
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 24 June 2020
Sec. Infectious Agents and Disease
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.01402
 Yu-Shan Huang1,2
Yu-Shan Huang1,2 Liang-Chuan Lai3,4
Liang-Chuan Lai3,4 Yu-An Chen4
Yu-An Chen4 Kuan-Yin Lin5
Kuan-Yin Lin5 Yi-Hsuan Chou5
Yi-Hsuan Chou5 Hsiu-Chi Chen6
Hsiu-Chi Chen6 Shu-Sheng Wang7
Shu-Sheng Wang7 Jann-Tay Wang1*
Jann-Tay Wang1* Shan-Chwen Chang1
Shan-Chwen Chang1Background: The prevalence of colonization with multidrug-resistant organisms (MDROs) among healthy adults in the community is largely unknown. This study investigated the colonization rate of multidrug-resistant Enterobacteriaceae, methicillin-resistant Staphylococcus aureus (MRSA), and vancomycin-resistant enterococci (VRE) in the community in Taiwan, and compared the gut microbiota between MDRO carriers and non-carriers.
Methods: This prospective cohort study was conducted from March 2017 to February 2018 at the Hsin-Chu and Jin-Shan branches of National Taiwan University Hospital. Nasal swabs and stool samples were obtained from healthy adults attending a health examination to screen for MDROs. Bacteria isolates of MDROs were tested for antibiotic susceptibility and resistant genes. Relevant data were collected using a standardized questionnaire to evaluate the risk factors for MDROs carriage, and 16S rRNA metagenomics sequencing was performed to analyze gut microbiota.
Results: Among 187 participants, 4.6% (8/174) carried MRSA and 41.4% (77/186) carried third-generation cephalosporin-resistant (3GC-R) Escherichia coli or Klebsiella pneumoniae. The carriage rate of AmpC beta-lactamases and ESBL-producing strains were 16.1 and 27.4%, respectively. No carbapenem-resistant Enterobacteriaceae (CRE) or VRE were detected. The dominant resistant gene of E. coli isolates was CTX-M-type (73%), while that of K. pneumoniae was AmpC beta-lactamases (80%). In the multivariate analysis, the significant risk factors for carrying 3GC-R E. coli or K. pneumoniae were being an employee of technology company A [adjusted odds ratio (aOR) 4.127; 95% confidence interval (CI) 1.824–9.336; p = 0.001], and traveling to Southeast Asia in the past year (aOR 6.545; 95% CI 1.071–40.001; p = 0.042). The gut microbiota analysis showed that the phylum Proteobacteria and the family Enterobacteriaceae were significantly more abundant in 3GC-R E. coli and K. pneumoniae carriers.
Conclusion: A high rate of Taiwanese adults in the community carried 3GC-R Enterobacteriaceae, while no CRE or VRE colonization was noted. Compared with non-carriers, an expansion of Enterobacteriaceae in gut microbiota was found among 3GC-R Enterobacteriaceae carriers.
The wide use of antibiotics in health care institutes and agriculture has led to the emergence of resistant pathogens (Spellberg et al., 2008; Heuer et al., 2009). As highly resistant bacteria spread worldwide, however, newer therapeutic agents are lacking. For infections caused by multidrug-resistant organisms (MDROs), effective treatment is often limited, and this is associated with additional morbidity and mortality and increased medical costs (Founou et al., 2017).
The burden of antibiotic resistance varies geographically, and is an especially great threat in Asia (Lai et al., 2014). In Taiwan, the multicenter surveillance programs for MDROs monitoring in the hospitals had revealed an increase in the prevalence of vancomycin-resistant enterococci (VRE) and carbapenem-resistant Gram-negative bacteria (Tseng et al., 2011). In contrast to the understanding of the epidemiology of MDROs in hospitals, the prevalence of human carriage of these resistant bacteria in the community is largely unknown. Prior studies reported that the colonization rate of methicillin-resistant Staphylococcus aureus (MRSA) was 3.8% and 7.8% among Taiwanese adults and children in the community, respectively (Wang et al., 2009; Chen et al., 2011). Another study surveyed Escherichia coli from different sources in the community setting and found that the prevalence of extended-spectrum β-lactamases (ESBL)-producers increased from 4.0 to 10.7% within 8 years (Wang et al., 2015). However, at present, data on the fecal carriage rate of VRE and carbapenem-resistant Enterobacteriaceae (CRE) among healthy adults in the community are still limited. It also remains unknown whether an increasing and high prevalence of MDROs in hospitals is associated with a high fecal carriage rate of these pathogens in the community.
Resistance to intestinal colonization by MDROs relies on the undisrupted commensal gut microbiota (Buffie and Pamer, 2013). It has been shown that fecal microbiota transplantation successfully eradicates colonization with MDROs in hospitalized patients (Crum-Cianflone et al., 2015). A recent study targeting VRE found that normal gut microbiota in the colon can secrete a lantibiotic to reduce VRE colonization (Kim et al., 2019). As the interaction between commensal gut microbiota and resistant pathogens were being explored, studies focusing on fecal carriage of Gram-negative bacteria and its correlation with changes in gut microbiota remained scarce.
This study aimed to investigate the prevalence of colonization with MDROs, including MRSA, third-generation cephalosporin- or carbapenem-resistant E. coli and Klebsiella pneumoniae, and VRE, among healthy adults in the community. Furthermore, differences in the gut microbiome composition between MDRO carriers and non-carriers were analyzed.
This prospective cohort study was conducted at two regional hospitals (Hsin-Chu and Jin-Shan branches of National Taiwan University Hospital) in Northern Taiwan between March 2017 and February 2018. During the study period, healthy adults aged 20 years or older who attended mandatory health examination as a part of the workplace health promotion program at the two participating hospitals were invited to participate. Those who met any of the following criteria before formal enrollment were excluded: (1) being hospitalized in an acute care hospital within the past 90 days, (2) resident of a nursing home or long-term care facility, (3) received intravenous therapy, oral, or intravenous chemotherapy within the past 30 days, or (4) attended a hemodialysis clinic within the past 30 days. A total of 187 healthy adults were enrolled. After enrollment, all participants were requested to fill out a questionnaire and provide a fresh stool sample. In addition, the investigator and a well-trained study assistant took a nasal swab from each enrolled participant.
This study was approved by the Research Ethics Committee of National Taiwan University Hospital (registration No. NTUH-201711066RINA). All patients provided written informed consent before enrollment to provide a nasal swab, stool sample, and clinical data for research.
A standardized questionnaire was used to collect the participants’ demographic data, including age, sex, dietary habits, educational degree, economic status, number of household members, presence of any household member who was a health care worker, presence of any household member who was younger than 7 years old, presence of chronic diseases, smoking habits, hospitalizations within the previous year, history of caring for inpatients within the past year, outpatient clinic visits within the past year, use of antibiotics within the past year, parenteral drug use, dialysis treatment within the past year, history of animal contact, and travel history within the past year.
