- Department of Pharmacology and Toxicology, College of Animal Science and Veterinary Medicine, Henan Agricultural University, Zhengzhou, China
ICEHpa1 was identified in the genome of a serovar 8 Haemophilus parasuis ST288 isolate YHP170504 from a case of swine lower respiratory tract infection. The aim of the present study was to characterize the integrative conjugative element ICEHpa1 and its multiresistance region. Susceptibility testing was determined by broth microdilution and the complete ICEHpa1 was identified by WGS analysis. The full sequence of ICEHpa1 was analyzed with bioinformatic tools. The presence of ICEHpa1, its circular intermediate and integration site were confirmed by PCR and sequence analysis. Transfer of ICEHpa1 was confirmed by conjugation. ICEHpa1 has a size of 68,922 bp with 37.42% GC content and harbors 81 genes responsible for replication and stabilization, transfer, integration, and accessory functions, as well as seven different resistance genes [blaRob–3, tet(B), aphA1, strA, strB, aac(6)′-Ie-aph(2′)-Ia, and sul2]. Conjugation experiments showed that ICEHpa1 could be transferred to H. parasuis V43 with frequencies of 6.1 × 10–6. This is the first time a multidrug-resistance ICE has been reported in H. parasuis. Seven different resistance genes were located on a novel integrative conjugative element ICEHpa1, which suggests that the ICEHpa1 is capable of acquiring foreign genes and serving as a carrier for various resistance genes.
Introduction
The gram-negative bacterium Haemophilus parasuis is the causative agent of Glasser’s disease characterized by polyarthritis, fibrinous polyserositis, and meningitis in swine (Oliveira and Pijoan, 2004). The H. parasuis infection may cause great economic losses to the global pig industry (Oliveira et al., 2001).
More and more attention has been drawn to the antimicrobial resistance in bacteria from food-producing animals. In H. parasuis, the resistant genes are usually located on small plasmids, in which mob genes (mobA, mobB, mobC, mobA-like, mobC-like, and mobA-L) and ISApl1 are usually identified flanking the resistant genes (Lancashire et al., 2005; Chen et al., 2010; Yang et al., 2013; Li et al., 2015; Moleres et al., 2015). However, no other mobile genetic elements [transposons, integrons, and integrative and conjugative elements (ICEs)] have been found to be associated with the resistant genes in H. parasuis. ICEs are self-transmissible mobile elements that are widespread among different bacteria (Burrus et al., 2002; Burrus and Waldor, 2004; Lei et al., 2016). ICEs are composed of a set of core genes that are responsible for replication, maintenance, conjugation, recombination, and regulation, with other accessory modules, such as antimicrobial resistance genes (Robinson et al., 2013; Johnson and Grossman, 2015). In addition, ICEs usually have a single insertion site, which is often in the 5′ or 3′ end of a tRNA or other highly conserved genes such as the gene prfC, in the chromosome of their host (Mulvey et al., 2001; Johnson and Grossman, 2015). ICEs, as vehicles for active DNA exchange among different bacteria, contain some specific genes or sites needed for processing their DNA for transfer. Most of these genes are not expressed when the ICE is integrated in the chromosome; however, expression of the genes needed for excision, integration, and conjugation is induced under certain conditions, and the ICE may excise from the host chromosome to form a dsDNA circular intermediate. Some ICE-encoded proteins assemble into a mating pore that is responsible for transferring the ICE. The new host may recognize the origin of transfer (oriT), process the ICE dsDNA to generate a linear ssDNA-protein (T-DNA) through the ICE-encoded relaxase, and pump the T-DNA into the recipient. Then the ICE was recombined into the new host chromosome through an ICE-encoded integrase (Toleman and Walsh, 2011; Johnson and Grossman, 2015; Wright et al., 2015).
Quite a few ICEs have been identified in Pasteurellaceae, such as Haemophilus influenzae, Pasteurella multocida, Mannheimia haemolytica, and Actinobacillus pleuropneumoniae (Juhas et al., 2007; Brenner et al., 2012; Eidam et al., 2015; Bossé et al., 2016; Li Y. et al., 2018). However, no complete multidrug-resistance ICE in H. parasuis has been described in detail to date. In this study, we identified ICEHpa1, a novel ICE carrying multiple resistance genes, in the chromosome of a serovar 8 H. parasuis ST288 isolate YHP170504, in a feedlot from Henan, China, in 2017.
Materials and Methods
Bacterial Strains and Susceptibility Testing
The strain YHP170504 was obtained from a case of swine lower respiratory tract infection in a feedlot from Henan, China, in 2017. Owing to the unavailability of an approved method for H. parasuis, MICs of H. parasuis isolates were determined using broth microdilution method following CLSI standard (Clinical and Laboratory Standards Institute [CLSI], 2018) for A. pleuropneumoniae. The antimicrobial agents tested were oxytetracycline, doxycycline, ampicillin, amoxicillin, ceftiofur, cefquinome, enrofloxacin, streptomycin, gentamicin, tilmicosin, tylosin, florfenicol, sulfamethoxazole/trimethoprim (19/1), lincomycin, and colistin. A. pleuropneumoniae ATCC27090 and Escherichia coli ATCC 25922 were used as control strains.
