
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 31 July 2020
Sec. Fungi and Their Interactions
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.01824
A correction has been applied to this article in:
Corrigendum: Characterization of cmcp Gene as a Pathogenicity Factor of Ceratocystis manginecans
Ceratocystis manginecans causes mango wilt with significant economic losses. In the infection court, cerato-platanin (CP) family proteins (CPPs) are believed to involve in pathogenesis but has not been determined in C. manginecans. To confirm this function, a CP protein (CmCP) of C. manginecans was characterized in this study. A protoplast of C. manginecans was prepared by treating its mycelia with driselase and lysing enzymes. The cmcp gene was edited using CRISPR/Cas-U6-1 expression vectors in 60% PEG and 50 μg/mL hygromycin B in the medium, resulting in mutants with cmcp deletion (Δcmcp). A complemented mutant (Δcmcp-C) was obtained by transforming cmcp to Δcmcp. Both Δcmcp and Δcmcp-C were characterized by comparing them with a wild-type strain on morphology, mycelial growth, conidial production and pathogenicity. Additionally, cmcp was transformed and expressed in Pichia pastoris, and the derived recombinant protein CmCP caused a severe necrosis on Nicotiana tabacum leaves. CmCP-treated plant leaves showed symptoms of hypersensitive response including electrolyte leakage, reactive oxygen species generation and overexpression of defense-related genes PR-1, PAD3, ERF1, HSR203J, and HIN1. All those results suggested that cmcp gene was required for the growth development of C. manginecans and functioned as a major pathogenicity factor in mango infection.
Ceratocystis spp. attack a wide range of economically important plants, causing cankers, lethal, wilt-type diseases, and black rot of storage roots on many plants. More than 30 plant species of plants representing 14 families are their hosts (Baker et al., 2003; Johnson et al., 2005). Ceratocystis fimbriata was firstly reported on mango in Brazil (Viégas, 1960). Subsequently, several other pathogenic species were reported in the world, including C. manginecans, C. acaciivora, and C. omanensis (Tarigan et al., 2011; Van Wyk et al., 2011). Among them, C. manginecans is considered as an important pathogen of mango tree and several other plants (Fourie et al., 2016). Mango wilt caused by C. manginecans is a serious vascular disease and has caused significantly economic losses to mango industry (Van Wyk et al., 2007; Al Adawi et al., 2013; Zhang et al., 2017).
In the infection course of Ceratocystis spp., cerato-platanin (CP) plays a major role, which is a phytotoxic protein secreted by pathogens such as C. fimbriata f. sp. platani, and induces cell death of tobacco (Pazzagli et al., 1999, 2006). CP is the first member of the cerato-platanin family. Cerato-populin (Pop1) from C. populicola is the second type of CP protein secreted by Ceratocystis species (Comparini et al., 2009). Up to date, genes codifying for cerato-platanin family proteins (CPPs) have been found in more than 50 fungal genomes (Chen et al., 2013). CPPs are a group of small, secreted, and cysteine-rich proteins. Previous studies have shown that CPPs in fungi are involved in the growth and development of fungi, and the interaction with host plants (Gaderer et al., 2014; Pazzagli et al., 2014). They function as effectors or elicitor molecules found in ascomycetes and basidiomycetes (Wösten, 2001; Pazzagli et al., 2014; Chen et al., 2015). For example, the Epl-1 protein from Trichoderma harzianum involves in mycoparasitism, plant resistance induction and self-cell wall protection (Gomes et al., 2015); the HaLP2 protein from Heterobasidion annosum induces cell death, autofluorescence and expression of defense genes in host plants (Chen et al., 2015); the elicitor Sm1 from Trichoderma virens induces plant defense response and autofluorescence (Djonović et al., 2006); and both SP1 and BcSPl1 induce the production of reactive oxygen species (ROS) and the expression of defense genes of host plants (Wilson et al., 2002; Frías et al., 2011).
CPPs are a well conserved family with a 70% similarity at some conserved motifs (Chen et al., 2013). However, sequence identity is only 13% and sequence similarity is about 40% in the representative members of CP family on protein analysis (Pazzagli et al., 2014). Functions of some genes encoding for CPPs have been confirmed in several fungi, which vary greatly depending on their taxon. For instance, the mpg1 associates with virulence in Magnaporthe grisea (Jeong et al., 2010). The bcspl1 gene contributes to pathogenicity of Botrytis cinerea, and BcSpl1 associates with the plant plasma membrane causing cell shrinkage and chloroplast disorganization (Frías et al., 2011, 2014). On the contrary, the cu gene does not directly affect virulence of Ophiostoma ulmi (Bowden et al., 1996). The sp1 gene of Leptosphaeria maculans is not crucial for pathogenicity on Brassica napus cotyledons (Wilson et al., 2002). In addition, the mpg1 and cu genes both associated with “easily wettable” phenotype, which causes by a loss of surface hydrophobicity (Stringer et al., 1991; Bell-Pedersen et al., 1992). Currently, little is known about the function of the gene encoding for CPPs in C. manginecans.
As shown in the above studies, gene knock out has been frequently used for studying the function of a gene of interest. Recently, the clustered regularly interspaced short palindromic repeat (CRISPR) system is a promising approach for efficient and precise genome modification (Cong et al., 2013). It has been successfully applied to genome editing in various organisms and cell types, including animals, plants, insects, and bacteria (Carroll, 2014; Cong and Zhang, 2014). This advanced technology has been used on a growing number of filamentous fungi, including several agriculturally important pathogens (Kück and Hoff, 2010; Jiang et al., 2013), such as Magnaporthe grisea (Synonyms: Pyricularia oryzae) (Arazoe et al., 2015), Ustilago maydis (Schuster et al., 2015), and Alternaria alternata (Wenderoth et al., 2017). However, less report has been documented in genetic manipulation methods of Ceratocystis spp. The purpose of this study was to establish a system of mutation in C. manginecans using CRISPR/Cas technology and gene knockout method and characterize the function of cmcp gene based on this system.
Ceratocystis manginecans strain MG-1-10 was previously isolated from mango (Mangifera indica) and maintained in the Laboratory of Seed Disease Testing and Control (BKL-SDTC), China Agricultural University, and was used as a wild-type parental strain for this study. The fungus was cultured and maintained on malt yeast extract agar (MYEA) (Baker et al., 2003). Tobacco (Nicotiana tabacum) plants were maintained at conditions of controlled humidity, temperature, and photoperiod in a growth chamber.
The homologous protein of CmCP in Ceratocystis manginecans was predicted using BLASTP 2.9 and HMMER 3.1b2 based on sequence similarity and structure similarity (Camacho et al., 2009; Potter et al., 2018). Total RNA was extracted from the mycelium of C. manginecans strain MG-1-10 using the Eastep® Super Total RNA Extraction Kit (Promega, Madison, WI, United States). cDNA was reversely transcribed from the RNA using PrimaScriptTM RT reagent Kit with gDNA Eraser (Takara, Beijing, China). SIGNALP 4.0 (Petersen et al., 2011) were used for signal peptide prediction. The cp orthologs gene (cmcp) of strain MG-1-10 and its CDS sequence encoding the mature protein were isolated using the primer pair AM-F and AM-R. A 360-bp fragment from cmcp, carrying almost the whole cerato-platanin open reading frame, from the end of the predicted signal peptide to the stop codon, was obtained by PCR from strain MG-1-10 cDNA with primers cp-F and cp-R. Other CPPs sequences were obtained from the GenBank database1. Amino acid sequence alignments were generated using ClustalW in MAGA 5.0 (Tamura et al., 2011). Phylogenetic analysis was conducted online using IQ-TREE with the maximum-likelihood method (Nguyen et al., 2015).
