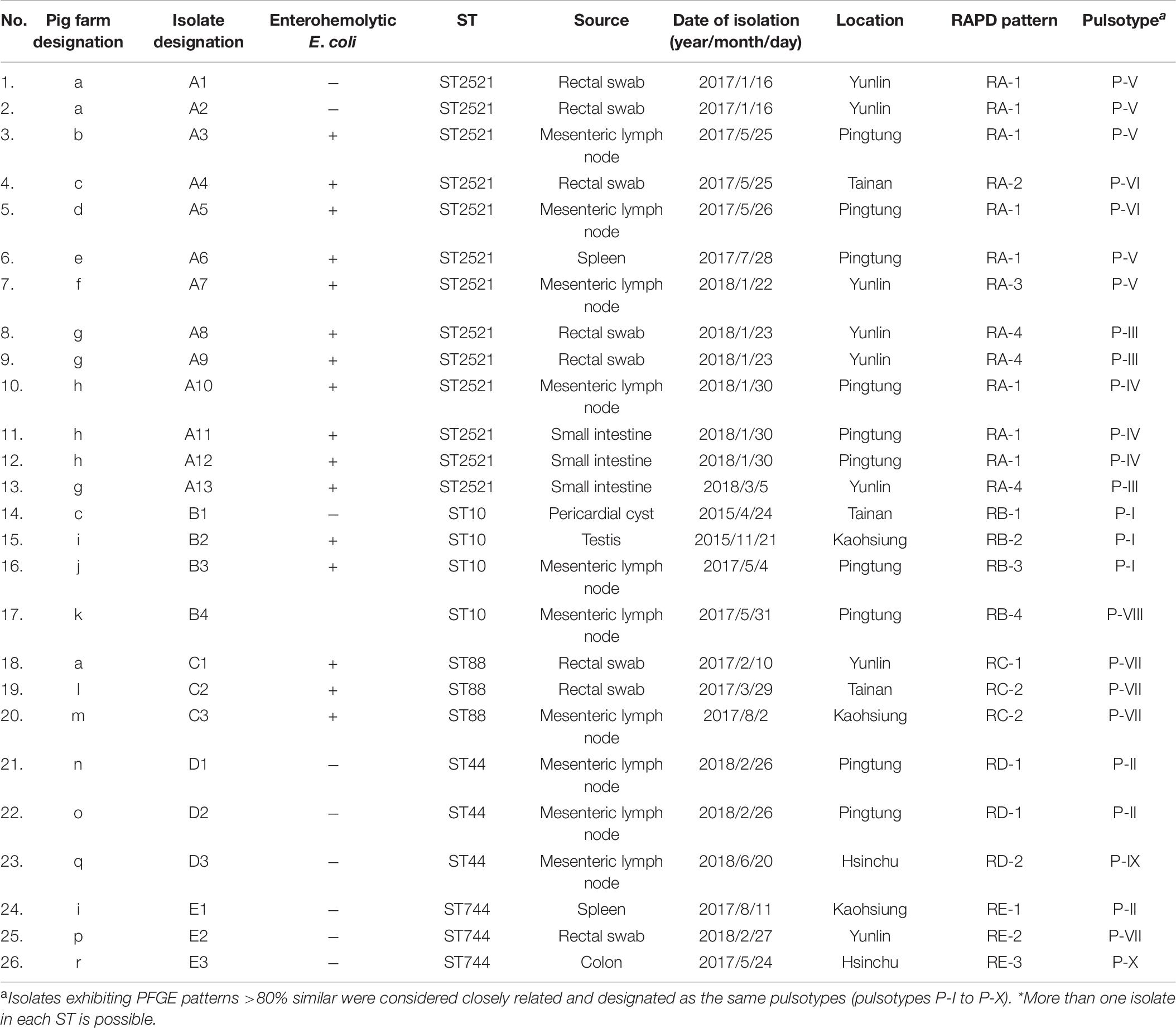
Table 6. Characteristics of the 26 porcine mcr-1-carrying E. coli isolates exhibiting five major sequence types (STs)*.
