- 1Department of Virology, Faculty of Medicine, Iran University of Medical Sciences, Tehran, Iran
- 2Student Research Committee, Iran University of Medical Sciences, Tehran, Iran
- 3Wellman Center for Photomedicine, Massachusetts General Hospital, Boston, MA, United States
- 4Department of Dermatology, Harvard Medical School, Boston, MA, United States
- 5Laser Research Centre, Faculty of Health Science, University of Johannesburg, Doornfontein, South Africa
- 6Student Research Committee, Mashhad University of Medical Sciences, Mashhad, Iran
- 7Department of Medical Biotechnology, Faculty of Medicine, Mashhad University of Medical Sciences, Mashhad, Iran
- 8Department of Regenerative Medicine, Cell Science Research Center, Royan Institute for Stem Cell Biology and Technology, ACECR, Tehran, Iran
- 9Department of Gynecology and Obstetrics, Mahdieh Hospital, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 10Department of Microbiology, Faculty of Medicine, Kermanshah University of Medical Sciences, Kermanshah, Iran
- 11Infectious and Tropical Diseases Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
- 12Immunology Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
- 13Research Center for Biochemistry and Nutrition in Metabolic Diseases, Kashan University of Medical Sciences, Kashan, Iran
Covalent closed circular RNAs (circRNAs) can act as a bridge between non-coding RNAs and coding messenger RNAs. CircRNAs are generated by a back-splicing mechanism during post-transcriptional processing and are abundantly expressed in eukaryotic cells. CircRNAs can act via the modulation of RNA transcription and protein production, and by the sponging of microRNAs (miRNAs). CircRNAs are now thought to be involved in many different biological and pathological processes. Some studies have suggested that the expression of host circRNAs is dysregulated in several types of virus-infected cells, compared to control cells. It is highly likely that viruses can use these molecules for their own purposes. In addition, some viral genes are able to produce viral circRNAs (VcircRNA) by a back-splicing mechanism. However, the viral genes that encode VcircRNAs, and their functions, are poorly studied. In this review, we highlight some new findings about the interaction of host circRNAs and viral infection. Moreover, the potential of VcircRNAs derived from the virus itself, to act as biomarkers and therapeutic targets is summarized.
Introduction
Single-stranded circular RNAs (circRNAs) belong to the non−coding RNA family. Unlike linear RNAs, they are take the form of a covalently closed continuous loop with neither 5′ capping nor 3′ polyadenylation, and are formed by a back-splicing process (Guo et al., 2014; Holdt et al., 2018). Sanger et al. (1976) originally discovered the presence of circRNAs in a viroid-infected plant using electron microscopy in 1970. Later, the presence of circRNA was detected in the hepatitis D virus (HDV) and in yeast mitochondria (Arnberg et al., 1980). The first study to confirm the presence of circRNAs in human cells by Nigro et al. (1991), reported the detection of circular transcripts derived from the tumor suppressor gene DCC in several human tumor cell lines. Recently, with the advent of new sequencing technologies, such as next-generation sequencing (NGS), a growing number of circRNAs have been reported (Hanan et al., 2017; Wang et al., 2017; Zhang Z.-C. et al., 2018; Zaiou, 2019), and have now become a “hot topic.”
Since some circRNAs have the ability to encode proteins, they have therefore been suggested as a crucial bridge between non-coding RNAs and coding RNAs (Braicu et al., 2019). To date, the function(s) of several circRNAs have been identified, including the sponging of microRNA (miRNA), regulation of RNA transcription and protein production, and the translation of proteins and peptides (Li J. et al., 2015). Considering the multifunctional nature of circRNAs, they may be involved in many biological and pathological processes, which could influence the progression of diseases such as cancer and viral infections. In cancer it has been suggested that circRNAs could affect the malignant phenotype through regulating cancer-related pathways, and could exert either an anti-cancer activity or a pro-cancer activity. Therefore, these molecules could either act as tumor suppressors or alternatively as oncogenes depending on the tumor type and stage (Wang et al., 2017), and could therefore serve as a therapeutic target in the treatment of cancer.
Moreover, the unique covalent closed-loop structure of circRNAs makes them insensitive to the enzyme activity of ribonucleases (Harland and Misher, 1988). The expression levels of circRNAs are generally lower than those of messenger RNAs (mRNAs) (Jeck et al., 2013; Guo et al., 2014; Shang et al., 2019). However, circRNAs mostly act in a tissue and cell type-specific manner, and have been found to be stably expressed in several biological materials including saliva, tissue, blood, and exosomes. Therefore, circRNAs could be potential biomarkers in the diagnosis and prognosis of several different diseases (Zhang Z. et al., 2018; Naeli et al., 2019). Up to now, only relatively few studies have been conducted on the interaction between viruses and host circRNAs; however, it has been reported that the expression patterns of host circRNAs are altered in virus-infected cells and patients compared to the control groups (Cui et al., 2018; Shi et al., 2018; Zheng et al., 2018; Yu T. et al., 2019). Therefore, it has been proposed that viruses are likely to use these molecules for their own progression. It has also been observed that some viral genes are able to produce viral circRNA (VcircRNA) molecules through back-splicing (Toptan et al., 2018; Ungerleider et al., 2019; Zhao et al., 2019b), but the viral genes that can encode circRNAs and their functions are poorly studied. In this review, we will first briefly discuss the biogenesis, and function of circRNAs, and then highlight some new findings concerning the interaction of host circRNAs with viral infections. Lastly, circRNAs derived from the virus genome and their potential as biomarkers or therapeutic targets for viral diseases are summarized.
The Biogenesis of circRNAs
In eukaryotic cells, the splicing of precursor mRNA (pre-mRNA) is catalyzed by spliceosomes. The spliceosome is an assembly of small nuclear ribonucleoproteins (snRNPs) and other protein factors that act to join together exons, and remove introns (Valadkhan, 2005). During splicing, if the upstream 5′ splice-site (donor site) is joined to the downstream 3′ splice site (acceptor site) this leads to the production of linear (m)RNAs. However the spliceosome can also generate circRNAs during the processing of pre-mRNA, pre-transfer ribonucleic acid (tRNA), and pre-ribosomal ribonucleic acid (rRNA) molecules (Zhang et al., 2017). These loop structures can originate from exons or from introns in either a single sequence or two different sequences (Xin et al., 2017). CircRNAs are considered to be a highly stable class of long non-coding RNAs, and are believed to be an important bridge between non-coding RNAs and coding RNAs (Braicu et al., 2019). CircRNAs are generally divided into three categories based on their components. Firstly, exonic circRNAs (ecircRNAs) are exclusively composed of exons, and represent the largest group of circRNAs. Secondly, intronic circRNAs (ciRNA) are exclusively composed of introns. Thirdly, exon-intron circRNAs (elciRNA) contain both exon and intron related sequences (Figure 1) (Wang et al., 2019). Intergenic circRNA is another non-exonic circRNA identified by a circRNA identifier (CIRI). This integrated circRNA is formed by two intronic circRNA fragments (ICFs) flanked by GT-AG splicing signals acting as the splice donor (SD) and splice acceptor (SA) sites of the circular junction (Geng et al., 2018). Studies using different techniques have shown that ecircRNAs are localized within the cytoplasm (Jeck and Sharpless, 2014). On the other hand, ciRNA and elciRNA are predominantly localized within the nucleus (Meng et al., 2017). The “direct back-splicing” and “lariat formation” are the two main mechanisms that can lead to the conversion of pre-mRNAs into circRNAs (Qu et al., 2015; Yang et al., 2017; Braicu et al., 2019). In the back splicing process, the downstream 5′ donor site is linked to the upstream 3′ acceptor site resulting in the production of circRNAs (Kristensen et al., 2019; Zhao et al., 2019c). Interestingly, multiple circRNAs can be generated through two different types of back-splicing, including alternative 5′ back-splicing (A5BS), and alternative 3′ back-splicing (A3BS). A5BS occurs when two or more 5′ downstream back-splice sites bind to the same upstream 3′ back-splice site in a reverse orientation. Similarly, A3BS occurs when two or more upstream 3′ back-splice sites bind to the same downstream 5′ back-splice site (Qu et al., 2015; Dong et al., 2018).
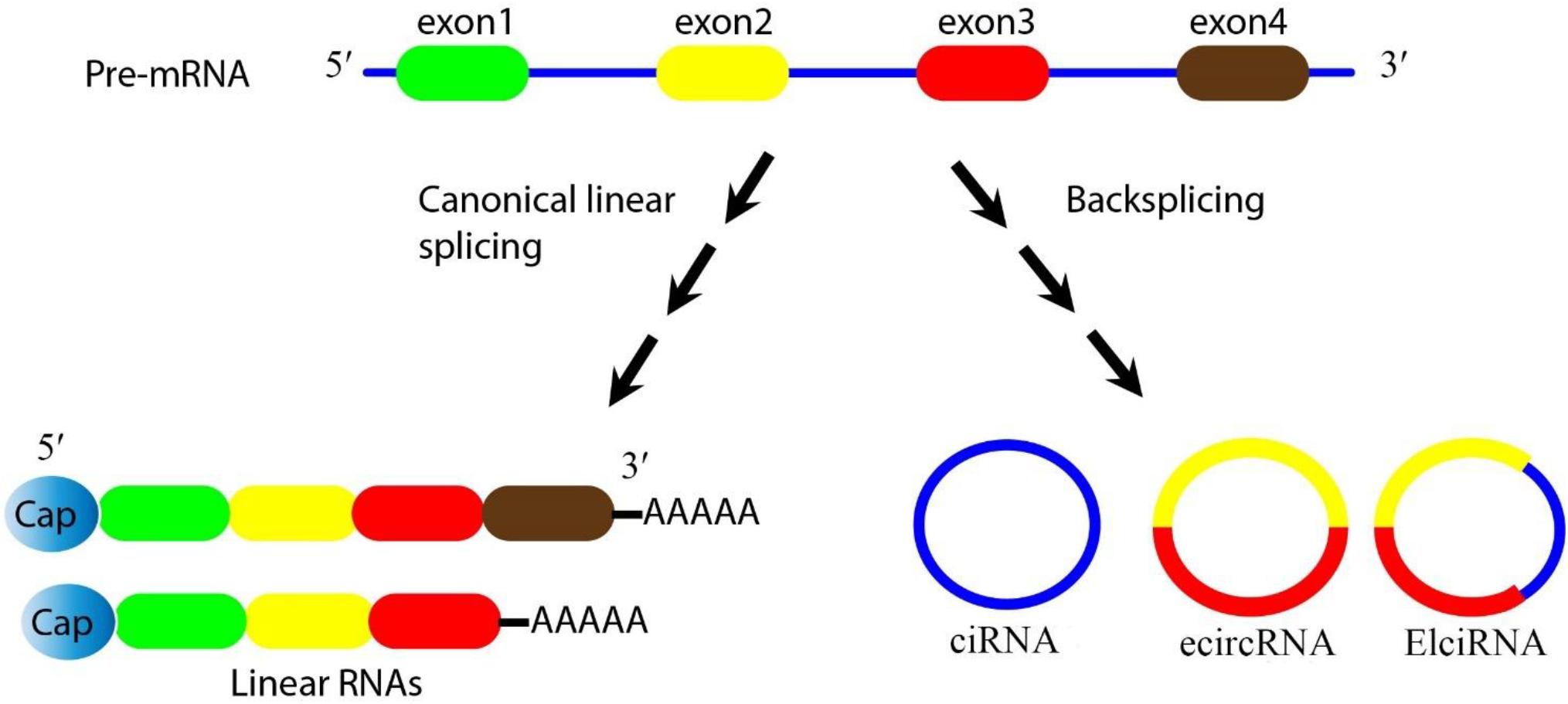
Figure 1. Biogenesis of circRNAs. Pre-mRNA can be processed through either canonical splicing to produce linear RNAs (left) or backsplicing to generate circRNAs (right). CircRNAs are mainly divided into three categories based on their components. Exonic circRNAs (ecircRNAs) are exclusively composed of exons and represent the largest group of circRNAs, intronic circRNA (ciRNA) are exclusively composed of introns, and exon-intron circRNA (elciRNA) are exon-intron sequences. In this figure, arrows show the splicing events for generating the products, blue lines show introns, and colored lines (red/yellow) show exons.