Vectors for replacing the cmcp gene were constructed as described by Zheng et al. (2014) and Arazoe et al. (2015) with slight modifications. Briefly, primer pair Cf12-F and Cf12-R was used to amplify a 464-bp region of the cmcp gene of genomic DNA of strain MG-1-10. Plasmids PtrpChptA-PItk, pCRISPR/Cas-U6-1 were obtained from other laboratories (Zheng et al., 2014; Arazoe et al., 2015). Deletion vectors for targeting genes were generated as follows. The sequence of 1.7 kb downstream was amplified by thermal asymmetric interlaced polymerase chain reaction (TAIL-PCR) using the Genome Walking Kit (Takara, Beijing, China). Primer pairs M1 + M2 and M3 + M4 were used to amplify a segment of 1.0 kb upstream and 1.0 kb downstream, respectively, of the cmcp gene. Similarly, primer pair H1 and H2 was used to amplify a 3.5-kb sequence of HPH-hsv-tk DNA from plasmid PtrpChptA-PItk. Additionally, three fragments, including 1.0-upstream, 3.5- HPH-hsv-tk, and 1.0-downstream, were combined into a pBluescript SK(−) plasmid using In-Fusion® HD Cloning Kit (Takara, Beijing, China). The recombinant plasmid was cloned into Escherichia coli DH5α. With the recombinant plasmid being a template, a 5.5 kb size of the replacement fragment was obtained by PCR amplification using the primer pair M1 and M4. For the pCRISPR/Cas-U6-1 expression vectors of cmcp gene, sense and antisense oligonucleotides of target genes were designed using the web-based service CRISPRdirect2, and were annealed according to the procedures previously reported (Ran et al., 2013). The annealed oligonucleotides were inserted into plasmid pCRISPR/Cas-U6-1 by Golden Gate cloning, as previously reported (Sakuma et al., 2014; Arazoe et al., 2015). The recombinant plasmid (pCRISPR/Cas-U6-1-SgRNAcmcp) was cloned into E. coli DH5α. The recombinant plasmid pCRISPR/Cas-U6-1-SgRNAcmcp was extracted using the TIANpure Mini Plasmid Kit (TIANGE, Beijing, China). The deletion vectors and pCRISPR/Cas-U6-1-SgRNAcmcp expression vectors of the cmcp gene were confirmed by PCR and DNA sequencing.
The experiment was conducted following the procedures of Royer et al. (1991) with some modifications. In the optimized procedure of protoplast preparation, eight mycelial plugs (5 mm in diameter) taken from the margin of a 7-day-old colony of C. manginecans MG-1-10 were added into a 300-mL flask containing 100 mL complete medium (CM) (Royer et al., 1991). After incubation on a rotary shaker at 100 rpm for 24–30 h at 25°C, mycelia in CM were filtered, washed with 0.5 M MgSO4 and treated with enzyme solution, which was prepared by mixing 2% lysing enzymes (Sigma, Beijing, China) and 2.5% driselase (Sigma, Beijing, China) and dissolving them in 0.5 M MgSO4. After 3–6 h at 25°C, the enzyme solution was filtered through three layers of Miracloth (Millipore, Beijing, China) to eliminate mycelial residues. The protoplasts in the filtrate were washed with 0.6 M KCl and STC (1 M sorbitol, 25 mM Tris-HCl pH 7.5 and 50 mM CaCl2) solutions, and then resuspended in STC at a concentration of 5 × 107 protoplasts/mL.
For an improved method of transformation, DNA fragments of deletion vector (5–10 μg), pCRISPR/Cas-U6-1-SgRNAcmcp plasmid mixture (5–10 μg) and heparin (5 μL, 5 mg/mL) were added into 200 μL protoplasts and incubated on ice for 30 min, an aliquot of 2.5-mL PTC (25 mM Tris-HCl pH 7.5, 50 mM CaCl2 and 60% PEG 4000) was mixed with the suspension, which was incubated at room temperature for 20 min. The protoplast suspension was diluted with 20 mL STC, and then centrifuged at 4000 rpm for 20 min. The resulting pellet was resuspended in 1 mL OCM [0.5% yeast extract, 0.5% malt extract, 0.132% (NH4)2SO4, 0.6 M sucrose]. After 6 h incubation at 25°C, this transformation mixture was spread on OCMA (OCM plus 1.6% agar) flat plates. After 24–36 h, the plates were overlaid with 12 mL of selective agar (2% malt extract, 0.2% yeast extract, 1% agarose in water containing 50 μg/mL of hygromycin B) and continued to be incubated. Transformants were obtained after 3–7 days post-transformation and transferred to fresh MYEA plates with 50 μg/mL hygromycin B (Supplementary Figure S1). The putative transformants were purified by the single-spore isolation method and confirmed by PCR and quantitative real-time PCR (qRT-PCR) with corresponding primers (Supplementary Table S1). Primer pair M5 and M6 was used to amplify the fragment of the connecting area to confirm that the HPH-hsv-tk was inserted into the sites where cmcp is normally located in the genome. The results of qRT-PCR were analyzed with the 2−ΔΔCt method using the 18S rRNA gene as endogenous control (Baccelli et al., 2012).
To confirm that the loss of pathogenesis was due to the deletion of cmcp gene, Δcmcp was complemented with a full-length cmcp gene (Δcmcp-C). The cmcp gene was amplified from the genomic DNA of strain MG-1-10 using primer pair Cf12-F and Cf12-R (Supplementary Table S1). Protoplast transformation of Δcmcp was conducted as described above except that 25 μg/mL F2dU (which supports the growth of the complemented strains Δcmcp-C but not the growth of Δcmcp) was used as a selection agent as previously described (Zheng et al., 2014).
The wild-type C. manginecans strain MG-1-10, its cmcp deletion mutants (Δcmcp), and the complemented strains (Δcmcp-C) were routinely cultured on MYEA at 25°C for 7 days. To test mycelial growth on MYEA plates, colony diameter was measured after incubation at 25°C for 12 days in an incubator. Each plate was inoculated with a 5-mm-diameter mycelial plug taken from the edge of a 7-day-old colony. There were three replicated plates per treatment, and the colony diameter was perpendicularly measured. Sporulation was counted using a hemocytometer. The procedure of “easily wettable” phenotype was described by Talbot et al. (1993), which was measured in rodA– or Eas– mutants. Three biological replicates were used per experiment.
The wild-type C. manginecans strain MG-1-10, Δcmcp and Δcmcp-C were inoculated on healthy mango branches. A small hole was created on those branches using a cork borer, followed by inoculation with a 5-mm-diameter mycelial plug taken from the edge of a 7-day-old culture. There were three biological replicates per treatment. After 10 days of incubation, lesion size was measured. Pathogen-free MYEA plugs were used as a blank control. This experiment was done three times.
The CDS of cmcp gene (without signal peptide) was cloned in the NcoI and XhoI restriction sites of plasmid pPICZ-αAM. The recombinant protein coded by this constructed PCR product contained the Pichia α-factor signal sequence at the N-terminus of the cmcp sequence and the c-myc and 6 × His epitopes at the C-terminus. The resulting plasmid, pPICZ-αAM-cmcp, was linearized with SacI-HF restriction enzyme (NEB, United States) and then transformed into P. pastoris X33 strain by electroporation. One of the transformants expressing cmcp (X33-pPICZ-αAM-cmcp) and one transformant of empty vector or EV (X33-pPICZ-αAM, without cmcp insert) confirmed by colony PCR were chosen for all subsequent work.
The supernatant of a culture of the selected transformant, induced for 6 days with 0.5% methanol at 28°C with 250 rpm in BMMY (0.3% K2HPO4⋅3H2O, 1.18% KH2PO4, 1.34%YNB, 0.4 μg/mL Biotin), was the starting material in the purification of the recombinant protein. CmCP was purified with the aid of the 6 × His tag and carried out with Ni-NTA His∙Bind®Resin prepacked column (Millipore, Beijing, China) according to the manufacturer’s instructions. Prior to loading to the column, yeast culture supernatant was adjusted to pH 7.4 by addition of binding buffer (20 mM Na3PO4, 0.5 M NaCl) and flow through the manufacturer’s instructions at 10 ml min–1. After treating with a washing buffer (20 mM Na3PO4, 0.5 M NaCl, 50 mM iminazole, pH 7.3), the protein was eluted by an elution buffer (20 mM Na3PO4, 0.5 M NaCl, 500 mM iminazole, pH 7.3), and concentrated by an ultrafiltration tube. The protein solution containing CmCP was confirmed by Western blot (Chen et al., 2015), which was stored at 80°C for later use. Western blot was performed using anti-c-myc antibody produced in rabbit (Sigma, Beijing, China). As a negative control, EV was determined by using the same method as described above. Protein concentrations were determined using BCA Protein Assay Kit (ComWin, Beijing, China), with BSA as a standard. Primers and oligonucleotides used in this study are listed on Supplementary Table S1.
