
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 22 July 2020
Sec. Systems Microbiology
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.01614
Plant based by-products (BP) produced from food and bioethanol industries are human inedible, but can be recycled into the global food chain by ruminant livestock. However, limited data is available on the methanogenesis potential associated with supplementing a solely BP formulated concentrate to a pastoral based diet. Therefore the objective of this in vitro study was to investigate the effects of BP inclusion rate (in a formulated concentrate) to a pasture based diet on dietary digestibility, rumen fermentation patterns, methane production and the prokaryotic microbial community composition. Diets consisted of perennial ryegrass and one of two supplementary concentrates, formulated to be isonitrogenous (16% CP) and isoenergetic (12.0 MJ/ME/kg), containing either 35% BP, barley and soybean meal (BP35) or 95% BP (BP95) offered on a 50:50 basis, however, starch, NDF and fat content varied. The BPs, included in equal proportions on a DM basis, were soyhulls, palm kernel expeller and maize dried distillers grains. The BP35 diet had greater (P < 0.05) digestibility of the chemical constituents DM, OM, CP, NDF, ADF. Greater total VFA production was seen in the BP35 diet (P < 0.05). Daily methane production (mmol/day; +22.7%) and methane output per unit of total organic matter digested (MPOMD; +20.8%) were greatest in the BP35 diet (P < 0.01). Dietary treatment influenced microbial composition (PERMANOVA; P = 0.023) with a greater relative abundance of Firmicutes (adj P < 0.01) observed in the BP35. The Firmicutes:Bacteroidetes ratio was significantly reduced in the BP95 diet (P < 0.01). The relative proportions of Proteobacteria (adj P < 0.01), Succinivibrionaceae (adj P < 0.03) and Succinivibrio (adj P = 0.053) increased in the BP95 diet. The abundance of Proteobacteria was found to be negatively associated with daily methane production (rs, −0.71; P < 0.01) and MPOMD (rs, −0.65; P < 0.01). Within Proteobacteria, the relationship of methane production was maintained with the mean abundance of Succinivibrio (rs, −0.69; P < 0.01). The abundance of the Firmicutes phyla was found to be positively correlated with both daily methane production (rs, 0.79; P < 0.001) and MPOMD (rs, 0.75; P < 0.01). Based on in vitro rumen simulation data, supplementation of an exclusively BP formulated concentrate was shown to reduce daily methane output by promoting a favorable alteration to the rumen prokaryotic community.
The processing of crops for the global food, oil and ethanol industries generates a source of residual by-product (BP) plant matter which can be utilized as animal feed (Mirzaei-Aghsaghali and Maheri-Sis, 2008). Such BPs contain little economic value as edible food for human consumption due to safety, quality and digestibility considerations (Grasser et al., 1995). However, BPs contain digestible nutrients that can be used as feed by the livestock industry. In particular, ruminant animals, through microbial fermentation, have the ability to digest cellulose and utilize highly fibrous feed sources (Sasson et al., 2017) which is often typical of such BPs. As a result, ruminants have the unique ability to convert these human inedible low value raw ingredients, into high quality animal dairy and meat proteins for human consumption.
Grazed pasture is usually the most economical feed source for ruminants in temperate regions such as Ireland (Finneran et al., 2012) although strategic concentrate usage is common throughout the ruminant production cycle (O’Brien et al., 2018) in order to overcome deficiencies in nutrient supply. Concentrates are utilized to supplement the diet of dairy cows during bad weather conditions and poor grass growth in order to ensure adequate energy intake to support milk production and quality (McGrath, 2013). Feeding concentrates is also a key component of beef production systems, especially during the winter finishing period (McGee, 2005). Currently, near 10% of the global area dedicated to cereal production, is used to produce grain for ruminant diets (Mottet et al., 2017). The inefficiency with which human edible feed is converted into ruminant derived protein and energy (Wilkinson, 2011) coupled with the need to feed a growing global population, has therefore brought into question the strategy of supplementing ruminant diets with human edible sources of feed.
Replacing human edible cereals with BP feeds, offers a more sustainable and efficient use of supplemented feed in pastoral based ruminant production systems (Wilkinson, 2011). However, the nutritional value between individual BPs can vary (Lee et al., 2003; Kim et al., 2013) and therefore must be accounted for when considering their inclusion in dietary formulations. Nonetheless, members of our group have shown the formulation of concentrates, formulated solely from dried distillers grains and solubles (DDGS), palm kernel expeller (PKE) and soyhulls (SH) supplemented to a pasture based dairy system, to produce a similar level of milk output, to grain formulated concentrates (Whelan et al., 2016; Condren et al., 2019). However, data on the effects of this BP concentrate on the rumen microbiome and methane production is needed to further assess the sustainability of the formulation.
Both the microbial population residing in the rumen (Morgavi et al., 2010; Henderson et al., 2015; Tapio et al., 2017) and enteric methane production (Beauchemin et al., 2008, 2020; Martin et al., 2010) has been shown to be altered by the composition of the diet offered to ruminants. Indeed, certain BP feeds are known to be lower in starch and higher in fiber than conventional cereal and oilseed grains (Ertl et al., 2015) leading to shifts in ruminal fermentation patterns and digestion end products. Thus, this highlights the importance of investigating, in tandem, the effects of replacing grain based concentrates with formulations derived from BPs, on the rumen microbiome and methane production.
On an individual BP bases, the high lipid content of DDGS have been shown to reduce methane output in beef and dairy cattle, in comparison to cereal and soybean meal based diets (McGinn et al., 2009; Benchaar et al., 2013; Hünerberg et al., 2013). Indeed, the lipid proportion of DDGS has been shown to decrease the abundance of members of the fibrolytic microbial community in some (Ramirez et al., 2012; Castillo-Lopez et al., 2014) but not all studies (Castillo-Lopez et al., 2017). Additionally, PKE has shown methane abatement potential through reductions to the methanogen population in vivo (Abubakr et al., 2014) and production of methane in vitro (Lee et al., 2003; Kim et al., 2013).
Therefore, this experiment was designed to evaluate, the effects of supplementing a solely BP formulated concentrate, with increased dietary fiber and fat, on rumen fermentation parameters, methane production and the microbial community composition with perennial ryegrass pasture as the main forage source in vitro.
All procedures described in this experiment were approved by the animal research ethics committee (AREC) at University College Dublin (UCD) and conducted under the European directive 2010/63/EU and S.I. No. 543 of 2012. Each person who carried out procedures on experimental animals, during the course of this experiment, was licensed to do so by means of individual authorisation from the Health Product Regulatory Authority (HPRA). Two rumen simulation technique (RUSITEC) systems with eight vessels/system (Sanshin Industrial Co. Ltd., Yokohama, Japan), were used to simulate the rumen environment. Each fermenter had a nominal volume of 850 ml with the general incubation procedure described by Czerkawski and Breckenridge (1977).
Dietary treatments consisted of a perennial ryegrass (Lolium perenne) based pasture (PRG) and one of two supplementary concentrates containing either barley, soybean meal and 35% BP (BP35) or solely 95% BP (BP95) offered on a 50:50 basis (Table 1). The BPs, included in equal proportions on a dry matter (DM) basis, were SHs, PKE and maize DDGS. The PRG component of each treatment was harvested in July 2015 from the grazing platform at UCD Lyons Estate Farm, Celbridge, Kildare, Ireland (53°17 ′56 ′ ′N, 6°32 ′18 ′ ′W). Samples were cut to a height of 4 cm from the same pasture using a handheld shears (Gardena Accu 90; Gardena GmbH, Ulm, DE) and subsequently stored at −20°C. Concentrates were formulated to be isonitrogenous [16% crude protein (CP)] and isoenergetic (12.0 MJ/ME/kg), however, starch, NDF and fat content varied. The BP35 concentrate was targeted to contain 280 g of starch and 270 g of neutral detergent fiber (NDF) per kg of DM with the BP95 containing 30 g of starch and 500 g of NDF per kg of DM, respectively. A crude fat content, based on ether extract (EE), was targeted at 2.51 and 5.6% for the BP35 and BP95 diets, respectively.
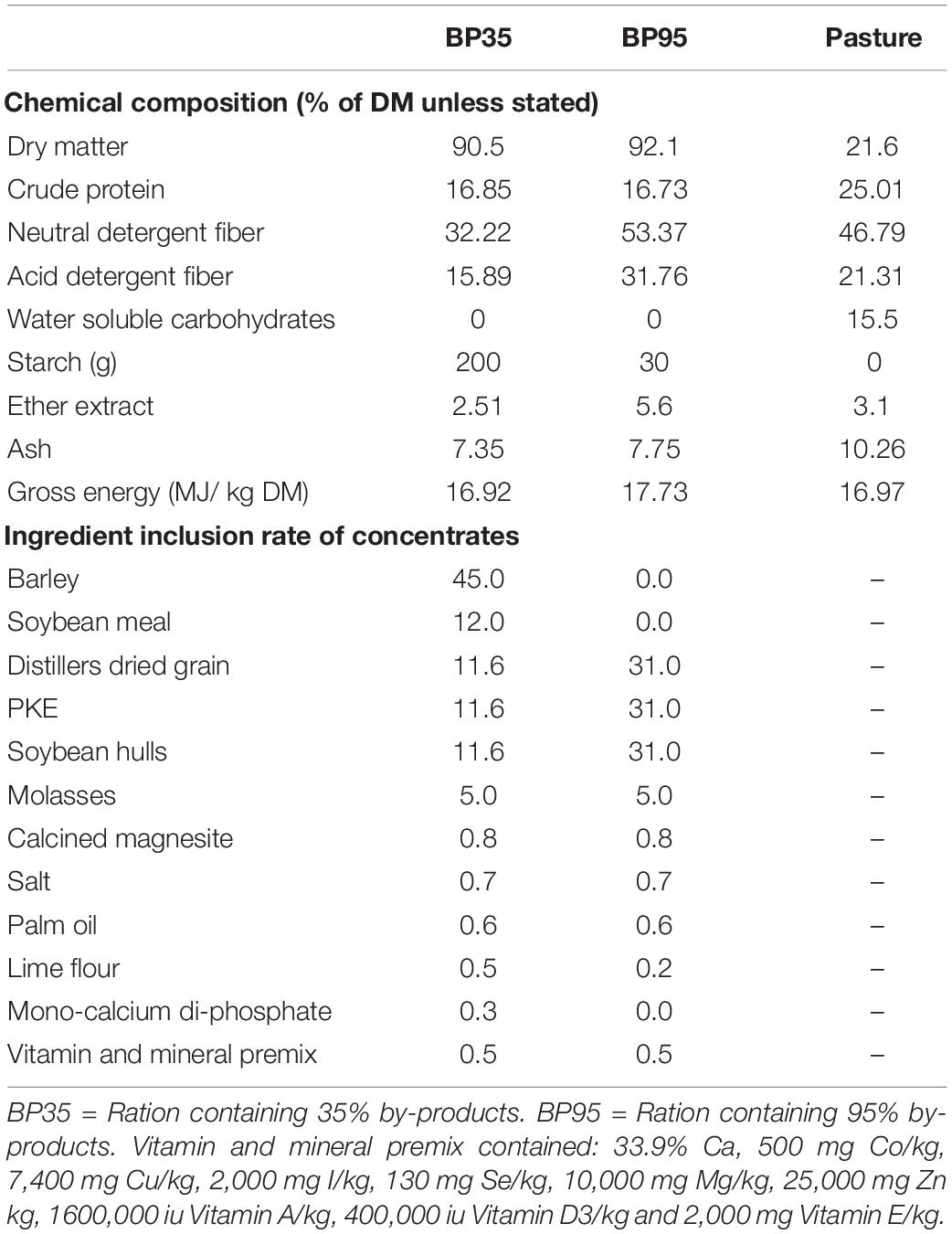
Table 1. Chemical composition of concentrates and grass along with ingredient inclusion rate of concentrates fed during the experiment.