WGS and Analysis
Total genomic DNA of strain YHP170504 was extracted using the TIANamp Bacteria DNA Kit (TIANGEN, Beijing, China) and subjected to WGS using Illumina Nextseq 500 and the Oxford Nanopore Technologies (ONT) MinION platforms. Sequencing reads including short-read and long-read data were assembled using Unicycler 0.4.4 with the hybrid strategy (Wick et al., 2017; Li R. et al., 2018). The complete sequence of ICEHpa1 was initially annotated using the RAST server1 and corrected manually. Comparison analysis was conducted using the genome comparison visualizer Easyfig.
Confirmation of the Circular Extrachromosomal Form of the ICE and Conjugal Transfer of ICEHpa1
Chromosomal insertion sites were confirmed by PCR in YHP170504 and transconjugants. For the 5′ junction, primers F1 and R1 were designed to amplify a 515-bp fragment from upstream of the ICEHpa1 insertion to a sequence within the 5′ end of ICEHpa1. For the 3′ insertion, primers F2 and R2 were designed to amplify a 458 bp fragment from within the 3′ end of ICEHpa1 insertion to a sequence of the downstream of the ICEHpa1 insertion. To confirm the extrachromosomal circular form of the ICE, outward-facing primers ICE-out-F and ICE-out-R were used.
To investigate self-transfer ability of the ICEHpa1, the conjugation assay was performed using the H. parasuis strain YHP170504 as the donor and H. parasuis V43 (rifampicin resistance) as the recipient. The serovar 4 H. parasuis ST170 isolate V43 was from the strain collection of our laboratory, and the rifampicin-resistant mutant of this strain was generated by selection on Tryptic Soy Agar (TSA) plates supplemented with 10% fetal bovine serum, 10 mg/L nicotinamide adenine dinucleotide (NAD), and increasing rifampicin concentration. For the conjugation assay, overnight cultures of donor and recipient strains grown in Tryptic Soy Broth supplemented with 10% fetal bovine serum and 10 mg/L NAD were mixed (1:5) and incubated for 4 h at 37°C. Bacterial cultures were spread on TSA plates supplemented with 10% fetal bovine serum, 10 mg/L NAD, oxytetracycline (8 mg/L), and rifampicin (100 mg/L), incubated at 37°C for 24 h. Then the transconjugants were screened on the plates. The conjugation frequency was calculated as the number of transconjugants per donor. All the transconjugants were confirmed with PCR using the primers virB4-F and virB4-R, susceptibility testing, and MLST. All the primers used are listed in Table 1.
Serotyping and Multilocus Sequence Typing (MLST)
The strain YHP170504, V43, and the transconjugant V43::ICEHpal1 were typed by serotyping and MLST. Serovars of the strains were determined using the primers Howell previously described (Howell et al., 2015). Seven housekeeping genes (atpD, infB, mdh, rpoB, 6pgd, g3pd, and frdB) were amplified and sequenced as described previously (Mullins et al., 2013), after registration of sequences at https://pubmlst.org/hparasuis/ for assignment of allele numbers and STs; data were analyzed using software available on the website.
Evolutionary Analyses of the Integrase
The integrase, an important core gene of ICE, is needed for both integration and excision. Evolutionary analyses were conducted in MEGA7 (Kumar et al., 2016) and the analysis involved 11 integrases complete sequences of eight ICEs from Pasteurellaceae. Initial trees for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood approach and then selecting the topology with superior log-likelihood value (Kumar et al., 2016).
Nucleotide Sequence Accession Number
The complete sequences of the chromosome and the ICEHpa1 in H. parasuis YHP170504 have been submitted to GenBank with the following accession numbers: CP054198 and MN844034.
Results and Discussion
The strain YHP170504 was serotyped as serovar 8, belonging to ST288. Also, it exhibited high MICs of oxytetracycline (64 mg/L), doxycycline (8 mg/L), ampicillin (64 mg/L), amoxicillin (64 mg/L), gentamicin (256 mg/L), streptomycin (128 mg/L), sulfamethoxazole/trimethoprim (513/27 mg/L), and enrofloxacin (8 mg/L).
The chromosome of H. parasuis YHP170504 is 2,520,015 bp long with a GC content of 39.64%. Sequence analysis showed that the genome harbors seven resistance genes including the β-lactamase-encoding gene blaRob–3, tetracycline resistance gene tet(B), the aminoglycoside resistance genes (aphA1, strA, and strB), aminoglycoside and fluoroquinolone resistance gene [aac(6)′-Ie-aph(2′)-Ia], and sulfonamide resistance gene sul2. WGS analysis showed that all seven resistance genes were located on a novel integrative conjugative element, designated as ICEHpa1 (Figure 1A) according to the nomenclature of ICEs2.