Nasal swabs were performed in both nares using the BBLTM CultureSwabTM (Becton, Dickinson and Company, Franklin Lakes, NJ, United States). After collection, each nasal swab was plated onto a blood agar plate. Isolates identified as S. aureus based on colony morphology and biochemical reactions were subcultured on CHROMID MRSA agar (BioMerieux, Marcy-l’Étoile, France) and incubated at 35°C in ambient air for 24 h to screen for MRSA. Fecal samples were collected into a clean container. A small amount of each stool sample was spread on a blood agar plate after collection, and the rest of the part was stored at −80°C as soon as possible. Cefotaxime (30 μg), ertapenem (10 μg), and imipenem (10 μg) disks were then applied to the blood agar plate and incubated at 35°C in ambient air for 24 h to screen for third-generation cephalosporin-resistant (3GC-R) Enterobacteriaceae or CRE. Isolates that grew within the inhibition zone of the cefotaxime and were identified as E. coli or K. pneumoniae by biochemical tests were subcultured on CHROMID ESBL agar (BioMerieux) to screen for ESBL production or not (Wickramasinghe et al., 2012; Ko et al., 2013; Blom et al., 2016). All 3GC-R E. coli or K. pneumoniae, including phenotypically selected ESBL and non-ESBL, isolates were further be tested genotypically by performing polymerase chain reaction (PCR) for the detection of AmpC β-lactamases or ESBL genes. Isolates that grew within the inhibition zone of the ertapenem and imipenem disks and were identified as E. coli or K. pneumoniae by biochemical tests were collected for further confirmation. A small amount of stool was also spread on CHROMID VRE agar (BioMerieux) to screen for VRE. Species identification of all presumed MRSA, third-generation cephalosporin- or carbapenem-resistant E. coli and K. pneumoniae, and VRE isolates were processed using the Vitek-2 bacterial identification system (BioMerieux).
The minimal inhibitory concentration of each isolate of S. aureus, E. coli, or K. pneumoniae to various drugs was determined by broth microdilution using Sensititre antimicrobial susceptibility testing system with customized MRSA plate format (plate code: NHRIGP9, including cefoxitin screen test and oxacillin) and commercialized ESBL plate format (plate code: ESB1F) (Trek Diagnostic Systems, East Grinstead, United Kingdom). The interpretation of the results was performed according to Clinical and Laboratory Standards Institute 2016 criteria (CLSI, 2016). The tested drugs included oxacillin, clindamycin, erythromycin, gentamicin, ciprofloxacin, tetracycline, rifampin (rifampicin), trimethoprim-sulfamethoxazole, vancomycin, teicoplanin, daptomycin, linezolid, and tigecycline for Gram-positive bacteria, and cefoxitin, cefotaxime, ceftazidime, cefepime, gentamicin, amikacin, ciprofloxacin, imipenem, and meropenem for Gram-negative bacteria. If the Gram-negative isolates were carbapenem-resistant, the susceptibility to tigecycline and colistin was further determined. For S. aureus isolates, we used oxacillin MIC to detect the oxacillin resistance. For E. coli and K. pneumoniae isolates, the growth at or above the centration of cefotaxime 1 μg/mL indicated ESBL production, and an imipenem or meropenem MIC of ≥2 μg/mL indicated carbapenemase production (CLSI, 2016).
For MRSA, multilocus sequence typing (MLST) was performed as described by Enright et al. (2000). For Enterobacteriaceae, multiplex PCR, which follows previously published protocols (Monstein et al., 2007; Queenan and Bush, 2007), was used to determine the presence of the genes encoding AmpC β-lactamases and ESBLs. Pulsed-field gel electrophoresis (PFGE) was used to determine the genetic relatedness among MRSA and 3GC-R E. coli or K. pneumoniae. For interpretation of the PFGE banding patterns, unweighted-pair group method using average linkages (UPGMA) dendrograms were constructed from the original data. Isolates that exhibited a similarity of 80% or greater of their banding patterns were considered to belong to the same cluster if more than three isolates were present.
Stool genomic DNA was extracted using QIAamp DNA stool MiniKits (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The V3-V4 region of the 16S rRNA genes was amplified using universal primers linked with indices and sequencing adaptors. The amplicons were sequenced on an Illumina MiSeq platform (San Diego, CA, United States) to obtain 300-bp paired-end reads and for taxonomic assignment. The raw sequence quality was first estimated by total read counts and Q30, and then merged with Illumina Paired-End reAd mergeR (PEAR, version 0.9.8), generating effective reads. The effective reads were then denoised by QIIME (version 1.9.1), and clustered into operational taxonomic units (OTUs) with ≥ 97% sequence homology. The SILVA rRNA database (release 132) was used for taxonomy assignment of clustered OTUs. The rarefied OTU table generated by QIIME was used for calculating alpha-diversity with the Shannon diversity index and beta-diversity with weighted and unweighted UniFrac distance matrices. The UniFrac distance matrices were then used to perform principal coordinate analysis (PCoA) to compare bacterial composition differences between sample groups. The linear discriminant analysis (LDA) effect size (LEfSe) algorithm was used to identify statistically significant bacterial taxa at different taxonomic levels for MDRO carriers and non-carriers. The threshold used to consider a discriminative feature for the logarithmic LDA score was set to 3.0.
Categorical variables were described as proportions and compared using the Chi-squared test or Fisher’s exact test if the estimated number was less than 10. The continuous variables were described as mean ± standard deviation (SD) and compared using the Mann–Whitney U test. Univariate and multivariate logistic regression models were used to assess factors associated with the carriage of drug-resistant pathogens. All p-values were two-sided, and a p-value < 0.05 was considered statistically significant. Statistical analyses were performed using SPSS software version 25.0 (SPSS Inc., Chicago, IL, United States).
One hundred and eighty-seven healthy adults from the community participated this study. Among whom, 174 provided nasal swabs and 186 stool samples were collected. The demographic data for all participants are presented in Table 1. The average age of the participants was 46 years, and 81 (43.3%) were male. Among all participants, 24.1% (45/187) were employees of technology company A; the other 75.9% were attendees of a health examination program who joined individually. The major products of the technology company A were the integrated circuit chip packages.
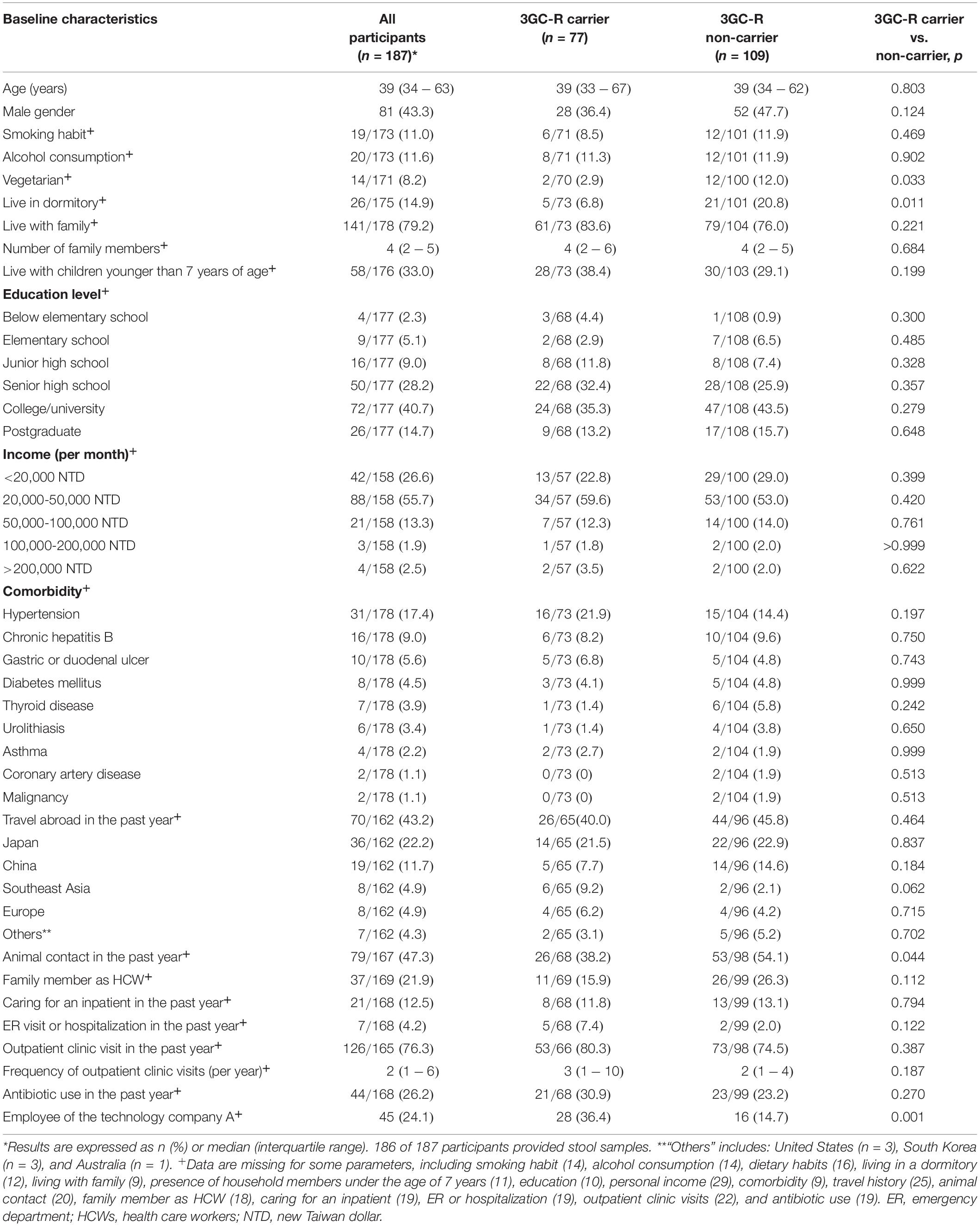
Table 1. Baseline demographics of the participants with or without fecal carriage of third-generation cephalosporin-resistant (3GC-R) E. coli or K. pneumoniae.
