- Department of Biotechnology and Life Sciences, University of Insubria, Varese, Italy
Enzymes are increasingly applied as biocatalysts for fulfilling industrial needs in a variety of applications and there is a bursting of interest for novel therapeutic proteins. Consequently, developing appropriate expression platforms for efficiently producing such recombinant proteins represents a crucial challenge. It is nowadays widely accepted that an ideal ‘universal microbial host’ for heterologous protein expression does not exist. Indeed, the first-choice microbes, as Escherichia coli or yeasts, possess known intrinsic limitations that inevitably restrict their applications. In this scenario, bacteria belonging to the Streptomyces genus need to be considered with more attention as promising, alternative, and versatile platforms for recombinant protein production. This is due to their peculiar features, first-of-all their natural attitude to secrete proteins in the extracellular milieu. Additionally, streptomycetes are considered robust and scalable industrial strains and a wide range of tools for their genetic manipulation is nowadays available. This mini-review includes an overview of recombinant protein production in streptomycetes, covering nearly 100 cases of heterologous proteins expressed in these Gram-positives from the 1980s to December 2019. We investigated homologous sources, heterologous hosts, and molecular tools (promoters/vectors/signal peptides) used for the expression of these recombinant proteins. We reported on their final cellular localization and yield. Thus, this analysis might represent a useful source of information, showing pros and cons of using streptomycetes as platform for recombinant protein production and paving the way for their more extensive use in future as alternative heterologous hosts.
Introduction
Nowadays, we witness the increasing application of enzymes in industrial sectors, including food, detergent, and textile manufactures (Trono, 2019) and the bursting of interest in proteins for therapeutic and diagnostic purposes (Tripathi and Shrivastava, 2019). Developing efficient bioprocessing strategies for protein production is consequently of utmost importance. Most of valuable industrial enzymes and therapeutic proteins are recombinant versions, produced by heterologous platforms (Adrio and Demain, 2014). However, an ideal ‘universal host’ for protein heterologous expression does not exist. Those microbes (as Escherichia coli or yeasts) that are still considered the first-choices to this purpose possess intrinsic limitations inevitably restricting their use. Production of heterologous proteins in E. coli is limited by self-cytotoxicity, incorrect folding, aggregation into inclusion bodies, and/or lack of secretion (Adrio and Demain, 2014). In yeasts, recombinant protein production is often associated with hyper-glycosylation and product retention within the periplasmic space (Vieira Gomes et al., 2018).
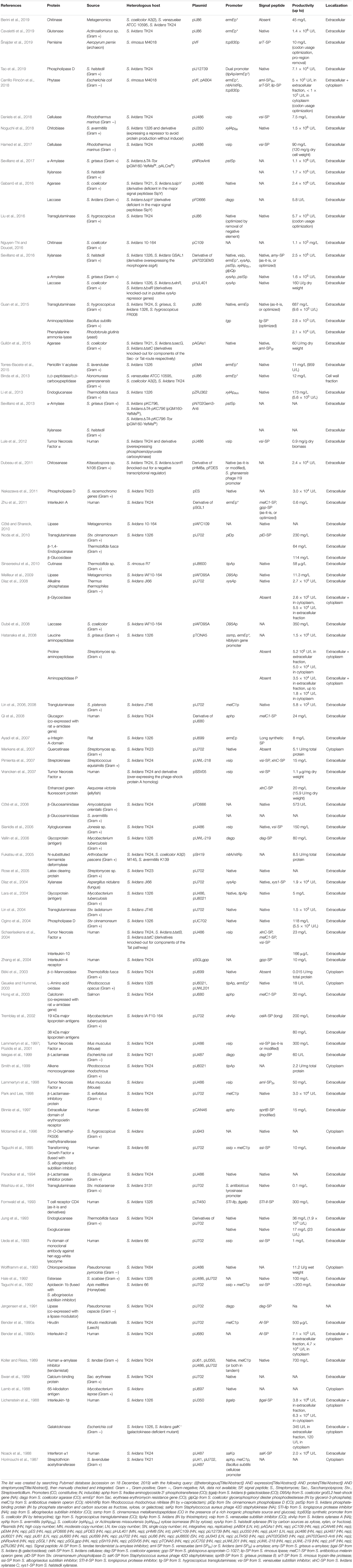
In this scenario, bacteria belonging to the Streptomyces genus might represent a promising alternative platform for recombinant protein production. Streptomycetes are Gram-positive, aerobic bacteria, characterized by a mycelial life style and commonly found in soils, where they secrete multiple hydrolytic enzymes to degrade complex organic substrates. This natural secretion capacity represents their most attractive feature for recombinant protein production. Secretion may prevent local accumulation of the overexpressed recombinant proteins, reducing toxicity to host cells and promoting correct folding (Anné et al., 2012). It facilitates downstream recovery decreasing production costs (Hamed et al., 2018). In addition, streptomycetes are characterized by low endogenous proteolytic activity; they grow relatively fast and in inexpensive media; they do not produce pyrogenic lipopolysaccharides and endotoxins; they are not pathogenic; and they might express G + C-rich genes without codon usage optimization (Anné et al., 2012; Sevillano et al., 2013). Thanks to the extensive fermentation knowhow deriving from their use as antibiotic producers (Ndlovu et al., 2015), streptomycetes are robust and scalable industrial strains, and a wide range of tools for their genetic manipulation have recently become available (Kieser et al., 2000). Notwithstanding these potential advantages, nowadays their use is not so common as it could be expected. To investigate this aspect, in this mini-review we cover – to the best of our knowledge – all studies published from 1980s to December 2019, in which streptomycetes were used as heterologous hosts for recombinant protein production. Table 1 reports these 94 cases of proteins expressed in streptomycetes. Figure 1 highlights the main results emerging from the analysis of Table 1 in terms of protein class, homologous source, heterologous host, and molecular tools.
Figure 1. Distribution of the 94 recombinant proteins heterologously produced by streptomycetes and listed in Table 1, according to: use/family of the recombinant protein (A), homologous source (B), heterologous host (C), plasmid (D), and promoter (E) used for heterologous production. (C–E) When for the same protein, multiple hosts and/or plasmids, and/or promoters were used, these different conditions were counted separately.
What Are the Recombinant Proteins Produced in Streptomycetes?
