- 1Plant and Environmental Protection Sciences, University of Hawai‘i at Mānoa, Honolulu, HI, United States
- 2Istituto per la Protezione Sostenibile delle Piante, Consiglio Nazionale delle Ricerche, Bari, Italy
Based on high-throughput sequencing (HTS) data, the existence of viroid-like RNAs (Vd-LRNAs) associated with fig trees grown in the Hawaiian Islands has been predicted. One of these RNAs has been characterized as a circular RNA ranging in size from 357 to 360 nucleotides. Structural and biochemical features of this RNA, tentatively named fig hammerhead viroid-like RNA (FHVd-LR), markedly resemble those previously reported for several viroids and viroid-like satellite RNAs (Vd-LsatRNAs), which are non-protein-coding RNAs infecting their hosts autonomously and in combination with a helper virus, respectively. The full-length sequence of FHVd-LR variants was determined by RT-PCR, cloning, and sequencing. Despite a low global sequence identity with known viroids and Vd-LsatRNAs, FHVd-LR contains a hammerhead ribozyme (HRz) in each polarity strand. Northern blot hybridization assays identified the circular and linear forms of both polarity strands of FHVd-LR and showed that one strand, assigned the (+) polarity, accumulates at higher levels than the (−) polarity strand in vivo. The (+) polarity RNA assumes a rod-like secondary structure of minimal free energy with the conserved domains of the HRzs located in opposition to each other, a feature typical of several viroids and Vd-LRNAs. The HRzs of both FHVd-LR polarity strands were shown to be active in vitro during transcription, self-cleaving the RNAs at the predicted sites. These data, together with the sequence variability observed in the cloned and sequenced full-length variants, indicate that FHVd-LR is a novel viroid or Vd-LsatRNA. According to HTS data, the coexistence of FHVd-LR of different sizes in the same host cannot be excluded. The relationships of FHVd-LR with previously reported viroids and Vd-LsatRNAs, and the need to perform bioassays to conclusively clarify the biological nature of this circular RNA, are discussed.
Introduction
Viroids are small, infectious, non-protein-coding circular RNAs, so far identified only in plants (Kovalskaya and Hammond, 2014; Flores et al., 2015; Gago-Zachert, 2016). Other RNAs resembling viroids from a structural point of view, but differing from them at the biological level, or not conclusively characterized in this respect, have also been reported from plants and designated as viroid-like RNAs (Vd-LRNAs). Viroid-like satellite RNAs (Vd-LsatRNAs) and retroviroid-like elements are Vd-LRNAs markedly diverging from viroids at the biological level. While viroids replicate and systemically infect their host plants autonomously (in the absence of any helper virus), the infectivity of Vd-LsatRNAs depends on a coinfecting helper virus (Navarro et al., 2017), Vd-LsatRNAs are encapsidated by the helper virus capsid proteins and have been also named virusoids (Symons and Randles, 1999). Both viroids and Vd-LsatRNAs differ from carnation small Vd-LRNA, which is non-infectious and has a DNA counterpart integrated in the genome of a plant pararetrovirus (Daròs and Flores, 1995; Vera et al., 2000) or in the plant genome (Hegedus et al., 2004). For this reason, carnation small Vd-LRNA is considered a retroviroid-like element.
Viroids replicating in the nucleus and the chloroplast have been classified into the families Pospiviroidae and Avsunviroidae, respectively (Di Serio et al., 2014). Members of the two families also differ in structural and other functional features. In viroids of the family Pospiviroidae, one RNA polarity strand generates circular forms that assume a rod-like or quasi-rod-like secondary structure of minimal free energy containing a central conserved region (CCR) and other conserved motifs. The CCR is involved in the replication of nuclear viroids through an asymmetric rolling-circle mechanism (Flores et al., 2014). Viroids of the family Avsunviroidae lack the CCR and other typical structural motifs conserved in nuclear viroids. Instead, they assume rod-like, quasi-rod-like, or branched conformation and contain hammerhead ribozymes (HRzs) (Di Serio et al., 2018b). HRzs are inactive in the most stable viroid RNA conformation, but they assume an active structure responsible for RNA self-cleavage (without the catalytic contribution of any protein) during replication (Hutchins et al., 1986). In contrast to nuclear viroids, members of the family Avsunviroidae replicate through a symmetric rolling-circle mechanism, in which self-cleaved oligomeric RNAs of both polarity strands are circularized (Flores et al., 2014). HRzs with similar functional roles during replication also have been identified in several Vd-LsatRNAs (Navarro et al., 2017). However, in most of them the HRz is present in only one polarity, with the other polarity containing a different ribozyme (named paperclip) or no ribozyme at all. In accord with the presence of ribozymes in one or both polarity strands, asymmetric or symmetric rolling-circle replication mechanisms have been proposed for Vd-LsatRNAs, respectively (Navarro et al., 2017). Among Vd-LsatRNAs, only those of lucerne transient streak virus (LTSV, genus Sobemovirus), cereal yellow dwarf virus-RPV (genus Polerovirus), and two cherry small circular viroid-like RNAs (cscRNAs, likely associated with a mycovirus) contain two HRzs in both polarity strands (Di Serio et al., 1997, 2006; Navarro et al., 2017).
In tissues infected by viroids or Vd-LsatRNAs, the two polarity strands of the infectious RNAs are detectable, but generally one strand accumulates at a higher level and, by convention, is designated the (+) strand. Akin to viruses, viroids and Vd-LsatRNAs have the typical features of quasispecies (Codoñer et al., 2006; Flores et al., 2014), which is consistent with the observation that these infectious agents accumulate in their hosts as populations of closely related sequence variants, among which one (the master sequence) or a few may prevail depending on the selection pressures imposed by the host and environment (Di Serio et al., 2017).
Edible fig (Ficus carica L.) trees are known to be natural hosts of several viruses (Elbeaino et al., 2010, 2012; Laney et al., 2012; Ale-Agha and Rakhshandehroo, 2014; Minafra et al., 2017). Viroids of the family Pospiviroidae, including hop stunt viroid, citrus exocortis viroid, and a viroid resembling apple dimple fruit viroid, have also been reported in fig trees (Yakoubi et al., 2007; Chiumenti et al., 2014). The latter viroid was initially identified in fig by high-throughput sequencing (HTS), a powerful technology that has been applied to investigate the virome of several plant species (Hadidi et al., 2016; Villamor et al., 2019). Starting with HTS data, we report the identification of Vd-LRNAs containing HRzs in fig. One of these RNAs has been molecularly characterized, and its relationships with previously reported viroids and Vd-LsatRNAs are discussed.
Materials and Methods
Plant Material, RNA Isolation, and HTS
A fig plant from a commercial nursery on the island of Kauai, Hawaii, displaying symptoms of severe mosaic and leaf distortion, was analyzed by HTS. Nucleic acid preparations enriched in double-stranded RNAs (dsRNAs) were obtained from root tissue as described in Navarro and Di Serio (2018) and used as a source for generating a random-amplified cDNA library (Melzer et al., 2010). Sequencing of the cDNA library was performed using a 454 GS FLX Titanium platform (Roche, Branford, CT, United States) at the University of Hawaii’s Advanced Studies in Genomics, Proteomics and Bioinformatics (ASGPB) laboratory.
In 2018 and 2019, samples composed of mixed leaves, petioles, and green bark from twelve fig trees displaying symptoms resembling those of the Kauai fig sample were collected from three locations on the island of Oahu (Hawaii). Total nucleic acids were extracted as described by Li et al. (2008) and tested by RT-PCR (see below). Nucleic acids enriched in highly structured RNAs were obtained from young leaf, bark, and petiole tissues of the fig trees with buffer-saturated phenol and partitioning the nucleic acids by chromatography on non-ionic cellulose CF-11 (Whatman, Maidstone, United Kigdom) as described previously (Pallás et al., 1987).
Bioinformatic Analysis
Analyses of HTS data were performed as described earlier (Olmedo-Velarde et al., 2019). Briefly, after trimming, quality control, and de novo assembly using Trinity (Grabherr et al., 2011), Velvet (Zerbino and Birney, 2008), and Unicycler (Wick et al., 2017), all the assembled contigs were sorted by length. Duplicate contigs and those <100 nucleotides (nt) were discarded using Geneious v.10.1.31. The remaining contigs were screened for viral and viroid sequence homology using BlastX and BlastN, respectively2. Alignments of HTS reads with a sequence reference and reassembling of selected reads were performed using Bowtie (Langmead et al., 2009) and Phrap (Machado et al., 2011), respectively, and then implemented by the MacVector Assembler platform (17.5.3, MacVector, Inc., Apex, NC, United States). Multiple alignments of nucleotide sequences were performed using Clustal Omega (Sievers et al., 2011). RNAfold software (Lorenz et al., 2011) was used to predict the secondary structure of minimal free energy of the RNAs.
