- 1Department of Isotope Biogeochemistry, Helmholtz Centre for Environmental Research – UFZ, Leipzig, Germany
- 2Chair of Geobiotechnology at TU Berlin, Berlin, Germany
Dehalococcoides mccartyi strain BTF08 has the unique property to couple complete dechlorination of tetrachloroethene and 1,2-dichloroethane to ethene with growth by using the halogenated compounds as terminal electron acceptor. The genome of strain BTF08 encodes 20 genes for reductive dehalogenase homologous proteins (RdhA) including those described for dehalogenation of tetrachloroethene (PceA, PteA), trichloroethene (TceA) and vinyl chloride (VcrA). Thus far it is unknown under which conditions the different RdhAs are expressed, what their substrate specificity is and if different reaction mechanisms are employed. Here we found by proteomic analysis from differentially activated batches that PteA and VcrA were expressed during dechlorination of tetrachloroethene to ethene, while TceA was expressed during 1,2-dichloroethane dehalogenation. Carbon and chlorine compound-specific stable isotope analysis suggested distinct reaction mechanisms for the dechlorination of (i) cis-dichloroethene and vinyl chloride versus (ii) tetrachloroethene. This differentiation was observed independent of the expressed RdhA proteins. Differently, two stable isotope fractionation patterns were observed for 1,2-dichloroethane transformation, for cells with distinct RdhA inventories. Conclusively, we could link specific RdhA expression with functions and provide an insight into the apparently substrate-specific reaction mechanisms in the pathway of reductive dehalogenation in D. mccartyi strain BTF08. Data are available via ProteomeXchange with identifiers PXD018558 and PXD018595.
Introduction
Chlorinated organic contaminants, such as tetrachloroethene (PCE), belong to the most frequent groundwater contaminants entering the environment through accidents, improper handling and waste disposal (Carter et al., 2008). Thus far, only microorganisms belonging to the class Dehalococcoidia can catalyze complete dehalogenation of PCE to the non-toxic ethene and have been shown to be key players for in situ bioremediation of chlorinated solvents (Löffler et al., 2013). Dehalococcoides mccartyi strain BTF08 harbors the unique property to couple all reductive dehalogenation steps starting from PCE down to ethene to growth referred to as organohalide respiration (Cichocka et al., 2010; Kaufhold et al., 2013). Further, strain BTF08 is capable of dehalogenating vicinal halogenated alkanes to ethene (Schmidt et al., 2014). Its genome contains 20 reductive dehalogenase homologous gene clusters (rdhAB) wherefrom 16 rdhAB genes belong to classified orthologue groups being conserved throughout cultivated D. mccartyi strains (Hug et al., 2013). The rdhAB genes in strain BTF08 include orthologues of pceA, tceA, and vcrA, proposed to be involved in PCE, TCE, and vinyl chloride (VC) dehalogenation, respectively, while the other rdhA genes can currently not be functionally assigned. Thus, D. mccartyi strain BTF08 possesses all RdhA encoding genes required for complete reductive dehalogenation of PCE to ethene (Pöritz et al., 2013). Functional annotation of the rdhA genes in strain BTF08 has been done on the basis of their affiliation to orthologue groups containing biochemically characterized orthologues in other D. mccartyi strains (Hug et al., 2013), but the biochemical activity has not been confirmed experimentally in D. mccartyi strain BTF08.
Among all RdhA several conserved features were identified, including a twin-arginine signal motif for cytoplasmic membrane translocation, two iron-sulfur cluster binding motifs as well as a motif for a corrinoid-containing co-factor (Magnuson et al., 2000; Holscher et al., 2004). Based on investigation with the corrinoid cyanocobalamin (Vitamin B12), a super-reduced Co(I) species was shown to be involved in the catalytic activity of RdhA (Miller et al., 1996; Schumacher et al., 1997; Middeldorp et al., 1999; Parthasarathy et al., 2015; Kunze et al., 2017). Reactions with the pure corrinoid were proposed to take place as (i) inner sphere single electron transfer (SET) (Cooper et al., 2015; Payne et al., 2015), (ii) inner sphere two electron transfer (Kliegman and McNeill, 2008) or (iii) long range single electron transfer (Bommer et al., 2014; Figure 1). Studies on structurally characterized RdhA proposed either long range SET for PceA from Sulfurospirillum multivorans (Bommer et al., 2014) and VcrA from Dehalococcoides mccartyi strain VS (Parthasarathy et al., 2015) or inner sphere SET for NpRdhA from Nitratireductor pacificus (Payne et al., 2015). Inner sphere two electron transfer was recently proposed in studies investigating biotic and abiotic dehalogenation of halogenated ethenes using compound-specific stable isotope analysis (CSIA) (Heckel et al., 2017, 2018; Lihl et al., 2019).
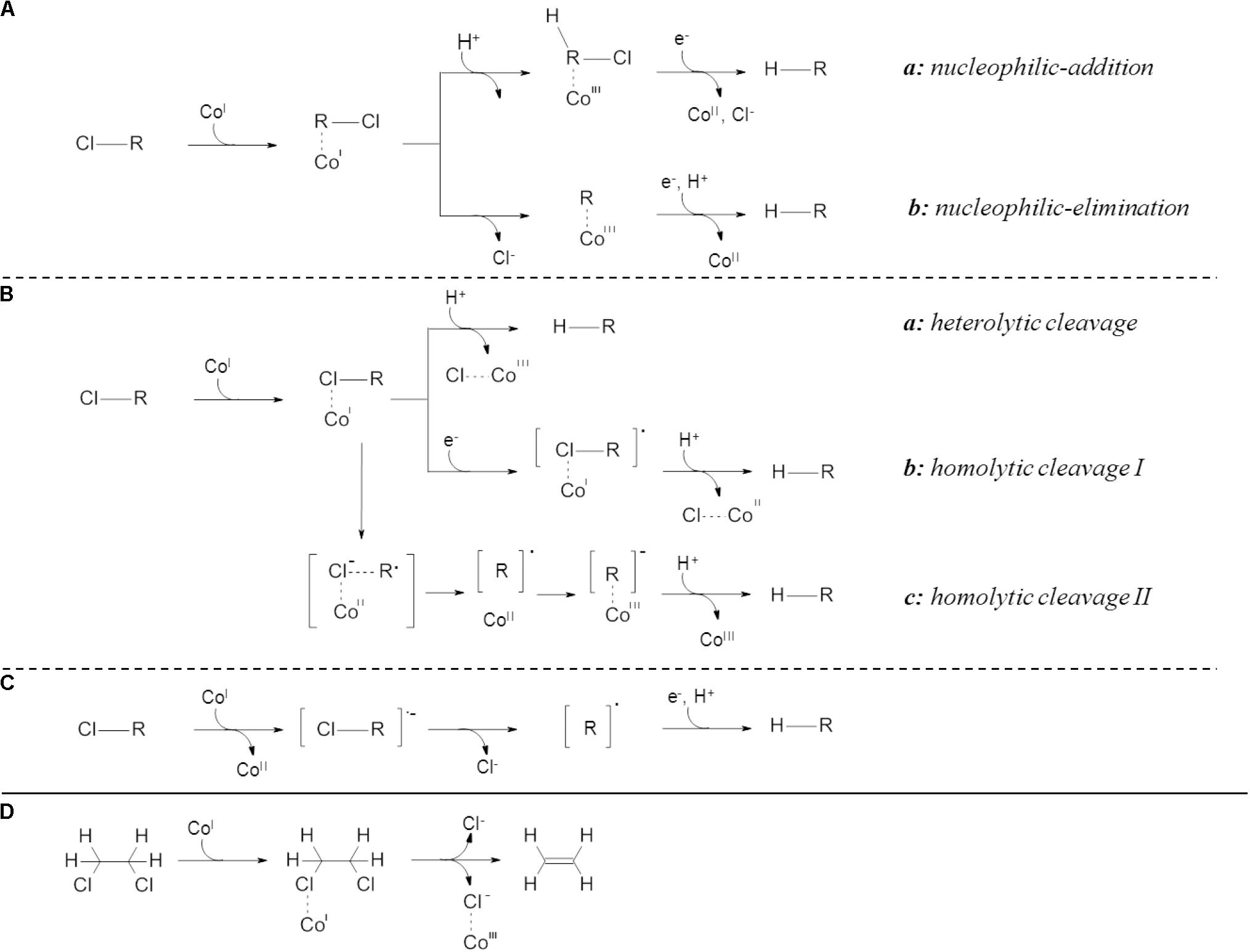
Figure 1. Proposed mechanisms for reductive dehalogenation. (A) inner sphere two electron transfers, (B) inner sphere single electron transfers displaying different modes of bond cleavage, (C) outer sphere single electron transfer and (D) inner sphere single electron transfer for vicinally substituted compounds. Inner sphere two electron transfer (A) is further divided in nucleophilic-addition reaction (a), characterized by initial protonation followed by carbon-halogen bond cleavage, and nucleophilic elimination (b), possessing initial carbon-halogen bond cleavage with subsequent protonation of the remaining carbon (Heckel et al., 2018; Lihl et al., 2019). Inner sphere single electron transfers (B) are described as heterolytic (a) or homolytic (b,c) cleavage (Cooper et al., 2015; Payne et al., 2015). Two distinct mechanisms of homolytic cleavage are proposed leading either to abstraction of a cobalt-halogen complex (Cooper et al., 2015; Payne et al., 2015) (b) or association of the carbon backbone to the cobalt (Cooper et al., 2015; Payne et al., 2015) (c). Outer sphere single electron transfers (C) do not involve a carbon or halogen to cobalt interaction (Kunze et al., 2017). Inner sphere single electron transfer for vicinally substituted compounds (D) was proposed for concerted halogen removal (Payne et al., 2015). Displayed are the hydrocarbon backbone [R] and the oxidation states of the cobalt within the cobamid cofactor [CoI–III].