To study in vivo effect of CmCP on N. tabacum plant, the protein expressed and purified from X33-pPICZ-αAM-cmcp culture medium at concentrations of 60, 12, 2.4, 0.48, and 0.1 μM were infiltrated into N. tabacum leaves. The products expressed and purified from EV culture medium and H2O were used for controls. The purified protein was infiltrated to a healthy leaf of 5-week-old N. tabacum with the aid of a 1-mL syringe without needle. Ten plants per treatment and three biological replicates were used. The treated plants were incubated in a growth chamber under controlled conditions (16-h light/8-h dark period at 22°C with 80% relative humidity).
Hypersensitive response (HR) comprises a series of characteristic symptoms that include the induction of ROS and electrolyte leakage (Frías et al., 2011). The generation of H2O2 and O2– was evaluated on CmCP-infiltrated leaves using 3,3′-diaminobenzidine (DAB). CmCP protein solution and the products expressed and purified from EV were infiltrated into N. tabacum leaves with a syringe using a half leaf method. Thirty hours later, the leaves were cut and treated with DAB (1 mg/mL, pH 3.8) for 12 h, followed by visualization of DAB deposits. Treated leaves were incubated in ethanol to eliminate chlorophyll prior to photographing. Three biological replicates were employed for each experiment.
To assay electrolyte leakage, N. tabacum leaves were infiltrated with CmCP protein solution and the products expressed and purified from EV. After 30 h, 5 leaf disks of 9 mm diameter were cut and submerged into 50 mL water at 28°C on a shaker at 80 rpm. Three hours later, the conductivity was measured with a conductivity meter (Mettleer-Toledo, Shanghai, China). The leaves were treated with boiling water for 20 min, and the conductivity was measured using the same method as described above. Relative conductivity is the ratio of the two results. Three biological replicates were employed in each trial.
For the expression of tobacco defense genes upon CmCP or the products expressed and purified from EV infiltration, the infiltrated area of the N. tabacum leaves was excised. All the treated leaves were collected at 6, 12, and 24 h post-infiltration (hpi), followed by RNA isolation. cDNA was reversely transcribed from the RNA using PrimaScriptTM RT reagent Kit with gDNA Eraser (Takara, Beijing, China). Three biological replicates were performed for each time point and three technical replicates were carried out by qRT-PCR. All qRT-PCR reactions were performed with specific primers (Supplementary Table S1) on an ABI 7500 Fast real-time detection system (Applied Biosystems, Beijing, China). Expression level was analyzed with the 2−ΔΔCt method compared to infiltration with EV-derived protein (set to 1) and using Tac9 and EF1α (Eeongation factor 1 alpha) as reference genes. The induction of five pathogenesis-related genes was studied upon CmCP infiltration: HSR203J and HIN1, which are considered markers of HR in tobacco (Pontier et al., 1994, 2001), and PR-1, which is a pathogenesis-related gene under the control of the transcription coactivator NPR1, the master regulator of systemic acquired resistance (SAR) (Spoel et al., 2009), and PAD3, which is a camalexin biosynthesis pathway related gene (Chen et al., 2015), and ERF1, which is a marker gene of ET-mediated signaling pathway (Chen et al., 2015).
Bioinformatics analysis showed that there was no homologous protein of CmCP in C. manginecans. The CDS sequences of cmcp were 405 bp, encoding a protein of 134 amino acids, and contained the cerato-platanin domain. Four cysteine residues putatively involved in disulfide bridge formation were highly conserved. A signal peptide was predicted for CmCP in the first 14 amino acids. Phylogenetic analysis indicated that orthologs of CmCP were widely present in fungi, including some important plant pathogens (Figure 1A). The similarity was 95% between putative mature protein and cerato-platanin (Figure 1B).
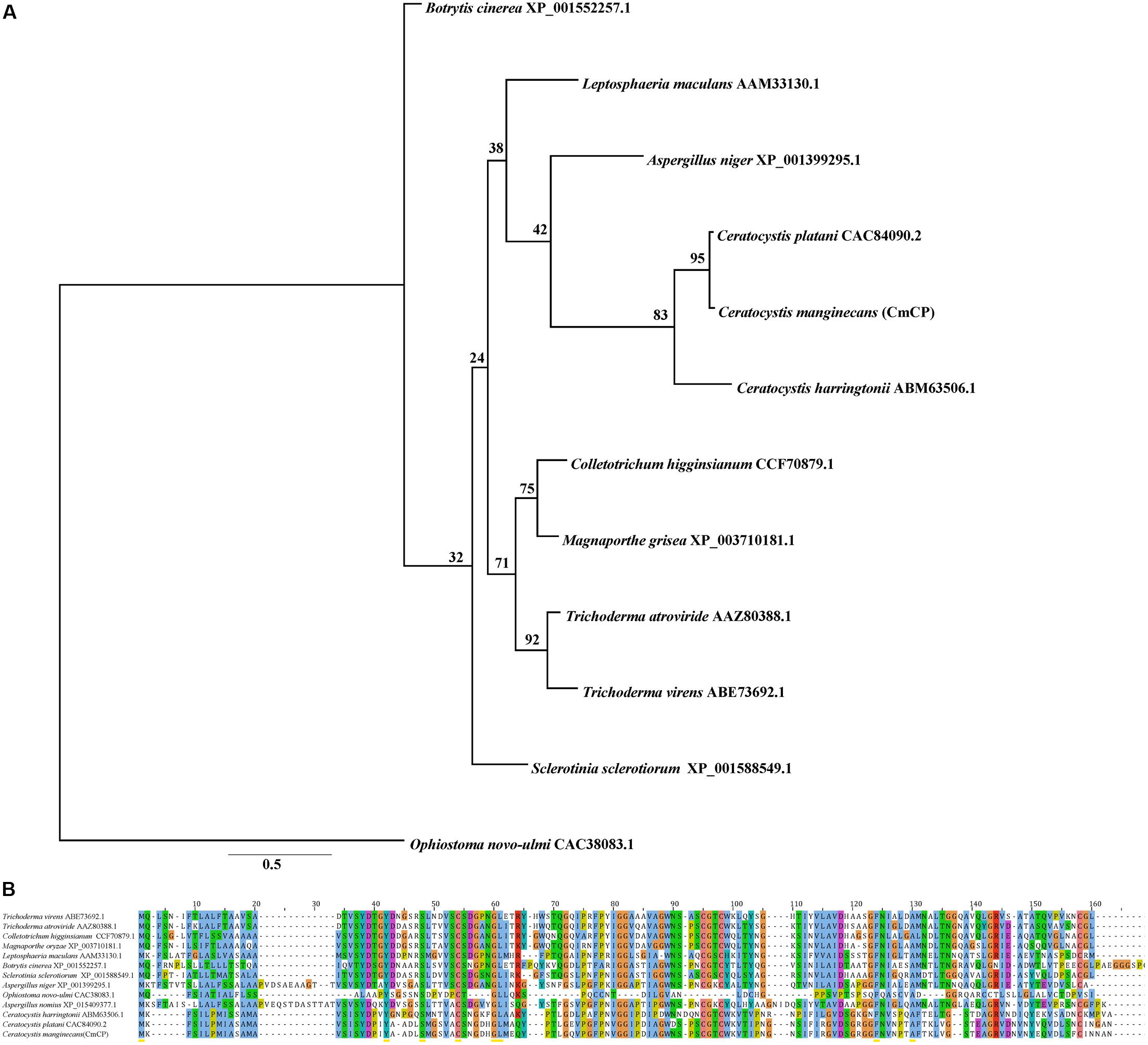
Figure 1. Analysis of the cerato-platanin protein CmCP. (A) Evolutionary relationship of CmCP and its homologs from other fungi determined with the maximum-likelihood algorithm. (B) Alignment of CmCP and its homologs with ClustalW by MAGA 5.0. Cerato-platanin protein were from: Trichoderma virens (GenBank accession number: ABE73692.1), Trichoderma atroviride (AAZ80388.1), Colletotrichum higginsianum (CCF70879.1), Magnaporthe oryzae (XP_003710181.1), Leptosphaeria maculans (AAM33130.1), Botrytis cinerea (XP_001552257.1), Sclerotinia sclerotiorum (XP_001588549.1), Aspergillus niger (XP_001399295.1), Ophiostoma novo-ulmi (CAC38083.1), Aspergillus nomius (XP_015409377.1), Ceratocystis harringtonii (ABM63506.1), Ceratocystis platani (CAC84090.2).
