
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 30 July 2020
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.01747
 Heba H. Mostafa1
,2
†
Heba H. Mostafa1
,2
† Andrew Cameron1
†
Andrew Cameron1
† Samantha M. Taffner1
Samantha M. Taffner1
 Jun Wang1
Jun Wang1
 Adel Malek1
Adel Malek1
 Ghinwa Dumyati3
Ghinwa Dumyati3
 Dwight J. Hardy1
,4
Dwight J. Hardy1
,4
 Nicole D. Pecora1
,4
*
Nicole D. Pecora1
,4
*Extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae pose significant treatment and infection prevention challenges. Escherichia coli sequence type (ST) 131 associated with the blaCTX-M-15 gene has been the dominant lineage of ESBL-producing E. coli in the US and worldwide. In this study, our objective was to determine the β-lactamase profile, means of dissemination, prevalence, and the clonal identity of ESBL-producing E. coli in our region of Western New York. Whole-genome SNP-based phylogenomics was used to assess 89 ceftriaxone-resistant (CTR) E. coli. Isolates were collected from both inpatients and outpatients and from urine and sterile-sites over a 2 month period in 2017 or throughout the year, respectively. ST131 was the predominant ST (46.0%), followed by ST38 (15.7%). The blaCTX-M-15 gene was commonly found in 53.7% of ST131 isolates, whereas the blaCTX-M-27 gene was found in 26.8% of ST131, though was significantly associated with ST38, and was found in 71.4% of those strains. When compared to ST131, ST38 E. coli exhibited increased frequency of resistance to nitrofurantoin and decreased frequency of resistance to ciprofloxacin and ampicillin-sulbactam. Using Nanopore long-read sequencing technology, an analysis of the ESBL genetic context showed that the blaCTX-M-15 gene was chromosomal in 68.2% of ST131, whereas the blaCTX-M-27 gene was plasmid-borne in all ST131 and 90% of ST38 isolates. Notably, the blaCTX-M-27 gene in ST38 resided on highly-related (99.0–100.0% identity and 65.0–98.0% query coverage) conjugative IncF plasmids of distinct plasmid multi-locus sequence types (pMLSTs) from those in ST131. Furthermore, ST131 and ST38 were found to harbor different antibiotic resistance gene and virulence factor profiles. These findings raise the possibility of an emerging ESBL-producing E. coli lineage in our region.
Extended-spectrum β-lactamase (ESBL)-producing organisms are responsible for ~26,000 drug-resistant infections and ~1,700 deaths per year in the US, where they are categorized as a serious and increasing threat within the Centers for Disease Control and Prevention’s (CDC) 2019 Antibiotic Resistance Threat Report (CDC, 2019). Among hospitalized patients, ESBL-producers may account for up to 11.6 and 16.1% of Escherichia coli causing urinary tract infection (UTI) and bloodstream infections (BSIs), respectively (Mendes et al., 2019). At present, CTX-M β-lactamases are the prevailing family of ESBLs and include more than 150 genes (Zhao and Hu, 2013). They may have originated as chromosomally-encoded enzymes in Kluyvera spp. before spreading to Escherichia, Klebsiella, and other enteric bacteria (Rossolini et al., 2008). Documented mechanisms of mobilization include capture by the insertion elements ISEcp1 and ISCR1, as well as bacteriophages (Poirel et al., 2008). While chromosomal integration is reported, CTX-M β-lactamase genes are more frequently associated with IncF plasmids (Doumith et al., 2012; Stoesser et al., 2016).
The most frequently reported gene, blaCTX-M-15, is associated with uropathogenic E. coli (UPEC) sequence type 131 (ST131), the predominant lineage of extra-intestinal pathogenic E. coli (ExPEC) worldwide (Nicolas-Chanoine et al., 2014; Pitout and DeVinney, 2017; Birgy et al., 2020). The successful emergence of this clone is attributed to the acquisition of antimicrobial resistance (AMR), specifically to fluoroquinolones (Nicolas-Chanoine et al., 2014; Shaik et al., 2017). In the US, the blaCTX-M-15 gene has also been frequently reported in the context of ST131 (often carried on IncF plasmids; Chandramohan and Revell, 2012; Banerjee et al., 2013; Doi et al., 2013; Chen et al., 2014; Kanamori et al., 2017). However, hinting that the epidemiology may be changing, the 2016 SENTRY Antimicrobial Surveillance Study demonstrated that the blaCTX-M-27 gene (17.3%) is also significant in E. coli UTI and BSI isolates, compared to the blaCTX-M-15 gene (55.5%; Mendes et al., 2019). Similarly, ST131 carrying the blaCTX-M-27 gene is also a frequent minority around the globe (Livermore et al., 2007; Cao et al., 2011; Dahbi et al., 2013; Matsumura et al., 2013; Seiffert et al., 2013; Roer et al., 2017; Guiral et al., 2019; Peirano and Pitout, 2019; Birgy et al., 2020). The relative advantages of one CTX-M family gene vs. another are not clear, though the blaCTX-M-27 gene may confer additional activity against ceftazidime (Bonnet et al., 2003).
The genetic context of blaCTX-M-15 and other CTX-M family genes in ESBL-producing E. coli in the US remains relatively undefined. In this study, we used bacterial whole-genome sequencing (WGS) to investigate the genomic epidemiology of 89 ceftriaxone-resistant (CTR) E. coli with respect to clonality, susceptibility, multi-drug resistance (MDR), and β-lactamase profiles. One of our goals was to compare the prevailing clonal types and ARGs between urine and sterile-site isolates from both inpatients and outpatients. Plasmid CTX-M gene context was further examined for all isolates using long-read sequencing. Complete plasmid sequences and chromosomal integration sites were mapped. Thus, this study represents a detailed snapshot of the genomic landscape, including the apparatus of horizontal transmission, of ESBL-producing E. coli isolated in our region of Western New York (NY).
This study was performed under University of Rochester Medical Center (URMC) IRB protocol RSRB00068143. Eighty-nine CTR E. coli isolates collected as part of routine clinical care at the URMC Clinical Microbiology laboratory in Rochester, New York were selected for this project. The laboratory provides diagnostic services to a population of ~0.5 million people in Western NY and services several area hospitals, urgent cares, nursing homes, and outpatient practices. To identify potential ESBL-producing organisms, we selected unique patient isolates for WGS based on CTR. Urine CTR E. coli were collected at convenience during the months of October and November in 2017 (53 isolates), and CTR E. coli derived from sterile-site infections were collected throughout 2017 (36 isolates: 28 BSIs, 4 bone, 2 peritoneal fluid, 1 joint fluid, and 1 drain). In their respective timeframes, this captured 53/67 (79.1%) of unique patient CTR urine isolates and 36/36 (100%) CTR sterile-site isolates. Initial E. coli identification was performed with MALDI-TOF (Vitek MS v3.0; bioMérieux Inc., Durham, NC). Antibiotic susceptibility (including ESBL production) was assessed with Vitek 2 (bioMérieux Inc.; AST-GN70 test card). Phenotypic ESBL production assessed by Vitek2 is indicated by “+” (Data S1 – Antibiogram). Cefazolin susceptibility was determined by Kirby-Bauer disk diffusion for isolates from sterile-sites and is indicated with “/KB” in Data S1 – Antibiogram. Kirby-Bauer zone of inhibition diameter is indicated in millimeters. Susceptibility was interpreted with CLSI standard M100 (CLSI, 2019).
E. coli isolates were archived at −80°C in trypticase soy broth (TSB) with 20% glycerol and maintained at 35°C on blood agar (BD BBL trypticase soy agar with 5% Sheep Blood; BD). Bacterial DNA was extracted with the MagNA Pure Compact System (Roche, Indianapolis, IN). DNA was quantified with the QuantiFluor dsDNA system (Promega, Madison, WI).
DNA library preparation was performed according to the manufacturer’s protocol (Nextera XT DNA Library Preparation Kit; Illumina, San Diego, CA), purified using Agencourt AMPure XP beads (Beckman Coulter Inc., Indianapolis, IN), and quality-checked using the Agilent 4,200 TapeStation System (Agilent; Santa Clara, CA). Purified PCR products were normalized using Nextera XT Library Normalization Beads (Illumina). Normalized samples were pooled and quantified using the Qubit ssDNA Assay kit (Invitrogen). Library pools were loaded with 2.45 ng ssDNA and 20 μl PhiX control DNA (20 pM). Paired-end sequencing was performed with MiSeq Reagent v3 600-cycle kits on the MiSeq instrument (Illumina).
Plasmids were sequenced on the MinION platform (Oxford Nanopore Technologies; Cambridge, MA). Briefly, for each of the 89 strains, 100 ml of Luria-Bertani (LB) broth was inoculated and incubated for ~18 h at 37°C, and plasmids were purified using the QIAfilter Plasmid Midi Kit (Qiagen, Germantown, MD). Plasmid DNA was purified (Agencourt AMPure XP beads), and quantified (QuantiFluor dsDNA), and adjusted to 400 ng in a total volume of 7.5 μl molecular biology-grade H2O. Sequencing library preparation (Rapid Barcoding Sequencing kit; Oxford Nanopore Technologies) was performed as indicated with the following alterations: Fragmentation Mix RB01-12 volume was reduced to 1.5 μl and was incubated for 20 s at 30°C. Base-calling and de-multiplexing was performed using Albacore v2.3.1 (Oxford Nanopore Technologies) and Illumina-Nanopore hybrid read assemblies were generated by Unicycler (Wick et al., 2017). Plasmids were denoted as “circular” (complete) or “uncircularized” (i.e., incomplete or fragments). BLASTn was used to identify plasmids, plasmid multi-locus sequence typing (pMLST), and virulence and antimicrobial resistance genes (ARGs), which were also assessed with Center for Genomic Epidemiology (CGE) tools (including ResFinder and FimTyper; Thomsen et al., 2016) and with ABRicate (Seemann, 2017). Plasmids were annotated using RAST (Aziz et al., 2008) and aligned with Mauve (Darling et al., 2004). In some instances, genomic sequences recovered from long-read plasmid sequencing were used to create hybrid assemblies with Illumina reads.
