
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 22 July 2020
Sec. Microbiotechnology
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.01742
This article is part of the Research TopicBridging Microbial Activities and Engineering Performance: Recent Advances in Microbial Metabolism and Interactions in Engineered EnvironmentsView all 8 articles
 Thomas Eng1,2
Thomas Eng1,2 Robin A. Herbert1,2,3,4
Robin A. Herbert1,2,3,4 Uriel Martinez2,5
Uriel Martinez2,5 Brenda Wang1
Brenda Wang1 Joseph C. Chen5
Joseph C. Chen5 James B. Brown2,3,6,7.8
James B. Brown2,3,6,7.8 Adam M. Deutschbauer3,4
Adam M. Deutschbauer3,4 Mina J. Bissell2
Mina J. Bissell2 Jenny C. Mortimer1,2,3*
Jenny C. Mortimer1,2,3* Aindrila Mukhopadhyay1,2,3*
Aindrila Mukhopadhyay1,2,3*The rhizosphere microbiome (rhizobiome) plays a critical role in plant health and development. However, the processes by which the constituent microbes interact to form and maintain a community are not well understood. To investigate these molecular processes, we examined pairwise interactions between 11 different microbial isolates under select nutrient-rich and nutrient-limited conditions. We observed that when grown with media supplemented with 56 mM glucose, two microbial isolates were able to inhibit the growth of six other microbes. The interaction between microbes persisted even after the antagonistic microbe was removed, upon exposure to spent media. To probe the genetic basis for these antagonistic interactions, we used a barcoded transposon library in a proxy bacterium, Pseudomonas putida, to identify genes which showed enhanced sensitivity to the antagonistic factor(s) secreted by Acinetobacter sp. 02. Iron metabolism-related gene clusters in P. putida were implicated by this systems-level analysis. The supplementation of iron prevented the antagonistic interaction in the original microbial pair, supporting the hypothesis that iron limitation drives antagonistic microbial interactions between rhizobionts. We conclude that rhizobiome community composition is influenced by competition for limiting nutrients, with implications for growth and development of the plant.
Microbial communities are being increasingly appreciated for their impact on larger biological systems, such as in relation to human health (e.g., the gut microbiome) (Chu et al., 2016) or crop productivity (e.g., the rhizobiome) (Mendes et al., 2018). Recent studies suggest that antagonism between microbes can influence host fitness in various systems (Mendes et al., 2018; Zhao et al., 2018). However, while some direct chemical interactions between hosts and their microbiome are known (Stringlis et al., 2018), it is unclear how beneficial microbial communities are assembled and maintained throughout host development. A better understanding of microbial communities and their relation to the plant root is of specific interest to understand plant development, biogeochemical carbon cycling, and applications in agriculture.
The environmental milieu of the plant root is distinct from that of bulk soil. Photosynthetic bioproducts are exuded through the roots, modifying the rhizosphere through the accumulation of sugars and secondary plant metabolites. The composition of root exudate is complex (Chaparro et al., 2013; Kawasaki et al., 2016); however, simple sugars (such as D-glucose) have often been detected as major components of the root exudate in terrestrial plants such as the model dicot Arabidopsis thaliana (Okubo et al., 2016). Changes in exudate profile correlate with compositional and functional changes in the rhizobiome (Chaparro et al., 2013).
While many isolates have been identified across ecologically distinct rhizobiomes, isolates from several major phyla tend to dominate any given rhizobiome (Vorholt, 2012; Bai et al., 2015). The formation of the rhizobiome is likely a deterministic process, in which microbes both synergize and antagonize each other in competition for limited resources from root exudate (Hassani et al., 2018). A population equilibrium, where several microbial phyla dominate, is eventually established. For example, specific microbes could be growth-limited by the plant through the secretion of iron-sequestering siderophores or antibiotics (Joshi et al., 2006; van der Meij et al., 2017). Alternatively, microbial inhibitory mechanisms such as secretion systems capable of puncturing neighboring cell membranes, or the secretion of antimicrobial compounds, have also been described (Nester, 2014; Alteri and Mobley, 2016). However, the specific environmental cues that prime a microbial response to limit the growth of other microbes remain poorly described.
In this study, we sought to understand if microbes from a model rhizobiome compete with each other under defined glucose supplementation conditions. We chose 11 rhizobacteria (Table 1 and Supplementary Figure S1) representative of phyla previously detected as enriched within the roots of A. thaliana relative to bulk soil (Lundberg et al., 2012; Levy et al., 2017). These microbes are of general interest because they are well characterized and can improve plant health in the presence of specific environmental stressors (Herbert et al., 2019). We hypothesized that microbe-microbe interactions could be readily detected by tracking the frequency by which microbes inhibited the growth of their neighboring species. Out of 37 pairwise microbial competition assays in this format, we detected three microbes which either blocked colony formation of many microbes or specific microbial isolates. To examine the molecular mechanism underlying an inhibitory interaction, we used a pooled transposon mutant library (Wetmore et al., 2015) built in the soil microbe Pseudomonas putida KT2440 to identify genes required for growth in Acinetobacter sp. 02 spent medium, and validated our findings back in the reconstituted rhizobiome microbe pair. A summary of our experimental approach is described in Figure 1.
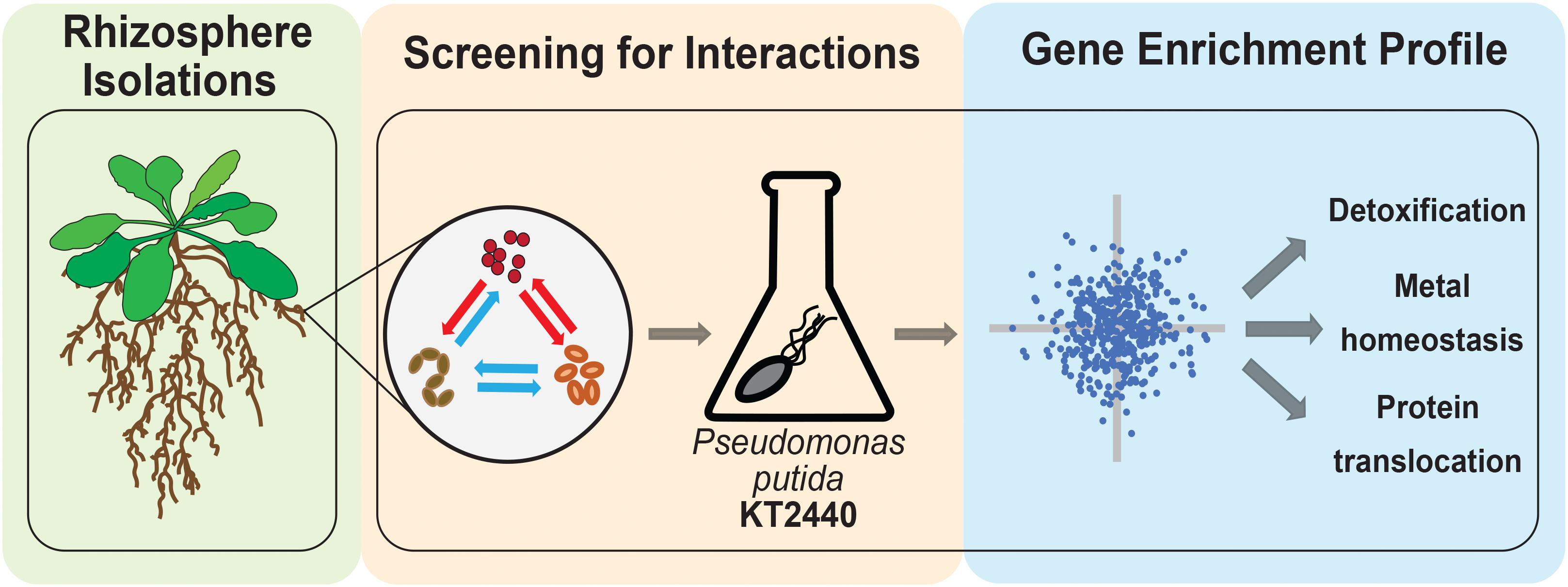
Figure 1. Workflow for bacterial chemical genomic screen. Rhizobacteria mediate complex interactions in the rhizosphere (green-shaded box). After isolation, rhizobacteria are screened for potential interactions against one another and with a random barcode transposon sequencing (RB-TnSeq) library of a model microorganism (orange-shaded box). Finally, RB-TnSeq data are analyzed and used to characterize and validate microbe-microbe interactions (blue-shaded box).