Figure 1 shows the results of colonization with target pathogens. Overall, the prevalences of colonization with methicillin-sensitive S. aureus, MRSA, and 3GC-R E. coli or K. pneumoniae were 20.1% (35/174), 4.6% (8/174), and 41.4% (77/186), respectively. Using ESBL screening media, 72 participants colonized with 3GC-R E. coli or K. pneumoniae showed positivity results of ESBL screening. However, based on PCR methods, 16.1% (30/186) of the participants carried AmpC β-lactamase gene-positive isolates and 27.4% (51/186) carried ESBL-positive isolates, respectively. No fecal carriage of CRE or VRE was found. A significantly higher rate of colonization with ESBL-producing E. coli or K. pneumoniae was found among employees of technology company A than among the other participants (56.8 vs. 18.3%, respectively; p < 0.001), whereas the colonization rates of MRSA and AmpC β-lactamases-producing E. coli or K. pneumoniae were similar between the two groups (4.5 vs. 4.6%, p > 0.999 and 15.9 vs.16.2%, p = 0.286, respectively).
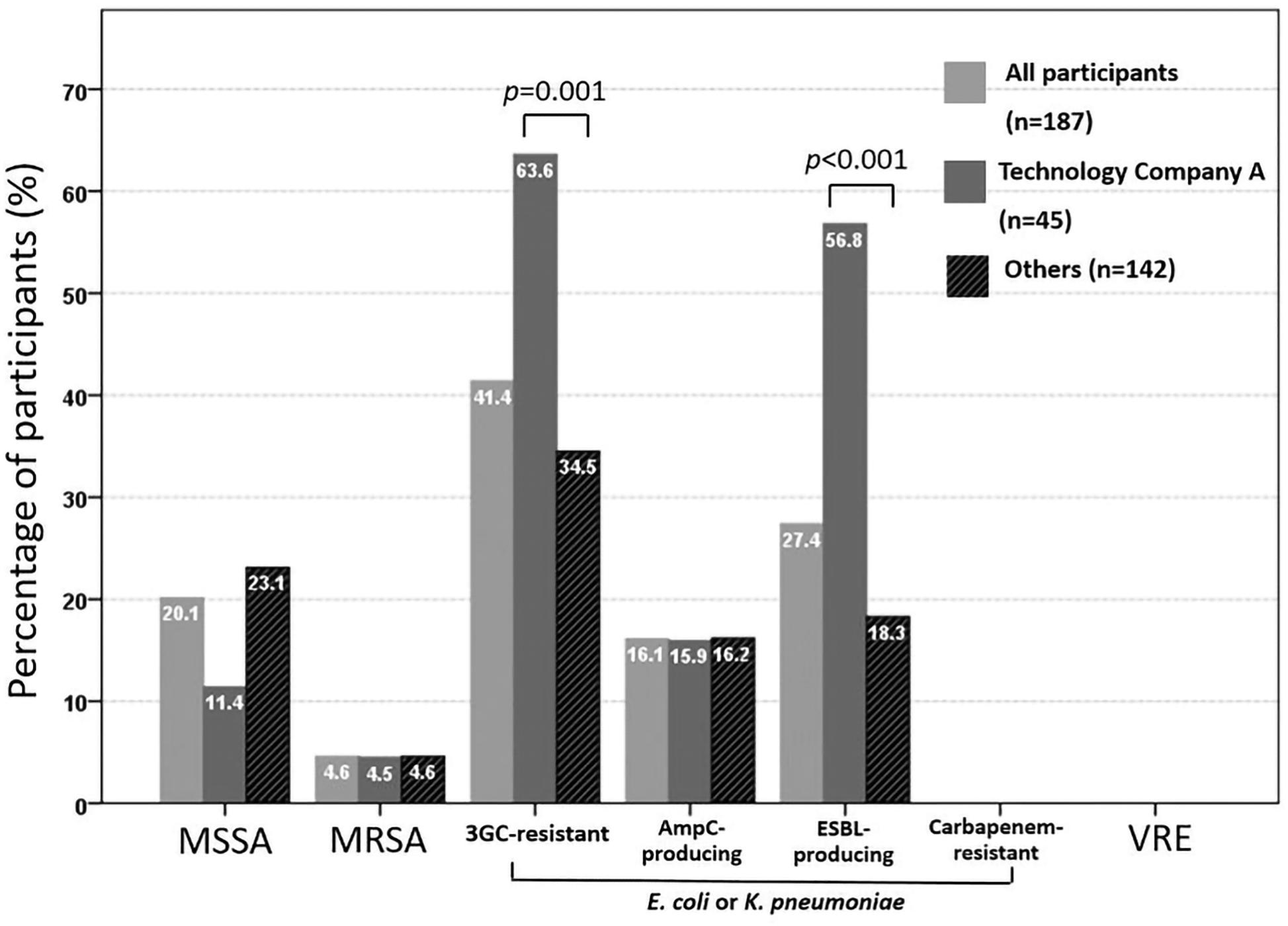
Figure 1. Rate of fecal colonization with third-generation cephalosporin (3GC)-resistant or carbapenem-resistant E. coli or K. pneumoniae and vancomycin-resistant enterococci (VRE) among 186 participants, and nasal colonization with methicillin-resistant Staphylococcus aureus (MRSA) and methicillin-sensitive Staphylococcus aureus (MSSA) among 174 participants.
Comparisons of the demographics data between participants with fecal carriage of 3GC-R E. coli or K. pneumoniae (n = 77) and those without (n = 109) are shown in Table 1. In the multivariate logistic regression, the significant risk factors for carrying 3GC-R were being an employee of technology company A [adjusted odds ratio (aOR) 4.127; 95% confidence interval (CI) 1.824–9.336; p = 0.001] and traveling to Southeast Asia in the past year (aOR 6.545, 95% CI 1.071–40.001; p = 0.042) (Table 2). In the same regression model, these two factors were also significantly associated with carrying ESBL-producing E. coli or K. pneumoniae. Among the employee of technology company A, a significant higer proportion of antibiotic use in the past year was found in 3GC-R E. coli or K. pneumoniae carriers than non-carriers [10/28 (35.7%) vs. 1/16 (6.3%), p = 0.036] (Supplementary Table 2).