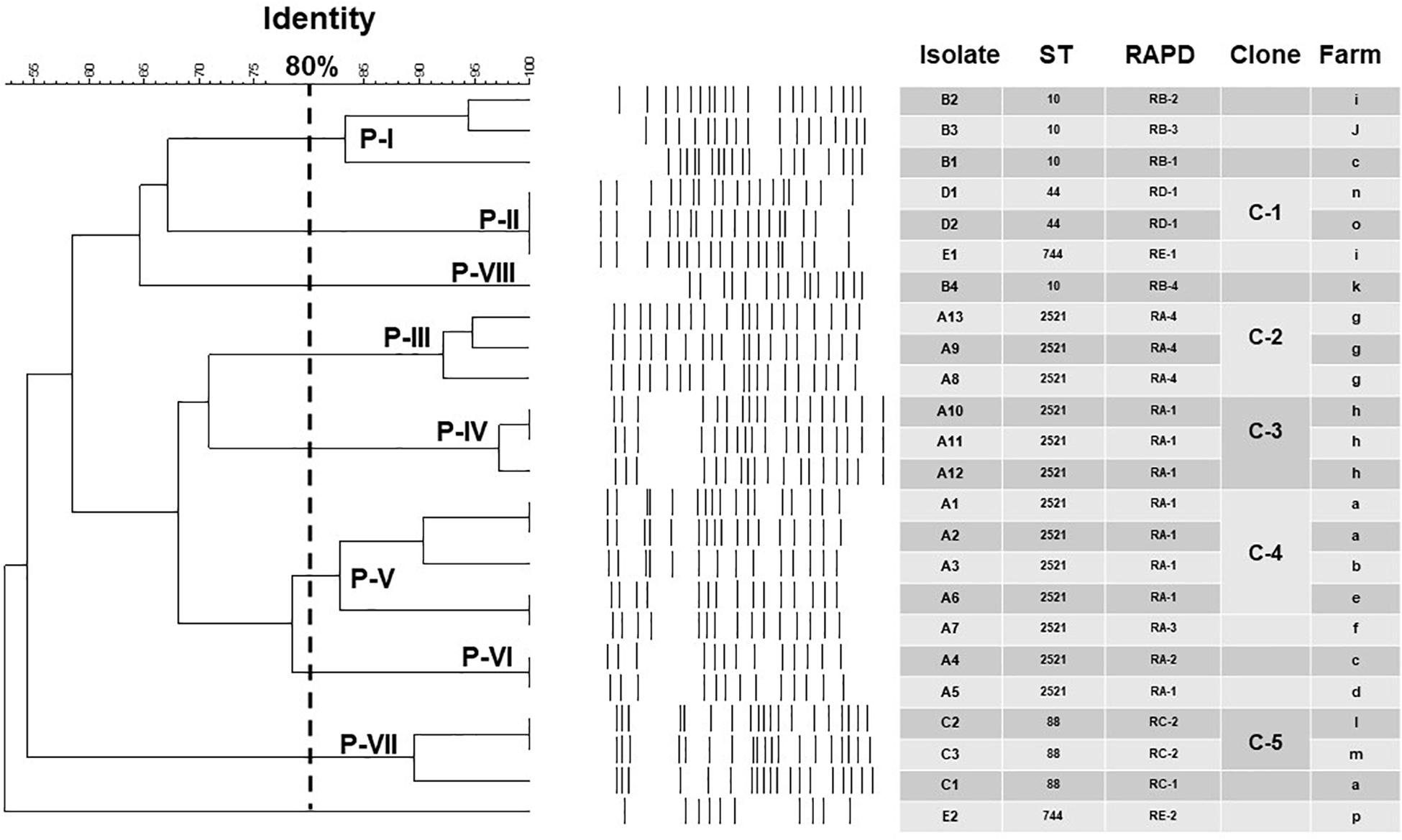
Figure 4. Epidemiological and molecular characteristics of the 24 porcine E. coli isolates harboring mcr-1 and displaying five major sequence types (STs) (more than one isolate in each ST). Not included are two porcine mcr-1-carrying E. coli isolates (D3 and E3; ST44 and ST744, respectively), different RAPD patterns (RD-2 and RE-3), and different pulsotypes (Clones P-IX and P-X) from the ST44 and ST744 isolates from northern Taiwan (Hsinchu) (see Table 6).