RT-PCR and Cloning
Total nucleic acids were reverse transcribed using random hexamers and Superscript IV reverse transcriptase (Invitrogen, Thermo Fisher Scientific, Waltham, MA, United States), following the manufacturer’s instructions. Two microliters of the cDNA reaction served as template for PCR amplification using 0.4 units of Phusion High-Fidelity DNA polymerase (Thermo Fisher Scientific, Waltham, MA, United States) in a 20-μl mixture containing 1 × reaction buffer, 0.25 mM of dNTPs, and 0.5 μM of two FHVd-LR-specific primers (Table 1) and using the following cycling conditions: initial denaturation at 98°C for 30 s, followed by 33 cycles at 98°C for 15 s, 59°C for 15 s, 72°C for 15 s, and a final extension step at 72°C for 7 min. After agarose gel purification, an adenine-residue overhang was added at the 5′ end of the amplicons using GoTaq DNA polymerase (Promega, Madison, WI, United States). Amplification products of the expected size (monomeric and dimeric) were then cloned into a pGEMT-Easy vector (Promega, Madison, WI, United States) and sequenced by Sanger Sequencing Custom Service (Macrogen, Amsterdam, Netherlands).
Northern Blot Hybridization
Nucleic acid preparations enriched in highly structured RNAs were subjected to double polyacrylamide gel electrophoresis (PAGE) (Flores et al., 1985). Briefly, the nucleic acids were separated by two consecutive 5% PAGE, the first under non-denaturing conditions (TAE buffer: 40 mM Tris, 20 mM sodium acetate, 1 mM EDTA, pH 7.2) and the second under denaturing conditions (8 M urea and 0.25 × TBE buffer). After staining the second gel with ethidium bromide, the nucleic acids were electroblotted to a nylon membrane (Hybond-N, Amersham, Little Chalfont, United Kingdom) in 0.5 × TBE buffer and immobilized by UV cross-linking. The membranes were then hybridized with DIG-labeled riboprobes complementary to (+) or (−) polarity strands of FHVd-LR as described previously (Hajizadeh et al., 2012). The hybridization signals were revealed with an anti-DIG alkaline phosphatase conjugate and the chemiluminescence substrate CSPD (Roche Applied Science) and visualized with a ChemiDoc Touch Imaging system (Bio-Rad, Hercules, CA, United States). The riboprobes were generated by in vitro transcription from linearized plasmids containing the full-length cDNA of FHVd-VL (see below) using a commercial Dig-labeling kit (Roche Diagnostics GmbH, Germany).
Analyses of RNA Self-Cleavage
Monomeric and dimeric transcripts of both polarity strands were obtained by in vitro transcription of plasmids containing monomeric and head-to-tail dimeric FHVd-LR cDNA sequences in both orientations. Recombinant plasmids were linearized by digestion with the appropriate restriction enzyme (SalI or NcoI) and, after phenol–chloroform extraction and ethanol precipitation, used as a template for the in vitro transcription with T7 or SP6 RNA polymerase (Forster et al., 1990). The transcription reactions containing the primary transcripts and their self-cleavage products were separated in 5% PAGE containing 8 M urea and 1 × TBE (89 mM Tris, 89 mM boric acid, 2.5 mM EDTA, pH 8.3), stained with ethidium bromide and UV visualized.
The 5′ terminal sequence of the 3′ self-cleavage product of the FHVd-LR monomeric transcripts of both polarity strands was determined by 5′ RACE experiments. Briefly, 3′ RNA fragments were eluted from the denaturing 5% PAGE by phenol–chloroform extraction and ethanol precipitation. The eluted RNA was reverse transcribed (as described above) using FHVd-4R and FHVd-5F (see Table 1) for the (+) and (−) polarity RNA fragment, respectively. Following the addition of a poly(dG) tail, the tailed cDNAs were PCR amplified using GoTaq DNA polymerase (Promega, Madison, WI, United States) with a 5′ RACE primer (Table 1) and the same primer used for the cDNA synthesis. PCR amplicons were gel-purified, cloned, and sequenced as reported above.
Results
A cDNA library was generated from dsRNAs extracted from a fig tree with symptoms of mosaic and leaf distortion, grown on Kauai, Hawaii. HTS of such a library produced 262,700 reads with an average size of approximately 500 base pair (bp), which were filtered for quality and de novo assembled, generating 1,183 contigs (size 100 to 17,165 nt). BlastX and BlastN searches against the viral database revealed contigs with significant sequence identity to several viruses, including badnaviruses, closteroviruses, emaraviruses, endornaviruses, totiviruses, trichoviruses, and umbraviruses (Olmedo-Velarde et al., unpublished data). Moreover, BlastN analysis revealed two contigs of 462 and 674 nt that shared sequence similarity with some viroids and Vd-LRNAs. In particular, a short region of 40–50 nt of both contigs shared 84–87% sequence identity with HRzs contained in eggplant latent viroid (Fadda et al., 2003), grapevine hammerhead Vd-LRNA (Wu et al., 2012), and cscRNAs (Di Serio et al., 1997, 2006). Interestingly, each contig consisted of a partial direct repeat of a monomeric sequence of 358 and 390 nt, respectively (Supplementary Figure S1), thus suggesting a possible multimeric or circular nature of the corresponding RNAs. The monomeric sequences of the two contigs shared about 84% sequence identity with each other. More specifically, they shared almost identical sequences spanning a region of 260 nt, while the remaining part of the molecules (about 120 nt) largely diverged (Supplementary Figure S2). Despite the sequence diversity, the secondary structure of lowest free energy of both RNAs was of the rod-like class, with the largely divergent sequences located in the terminal right regions (Supplementary Figure S2). The two RNAs contained HRzs in both polarity strands (see below), thus displaying typical structural features previously reported for members of the family Avsunviroidae and in some Vd-LsatRNAs (Di Serio et al., 2018b; Navarro et al., 2017). Although these data strongly suggested the possible existence of at least two novel Vd-LRNAs in fig, it was not possible to confirm these preliminary results because the original fig source from Kauai was destroyed. In an attempt to identify additional isolates containing similar Vd-LRNAs, specific primers were designed to test other fig trees by RT-PCR.
Identification of a Novel Fig Viroid-Like RNA on the Island of Oahu
An RT-PCR-based preliminary survey was performed on the island of Oahu, Hawaii, using primers FHVd-1F and FHVd-2R (Table 1) derived from two adjacent regions of a sequence common to the two contigs and, therefore, expected to amplify full-length cDNAs of the potential circular RNAs (Candresse et al., 1998). Three out of the twelve fig trees examined, all grown in the same location on Oahu, generated amplification products of about 360 bp. Cloning and sequencing of the amplicons showed sequence variants ranging in size from 357 to 359 nt, which differed from each other in a few positions (Figure 1), The conserved nucleotides reported in almost all natural HRzs were found in both polarity strands of the cloned RNA variants (Figure 1), and BlastN searches confirmed that only the short-sequence fragment corresponding to the ribozyme domain matched with some members of the family Avsunviroidae and with csc-RNAs. Importantly, the sequenced variants from Oahu and the short Vd-LRNA from the fig tree on the island of Kauai shared high identity (96%), with only 14 polymorphic positions observed in a pairwise alignment between them (Supplementary Figure S3). Altogether, these data suggested that the RNA identified from Oahu was closely related to those found by HTS in the Kauai sample and could represent a novel Vd-LRNA, hereafter named fig hammerhead viroid-like RNA (FHVd-LR).

Figure 1. Multiple-sequence alignment of full-length cDNAs of fig hammerhead viroid-like RNA (FHVd-LR) variants amplified with primer pairs FHVd-1F/2R (in red), FHVd-3F/4R (in blue), and FHVd-5F/6R (in green). Names of the sequence variants and primers (horizontal arrows) used to amplify the corresponding cDNAs are reported with the same colors; the numbers of clones containing the same variant are indicated in brackets. The reference variant (FHVd-LR_A1) reported at the top is identical to the variant FHVd-LR_B6 and, being the most frequently sequenced (found in 13 independent clones), also corresponds to the master sequence in the population. Nucleotide identity and gaps with respect to the FHVd-LR_A1 variant are indicated by dots and dashes, respectively. The light blue background highlights mutations. Regions involved in the formation of (+) and (−) hammerhead structures are delimited by flags; arrows and bars indicate the self-cleavage sites and the nucleotides conserved in most natural hammerhead structures, respectively; filled (black) and open symbols refer to (+) and (−) polarity, respectively. Nucleotide positions in the multiple alignments are reported on the left.