Dietary treatments were randomly allocated to sixteen fermentation vessels and added to each vessel in nylon bags (100-μm pore size). Samples of PRG were removed from storage (−20°C) and immediately chopped to 1–2 cm in length with a bowl chopper before being added to each nylon bag and subsequently being refrozen and stored (−20°C). For each treatment, 10 g DM of either concentrate under investigation and the fresh weight equivalent of 10 g DM of PRG were added to separate nylon bags.
Rumen inoculum was obtained from three ruminally cannulated beef heifers of continental breed crosses, offered grass silage (18% DM, 13.37% CP, 51.5% NDF and 37.39% acid detergent fiber (ADF)) and concentrate (86% DM, 16% CP) in a 50:50 ratio, in order to support estimated maintenance energy requirements (Agriculture Food Research Council, 1993). Rumen fluid collection took place before morning feeding at 09:00.
A single in vitro incubation period of 14 days, using the first 10 days for microbial adaptation and fermentation stabilization (Jaurena et al., 2005) and the last 4 for sampling was implemented using the RUSITEC. On Day 1 of the experiment, solid digesta and rumen fluid were collected from the three ruminally cannulated heifers, with the fluid strained through four layers of cheesecloth. The samples were pooled, flushed with carbon dioxide (CO2) and incubated at 39°C before being transferred to the RUSITEC vessels within 30 min of collection. Each vessel was inoculated with 500 ml of rumen fluid and 350 ml of artificial saliva (McDougall, 1948). Nylon bags containing PRG were removed from storage (−20°C), on the day prior to incubation, and allowed to thaw for 24 h. Treatments were added to each vessel in individual nylon bags (n = 2) containing PRG and the BP concentrate under investigation. In addition, a third nylon bag, containing 70 g of rumen solid digesta, was added to each vessel. This resulted in each vessel containing a total of three nylon bags (PRG, concentrate and rumen digesta) at the beginning of the experiment. Artificial saliva was prepared daily and was continuously infused at a rate of 640 ml/d (dilution rate of 3.33%/h) using the Watson-Marlow 500 series peristaltic pump (Watson-Marlow Fluid Technology Group, Cornwall, United Kingdom). The displaced effluent and fermentation gasses from each vessel were collected into effluent bottles and gas collection bags, respectively. On day 2, each vessel was opened and nylon bags containing rumen digesta solids and the concentrates under investigation were removed, squeezed and washed in artificial saliva. The liquid fractions of the washings were returned to each vessel along with two new nylon bags, containing PRG and the concentrate under examination. Throughout the 14-day period, nylon bags containing PRG were incubated for a 48 h period after which they were replaced by a new nylon bag containing forage. Nylon bags containing either concentrate under investigation were replaced every 24 h by new bags containing the same concentrate. During the adaptation period effluent outflow and gas production were measured, as well as pH of the fermentation vessels.
Dry matter degradation, gas production and outflow of fermentation products were measured on days 11, 12, 13, and 14. Overflow vessels were kept in cold water (2°C) to stabilize fermentation products. A 4 ml sample of outflow liquor, from each vessel, was preserved in 1 ml of 50% trichloroacetic acid (TCA), and stored at −20°C for volatile fatty acid (VFA) and ammonia (NH3-N) analysis.
After removal from fermenters, nylon bags were rinsed with cold water. Feed residues in the nylon bags were washed in a domestic washing machine using the cold rinse cycle in the absence of detergent (30 min) to remove the loosely attached bacteria. The feed residues were dried in a 55°C forced air oven and weighed. Feed dry matter digestibility (DMD) was calculated as the amount of material that had disappeared from the nylon bags after 24 and 48 h of incubation, for concentrates and forage respectively. Chemical composition of the dried incubation residues was determined to calculate digestibility of feed components.
Gas was measured using gas reusable polyethylene bags fitted with one way valves. These were placed on each outflow vessel to measure gas production including methane percentage. Gas volume was quantified by manual expulsion through the dual diaphragm DC-1 dry gas test meter (Sinagawa Corp. Tokyo, Japan) with the percentage of methane estimated using the infra-red ADC SB2000 wall mounted analyser (ADC Gas Analysis Ltd., Hoddeston, United Kingdom) (O’Connor et al., 2019a, b). Daily calibrations of the ADC SB2000 were conducted with a 10% methane span gas.
The DM content (g/kg) of the feed and feed residue samples were determined after drying the samples at 55°C for 72 h in a forced air oven. Dried feed and feed residue samples were then ground in a hammer mill fitted with a 1-mm screen for subsequent chemical analysis (Lab Mill, Christy Turner, Suffolk, United Kingdom). Ash concentrations (g/kg DM) were determined by complete combustion in a muffle furnace (Nabertherm, GmbH, Lilienthal, Germany) at 550°C for 4 h. The nitrogen concentration (g/kg DM) of feed was determined using a LECO FP 528 instrument (Leco Instruments UK Ltd., Stockport, United Kingdom). The nitrogen concentration of the feed was multiplied by 6.25, to determine CP concentrations (g/kg DM). Determination of NDF and ADF concentrations were determined by the method of Van Soest et al. (1991) using the ANKOM220 Fiber Analyzer (ANKOM Technology, Macedon, NY, United States). Grass and concentrate samples were analyzed for NDF with sodium sulphite and with a heat stable amylase included for concentrate samples only. Both NDF and ADF are expressed inclusive of residual ash (g/kg DM). Gross energy was determined on pelletized samples using a bomb calorimeter (Parr Instrument Company, Moline, IL, United States). The EE content was determined using Soxtec instruments (Tecator, Höganäs, SE) and light petroleum ether. The chemical composition of the concentrates and PRG are presented in Table 1. Samples of vessel effluent were thawed for 16 h to 4°C before being centrifuged for 10 min (1600 g; 4°C). A 1 ml sample of supernatant was drawn off and diluted one in five with distilled water (dH2O) and centrifuged for 15 min (1600 g; 4°C). Following this 200 and 250 μl of supernatant was drawn off into separate test tubes for NH3-N and VFA analysis, respectively. Concentrations of NH3-N were determined using the phenol-hypochlorite method of Weatherburn (1967) using a Shimadzu UV/Vis UVmini-1240 spectrophotometer (Shimadzu UK Ltd., Wolverton Mill South, Milton Keynes, United Kingdom). For VFA analysis, the 250 μl of supernatant was diluted with 3.75 ml of dH2O and 1 ml of internal standard (0.5 g 3-metyl-n-valeric acid in one liter of 0.15 M oxalic acid). Following this, diluted VFA samples were centrifuged for 5 min (260 g; 21°C) followed by filtration through a 0.45 μm filter (Cronus Syringe filter PTFE 13 mm; SMI-LabHut Ltd., Maisemore, Gloucester, United Kingdom) into a 4 ml GC vial (Thermo Scientific, Langerwehe, Germany) and stored (−20°C) until analysis was conducted. VFAs were quantified by injecting 1 μl, via an auto sampler, onto a 25 m × 0.53 mm i.d. megabore Varian gas chromatograph (GC) 3800 column, [coating CP-Wax 58 (FFAP) – CB (no. CP7614)] (Varian, Middelburg, Netherlands). An initial injector temperature of 75°C, immediately rose to 95°C, followed by a rate of temperature increase of 3°C/min up to 200°C (held for 50 s). Nitrogen was used as a carrier gas. The pressure of the column was held at 2.3 psi and the column rate was 8.1 ml/min. Estimates of hydrogen (H) recovery within each vessel were calculated based on the concentration of individual VFAs as described by Marty and Demeyer (1973) with the exclusion of hydrogen gas (H2).
On the final day of incubation, prior to opening each vessel, a 15 ml sample of fluid was collected via an aspiration port at the top of each RUSITEC vessel. Fluid samples were collected in individual disposable sterile syringes and deposited in 15 ml falcon tubes. Immediately after collection, samples of vessel effluent were snap frozen in liquid nitrogen and subsequently stored at −80°C. Under liquid nitrogen, samples were ground to a fine powder using a pestle and mortar. Microbial DNA was extracted from approximately 250 mg of ground sample using the repeated bead beating and column purification method (Yu and Morrison, 2004). DNA quality was assessed on 0.8% agarose gels with the concentration of extracted DNA quantified on the Nanodrop 1000 spectrophotometer.
Microbial DNA extractions were also performed on the ZymoBIOMICSTM Microbial Community Standard (MC) (Zymo Research Corp., Irvine, CA, United States) to assess DNA extraction performance. Extractions were performed on the ZymoBIOMICSTM MC in triplicate and were treated as an internal control and subject to the same library preparation and sequencing regime as samples collected from the RUSITEC systems.
Using 12.5 ng of individual microbial DNA obtained from the RUSITEC vessels, 16 amplicon libraries were generated by performing two rounds of PCR amplification as outlined in the Illumina Miseq 16S Sample Preparation Guide with minor modifications to cycle length, as outlined by McGovern et al. (2018a). An additional four amplicon libraries were generated to assess sequencing run performance and library preparation. Three amplicon libraries were generated from the extractions performed on the ZymoBIOMICSTM MC with a final library generated using the ZymoBIOMICSTM Microbial Community DNA Standard (DS) (Zymo Research Corp., Irvine, CA, United States) to assess library preparation and sequencing run performance. This resulted in a total of 20 amplicon libraries being generated (RUSITEC n = 16; MC n = 3; DS n = 1).
The first round of PCR amplification, targeting the V4 hypervariable region of the 16S rRNA gene, was performed using the 515F/806R primers (Caporaso et al., 2011), designed with Nextera over hang adapters, and 2x KAPA Hifi HotStart ReadyMix DNA polymerase (Roche Diagnositics, West Sussex, United Kingdom). Cycle conditions were as follows: 95°C for 3 min, 20 cycles at 95°C for 30 s, 55°C for 30 s, 72°C for 30 s and then 72°C for 5 min.
