
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol. , 07 July 2020
Sec. Microbial Physiology and Metabolism
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.01505
This article is part of the Research Topic Role of Sigma Factors of RNA Polymerase in Bacterial Physiology View all 7 articles
Sensing and responding to environmental cues is critical for the adaptability and success of the food-borne bacterial pathogen Listeria monocytogenes. A supramolecular multi-protein complex known as the stressosome, which acts as a stress sensing hub, is responsible for orchestrating the activation of a signal transduction pathway resulting in the activation of σB, the sigma factor that controls the general stress response (GSR). When σB is released from the anti-sigma factor RsbW, a rapid up-regulation of the large σB regulon, comprised of ≥ 300 genes, ensures that cells respond appropriately to the new environmental conditions. A diversity of stresses including low pH, high osmolarity, and blue light are known to be sensed by the stressosome, resulting in a generalized increase in stress resistance. Appropriate activation of the stressosome and deployment of σB are critical to fitness as there is a trade-off between growth and stress protection when the GSR is deployed. We review the recent developments in this field and describe an up-to-date model of how this sensory organelle might integrate environmental signals to produce an appropriate activation of the GSR. Some of the outstanding questions and challenges in this fascinating field are also discussed.
The firmicute Listeria monocytogenes is a remarkably robust bacterium with a capacity to grow and survive over a wide range of challenging environmental conditions. It is unusual among food-borne pathogens in being able to grow at refrigeration temperatures and it is very tolerant to high salt concentrations, being able to grow in media containing over 1.5 M NaCl. Additionally, it has an effective protective response against low pH, designated the adaptive acid tolerance response, which allows it to survive at pH values as low as 3.0 for extended periods (O’Byrne and Karatzas, 2008; Dorey et al., 2019b). These traits, combined with the almost ubiquitous occurrence of this microorganism, can allow it to persist in the human food-chain and occasionally establish infections in immunocompromised individuals, elderly people and pregnant women (NicAogáin and O’Byrne, 2016). When they arise, infections can be life-threatening, and outbreaks are associated with high mortality rates, typically 20–30% (Lecuit, 2007).
While many factors contribute to the phenotypic robustness of this pathogen the general stress response (GSR) plays a central role (Gandhi and Chikindas, 2007; Hecker et al., 2007; O’Byrne and Karatzas, 2008; Dorey et al., 2019b). This response is characterized by a general reprogramming of cellular transcription mediated by an alternative sigma factor called SigB (σB), first identified in L. monocytogenes just over two decades ago (Becker et al., 1998; Wiedmann et al., 1998). Homologs of σB are found in most Gram-positive bacteria (Hecker et al., 2007).
In this mini-review, we discuss the recent developments in our understanding of how σB contributes to stress tolerance and how its activity is regulated in response to stress. We explore its contribution to virulence and analyze the resource implications for the cell of deploying the GSR. We highlight some of the key research questions that remain to be answered in this important field.
The robustness of L. monocytogenes is modulated in part by σB, an alternative sigma factor responsible for the upregulation of approximately 300 genes in L. monocytogenes (Milohanic et al., 2003; Wemekamp-Kamphuis et al., 2004; Chatterjee et al., 2006; Abram et al., 2008a, b; Raengpradub et al., 2008; Ollinger et al., 2009; Toledo-Arana et al., 2009; Oliver et al., 2010; Shin et al., 2010b; Chaturongakul et al., 2011; Palmer et al., 2011; Ribeiro et al., 2014; Liu et al., 2017; Cortes et al., 2020), including several non-coding sRNA (Nielsen et al., 2008; Toledo-Arana et al., 2009). The σB regulon, which is not the primary focus of this mini-review, has recently been systematically reviewed by Liu et al., 2019. A subset of approximately 60 genes, identified across strains of L. monocytogenes belonging to different lineages, constitute the σB core regulon (Oliver et al., 2010). Genes comprising the σB regulon are involved in carbohydrate metabolism (Abram et al., 2008b; Tapia et al., 2020), cell envelope modification (Abram, 2007; Tiensuu et al., 2013), pH homeostasis (Cotter et al., 2005; Karatzas et al., 2010, 2012), osmoregulation (Fraser et al., 2003; Cetin et al., 2004; Wemekamp-Kamphuis et al., 2004; Abram et al., 2008a), regulation of amino acids biosynthesis (Marinho et al., 2019), flagellar biosynthesis (Raengpradub et al., 2008; Toledo-Arana et al., 2009), quorum sensing (Marinho et al., 2020), and antibiotic resistance (Begley et al., 2006). These mechanisms under σB control have been previously reviewed (O’Byrne and Karatzas, 2008; NicAogáin and O’Byrne, 2016; Dorey et al., 2019b; Liu et al., 2019), and they contribute to the survival of L. monocytogenes under a broad range of lethal stresses (Cole et al., 1990; Ferreira et al., 2001; Sue et al., 2003; Wemekamp-Kamphuis et al., 2004; Begley et al., 2005, 2006; Giotis et al., 2008; Palmer et al., 2009; Shin et al., 2010a; Dowd et al., 2011; Feehily et al., 2012, 2013, 2014; O’Donoghue et al., 2016; Curtis et al., 2017; Bourke et al., 2019; Williams et al., 2019). Activation of σB by one stress often triggers cross protection against other types of stress in L. monocytogenes (Begley et al., 2002; Bergholz et al., 2012; Pittman et al., 2014), indicating that a large fraction of the σB regulon is activated simultaneously. However, many σB-dependent genes are differentially expressed under different growth conditions (Toledo-Arana et al., 2009), suggesting the involvement of additional transcriptional regulators to achieve condition-specific gene expression.
To sense environmental changes L. monocytogenes relies on a 1.8 MDa supramolecular apparatus designated the stressosome (Figure 1). This stress-sensing organelle is found in members of the proteobacteria, the firmicutes, the actinobacteria, the cyanobacteria, and in the Bacteroides and Deinococcus groups (Pané-Farré et al., 2005). In Bacillus subtilis, the stressosome is composed of RsbRA and its paralogs (RsbRB, RsbRC, RsbRD, and YtvA), RsbS and RsbT forming a pseudo-icosahedral core with turrets on its surface (Chen et al., 2003; Marles-Wright and Lewis, 2008; Martinez et al., 2010; Pané-Farré et al., 2017), the presence of which was later confirmed in L. monocytogenes. The L. monocytogenes stressosome is composed of RsbR (Lmo0899) and its paralogs RsbR2 (Lmo0161), RsbL (Lmo0799), RsbR3 (Lmo1642), RsbS and RsbT (Impens et al., 2017). The C-terminal domains of RsbS and RsbR fold into Sulfate Transporter and Anti-Sigma (STAS) factor antagonist domains and self-assemble into the stressosome’s core (Aravind and Koonin, 2000). RsbR N-terminal domains, the putative sensory elements of the stressosome, fold into a non-heme globin like structure and associate in dimers (Murray et al., 2005), forming turrets at the complex surface. Pull-down experiments revealed that the RsbR N-terminal domain in L. monocytogenes can bind to the small membrane-spanning peptide Prli42, which has been suggested to anchor the stressosome to the cell membrane and to contribute to oxidative stress sensing (Impens et al., 2017). In the same study, the remaining RsbR paralogs were also found associated with the stressosome, the exception being Lmo1842, which was not detected, perhaps consistent with the low transcription levels of the corresponding gene (Wurtzel et al., 2012; Bécavin et al., 2017). In a recent study, in vitro assembly of the L. monocytogenes stressosome proteins purified from Escherichia coli, revealed that it has an icosahedral shape with a 2:1:1 RsbR:RsbS:RsbT stoichiometry and an hexagonal basic structural subunit composed of two dimers of RsbR and one dimer of RsbS (Figure 1A), where the dimeric interfaces form a rigid structure that is responsible for the stressosome integrity (Williams et al., 2019). While the current understanding of the stressosome structure has been thoroughly reviewed in a number of studies (Marles-Wright and Lewis, 2010; Pané-Farré et al., 2017; Tiensuu et al., 2019), there are no structural models available yet that include all RsbR paralogs.