Primary and Proposed Secondary Structure of FHVd-LR
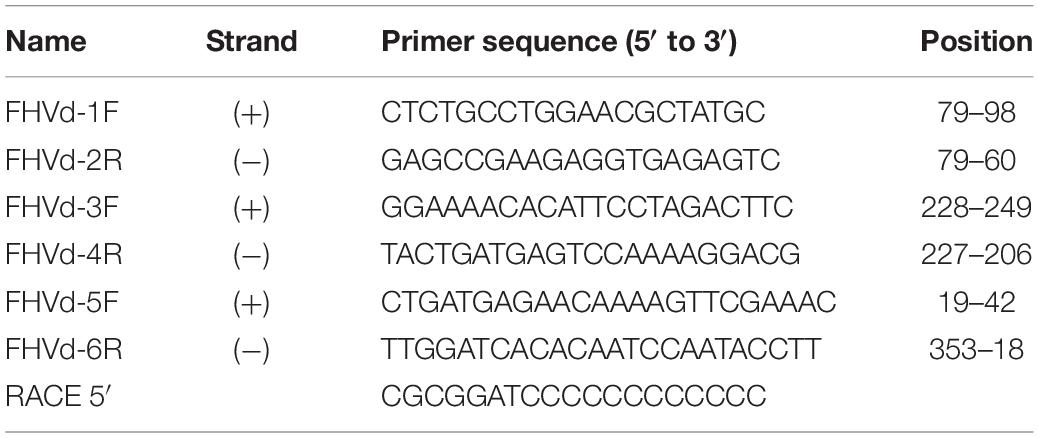
To further assess the nucleotide sequence composition in the region covered by the first primer set and the possible circularity of the RNA, full-length cDNAs were also amplified and cloned using two additional pairs of adjacent primers (FHVd-3F/4R and FHVd-5F/6R, Table 1). Sequencing of 10 and 8 clones of the RT-PCR amplicons generated with the two respective primer sets revealed variants ranging in size from 356 to 360 nt. They showed high sequence identity (97.8–100%) between them and to those obtained previously using the primer set FHVd-1F/2R. A total of nine different sequence variants were annotated in GenBank (with the accession numbers from MT57734 to MT577542). The most frequently sequenced variant was FHVd-LR_A1, which differed from the others in up to six positions and will be considered as the reference variant for this new Vd-LRNA (Figure 1). These results support the circular and quasispecies nature of the RNA. Multiple-sequence alignments also revealed the absence of nucleotide variability in the region covered by FHVd-1F/2R, while only two polymorphic positions were found in the regions targeted by the other two primer pairs (FHVd-3F/4R and FHVd-5F/6R). Interestingly, 9 out of 15 changes mapped in a region covering almost 100 nt, between positions 265 and 358 of the multiple sequence alignment (Figure 1). FHVd-LR was composed of 22.6, 28.2, 24.6, and 24.6% of A, U, C, and G, respectively. Therefore, its G + C content (49.2%) resembled that of most viroids (ranging from 52.2% to 61.6%) and Vd-LsatRNAs (ranging from 50.0 to 63.6%). However, the G + C content differed from avocado sunblotch viroid (ASBVd) (Symons, 1981), which is unique in its low G + C content of about 38%. The existence of a DNA counterpart of FHVd-LR, and therefore its potential retroviroid-like nature, was excluded based on the negative results of amplification by PCR without previous reverse transcription using nucleic acid preparations from a FHVd-LR-positive tree and different primer pairs (Figure 2).
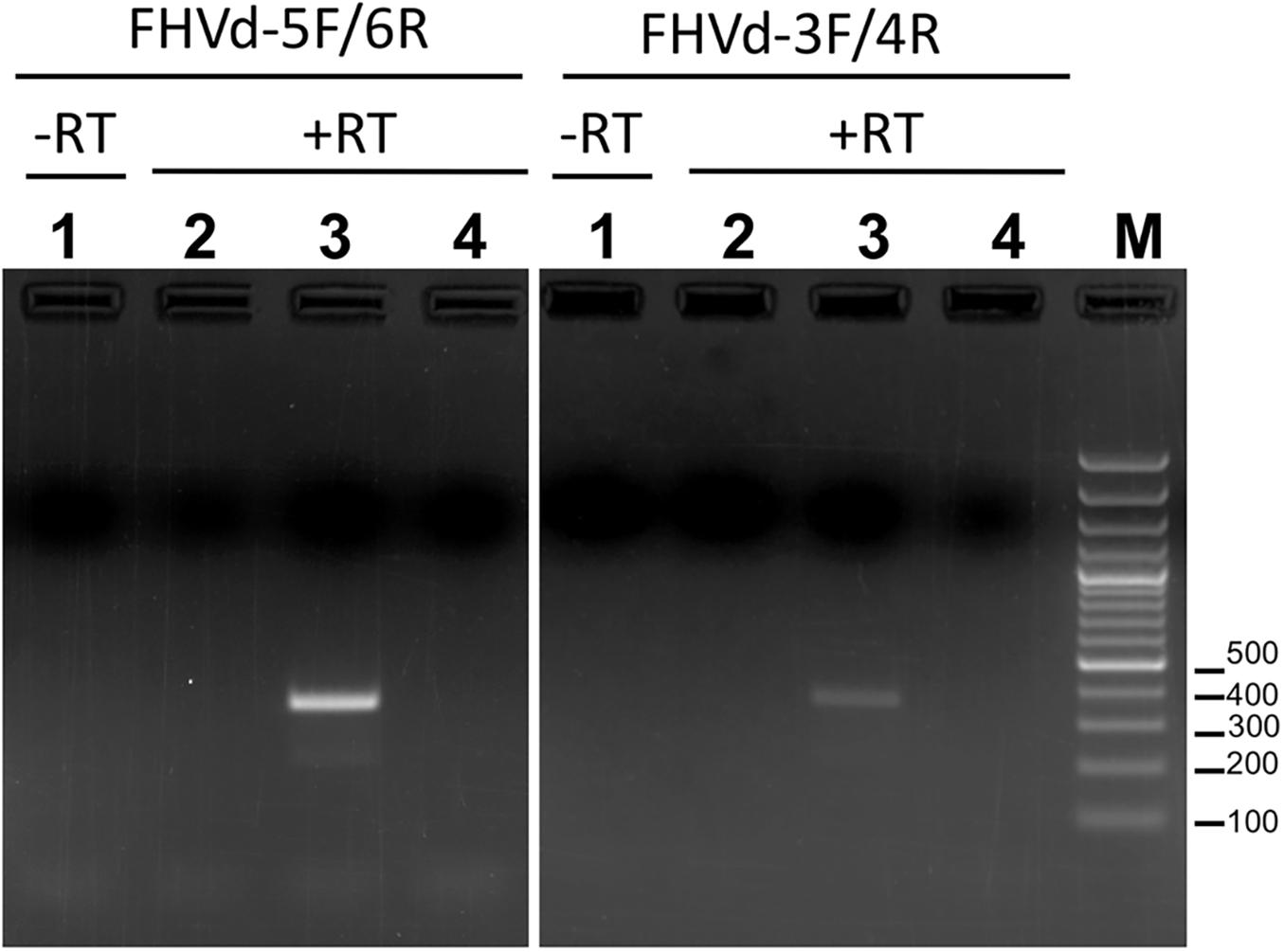
Figure 2. Amplification assays using RNA preparations from FHVd-LR-positive and -negative samples, and primer pairs FHVd-5F/6R (left) or FHVd-3F/4R (right). Lane 1, amplification was performed in the absence of reverse transcription (−RT) using an FHVd-LR-positive sample. Lanes 2 and 3, amplifications were performed after reverse transcription (+RT) using RNA preparations from FHVd-LR-negative or -positive samples, respectively. Lane 4 is a non-template control. The same FHVd-LR-positive sample was tested in lanes 1 and 3.
When the secondary structure of minimal free energy was calculated, FHVd-LR variants assumed a rod-like conformation with about 70% paired residues (Figure 3). In such a structure, the conserved sequences in the natural HRzs were located in a central region and opposite to each other (Figure 3), as previously observed in most members of the family Avsunviroidae and several Vd-LsatRNAs. The proposed rod-like secondary structure of FHVd-LR was also supported by most of the heterogeneity found in the sequenced variants consisting of changes in loops or compensatory mutations, which did not result in major modifications of the proposed conformation (Figure 3). It is interesting that the nucleotide changes were asymmetrically distributed in the proposed structure, with most of them mapping at the terminal right domain of the molecule and covering about 120 nt.