The prevalence of the mcr-1 gene in porcine E. coli isolates from different geographical regions of Taiwan may be unrelated to a specific clonal population. The strains may carry different plasmids encoding the mcr-1 gene. The MLST of 32 mcr-1-positive E. coli isolates from 18 retail meat samples and 14 human samples in Taiwan (2010–2015) revealed 18 distinct STs, including ST38 (n = 8), ST117 (n = 5), and two each of ST701, ST744, and ST428. The STs of the remaining isolates were distinct (Kuo et al., 2016). ST744 was also documented in this study, in addition to many other STs. For example, ST2521 was the most common ST, documented since 2017, but it had never been observed in pigs or humans (Kuo et al., 2016; Hadjadj et al., 2017; Kong et al., 2017; Lai et al., 2018; Wang et al., 2020). Our results suggest that further investigation is required to assess the clinical significance of ST2521 among mcr-1 positive E. coli isolates. Our findings also indicate that mcr-1-positive E. coli isolates from different geographical regions of Taiwan may have variable STs. The RAPD patterns showed no evidence of clonal dissemination of the mcr-1-positive isolates between humans and pigs. In contrast, we identified five clone isolates from the MLST results, RAPD patterns, and pulsotypes. One clone was from sick pigs on different farms and also sick pigs on the same farm. This may indicate intra- and inter-farm spread of porcine E. coli harboring mcr-1. Thus, active and regular screening of mcr-1-containing E. coli isolates from humans and animals is imperative.
A study from China reported 16 different STs among 64 blaNDM–5- and mcr-1-carrying E. coli isolates from a commercial swine farm (Kong et al., 2017). The five main STs were ST48 (n = 12), ST4463 (n = 8), ST54 (n = 7), ST410 (n = 6), and ST165 (n = 6). We did not observe any of these STs in our study. Another Chinese study described 40 distinct STs among 58 mcr-1-positive isolates, indicating considerable diversity among the mcr-1-positive isolates from different geographic locations (ST48 and ST10 are widespread) (Yang et al., 2017). In France, six STs (ST4015, ST3997, ST10, ST93, ST48, and ST648) were detected among 25 mcr-1-carrying E. coli isolates from humans, pigs, and chickens (Hadjadj et al., 2017). In the latter study, the ST4015, ST4704, and ST93 porcine mcr-1-carrying E. coli isolates were also different.
Animal-to-human transmission remains a serious concern. A study conducted in Laos reported two cases of colistin-resistant E. coli; one in a boy with no recent history of antibiotic usage and another in a pig that belonged to the boy’s family. Both isolates belonged to the same novel ST and displayed the same virulence and PFGE patterns (Olaitan et al., 2015). The boy normally fed the pig without protective equipment (e.g., boots). The presence of the same ST4015 mcr-1-carrying E. coli strain in the boy and pig indicates possible horizontal transmission of colistin-resistant E. coli. Another study described ST648 in two travelers and ST3997 in two villagers, both of which may represent inter-human transmission (Hadjadj et al., 2017). However, the clonality of the 39 E. coli isolates harboring the mcr-1 gene was diverse, and 18 different STs were detected (though some STs appeared more than once). Previously, we described six mcr-1-carrying E. coli isolates from patients with bacteremia; two were ST69, but the rest only occurred once (ST1196, ST361, ST1463, and ST1011) (Lai et al., 2018). The human mcr-1-positive E. coli isolates had different pulsotypes, and the STs were different from the porcine mcr-1-carrying E. coli isolates. Although we failed to detect mutual transmission between humans and pigs in Taiwan, we cannot exclude its occurrence. Further surveillance is necessary to identify potential transmissions of the mcr-1-carrying E. coli between animals and humans.