A pipeline written in Python, sqlite3, BASH, JavaScript, D3 (Bostock et al., 2011), JQuery, HTML, and Bootstrap was used for genomic DNA read quality control, genomic sequence assembly, mapping, SNP calling, and phylogenomics. Briefly, human DNA reads were removed using Bowtie2 v2.3.5.1 (Langmead and Salzberg, 2012). Trimmomatic v0.39 (Bolger et al., 2014) was used for adaptor removal and read quality control (with altered criteria: SLIDINGWINDOW: 5:20; and MINLEN: 50). Assemblies were built using SPAdes v3.13.0 (Bankevich et al., 2012). Genome assembly quality was assessed by Quast (Gurevich et al., 2013). Genomes were aligned against E. coli MG1655 (GenBank: NC_000913.3) using Bowtie2 (Langmead and Salzberg, 2012). BLASTn was used to determine multi-locus sequence typing (MLST; Jolley and Maiden, 2010). ARGs and virulence factors were determined as above. Phage sequences were detected with PHASTER (Arndt et al., 2016). E. coli phylogroups were determined with ClermonTyper (Beghain et al., 2018).
A modified CFSAN SNP Pipeline was used for reference-based SNP calling and phylogenetic analysis (Davis et al., 2015). The “call_sites” function uses samtools (Li et al., 2009) and varscan (settings: min-var-freq:0.90; min-coverage:12; min-avg-qual:20; Koboldt et al., 2012) to find high-confidence SNPs between the reference sequence and mapped reads. SNPs that occur inside phages, mobile elements, and transposons were removed using a custom RAST-annotated gff file (Aziz et al., 2008). Maximum likelihood trees were built using a concatenated SNP fasta file for each analyzed genome (settings: call_consensus, minConsFreq: 0.9). Trees were produced with FastTree (Price et al., 2010). Plasmid content was determined with Roary v3.11.2 (Page et al., 2015) and visualized with hierarchical clustering using Morpheus (Broad Institute, Cambridge, MA). Circular alignment plots were produced with Circos (Krzywinski et al., 2009). Linear alignment gene diagrams were produced with EasyFig (Sullivan et al., 2011).
Statistical calculations were performed in Prism 8 (GraphPad Software, San Diego, CA). Multiple t-tests (Holm-Sidak method) were used for comparison of AMR frequencies. Otherwise, Fisher’s exact test was used, including for gene content comparisons between ST131 and ST38. Unless otherwise indicated, data shown represent the mean ± SEM.
Surveying over 5 years (2013–2018), the average frequency (mean ± SEM) of CTR E. coli in sterile-site and urine isolates was 5.6 ± 1.2% and 3.7 ± 0.1%, respectively. Likewise, ESBL-producers were detected at frequencies of 4.8 ± 1.3% and 3.0% ± 0.1% for sterile-site and urine isolates (Figure 1A). The majority of CTR E. coli were isolated from urine specimens (90.3%; Figure 1B). The sequenced CTR E. coli originated across a geographic distribution in Western NY that approximated the patient population served by our laboratory (Figure 1C). Isolates from inpatients accounted for 53.9% of the overall study set. Of sterile-site isolates, 94.4% were from inpatients vs. 26.4% of urine isolates. The cumulative antibiogram for CTR E. coli isolated from urine indicated the following resistance frequencies (mean ± SEM): ampicillin (99.9 ± 0.007%), ampicillin-sulbactam (78.4 ± 0.9%), amikacin (0.4 ± 0.1%), aztreonam (69.5 ± 1.0%), ciprofloxacin (65.6 ± 1.0%), nitrofurantoin (13.3 ± 0.8%), cefepime (35.0 ± 1.2%), gentamicin (28.7 ± 1.1%), trimethoprim-sulfamethoxazole (48.6 ± 1.2%), tigecycline (0%), tobramycin (32.2 ± 1.1%), and piperacillin-tazobactam (12.7 ± 0.8%). The resistance frequencies for CTR E. coli isolated from sterile-sites were: ampicillin (94.3 ± 3.2%), ampicillin-sulbactam (67.9 ± 6.5%), amikacin (0%), aztreonam (75.5 ± 6.0%), ciprofloxacin (71.7 ± 6.2%), nitrofurantoin (17.0 ± 5.2%), cefepime (22.6 ± 5.8%), gentamicin (20.7 ± 5.6%), trimethoprim-sulfamethoxazole (56.6 ± 6.8%), tigecycline (1.9 ± 1.8%), tobramycin (20.7 ± 5.6%), and piperacillin-tazobactam (37.7 ± 2.6%). Resistance frequencies of the total E. coli in our hospital system and community vs. the sequenced isolates are depicted in Figure 1D. For urine isolates, the frequency of ampicillin resistance was different between the larger collection of CTR E. coli vs. sequenced E. coli (99.8 ± 0.007 vs. 94.3 ± 3.2%, p < 0.000001). Otherwise, there were no significant differences observed for any other drug tested when compared to urine isolates selected for sequencing. Likewise, there were no significant differences in the antibiogram between the sequenced sterile-site CTR E. coli and the larger collection of sterile-site E. coli. Antibiotic resistance profiles of sequenced isolates are summarized in supplemental data (Data S1 – Antibiogram).
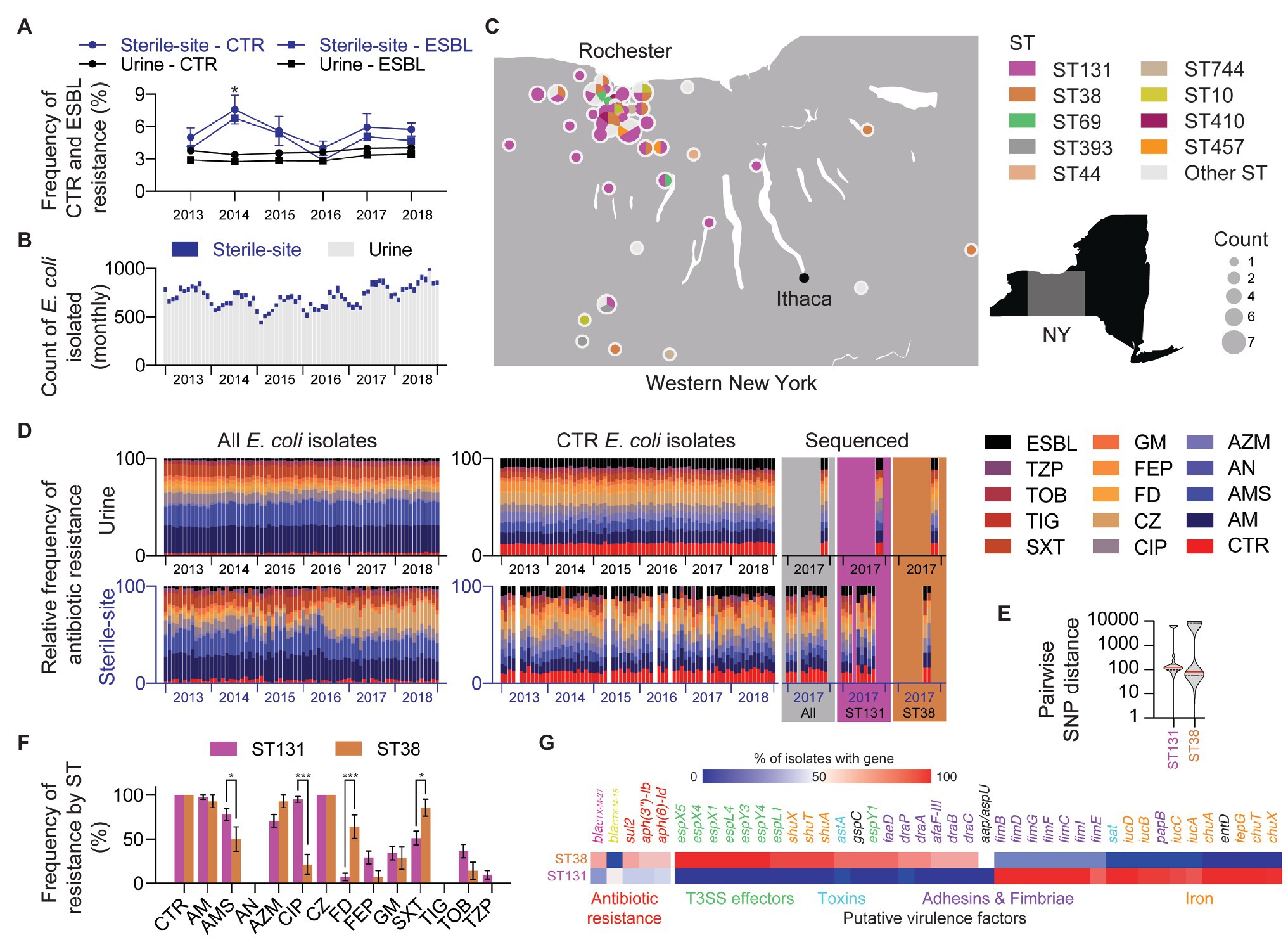
Figure 1. Incidence of ceftriaxone-resistant (CTR) and extended-spectrum β-lactamase (ESBL)-producing Escherichia coli in Western New York (NY). (A) Yearly frequency of CTR and ESBL-producing E. coli in sterile-site and urine E. coli. Mean percentage with error bars showing SEM. Statistical test: t-test (Holm-Sidak method). Significance: * p ≤ 0.05. (B) Total monthly count of sterile-site and urine E. coli isolated between 2013 and 2018. Data include only one isolate per patient. (C) Geographic distribution and prevalence of E. coli sequence types (STs). (D) Antimicrobial resistance (AMR) profiles of sterile-site and urine E. coli isolated between 2013 and 2018. Relative frequency is shown for monthly profiles for all isolates (left), CTR isolates (right), and whole-genome sequenced (WGS) CTR isolates (colored panels). CTR, ceftriaxone; AM, ampicillin; AMS, ampicillin-sulbactam; AN, amikacin; AZM, aztreonam; CIP, ciprofloxacin; CZ, ceftizoxime; FD, nitrofurantoin; FEP, cefepime; GM, gentamicin; SXT, trimethoprim-sulfamethoxazole; TIG, tigecycline; TOB, tobramycin; TZP, piperacillin-tazobactam; ESBL, Extended spectrum β-lactamase. (E) Violin plot showing distribution of intraclade pairwise SNP distances for ST131 and ST38. (F) Differences in AMR rates between sterile-site and urine E. coli ST131 and ST38 STs. Statistical test: multiple t-tests (Holm-Sidak method). Significance: *p ≤ 0.05 and ***p ≤ 0.001. (G) ARGs and putative virulence factors associated with ST131 and ST38. Color indicates percentage of ST131 or ST38 isolates encoding genes listed. Genes are listed only if mean ST131 and ST38 percentages differ by >30% (for ARGs) and >50% (for putative virulence factors).