The two different types of the lariat formation mechanism, are “exon skipping” and “intron−pairing−driven” circularization (Wang et al., 2015). In intron−pairing−driven circularization, the 5′ end of the pre-mRNA is cleaved by the U1 small nuclear RNA (snRNA), and the 5′- and 2′-bases between the guanidine and adenosine are ligated together. The processed intronic lariat is retained in a circular form within the nucleus (Konarska et al., 1985; Hansen et al., 2013; Braicu et al., 2019). Exon skipping occurs naturally only at a low frequency (Goyenvalle et al., 2004). In exon skipping a “hetero-lariat” is formed that contains both introns and exons (Suzuki et al., 2016; Zhu et al., 2017; Braicu et al., 2019). This process could continue until all the introns have been removed, and only circRNAs containing exonic sequences are formed (Yang et al., 2017).
Some studies have shown that various viruses can encode circRNAs by alternative splicing of the viral mRNAs (Bodescot and Perricaudet, 1986; Purcell and Martin, 1993). Thus, viruses can produce VcircRNAs (Zhao et al., 2019b) using a back-splicing mechanism (Toptan et al., 2018). However, only few studies have been performed on this topic and more experimental studies are needed.
The Functions of circRNAs
Considering the great diversity of circRNAs and their many individual targets, it is to be expected that circRNAs will play several key roles within the cell, but not all of them have yet been well characterized. However, some cellular and molecular roles of circRNAs have been identified (Ruskin and Green, 1990; Vicens and Westhof, 2014). As mentioned above, circRNAs can be derived from introns (intronic circRNA or ciRNA), exons (exonic circRNA or ecircRNA) and exon-intron sequences (exon-intron circRNA or elciRNA) (Zhang et al., 2013; Zhang et al., 2014; Chen et al., 2015; Li Z. et al., 2015). Some elciRNAs and ciRNAs are found in the nucleus and are likely to play a regulatory function in gene transcription, whereas ecircRNAs are predominantly found in the cytoplasm, and are involved in post-transcriptional regulation (Haddad and Lorenzen, 2019). The structural examination of some circRNAs has shown that they contain N6-methyladenosine modifications or an internal ribosome entry site (IRES), and have the potential to be efficiently translated into proteins (Yu and Kuo, 2019). Depending on the type of protein produced, they could play important roles within the cell. For example, Yang et al. (2019) found that circ-FBXW7 was abundantly expressed in the normal human brain tissue, and reported that circ-FBXW7 encodes the FBXW7-185aa protein. The FBXW7-185aa protein inhibits cell proliferation and tumorigenesis, while its down-regulation is associated with the induction of a malignant phenotype in cancer cells (Yang et al., 2019). Additionally, circRNAs may also affect the activity of proteins, and are probably involved in the modulation of protein–protein interactions, protein binding, and protein sorting (Du et al., 2017). For example, circRNA CDR1 interacts strongly with argonaute (AGO) proteins, which are essential components of the RISC complex (Memczak et al., 2013).
CircRNAs could also regulate the activity of miRNAs via a novel mechanism (Kulcheski et al., 2016). Endogenous circRNAs can function as miRNA sponges, to negatively regulate the activity of miRNAs by removing them from their site of action (Hansen et al., 2013; Memczak et al., 2013). Since miRNAs control many biological events, circRNAs could influence these processes through a miRNA sponging effect (Kulcheski et al., 2016). It has been shown that several miRNA sponges may be associated with human disease, such as Alzheimer’s, Hirschsprung’s, diabetes, osteoarthritis, and several types of cancer (Lukiw, 2013; Wang et al., 2016; Zhao et al., 2016; Han et al., 2017; Peng et al., 2017; Zhao et al., 2017; Zhou and Yu, 2017; Zhou Z.-B. et al., 2018; Kristensen et al., 2018).
For example, it has been shown that some circRNAs can contribute to cancer progression by sponging tumor-suppressive miRNAs, or alternatively suppress the cancer phenotype via sponging of oncogenic miRNAs. It has generally been observed that tumor suppressor circRNAs are down-regulated in cancers, while oncogenic circRNAs are up-regulated. For instance, Hao et al. (2019) reported that circ_0007534 was over-expressed in patients with pancreatic ductal adenocarcinoma. They also found that ectopic expression of circ_0007534 caused increased proliferation, invasion, and migration in the PDAC cell line. This oncogenic effect of circ_0007534 was mediated by sponging of miR-892b and miR-625 (Hao et al., 2019). Circ_0026344 was found to be down-regulated in colorectal cancer, while the expression level of miR-31 and miR-21 was increased in colorectal cancer tissue. Further analysis demonstrated that over-expression of circ_0026344 decreased tumor growth and increased apoptosis in cell lines, through sponging of miR-21 and miR-31 (Yuan et al., 2018).
However, up to now only a few studies have been performed on the function of circRNAs in viral infections, and the role they may play in inhibiting or enhancing virus replication is not well understood. A summary of the circRNA functions is shown in Figure 2.
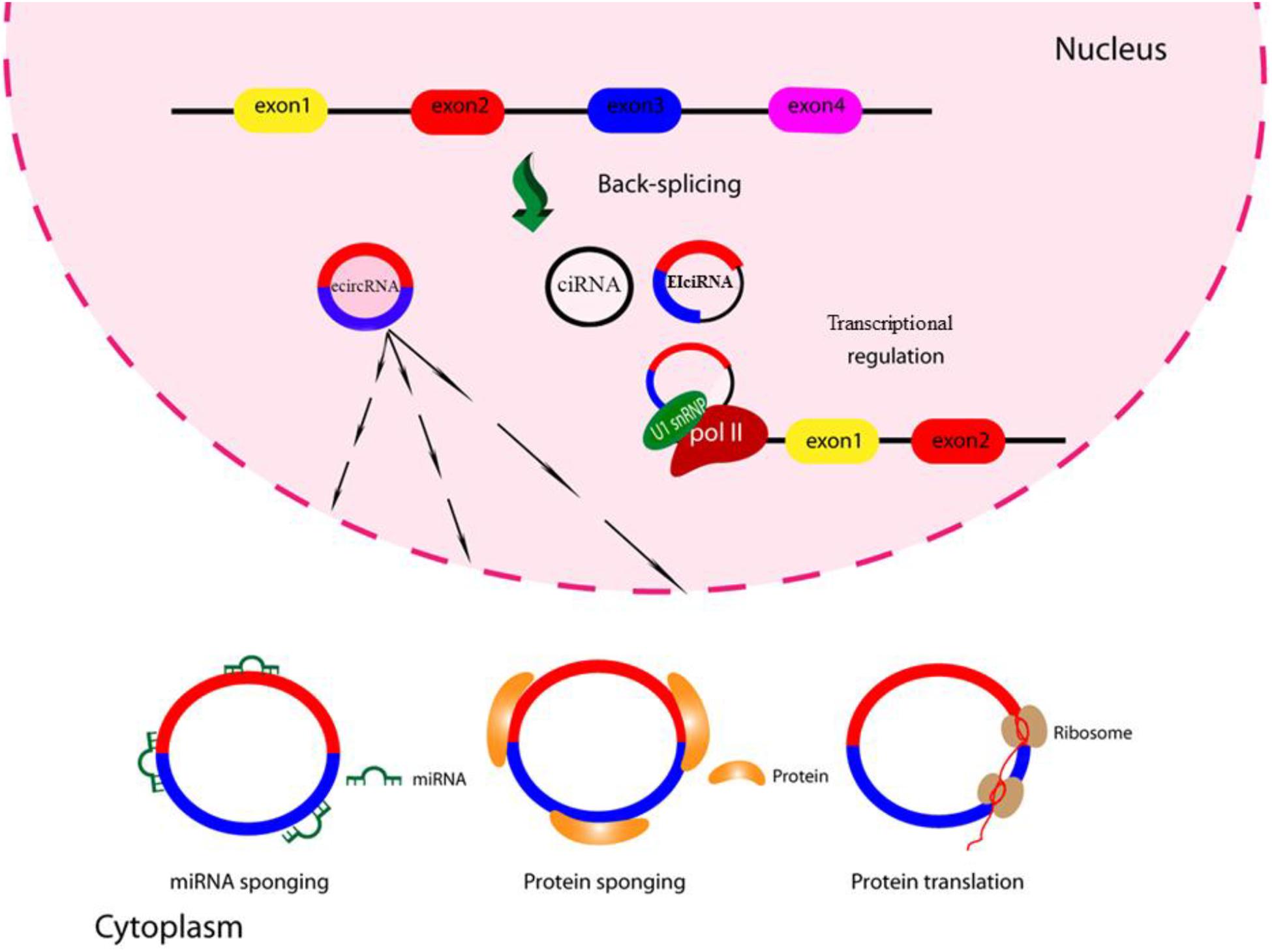
Figure 2. Schematic of circRNA functions. CircRNAs might function as miRNA sponges by competing for the binding of miRNA sequences, lessening the impact of miRNA-mediated regulation of gene expression. CircRNAs might function as protein sponges. Some circRNAs might control the expression of proteins by sequestering mRNA translation start sites. CircRNAs might be translated to create functional proteins.
Host circRNAs and Viral Infections
As discussed above, it has been shown that the deregulation of circRNA expression in cancer cells, i.e., down-regulation or up-regulation, can contribute to the malignancy of cancer cells (Wu et al., 2019). However, it is not yet clear whether the aberrant expression of circRNAs in virus-infected cells, ultimately contributes to viral replication or not. It has been demonstrated that viral infections can remodel the entire transcriptome landscape of host cells (Park et al., 2015; Hu et al., 2016). One important effect is to improve the viral replication cycle through modifying transcriptomic responses related to antiviral immunity and cellular apoptosis. This common strategy has been observed in many different viral infections (Gao et al., 2017a; Martin et al., 2017; Wang et al., 2017). HSV-1 infection, in addition to altering the expression of cellular genes, can affect RNA processing in the host cells leading to changes in alternative polyadenylation and splicing in the host transcriptome (Zheng et al., 2017; Shi et al., 2018). An interaction between viruses and circRNAs was observed in a study by Li et al. (2017). They reported that the transcription factor NF90/NF110 (derived from interleukin enhancer binding factor, ILF3) could regulate the biogenesis and function of circRNAs. Upon viral infection, these factors led to the down-regulation of circRNA levels (Li et al., 2017).
Recently, Shi et al. (2018) examined the profile of the circRNA transcriptome in HSV-infected cells. In this study, the aberrant circRNA expression profile in the HSV-1-infected KMB17 cell line was investigated using deep RNA sequencing. The results showed that the expression levels of 536 separate circRNAs were significantly dysregulated after HSV-1 infection, and of these, 348 circRNAs were down-regulated and 188 were up-regulated. The expression levels of five circRNAs (circRNA7231, circRNA3683, circRNA3046, circRNA6783, and circRNA7752) were measured by real time quantitative reverse transcription (RT-qPCR), and it was found that their expression was significantly increased after infection with HSV-1 (Shi et al., 2018). However, the function and interaction of these circRNAs with the HSV-1 virus, and whether they could be used as diagnostic biomarkers or therapeutic targets is still unclear.
Cervical cancer is the fourth most common cancer among women throughout the world (Bhadelia, 2019). Persistent infection with the high risk type of human papillomaviruses (HPV-16 and HPV-18) is the main cause of the progression of papilloma lesions to cervical cancer, and is found in more than 70% of cases (Ghittoni et al., 2010). HPV E6 and E7 oncoproteins play a critical role in the transformation of cervical cells by interfering with the p53 and pRb pathways, respectively (Burroni et al., 2013). In addition, HPV E6 and E7 oncoproteins can modulate the expression of cellular miRNAs, which may contribute to the tumorigenesis of cervical cancer cells (Nahand et al., 2019). Since circRNAs act as sponges for miRNAs, the virus is likely to indirectly alter the expression level of miRNAs by affecting circRNAs. Current studies suggest that circRNAs have the potential to be used as diagnostic biomarkers in cervical cancer (Chaichian et al., 2019). Gao et al. (2017b) demonstrated that hsa_circ_0018289 was over-expressed in cervical cancer cells, and knockdown of this circRNA could suppress the migration, proliferation, and invasion of cervical cancer cells. They suggested hsa_circ_0018289 may play an oncogenic role in the tumorigenesis of cervical cancer (Gao et al., 2017b).