50 (out of 94) proteins listed in Table 1 are enzymes with potential industrial/environmental applications (Figure 1A). The most represented class is that of glycosyl hydrolases (23 proteins), including: (i) (hemi)cellulases, for lignocellulose saccharification and biofuel production; (ii) chitinases, for generating value-added chitin-derivatives as chitosan or biopesticides (Berini et al., 2018a); and (iii) amylases for starch processing. The lipase/esterase group (8 proteins) with applications in detergent, food, and biofuel industries, and the oxidoreductase class (7), including laccases and peroxidases for bioremediation (Berini et al., 2018b), follow. Interesting examples are the phospholipase D from Streptomyces racemochromogenes, for producing phosphatidyl derivatives from lecithin with emulsifying properties for food and cosmetics (Nakazawa et al., 2011), and the cutinase from Thermobifida sp. with polyester-degrading activity in bioplastic recycle (Sinsereekul et al., 2010). Dubé et al. (2008) produced in Streptomyces lividans up to 350 mg/L of Streptomyces coelicolor small laccase, a thermostable enzyme decolorizing synthetic dyes that is considered promising for pollutant degradation in urban or industrial wastewaters. Finally, Table 1 and Figure 1A include transferases (6 proteins) for food processing, proteases/peptidases (5) for feed and detergent industries, and phosphatases (2), including a phytase used as supplement for animal nutrition (Carrillo Rincón et al., 2018). Additionally, Torres-Bacete et al. (2015) expressed a novel Penicillin V acylase for producing semisynthetic penicillins, whereas Rose et al. (2005) a latex clearing protein for bioconversion of rubber wastes. Unfortunately, only few of these studies reported a comparison of protein expression yield between streptomycetes and other microbial hosts. Hamed et al. (2017) succeed in producing 90 mg/L of a thermostable cellulase from the bacteroidetes Rhodotermus marinus using S. lividans TK24 as host; the same protein could not be produced in E. coli. Very recently, Šnajder et al. (2019) reported the first and so far the only case of expression of an archaeal thermozyme (pernisine) in Streptomyces rimosus. The homologous host – the hyperthermophilic Aeropyrum pernix – was uncultivable in industrial fermentation facilities. The protein productivity (10 mg/L) in this case was comparable to that achieved in E. coli, but with the advantage of simplified downstream processes due to protein secretion in the streptomycete (Šnajder et al., 2019). Similarly, the Streptomyces halstedii phospholipase expression was approximately 60 and 30 times higher in S. lividans TK24 than in E. coli and Pichia pastoris, respectively (Tao et al., 2019). Sianidis et al. (2006) and Sinsereekul et al. (2010) reported that their attempts to express a xyloglucanase from Jonesia sp. and a cutinase from Thermobifida sp. failed, respectively, in E. coli and B. subtilis, and E. coli and P. pastoris. Finally, Díaz et al. (2004) produced in S. lividans JI66 a xylanase from Aspergillus nidulans with a yield 3- and 19-fold higher than in lactic bacteria and Saccharomyces cerevisiae, respectively. Despite these successes at laboratory level, we are indeed unaware of any further scaling up at industrial level of recombinant enzyme production from streptomycetes. We can suppose that this is probably due to an overall limited protein productivity in streptomycetes that rarely reaches the g/L production level usually required for industrial application. As reported in Table 1, only in the case of a chitinase (Nguyen-Thi and Doucet, 2016), the protein productivity was more than 1 g/L. These results point out the crucial need to overcome intrinsic bottlenecks in protein productivity in streptomycetes, by redesigning their regulatory networks and secretion pathways by system biology, as recently proposed by Kim et al. (2020).
In Table 1, 21 are the recombinant proteins curing human diseases (Figure 1A), including those for treating cancer (interleukin, interferon, Tumor Necrosis Factor Alpha-TNF-α), cardiovascular pathologies (streptokinase, hirudin), and metabolic or auto-immune disorders (glucagon, phenylalanine ammonia-lyase, tendamistat). Recently, S. lividans TK24 was used for producing an Actinoallomurus A8-sourced glutenase, a promising candidate for oral enzymatic management of gluten toxicity (Cavaletti et al., 2019). Streptomycetes were also used to express 8 ‘target’ proteins, as antigens from Mycobacterium tuberculosis (Vallin et al., 2006) or the α-integrin A-domain for screening ligands for treating inflammatory disorders (Ayadi et al., 2007), and few diagnostic proteins (2) as the T Cell receptor CD4 for diagnosis of HIV infection (Fornwald et al., 1993). Biopharmaceutical production of proteins in streptomycetes is generally acceptable to the Food and Drug Administration and European Medicine Agency since these bacteria have been used for decades in industrial manufacturing of antibiotics, immunomodulating and antitumor drugs, and nutraceuticals (Marinelli and Marcone, 2011). Additionally, these naturally soil-inhabiting bacteria are recognized as useful components of natural ecosystem and they are considered safer than other microorganisms for agricultural use (Berini et al., 2019). Interestingly, besides the proteins listed in Table 1, S. lividans was employed by Cangene Corporation (now part of Emergent BioSolutions) for the recombinant production of the macrophage-colony stimulating factor Leucotropin™, a therapeutic agent that successfully completed Phase III trials for treating Hodgkin’s and non-Hodgkin’s lymphoma (Vrancken and Anné, 2009). To our best knowledge, this is the only reported case of a therapeutic protein production in streptomycetes that reached the clinical phases.
Finally, Table 1 includes 7 proteins without any direct industrial/therapeutic application: they were produced in streptomycetes for studying biochemical/functional properties and/or mode of action, as in case of the novel N-substitute formamide deformylase from Arthrobacter pascens involved in the metabolism of isonitriles (Fukatsu et al., 2005). Another example is VanYn, a D,D-dipeptidase/D,D-carboxypeptidase identified as the sole resistant determinant in the glycopeptide producer Nonomuraea gerenzanensis (Binda et al., 2013; Dalmastri et al., 2016). VanYn expression in Streptomyces venezuelae allowed a higher production than in E. coli (Binda et al., 2012), and contributed to elucidating cell wall turnover during antibiotic production (Marcone et al., 2010a, 2014).
Where Do Recombinant Proteins Expressed in Streptomycetes Come From?
71 of the proteins listed in Table 1 derive from prokaryotes and 23 from eukaryotes (Figure 1B). Most of prokaryote-sourced proteins come from Gram-positive bacteria: 49 are from Streptomyces or Streptoverticillium spp., or other actinomycetes as Nonomuraea, Kitasatospora, or Thermobifida spp. This is not surprising, as heterologous expression is facilitated when the host is phylogenetically related to the homologous producer, due to the similar metabolic and genetic background (Binda et al., 2013). Streptomycetes (DNA G + C > 60%) offer an optimized codon usage for high G + C content genes and they represent a complementary tool versus E. coli (DNA G + C ca. 51%). For instance, chitinases, usually produced by soil-inhabitant actinomycetes, were successfully produced in streptomycetes (Berini et al., 2019). Cloning a S. coelicolor chitinase in S. lividans 10–164 resulted in 486-fold production improvement compared to E. coli, allowing gram-scale production for converting crystalline chitin in N-acetylglucosamine (Nguyen-Thi and Doucet, 2016). 9 additional recombinant proteins derive from the firmicutes Bacillus subtilis and Streptococcus equisimilis, and other 9 from the Gram-negative Escherichia, Thermus, and Pseudomonas spp. (Figure 1B). The thermostable cellulase from the bacteroidetes Rhodotermus marinus (Hamed et al., 2017) and the archaeal thermozyme (pernisine) (Šnajder et al., 2019), described above, complete the list of the prokaryote proteins.
Streptomycetes were successfully used for expressing metagenome-sourced bacterial enzymes (Berini et al., 2017). 2 lipases from enriched fed-batch bioreactors (Meilleur et al., 2009; Côté and Shareck, 2010) and 1 chitinase (named 53D1) from agricultural soil (Berini et al., 2019) were produced in different Streptomyces strains. In case of 53D1, the protein was secreted (45 mg/L) into the culture broth by S. coelicolor A3(2), with a clear improvement over its expression in E. coli, where the protein was mostly accumulated as inactive into inclusion bodies (Cretoiu et al., 2015). Enough 53D1 was produced in the streptomycete to test its activity as biopesticide (Berini et al., 2019).
The heterogeneity of eukaryote sources of the recombinant proteins expressed in streptomycetes confirms their versatility (Table 1 and Figure 1B). The homologous producers of the eukaryote proteins listed in Table 1 span from filamentous fungi or yeasts (2), to invertebrates (insect, leech, and jellyfish; 3) or vertebrates (fish and mammals; 18). Notably, 14 human proteins were produced in these hosts. A chronological analysis indicates that eukaryote protein expression in streptomycetes was more frequent in the 1990s, becoming after that rarer. The last example of eukaryote protein produced in S. lividans TK24 dated back to 2012 (Lule et al., 2012). This is probably due to recent developments in using engineered yeasts, and mammalian and insect cell lines for manufacturing high-value eukaryote proteins, especially those requiring post-translational modifications (Hunter et al., 2019).
Which Is the Best Promoter/Vector/Host System for Recombinant Protein Production in Streptomycetes?