WGS MLST determined that ST131 was the most frequent (46.0%, 41/89) and ST38 was the second-most frequent (15.7%, 14/89) ST. These were followed by ST69 (4.5%, 4/89) and ST10 (3.4%, 3/89; Data S1 – Genomes). Excluding ST131 and ST38, the remaining sequenced isolates (35/89) belonged to 25 other STs. Diversity of STs was similar between sterile-site and urine isolates with 16 vs. 17 different STs identified, respectively. Among ST131 isolates, 68.3% were isolated from inpatients (including long-term care facilities), whereas 35.7% of ST38 were isolated from inpatients. Although not significantly different, 41.5% of ST131 were sterile-site isolates compared to 21.4% of ST38 (Table 1). All ST131 isolates were phylogroup B2, and all ST38 isolates were phylogroup D.
SNP-based phylogenetic clustering revealed that the median SNP distance between ST131 and ST38 was 24,641 SNPs ± 3.0 SNPs. The median SNP distance among ST131 isolates was 122 SNPs (range: 0–6,792 SNPs) and 82 SNPs (range: 0–11,470 SNPs) for ST38 isolates (Figure 1E and Data S1 – SNP distance).
Compared to ST131, ST38 isolates from urine demonstrated increased frequency of resistance to nitrofurantoin (70.0 ± 15.3 vs. 4.1 ± 4.1%, p < 0.000001) and trimethoprim-sulfamethoxazole (90.0 ± 10.0 vs. 41.6 ± 10.2%, p = 0.00003; Figure 1F). For nine ST38 strains with intermediate or resistant nitrofurantoin phenotypes, Q67STOP (eight isolates) and Q26STOP (one isolate) truncations were detected in the nitroreductase gene nfsA (data not shown). Likewise, mutations in nfsA (L60STOP, Q26STOP, and Q113STOP) were detected in three nitrofurantoin-resistant ST131 isolates (data not shown). Increased frequency of trimethoprim-sulfamethoxazole resistance correlated with the presence of sul1, sul2, and dfrA genes, which, when considered together, were more frequent in ST38 than in ST131 (69.1 vs. 28.7%, p < 0.0001; Figure 1G). ST38 isolates from urine were also more often susceptible to ampicillin-sulbactam compared to ST131 (40.0 ± 16.3 vs. 79.1 ± 8.4%, p = 0.0006). However, blaTEM and blaOXA-1 genes were also absent in both ST38 and ST131 isolates susceptible to ampicillin-sulbactam (Data S1 – Genomes).
ST131 isolates were more frequently resistant to ciprofloxacin than ST38 strains regardless of urine vs. sterile-site specimen type (urine: 95.8 ± 4.1 vs. 30.0 ± 15.2%, p < 0.000001; sterile-site: 94.1 ± 5.8% vs. none detected, p = 0.000004). Mutations known to be associated with quinolone resistance were detected in ST131 isolates, including in gyrA (D87N, S83L, and A828S), and in the parC gene (S80I, E84V; Data S1 – ST131; Sorlozano et al., 2007; Phan et al., 2015; Kanamori et al., 2017; Nicolas-Chanoine et al., 2017). Fluoroquinolone resistance correlated with ST131 clades. URMC_16, URMC_22, and URMC_112 demonstrated full or intermediate susceptibility to fluoroquinolones, harbored single gyrA mutations, and were found to be fimH41, all consistent with Clade A (Data S1 – ST131). The remaining ST131 isolates were all resistant to ciprofloxacin and were characterized as fimH30 with one exception (fimH34, URMC_111), placing them within ST131 Clade C (Data S1 – ST131). Three ciprofloxacin-resistant ST38 isolates had gyrA (S83L) and parC (S80I) gene mutations.
Profiling for putative virulence factors revealed differences in gene content between ST131 and ST38 isolates (Figure 1G). The majority of ST131 isolates carried genes encoding the secreted autotransporter toxin (sat: ST131, 100% vs. ST38, 7.1%; p < 0.0001; Guignot et al., 2007), genes for iron acquisition (e.g., iucC: ST131, 92.6% vs. ST38, 7.1%; p < 0.0001), and genes for Type 1 fimbriae biogenesis (e.g., fimI, fimC, fimD, fimF, and fimG: ST131, 100% vs. ST38, 21.4%; p < 0.0001). Additionally, cytotoxic necrotizing factor (cnf1: ST131, 29.3% vs. ST38, 0%; p = 0.02) and the plasmid-encoded enterotoxin (senB: ST131, 46.3% vs. ST38, 7.1%; p = 0.01; Nataro et al., 1995) were also enriched in ST131 isolates, compared to ST38. In contrast, ST38-enriched genes included determinants characteristic of enteroaggregative E. coli (EAEC; Lima et al., 2013; Chattaway et al., 2014; Havt et al., 2017), such as genes for aggregative heat stable toxin (astA: ST131, 0% vs. ST38, 92.9%; p < 0.0001; Paiva de Sousa and Dubreuil, 2001), non-fimbrial adhesin (afaF: ST131, 0% vs. ST38, 71.4%; p < 0.0001; Korotkova et al., 2008), iron transport (shuA, shuT, shuX: ST131, 0% vs. ST38, 92.9%; p < 0.0001), and anti-aggregation protein (aap: ST131, 0% vs. ST38, 50.0%; p < 0.0001; Sheikh et al., 2002).
Among the 89 sequenced isolates, the blaCTX-M-15 gene was found in 30/89 (34%) isolates (ST131, 22/41 vs. ST38, 1/14; p = 0.004; Figure 2A and Data S1 – Genomes). The blaCTX-M-27 gene was nearly as common and was found in 27/89 (30.3%) isolates. Whereas the blaCTX-M-15 gene was mainly associated with ST131, the blaCTX-M-27 gene was associated with both ST131 and ST38, accounting, respectively, for 26.8 and 64.3% of those isolates (ST131, 11/41 vs. ST38, 10/14; p = 0.005; Figure 2B and Table 1). Less frequent blaCTX-M genes included the blaCTX-M-14 gene (7/89), the blaCTX-M-55 gene (6/89), and the blaCTX-M-1 gene (4/89). Non-CTX-M family β-lactamase genes were found in 9/89 isolates, including the blaCMY-2 gene (8/89), the blaSHV-12 gene (1/89), and a blaKPC-3 gene (1/89). Of 21/41 ST131 strains, the blaCTX-M-15 gene correlated with fimH30 and ciprofloxacin resistance, placing those strains in clade C2 (H30-Rx; Price et al., 2013; Petty et al., 2014; Stoesser et al., 2016). Of the remaining ST131 isolates, 12.2% (5/41) were Clade C1 (H30-R), 26.8% (11/41) were clade C1-M27 (H30-R), 7.3% (3/41) were clade A, and one isolate (URMC_111, fimH34) was of an undefined clade (Data S1 – ST131). Within the most common MLST, ST131, the blaCTX-M-15 gene was not more likely to be found among inpatients than the blaCTX-M-27 gene (63.6 vs. 72.7%; p = not significant).