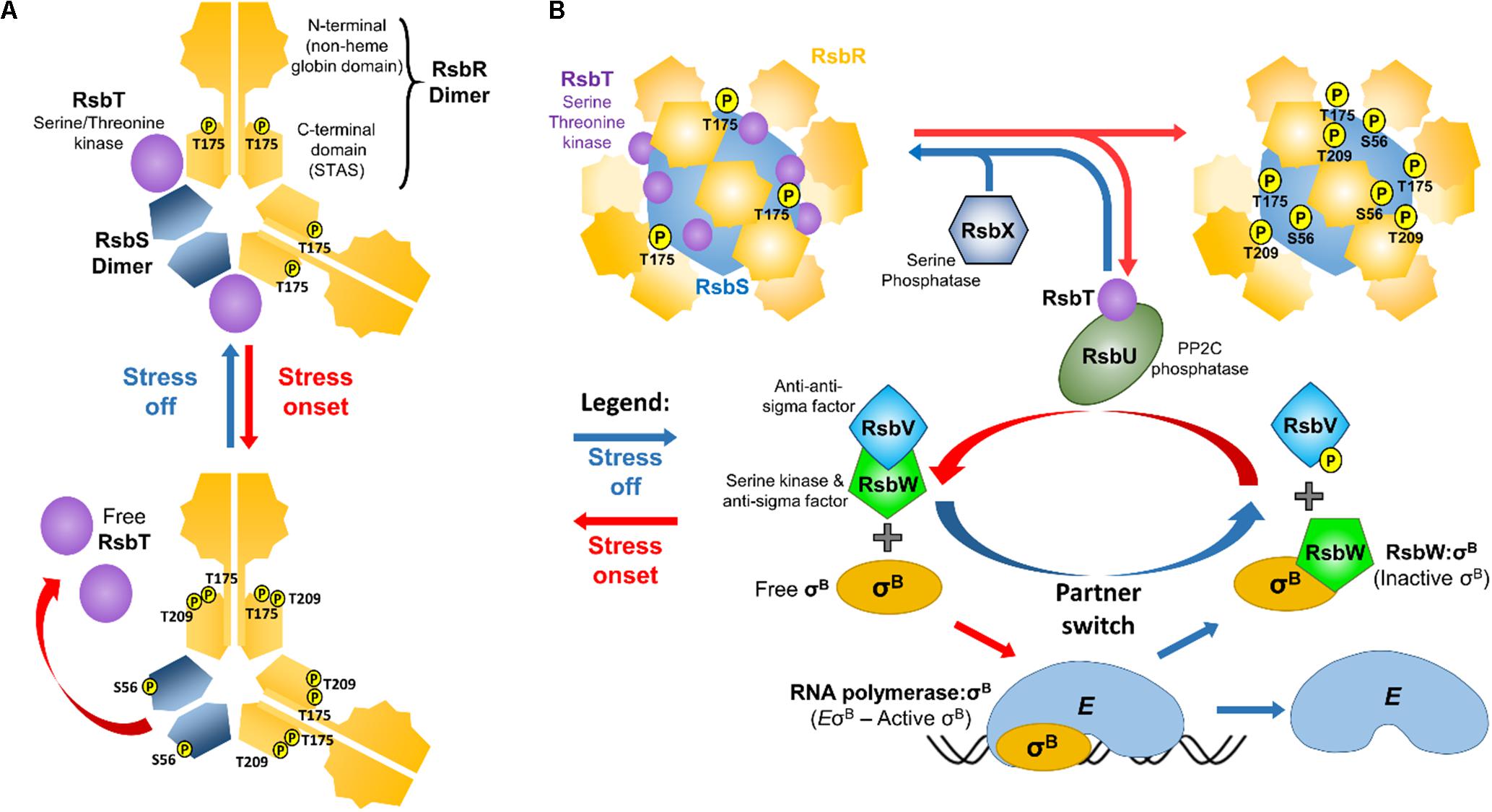
Figure 1. Schematic representation of the oB regulatory pathway in L. monocytogenes. (A) The repeating hexagonal subunits of the stressosome that are composed of two dimers of RsbR, one dimer of RsbS, and two monomers of RsbT (Williams et al., 2019). It is hypothesized that the N-terminal turrets formed by the RsbR dimers detect stress signals, triggering conformational changes that propagate into the STAS domains, initiating the kinase activity of RsbT. This results in the phosphorylation of RsbR Thr209 and Thr175 and RsbS Ser56, which in turn leads to the release of RsbT from the stressosome. (B) Free RsbT interacts with and activates the RsbU phosphatase, which acts on phosphorylated RsbV. Simultaneously, the anti-sigma factor RsbW that binds and prevents σB from interacting with the RNA polymerase (E), releases σB and preferentially interacts with non-phosphorylated RsbV. σB can then interact with RNA polymerase forming the holoenzyme EσB. Once stress is removed, RsbX dephosphorylates the stressosome (except for RsbR Thr175 which remains phosphorylated; Misra et al., 2011), resulting in the sequestration of RsbT back into the stressosome and inactivating the signal transduction.
In B. subtilis two residues in the RsbRA STAS domain (Thr171 and Thr205) and one in RsbS (Ser59), can be phosphorylated through the action of the serine/threonine kinase RsbT (Kim et al., 2004), and these residues are all conserved in RsbR and RsbS of L. monocytogenes (Ferreira et al., 2004) (Figure 1). In contrast to B. subtilis, where all RsbRA paralogs possess phosphorylatable residues, in L. monocytogenes only RsbR has these conserved threonines (Thr175 and Thr209). In B. subtilis RsbRA Thr171 (Lm Thr175) is constitutively phosphorylated (Kim et al., 2004). Indeed, L. monocytogenes RsbR Thr175 was also found phosphorylated in the absence of stress, but not Lm RsbR Thr209 nor Lm RsbS Ser56 (Misra et al., 2011). B. subtilis RsbRA Thr205 was found to be phosphorylated only under extreme conditions (Eymann et al., 2011). Bs RsbS Ser59 phosphorylation rate seems dependent on Bs RsbRA Thr171 and Thr205 phosphorylation (Chen et al., 2004). Amino acid substitutions of L. monocytogenes Thr175 and/or Thr209 to Ala resulted in reduced σB activity and consequently reduced survival in acidic conditions (He et al., 2019).
Following the release of RsbT and consequent activation of σB, a negative feedback mechanism controlled by the phosphatase RsbX allows the stressosome to be reset to its non-stressed dephosphorylated state, which leads to the recapture of RsbT (Voelker et al., 1997; Chen et al., 2004; Eymann et al., 2011). Deletion of rsbX produces a constitutive σB activation and consequently increased survival in acidic conditions (Xia et al., 2016) and a reduced competitiveness against a WT strain, a consequence of the reduced growth rate associated with increased σB activity (Guerreiro et al., 2020) (see section “σB Deployment Is a Double-Edged Sword”).
From the plethora of stresses that result in σB activation, only the blue-light sensing mechanism is well understood. Light is sensed by the phototropin RsbL in L. monocytogenes (Ondrusch and Kreft, 2011; Tiensuu et al., 2013; O’Donoghue et al., 2016; Dorey et al., 2019a) and by YtvA in B. subtilis (Gaidenko et al., 2006; Ávila-Pérez et al., 2010), both of which have N-terminal light-oxygen-voltage (LOV) domains that associate with a flavin mononucleotide (FMN) (Losi et al., 2002; Ondrusch and Kreft, 2011). Like other RsbR paralogs, RsbL/YtvA associate in homodimers (Buttani et al., 2007; Möglich and Moffat, 2007; Jurk et al., 2010). After blue-light absorption, the FMN forms a covalent adduct with the Cys56 in L. monocytogenes RsbL and Cys62 in B. subtilis YtvA (Avila-Pérez et al., 2006; Gaidenko et al., 2006; O’Donoghue et al., 2016). The adduct produces a local structural rearrangement in RsbL, propagating into the stressosome core and activating the signal transduction (Salomon et al., 2001; Crosson and Moffat, 2002). Once blue-light is removed, the covalent adduct decays to its ground state (τ1/2 = 95 min), resetting the protein to its non-stressed state (Chan et al., 2013). Interestingly, L. monocytogenes does not activate σB when exposed to blue-light at 37°C, suggesting that the bond between FMN and residue Cys56 may not form at this temperature (Dorey et al., 2019a). Indeed Chan et al. reported that FMN:RsbL association is reduced as the temperature increases above 26°C. Presumably the absence of an evolutionary pressure to detect light at 37°C, when the pathogen is most likely within the dark confines of a host, produced this temperature-dependent light sensing phenotype.
It is hypothesized that the N-terminal domains of RsbR and the other paralogs are also responsible for the stress signal integration into the stressosome, however, neither the mechanisms involved nor the stress signals being detected are known at present. In B. subtilis nutritional stress is sensed through RsbP and RsbQ and integrated into the signal transduction pathway regulating σB downstream of the stressosome via RsbV (Vijay et al., 2000). In L. monocytogenes homologs of RsbPQ are not present, and nutritional starvation is detected by the stressosome through RsbR instead (Chaturongakul and Boor, 2004; Chaturongakul and Boor, 2006; Martinez et al., 2010).
The stressosome, along with proteins that integrate the signal transduction responsible for σB regulation, are encoded in the sigB operon (rsbR, rsbS, rsbT, rsbU, rsbV, rsbW, sigB, and rsbX). Once stress is sensed, RsbT is released from the stressosome core and is then free to initiate a signal cascade by associating with RsbU, which in turn directs its phosphatase activity toward phosphorylated RsbV (Yang et al., 1996). The anti-sigma factor RsbW, which binds to σB and blocks its interaction with RNA polymerase (RNApol), has a higher affinity for the dephosphorylated form of RsbV than for σB. RsbV–RsbW interaction restores the phosphorylated state of RsbV through the kinase activity of RsbW, which in turn promotes the reassociation of RsbW with σB, thereby establishing another negative feedback loop (Yang et al., 1996). Once dissociated from RsbW, σB interacts with RNApol resulting in the upregulation of the σB regulon. It has been proposed that the signal transduction cascade in L. monocytogenes can be inferred from the well-studied B. subtilis, since both species share a high level of conservation (Ferreira et al., 2004). Many studies of σB regulation in L. monocytogenes have confirmed that the signal transduction pathways likely function in a very similar way between these two microorganisms (Chaturongakul and Boor, 2004, 2006; Cosgrave, 2010; Shin et al., 2010a; Utratna et al., 2014; O’Donoghue,, 2016; Guerreiro et al., 2020; Hsu et al., 2020).