One study evaluated the circRNA expression profile in cervical cancer cells infected with HPV16 E7. Using a microarray approach, they found that 526 separate circRNAs had significantly altered expression levels (174 down-regulated 352 up-regulated) (Zheng et al., 2018). In order to confirm these results, eight circRNAs with different expression levels were measured using the qRT-PCR method, and it was found that the results of both methods were in agreement. The finding that the expression pattern of circRNAs in HPV-16 E7 infected cells was altered (Zheng et al., 2018), provided new insights into potential therapeutic targets and candidate biomarkers in cervical cancer associated with HPV.
MicroRNAs are involved in many cellular biological processes, including inflammation, cellular differentiation, apoptosis, and also in viral infection (Hagen and Lai, 2008; Hulsmans and Holvoet, 2013; Sadri et al., 2019). Transmissible gastroenteritis virus (TGEV) is a member of the Coronavirus family with a positive-sense single-stranded RNA genome, which emerged as a cause of severe gastroenteritis in pigs (Laude et al., 1990). TGEV can activate the NF-κB pathway resulting in severe inflammation in the epithelial cells of porcine intestines (Ma et al., 2014). Ma et al. (2018) analyzed the expression profile of circRNAs, miRNAs, and mRNAs in TGEV-infected IPEC-J2 (intestinal porcine epithelial cell-jejunum 2) cell line using NGS technology. They observed that miR-22 was over-expressed in TGEV infected cells, and suggested that ssc_circ_009380 by sponging of miR-22 was able to trigger NF-κB pathway activity. It is known that miR-22 exerts its anti-inflammatory effects via targeting IL-6, CCL5 and DDX58 (Ma et al., 2018). Moreover, it was demonstrated that miR-22 could attenuate myocardial ischemia-reperfusion injury in rats via an anti-inflammatory mechanism (Yang et al., 2016).
The hepatitis B virus (HBV) belongs to the Hepadnaviridae family, and between 5 and 10% of HBV-infected patients develop a chronic liver infection about 6 months after the acute infection (Mayerat et al., 1999). Chronic HBV infection can cause serious liver diseases, such as cirrhosis and hepatocellular carcinoma (HCC) (Sinn et al., 2015). The progression of HBV infection in many individuals is limited by the host immune response. However, in others the infections becomes chronic due to comparatively weak T-cell responses against HBV, suppression of NK cell function by several immunomodulatory cytokines, and ineffective adaptive responses against HBV viral infected cells (Mayerat et al., 1999; Tan et al., 2015). Other factors, such as alcohol, aflatoxin, gut microbiota, mannose-binding lectin (MBL) genotype, and also miRNAs, can affect the progression of chronic hepatitis B (Ohnishi et al., 1982; Handin et al., 2003; Chong et al., 2005; Sagnelli et al., 2018; Sadri Nahand et al., 2019). Evidence suggests that some circRNAs are associated with the carcinogenesis process of HCC (Fu et al., 2018). Zhu et al. (2018) showed that hsa_circ_0067934 by sponging of miR−1324, could alter the FZD5/Wnt/β−catenin signaling pathway, which was involved in migration, proliferation, and invasion of HCC cells. However, the role of HBV infection in altering the expression of circRNAs, which may then contribute to liver disease, is poorly characterized. Yang et al. (2016, 2017, 2019) investigated the deregulation of circRNAs in hepatitis B-related HCC, and found that circRNA_100338 was significantly over-expressed in HCC tissue. They also found that circRNA_100338 could act as a miR-141-3p sponge in HCC tissue, and suggested that circRNA_100338 could be a new biomarker for the diagnosis of HBV-associated HCC (Huang et al., 2017). Yu J. et al. (2019) examined the expression of circRNAs in plasma and HCC tissues from infected patients by microarray and qRT-PCR techniques. They reported that there was a positive correlation between the expression levels of hsa_circ_0139897, hsa_circ_0000976, and hsa_circ_0007750 in the patient samples, and they were significantly increased after hepatectomy (Yu J. et al., 2019). Data concerning the role of circRNAs as diagnostic or therapeutic biomarkers in HBV-HCC is limited and needs further investigation.
Autophagy is a form of programmed cell death, and is a highly conserved cellular process designed to dispose of damaged cellular organelles or protein aggregates (Pourhanifeh et al., 2020). Autophagy is activated in many viral infections such as HSV-1, HBV, and influenza A virus (IAV). Autophagy has been reported to either inhibit or stimulate the replication of some viruses, and plays a critical role in modulating cell survival (Ahmad et al., 2018). Additionally, it has been observed that viral proteins and noncoding RNAs produced during viral infections, can regulate autophagy in the host cells, which may contribute to escape from the immune system, release of viruses from cells, and viral replication (Kudchodkar and Levine, 2009; Fu et al., 2015; Wu et al., 2016). circRNAs have the ability to either attenuate or activate autophagy depending on the conditions (Du et al., 2018; Zhou L.-Y. et al., 2018), and therefore viruses may also affect autophagy by altering the expression of circRNA. Recently, it was reported that the expression level of circ-GATAD2A was up-regulated during infection of A549 cells with IAV. Furthermore, after the knockdown of circ-GATAD2A within the cells, H1N1 replication was suppressed and autophagy was promoted. In agreement, the up-regulation of circ-GATAD2A in A549 cells-infected with H1N1 promoted virus replication and also inhibited autophagy. Overall, the results of this study suggested that this circRNA could promote H1N1 replication by suppressing autophagy (Yu T. et al., 2019), and this finding might be useful for therapeutic purposes.
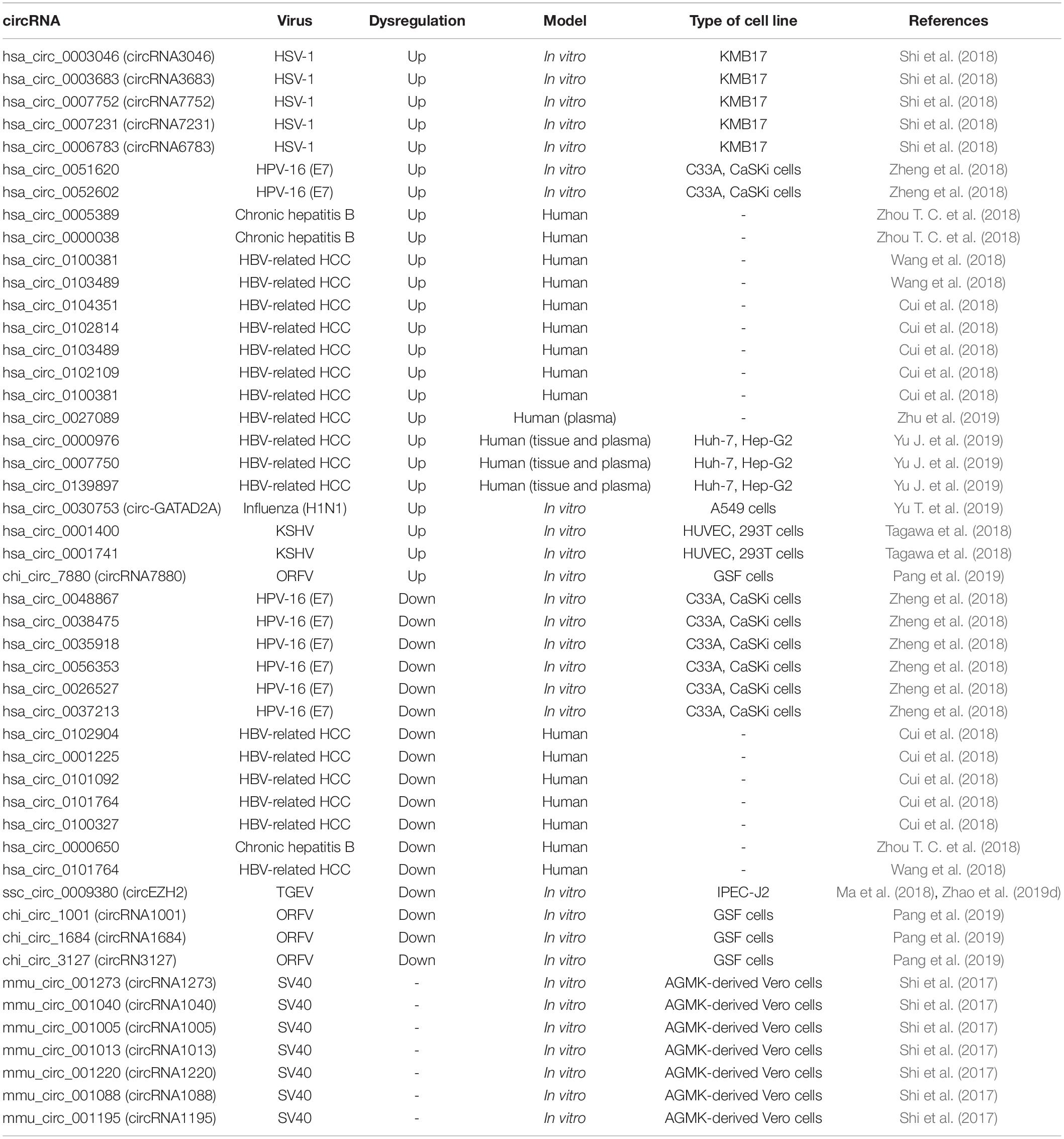
The role of circRNAs in viral replication has only so far been studied to a limited extent, however, given the significant changes observed in the circRNA expression profile during viral infections, we believe that viruses are likely to use this effect to their advantage. Reports of the deregulation of circRNAs during viral infections are shown in Table 1.
Virus-Encoded circRNAs
Viral infections in both humans and animals have been reported to occur with increased frequency in recent years. There are several types of viral disease, according to the underlying virus. HDV was the first human viral pathogen to be found to possess a circRNA within the genome (Kos et al., 1986). This circRNA can encode only one protein, hepatitis delta antigen (HDAg) (Farci, 2003). The Herpesviridae is a large family of DNA viruses, which can be categorized into three subfamilies: Alphaherpesvirinae, e.g., herpes simplex virus (HSV)-1, 2, varicella-zoster virus (VZV), Betaherpesvirinae, e.g., cytomegalovirus (CMV), human herpes virus (HHV)-6, 7, and Gammaherpesvirinae, e.g., Epstein Barr virus (EBV) and Kaposi’s sarcoma-associated herpes virus (KSHV). Almost all herpes viruses remain latent after the primary infection has subsided and are later reactivated under certain conditions (Pellett and Roizman, 2013). The viral replication and the expression of viral genes in the latent phase are limited, and this accounts for the ability of the virus to escape from the host immune response (Grinde, 2013). HPV is another viral infection that is passed between people through skin-to-skin contact. As discussed earlier, persistent infection with HPV-16 and HPV-18 is the main cause of the papilloma lesions that are precursors to cervical cancer, and is found in more than 70% of cases (Ghittoni et al., 2010). Anal squamous cell carcinoma (ASCC) is a rare malignancy, but high-risk strains of HPV have been implicated in 70–90% of ASCC cases (Martin et al., 2018).
Hepatitis D virus produces its circRNA molecules by using the cellular machinery and the rolling-circle mechanism. The connection of both ends of the RNA molecules by formation of 3′–5′ or 2′–5′ phosphodiester bonds leads to the formation of circRNA molecules (Reid and Lazinski, 2000; Eger et al., 2018). Two advantages of the circularization process of the HDV genome or viral encoded circRNAs could be suggested: (a) escape from recognition via innate immune pattern recognition receptors such as RIG-I and MDA-5; and (b) protection against degradation by intracellular exonucleases (Eger et al., 2018).