Compound-specific stable isotope analysis emerged as powerful tool to evaluate reaction mechanisms (Elsner et al., 2005; Hirschorn et al., 2007; Schmidt et al., 2014), allowing to relate isotope fractionation patterns with distinct reactions and characterization of the rate-determining step (Vogt et al., 2008). Recently, the absence of chlorine isotope fractionation was shown for outer-sphere SET on halogenated ethenes (Heckel et al., 2017). Additionally, a dichotomy in the reaction mechanisms during TCE-dehalogenation was observed in dependence on the electron acceptor used in the previous cultivation for several D. mccartyi containing mixed cultures and Geobacter lovleyi strain KB-1 (Lihl et al., 2019). The involved electron transfer reactions were proposed to be inner sphere two electron transfers (Figure 1A). However, this was not linked to specific microorganisms or dehalogenases (Lihl et al., 2019). Thus, we hypothesized the presence of distinct, electron-acceptor specific reaction mechanisms during corrinoid-dependent dechlorination at their respective RdhAs.
Here we aimed to characterize the reaction mechanisms during dehalogenation of PCE, cDCE, VC, and 1,2-dichloroethane (1,2-DCA) using dual element (C/Cl) CSIA. Furthermore we aimed to investigate the RdhA inventory present in the underlying dehalogenation processes by experiments.
Materials and Methods
Cultivation of Dehalococcoides mccartyi Strain BTF08
Dehalococcoides mccartyi strain BTF08 was pre-cultivated for at least 4 transfers in 100 mL mineral medium (final pH 6.8) in 240 mL serum bottles and in total 1.2 mM PCE, cis-DCE, 1,2-DCA or VC as electron acceptor prior to transfer 0 (Supplementary Figure S1A) as described elsewhere (Schmidt et al., 2014; Franke et al., 2017). Chlorinated solvents were added stepwise with initially approximately 200 μM, and subsequently 400 and 600 μM, added after the previous dose was converted. Cultivation in transfers 0, 1, and 2 was conducted using 100 mL mineral salt medium in 240 mL serum bottles (Supplementary Figures S1B,C). Upscaling of cultivation (transfer 1, Supplementary Figure S1D) was done with 500 mL mineral salt media in 1 L serum bottles. All cultivation bottles were flushed with N2 and CO2 (70/30%), closed with Teflon-lined butyl rubber stoppers and aluminum crimp caps and sterilized for 40 min at 120°C. Triplicate bottles were amended with 500 μM PCE, cis-DCE, 1,2-DCA or VC, and with another 1 mM of the respective electron acceptor after the first dose was completely consumed. The bottles were inoculated with 5% (v/v) of transfer 0 grown on PCE, cis-DCE, 1,2-DCA or VC, respectively. Hydrogen as electron donor was added at 0.5 bar overpressure. Cultures were incubated in the dark without shaking at 20°C.
For proteomic analyses, 30 mL of the culture were processed for LC-MS/MS analysis after each dose of electron acceptor (first: 0.5 mM, second: 1 mM) was completely dehalogenated. Cultures were then transferred to fresh medium with an inoculum of 10% (v/v) and sampling was repeated (Supplementary Figures S1B,C).
Isotope Fractionation Experiments and Activity Assays
Isotope fractionation experiments and activity assays were conducted using resting cells. Cells were harvested from 400 mL culture (Supplementary Figure S1D) by anoxic centrifugation at 16°C and 5000 × g for 1 h. Half of the supernatant was removed anoxically and the residual fraction was centrifuged again under the conditions stated above. Under anoxic conditions, the supernatant was removed completely, the pellet was resuspended in 1 M potassium acetate buffer pH 6.8 and transferred to a sterile anoxic glass vial crimp closed with Teflon-lined butyl rubber stoppers.
Fractionation experiments and activity assays were conducted anoxically using 200 mM potassium acetate buffer pH 6.8, 1 mM methyl viologen and 0.2 mM titanium-III-citrate in 10 mL glass vials, yielding in total 3.9 mL reaction mixture, and crimp closed with Teflon-lined butyl rubber stoppers. 50 μL of the 100 mM electron acceptor stock solution in ethanol were added and the mixture was shaken for 30 min at 20°C in the dark. Another 2 mM titanium-III-citrate and 1 mL of the resuspended pellet were added per vial to start the dehalogenation reaction. Nine vials were amended per set (Supplementary Figure S1E), with one of the electron acceptors PCE, cis-DCE, VC or 1,2-DCA, and one of the four resting cell suspensions (derived from cultures amended with one of the electron acceptors, PCE, cis-DCE, VC or 1,2-DCA). Additionally, four vials were kept as abiotic control without addition of cells but 1 mL anoxic desalted and bi-distillated water. Vials were shaken during incubation at 30°C in the dark and dehalogenation progress was determined by analysis of electron acceptor and product concentrations in the culture headspace via GC-FID. Vials were sacrificed at different extents of dehalogenation, described as Ct/C0 (where Ct is the residual electron acceptor concentration at the sampling time t and C0 the initial concentration) ranging between 0.1 and 0.9, by addition of 0.5 mL of acidic sodium sulfate solution (280 g⋅L–1, pH 1) as described elsewhere (Franke et al., 2017). Sacrificed samples were analyzed using gas chromatography with isotope ratio mass spectrometry.
Concentration Analysis by Gas Chromatography-Flame Ionization Detection (GC-FID)
Concentrations of PCE, cDCE, 1,2-DCA, VC and the product ethene were determined by analysis of headspace samples via gas chromatography with FID (Varian Chrompack CP-3800, Middleburg, the Netherlands) equipped with a GS-Q-column (30 m × 0.53 mm, J&W Scientific, Waldbronn, Germany) as described previously (Schmidt et al., 2014).
Analysis of Carbon Isotope Ratios by Gas Chromatography – Isotope Ratio Mass Spectrometry
Carbon isotope ratios were determined via compound-specific stable isotope analyses using GC-isotope ratio mass spectrometry (IRMS). The organic mixture was separated by gas chromatography and baseline-separated compounds were chemically converted into the analyte gas CO2 for 13C/12C analysis (Elsner et al., 2012). Carbon isotope fractionation was determined in triplicate by injection into the gas chromatograph (Agilent 6890, Palo Alto, CA, United States) using a CP-PoraBOND column (50 m × 0.32 mm, 5 μm inner diameter, J&W Scientific, Germany). One mL of the reaction mixture was transferred to a He-flushed 10 mL glass vial wherefrom 1 mL headspace sample was injected via autosampler (TriPlus RSH, Thermo Scientific, Germany) with a split ratio of 1:5. For chromatographic separation a temperature gradient program was used, starting from 30°C (held for 10 min) followed by a 20°C ⋅ min–1 gradient to 250°C (held for 5 min) with 2 mL ⋅ min–1 flow and an injector temperature of 280°C (Franke et al., 2017). Carbon isotope ratios were determined at the IRMS (MAT 235, Thermo Scientific, Germany) relative to the laboratory reference gas [CO2, calibrated against Vienna Pee Dee Belemnite standard V-PDB, IAEA Vienna, Austria (Coplen et al., 2006)]. The overall analytical uncertainty was <0.5%.
Analysis of Chlorine Isotope Ratios by Gas Chromatography – Multi Collector-Inductively Coupled Plasma Mass Spectrometry
For chlorine isotope analysis a multi collector-inductively coupled plasma mass spectrometer was coupled to gas chromatography (GC-MC-ICPMS) (Horst et al., 2017). The MC-ICPMS, a Neptune (Thermo Fisher Scientific, Germany), was equipped with a gas chromatograph Trace 1310 (Thermo Scientific, Germany) coupled to FID. Triplicate samples were analyzed by manual injection of 0.05–1 mL headspace with a split ratio of 1:10 with injector kept at 250°C and a carrier gas flow of 2 mL⋅min–1. For the analysis, a Zebron ZB-1 capillary column (60 m × 0.32 mm i.d., 1 μm film thickness; Phenomenex Inc.) was utilized isothermal at 100°C for 1,2-DCA, VC and cis-DCE and 120°C for PCE analysis. The separated compounds were transferred to the plasma via a Thermo Electron TransferLine (AE2080, Aquitaine Electronique, France) heated to 250°C using an auxiliary helium flow of 5 mL⋅min–1. Instrument tuning and preparation was performed daily prior to the measurements (Horst et al., 2017). Chlorine isotope ratios were determined relative to the laboratory standards [methyl chloride, TCE-2, and TCE-6 (Horst et al., 2017; Renpenning et al., 2018)]. The overall analytical uncertainty was <0.5%.