Bacterial populations display random fluctuations in the expression of individual genes, metabolite pools, and macromolecular concentrations that generate heterogeneity within the population (Cai et al., 2006; Levine et al., 2013). These differences can give rise to a “bet hedging” survival strategy, where some cells are better prepared for environmental changes and hence have a higher chance of survival under unfavorable conditions. Emerging single-cell analytical methods are increasingly being used to further investigate how σB activity is regulated at the single-cell level. After exposing B. subtilis to mycophenolic acid (MPA), an inhibitor of GTP synthesis that indirectly triggers energy stress, σB activation was studied using fluorescent protein reporters and time-lapse microscopy (Locke et al., 2011). A series of stochastic pulses of σB activity was observed in individual cells, with an increased frequency of pulses observed with increasing MPA concentrations. These observations could be explained by fluctuations (noise) in the concentration of some of the key components of σB regulatory circuit. A minimal mathematical model of the circuit, where fluctuations in the RsbQP phosphatase/RsbW kinase ratio cause sudden increases in σB activation, exhibited a similar behavior to the experimental observations (Locke et al., 2011). Surprisingly, when a microfluidic-based strategy was used to study σB activation, the results obtained were somewhat different from those obtained by Locke et al. (Cabeen et al., 2017). In this case, the amplitude of the response increased with the magnitude of the stress, but the frequency of σB activation remained unchanged (no stochastic pulses were observed). When bacteria were exposed to environmental stresses (osmotic stress and ethanol), a single pulse of activation of σB was observed, whose amplitude depended on the rate at which the stress increased (Young et al., 2013) or its magnitude (Cabeen et al., 2017). However, strains producing only one of the four RsbR paralogs present in B. subtilis displayed repeated stress-activation peaks in single cells, resembling the stochastic activation of σB reported previously (Cabeen et al., 2017). Pulsing activity of σB has also been observed during biofilm development, allowing mutually exclusive cell states to co-exist in the same regions of the biofilm and enabling the formation of simple spatial patterns (Nadezhdin et al., 2020). The presence of positive and negative feedback loops within the σB activation pathway contributes to the generation of noise, with a positive feedback loop amplifying the fluctuations and negative feedback loop, once RsbW is activated, that terminates the pulsing (Nadezhdin et al., 2020). Differences in the experimental approach might affect σB dynamics differently, causing distinct responses. Future studies will probably need to refine the mathematical models used to predict the activation patterns of σB in order to resolve the observed experimental discrepancies.
In L. monocytogenes heterogeneous activation of σB was observed when cells were subjected to osmotic shock, with an increased proportion of cells having an active σB as the magnitude of the stress was increased (Utratna et al., 2012). A similar stochastic behavior of σB was also observed in another study under similar stress conditions (Guldimann et al., 2017), further supporting the idea of a bet-hedging survival strategy in L. monocytogenes.
To establish an infection L. monocytogenes needs to survive under the harsh conditions of the gastrointestinal (GI) tract, including the acidic conditions of the stomach, osmotic stress in the small intestine, and the presence of bile salts in the duodenum (Sleator et al., 2009; Gaballa et al., 2019; Tiensuu et al., 2019). Survival in the presence of these stresses is partially dependent on σB, as an intragastrically inoculated ΔsigB strain exhibits attenuated virulence (Garner et al., 2006; Oliver et al., 2010). σB regulates the glutamate decarboxylase (GAD) system (Wemekamp-Kamphuis et al., 2002; Cotter et al., 2001a, b, 2005), bile resistance genes such as bilE (Fraser et al., 2003; Sleator et al., 2005), bsh (Sue et al., 2003; Zhang et al., 2011), pva (Begley et al., 2005), and also controls opuC, gbu, and betL to help the bacteria cope with osmotic stress (Fraser et al., 2003; Sue et al., 2003; Cetin et al., 2004; Raengpradub et al., 2008).
A growing body of evidence points toward a complex two-way regulatory network between σB and the master regulator of virulence, PrfA (Gaballa et al., 2019; Tiensuu et al., 2019). σB is also responsible for the regulation of the RNA chaperone Hfq which also plays a role in virulence and osmotic stress resistance (Christiansen et al., 2004). The activity of PrfA is crucial for the expression of genes that are important for pathogenesis, including the genes from the Listeria Pathogenicity Island 1 (LIPI-1) and the inlAB loci (de las Heras et al., 2011). One of the three promoters that drive prfA transcription, P2, is a σB-dependent promoter (Nadon et al., 2002). Under certain forms of stress, transcription from the P2 promoter is enhanced, demonstrating a role for σB in prfA expression (Kazmierczak et al., 2006). There is also a transcriptional overlap between σB and PrfA regulons, with a group of genes being under the control of both systems (Milohanic et al., 2003). Significantly, it has been shown that σB plays a crucial role in limiting the availability of branched chain amino acids (BCAA) in L. monocytogenes, raising the possibility that σB might influence PrfA activity via CodY, a global transcription regulator and sensor of BCAA (Marinho et al., 2019). When BCAA availability is low, as they are inside the mammalian host cell, CodY plays a direct role in the transcriptional activation of prfA (Lobel et al., 2015). A genome wide analysis of the CodY regulon identified sigB as one of the genes that is also directly regulated by CodY, indicating that CodY may promote prfA transcription by at least two different mechanisms: directly via binding to the prfA gene and indirectly by relieving sigB repression (Lobel and Herskovits, 2016). However, in vitro binding of CodY to the 5’ coding region of prfA is very weak, suggesting that other indirect mechanisms are likely to be involved in CodY-mediated prfA activation (Biswas et al., 2020).
Unlike most Gram-positive bacteria, L. monocytogenes has the ability to synthesize glutathione (GSH) (Gopal et al., 2005), and is also capable of utilizing exogenous GSH (Portman et al., 2017). It has been shown that GSH allosterically activates PrfA, causing a conformational change that increases binding of PrfA to DNA, promoting the transcription of virulence genes accordingly (Reniere et al., 2015; Hall et al., 2016). The expression of GSH reductase (lmo1433), an enzyme that contributes to oxidative stress resistance by reducing GSH disulfide to GSH, is positively regulated by σB (Kazmierczak et al., 2003). These observations could imply that σB can indirectly contribute to PrfA activation by maintaining the intracellular GSH levels high through the expression of GSH reductase. This multi-layered regulatory network plays a major role in modifying gene expression in response to environmental stress in L. monocytogenes and is central to this pathogen’s remarkable adaptive capacity.
In addition to conferring stress resistance, the activation of σB also results in reduced growth in L. monocytogenes (Figure 2) (Brøndsted et al., 2003; Chaturongakul and Boor, 2004; Abram, 2007; Cosgrave, 2010; Zhang et al., 2013; O’Donoghue et al., 2016; Curtis et al., 2017; Marinho et al., 2019; Sæbø et al., 2019; Guerreiro et al., 2020). It has been hypothesized that living organisms often limit their growth in exchange for increased survival, when conditions are unfavorable due to nutrient limitation (Nyström, 2004). Recently, we have shown that L. monocytogenes σB-defective strains exhibit a decreased acid tolerance but have increased growth rates and a competitiveness advantage under mild heat stress (Guerreiro et al., 2020). This growth advantage allows strains with reduced σB activity to overtake the WT in mixed strain competition experiments. The reason for this growth advantage is not clear at present but three hypotheses seem worth considering. First, competition of different sigma factors for the same allosteric site of the RNApol to produce an active holoenzyme (Eσ) could redirect transcription away from growth-related functions (Figure 2). In L. monocytogenes the availability of σB to form EσB is ultimately governed by the signal transduction leading to the release of σB from RsbW. As more σB is released from RsbW the competition with other sigma factors increases (Figure 2). This potentially impacts the housekeeping σA, which is responsible for the transcription of growth related genes (Österberg et al., 2011). Indeed mathematical models support this type of competition (Mauri and Klumpp, 2014). Whether σB has a higher affinity for RNApol than σA or a displacement mechanism exists, as has been shown for B. subtilis σE and σK during sporulation (Ju et al., 1999), are still unknown. Second, σB activation may result in the depletion of energy resources to the extent that it has a negative impact on growth. Indeed, exposure to different types of stress results in the reduction of the ATP pool in several bacteria (Antonietti and Ferrini, 1986; Hecker et al., 1989; Antonietti and Tomaselli, 1991). Additionally, ΔsigB mutants exhibit higher intracellular ATP levels compared to a WT strain after the exposure to osmotic stress (Xia et al., 2016). In contrast, an ΔrsbX mutant has lower ATP levels, as a result of the over-activation of σB (Xia et al., 2016). Third, it is conceivable that σB specifically reduces growth as part of an overall damage mitigation strategy in the face of stress. We recently showed that the σB-dependent sRNA, Rli47, blocks isoleucine biosynthesis in L. monocytogenes through a direct interaction with ilvA mRNA. This interaction results in restricted growth under conditions where isoleucine is limited and suggests a possible role for σB in controlling growth under those conditions (Marinho et al., 2019). Further studies will be needed to tease these possibilities out fully but it is already clear that σB has an important impact on fitness and is likely to be subjected to a strong selective pressure. Indeed this may well explain the complexity of the regulatory system controlling σB activity; deciding precisely when and to what extent σB should be deployed is critical to resource allocation in times of stress and this ultimately determines fitness (Figure 2).