The innate immune system, the first line of host defense, is very important against pathogens. It contains pattern recognition receptors which respond to some specific structures that are typical of pathogens (Mogensen, 2009). Protein kinase R (PKR) is one of the receptors that recognize long dsRNAs (>33 bp) in the cytoplasm, and inhibits synthesis of protein. Although PKR should be readily activatable if needed, it should remain in an inactive state to prevent autoimmunity and inappropriate reactions. Studies have shown that activation of PKR can be inhibited by binding to the adenovirus small-noncoding VAI RNA or short (16–33 bp) dsRNAs. Liu C. X. et al. (2019) found that endogenous circRNAs could bind to PKR (Kitajewski et al., 1986; Zheng and Bevilacqua, 2004). Importantly, by comparing the binding profiles of circular and linear RNAs with the same base sequence, they found that circRNAs could bind more strongly to PKR than linear RNAs. This result suggested that the secondary structures of circRNAs and linear RNAs are different. In fact, structural mapping showed that circRNAs inside cells could form stable secondary structures which contained short (16–26 bp) imperfect duplexes, whereas the linear RNAs were folded into unstable and more dynamic structures (Liu C. X. et al., 2019).
As mentioned above, alternative splicing is a key post-transcriptional mechanism involved in the production of circRNA molecules from pre-mRNAs (Eger et al., 2018). One way that the virus could produce multiple products from a single gene, is alternative splicing. Since this mechanism occurs in the nucleus, DNA viruses (e.g., Adenoviridae, Herpesviridae, Papillomaviridae, Polyomaviridae, and Hepadnaviridae) (Ge and Manley, 1990; Tormanen et al., 2006; Verma and Swaminathan, 2008; Mole et al., 2009; Li et al., 2010; Guan et al., 2011) and some RNA viruses (e.g., Retroviridae, Bornaviridae, and Orthomyxoviridae) (Hope, 1999; Tomonaga et al., 2000; Robb and Fodor, 2012) that all replicate within the nucleus, may be capable of producing circRNAs. Recently, the RNA-sequencing technique (RNA-seq) has been used to identify the expression of circRNAs, and many results have been confirmed by RT-qPCR (Li and Han, 2019). RNA-seq was applied to evaluate the expression level of RNAs, and to survey the overall RNA population, including long noncoding RNA (lncRNA), small RNA, rRNA, and circRNA (Wang et al., 2009; Ingolia et al., 2012). The following methods have employed for different purposes in the RNA-seq experiments: (a) RiboMinus treatment was used for the accurate comparison between circRNA and mRNA expression levels; (b) RNase R and RiboMinus treatment was used for the comprehensive profiling of circRNAs; and (c) poly(A) enrichment was used to increase the concentrations of lncRNA and mRNA (Ji et al., 2019).
According to the different expression patterns of EBV genes, at least three distinct latent phases (I, II, and III) have been identified (Münz, 2015). In addition to its coding RNAs, the EBV virus can also express non-coding RNAs. It has been shown that the intronic regions of the Bam HI-A region rightward transcript (BART) gene of EBV is capable of expressing many non-coding RNAs (ncRNA), including the small non-coding EBV-encoded RNA 1 (EBER1) and EBER2 (Fok et al., 2006; Verhoeven et al., 2019). Unlike EBV, differential gene expression in various types of Karposi’s sarcoma tumor has been described (Dittmer and Damania, 2013). During latent infection with KSHV a number of non-coding RNAs have been detected, including miRNAs and the polyadenylated nuclear RNA (PAN) (Samols et al., 2007; Conrad, 2016; Toptan et al., 2018). PAN transcripts are important for KSHV replication and are abundantly expressed during lytic replication, but are found only in low levels during the latent phase (Conrad, 2016). The viral genes encoding circRNAs in many viruses have not yet been fully investigated.
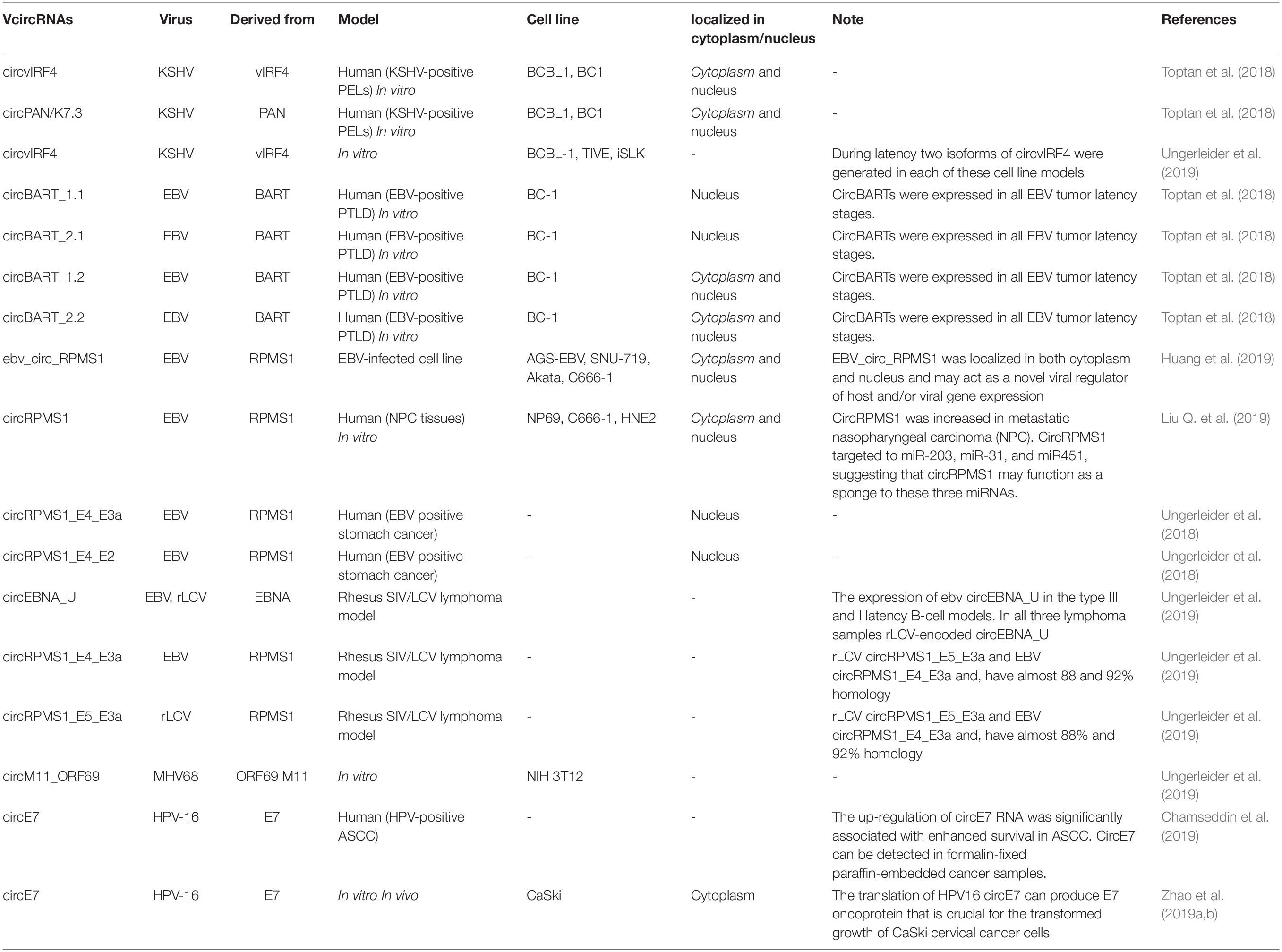
Toptan et al. (2018), for the first time investigated the VcircRNAs encoded by EBV and KSHV in tumor samples and cell lines. In this study RNA-seq was used to identify VcircRNAs after treatment with RNase R. RNase R-resistant RNA sequencing showed that both EBV and KSHV could encode several circRNAs. They observed that in EBV-positive PTLD samples, there were several EBV-derived circRNAs (circBARTs). These were both exon-intron and exon-only circRNAs, formed by back-splicing of BART transcripts. Similarly, circBARTs derived from BART are expressed in all types of latent EBV infection. The authors found that circBARTs were found in all tumors associated with EBV, and suggested that circBARTs could play a role in the reproductive ability of EBV-positive tumor cells. Furthermore, the sequencing of circRNAs within KSHV-infected primary effusion lymphoma (PEL) cells showed that KSHV circRNAs (including circvIRF4 and circPAN/K7.3) were expressed in these cells, and were found to originate from the vIRF4 locus and the PAN region, respectively (Toptan et al., 2018). These VcircRNAs may provide a novel approach for diagnostic biomarkers and therapy for KSHV and EBV associated malignancies.
Huang et al. (2019) used RNA-seq to investigate whether EBV is able to encode circRNAs in EBV-infected cell lines, including Akata (latency I), AGS-EBV (latency I), SNU-719 (latency I), C666-1 (latency II), and EBV-positive nasopharyngeal carcinoma (NPC) tissue samples. They found that ebv-circRPMS1 originated from exons 2–4 of the RPMS1 gene by back-splicing (Figure 3) (Huang et al., 2019). In another study, the role of EBV-encoded circRPMS1 in NPC tumorigenesis was examined (Liu Q. et al., 2019), and it was found that circRPMS1 expression was correlated with a shorter survival time, and was further up-regulated in metastatic NPC samples. Moreover, knockdown of circRPMS1 led to inhibition of proliferation and invasion of NPC cells infected with EBV, and induced apoptosis in these cells. Further investigation suggested that circRPMS1 was most probably involved in inducing the epithelial-mesenchymal transition (EMT) in NPC cells and encouraged oncogenesis via sponging of miR-31, miR-451, and miR-203. Down-regulation of this circRNA inhibited the aggressiveness and slowed down the EMT of NPC cells (Liu Q. et al., 2019). These studies suggest that circRPMS1 may be a potential therapeutic target for EBV-associated NPC.