Evaluation of Isotopic Data
Stable isotope composition was reported in δ-notation (%) relative to an international standard (Coplen et al., 2006; Elsner, 2010; Renpenning et al., 2015). The calculation of carbon and chlorine isotope enrichment factors εX (where X represents the analyzed element, i.e., C or Cl, respectively) was accomplished according to the Rayleigh-Equation (1) (Mariotti et al., 1981). The concentrations at different time points (t) of the reaction and the starting concentration (0) are represented as Ct and C0, respectively, whereas R = 1 + δ (Coplen et al., 2006).
The slope derived from the dual-element plot (m2D) equals the Λ-value and approximately conforms to the ratio of the enrichment factors of the compared elements (εC/εCl) according to Eq. 2.
Based on the slope of the regression a two-tailed T-test was uses to calculate the 95% confidence interval (data evaluation in Microsoft Excel).
Apparent Kinetic Isotope Effects
The previously calculated bulk enrichment factor εXbulk describes the isotopic fractionation of the whole molecule. To determine position-specific kinetic isotope effects intra-molecular competition and non-reacting positions have to be taken into account (Elsner et al., 2005). The AKIE values were determined for PCE, cDCE, 1,2-DCA, and VC according to Eq. (3) (Elsner et al., 2005). The number of carbon or chlorine atoms is described by n, the number of atoms at the reactive position by x, whereas z takes the number of indistinguishable reactive sites into consideration. The following values were used for carbon (n = 2 for all cases): PCE: x = 1, z = 4; cis-DCE: x = 1, z = 2; VC: x = 1, z = 1; 1,2-DCA: x = 2, z = 2 (concerted) or x = 1, z = 2 (stepwise). For chlorine, the following values were used: PCE: n = 4, x = 1, z = 4; cis-DCE: n = 2, x = 1, z = 2; VC: n = 1, x = 1, z = 1; 1,2-DCA: n = 2 and x = 2, n = 2 (concerted) or x = 1, z = 2.
Cell Harvesting and Sample Preparation for Proteomic Analysis
Dehalococcoides mccartyi strain BTF08 cultures cultivated with different halogenated electron acceptors PCE, cDCE, 1,2-DCA or VC, were harvested by centrifugation of 30 mL culture at 10,000 × g for 45 min at 16°C. Cell pellets were washed with 100 mM ammonium bicarbonate buffer pH 7.9, centrifuged at 10,000 × g for 30 min at 16°C and subsequent removal of the supernatant. The cell pellet was re-suspended in 30 μL 50 mM ammonium bicarbonate buffer pH 7.9 and the cells were disrupted by three cycles of freezing, using liquid nitrogen, and thawing at 40°C while shaking at 750 rpm. Proteins were reduced using 50 mM dithiothreitol for 1 h at 30°C (400 rpm) and alkylated by 130 mM 2-iodoacetamide for 1 h at 22°C and 400 rpm prior to trypsin digest overnight through 0.6 μg trypsin (Sequencing Grade Modified Trypsin, Promega) at 37°C at 400 rpm. Trypsin digest was stopped by 1 μL 100% formic acid and followed by centrifugation at 13,000 rpm (Centrifuge 5430, rotor FA-45-24-11, Eppendorf, Germany) for 20 min. The pellet was discarded, the supernatant concentrated to 10 μL by vacuum centrifugation at 20 mbar and stored until further processing at −20°C.
Peptides for LC-MS/MS analysis were desalted before injection using ZipTip-μC18 material (Merck Millipore, Germany). Solvents were evaporated under vacuum and samples re-suspended in 0.1% formic acid prior to LC-MS/MS analysis.
LC-MS/MS and Proteomic Data Analysis
Proteomic analysis was performed via nano-liquid chromatography-tandem mass spectrometry (nLC-MS/MS) using an Orbitrap Fusion (Thermo Fisher Scientific, Bremen, Germany) coupled with a nano ultra-performance liquid chromatography system (Dionex Ultimate 3000RSLC; Thermo Scientific, Germany) and an Acclaim PepMap 100 C18 LC-Column (Thermo Fisher Scientific) as described previously (Goris et al., 2016). For liquid chromatography 0.1% formic acid (eluent A) and 80% acetonitrile with 0.08% formic acid (eluent B) were used and 3 μL of sample was injected. With a constant flow rate of 300 nL⋅min–1 4% eluent A was hold for 6 min prior to increase to 55% eluent A over 120 min being further increase to 90% within 1 min, hold for 4 min and decreased back to 4% eluent A within 1 min.
Subsequent peptide analysis was conducted using Proteome Discoverer (v2.2, Thermo Fisher Scientific, Germany) with the D. mccartyi strain BTF08 genome as search database (UniProt Proteome ID: UP000011727) (Pöritz et al., 2013). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [1] partner repository with the dataset identifiers PXD018558 and PXD018595.
Classification Into RdhA Orthologous Groups
Determination of the orthologous groups to which the identified RdhA proteins belong was done via sequence alignment of the data used by Hug et al. (2013) and the proteome of D. mccartyi strain BTF08 (UniProt Proteome ID: UP000011727) (Hug et al., 2013; Pöritz et al., 2013) followed by phylogenetic analysis of this alignment. The alignment and phylogenetic tree (Minimum Evolution Tree with 100 bootstrap repetitions) were performed using Mega X (Kumar et al., 2016).
Results
Dehalogenation Activity in Growing Cultures Correlates With Electron Acceptor Change
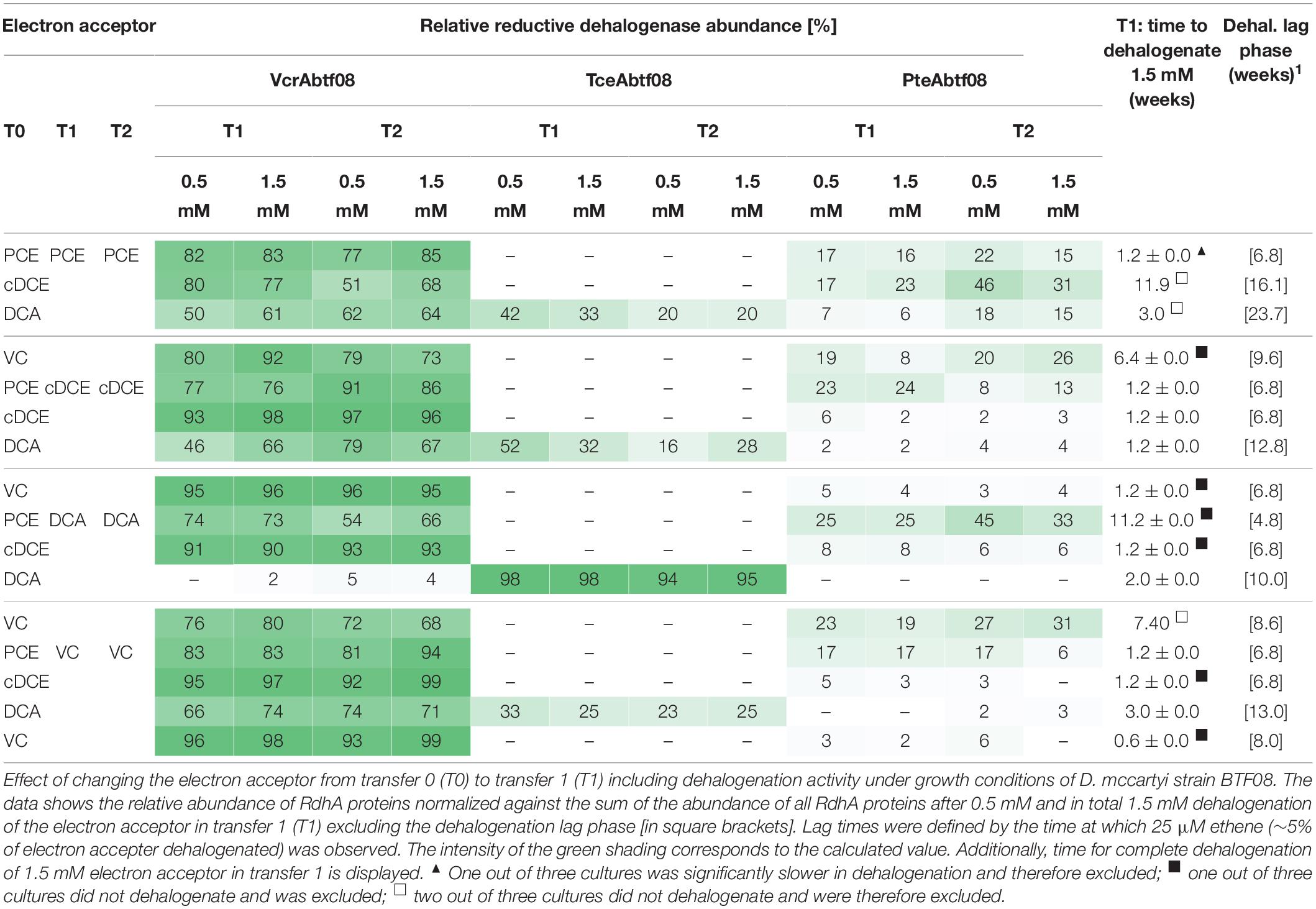
Dehalococcoides mccartyi strain BTF08 was cultivated on PCE, cDCE, VC or 1,2-DCA as electron acceptor after growth for at least four transfers with one constant electron acceptor, either PCE, cDCE, VC or 1,2-DCA (Supplementary Figures S1A–C). D. mccartyi strain BTF08 adapted fast, with no obvious delay in dehalogenation compared to controls maintained on the same substrate, when the electron acceptor was switched in most cases (Table 1). Delayed dechlorination to ethene (as indicated by the dehalogenation lag time) was observed when switching from 1,2-DCA, cDCE or VC to PCE. The dechlorination activities (the time to dehalogenate 1.5 mM organohalide, see Table 1) were highest with cDCE and VC regardless of the electron acceptor used for pre-cultivation, whereas those with PCE and 1,2-DCA showed stronger variations. Dechlorination activity, however, was decreased when switching from 1,2-DCA to VC as well as from PCE or VC to 1,2-DCA.