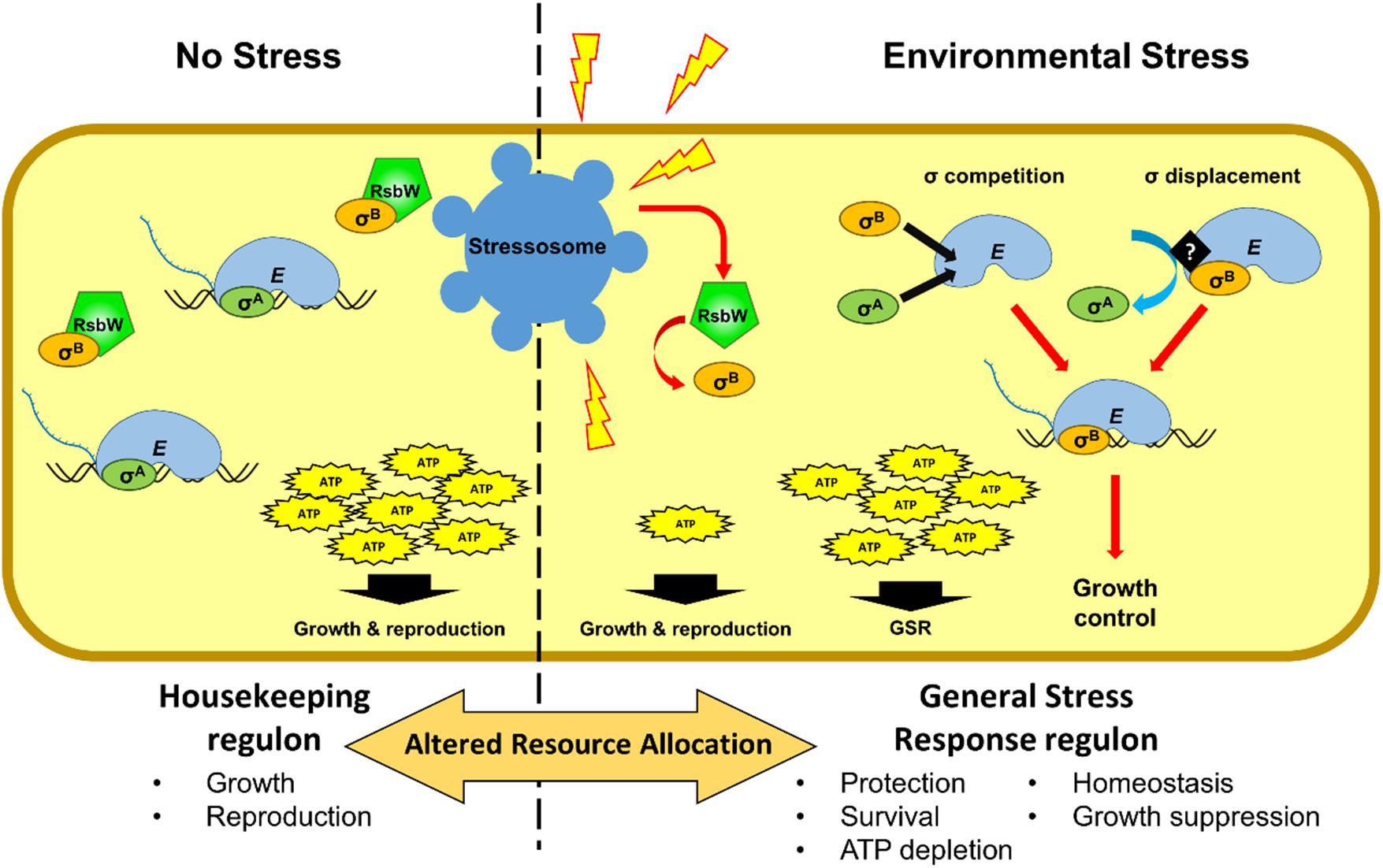
Figure 2. Schematic representation of the alterations in resource allocation that occur during the GSR. Cell growth largely depends on the housekeeping sigma factor σA in the absence nutrient limitations or stressful conditions. Under these conditions (no stress), most of the transcriptional machinery is dedicated to the transcription of housekeeping genes that preceded by σA promoters. In the absence of stress, σB is sequestered by the anti-sigma factor RsbW. At the onset of stress σB is released from its anti-sigma factor RsbW, resulting in competition between σB and σA and the displacement of σA from a proportion of the RNA polymerase pool. It is possible that the interaction of σB with RNA polymerase is specifically regulated as has been described in other species. Consequently, genes under σA control that are associated with growth functions are downregulated and σB dependent genes (the GSR regulon) are upregulated. The energy resources needed to maintain the general stress response reduces the availability of ATP for growth and reproduction. σB may specifically regulate growth rate to allow for improved maintenance and repair, thereby increasing the likelihood of survival.
It is over 20 years since the σB system has been discovered in L. monocytogenes and its role in controlling the GSR and the many stress-related phenotypes associated with loss-of-function have been well described. However, there is still much to learn about how its activity is regulated.
Probably the biggest challenge facing the field, and this is also true in Bacillus, is that there is very little understanding of what stress signals are detected and how these signals are integrated into the σB regulatory pathway via the stressosome. The only exception to this is the mechanism that allows photons of blue light to be detected by the stressosome protein RsbL (Chan et al., 2013; O’Donoghue et al., 2016). It is clear that acid and salt and growth-phase all trigger the activation of σB (Utratna et al., 2011) but the nature of the stress signal detected in each case is unknown, neither is the sensory mechanism known. It is thought that, like RsbL, the N-terminal domains of RsbR or its paralogs (RsbR2, 3, and 4), which are predicted to form turret-like structures that protrude from the surface of the stressosome, are likely to play an important role in signal integration. Whether multiple distinct signals can be detected (possible by virtue of the distinct N-terminal domains of RsbR and its paralogs), or whether a single generic stress-associated signal is detected is still unknown at present. In the case of oxidative stress, it has been proposed that the membrane-spanning miniprotein Prli42 might transduce signals directly to the stressosome through its interaction with RsbR, but the mechanism involved has not been elucidated (Impens et al., 2017).
Although some structural information is available for the stressosome (Williams et al., 2019), high resolution crystal structures of individual components combined with cryo-electron microscopic images of native stressosomes (as opposed to in vitro reconstituted stressosomes) will be required to build a clear picture of what the in vivo structure of the stressosome is like. Information on subcellular localization and assembly dynamics will also be useful to build a model of where in the cell stress sensing occurs and whether stressosomes are structurally homogeneous in vivo or whether different stoichiometries can produce functional differences between them. Single-cell time-resolved approaches will be necessary to see whether structural or stoichiometric differences in stressosomes between cells might contribute to heterogeneity in σB activity observed within populations subjected to stress. The extent to which individual L. monocytogenes cells engage in bet-hedging in response to stressful environmental conditions remains to be fully explored.
Finally the role of the GSR in modulating the virulence of L. monocytogenes is still an open question. There are multiple lines of evidence suggesting regulatory crosstalk between σB and PrfA and these need to be explored further (Gaballa et al., 2019; Tiensuu et al., 2019). While σB plays an essential role during the GI stage of the infectious cycle, it is less important during the systemic stages, where PrfA appears to be the dominant regulator. Both regulators are modulated by complex multi-layered control circuitry, highlighting the importance to fitness of deploying these systems only when the prevailing conditions are suitable. We have seen clear evidence that there is a significant burden on resources associated with deploying the GSR (Guerreiro et al., 2020) and a similar cost has been reported for inappropriate activation of PrfA (Bruno and Freitag, 2010). Clarification of the nature of the regulatory crosstalk between these systems will give new insights into the biology of this human pathogen as well as suggesting new approaches to control it.
All three authors contributed to researching, writing, and editing this mini-review.
This project has received funding from the European Union’s Horizon 2020 Research and Innovation Program under Marie Skłodowska-Curie grant agreement no. 721456.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abram, F. (2007). Responses to Stress in Listeria Monocytogenes: Using Proteomics to Investigate the Role of the Alternative Sigma Factor, Sigma B. Doctoral Thesis, National University of Ireland, Galway.
Abram, F., Starr, E., Karatzas, K. A., Matlawska-Wasowska, K., Boyd, A., Wiedmann, M., et al. (2008a). Identification of components of the sigma B regulon in Listeria monocytogenes that contribute to acid and salt tolerance. Appl. Environ. Microbiol. 74, 6848–6858. doi: 10.1128/aem.00442-08
Abram, F., Su, W. L., Wiedmann, M., Boor, K. J., Coote, P., Botting, C., et al. (2008b). Proteomic analyses of a Listeria monocytogenes mutant lacking σB identify new components of the σB regulon and highlight a role for σB in the utilization of glycerol. Appl. Environ. Microbiol. 74, 594–604. doi: 10.1128/aem.01921-07
Antonietti, R., and Ferrini, C. (1986). An automatic method for the measurement of the effect of short stresses on the microbial ATP pool. J. Microbiol. Methods 6, 21–25. doi: 10.1016/0167-7012(86)90028-x
Antonietti, R., and Tomaselli, L. (1991). Effects of changes in extracellular pH on ATP pools of some prokaryotic and eukaryotic microorganisms. Verh. Int. Ver. Limnol. 24, 2618–2620. doi: 10.1080/03680770.1989.11900036
Aravind, L., and Koonin, E. V. (2000). The STAS domain—a link between anion transporters and antisigma-factor antagonists. Curr. Biol. 10, R53–R55.