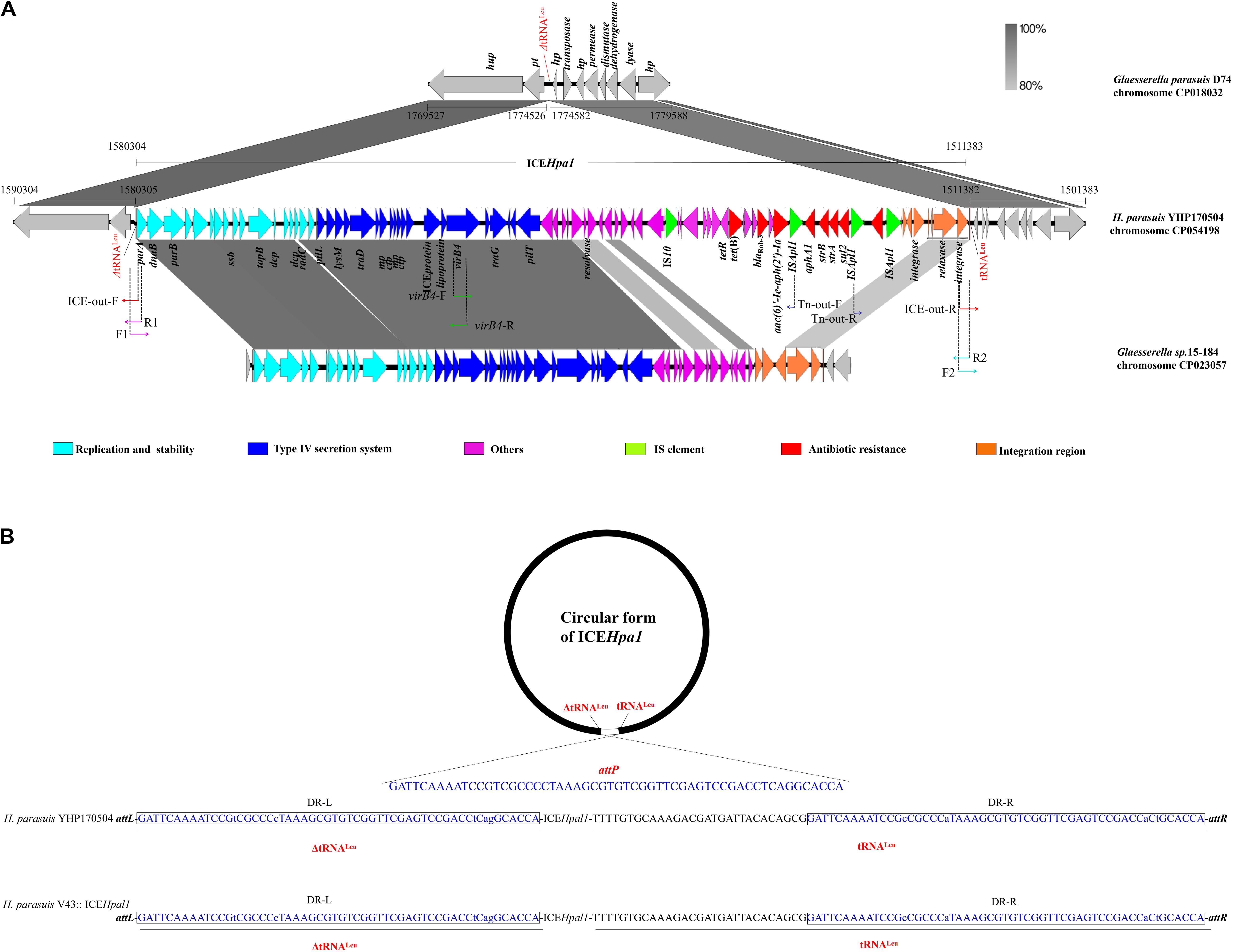
Figure 1. (A) Schematic representation of ICEHpa1 and its border regions. Genes are presented as broad arrows, with the arrowhead indicating the direction of transcription. Linear comparison of ICEHpa1 and its border regions with the homologous region of the genome of Glaesserella parasuis (H. parasuis) strain D74 (CP018032), and Glaesserella sp. (H. parasuis) strain 15–184 (CP023057). Thin arrows represent the orientation of each primer and relative positions of the primers along the tested linear sequence. mp, gene encoding membrane protein; ctp, gene encoding conjugal transfer protein. (B) Site-specific integration of ICEHpa1 into the tRNALeu of donor and recipient strain. The sequences of the tRNALeu are shown in the orientation that matches the orientation of the ICEHpa1 sequence. Two tRNALeu copies (underlined, a truncated copy and an intact copy), the sequences involved in the crossover (attP sequence, 56 bp), and the resulting imperfect direct repeats (with lowercase letters indicating the bases that differ) located on the left termini (DR-L, in the closed boxes) and on the right termini (DR-R, in the closed boxes) of the inserted ICEHpa1 are also shown.
ICEHpa1 (68,922 bp, corresponding to bases 1,511,383–1,580,304 in GenBank accession number CP054198 or bases 5938–74,859 in GenBank accession number MN844034), a novel ICE with a lower GC content (37.42%), differing from the GC content (39.64%) of entire genome of H. parasuis YHP170504, was integrated into the tRNALeu, a common insertion site for ICEs in other species of the family Pasteurellaceae (Juhas et al., 2007; Brenner et al., 2012; Eidam et al., 2015; Bossé et al., 2016). WGS analysis showed that the ICEHpa1 was flanked by two tRNALeu copies (a truncated copy and an intact copy, Figure 1A). The presence of an extrachromosomal circular form of ICEHpa1 was confirmed in YHP170504 using primers ICE-out-F and ICE-out-R. Sequencing of the closed circular form of ICEHpa1 showed that the junction (attP sequence) sequence was formed by 56 bp imperfect direct repeats (Figure 1B).