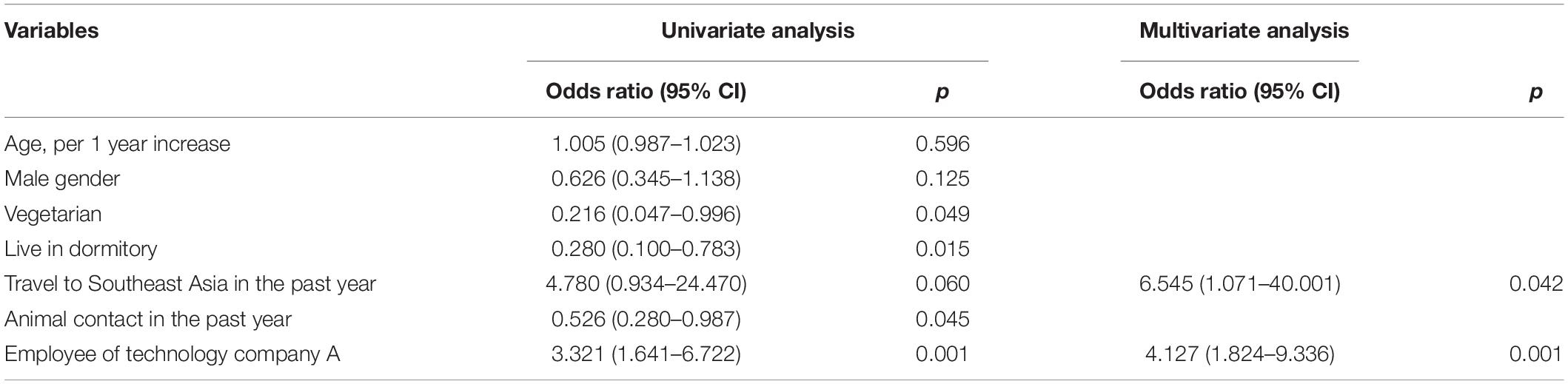
Table 2. Associated risk factors of colonization with third-generation cephalosporin-resistant E. coli or K. pneumoniae.
In total, 74 3GC-R E. coli and 15 3GC-R K. pneumoniae were isolated from the stool samples of 77 participants. The susceptibilities of these isolates to piperacillin/tazobactam, cefepime, and ciprofloxacin were 96.6, 87.6, and 70.8%, respectively (Figure 2). The antibiotic susceptibility rate of E. coli isolates from employees of technology company A was similar to that of the other participants; however, a lower susceptibility to cefepime was found among the E. coli isolated from employees of technology company A (Supplementary Figure 1).
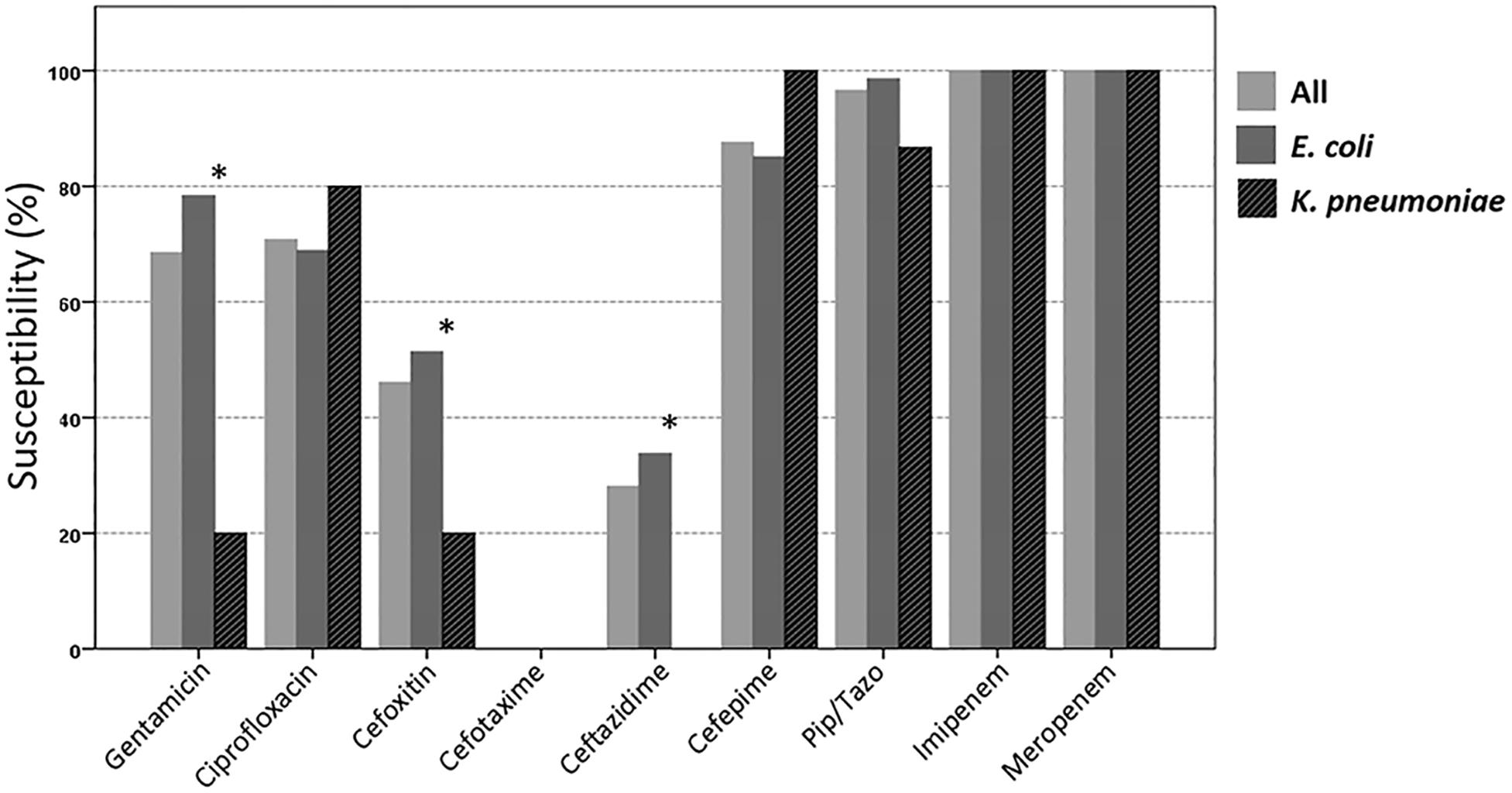
Figure 2. Susceptibilities of the 89 third-generation cephalosporin-resistant E. coli or K. pneumoniae isolates to different antimicrobial agents. ∗The susceptibility is significantly lower among K. pneumoniae than among E. coli (p < 0.05; Pip/Tazo, piperacillin/tazobactam).
Among all 3GC-R isolates, 57 (64%) carried CTX-M-type genes, 46 (52%) non-ESBL TEM genes (all were TEM-1), 32 (36%) AmpC β-lactamase genes, 14 (16%) non-ESBL SHV-type genes, and 1 (1%) OXA-10-type gene. Two E. coli isolates carried both CTX-M and AmpC genes. The prevalence of CTX-M genes was significantly higher in E. coli than in K. pneumoniae (73 vs. 20%, respectively; p < 0.001). By contrast, AmpC genes were dominant in K. pneumoniae, but not in E. coli (80 vs. 27%, respectively; p < 0.001).
For the eight MRSA isolates, MLST was performed, and five sequence types were identified: ST 59 (n = 3), ST 508 (n = 2), ST 8 (n = 1), ST 630 (n = 1), and ST 2846 (n = 1). The susceptibilities of these MRSA isolates to different antimicrobial agents are shown in Supplementary Figure 2.
