
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CORRECTION article
Front. Microbiol. , 29 June 2020
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.01323
This article is a correction to:
Characterization of Milkisin, a Novel Lipopeptide With Antimicrobial Properties Produced By Pseudomonas sp. UCMA 17988 Isolated From Bovine Raw Milk
 Margot Schlusselhuber1*
Margot Schlusselhuber1* Justine Godard1
Justine Godard1 Muriel Sebban2
Muriel Sebban2 Benoit Bernay3
Benoit Bernay3 David Garon1
David Garon1 Virginie Seguin1
Virginie Seguin1 Hassan Oulyadi2
Hassan Oulyadi2 Nathalie Desmasures1
Nathalie Desmasures1A Corrigendum on
Characterization of Milkisin, a Novel Lipopeptide With Antimicrobial Properties Produced By Pseudomonas sp. UCMA 17988 Isolated From Bovine Raw Milk
by Schlusselhuber, M., Godard, J., Sebban, M., Bernay, B., Garon, D., Seguin, V., et al. (2018). Front. Microbiol. 9:1030. doi: 10.3389/fmicb.2018.01030
In the original article, there was a mistake in Figure 4, Figure 5, Figure 6, Figure 8, and Supplementary Table S1 as published. Indeed, the quasi–identical positions of different amino acids signals in NMR spectra (Murphy's Law), lead to a mis-interpretation of the sequence of amino acids of milkisin that has to be corrected. Thus, NMR spectra figures and Supplementary Table S1 were corrected as well as Figures 4 and 8. The corrected figures and Supplementary Table S1 appear below.
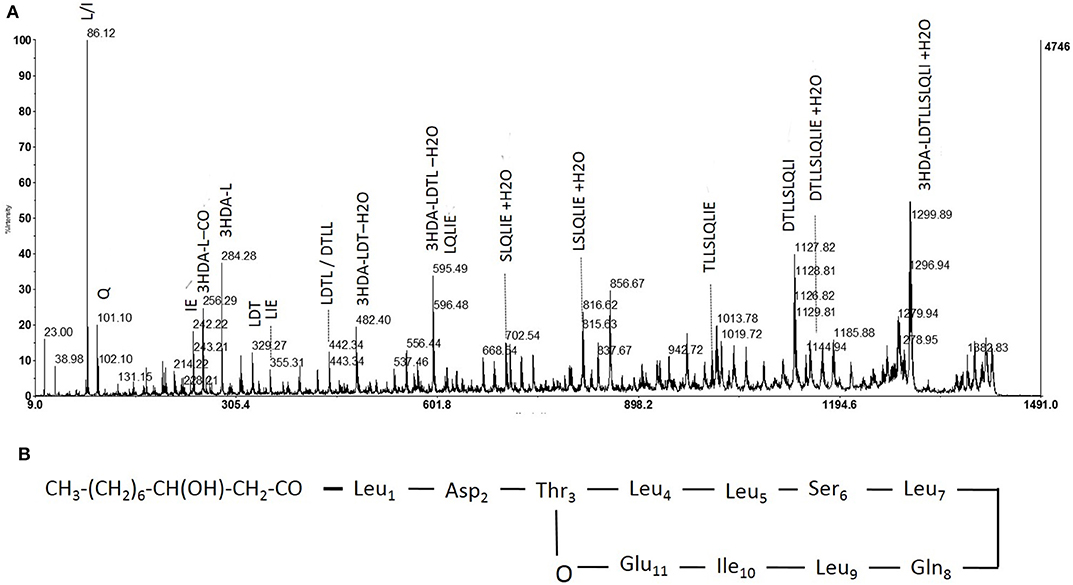
Figure 4. Fragmentation pattern of lipopeptide m/z 1409 and proposed structure. (A) Product ions obtained by fragmentation using MALDI-TOF. (B) Determined structure of purified lipopeptide (m/z 1409) based on mass spectrometric and NMR analysis.