Amplicons were purified using the MinElute PCR Purification Kit (Qiagen, Manchester, United Kingdom). Following purification, amplicons were subject to a second round of PCR to permit attachment of dual indices and Illumina sequencing adapters using the Nextera XT indexing kit (Illumina, San Diego, CA, United States). Cycle conditions for the second round of PCR were 95°C for 3 min, 8 cycles at 95°C for 30 s, 55°C for 30 s, 72°C for 30 s and then 72°C for 5 min followed by an additional PCR purification with the MinElute PCR Purification Kit (Qiagen, Manchester, United Kingdom). Confirmation of amplicon generation was conducted visually on a 2% agarose gel. Amplicons were pooled together in equal concentration and subject to gel purification using the QIAquick Gel Extraction Kit (Qiagen, Manchester, United Kingdom) to remove adapter primers and further purified to remove any residues of agarose using the MinElute PCR purification kit (Qiagen, Manchester, United Kingdom).
Pooled sample purity and quantity was analyzed on the Nanodrop 1000 with further validation on the Qubit fluorometer and using the KAPA SYBR FAST qPCR universal kit with Illumina Primer Premix (Roche Diagnositics, West Sussex, United Kingdom). Following this, the library pool was diluted and denatured as per the Illumina Miseq 16S Sample Preparation Guide with sequencing conducted on the Illumina MiSeq using the 500 cycle version 2 MiSeq reagent kit (Illumina, San Diego, CA, United States).
Amplicon sequence data was processed in R (version 3.5.2) using DADA2 (version 1.11.3) and submitted to the pipeline as previously described (Callahan et al., 2016) with minor modifications. Read quality was determined based on the visualization of Q scores with the aim to ensure mean Q scores of >30 were upheld for forward and reverse reads. To achieve this, forward reads were trimmed to a length of 240 bp and reverse reads trimmed to 200 bp. The removal of primer sequences was conducted using the trimLeft function. Identical sequences were combined using the dereplication function followed by the merging of forward and reverse reads. Following this an amplicon sequence variant (ASV) table was constructed after which chimeric sequences were removed. The assignTaxonomy function was used to assign taxonomy to each ASV using the RefSeq + RDP database (NCBI RefSeq 16S rRNA database supplemented by RDP) downloaded from the DADA2 website1. A bootstrapping threshold of 80 was applied with the addition of the minBoot = 80 function. Sample metadata, sequence taxonomy and ASVs were combined into a phyloseq object using phyloseq (version 1.24.2) (McMurdie and Holmes, 2013) for further analysis. Predictions of metabolic pathways for each sample, based on the generated ASVs, were conducted using CowPI (Wilkinson et al., 2018).
Data were checked for normality and homogeneity of variance by histograms, qqplots, and formal statistical tests as part of the UNIVARIATE procedure of SAS (version 9.1.3; SAS Institute). Data that were not normally distributed were transformed by raising the variable to the power of lambda. The appropriate lambda value was obtained by conducting a Box-Cox transformation analysis using the TRANSREG procedure of SAS. In-vitro data were analyzed using a mixed model ANOVA (PROC MIXED). Fixed effects in the model included BP concentrate (BP35 or BP95). Fermentation vessel was included as a random effect. Differences between treatments were determined by F-tests using Type III sums of squares. The PDIFF command incorporating the Tukey test was applied to evaluate pairwise comparisons between treatment means.
The generated ASV table and sequence taxonomies were analyzed in R (version 3.4.2). Of the 16 samples sequenced from the dietary treatments, one sample from the BP35 group was excluded from the analysis due to having a substantially lower sequencing depth compared to all other samples leaving a total of 15 samples for analysis (BP35 n = 7; BP95 n = 8). The relative abundance of taxa was calculated for each sample at the genus level in phyloseq. A PERMANOVA test, based on 9,999 permutations and a significance level of P < 0.05, was carried using the R package vegan (Oksanen et al., 2019) (version 2.5.4) implemented through microbiome (Leo et al., 2017) (version 1.0.2) to investigate differences in the community structure amongst samples based on treatment at the level of genus. The Wilcoxon rank sum test, with Benjamini Hochberg (BH) correction for false discovery rate (FDR) was implemented for identification of significant treatment differences in relative abundance of taxa based on adjusted P values (adj P) of <0.05. The ZymoBIOMICSTM MC standard contains eight bacteria and two yeast. As a result the mean relative abundance of the eight most abundant ASVs, within the three MC controls, was used to analyze performance of the DNA extraction method. Spearman’s rank correlation coefficient was used to determine consistency of the internal positive controls and correlations between microbial abundances and production data. A Student’s t-test was used to compare diversity metrics for microbial community structure comparisons and the ratio of Firmicutes:Bacteroidetes. Statistical analysis of predicted pathways was carried out using STAMP (version 2.1.3; Parks et al., 2014). Comparison of predicted pathways was conducted using the proportion of reads that were annotated to each individual metabolic pathway as a percentage of the total reads (read relative abundance). Statistical analysis of read relative abundance was conducted using a Student’s t-test, with BH correction for FDR (adj P < 0.05).
The disappearance of DM and its chemical constituents are given in Table 2. The BP35 diet had greater (P < 0.05) digestibility of the chemical constituents DM, organic matter (OM), CP, NDF, ADF compared to the BP95 diet. The fermentation parameters including VFA production per day and molar proportions for each dietary treatment are presented in Table 3. No significant difference was recorded in pH between treatments (P = 0.132). The BP35 diet had greater total VFA production than BP95 (P < 0.05). Diet did not alter (P > 0.10) the VFA proportions of either acetic or propionic acid, nor acetate:proprionate ratio. The proportion of butyric acid was greatest in the BP35 diet compared to the BP95 (P < 0.01). There was no effect of diet type on NH3-N output per day (P = 0.429).
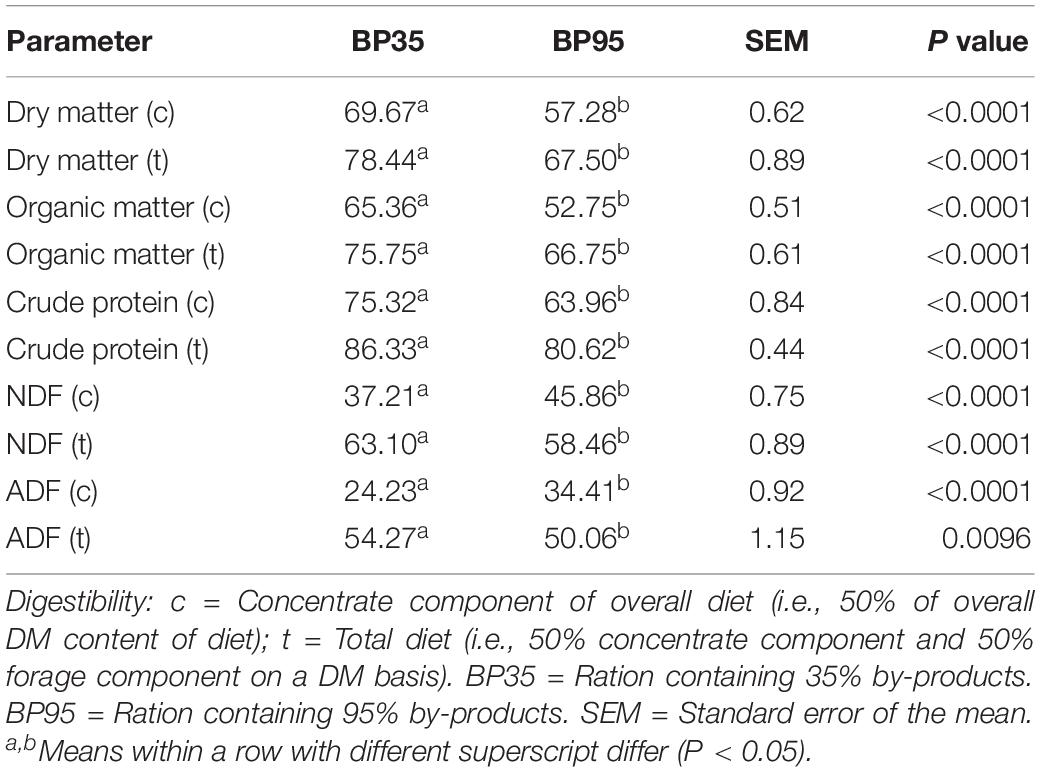
Table 2. Mean effects of by-product inclusion rate on disappearance of feed and its components in the rumen simulation technique (RUSITEC) system.
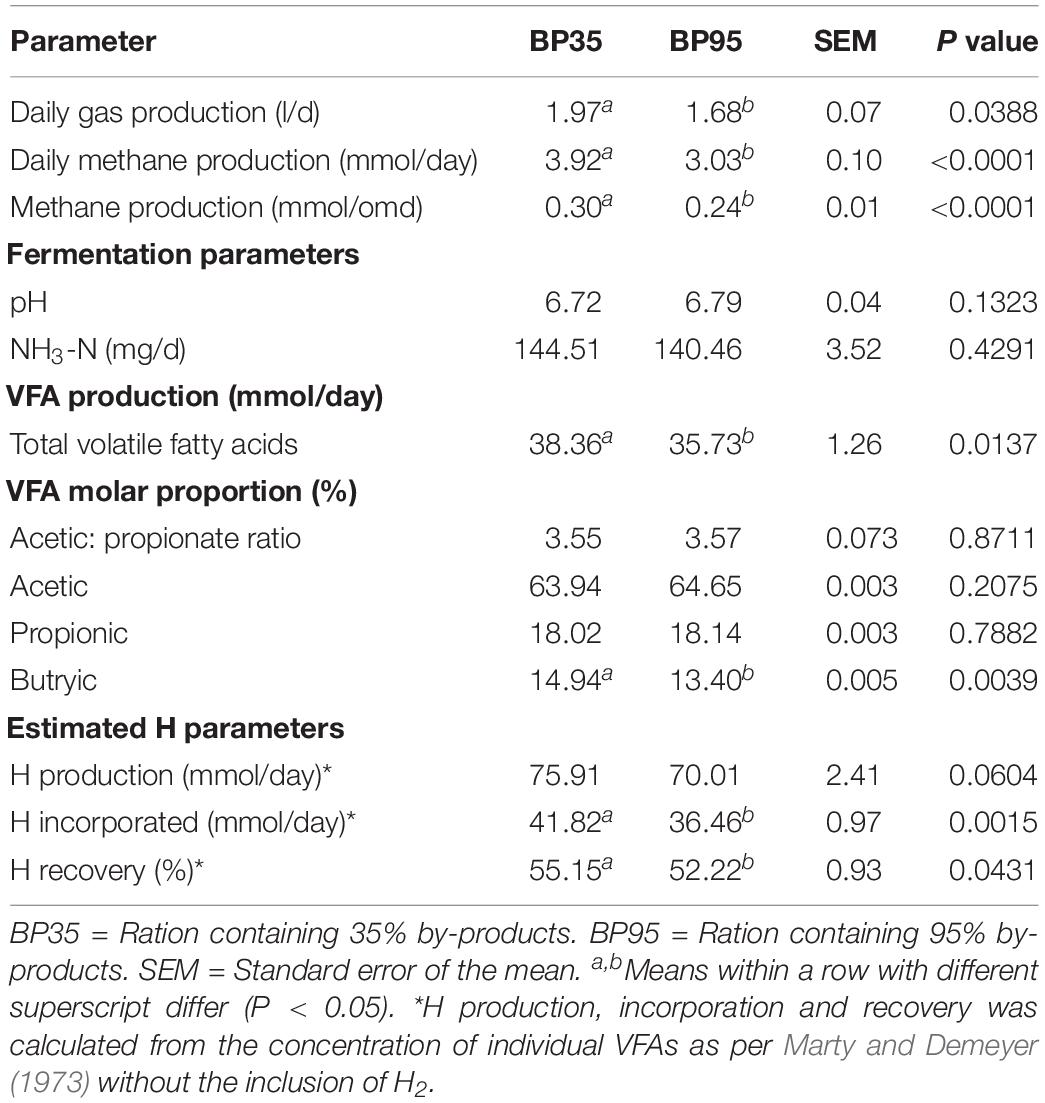
Table 3. Mean effects of by-product inclusion rate on pH and fermentation pattern in the rumen simulation technique (RUSITEC) system.