PCR assays and susceptibility testing confirmed the presence and activity of the ICEHpa1-associated resistance genes in the H. parasuis V43. The transconjugant H. parasuis V43::ICEHpal1 showed, in comparison with H. parasuis V43, increased MICs of oxytetracycline (from < 0.5 to 32 mg/L), doxycycline (from < 0.5 to 8 mg/L), ampicillin (from 1 to 64 mg/L), amoxicillin (from 1 to 64 mg/L), gentamicin (from < 0.5 to 128 mg/L), streptomycin (from 2 to 128 mg/L), sulfamethoxazole/trimethoprim (from 1.9/0.1 to 513/27 mg/L), and enrofloxacin (from < 0.5 to 4 mg/L). Also, the transconjugant H. parasuis V43::ICEHpal1 showed the same serovar and ST as H. parasuis V43. Although only traD, traG, virB4, pilT, and pilL genes encoding components of the type IV secretion system were identified, many other conjugal transfer proteins and membrane proteins were present in ICEHpa1, which may also be involved in the conjugal transfer and responsible for the host specificity of the ICE. The ICEHpa1 could be transferred to H. parasuis V43 at a low frequency of conjugation, with 6.1 × 10–6 transconjugants per donor. Our results revealed that ICEHpa1 has the self-transmissible capacity to facilitate the dissemination of the resistance genes.
Sequence analysis indicated that the insertion point of ICEHpa1 in the transconjugant was located in a tRNALeu. Also, the tRNALeu locus, in which the ICEHpa1 was inserted in the H. parasuis V43, showed the same sequence as the one in the H. parasuis YHP170504 (Figure 1B). The tRNALeu into which ICEHpa1 was inserted in H. parasuis YHP170504 was flanked on one side by a peptide transporter and on the other side by a hypothetical protein. A comparison between the genome of H. parasuis YHP170504 and another similar genome of Glaesserella parasuis (H. parasuis) D74 showed that the genome fragment flanking the left border and right border regions of the ICEHpa1 in YHP170504 had high identity to the D74, but a 55-bp truncated tRNALeu was replaced by the ICEHpa1 insertion in YHP170504. Similarly, a peptide transporter and a hypothetical protein were found immediately up- and downstream of the tRNALeu in Glaesserella parasuis D74. The result revealed the strain Glaesserella parasuis D74 may serve as a potential recipient for acquiring ICEHpa1.
A total of 81 genes flanked by two tRNALeu copies were identified within ICEHpa1, of which 17 coded for the putative replication, 20 for type IV secretion system (T4SS), 7 for integration, 4 for transposases of IS elements (an IS10 and three copies of ISApl1), and 7 for resistance genes including the blaRob–3, tet(B), aphA1, strA, strB, aac(6)′-Ie-aph(2′)-Ia, and sul2. Comparative sequence analysis (Figure 1A) showed that the complete sequence of ICEHpa1 shared only 63% identity (the highest rate of match) with the region of Glaesserella sp. (H. parasuis) 15–184 chromosome, differing clearly from the previous reports about the ICEs from the other Pasteurellaceae species (Juhas et al., 2007; Brenner et al., 2012; Eidam et al., 2015; Bossé et al., 2016; Li Y. et al., 2018).
ICEHpa1 contains five components (replication and stabilization, T4SS, antimicrobial resistance region, integration and accessory region) (Figure 1A), whose replication and T4SS region shared 98.18% identity with the corresponding region of Glaesserella sp. 15–184 chromosome. The accessory region and integration region (including two integrase genes and a relaxase gene) exhibited only partial homology to corresponding region of Glaesserella sp. 15–184 chromosome (69% coverage with 96.41% identity and 61% coverage with 85.05% identity, respectively) (Figure 1A). Similar to ICEPmu1 in P. multocida (Brenner et al., 2012), two integrases were found in ICEHpa1. Identity of 47.35% (93% coverage) was seen when the amino acid sequences of these two integrases were aligned. Comparative sequence analysis revealed that these two integrases are tyrosine recombinases of the Xer family, which are responsible for the integration by site-specific recombination. The integrase 1 in ICEHpa1, belonging to tyrosine recombinase XerD, shared 93.98% (98% coverage) amino acid identity to integrase (Pmu_02700) of ICEPmu1 (CP003022). The integrase 2 in ICEHpa1, belonging to tyrosine recombinase XerC, shared 100% (100% coverage) amino acid identity to integrase of Glaesserella parasuis strain F9 (KEZ23006.1). However, both of them differed from the integrases reported in ICEs from Pasteurellaceae according to the maximum-likelihood tree obtained by using MEGA 7 software (Figure 2). Other experiments are necessary to show which of them or if both are responsible for the integration of ICEHpa1.