Analysis of the Expression of RdhA Proteins in vivo in Response to Electron Acceptor Change
The expression of RdhA proteins (Table 1) was analyzed in response to the electron acceptor present. Seven RdhA proteins were detected and assigned to orthologous groups (OG) (Supplementary Figure S4). The RdhA_ btf08_185 (NCBI: AGG07294, OG5) and RdhA_btf08_1407 (AGG08472, OG8) correspond to the TCE reductive dehalogenase (TceAbtf08) and VC reductive dehalogenase (VcrAbtf08), respectively, previously described (Müller et al., 2004; Pöritz et al., 2013). RdhA_btf08_1393 (AGG08458, OG16) was present in increased abundance if PCE was used as electron acceptor suggesting that it acts as a PCE reductive dehalogenase. As RdhA_btf08_1393 shares 99% amino acid sequence similarity to an identified PteA from D. mccartyi strain 11a5 (Zhao et al., 2016) it will be referred to as PteAbtf08. Thus, RdhA_btf08_1393 (PteAbtf08) and not the expected RdhA_btf08_1454 (PceAbtf08, AGG08519), also encoded in the genome (Pöritz et al., 2013), was associated with PCE dechlorination. Minor abundant (less than 2% of the expressed RdhA) and not functionally identified were RdhA_btf08_1481 (AGG08546, OG10), RdhA_btf08_1497 (AGG08562, OG15), RdhA_btf08_1488 (AGG08553, OG17), and RdhA_btf08_121 (AGG07230, OG23).
PteAbtf08 had a relative high abundance in cultures with PCE as electron acceptor (Table 1, 2). PteAbtf08 functionality was confirmed as its expression increased if PCE was used as electron acceptor, after transfer of cells pre-grown on cDCE or 1,2-DCA. PteAbtf08 relative abundance decreased if the electron acceptor was switched from PCE to cDCE or VC (Table 1). Expression of PteAbtf08 increased once electron acceptor was switched from PCE or VC to 1,2-DCA, although with apparently lower rates for 1,2-DCA dehalogenation (Table 1). VcrAbtf08 was dominantly expressed in presence of halogenated ethenes but not for the cultures maintained on 1,2-DCA (VcrAbtf08 ≤ 5%). After transfer from cultures with 1,2-DCA to halogenated ethenes, VcrAbtf08 expression was increasing with each dose of the respective halogenated ethene. No change in expression was visible after the next transfer anymore. Strikingly, TceAbtf08 was expressed as almost the only RdhA if 1,2-DCA was the sole electron acceptor during cultivation. The abundance of TceAbtf08 decreased when the electron acceptor was changed to one of the halogenated ethenes. Interestingly, TceAbtf08 was not detected in cultures with 1,2-DCA, previously cultivated on the chlorinated ethenes, however, PteAbtf08 and VcrAbtf08 were detected.
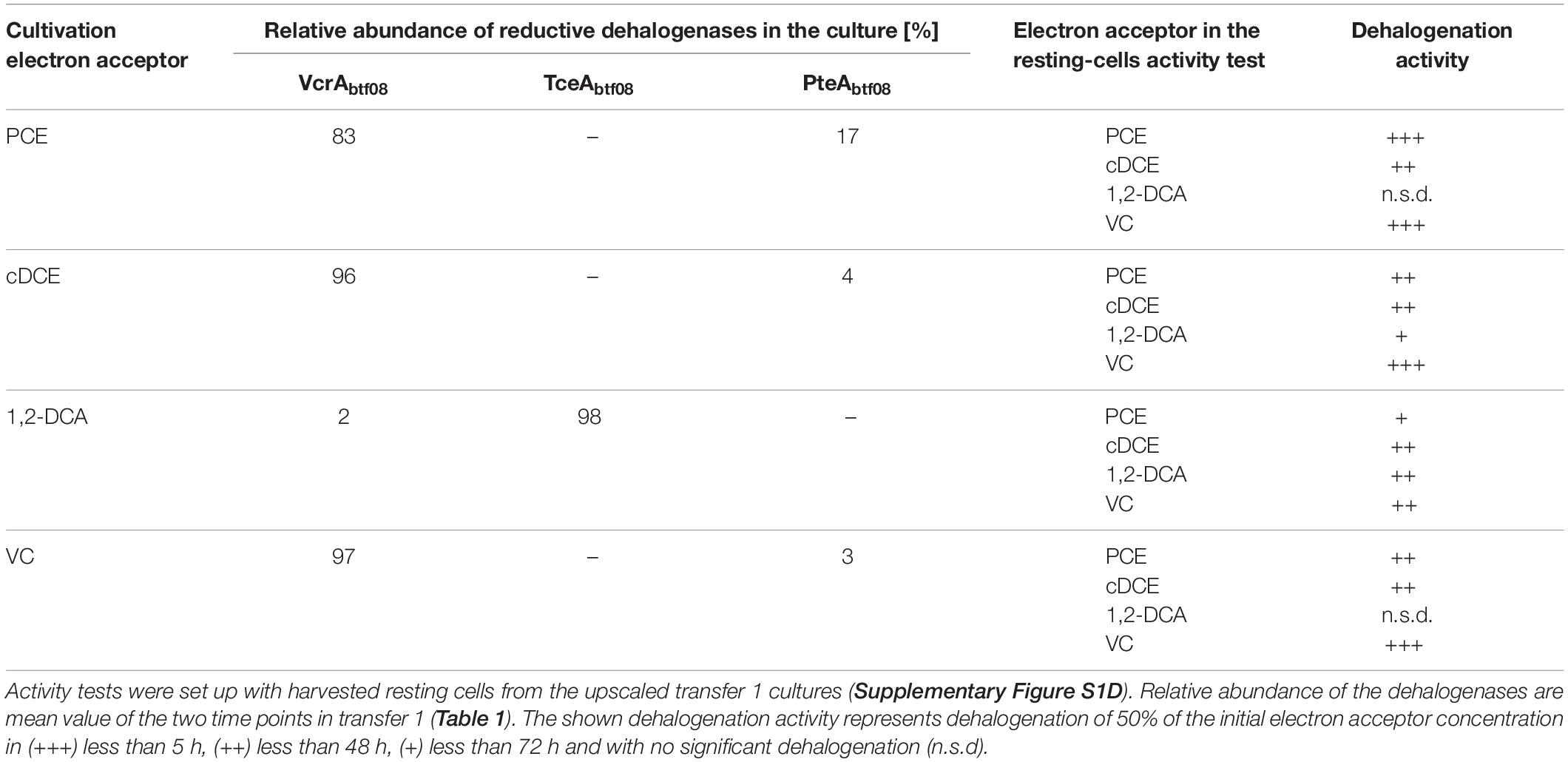
Substrate Specificity of Differently Induced Cells and Related RdhA Inventory
Subsequently, substrate specificity of the RdhAs was tested using resting cells and the artificial electron donor methyl viologen. Resting cells contained the above described RdhA inventory (Table 2) and were derived from D. mccartyi strain BTF08 cultures which were cultivated for several transfers with a single specific electron acceptor (Supplementary Figure S1E). Cells with almost exclusively TceAbtf08 present, dechlorinated 1,2-DCA, cDCE, and VC effectively and PCE at lower rates completely to ethene (data not shown). 1,2-DCA was dechlorinated in cases when TceAbtf08 was absent only if cells were cultivated with cDCE, albeit at lower rates (VcrAbtf08 was present as almost exclusive RdhA). In cells cultivated with PCE or VC as electron acceptor 1,2-DCA was not dehalogenated. All tested halogenated ethenes were dechlorinated in cells when VcrAbtf08 (>96%) was dominantly present.