S. lividans strains are by far the most frequently used heterologous hosts, employed for producing 91 proteins listed in Table 1. 31 proteins were expressed in the parental S. lividans 66 (also named JI66 or 1326), whereas 37 in its derivative TK24, which is a two-plasmid-free mutant carrying streptomycin resistance mutation (str-6, SLP2–, SLP3–) (Kieser et al., 2000) (Figure 1C). 1 additional protein was produced in TK64, carrying the same mutations as TK24 plus the pro-2 mutation, and 1 in TK54, characterized by his-2, leu-2, and spc-1 mutations. The use of S. lividans TK24 has the following advantages: (i) low level of extracellular protease activity, (ii) poorly active restriction-modification system of exogenous DNA, (iii) known biochemistry/genetic background due to its high similarity to the model organism S. coelicolor A3(2) (Daniels et al., 2018). Other S. lividans used as hosts were the plasmid-free mutants S. lividans TK23 (for 3 proteins), carrying spectinomycin resistance mutation (spc-1, SLP2–, SLP3–), and its derivative JT46 (2 proteins) mutated in rec-46 gene to reduce inter-plasmid recombination (Kieser et al., 2000). 4 proteins were produced in S. lividans TK21, which lacks only SLP2 plasmid. Ad hoc constructed S. lividans hosts were derivatives of S. lividans 66 or TK24, as the pleiotropic mutant S. lividans 10–164 (Hurtubise et al., 1995) defective in cellobiose and xylobiose uptake and used for producing a metagenome-sourced lipase (Meilleur et al., 2009; Côté and Shareck, 2010), and S. lividans galK– (galactokinase-deficient mutant) for the production of E. coli galactokinase (Lichenstein et al., 1988). S. lividans GSAL1, used for the production of a xylanase and a α-amylase, overexpresses the morphogene ssgA, which pleiotropically controls growth and cell division. ssgA overexpression markedly enhances septation in vegetative hyphae, leading to fragmented growth and to wider hyphae, a phenotype that apparently favors protein production and secretion (Sevillano et al., 2016). Other streptomycetes employed as hosts were S. coelicolor A3(2) and its derivative M145 (3 proteins), Streptomyces griseus (3), S. rimosus (3), Streptomyces hygroscopicus (3), S. venezuelae (2), and Streptomyces avermitilis (1) (Table 1 and Figure 1C). Although less frequently used than S. lividans, in certain cases these alternative streptomycetes permitted the production of proteins poorly or not at all expressed in S. lividans (Binda et al., 2013; Berini et al., 2019), thus indicating that expanding the range of streptomycete hosts might be promising.
As regards to vectors, the mostly used are high copy number replicative ones (in 93 cases) (Table 1 and Figure 1D) as for examples pIJ702 (25 proteins), pIJ486 (14), and pIJ86 (7 proteins). pIJ702 vector, which carries thiostrepton resistance (tsrR) and tyrosinase production (mel+) markers, is the derivative of pIJ350, a non-conjugative broad host range vector (Kieser et al., 2000). pIJ486 (tsrR) derived from pIJ101, which contains the promoterless neo gene (kanamycin resistance) and lacks both the transfer function and the sti locus that usually confers ‘strong incompatibility’. Removing the sti locus increases the chance that different plasmids can be retained at similar copy numbers (Deng et al., 1988; Kieser et al., 2000). The more recent pIJ86 carries apramycin resistance marker (aprR) and it is a conjugative vector used for the strong constitutive expression of proteins under erythromycin promoter (ermE∗ promoter) from Saccharopolyspora erythraea. Recent works (Sevillano et al., 2013, 2017) described new replicative high copy number ‘marker-free’ systems, which allowed the production of high levels of proteins without using antibiotics as selection markers. One example is based on the presence of a toxin gene localized in the genome and of an anti-toxin gene located on the expression plasmid of the yefM/yoeBsl operon from S. lividans (Sevillano et al., 2013). Only for 5 proteins, replicative moderate or low copy number vectors were used. For instance, the moderate copy number pIJ12739 was constructed for the expression of the phospholipase D from S. halstedii in S. lividans TK24, following the same approach previously described by Fernández-Martínez and Bibb (2014) to produce a dual-promoter expression vector (Tao et al., 2019). The low copy number pIJ943 was used for producing the 31-O-demethyl-FK506 methyltransferase in S. lividans (Motamedi et al., 1996). For only 3 proteins, integrative vectors were employed such as pAB04 – low copy number plasmid used for producing a phytase (Carrillo Rincón et al., 2018), or pIJ8600 – single copy number vector employed for the expression of the cutinase from the Gram-positive Thermobifida sp. in S. rimosus R7 (Sinsereekul et al., 2010). Although less explored, integrative vectors might present some advantages. When the integrative single copy number pHM8a plasmid was used for expressing a chitosanase, productivity was comparable to that achieved with replicative multicopy pFDES plasmid, but with the advantage of not requiring antibiotic addition for selection (Dubeau et al., 2011). Interestingly, this last work is the only one, among those cited in this mini-review, which allowed a direct comparison on the effect of different vectors on protein yield. Most of the studies were driven by an empirical case-by-case approach to optimize the tools for a specific protein production, making difficult to draw final conclusions on which is the preferable vector system to be used.
In 20 cases (out of 94), the heterologous protein genes were cloned under the control of their native promoters, but more frequently streptomycete (or other actinomycete) heterologous promoters were used (Table 1 and Figure 1C). The heterologous promoters can be constitutive (e.g., vsip from S. venezuelae; dagp from S. coelicolor; ermE∗p from Sac. erythraea; ssip from Streptomyces albogriseolus; aphp from Streptomyces fradiae) or inducible (e.g., xysAp from S. halstedii, induced by xylane; pstSp from S. lividans, by phosphate starvation and different carbon sources; tcp830p from S. coelicolor, by tetracycline; tipAp from S. lividans, by thiostrepton). Constitutive promoters were more frequently used than inducible ones (50 vs. 24 cases, respectively). If in E. coli, a balance between the vector copy number and the promoter strength is needed for controlling protein production and slowing down inclusion body formation (Adrio and Demain, 2014), in streptomycetes this problem is overcome by protein secretion. On the other hand, in streptomycetes, constitutive expression may cause a growth rate reduction negatively impacting on protein productivity: in these cases, inducible expression could be advantageous, although weak points of an inducible system remain as low level of expression, a narrow host range, and the need of an expensive inducer (Herai et al., 2004). As in the case of vectors, only very few studies systemically compared the effect of different promoters on protein yield. Sevillano et al. (2016) investigated the expression of a xylanase from S. halstedii cloning the gene under the control of six strong promoters, including two commonly used (vsip and ermE∗p) and four recently identified. Two belonging to the last group (xysAp and pstSp) performed better than those considered the golden standards, confirming that there is room for developing new tools for improving protein expression in streptomycetes.