This was the first report on an improved CRISPR/Cas and homologous recombination system in C. manginecans, which generated cmcp gene deletion mutants. For the preparation of Ceratocystis protoplast, a large number of high-quality protoplasts were obtained from the mycelium treated with enzymes. For the plasmid transfection of Ceratocystis, Δcmcp and Δcmcp-C were selected on MYEA plates amended with 50 μg/mL hygromycin B and 25 μg/mL F2dU, respectively.
Deletion vectors and pCRISPR/Cas-U6-1-SgRNAcmcp expression vectors of cmcp gene were successfully constructed and confirmed by sequencing. Gene-deletion mutants were generated using a CRISPR/Cas system-based homology recombination strategy (Figure 2A). The expected size of the whole transformation fragment was 5500 bp. Using this transformation system (CRISPR/Cas System based homology recombination), 56 hygromycin-resistant transformants were recovered after 3–7 days and genetically purified by single-spore isolation. No colonies were observed on control (no vector DNA fragments) plates. The primers M5 and M6 amplified 5929 bp fragments from the cmcp deletion mutants but amplified 2893 bp fragments from the parental wild-type strain MG-1-10 (Figure 2B). The primes M7 and M8 amplified 464 bp fragments on cmcp gene of the wild-type strain MG-1-10 but did not amplify any fragments from the cmcp deletion mutants. In addition, the same fragments amplified from the wild-type strain MG-1-10 were amplified in Δcmcp-C strains (Figure 2B).
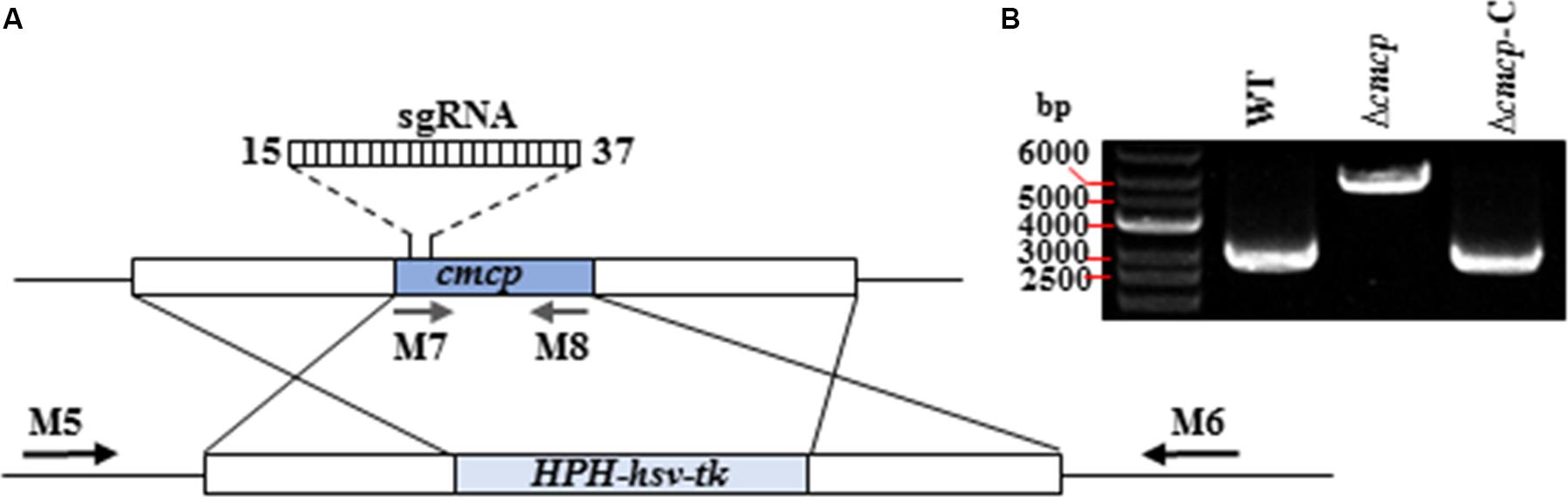
Figure 2. Generation and identification of Ceratocystis manginecans mutants with the cmcp gene deleted. (A) Schematic diagram of replacing cmcp gene. The gene replacement cassette HPH-hsv-tk contains hygromycin B-resistant gene and herpes simplex virus thymidine kinase gene. Binding sites for primers (M5, M6, M7, and M8) used in polymerase chain reaction (PCR) are indicated by arrows (see Supplementary Table S1 for primer sequences). (B) PCR amplicons for Δcmcp transformant and Δcmcp-C strain, using primer pair M5/M6. A 5.9 Kb amplified fragment indicates the HPH–hsv-tk was inserted into the sites where cmcp is normally located on the genome. WT, wild-type C. manginecans strain MG-1-10; Δcmcp, cmcp-deletion mutant derived from MG-1-10; Δcmcp-C, cmcp-complemented strain of Δcmcp mutant.
In qRT-PCR analyses, the wild-type strain MG-1-10, Δcmcp and Δcmcp-C were used to quantify the expression of cmcp gene using the 18S gene as endogenous control. Normal expression of cmcp gene was detected on the wild-type parental strain MG-1-10 and Δcmcp-C strains but no expression was detected of cmcp gene in Δcmcp mutants. Those results showed that cmcp gene were successfully knocked out or complemented in the mutants. The transformation efficiency was higher than 80%.
To examine the effect of cmcp on mycelial growth and conidial production, colony diameter was measured, and the number of conidia was counted after incubation at 25°C for 12 days. Δcmcp and MG-1-10 strains reached an average diameter of 5.18 and 6.15 cm, respectively. Δcmcp grew significantly slower than the wild-type strain MG-1-10 on MYEA plates (Figures 3A,B). In addition, conidial production was significantly reduced in Δcmcp compared to MG-1-10 (Figure 3C). After 12 days of incubation on MYEA plates, the number of conidia was 44 × 106 spores/mL and 27 × 106 spores/mL for MG-1-10 and Δcmcp strains, respectively. The Δcmcp-C strain showed similar colony diameter and conidial production as the wild-type strain. Therefore, cmcp gene might involve in the growth and sporulation of C. manginecans.
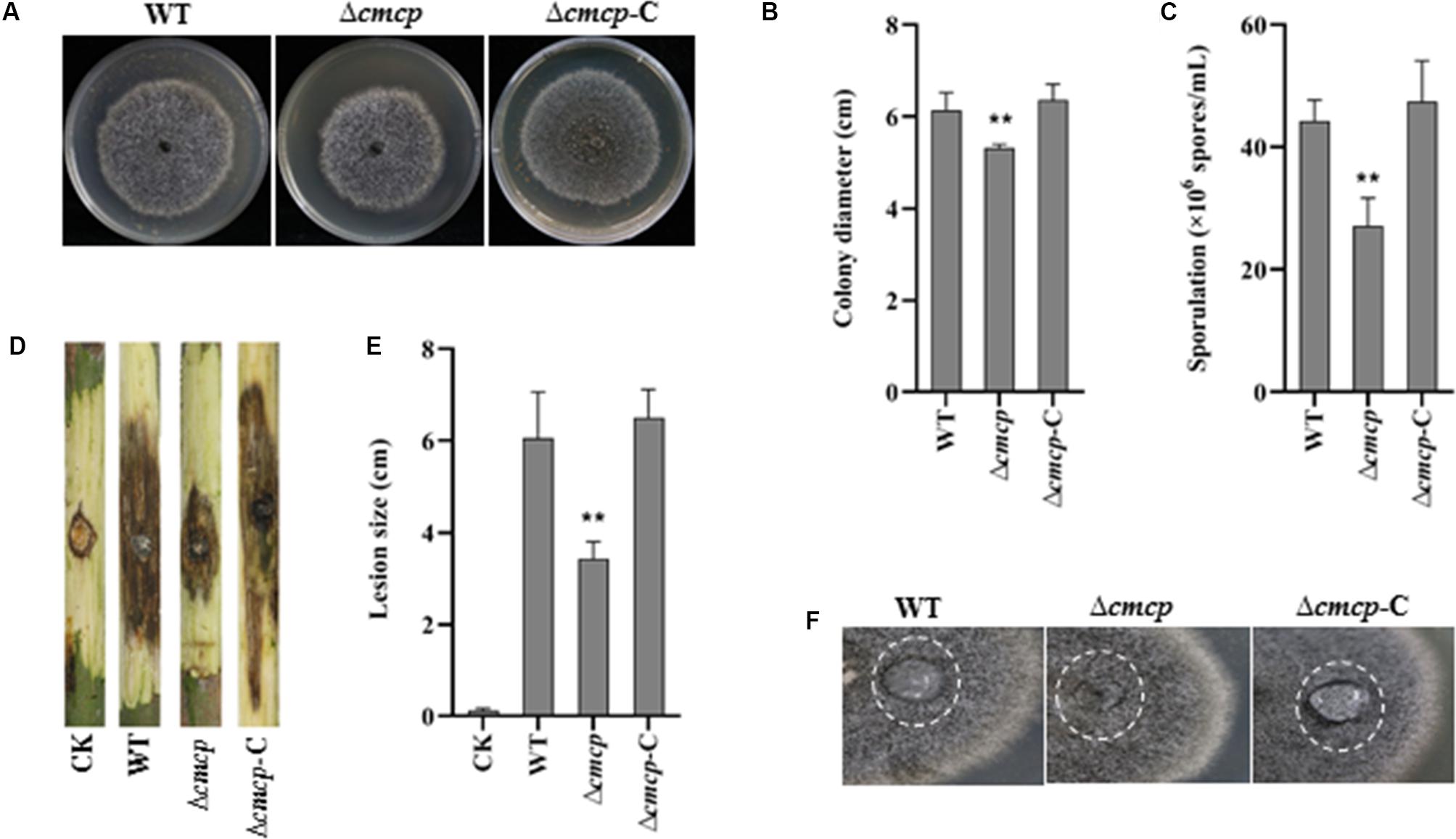
Figure 3. Biological function and virulence of cmcp gene in Ceratocystis manginecans. Wild-type (WT) C. manginecans strain MG-1-10, its cmcp-deletion mutant Δcmcp, and cmcp-complemented strain Δcmcp-C were incubated on malt yeast extract agar for 12 days at 25°C, resulting in (A) culture morphology; (B) colony size (n = 4 per plot, **P < 0.01, ±SD); (C) sporulation (n = 4 per plot, **P < 0.01, ±SD); (D,E) virulence on mango branches (n = 4 per plot, **P < 0.01, ±SD); and (F) “easily wettable” phenotype shown in a white circle. Three biological replicates were employed for each experiment.