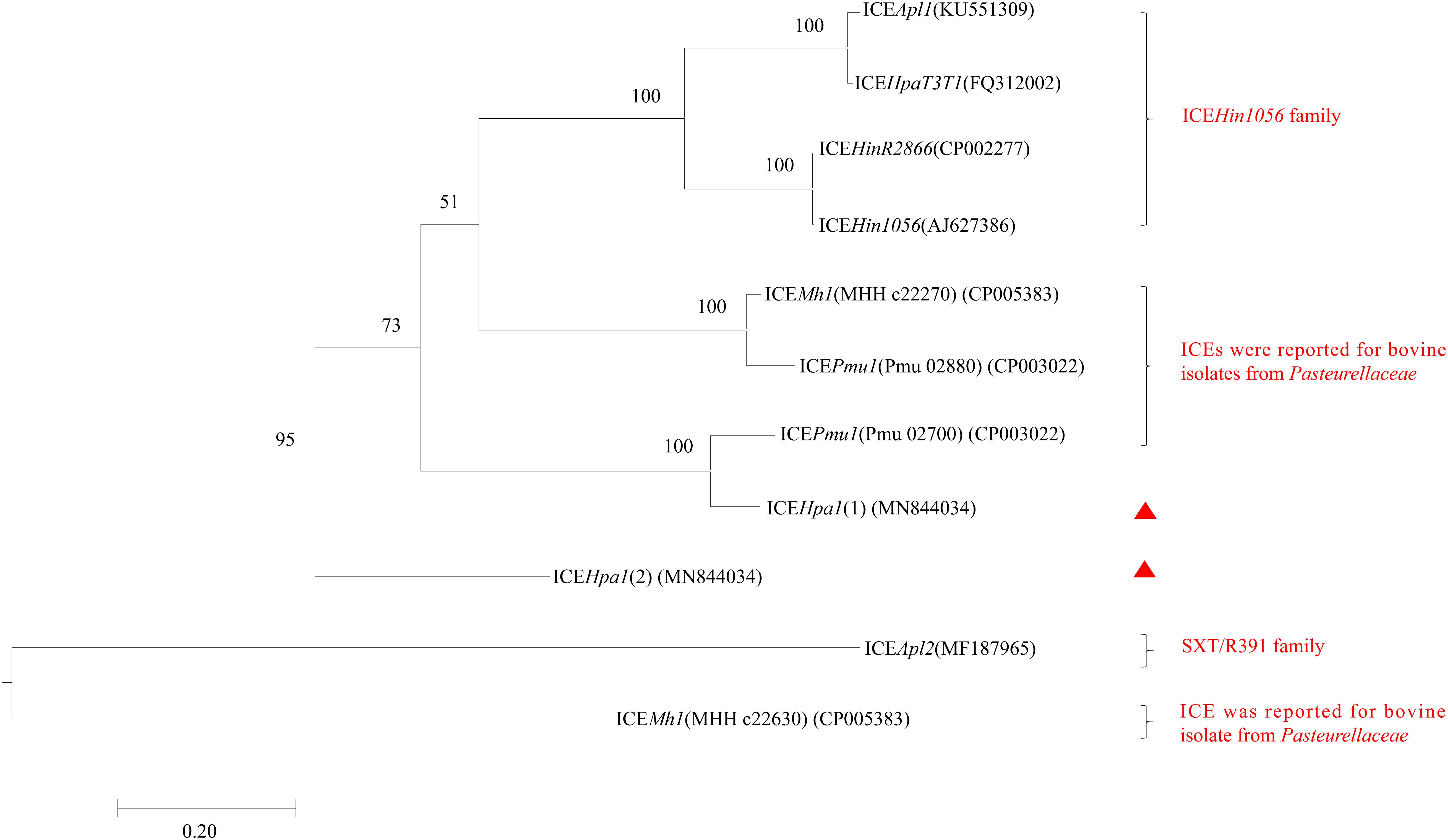
Figure 2. Maximum-likelihood phylogenetic tree based on 11 integrases complete sequences of 8 ICEs from Pasteurellaceae by using MEGA7. Two integrases of ICEHpa1 are indicated by red triangles. The percentage of trees in which the associated taxa clustered together is shown next to the branches. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. All positions containing gaps and missing data were eliminated. Bootstrap analysis was performed with 1000 replications. Bar, 0.2 substitution per nucleotide position.