In 30 out of the 94 proteins, the presence of native signal peptides (SP) guaranteed secretion in the heterologous hosts, while in 2 cases proteins expressed with their native SP accumulated into the cytoplasm and in 1 case the enzyme was recovered from the cell wall fraction (Table 1). In streptomycetes, the Sec pathway constitutes the main secretion system (Anné et al., 2012). Accordingly, proteins to be secreted have N-terminal hydrophobic SP, followed by a longer hydrophobic H-domain and a C-terminal part containing at the end three amino acids which form the signal peptidase recognition site. Other minor secretion systems were reported, including the twin-arginine dependent translocation (Tat) pathway (Anné et al., 2012). The Tat machinery exports fully folded proteins across the cytoplasmic membrane: SPs that target proteins to this pathway resemble Sec SPs, but contain a conserved twin-arginine motif in the N-region (Valverde et al., 2018). A comparison between the efficiency of these two pathways for recombinant protein production showed that replacing Sec-dependent SP with Tat-dependent SP drastically reduced protein expression (Schaerlaekens et al., 2004). When native SPs were absent or not functional, heterologous genes were fused to SP encoding sequences from genes for highly expressed/secreted endogenous Streptomyces proteins (Anné et al., 2016), such as the one from the subtilisin inhibitor (vsi) of S. venezuelae CBS762.70 (Van Mellaert et al., 1998). Other SP sequences, frequently used in Streptomyces expression-systems are also listed in Table 1. They derived from the genes for the trypsin-like protease (srT) from S. rimosus, for the α-amylase from Streptomyces tendae, S. griseus, S. lividans, or Streptomyces limoseus, for the melanin operon gene (melC1) from Streptomyces antibioticus, for the subtilisin inhibitor (ssi) from S. albogriseolus. The final result is that in 77 out of the 94 proteins listed in Table 1, the recombinant proteins were completely secreted with productivities up to 100s of mg/L (Guan et al., 2015). In the few cases (8) where proteins were accumulated into cytoplasm, their productivity was generally low. 7 proteins were found produced either inside or outside the cells, whereas VanYn was localized in the cell wall fraction where it plays its physiological role in antibiotic resistance (Marcone et al., 2010a; Binda et al., 2012, 2013).
Conclusion
From the analysis of the literature in the last four decades, it emerges that, although promising, streptomycetes have been used for heterologous protein production less than their potential indicates to do. Notwithstanding their efficient protein secretion machine – which definitively facilitates downstream operations and protein purification – the mycelial lifestyle of these bacteria has probably discouraged scientists to use them more frequently. In liquid media, streptomycetes grow as mycelial pellets consisting of cells in different physiological states, and cultures are not homogenous and might become very viscous. In this regard, combining different specific mutations as ssgA for improving disperse growth (Sevillano et al., 2016), and galK for generating auxotrophic mutants not requiring antibiotic-dependent selection (Lichenstein et al., 1988) might facilitate upstream processes. Additionally, formulation of novel cultivation media – replacing those used for antibiotic biosynthesis – could facilitate protein downstream (Binda et al., 2013; Berini et al., 2019). Another aspect probably limiting their application is that streptomycetes cannot be genetically manipulated by the methods commonly used for E. coli and S. cerevisiae. They need ad hoc protocols based on intergeneric conjugation or protoplast transformation (Kieser et al., 2000; Marcone et al., 2010b, c). With time, these protocols have become available and, as reported in this review, nowadays we can count on a large variety of vectors, promoters, and SP sequences. What is still missing is the systematic and critical comparison of the available toolkits. Optimization of protein production is still conducted following a case-by-case – and somehow random – approach. Finally, an important issue is the intrinsic low protein productivity of streptomycetes in comparison with the mostly used E. coli and yeasts. Further improvements, in this sense, are urgently needed and may derive from system and synthetic biology approaches, still poorly applied to streptomycetes. Indeed, progresses in system biology and -omics technologies may shed light on the interplay of elements involved in protein expression, thus helping in the rational improvement of both expression platforms and fermentation conditions, finalized at reducing the metabolic burden due to heterologous protein production. A demonstration is present in the pioneering work conducted by Muhamadali et al. (2015) on a S. lividans strain producing the murine TNF-α, where heterologous protein expression determined profound changes in the metabolomics of the streptomycete causing an overflow of organic acids and sugars. In post-genomic era, a further ambitious goal is applying synthetic biology approaches for building a Streptomyces ‘super host’ with metabolic networks rewired to facilitate heterologous protein expression. Synergic application of genome minimization strategies (i.e., systematic removal of those elements – as secondary metabolites or proteases – that can hamper protein production) and engineering of translation and transcription machineries, might help reaching this goal (Kim et al., 2020). To this end, it is encouraging considering that performing Streptomyces ‘super hosts’ have been already constructed for the heterologous production of antibiotics (Gomez-Escribano and Bibb, 2011; Myronovskyi et al., 2018). We believe that integrating these tools could help in improving streptomycetes as robust producers of recombinant proteins, increasing their competitiveness to other platforms and stimulating their large-scale application as cell factories.
Author Contributions
FB and EB collected the data and analyzed them. FB, FM, and EB co-wrote the review. FB prepared the figure and the table. EB coordinated the work. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grant ‘Fondo di Ateneo per la Ricerca’ 2017 and 2018 to FM, and Consorzio Italbiotec to EB.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
CIB (Consorzio Interuniversitario per le Biotecnologie) fellowship to FB and FEMS (Federation of European Microbiological Societies) Research and Training Grant support to FB and EB are acknowledged.
References
Adrio, J. L., and Demain, A. L. (2014). Microbial enzymes: tools for biotechnological processes. Biomolecules 4, 117–139. doi: 10.3390/biom4010117