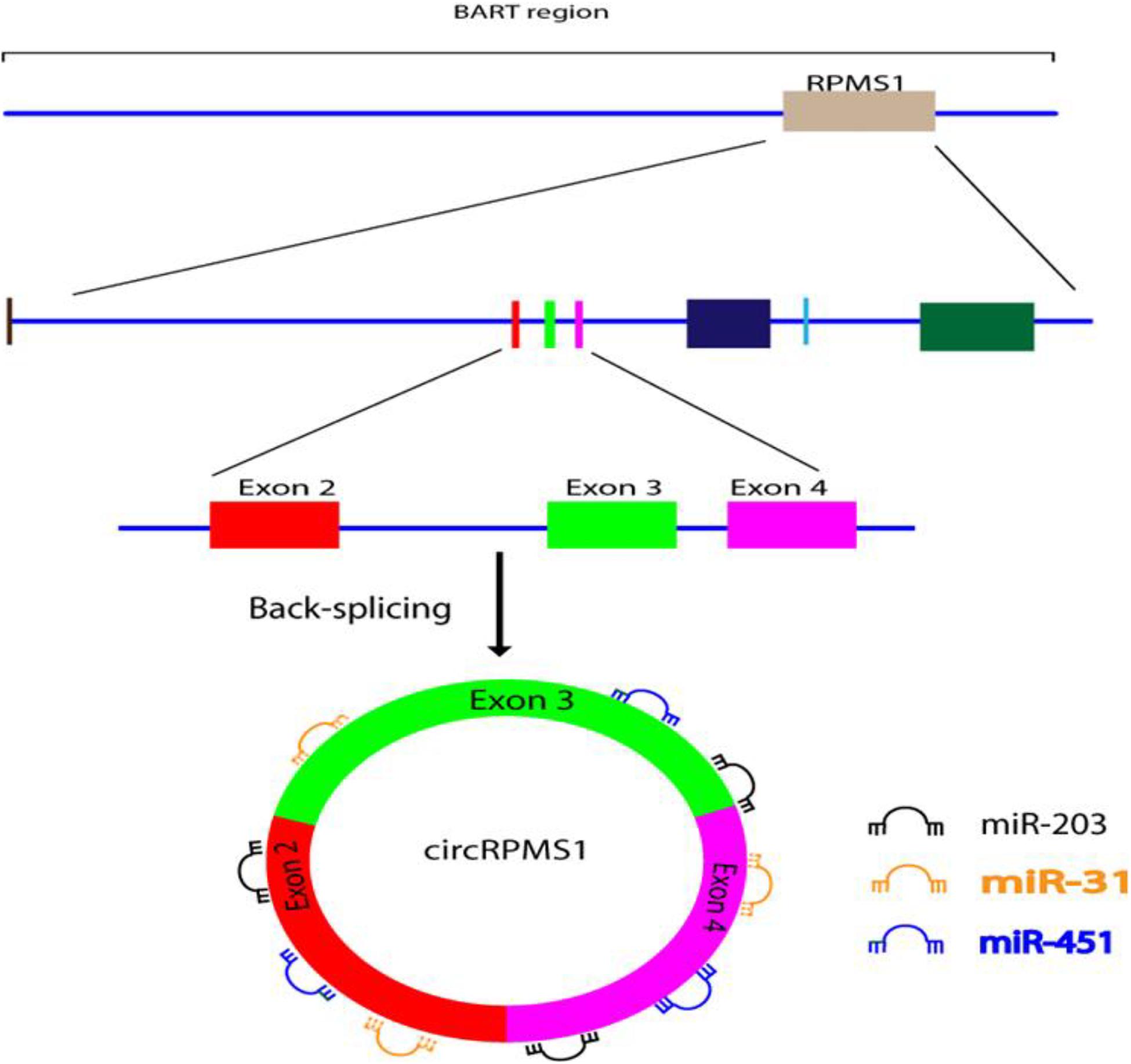
Figure 3. Formation and function of ebv-circRPMS1in EBV-infected NPC cells. Genomic location of the RPMS1 gene in EBV-BART region and ebv-circRPMS1 that originated from exon 2–4 of the RPMS1 gene by back-splicing. CircRPMS1 may be involved in the oncogenesis of NPC via sponging of miR-31, miR-451, and miR-203.
Alternative splicing and polyadenylation regulate HPV-RNA processing during the primary and later-stages of HPV infection. The polycistronic HPV mRNA transcripts undergo alternative splicing using various SD and acceptor sites (Johansson and Schwartz, 2013). In the HPV-16 genome, splicing between SD site 226 (SD226) and splice acceptor site 409 (SA409) or SA526 leads to the production of E7 mRNAs, and also several truncated E6 transcripts (e.g., E6∗I). On the other hand, preservation of the intron between SD226 and SA409 generates mRNAs that express E6 (Sedman et al., 1991; Tang et al., 2006; Johansson and Schwartz, 2013). Recently, circRNAs derived from oncogenic HPV subtypes (HPV-16 and 18) were analyzed using the cancer genome atlas (TCGA) RNA-Seq data. The presence of back-splicing in the HPV-16 integrated cell lines (SiHa, CaSki, and UPCI:SCC154 cells) was investigated by inverse PCR. After treatment with RNase R, HPV-E7 derived circRNA (circE7) was detected in all these cell lines. Moreover, they detected circE7 in cells that had been transformed with HPV-16 using both Northern blotting and inverse RT-PCR, but the analogous HPV-18 circE7 could not be robustly detected using these techniques. Several lines of evidence have suggested that circE7 could be translated into the E7 oncoprotein. The knockdown of circE7 led to a decreased level of E7 oncoprotein in CaSki cells, and limited the growth of cancer cells in vitro and tumor xenografts in vivo (Zhao et al., 2019b).
Chamseddin et al. (2019) compared the role of circE7 as a diagnostic biomarker in ASCC with already established biomarkers (PD-L1, HPV-ISH, and GLUT1), and the relationship between these biomarkers and the clinical outcome in ASCC patients. The circE7 levels were evaluated by RT-qPCR showing that the high circE7 group (top 50% of patients) had the best overall survival rate, and this was also correlated with low PD-L1 expression, positive HPV-ISH, and a better tumor stage (Chamseddin et al., 2019). Overall, studies have shown that patients with HPV-16 positive tumors with a high level of circE7 show improved survival compared to HPV-16 positive tumors with a low level of circE7 (Chamseddin et al., 2019; Zhao et al., 2019b). Due to its importance, more attention should be given to role of circE7 as a biomarker in cervical cancer. A number of other studies are summarized in Table 2.
Conclusion
The discovery of non-coding RNAs (such as lncRNA, miRNA, and circRNA) has provided better understanding of the mechanisms involved in many physiological and pathological processes, and has made them the focal point of studies concerned with their role in cancer, infectious disease (e.g., viral infections), and autoimmune diseases. It was previously thought that circRNAs were only random errors that occurred during transcription, but today it is known that they are fundamentally involved in the regulation of gene expression, and can affect many biological and pathological processes. However, the entire scope of their functions is still unclear. In addition, it has been observed that the expression of circRNAs is altered under different physiological and pathological conditions, and major changes are significantly associated with the progression of several diseases, and may serve as a promising diagnostic biomarker for diseases including viral infections. Recent studies have confirmed that some host circRNAs are deregulated in viral infections, and suggest that the virus uses this cellular mechanism to its advantage. The fact that members of different viral families are capable of encoding circRNAs, promises new advances in the scientific understanding of the diagnosis of viral diseases. Unfortunately, knowledge about the function, mechanism of formation, and transportation of VcircRNAs, as well as identification of the viral genes that encode circRNAs, is still very limited. However, more research into the function of VcircRNAs is crucial, since distinguishing between the roles of host circRNAs and VcircRNAs in viral infection might provide unique insight in the development of novel therapeutic strategies. In the future, the potential application of VcircRNAs can be probably divided into two categories: one being their use as novel biomarkers for prognosis and diagnosis of viral diseases. The other is the development of VcircRNA-based therapeutic approaches which could be useful for deadly viruses, e.g., Ebola virus and human immunodeficiency virus (HIV), or for viruses which are highly contagious and spread rapidly, e.g., novel coronavirus SARS-CoV2. Furthermore, VcircRNA-based vaccines might represent a highly versatile platform for development of vaccines against other viruses. Therefore, further research on circRNAs (especially VcircRNA) could open a new window of opportunity for the treatment of deadly diseases, preventing thousands of deaths and saving a lot of money.
Author Contributions
HM and HB contributed in conception, design, statistical analysis, and drafting of the manuscript. JN, SJ, MJ, MM-T, MV, AK, and MM contributed in data collection and manuscript drafting. MH critically revised the manuscript. All authors approved the final version for submission.
Funding
MH was supported by US NIH Grants R01AI050875 and R21AI121700.
Conflict of Interest
MH declares the following potential conflicts of interest. Scientific Advisory Boards: Transdermal Cap Inc., Cleveland, OH; BeWell Global Inc., Wan Chai, Hong Kong; Hologenix Inc., Santa Monica, CA; LumiThera Inc., Poulsbo, WA; Vielight, Toronto, Canada; Bright Photomedicine, São Paulo, Brazil; Quantum Dynamics LLC, Cambridge, MA; Global Photon Inc., Bee Cave, TX; Medical Coherence, Boston, MA; NeuroThera, Newark, DE; JOOVV Inc., Minneapolis-St. Paul, MN; AIRx Medical, Pleasanton, CA; FIR Industries Inc., Ramsey, NJ; UVLRx Therapeutics, Oldsmar, FL; Ultralux UV Inc., Lansing MI; Illumiheal & Petthera, Shoreline, WA; MB Lasertherapy, Houston, TX; ARRC LED, San Clemente, CA; Varuna Biomedical Corp. Incline Village, NV; Niraxx Light Therapeutics Inc., Boston, MA; Consulting; Lexington Int, Boca Raton, FL; USHIO Corp, Japan; Merck KGaA, Darmstadt, Germany; Philips Electronics Nederland B.V., Eindhoven, Netherlands; Johnson & Johnson Inc., Philadelphia, PA; Sanofi-Aventis Deutschland GmbH, Frankfurt am Main, Germany. Stockholdings: Global Photon Inc., Bee Cave, TX; Mitonix, Newark, DE.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
A3BS, alternative 3′ back-splicing; A5BS, alternative 5′ back-splicing; AGO, argonaute; ASCC, anal squamous cell carcinoma; BART, Bam HI-A region rightward transcript; circBART, EBV-derived circRNA; circE7, HPV-E7 derived circRNA; circRNAs, covalent closed circular RNAs or circular RNAs; CMV, cytomegalovirus; EBER, EBV-encoded RNA; EBV, Epstein Barr virus; EMT, epithelial-mesenchymal transition; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; HDAg, hepatitis delta antigen; HDV, hepatitis D virus; HHV, human herpesvirus; HPV, human papillomavirus; HSV, herpes simplex virus; IAV, influenza A virus; IRES, internal ribosome entry site; KSHV, Kaposi’s sarcoma-associated herpesvirus; lncRNA, long noncoding RNA; MBL, mannose-binding lectin; NGS, next-generation sequencing; NPC, nasopharyngeal carcinoma; PAN, polyadenylated nuclear RNA; PEL, primary effusion lymphoma; RNA-seq, RNA-sequencing; rRNA, ribosomal ribonucleic acid; SA409, splice acceptor 409; SD226, splicing between splice donor 226; snRNA, small nuclear RNA; snRNPs, small nuclear ribonucleoproteins; TGEV, transmissible gastroenteritis virus; tRNA, transfer ribonucleic acid; VcircRNAs, virus-encoded circRNAs; VZV, varicella-zoster virus.
References
Ahmad, L., Mostowy, S., and Sancho Shimizu, V. (2018). Autophagy-virus interplay: from cell biology to human disease. Front. Cell. Dev. Biol. 6:155. doi: 10.3389/fcell.2018.00155
Arnberg, A. C., Van Ommen, G. J., Grivell, L. A., Van Bruggen, E. F., and Borst, P. (1980). Some yeast mitochondrial RNAs are circular. Cell 19, 313–319. doi: 10.1016/0092-8674(80)90505-x
Bhadelia, A. (2019). Evidence for the path to cervical cancer elimination. Lancet Global Health 8, e155–e156. doi: 10.1016/s2214-109x(19)30523-6
Bodescot, M., and Perricaudet, M. (1986). Epstein-Barr virus mRNAs produced by alternative splicing. Nucleic Acids Res. 14, 7103–7114. doi: 10.1093/nar/14.17.7103
Braicu, C., Zimta, A.-A., Gulei, D., Olariu, A., and Berindan-Neagoe, I. (2019). Comprehensive analysis of circular RNAs in pathological states: biogenesis, cellular regulation, and therapeutic relevance. Cell. Mol. Life Sci. 76, 1559–1577. doi: 10.1007/s00018-019-03016-5
Burroni, E., Bisanzi, S., Sani, C., Puliti, D., and Carozzi, F. (2013). Codon 72 polymorphism of p53 and HPV type 16 E6 variants as risk factors for patients with squamous epithelial lesion of the uterine cervix. J. Med. Virol. 85, 83–90. doi: 10.1002/jmv.23417
Chaichian, S., Shafabakhsh, R., Mirhashemi, S. M., Moazzami, B., and Asemi, Z. (2019). Circular RNAs: a novel biomarker for cervical cancer. J. Cell. Physiol. 235, 718–724. doi: 10.1002/jcp.29009
Chamseddin, B. H., Lee, E. E., Kim, J., Zhan, X., Yang, R., Murphy, K. M., et al. (2019). Assessment of circularized E7 RNA, GLUT1, and PD-L1 in anal squamous cell carcinoma. Oncotarget 10:5958. doi: 10.18632/oncotarget.27234
Chen, I., Chen, C. Y., and Chuang, T. J. (2015). Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip. Rev. RNA 6, 563–579. doi: 10.1002/wrna.1294
Chong, W. P., To, Y. F., Ip, W. K., Yuen, M. F., Poon, T. P., Wong, W. H., et al. (2005). Mannose-binding lectin in chronic hepatitis B virus infection. Hepatology 42, 1037–1045.