To check the hydrophobicity of the Δcmcp, water was placed on the top of fungal cultures. After 12 h, water drop was still suspended on the culture surface of the wild-type strain and Δcmcp-C but had soaked into the surface of all the Δcmcp mutants, leaving a pronounced water-soaked mark (Figure 3F). Therefore, the gene product of cmcp contributed to cell surface hydrophobicity of the cell surface of mycelium of C. manginecans.
The virulence of the wild-type strain and the Δcmcp mutants was studied using mycelial inoculation. After 10 days of incubation, both wild-type strain, Δcmcp-C and Δcmcp caused brown or dark lesions on mango branches and many perithecia were observed on the inoculated site. However, virulence of the Δcmcp mutants was significantly reduced (Figures 3D,E). At 10 days post-inoculation, the lesion size caused by wild-type strain and Δcmcp-C were 6.05 and 6.5 cm respectively, but Δcmcp produced much significantly smaller lesions on mango branches (P < 0.01). No symptom was observed in control branches. Thus, cmcp was a major pathogenicity factor in the development of mango wilt.
The cmcp gene was expressed in P. pastoris X33 using the vector pPICZ-αAM-cmcp, and the recombinant protein was successfully obtained from yeast culture supernatant (Figure 4A). After being applied to N. tabacum leaves, CmCP significantly induced plant cell death at concentrations of 60 and 12 μM but was not effective at 2.4 μM and lower protein concentrations. The necrotic symptoms started to appear at 24 hpi and continued to develop to a larger and visible lesion for several days. A typical symptom at 5 days after infiltration was shown on Figure 4B. Control infiltrations with H2O or the products expressed and purified from EV did not produce any lesions on N. tabacum leaves. Therefore, cell death of plant leaves was specifically induced by CmCP, and this activity was dose dependent.
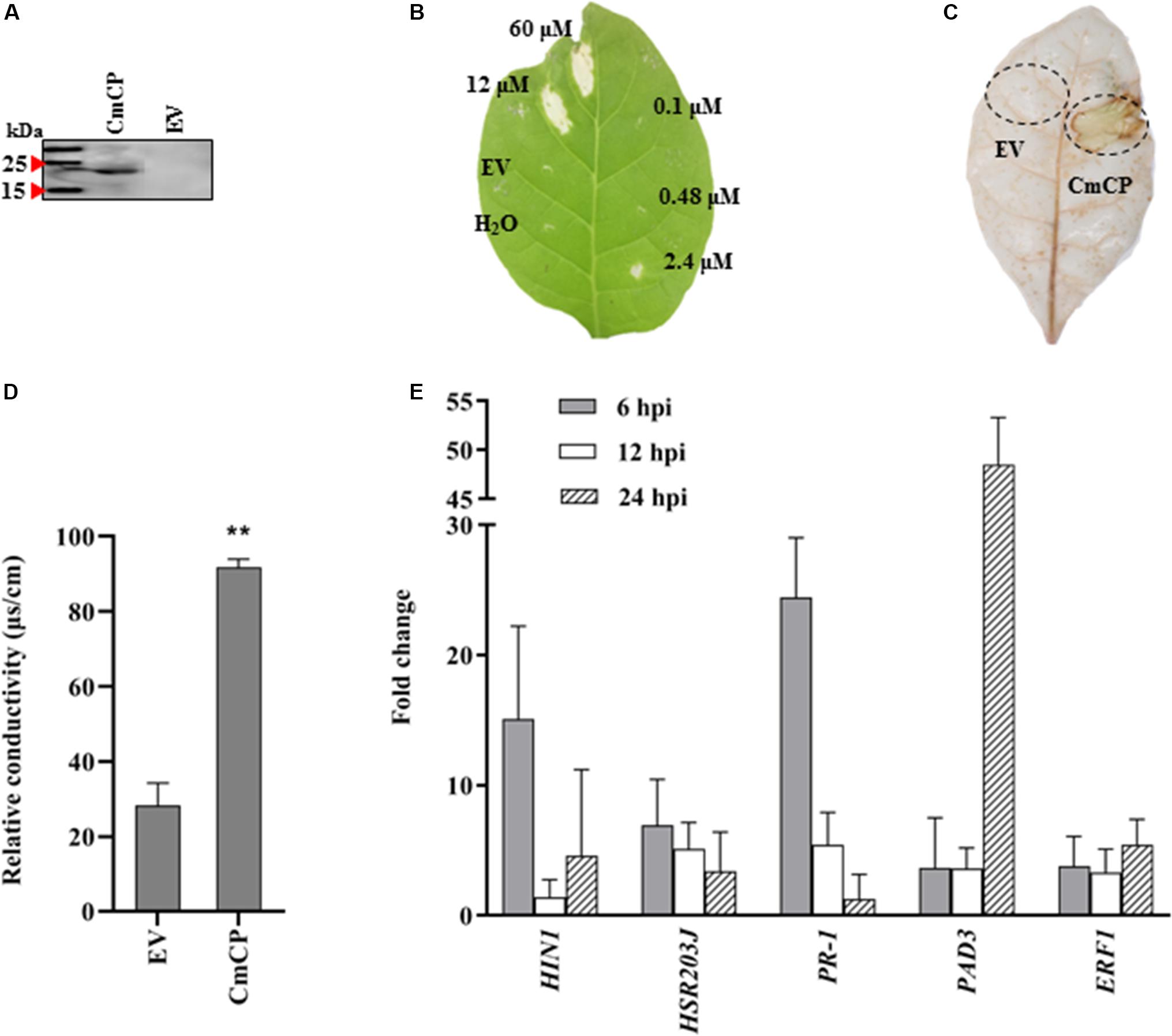
Figure 4. Induction of hypersensitive response (HR) on tobacco (Nicotiana tabacum) leaves with CmCP protein. (A) Western-blot analysis of the purified recombinant CmCP. (B) Tabaco leaves infiltrated with water, EV, and CmCP protein at 0.1, 0.48, 2.4, 12, and 60 μM and incubated for 5 days. (C) Reactive oxygen species (ROS) in tabaco leaves infiltrated with 12 μM CmCP or EV for 30 h post inoculation (hpi), detected using 3,3′-diaminobenzidine. (D) Electrolyte leakage of tabaco leaves infiltrated with 12 μM CmCP or EV for 30 hpi (n = 3 per plot, **P < 0.01, ±SD). (E) Expression of defense genes induced by CmCP protein in tabaco leaves detected by quantitative polymerase chain reaction (n = 3, **P < 0.01, ±SD). The y-axis represents normalized fold (2−ΔΔCt) compared to infiltrate with EV (set to 1). HIN1 and HSR203J are markers of HR; PR-1 is pathogenesis-related gene; PAD3 is phytoalexin-deficiency gene; and ERF1 is ethylene response factor 1. EV: the products expressed and purified from empty vector X33-pPICZ-αAM. Three biological replicates were employed for each experiment.