Anné, J., Economou, A., and Bernaerts, K. (2016). “Protein secretion in Gram-Positive bacteria: from multiple pathways to biotechnology,” in Protein and Sugar Export and Assembly in Gram-Positive Bacteria. Current Topics in Microbiology and Immunology, eds F. Bagnoli and R. Rappuoli (Cham: Springer), 267–308. doi: 10.1007/82_2016_49
Anné, J., Maldonado, B., Van Impe, J., Van Mellaert, L., and Bernaerts, K. (2012). Recombinant protein production and streptomycetes. J. Biotechnol. 158, 159–167. doi: 10.1016/j.jbiotec.2011.06.028
Ayadi, D. Z., Chouayekh, H., Mhiri, S., Zerria, K., Fathallah, D. M., and Bejar, S. (2007). Expression by Streptomyces lividans of the rat alpha integrin CD11b A-domain as a secreted and soluble recombinant protein. J. Biomed. Biotechnol. 2007:54327. doi: 10.1155/2007/54327
Béki, E., Nagy, I., Vanderleyden, J., Jäger, S., Kiss, L., Fülöp, L., et al. (2003). Cloning and heterologous expression of a beta-D-mannosidase (EC 3.2.1.25)-encoding gene from Thermobifida fusca TM51. Appl. Environ. Microbiol. 69, 1944–1952. doi: 10.1128/aem.69.4.1944-1952.2003
Bender, E., Koller, K. P., and Engels, J. (1990a). Secretory synthesis of human interleukin-2 by Streptomyces lividans. Gene 86, 227–232. doi: 10.1016/0378-1119(90)90283-W
Bender, E., Vogel, R., Koller, K. P., and Engels, J. (1990b). Synthesis and secretion of hirudin by Streptomyces lividans. Appl. Microbiol. Biotechnol. 34, 203–207. doi: 10.1007/BF00166781
Berini, F., Casartelli, M., Montali, A., Reguzzoni, M., Tettamanti, G., and Marinelli, F. (2019). Metagenome-sourced microbial chitinases as potential insecticide proteins. Front. Microbiol. 10:1358. doi: 10.3389/fmicb.2019.01358
Berini, F., Casciello, C., Marcone, G. L., and Marinelli, F. (2017). Metagenomics: novel enzymes from non-culturable microbes. FEMS Microbiol. Lett. 364:fnx211. doi: 10.1093/femsle/fnx211
Berini, F., Katz, C., Gruzdev, N., Casartelli, M., Tettamanti, G., and Marinelli, F. (2018a). Microbial and viral chitinases: attractive biopesticides for integrated pest management. Biotechnol. Adv. 36, 818–838. doi: 10.1016/j.biotechadv.2018.01.002
Berini, F., Verce, M., Ausec, L., Rosini, E., Tonin, F., Pollegioni, L., et al. (2018b). Isolation and characterization of a heterologously expressed bacterial laccase from the anaerobe Geobacter metallireducens. Appl. Microbiol. Biotechnol. 102, 2425–2439. doi: 10.1007/s00253-018-8785-z
Binda, E., Marcone, G. L., Berini, F., Pollegioni, L., and Marinelli, F. (2013). Streptomyces spp. as efficient expression system for a D,D-peptidase/D,D-carboxypeptidase involved in glycopeptide antibiotic resistance. BMC Biotechnol. 13:24. doi: 10.1186/1472-6750-13-24
Binda, E., Marcone, G. L., Pollegioni, L., and Marinelli, F. (2012). Characterization of VanYn, a novel D,D-peptidase/D,D-carboxypeptidase involved in glycopeptide antibiotic resistance in Nonomuraea sp. ATCC 39727. FEBS J. 279, 3203–3213. doi: 10.1111/j.1742-4658.2012.08706.x
Binnie, C., Jenish, D., Cossar, D., Szabo, A., Trudeau, D., Krygsman, P., et al. (1997). Expression and characterization of soluble human erythropoietin receptor made in Streptomyces lividans 66. Protein Expr. Purif. 11, 271–278. doi: 10.1006/prep.1997.0787
Carrillo Rincón, A. F., Magdevska, V., Kranjc, L., Fujs, Š., Müller, R., and Petković, H. (2018). Production of extracellular heterologous proteins in Streptomyces rimosus, producer of the antibiotic oxytetracycline. Appl. Microbiol. Biotechnol. 102, 2607–2620. doi: 10.1007/s00253-018-8793-z
Cavaletti, L., Taravella, A., Carrano, L., Carenzi, G., Sigurtà, A., Solinas, N., et al. (2019). E40, a novel microbial protease efficiently detoxifying gluten proteins, for the dietary management of gluten intolerance. Sci. Rep. 9:13147. doi: 10.1038/s41598-019-48299-7
Côté, A., and Shareck, F. (2010). Expression and characterization of a novel heterologous moderately thermostable lipase derived from metagenomics in Streptomyces lividans. J. Ind. Microbiol. Biotechnol. 37, 883–891. doi: 10.1007/s10295-010-0735-4
Côté, N., Fleury, A., Dumont-Blanchette, E., Fukamizo, T., Mitsutomi, M., and Brzezinski, R. (2006). Two exo-beta-D-glucosaminidases/exochitosanases from actinomycetes define a new subfamily within family 2 of glycoside hydrolases. Biochem. J. 394, 675–686. doi: 10.1042/BJ20051436
Cretoiu, M. S., Berini, F., Kielak, A. M., Marinelli, F., and van Elsas, J. D. (2015). A novel salt-tolerant chitobiosidase discovered by genetic screening of a metagenomic library derived from chitin-amended agricultural soil. Appl. Microbiol. Biotechnol. 99, 8199–8215. doi: 10.1007/s00253-015-6639-5
Dalmastri, C., Gastaldo, L., Marcone, G. L., Binda, E., Congiu, T., and Marinelli, F. (2016). Classification of Nonomuraea sp. ATCC 39727, an actinomycete that produces the glycopeptide antibiotic A40926, as Nonomuraea gerenzanensis sp. nov. Int. J. Syst. Evol. Microbiol. 66, 912–921. doi: 10.1099/ijsem.0.000810
Daniels, W., Bouvin, J., Busche, T., Rückert, C., Simoens, K., Karamanou, S., et al. (2018). Transcriptomic and fluxomic changes in Streptomyces lividans producing heterologous protein. Microb. Cell Fact. 17, 2–19. doi: 10.1186/s12934-018-1040-6
Deng, Z., Kieser, T., and Hopwood, D. (1988). “Strong incompatibility” between derivatives of the Streptomyces multi-copy plasmid pIJ101. Mol. Gen. Genet. 214, 286–294. doi: 10.1007/BF00337723
Díaz, M., Adham, S. A., Ramón, D., Gil, J. A., and Santamaría, R. I. (2004). Streptomyces lividans and Brevibacterium lactofermentum as heterologous hosts for the production of X22 xylanase from Aspergillus nidulans. Appl. Microbiol. Biotechnol. 65, 401–406. doi: 10.1007/s00253-004-1633-3
Díaz, M., Ferreras, E., Moreno, R., Yepes, A., Berenguer, J., and Santamaria, R. (2008). High-level overproduction of Thermus enzymes in Streptomyces lividans. Appl. Microbiol. Biotechnol. 79, 1001–1008. doi: 10.1007/s00253-008-1495-1
Dubé, E., Shareck, F., Hurtubise, Y., Daneault, C., and Beaurengard, M. (2008). Homologous cloning, expression, and characterisation of a laccase from Streptomyces coelicolor and enzymatic decolourisation of an indigo dye. Appl. Microbiol. Biotechnol. 79, 597–603. doi: 10.1007/s00253-008-1475-5
Dubeau, M. P., Guay, I., and Brzezinski, R. (2011). Modification of genetic regulation of a heterologous chitosanase gene in Streptomyces lividans TK24 leads to chitosanase production in the absence of chitosan. Microb. Cell Fact. 10:7. doi: 10.1186/1475-2859-10-7
Fernández-Martínez, L. T., and Bibb, M. J. (2014). Use of the meganuclease I-SceI of Saccharomyces cerevisiae to select for gene deletions in actinomycetes. Sci. Rep. 4:7100. doi: 10.1038/srep07100
Fornwald, J. A., Donovan, M. J., Gerber, R., Keller, J., Taylor, D. P., Arcuri, E. J., et al. (1993). Soluble forms of the human T cell receptor CD4 are efficiently expressed by Streptomyces lividans. Nat. Biotechnol. 11, 1031–1036. doi: 10.1038/nbt0993-1031