We observed a high VME (16.7%) in the VITEK 2 susceptibility testing of colistin compared to the BMD method. Our findings are contradictory to a previous study in human bacteremic E. coli isolates (no VME) and indicate that the VITEK 2 method is a low-sensitivity tool for identifying colistin resistance in porcine E. coli isolates (Lai et al., 2019). Gentamicin is frequently used to treat colibacillosis in pigs, especially in neonatal piglets (via intramuscular or oral administration). Of the 90 E. coli isolates, the resistance to gentamicin was 55.6% (50/90). High levels of gentamicin resistance were reported in E. coli isolates from sick pigs in several countries – 32.7% in the United States (Jiang et al., 2019), 46% in Belgium, 45% in Poland, 20% in Spain, and 77% in Korea (Lee et al., 2009; Luppi, 2017). Plasmid-mediated antimicrobial resistance genes are transmissible and cross-resistance between gentamicin and other aminoglycosides such as apramycin (a veterinary drug) has been described (Jensen et al., 2006). However, the prevalence of antimicrobial resistance genes in aminoglycosides requires further investigation.
This study has several limitations. First, a small number of porcine E. coli isolates were evaluated and all were from sick pigs. This might limit the power of intra- and inter-farm spreading analysis of porcine E. coli isolates harboring mcr-1. The inclusion of more E. coli isolates from healthy and sick pigs raised on different farms may better describe the prevalence of mcr-1-harboring E. coli in swine and porcine farms. Second, the mechanisms mediating colistin resistance among the mcr-1-negative colistin-intermediate E. coli isolates were not investigated. Third, we investigated mcr-1 to mcr-5, but at least nine mcr genes have been reported. The roles of mcr-6 to mcr-9 should be investigated.
In conclusion, the occurrence of mcr-1-positive E. coli isolates in sick pigs has continuously increased in Taiwan. Regular screening for the mcr-1 gene in E. coli in sick pigs and their environment must be performed to prevent the spread of these resistant organisms.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
This study was approved by the Research Ethics Committee of National Taiwan University Hospital (201609066RINB).
S-CH, C-CL, and P-RH designed the experiments. S-CH, M-TC, and C-HL executed the lab experiments. S-CH, C-CL, Y-TH, C-HL, and C-NL analyzed the data. S-CH, C-CL, and P-RH prepared the manuscript. S-CH, C-CL, Y-TH, C-HL, M-TC, C-NL, and P-RH read and approved the final version of the manuscript.
This work was supported by grants from the Centers for Disease Control and Prevention, Minister of Health and Welfare, Executive Yuan, Taiwan (MOHW106-CDC-C-114-114701). The funder was not involved in the study design, data collection, analysis, or interpretation, writing the manuscript, or the decision to submit this work for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Brennan, E., Martins, M., McCusker, M. P., Wang, J., Alves, B. M., Hurley, D., et al. (2016). Multidrug-resistant Escherichia coli in bovine animals, Europe. Emerg. Infect. Dis. 22, 1650–1652. doi: 10.3201/eid2209.160140
Chapman, T. A., Wu, X. Y., Barchia, I., Bettelheim, K. A., Driesen, S., Trott, D., et al. (2006). Comparison of virulence gene profiles of Escherichia coli strains isolated from healthy and diarrheic swine. Appl. Environ. Microbiol. 72, 4782–4795. doi: 10.1128/AEM.02885-05
Chen, S. Y., Liao, C. H., Wang, J. L., Chiang, W. C., Lai, M. S., Chie, W. C., et al. (2014). Method-specific performance of vancomycin MIC susceptibility tests in predicting mortality of patients with methicillin-resistant Staphylococcus aureus bacteraemia. J Antimicrob Chemother 69, 211–218. doi: 10.1093/jac/dkt340
Chiou, C. S., Chen, Y. T., Wang, Y. W., Liu, Y. Y., Kuo, H. C., Tu, Y. H., et al. (2017). Dissemination of mcr-1-carrying plasmids among colistin-resistant Salmonella strains from humans and food-producing animals in Taiwan. Antimicrob. Agents Chemother. 61:e00338-17. doi: 10.1128/AAC.00338-17
Clinical and Laboratory Standards Institute [CLSI] (2020). Performance Standards for Antimicrobial Susceptibility Testing, 30th Edn. Wayne, PA: CLSI.
Hadjadj, L., Riziki, T., Zhu, Y., Li, J., Diene, S. M., and Rolain, J. M. (2017). Study of mcr-1 gene-mediated colistin resistance in Enterobacteriaceae isolated from humans and animals in different countries. Genes 8:E394. doi: 10.3390/genes8120394
Hsueh, P. R., Teng, L. J., Chen, C. Y., Chen, W. H., Yu, C. J., Ho, S. W., et al. (2002). Pandrug-resistant Acinetobacter baumannii causing nosocomial infections in a university hospital, Taiwan. Emerg. Infect. Dis. 8, 827–832.