Reactive oxygen species and electrolyte leakage were assayed in plants cells infiltrated with CmCP product. The generation of H2O2 and O2– was evaluated with DAB. DAB staining results showed that an increase in brown DAB precipitate was observed in the area where CmCP had been infiltrated, while control infiltrations with the products expressed and purified from EV did not show any chemical reactions in the leaves (Figure 4C). The production of superoxide anion in tobacco leaves increased at 30 hpi on tobacco leaves with CmCP treatment. Therefore, infiltration of N. tabacum leaves with CmCP induced ROS of the treated tissue.
A serious electrolyte leakage was detected at 30 hpi on tobacco leaves with CmCP infiltration. The relative conductivity of CmCP-infiltrated area was significantly higher than the control area infiltrated with the products expressed and purified from EV (Figure 4D). Thus, infiltration of N. tabacum leaves with CmCP caused electrolyte leakage of the treated tissue.
The expression of five genes related to plant defenses was examined after N. tabacum leaves infiltrated with 12 μM CmCP. qRT-PCR results (Figure 4E) showed that in N. tabacum leaves, PR-1 (pathogenesis-related gene under the control of the transcription coactivator NPR1), HSR203J and HIN1 (marker genes of HR), PAD3 (phytoalexin biosynthesis pathway) and ERF1 (Ethylene-mediated signaling pathway) were significantly up-regulated. In the early phase at 6 hpi, transcript levels of PR-1, HIN1 and HSR203J reached the maximum at the detecting time point. Relative to reference genes, the transcription of PR-1 was more than 24 folds above control (the products expressed and purified from EV treated leaves) relative to reference genes; and the transcription of HIN1 was >15 folds above control. After infiltration for 24 h, PAD3 expression reached the highest level, and its transcription increased by >40 folds above control. Therefore, CmCP induced the expression of all five genes related to plant defenses.
We have demonstrated that cmcp gene was required for growth development of C. manginecans and played a key role in pathogenicity of the fungus. The latter was confirmed by the application of CmCP protein that induced necrotic symptoms and HR in N. tabacum, including electrolyte leakage, reactive oxygen species generation and overexpression of defense-related genes. We have also demonstrated that an improved CRISPR/Cas system was an effective tool in studying Ceratocystis.
Because Ophiostoma is a genus genetically close to Ceratocystis, we adapted the method of transformation-mediated gene disruption used for Ophiostoma novo-ulmi, cu– (Royer et al., 1991; Bowden et al., 1996) with improvement. The results were promising. This method showed a high efficiency (80%) in transformation of Ceratocystis. Our improvements included (1) using driselase and lysing enzymes instead of NovoZym234 enzyme and 2-mercaptoethanol (strong toxicity) to digest the mycelium (Royer et al., 1991) in protoplast preparation; (2) Because the growth of C. manginecans was extremely slow, we used plating the transformation mixture on rather than in the medium (Talbot et al., 1993); and (3) using CRISPR/Cas-U6-1 expression vectors, 60% PEG and 50 μg/mL of hygromycin B (Parsons et al., 1987; Leung et al., 1990; Arazoe et al., 2015). We have shown that using conventional homologous recombination method (not adding pCRISPR/Cas-U6-1-SgRNAcmcp expression vectors) and different promoters including 35S and TrpC was unsuccessful. Therefore, we suggested that the improved CRISPR/Cas system we established was an efficient gene editing protocol for Ceratocystis.
CPPs play important roles in virulence and mycelial growth and spore formation of fungi (Jeong et al., 2010; Marcos et al., 2011; De O Barsottini et al., 2013; Baccelli, 2015). In this study, we have demonstrated that the virulence of cmcp deletion mutants were significantly reduced, as shown in M. grisea and B. cinera (Jeong et al., 2010; Frías et al., 2011). We also found that cmcp deletion mutants possessed a water-soaked phenotype, which was in agreement with others suggesting that CPPs contribute to cell surface hydrophobicity of aerial hyphae of certain plant pathogenic fungi, such as M. grisea and Ophiostoma novo-ulmi (Talbot et al., 1993; Bowden et al., 1996; Pazzagli et al., 2014). In M. grisea, mpg1 may involve in attachment, infection court preparation, or topological signaling during infection on plant surface (Mendgen and Deising, 1993; Talbot et al., 1993). In contrast, cmcp expression affected mycelial growth and conidial production. Therefore, those phenotypes may be related to the reduced pathogenicity of cmcp deletion mutants of C. manginecans.
CPPs constitute a well conserved protein family (Chen et al., 2013). However, functions of these genes encoding for CPPs vary greatly depending on the fungal taxon. On the one hand, not all genes encoding CPPs are involved in the pathogenicity of fungi, as confirmed with sp1 gene in Leptosphaeria maculans (Wilson et al., 2002) and cu gene in Ophiostoma novo-ulmi (Bowden et al., 1996). On the other hand, the growth and overall phenotype are not affected by epl1 and epl2 genes in Trichoderma atroviride (Frischmann et al., 2013) and bcsp1 in B. cinera (Frías et al., 2011). This may be related to the difference of CPPs in the interaction between pathogen and plant cells, the differences of plant species, or the intrinsic differences in the proteins. cp is a single-copy gene in C. platanin (Baccelli et al., 2012), which is different with some fungal species containing multiple copies of CPPs homologs (Luti et al., 2020). We have found that there was one predicted cp gene in C. manginecans. It seems that the number of cp gene copy is dependent on fungal species.
By expressing cmcp in P. pastoris, the function of obtained CmCP product can be examined on N. tabacum leaves. Results showed that CmCP-induced necrosis was a dose-dependent activity, and it also induced the expression of defense genes in plant cells. Similar reports have been documented on CPPs from C. platanin (Pazzagli et al., 1999), B. cinerea (Frías et al., 2011), and M. grisea (Yang et al., 2009). CPPs act as microbe/pathogen-associated molecular patterns (MAMPs/PAMPs) (Gaderer et al., 2014; Baccelli, 2015) and induce plant defense responses related to HR, including electrolyte leakage (Frías et al., 2011), reactive oxygen species generation (Djonović et al., 2006) and defenses gene overexpression (Buensanteai et al., 2010; Frías et al., 2011), which supports our results.
We have shown that HSR203J and HIN1 were responsible for HR-induced cell death (Pontier et al., 1994, 2001); PR-1 is a pathogenesis-related gene associated with salicylic acid-mediated signaling pathways (Spoel et al., 2009); ERF1 functions via ethylene-mediated signaling pathway (Chen et al., 2015); and PAD3 is a phytoalexin-related gene (Chen et al., 2015). The expression of all these gene were induced by CmCP. Thus, CmCP may be related to the activation of phytoalexin, and participate in the SA/ET-mediated signaling pathways after infection plant cell. PR1 is a common host protein involved in host defense, and may be a ‘universal’ target to be attacked by pathogen-secreted proteins (Breen et al., 2016; Yang et al., 2017). SsCP1 interacted and targeted plant PR1 and contributes to virulence of Sclerotinia sclerotiorum (Yang et al., 2017). Therefore, CmCP can be an effector involved in the process of pathogen-plant interaction, and PR1 should the target of CmCP.
Although CPPs are well-known to act as elicitors or effectors in ascomycete and basidiomycete fungi (Pazzagli et al., 2014), other additional functions should be illustrated. In addition, the low similarity of genes encoding for CPPs leads to the functions of these genes vary greatly in different fungi. Thus, the expression pattern, subcellular localization and structure of CmCP, and its interaction with the host should be further confirmed.
All datasets generated for this study are included in the article/Supplementary Material.
ZZ performed the majority of the experiments, data analysis and preparation of the manuscript. JL provided guidance and designed the experiments. LL, JH, and YL contributed in suggestions in research process as well as in revising the manuscript. All the authors participated in the project proposal and approved the final version of the manuscript.