The multiresistance region (Figure 3A) contains three segments harboring seven different resistance genes. The first segment harboring tet(B) was characterized by a truncated transposon Tn10, which shows 99% identity with the corresponding region of the Tn10 transposon. The second segment carries two resistance genes, blaRob–3 and aac(6)′-Ie-aph(2′)-Ia. Notably, although this region showed 99% identity with the corresponding region of the H. parasuis pQY431 complete sequence, its blaRob gene significantly differed from its counterpart in H. parasuis pQY431. Compared with the BlaRob protein encoded by blaRob–1 from the H. parasuis pQY431, two alanine residues [leucine (L) and threonine (T)] were added between positions 16 and 17 in the BlaRob from ICEHpa1. The BlaRob from ICEHpa1 shared 100% amino acid identity with the BlaRob–3 from Moraxella pluranimalium CCUG 54913 (NG059331). Resistance to β-lactam antibiotics in H. parasuis is conferred by two potential mechanisms: mutations in the ftsI gene encoding PBP3 and/or production of β-lactamases. WGS analysis showed that no point mutation was found in the ftsI gene and no other bla gene was detected in H. parasuis YHP170504 except blaRob–3, which suggests that the blaRob–3 gene conferred resistance to ampicillin and amoxicillin. To our knowledge, this is the first report of blaRob–3 gene in Pasteurellaceae species. The third segment harboring a resistance module aphA1-strB-strA-sul2, which is flanked by two ISApl1 elements oriented in the same direction, was regarded as a putative small transposon designated Tn6742 (Figure 3B). To confirm the excision and cyclization of this structure, PCR was conducted using the primers, Tn-out-F and Tn-out-R. The result showed this structure can be looped out, which indicated ISApl1 might accelerate the dissemination of the module aphA1-strB-strA-sul2. ISApl1 has been reported to produce a 2 bp direct duplication GG at its integration site (Tegetmeyer et al., 2008). The 2 bp direct duplication GG was detected upstream of the left-hand copy and downstream of the right-hand copy of ISApl1, which suggested that the transposon Tn6742 was reassembled into the host chromosome via ISApl1-mediated insertion rather than homologous recombination. However, the formation of a similar structure ISApl1-aphA1-strB-strA-sul2-ISCR21-ΔISCR2-floR-ISCR2-erm42-ISApl1 in ICEPmu1 from P. multocida was proved to insert the chromosome by producing a 2-bp direct duplication GT rather than GG (Brenner et al., 2012). In addition, all the four resistance genes aphA1, strB, strA, and sul2 oriented in the same direction, which is opposite the direction of the two ISApl1. Compared with that, the four genes and two ISApl1, all oriented in the same direction, were identified in ICEPmu1 from P. multocida (Brenner et al., 2012). Additional experiments are necessary to show the role of the third ISApl1 in the transmission of these resistance genes.
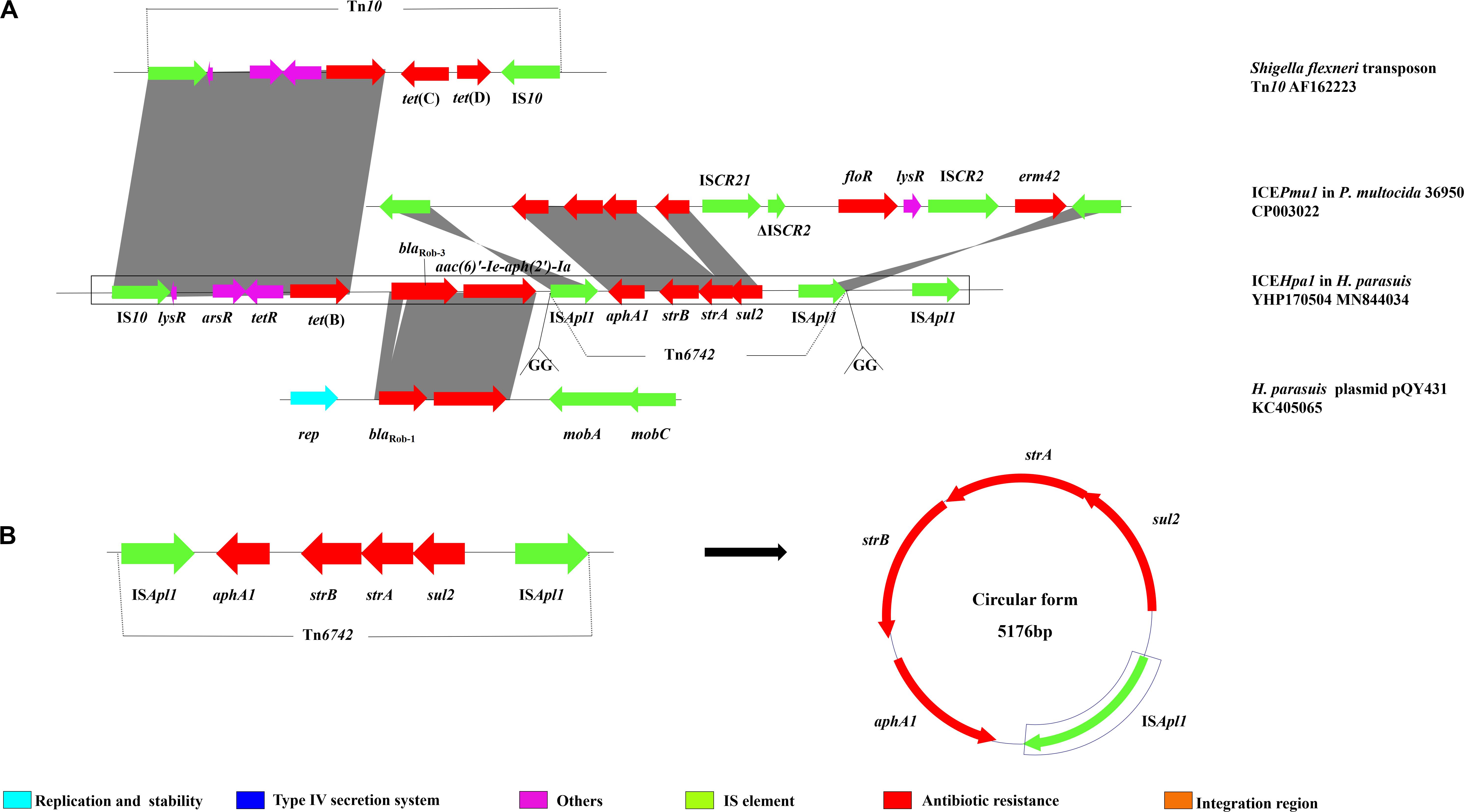
Figure 3. (A) Schematic representation the resistance gene region of ICEHpa1. Genes are presented as arrows, with the arrowhead indicating the direction of transcription. Linear comparison of resistance gene region of ICEHpa1 with the homologous region of the transposon Tn10 of Shigella flexneri (AF162223), plasmid pQY431 of H. parasuis (KC405065), and ICEPmu1 of P. multocida 36950 (CP003022). (B) The circular form of the ISApl1-aphA1-strB-strA-sul2-ISApl1 segment in ICEHpa1.
