
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
OPINION article
Front. Microbiol. , 18 August 2020
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.01820
This article is part of the Research Topic Counteracting the Global Emergence of Antimicrobial Resistance in Biomaterial-Associated Infections: Current Research and New Breakthroughs View all 5 articles
 Ivan Brukner1,2,3*
Ivan Brukner1,2,3* Matthew Oughton1,2,3*
Matthew Oughton1,2,3*Most patients with an acute bacterial infection have immune defense mechanisms to respond to the pathogen and destroy both resistant and non-resistant bacterial cells, facilitating the elimination of the pathogen. However, a growing population of patients–those with an immune deficiency—may lack some, or many aspects of those defense mechanisms, and thus require effective antibiotic therapy to survive or reduce their time required for recovery. Antibiotic Susceptibility Testing (AST) can help in choosing the most appropriate option. However, misguiding AST results can lead to poor and even fatal outcomes (Zilberberg et al., 2017; Jiang et al., 2018; Karve et al., 2018; Band and Weiss, 2019; Eliakim-Raz et al., 2019; Peeters et al., 2019; Rodriguez-Gomez et al., 2019; Uppsala University, 2019). There is a consensus among some medical researchers that these tests need improvement (Valsesia et al., 2015; Karve et al., 2018; Martin et al., 2018; Band and Weiss, 2019; Kahlmeter et al., 2019; Lerminiaux and Cameron, 2019; Nicoloff et al., 2019; Peeters et al., 2019). To improve the clinical value of AST, we challenge and propose a change of the two most dogmatic steps of the clinical tests: standardization of bacterial inoculation size (McFaland, 1907; Smith and Kirby, 2018) and the use of a limited number of bacterial colonies (Kao et al., 2014; Qin et al., 2018; Maciel et al., 2020; Montealegre et al., 2020). Problems with AST can be resolved by replacing the current approach that relies on selection of a small number of colonies with a population-level approach.
Two leading organizations that set standards for AST, the European Committee on Antimicrobial Susceptibility Testing (EUCAST) and the Clinical and Laboratory Standards Institute (CLSI), acknowledge that there are challenges associated with the interpretation of their recommended tests (Kahlmeter et al., 2019). They have used different strategies to deal with these challenges. One such strategy, implemented by EUCAST, was to coin the term “technical uncertainty.” Use of this term was intended to attenuate small, uncontrolled technical factors from causing significant discrepancies in interpretations (Kahlmeter et al., 2019). On the contrary, CLSI acknowledged the inherent uncertainty of the test, without insisting it was solely due to technical factors (Kahlmeter et al., 2019). Future changes in regulation and standardization of AST would need to be convergent and to facilitate clinical interpretation of the results.
The response to antibiotics within bacterial populations is inherently variable due to the unknown genetic complexity of the population, as previously suggested by some authors (Qin et al., 2018; Kahlmeter et al., 2019; Mouton et al., 2019; Nicoloff et al., 2019). Present formats of AST are designed to detect the susceptibility of the most prevalent bacteria, but not necessarily the most resistant. Other bacterial subtypes with higher resistance than the majority of the bacterial population may be present at lower frequencies in clinical samples, but remain unanalyzed (Martin et al., 2018; Qin et al., 2018; Maciel et al., 2020; Montealegre et al., 2020). Unfortunately, diversity in susceptibility remains undetected due to the current standard operating procedures of AST. As a common example Figure 1 demonstrates typical but contradictory logical reasoning between antibiotic screening results and AST, which can be reproduced in any clinical microbiology laboratory. What is the cumulative probability (Pc) of getting “resistant result” from hetero-resistant samples with different relative frequencies of resistant (Rf), vs. sensitive bacterial cells present in the sample? Cumulative binomial probability (probability of getting a result indicating resistance, or Pc, in Figure 1A), refers to the probability that the value of a binomial random variable falls within a specified range. The table from Figure 1A demonstrates that even with a Rf of 0.25, the chances of detecting resistance by selecting only 5 colonies/cells is only 0.76. If the resistant cells are 10x less frequent (Rf = 0.025), the probability drops to 0.12, with further decrease as the frequency of resistant cells continues to drop. However, by the application of a plate screening method, (see below), where 10,000–100,000 cells can be inoculated and screened on the same plate, the cumulative probability of getting resistant cells is quite good, even if relative frequencies are only 0.00025 of resistant vs. sensitive cells. Ultimately, a combination of species-specific bacterial qPCR detection combined with a brief incubation in the presence of antibiotic permits the detection of even rare resistant bacterial cells. A common clinical example is the urine culture (see Figure 1B), declared as positive for E. coli (more than 106 colony forming units per mL of urine). AST, when performed routinely, determines the E.coli to be sensitive on ciprofloxacin. However, when the same sample is inoculated directly on agar containing 10 microgram/mL of ciprofloxacin, (MacConkey Agar with Ciprofloxacin, Hardy Diagnostics, VWR, Canada), resistance colonies were clearly detected (Figure 1B). These examples are indicating that AST methodology may yield inaccurate results for pathogens exhibiting hetero resistance.