This work was funded by the National Natural Science Foundation of China, project number 31571958.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01824/full#supplementary-material
FIGURE S1 | Wild-type Ceratocystis manginecans strain MG-1-10 grown on agar plates amended with different concentrations of hygromycin B (A–L): 0, 2.5, 5, 10, 15, 20, 30, 50, 100, 150, 200, and 250 μg/mL.
Al Adawi, A. O., Al Jabri, R. M., Deadman, M. L., Barnes, I., Wingfield, B., Wingfield, M. J., et al. (2013). The mango sudden decline pathogen, Ceratocystis manginecans, is vectored by Hypocryphalus mangiferae (Coleoptera: Scolytinae) in Oman. Infect. Genet. Evol. 135, 243–251. doi: 10.1007/s10658-012-0081-7
Arazoe, T., Miyoshi, K., Yamato, T., Ogawa, T., Ohsato, S., Arie, T., et al. (2015). Tailor-made CRISPR/Cas system for highly efficient targeted gene replacement in the rice blast fungus. Biotechnol. Bioeng. 112, 2543–2549. doi: 10.1002/bit.25662
Baccelli, I. (2015). Cerato-platanin family proteins: one function for multiple biological roles? Front. Plant Sci. 5:769. doi: 10.3389/fpls.2014.00769
Baccelli, I., Comparini, C., Bettini, P. P., Martellini, F., Ruocco, M., Pazzagli, L., et al. (2012). The expression of the cerato-platanin gene is related to hyphal growth and chlamydospores formation in Ceratocystis platani. FEMS Microbiol. Lett. 327, 155–163. doi: 10.1111/j.1574-6968.2011.02475.x
Baker, C. J., Harrington, T. C., Ulrike, K., and Alfenas, A. C. (2003). Genetic variability and host specialization in the Latin American clade of Ceratocystis fimbriata. Phytopathology 93, 1274–1284. doi: 10.1094/PHYTO.2003.93.10.1274
Bell-Pedersen, D., Dunlap, J. C., and Loros, J. J. (1992). The Neurospora circadian clock-controlled gene, ccg-2, is allelic to eas and encodes a fungal hydrophobin required for formation of the conidial rodlet layer. Genes Dev. 6, 2382–2394. doi: 10.1101/gad.6.12a.2382
Bowden, C. G., Smalley, E., Guries, R. P., Hubbes, M., and Horgen, P. A. (1996). Lack of association between cerato-ulmin production and virulence in Ophiostoma novo-ulmi. Mol. Plant Microbe Interact. 9, 556–564. doi: 10.1094/MPMI-9-0556
Breen, S., Williams, S. J., Winterberg, B., Kobe, B., and Solomon, P. S. (2016). Wheat PR-1 proteins are targeted by necrotrophic pathogen effector proteins. Plant J. 88, 13–25. doi: 10.1111/tpj.13228
Buensanteai, N., Mukherjee, P. K., Horwitz, B. A., Cheng, C., Dangott, L. J., Kenerley, C. M., et al. (2010). Expression and purification of biologically active Trichoderma virens proteinaceous elicitor Sm1 in Pichia pastoris. Protein Expr. Purif. 72, 131–138. doi: 10.1016/j.pep.2010.03.006
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinf. 10:421. doi: 10.1186/1471-2105-10-421
Carroll, D. (2014). Genome engineering with targetable nucleases. Annu. Rev. Biochem. 83, 409–439. doi: 10.1146/annurev-biochem-060713-035418
Chen, H., Kovalchuk, A., Kerio, S., and Asiegbu, F. O. (2013). Distribution and bioinformatic analysis of the cerato-platanin protein family in Dikarya. Mycologia 105, 1479–1488. doi: 10.3852/13-115
Chen, H., Quintana, J., Kovalchuk, A., Ubhayasekera, W., and Asiegbu, F. O. (2015). A cerato-platanin-like protein HaCPL2 from Heterobasidion annosum sensu stricto induces cell death in Nicotiana tabacum and Pinus sylvestris. Fungal Genet. Biol. 84, 41–51. doi: 10.1016/j.fgb.2015.09.007
Comparini, C., Carresi, L., Pagni, E., Sbrana, F., Sebastiani, F., Luchi, N., et al. (2009). New proteins orthologous to cerato-platanin in various Ceratocystis species and the purification and characterization of cerato-populin from Ceratocystis populicola. Appl. Microbiol. Biotechnol. 84, 309–322. doi: 10.1007/s00253-009-1998-4
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. doi: 10.1126/science.1231143
Cong, L., and Zhang, F. (2014). “Genome engineering using CRISPR-Cas9 system,” in Chromosomal Mutagenesis ed. S. Pruett-Miller (New York, FL: Humana Press) 197–217.
De O Barsottini, M. R., De Oliveira, J. F., Adamoski, D., Teixeira, P. J., do Prado, P. F., et al. (2013). Functional diversification of cerato-platanins in Moniliophthora perniciosa as seen by differential expression and protein function specialization. Mol. Plant Microbe Interact. 26, 1281–1293. doi: 10.1094/mpmi-05-13-0148-r
Djonović, S., Pozo, M. J., Dangott, L. J., Howell, C. R., and Kenerley, C. M. (2006). Sm1, a proteinaceous elicitor secreted by the biocontrol fungus Trichoderma virens induces plant defense responses and systemic resistance. Mol. Plant Microbe Interact. 19, 838–853. doi: 10.1046/j.1365-3164.2002.00305.x
Fourie, A., Wingfield, M. J., Wingfield, B. D., Thu, P. Q., and Barnes, I. (2016). A possible centre of diversity in South East Asia for the tree pathogen, Ceratocystis manginecans. Infect. Genet. Evol. 41, 73–83. doi: 10.1016/j.meegid.2016.03.011
Frías, M., Brito, N., González, M., and González, C. (2014). The phytotoxic activity of the cerato-platanin BcSpl1 resides in a two-peptide motif on the protein surface. Mol. Plant Pathol. 15, 342–351. doi: 10.1111/mpp.12097
Frías, M., González, C., and Brito, N. (2011). BcSpl1, a cerato-platanin family protein, contributes to Botrytis cinerea virulence and elicits the hypersensitive response in the host. New Phytol. 192, 483–495. doi: 10.2307/41320501
Frischmann, A., Neudl, S., Gaderer, R., Bonazza, K., Zach, S., Gruber, S., et al. (2013). Self-assembly at air/water interfaces and carbohydrate-binding properties of the small secreted protein EPL1 from the fungus Trichoderma atroviride. J. Biol. Chem. 288, 4278–4287. doi: 10.1074/jbc.M112.427633
Gaderer, R., Bonazza, K., and Seidl-Seiboth, V. (2014). Cerato-platanins: a fungal protein family with intriguing properties and application potential. Appl. Microbiol. Biotechnol. 98, 4795–4803. doi: 10.1007/s00253-014-5690-y
Gomes, E. V., Costa, M. D. N., De Paula, R. G., de Azevedo, R. R., da Silva, F. L., Noronha, E. F., et al. (2015). The cerato-platanin protein Epl-1 from Trichoderma harzianum is involved in mycoparasitism, plant resistance induction and self cell wall protection. Sci Rep 5:17998. doi: 10.1038/srep17998
Jeong, J. S., Mitchell, T. K., and Dean, R. A. (2010). The Magnaporthe grisea snodprot1 homolog, MSP1, is required for virulence. FEMS Microbiol. Lett. 273, 157–165. doi: 10.1111/j.1574-6968.2007.00796.x
Jiang, D., Zhu, W., Wang, Y., Sun, C., Zhang, K. Q., Yang, J., et al. (2013). Molecular tools for functional genomics in filamentous fungi: recent advances and new strategies. Biotechnol. Adv. 31, 1562–1574. doi: 10.1016/j.biotechadv.2013.08.005
Johnson, J. A., Harrington, T. C., and Cengelbrecht, C. J. B. (2005). Phylogeny and taxonomy of the North American clade of the Ceratocystis fimbriata complex. Mycologia 97, 1067–1092. doi: 10.2307/3762286
Kück, U., and Hoff, B. (2010). New tools for the genetic manipulation of filamentous fungi. Appl. Microbiol. Biotechnol. 86, 51–62. doi: 10.1007/s00253-009-2416-7
Leung, H., Lehtinen, U., Karjalainen, R., Skinner, D., Tooley, P., Leong, S., et al. (1990). Transformation of the rice blast fungus Magnaporthe grisea to hygromycin B resistance. Curr. Genet. 17, 409–411. doi: 10.1007/bf00334519