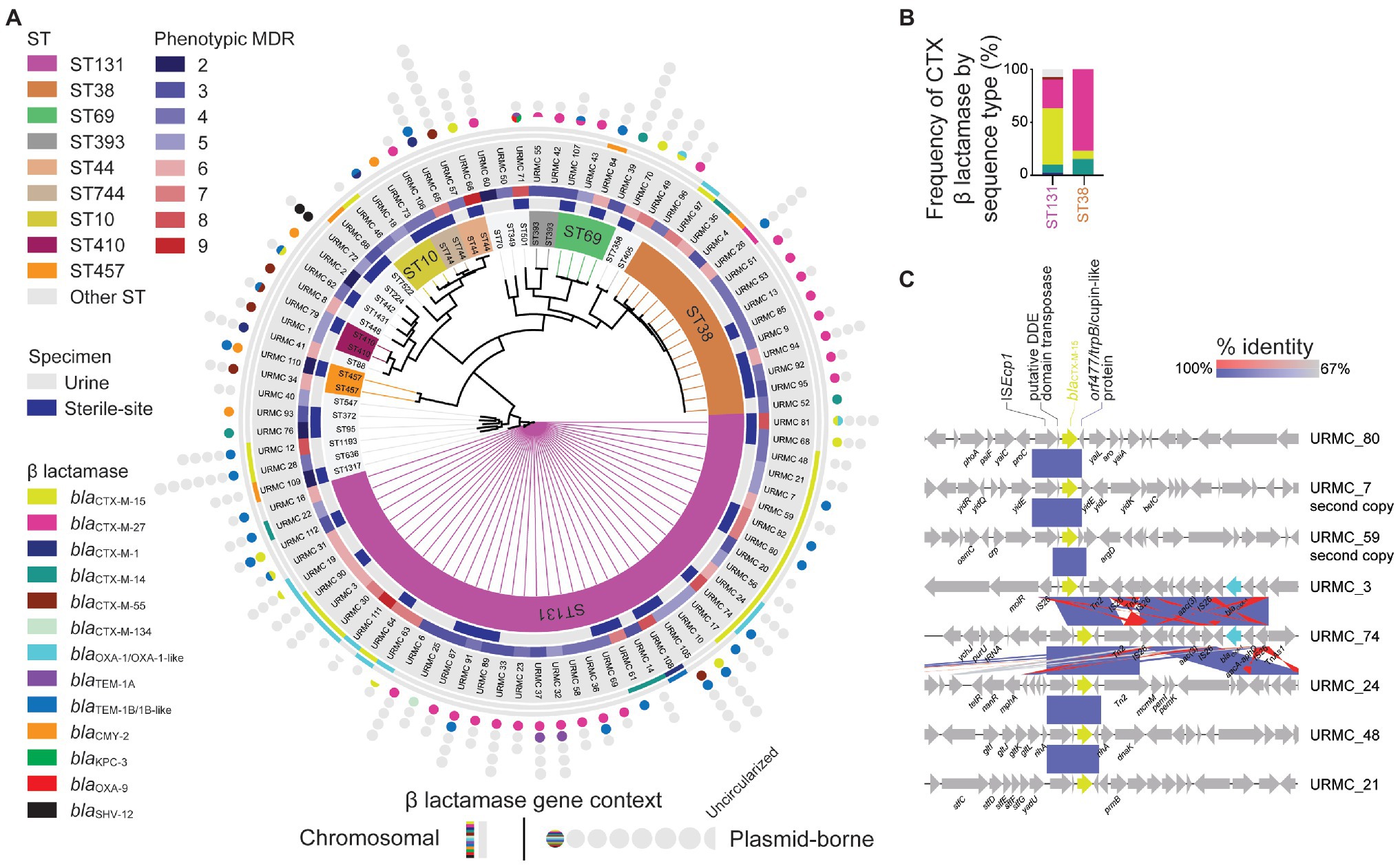
Figure 2. WGS SNP-based phylogenomic tree and ST distribution of sequenced CTR E. coli. (A) Isolates (89 total) selected for WGS included those from urine (53 isolates) and sterile-sites (36 total: 28 blood, 4 bone, 2 peritoneal fluid, 1 joint fluid, and 1 drain) specimens. Detected ARGs are displayed as chromosomal (lines) or plasmid-borne (circles). Circles indicate the number of plasmids identified in each E. coli isolate. Half-circles indicate incomplete sequences. (B) Relative frequency of β-lactamase genes detected in E. coli ST131 and ST38 STs. (C) Gene context schematic of representative blaCTX-M-15 gene chromosomal insertion sites.
Among the 30 isolates with the blaCTX-M-15 gene, 15/22 ST131 isolates had the gene integrated into the chromosome. The blaCTX-M-15 gene was chromosomal in 4/8 isolates of other STs, one of which was ST38 (Figure 2A and Data S1 – Genomes). For most chromosomal insertions, alignment (with 100% identity) of the immediate blaCTX-M-15 context revealed the presence of the well-known ISEcp1-flanked blaCTX-M-15-orf477 arrangement (Rodríguez et al., 2004). The blaCTX-M-15 gene insertions were also typically flanked by other IS element insertions and scars (Figure 2C).
Among the ST131 isolates with chromosomal blaCTX-M-15, the gene was frequently located in one of two sites, either: (1) between genes encoding shikimate kinase (aro) and pyrroline-5-carboxylate-reductase (proC; 6/15 isolates); or (2) inserted adjacent to a molybdate metabolism regulator gene (molR; 4/15 isolates). In the remaining 5/15 ST131 isolates with chromosomal integration, the blaCTX-M-15 gene was inserted in unique sites. In two instances, long-read hybrid assembly using chromosomal reads obtained during plasmid sequencing also identified one additional chromosomal copy of the blaCTX-M-15 gene inserted into unique locations (in URMC_7 and URMC_59; Figure 2C).
In 6/11 isolates, the plasmid-encoded blaCTX-M-15 genes were carried on IncF-type plasmids of a variety of pMLSTs (Figure 3B). Two additional plasmids (URMC_62_p_96678 and URMC_112_p_99275) were typed as IncY (Data S1, Plasmids). In general, blaCTX-M-15 plasmids shared little synteny with each other or with blaCTX-M-27 plasmids (Figures 3A,B). The plasmids varied in their carriage and arrangement of ARGs. Most (9/11) carried MDR regions with up to 10 ARGs, typically including a Class 1 integron (Data S2). Interestingly, URMC_112_p_99275 carried the blaCTX-M-15 gene in an intact phage (Escherichia phage RCS47; Genbank: NC_042128).
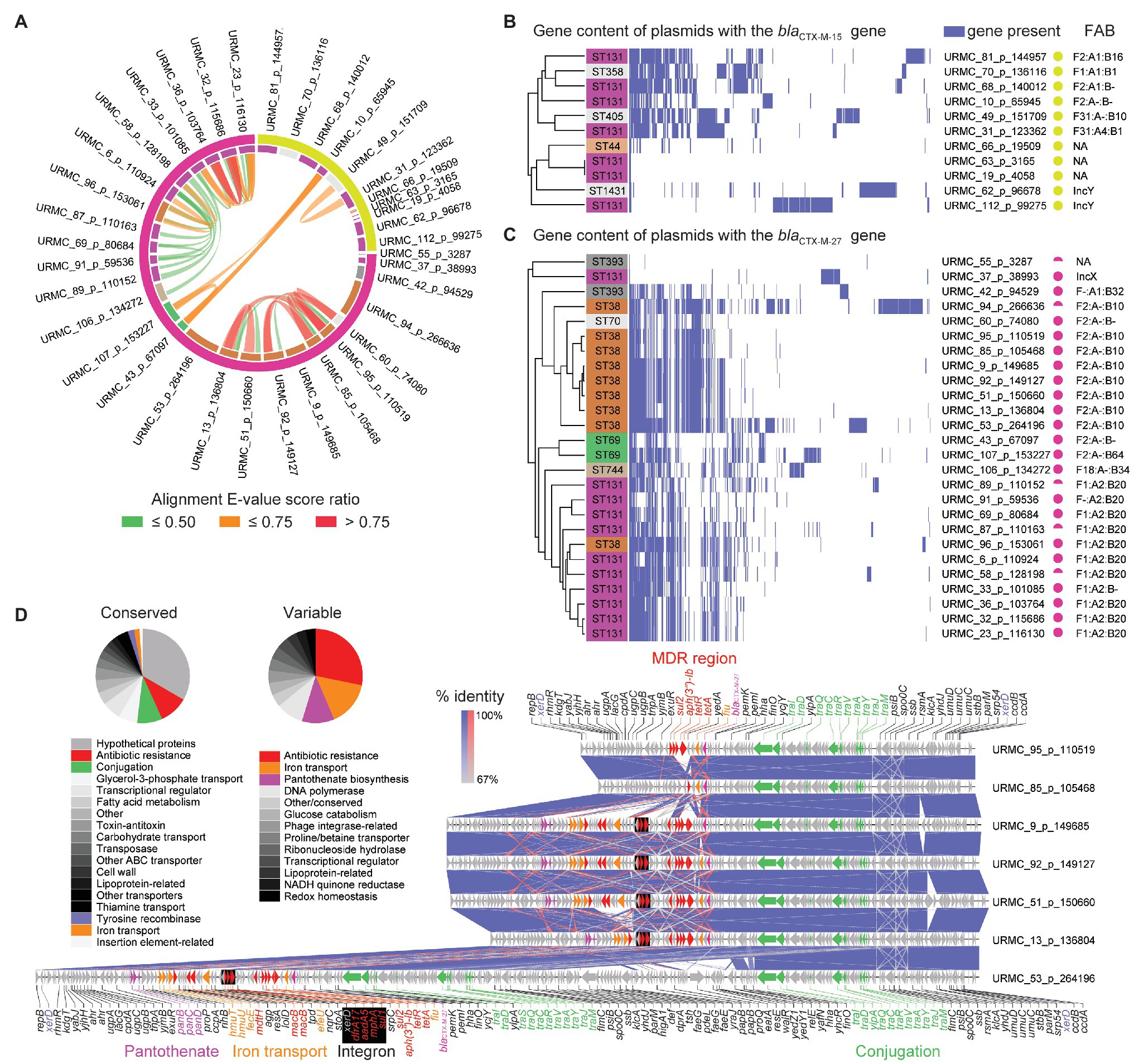
Figure 3. Plasmids harboring the blaCTX-M-15 and blaCTX-M-27 β-lactamase genes. (A) Circular alignment plot of plasmids with either blaCTX-M-15 or blaCTX-M-27 genes showing E. coli ST (inner ring; color key consistent with Figure 2A) and β-lactamase (outer ring). Ribbons colored by alignment E-value score ratio. (B) Gene content of all plasmids harboring the blaCTX-M-15 gene. Detected genes shown in blue with hierarchical clustering. (C) Gene content of all plasmids harboring the blaCTX-M-27 gene. Detected genes shown in blue with hierarchical clustering. (D) Gene schematic and linear alignment of complete plasmids encoding the blaCTX-M-27 gene from ST38 (with genes shown as arrows). Arrows are colored to show antibiotic resistance genes (red), conjugation-associated genes (purple), and other/hypothetical genes (gray). Pie charts show relative frequencies of annotated genes by functional characteristics found in conserved (left) and variable (right) regions in plasmids with the blaCTX-M-27 gene.