We developed a rapid and reproducible assay to monitor microbe-microbe interactions based on cell viability as an alternative to technically involved methods, such as metagenomic RNAseq or microbial capture in microdroplets (Jackman et al., 2019; Mutha et al., 2019). We adapted an established agar plate based assay used to monitor cell-cell interactions in Saccharomyces cerevisiae (Abelson et al., 1990; Liu et al., 2019). When two microbial isolates are grown together (Figure 2A), it should be possible to observe one of four types of interactions based on growth. First, there could be no detectable interaction between the two microbes. Second, both strains could still form colonies, but exhibit morphological changes or exhibit synergistic, improved growth. Third, one of the microbes could fail to grow in the presence of the other microbe. Finally, both microbes could be growth-inhibited when co-cultivated. Bioinformatic analysis of these microbial genomes (Weber et al., 2015) to identify putative secreted gene clusters indicated that many of these species had the capacity to produce a range of secreted molecules, but it was unclear what conditions would be needed to induce production, or if any given microbe would be sensitive to a given secreted molecule (Supplementary Table S1). A representative agar plate is shown in Figure 2B, where Acinetobacter sp. 02 is the primary species and tested for interactions with six secondary species. Finally, we modified this method with a staggered plating regime (see section “Materials and Methods”) to determine if the continued presence of cells was required for an antagonistic interaction with a staggered plating regime (see section “Materials and Methods”).
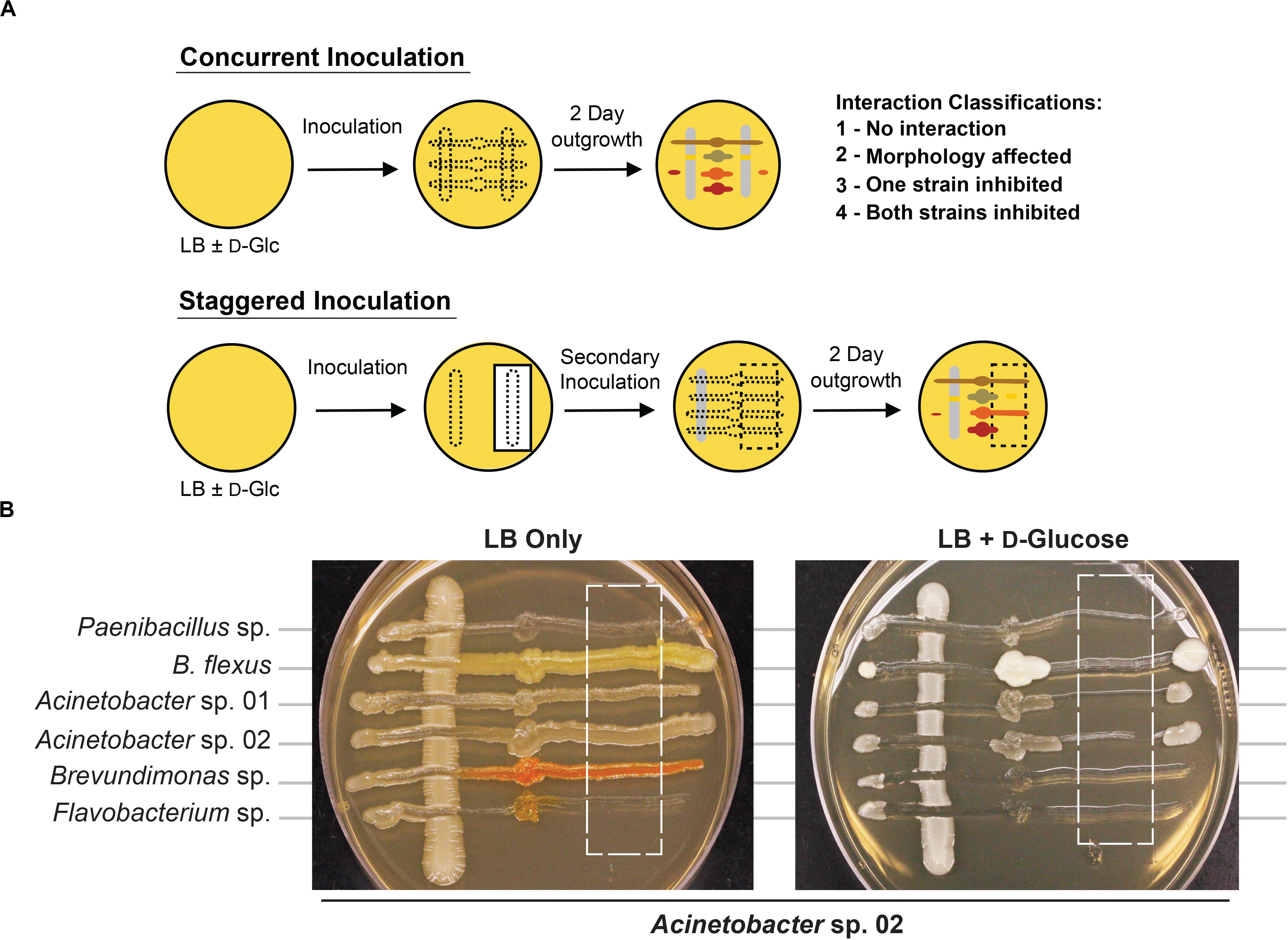
Figure 2. Methodology for microbial interaction screen. (A) Multiple rhizobacteria are used to inoculate LB plates, either concurrently or staggered such that the vertical, “primary” species was grown for 2 days before the introduction of a secondary species. For staggered inoculation assays, half of the primary inoculum was grown on top of a nitrocellulose membrane that was removed prior to secondary inoculation. (B) Representative images of staggered inoculation assays with Acinetobacter sp. 02 grown in co-culture with six rhizobacterial species. Dashed lines indicate the location of a 0.44-μm nitrocellulose membrane prior to removal. Brightness and contrast have been uniformly edited to increase visibility.
When the primary and secondary microbes were inoculated at the same time, we detected two pairwise interaction types. The majority of microbes tested for interactions showed no detectable change in growth when co-cultured (Figure 3A). The following six isolates were screened for interactions (Agrobacterium rhizogenes, Acinetobacter sp. 02, Arthrobacter sp., Flavobacterium sp., Paenibacillus sp., and Ralstonia sp.) against seven others (A. rhizogenes, Acinetobacter sp. 01, Acinetobacter sp. 02, Bacillus flexus, Brevundimonas sp., Flavobacterium sp., and Paenibacillus sp.; n ≥ 3) on solid lysogeny broth (LB) (“limited glucose”). However, both Acinetobacter sp. 02 and Flavobacterium sp. specifically inhibited the growth of Brevundimonas sp. under the concurrent growth regimen (Figure 3A).