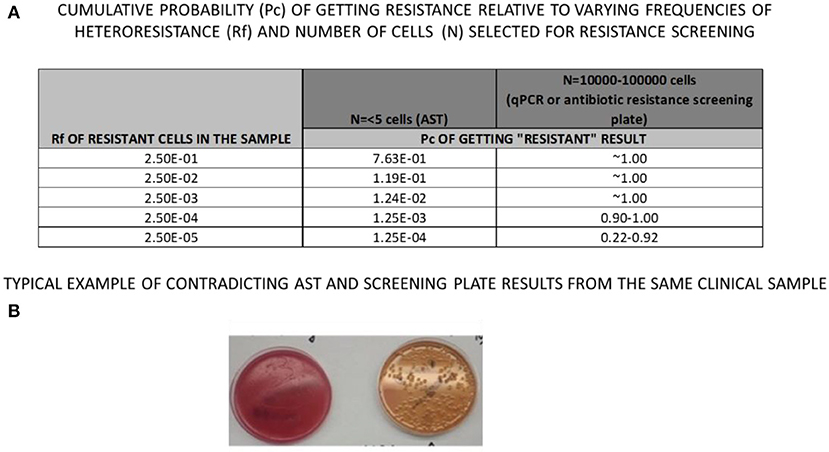
Figure 1. Same clinical sample was loaded on (A) blood Agar plate (left) forming confluent culture (more then 10,000,000 cells per ml) and on (B) MacConkey, Cipro-screening plate (right), revealing 200 resistant Colony Forming Units, CFU), contrary to Antibiotic Susceptibility test which reported only CIPRO-sensitive culture.
To predict response to antibiotic therapy, these undetected distribution-outliers—characterized by higher antibiotic resistance—should be considered.
As a reminder of standard operating procedures of AST in clinical microbiology, the first step of testing is to culture bacteria from the original sample on primary inoculum plates. The second step is to select a few individual colonies from the primary plate, prepare a standardized inoculum for AST (Kirby-Bauer plate), and determine the susceptibility result after overnight (16–18 h) incubation. Contrary to this approach, inoculating the original sample directly on antibiotic screening agar plate (Figure 1B) reveals antimicrobial resistance present in some bacteria from the inoculum. Therefore, antibiotic-resistant bacteria that were not detectable by conventional AST, were present at very low frequencies (≤1 in 10,000 colonies). This discrepancy is rooted in inoculum standardization and reduction of sample diversity when selecting individual colonies during these measurements.
There is a consensus among researchers that inoculum size has a dramatic impact on the test result. Standard tests use an inoculum size of 0.5 McFarland units (a measure of turbidity, ~108 CFU/mL) (McFaland, 1907). The rationale for this decision was to eliminate variability due to inoculum size. However, bacterial abundance is inherently variable in each clinical sample. Furthermore, the higher the bacterial inoculum concentration is, the more likely the bacterial population will develop resistance. To illustrate, think of two hypothetical patients with the similar infections (same bacterial species). The first patient, X, has 100 times the burden of infection (bacteria per volume or mass of biological fluid/tissue) compared to the second patient, Y. With the current AST, samples from patients X and Y tested multiple times would, over time, produce the same susceptibility result. However, patient X's infection would have a higher propensity for developing resistance than patient Y's. Moreover, patient X's treatment would be more likely to fail due to this difference. Therefore, standardizing inoculum size will cause a loss of clinically valuable information if we are aiming to develop a new generation of AST.
The major limitation with current AST design is the need to select only a few isolated colonies. This practice limits the test to a small number of isolates from an often-complex bacterial population. Only progenitors of a few individual bacterial cells will be standardized to produce an inoculum size of 0.5 McFarland units. This critical step in sample processing typically reduces bacterial diversity by up to 106 and leaves the result of the test to “pure” sampling chance, where statistics are not in favor of detecting resistance. Altogether, standardization of sample size and reduction of sample diversity both reduce the clinical value and predictive power of AST.
The predictive power of AST can improve, though, if bacterial species are analyzed in the context of the original clinical sample, reflecting the patient-specific burden of infection. A new generation of nucleic acid amplification tests (NAATs) can measure species-specific growth rates of bacteria in the original sample in response to different antibiotics (Rolain et al., 2004; Maxson et al., 2018). These tests require ~2–4 h of in vitro incubation, which would be a significant improvement in turnaround time (Luo et al., 2018). Furthermore, the current operative cost of these PCR-based tests can be below equivalent cost related to traditional plate-based tests.