Avila-Pérez, M., Hellingwerf, K. J., and Kort, R. (2006). Blue light activates the σB-dependent stress response of Bacillus subtilis via YtvA. J. Bacteriol. 188, 6411–6414. doi: 10.1128/jb.00716-06
Ávila-Pérez, M., van der Steen, J. B., Kort, R., and Hellingwerf, K. J. (2010). Red light activates the σB-mediated general stress response of Bacillus subtilis via the energy branch of the upstream signaling cascade. J. Bacteriol. 192, 755–762. doi: 10.1128/jb.00826-09
Bécavin, C., Koutero, M., Tchitchek, N., Cerutti, F., Lechat, P., Maillet, N., et al. (2017). Listeriomics: an interactive web platform for systems biology of Listeria. MSystems 2:e186-e116.
Becker, L. A., Cetin, M. S., Hutkins, R. W., and Benson, A. K. (1998). Identification of the gene encoding the alternative sigma factor σB from Listeria monocytogenes and its rrole in osmotolerance. J. Bacteriol. 180, 4547–4554. doi: 10.1128/jb.180.17.4547-4554.1998
Begley, M., Gahan, C. G., and Hill, C. (2002). Bile stress response in Listeria monocytogenes LO28: adaptation, cross-protection, and identification of genetic loci involved in bile resistance. Appl. Environ. Microbiol. 68, 6005–6012. doi: 10.1128/aem.68.12.6005-6012.2002
Begley, M., Hill, C., and Ross, R. P. (2006). Tolerance of Listeria monocytogenes to cell envelope-acting antimicrobial agents is dependent on SigB. Appl. Environ. Microbiol. 72, 2231–2234. doi: 10.1128/aem.72.3.2231-2234.2006
Begley, M., Sleator, R. D., Gahan, C. G., and Hill, C. (2005). Contribution of three bile-associated loci, bsh, pva, and btlB, to gastrointestinal persistence and bile tolerance of Listeria monocytogenes. Infect. immun. 73, 894–904. doi: 10.1128/iai.73.2.894-904.2005
Bergholz, T. M., Bowen, B., Wiedmann, M., and Boor, K. J. (2012). Listeria monocytogenes shows temperature-dependent and-independent responses to salt stress, including responses that induce cross-protection against other stresses. Appl. Environ. Microbiol. 78, 2602–612.
Biswas, R., Sonenshein, A. L., and Belitsky, B. R. (2020). Genome-wide identification of Listeria monocytogenes CodY-binding sites. Mol. Microbiol. 113:841–858. doi: 10.1111/mmi.14449
Bourke, P., O’Byrne, C., Boehm, D., Cullen, P. J., Keener, K., Bourke, P., et al. (2019). The effect of atmospheric cold plasma on bacterial stress responses and virulence using Listeria monocytogenes knockout mutants. Front. Microbiol. 10:2841. doi: 10.3389/fmicb.2019.02841
Brøndsted, L., Kallipolitis, B. H., Ingmer, H., and Knöchel, S. (2003). kdpE and a putative RsbQ homologue contribute to growth of Listeria monocytogenes at high osmolarity and low temperature. FEMS Microbiol. Lett. 219, 233–239. doi: 10.1016/s0378-1097(03)00052-1
Bruno, J. C.Jr, and Freitag, N. E. (2010). Constitutive activation of PrfA tilts the balance of Listeria monocytogenes fitness towards life within the host versus environmental survival. PLoS One 5, e15138. doi: 10.1371/journal.pone.0015138
Buttani, V., Losi, A., Eggert, T., Krauss, U., Jaeger, K. E., Cao, Z., et al. (2007). Conformational analysis of the blue-light sensing protein YtvA reveals a competitive interface for LOV–LOV dimerization and interdomain interactions. Photochem. Photobiol. Sci. 6, 41–49. doi: 10.1039/b610375h
Cabeen, M. T., Russell, J. R., Paulsson, J., and Losick, R. (2017). Use of a microfluidic platform to uncover basic features of energy and environmental stress responses in individual cells of Bacillus subtilis. PLoS Genet 13:e1006901. doi: 10.1371/journal.pgen.1006901
Cai, L., Friedman, N., and Xie, X. S. (2006). Stochastic protein expression in individual cells at the single molecule level. Nature 440, 358–362. doi: 10.1038/nature04599
Cetin, M. S., Zhang, C., Hutkins, R. W., and Benson, A. K. (2004). Regulation of transcription of compatible solute transporters by the general stress sigma factor, σB, in Listeria monocytogenes. J. Bacteriol. 186, 794–802. doi: 10.1128/jb.186.3.794-802.2004
Chan, R. H., Lewis, J. W., and Bogomolni, R. A. (2013). Photocycle of the LOV−STAS protein from the pathogen Listeria monocytogenes. Photochem. Photobiol. 89, 361–369. doi: 10.1111/php.12004
Chatterjee, S. S., Hossain, H., Otten, S., Kuenne, C., Kuchmina, K., Machata, S., et al. (2006). Intracellular gene expression profile of Listeria monocytogenes. Infect. Immun. 74, 1323–1338. doi: 10.1128/iai.74.2.1323-1338.2006
Chaturongakul, S., and Boor, K. J. (2004). RsbT and RsbV contribute to σB-dependent survival under environmental, energy, and intracellular stress conditions in Listeria monocytogenes. Appl. Environ. Microbiol. 70, 5349–5356. doi: 10.1128/aem.70.9.5349-5356.2004
Chaturongakul, S., and Boor, K. J. (2006). σB activation under environmental and energy stress conditions in Listeria monocytogenes. Appl. Environ. Microbiol. 72, 5197–5203. doi: 10.1128/aem.03058-05
Chaturongakul, S., Raengpradub, S., Palmer, M. E., Bergholz, T. M., Orsi, R. H., Ollinger, J., et al. (2011). Transcriptomic and phenotypic analyses identify coregulated, overlapping regulons among PrfA, CtsR, HrcA, and the alternative sigma factors σB, σC, σH, and σL in Listeria monocytogenes. Appl. Environ. Microbiol. 77, 187–200. doi: 10.1128/aem.00952-10
Chen, C. C., Lewis, R. J., Harris, R., Yudkin, M. D., and Delumeau, O. (2003). A supramolecular complex in the environmental stress signalling pathway of Bacillus subtilis. Mol. Microbiol. 49, 1657–1669. doi: 10.1046/j.1365-2958.2003.03663.x
Chen, C.-C., Yudkin, M. D., and Delumeau, O. (2004). Phosphorylation and RsbX-dependent dephosphorylation of RsbR in the RsbR-RsbS complex of Bacillus subtilis. J. Bacteriol. 186, 6830–6836. doi: 10.1128/jb.186.20.6830-6836.2004
Christiansen, J. K., Larsen, M. H., Ingmer, H., Søgaard-Andersen, L., and Kallipolitis, B. H. (2004). The RNA-binding protein Hfq of Listeria monocytogenes: role in stress tolerance and virulence. J. Bacteriol. 186, 3355–3362. doi: 10.1128/jb.186.11.3355-3362.2004
Cole, M., Jones, M., and Holyoak, C. (1990). The effect of pH, salt concentration and temperature on the survival and growth of Listeria monocytogenes. J. Appl. Bacteriol. 69, 63–72. doi: 10.1111/j.1365-2672.1990.tb02912.x
Cortes, B. W., Naditz, A. L., Anast, J. M., and Schmitz-Esser, S. (2020). Transcriptome sequencing of Listeria monocytogenes reveals major gene expression changes in response to lactic acid stress exposure but a less pronounced response to oxidative stress. Front. Microbiol. 10:3110. doi: 10.3389/fmicb.2019.03110
Cosgrave, E. (2010). An Investigation into the Roles of RsbV and RsbW in the Regulation of Sigma B activity in the Human Pathogen Listeria monocytogenes. Doctoral Thesis, National University of Ireland, Galway.
Cotter, P. D., Gahan, C. G., and Hill, C. (2001a). A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol. Microbiol. 40, 465–475. doi: 10.1046/j.1365-2958.2001.02398.x
Cotter, P. D., O’Reilly, K., and Hill, C. (2001b). Role of the glutamate decarboxylase acid resistance system in the survival of Listeria monocytogenes LO28 in low pH foods. J. Food Protect. 64, 1362–1368. doi: 10.4315/0362-028x-64.9.1362
Cotter, P. D., Ryan, S., Gahan, C. G., and Hill, C. (2005). Presence of GadD1 glutamate decarboxylase in selected Listeria monocytogenes strains is associated with an ability to grow at low pH. Appl. Environ. Microbiol. 71, 2832–2839. doi: 10.1128/aem.71.6.2832-2839.2005
Crosson, S., and Moffat, K. (2002). Photoexcited structure of a plant photoreceptor domain reveals a light-driven molecular switch. Plant Cell 14, 1067–1075. doi: 10.1105/tpc.010475
Curtis, T. D., Takeuchi, I., Gram, L., and Knudsen, G. M. (2017). The influence of the Toxin/Antitoxin mazEF on growth and survival of Listeria monocytogenes under stress. Toxins 9:31. doi: 10.3390/toxins9010031
de las Heras, A., Cain, R. J., Bielecka, M. K., and Vázquez-Boland, J. A. (2011). Regulation of Listeria virulence: PrfA master and commander. Curr. Opin. Microbiol. 14, 118–127. doi: 10.1016/j.mib.2011.01.005
Dorey, A. L., Lee, B.-H., Rotter, B., and O’Byrne, C. P. (2019a). Blue light sensing in Listeria monocytogenes is temperature-dependent and the transcriptional response to it is predominantly SigB-dependent. Front. Microbiol. 10:2497. doi: 10.3389/fmicb.2019.02497
Dorey, A., Marinho, C., Piveteau, P., and O’Byrne, C. (2019b). Role and regulation of the stress activated sigma factor sigma B (σB) in the saprophytic and host-associated life stages of Listeria monocytogenes. Adv. Appl. Microbiol. 106, 1–48. doi: 10.1016/bs.aambs.2018.11.001
Dowd, G. C., Joyce, S. A., Hill, C., and Gahan, C. G. (2011). Investigation of the mechanisms by which Listeria monocytogenes grows in porcine gallbladder bile. Infect. Immun. 79, 369–379.