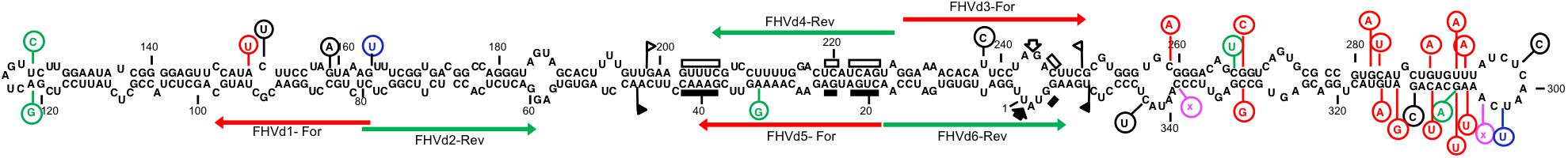
Figure 3. Primary and predicted secondary structures of lowest free energy of the fig hammerhead viroid-like RNA variant A1 (FHVd-LR_A1) collected from the island of Oahu. Regions involved in the formation of (+) and (−) hammerhead structures are delimited by flags; arrows and bars indicate the self-cleavage sites and the nucleotides conserved in most natural hammerhead structures, respectively; filled (black) and open symbols refer to (+) and (−) polarity, respectively. Mutated positions in other FHVd-LR variants are indicated in circles, with compensatory mutations, insertions, and deletions reported in green, blue, and pink, respectively. Nucleotide changes with respect to the short VL-RNA from the island of Kauai are in red. Horizontal arrows delimit the regions targeted by primer pairs used in the amplification steps of the cloning and sequencing strategy.
As reported above, FHVd-LR and the short Vd-LRNA from Kauai shared high sequence identity. Interestingly, most nucleotide changes in the latter RNA mapped in the terminal right domain of the secondary structure proposed for FHVd-LR and were compensatory mutations or covariations preserving the rod-like conformation (Figure 3). These findings highlighted an additional structural parallelism between the variants from the Oahu samples, which we confirmed by cloning and sequencing, and the Vd-LRNAs from the Kauai isolate that were detected only in silico due to destruction of the original tree.
Circularity and Accumulation Levels of Both Polarity Strands of FHVd-LR
The presence of FHVd-LR in fig and evidence of its circularity were further inferred by northern blot hybridization. Nucleic acid preparations enriched in highly structured RNAs from fig trees that tested positive and negative in the RT-PCR assays were loaded side by side in a non-denaturing PAGE followed by a denaturing PAGE (double PAGE), a method specifically developed to separate circular from linear RNAs of the same size (Flores et al., 1985). Moreover, to test whether both polarity strands of FHVd-LR generated circular forms and to ascertain whether one of them accumulated at a higher level in vivo, equalized riboprobes specific for detecting each FHVd-LR polarity strand were used separately in parallel hybridization experiments. As expected, the circular and linear forms of FHVd-LR were only detected in the samples from the fig trees that previously tested positive by RT-PCR, thus confirming the association of this RNA with some fig plants only (Figure 4). Circular forms of both FHVd-LR polarity strands were detected. Based on the intensity of the hybridization signals, one polarity strand accumulated at higher level in vivo (Figure 4). Therefore, this strand, corresponding to the sequences in Figures 1, 2, was considered as the (+) FHVd-LR polarity. The possibility that one probe could cross-hybridize with the strand of the same polarity due to the self-complementarity of the FHVd-LR sequence was excluded by northern blot assays showing that this did not happen under the experimental conditions used (Supplementary Figure S4).
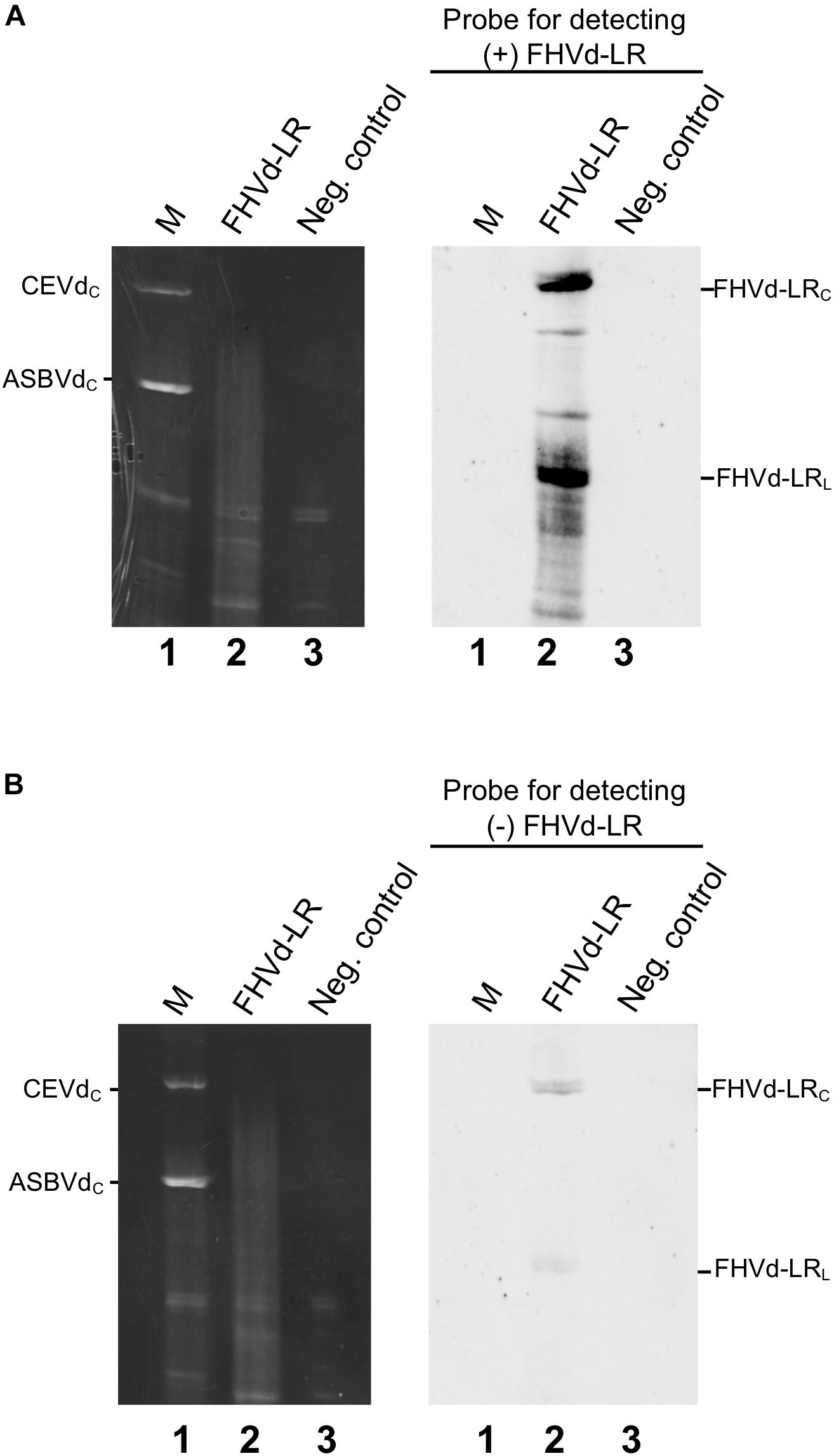
Figure 4. Northern blot hybridization assays to detect circular RNAs and identify the most abundant FHVd-LR strand denoted (+) polarity strand. Fig RNA preparations separated by double PAGE (left) were transferred to nylon membranes and hybridized with equalized full-length digoxigenin-labeled riboprobes for detecting the (+) and (−) FHVd-LR strands (A,B), respectively. The second (denaturing) gel of the double PAGE, stained with ethidium bromide, and the northern blot results are respectively reported on the left and on the right of each panel. Lane 1 corresponds to RNA preparations from Gynura aurantiaca and avocado infected by citrus exocortis viroid (CEVd, 371 nt) and avocado sunblotch viroid (ASBVd, 246 nt) respectively, used as molecular markers (M); lane 2 is an RNA preparation from a fig tree that tested positive to FHVd-LR by RT-PCR assay; lane 3 is an RNA preparation from FHVd-LR fig tree that tested negative to FHVd-LR and used as negative control. Identical aliquots of the same RNA preparations were loaded in the gels shown in panels (A,B). The positions of circular forms of CEVd and ASBVd (CEVdc and ASBVdc), visible in the gel stained with ethidium bromide, are reported on the left; the positions of circular and linear forms of FHVd-LR detected by the specific probes for the (+) and (−) polarity strands [panels (A,B), respectively] are indicated on the right. Bands between the circular and linear forms could correspond to artifacts generated by UV-induced cross-links during the visualization of the first non-denaturing gel of the double PAGE, as observed previously for some viroids (Hernández et al., 2006; Serra et al., 2018).