Average daily gas and methane production was altered between the treatment groups (P < 0.05). A greater mean daily methane output (mmol/day) was observed in the BP35 compared to the BP95 diet (P < 0.01) (Figure 1). Increased methane output with the BP35 diet persisted when expressed per unit of total organic matter digested (OMD; P < 0.001) (Figure 2). Daily gas production (l/day) was greatest in the BP35 diet (P < 0.05). Based on VFA analysis, there was a tendency for an increase in the production of theoretical H in the BP35 diet (P = 0.06). A reduction in the H incorporation (P < 0.01) and recovery (P < 0.05) was observed in the BP95 group.
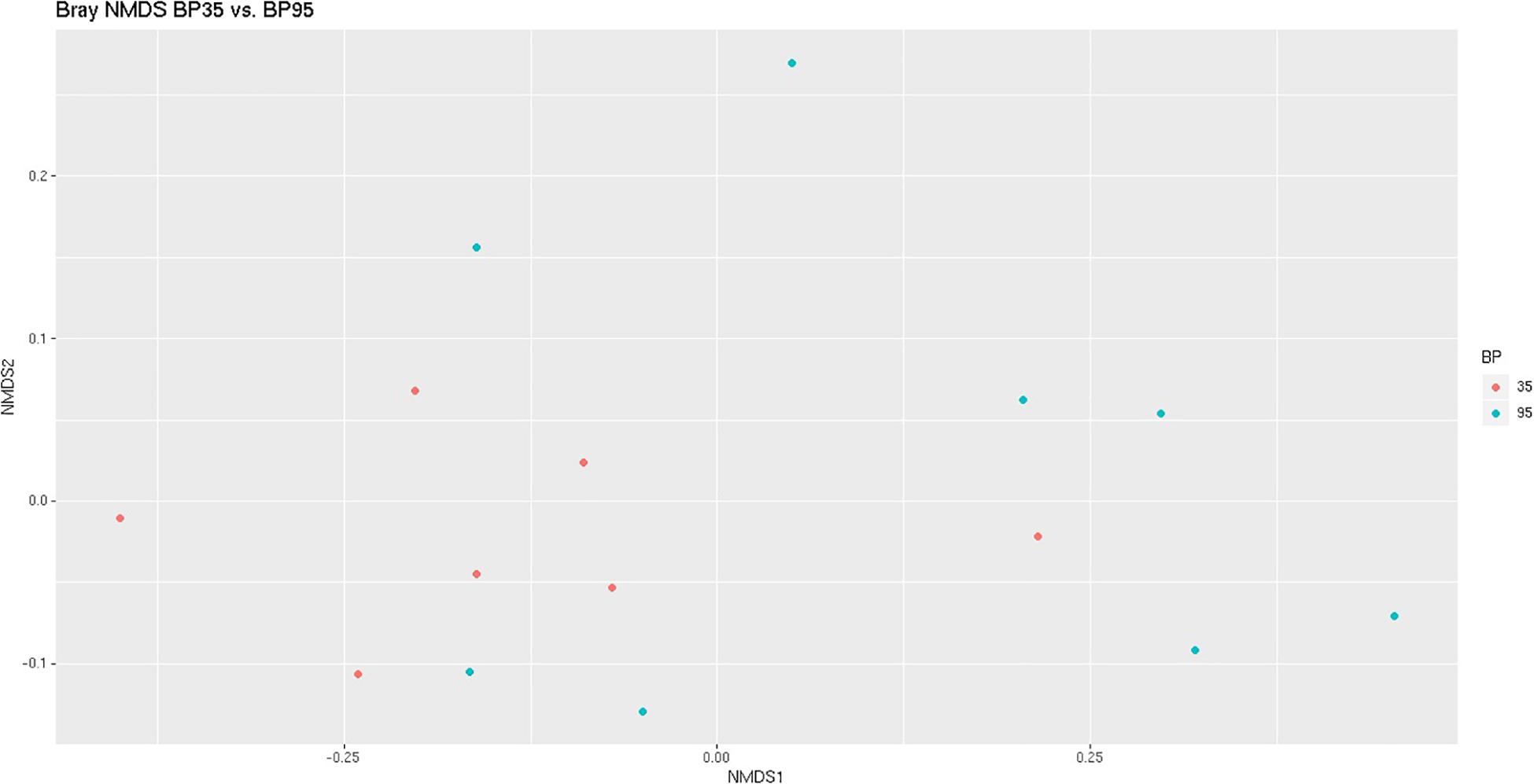
Figure 1. Bray Curtis NMDS plot highlighting differences in the community composition between diets. Different color dots represent samples obtained from rumen simulation technique (RUSITEC) systems incubated with different diets. Red = Concentrate formulated with 35% by-product (BP35), Blue = Concentrate formulated with 95% by-product (BP95).
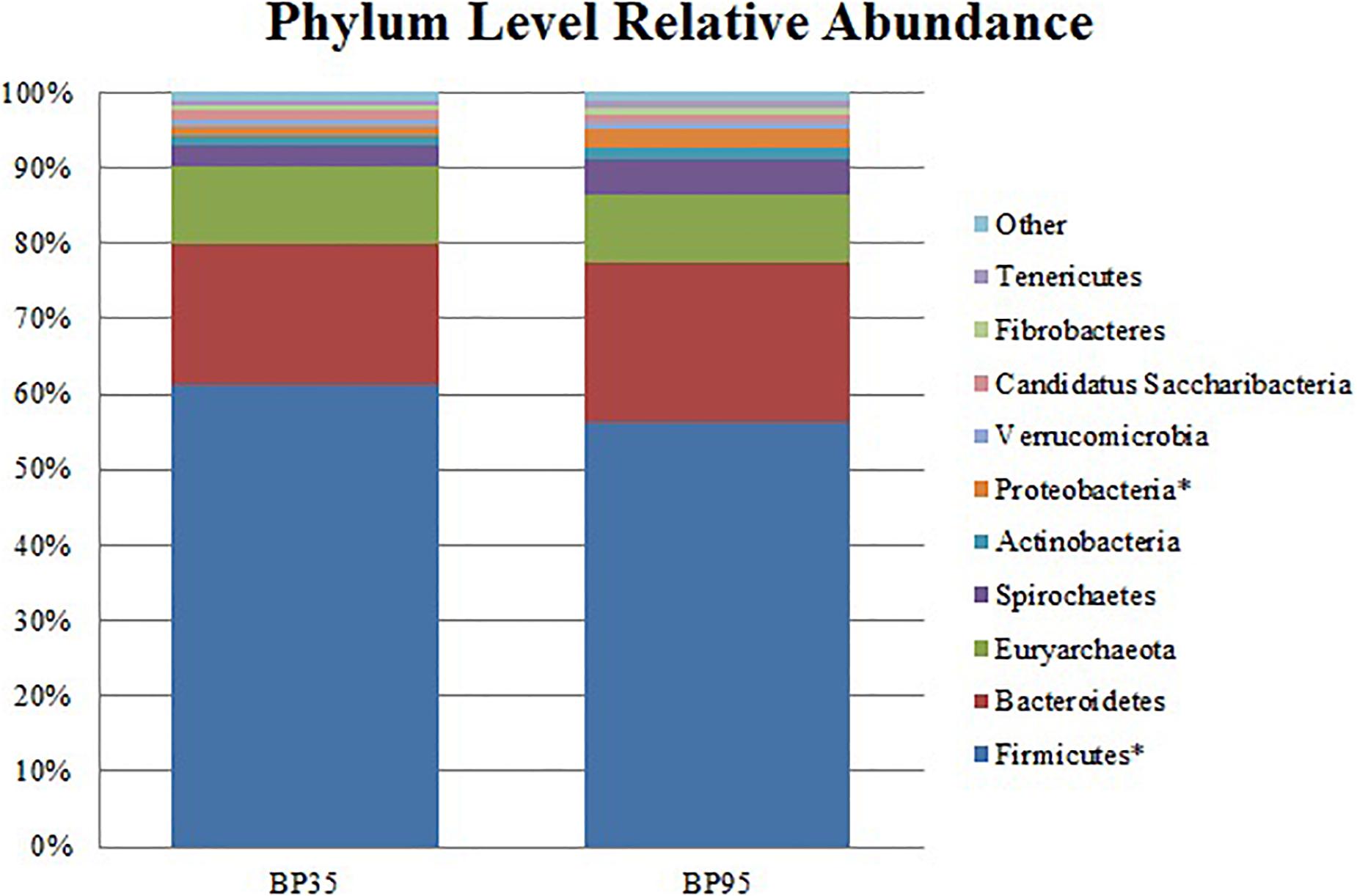
Figure 2. Stack plot comparing the differences, between diets, in the relative abundances of the ten most abundant microbes at the phylum level. Asterisk (∗) signifies phyla with a significantly different relative abundance between diets. BP35 = Concentrate formulated with 35% by-product. BP95 = Concentrate formulated with 95% by-product.
After quality filtering, merging and removal of chimeric sequences, a total of 1,411,464 reads were generated with a mean of 70,573.2 ± 16,920.78 reads per sample. The eight most abundant ASVs in the MC control accounted for 97.78 ± 0.24% of sequences. The mean correlation coefficient between the composition of libraries generated from the MC standards, and the theoretical composition of the ZymoBIOMICSTM standard was (rs, 0.79; P < 0.03). In addition, a strong positive correlation between the DS standard and the theoretical composition of the ZymoBIOMICSTM standard was observed (rs, 0.95; P < 0.001). Performance of the extraction method, library preparation and the sequencing run were deemed adequate based on correlations of our internal controls, and that of theoretical composition of the ZymoBIOMICSTM standards above that previously reported by members of our group (McGovern et al., 2018b).