Eymann, C., Schulz, S., Gronau, K., Becher, D., Hecker, M., and Price, C. W. (2011). In vivo phosphorylation patterns of key stressosome proteins define a second feedback loop that limits activation of Bacillus subtilis σB. Mol. Microbiol. 80, 798–810. doi: 10.1111/j.1365-2958.2011.07609.x
Feehily, C., Finnerty, A., Casey, P. G., Hill, C., Gahan, C. G., O’Byrne, C. P., et al. (2014). Divergent evolution of the activity and regulation of the glutamate decarboxylase systems in Listeria monocytogenes EGD-e and 10403S: roles in virulence and acid tolerance. PloS one 9:e112649. doi: 10.1371/journal.pone.0112649
Feehily, C., O’Byrne, C. P., and Karatzas, K.-A. G. (2012). Listeria monocytogenes has a functional γ-aminobutyrate (GABA) shunt: role in acid tolerance and succinate biosynthesis. Appl. Environ. Microbiol. 79, 74–80.
Feehily, C., O’Byrne, C. P., and Karatzas, K. A. G. (2013). Functional γ-aminobutyrate shunt in Listeria monocytogenes: role in acid tolerance and succinate biosynthesis. Appl. Environ. Microbiol. 79, 74–80. doi: 10.1128/aem.02184-12
Ferreira, A., Gray, M., Wiedmann, M., and Boor, K. J. (2004). Comparative genomic analysis of the sigB operon in Listeria monocytogenes and in other gram-positive bacteria. Curr. Microbiol. 48, 39–46. doi: 10.1007/s00284-003-4020-x
Ferreira, A., O’Byrne, C. P., and Boor, K. J. (2001). Role of σB in heat, ethanol, acid, and oxidative stress resistance and during carbon starvation in Listeria monocytogenes. Appl. Environ. Microbiol. 67, 4454–4457. doi: 10.1128/aem.67.10.4454-4457.2001
Fraser, K. R., Sue, D., Wiedmann, M., Boor, K., and O’Byrne, C. P. (2003). Role of σB in regulating the compatible solute uptake systems of Listeria monocytogenes: osmotic induction of opuC is σB dependent. Appl. Environ. Microbiol. 69, 2015–2022. doi: 10.1128/aem.69.4.2015-2022.2003
Gaballa, A., Guariglia-Oropeza, V., Wiedmann, M., and Boor, K. J. (2019). Cross talk between SigB and PrfA in Listeria monocytogenes facilitates transitions between extra-and intracellular environments. Microbiol. Mol. Biol. Rev. 83:e 34-19.
Gaidenko, T. A., Kim, T.-J., Weigel, A. L., Brody, M. S., and Price, C. W. (2006). The blue-light receptor YtvA acts in the environmental stress signaling pathway of Bacillus subtilis. J. Bacteriol. 188, 6387–6395. doi: 10.1128/jb.00691-06
Gandhi, M., and Chikindas, M. L. (2007). Listeria: a foodborne pathogen that knows how to survive. Int. J. Food Microbiol. 113, 1–15. doi: 10.1016/j.ijfoodmicro.2006.07.008
Garner, M., Njaa, B., Wiedmann, M., and Boor, K. (2006). Sigma B contributes to Listeria monocytogenes gastrointestinal infection but not to systemic spread in the guinea pig infection model. Infect. Immun. 74, 876–886. doi: 10.1128/iai.74.2.876-886.2006
Giotis, E. S., Julotok, M., Wilkinson, B. J., Blair, I. S., and McDowell, D. A. (2008). Role of sigma B factor in the alkaline tolerance response of Listeria monocytogenes 10403S and cross-protection against subsequent ethanol and osmotic stress. J. Food Prot. 71, 1481–1485. doi: 10.4315/0362-028x-71.7.1481
Gopal, S., Borovok, I., Ofer, A., Yanku, M., Cohen, G., Goebel, W., et al. (2005). A multidomain fusion protein in Listeria monocytogenes catalyzes the two primary activities for glutathione biosynthesis. J. Bacteriol. 187, 3839–3847. doi: 10.1128/JB.187.11.3839-3847.2005
Guerreiro, D. N., Wu, J., Dessaux, C., Oliveira, A. H., Tiensuu, T., Gudynaite, D.,, et al. (2020). Mild stress conditions during laboratory culture promote the proliferation of mutations that negatively affect Sigma B activity in Listeria monocytogenes. J. Bacteriol. 202(9):e751–e719..
Guldimann, C., Guariglia-Oropeza, V., Harrand, S., Kent, D., Boor, K. J., Wiedmann, M., et al. (2017). Stochastic and differential activation of σB and PrfA in Listeria monocytogenes at the single cell level under different environmental stress conditions. Front. Microbiol. 8:348. doi: 10.3389/fmicb.2017.00348
Hall, M., Grundström, C., Begum, A., Lindberg, M. J., Sauer, U. H., Almqvist, F., et al. (2016). Structural basis for glutathione-mediated activation of the virulence regulatory protein PrfA in Listeria. Proc Natl Acad Sci U.S.A. 113, 14733–14738. doi: 10.1073/pnas.1614028114
He, K., Xin, Y. P., Shan, Y., Zhang, X., Song, H. H., Fang, W. H., et al. (2019). Phosphorylation residue T175 in RsbR protein is required for efficient induction of sigma B factor and survival of Listeria monocytogenes under acidic stress. J. Zhejiang Univ.Sci. B 20, 660–669. doi: 10.1631/jzus.b1800551
Hecker, M., Pané-Farré, J., and Uwe, V. (2007). SigB-dependent general stress response in Bacillus subtilis and related gram-positive bacteria. Annu. Rev. Microbiol. 61, 215–236. doi: 10.1146/annurev.micro.61.080706.093445
Hecker, M., Völker, U., and Heim, C. (1989). RelA-independent (p) ppGpp accumulation and heat shock protein induction after salt stress in Bacillus subtilis. FEMS Microbiol. Lett. 58, 125–128. doi: 10.1111/j.1574-6968.1989.tb03031.x
Hsu, C.-Y., Cairns, L., Hobley, L., Abbott, J., O’Byrne, C., Stanley-Wall, N. R., et al. (2020). Genomic differences between Listeria monocytogenes EGDe isolates reveals crucial roles for SigB and wall rhamnosylation in biofilm formation. J. Bacteriol. 202(7):e692–e619
Impens, F., Rolhio, N., Radoshevich, L., Bécavin, C., Duval, M., Mellin, J., et al. (2017). N-terminomics identifies Prli42 as a membrane miniprotein conserved in Firmicutes and critical for stressosome activation in Listeria monocytogenes. Nat. Microbiol. 2:17005.