Fukatsu, H., Herai, S., Hashimoto, Y., Maseda, H., Higashibata, H., and Kobayashi, M. (2005). High-level expression of a novel amine-synthesizing enzyme, N-substituted formamide deformylase, in Streptomyces with a strong protein expression system. Protein Expr. Purif. 40, 212–219. doi: 10.1016/j.pep.2004.11.013
Gabarró, M. V., Gullón, S., Vicente, R. L., Carminal, G., Mellado, R. P., and López-Santín, J. (2016). A Streptomyces lividans SipY deficient strain as a host for protein production: standardization of operational alternatives for model proteins. J. Chem. Technol. Biotechnol. 92, 217–223. doi: 10.1002/jctb.4933
Geueke, B., and Hummel, W. (2003). Heterologous expression of Rhodococcus opacus L-amino acid oxidase in Streptomyces lividans. Protein Expr. Purif. 28, 303–309. doi: 10.1016/S1046-5928(02)00701-5
Gomez-Escribano, J. P., and Bibb, M. J. (2011). Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb. Biotechnol. 4, 207–215. doi: 10.1111/j.1751-7915.2010.00219.x
Guan, C., Cui, W., He, X., Hu, X., Xu, J., and Du, G. (2015). Construction and development of a novel expression system of Streptomyces. Protein Expr. Purif. 113, 17–22. doi: 10.1016/j.pep.2015.04.009
Gullón, S., Vicente, R. L., Valverde, J. R., Marín, S., and Mellado, R. P. (2015). Exploring the feasibility of the Sec route to secrete proteins using the Tat route in Streptomyces lividans. Mol. Biotechnol. 57, 931–938. doi: 10.1007/s12033-015-9883-0
Hale, V., McGrew, M., Carlson, B., and Schottel, J. L. (1992). Heterologous expression and secretion of a Streptomyces scabies esterase in Streptomyces lividans and Escherichia coli. J. Bacteriol. 174, 2431–2439. doi: 10.1128/JB.174.8.2431-2439.1992
Hamed, M. B., Anné, J., Karamanou, S., and Economou, A. (2018). Streptomyces protein secretion and its application in biotechnology. FEMS Microbiol. Lett. 365:fny250. doi: 10.1093/femsle/fny250
Hamed, M. B., Karamanou, S., Ólafsdottir, S., Basílio, J. S. M., Simoens, K., Tsolis, K. C., et al. (2017). Large-scale production of a thermostable Rhodothermus marinus cellulase by heterologous secretion from Streptomyces lividans. Microb. Cell Fact. 16:232. doi: 10.1186/s12934-017-0847-x
Hatanaka, T., Onaka, H., Arima, J., Uraji, M., Uesugi, Y., Usuki, H., et al. (2008). pTONA5: a hyperexpression vector in streptomycetes. Protein Expr. Purif. 62, 244–248. doi: 10.1016/j.pep.2008.09.001
Herai, S., Hashimoto, Y., Higashibata, H., Maseda, H., Ikeda, H., Omura, S., et al. (2004). Hyper-inducible expression system for streptomycetes. Proc. Natl. Acad. Sci. U.S.A. 101, 14031–14035. doi: 10.1073/pnas.0406058101
Hong, B., Wu, B., and Li, Y. (2003). Production of C-terminal amidated recombinant salmon calcitonin in Streptomyces lividans. Appl. Biochem. Biotechnol. 110, 113–123. doi: 10.1385/abab:110:2:113
Horinouchi, S., Furuya, K., Nishiyama, M., Suzuki, H., and Beppu, T. (1987). Nucleotide sequence of the streptothricin acetyltransferase gene from Streptomyces lavendulae and its expression in heterologous hosts. J. Bacteriol. 169, 1929–1937. doi: 10.1128/jb.169.5.1929-1937.1987
Hunter, M., Yuan, P., Vavilala, D., and Fox, M. (2019). Optimization of protein expression in mammalian cells. Curr. Protoc. Protein Sci. 95:e77. doi: 10.1002/cpps.77
Hurtubise, Y., Shareck, F., Kluepfel, D., and Morosoli, R. (1995). A cellulase/xylanase-negative mutant of Streptomyces lividans 1326 defective in cellobiose and xylobiose uptake is mutated in a gene encoding a protein homologous to ATP-binding proteins. Mol. Microbiol. 17, 367–377. doi: 10.1111/j.1365-2958.1995.mmi_17020367.x
Isiegas, C., Parro, V., and Mellado, R. P. (1999). Streptomyces lividans as a host for the production and secretion of Escherichia coli TEM beta-lactamase. Lett. Appl. Microbiol. 28, 321–326. doi: 10.1046/j.1365-2672.1999.00519.x
Jørgensen, S., Skov, K. W., and Diderichsen, B. (1991). Cloning, sequence, and expression of a lipase gene from Pseudomonas cepacia: lipase production in heterologous hosts requires two Pseudomonas genes. J. Bacteriol. 173, 559–567. doi: 10.1128/jb.173.2.559-567.1991
Jung, E. D., Lao, G., Irwin, D., Barr, B. K., Benjamin, A., and Wilson, D. B. (1993). DNA sequences and expression in Streptomyces lividans of an exoglucanase gene and an endoglucanase gene from Thermomonospora fusca. Appl. Environ. Microbiol. 59, 3032–3043. doi: 10.1128/aem.59.9.3032-3043.1993
Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F., and Hopwood, D. A. (2000). Practical Streptomyces Genetics. Norwich: John Innes Foundation.
Kim, K., Choe, D., Lee, D. H., and Cho, B. K. (2020). Engineering biology to construct microbial chassis for the production of difficult-to-express proteins. Int. J. Mol. Sci. 21:990. doi: 10.3390/ijms21030990
Koller, K. P., and Riess, G. (1989). Heterologous expression of the alpha-amylase inhibitor gene cloned from an amplified genomic sequence of Streptomyces tendae. J. Bacteriol. 171, 4953–4957. doi: 10.1128/jb.171.9.4953-4957.1989
Lamb, F. I., Kingston, A. E., Estrada, I., and Colston, M. J. (1988). Heterologous expression of the 65-kilodalton antigen of Mycobacterium leprae and murine T-cell responses to the gene product. Infect. Immun. 56, 1237–1241. doi: 10.1128/iai.56.5.1237-1241.1988
Lammertyn, E., Desmyter, S., Schacht, S., Van Mellaert, L., and Anné, J. (1998). Influence of charge variation in the Streptomyces venezuelae alpha-amylase signal peptide on heterologous protein production by Streptomyces lividans. Appl. Microbiol. Biotechnol. 49, 424–430. doi: 10.1007/s002530051193
Lammertyn, E., Van Mellaert, L., Schacht, S., Dillen, C., Sablon, E., Van Broekhoven, A., et al. (1997). Evaluation of a novel subtilisin inhibitor gene and mutant derivatives for the expression and secretion of mouse tumor necrosis factor alpha by Streptomyces lividans. Appl. Environ. Microbiol. 63, 1808–1813. doi: 10.1128/aem.63.5.1808-1813.1997
Lara, M., Servín-González, L., Singh, M., Moreno, C., Cohen, I., Nimtz, M., et al. (2004). Expression, secretion, and glycosylation of the 45- and 47-kDa glycoprotein of Mycobacterium tuberculosis in Streptomyces lividans. Appl. Environ. Microbiol. 70, 679–685. doi: 10.1128/aem.70.2.679-685.2004
Li, J. X., Zhao, L. M., Wu, R. J., Zheng, Z. J., and Zhang, R. J. (2013). High-level overproduction of Thermobifida enzyme in Streptomyces lividans using a novel expression vector. Int. J. Mol. Sci. 14, 18629–18639. doi: 10.3390/ijms140918629
Lichenstein, H., Brawner, M. E., Miles, L. M., Meyers, C. A., Young, P. R., and Simon, P. L. (1988). Secretion of interleukin-1 beta and Escherichia coli galactokinase by Streptomyces lividans. J. Bacteriol. 170, 3924–3929. doi: 10.1128/jb.170.9.3924-3929.1988
Lin, S. J., Hsieh, Y. F., Lai, L. A., Chao, M. L., and Chu, W. S. (2008). Characterization and large-scale production of recombinant Streptoverticillium platensis transglutaminase. J. Ind. Microbiol. Biotechnol. 35, 981–990. doi: 10.1007/s10295-008-0373-2