Across all samples, the relative abundance of bacteria (91.05%) was greater in comparison to archaea (8.95%). At the phylum level, Firmicutes dominated preceded by Bacteroidetes and Euryarchaeota. At the genus level, 171 different genera of bacteria and archaea were identified, however, only those with a relative abundance greater than 0.01% are reported. Methanobrevibacter (11.42%) was the most abundant microbe followed by Prevotella (10.32%). Other highly abundant bacteria included Lactobacillus, Streptococcus, Treponema and Butyrivibrio. After Methanobrevibacter, other highly abundant methanogens included Methanosphaera, Methanobacterium, Methanomassiliicoccus, Methanimicrococcus and Methanomicrobium.
A NMDS plot of sample data generated using Bray Curtis dissimilarity analysis (Figure 1) displayed a moderate degree of clustering of samples based on diet, particularly BP35, with respect to the microbial communities. Based on the results of a PERMANOVA test, BP inclusion rate was deemed to have altered the microbial composition between dietary treatments (P = 0.023).
Differences in the alpha diversity of the microbial community structure, between the groups, was not observed when comparisons were conducted using the Shannon Index (P = 0.76). However, there was a tendency for a decrease in microbial diversity in the BP95 when diversity investigations were conducted with the Simpson metric (0.989 vs. 0.991; P = 0.08).
Comparisons of the ten and fifteen most abundant microbes classified at the phylum and family level are displayed in Figures 2, 3, respectively. A dietary effect was observed at the phylum level (Table 4), with a greater relative abundance of Firmicutes (adj P < 0.01) observed in the BP35 with an increased proportion of Proteobacteria in BP95 diet (adj P < 0.01). In addition, there was a tendency for an increased relative abundance of the Bacteroidetes phylum in the BP95 diet (adj P = 0.084). The increased proportion of bacteria, belonging to the phyla Proteobacteria, persisted at lower taxonomic ranks. For example, an increased abundance of Succinivibrionaceae (adj P < 0.03) at the family level with a strong tendency toward a statistically significant rise in the relative abundance of Succinivibrio (adj P = 0.053) at the genus level (Table 5), was observed in the BP95 diet. The Firmicutes:Bacteroidetes ratio was reduced in the BP95 diet (P < 0.01).
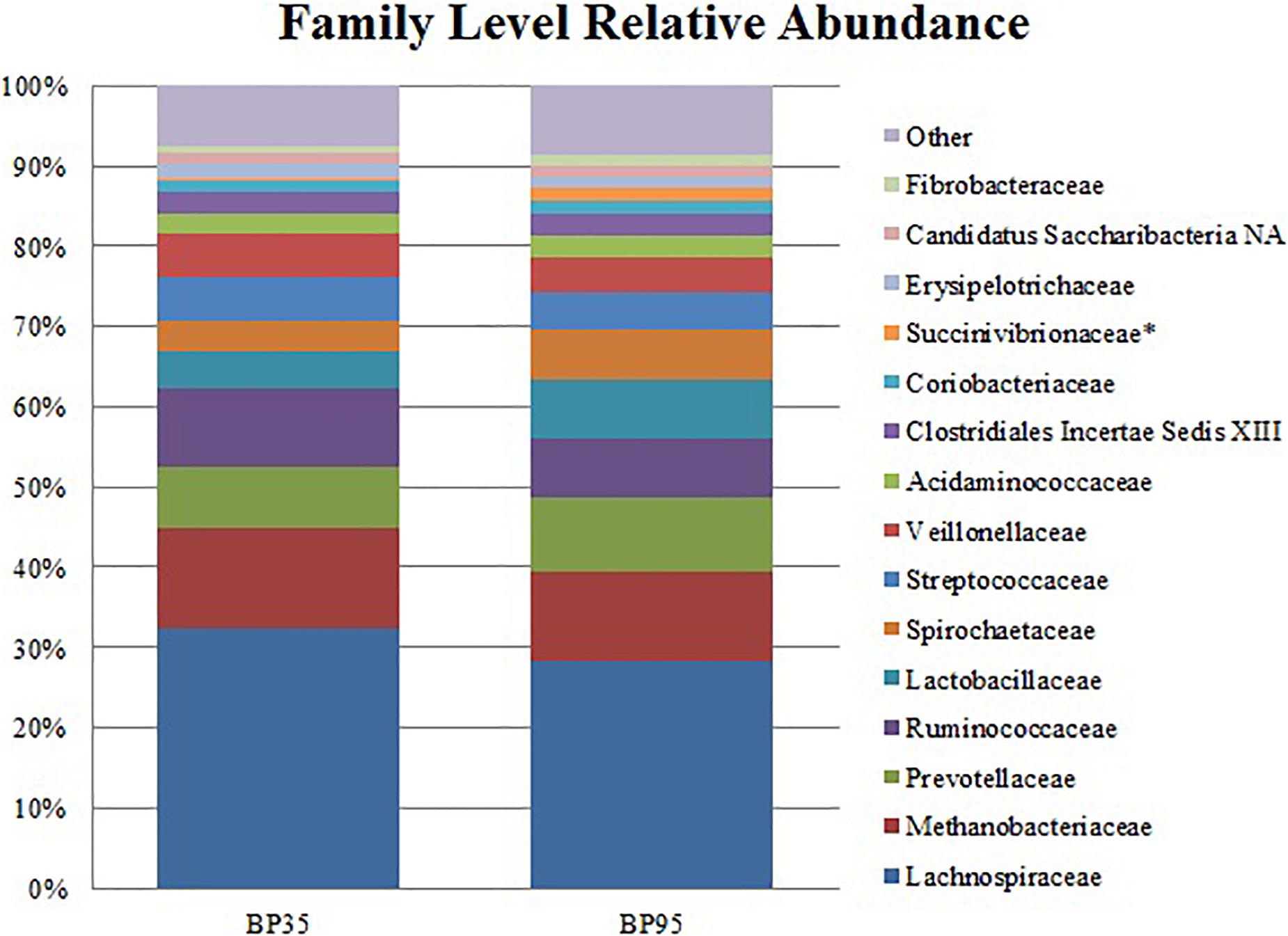
Figure 3. Stack plot comparing the differences, between diets, in the relative abundances of the fifteen most abundant microbes at the family level. Asterisk (∗) signifies families with a significantly different relative abundance between diets. BP35 = Concentrate formulated with 35% by-product. BP95 = Concentrate formulated with 95% by-product.
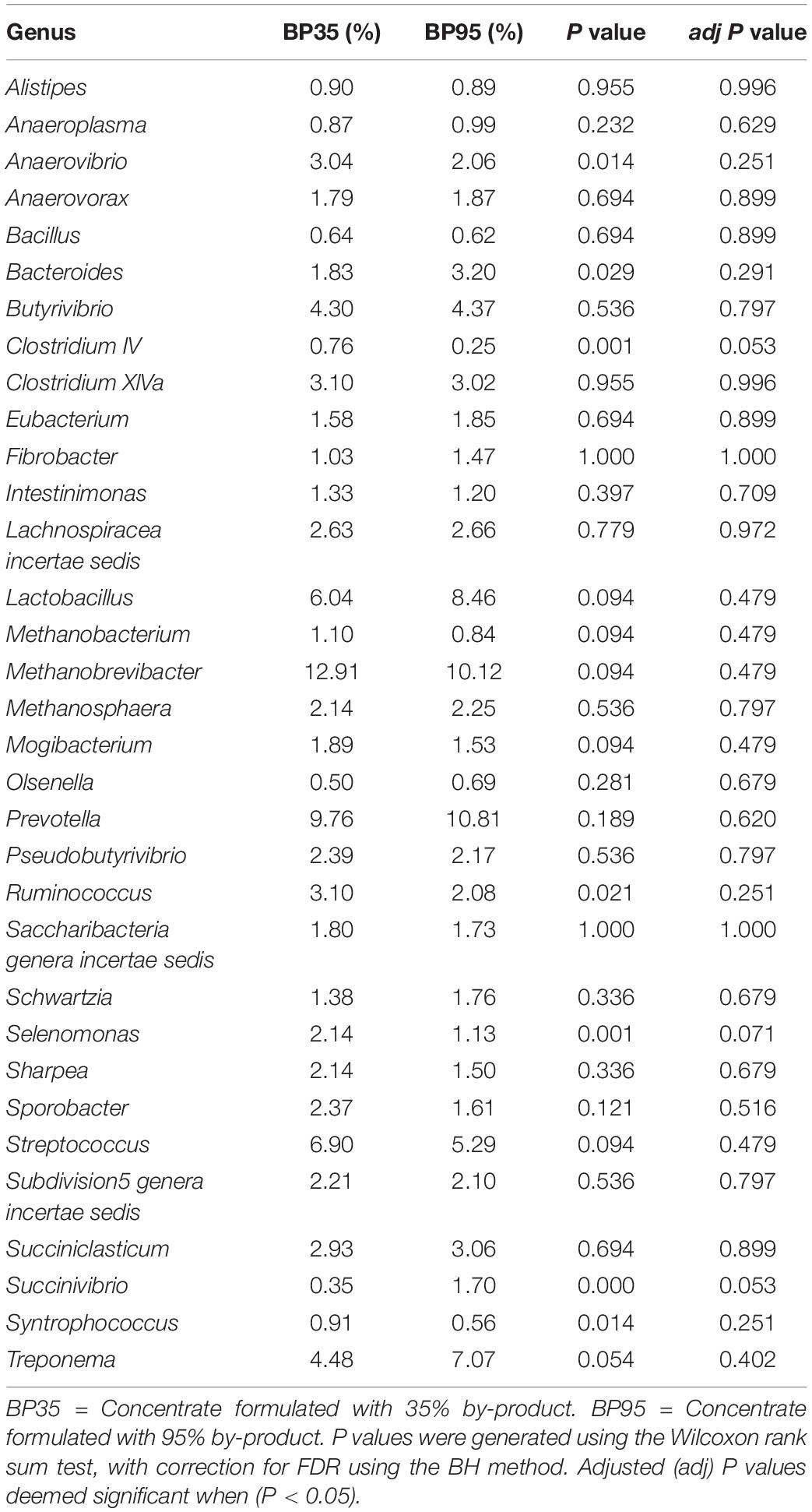
Table 5. Mean relative abundance of the 33 most abundant genera accounting for >90% of the relative abundance across both diets.
BP inclusion rate affected the abundance of Selenomonas (adj P = 0.071) and Clostridium IV (adj P = 0.053) with both genera tending to have a greater abundance in the BP35 dietary treatment (Table 4). Although numeric differences were apparent in the average relative abundances, BP inclusion rate had no significant influence on the abundance of methanogens when calculated relative to all members of the microbial communities.