Hammerhead Ribozymes of FHVd-LR Are Active During Transcription
Fig hammerhead viroid-like RNA HRzs of both polarity strands were composed of three hairpins, two of them closed by short loops, which are located around the central core containing the predicted self-cleavage site (Figure 5A). In the (−) HRz, this site is preceded by a GUC trinucleotide, as in most HRzs of other viroids and Vd-LRNAs (Hutchins et al., 1986; Navarro et al., 2017). In contrast, a GUA trinucleotide was found at the same position in the (+) HRz of FHVd-LR, a situation previously reported only in the (−) HRz of the Vd-LsatRNAs of velvet tobacco mottle virus and LTSV (Forster et al., 1988; Collins et al., 1998) and in the (+) HRz of cscRNAs (Di Serio et al., 2006). The relevance in vivo of the FHVd-LR HRzs was confirmed by the limited sequence variability observed in these catalytic domains in the FHVd-LR variants (Figures 1, 3). Indeed, the single mutation detected in the (+) HRz mapped in the loop that closes the hairpin II, while that found in the (−) HRz was a compensatory mutation located in the stem II (Figure 5A). Therefore, both mutations preserved the typical hammerhead structures.
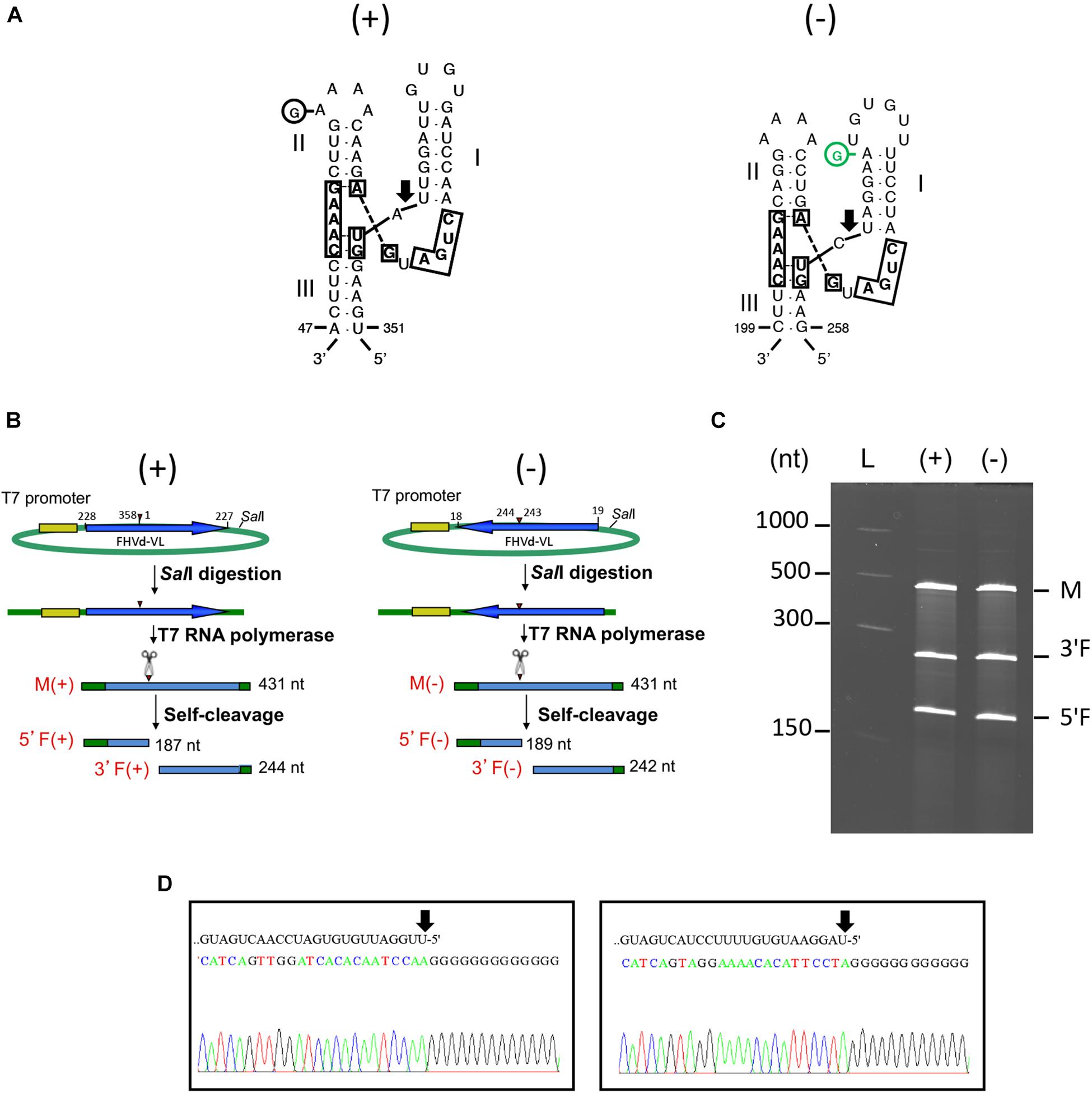
Figure 5. (A) Primary and Y-shaped secondary structure of hammerhead ribozymes (HRzs) of (+) and (−) FHVd-LR are presented considering the existing X-ray crystallography data on this class of ribozymes (Pley et al., 1994). Stems I and II are closed by loops. The nucleotides of the catalytic core conserved in most natural hammerhead structures are boxed. The cleavage site of each ribozyme is indicated by an arrow. Nucleotides in the (+) and (−) polarity are numerated considering their positions in the variant FHVd-LR_A1 (+). Mutations are indicated in circles, with the compensatory mutation reported in green. (B) Schematic representation of the DNA templates and monomeric RNA products generated by in vitro transcription. Plasmids containing the monomeric sequence of FHVd-LR in opposite orientations were linearized with SalI and transcribed with T7 RNA polymerase to produce monomeric transcripts (M) of opposite polarity and the respective 5′ and 3′ fragments (5′F and 3′F, respectively) derived from the HRz self-cleaving activity. In green, plasmid sequences; in yellow, polymerase promoter; in blue, FHVd-LR sequence, with the arrows indicating the (+) orientation; arrowheads and scissors mark the position of the self-cleavage sites. Numbers on the right of each RNA fragment indicate its expected size. (C) Analyses by PAGE of the in vitro transcription of the plasmids containing (+) and (−) monomeric FHVd-LR cDNA. L, RNA ladder with sizes indicated on the left; see panel (B) for M, 3′F and 5′F abbreviations. (D) Determination of self-cleavage site by 5′ RACE of 3′F fragment resulting from the HRz-mediated cleavage of FHVd-LR monomeric transcripts [generated as reported in panels (B,C)]. Sequencing electropherograms of 5′ RACE products of the (+) and (−) 3′F fragments are shown on the left and right, respectively, with the self-cleaved RNA sequence reported on the top and the 5′ terminal nucleotide indicated by the arrow.
In vitro transcription of recombinant plasmids containing monomeric and dimeric head-to-tail constructs of FHVd-LR indicated that HRzs of both polarity strands were active, generating RNA fragments of sizes consistent with those expected and in accord with the self-cleavage of transcripts at the predicted sites (Figures 5B,C and Supplementary Figure S5). Moreover, the self-cleavage site of both polarity strands was further confirmed by 5′ RACE experiments, showing that the 3′ fragments generated by the ribozymes in each polarity strand had the expected 5′ terminal termini (Figure 5D).
Discussion
Analyses of HTS data from a library prepared from dsRNAs allowed us to predict the existence of two novel Vd-LRNAs in a fig tree grown on the island of Kauai, Hawaii. The two potential small circular RNAs contained HRzs in both polarity strands and had closely related sequences of 358 nt and 390 nt that mainly diverged in a specific region of about 120 nt. Discrimination between the long and short Vd-LRNAs in silico was possible due to the availability of long reads, up to 382 nt, which covered common and divergent sequences of the two de novo assembled contigs. This finding suggests that HTS technologies providing long sequence reads, like the 454 platform used in the present study, increase the chances of detecting coexisting but slightly divergent sequence variants of Vd-LRNAs.