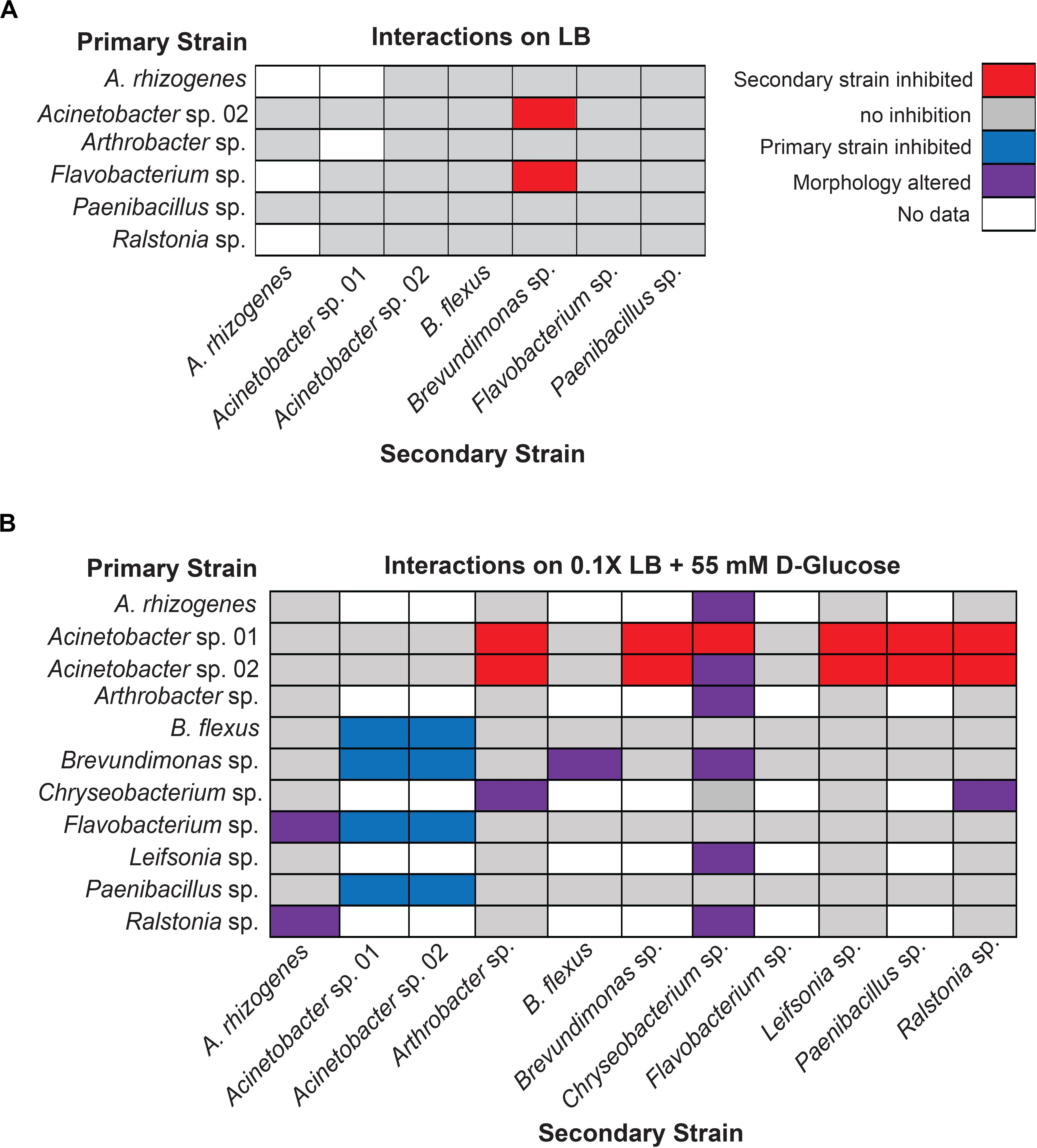
Figure 3. Results of concurrent inoculation interaction screen. Interactions observed on (A) LB or (B) 0.1× LB supplemented with 55 mM D-glucose. Rows represent primary, vertically streaked species and columns represent secondary, horizontally streaked species. The same color legend is used for both (A) and (B).
Increasing the concentration of available glucose to 55 mM in the media allowed us to detect several additional cases of antagonistic interactions. We examined a larger set of pairwise microbial interactions by competing a set of six (Acinetobacter sp. 01, Acinetobacter sp. 02, B. flexus, Brevundimonas sp., Flavobacterium sp., and Paenibacillus sp.; n ≥ 3 replicates) against the same set, plus the following five microbes: A. rhizogenes, Arthrobacter sp., Chryseobacterium sp., Leifsonia sp., and Ralstonia sp. (n ≥ 2).
Under the glucose-supplemented media conditions, additional inhibitory interactions were observed. Specifically, Acinetobacter sp. 01 inhibited the growth of seven different microbes: Arthrobacter sp., Brevundimonas sp., Chryseobacterium sp., Flavobacterium sp., Leifsonia sp., Paenibacillus sp., and Ralstonia sp. (Figure 3B). Acinetobacter sp. 02 inhibited a similar set of microbes as Acinetobacter sp. 01 (Figure 3B), with the exception of Chryseobacterium sp. In contrast, Flavobacterium sp., which was able to inhibit the growth of Brevundimonas sp. when grown on LB media (Figure 3A), had no effect when cultivated on glucose-supplemented conditions (Figure 3B). However, there were a number of interactions, e.g., between B. flexus and either of the Acinetobacter spp., wherein the order of inoculation could have a deterministic effect.
We also observed a range of morphological changes in the concurrent inoculation assay under glucose-supplemented conditions. Examples include Chryseobacterium sp. changing colony morphology and color in the co-culture assay when grown with 6 of the 11 isolates tested (Figure 3B and Supplementary Figure S2). Ralstonia sp. appeared to rapidly overtake cells of both A. rhizogenes or Chryseobacterium sp., but not other species such as Arthrobacter sp. or Leifsonia sp., consistent with our definition of a non-inhibitory microbial interaction (Supplementary Figure S3). B. flexus also appeared to grow more rapidly along Brevundimonas sp. (n = 4 out of 6). Finally, Flavobacterium sp. formed an irregular border when co-cultured with A. rhizogenes (Figure 3B). Together, these results suggest a variety of interactions may occur between different rhizobacterial pairs and the identification of three of the four types of potential microbial interactions.
Having identified three microbes with clear growth-inhibitory phenotypes in our concurrent inoculation assay (Flavobacterium sp. and two isolates of Acinetobacter spp.), we next sought to understand the mechanism underlying these interactions. Of the three, both Flavobacterium sp. and Acinetobacter sp. 02 were able to inhibit growth of other microbes at a distance. We hypothesized that these strains could be secreting an environment-modifying molecule (e.g., an antimicrobial compound) or depleting a nutrient. This is in contrast to microbial type VI secretion systems, which requires direct cell contact to lyse competitors (Alteri and Mobley, 2016). In addition, while we did not detect any cell-cell interactions using Paenibacillus sp. in the concurrent inoculation assay (Figure 3), evidence from the literature suggested that members of the genus can produce growth-inhibitory antimicrobials (Shaheen et al., 2011; Meng et al., 2018). Therefore, we chose these three microbes to test whether they could alter the growth media such that it was inhibitory to other microbes.
We first used the staggered inoculation regime to test for microbial interactions. Acinetobacter sp. 02, Flavobacterium sp., and Paenibacillus sp. were grown for 2 days on a sterile nitrocellulose membrane placed on top of the solid agar media. After 2 days, the membrane and cells were removed, resulting in spent solid media. Alternatively, the microbes were plated directly onto solid media (refer to Figure 2 for diagram and representative plate). The second microbe was then streaked onto the plate (Figure 2). Being able to directly assess growth conditions on the same agar plate strengthens our ability to detect microbial interactions in spent agar media. Flavobacterium sp. and Paenibacillus sp. grew poorly on the nitrocellulose membrane under nutrient-poor conditions (data not shown), and so standard LB media was used for these assays instead. Following removal of Acinetobacter sp. 02, Flavobacterium sp. was still inhibited (Figure 4A), the only case in which this antagonism of Flavobacterium sp. was observed. As a primary strain, Flavobacterium sp. pretreatment inhibited the growth of both Brevundimonas sp., as well as freshly plated Flavobacterium sp. itself, however, this antagonism was not observed on the spent solid media (Figure 4A). Similarly, Paenibacillus sp. was inhibitory against B. flexus in the presence of Paenibacillus sp. cells only (Figure 4A). No other interactions between other microbe pairs in the staggered assay were detected.