Phylogenetic analysis of all 3GC-R E. coli and K. pneumoniae isolates from the participants was performed using PFGE, and the results revealed great genetic diversity (Supplementary Figures 3A,B). Because of the significantly higher rate of colonization with ESBL-producers among employees of technology company A compared with the other participants, PFGE was performed with the 27 E. coli and three K. pneumoniae isolates from employees of technology company A. Only one small cluster of three E. coli isolates sharing ≥80% similarity in the PFGE pattern was found (Supplementary Figures 4A,B). For MRSA isolates, PFGE showed diverse pulsotypes (Supplementary Figure 5).
From participants reporting no exposure to antibiotics within the past year, 20 carrying 3GC-R E. coli or K. pneumoniae (3GC-R carriers) were randomly selected and matched to 60 non-carriers by age and sex for gut microbiota analysis. The demographic data of the 20 3GC-R carriers and 60 non-carriers are presented in Supplementary Table 1.
The diversity within each group was measured by alpha-diversity using the Shannon diversity index, and no significant differences were found between 3GC-R carriers and non-carriers (p = 0.272) (Figure 3A). The diversity between the two groups (beta-diversity analysis) was compared using weighted and unweighted UniFrac analysis. The PCoA revealed that the gut microbiota of 3GC-R carriers was distinct from those of non-carriers using the unweighted UniFrac distance (p = 0.020), whereas the weighted UniFrac analysis was unable to differentiate carriers from non-carriers (p = 0.055) (Figure 3B).
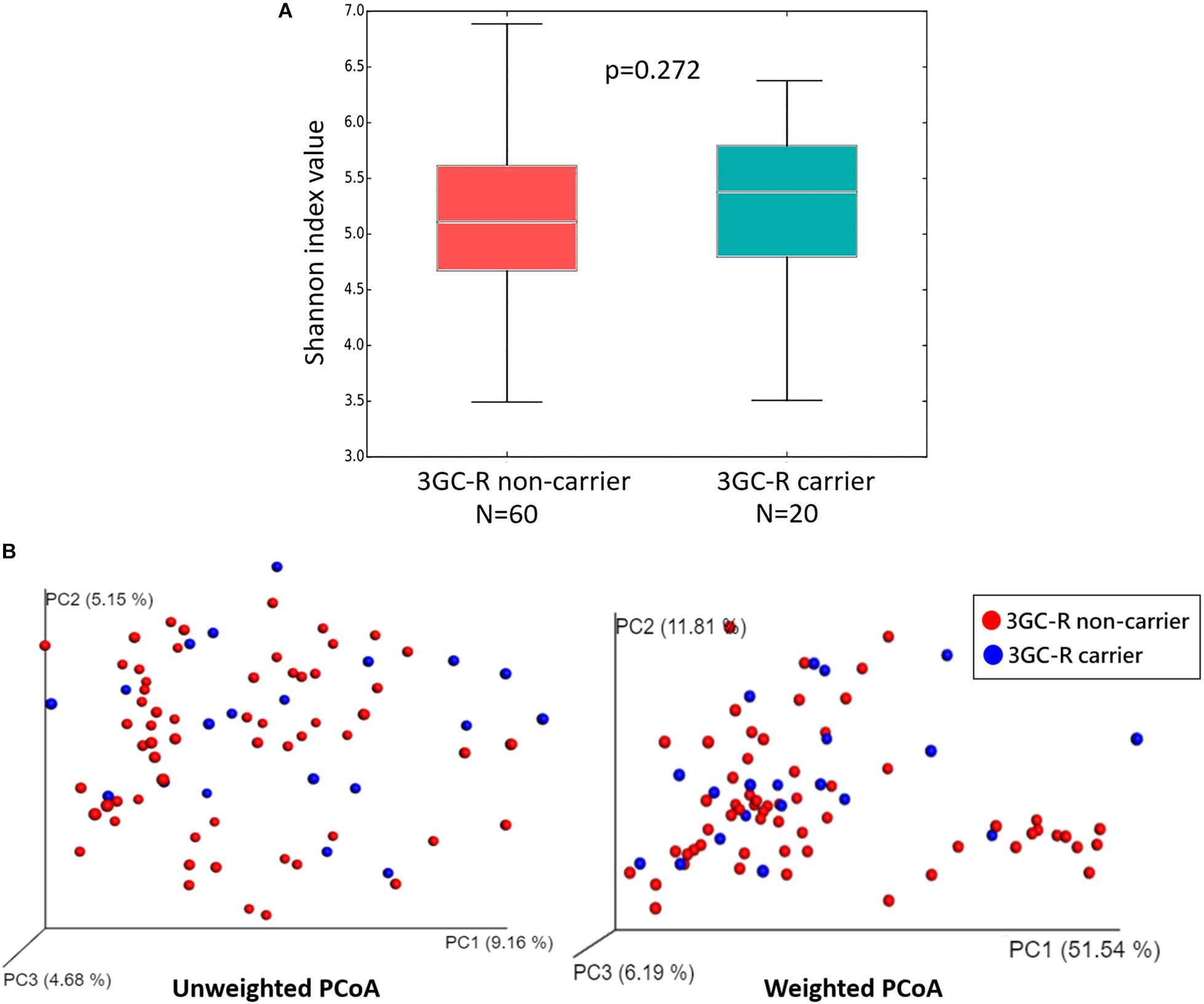
Figure 3. (A) Box plot of the alpha-diversity index measured based on the Shannon diversity index for third-generation cephalosporin-resistant (3GC-R) E. coli or K. pneumoniae carriers (blue) and non-carriers (red). Statistical testing showed no differences for the Shannon diversity index (p = 0.272). (B) PCoA plots of bacterial beta-diversity for 3GC-R carriers (blue) and non-carriers (red) based on the unweighted and weighted UniFrac distance.
The taxa that differ significantly in abundance between 3GC-R carriers and non-carriers were discriminated by the LEfSe algorithm. The results of the LEfSe analysis revealed that the phylum Proteobacteria, as well as the family Enterobacteriaceae and the species E. coli, K. pneumoniae, and Klebsiella aerogenes were significantly more abundant in 3GC-R carriers than in non-carriers. Bacteroides vulgatus was also more abundant in 3GC-R carriers, while non-carriers had a higher abundance of Prevotella massiliensis and the family Pseudomonadaceae (Figures 4A,B).