The abundance of the Firmicutes phylum had a strong positive correlation with the total DM digestibility (DMD) (rs, 0.84; P < 0.001), CP digestibility (CPD) (rs, 0.83; P < 0.001), NDF digestibility (NDFD) (rs, 0.86; P < 0.0001) and ADF digestibility (ADFD) (rs, 0.80; P < 0.001). Total (rs, 0.63; P < 0.02) and all individual VFA production levels had a positive correlation with the abundance of Firmicutes.
The relative abundance of Proteobacteria had a strong negative correlation with all digestibility parameters [total DMD (rs, −0.88; P < 0.0001), OMD (rs, −0.63; P < 0.02), CPD (rs, −0.86; P < 0.0001), NDFD (rs, −0.89; P < 0.0001) and ADFD (rs, −0.86; P < 0.0001)]. A moderate negative relationship was observed for the relative abundance of Proteobacteria with total VFA (rs, −0.60; P < 0.02) and propionate (rs, −0.58; P < 0.03) production. A strong negative relationship with butyrate (rs, −0.76; P < 0.01) and a near significant association with acetate (rs, −0.51; P = 0.052) production, was also observed. The relative abundance of Succinivibrionaceae had a negative correlation with all digestibility parameters. Total VFA production had a significant negative association (rs, −0.52; P < 0.05) with Succinivibrionaceae with both propionate (rs, −0.53; P < 0.05) and butyrate (rs, −0.68; P < 0.01) production observed to a have a moderately negative association.
The associations between members of the microbial community and methane production were investigated in an effort to better understand the microbial factors resulting in the divergence of methane production. At lower taxonomic levels methanogen abundance was found to be associated with methane production. For example a strong positive relationship between Methanobrevibacter and daily methane output (rs, 0.63; P < 0.02) was observed. In addition, there was a tendency for a similar correlation between Methanobacterium and methane output (rs, 0.46; P = 0.081).
Within the bacterial proportion, the abundance of Proteobacteria was found to be negatively associated with methane production when expressed on a daily basis (rs, −0.71; P < 0.01) or per unit of OMD (rs, −0.65; P < 0.01). At lower taxonomic levels, the relationship of members of the Proteobacteria phylum with methane production was maintained for the mean abundance of Succinivibrio (rs, −0.68; P < 0.01). The abundance of the Firmicutes phyla was found to be positively correlated with both daily methane production (rs, 0.79; P < 0.001) and output per unit of OMD (rs, 0.75; P < 0.01). Similarly this relationship was maintained for the genera Selenomonas (rs, 0.82; P < 0.001) and Clostridium IV (rs, 0.73; P < 0.01) with daily methane output.
Findings from this in vitro study have shown a 22.7% reduction in methane output with the supplementation of a concentrate formulated exclusively from DDGS, PKE and SHs to a pasture based diet, in comparison to a barley and soybean meal based concentrate formulation. The observed differences in all digestibility parameters associated with the BP95 diet, most likely originated from the varied supply of fiber (NDF), starch, and fat between the diets (Table 1). Indeed, fiber has a lower digestibility in comparison to starch (Getachew et al., 2004) with some components of our BP mixture previously shown to be moderately degradable (Ipharraguerre and Clark, 2003; Schingoethe et al., 2009; Wang et al., 2018). In addition, fat supplementation is known to reduce fiber degradation in the rumen (Patra, 2013) further explaining the reduced digestibility observed in the high fiber BP95 diet. Furthermore, the limited fermentation of lipids by ruminal microbes (Nagaraja et al., 1997) likely contributed to the reduced diet digestibility, VFA and daily methane production in the BP95 diet, as fat replaced fermentable OM. However, the proportions of individual VFAs were similar across both diets, with the exception of butyric acid.
Findings from this study are in agreement with the proposed relationship between the tandem reduction in methane production and H recovery (Ungerfeld, 2015). However, some 45% of H was unaccounted for in our analysis, which may have been redirected to other H sinks not investigated in this study, such as H2 and additional fermentation end products beyond the predominant VFAs and methane, as proposed by previous authors (Machmüller et al., 1998; Dohme et al., 2000; Guyader et al., 2017).
While differences in fat, starch and NDF content between the two diets is accepted to have influenced daily methane production, it is more precise to estimate methane output, in vitro, per unit of substrate digested (Yáñez-Ruiz et al., 2016). After correcting methane output for OMD, dietary ranking was consistent with a 20.8% reduction in output detected in the BP95 diet. As methane is a product of microbial fermentation (Tapio et al., 2017) differences in the components of the diets likely altered the composition of the microbial communities, which may have contributed to the observed variation in methane output per OMD between the diets.
An increase in the proportion of dietary unsaturated fat has been shown to increase the abundance of Succinivibrionacea and Succinivibrio in cattle and sheep (Huws et al., 2015; Lyons et al., 2017). Therefore the increased abundance of some members of the Succinivibrionacea family in the BP95 diet may be due to the elevated supply of lipids, some of which may have been unsaturated and originated from DDGS (Castillo-Lopez et al., 2014; Xu et al., 2014). In addition, the non-structural carbohydrate component of SHs is high in pectin (Makkar, 2012) which may have benefitted some members of the Succinivibrionacea family capable of degrading pectin (Stewart et al., 1997).
Bacteria belonging to the phylum, Firmicutes, such as members of the Lachnospiraceae and Ruminococcaceae family are known degraders of cellulose and hemicellulose (Krause et al., 2003; Dai et al., 2015; Huws et al., 2016; Mayorga et al., 2016) while others are accepted producers of CO2 and H2 (Marounek and Dušková, 1999; Rooke et al., 2014). In agreement with the literature, the abundance of Firmicutes was strongly correlated with NDF digestibility and daily gas production. In addition, it is likely the strong relationship between the abundance of Firmicutes and methane output (daily and per unit of OMD) was as a result of the fibrolytic capabilities of members of the phyla.
The observed reduction in the Firmicutes:Bacteroidetes ratio in the BP95 is in agreement with the effects of DDGS dietary inclusion, as presented by others (Callaway et al., 2010; Ramirez et al., 2012). Equally, the Firmicutes phylum is likely to have been impacted by the increased fat content in the BP95 diet, with some members of the phylum found to be adversely impacted by lipids (Maia et al., 2007). SHs have also been shown to reduce the abundance of Firmicutes in vitro (Wang et al., 2018). As a result, dietary related effects to the abundance of the Firmicutes phyla most likely resulted in a varied supply of CO2 and H2 to hydrogenotrophic methanogens. Finally, the abundance of Selenomonas likely benefited from the higher starch content observed in the BP35 with previous studies showing an elevated abundance of the bacteria in high grain diets (Fernando et al., 2010).
In this study, a high relative abundance of archaea was observed across both dietary groups at all taxonomic levels, in comparison to other RUSITEC and in vivo experiments, utilizing the same primers to simultaneously target bacterial and archaeal populations (Duarte et al., 2017; Bowen et al., 2020). However, archaea have previously been reported as having a relative abundance of greater than 7% in feed restricted animals (McCabe et al., 2015) and thus similar to the proportion observed in this study. In addition, comparisons between other RUSITEC experiments are difficult due to the separate reporting of archaeal and bacterial relative abundances by some groups (Belanche et al., 2016a, b). Furthermore, sampling time may have had an effect on the abundance of methanogens, with both the abundance of bacteria and archaea shown to peak 2–4 h after the introduction of feed to the RUSITEC (Belanche et al., 2013). As samples were collected 24 h after feeding, potentially a greater post feeding decrease in bacteria, relative to the archaea, may have occurred resulting in the high relative abundance of archaea observed in our study.
Both PKE and DDGS have previously been shown to negatively affect the methanogen population in goats (Abubakr et al., 2014) and cattle (Zhou et al., 2012). However, in this study the relative abundance of the methanogen proportion of the microbial community was not affected by diet. McrA gene expression has previously been shown to be associated with methane emissions (Shi et al., 2014) and is noted as a more credible methanogenesis biomarker than 16S rRNA based methods (Wilkins et al., 2015). Therefore the effect of diet on abundance of Firmicutes may have resulted in a reduced supply of H2 and CO2, and subsequently influenced the expression of methanogenesis pathways resulting in the reduced methane production observed in the BP95 diet.
Findings from this in vitro study highlight the methane abatement potential of a low starch but high fat and fiber formulated concentrate, for grass based ruminant production systems. Data generated from our metataxonomic approach, suggests the rumen prokaryotic composition to be altered in favor of a potential reduction in the production of methanogenesis substrates, with the supplementation of our 95% BP formulated concentrate. However, confirmation in vivo will be required to confirm the methane abatement potential, of concentrates formulated solely from SHs, PKE and DDGS to act a cost effective dietary mitigation strategy for pasture based livestock production.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA610813.
The animal study was reviewed and approved by The animal research ethics committee (AREC) at University College Dublin (UCD).
AK, TB, and JH conceived and designed the experiments. PS and JH performed the experiments. PS, JH, and AK analyzed the data. AK, JH, SW, DK, and TB contributed reagents, materials, and analysis tools. PS, AK, SW, DK, TB, and JH interpreted results and wrote the manuscript. All authors contributed to the article and approved the submitted version.
The RUSITEC experiment was funded by the Irish Government under the National Development Plan 2007–2013 through the Department of Agriculture Food and the Marine Research Stimulus Fund: RSF 11/S/122: FEFAN. The molecular rumen microbiome work was funded by the FACCE ERA-GAS “RumenPredict” grant (2017). PS is funded by a Walsh Scholarship (RMIS 0364).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abubakr, A., Alimon, A. R., Yaakub, H., Abdullah, N., and Ivan, M. (2014). Effect of feeding palm oil by-products based diets on total bacteria, cellulolytic bacteria and methanogenic archaea in the rumen of goats. PLoS One 9:e95713. doi: 10.1371/journal.pone.0095713
Agriculture Food Research Council (1993). Energy and Protein Requirements of Ruminants. An Advisory Manual Prepared by the AFRC Technical Committee on Responses to Nutrients. Wallingford: CAB International.
Beauchemin, K. A., Kreuzer, M., O’Mara, F., and McAllister, T. A. (2008). Nutritional management for enteric methane abatement: a review. Aust. J. Exp. Agric. 48, 21–27.
Beauchemin, K. A., Ungerfeld, E. M., Eckard, R. J., and Wang, M. (2020). Fifty years of research on rumen methanogenesis: lessons learned and future challenges for mitigation. Animal 14, 2–16.
Belanche, A., Lee, M. R. F., Moorby, J. M., and Newbold, C. J. (2013). Comparison of ryegrass and red clover on the fermentation pattern, microbial community and efficiency of diet utilisation in the rumen simulation technique (Rusitec). Anim. Prod. Sci. 53, 1052–1064.