Stable Isotope Fractionation Patterns Suggest Distinct Dehalogenation Mechanisms
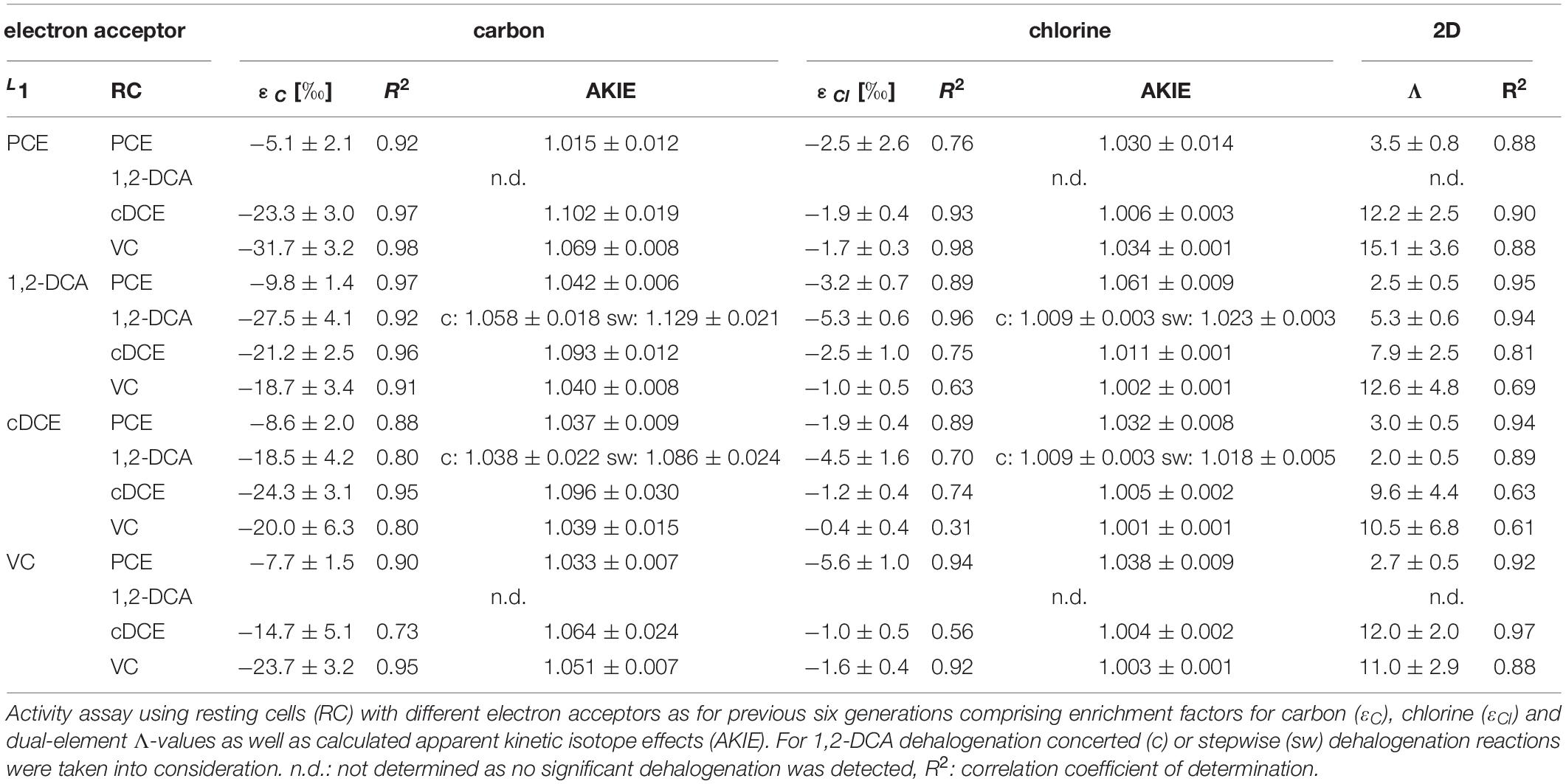
The underlying reaction mechanisms for dechlorination of PCE, cDCE, DCA, and VC were characterized by dual carbon and chlorine stable isotope analysis. Stable carbon isotope analysis of cDCE-, DCA- and VC-dechlorination show stronger isotope fractionation compared to PCE-dechlorination (Supplementary Figure S2), independently of the electron acceptor used for cultivation of cells and related RdhA inventory (Table 2). Stable chlorine isotope analysis revealed stronger chlorine isotope fractionation for PCE or DCA compared to cDCE or VC (Supplementary Figure S3). For comparison of the reaction mechanisms, dual-element fractionation patterns were evaluated (Figure 2) and Λ-values, relating the carbon to chlorine isotope fractionation, were calculated. Two main patterns could be observed for halogenated ethenes, (i) relatively strong carbon versus chlorine isotope fractionation for cDCE and VC and (ii) lower carbon vs. chlorine isotope fractionation for PCE. 1,2-DCA dehalogenation showed strong chlorine isotope fractionation either vs. (i) strong carbon isotope fractionation (>98% TceAbtf08) or vs. (ii) lower carbon isotope fractionation 1,2-DCA (>96% VcrAbtf08).
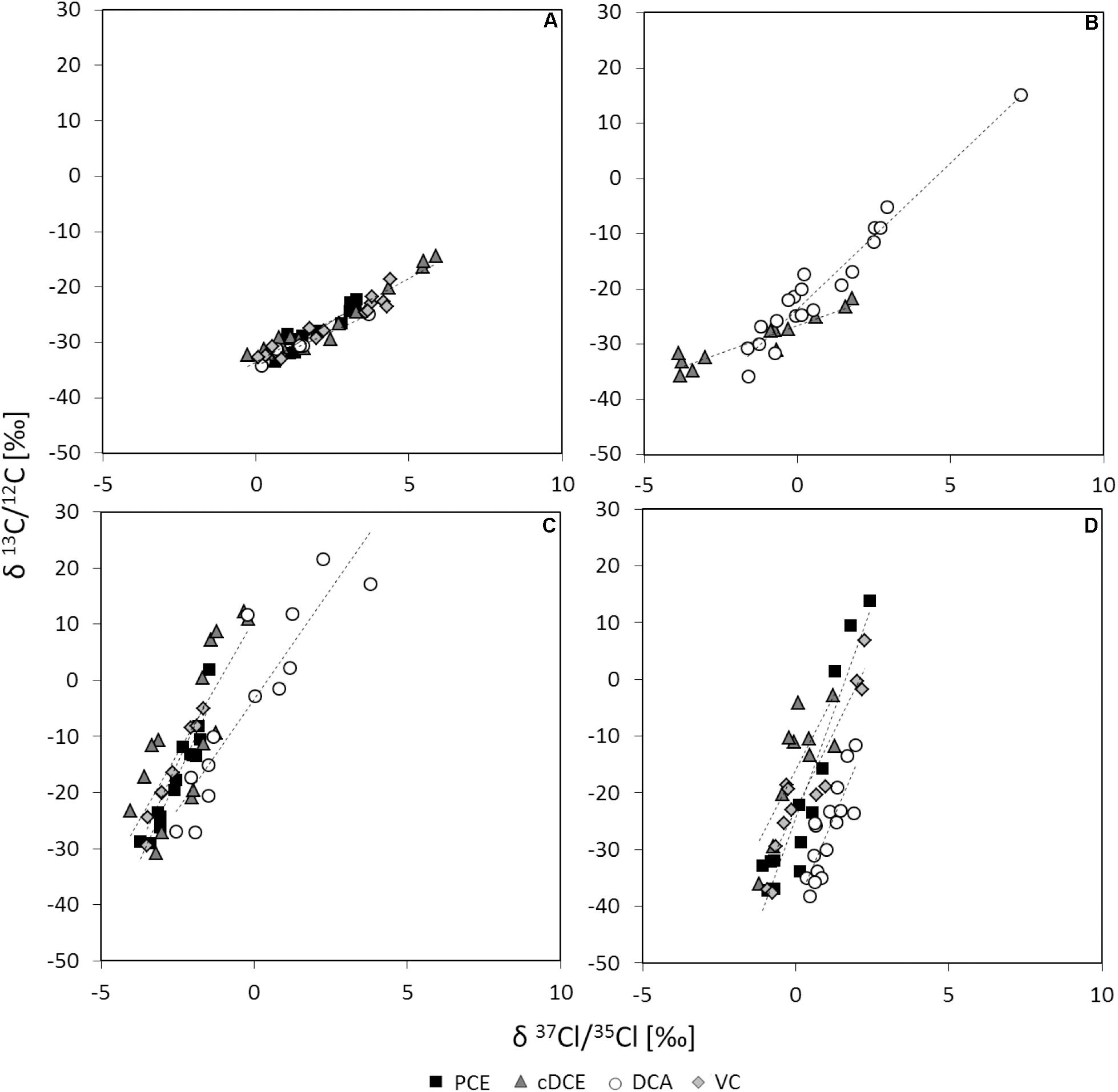
Figure 2. Double-element-plots for stable carbon and chlorine isotope fractionation. Activity assays using resting cells of D. mccartyi strain BTF08 with electron acceptors (A) PCE, (B) 1,2-DCA, (C) cDCE or (D) VC in dependence of the electron acceptors used for growth in upscaled transfer 1. Linear fits are represented by dashed lines.