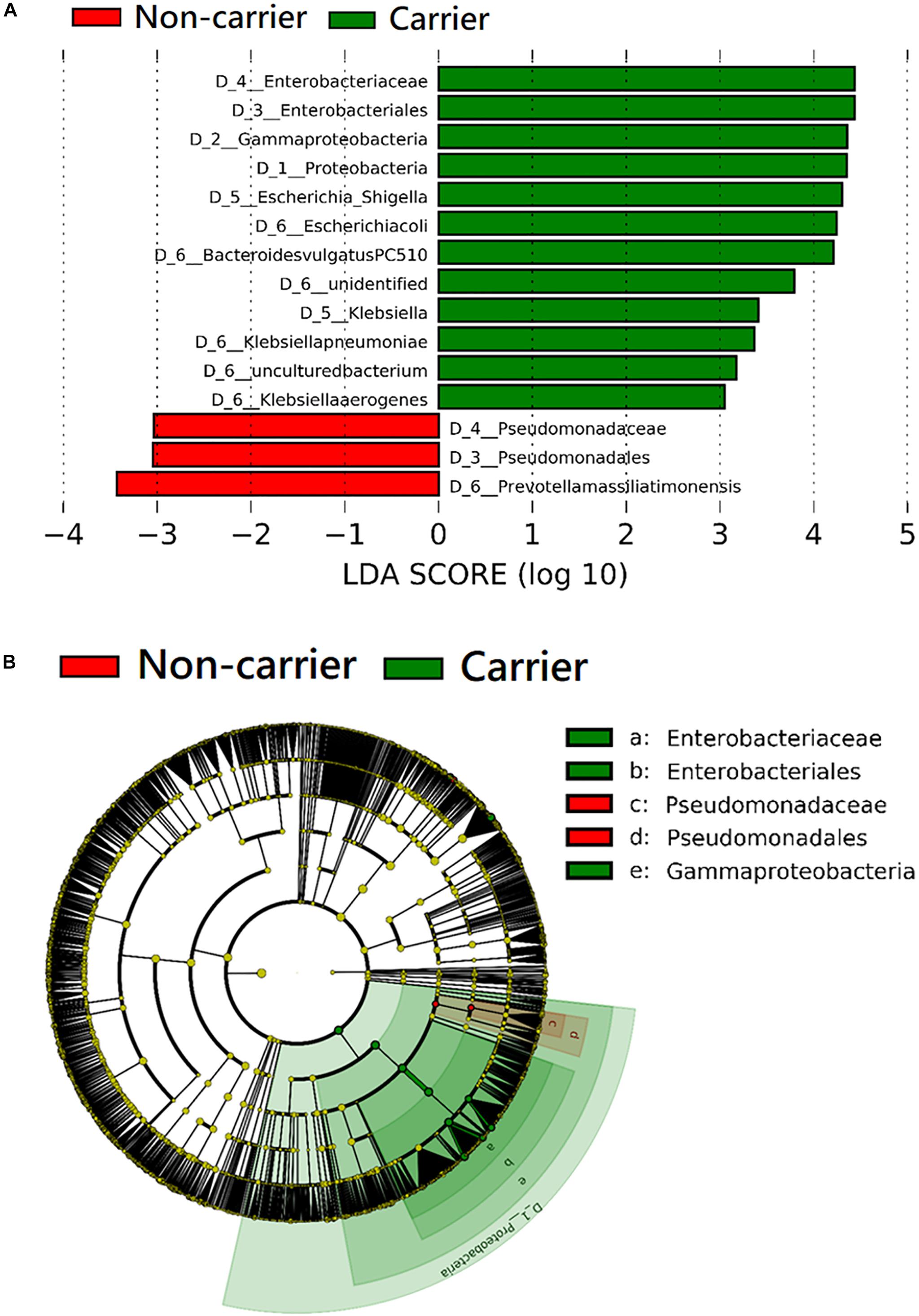
Figure 4. Comparison of the composition of fecal microbiota between third-generation cephalosporin-resistant E. coli or K. pneumoniae carriers (n = 20) and non-carriers (n = 60) by linear discriminant analysis (LDA) effect size (LEfSe). (A) Histogram of the LDA scores shows the most differentially abundant taxa in the two groups. (B) The cladogram shows the significantly overrepresented bacterial taxa in each group.
In this cohort of healthy adults in the community, the rate of colonization with 3GC-R Enterobacteriaceae was 41.4%. No CRE or VRE colonization was noted, and the nasal carriage rate of MRSA was 4.6%, which remained stationary compared with previous reports in Taiwan. The gut microbiota analysis revealed an increased Enterobacteriaceae abundance in 3GC-R E. coli or K. pneumoniae carriers when compared with non-carriers.
In this study, 27.4% of healthy adults carried ESBL-producing E. coli or K. pneumoniae. The results are concordant with the high prevalence of ESBL-producing Enterobacteriaceae colonization reported among healthy individuals in Southeast Asia (Karanika et al., 2016; Piewngam et al., 2019). However, a study conducted in Southern Taiwan reported a low (1.9%) prevalence of ESBL-producing E. coli fecal carriage among healthy adults (Wu et al., 2019). This discrepancy might result from differences in local epidemiology, the proportion of participants with exposure to antibiotics, and the fact that K. pneumoniae was also included in our study. In Taiwan, a longitudinal study investigating 3481 E. coli from outpatients and patients visiting emergency rooms reported that non-susceptibility to cefotaxime reached 21.1% in 2012 (Wang et al., 2015). Another single-center study reported that 19.7% of E. coli isolates causing community-onset bacteremia were 3GC-R (Lin et al., 2019). Accordingly, a high prevalence of colonization with 3GC-R Enterobacteriaceae in the community is expected, and more extensive surveys are needed to understand the burden across Taiwan.
Several factors have been linked to increased fecal carriage of 3GC-R or ESBL-producing Enterobacteriaceae, including antibiotic use, international travel, hospitalization, dietary habits, and animal contact (Meyer et al., 2012; Leistner et al., 2013; Hamprecht et al., 2016; Otter et al., 2019). In our analysis, traveling to Southeast Asia in the past year and working at a specific company were associated with colonization by 3GC-R and ESBL-producing Enterobacteriaceae. The subgroup analysis of employees of technology company A showed that 3GC-R carriers were more likely to have antibiotics exposure in the past year. In addition, we found a higher carriage rate of ESBL-producers, but not AmpC-producers, among employees of technology company A, which indicated that the high rate of 3GC-R colonization in the community might be attributed to the spread of ESBL-producers in particular populations.
The VRE colonization rate of healthy adults is inconsistent in the literature, ranging from 0 to 21% (Balzereit-Scheuerlein and Stephan, 2001; Kolar et al., 2006; Hannaoui et al., 2016; Decker et al., 2018). Decker et al. (2018) examined 800 health care personnel and found no VRE colonization. Studies investigating CRE colonization among healthy adults in the community were scarce. One study from India reported no urine colonization of imipenem- or meropenem-resistant Enterobacteriaceae in 433 healthy individuals (Lohiya et al., 2015). In our study, we did not find VRE or CRE colonization among healthy adults, despite the increased prevalence of both pathogens in hospitals in Taiwan (Wang et al., 2013; Jean et al., 2018). Our results suggest that in the community, the spread of VRE or CRE occurs mainly in health care facilities such as nursing homes (Lee et al., 2017).
How commensal gut microbiota react with colonization by specific pathogens remains unclear. To date, only two studies in rural areas have investigated the composition of gut microbiota in participants with or without colonization by ESBL-producing Enterobacteriaceae (Gosalbes et al., 2015; Piewngam et al., 2019), and both studies included fewer cases than did our study. We showed that the gut microbiota of 3GC-R Enterobacteriaceae carriers was characterized by an increased relative abundance of Proteobacteria and Enterobacteriaceae; similar results had been described in CRE-carriers (Korach-Rechtman et al., 2019). The expansion of Enterobacteriaceae in gut microbiota is regarded as a signature of dysbiosis, which could be induced by intestinal inflammation or treatment with antibiotics (Shin et al., 2015; Litvak et al., 2017). In patients undergoing liver transplantation, the pre-transplant dysbiosis of gut microbiota has been shown to increase the likelihood of subsequent colonization by MDROs (Annavajhala et al., 2019). Our findings imply that dysbiosis gut microbiota in healthy adults is associated with colonization by drug-resistant Enterobacteriaceae. In humans, the domination of Proteobacteria or a high relative abundance of carbapenemase-producing K. pneumoniae in the gut microbiota has been found to increase the risk of subsequent Gram-negative rod bacteremia (Taur et al., 2012; Shimasaki et al., 2019). However, whether an increased level of Enterobacteriaceae in the gut microbiota would persist through the progression from asymptomatic colonization to invasive infection of drug-resistant Enterobacteriaceae remains unclear.
This study has several limitations. First, the number of cases was small and we included only E. coli and K. pneumoniae. The prevalence of colonization with other ESBL-producing Enterobacteriaceae was not evaluated. Second, the demographic data collected by the questionnaire might be imprecise and lead to recall bias. Third, the type of antibiotics taken by the participants was not documented, so we were unable to evaluate the impact of specific classes of antibiotics on the colonization of MDROs. Fourth, the number of participants from technology company A was small, which limited further statistical analysis for associated risk factors of MDRO colonization among these individuals. Lastly, the database we used in the gut microbiome analysis was unable to identify the antibiotic susceptibility profiles of each species, thus we could not know whether the increased abundance of E. coli in 3GC-R carriers was caused by the increase in resistant strains. It was still unknown whether subjects with MDRO fecal colonization would have an increased abundance of MDROs in their gut microbiota.