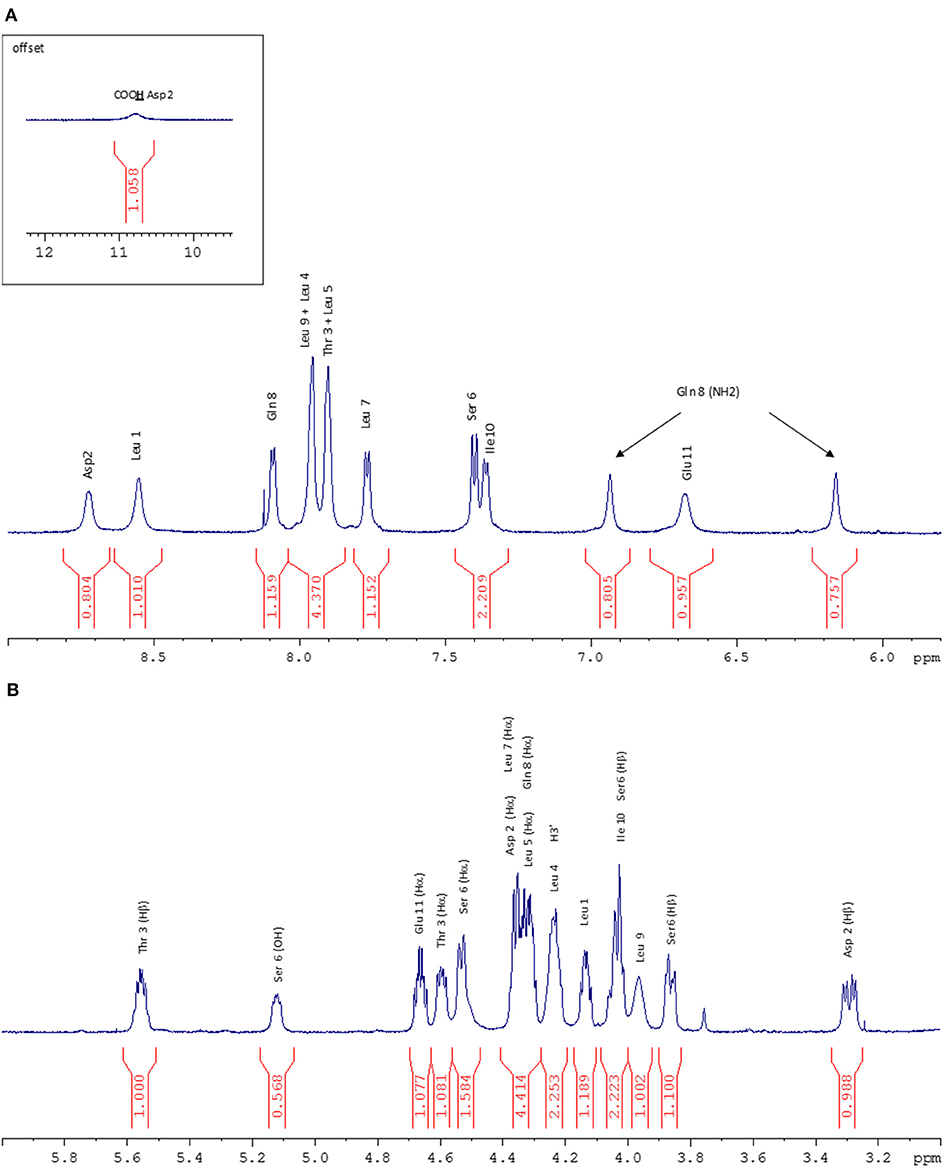
Figure 5. (A) NH zone of the 1H NMR spectrum (600 MHz, 298 K, Acetone-d6). (B) Hα zone of the 1H NMR spectrum (600 MHz, 298 K, Acetone-d6).
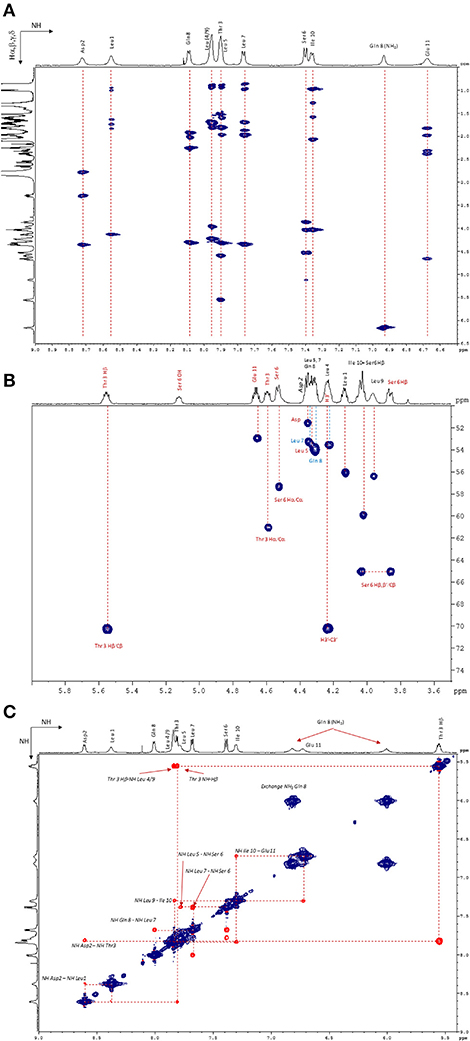
Figure 6. (A) NH/Hα, β, and γ zone of the 1H−1H TOCSY NMR spectrum (600 MHz, 298 K, Acetone-d6). (B) Hα/Cα zone of the 1H–13C HSQC NMR spectrum (600 MHz, 298 K, Acetone-d6). (C) NH/NH zone of the 1H–1H NOESY NMR spectrum (600 MHz, 323 K, Acetone-d6).