In the absence of additional material from the original plant isolate, in silico data were used to design specific primers and perform a preliminary RT-PCR survey of figs on the island of Oahu, Hawaii. This analysis resulted in the identification of several fig trees associated with FHVd-LR, a small circular RNA with HRz in both polarity strands closely related to the Vd-LRNAs from Kauai. The presence of FHVd-LR only in some fig trees from Oahu, but not in others, was confirmed by northern blot hybridization. Northern blotting also showed that linear and circular forms of both polarity strands of this RNA exist in vivo, with one strand accumulating at a higher level than the other. Further experiments showed that this RNA has no DNA counterpart and that it accumulates in infected tissues as a population of sequence variants differing from each other in a few positions. These are typical features of quasispecies previously reported for other RNA replicons, including viruses, viroids, and VL-sat RNAs (Biebricher and Eigen, 2006; Codoñer et al., 2006; Flores et al., 2014).
Hammerhead ribozyme-mediated self-cleavage of both polarity strands of FHVd-LR during transcription was demonstrated and correspondence between the predicted and actual self-cleavage sites confirmed by RACE experiments. Interestingly, the nucleotide changes observed in the HRzs of both polarity strands of FHVd-LR did not impair the formation of the stable hammerhead structures, thus upholding the relevance in vivo of these ribozymes. Altogether, these data strongly support that FHVd-LR is a novel Vd-LRNA that likely replicates through a symmetric rolling-circle mechanism as previously proposed for some viroids and Vd-LsatRNAs (Flores et al., 2011). However, these structural and biochemical features did not provide any indication on whether FHVd-LR is a viroid or a Vd-LsatRNA. Indeed, HRzs in both polarity strands have been reported in three Vd-LsatRNAs and in all viroids classified in the family Avsunviroidae. HRzs were also found in both polarity strands of two Vd-LRNAs whose nature remains unclear (grapevine hammerhead Vd-LRNA and cscRNAs, which possibly are a viroid and two Vd-LsatRNAs of a mycovirus, respectively) (Alioto et al., 2003; Wu et al., 2012; Minoia et al., 2014).
Fig hammerhead viroid-like RNA assumes a rod-like conformation of minimal free energy, the relevance in vivo of which is supported by the variability observed in the FHVd-LR population accumulating in fig tissues. Indeed, most nucleotide changes detected in the FHVd-LR sequence variants mapped at loops or were compensatory mutations or covariations preserving the proposed structure. Rod-like structures have been proposed for most viroids and Vd-LsatRNAs. In members of the family Avsunviroidae, the G + C content, morphology, and thermodynamic stability of the HRzs and the secondary structure are of taxonomic relevance (Di Serio et al., 2018b). When these structural elements are taken into consideration, FHVd-LR seems to diverge from all members of the family Avsunviroidae. Among them, only ASBVd adopts a rod-like conformation in silico, in vitro, and in vivo (Symons, 1981; López-Carrasco and Flores, 2017). However, with a 38% G + C content and thermodynamically unstable HRzs due to a short stem III (Forster et al., 1988), this viroid appears different from FHVd-LR, which has about 50% G + C and stable HRzs. In contrast, the remaining members of the family Avsunviroidae, which share with FHVd-LR a high G + C content and stable HRzs (Di Serio et al., 2018b; Serra et al., 2018), assume branched or bifurcated conformations. Actually, among all known Vd-LRNAs, only the Vd-LsatRNA of LTSV and the two Vd-LRNAs from cherry (cscRNAs) resemble FHVd-LR in their rod-like secondary structure, high G + C content (about 50%), and stable HRzs in both polarity strands (Forster and Symons, 1990; Di Serio et al., 2006). Additionally, akin to the HRzs of FHVd-LR, the (+) and (−) HRzs of the two cscRNAs and the (−) HRz of VdL-satRNA of LTSV also contain an atypical adenine residue preceding the self-cleavage site.
In this context, note that the in silico data reported here supported the coexistence of at least two Vd-LRNAs of different size in the fig sample from Kauai, a hypothesis that was not conclusively tested due to the destruction of the original isolate. Attempted amplifications by RT-PCR, using different primer pairs, of Vd-LRNAs larger than FHVd-LR in the Oahu isolates have so far been unsuccessful, suggesting the absence of larger coinfecting Vd-LRNAs. Moreover, the two cscRNAs coinfecting cherry trees were not simultaneously observed in all the studied isolates, with some of them being infected by only one (Minoia et al., 2014).
Although the FHVd-LR structural features are closer to the VL-satRNA of LTSV and to cscRNAs than to viroids, we are unable to predict the biological nature of this novel Vd-LRNA from fig. Only bioassays may answer the specific question of whether FHVd-LR is able to replicate autonomously and systemically infect fig (Di Serio et al., 2018a). In this respect, experiments to test the transmission of FHVd-LR to virus-free fig plants by grafting and slash-inoculation of RNA preparations enriched in Vd-LRNAs and of in vitro-generated dimeric head-to tail transcripts are ongoing. Slash-inoculated plants were negative by RT-PCR and northern blot assays one and two months post-inoculation, but such bioassays in woody hosts may require longer times for systemic infection (Di Serio et al., 2018a).
Although all viruses identified by HTS as potentially infecting the original fig tree from Kauai belong to viral taxa not containing helper viruses of Vd-LsatRNAs, the association of FHVd-LR with a plant virus (known or unknown) cannot be ruled out; further studies are needed. The association of FHVd-LR with a virus infecting a fungus or another organism associated with fig cannot be excluded either. It has been in fact reported that HTS applied to cDNA libraries from plant tissues may detect viruses (and possibly Vd-LRNAs) infecting other transiently plant-associated organisms (Hily et al., 2018).
Conclusion
In conclusion, HTS allowed the initial identification of two novel Vd-LRNAs associated with fig, providing the necessary information for molecular studies to confirm the existence, self-cleaving activity mediated by HRzs, circularity of both polarity strands, and quasispecies nature of FHVd-LR. The data reported here make available detection methods to further investigate the biology and epidemiology of this and possibly other coinfecting novel Vd-LRNAs in fig trees.
Data Availability Statement
The datasets presented in this study can be found in NCBI database GenBank, accession numbers from MT57734 to MT577542.
Author Contributions
FD and MM supervised the project. AO-V and BN performed the experiments. AO-V, MM, JH, FD, and BN conceived the study and analyzed the data. FD, AO-V, and BN wrote the manuscript. All authors revised the manuscript.
Funding
This work was supported by USDA NIFA Hatch project HAW09030-H managed by the College of Tropical Agriculture and Human Resources at the University of Hawai‘i at Mānoa. This project has also received funding from the European Union’s Horizon 2020 Research and Innovation Program under the Marie Skłodowska-Curie grant agreement no. 734736. This article reflects only the author’s view, and the agency is not responsible for any use that may be made of the information it contains.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We sincerely thank Professor Ricardo Flores for critical reading of the manuscript and for suggestions. We would also like to thank the reviewers and the editor for their comments contributing to improve the article.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01903/full#supplementary-material
FIGURE S1 | Contigs of different size consisting of direct repeats (green arrows) generated by overlapping reads (in pink) obtained by HTS of a cDNA library generated from dsRNA extracted from a fig tree grown on the island of Kauai, Hawaii.
FIGURE S2 | (A) Pairwise comparison between a short and a long viroid-like RNA (Vd-LRNA) identified in silico in a fig sample from the island of Kauai. Positions with identical nucleotides are denoted by asterisks. (B) Primary and predicted secondary structure of the free energy of the long and short Vd-LRNA variants from Kauai. Differences between both RNA molecules are denoted in red. Regions involved in the formation of (+) and (−) hammerhead structures are delimited by flags; arrows and bars indicate the self-cleavage sites and the nucleotides conserved in most natural hammerhead structures, respectively; filled (black) and open symbols refer to (+) and (−) polarity, respectively.
FIGURE S3 | Pairwise comparison between the short viroid-like RNA (Vd-LRNA) from island of Kauai and FHVd-LR (master sequence) from the island of Oahu. The sequence identity is 96.1%. Positions with identical nucleotides are denoted by asterisks.
FIGURE S4 | Northern blot hybridization with equalized full-length digoxigenine-labeled riboprobes for detecting FHVd-LR (+) or (−) strands (left and right panel, respectively). In each panel, lanes 1 and 2 correspond to equal amounts of dimeric FHVd-LR (+) and (−) transcripts, respectively. Positions of the FHVd-LR (+) and (−) linear monomeric forms (m) are indicated by an arrow. Position and size (in nt) of the RNAs used as molecular markers are reported on the left.