In conclusion, our study shows a high prevalence of fecal carriage of 3GC-R E. coli or K. pneumoniae among healthy adults in the community, which should be considered in the empiric management of community-associated infection when involvement of Enterobacteriaceae is suspected. Dysbiosis of the gut microbiota presented as the expansion of Enterobacteriaceae was found in carriers of 3GC-R Enterobacteriaceae. Further studies are needed to investigate the causal relationship between changes in the gut microbiota and fecal colonization with antibiotic-resistant Enterobacteriaceae.
The datasets generated for this study can be found in the Sequence Read Archive (SRA) of NCBI with the following link: https://www.ncbi.nlm.nih.gov/sra/PRJN628533.
The studies involving human participants were reviewed and approved by the Research Ethics Committee of National Taiwan University Hospital. The patients/participants provided their written informed consent to participate in this study.
Y-SH and J-TW: conceived and designed the analysis. Y-SH, K-YL, Y-HC, H-CC, and S-SW: acquisition of the data. Y-SH, Y-AC, L-CL, and J-TW: analysis and interpretation of the data. Y-SH: drafted the manuscript. J-TW: critical revision. S-CC: supervised the work. All authors contributed to the article and approved the submitted version.
The study was supported by the National Taiwan University Hospital Hsinchu Branch under Grant 106-HCH016 and Ministry of Science and Technology of Taiwan under Grant MOST 106-2314-B-002-228. The sponsors had no role in the study design, data collection and analysis, manuscript preparation, or the decision to submit for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to acknowledge the services provided by the Medical Microbiota Center of the First Core Laboratory, National Taiwan University College of Medicine.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01402/full#supplementary-material
Annavajhala, M. K., Gomez-Simmonds, A., Macesic, N., Sullivan, S. B., Kress, A., Khan, S. D., et al. (2019). Colonizing multidrug-resistant bacteria and the longitudinal evolution of the intestinal microbiome after liver transplantation. Nat. Commun. 10:4715. doi: 10.1038/s41467-019-12633-4
Balzereit-Scheuerlein, F., and Stephan, R. (2001). Prevalence of colonisation and resistance patterns of vancomycin-resistant enterococci in healthy, non-hospitalised persons in Switzerland. Swiss Med. Wkly. 131, 280–282.
Blom, A., Ahl, J., Mansson, F., Resman, F., and Tham, J. (2016). The prevalence of ESBL-producing Enterobacteriaceae in a nursing home setting compared with elderly living at home: a cross-sectional comparison. BMC Infect. Dis. 16:111. doi: 10.1186/s12879-016-1430-5
Buffie, C. G., and Pamer, E. G. (2013). Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol. 13, 790–801. doi: 10.1038/nri3535
Chen, C. J., Hsu, K. H., Lin, T. Y., Hwang, K. P., Chen, P. Y., and Huang, Y. C. (2011). Factors associated with nasal colonization of methicillin-resistant Staphylococcus aureus among healthy children in Taiwan. J. Clin. Microbiol. 49, 131–137. doi: 10.1128/JCM.01774-10
CLSI (2016). Performance Standards for Antimicrobial Susceptibility Testing. 26th Informational Supplement M100-S26. Wayne, PA: Clinical and Laboratory Standards Institute (CLSI).
Crum-Cianflone, N. F., Sullivan, E., and Ballon-Landa, G. (2015). Fecal microbiota transplantation and successful resolution of multidrug-resistant-organism colonization. J. Clin. Microbiol. 53, 1986–1989. doi: 10.1128/JCM.00820-15
Decker, B. K., Lau, A. F., Dekker, J. P., Spalding, C. D., Sinaii, N., Conlan, S., et al. (2018). Healthcare personnel intestinal colonization with multidrug-resistant organisms. Clin. Microbiol. Infect. 24, 82.e81–82.e84. doi: 10.1016/j.cmi.2017.05.010
Enright, M. C., Day, N. P., Davies, C. E., Peacock, S. J., and Spratt, B. G. (2000). Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38, 1008–1015.
Founou, R. C., Founou, L. L., and Essack, S. Y. (2017). Clinical and economic impact of antibiotic resistance in developing countries: a systematic review and meta-analysis. PLoS One 12:e0189621. doi: 10.1371/journal.pone.0189621
Gosalbes, M. J., Vázquez-Castellanos, J. F., Angebault, C., Woerther, P.-L., Ruppé, E., Ferrús, M. L., et al. (2015). Carriage of enterobacteria producing extended-spectrum β-lactamases and composition of the gut microbiota in an Amerindian community. Antimicrob. Agents Chemother. 60, 507–514. doi: 10.1128/AAC.01528-15
Hamprecht, A., Rohde, A. M., Behnke, M., Feihl, S., Gastmeier, P., Gebhardt, F., et al. (2016). Colonization with third-generation cephalosporin-resistant Enterobacteriaceae on hospital admission: prevalence and risk factors. J. Antimicrob. Chemother. 71, 2957–2963. doi: 10.1093/jac/dkw216
Hannaoui, I., Barguigua, A., Serray, B., El Mdaghri, N., Timinouni, M., Ait Chaoui, A., et al. (2016). Intestinal carriage of vancomycin-resistant enterococci in a community setting in Casablanca, Morocco. J. Glob. Antimicrob. Resist. 6, 84–87. doi: 10.1016/j.jgar.2016.03.008
Heuer, O. E., Kruse, H., Grave, K., Collignon, P., Karunasagar, I., and Angulo, F. J. (2009). Human health consequences of use of antimicrobial agents in aquaculture. Clin. Infect. Dis. 49, 1248–1253. doi: 10.1086/605667
Jean, S.-S., Lee, N.-Y., Tang, H.-J., Lu, M.-C., Ko, W.-C., and Hsueh, P.-R. (2018). Carbapenem-resistant Enterobacteriaceae infections: Taiwan aspects. Front. Microbiol. 9:2888. doi: 10.3389/fmicb.2018.02888
Karanika, S., Karantanos, T., Arvanitis, M., Grigoras, C., and Mylonakis, E. (2016). Fecal Colonization with extended-spectrum beta-lactamase-producing Enterobacteriaceae and risk factors among healthy individuals: a systematic review and metaanalysis. Clin. Infect. Dis. 63, 310–318. doi: 10.1093/cid/ciw283
Kim, S. G., Becattini, S., Moody, T. U., Shliaha, P. V., Littmann, E. R., Seok, R., et al. (2019). Microbiota-derived lantibiotic restores resistance against vancomycin-resistant Enterococcus. Nature 572, 665–669. doi: 10.1038/s41586-019-1501-z
Ko, Y. J., Moon, H. W., Hur, M., Park, C. M., Cho, S. E., and Yun, Y. M. (2013). Fecal carriage of extended-spectrum beta-lactamase-producing Enterobacteriaceae in Korean community and hospital settings. Infection 41, 9–13. doi: 10.1007/s15010-012-0272-3
Kolar, M., Pantucek, R., Vagnerova, I., Sauer, P., Kesselova, M., Cekanova, L., et al. (2006). Prevalence of vancomycin-resistant enterococci in hospitalized patients and those living in the community in the Czech Republic. New Microbiol. 29, 121–125.