Belanche, A., Kingston-Smith, A. H., and Newbold, C. J. (2016a). An integrated multi-omics approach reveals the effects of supplementing grass or grass hay with vitamin E on the rumen microbiome and its function. Front. Microbiol. 7:905. doi: 10.3389/fmicb.2016.00905
Belanche, A., Pinloche, E., Preskett, D., and Newbold, C. J. (2016b). Effects and mode of action of chitosan and ivy fruit saponins on the microbiome, fermentation and methanogenesis in the rumen simulation technique. FEMS Microbiol. Ecol. 92:fiv160. doi: 10.1093/femsec/fiv160
Benchaar, C., Hassanat, F., Gervais, R., Chouinard, P. Y., Julien, C., Petit, H. V., et al. (2013). Effects of increasing amounts of corn dried distillers grains with solubles in dairy cow diets on methane production, ruminal fermentation, digestion, N balance, and milk production. J. Dairy Sci. 96, 2413–2427. doi: 10.3168/jds.2012-6037
Bowen, J. M., Cormican, P., Lister, S. J., McCabe, M. S., Duthie, C. A., Roehe, R., et al. (2020). Links between the rumen microbiota, methane emissions and feed efficiency of finishing steers offered dietary lipid and nitrate supplementation. PLoS One 15:e0231759. doi: 10.1371/journal.pone.0231759
Callahan, B. J., Mcmurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Callaway, T. R., Dowd, S. E., Edrington, T. S., Anderson, R. C., Krueger, N., Bauer, N., et al. (2010). Evaluation of bacterial diversity in the rumen and feces of cattle fed different levels of dried distillers grains plus solubles using bacterial tag-encoded FLX amplicon pyrosequencing. J. Anim. Sci. 88, 3977–3983. doi: 10.2527/jas.2010-2900
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108, 4516–4522. doi: 10.1073/pnas.1000080107
Castillo-Lopez, E., Jenkins, C. J. R., Aluthge, N. D., Tom, W., Kononoff, P. J., and Fernando, S. C. (2017). The effect of regular or reduced-fat distillers grains with solubles on rumen methanogenesis and the rumen bacterial community. J. Appl. Microbiol. 123, 1381–1395. doi: 10.1111/jam.13583
Castillo-Lopez, E., Ramirez Ramirez, H. A., Klopfenstein, T. J., Anderson, C. L., Aluthge, N. D., Fernando, S. C., et al. (2014). Effect of feeding dried distillers grains with solubles on ruminal biohydrogenation, intestinal fatty acid profile, and gut microbial diversity evaluated through DNA pyro-sequencing. J. Anim. Sci. 92, 733–743. doi: 10.2527/jas.2013-7223
Condren, S. A., Kelly, A. K., Lynch, M. B., Boland, T. M., Whelan, S. J., Grace, C., et al. (2019). The effect of by-product inclusion and concentrate feeding rate on milk production and composition, pasture dry matter intake, and nitrogen excretion of mid-late lactation spring-calving cows grazing a perennial ryegrass-based pasture. J. Dairy Sci. 102, 1247–1256. doi: 10.3168/jds.2018-14970
Czerkawski, J. W., and Breckenridge, G. (1977). Design and development of a long-term rumen simulation technique (Rusitec). Br. J. Nutr. 38, 371–384. doi: 10.1079/bjn19770102
Dai, X., Tian, Y., Li, J., Su, X., Wang, X., Zhao, S., et al. (2015). Metatranscriptomic analyses of plant cell wall polysaccharide degradation by microorganisms in the cow rumen. Appl. Environ. Microbiol. 81, 1375–1386. doi: 10.1128/aem.03682-14
Dohme, F., Machmüller, A., Wasserfallen, A., and Kreuzer, M. (2000). Comparative efficiency of various fats rich in medium-chain fatty acids to suppress ruminal methanogenesis as measured with RUSITEC. Can. J. Anim. Sci. 80, 473–484. doi: 10.4141/a99-113
Duarte, A. C., Holman, D. B., Alexander, T. W., Durmic, Z., Vercoe, P. E., and Chaves, A. V. (2017). The type of forage substrate preparation included as substrate in a RUSITEC system affects the ruminal microbiota and fermentation characteristics. Front. Microbiol. 8:704. doi: 10.3389/fmicb.2017.00704
Ertl, P., Knaus, W., Metzler-Zebeli, B. U., Klevenhusen, F., Khiaosa-Ard, R., and Zebeli, Q. (2015). Substitution of common concentrates with by-products modulated ruminal fermentation, nutrient degradation, and microbial community composition in vitro. J. Dairy Sci. 98, 4762–4771. doi: 10.3168/jds.2014-9063
Fernando, S. C., Purvis, H. T., Najar, F. Z., Sukharnikov, L. O., Krehbiel, C. R., Nagaraja, T. G., et al. (2010). Rumen microbial population dynamics during adaptation to a high-grain diet. Appl. Environ. Microbiol. 76, 7482–7490. doi: 10.1128/aem.00388-10
Finneran, E., Crosson, P., O’Kiely, P., Shalloo, L., Forristal, D., and Wallace, M. (2012). Stochastic simulation of the cost of home-produced feeds for ruminant livestock systems. J. Agric. Sci. 150, 123–139. doi: 10.1017/s002185961100061x
Getachew, G., Robinson, E. J., DePeters, H., and Taylor, S. (2004). Relationships between chemical composition, dry matter degradation and in vitro gas production of several ruminant feeds. Anim. Feed Sci. Technol. 111, 57–71. doi: 10.1016/s0377-8401(03)00217-7
Grasser, L. A., Fadel, J. G., Garnett, I., and Depeters, E. J. (1995). Quantity and economic importance of nine selected by-products used in California dairy rations. J. Dairy Sci. 78, 962–971. doi: 10.3168/jds.s0022-0302(95)76711-x
Guyader, J., Ungerfeld, E. M., and Beauchemin, K. A. (2017). Redirection of metabolic hydrogen by inhibiting methanogenesis in the rumen simulation technique (RUSITEC). Front. Microbiol. 8:393. doi: 10.3389/fmicb.2017.00393
Henderson, G., Cox, F., Ganesh, S., Jonker, A., Young, W., Abecia, L., et al. (2015). Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci. Rep. 5:14567.
Hünerberg, M., McGinn, S. M., Beauchemin, K. A., Okine, E. K., Harstad, O. M., and McAllister, T. A. (2013). Effect of dried distillers’ grains with solubles on enteric methane emissions and nitrogen excretion from finishing beef cattle. Can. J. Anim. Sci. 93, 373–385. doi: 10.4141/cjas2012-151
Huws, S. A., Edwards, J. E., Creevey, C. J., Rees Stevens, P., Lin, W., Girdwood, S. E., et al. (2016). Temporal dynamics of the metabolically active rumen bacteria colonizing fresh perennial ryegrass. FEMS Microbiol. Ecol. 92:fiv137. doi: 10.1093/femsec/fiv137
Huws, S. A., Kim, E. J., Cameron, S. J., Girdwood, S. E., and Davies, L. (2015). Characterization of the rumen lipidome and microbiome of steers fed a diet supplemented with flax and echium oil. Microb. Biotechnol. 8, 331–341. doi: 10.1111/1751-7915.12164
Ipharraguerre, I. R., and Clark, J. H. (2003). Soyhulls as an alternative feed for lactating dairy cows: a review. J. Dairy Sci. 86, 1052–1073. doi: 10.3168/jds.s0022-0302(03)73689-3
Jaurena, G., Moorby, J. M., and Davies, D. R. (2005). Efficiency of microbial protein synthesis on red clover and ryegrass silages supplemented with barley by rumen simulation technique (RUSITEC). Anim. Feed Sci. Technol. 118, 79–91. doi: 10.1016/j.anifeedsci.2004.09.008
Kim, S. H., Mamuad, L. L., Jeong, C. D., Choi, Y. J., Lee, S. S., Ko, J. Y., et al. (2013). In vitro evaluation of different feeds for their potential to generate methane and change methanogen diversity. Asian Australas. J. Anim. Sci. 26, 1698–1707. doi: 10.5713/ajas.2013.13260
Krause, D. O., Denman, S. E., Mackie, R. I., Morrison, M., Rae, A. L., Attwood, G. T., et al. (2003). Opportunities to improve fiber degradation in the rumen: microbiology, ecology, and genomics. FEMS Microbiol. Rev. 27, 663–693. doi: 10.1016/s0168-6445(03)00072-x
Lee, H. J., Lee, S. C., Kim, J. D., Oh, Y. G., Kim, B. K., Kim, C. W., et al. (2003). Methane production potential of feed ingredients as measured by in vitro gas test. Asian Australas. J. Anim. Sci. 16, 1143–1150. doi: 10.5713/ajas.2003.1143
Leo, L., Sudarshan, R., Shetty, K., Tweed, J., Vallin, H., Scollan, N. D., et al. (2017). Tools for Microbiome Analysis in R. Version 1.9.19.
Lyons, T., Boland, T., Storey, S., and Doyle, E. (2017). Linseed oil supplementation of lambs’ diet in early life leads to persistent changes in rumen microbiome structure. Front. Microbiol. 8:1656. doi: 10.3389/fmicb.2017.01656
Machmüller, A., Ossowski, D. A., Wanner, M., and Kreuzer, M. (1998). Potential of various fatty feeds to reduce methane release from rumen fermentation in vitro (Rusitec). Anim. Feed Sci. Technol. 71, 117–130. doi: 10.1016/s0377-8401(97)00126-0
Maia, M. R., Chaudhary, L. C., Figueres, L., and Wallace, R. J. (2007). Metabolism of polyunsaturated fatty acids and their toxicity to the microflora of the rumen. Antonie Van Leeuwenhoek 91, 303–314. doi: 10.1007/s10482-006-9118-2
Makkar, H. P. S. (2012). Biofuel Co-products as Livestock Feed-Opportunities and Challenges. Rome: Food and Agriculture Organization.
Marounek, M., and Dušková, D. (1999). Metabolism of pectin in rumen bacteria Butyrivibrio fibrisolvens and Prevotella ruminicola. Lett. Appl. Microbiol. 29, 429–433. doi: 10.1046/j.1472-765x.1999.00671.x
Martin, C., Morgavi, D. P., and Doreau, M. (2010). Methane mitigation in ruminants: from microbe to the farm scale. Animal 4, 351–365. doi: 10.1017/s1751731109990620
Marty, R. J., and Demeyer, D. I. (1973). The effect of inhibitors of methane production of fermentation pattern and stoichiometry in vitro using rumen contents from sheep given molasses. Br. J. Nutr. 30, 369–376. doi: 10.1079/bjn19730041
Mayorga, O. L., Kingston-Smith, A. H., Kim, E. J., Allison, G. G., Wilkinson, T. J., Hegarty, M. J., et al. (2016). Temporal metagenomic and metabolomic characterization of fresh perennial ryegrass degradation by rumen bacteria. Front. Microbiol. 7:1854. doi: 10.3389/fmicb.2016.01854
McCabe, M. S., Cormican, P., Keogh, K., O’Connor, A., O’Hara, E., Palladino, R. A., et al. (2015). Illumina MiSeq phylogenetic amplicon sequencing shows a large reduction of an uncharacterised Succinivibrionaceae and an increase of the Methanobrevibacter gottschalkii clade in feed restricted cattle. PLoS One 10:e0133234. doi: 10.1371/journal.pone.0133234
McDougall, E. I. (1948). Studies on ruminant saliva. 1. The composition and output of sheep’s saliva. Biochem. J. 43, 99–109. doi: 10.1042/bj0430099
McGee, M. (2005). “Recent developments in feeding beef cattle on grass silage-based diets. Silage production and utilisation,” in Proceedings of the XIVth International Silage Conference, a Satellite Workshop of the XXth International Grassland Congress, July 2005, (Belfast: Wageningen Academic).