The 26 blaCTX-M-27 β-lactamase genes identified here were borne on plasmids in 11/11 ST131 isolates (clade C1-M27) and 9/10 ST38 isolates (Figure 3C). With the exception of URMC_37_p_38993 (IncX), the 11 ST131 plasmids that encoded the blaCTX-M-27 gene were of various IncF-types, with pMLST IncF[F1:A2:B20] being found in 9/11 (Figure 3C and Data S1 – Plasmids). Five of these ST131 plasmids had high homology (101,085–116,130 bp, >99% identity) to the multi-replicon plasmid pH105 (134,899 bp; Genbank: CP021871) recently characterized in a vaginal ST131 isolate with the blaCTX-M-27 gene in Germany (Ghosh et al., 2017). These also carried ARGs acting against aminoglycosides (aadA5, streptomycin/spectinomycin resistance; aph(6)-Id, tobramycin and amikacin resistance; aph(3″)-Ib, streptomycin resistance; and ant(3″)-Ia, streptomycin and spectinomycin resistance). Other ARGs detected in the pH105-like plasmids identified here included genes conferring resistance to macrolides [mph(A)], tetracyclines [tet(A)], sulfonamides (sul1, sul2), and trimethoprim (dfrA17). The MDR regions also encoded a chromate resistance gene (chrA), a Class I integron, and were scarred with transpositions of ISs. Typically, the blaCTX-M-27 gene was flanked by IS903B and IS26. The other 5 IncF plasmids in ST131 had a variety of close hits in Genbank (Data S1 – Plasmids), with 4/5 bearing no other ARGs other than the blaCTX-M-15 gene.
The nine ST38 isolates with IncF plasmids carrying the blaCTX-M-27 gene were highly related (99.0–100.0% identity and 65.0–98.0% query coverage) and ranged in size from 99,138 to 266,636 bp. The majority (8/9) were typed as IncF[F2:A-:B10]. The remaining 1/9 was IncF[F1:A2:B20]. In general, the ST38 plasmids with the blaCTX-M-27 gene had a conserved ~40-kb conjugation-associated tra region and a single complex MDR region (Figure 3D). The MDR region frequently harbored a Class 1 integron stacked with multiple AMR determinants, other ARGs, and genes involved in heavy metal resistance (e.g., srpC putative chromate transporter). The ARGs were typically flanked by IS26 and IS15D1. IS associated with the blaCTX-M-27 gene included IS903B and a fragment of ISEcp1, into which IS26 had inserted. URMC_53_p_264196 was larger (>200-kb) due to a duplication of the tra gene region and additional gene content, including a putative enteroaggregative virulence factor (e.g., astA heat-stable enterotoxin 1; Figure 3D). In URMC_60_p_74080, the MDR region was not present, and the blaCTX-M-27 gene was the only ARG on the plasmid.
Our data indicated that the blaCTX-M-15 gene was the predominant ESBL in our region, but also that the blaCTX-M-27 gene constituted a large minority, being highly represented in ST131 (26.8%) and in ST38 (64.2%) isolates. Prior studies conducted in the US have indicated that the blaCTX-M-15 gene is the predominant ESBL, and is frequently carried by ST131, the most widely established extraintestinal clone (Johnson et al., 2012).
In our clade C2 (H30-Rx) ST131 isolates, two integration sites accounted for 10/15 chromosomally integrated blaCTX-M-15 genes. Both of these integration sites have been reported for ESBL E. coli collected in other studies (i.e., Genbank: NZ_CP018979 and NZ_CP018991.1). Indicating that these groups did not represent recent local clonal transmission, they were separated by >50 SNPs. The blaCTX-M-15 gene was carried on a diverse group of plasmids in the clade C2 group, all with unique pMLSTs (Figure 3B).
Others have shown the increasing prevalence of blaCTX-M-27 in ST131 (Matsumura et al., 2015). In these studies and others, the blaCTX-M-27 gene has been associated with fimH30 and fluoroquinolone resistance as part of clade C1-M27. Here, we found the same, as well as noting that the blaCTX-M-27 gene was often embedded in plasmids of pMLST IncF[F1:A2:B20] which was almost exclusively restricted to the C1-M27 clade, as reported by others (Ghosh et al., 2017; Kondratyeva et al., 2020). Interestingly, An IncF[F1:A2:B20] plasmid was also found in a single ST38 strain (URMC_96; Figure 3C).
Among ST38 strains in our study set, the blaCTX-M-27 gene was borne on plasmids with few close homologs in Genbank, and which were distinct from those in ST131. These were among the most novel of all the plasmids described in this study. The closest homologue was the IncF plasmid p7_2.1 (Genbank: CP023821), which shared >99% identity over a query coverage range of 33–61% with the ST38 plasmids that carried the blaCTX-M-27 gene (Data S1 – Plasmids). The p7_2.1 plasmid was identified in a Swedish study of stool isolates but does not harbor the blaCTX-M-27 gene. Another ST38 blaCTX-M-27-carrying plasmid (URMC_96_p153061) matched closely (99% identity and 80% query coverage) to plasmid p4_4.1 from the same study (GenBank: CP023827.1), but also had 100% identity (34% query coverage) to pDA33137-178 from a ST44 isolate (Nicoloff et al., 2019). In our study, IncF plasmids in ST38 were commonly (8/9) IncF[F2:A-:B10], where the blaCTX-M-27 gene was closely associated with IS903B and IS26. The latter has been reported to help drive the dissemination of some CTX-M β-lactamase genes (e.g., blaCTX-M-14; Zhao and Hu, 2013). FAB pMLST assessment of ST38 plasmids has not been widely done, though IncF[F2:A-:B10] has been shown in one study to account for 1/12 isolates of a collection of ST38, where IncF[F1:A-:B33] was more commonly observed (4/12; Shaik et al., 2017).
While ST131 is a well-known epidemic lineage, ST38 has historically been found less frequently in surveillance studies, though that may be changing. For example, in the US, 4.8% of E. coli causing UTIs and 6.2% of BSI isolates were typed as ST38 in a recent nationwide surveillance study (Mendes et al., 2019). Furthermore, after selecting isolates with elevated MICs for ceftriaxone, aztreonam, ceftazidime, and/or imipenem/meropenem, ST38 comprised 11.6% of UTI and 16.1% of BSI isolates (Mendes et al., 2019). Studies from Europe, the Middle East, and Asia have also found ST38 to be a significant minority among ESBL-producing E. coli (Alghoribi et al., 2015; Hertz et al., 2016; Peirano and Pitout, 2019; Sepp et al., 2019). By comparison, our study found that ST38 comprised 15.7% of total CTR isolates (urine: 18.8%; sterile-site: 11.1%). Human clinical ST38 isolates have previously been associated with blaCTX-M-9, blaCTX-M-14, blaNDM-1, and increasingly, with blaOXA-48 genes (Suzuki et al., 2009; Kim et al., 2011; Mshana et al., 2011; Poirel et al., 2011; van der Bij et al., 2011; Brodrick et al., 2017), but only sporadically with the blaCTX-M-27 gene (Flament-Simon et al., 2020; Yasir et al., 2020). Other molecular epidemiological studies have not, to our knowledge, detected this association (Day et al., 2019), indicating that carriage of the blaCTX-M-27 gene by ST38 isolates may be emerging.
Published genomic comparisons of ST131 with ST38 suggest that the latter harbors fewer UPEC-associated virulence genes, though the two have similar in vitro adhesion, invasion, and serum resistance phenotypes (Shaik et al., 2017). Elsewhere, ST38 has been described as an ExPEC or a UPEC/EAEC hybrid (Chattaway et al., 2014; Phan et al., 2015; Muenzner et al., 2016). Others have suggested that EAEC attributes may increase the potential of such strains to cause UTIs (Boll et al., 2013). The ST38 isolates identified in this study did not harbor genes encoding for aggregative adherence fimbriae (AAF) nor for AggR, the transcription factor that regulates AAF biogenesis (Boll et al., 2012). While profiling virulence factors in silico is limited by the quality and quantity of available databases, the ST38 isolates in this study did harbor some putative virulence factors that may be associated with aggregation and dispersion in EAEC. For example, ST38 harbored afaF-III (Afa/Dr. adhesin family; Muenzner et al., 2016). The pathogenicity and clinical pertinence of E. coli expressing Afa/Dr. adhesins in UTIs are well established (Servin, 2014).
In this study, ST131 exhibited increased frequency of resistance to fluoroquinolones and ampicillin-sulbactam compared to ST38, which was often non-susceptible to nitrofurantoin. Increased frequency of fluoroquinolone resistance in ST131 vs. ST38 has been previously described (Alghoribi et al., 2015; Gauthier et al., 2018; Guiral et al., 2019) and is a hallmark of clade C strains. Only 3/41 ST131 isolates in this study were susceptible (S or I) to fluoroquinolones, all of which were clade A. To the best of our knowledge, the increased frequency of nitrofurantoin resistance in ST38 has not been reported. Nonsense mutations in the nitroreductase genes nfsA and nfsB are associated with nitrofurantoin resistance and were found in all nonsusceptible strains of both ST38 and ST131 (Sandegren et al., 2008; Shanmugam et al., 2016).