Jensen, V. F., Jakobsen, L., Emborg, H. D., Seyfarth, A. M., and Hammerum, A. M. (2006). Correlation between apramycin and gentamicin use in pigs and an increasing reservoir of gentamicin-resistant Escherichia coli. J. Antimicrob. Chemother. 58, 101–107. doi: 10.1093/jac/dkl201
Jiang, F., Wu, Z., Zheng, Y., Frana, T. S., Sahin, O., Zhang, Q., et al. (2019). Genotypes and antimicrobial susceptibility profiles of hemolytic Escherichia coli from diarrheic piglets. Foodborne Pathog. Dis. 16, 94–103. doi: 10.1089/fpd.2018.2480
Kempf, I., Jouy, E., and Chauvin, C. (2016). Colistin use and colistin resistance in bacteria from animals. Int. J. Antimicrob. Agents 48, 598–606. doi: 10.1016/j.ijantimicag.2016.09.016
Khan, S. B., Zou, G., Cheng, Y. T., Xiao, R., Li, L., Wu, B., et al. (2017). Phylogenetic grouping and distribution of virulence genes in Escherichia coli along the production and supply chain of pork around Hubei, China. J. Microbiol. Immunol. Infect. 50, 382–385. doi: 10.1016/j.jmii.2016.03.006
Kong, L. H., Lei, C. W., Ma, S. Z., Jiang, W., Liu, B. H., Wang, Y. X., et al. (2017). Various sequence types of Escherichia coli isolates coharboring blaNDM-5 and mcr-1 genes from a commercial swine farm in China. Antimicrob. Agents Chemother. 61:e02167-16. doi: 10.1128/AAC.02167-16
Kuo, S. C., Huang, W. C., Wang, H. Y., Shiau, Y. R., Cheng, M. F., and Lauderdale, T. L. (2016). Colistin resistance gene mcr-1 in Escherichia coli isolates from humans and retail meats, Taiwan. J. Antimicrob. Chemother. 71, 2327–2329. doi: 10.1093/jac/dkw122
Lai, C. C., Chen, Y. S., Lee, N. Y., Tang, H. J., Lee, S. S., Lin, C. F., et al. (2019). Susceptibility rates of clinically important bacteria collected from intensive care units against colistin, carbapenems, and other comparative agents: results from Surveillance of Multicenter Antimicrobial Resistance in Taiwan (SMART). Infect. Drug Resist. 12, 627–640. doi: 10.2147/IDR.S194482
Lai, C. C., Chuang, Y. C., Chen, C. C., and Tang, H. J. (2017). Coexistence of MCR-1 and NDM-9 in a clinical carbapenem-resistant Escherichia coli isolate. Int. J. Antimicrob. Agents 49, 517–518. doi: 10.1016/j.ijantimicag.2017.02.001
Lai, C. C., Lin, Y. T., Lin, Y. T., Lu, M. C., Shi, Z. Y., Chen, Y. S., et al. (2018). Clinical characteristics of patients with bacteraemia due to the emergence of mcr-1-harboring Enterobacteriaceae in humans and pigs in Taiwan. Int. J. Antimicrob. Agents 52, 651–657. doi: 10.1016/j.ijantimicag.2018.08.015
Lee, S. I., Rayamahji, N., Lee, W. J., Cha, S. B., Shin, M. K., Roh, Y. M., et al. (2009). Genotypes, antibiogram, and pulsed-field gel electrophoresis profiles of Escherichia coli strains from piglets in Korea. J. Vet. Diagn. Invest. 21, 510–516. doi: 10.1177/104063870902100413
Lin, Y. C., Kuroda, M., Suzuki, S., and Mu, J. J. (2018). Emergence of an Escherichia coli strain co-harboring mcr-1 and blaNDM-9 from urinary tract infection in Taiwan. J. Glob. Antimicrob. Resist. 16, 286–290. doi: 10.1016/j.jgar.2018.10.003
Liu, J. Y., Liao, T. L., Huang, W. C., Liu, Y. M., Wu, K. M., Lauderdale, T. L., et al. (2018). Increased mcr-1 in pathogenic Escherichia coli from diseased swine, Taiwan. J. Microbiol. Immunol. Infect. doi: 10.1016/j.jmii.2018.10.011 [Epub ahead of print].