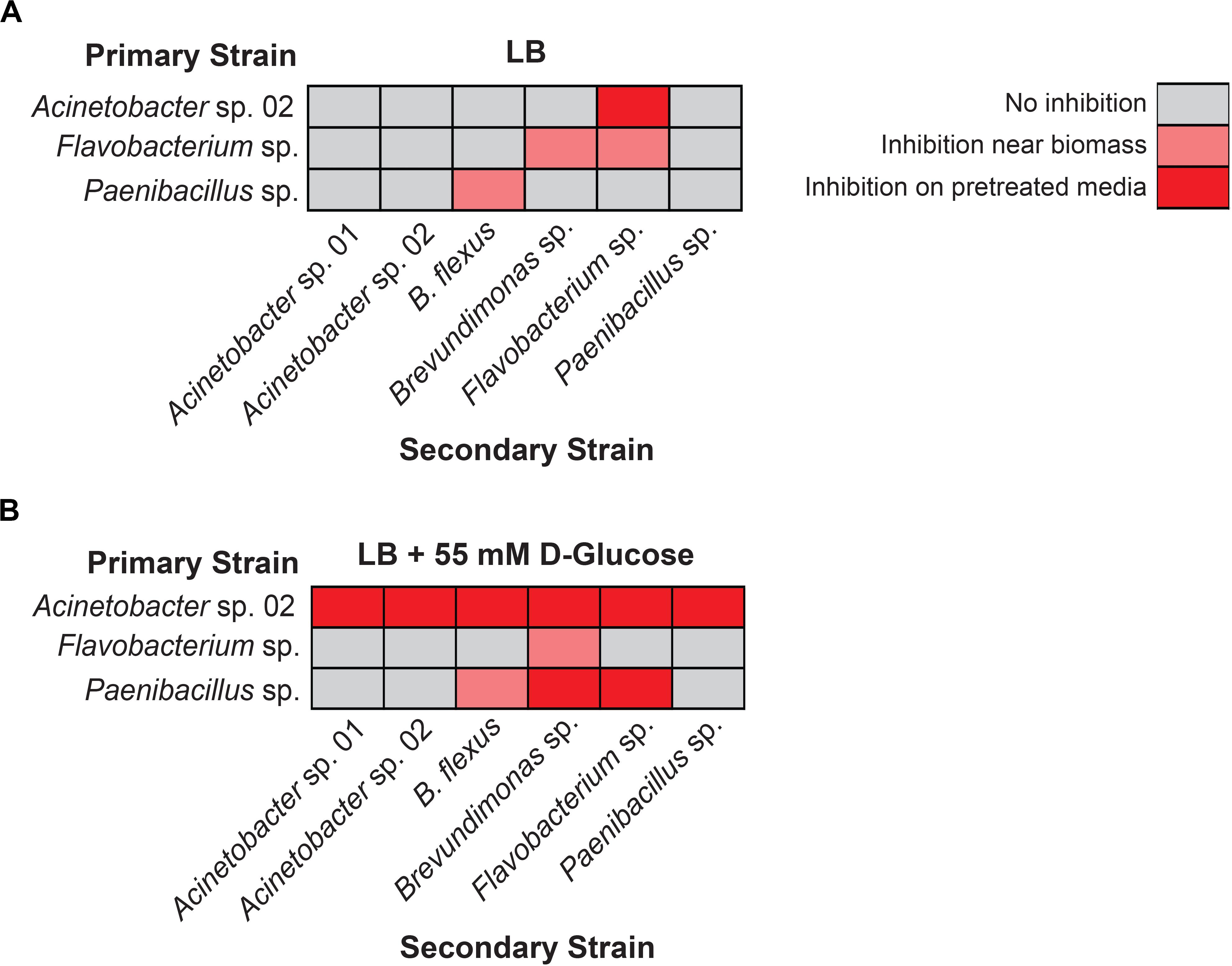
Figure 4. Results of staggered inoculation interaction screen. Interactions observed on (A) LB or (B) LB supplemented with 55 mM D-glucose. Rows represent primary, vertically streaked species and columns represent secondary, horizontally streaked species. n ≥ 3. Where interactions were variable by colony, the most frequently observed interaction is displayed.
Next, we repeated the staggered inoculation assay with glucose-supplemented media to determine if the spent solid media was now more or less toxic. We observed that in this experiment, Acinetobacter sp. 02 was inhibitory to all six tested microbes both with and without the cells present (see representative plate in Figures 2B, 4B). Flavobacterium sp. inhibited the growth of Brevundimonas sp. and no other (Figure 4B). Finally, spent solid media from Paenibacillus sp. was inhibitory to both Brevundimonas sp. and Flavobacterium sp. (Figure 4B). This data suggests that specific microbial interactions do not require direct cell-cell contact, and can persist even after a microbe is removed from the environment. These interactions are differentially mediated by D-glucose concentration/availability. These observations support our hypothesis that growth inhibition arises through depletion of an essential nutrient or the secretion of an antimicrobial compound.
Having established that Acinetobacter sp. 02 modified its environment such that it was inhibitory to a range of different microbes, we sought to identify the secreted factor(s) that conferred this growth inhibitory phenotype. Since genetic tools are not yet available for these recently isolated microbes, we used P. putida KT2440 as a proxy. P. putida KT2440 is an established soil microbe found in similar environments as many microbes in our representative rhizobiome (Molina, 2000). Wild-type P. putida KT2440 is not sensitive to the Acinetobacter sp. 02 supernatant (Supplementary Figure S4). Accordingly, we hypothesized that individual genes in P. putida KT2440 which are responsive to the potential antagonistic agent/condition present in the media would provide insight into the phenotype being observed in the rhizobacterial pairs. Using a barcoded transposon library for parallel fitness assays is an ideal method for the rapid identification of such mutants and is commonly referred to as random barcode transposon sequencing (RB-TnSeq) (Wetmore et al., 2015). This library has been generated for P. putida KT2440 and contains ∼100,000 unique transposon mutants with coverage of most non-essential genes (Price et al., 2019; Thompson et al., 2019). By growing these mutants in a pooled format, P. putida mutants that are sensitive to the toxic agents in the supernatant will be outcompeted by more fit strains, and the absolute abundance of each mutant can be determined using Illumina sequencing specific to each barcoded transposon mutant. Analysis of transposon abundances are used to implicate gene functions that are correlated with resistance/susceptibility to the secreted molecule (Figure 5A).
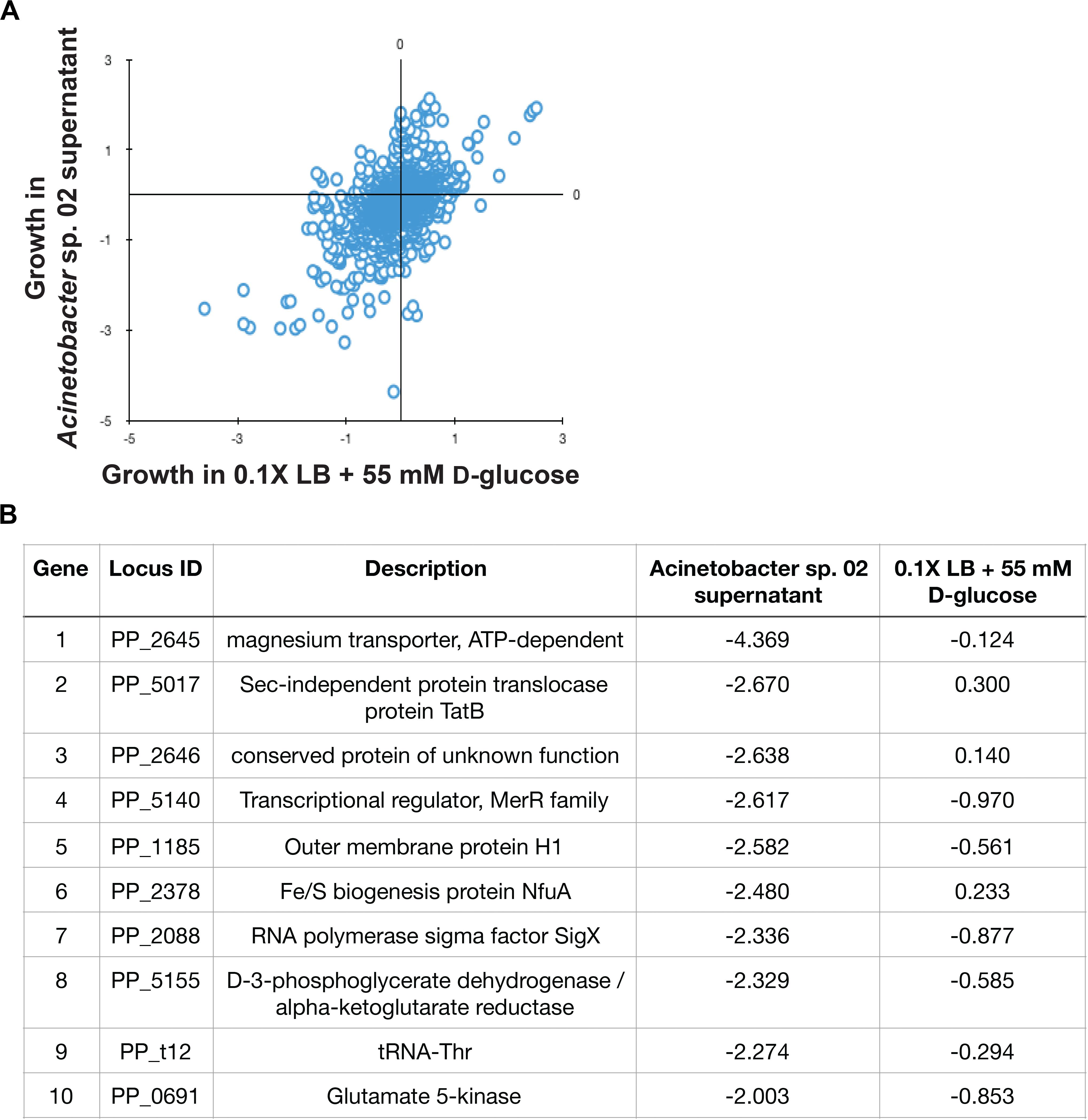
Figure 5. Pseudomonas putida KT2440 RB-TnSeq results. (A) Scatterplot of gene fitness values from P. putida KT2440 grown in Acinetobacter sp. 02 supernatant vs. control media. (B) The top 10 depleted genes in library populations grown in supernatant and relevant log2-fold decreases relative to the population at Time 0.