If ST38 is emerging as a prominent ESBL lineage, then concurrent resistance to nitrofurantoin is concerning because this drug has thus far remained useful for fluoroquinolone-resistant and ESBL-producing organisms (Hertz et al., 2016; Tulara, 2018). Vice-versa, the emergence of this lineage may be influenced by the reduction of fluoroquinolone use. With respect to the observed differences for ampicillin-sulbactam, others have noted that blaCTX-M-15 genes (or isolates harboring these determinants) are associated with increased frequency of resistance compared to isolates carrying the blaCTX-M-27 gene (Faheem et al., 2013; Matsumura et al., 2015). This is consistent with our results given the respective preponderance of these enzymes in ST131 vs. ST38. This observation may also be related to the presence of other β-lactamases, as isolates of both ST131 and ST38 were more often resistant to ampicillin-sulbactam if they also harbored broad spectrum β-lactamase genes such as blaTEM and blaOXA.
Limitations of this study include the regional nature and narrow timeframe of isolate collection. Furthermore, while our sequenced E. coli may have similar phenotypic susceptibility to both current and past isolates, they may not be representative of the ARGs, STs, and mobile genetic elements found in the community across time. The findings in this study raise several questions. For example, is there a fitness advantage for isolates carrying the blaCTX-M-27 gene? This gene may confer greater resistance to ceftazidime (Kuroda et al., 2012). What explains the almost exclusive association between ST38 and the blaCTX-M-27 gene, while ST131 is associated with both the blaCTX-M-27 and blaCTX-M-15 genes? Is the spread of blaCTX-M-27 in our region associated with human carriage from areas of high prevalence or an isolated clonal outbreak? Recent surveillance of food-producing animals in the US showed that cattle and turkey E. coli frequently carried the blaCTX-M-27 gene (Tadesse et al., 2018). Salmonella spp. from food-producing animals have also been shown to carry the blaCTX-M-27 gene (Zhang et al., 2016), though the plasmids in our study bore little resemblance to publically available sequences from Salmonella (data not shown). Establishing a link between these observations highlights the need for more extensive and longitudinal “One Health” surveillance studies. In conclusion, although this work may serve as a window through which to view national epidemiological trends, additional surveillance is needed to confirm the emergence of ST38 and its association with the blaCTX-M-27 gene.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
HM and NP designed the study. HM and NP selected clinical isolates. HM isolated genomic DNA. JW performed sequencing. ST performed and managed bioinformatics analyses and pipelines. HM, AM, and AC analyzed sequence data. HM and AC performed statistical analyses. HM, AC, and NP analyzed overall data/results and wrote the first draft of the manuscript. NP provided funding and resources. NP, GD, and DH provided technical expertise. All authors participated in editing and reviewing the manuscript and approved the final manuscript. All authors contributed to the article and approved the submitted version.
Funding from the Department of Pathology and Laboratory Medicine, University of Rochester Medical Center supported this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank UR Medicine Central Laboratory Clinical Microbiology for specimen collection and antimicrobial susceptibility testing. Ruichao Li and Chen Sheng (Hong Kong PolyU Shen Zhen Research Institute, Shenzhen, PR China) provided technical advice for Nanopore sequencing. Dr. Steven Gill (URMC) and his group provided technical assistance with plasmid extractions. We also thank Dr. Paul Dunman (URMC) for critical reading of the manuscript.
The Supplementary Material for this article can be found online at:
https://www.frontiersin.org/articles/10.3389/fmicb.2020.01747/full#supplementary-material.
Alghoribi, M. F., Gibreel, T. M., Farnham, G., Al Johani, S. M., Balkhy, H. H., and Upton, M. (2015). Antibiotic-resistant ST38, ST131 and ST405 strains are the leading uropathogenic Escherichia coli clones in Riyadh, Saudi Arabia. J. Antimicrob. Chemother. 70, 2757–2762. doi: 10.1093/jac/dkv188
Arndt, D., Grant, J. R., Marcu, A., Sajed, T., Pon, A., Liang, Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21. doi: 10.1093/nar/gkw387
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Banerjee, R., Robicsek, A., Kuskowski, M. A., Porter, S., Johnston, B. D., Sokurenko, E., et al. (2013). Molecular epidemiology of Escherichia coli sequence type 131 and its H30 and H30-Rx subclones among extended-spectrum-beta-lactamase-positive and -negative E. coli clinical isolates from the Chicago region, 2007 to 2010. Antimicrob. Agents Chemother. 57, 6385–6388. doi: 10.1128/AAC.01604-13
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Beghain, J., Bridier-Nahmias, A., Le Nagard, H., Denamur, E., and Clermont, O. (2018). ClermonTyping: an easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb. Genom. 4:e000192. doi: 10.1099/mgen.0.000192
Birgy, A., Madhi, F., Jung, C., Levy, C., Cointe, A., Bidet, P., et al. (2020). Diversity and trends in population structure of ESBL-producing Enterobacteriaceae in febrile urinary tract infections in children in France from 2014 to 2017. J. Antimicrob. Chemother. 75, 96–105. doi: 10.1093/jac/dkz423
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Boll, E. J., Struve, C., Boisen, N., Olesen, B., Stahlhut, S. G., and Krogfelt, K. A. (2013). Role of enteroaggregative Escherichia coli virulence factors in uropathogenesis. Infect. Immun. 81, 1164–1171. doi: 10.1128/IAI.01376-12
Boll, E. J., Struve, C., Sander, A., Demma, Z., Nataro, J. P., McCormick, B. A., et al. (2012). The fimbriae of enteroaggregative Escherichia coli induce epithelial inflammation in vitro and in a human intestinal xenograft model. J. Infect. Dis. 206, 714–722. doi: 10.1093/infdis/jis417
Bonnet, R., Recule, C., Baraduc, R., Chanal, C., Sirot, D., De Champs, C., et al. (2003). Effect of D240G substitution in a novel ESBL CTX-M-27. J. Antimicrob. Chemother. 52, 29–35. doi: 10.1093/jac/dkg256
Bostock, M., Ogievetsky, V., and Heer, J. (2011). D(3): data-driven documents. IEEE Trans. Vis. Comput. Graph. 17, 2301–2309. doi: 10.1109/TVCG.2011.185
Brodrick, H. J., Raven, K. E., Kallonen, T., Jamrozy, D., Blane, B., Brown, N. M., et al. (2017). Longitudinal genomic surveillance of multidrug-resistant Escherichia coli carriage in a long-term care facility in the United Kingdom. Genome Med. 9:70. doi: 10.1186/s13073-017-0457-6
Cao, X., Cavaco, L. M., Lv, Y., Li, Y., Zheng, B., Wang, P., et al. (2011). Molecular characterization and antimicrobial susceptibility testing of Escherichia coli isolates from patients with urinary tract infections in 20 Chinese hospitals. J. Clin. Microbiol. 49, 2496–2501. doi: 10.1128/JCM.02503-10
Chandramohan, L., and Revell, P. A. (2012). Prevalence and molecular characterization of extended-spectrum-beta-lactamase-producing Enterobacteriaceae in a pediatric patient population. Antimicrob. Agents Chemother. 56, 4765–4770. doi: 10.1128/AAC.00666-12
Chattaway, M. A., Jenkins, C., Ciesielczuk, H., Day, M., DoNascimento, V., Day, M., et al. (2014). Evidence of evolving extraintestinal enteroaggregative Escherichia coli ST38 clone. Emerg. Infect. Dis. 20, 1935–1937. doi: 10.3201/eid2011.131845
Chen, L. F., Freeman, J. T., Nicholson, B., Keiger, A., Lancaster, S., Joyce, M., et al. (2014). Widespread dissemination of CTX-M-15 genotype extended-spectrum-beta-lactamase-producing enterobacteriaceae among patients presenting to community hospitals in the southeastern United States. Antimicrob. Agents Chemother. 58, 1200–1202. doi: 10.1128/AAC.01099-13
CLSI (2019). “Performance standards for antimicrobial susceptibility testing” in CLSI supplement M100. 29th Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
Dahbi, G., Mora, A., Lopez, C., Alonso, M. P., Mamani, R., Marzoa, J., et al. (2013). Emergence of new variants of ST131 clonal group among extraintestinal pathogenic Escherichia coli producing extended-spectrum beta-lactamases. Int. J. Antimicrob. Agents 42, 347–351. doi: 10.1016/j.ijantimicag.2013.06.017
Darling, A. C., Mau, B., Blattner, F. R., and Perna, N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. doi: 10.1101/gr.2289704
Davis, S., Pettengill, J. B., Luo, Y., Payne, J., Shpuntoff, A., Rand, H., et al. (2015). CFSAN SNP pipeline: an automated method for constructing SNP matrices from next-generation sequence data. PeerJ Comput. Sci. 1:e20. doi: 10.7717/peerj-cs.20