FIGURE S5 | (A) Schematic representation of plasmids containing head-to-tail dimeric constructs of FHVd-LR and of the products generated by in vitro transcription. Plasmids containing the dimeric sequence of FHVd-LR in opposite orientations were linearized with SalI or NcoI and transcribed with T7 or SP6 RNA polymerase, respectively. Transcription of these templates is expected to produce complete dimeric transcripts (D), fragments longer than a monomer (5′F-M and 3′F-M) generated by the self-cleavage of one hammerhead ribozyme (HRz), monomeric RNAs (M) and fragments (5′F and 3′F) generated by the self-cleavage of both HRzs. In green, plasmid sequences; in yellow, polymerase promoter; in blue, FHVd-LR sequence, with the arrows indicating the (+) orientation; arrowheads and scissors mark the positions of the self-cleavage sites. Numbers on the left of each RNA fragment indicate its expected size. (B) Analysis by PAGE of the in vitro transcription of plasmids containing (+) and (−) dimeric FHVd-LR cDNA; L1 and L2, RNA ladders with sizes indicated on the left and on the right, respectively; for other symbols see panel (A).
Footnotes
References
Ale-Agha, G. N., and Rakhshandehroo, F. (2014). Detection and molecular variability of fig fleck-associated virus and fig cryptic virus in Iran. J. Phytopathol. 162, 417–425. doi: 10.1111/jph.12204
Alioto, D., Zaccaria, F., Covelli, L., Di Serio, F., Ragozzino, A., and Milne, R. G. (2003). Light and electron microscope observations on chlorotic rusty spot, a disorder of cherry in Italy. J. Plant Pathol. 85, 215–218. doi: 10.4454/jpp.v85i3.1033
Biebricher, C. K., and Eigen, M. (2006). What is a quasispecies? Curr. Top. Microbiol. 229, 1–31. doi: 10.1007/3-540-26397-7_1
Candresse, T., Hammond, R. W., and Hadidi, A. (1998). “Detection and identification of plant viruses and viroids using polymerase chain reaction (PCR),” in Plant Virus Disease Control, eds A. Hadidi, R. K. Khetarpal, and H. Koganezawa (St Paul: APS Press), 399–416.
Chiumenti, M., Torchetti, E. M., Di Serio, F., and Minafra, A. (2014). Identification and characterization of a viroid resembling apple dimple fruit viroid in fig (Ficus carica L.) by next generation sequencing of small RNAs. Virus Res. 188, 54–59. doi: 10.1016/j.virusres.2014.03.026
Codoñer, F. M., Daròs, J. A., Sole, R. V., and Elena, S. F. (2006). The fittest versus the flattest: experimental confirmation of the quasispecies effect with subviral pathogens. PLoS Pathog. 2:0020136. doi: 10.1371/journal.ppat.0020136
Collins, R. F., Gellatly, D. L., Sehgal, O. P., and Abouhaidar, M. G. (1998). Self-cleaving circular RNA associated with rice yellow mottle virus is the smallest viroid-like RNA. Virology 241, 269–275. doi: 10.1006/viro.1997.8962
Daròs, J. A., and Flores, R. (1995). Identification of a retroviroid-like element from plants. Proc. Natl. Acad. Sc.i U.S.A. 92, 6856–6860. doi: 10.1073/pnas.92.15.6856
Di Serio, F., Ambrós, S., Sano, T., Flores, R., and Navarro, B. (2018a). Viroid diseases in pome and stone fruit trees and Koch’s postulates: a critical assessment. Viruses 10:612. doi: 10.3390/v10110612
Di Serio, F., Daròs, J. A., Ragozzino, A., and Flores, R. (1997). A 451-nucleotide circular RNA from cherry with hammerhead ribozymes in its strands of both polarities. J. Virol. 71, 6603–6610. doi: 10.1128/jvi.71.9.6603-6610.1997
Di Serio, F., Daròs, J. A., Ragozzino, A., and Flores, R. (2006). Close structural relationship between two hammerhead viroid-like RNAs associated with cherry chlorotic rusty spot disease. Arch. Virol. 151, 1539–1549. doi: 10.1007/s00705-006-0732-0
Di Serio, F., Flores, R., Verhoeven, J. T., Li, S. F., Pallás, V., Randles, J. W., et al. (2014). Current status of viroid taxonomy. Arch. Virol. 159, 3467–3478. doi: 10.1007/s00705-014-2200-6
Di Serio, F., Li, S. F., Matousek, J., Owens, R. A., Pallás, V., Randles, J. W., et al. (2018b). ICTV virus taxonomy profile: Avsunviroidae. J. Gen. Virol. 99, 611–612. doi: 10.1099/jgv.0.001045
Di Serio, F., Navarro, B., and Flores, R. (2017). “Origin and evolution of viroids,” in Viroids and Satellites, eds A. Hadidi, R. Flores, J. W. Randles, and P. Palukaitis (Cambridge: Academic Press), 125–134. doi: 10.1016/b978-0-12-801498-1.00012-7
Elbeaino, T., Abou Kubaa, R., Ismaeil, E., Mando, J., and Digiaro, M. (2012). Viruses and hop stunt viroid of fig trees in Syria. J. Plant Pathol. 94, 687–691. doi: 10.4454/JPP.V94I3.002
Elbeaino, T., Digiaro, M., Heinoun, K., De Stradis, A., and Martelli, G. P. (2010). Fig mild mottle-associated virus, a novel closterovirus infecting fig. J. Plant Pathol. 92, 165–172. doi: 10.4454/jpp.v92i1.26
Fadda, Z., Daròs, J. A., Flores, R., and Durán-Vila, N. (2003). Identification in eggplant of a variant of citrus exocortis viroid (CEVd) with a 96 nucleotide duplication in the right terminal region of the rod-like secondary structure. Virus Res. 97, 145–149. doi: 10.1016/j.virusres.2003.08.002
Flores, R., Durán-Vila, N., Pallás, V., and Semancik, J. S. (1985). Detection of viroid and viroid-like RNAs from grapevine. J. Gen. Virol. 66, 2095–2102. doi: 10.1099/0022-1317-66-10-2095
Flores, R., Gago-Zachert, S., Serra, P., Sanjuán, R., and Elena, S. F. (2014). Viroids: survivors from the RNA world? Annu. Rev. Microbiol. 68, 395–414. doi: 10.1146/annurev-micro-091313-103416
Flores, R., Grubb, D., Elleuch, A., Nohales, M. A., Delgado, S., and Gago, S. (2011). Rolling-circle replication of viroids, viroid-like satellite RNAs and hepatitis delta virus: variations on a theme. RNA Biol. 8, 200–206. doi: 10.4161/rna.8.2.14238
Flores, R., Minoia, S., Carbonell, A., Gisel, A., Delgado, S., López-Carrasco, A., et al. (2015). Viroids, the simplest RNA replicons: how they manipulate their hosts for being propagated and how their hosts react for containing the infection. Virus Res. 209, 136–145. doi: 10.1016/j.virusres.2015.02.027
Forster, A. C., Davies, C., Hutchins, C. J., and Symons, R. H. (1990). Characterization of self-cleavage of viroid and virusoid RNAs. Methods Enzymol. 181, 583–607. doi: 10.1016/0076-6879(90)81153-l
Forster, A. C., Davies, C., Sheldon, C. C., Jeffries, A. C., and Symons, R. H. (1988). Self-cleaving viroid and newt RNAs may only be active as dimers. Nature 334, 265–267. doi: 10.1038/334265a0
Forster, A. C., and Symons, R. H. (1990). Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell 49, 211–220. doi: 10.1016/0092-8674(87)90562-9
Gago-Zachert, S. (2016). Viroids, infectious long non-coding RNAs with autonomous replication. Virus Res. 212, 12–24. doi: 10.1016/j.virusres.2015.08.018
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Hadidi, A., Flores, R., Candresse, T., and Barba, M. (2016). Next-generation sequencing and genome editing in plant virology. Front. Microbiol. 7:1325. doi: 10.3389/fmicb.2016.01325
Hajizadeh, M., Navarro, B., Bashir, N. S., Torchetti, E. M., and Di Serio, F. (2012). Development and validation of a multiplex RT-PCR method for simultaneous detecting five grapevine viroids. J. Virol. Methods 179, 62–69. doi: 10.1016/j.jviromet.2011.09.022
Hegedus, K., Dallmann, G., and Balazs, E. (2004). The DNA form of a retroviroid-like element is involved in recombination events with itself and with the plant genome. Virology 325, 277–286. doi: 10.1016/j.virol.2004.04.035