Apparent kinetic isotope effects (AKIE) were calculated for the substrate to confirm the difference in reaction of the chlorinated ethenes (Table 3). Relative AKIEC/AKIECl showed a similar trend with 0.98 to 1.00 for PCE vs. 1.04 to 1.10 for cis-DCE and VC. Further, the AKIE for 1,2-DCA was evaluated to investigate whether concerted or stepwise removal of chlorine takes place during dehalogenation (Table 3). For carbon stable isotope analysis, AKIE calculated for concerted 1,2-DCA dechlorination did not differ significantly in comparison to dechlorination of PCE and VC. Chlorine AKIE were similar to those calculated for cDCE and VC but significantly lower than for PCE. Calculated carbon AKIE for stepwise dehalogenation of 1,2-DCA (1.086 to 1.129) were similar to those calculated for cDCE (1.064 to 1.102) and significantly higher than those for PCE (1.015 to 1.042) or VC (1.039 to 1.069). Chlorine AKIE for stepwise 1,2-DCA dehalogenation (1.018 to 1.023) were significantly higher than those for cDCE and VC (1.001 to 1.011) but lower than for PCE dehalogenation (1.030 to 1.061). For stepwise dechlorination of 1,2-DCA all AKIE exceeded the theoretical Streitwieser limit for C-Cl-bond cleavage, KIEC = 1.057 and KIECl = 1.013 (Elsner et al., 2005). Stepwise dehalogenation of 1,2-DCA is therefore considered unlikely, agreeing with previous studies (Fletcher et al., 2011; Palau et al., 2017). Overall, the high observed AKIEC for cDCE and VC fit with previous observed isotope fractionation during dechlorination by various D. mccartyi strains and D. mccartyi containing consortia (Fletcher et al., 2011).
Discussion
Expression analysis of strain BTF08 under different conditions led to different conclusions of the functional activity of the reductive dehalogenases as assumed previously. For example TceAbtf08 sharing 96% amino acid sequence identity TceA195 (Seshadri et al., 2005; Pöritz et al., 2013), was previously described to be capable of cDCE, VC, TCE, and 1,2-DCA dehalogenation (Magnuson et al., 2000). However, in D. mccartyi strain BTF08 TceAbtf08 dehalogenated 1,2-DCA, cDCE and VC as well as PCE to ethene, albeit at lower rates. Thus, TceAbtf08 is presumably capable of complete dehalogenation of PCE to ethene. Interestingly, changing the electron acceptor from halogenated ethenes to 1,2-DCA did not lead to expression of TceAbtf08 although dehalogenation of 1,2-DCA was observed. PCR analysis showed that the gene encoding for TceAbtf08 was still present in the samples, excluding the possibility that it might have been lost during transfers (data not shown). VcrAbtf08, highly expressed after 1,2-DCA addition, could have catalyzed the dehalogenation of 1,2-DCA in our experiments, similar to observation for strain VS (Parthasarathy et al., 2015). For dehalogenation of PCE to ethene TceAbtf08 was not expressed, indicating that it is not needed for this reaction sequence, in contrast to a previous proposal based on genome analysis (Pöritz et al., 2013). PCE was completely dechlorinated to ethene in the presence of PteAbtf08 and VcrAbtf08. The tetrachloroethene reductive dehalogenase PteAbtf08 was expressed in D. mccartyi strain BTF08 in the presence of PCE. This was noteworthy, as pceA was described in the genome of strain BTF08 (Pöritz et al., 2013) and PceAbtf08 was thought to catalyze PCE dehalogenation. However, it was not detected within in this study. Consequently, in strain BTF08, PteAbtf08 is proposed to dechlorinate PCE to TCE, and VcrAbtf08 presumably dehalogenates TCE, cDCE, and VC to ethene. VcrA from D. mccartyi strain VS was described capable of 1,2-DCA (Parthasarathy et al., 2015) and TCE (Müller et al., 2004) dehalogenation, albeit TCE dehalogenation proceeds at significantly lower dehalogenation rates compared to cDCE or VC dehalogenation (Müller et al., 2004).
Based on dual-element CSIA results, the dehalogenation mechanism of the chlorinated ethenes was observed to be dependent on the electron acceptor present in the activity assay. The specific reductive dehalogenases present apparently did not affect mechanism in D. mccartyi strain BTF08. This is in contrast to recent findings of Lihl et al. (2019), presenting mechanistic division for TCE dehalogenation if previously cultivated on (i) cDCE or VC (Figure 1Aa) or (ii) TCE or PCE (Figure 1Ab). In this previous study, the presence of rdhA genes was analyzed in the investigated consortia, and not for the proteins present. Nevertheless, even when similar rdhA genes were dominant, different isotope fractionation patterns were observed (e.g., KB-1 RF and KB-1/VC with dominant vcrA but Λ = 2.7 ± 0.2 and 18.2 ± 4.4). Contrarily, dehalogenases without close sequence similarity led to similar dual-element patterns (e.g., KB-1 RF with vcrA and Donna II with tceA with Λ = 2.7 ± 0.2 and 2.3 ± 0.1, respectively) (Lihl et al., 2019). Thus, investigation of the actual RdhA expressed is essential.
In our study, similar carbon and chlorine isotope fractionation patterns were observed for dechlorination of cDCE and VC, suggesting involvement of a similar reaction mechanism, in accordance with the dechlorination of both, cDCE and VC, by VcrAbtf08. However, different carbon and chlorine isotope fractionation patterns were observed during dehalogenation of PCE or 1,2-DCA suggesting additional, distinct, dechlorination mechanisms for these two electron acceptors. Studies on the recently isolated PceA from Sulfurospirillum multivorans (Bommer et al., 2014) and the VcrA from D. mccartyi strain VS (Parthasarathy et al., 2015) proposed long range SET (Figure 1C) whereas inner sphere SET was proposed either by homolytic or heterolytic bond cleavage for NpRdhA from Nitratireductor pacificus (Payne et al., 2015) and by homolytic bond cleavage for D. mccartyi strain CBDB1 (Cooper et al., 2015; Figure 1Ba,b). Inner sphere nucleophilic substitutions involving covalent cobalt-carbon-intermediates were postulated for halogenated alkenes on the basis of mass spectrometry and isotope fractionation (Heckel et al., 2017, 2018; Figure 1A).
The pronounced carbon isotope fractionation relative to chlorine isotope fractionation for cDCE and VC suggests a nucleophilic substitution-addition reaction via a Co(III)-alkyl-complex (Figure 1Aa). As PCE dehalogenation led to a more pronounced chlorine isotope fractionation relative to carbon isotope fractionation, a nucleophilic substitution-elimination mechanism via a Co(III)-vinyl-complex intermediate was proposed (Figure 1Ab). This difference in reaction between PCE vs. cDCE and VC was also predicted by computational modeling for the reductive dechlorination of chlorinated ethenes with cobalamin (Ji et al., 2017). Heckel et al. (2018) showed that outer sphere SET does not result in chlorine isotope fractionation for halogenated ethenes. Therefore, this mechanism may be excluded for dechlorination of PCE, cDCE and VC in D. mccartyi strain BTF08, as we observed significant chlorine isotope fractionation. However, chlorine isotope fractionation was reported previously for PCE dehalogenation in S. multivorans (Renpenning et al., 2014) for which long range SET is currently proposed (Kunze et al., 2017). Recent DFT (density function theory) modeling led to the proposition of a concerted chlorine removal and hydrogen insertion from a single [cobalt…halogen…carbon] transition state, either in inner sphere SET or by long range SET (Johannissen et al., 2017). Contradictorily to the studies by Heckel et al. (2018), the herein determined isotope fractionation pattern may be also explained in dependence of whether the insertion or the abstraction is taking place as rate-limiting step.