Conrad, N. K. (2016). New insights into the expression and functions of the Kaposi’s sarcoma-associated herpesvirus long noncoding PAN RNA. Virus Res. 212, 53–63. doi: 10.1016/j.virusres.2015.06.012
Cui, S., Qian, Z., Chen, Y., Li, L., Li, P., and Ding, H. (2018). Screening of up-and downregulation of circRNAs in HBV-related hepatocellular carcinoma by microarray. Oncol. Lett. 15, 423–432.
Dittmer, D. P., and Damania, B. (2013). Kaposi sarcoma associated herpesvirus pathogenesis (KSHV)—an update. Curr. Opin. Virol. 3, 238–244. doi: 10.1016/j.coviro.2013.05.012
Dong, R., Ma, X.-K., Li, G.-W., and Yang, L. (2018). CIRCpedia v2: an updated database for comprehensive circular RNA annotation and expression comparison. Genomics Proteomics Bioinformatic 16, 226–233. doi: 10.1016/j.gpb.2018.08.001
Du, W. W., Yang, W., Li, X., Awan, F. M., Yang, Z., Fang, L., et al. (2018). A circular RNA circ-DNMT1 enhances breast cancer progression by activating autophagy. Oncogene 37, 5829–5842. doi: 10.1038/s41388-018-0369-y
Du, W. W., Zhang, C., Yang, W., Yong, T., Awan, F. M., and Yang, B. B. (2017). Identifying and characterizing circRNA-protein interaction. Theranostics 7, 4183–4191. doi: 10.7150/thno.21299
Eger, N., Schoppe, L., Schuster, S., Laufs, U., and Boeckel, J.-N. (2018). Circular RNA splicing. Adv. Exp. Med. Biol. 1087, 41–52. doi: 10.1007/978-981-13-1426-1_4
Farci, P. (2003). Delta hepatitis: an update. J. Hepatol. 39, 212–219. doi: 10.1016/s0168-8278(03)00331-3
Fok, V., Friend, K., and Steitz, J. A. (2006). Epstein-Barr virus noncoding RNAs are confined to the nucleus, whereas their partner, the human La protein, undergoes nucleocytoplasmic shuttling. J. Cell Biol. 173, 319–325. doi: 10.1083/jcb.200601026
Fu, L., Jiang, Z., Li, T., Hu, Y., and Guo, J. (2018). Circular RNA s in hepatocellular carcinoma: functions and implications. Cancer Med. 7, 3101–3109.
Fu, Y., Xu, W., Chen, D., Feng, C., Zhang, L., Wang, X., et al. (2015). Enterovirus 71 induces autophagy by regulating has-miR-30a expression to promote viral replication. Antiviral Res. 124, 43–53. doi: 10.1016/j.antiviral.2015.09.016
Gao, F.-X., Wang, Y., Zhang, Q.-Y., Mou, C.-Y., Li, Z., Deng, Y.-S., et al. (2017a). Distinct herpesvirus resistances and immune responses of three gynogenetic clones of gibel carp revealed by comprehensive transcriptomes. BMC Genomics 18:561. doi: 10.1186/s12864-017-3945-6
Gao, Y. L., Zhang, M. Y., Xu, B., Han, L. J., Lan, S. F., Chen, J., et al. (2017b). Circular RNA expression profiles reveal that hsa_circ_0018289 is up-regulated in cervical cancer and promotes the tumorigenesis. Oncotarget 8, 86625–86633. doi: 10.18632/oncotarget.21257
Ge, H., and Manley, J. L. (1990). A protein factor, ASF, controls cell-specific alternative splicing of SV40 early pre-mRNA in vitro. Cell 62, 25–34. doi: 10.1016/0092-8674(90)90236-8
Geng, Y., Jiang, J., and Wu, C. (2018). Function and clinical significance of circRNAs in solid tumors. J. Hematol. Oncol. 11:98.
Ghittoni, R., Accardi, R., Hasan, U., Gheit, T., Sylla, B., and Tommasino, M. (2010). The biological properties of E6 and E7 oncoproteins from human papillomaviruses. Virus Genes 40, 1–13. doi: 10.1007/s11262-009-0412-8
Goyenvalle, A., Vulin, A., Fougerousse, F., Leturcq, F., Kaplan, J.-C., Garcia, L., et al. (2004). Rescue of dystrophic muscle through U7 snrna-mediated exon skipping. Science 306, 1796–1799. doi: 10.1126/science.1104297
Grinde, B. (2013). Herpesviruses: latency and reactivation–viral strategies and host response. J. Oral. Microbiol. 5:22766. doi: 10.3402/jom.v5i0.22766
Guan, W., Huang, Q., Cheng, F., and Qiu, J. (2011). Internal polyadenylation of the parvovirus B19 precursor mRNA is regulated by alternative splicing. J. Biol. Chem. 286, 24793–24805. doi: 10.1074/jbc.m111.227439
Guo, J. U., Agarwal, V., Guo, H., and Bartel, D. P. (2014). Expanded identification and characterization of mammalian circular RNAs. Genome. Biol. 15:409.
Haddad, G., and Lorenzen, J. M. (2019). Biogenesis and function of circular RNAs in health and in disease. Front. Pharmacol. 10:428. doi: 10.3389/fphar.2019.00428
Hagen, J. W., and Lai, E. C. (2008). microRNA control of cell-cell signaling during development and disease. Cell Cycle 7, 2327–2332. doi: 10.4161/cc.6447
Han, D., Li, J., Wang, H., Su, X., Hou, J., Gu, Y., et al. (2017). Circular RNA circMTO1 acts as the sponge of microrna-9 to suppress hepatocellular carcinoma progression. Hepatology 66, 1151–1164. doi: 10.1002/hep.29270
Hanan, M., Soreq, H., and Kadener, S. (2017). CircRNAs in the brain. RNA Biol. 14, 1028–1034. doi: 10.1080/15476286.2016.1255398
Handin, R. I., Lux, S. E., and Stossel, T. P. (2003). Blood: Principles and Practice of Hematology, Vol. 1. (Philadelphia, PA: Lippincott Williams & Wilkins), 2304.
Hansen, T. B., Jensen, T. I., Clausen, B. H., Bramsen, J. B., Finsen, B., Damgaard, C. K., et al. (2013). Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388. doi: 10.1038/nature11993
Hao, L., Rong, W., Bai, L., Cui, H., Zhang, S., Li, Y., et al. (2019). Upregulated circular RNA circ_0007534 indicates an unfavorable prognosis in pancreatic ductal adenocarcinoma and regulates cell proliferation, apoptosis, and invasion by sponging miR-625 and miR-892b. J. Cell. Biochem. 120, 3780–3789. doi: 10.1002/jcb.27658
Harland, R., and Misher, L. (1988). Stability of RNA in developing Xenopus embryos and identification of a destabilizing sequence in TFIIIA messenger RNA. Development 102, 837–852.
Holdt, L. M., Kohlmaier, A., and Teupser, D. (2018). Circular RNAs as therapeutic agents and targets. Front. Physiol. 9:1262. doi: 10.3389/fphys.2018.01262
Hope, T. J. (1999). The ins and outs of HIV Rev. Arch. Biochem. Biophys. 365, 186–191. doi: 10.1006/abbi.1999.1207
Hu, B., Li, X., Huo, Y., Yu, Y., Zhang, Q., Chen, G., et al. (2016). Cellular responses to HSV-1 infection are linked to specific types of alterations in the host transcriptome. Sci. Rep. 6:28075.
Huang, J.-T., Chen, J.-N., Gong, L.-P., Bi, Y.-H., Liang, J., Zhou, L., et al. (2019). Identification of virus-encoded circular RNA. Virology 529, 144–151. doi: 10.1016/j.virol.2019.01.014
Huang, X.-Y., Huang, Z.-L., Xu, Y.-H., Zheng, Q., Chen, Z., Song, W., et al. (2017). Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141-3p pathway in hepatitis B-related hepatocellular carcinoma. Sci. Rep. 7:5428.
Hulsmans, M., and Holvoet, P. (2013). MicroRNA-containing microvesicles regulating inflammation in association with atherosclerotic disease. Cardiovasc. Res. 100, 7–18. doi: 10.1093/cvr/cvt161
Ingolia, N. T., Brar, G. A., Rouskin, S., Mcgeachy, A. M., and Weissman, J. S. (2012). The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 7, 1534–1550. doi: 10.1038/nprot.2012.086
Jeck, W. R., and Sharpless, N. E. (2014). Detecting and characterizing circular RNAs. Nat. Biotechnol. 32, 453–461. doi: 10.1038/nbt.2890
Jeck, W. R., Sorrentino, J. A., Wang, K., Slevin, M. K., Burd, C. E., Liu, J., et al. (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19, 141–157. doi: 10.1261/rna.035667.112
Ji, P., Wu, W., Chen, S., Zheng, Y., Zhou, L., Zhang, J., et al. (2019). Expanded expression landscape and prioritization of circular RNAs in mammals. Cell Rep. 26, 3444–3460.e5. doi: 10.1016/j.celrep.2019.02.078
Johansson, C., and Schwartz, S. (2013). Regulation of human papillomavirus gene expression by splicing and polyadenylation. Nat. Rev. Microbiol. 11, 239–251. doi: 10.1038/nrmicro2984
Kitajewski, J., Schneider, R. J., Safer, B., Munemitsu, S. M., Samuel, C. E., Thimmappaya, B., et al. (1986). Adenovirus VAI RNA antagonizes the antiviral action of interferon by preventing activation of the interferon-induced eIF-2 alpha kinase. Cell 45, 195–200. doi: 10.1016/0092-8674(86)90383-1
Konarska, M. M., Grabowski, P. J., Padgett, R. A., and Sharp, P. A. (1985). Characterization of the branch site in lariat RNAs produced by splicing of mRNA precursors. Nature 313, 552–557. doi: 10.1038/313552a0
Kos, A., Dijkema, R., Arnberg, A., Van Der Meide, P., and Schellekens, H. (1986). The hepatitis delta (δ) virus possesses a circular RNA. Nature 323, 558–560. doi: 10.1038/323558a0
Kristensen, L., Hansen, T., Venø, M., and Kjems, J. (2018). Circular RNAs in cancer: opportunities and challenges in the field. Oncogene 37, 555–565. doi: 10.1038/onc.2017.361
Kristensen, L. S., Andersen, M. S., Stagsted, L. V., Ebbesen, K. K., Hansen, T. B., and Kjems, J. (2019). The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 20, 675–691.
Kulcheski, F. R., Christoff, A. P., and Margis, R. (2016). Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J. Biotechnol. 238, 42–51. doi: 10.1016/j.jbiotec.2016.09.011
Laude, H., Rasschaert, D., Delmas, B., Godet, M., Gelfi, J., and Charley, B. (1990). Molecular biology of transmissible gastroenteritis virus. Vet. Microbiol. 23, 147–154. doi: 10.1016/0378-1135(90)90144-k
Li, H. C., Huang, E. Y., Su, P. Y., Wu, S. Y., Yang, C. C., Lin, Y. S., et al. (2010). Nuclear export and import of human hepatitis B virus capsid protein and particles. PLoS. Pathog. 6:e1001162. doi: 10.1371/journal.ppat.1001162
Li, J., Yang, J., Zhou, P., Le, Y., Zhou, C., Wang, S., et al. (2015). Circular RNAs in cancer: novel insights into origins, properties, functions and implications. Am. J. Cancer Res. 5, 472–480.
Li, S., and Han, L. (2019). Circular RNAs as promising biomarkers in cancer: detection, function, and beyond. Genome Med. 11:15.