In the RB-TnSeq data, insertions in 10 genes had a statistically significant fitness defect in the spent media, relative to the control conditions. Interestingly, many of these genes with phenotypes are involved in metal ion transport or metabolism. Both genes in a two-gene operon, PP_2645 and PP_2646, were associated with sensitivity to the Acinetobacter sp. 02 supernatant. PP_2645 shows sequence similarity to ATP-dependent magnesium transporters (Figure 5B) while PP_2646 remains uncharacterized (Figure 5B). Mutants in the metal responsive transcriptional regulator, PP_5140 [merR (Miller et al., 2009)] and a metal-responsive outer membrane protein PP_1185 were also sensitive to this supernatant (Figure 5B). Moreover, we also recovered mutants in PP_2378, a candidate Fe/S related gene (nfuA) (Figure 5B). A comparison with an additional biological replicate of the control condition also implicated two other metal ion related genes, PP_3244 and PP_5139, which may be related to magnesium and cadmium transport. Both full, gene-for-gene comparisons can be found in Supplementary Data Sheet 1. Together, these gene targets suggested that the Acinetobacter sp. 02 supernatant was growth-limiting due to the absence or inactivation of an essential metal cofactor such as magnesium or iron.
Next, we tested whether Acinetobacter sp. 02 could be reducing iron availability in the media, for example by sequestering metal ions with a siderophore like pyoverdine (Drehe et al., 2018). Elemental iron is a limiting and essential nutrient in all cells (Cairo et al., 2006). We used an established genomic analysis software package, antibiotics and secondary metabolite analysis shell (antiSMASH) (Weber et al., 2015), to determine if putative gene clusters encoding bioactive small molecules were present in these microbial genomes. Our bioinformatics analysis (Supplementary Table S1) indicated that several of the representative microbes had the capacity to produce siderophores, and Acinetobacter sp. 02 encodes a putative siderophore that was weakly similar (22% identity) to vicibactin (Wright et al., 2013; Supplementary Table S1). We tested this hypothesis by repeating our concurrent inoculation assay with Acinetobacter sp.02 and several secondary strains, but examined conditions where cells were grown in nutrient-rich, dilute glucose medium (LB) with or without supplemental iron. LB media contains approximately 9.6 μM iron from yeast extract and tryptone. Consistent with our predictions from our functional genomics analysis, iron supplementation of the media restored growth of Brevundimonas sp. and Leifsonia sp. (Figures 6A,B). The effect of iron supplementation was dose-dependent, as 10 and 100 μM FeCl3 were sufficient to improve Brevundimonas sp. and Leifsonia sp. growth in the presence of Acinetobacter sp. 02, but 1 μM FeCl3 was not (Supplementary Figure S5).
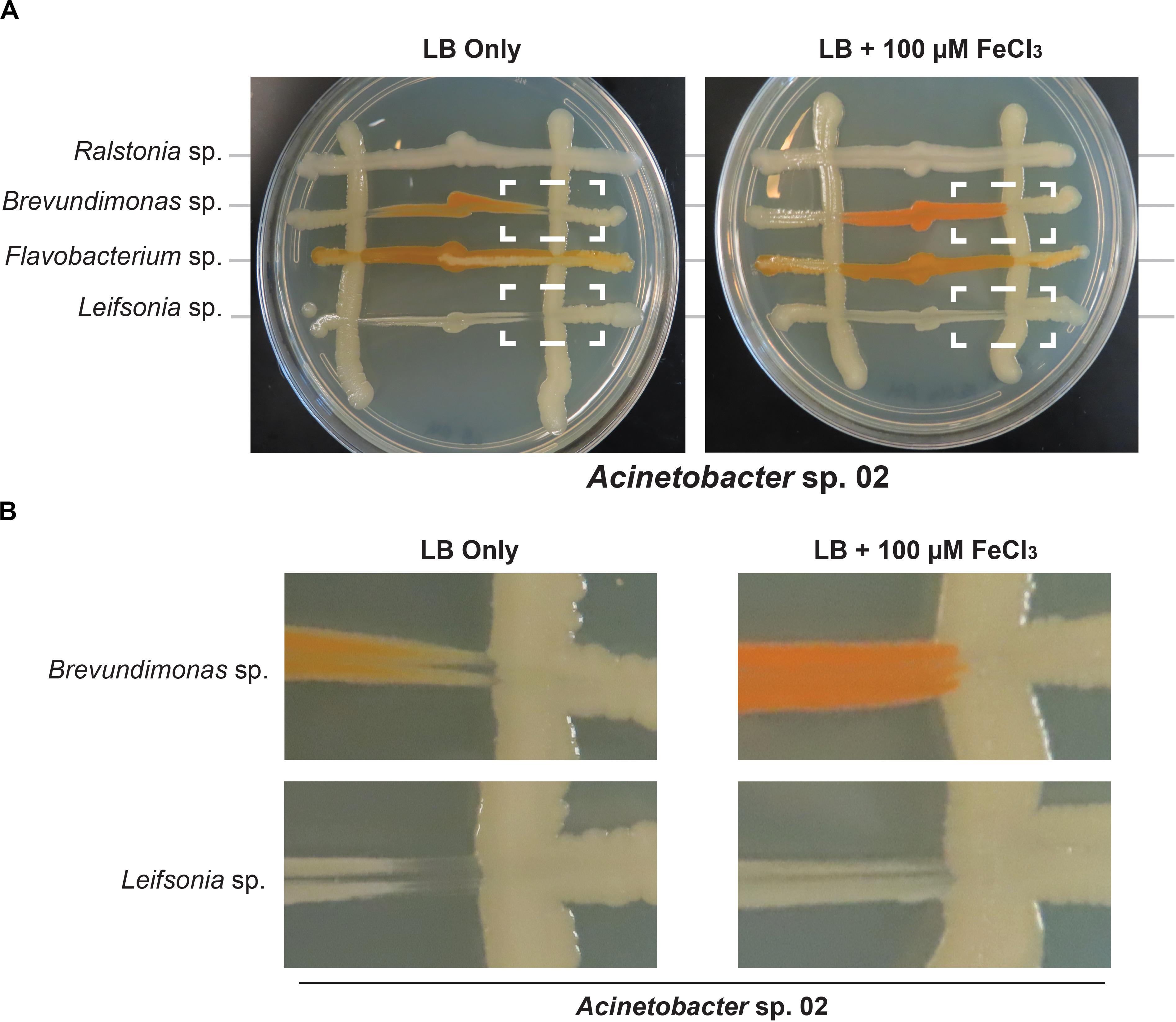
Figure 6. Iron supplementation in Acinetobacter sp. 02 concurrent interaction assay. (A) Acinetobacter colonies were streaked vertically on LB with or without 100 μM supplemental FeCl3 followed by the perpendicular inoculation of the secondary species indicated. Dashed boxes indicate interactions that change depending on iron supplementation. n ≥ 3. (B) A magnified view of the region indicated with dashed white boxes from (A).
Microbes interact with plant roots and each other to develop the specific architecture of the root-associated microbial communities. However, the relationship between the root and its associated microbes is complex and remains to be elucidated. In this report we dissect the pairwise interactions which may occur between several representative bacterial members of the Arabidopsis root microbiome. We tested predictions generated from analyzing the behavior of a proxy soil microbe in response to a complex spent growth media using a systems biology approach. This analysis generated predictive insights on the potential growth inhibitory molecule in the Acinetobacter sp. 02 supernatant, which in turn helped our understanding of microbial interactions between two genetically unmodified rhizobacterial species. From this analysis, we provide evidence that competition for iron in the culture media can lead to physiological changes in Acinetobacter sp. 02 such that it acts to limit competition from other microbes in its vicinity. Our work sheds light on the context by which individual microbes may respond to the specialized nutrient cues from the environment near the root, as the broad growth inhibitory activity of Acinetobacter sp. 02 is dependent on specific growth conditions. Moreover, the founder effects we observed in this study could confound higher degree microbial interaction studies if the order of species arrival in an environment is not carefully controlled.