1,2-DCA dehalogenation in the presence of solely TceAbtf08 showed both pronounced chlorine and carbon isotope effects which cannot be explained with a reaction via nucleophilic substitution. 1,2-DCA was previously shown to be dechlorinated via dihaloelimination presumably following a concerted carbon-chlorine bond cleavage (Franke et al., 2017). An outer sphere SET (Figure 1C; Heckel et al., 2017) as well as an inner sphere SET are possible only if two electrons can be transferred almost concurrently and the remaining alkyl-radical can be stabilized sufficiently within the active site of the RdhA. Alternatively, a Co-halogen-bond formation with concomitant removal of vicinal halogens excludes the need for radical stabilization as proposed for NpRdhA (Payne et al., 2015) and is therefore assumed most likely (Figure 1D). Dual-element stable isotope patterns for 1,2-DCA dehalogenation are in close proximity to those determined for PCE dehalogenation, pointing toward an initial C-Cl-bond breakage. In our study, two main patterns are observed corresponding with previous observations for a Dehalogenimonas-containing microcosms (Λ = 1.89 ± 0.02) and Dehalococcoides-containing cultures (Λ = 6.8 ± 0.2; 6.9 ± 1.2) (Palau et al., 2017; Lihl et al., 2019). The differences in the Λ-values for 1,2-DCA dehalogenation in dependence on the most abundant RdhA can presumably be explained by (i) different underlying dehalogenation mechanisms or (ii) a different stabilization of intermediates within, the most likely different, active site of both RdhA. Latter assumption fits with differences between TceAbtf08 and VcrAbtf08, sharing 38% amino acid sequence identity, only (data not shown).
Isotope analyses allowed to characterize the underlying reaction, indicating distinct mechanisms for (i) PCE, (ii) cDCE and VC, and (iii) 1,2-DCA dechlorination. In accordance, our findings indicate that only two reductive dehalogenases, PteAbtf08 and VcrAbtf08, are needed for complete PCE dehalogenation to ethene in strain BTF08. TceAbtf08 was furthermore presumably identified capable of complete PCE dehalogenation to ethene, as well as for 1,2-DCA dehalogenation, although solely expressed if 1,2-DCA was present. Our findings support the previous proposed diversity in reaction mechanism for RdhA and reinforce CSIA as a powerful tool to elucidate biochemical reaction mechanisms. Furthermore, our findings contribute to a pronounced understanding of the underlying dehalogenation processes in anoxic zones, necessary to achieve applicability from laboratory micro-scale experiments to field site remediation approaches.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: http://www.proteomexchange.org/, PXD018558 and PXD018595.
Author Contributions
IN and SF developed the overall concept. IN, LA, and SF conceived the experimental study. SF conducted the cultivation, activity tests, isotope fractionation experiments, proteomics sample preparation, analyzed and evaluated the data for activity tests and isotope fractionation, evaluated proteomics data, and developed the first draft. LA and KS conducted phylogenetic analysis. All authors edited and approved the manuscript.
Funding
This work was supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) Project FOR1530 (NI 1323/1-2 and AD 178/5-2).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Matthias Schmidt, Julian Renpenning, and Steffen Kümmel for providing technical assistance, support in data evaluation, and helpful discussions concerning isotope analysis. We acknowledge Benjamin Scheer for technical support in nLC-MS/MS analysis as well as Felicitas Ehme, Florian Tschernikl, and Paul-Georg Richter for assistance in sample preparation. We acknowledge the Centre for Chemical Microscopy (ProVIS) at the Helmholtz Centre for Environmental Research supported by European regional Development Funds (EFRE – Europe funds Saxony) for access to chlorine stable isotope and LC-MS/MS analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01507/full#supplementary-material
References
Bommer, M., Kunze, C., Fesseler, J., Schubert, T., Diekert, G., and Dobbek, H. (2014). Structural basis for organohalide respiration. Science 346, 455–458. doi: 10.1126/science.1258118
Carter, J. M., Lapham, W. W., and Zogorski, J. S. (2008). Occurrence of volatile organic compounds in aquifers of the United States. JAWRA 44, 399–416. doi: 10.1111/j.1752-1688.2008.00170.x
Cichocka, D., Nikolausz, M., Haest, P. J., and Nijenhuis, I. (2010). Tetrachloroethene conversion to ethene by a Dehalococcoides-containing enrichment culture from Bitterfeld. FEMS Microbiol. Ecol. 72, 297–310. doi: 10.1111/j.1574-6941.2010.00845.x
Cooper, M., Wagner, A., Wondrousch, D., Sonntag, F., Sonnabend, A., Brehm, M., et al. (2015). Anaerobic microbial transformation of halogenated aromatics and fate prediction using electron density modeling. Environ. Sci. Technol. 49, 6018–6028. doi: 10.1021/acs.est.5b00303
Coplen, T. B., Brand, W. A., Gehre, M., Groning, M., Meijer, H. A., Toman, B., et al. (2006). After two decades a second anchor for the VPDB delta13C scale. Rapid. Commun. Mass. Spectrom. 20, 3165–3166. doi: 10.1002/rcm.2727
Elsner, M. (2010). Stable isotope fractionation to investigate natural transformation mechanisms of organic contaminants: principles, prospects and limitations. J. Environ. Monit. 12, 2005–2031.
Elsner, M., Jochmann, M. A., Hofstetter, T. B., Hunkeler, D., Bernstein, A., Schmidt, T. C., et al. (2012). Current challenges in compound-specific stable isotope analysis of environmental organic contaminants. Anal. Bioanal. Chem. 403, 2471–2491. doi: 10.1007/s00216-011-5683-y
Elsner, M., Zwank, L., Hunkeler, D., and Schwarzenbach, R. P. (2005). A new concept linking observable stable isotope fractionation to transformation pathways of organic pollutants. Environ. Sci. Technol. 39, 6896–6916. doi: 10.1021/es0504587
Fletcher, K. E., Nijenhuis, I., Richnow, H. H., and Löffler, F. E. (2011). Stable carbon isotope enrichment factors for cis-1,2-dichloroethene and vinyl chloride reductive dechlorination by Dehalococcoides. Environ. Sci. Technol. 45, 2951–2957. doi: 10.1021/es103728q
Franke, S., Lihl, C., Renpenning, J., Elsner, M., and Nijenhuis, I. (2017). Triple-element compound-specific stable isotope analysis of 1,2-dichloroethane for characterization of the underlying dehalogenation reaction in two Dehalococcoides mccartyi strains. FEMS Microbiol. Ecol. 93:fix137.
Goris, T., Schiffmann, C. L., Gadkari, J., Adrian, L., von Bergen, M., Diekert, G., et al. (2016). Proteomic data set of the organohalide-respiring Epsilonproteobacterium Sulfurospirillum multivorans adapted to tetrachloroethene and other energy substrates. Data Brief 8, 637–642. doi: 10.1016/j.dib.2016.06.022
Heckel, B., Cretnik, S., Kliegman, S., Shouakar-Stash, O., McNeill, K., and Elsner, M. (2017). Reductive outer-sphere single electron transfer is an exception rather than the rule in natural and engineered chlorinated ethene dehalogenation. Environ. Sci. Technol. 51, 9663–9673. doi: 10.1021/acs.est.7b01447
Heckel, B., McNeill, K., and Elsner, M. (2018). Chlorinated ethene reactivity with vitamin B12 is governed by cobalamin chloroethylcarbanions as crossroads of competing pathways. ACS Catalysis 8, 3054–3066. doi: 10.1021/acscatal.7b02945
Hirschorn, S. K., Grostern, A., Lacrampe-Couloume, G., Edwards, E. A., Mackinnon, L., Repta, C., et al. (2007). Quantification of biotransformation of chlorinated hydrocarbons in a biostimulation study: added value via stable carbon isotope analysis. J. Contam. Hydrol. 94, 249–260. doi: 10.1016/j.jconhyd.2007.07.001
Holscher, T., Krajmalnik-Brown, R., Ritalahti, K. M., Von Wintzingerode, F., Gorisch, H., Löffler, F. E., et al. (2004). Multiple nonidentical reductive-dehalogenase-homologous genes are common in Dehalococcoides. Appl. Environ. Microbiol. 70, 5290–5297. doi: 10.1128/aem.70.9.5290-5297.2004
Horst, A., Renpenning, J., Richnow, H. H., and Gehre, M. (2017). Compound specific stable chlorine isotopic analysis of volatile aliphatic compounds using gas chromatography hyphenated with multiple collector inductively coupled plasma mass spectrometry. Anal. Chem. 89, 9131–9138. doi: 10.1021/acs.analchem.7b01875
Hug, L. A., Maphosa, F., Leys, D., Loffler, F. E., Smidt, H., Edwards, E. A., et al. (2013). Overview of organohalide-respiring bacteria and a proposal for a classification system for reductive dehalogenases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368:20120322. doi: 10.1098/rstb.2012.0322
Ji, L., Wang, C. C., Ji, S. J., Kepp, K. P., and Paneth, P. (2017). Mechanism of cobalamin-mediated reductive dehalogenation of chloroethylenes. ACS Catalysis 7, 5294–5307. doi: 10.1021/acscatal.7b00540
Johannissen, L. O., Leys, D., and Hay, S. (2017). A common mechanism for coenzyme cobalamin-dependent reductive dehalogenases. Phys. Chem. Chem. Phys. 19, 6090–6094. doi: 10.1039/c6cp08659d
Kaufhold, T., Schmidt, M., Cichocka, D., Nikolausz, M., and Nijenhuis, I. (2013). Dehalogenation of diverse halogenated substrates by a highly enriched Dehalococcoides-containing culture derived from the contaminated mega-site in Bitterfeld. FEMS Microbiol. Ecol. 83, 176–188. doi: 10.1111/j.1574-6941.2012.01462.x
Kliegman, S., and McNeill, K. (2008). Dechlorination of chloroethylenes by cob(I)alamin and cobalamin model complexes. Dalton Trans. 32, 4191–4201.