McGinn, S. M., Chung, K. A., Beauchemin, A. D., Iwaasa, S., and Grainger, C. (2009). Use of corn distillers’ dried grains to reduce enteric methane loss from beef cattle. Can. J. Anim. Sci. 89, 409–413. doi: 10.4141/cjas08133
McGovern, E., Kenny, D. A., McCabe, M. S., Fitzsimons, C., McGee, M., Kelly, A. K., et al. (2018a). 16S rRNA sequencing reveals relationship between potent cellulolytic genera and feed efficiency in the rumen of bulls. Front. Microbiol. 9:1842. doi: 10.3389/fmicb.2018.01842
McGovern, E., Waters, S. M., Blackshields, G., and Mccabe, M. S. (2018b). Evaluating established methods for rumen 16S rRNA amplicon sequencing with mock microbial populations. Front. Microbiol. 9:1365. doi: 10.3389/fmicb.2018.01365
McGrath, O. (2013). Mid-Season Nutrition of Dairy Cows. Teagasc Advisory Services Article 17. Dublin: Teagasc.
McMurdie, P. J., and Holmes, H. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217
Mirzaei-Aghsaghali, A., and Maheri-Sis, N. (2008). Nutritive value of some agro-industrial by-products for ruminants-A review. World J. Zool. 3, 40–46.
Morgavi, D. P., Forano, E., Martin, C., and Newbold, C. J. (2010). Microbial ecosystem and methanogenesis in ruminants. Animal 4, 1024–1036. doi: 10.1017/s1751731110000546
Mottet, A., de Haan, C., Falcucci, A., Tempio, G., Opio, C., and Gerber, P. (2017). Livestock: on our plates or eating at our table? A new analysis of the feed/food debate. Glob. Food Sec. 14, 1–8. doi: 10.1016/j.gfs.2017.01.001
Nagaraja, T. G., Newbold, C. J., Van Nevel, C. J., and Demeyer, D. I. (1997). “Manipulation of ruminal fermentation,” in The Rumen Microbial Ecosystem, eds P. N. Hobson, and C. S. Stewart, (Berlin: Springer), 523–632. doi: 10.1007/978-94-009-1453-7_13
O’Brien, D., Moran, B., and Shalloo, L. (2018). A national methodology to quantify the diet of grazing dairy cows. J. Dairy Sci. 101, 8595–8604. doi: 10.3168/jds.2017-13604
O’Connor, A., McGee, M., Moloney, A., Boland, T., and O’Kiely, P. (2019a). Digestion and nitrogen metabolism in beef cattle and in vitro rumen fermentation of autumn grass differing in fertilizer nitrogen application rate. Grass Forage Sci. 74, 535–547.
O’Connor, A., Moloney, A. P., O’Kiely, P., Boland, T. M., and McGee, M. (2019b). Effects of fertiliser nitrogen rate to spring grass on apparent digestibility, nitrogen balance, ruminal fermentation and microbial nitrogen production in beef cattle and in vitro rumen fermentation and methane output. Anim. Feed Sci. Technol. 254:114198. doi: 10.1016/j.anifeedsci.2019.06.002
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O’Hara, R. B., et al. (2019). vegan: Community Ecology Package. R package.
Parks, D. H., Tyson, G. W., Hugenholtz, P., and Beiko, R. G. (2014). STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30, 3123–3124. doi: 10.1093/bioinformatics/btu494
Patra, A. K. (2013). The effect of dietary fats on methane emissions, and its other effects on digestibility, rumen fermentation and lactation performance in cattle: a meta-analysis. Livest. Sci. 155, 244–254. doi: 10.1016/j.livsci.2013.05.023
Ramirez, H. R., Nestor, K., Tedeschi, L. O., Callaway, T. R., Dowd, S. E., Fernando, S. C., et al. (2012). The effect of brown midrib corn silage and dried distillers’ grains with solubles on milk production, nitrogen utilization and microbial community structure in dairy cows. Can. J. Anim. Sci. 92, 365–380. doi: 10.4141/cjas2011-133
Rooke, J. A., Wallace, R. J., Duthie, C.-A., Mckain, N., De Souza, S. M., Hyslop, J. J., et al. (2014). Hydrogen and methane emissions from beef cattle and their rumen microbial community vary with diet, time after feeding and genotype. Br. J. Nutr. 112, 398–407. doi: 10.1017/s0007114514000932
Sasson, G., Ben-Shabat, S. K., Seroussi, E., Doron-Faigenboim, A., Shterzer, N., Yaacoby, S., et al. (2017). Heritable bovine rumen bacteria are phylogenetically related and correlated with the cow’s capacity to harvest energy from its feed. mBio 8:e00703-17. doi: 10.1128/mBio.00703-17
Schingoethe, D. J., Kalscheur, K. F., Hippen, A. R., and Garcia, A. D. (2009). Invited review: the use of distillers products in dairy cattle diets. J. Dairy Sci. 92, 5802–5813. doi: 10.3168/jds.2009-2549
Shi, W. B., Moon, C. D., Leahy, S. C., Kang, D. W., Froula, J., Kittelmann, S., et al. (2014). Methane yield phenotypes linked to differential gene expression in the sheep rumen microbiome. Genome Res. 24, 1517–1525. doi: 10.1101/gr.168245.113
Stewart, C. S., Flint, H. J., and Bryant, M. P. (1997). “The rumen bacteria,” in The Rumen Microbial Ecosystem, eds P. N. Hobson, and C. S. Stewart, (Berlin: Springer), 10–72. doi: 10.1007/978-94-009-1453-7_2
Tapio, I., Snelling, T. J., Strozzi, F., and Wallace, R. J. (2017). The ruminal microbiome associated with methane emissions from ruminant livestock. J. Anim. Sci. Biotechnol. 8:7.
Ungerfeld, E. M. (2015). Shifts in metabolic hydrogen sinks in the methanogenesis-inhibited ruminal fermentation: a meta-analysis. Front. Microbiol. 6:37. doi: 10.3389/fmicb.2015.00037
Van Soest, P. V., Robertson, J. B., and Lewis, B. A. (1991). Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 74, 3583–3597. doi: 10.3168/jds.s0022-0302(91)78551-2
Wang, K., Nan, X., Chu, K., Tong, J., Yang, L., Zheng, S., et al. (2018). Shifts of hydrogen metabolism from methanogenesis to propionate production in response to replacement of forage fiber with non-forage fiber sources in diets in vitro. Front. Micrbiol. 9:2764. doi: 10.3389/fmicb.2018.02764
Weatherburn, M. W. (1967). Phenol-hypochlorite reaction for determination of ammonia. Anal. Chem. 39, 971–974. doi: 10.1021/ac60252a045
Whelan, S. J., Carey, W., Boland, T. M., Lynch, M. B., Kelly, A. K., Rajauria, G., et al. (2016). The effect of by-product inclusion level on milk production, nutrient digestibility and excretion, and rumen fermentation parameters in lactating dairy cows offered a pasture-based diet. J. Dairy Sci. 100, 1055–1062. doi: 10.3168/jds.2016-11600
Wilkins, D., Lu, X. Y., Shen, Z., Chen, J., and Lee, P. K. (2015). Pyrosequencing of mcrA and archaeal 16S rRNA genes reveals diversity and substrate preferences of methanogen communities in anaerobic digesters. Appl. Environ. Microbiol. 8, 604–613. doi: 10.1128/aem.02566-14
Wilkinson, J. M. (2011). Re-defining efficiency of feed use by livestock. Animal 5, 1014–1022. doi: 10.1017/s175173111100005x
Wilkinson, T. J., Huws, S. A., Edwards, J. E., Kingston-Smith, A. H., Siu-Ting, K., Hughes, M., et al. (2018). CowPI: a rumen microbiome focussed version of the PICRUSt functional inference software. Front. Microbiol. 9:1095. doi: 10.3389/fmicb.2018.01095
Xu, L., Jin, Y., He, M. L., Li, C., Beauchemin, K. A., and Yang, W. Z. (2014). Effects of increasing levels of corn dried distillers grains with solubles and monensin on ruminal biohydrogenation and duodenal flows of fatty acids in beef heifers fed high-grain diets. J. Anim. Sci. 92, 1089–1098. doi: 10.2527/jas.2013-6668
Yáñez-Ruiz, D. R., Bannink, A., Dijkstra, J., Kebreab, E., Morgavi, D. P., O’Kiely, P., et al. (2016). Design, implementation and interpretation of in vitro batch culture experiments to assess enteric methane mitigation in ruminants—a review. Anim. Feed Sci. Technol. 216, 1–18. doi: 10.1016/j.anifeedsci.2016.03.016
Yu, Z., and Morrison, M. (2004). Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques 36, 808–812. doi: 10.2144/04365st04
Zhou, M., Hünerberg, M., Beauchemin, K. A., McAllister, T. A., Okine, E. K., and Guan, L. L. (2012). Individuality of ruminal methanogen/protozoa populations in beef cattle fed diets containing dried distillers’ grain with solubles. Acta Agric. Scand. A Anim. Sci. 62, 273–288. doi: 10.1080/09064702.2013.788206
Keywords: rumen microbiome, by-products, RUSITEC, methane, 16S rRNA
Citation: Smith PE, Waters SM, Kenny DA, Boland TM, Heffernan J and Kelly AK (2020) Replacing Barley and Soybean Meal With By-products, in a Pasture Based Diet, Alters Daily Methane Output and the Rumen Microbial Community in vitro Using the Rumen Simulation Technique (RUSITEC). Front. Microbiol. 11:1614. doi: 10.3389/fmicb.2020.01614
Received: 02 April 2020; Accepted: 19 June 2020;
Published: 22 July 2020.
Edited by:
Antonio Faciola, University of Florida, United StatesReviewed by:
Gabriel O. Ribeiro, University of Saskatchewan, CanadaCopyright © 2020 Smith, Waters, Kenny, Boland, Heffernan and Kelly. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sinéad M. Waters, U2luZWFkLldhdGVyc0B0ZWFnYXNjLmll
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.