Korach-Rechtman, H., Hreish, M., Fried, C., Gerassy-Vainberg, S., Azzam, Z. S., Kashi, Y., et al. (2019). Intestinal dysbiosis in carbapenem-resistant Enterobacteriaceae carriers. bioRxiv [Preprint]. doi: 10.1101/855718
Lai, C. C., Lee, K., Xiao, Y., Ahmad, N., Veeraraghavan, B., Thamlikitkul, V., et al. (2014). High burden of antimicrobial drug resistance in Asia. J. Glob. Antimicrob. Resist. 2, 141–147. doi: 10.1016/j.jgar.2014.02.007
Lee, C. M., Lai, C. C., Chiang, H. T., Lu, M. C., Wang, L. F., Tsai, T. L., et al. (2017). Presence of multidrug-resistant organisms in the residents and environments of long-term care facilities in Taiwan. J. Microbiol. Immunol. Infect. 50, 133–144. doi: 10.1016/j.jmii.2016.12.001
Leistner, R., Meyer, E., Gastmeier, P., Pfeifer, Y., Eller, C., Dem, P., et al. (2013). Risk factors associated with the community-acquired colonization of extended-spectrum beta-lactamase (ESBL) positive Escherichia Coli. an exploratory case-control study. PLoS One 8:e74323. doi: 10.1371/journal.pone.0074323
Lin, W. P., Huang, Y. S., Wang, J. T., Chen, Y. C., and Chang, S. C. (2019). Prevalence of and risk factor for community-onset third-generation cephalosporin-resistant Escherichia coli bacteremia at a medical center in Taiwan. BMC Infect. Dis. 19:245. doi: 10.1186/s12879-019-3880-z
Litvak, Y., Byndloss, M. X., Tsolis, R. M., and Bäumler, A. J. (2017). Dysbiotic Proteobacteria expansion: a microbial signature of epithelial dysfunction. Curr. Opin. Microbiol. 39, 1–6. doi: 10.1016/j.mib.2017.07.003
Lohiya, A., Kant, S., Kapil, A., Gupta, S. K., Misra, P., and Rai, S. K. (2015). Pattern of antibiotic resistance among community derived isolates of Enterobacteriaceae using urine sample: a study from Northern India. J. Clin. Diagn. Res. 9, Lc15–Lc19. doi: 10.7860/jcdr/2015/14230.6254
Meyer, E., Gastmeier, P., Kola, A., and Schwab, F. (2012). Pet animals and foreign travel are risk factors for colonisation with extended-spectrum β-lactamase-producing Escherichia coli. Infection 40, 685–687. doi: 10.1007/s15010-012-0324-8
Monstein, H. J., Ostholm-Balkhed, A., Nilsson, M. V., Nilsson, M., Dornbusch, K., and Nilsson, L. E. (2007). Multiplex PCR amplification assay for the detection of blaSHV, blaTEM and blaCTX-M genes in Enterobacteriaceae. Acta Pathol. Microbiol. Immunol. Scand. 115, 1400–1408. doi: 10.1111/j.1600-0463.2007.00722.x
Otter, J. A., Natale, A., Batra, R., Tosas Auguet, O., Dyakova, E., Goldenberg, S. D., et al. (2019). Individual- and community-level risk factors for ESBL Enterobacteriaceae colonization identified by universal admission screening in London. Clin. Microbiol. Infect. 25, 1259–1265. doi: 10.1016/j.cmi.2019.02.026
Piewngam, P., Quinones, M., Thirakittiwatthana, W., Yungyuen, T., Otto, M., and Kiratisin, P. (2019). Composition of the intestinal microbiota in extended-spectrum beta-lactamase-producing Enterobacteriaceae carriers and non-carriers in Thailand. Int. J. Antimicrob. Agents 53, 435–441. doi: 10.1016/j.ijantimicag.2018.12.006
Queenan, A. M., and Bush, K. (2007). Carbapenemases: the versatile beta-lactamases. Clin. Microbiol. Rev. 20, 440–458. doi: 10.1128/cmr.00001-07
Shimasaki, T., Seekatz, A., Bassis, C., Rhee, Y., Yelin, R. D., Fogg, L., et al. (2019). Increased relative abundance of Klebsiella pneumoniae carbapenemase-producing Klebsiella pneumoniae within the gut microbiota is associated with risk of bloodstream infection in long-term acute care hospital patients. Clin. Infect. Dis. 68, 2053–2059. doi: 10.1093/cid/ciy796
Shin, N.-R., Whon, T. W., and Bae, J.-W. (2015). Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 33, 496–503. doi: 10.1016/j.tibtech.2015.06.011
Spellberg, B., Guidos, R., Gilbert, D., Bradley, J., Boucher, H. W., Scheld, W. M., et al. (2008). The epidemic of antibiotic-resistant infections: a call to action for the medical community from the infectious diseases society of America. Clin. Infect. Dis. 46, 155–164. doi: 10.1086/524891
Taur, Y., Xavier, J. B., Lipuma, L., Ubeda, C., Goldberg, J., Gobourne, A., et al. (2012). Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin. Infect. Dis. 55, 905–914. doi: 10.1093/cid/cis580
Tseng, S. H., Lee, C. M., Lin, T. Y., Chang, S. C., and Chang, F. Y. (2011). Emergence and spread of multi-drug resistant organisms: think globally and act locally. J. Microbiol. Immunol. Infect. 44, 157–165. doi: 10.1016/j.jmii.2011.03.001
Wang, J. T., Chang, S. C., Chang, F. Y., Fung, C. P., Chuang, Y. C., Chen, Y. S., et al. (2015). Antimicrobial non-susceptibility of Escherichia coli from outpatients and patients visiting emergency rooms in Taiwan. PLoS One 10:e0144103. doi: 10.1371/journal.pone.0144103
Wang, J.-T., Chang, S.-C., Wang, H.-Y., Chen, P.-C., Shiau, Y.-R., Lauderdale, T.-L., et al. (2013). High rates of multidrug resistance in Enterococcus faecalis and E. faecium isolated from inpatients and outpatients in Taiwan. Diagn. Microbiol. Infect. Dis. 75, 406–411. doi: 10.1016/j.diagmicrobio.2013.01.004
Wang, J. T., Liao, C. H., Fang, C. T., Chie, W. C., Lai, M. S., Lauderdale, T. L., et al. (2009). Prevalence of and risk factors for colonization by methicillin-resistant Staphylococcus aureus among adults in community settings in Taiwan. J. Clin. Microbiol. 47, 2957–2963. doi: 10.1128/JCM.00853-09
Wickramasinghe, N. H., Xu, L., Eustace, A., Shabir, S., Saluja, T., and Hawkey, P. M. (2012). High community faecal carriage rates of CTX-M ESBL-producing Escherichia coli in a specific population group in Birmingham, UK. J. Antimicrob. Chemother. 67, 1108–1113. doi: 10.1093/jac/dks018
Wu, P. C., Wang, J. L., Hsueh, P. R., Lin, P. H., Cheng, M. F., Huang, I. F., et al. (2019). Prevalence and risk factors for colonization by extended-spectrum beta-lactamase-producing or ST 131 Escherichia coli among asymptomatic adults in community settings in Southern Taiwan. Infect. Drug Resist. 12, 1063–1071. doi: 10.2147/idr.S201086
Keywords: multidrug-resistant organisms, extended-spectrum β-lactamases, colonization, community, gut microbiota, third generation cephalosporin
Citation: Huang Y-S, Lai L-C, Chen Y-A, Lin K-Y, Chou Y-H, Chen H-C, Wang S-S, Wang J-T and Chang S-C (2020) Colonization With Multidrug-Resistant Organisms Among Healthy Adults in the Community Setting: Prevalence, Risk Factors, and Composition of Gut Microbiome. Front. Microbiol. 11:1402. doi: 10.3389/fmicb.2020.01402
Received: 27 February 2020; Accepted: 29 May 2020;
Published: 24 June 2020.
Edited by:
Daniel Pletzer, University of Otago, New ZealandReviewed by:
Sabrina John Moyo, University of Bergen, NorwayCopyright © 2020 Huang, Lai, Chen, Lin, Chou, Chen, Wang, Wang and Chang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jann-Tay Wang, d2FuZy5qdDE5NjhAZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.