Competition for essential metals beyond iron represents a general mechanism likely to affect rhizobiome assembly. Understanding the microbial response to extracellular small molecules is an active field of research (Chandrangsu et al., 2017); extending this analysis to plant-microbial communities has the potential to help reveal a greater understanding of complex interactions. Both plant and animal hosts have been observed to engage in nutritional immunity, wherein hosts sequester essential metals in order to limit the growth of potential opportunistic pathogens. Therefore, the ability of these bacteria to compete for metals, both in relation to their host and other microbes, could be general mechanisms that influence host colonization. For instance, iron and zinc homeostasis are essential for colonization of mammalian host tissues by the pathogen Acinetobacter baumannii (Hood et al., 2012; Lonergan et al., 2019). Similarly, root-associated Acinetobacter sp. have been observed to sequester metals such as copper (Rojas-Tapias et al., 2012, 2014), thereby affecting plant growth. In our system, we deduced that iron was a key driver of microbial antagonism; iron has been shown to be a key factor in shaping both plant exudates and the microbiome (Butaitë et al., 2017; Voges et al., 2019). If we consider the context of the root, Arabidopsis roots secrete coumarins under alkaline conditions, where iron is less bioavailable. Coumarins can both mobilize iron and act as selective antimicrobials (Voges et al., 2019). Secretion of coumarins is regulated by the transcription factor MYB72 (Stringlis et al., 2018), the expression of which can be induced by certain (coumarin-resistant) rhizobacteria (Stringlis et al., 2018). With both direct and indirect effects of iron limitation in plant/microbe communities described, identifying interactions that occur between microbes in the community can clarify the interplay between plant root and microbial community.
We had also expected to identify antibiotic compounds as active growth inhibitors in these pairwise microbe interaction assays, as our bioinformatic analysis had predicted the existence of several candidate antibiotic gene clusters in their genomes. Our functional genomics assay of Acinetobacter sp. 02 supernatant with the P. putida RB-TnSeq library did not implicate efflux pump gene clusters, which are a common resistance mechanism to protect against inhibitory small molecules (Mukhopadhyay, 2015; Eng et al., 2018). As the microbes tested in this study are representative of a synthetic model microbiome, we speculate that activation of such gene clusters would require the presence of a pathogenic or otherwise invasive microbial species. Introducing chemical or physical stressors, such as changes in temperature or humidity, DNA damaging agents, or plant hormones, could reveal new interactions between otherwise stable populations.
The methodology developed in this study enabled the examination of interactions between relevant rhizobacterial strains and revealed the role of nutrient limitation. This approach can be expanded to a much larger number of microbes and also higher order (greater than pairs) interactions. While metagenomic sequencing has identified the correlating microbial association networks present in the plant-microbe holobiont (Nguyen and Bruns, 2015; Agler et al., 2016; Marupakula et al., 2016), our study could provide the evidence to establish causal relationships that have been implicated in these high-throughput studies.
Rhizobacterial strains (Table 1) and P. putida KT2440 were maintained in glycerol stocks stored at −80°C. Microbes were routinely cultivated on lysogeny broth (LB; 10 g/L tryptone, 5 g/L yeast extract, and 5 g/L NaCl) with 10 g/L agar. For nutrient-rich microbe-microbe interaction experiments, 1× LB was used with or without supplementation with 10 g/L glucose. For nutrient-poor experiments, 0.1× LB was prepared by dilution with sterile water, then supplemented with glucose (final concentration: 1 g/L tryptone, 0.5 g/L yeast extract, 0.5 g/L NaCl, 10 g/L glucose). All reagents were purchased from BD Biosciences (San Jose, CA, United States) and were of molecular biology grade. Single colonies were obtained by streaking the desired microbe onto LB agar and incubating for 1–3 days at 30°C, depending on the species. Liquid cultures were inoculated using single colonies from these plates. To verify taxonomy, strain identification was confirmed by analysis of 16S ribosomal sequences using the following two primers for amplification followed by Sanger sequencing: 27F: 5′-AGAGTTTGATCMTGGCTCAG-3′; 1510R: 5′-GGTTACCTTGTTACGACTT-3′. Standard protocols for PCR were followed using Q5 polymerase (New England Biolabs, Ipswich, MA, United States). PCR was performed using manufacturer’s guidelines, and the annealing temperature was set to 50°C for 30 cycles and a 120 s extension step at 72°C. Phylogenetic trees were generated using NCBI taxonomy IDs and visualized using iTOLv4 (Letunic and Bork, 2019).
Microbial genomes in this study were analyzed using bacterial antiSMASH 3.0 (Weber et al., 2015). Genomes were inputted into the algorithm using the appropriate NCBI TaxonID with the following activated parameters: default strictness; KnownClusterBlast; ActiveSiteFinder; SubClusterBlast.
For both the concurrent and staggered inoculation assays, microbe-microbe interactions were characterized based on inhibition of colony formation on an agar plate (Figure 1). Due to the order in which primary and secondary strains were applied to the agar plate, regions were formed for each secondary strain in which plated cells were inoculated alone or mixed with other microbial species (see Figure 2). Microbial growth was inspected every 24 h for the appearance of colonies or when many viable cells were present, bacterial lawns. If both microbes showed similar colony formation when comingled or free from the presence of a second microbe, there was no observed interaction. Where we observed apparent reductions in growth of one or both microbes, these interactions were classified as inhibitory. We further defined a category for morphological changes wherein both microbes could grow in each other’s presence, but had a change in colony or lawn formation. With few exceptions, plate-based assays were repeated with >3 biological replicates over many weeks, using different batches of prepared solid agar media. Experiments were typically started using single colonies streaked out from glycerol stocks no longer than 5 days prior.
Agar (1% w/v) plates were prepared containing either 1× or 0.1× LB + 55 mM D-glucose as previously described (Rigali et al., 2008; Nguyen et al., 2011). A representative plate is shown in Figure 2. The concentration of iron in LB was calculated using specifications provided in the “Bionutrients Technical Manual Vol. 3” supplied by the manufacturer (BD Biosciences). For assays testing the role of iron availability, plates were supplemented with 1, 10, or 100 μM FeCl3. Single colonies of a candidate microbe, designated the primary species, were first streaked along a plate in two parallel, vertical lines. Immediately afterward, colonies from 4 to 6 secondary candidate microbes were blotted at a small point roughly equidistant from each streak of the primary species. A sterile toothpick was then used to streak the secondary species in a line perpendicular to the primary species such that they intersect and mix. Plates were then incubated at 30°C, imaged daily for 1–3 days using a Canon 550D camera, and images were saved as JPEG files for further analysis. The brightness was adjusted uniformly to maximize visibility. This experimental design allowed for observation of both direct cell-cell contact, as well as effects that happen at a distance. Antagonism between microbes was defined as a visible zone of growth inhibition in one or both species as a result of their co-culture.
A sterile 0.44-μm nitrocellulose membrane (1 × 5 cm) was applied to one half of the agar plate, prepared as described above. The primary species was inoculated vertically along the plate, with one streak on top of this membrane, and one directly onto the agar surface. Plates were incubated for 2 days at 30°C, after which the nitrocellulose membrane was removed along with any associated bacteria. At this point, the inoculation of secondary species, imaging, and analysis proceeded as in the concurrent growth assays described above.
To test if a growth-inhibitory compound was present in liquid culture media, Acinetobacter sp. 02 was inoculated into 5 mL of 0.1× LB + 55 mM D-glucose and grown for 3 days (200 RPM, 30°C). Spent medium was obtained by centrifuging the saturated culture and filtering the supernatant at 0.22 μm. Spent medium was kept at room temperature until use, which was no longer than 1 month. Either WT P. putida KT2440 or a P. putida KT2440 pooled transposon library [described in Rand et al. (2017)] was inoculated into this spent medium. Cultures were incubated at 30°C shaking at 200 RPM and samples for fitness analysis were taken 3 h post-inoculation.