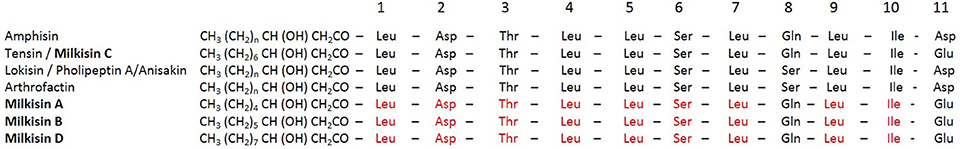
Figure 8. Alignment of lipopeptide milkisin isoforms produced by Pseudomonas sp. UCMA 17988 with members of amphisin group.
In the original article, there was error. As previously mentioned, the quasi–identical positions of different amino acids signals in NMR spectra (Murphy's Law), lead to a mis-interpretation of the sequence of amino acids of milkisin that has to be corrected. The corrections have been made to the Abstract and the Results section, Extraction and Structural Analysis of Biosurfactants sub-section respectively:
“Biosurfactants such as lipopeptides are amphiphilic compounds produced by microorganisms such as bacteria of the genera of Pseudomonas and Bacillus. Some of these molecules proved to have interesting antimicrobial, antiviral, insecticide and/or tensio-active properties that are potentially useful for the agricultural, chemical, food, and pharmaceutical industries. Raw milk provides a physicochemical environment that is favorable to the multiplication of a broad spectrum of microorganisms. Among them, psychrotrophic bacterial species, especially members of the genus Pseudomonas, are predominant and colonize milk during cold storage and/or processing. We isolated the strain Pseudomonas sp. UCMA 17988 from raw cow milk, with antagonistic activity against Listeria monocytogenes, Staphylococcus aureus, and Salmonella enterica Newport. Antimicrobial molecules involved in the antagonistic activity of this strain were characterized. A mass spectrometry analysis highlighted the presence of four lipopeptides isoforms. The major isoform (1409 m/z), composed of 10 carbons in the lipidic chain, was named milkisin C. The three other isoforms detected at 1381, 1395, and 1423 m/z, that are concomitantly produced, were named milkisin A, B and D, respectively. The structure of milkisin, as confirmed by NMR analyses, is closely related to amphisin family. Indeed, the peptidic chain was composed of 11 amino acids, 9 of which are conserved among the family. In conclusion, Pseudomonas sp. UCMA 17988 produces new members of the amphisin family which are responsible for the antagonistic activity of this strain.”
“Hereafter, collected fractions were analyzed by MALDI-TOF mass spectrometry. The spectra also highlighted the presence of four molecules (Figure 4). Two intense pseudomolecular ions [M+H]+ at m/z 1409.49 and 1395.48 were observed. Corresponding sodium and potassium adducts were also found at m/z [M+Na]+ at 1417 and 1431.60 and m/z [M+K]+ at 1433.57 and 1447.57 respectively. The two other molecules could be detected through their potassium and/or sodium adducts (m/z [M+Na]+ at 1403; m/z [M+K]+ at 1419.57 and 1461.58). The presence of four molecules harboring a difference of 14 Da was consistent with the presence of lipopeptides isoforms. Indeed, such difference is typical of the addition or substitution of a methyl group in the fatty acid chain. A MS/MS analysis was performed on the dominant [M+H]+ ion (m/z at 1409.49). The fragmentation spectrum obtained is shown in Figure 5. Fragments were assigned by subtraction of the different peaks between them. The ions with 284 Da and 86 Da masses were observed and respectively correspond to fragment 3-hydroxy-fatty acid-Leu/Ile and immonium ion of leucine or isoleucine. The following linear sequence was proposed: 3HDA-Leu/Ile1-Asp2-Thr3-Leu/Ile4-Leu/Ile5-Ser6-Leu/Ile7-Gln8-Leu/Ile9-Leu/Ile10-Glu11 with cyclisation between Thr3 and Glu11.”
The authors apologize for these errors and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
Keywords: antimicrobial activity, Pseudomonas, milkisin, amphisin, lipopeptide
Citation: Schlusselhuber M, Godard J, Sebban M, Bernay B, Garon D, Seguin V, Oulyadi H and Desmasures N (2020) Corrigendum: Characterization of Milkisin, a Novel Lipopeptide With Antimicrobial Properties Produced By Pseudomonas sp. UCMA 17988 Isolated From Bovine Raw Milk. Front. Microbiol. 11:1323. doi: 10.3389/fmicb.2020.01323
Received: 11 March 2020; Accepted: 25 May 2020;
Published: 29 June 2020.
Edited and reviewed by: Jørgen Johannes Leisner, University of Copenhagen, Denmark
Copyright © 2020 Schlusselhuber, Godard, Sebban, Bernay, Garon, Seguin, Oulyadi and Desmasures. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Margot Schlusselhuber, bWFyZ290LnNjaGx1c3NlbGh1YmVyQHVuaWNhZW4uZnI=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.