Li, X., Liu, C. X., Xue, W., Zhang, Y., Jiang, S., Yin, Q. F., et al. (2017). Coordinated circRNA Biogenesis and Function with Nf90/Nf110 in Viral Infection. Mol. Cell 67, 214–227.e7. doi: 10.1016/j.molcel.2017.05.023
Li, Z., Huang, C., Bao, C., Chen, L., Lin, M., Wang, X., et al. (2015). Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 22, 256–264. doi: 10.1038/nsmb.2959
Liu, C. X., Li, X., Nan, F., Jiang, S., Gao, X., Guo, S. K., et al. (2019). Structure and Degradation of Circular RNAs Regulate PKR Activation in Innate Immunity. Cell 177, 865–880.e21. doi: 10.1016/j.cell.2019.03.046
Liu, Q., Shuai, M., and Xia, Y. (2019). Knockdown of EBV-encoded circRNA circRPMS1 suppresses nasopharyngeal carcinoma cell proliferation and metastasis through sponging multiple miRNAs. Cancer Manag. Res. 11, 8023–8031. doi: 10.2147/cmar.s218967
Lukiw, W. (2013). Circular RNA (circRNA) in Alzheimer’s disease (AD). Front. Genet. 4:307. doi: 10.3389/fgene.2013.0030
Ma, R., Zhang, Y., Liu, H., and Ning, P. (2014). Proteome profile of swine testicular cells infected with porcine transmissible gastroenteritis coronavirus. PLoS One 9:e110647. doi: 10.1371/journal.pone.0110647
Ma, X., Zhao, X., Zhang, Z., Guo, J., Guan, L., Li, J., et al. (2018). Differentially expressed non-coding RNAs induced by transmissible gastroenteritis virus potentially regulate inflammation and NF-kappaB pathway in porcine intestinal epithelial cell line. BMC Genomics 19:747. doi: 10.1186/s12864-018-5128-5
Martin, D., Balermpas, P., Winkelmann, R., Rödel, F., Rödel, C., and Fokas, E. (2018). Anal squamous cell carcinoma–State of the art management and future perspectives. Cancer Treat. Rev. 65, 11–21. doi: 10.1016/j.ctrv.2018.02.001
Martin, K. M., Barandoc-Alviar, K., Schneweis, D. J., Stewart, C. L., Rotenberg, D., and Whitfield, A. E. (2017). Transcriptomic response of the insect vector, Peregrinus maidis, to Maize mosaic rhabdovirus and identification of conserved responses to propagative viruses in hopper vectors. Virology 509, 71–81. doi: 10.1016/j.virol.2017.05.019
Mayerat, C., Mantegani, A., and Frei, P. (1999). Does hepatitis B virus (HBV) genotype influence the clinical outcome of HBV infection? J. Viral. Hepat. 6, 299–304. doi: 10.1046/j.1365-2893.1999.00174.x
Memczak, S., Jens, M., Elefsinioti, A., Torti, F., Krueger, J., Rybak, A., et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338. doi: 10.1038/nature11928
Meng, X., Li, X., Zhang, P., Wang, J., Zhou, Y., and Chen, M. (2017). Circular RNA: an emerging key player in RNA world. Brief. Bioinformatics 18, 547–557.
Mogensen, T. H. (2009). Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 22, 240–273. doi: 10.1128/cmr.00046-08
Mole, S., Milligan, S. G., and Graham, S. V. (2009). Human papillomavirus type 16 E2 protein transcriptionally activates the promoter of a key cellular splicing factor, SF2/ASF. J. Virol. 83, 357–367. doi: 10.1128/jvi.01414-08
Naeli, P., Pourhanifeh, M. H., Karimzadeh, M. R., Shabaninejad, Z., Movahedpour, A., Tarrahimofrad, H., et al. (2019). Circular RNAs and gastrointestinal cancers: epigenetic regulators with a prognostic and therapeutic role. Crit. Rev. Oncol. Hematol. 145:102854. doi: 10.1016/j.critrevonc.2019.102854
Nahand, J. S., Taghizadeh-Boroujeni, S., Karimzadeh, M., Borran, S., Pourhanifeh, M. H., Moghoofei, M., et al. (2019). microRNAs: new prognostic, diagnostic, and therapeutic biomarkers in cervical cancer. J. Cell. Physiol. 234, 17064–17099. doi: 10.1002/jcp.28457
Nigro, J. M., Cho, K. R., Fearon, E. R., Kern, S. E., Ruppert, J. M., Oliner, J. D., et al. (1991). Scrambled exons. Cell 64, 607–613. doi: 10.1016/0092-8674(91)90244-s
Ohnishi, K., Iida, S., Iwama, S., Goto, N., Nomura, F., Takashi, M., et al. (1982). The effect of chronic habitual alcohol intake on the development of liver cirrhosis and hepatocellular carcinoma: relation to hepatitis B surface antigen carriage. Cancer 49, 672–677. doi: 10.1002/1097-0142(19820215)49:4<672::aid-cncr2820490415>3.0.co;2-#
Pang, F., Zhang, M., Yang, X., Li, G., Zhu, S., Nie, X., et al. (2019). Genome-wide analysis of circular RNAs in goat skin fibroblast cells in response to Orf virus infection. PeerJ 7:e6267. doi: 10.7717/peerj.6267
Park, S.-J., Kumar, M., Kwon, H.-I., Seong, R.-K., Han, K., Song, J.-M., et al. (2015). Dynamic changes in host gene expression associated with H5N8 avian influenza virus infection in mice. Sci. Rep. 5:16512.
Peng, L., Chen, G., Zhu, Z., Shen, Z., Du, C., Zang, R., et al. (2017). Circular RNA ZNF609 functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-150-5p in Hirschsprung’s disease. Oncotarget 8, 808–818. doi: 10.18632/oncotarget.13656
Pourhanifeh, M. H., Mahjoubin-Tehran, M., Karimzadeh, M. R., Mirzaei, H. R., Razavi, Z. S., Sahebkar, A., et al. (2020). Autophagy in cancers including brain tumors: role of MicroRNAs. Cell Commun. Signal. 18:88.
Purcell, D., and Martin, M. A. (1993). Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J. Virol. 67, 6365–6378. doi: 10.1128/jvi.67.11.6365-6378.1993
Qu, S., Yang, X., Li, X., Wang, J., Gao, Y., Shang, R., et al. (2015). Circular RNA: a new star of noncoding RNAs. Cancer Lett. 365, 141–148. doi: 10.1016/j.canlet.2015.06.003
Reid, C. E., and Lazinski, D. W. (2000). A host-specific function is required for ligation of a wide variety of ribozyme-processed RNAs. Proc. Natl. Acad. Sci. U.S.A. 97, 424–429. doi: 10.1073/pnas.97.1.424
Robb, N. C., and Fodor, E. (2012). The accumulation of influenza A virus segment 7 spliced mRNAs is regulated by the NS1 protein. J. Gen. Virol 93, 113–118. doi: 10.1099/vir.0.035485-0
Ruskin, B., and Green, M. R. (1990). RNA lariat debranching enzyme as tool for analyzing RNA structure. Methods Enzymol. 181, 180–188. doi: 10.1016/0076-6879(90)81120-j
Sadri, J. N., Bokharaei-Salim, F., Karimzadeh, M., Moghoofei, M., Karampoor, S., Mirzaei, H., et al. (2019). MicroRNAs and exosomes: key players in HIV pathogenesis. HIV Med. 21, 246–278. doi: 10.1111/hiv.12822
Sadri Nahand, J., Bokharaei-Salim, F., Salmaninejad, A., Nesaei, A., Mohajeri, F., Moshtzan, A., et al. (2019). microRNAs: key players in virus-associated hepatocellular carcinoma. J. Cell. Physiol. 234, 12188–12225. doi: 10.1002/jcp.27956
Sagnelli, E., Potenza, N., Onorato, L., Sagnelli, C., Coppola, N., and Russo, A. (2018). Micro-RNAs in hepatitis B virus-related chronic liver diseases and hepatocellular carcinoma. World J. Hepatol. 10, 558–570. doi: 10.4254/wjh.v10.i9.558
Samols, M. A., Skalsky, R. L., Maldonado, A. M., Riva, A., Lopez, M. C., Baker, H. V., et al. (2007). Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathog. 3:e65. doi: 10.1371/journal.ppat.0030065
Sanger, H. L., Klotz, G., Riesner, D., Gross, H. J., and Kleinschmidt, A. K. (1976). Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. U.S.A 73, 3852–3856. doi: 10.1073/pnas.73.11.3852
Sedman, S., Barbosa, M., Vass, W., Hubbert, N., Haas, J., Lowy, D., et al. (1991). The full-length E6 protein of human papillomavirus type 16 has transforming and trans-activating activities and cooperates with E7 to immortalize keratinocytes in culture. J. Virol. 65, 4860–4866. doi: 10.1128/jvi.65.9.4860-4866.1991
Shang, Q., Yang, Z., Jia, R., and Ge, S. (2019). The novel roles of circRNAs in human cancer. Mol. Cancer 18:6.
Shi, J., Hu, N., Li, J., Zeng, Z., Mo, L., Sun, J., et al. (2017). Unique expression signatures of circular RNAs in response to DNA tumor virus SV40 infection. Oncotarget 8, 98609–98622. doi: 10.18632/oncotarget.21694
Shi, J., Hu, N., Mo, L., Zeng, Z., Sun, J., and Hu, Y. (2018). Deep RNA Sequencing reveals a repertoire of human fibroblast circular RNAs associated with cellular responses to herpes simplex virus 1 infection. Cell. Physiol. Biochem. 47, 2031–2045. doi: 10.1159/000491471
Sinn, D. H., Lee, J., Goo, J., Kim, K., Gwak, G. Y., Paik, Y. H., et al. (2015). Hepatocellular carcinoma risk in chronic hepatitis B virus-infected compensated cirrhosis patients with low viral load. Hepatology 62, 694–701. doi: 10.1002/hep.27889
Suzuki, H., Aoki, Y., Kameyama, T., Saito, T., Masuda, S., Tanihata, J., et al. (2016). Endogenous multiple exon skipping and back-splicing at the DMD mutation hotspot. Int. J. Mol. Sci. 17:1722. doi: 10.3390/ijms17101722
Tagawa, T., Gao, S., Koparde, V. N., Gonzalez, M., Spouge, J. L., Serquiña, A. P., et al. (2018). Discovery of Kaposi’s sarcoma herpesvirus-encoded circular RNAs and a human antiviral circular RNA. Proc. Natl. Acad. Sci. U.S.A. 115, 12805–12810. doi: 10.1073/pnas.1816183115
Tan, A., Koh, S., and Bertoletti, A. (2015). Immune response in hepatitis B virus infection. Cold Spring Harb. Perspect. Med. 5:a021428.
Tang, S., Tao, M., Mccoy, J. P., and Zheng, Z.-M. (2006). The E7 oncoprotein is translated from spliced E6∗ I transcripts in high-risk human papillomavirus type 16-or type 18-positive cervical cancer cell lines via translation reinitiation. J. Virol. 80, 4249–4263. doi: 10.1128/jvi.80.9.4249-4263.2006
Tomonaga, K., Kobayashi, T., Lee, B. J., Watanabe, M., Kamitani, W., and Ikuta, K. (2000). Identification of alternative splicing and negative splicing activity of a nonsegmented negative-strand RNA virus. Borna disease virus. Proc. Natl. Acad. Sci. U.S.A. 97, 12788–12793.
Toptan, T., Abere, B., Nalesnik, M. A., Swerdlow, S. H., Ranganathan, S., Lee, N., et al. (2018). Circular DNA tumor viruses make circular RNAs. Proc. Natl. Acad. Sci. U.S.A. 115, E8737–E8745.
Tormanen, H., Backstrom, E., Carlsson, A., and Akusjarvi, G. (2006). L4-33K, an adenovirus-encoded alternative RNA splicing factor. J. Biol. Chem. 281, 36510–36517. doi: 10.1074/jbc.m607601200
Ungerleider, N., Concha, M., Lin, Z., Roberts, C., Wang, X., Cao, S., et al. (2018). The Epstein Barr virus circRNAome. PLoS Pathog. 14:e1007206. doi: 10.1371/journal.ppat.1007206
Ungerleider, N. A., Jain, V., Wang, Y., Maness, N. J., Blair, R. V., Alvarez, X., et al. (2019). Comparative analysis of gammaherpesvirus circular RNA repertoires: conserved and unique viral circular RNAs. J. Virol. 93:e01952-18.