Ju, J., Mitchell, T., Peters, H., and Haldenwang, W. (1999). Sigma factor displacement from RNA polymerase during Bacillus subtilis sporulation. J. Bacteriol. 181, 4969–4977. doi: 10.1128/jb.181.16.4969-4977.1999
Jurk, M., Dorn, M., Kikhney, A., Svergun, D., Gärtner, W., Schmieder, P., et al. (2010). The switch that does not flip: the blue-light receptor YtvA from Bacillus subtilis adopts an elongated dimer conformation independent of the activation state as revealed by a combined AUC and SAXS study. J. Mol. Biol. 403, 78–87. doi: 10.1016/j.jmb.2010.08.036
Karatzas, K.-A. G., Brennan, O., Heavin, S., Morrissey, J., and O’Byrne, C. P. (2010). Intracellular accumulation of high levels of γ-aminobutyrate by Listeria monocytogenes 10403S in response to low pH: uncoupling of γ-aminobutyrate synthesis from efflux in a chemically defined medium. Appl. Environ. Microbiol. 76, 3529–3537. doi: 10.1128/aem.03063-09
Karatzas, K.-A. G., Suur, L., and O’Byrne, C. P. (2012). Characterization of the intracellular glutamate decarboxylase system: analysis of its function, transcription, and role in the acid resistance of various strains of Listeria monocytogenes. Appl. Environ. Microbiol. 78, 3571–3579. doi: 10.1128/aem.00227-12
Kazmierczak, M. J., Mithoe, S. C., Boor, K. J., and Wiedmann, M. (2003). Listeria monocytogenes sigma B regulates stress response and virulence functions. J. Bacteriol. 185, 5722–5734 doi: 10.1128/jb.185.19.5722-5734.2003
Kazmierczak, M. J., Wiedmann, M., and Boor, K. J. (2006). Contributions of Listeria monocytogenes sigmaB and PrfA to expression of virulence and stress response genes during extra- and intracellular growth. Microbiology 152, 1827–1838. doi: 10.1099/mic.0.28758-0
Kim, T.-J., Gaidenko, T. A., and Price, C. W. (2004). In vivo phosphorylation of partner switching regulators correlates with stress transmission in the environmental signaling pathway of Bacillus subtilis. J. Bacteriol. 186, 6124–6132. doi: 10.1128/jb.186.18.6124-6132.2004
Lecuit, M. (2007). Human listeriosis and animal models. Microbes Infect. 9, 1216–1225. doi: 10.1016/j.micinf.2007.05.009
Levine, J. H., Lin, Y., and Elowitz, M. B. (2013). Functional roles of pulsing in genetic circuits. Science 342, 1193–1200. doi: 10.1126/science.1239999
Liu, Y., Orsi, R. H., Gaballa, A., Wiedmann, M., Boor, K. J., and Guariglia-Oropeza, V. (2019). Systematic review of the Listeria monocytogenes σB regulon supports a role in stress response, virulence and metabolism. Future Microbiol. 14, 801–828. doi: 10.2217/fmb-2019-0072
Liu, Y., Orsi, R. H., Boor, K. J., Wiedmann, M., and Guariglia-Oropeza, V. (2017). Home alone: elimination of all but one alternative sigma factor in Listeria monocytogenes allows prediction of new roles for σB. Front. Microbiol. 8:1910. doi: 10.3389/fmicb.2017.01910
Lobel, L., and Herskovits, A. A. (2016). Systems level analyses reveal multiple regulatory activities of CodY controlling metabolism, motility and virulence in Listeria monocytogenes. PLoS Genet 12:e1005870. doi: 10.1371/journal.pgen.1005870
Lobel, L., Sigal, N., Borovok, I., Belitsky, B. R., Sonenshein, A. L., and Herskovits, A. A. (2015). The metabolic regulator CodY links Listeria monocytogenes metabolism to virulence by directly activating the virulence regulatory gene prfA. Mol. Microbiol. 95, 624–644. doi: 10.1111/mmi.12890
Locke, J. C., Young, J. W., Fontes, M., Hernandez Jimenez, M. J., and Elowitz, M. B. (2011). Stochastic pulse regulation in bacterial stress response. Science 334, 366–369. doi: 10.1126/science.1208144
Losi, A., Polverini, E., Quest, B., and Gärtner, W. (2002). First evidence for phototropin-related blue-light receptors in prokaryotes. Biophys. J. 82, 2627–2634. doi: 10.1016/s0006-3495(02)75604-x
Marinho, C. M., Dos Santos, P. T., Kallipolitis, B. H., Johansson, J., Ignatov, D., and Guerreiro, D. N. (2019). The σB-dependent regulatory sRNA Rli47 represses isoleucine biosynthesis in Listeria monocytogenes through a direct interaction with the ilvA transcript. RNA Biol. 16, 1424–1437. doi: 10.1080/15476286.2019.1632776
Marinho, C. M., Garmyn, D., Ga, L., Brunhede, M. Z., O’Byrne, C., and Piveteau, P. (2020). Investigation of the roles of AgrA and σB regulators in Listeria monocytogenes adaptation to roots and soil. FEMS Microbiol. Lett. 367:fnaa036.
Marles-Wright, J., and Lewis, R. J. (2008). The Bacillus subtilis stressosome: a signal integration and transduction hub. Commun. Integr. Biol. 1, 182–184.
Marles-Wright, J., and Lewis, R. J. (2010). The stressosome: molecular architecture of a signalling hub. Biochem. Soc. Trans. 38, 928–933. doi: 10.1042/BST0380928
Martinez, L., Reeves, A., and Haldenwang, W. (2010). Stressosomes formed in Bacillus subtilis from the RsbR protein of Listeria monocytogenes allow σB activation following exposure to either physical or nutritional stress. J. Bacteriol. 192, 6279–6286. doi: 10.1128/jb.00467-10
Mauri, M., and Klumpp, S. (2014). A model for sigma factor competition in bacterial cells. PLoS comput. Biol. 10:e1003845. doi: 10.1371/journal.pcbi.1003845
Milohanic, E., Glaser, P., Coppée, J. Y., Frangeul, L., Vega, Y., Vázquez-Boland, J. A., et al. (2003). Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol. Microbiol. 47, 1613–1625. doi: 10.1046/j.1365-2958.2003.03413.x
Misra, S. K., Milohanic, E., Aké, F., Mijakovic, I., Deutscher, J., Monnet, V., et al. (2011). Analysis of the serine/threonine/tyrosine phosphoproteome of the pathogenic bacterium Listeria monocytogenes reveals phosphorylated proteins related to virulence. Proteomics 11, 4155–4165. doi: 10.1002/pmic.201100259
Möglich, A., and Moffat, K. (2007). Structural basis for light-dependent signaling in the dimeric LOV domain of the photosensor YtvA. J. Mol. Biol. 373, 112–126. doi: 10.1016/j.jmb.2007.07.039
Murray, J. W., Delumeau, O., and Lewis, R. J. (2005). Structure of a nonheme globin in environmental stress signaling. Proc. Natl. Acad. Sci. U.S.A. 102, 17320–17325. doi: 10.1073/pnas.0506599102
Nadezhdin, E., Murphy, N., Dalchau, N., Phillips, A., and Locke, J. C. (2020). Stochastic pulsing of gene expression enables the generation of spatial patterns in Bacillus subtilis biofilms. Nat. Commun 11, 1–12.
Nadon, C. A., Bowen, B. M., Wiedmann, M., and Boor, K. J. (2002). Sigma B contributes to PrfA-mediated virulence in Listeria monocytogenes. Infect. Immun. 70, 3948–3952. doi: 10.1128/iai.70.7.3948-3952.2002
NicAogáin, K., and O’Byrne, C. P. (2016). The role of stress and stress adaptations in determining the fate of the bacterial pathogen Listeria monocytogenes in the food chain. Front. Microbiol. 7:1865. doi: 10.3389/fmicb.2016.01865
Nielsen, J. S., Olsen, A. S., Bonde, M., Valentin-Hansen, P., and Kallipolitis, B. H. (2008). Identification of a σB-dependent small noncoding RNA in Listeria monocytogenes. J. Bacteriol. 190, 6264–6270. doi: 10.1128/jb.00740-08
Nyström, T. (2004). MicroReview: growth versus maintenance: a trade−off dictated by RNA polymerase availability and sigma factor competition? Mol. Microbiol. 54, 855–862. doi: 10.1111/j.1365-2958.2004.04342.x
O’Byrne, C. P., and Karatzas, K. A. (2008). The role of sigma B (σB) in the stress adaptations of Listeria monocytogenes: overlaps between stress adaptation and virulence. Adv. Appl. Microbiol. 65, 115–140. doi: 10.1016/s0065-2164(08)00605-9
O’Donoghue, B. (2016). A Molecular Genetic Investigation into Stress Sensing in the Food-Borne Pathogen Listeria monocytogenes: Roles for RsbR and its Paralogues. Doctoral Thesis, National University of Ireland, Galway.
O’Donoghue, B., NicAogáin, K., Bennett, C., Conneely, A., Tiensuu, T., Johansson, J., et al. (2016). Blue-light inhibition of Listeria monocytogenes growth is mediated by reactive oxygen species and is influenced by σB and the blue-light sensor Lmo0799. Appl. Environ. Microbiol. 82, 4017–4027. doi: 10.1128/aem.00685-16
Oliver, H., Orsi, R., Wiedmann, M., and Boor, K. (2010). Listeria monocytogenes σB has a small core regulon and a conserved role in virulence but makes differential contributions to stress tolerance across a diverse collection of strains. Appl. Environ. Microbiol. 76, 4216–4232. doi: 10.1128/aem.00031-10
Ollinger, J., Bowen, B., Wiedmann, M., Boor, K. J., and Bergholz, T. M. (2009). Listeria monocytogenes σB modulates PrfA-mediated virulence factor expression. Infect. immun. 77, 2113–2124. doi: 10.1128/iai.01205-08
Ondrusch, N., and Kreft, J. (2011). Blue and red light modulates SigB-dependent gene transcription, swimming motility and invasiveness in Listeria monocytogenes. PloS one 6:e16151. doi: 10.1371/journal.pone.0016151
Österberg, S., Peso-Santos, T. D., and Shingler, V. (2011). Regulation of alternative sigma factor use. Annu. Rev. Microbiol. 65, 37–55.
Palmer, M. E., Chaturongakul, S., Wiedmann, M., and Boor, K. J. (2011). The Listeria monocytogenes σB regulon and its virulence-associated functions are inhibited by a small molecule. MBio 2, e241–e211.