Hernández, C., Di Serio, F., Ambrós, S., Daròs, J. A., and Flores, R. (2006). An element of the tertiary structure of Peach latent mosaic viroid RNA revealed by UV irradiation. J. Virol. 80, 9336–9340. doi: 10.1128/JVI.00630-06
Hily, J. M., Candresse, T., Garcia, S., Vigne, E., Tannière, M., Komar, V., et al. (2018). High-throughput sequencing and the viromic study of grapevine leaves: from the detection of grapevine-infecting viruses to the description of a new environmental Tymovirales member. Front. Microbiol. 9:1782. doi: 10.3389/fmicb.2018.01782
Hutchins, C. J., Rathjen, P. D., Forster, A. C., and Symons, R. H. (1986). Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res. 14, 3627–3640. doi: 10.1093/nar/14.9.3627
Kovalskaya, N., and Hammond, R. W. (2014). Molecular biology of viroid-host interactions and disease control strategies. Plant Sci. 228, 48–60. doi: 10.1016/j.plantsci.2014.05.006
Laney, A. G., Hassan, M., and Tzanetakis, I. E. (2012). An integrated badnavirus is prevalent in fig germplasm. Phytopathology 102, 1182–1189. doi: 10.1094/PHYTO-12-11-0351
Langmead, B., Trapnell, C., Pop, M., and Salzberg, S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. doi: 10.1186/gb-2009-10-3-r25
Li, R., Mock, R., Huang, Q., Abad, J., Hartung, J., and Kinard, G. (2008). A reliable and inexpensive method of nucleic acid extraction for the PCR-based detection of diverse plant pathogens. J. Virol. Methods 154, 48–55. doi: 10.1016/j.jviromet.2008.09.008
López-Carrasco, A., and Flores, R. (2017). The predominant circular form of avocado sunblotch viroid accumulates in planta as a free RNA adopting a rod-shaped secondary structure unprotected by tightly bound host proteins. J. Gen. Virol. 98, 1913–1922. doi: 10.1099/jgv.0.000846
Lorenz, R., Bernhart, S. H., Höner, Z., Siederdissen, C., Tafer, H., Flamm, C., et al. (2011). ViennaRNA Package 2.0. Algorithms Mol. Biol. 6:26. doi: 10.1186/1748-7188-6-26
Machado, M., Magalhães, W. C., Sene, A., Araújo, B., Faria-Campos, A. C., Chanock, S. J., et al. (2011). Phred-Phrap package to analyses tools: a pipeline to facilitate population genetics re-sequencing studies. Investig. Genet. 2:3. doi: 10.1186/2041-2223-2-3
Melzer, M. J., Borth, W. B., Sether, D. M., Ferreira, S., Gonsalves, D., and Hu, J. S. (2010). Genetic diversity and evidence for recent modular recombination in Hawaiian Citrus tristeza virus. Virus Genes 40, 111–118. doi: 10.1007/s11262-009-0409-3
Minafra, A., Savino, V., and Martelli, G. P. (2017). Virus diseases of fig and their control. Acta Horticulturae 1173, 237–244. doi: 10.17660/actahortic.2017.1173.41
Minoia, S., Navarro, B., Covelli, L., Barone, M., Garcia-Becedas, M. T., Ragozzino, A., et al. (2014). Viroid-like RNAs from cherry trees affected by leaf scorch disease: further data supporting their association with mycoviral double-stranded RNAs. Arch. Virol. 159, 589–593. doi: 10.1007/s00705-013-1843-z
Navarro, B., and Di Serio, F. (2018). “Double-stranded RNA-enriched preparations to identify viroids by next-generation sequencing,” in Viral Metagenomics: Methods and Protocols. Methods in Molecular Biology, Vol. 1746, eds V. Pantaleo and M. Chiumenti (New York, NY: Humana Press), 37–43. doi: 10.1007/978-1-4939-7683-6_3
Navarro, B., Rubino, L., and Di Serio, F. (2017). “Small circular satellite RNAs,” in Viroids and Satellites, eds A. Hadidi, R. Flores, J. W. Randles, and P. Palukaitis (Cambridge: Academic Press), 659–669. doi: 10.1016/b978-0-12-801498-1.00061-9
Olmedo-Velarde, A., Park, A. C., Sugano, J., Uchida, J. Y., Kawate, M., Borth, W. B., et al. (2019). Characterization of ti ringspot-associated virus, a novel Emaravirus associated with an emergent ringspot disease of Cordyline fruticose. Plant Dis. 103, 2345–2352. doi: 10.1094/PDIS-09-18-1513-RE
Pallás, V., Navarro, A., and Flores, R. (1987). Isolation of a viroid-like RNA from hop different from hop stunt viroid. J. Gen. Virol. 68, 3201–3205. doi: 10.1099/0022-1317-68-12-3201
Pley, H. W., Flaherty, K. M., and McKay, D. B. (1994). Three-dimensional structure of a hammerhead ribozyme. Nature 372, 68–74. doi: 10.1038/372068a0
Serra, P., Messmer, A., Sanderson, D., James, D., and Flores, R. (2018). Apple hammerhead viroid-like RNA is a bona fide viroid: autonomous replication and structural features support its inclusion as a new member in the genus Pelamoviroid. Virus Res. 249, 8–15. doi: 10.1016/j.virusres.2018.03.001
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
Symons, R. H. (1981). Avocado sunblotch viroid: primary sequence and proposed secondary structure. Nucleic Acids Res. 9, 6527–6537. doi: 10.1093/nar/9.23.6527
Symons, R. H., and Randles, J. W. (1999). “Encapsidated circular viroid-like satellite RNAs (virusoids) of plants,” in Satellites and Defective Viral RNAs, eds P. K. Vogt and A. O. Jackson (Berlin: Springer-Verlag).
Vera, A., Daròs, J. A., Flores, R., and Hernández, C. (2000). The DNA of a plant retroviroid-like element is fused to different sites in the genome of a plant pararetrovirus and shows multiple forms with sequence deletions. J. Virol. 74, 10390–10400. doi: 10.1128/jvi.74.22.10390-10400.2000
Villamor, D. E. V., Ho, T., Al Rwahnih, M., Martin, R. R., and Tzanetakis, I. E. (2019). High throughput sequencing for plant virus detection and discovery. Phytopathology 109, 716–725. doi: 10.1094/PHYTO-07-18-0257-RVW
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Wu, Q., Wang, Y., Cao, M., Pantaleo, V., Burgyan, J., Li, W. X., et al. (2012). Homology-independent discovery of replicating pathogenic circular RNAs by deep sequencing and a new computational algorithm. Proc. Natl. Acad. Sci. U.S.A. 109, 3938–3943. doi: 10.1073/pnas.1117815109
Yakoubi, S., Elleuch, A., Besaies, N., Marrakchi, M., and Fakhfakh, H. (2007). First report of Hop stunt viroid and Citrus exocortis viroid on fig with symptoms of fig mosaic disease. J. Phytopathol. 155, 125–128. doi: 10.1111/j.1439-0434.2007.01205.x
Keywords: circular RNA, infectious RNAs, Ficus carica, next generation sequencing, non-coding RNAs, ribozyme, viroid, virusoid
Citation: Olmedo-Velarde A, Navarro B, Hu JS, Melzer MJ and Di Serio F (2020) Novel Fig-Associated Viroid-Like RNAs Containing Hammerhead Ribozymes in Both Polarity Strands Identified by High-Throughput Sequencing. Front. Microbiol. 11:1903. doi: 10.3389/fmicb.2020.01903
Received: 11 June 2020; Accepted: 20 July 2020;
Published: 18 August 2020.
Edited by:
John Wesley Randles, The University of Adelaide, AustraliaReviewed by:
Peter Palukaitis, Seoul Women’s University, South KoreaZhixiang Zhang, State Key Laboratory for Biology of Plant Diseases and Insect Pests (CAAS), China
Copyright © 2020 Olmedo-Velarde, Navarro, Hu, Melzer and Di Serio. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael J. Melzer, bWVsemVyQGhhd2FpaS5lZHU=; Francesco Di Serio, ZnJhbmNlc2NvLmRpc2VyaW9AaXBzcC5jbnIuaXQ=
†These authors have contributed equally to this work
 Alejandro Olmedo-Velarde
Alejandro Olmedo-Velarde Beatriz Navarro
Beatriz Navarro John S. Hu
John S. Hu Michael J. Melzer
Michael J. Melzer Francesco Di Serio
Francesco Di Serio