In conclusion, the current evidence suggests that the present methodology of AST should be reconsidered. We must move forward, taking advantage of the new technology available, and abandon inoculum size standardization and the use of pure cultures. This might change the rules both for industry and the public sector but will ultimately benefit patients in need of antibiotic treatment, especially those that are immunocompromised.
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
This work was promoted by the Lady Davis Institute, Jewish General Hospital, Montreal, QC, Canada.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Unfortunately, the preservation the current concept of AST is continuing misleading antibiotic therapy with fatal consequences among immunocompromised patients every day. Our attempt/s to get funds and resolve technical issues of AST, using CIHR peer review granting system (https://cihr-irsc.gc.ca/e/13533.html) failed in 2019/2020. Hereby, we would like to acknowledge Justine Dees for significant writing and editing assistance. Supportive discussions with Drs. S. Eintracht, A. Papadakis, (JGH, Montreal), S. Leveque, (U. of Sherbrooke, Sherbrooke), F. Rallu, (SJH, Montreal), and E. Levy (Micra, NY), were very appreciated. Talking with Coleen Delisle, Anastasia Giannakakis, and Maria Pastras was clarifying the changes of routine clinical work. Dedicated to the memory of Olga Brukner, who's recurrent urosepsis was not resolved with meropenem therapy, despite multiple AST results of meropenem-sensitive Pseudomonas aeruginosa. For more information about population-based AST via qPCR, please write to aXZhbi5icnVrbmVyMkBtY2dpbGwuY2E= or bWF0dGhldy5vdWdodG9uQG1jZ2lsbC5jYQ==.
Band, V. I., and Weiss, D. S. (2019). Heteroresistance: a cause of unexplained antibiotic treatment failure? PLoS Pathog. 15:e1007726. doi: 10.1371/journal.ppat.1007726
Eliakim-Raz, N., Babitch, T., Shaw, E., Addy, I., Wiegand, I., Vank, C., et al. (2019). Risk factors for treatment failure and mortality among hospitalized patients with complicated urinary tract infection: a multicenter retrospective cohort study (RESCUING study group). Clin. Infect. Dis. 68, 29–36. doi: 10.1093/cid/ciy418
Jiang, P., Liss, M. A., and Szabo, R. J. (2018). Targeted antimicrobial prophylaxis does not always prevent sepsis after transrectal prostate biopsy. J. Urol. 200, 361–368. doi: 10.1016/j.juro.2018.03.078
Kahlmeter, G., Giske, C. G., Kirn, T. J., and Sharp, S. E. (2019). Point-counterpoint: differences between the European Committee on Antimicrobial Susceptibility Testing and Clinical and Laboratory Standards Institute Recommendations for Reporting Antimicrobial Susceptibility Results. J. Clin. Microbiol. 57:e01129–19. doi: 10.1128/JCM.01129-19
Kao, C. Y., Lee, A. Y., Huang, A. H., Song, P. Y., Yang, Y. J., Sheu, S. M., et al. (2014). Heteroresistance of Helicobacter pylori from the same patient prior to antibiotic treatment. Infect. Genet. Evol. 23, 196–202. doi: 10.1016/j.meegid.2014.02.009
Karve, S., Ryan, K., Peeters, P., Baelen, E., Rojas-Farreras, S., Potter, D., et al. (2018). The impact of initial antibiotic treatment failure: real-world insights in patients with complicated urinary tract infection. J. Infect. 76, 121–131. doi: 10.1016/j.jinf.2017.11.001
Lerminiaux, N. A., and Cameron, A. D. S. (2019). Horizontal transfer of antibiotic resistance genes in clinical environments. Can. J. Microbiol. 65, 34–44. doi: 10.1139/cjm-2018-0275
Luo, J., Yu, J., Yang, H., and Wei, H. (2018). Parallel susceptibility testing of bacteria through culture-quantitative PCR in 96-well plates. Int. J. Infect. Dis. 70, 86–92. doi: 10.1016/j.ijid.2018.03.014
Maciel, J. F., Gressler, L. T., da Silveira, B. P., Dotto, E., Balzan, C., Matter, L. B., et al. (2020). Caution at choosing a particular colony-forming unit from faecal Escherichia coli: it may not represent the sample profile. Lett. Appl. Microbiol. 70, 130–136. doi: 10.1111/lam.13252
Martin, L. W., Robson, C. L., Watts, A. M., Gray, A. R., Wainwright, C. E., Bell, S. C., et al. (2018). Expression of Pseudomonas aeruginosa antibiotic resistance genes varies greatly during infections in cystic fibrosis patients. Antimicrob. Agents Chemother. 62:18. doi: 10.1128/AAC.01789-18