Day, M. J., Hopkins, K. L., Wareham, D. W., Toleman, M. A., Elviss, N., Randall, L., et al. (2019). Extended-spectrum β-lactamase-producing Escherichia coli in human-derived and foodchain-derived samples from England, Wales, and Scotland: an epidemiological surveillance and typing study. Lancet Infect. Dis. 19, 1325–1335. doi: 10.1016/S1473-3099(19)30273-7
Doi, Y., Park, Y. S., Rivera, J. I., Adams-Haduch, J. M., Hingwe, A., Sordillo, E. M., et al. (2013). Community-associated extended-spectrum beta-lactamase-producing Escherichia coli infection in the United States. Clin. Infect. Dis. 56, 641–648. doi: 10.1093/cid/cis942
Doumith, M., Dhanji, H., Ellington, M. J., Hawkey, P., and Woodford, N. (2012). Characterization of plasmids encoding extended-spectrum beta-lactamases and their addiction systems circulating among Escherichia coli clinical isolates in the UK. J. Antimicrob. Chemother. 67, 878–885. doi: 10.1093/jac/dkr553
Faheem, M., Rehman, M. T., Danishuddin, M., and Khan, A. U. (2013). Biochemical characterization of CTX-M-15 from Enterobacter cloacae and designing a novel non-β-lactam-β-lactamase inhibitor. PLoS One 8:e56926. doi: 10.1371/annotation/049bf1aa-d866-471f-95c1-5939d4461f8c
Flament-Simon, S. C., Garcia, V., Duprilot, M., Mayer, N., Alonso, M. P., Garcia-Menino, I., et al. (2020). High prevalence of ST131 subclades C2-H30Rx and C1-M27 among extended-spectrum beta-lactamase-producing Escherichia coli causing human extraintestinal infections in patients from two hospitals of Spain and France during 2015. Front. Cell. Infect. Microbiol. 10:125. doi: 10.3389/fcimb.2020.00125
Gauthier, L., Dortet, L., Cotellon, G., Creton, E., Cuzon, G., Ponties, V., et al. (2018). Diversity of carbapenemase-producing Escherichia coli isolates in France in 2012-2013. Antimicrob. Agents Chemother. 62, e00266–e00218. doi: 10.1128/AAC.00266-18
Ghosh, H., Bunk, B., Doijad, S., Schmiedel, J., Falgenhauer, L., Sproer, C., et al. (2017). Complete genome sequence of blaCTX-M-27-encoding Escherichia coli strain H105 of sequence type 131 lineage C1/H30R. Genome Announc. 5, e00736–e00717. doi: 10.1128/genomeA.00736-17
Guignot, J., Chaplais, C., Coconnier-Polter, M. H., and Servin, A. L. (2007). The secreted autotransporter toxin, Sat, functions as a virulence factor in Afa/Dr diffusely adhering Escherichia coli by promoting lesions in tight junction of polarized epithelial cells. Cell. Microbiol. 9, 204–221. doi: 10.1111/j.1462-5822.2006.00782.x
Guiral, E., Quiles, M. G., Muñoz, L., Moreno-Morales, J., Alejo-Cancho, I., Salvador, P., et al. (2019). Emergence of resistance to quinolones and β-lactam antibiotics in enteroaggregative and enterotoxigenic Escherichia coli causing traveler’s diarrhea. Antimicrob. Agents Chemother. 63, e01745–e01718. doi: 10.1128/AAC.01745-18
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Havt, A., Lima, I. F., Medeiros, P. H., Clementino, M. A., Santos, A. K., Amaral, M. S., et al. (2017). Prevalence and virulence gene profiling of enteroaggregative Escherichia coli in malnourished and nourished Brazilian children. Diagn. Microbiol. Infect. Dis. 89, 98–105. doi: 10.1016/j.diagmicrobio.2017.06.024
Hertz, F. B., Nielsen, J. B., Schonning, K., Littauer, P., Knudsen, J. D., Lobner-Olesen, A., et al. (2016). Population structure of drug-susceptible, -resistant and ESBL-producing Escherichia coli from community-acquired urinary tract. BMC Microbiol. 16:63. doi: 10.1186/s12866-016-0681-z
Johnson, J. R., Urban, C., Weissman, S. J., Jorgensen, J. H., Lewis, J. S., Hansen, G., et al. (2012). Molecular epidemiological analysis of Escherichia coli sequence type ST131 (O25: H4) and blaCTX-M-15 among extended-spectrum-β-lactamase-producing E. coli from the United States, 2000 to 2009. Antimicrob. Agents Chemother. 56, 2364–2370. doi: 10.1128/AAC.05824-11
Jolley, K. A., and Maiden, M. C. (2010). BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinform. 11:595. doi: 10.1186/1471-2105-11-595
Kanamori, H., Parobek, C. M., Juliano, J. J., Johnson, J. R., Johnston, B. D., Johnson, T. J., et al. (2017). Genomic analysis of multidrug-resistant Escherichia coli from North Carolina community hospitals: ongoing circulation of CTX-M-producing ST131-H30Rx and ST131-H30R1 strains. Antimicrob. Agents Chemother. 61, e00912–e00917. doi: 10.1128/AAC.00912-17
Kim, J., Bae, I. K., Jeong, S. H., Chang, C. L., Lee, C. H., and Lee, K. (2011). Characterization of IncF plasmids carrying the blaCTX-M-14 gene in clinical isolates of Escherichia coli from Korea. J. Antimicrob. Chemother. 66, 1263–1268. doi: 10.1093/jac/dkr106
Koboldt, D. C., Zhang, Q., Larson, D. E., Shen, D., McLellan, M. D., Lin, L., et al. (2012). VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22, 568–576. doi: 10.1101/gr.129684.111
Kondratyeva, K., Salmon-Divon, M., and Navon-Venezia, S. (2020). Meta-analysis of pandemic Escherichia coli ST131 plasmidome proves restricted plasmid-clade associations. Sci. Rep. 10:36. doi: 10.1038/s41598-019-56763-7
Korotkova, N., Yarova-Yarovaya, Y., Tchesnokova, V., Yazvenko, N., Carl, M. A., Stapleton, A. E., et al. (2008). Escherichia coli DraE adhesin-associated bacterial internalization by epithelial cells is promoted independently by decay-accelerating factor and carcinoembryonic antigen-related cell adhesion molecule binding and does not require the DraD invasin. Infect. Immun. 76, 3869–3880. doi: 10.1128/IAI.00427-08
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Kuroda, H., Yano, H., Hirakata, Y., Arai, K., Endo, S., Kanamori, H., et al. (2012). Molecular characteristics of extended-spectrum beta-lactamase-producing Escherichia coli in Japan: emergence of CTX-M-15-producing E. coli ST131. Diagn. Microbiol. Infect. Dis. 74, 201–203. doi: 10.1016/j.diagmicrobio.2012.06.011
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Lima, I. F., Boisen, N., Quetz Jda, S., Havt, A., de Carvalho, E. B., Soares, A. M., et al. (2013). Prevalence of enteroaggregative Escherichia coli and its virulence-related genes in a case-control study among children from North-Eastern Brazil. J. Med. Microbiol. 62, 683–693. doi: 10.1099/jmm.0.054262-0
Livermore, D. M., Canton, R., Gniadkowski, M., Nordmann, P., Rossolini, G. M., Arlet, G., et al. (2007). CTX-M: changing the face of ESBLs in Europe. J. Antimicrob. Chemother. 59, 165–174. doi: 10.1093/jac/dkl483
Matsumura, Y., Johnson, J. R., Yamamoto, M., Nagao, M., Tanaka, M., Takakura, S., et al. (2015). CTX-M-27-and CTX-M-14-producing, ciprofloxacin-resistant Escherichia coli of the H30 subclonal group within ST131 drive a Japanese regional ESBL epidemic. J. Antimicrob. Chemother. 70, 1639–1649. doi: 10.1093/jac/dkv017
Matsumura, Y., Yamamoto, M., Nagao, M., Ito, Y., Takakura, S., Ichiyama, S., et al. (2013). Association of fluoroquinolone resistance, virulence genes, and IncF plasmids with extended-spectrum-beta-lactamase-producing Escherichia coli sequence type 131 (ST131) and ST405 clonal groups. Antimicrob. Agents Chemother. 57, 4736–4742. doi: 10.1128/AAC.00641-13
Mendes, R. E., Jones, R. N., Woosley, L. N., Cattoir, V., and Castanheira, M. (2019). Application of next-generation sequencing for characterization of surveillance and clinical trial isolates: analysis of the distribution of beta-lactamase resistance genes and lineage background in the United States. Open Forum Infect. Dis. 6, S69–S78. doi: 10.1093/ofid/ofz004
Mshana, S. E., Imirzalioglu, C., Hain, T., Domann, E., Lyamuya, E. F., and Chakraborty, T. (2011). Multiple ST clonal complexes, with a predominance of ST131, of Escherichia coli harbouring blaCTX-M-15 in a tertiary hospital in Tanzania. Clin. Microbiol. Infect. 17, 1279–1282. doi: 10.1111/j.1469-0691.2011.03518.x
Muenzner, P., Tchoupa, A. K., Klauser, B., Brunner, T., Putze, J., Dobrindt, U., et al. (2016). Uropathogenic E. coli exploit CEA to promote colonization of the urogenital tract mucosa. PLoS Pathog. 12:e1005608. doi: 10.1371/journal.ppat.1005608
Nataro, J. P., Seriwatana, J., Fasano, A., Maneval, D. R., Guers, L. D., Noriega, F., et al. (1995). Identification and cloning of a novel plasmid-encoded enterotoxin of enteroinvasive Escherichia coli and Shigella strains. Infect. Immun. 63, 4721–4728. doi: 10.1128/IAI.63.12.4721-4728.1995
Nicolas-Chanoine, M. H., Bertrand, X., and Madec, J. Y. (2014). Escherichia coli ST131, an intriguing clonal group. Clin. Microbiol. Rev. 27, 543–574. doi: 10.1128/CMR.00125-13