Lin, Y. S., Chao, M. L., Liu, C. H., and Chu, W. S. (2004). Cloning and expression of the transglutaminase gene from Streptoverticillium ladakanum in Streptomyces lividans. Process Biochem. 39, 591–598. doi: 10.1016/S0032-9592(03)00134-1
Lin, Y. S., Chao, M. L., Liu, C. H., Tseng, M., and Chu, W. S. (2006). Cloning of the gene coding for transglutaminase from Streptomyces platensis and its expression in Streptomyces lividans. Process Biochem. 41, 519–524. doi: 10.1016/j.procbio.2005.09.009
Liu, S., Wang, M., Du, G., and Chen, J. (2016). Improving the active expression of transglutaminase in Streptomyces lividans by promoter engineering and codon optimization. BMC Biotechnol. 16:75. doi: 10.1186/s12896-016-0304-7
Lule, I., Maldonado, B., D’Huys, P. J., Van Mellaert, L., Van Impe, J., Bernaerts, K., et al. (2012). On the influence of overexpression of phosphoenolpyruvate carboxykinase in Streptomyces lividans on growth and production of human tumor necrosis factor-alpha. Appl. Microbiol. Biotechnol. 96, 367–372. doi: 10.1007/s00253-012-4182-1
Marcone, G. L., Beltrametti, F., Binda, E., Carrano, L., Foulston, L., Hesketh, A., et al. (2010a). Novel mechanism of glycopeptide resistance in the A40926 producer Nonomuraea sp. ATCC 39727. Antimicrob. Agents Chemother. 54, 2465–2472. doi: 10.1128/aac.00106-10
Marcone, G. L., Binda, E., Carrano, L., Bibb, M., and Marinelli, F. (2014). Relationship between glycopeptide production and resistance in the actinomycete Nonomuraea sp. ATCC 39727. Antimicrob. Agents Chemother. 58, 5191–5201. doi: 10.1128/aac.02626-14
Marcone, G. L., Carrano, L., Marinelli, F., and Beltrametti, F. (2010b). Protoplast preparation and reversion to the normal filamentous growth in antibiotic-producing uncommon actinomycetes. J. Antibiot. 63, 83–88. doi: 10.1038/ja.2009.127
Marcone, G. L., Foulston, L., Binda, E., Marinelli, F., Bibb, M., and Beltrametti, F. (2010c). Methods for the genetic manipulation of Nonomuraea sp. ATCC 39727. J. Ind. Microbiol. Biotechnol. 37, 1097–1103. doi: 10.1007/s10295-010-0807-5
Marinelli, F., and Marcone, G. L. (2011). “Small molecules - microbial secondary metabolites,” in Comprehensive Biotechnology, Second Edn, ed. M.-Y. Murray (Amsterdam: Elsevier), 285–297. doi: 10.1016/b978-0-08-088504-9.00539-0
Meilleur, C., Hupé, J. F., Juteau, P., and Shareck, F. (2009). Isolation and characterization of a new alkali-thermostable lipase cloned from a metagenome library. J. Ind. Microbiol. Biotechnol. 36, 853–861. doi: 10.1007/s10295-009-0562-7
Merkens, H., Sielker, S., Rose, K., and Fetzner, S. (2007). A new monocupin quercetinase of Streptomyces sp. FLA: identification and heterologous expression of the queD gene and activity of the recombinant enzyme towards different flavonols. Arch. Microbiol. 187, 475–487. doi: 10.1007/s00203-007-0215-z
Motamedi, H., Shafiee, A., Cai, S. J., Streicher, S. L., Arison, B. H., and Miller, R. R. (1996). Characterization of methyltransferase and hydroxylase genes involved in the biosynthesis of the immunosuppressants FK506 and FK520. J. Bacteriol. 178, 5243–5248. doi: 10.1128/jb.178.17.5243-4248.1996
Muhamadali, H., Xu, Y., Ellis, D. I., Trivedi, D. K., Rattray, N. G. W., Bernaerts, K., et al. (2015). Metabolomics investigation of recombinant mTNFα production in Streptomyces lividans. Microb. Cell Fact. 14:157. doi: 10.1186/s12934-015-0350-1
Myronovskyi, M., Rosenkranzer, B., Nadmid, S., Pujic, P., Normand, P., and Luzhetskyy, A. (2018). Generation of a cluster-free Streptomyces albus chassis strains for improved heterologous expression of secondary metabolite clusters. Metab. Eng. 49, 316–324. doi: 10.1016/j.ymben.2018.09.004
Nakazawa, Y., Sagane, Y., Sakurai, S., Uchino, M., Sato, H., and Toeda, K. (2011). Large-scale production of phospholipase D from Streptomyces racemochromogenes and its application to soybean lecithin modification. Appl. Biochem. Biotechnol. 165, 1494–1506. doi: 10.1007/s12010-011-9370-4
Ndlovu, T. M., Ward, A. C., Glassey, J., Eskildsen, J., and Akay, G. (2015). Bioprocess intensification of antibiotic production by Streptomyces coelicolor A3(2) in micro-porous culture. Mater. Sci. Eng. C. 49, 799–806. doi: 10.1016/j.msec.2015.01.052
Nguyen-Thi, N., and Doucet, N. (2016). Combining chitinase C and N-acetylhexosaminidase from Streptomyces coelicolor A3(2) provides an efficient way to synthesize N-acetylglucosamine from crystalline chitin. J. Biotechnol. 220, 25–32. doi: 10.1016/j.jbiotec.2015.12.038
Noack, D., Geuther, R., Tonew, M., Breitling, R., and Behnke, D. (1988). Expression and secretion of interferon-alpha 1 by Streptomyces lividans: use of staphylokinase signals and amplification of a neo gene. Gene 68, 53–62. doi: 10.1016/0378-1119(88)90598-7
Noda, S., Ito, Y., Shimizu, N., Tanaka, T., Ogino, C., and Kondo, A. (2010). Over-production of various secretory-form proteins in Streptomyces lividans. Protein Expr. Purif. 73, 198–202. doi: 10.1016/j.pep.2010.05.011
Noguchi, Y., Kashiwagi, N., Uzura, A., Ogino, C., Kondo, A., Ikeda, H., et al. (2018). Development of a strictly regulated xylose-induced expression system in Streptomyces. Microb. Cell Fact. 17:151. doi: 10.1186/s12934-018-0991-y
Ogino, C., Kanemasu, M., Hayashi, Y., Kondo, A., Shimizu, N., and Tokuyama, S. (2004). Over-expression system for secretory phospholipase D by Streptomyces lividans. Appl. Microbiol. Biotechnol. 64, 823–828. doi: 10.1007/s00253-003-1552-8
Paradkar, A. S., Petrich, A. K., Leskiw, B. K., Aidoo, K. A., and Jensen, S. E. (1994). Transcriptional analysis and heterologous expression of the gene encoding beta-lactamase inhibitor protein (BLIP) from Streptomyces clavuligerus. Gene 144, 31–36. doi: 10.1016/0378-1119(94)90199-6
Park, H. U., and Lee, K. J. (1998). Cloning and heterologous expression of the gene for BLIP-II, a beta-lactamase-inhibitory protein from Streptomyces exfoliates SFM19. Microbiology 144, 2161–2167. doi: 10.1099/mic.0.26310-0
Pimienta, E., Ayala, J. C., Rodríguez, C., Ramos, A., Van Mellaert, L., Vallín, C., et al. (2007). Recombinant production of Streptococcus equisimilis streptokinase by Streptomyces lividans. Microb. Cell Fact. 6:20. doi: 10.1186/1475-2859-6-20
Pozidis, C., Lammertyn, E., Politou, A. S., Anné, J., Tsiftsoglou, A. S., Sianidis, G., et al. (2001). Protein secretion biotechnology using Streptomyces lividans: large-scale production of functional trimeric tumor necrosis factor alpha. Biotechnol. Bioeng. 72, 611–619. doi: 10.1002/1097-0290(20010320)72:6<611::aid-bit1026>3.0.co;2-0
Qi, X., Jiang, R., Yao, C., Zhang, R., and Li, Y. (2008). Expression, purification, and characterization of C-terminal amidated glucagon in Streptomyces lividans. J. Microbiol. Biotechnol. 18, 1076–1080.