Maxson, T., Blancett, C. D., Graham, A. S., Stefan, C. P., and Minogue, T. D. (2018). Rapid antibiotic susceptibility testing from blood culture bottles with species agnostic real-time polymerase chain reaction. PLoS ONE 13:e0209042. doi: 10.1371/journal.pone.0209042
McFaland J. Nephelometer: an instrument for media used for estimating the number of bacteria in suspensions used for calculating the opsonic index and for vaccines. J Am Med Assoc. (1907) 49:1176–8. doi: 10.1001/jama.1907.25320140022001f
Montealegre, M. C., Talavera Rodriguez, A., Roy, S., Hossain, M. I., Islam, M. A., Lanza, V. F., et al. (2020). High genomic diversity and heterogenous origins of pathogenic and antibiotic-resistant Escherichia coli in household settings represent a challenge to reducing transmission in low-income settings. mSphere 5:19. doi: 10.1128/mSphere.00704-19
Mouton, J. W., Meletiadis, J., Voss, A., and Turnidge, J. (2019). Variation of MIC measurements: the contribution of strain and laboratory variability to measurement precision-authors' response. J. Antimicrob. Chemother. 74, 1761–1762. doi: 10.1093/jac/dkz142
Nicoloff, H., Hjort, K., Levin, B. R., and Andersson, D. I. (2019). The high prevalence of antibiotic heteroresistance in pathogenic bacteria is mainly caused by gene amplification. Nat. Microbiol. 4, 504–514. doi: 10.1038/s41564-018-0342-0
Peeters, P., Ryan, K., Karve, S., Potter, D., Baelen, E., Rojas-Farreras, S., et al. (2019). The impact of initial antibiotic treatment failure: real-world insights in patients with complicated, health care-associated intra-abdominal infection. Infect. Drug Resist. 12, 329–343. doi: 10.2147/IDR.S184116
Qin, X., Zhou, C., Zerr, D. M., Adler, A., Addetia, A., Yuan, S., et al. (2018). Heterogeneous antimicrobial susceptibility characteristics in Pseudomonas aeruginosa isolates from cystic fibrosis patients. mSphere 3:17. doi: 10.1128/mSphere.00615-17
Rodriguez-Gomez, J., Perez-Nadales, E., Gutierrez-Gutierrez, B., Machuca, I., Martinez-Martinez, L., Rivera, F., et al. (2019). Prognosis of urinary tract infection caused by KPC-producing Klebsiella pneumoniae: the impact of inappropriate empirical treatment. J. Infect. 79, 245–252. doi: 10.1016/j.jinf.2019.06.014
Rolain, J. M., Mallet, M. N., Fournier, P. E., and Raoult, D. (2004). Real-time PCR for universal antibiotic susceptibility testing. J. Antimicrob. Chemother. 54, 538–541. doi: 10.1093/jac/dkh324
Smith, K. P., and Kirby, J. E. (2018). The inoculum effect in the era of multidrug resistance: minor differences in inoculum have dramatic effect on MIC determination. Antimicrob. Agents Chemother. 62:18. doi: 10.1128/AAC.00433-18
Uppsala University. Hard-to-Detect Antibiotic Resistance an Underestimated Clinical Problem. Uppsala: Uppsala University (2019).
Valsesia, G., Hombach, M., Maurer, F. P., Courvalin, P., Roos, M., and Bottger, E. C. (2015). The Resistant-population cutoff (RCOFF): a new concept for improved characterization of antimicrobial susceptibility patterns of non-wild-type bacterial populations. J. Clin. Microbiol. 53, 1806–1811. doi: 10.1128/JCM.03505-14
Zilberberg, M. D., Nathanson, B. H., Sulham, K., Fan, W., and Shorr, A. F. (2017). Carbapenem resistance, inappropriate empiric treatment and outcomes among patients hospitalized with Enterobacteriaceae urinary tract infection, pneumonia, and sepsis. BMC Infect. Dis. 17:279. doi: 10.1186/s12879-017-2383-z
Keywords: antibiotic susceptibility, antibiotic resisitance, bacterial growth, qPCR, microbiology, guide to therapy, laboratory results, immunocompromidsed patients
Citation: Brukner I and Oughton M (2020) A Fundamental Change in Antibiotic Susceptibility Testing Would Better Prevent Therapeutic Failure: From Individual to Population-Based Analysis. Front. Microbiol. 11:1820. doi: 10.3389/fmicb.2020.01820
Received: 17 May 2020; Accepted: 10 July 2020;
Published: 18 August 2020.
Edited by:
Thomas Dick, Hackensack Meridian Health, United StatesReviewed by:
Mariusz Stanislaw Grinholc, University of Gdańsk and Medical University of Gdańsk, PolandCopyright © 2020 Brukner and Oughton. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ivan Brukner, aWJydWtuZXJAamdoLm1jZ2lsbC5jYQ==; Matthew Oughton, bWF0dGhldy5vdWdodG9uQG1jZ2lsbC5jYQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.