A 1 mL aliquot of the P. putida RB-TnSeq library described previously was used to inoculate 25 mL of 0.1× LB + 55 mM D-glucose in a 250 mL baffled flask and grown, shaking at 200 RPM, at 30°C overnight. Spent liquid medium from Acinetobacter sp.02 was generated by growing Acinetobacter sp.02 in 25 mL 0.1× LB + 55 mM D-glucose for 3 days in a baffled shake flask at 200 RPM. After 3 days of growth, the culture was pelleted by centrifugation at 4000 rcf for 10 min, and the supernatant was recovered by sterile filtration through a 0.45 μm filter. The P. putida RB-TnSeq pooled mutant library was inoculated into either 0.1× LB + 55 mM D-glucose or the spent medium. Samples were taken as “Time 0”, and the pre-culture was used to inoculate 700 μL of 100% Acinetobacter sp.02 spent medium or control medium per well of a 48-well plate. Plates were sealed with a gas-permeable membrane and incubated at 30°C overnight, shaking at 200 RPM. Undiluted Acinetobacter sp. 02 spent media used here does not allow for the growth of P. putida KT2440; therefore, after a 24 h incubation we back-diluted the inoculated spent media 1:10 into fresh 0.1× LB + D-glucose in a 48-well plate. After an overnight outgrowth samples were pelleted and frozen at −80°C for barcode sequencing. We performed DNA barcode sequencing as previously described (Wetmore et al., 2015). The fitness of a strain is defined here as the normalized log2 ratio of barcode reads in the experimental sample to barcode reads in the time zero sample. The fitness of a gene is defined here as the weighted average of strain fitness for insertions in the central 10 to 90% of the gene.
All datasets generated for this study are included in the article/Supplementary Material.
AM, JM, TE, and RH conceived the study. RH, BW, and UM collected and analyzed the data. TE and AM helped to organize the data and project. JB, JC, MB, and AD contributed valuable reagents and technical expertise. JB, JM, MB, and AM provided funding. TE and RH wrote the first draft of the manuscript and prepared figures. All authors took part in editing the manuscript, read and approved the final version.
JB was employed by Arva Intelligence Inc.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was funded by a Laboratory Directed Research and Development (LDRD) grant at LBNL. TE, AM, and JM were also funded as part of the DOE Joint BioEnergy Institute (http://www.jbei.org) supported by the United States Department of Energy, Office of Science, through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the United States Department of Energy. JC was supported by the National Institute of General Medical Sciences under Award Number SC3GM096943.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01742/full#supplementary-material
FIGURE S1 | Phylogeny of rhizobacteria used in this study. Genus-level description of selected rhizobacteria and their representation within major bacterial phyla. The bold, solid lines in the phylogenetic tree indicate lineages represented by the isolates used in this study. The phylogenetic tree was generated using NCBI taxonomy IDs and visualized using iTOL V4.
FIGURE S2 | Representative images of Chryseobacterium sp. grown in co-culture with other rhizobacteria. Chryseobacterium biomass changes color and opacity when co-cultured with A. rhizogenes or Arthrobacter sp., but not when co-cultured with itself.
FIGURE S3 | Representative images of Ralstonia sp. growing along the biomass of other rhizobacteria.
FIGURE S4 | Pseudomonas putida KT2440 does not lose viability in Acinetobacter sp. 02 supernatant. A log phase culture of P. putida KT2440 was back-diluted into fresh 1/10× LB + glucose, or spent liquid media from Acinetobacter sp. 02. After 3 h of growth at 30°C, cultures were serially diluted and plated onto LB solid agar media to assess colony formation.
FIGURE S5 | The effect of iron on interactions between Acinetobacter sp. 02 and other rhizobacteria is dose dependent. n ≥ 3.
TABLE S1 | Summary of potential secondary metabolites produced by microbes used in this study as identified by antiSMASH.
antiSMASH, antibiotics and secondary metabolite analysis shell; JBEI, Joint BioEnergy Institute; JGI, Joint Genome Institute; LB, lysogeny broth; RB-TnSeq, random barcode transposon sequencing.
Abelson, J., Simon, M., Guthrie, C., and Fink, G. (1990). Guide to Yeast Genetics and Molecular Biology, Vol194, 1st Edn. Amsterdam: Elsevier.
Agler, M. T., Ruhe, J., Kroll, S., Morhenn, C., Kim, S.-T., Weigel, D., et al. (2016). Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol. 14:e1002352. doi: 10.1371/journal.pbio.1002352
Alteri, C. J., and Mobley, H. L. T. (2016). The versatile type VI secretion system. Microbiol. Spectr. 4:10.1128/microbiolsec.VMBF–0026–2015. doi: 10.1128/microbiolspec.VMBF-0026-2015
Bai, Y., Müller, D. B., Srinivas, G., Garrido-Oter, R., Potthoff, E., Rott, M., et al. (2015). Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528, 364–369. doi: 10.1038/nature16192
Butaitë, E., Baumgartner, M., Wyder, S., and Kümmerli, R. (2017). Siderophore cheating and cheating resistance shape competition for iron in soil and freshwater Pseudomonas communities. Nat. Commun. 8:414. doi: 10.1038/s41467-017-00509-4
Cairo, G., Bernuzzi, F., and Recalcati, S. (2006). A precious metal: iron, an essential nutrient for all cells. Genes Nutr. 1, 25–39. doi: 10.1007/BF02829934
Chandrangsu, P., Rensing, C., and Helmann, J. D. (2017). Metal homeostasis and resistance in bacteria. Nat. Rev. Microbiol. 15, 338–350. doi: 10.1038/nrmicro.2017.15
Chaparro, J. M., Badri, D. V., Bakker, M. G., Sugiyama, A., Manter, D. K., and Vivanco, J. M. (2013). Root exudation of phytochemicals in Arabidopsis follows specific patterns that are developmentally programmed and correlate with soil microbial functions. PLoS One 8:e55731. doi: 10.1371/journal.pone.0055731
Chu, H., Khosravi, A., Kusumawardhani, I. P., Kwon, A. H. K., Vasconcelos, A. C., Cunha, L. D., et al. (2016). Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science 352, 1116–1120. doi: 10.1126/science.aad9948
Drehe, I., Simonetti, E., and Ruiz, J. A. (2018). Contribution of the Siderophores Pyoverdine and Enantio-Pyochelin to Fitness in Soil of Pseudomonas protegens Pf-5. Curr. Microbiol. 75, 1560–1565. doi: 10.1007/s00284-018-1560-7
Eng, T., Demling, P., Herbert, R. A., Chen, Y., Benites, V., Martin, J., et al. (2018). Restoration of biofuel production levels and increased tolerance under ionic liquid stress is enabled by a mutation in the essential Escherichia coli gene cydC. Microb. Cell Fact. 17:159. doi: 10.1186/s12934-018-1006-8
Hassani, M. A., Durán, P., and Hacquard, S. (2018). Microbial interactions within the plant holobiont. Microbiome 6:58. doi: 10.1186/s40168-018-0445-0
Herbert, R. A., Eng, T., Martinez, U., Wang, B., Langley, S., Wan, K., et al. (2019). Rhizobacteria mediate the phytotoxicity of a range of biorefinery-relevant compounds. Environ. Toxicol. Chem. 38, 1911–1922. doi: 10.1002/etc.4501
Hood, M. I., Mortensen, B. L., Moore, J. L., Zhang, Y., Kehl-Fie, T. E., Sugitani, N., et al. (2012). Identification of an Acinetobacter baumannii zinc acquisition system that facilitates resistance to calprotectin-mediated zinc sequestration. PLoS Pathog. 8:e1003068. doi: 10.1371/journal.ppat.1003068
Jackman, C. M., Deans, K. W., Forney, L. J., and Lin, X. N. (2019). Microdroplet co-cultivation and interaction characterization of human vaginal bacteria. Integr. Biol. 11, 69–78. doi: 10.1093/intbio/zyz006
Joshi, F., Archana, G., and Desai, A. (2006). Siderophore cross-utilization amongst rhizospheric bacteria and the role of their differential affinities for Fe3+ on growth stimulation under iron-limited conditions. Curr. Microbiol. 53, 141–147. doi: 10.1007/s00284-005-0400-8