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Luppi, A. (2017). Swine enteric colibacillosis: diagnosis, therapy and antimicrobial resistance. Porc. Health Manag. 3:16. doi: 10.1186/s40813-017-0063-4
Meinersmann, R. J., Ladely, S. R., Plumblee, J. R., Cook, K. L., and Thacker, E. (2017). Prevalence of mcr-1 in the cecal contents of food animals in the United States. Antimicrob. Agents Chemother. 61:e02244-16. doi: 10.1128/AAC.02244-16
Nordmann, P., Assouvie, L., Prod’Hom, G., Poirel, L., and Greub, G. (2016). Screening of plasmid-mediated MCR-1 colistin-resistance from bacteraemia. Eur. J. Clin. Microbiol. Infect. Dis. 35, 1891–1892. doi: 10.1007/s10096-016-2739-0
Olaitan, A. O., Thongmalayvong, B., Akkhavong, K., Somphavong, S., Paboriboune, P., Khounsy, S., et al. (2015). Clonal transmission of a colistin-resistant Escherichia coli from a domesticated pig to a human in Laos. J. Antimicrob. Chemother. 70, 3402–3404.
Palmeira, J. D., Ferreira, H., Madec, J. Y., and Haenni, M. (2018). Draft genome of a ST443 mcr-1- and blaCTX-M-2-carrying Escherichia coli from cattle in Brazil. J. Glob. Antimicrob. Resist. 13, 269–270. doi: 10.1016/j.jgar.2018.05.010
Pulss, S., Semmler, T., Prenger-Berninghoff, E., Bauerfeind, R., and Ewers, C. (2017). First report of an Escherichia coli strain from swine carrying an OXA-181 carbapenemase and the colistin resistance determinant MCR-1. Int. J. Antimicrob. Agents 50, 232–236. doi: 10.1016/j.ijantimicag.2017.03.014
Rebelo, A. R., Bortolaia, V., Kjeldgaard, J. S., Pedersen, S. K., Leekitcharoenphon, P., Hansen, I. M., et al. (2018). Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro Surveill. 23:17-00672. doi: 10.2807/1560-7917.ES.2018.23.6.17-00672
Tenover, F. C., Arbeit, R. D., Goering, R. V., Mickelsen, P. A., Murray, B. E., Persing, D. H., et al. (1995). Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33, 2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995
Wang, C. H., Lin, J. C., Yu, C. M., and Wu, R. X. (2020). Emergence of multiple drug-resistant Escherichia coli harboring mcr-1 in immunocompetent patients from the community. J. Microbiol. Immunol. Infect. doi: 10.1016/j.jmii.2020.03.012 [Epub ahead of print].
Wirth, T., Falush, D., Lan, R., Colles, F., Mensa, P., Wieler, L. H., et al. (2006). Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60, 1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x
Keywords: Escherichia coli, sick pigs, colistin resistance, mcr-1, genotypes
Citation: Hsueh S-C, Lai C-C, Huang Y-T, Liao C-H, Chiou M-T, Lin C-N and Hsueh P-R (2020) Molecular Evidence for Intra- and Inter-Farm Spread of Porcine mcr-1-Carrying Escherichia coli in Taiwan. Front. Microbiol. 11:1967. doi: 10.3389/fmicb.2020.01967
Received: 16 May 2020; Accepted: 24 July 2020;
Published: 20 August 2020.
Edited by:
Patrick Rik Butaye, Ross University School of Veterinary Medicine, Saint Kitts and NevisReviewed by:
Chengming Wang, Auburn University, United StatesCopyright © 2020 Hsueh, Lai, Huang, Liao, Chiou, Lin and Hsueh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chao-Nan Lin, Y25saW42QG1haWwubnB1c3QuZWR1LnR3; Po-Ren Hsueh, aHNwb3JlbkBudHUuZWR1LnR3
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.