Luti, S., Sella, L., Quarantin, A., Pazzagli, L., and Baccelli, I. (2020). Twenty years of research on cerato-platanin family proteins: clues, conclusions, and unsolved issues. Fungal Biol. Rev. 34, 13–24. doi: 10.1016/j.fbr.2019.10.001
Marcos, F., Celedonio, G., and Nélida, B. (2011). BcSpl1, a cerato-platanin family protein, contributes to Botrytis cinerea virulence and elicits the hypersensitive response in the host. New Phytol. 192, 483–495. doi: 10.2307/41320501
Mendgen, K., and Deising, H. (1993). Infection structures of fungal plant pathogens:a cytological and physiological evaluation. New Phytol. 124, 193–213. doi: 10.1111/j.1469-8137.1993.tb03809.x
Nguyen, L. T., Schmidt, H. A., Von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effectivestochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Parsons, K. A., Chumley, F. G., and Valent, B. (1987). Genetic transformation of the fungal pathogen responsible for rice blast disease. Proc. Natl. Acad. Sci. U.S.A. 84, 4161–4165. doi: 10.1073/pnas.84.12.4161
Pazzagli, L., Cappugi, G., Manao, G., Camici, G., Santini, A., Scala, A., et al. (1999). Purification, characterization, and amino acid sequence of cerato-platanin, a new phytotoxic protein from Ceratocystis fimbriata f. sp. platani. J. Biol. Chem. 274, 24959–24964. doi: 10.1074/jbc.274.35.24959
Pazzagli, L., Pantera, B., Carresi, L., Zoppi, C., Pertinhez, T. A., Spisni, A., et al. (2006). Cerato-platanin, the first member of a new fungal protein family. Cell Biochem. Biophys. 44, 512–521. doi: 10.1385/cbb:44:3:512
Pazzagli, L., Seidl-Seiboth, V., Barsottini, M., Barsottini, M., Vargas, W. A., Scala, A., et al. (2014). Cerato-platanins: elicitors and effectors. Plant Sci. 228, 79–87. doi: 10.1016/j.plantsci.2014.02.009
Petersen, T. N., Brunak, S., Heijne, G. V., and Nielsen, H. H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786. doi: 10.1038/nmeth.1701
Pontier, D., Balague, C., Bezombes-Marion, I., Tronchet, M., Deslandes, L., Roby, D., et al. (2001). Identification of a novel pathogen-responsive element in the promoter of the tobacco gene HSR203J, a molecular marker of the hypersensitive response. Plant J. 26, 495–507. doi: 10.1046/j.1365-313x.2001.01049.x
Pontier, D., Godiard, L., Marco, Y., and Roby, D. (1994). HSR203J, a tobacco gene whose activation is rapid, highly localized and specific for incompatible plant/pathogen interactions. Plant J. 5, 507–521. doi: 10.1046/j.1365-313X.1994.5040507.x
Potter, S. C., Luciani, A., Eddy, S. R., Park, Y., Lopez, R., Finn, R. D., et al. (2018). HMMER web server: 2018 update. Nucleic Acids Res. 46, W200–W204. doi: 10.1093/nar/gky448
Ran, F. A., Hsu, P. D., Wright, J., Agarwala, V., Scott, D. A., Zhang, F., et al. (2013). Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308. doi: 10.5496/wjmg.v4.i3.69
Royer, J. C., Dewar, K., Hubbes, M., and Horgen, P. A. (1991). Analysis of a high frequency transformation system for Ophiostoma ulmi, the causal agent of Dutch elm disease. Mol. Gen. Genet. 225, 168–176. doi: 10.1007/bf00282655
Sakuma, T., Nishikawa, A., Kume, S., Chayama, K., and Yamamoto, T. (2014). Multiplex genome engineering in human cells using all-in-one CRISPR/Cas9 vector system. Sci. Rep. 4:5400. doi: 10.1038/srep05400
Schuster, M., Schweizer, G., Reissmann, S., and Kahmann, R. (2015). Genome editing in Ustilago maydis using the CRISPR-Cas system. Fungal Genet. Biol. 89, 3–9. doi: 10.1016/j.fgb.2015.09.001
Spoel, S. H., Mou, Z., Tada, Y., Spivey, N. W., Genschik, P., Dong, X., et al. (2009). Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell 137, 860–872. doi: 10.1016/j.cell.2009.03.038
Stringer, M. A., Dean, R. A., Sewall, T. C., and Timberlake, W. E. (1991). Rodletless, a new Aspergillus developmental mutant induced by directed gene inactivation. Genes Dev. 5, 1161–1171. doi: 10.1101/gad.5.7.1161
Talbot, N. J., Ebbole, D. J., and Hamer, J. E. (1993). Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell 5, 1575–1590. doi: 10.2307/3869740
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., Kumar, S., et al. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tarigan, M., Roux, J., Van Wyk, M., Tjahjono, B., and Wingfield, M. J. (2011). A new wilt and die-back disease of Acacia mangium associated with Ceratocystis manginecans and C. acaciivora sp. nov. in Indonesia. South Afr. J. Bot. 77, 292–304. doi: 10.1016/j.sajb.2010.08.006
Van Wyk, M., Al Adawi, A. O., Khan, I. A., Deadman, M. L., Jahwari, A. A. A., Wingfield, B. D., et al. (2007). Ceratocystis manginecans sp. nov., causal agent of a destructive mango wilt disease in Oman and Pakistan. Fungal Divers. 27, 213–230. doi: 10.1002/yea.1553
Van Wyk, M., Wingfield, B. D., Al Adawi, A. O., Rossetto, C. J., Ito, M. F., Wingfield, M. J., et al. (2011). Two new Ceratocystis species associated with mango disease in Brazil. Mycotaxon 117, 381–404. doi: 10.5248/117.381
Viégas, D. A. P. (1960). Seca da mangueira mango-blight. Bragantia 19, 163–182. doi: 10.1590/S0006-87051960000100011
Wenderoth, M., Pinecker, C., Vob, B., and Fischer, R. (2017). Establishment of CRISPR/Cas9 in Alternaria alternata. Fungal Genet. Biol. 101, 55–60. doi: 10.1016/j.fgb.2017.03.001
Wilson, L. M., Idnurm, A., and Howlett, B. J. (2002). Characterization of a gene (sp1) encoding a secreted protein from Leptosphaeria maculans, the blackleg pathogen of Brassica napus. Mol. Plant Pathol. 3, 487–493. doi: 10.1046/j.1364-3703.2002.00144.x
Wösten, H. A. B. (2001). Hydrophobins: multipurpose proteins. Annu. Rev. Microbiol. 55, 625–646. doi: 10.1146/annurev.micro.55.1.625
Yang, G., Tang, L., Gong, Y., Xie, J., Fu, Y., Jiang, D., et al. (2017). A cerato-platanin protein SsCP1 targets plant PR1 and contributes to virulence of Sclerotinia sclerotiorum. New Phytol. 217, 739–755. doi: 10.1111/nph.14842
Yang, Y. Y., Zhang, H. J., Li, G., Li, W., Wang, X., Song, F., et al. (2009). Ectopic expression of MgSM1, a Cerato-platanin family protein from Magnaporthe grisea, confers broad-spectrum disease resistance in Arabidopsis. Plant Biotechnol. J. 7, 763–777. doi: 10.1111/j.1467-7652.2009.00442.x
Zhang, Z. P., Li, Q., Luo, L. X., Hao, J. J., and Li, J. Q. (2017). First report of mango wilt caused by Ceratocystis fimbriata in Mangifera indica in China. Plant Dis. 101:1042. doi: 10.1094/pdis-10-16-1477-pdn
Keywords: mango wilt, CRISPR/Cas, cerato-platanin, virulence, hypersensitive response
Citation: Zhang Z, Li Y, Luo L, Hao J and Li J (2020) Characterization of cmcp Gene as a Pathogenicity Factor of Ceratocystis manginecans. Front. Microbiol. 11:1824. doi: 10.3389/fmicb.2020.01824
Received: 27 January 2020; Accepted: 10 July 2020;
Published: 31 July 2020.
Edited by:
Gustavo Henrique Goldman, University of São Paulo, BrazilReviewed by:
Changbin Chen, Institut Pasteur of Shanghai (CAS), ChinaCopyright © 2020 Zhang, Li, Luo, Hao and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianqiang Li, bGlqcTIzMUBjYXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.