Kawasaki, A., Donn, S., Ryan, P. R., Mathesius, U., Devilla, R., Jones, A., et al. (2016). Microbiome and exudates of the root and rhizosphere of brachypodium distachyon, a model for wheat. PLoS One 11:e0164533. doi: 10.1371/journal.pone.0164533
Letunic, I., and Bork, P. (2019). Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259. doi: 10.1093/nar/gkz239
Levy, A., Salas Gonzalez, I., Mittelviefhaus, M., Clingenpeel, S., Herrera Paredes, S., Miao, J., et al. (2017). Genomic features of bacterial adaptation to plants. Nat. Genet. 50, 138–150. doi: 10.1038/s41588-017-0012-9
Liu, X., Liu, Z., Dziulko, A. K., Li, F., Miller, D., Morabito, R. D., et al. (2019). iSeq 2.0: a modular and interchangeable toolkit for interaction screening in yeast. Cell Syst. 8, 338.e8–344.e8. doi: 10.1016/j.cels.2019.03.005
Lonergan, Z. R., Nairn, B. L., Wang, J., Hsu, Y.-P., Hesse, L. E., Beavers, W. N., et al. (2019). An Acinetobacter baumannii, zinc-regulated peptidase maintains cell wall integrity during immune-mediated nutrient sequestration. Cell Rep. 26, 2009.e6–2018.e6. doi: 10.1016/j.celrep.2019.01.089
Lundberg, D. S., Lebeis, S. L., Paredes, S. H., Yourstone, S., Gehring, J., Malfatti, S., et al. (2012). Defining the core Arabidopsis thaliana root microbiome. Nature 488, 86–90. doi: 10.1038/nature11237
Marupakula, S., Mahmood, S., and Finlay, R. D. (2016). Analysis of single root tip microbiomes suggests that distinctive bacterial communities are selected by Pinus sylvestris roots colonized by different ectomycorrhizal fungi. Environ. Microbiol. 18, 1470–1483. doi: 10.1111/1462-2920.13102
Mendes, L. W., Raaijmakers, J. M., de Hollander, M., Mendes, R., and Tsai, S. M. (2018). Influence of resistance breeding in common bean on rhizosphere microbiome composition and function. ISME J. 12, 212–224. doi: 10.1038/ismej.2017.158
Meng, J., Zhong, Z., and Qian, P.-Y. (2018). Paenialvin A-D, four peptide antibiotics produced by Paenibacillus alvei DSM 29. J. Antibiot. 71, 769–777. doi: 10.1038/s41429-017-0001-3
Miller, C. D., Pettee, B., Zhang, C., Pabst, M., McLean, J. E., and Anderson, A. J. (2009). Copper and cadmium: responses in Pseudomonas putida KT2440. Lett. Appl. Microbiol. 49, 775–783. doi: 10.1111/j.1472-765X.2009.02741.x
Molina, L. (2000). Survival of Pseudomonas putida KT2440 in soil and in the rhizosphere of plants under greenhouse and environmental conditions. Soil Biol. Biochem. 32, 315–321. doi: 10.1016/S0038-0717(99)00156-X
Mukhopadhyay, A. (2015). Tolerance engineering in bacteria for the production of advanced biofuels and chemicals. Trends Microbiol. 23, 498–508. doi: 10.1016/j.tim.2015.04.008
Mutha, N. V. R., Mohammed, W. K., Krasnogor, N., Tan, G. Y. A., Wee, W. Y., Li, Y., et al. (2019). Transcriptional profiling of coaggregation interactions between Streptococcus gordonii and Veillonella parvula by Dual RNA-Seq. Sci. Rep. 9:7664. doi: 10.1038/s41598-019-43979-w
Nester, E. W. (2014). Agrobacterium: nature’s genetic engineer. Front. Plant Sci. 5:730. doi: 10.3389/fpls.2014.00730
Nguyen, D., Joshi-Datar, A., Lepine, F., Bauerle, E., Olakanmi, O., Beer, K., et al. (2011). Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334, 982–986. doi: 10.1126/science.1211037
Nguyen, N. H., and Bruns, T. D. (2015). The microbiome of pinus muricata ectomycorrhizae: community assemblages, fungal species effects, and burkholderia as important bacteria in multipartnered symbioses. Microb. Ecol. 69, 914–921. doi: 10.1007/s00248-015-0574-y
Okubo, A., Matsusaka, M., and Sugiyama, S. (2016). Impacts of root symbiotic associations on interspecific variation in sugar exudation rates and rhizosphere microbial communities: a comparison among four plant families. Plant Soil 399, 345–356. doi: 10.1007/s11104-015-2703-2
Price, M. N., Ray, J., Iavarone, A. T., Carlson, H. K., Ryan, E. M., Malmstrom, R. R., et al. (2019). Oxidative pathways of deoxyribose and deoxyribonate catabolism. mSystems 4:e00297-18. doi: 10.1128/mSystems.00297-18
Rand, J. M., Pisithkul, T., Clark, R. L., Thiede, J. M., Mehrer, C. R., Agnew, D. E., et al. (2017). A metabolic pathway for catabolizing levulinic acid in bacteria. Nat. Microbiol. 2, 1624–1634. doi: 10.1038/s41564-017-0028-z
Rigali, S., Titgemeyer, F., Barends, S., Mulder, S., Thomae, A. W., Hopwood, D. A., et al. (2008). Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep. 9, 670–675. doi: 10.1038/embor.2008.83
Rojas-Tapias, D. F., Bonilla, R., and Dussán, J. (2014). Effect of inoculation and co-inoculation of Acinetobacter sp. RG30 and Pseudomonas putida GN04 on Growth, fitness, and copper accumulation of maize (Zea mays). Water Air Soil Pollut. 225:2232. doi: 10.1007/s11270-014-2232-2
Rojas-Tapias, D. F., Bonilla, R. R., and Dussán, J. (2012). Effect of inoculation with plant growth-promoting bacteria on growth and copper uptake by sunflowers. Water Air Soil Pollut. 223, 643–654. doi: 10.1007/s11270-011-0889-3
Shaheen, M., Li, J., Ross, A. C., Vederas, J. C., and Jensen, S. E. (2011). Paenibacillus polymyxa PKB1 produces variants of polymyxin B-type antibiotics. Chem. Biol. 18, 1640–1648. doi: 10.1016/j.chembiol.2011.09.017
Stringlis, I. A., Yu, K., Feussner, K., de Jonge, R., Van Bentum, S., Van Verk, M. C., et al. (2018). MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc. Natl. Acad. Sci. U.S.A. 115, E5213–E5222. doi: 10.1073/pnas.1722335115
Thompson, M. G., Blake-Hedges, J. M., Cruz-Morales, P., Barajas, J. F., Curran, S. C., Eiben, C. B., et al. (2019). Massively parallel fitness profiling reveals multiple novel enzymes in Pseudomonas putida lysine metabolism. mBio 10:e02577-18. doi: 10.1128/mBio.02577-18
van der Meij, A., Worsley, S. F., Hutchings, M. I., and van Wezel, G. P. (2017). Chemical ecology of antibiotic production by actinomycetes. FEMS Microbiol. Rev. 41, 392–416. doi: 10.1093/femsre/fux005
Voges, M. J. E. E. E., Bai, Y., Schulze-Lefert, P., and Sattely, E. S. (2019). Plant-derived coumarins shape the composition of an Arabidopsis synthetic root microbiome. Proc. Natl. Acad. Sci. U.S.A. 116, 12558–12565. doi: 10.1073/pnas.1820691116
Vorholt, J. A. (2012). Microbial life in the phyllosphere. Nat. Rev. Micro 10, 828–840. doi: 10.1038/nrmicro2910
Weber, T., Blin, K., Duddela, S., Krug, D., Kim, H. U., Bruccoleri, R., et al. (2015). antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 43, W237–W243. doi: 10.1093/nar/gkv437
Wetmore, K. M., Price, M. N., Waters, R. J., Lamson, J. S., He, J., Hoover, C. A., et al. (2015). Rapid quantification of mutant fitness in diverse bacteria by sequencing randomly bar-coded transposons. mBio 6:e00306-15. doi: 10.1128/mBio.00306-15
Wright, W., Little, J., Liu, F., and Chakraborty, R. (2013). Isolation and structural identification of the trihydroxamate siderophore vicibactin and its degradative products from Rhizobium leguminosarum ATCC 14479 bv. trifolii. Biometals 26, 271–283. doi: 10.1007/s10534-013-9609-3
Keywords: microbe-macroorganism interaction, iron depletion, RB-TnSeq, Pseudomonas putida, Acinetobacter, rhizobiota, composition and function
Citation: Eng T, Herbert RA, Martinez U, Wang B, Chen JC, Brown JB, Deutschbauer AM, Bissell MJ, Mortimer JC and Mukhopadhyay A (2020) Iron Supplementation Eliminates Antagonistic Interactions Between Root-Associated Bacteria. Front. Microbiol. 11:1742. doi: 10.3389/fmicb.2020.01742
Received: 12 February 2020; Accepted: 03 July 2020;
Published: 22 July 2020.
Edited by:
Marina G. Kalyuzhanaya, San Diego State University, United StatesReviewed by:
Naresh Singhal, The University of Auckland, New ZealandCopyright © 2020 Eng, Herbert, Martinez, Wang, Chen, Brown, Deutschbauer, Bissell, Mortimer and Mukhopadhyay. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jenny C. Mortimer, amNtb3J0aW1lckBsYmwuZ292; Aindrila Mukhopadhyay, YW11a2hvcGFkaHlheUBsYmwuZ292
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.