Valadkhan, S. (2005). snRNAs as the catalysts of pre-mRNA splicing. Curr. Opin. Chem. Biol. 9, 603–608. doi: 10.1016/j.cbpa.2005.10.008
Verhoeven, R., Tong, S., Mok, B. W.-Y., Liu, J., He, S., Zong, J., et al. (2019). Epstein-Barr virus BART long non-coding RNAs function as epigenetic modulators in nasopharyngeal carcinoma. Front. Oncol. 9:1120. doi: 10.3389/fonc.2019.01120
Verma, D., and Swaminathan, S. (2008). Epstein-Barr virus SM protein functions as an alternative splicing factor. J. Virol. 82, 7180–7188. doi: 10.1128/jvi.00344-08
Vicens, Q., and Westhof, E. (2014). Biogenesis of circular RNAs. Cell 159, 13–14. doi: 10.1016/j.cell.2014.09.005
Wang, H., Feng, C., Wang, M., Yang, S., and Wei, F. (2019). Circular RNAs: diversity of functions and a regulatory nova in oral medicine: a pilot review. Cell Transplant. 28, 819–830. doi: 10.1177/0963689719837917
Wang, K., Long, B., Liu, F., Wang, J.-X., Liu, C.-Y., Zhao, B., et al. (2016). A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur. Heart J. 37, 2602–2611. doi: 10.1093/eurheartj/ehv713
Wang, S., Cui, S., Zhao, W., Qian, Z., Liu, H., Chen, Y., et al. (2018). Screening and bioinformatics analysis of circular RNA expression profiles in hepatitis B-related hepatocellular carcinoma. Cancer Biomark 22, 631–640. doi: 10.3233/cbm-170910
Wang, X., Zhang, Y., Huang, L., Zhang, J., Pan, F., Li, B., et al. (2015). Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances. Int. J. Clin. Exp. Pathol. 8, 16020–16025.
Wang, Y., Mo, Y., Gong, Z., Yang, X., Yang, M., Zhang, S., et al. (2017). Circular RNAs in human cancer. Mol. Cancer 16:25.
Wang, Z., Gerstein, M., and Snyder, M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63. doi: 10.1038/nrg2484
Wu, Q., Li, P., Wu, M., and Liu, Q. (2019). Deregulation of circular RNAs in cancer from the perspectives of aberrant biogenesis, transport and removal. Front. Genet. 10:16. doi: 10.3389/fgene.2019.00016
Wu, S.-Y., Lan, S.-H., and Liu, H.-S. (2016). Autophagy and microRNA in hepatitis B virus-related hepatocellular carcinoma. World J. Gastroenterol. 22, 176–187.
Xin, Z., Ma, Q., Ren, S., Wang, G., and Li, F. (2017). The understanding of circular RNAs as special triggers in carcinogenesis. Brief. Funct. Genomics 16, 80–86.
Yang, J., Fan, Z., Yang, J., Ding, J., Yang, C., and Chen, L. (2016). microRNA-22 attenuates myocardial ischemia-reperfusion injury via an anti-inflammatory mechanism in rats. Exp. Ther. Med. 12, 3249–3255. doi: 10.3892/etm.2016.3777
Yang, Y., Gao, X., Zhang, M., Yan, S., Sun, C., Xiao, F., et al. (2019). Novel role of FBXW7 Circular RNA in repressing glioma tumorigenesis. J. Natl. Cancer Inst. 111:435. doi: 10.1093/jnci/djy116
Yang, Z., Xie, L., Han, L., Qu, X., Yang, Y., Zhang, Y., et al. (2017). Circular RNAs: regulators of cancer-related signaling pathways and potential diagnostic biomarkers for human cancers. Theranostics 7, 3106–3117. doi: 10.7150/thno.19016
Yu, C.-Y., and Kuo, H.-C. (2019). The emerging roles and functions of circular RNAs and their generation. J. Biomed. Sci. 26:29.
Yu, J., Ding, W. B., Wang, M. C., Guo, X. G., Xu, J., Xu, Q. G., et al. (2019). Plasma circular RNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma: a large-scale, multicenter study. Int. J. Cancer 146, 1754–1763. doi: 10.1002/ijc.32647
Yu, T., Ding, Y., Zhang, Y., Liu, Y., Li, Y., Lei, J., et al. (2019). Circular RNA GATAD2A promotes H1N1 replication through inhibiting autophagy. Vet. Microbiol. 231, 238–245. doi: 10.1016/j.vetmic.2019.03.012
Yuan, Y., Liu, W., Zhang, Y., Zhang, Y., and Sun, S. (2018). CircRNA circ_0026344 as a prognostic biomarker suppresses colorectal cancer progression via microRNA-21 and microRNA-31. Biochem. Biophys. Res. Commun. 503, 870–875. doi: 10.1016/j.bbrc.2018.06.089
Zaiou, M. (2019). Circular RNAs as potential biomarkers and therapeutic targets for metabolic diseases. Adv. Exp. Med. Biol. 1134, 177–191. doi: 10.1007/978-3-030-12668-1_10
Zhang, X.-O., Wang, H.-B., Zhang, Y., Lu, X., Chen, L.-L., and Yang, L. (2014). Complementary sequence-mediated exon circularization. Cell 159, 134–147. doi: 10.1016/j.cell.2014.09.001
Zhang, Y., Liang, W., Zhang, P., Chen, J., Qian, H., Zhang, X., et al. (2017). Circular RNAs: emerging cancer biomarkers and targets. J. Exp. Clin. Cancer Res. 36:152.
Zhang, Y., Zhang, X.-O., Chen, T., Xiang, J.-F., Yin, Q.-F., Xing, Y.-H., et al. (2013). Circular intronic long noncoding RNAs. Mol. Cell 51, 792–806. doi: 10.1016/j.molcel.2013.08.017
Zhang, Z., Yang, T., and Xiao, J. (2018). Circular RNAs: promising biomarkers for human diseases. EBioMedicine 34, 267–274. doi: 10.1016/j.ebiom.2018.07.036
Zhang, Z.-C., Guo, X.-L., and Li, X. (2018). The novel roles of circular RNAs in metabolic organs. Genes Dis. 5, 16–23. doi: 10.1016/j.gendis.2017.12.002
Zhao, J., Lee, E. E., Kim, J., Chamseddin, B., Yang, R., Xie, Y., et al. (2019a). Translation and transforming activity of a circular RNA from human papillomavirus. bioRxiv [Preprint]. doi: 10.1101/600056
Zhao, J., Lee, E. E., Kim, J., Yang, R., Chamseddin, B., Ni, C., et al. (2019b). Transforming activity of an oncoprotein-encoding circular RNA from human papillomavirus. Nat. Commun. 10:2300.
Zhao, X., Cai, Y., and Xu, J. (2019c). Circular RNAs: biogenesis, mechanism, and function in human cancers. Int. J. Mol. Sci. 20:3926. doi: 10.3390/ijms20163926
Zhao, X., Ma, X., Guo, J., Mi, M., Wang, K., Zhang, C., et al. (2019d). Circular RNA CircEZH2 suppresses transmissible gastroenteritis coronavirus-induced opening of mitochondrial permeability transition pore via targeting MiR-22 in IPEC-J2. Int. J. Biol.Sci. 15, 2051–2064. doi: 10.7150/ijbs.36532
Zhao, Y., Alexandrov, P., Jaber, V., and Lukiw, W. (2016). Deficiency in the ubiquitin conjugating enzyme UBE2A in Alzheimer’s disease (Ad) is linked to deficits in a natural circular miRNA-7 sponge (circRNA; ciRS-7). Genes 7:116. doi: 10.3390/genes7120116
Zhao, Z., Li, X., Jian, D., Hao, P., Rao, L., and Li, M. (2017). Hsa_circ_0054633 in peripheral blood can be used as a diagnostic biomarker of pre-diabetes and type 2 diabetes mellitus. Acta Diabetol. 54, 237–245. doi: 10.1007/s00592-016-0943-0
Zheng, S. R., Zhang, H. R., Zhang, Z. F., Lai, S. Y., Huang, L. J., Liu, J., et al. (2018). Human papillomavirus 16 E7 oncoprotein alters the expression profiles of circular RNAs in Caski cells. J. Cancer 9, 3755–3764. doi: 10.7150/jca.24253
Zheng, X., and Bevilacqua, P. C. (2004). Activation of the protein kinase PKR by short double-stranded RNAs with single-stranded tails. RNA 10, 1934–1945. doi: 10.1261/rna.7150804
Zheng, Y., Wang, Y., Ding, B., and Fei, Z. (2017). Comprehensive transcriptome analyses reveal that potato spindle tuber viroid triggers genome-wide changes in alternative splicing, inducible trans-acting activity of phased secondary small interfering RNAs, and immune responses. J. Virol. 91:e00247-17.
Zhou, B., and Yu, J.-W. (2017). A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting Tgf-β1. Biochem. Biophys. Res. Commun. 487, 769–775. doi: 10.1016/j.bbrc.2017.04.044
Zhou, L.-Y., Zhai, M., Huang, Y., Xu, S., An, T., Wang, Y.-H., et al. (2018). The circular RNA ACR attenuates myocardial ischemia/reperfusion injury by suppressing autophagy via modulation of the Pink1/FAM65B pathway. Cell Death Differ. 26, 1299–1315. doi: 10.1038/s41418-018-0206-4
Zhou, T. C., Li, X., Chen, L. J., Fan, J. H., Lai, X., Tang, Y., et al. (2018). Differential expression profile of hepatic circular Rna s in chronic hepatitis B. J. Viral Hepat. 25, 1341–1351. doi: 10.1111/jvh.12944
Zhou, Z.-B., Du, D., Huang, G.-X., Chen, A., and Zhu, L. (2018). Circular RNA Atp9b, a competing endogenous RNA, regulates the progression of osteoarthritis by targeting miR-138-5p. Gene 646, 203–209. doi: 10.1016/j.gene.2017.12.064
Zhu, K., Zhan, H., Peng, Y., Yang, L., Gao, Q., Jia, H., et al. (2019). Plasma hsa_circ_0027089 is a diagnostic biomarker for hepatitis B virus-related hepatocellular carcinoma. Carcinogenesis 41, 296–302.
Zhu, L.-P., He, Y.-J., Hou, J.-C., Chen, X., Zhou, S.-Y., Yang, S.-J., et al. (2017). The role of circRNAs in cancers. Biosci. Rep. 37:BSR20170750.
Keywords: circular RNA, VcircRNA, viral infection, biomarker, back-splicing, epigenetics
Citation: Nahand JS, Jamshidi S, Hamblin MR, Mahjoubin-Tehran M, Vosough M, Jamali M, Khatami A, Moghoofei M, Baghi HB and Mirzaei H (2020) Circular RNAs: New Epigenetic Signatures in Viral Infections. Front. Microbiol. 11:1853. doi: 10.3389/fmicb.2020.01853
Received: 17 May 2020; Accepted: 15 July 2020;
Published: 31 July 2020.
Edited by:
Fatah Kashanchi, George Mason University, United StatesReviewed by:
Walter Moss, Iowa State University, United StatesMarta Gòdia, Centre for Research in Agricultural Genomics (CRAG), Spain
Copyright © 2020 Nahand, Jamshidi, Hamblin, Mahjoubin-Tehran, Vosough, Jamali, Khatami, Moghoofei, Baghi and Mirzaei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hossein Bannazadeh Baghi, aGJhbm5hemFkZWhAdGJ6bWVkLmFjLmly; Hamed Mirzaei, aC5taXJ6YWVpMjAwMkBnbWFpbC5jb20=; bWlyemFlaS1oQGthdW1zLmFjLmly
†These authors have contributed equally to this work
 Javid Sadri Nahand1,2†
Javid Sadri Nahand1,2† Hamed Mirzaei
Hamed Mirzaei