Conclusion
In summary, a novel ICE was identified from a serovar 8 H. parasuis ST288 isolate. To our knowledge, this is the first time a multidrug-resistance ICE has been reported in H. parasuis. A total of seven different resistance genes were located on the ICEHpa1, which suggests that the ICEHpa1 may act as a reservoir for various resistance genes. Therefore, more research and effective surveillance is needed to monitor the dissemination of multidrug-resistance ICEs.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics Statement
This study was carried out in accordance with the guidelines of Henan Agricultural University Animal Ethics Committee. The owners of the farm animals from which samples were taken gave permission for their animals to be used in this study.
Author Contributions
G-ZH conceived and designed the experiments. H-RS, X-DC, X-KL, and K-FY produced the data. H-RS, Y-SP, D-DH, HW, LY, and G-ZH analyzed the data. H-RS and G-ZH wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was financed by the National Key Research and Development Program of China (2016YFD05101304).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Bossé, J. T., Li, Y., Roberto, F. C., Chaudhuri, R. R., Rogers, J., Holden, M. T. G., et al. (2016). ICEApl1, an integrative conjugative element related to ICEHin1056, identified in the pig pathogen Actinobacillus pleuropneumoniae. Front. Microbiol. 7:810. doi: 10.3389/fmicb.2016.00810
Brenner, M. G., Kristina, K., Sweeney, M. T., Elzbieta, B., Heiko, L., Rolf, D., et al. (2012). ICEPmu1, an integrative conjugative element (ICE) of Pasteurella multocida: analysis of the regions that comprise 12 antimicrobial resistance genes. Jo. Antimicrob. Chemother. 67, 84–90. doi: 10.1093/jac/dkr406
Burrus, V., Pavlovic, G., Decaris, B., and Guedon, G. (2002). Conjugative transposons: the tip of the iceberg. Mol. Microbiol. 46, 601–610. doi: 10.1046/j.1365-2958.2002.03191.x
Burrus, V., and Waldor, M. K. (2004). Shaping bacterial genomes with integrative an conjugative elements. Res. Microbiol. 155, 376–386. doi: 10.1016/j.resmic.01.012
Chen, L. P., Cai, X. W., Wang, X. R., Zhou, X. L., Wu, D. F., Xu, X. J., et al. (2010). Characterization of plasmid-mediated lincosamide resistance in a field isolate of Haemophilus parasuis. J. Antimicrob. Chemother. 65, 2256. doi: 10.1093/jac/dkq304
Clinical and Laboratory Standards Institute [CLSI] (2018). “Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals,” in CLSI supplement VET08, 4th Edn (Wayne, PA: CLSI).
Eidam, C., Poehlein, A., Leimbach, A., Michael, G. B., Kadlec, K., Liesegang, H., et al. (2015). Analysis and comparative genomics of ICEMh1, a novel integrative and conjugative element (ICE) of Mannheimia haemolytica. Jo. Antimicrob. Chemother. 70, 93–97. doi: 10.1093/jac/dku361
Howell, K. J., Peters, S. E., Wang, J., Hernandez-Garcia, J., Weinert, L. A., Luan, S. L., et al. (2015). Development of a multiplex PCR assay for rapid molecular serotyping of Haemophilus parasuis. J. Clin. Microbiol. 53, 3812–3821. doi: 10.1128/jcm.01991-15
Johnson, C. M., and Grossman, A. D. (2015). Integrative and Conjugative Elements (ICEs): what They Do and How They Work. Ann. Rev. Genet. 49, 577–601. doi: 10.1146/annurev-genet-112414-055018
Juhas, M., Power, P. M., Harding, R. M., Ferguson, D. J. P., Dimopoulou, I. D., Elamin, A. R. E., et al. (2007). Sequence and functional analyses of Haemophilus spp. genomic islands. Genome Biol. 8:R237. doi: 10.1186/gb-2007-8-11-r237
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lancashire, J. F., Terry, T. D., Blackall, P. J., and Jennings, M. P. (2005). Plasmid-encoded tet B tetracycline resistance in Haemophilus parasuis. Antimicrob. Agents Chemother. 49:1927. doi: 10.1128/AAC.49.5.1927-1931.2005
Lei, C. W., Zhang, A. Y., Wang, H. N., Liu, B. H., Yang, L. Q., and Yang, Y. Q. (2016). Characterization of SXT/R391 integrative and conjugative elements in Proteus mirabilis isolates from food-producing animals in China. Antimicrob. Agents Chemother. 60:1935. doi: 10.1128/AAC.02852-15