Rose, K., Tenberge, K. B., and Steinbüchel, A. (2005). Identification and characterization of genes from Streptomyces sp. strain K30 responsible for clear zone formation on natural rubber latex and poly(cis-1,4-isoprene) rubber degradation. Biomacromolecules 6, 180–188. doi: 10.1021/bm0496110
Schaerlaekens, K., Lammertyn, E., Geukens, N., De Keersmaeker, S., Anné, J., and Van Mellaert, L. (2004). Comparison of the Sec and Tat secretory pathways for heterologous protein production by Streptomyces lividans. J. Biotechnol. 112, 279–288. doi: 10.1016/j.jbiotec.2004.05.004
Sevillano, L., Díaz, M., and Santamaría, R. I. (2013). Stable expression plasmids for Streptomyces based on a toxin-antitoxin system. Microb. Cell Fact. 12:39. doi: 10.1186/1475-2859-12-39
Sevillano, L., Díaz, M., and Santamaría, R. I. (2017). Development of an antibiotic marker-free platform for heterologous protein production in Streptomyces. Microb. Cell Fact. 16:164. doi: 10.1186/s12934-017-0781-y
Sevillano, L., Vijgenboom, E., van Wezel, G. P., Díaz, M., and Santamaría, R. I. (2016). New approaches to achieve high level enzyme production in Streptomyces lividans. Microb. Cell Fact. 15:28. doi: 10.1186/s12934-016-0425-7
Sianidis, G., Pozidis, C., Becker, F., Vrancken, K., Sjoeholm, C., Karamanou, S., et al. (2006). Functional large-scale production of a novel Jonesia sp. xyloglucanase by heterologous secretion from Streptomyces lividans. J. Biotechnol. 121, 498–507. doi: 10.1016/j.jbiotec.2005.08.002
Sinsereekul, N., Wangkam, T., Thamchaipenet, A., Srikhirin, T., and Eurwilaichitr, L. (2010). Recombinant expression of BTA hydrolase in Streptomyces rimosus and catalytic analysis on polyesters by surface plasmon resonance. Appl. Microbiol. Biotechonol. 86, 1775–1784. doi: 10.1007/s00253-010-2465-y
Smith, T. J., Lloyd, J. S., Gallagher, S. C., Fosdike, W. L., Murrell, J. C., and Dalton, H. (1999). Heterologous expression of alkene monooxygenase from Rhodococcus rhodochrous B-276. Eur. J. Biochem. 260, 446–452. doi: 10.1046/j.1432-1327.1999.00179.x
Šnajder, M., Carrillo Rincón, A. F., Magdevska, V., Bahun, M., Kranjc, L., Paš, M., et al. (2019). Extracellular production of the engineered thermostable protease pernisine from Aeropyrum pernix K1 in Streptomyces rimosus. Microb. Cell Fact. 18:196. doi: 10.1186/s12934-019-1245-3
Swan, D. G., Cortes, J., Hale, R. S., and Leadlay, P. F. (1989). Cloning, characterization, and heterologous expression of the Saccharopolyspora erythraea (Streptomyces erythraeus) gene encoding an EF-hand calcium-binding protein. J. Bacteriol. 171, 5614–5619. doi: 10.1128/jb.171.10.5614-5619.1989
Taguchi, S., Maeno, M., and Momose, H. (1992). Extracellular production system of heterologous peptide driven by a secretory protease inhibitor of Streptomyces. Appl. Microbiol. Biotechnol. 36, 749–753. doi: 10.1007/BF00172187
Taguchi, S., Misawa, S., Yoshida, Y., and Momose, H. (1995). Microbial secretion of biologically active human transforming growth factor α fused to the Streptomyces protease inhibitor. Gene 159, 239–243. doi: 10.1016/0378.1119(95)00107-H
Tao, X., Zhao, M., Zhang, Y., Liu, M., Liu, Q., Wang, W., et al. (2019). Comparison of the expression of phospholipase D from Streptomyces halstedii in different hosts and its over-expression in Streptomyces lividans. FEMS Microbiol. Lett. 366:fnz051. doi: 10.1093/femsle/fnz051
Torres-Bacete, J., Hormigo, D., Torres-Gúzman, R., Arroyo, M., Castillón, M. P., García, L., et al. (2015). Overexpression of penicillin V acylase from Streptomyces lavendulae and elucidation of its catalytic residues. Appl. Environ. Microbiol. 81, 1225–1233. doi: 10.1128/AEM.02352-14
Tremblay, D., Lemay, J., Gilbert, M., Chapdelaine, Y., Dupont, C., and Morosoli, R. (2002). High-level heterologous expression and secretion in Streptomyces lividans of two major antigenic proteins from Mycobacterium tuberculosis. Can. J. Microbiol. 48, 43–48. doi: 10.1139/w01-133
Tripathi, N. K., and Shrivastava, A. (2019). Recent developments in bioprocessing of recombinant proteins: expression hosts and process development. Front. Bioeng. Biotechnol. 7:420. doi: 10.3389/fbioe.2019.00420
Trono, D. (2019). “Chapter 13. Recombinant enzymes in the food and pharmaceutical industries,” in Advances in Enzyme Technology, ed. A. Pandey (Amsterdam: Elsevier), 349–387. doi: 10.1016/B978-0-444-64114-4.00013-3
Ueda, Y., Tsumoto, K., Watanabe, K., and Kumagai, I. (1993). Synthesis and expression of a DNA encoding the Fv domain of an anti-lysozyme monoclonal antibody. HyHEL10, in Streptomyces lividans. Gene 129, 129–134. doi: 10.1016/0378-1119(93)90708-B
Vallin, C., Ramos, A., Pimienta, E., Rodríguez, C., Hernández, T., Hernández, I., et al. (2006). Streptomyces as host for recombinant production of Mycobacterium tuberculosis proteins. Tuberculosis 86, 198–202. doi: 10.1016/j.tube.2006.02.003
Valverde, J. R., Gullón, S., and Mellado, R. P. (2018). Modelling the metabolism of protein secretion through the Tat route in Streptomyces lividans. BMC Microbiol. 18:59. doi: 10.1186/s12866-018-1199-3
Van Mellaert, L., Lammertyn, E., Schacht, S., Proost, P., Van Damme, J., Wroblowski, B., et al. (1998). Molecular characterization of a novel subtilisin inhibitor protein produced by Streptomyces venezuelae CBS762.70. DNA Seq. 9, 19–30. doi: 10.3109/10425179809050021
Vieira Gomes, A. M., Souza Carmo, T., Silva Carvalho, L., Mendonça Bahia, F., and Parachin, N. S. (2018). Comparison of yeasts as hosts for recombinant protein production. Microorganisms 6:38. doi: 10.3390/microorganisms6020038
Vrancken, K., and Anné, J. (2009). Secretory production of recombinant proteins by Streptomyces. Future Microbiol. 4, 181–188. doi: 10.2217/17460913.4.2.181
Vrancken, K., De Keersmaeker, S., Geukens, N., Lammertyn, E., Anné, J., and Van Mallaert, L. (2007). pspA overexpression in Streptomyces lividans improves both Sec- and Tat-dependent protein secretion. Appl. Microbiol. Biotechnol. 73, 1150–1157. doi: 10.1007/s00253-006-0571-7
Washizu, K., Ando, K., Koikeda, S., Hirose, S., Matsuura, A., Takagi, H., et al. (1994). Molecular-cloning of the gene for microbial transglutaminase from Streptoverticillium and its expression in Streptomyces lividans. Biosci. Biotechnol. Biochem. 58, 82–87. doi: 10.1271/bbb.58.82
Wolfframm, C., Lingens, F., Mutzel, R., and van Pée, K. H. (1993). Chloroperoxidase-encoding gene from Pseudomonas pyrrocinia: sequence, expression in heterologous hosts, and purification of the enzyme. Gene 130, 131–135. doi: 10.1016/0378-1119(93)90356-8
Zhang, Y., Wang, W. C., and Li, Y. (2004). Cloning, expression, and purification of soluble human interleukin-4 receptor in Streptomyces. Protein Expr. Purif. 36, 139–145. doi: 10.1016/j.pep.2004.04.010
Keywords: streptomycetes, recombinant proteins, heterologous expression, industrial enzymes, therapeutic proteins
Citation: Berini F, Marinelli F and Binda E (2020) Streptomycetes: Attractive Hosts for Recombinant Protein Production. Front. Microbiol. 11:1958. doi: 10.3389/fmicb.2020.01958
Received: 27 May 2020; Accepted: 24 July 2020;
Published: 20 August 2020.
Edited by:
Sara Borin, University of Milan, ItalyReviewed by:
Aleš Berlec, Institut “Jožef Stefan” (IJS), SloveniaMingtao Huang, South China University of Technology, China
Copyright © 2020 Berini, Marinelli and Binda. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Francesca Berini, Zi5iZXJpbmlAdW5pbnN1YnJpYS5pdA==
 Francesca Berini
Francesca Berini Flavia Marinelli
Flavia Marinelli Elisa Binda
Elisa Binda