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kunze, C., Bommer, M., Hagen, W. R., Uksa, M., Dobbek, H., Schubert, T., et al. (2017). Cobamide-mediated enzymatic reductive dehalogenation via long-range electron transfer. Nat. Commun. 8:15858.
Lihl, C., Douglas, L. M., Franke, S., Perez de Mora, A., Meyer, A. H., Daubmeier, M., et al. (2019). Mechanistic dichotomy in bacterial trichloroethene dechlorination. Environ. Sci. Technol. 53, 4245–4254. doi: 10.1021/acs.est.8b06643
Löffler, F. E., Yan, J., Ritalahti, K. M., Adrian, L., Edwards, E. A., Konstantinidis, K. T., et al. (2013). Dehalococcoides mccartyi gen. nov., sp. nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov. and family Dehalococcoidaceae fam. nov., within the phylum Chloroflexi Int. J. Syst. Evol. Microbiol. 63, 625–635. doi: 10.1099/ijs.0.034926-0
Magnuson, J. K., Romine, M. F., Burris, D. R., and Kingsley, M. T. (2000). Trichloroethene reductive dehalogenase from Dehalococcoides ethenogenes: sequence of tceA and substrate range characterization. Appl. Environ. Microbiol. 66, 5141–5147. doi: 10.1128/aem.66.12.5141-5147.2000
Mariotti, A., Germon, J. C., Hubert, P., Kaiser, P., Letolle, R., Tardieux, A., et al. (1981). Experimental-determination of nitrogen kinetic isotope fractionation - some principles - illustration for the denitrification and nitrification processes. Plant Soil 62, 413–430. doi: 10.1007/bf02374138
Middeldorp, P. J. M., Luijten, M. L. G. C., van de Pas, B. A., van Eekert, M. H. A., Kengen, S. W. M., Schraa, G., et al. (1999). Anaerobic microbial reductive dehalogenation of chlorinated ethenes. Bioremed. J. 3, 151–169. doi: 10.1080/10889869991219280
Miller, E., Wohlfarth, G., and Diekert, G. (1996). Studies on tetrachloroethene respiration in Dehalospirillum multivorans. Arch. Microbiol. 166, 379–387. doi: 10.1007/s002030050399
Müller, J. A., Rosner, B. M., Von Abendroth, G., Meshulam-Simon, G., McCarty, P. L., and Spormann, A. M. (2004). Molecular identification of the catabolic vinyl chloride reductase from Dehalococcoides sp. strain VS and its environmental distribution. Appl. Environ. Microbiol. 70, 4880–4888. doi: 10.1128/aem.70.8.4880-4888.2004
Palau, J., Yu, R., Hatijah Mortan, S., Shouakar-Stash, O., Rosell, M., Freedman, D. L., et al. (2017). Distinct dual C-Cl isotope fractionation patterns during anaerobic biodegradation of 1,2-dichloroethane: potential to characterize microbial degradation in the field. Environ. Sci. Technol. 51, 2685–2694. doi: 10.1021/acs.est.6b04998
Parthasarathy, A., Stich, T. A., Lohner, S. T., Lesnefsky, A., Britt, R. D., and Spormann, A. M. (2015). Biochemical and EPR-spectroscopic investigation into heterologously expressed vinyl chloride reductive dehalogenase (VcrA) from Dehalococcoides mccartyi strain VS. J. Am. Chem. Soc. 137, 3525–3532. doi: 10.1021/ja511653d
Payne, K. A., Quezada, C. P., Fisher, K., Dunstan, M. S., Collins, F. A., Sjuts, H., et al. (2015). Reductive dehalogenase structure suggests a mechanism for B12-dependent dehalogenation. Nature 517, 513–516. doi: 10.1038/nature13901
Pöritz, M., Goris, T., Wubet, T., Tarkka, M. T., Buscot, F., Nijenhuis, I., et al. (2013). Genome sequences of two dehalogenation specialists - Dehalococcoides mccartyi strains BTF08 and DCMB5 enriched from the highly polluted Bitterfeld region. FEMS Microbiol. Lett. 343, 101–104. doi: 10.1111/1574-6968.12160
Renpenning, J., Horst, A., Schmidt, M., and Gehre, M. (2018). Online isotope analysis of 37Cl/35Cl universally applied for semi-volatile organic compounds using GC-MC-ICPMS. JAAS 33, 314–321. doi: 10.1039/c7ja00404d
Renpenning, J., Keller, S., Cretnik, S., Shouakar-Stash, O., Elsner, M., Schubert, T., et al. (2014). Combined C and Cl isotope effects indicate differences between corrinoids and enzyme (Sulfurospirillum multivorans PceA) in reductive dehalogenation of tetrachloroethene, but not trichloroethene. Environ. Sci. Technol. 48, 11837–11845. doi: 10.1021/es503306g
Renpenning, J., Kummel, S., Hitzfeld, K. L., Schimmelmann, A., and Gehre, M. (2015). Compound-specific hydrogen isotope analysis of heteroatom-bearing compounds via gas chromatography-chromium-based high-temperature conversion (Cr/HTC)-isotope ratio mass spectrometry. Anal. Chem. 87, 9443–9450. doi: 10.1021/acs.analchem.5b02475
Schmidt, M., Lege, S., and Nijenhuis, I. (2014). Comparison of 1,2-dichloroethane, dichloroethene and vinyl chloride carbon stable isotope fractionation during dechlorination by two Dehalococcoides strains. Water Res. 52, 146–154. doi: 10.1016/j.watres.2013.12.042
Schumacher, W., Holliger, C., Zehnder, A. J., and Hagen, W. R. (1997). Redox chemistry of cobalamin and iron-sulfur cofactors in the tetrachloroethene reductase of Dehalobacter restrictus. FEBS Lett. 409, 421–425. doi: 10.1016/s0014-5793(97)00520-6
Seshadri, R., Adrian, L., Fouts, D. E., Eisen, J. A., Phillippy, A. M., Methe, B. A., et al. (2005). Genome sequence of the PCE-dechlorinating bacterium Dehalococcoides ethenogenes. Science 307, 105–108. doi: 10.1126/science.1102226
Vogt, C., Cyrus, E., Herklotz, I., Schlosser, D., Bahr, A., Herrmann, S., et al. (2008). Evaluation of toluene degradation pathways by two-dimensional stable isotope fractionation. Environ. Sci. Technol. 42, 7793–7800. doi: 10.1021/es8003415
Keywords: compound-specific stable isotope analysis, nLC-MS/MS, Dehalococcoides mccartyi strain BTF08, reductive dehalogenation, 1, 2-dichloroethane, tetrachloroethene, cis-dichloroethene, vinyl chloride
Citation: Franke S, Seidel K, Adrian L and Nijenhuis I (2020) Dual Element (C/Cl) Isotope Analysis Indicates Distinct Mechanisms of Reductive Dehalogenation of Chlorinated Ethenes and Dichloroethane in Dehalococcoides mccartyi Strain BTF08 With Defined Reductive Dehalogenase Inventories. Front. Microbiol. 11:1507. doi: 10.3389/fmicb.2020.01507
Received: 23 March 2020; Accepted: 10 June 2020;
Published: 17 July 2020.
Edited by:
Mengyan Li, New Jersey Institute of Technology, United StatesReviewed by:
Yi Yang, Institute of Applied Ecology (CAS), ChinaUte Lechner, Martin Luther University of Halle-Wittenberg, Germany
Copyright © 2020 Franke, Seidel, Adrian and Nijenhuis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ivonne Nijenhuis, aXZvbm5lLm5pamVuaHVpc0B1ZnouZGU=
†Present address: Lorenz Adrian, Department of Environmental Biotechnology, Helmholtz Centre for Environmental Research – UFZ, Leipzig, Germany
 Steffi Franke1
Steffi Franke1 Katja Seidel
Katja Seidel Lorenz Adrian
Lorenz Adrian Ivonne Nijenhuis
Ivonne Nijenhuis