Nicolas-Chanoine, M. -H., Petitjean, M., Mora, A., Mayer, N., Lavigne, J. -P., Boulet, O., et al. (2017). The ST131 Escherichia coli H22 subclone from human intestinal microbiota: comparison of genomic and phenotypic traits with those of the globally successful H30 subclone. BMC Microbiol. 17:71. doi: 10.1186/s12866-017-0984-8
Nicoloff, H., Hjort, K., Levin, B. R., and Andersson, D. I. (2019). The high prevalence of antibiotic heteroresistance in pathogenic bacteria is mainly caused by gene amplification. Nat. Microbiol. 4, 504–514. doi: 10.1038/s41564-018-0342-0
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. doi: 10.1093/bioinformatics/btv421
Paiva de Sousa, C., and Dubreuil, J. D. (2001). Distribution and expression of the astA gene (EAST1 toxin) in Escherichia coli and Salmonella. Int. J. Med. Microbiol. 291, 15–20. doi: 10.1078/1438-4221-00097
Peirano, G., and Pitout, J. D. (2019). Extended-spectrum β-lactamase-producing Enterobacteriaceae: update on molecular epidemiology and treatment options. Drugs 79, 1529–1541. doi: 10.1007/s40265-019-01180-3
Petty, N. K., Ben Zakour, N. L., Stanton-Cook, M., Skippington, E., Totsika, M., Forde, B. M., et al. (2014). Global dissemination of a multidrug resistant Escherichia coli clone. Proc. Natl. Acad. Sci. U. S. A. 111, 5694–5699. doi: 10.1073/pnas.1322678111
Phan, M. D., Forde, B. M., Peters, K. M., Sarkar, S., Hancock, S., Stanton-Cook, M., et al. (2015). Molecular characterization of a multidrug resistance IncF plasmid from the globally disseminated Escherichia coli ST131 clone. PLoS One 10:e0122369. doi: 10.1371/journal.pone.0122369
Pitout, J. D., and DeVinney, R. (2017). Escherichia coli ST131: a multidrug-resistant clone primed for global domination. F1000Res. 6:195. doi: 10.12688/f1000research.10609.1
Poirel, L., Bernabeu, S., Fortineau, N., Podglajen, I., Lawrence, C., and Nordmann, P. (2011). Emergence of OXA-48-producing Escherichia coli clone ST38 in France. Antimicrob. Agents Chemother. 55, 4937–4938. doi: 10.1128/AAC.00413-11
Poirel, L., Naas, T., and Nordmann, P. (2008). Genetic support of extended-spectrum beta-lactamases. Clin. Microbiol. Infect. 14(Suppl. 1), 75–81. doi: 10.1111/j.1469-0691.2007.01865.x
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490
Price, L. B., Johnson, J. R., Aziz, M., Clabots, C., Johnston, B., Tchesnokova, V., et al. (2013). The epidemic of extended-spectrum-beta-lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H30-Rx. mBio 4, e00377–e00313. doi: 10.1128/mBio.00377-13
Rodríguez, M. M., Power, P., Radice, M., Vay, C., Famiglietti, A., Galleni, M., et al. (2004). Chromosome-encoded CTX-M-3 from Kluyvera ascorbata: a possible origin of plasmid-borne CTX-M-1-derived cefotaximases. Antimicrob. Agents Chemother. 48, 4895–4897. doi: 10.1128/AAC.48.12.4895-4897.2004
Roer, L., Hansen, F., Thomsen, M. C. F., Knudsen, J. D., Hansen, D. S., Wang, M., et al. (2017). WGS-based surveillance of third-generation cephalosporin-resistant Escherichia coli from bloodstream infections in Denmark. J. Antimicrob. Chemother. 72, 1922–1929. doi: 10.1093/jac/dkx092
Rossolini, G. M., D’Andrea, M. M., and Mugnaioli, C. (2008). The spread of CTX-M-type extended-spectrum beta-lactamases. Clin. Microbiol. Infect. 14(Suppl. 1), 33–41. doi: 10.1111/j.1469-0691.2007.01867.x
Sandegren, L., Lindqvist, A., Kahlmeter, G., and Andersson, D. I. (2008). Nitrofurantoin resistance mechanism and fitness cost in Escherichia coli. J. Antimicrob. Chemother. 62, 495–503. doi: 10.1093/jac/dkn222
Seemann, T. (2017). ABRicate: mass screening of contigs for antimicrobial resistance or virulence genes.
Seiffert, S. N., Hilty, M., Kronenberg, A., Droz, S., Perreten, V., and Endimiani, A. (2013). Extended-spectrum cephalosporin-resistant Escherichia coli in community, specialized outpatient clinic and hospital settings in Switzerland. J. Antimicrob. Chemother. 68, 2249–2254. doi: 10.1093/jac/dkt208
Sepp, E., Andreson, R., Balode, A., Bilozor, A., Brauer, A., Egorova, S., et al. (2019). Phenotypic and molecular epidemiology of ESBL-, AmpC-, and Carbapenemase-producing Escherichia coli in Northern and Eastern Europe. Front. Microbiol. 10:2465. doi: 10.3389/fmicb.2019.02465
Servin, A. L. (2014). Pathogenesis of human diffusely adhering Escherichia coli expressing Afa/Dr adhesins (Afa/Dr DAEC): current insights and future challenges. Clin. Microbiol. Rev. 27, 823–869. doi: 10.1128/CMR.00036-14
Shaik, S., Ranjan, A., Tiwari, S. K., Hussain, A., Nandanwar, N., Kumar, N., et al. (2017). Comparative genomic analysis of globally dominant ST131 clone with other epidemiologically successful extraintestinal pathogenic Escherichia coli (ExPEC) lineages. mBio 8, e01596–e01517. doi: 10.1128/mBio.01596-17
Shanmugam, D., Esak, S. B., and Narayanaswamy, A. (2016). Molecular characterisation of nfsA gene in nitrofurantoin resistant uropathogens. J. Clin. Diagn. Res. 10, DC05–DC09. doi: 10.7860/JCDR/2016/17280.7957
Sheikh, J., Czeczulin, J. R., Harrington, S., Hicks, S., Henderson, I. R., Le Bouguénec, C., et al. (2002). A novel dispersin protein in enteroaggregative Escherichia coli. J. Clin. Invest. 110, 1329–1337. doi: 10.1172/JCI16172
Sorlozano, A., Gutierrez, J., Jimenez, A., de Dios Luna, J., and Martinez, J. L. (2007). Contribution of a new mutation in parE to quinolone resistance in extended-spectrum-beta-lactamase-producing Escherichia coli isolates. J. Clin. Microbiol. 45, 2740–2742. doi: 10.1128/JCM.01093-07
Stoesser, N., Sheppard, A. E., Pankhurst, L., De Maio, N., Moore, C. E., Sebra, R., et al. (2016). Evolutionary history of the global emergence of the Escherichia coli epidemic clone ST131. mBio 7:e02162. doi: 10.1128/mBio.02162-15
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Suzuki, S., Shibata, N., Yamane, K., Wachino, J., Ito, K., and Arakawa, Y. (2009). Change in the prevalence of extended-spectrum-beta-lactamase-producing Escherichia coli in Japan by clonal spread. J. Antimicrob. Chemother. 63, 72–79. doi: 10.1093/jac/dkn463
Tadesse, D. A., Li, C., Mukherjee, S., Hsu, C. H., Bodeis Jones, S., Gaines, S. A., et al. (2018). Whole-genome sequence analysis of CTX-M containing Escherichia coli isolates from retail meats and cattle in the United States. Microb. Drug Resist. 24, 939–948. doi: 10.1089/mdr.2018.0206
Thomsen, M. C. F., Ahrenfeldt, J., Cisneros, J. L. B., Jurtz, V., Larsen, M. V., Hasman, H., et al. (2016). A bacterial analysis platform: an integrated system for analysing bacterial whole genome sequencing data for clinical diagnostics and surveillance. PLoS One 11:e0157718. doi: 10.1371/journal.pone.0157718
Tulara, N. K. (2018). Nitrofurantoin and fosfomycin for extended spectrum beta-lactamases producing Escherichia coli and Klebsiella pneumoniae. J. Global Infect. Dis. 10, 19–21. doi: 10.4103/jgid.jgid_72_17
van der Bij, A. K., Peirano, G., Goessens, W. H., van der Vorm, E. R., van Westreenen, M., and Pitout, J. D. (2011). Clinical and molecular characteristics of extended-spectrum-beta-lactamase-producing Escherichia coli causing bacteremia in the Rotterdam area, Netherlands. Antimicrob. Agents Chemother. 55, 3576–3578. doi: 10.1128/AAC.00074-11
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Yasir, M., Farman, M., Shah, M. W., Jiman-Fatani, A. A., Othman, N. A., Almasaudi, S. B., et al. (2020). Genomic and antimicrobial resistance genes diversity in multidrug-resistant CTX-M-positive isolates of Escherichia coli at a health care facility in Jeddah. J. Infect. Public Health 13, 94–100. doi: 10.1016/j.jiph.2019.06.011
Zhang, W. H., Lin, X. Y., Xu, L., Gu, X. X., Yang, L., Li, W., et al. (2016). CTX-M-27 producing Salmonella enterica serotypes Typhimurium and Indiana are prevalent among food-producing animals in China. Front. Microbiol. 7:436. doi: 10.3389/fmicb.2016.00436
Keywords: Escherichia coli, ceftriaxone, plasmids, bacterial genomics, mobile elements, extended-spectrum beta lactamases, nitrofurantoin
Citation: Mostafa HH, Cameron A, Taffner SM, Wang J, Malek A, Dumyati G, Hardy DJ and Pecora ND (2020) Genomic Surveillance of Ceftriaxone-Resistant Escherichia coli in Western New York Suggests the Extended-Spectrum β-Lactamase blaCTX-M-27 Is Emerging on Distinct Plasmids in ST38. Front. Microbiol. 11:1747. doi: 10.3389/fmicb.2020.01747
Received: 24 April 2020; Accepted: 03 July 2020;
Published: 30 July 2020.
Edited by:
Teresa M. Coque, Ramón y Cajal Institute for Health Research, SpainReviewed by:
Marie-Helene Nicolas-Chanoine, INSERM U1137 Infection, Antimicrobiens, Modélisation, Evolution, FranceCopyright © 2020 Mostafa, Cameron, Taffner, Wang, Malek, Dumyati, Hardy and Pecora. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nicole D. Pecora, bmljb2xlX3BlY29yYUB1cm1jLnJvY2hlc3Rlci5lZHU=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.