Li, B., Zhang, Y., Wei, J., Shao, D., Liu, K., Shi, Y., et al. (2015). Characterization of a novel small plasmid carrying the florfenicol resistance gene floR in Haemophilus parasuis. J. Antimicrob. Chemother. 70, 3159–3161. doi: 10.1093/jac/dkv230
Li, R., Xie, M., Dong, N., Lin, D., Yang, X., Wong, M. H. Y., et al. (2018). Efficient generation of complete sequences of MDR-encoding plasmids by rapid assembly of MinION barcoding sequencing data. Gigascience 7, 1–9. doi: 10.1093/gigascience/gix132
Li, Y., Li, Y., Crespo, R. F., Leanse, L. G., Langford, P. R., and Bosse, J. T. (2018). Characterization of the Actinobacillus pleuropneumoniae SXT-related integrative and conjugative element ICEApl2 and analysis of the encoded FloR protein: hydrophobic residues in transmembrane domains contribute dynamically to florfenicol and chloramphenicol efflux. J. Antimicrob. Chemother. 73, 57–65. doi: 10.1093/jac/dkx342
Moleres, J., Santos-López, A., Lázaro, I., Labairu, J., Prat, C., Ardanuy, C., et al. (2015). Novel blaROB-1-bearing plasmid conferring resistance to β-lactams in Haemophilus parasuis isolates from healthy weaning pigs. Appl. Environ. Microbiol. 81, 3255–3267. doi: 10.1128/AEM.03865-3814
Mullins, M. A., Register, K. B., Brunelle, B. W., Aragon, V., Galofré-Mila, N., Baylese, D., et al. (2013). A curated public database for multilocus sequence typing (MLST) and analysis of Haemophilus parasuis based on an optimized typing scheme. Vet. Microbiol. 162, 899–906. doi: 10.1016/j.vetmic.2012.11.019
Mulvey, M., Martin, M., Alfa, M., and Ng, R. M. K. (2001). Multiplex PCR for the detection of tetracycline resistant genes. Mol. Cell Probes. 15, 209–215. doi: 10.1006/mcpr.2001.0363
Oliveira, S., Galina, L., and Pijoan, C. (2001). Development of a PCR test to diagnose Haemophilus parasuis infections. J. Vet. Diagn. Invest. 13, 495–501. doi: 10.1177/104063870101300607
Oliveira, S., and Pijoan, C. (2004). Haemophilus parasuis: new trends on diagnosis epidemiology and control. Vet. Microbiol. 99, 1–12. doi: 10.1016/j.vetmic.2003.12.001
Robinson, E., Didelot, X., Hood, D., and Crook, D. (2013). Comparative phylogenetics of ICEHin1056 family reveals deep evolutionary associations of mobile genetic elements responsible for transfer of antibiotic resistance genes. Lancet 381(Suppl.1):S93. doi: 10.1016/S0140-6736(13)60533-4
Tegetmeyer, H. E., Jones, S. C. P., Langford, P. R., and Baltes, N. (2008). ISApl1, a novel insertion element of Actinobacillus pleuropneumoniae, prevents ApxIV-based serological detection of serotype 7 strain AP76. Vet. Microbiol. 128, 342–353. doi: 10.1016/j.vetmic.2007.10.025
Toleman, M. A., and Walsh, T. R. (2011). Combinatorial events of insertion sequences and ICE in Gram-negative bacteria. FEMS Microbiol. Rev. 35, 912–935. doi: 10.1111/j.1574-6976.2011.00294.x
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Wright, L. D., Johnson, C. M., Grossman, A. D., and Viollier, P. H. (2015). Identification of a single strand origin of replication in the integrative and conjugative element ICEBs1 of Bacillus subtilis. PLoS Genet. 11:e1005556. doi: 10.1371/journal.pgen.1005556
Keywords: multiresistance, H. parasuis, ICE, conjugation, transposon
Citation: Sun H-R, Cui X-D, Liu X-K, Li S-H, Yi K-F, Pan Y-S, Wu H, Yuan L, Hu G-Z and He D-D (2020) Molecular Characterization of a Novel Integrative Conjugative Element ICEHpa1 in Haemophilus parasuis. Front. Microbiol. 11:1884. doi: 10.3389/fmicb.2020.01884
Received: 24 April 2020; Accepted: 17 July 2020;
Published: 13 August 2020.
Edited by:
Axel Cloeckaert, Institut National de la Recherche Agronomique (INRA), FranceReviewed by:
Kristina Kadlec, Independent Researcher, Wunstorf, GermanyPatrick Boerlin, University of Guelph, Canada
Ruth M. Hall, The University of Sydney, Australia
Séamus Fanning, University College Dublin, Ireland
Copyright © 2020 Sun, Cui, Liu, Li, Yi, Pan, Wu, Yuan, Hu and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gong-Zheng Hu, eWFvbGlsYWJAMTYzLmNvbQ==; Dan-Dan He, OTk1MTI3ODE0MkBxcS5jb20=
†These authors have contributed equally to this work
 Hua-Run Sun†
Hua-Run Sun† Gong-Zheng Hu
Gong-Zheng Hu