Palmer, M. E., Wiedmann, M., and Boor, K. J. (2009). σB and σL contribute to Listeria monocytogenes 10403S response to the antimicrobial peptides SdpC and nisin. Foodborne pathog. Dis. 6, 1057–1065. doi: 10.1089/fpd.2009.0292
Pané-Farré, J., Lewis, R. J., and Stülke, J. (2005). The RsbRST stress module in bacteria: a signalling system that may interact with different output modules. J. Mol. Microbiol. Biotechnol. 9, 65–76. doi: 10.1159/000088837
Pané-Farré, J., Quin, M. B., Lewis, R. J., and Marles-Wright, J. (2017). “Structure and Function of the stressosome signalling hub” in Macromolecular Protein Complexes. Eds J. R. Harris and J. Marles-Wright (Cham: Springer International Publishing) 1–41. doi: 10.1007/978-3-319-46503-6_1
Pittman, J. R., Buntyn, J. O., Posadas, G., Nanduri, B., Pendarvis, K., Donaldson, J. R., et al. (2014). Proteomic analysis of cross protection provided between cold and osmotic stress in Listeria monocytogenes. J. Proteome Res. 13, 1896–1904. doi: 10.1021/pr401004a
Portman, J. L., Dubensky, S. B., Peterson, B. N., Whiteley, A. T., and Portnoy, D. A. (2017). Activation of the Listeria monocytogenes virulence program by a reducing environment. mBio 8:e1595-17. doi: 10.1128/mBio.01595-17
Raengpradub, S., Wiedmann, M., and Boor, K. J. (2008). Comparative analysis of the σB-dependent stress responses in Listeria monocytogenes and Listeria innocua strains exposed to selected stress conditions. Appl. Environ. Microbiol. 74, 158–171. doi: 10.1128/aem.00951-07
Reniere, M. L., Whiteley, A. T., Hamilton, K. L., John, S. M., Lauer, P., Brennan, R. G., et al. (2015). Glutathione activates virulence gene expression of an intracellular pathogen. Nature 517, 170–173. doi: 10.1038/nature14029
Ribeiro, V., Mujahid, S., Orsi, R. H., Bergholz, T. M., Wiedmann, M., Boor, K. J., et al. (2014). Contributions of σB and PrfA to Listeria monocytogenes salt stress under food relevant conditions. Int. J. Food Microbiol. 177, 98–108. doi: 10.1016/j.ijfoodmicro.2014.02.018
Sæbø, K. P., Sundaram, A. Y. M., Skjerdal, T., Wasteson, Y., Kijewski, A., Lindbäck, T., et al. (2019). Exposure to broad-spectrum visible light causes major transcriptomic changes in Listeria monocytogenes EGDe. Appl. Environ. Microbiol. 85:e1462-e1419.
Salomon, M., Eisenreich, W., Dürr, H., Schleicher, E., Knieb, E., Massey, V., et al. (2001). An optomechanical transducer in the blue light receptor phototropin from Avena sativa. Proc. Natl. Acad. Sci. U.S. A. 98, 12357–12361. doi: 10.1073/pnas.221455298
Shin, J.-H., Brody, M. S., and Price, C. W. (2010a). Physical and antibiotic stresses require activation of the RsbU phosphatase to induce the general stress response in Listeria monocytogenes. Microbiology 156, 2660–2669. doi: 10.1099/mic.0.041202-0
Shin, J.-H., Kim, J., Kim, S. M., Kim, S., Lee, J. C., Ahn, J. M., et al. (2010b). σB-dependent protein induction in Listeria monocytogenes during vancomycin stress. FEMS Microbiol. Lett. 308, 94–100. doi: 10.1111/j.1574-6968.2010.01998.x
Sleator, R. D., Watson, D., Hill, C., and Gahan, C. G. (2009). The interaction between Listeria monocytogenes and the host gastrointestinal tract. Microbiology 155, 2463–2475. doi: 10.1099/mic.0.030205-0
Sleator, R. D., Wemekamp−Kamphuis, H. H., Gahan, C. G., Abee, T., and Hill, C. A. (2005). PrfA−regulated bile exclusion system (BilE) is a novel virulence factor in Listeria monocytogenes. Mol. Microbiol. 55, 1183–1195. doi: 10.1111/j.1365-2958.2004.04454.x
Sue, D., Boor, K. J., and Wiedmann, M. (2003). σB-dependent expression patterns of compatible solute transporter genes opuCA and lmo1421 and the conjugated bile salt hydrolase gene bsh in Listeria monocytogenes. Microbiology 149, 3247–3256. doi: 10.1099/mic.0.26526-0
Tapia, N. C., Dorey, A. L., Gahan, C. G. M., den Besten, H. M. W., O’Byrne, C. P., and Abee, T. (2020). Different carbon sources result in differential activation of sigma B and stress resistance in Listeria monocytogenes. Int. J. Food Microbiol. 320:108504. doi: 10.1016/j.ijfoodmicro.2019.108504
Tiensuu, T., Andersson, C., Rydén, P., and Johansson, J. (2013). Cycles of light and dark co−ordinate reversible colony differentiation in Listeria monocytogenes. Mol. Microbiol. 87, 909–924. doi: 10.1111/mmi.12140
Tiensuu, T., Guerreiro, D. N., Oliveira, A. H., O’Byrne, C., and Johansson, J. (2019). Flick of a switch: regulatory mechanisms allowing Listeria monocytogenes to transition from a saprophyte to a killer. Microbiology 165, 819–833. doi: 10.1099/mic.0.000808
Toledo-Arana, A., Dussurget, O., Nikitas, G., Sesto, N., Guet-Revillet, H., Balestrino, D., et al. (2009). The Listeria transcriptional landscape from saprophytism to virulence. Nature 459, 950–956. doi: 10.1038/nature08080
Utratna, M., Cosgrave, E., Baustian, C., Ceredig, R. H., and O’Byrne, C. P. (2014). Effects of growth phase and temperature on activity within a Listeria monocytogenes population: evidence for RsbV-independent activation of at refrigeration temperatures. BioMed Res. Int. 2014: 641647.
Utratna, M., Cosgrave, E., Baustian, C., Ceredig, R., and O’Byrne, C. (2012). Development and optimization of an EGFP-based reporter for measuring the general stress response in Listeria monocytogenes. Bioengineered 3, 93–103. doi: 10.4161/bbug.19476
Utratna, M., Shaw, I., Starr, E., and O’Byrne, C. P. (2011). Rapid, transient, and proportional activation of σB in response to osmotic stress in Listeria monocytogenes. Appl. Environ. Microbiol. 77, 7841–7845. doi: 10.1128/aem.05732-11
Vijay, K., Brody, M. S., Fredlund, E., and Price, C. W. (2000). A. PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the σB transcription factor of Bacillus subtilis. Mol. Microbiol. 35, 180–188. doi: 10.1046/j.1365-2958.2000.01697.x
Voelker, U., Luo, T., Smirnova, N., and Haldenwang, W. (1997). Stress activation of Bacillus subtilis sigma B can occur in the absence of the sigma B negative regulator RsbX. J. Bacteriol. 179, 1980–1984. doi: 10.1128/jb.179.6.1980-1984.1997
Wemekamp-Kamphuis, H. H., Wouters, J. A., de Leeuw, P. P. L. A., Hain, T., Chakraborty, T., Abee, T., et al. (2004). Identification of sigma factor σB-controlled genes and their impact on acid stress, high hydrostatic pressure, and freeze survival in Listeria monocytogenes EGD-e. Appl. Environ. Microbiol. 70, 3457–3466. doi: 10.1128/aem.70.6.3457-3466.2004
Wemekamp-Kamphuis, H. H., Wouters, J. A., Sleator, R. D., Gahan, C. G., Hill, C., Abee, T., et al. (2002). Multiple deletions of the osmolyte transporters BetL, Gbu, and OpuC of Listeria monocytogenes affect virulence and growth at high osmolarity. Appl. Environ. Microbiol. 68, 4710–4716. doi: 10.1128/aem.68.10.4710-4716.2002
Wiedmann, M., Arvik, T. J., Hurley, R. J., and Boor, K. J. (1998). General stress transcription factor σB and its role in acid tolerance and virulence of Listeria monocytogenes. J. Bacteriol. 180, 3650–3656. doi: 10.1128/jb.180.14.3650-3656.1998
Williams, A. H., Redzej, A., Rolhion, N., Costa, T. R. D., Rifflet, A., Waksman, G., et al. (2019). The cryo-electron microscopy supramolecular structure of the bacterial stressosome unveils its mechanism of activation. Nat. Commun. 10:3005.
Wurtzel, O., Sesto, N., Mellin, J. R., Karunker, I., Edelheit, S., Bécavin, C., et al. (2012). Comparative transcriptomics of pathogenic and non−pathogenic Listeria species. Mol. Sys. Biol. 8:583. doi: 10.1038/msb.2012.11
Xia, Y., Xin, Y., Li, X., and Fang, W. (2016). To modulate survival under secondary stress conditions, Listeria monocytogenes 10403S employs RsbX To downregulate σB activity in the poststress recovery stage or stationary phase. Appl. Environ. Microbiol. 82, 1126–1135. doi: 10.1128/aem.03218-15
Yang, X., Kang, C. M., Brody, M. S., and Price, C. W. (1996). Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev. 10, 2265–2275. doi: 10.1101/gad.10.18.2265
Young, J. W., Locke, J. C., and Elowitz, M. B. (2013). Rate of environmental change determines stress response specificity. Proc Natl Acad Sci U.S.A. 110, 4140–4145. doi: 10.1073/pnas.1213060110
Zhang, Q., Feng, Y., Deng, L., Feng, F., Wang, L., Zhou, Q., and Luo, Q. (2011). SigB plays a major role in Listeria monocytogenes tolerance to bile stress. Int. J. Food Microbiol. 145, 238–243. doi: 10.1016/j.ijfoodmicro.2010.12.028
Keywords: Listeria monocytogenes, σB, stress response, virulence, stressosome, signal transduction
Citation: Guerreiro DN, Arcari T and O’Byrne CP (2020) The σB-Mediated General Stress Response of Listeria monocytogenes: Life and Death Decision Making in a Pathogen. Front. Microbiol. 11:1505. doi: 10.3389/fmicb.2020.01505
Received: 28 April 2020; Accepted: 10 June 2020;
Published: 07 July 2020.
Edited by:
Ilana Kolodkin-Gal, Weizmann Institute of Science, IsraelReviewed by:
Alexandre Leclercq, Institut Pasteur, FranceCopyright © 2020 Guerreiro, Arcari and O’Byrne. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Conor P. O’Byrne, Y29ub3Iub2J5cm5lQG51aWdhbHdheS5pZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.