- 1Shandong Province Key Laboratory of Agricultural Microbiology, Department of Plant Pathology, College of Plant Protection, Shandong Agricultural University, Tai’an, China
- 2State Key Laboratory of Crop Biology, Shandong Agricultural University, Tai’an, China
Phytopathogens deploy glycoside hydrolases (GHs) to disintegrate plant cell walls for nutrition and invasion. However, the pathogenic mechanisms of the majority of GHs in virulence remain unknown, especially in oomycetes. In this study, a Phytophthora sojae gene encodes a GH7 family cellobiohydrolase, named PsGH7a, was identified. PsGH7a was highly induced during the cyst germination and infection stages. PsGH7a is conserved in oomycetes, and shares a high amino acid sequence identity (>85%) within Phytophthora genus. The recombinant PsGH7a catalyzes the hydrolysis of β-1,4-glucan and avicel, which represent the major components of cellulose in plant cell wall. The mutation of catalytic residue Glu236 to alanine resulted in a lower catalytic activity. In addition, the PsGH7a promotes Phytophthora invasion, while the mutant can not. Notably, PsGH7a protein triggers hypersensitive cell death in diverse plants. PsGH7a knockout mutants were generated via CRISPR/Cas9 system, to investigate its biological function. Compared to wild-type strain P6497, the mutants showed reduced virulence on susceptible soybean, indicates PsGH7a is indispensable to P. sojae virulence.
Introduction
The battle between plants and microbes is the product of million-years of co-evolution. The front line of the plant defense is numerous physical barriers such as the cell walls, waxes and hairs (Hématy et al., 2009). A primary challenge for microbial pathogens is to penetrate the formidable and dynamic barrier of plant cell walls, which are constructed of cellulose, hemicellulose, pectin, and joined by complex distinct connection types (Somerville et al., 2004; Vorwerk et al., 2004; Wang et al., 2019).
Plant pathogens produce cell wall degrading enzymes (CWDEs) as part of their arsenal for nutrition and plant invasion (Faure, 2002; Martinez et al., 2004; Hashimoto et al., 2007; Hématy et al., 2009; Bakunina et al., 2013; van Wyk et al., 2017; Pluvinage et al., 2019). Phytopathogenic fungi and oomycetes are unique microbial pathogens that being able to break the intact physical surfaces of host plants (Soanes et al., 2007). Many plant-pathogenic fungi secrete a range of CWDEs to degrade the host cell wall, such as glycoside hydrolases, polysaccharide lyases, and esterases, even much more than that in Trichoderma reesei, which is known as a major industrial cellulase-producing fungus (Ma Y. A. et al., 2015; Gui et al., 2017; Le Mauff et al., 2019). For example, the genome of phytopathogenic fungi Magnaporthe grisea and Fusarium graminearum contains two to three times of genes encoding cellulases and xylanases as that in industrial fungus T. reesei (Martinez et al., 2008; King et al., 2011). The effects of the CWDEs usually support their direct contributions to invasion and disease. Emasculation of the endo-beta-1,4-xylanase xyn11A affected the virulence of Botrytis cinerea (Brito et al., 2006), and the mutation of pectate lyase CcpelA had a marked effect on the aggressiveness of Colletotrichum coccodes toward tomato fruits (Ben-Daniel et al., 2012). Oomycetes, from the kingdom Stramenopila (Baldauf, 2003; Yutin et al., 2008), encompass numerous phytopathogens such as Phytophthora, Pythium, Albugo, and downy mildews, which genomes encode abundant of CWDEs toward plant cell wall components (Tyler et al., 2006; Haas et al., 2009; Baxter et al., 2010). It is reported that Phytophthora sojae, the causal agent of soybean stem and root rot disease, manipulates plant immunity by protecting xyloglucanase XEG1 through its truncated paralogous PsXLP1 as a decoy (Ma et al., 2017). Nevertheless, the pathogenetic roles of the vast majority of CWDEs remain unknown, especially in oomycetes (Ma Z. et al., 2015).
The CWDEs produced by microorganisms were shown as multiple components, and their synergistic effect was reported (King et al., 2011; Payne et al., 2015). Some reports indicate that the glycoside hydrolases (GHs) is required for pathogen virulence on host, but the virulence mechanism are still unknown (Ma Z. et al., 2015; Baker et al., 2016; Ma et al., 2017; Fleming et al., 2017; Gui et al., 2017; Snarr et al., 2017; Le Mauff et al., 2019; Shen et al., 2020). Glycoside hydrolases are a widely distributed group of carbohydrate active enzymes (CAZy), which hydrolyze glycosidic bonds in glycosides, glycans and glycoconjugates (Henrissat and Davies, 2000). The primary action mode glycoside hydrolases is regarded as endoglucanases (EGs) act by cleaving β-1,4-glucosidic linkages in amorphous regions of cellulose chains, and cellobiohydrolases (CBHs) attach to the chain end of cello-oligosaccharides and then depolymerize these cellulosic fibers into disaccharide units (Payne et al., 2015). So far, the CAZy database1 classified glycoside hydrolases into 167 families based on predicted structures and sequence similarities (Turbe-Doan et al., 2019). Of these, GH7 family is somewhat enigmatic because it contains both CBHs and EGs in terms of the similar protein comformation, almost undoubtedly it provide the majority of hydrolytic turnover (Wood, 1985). This feature permits efficient hydrolysis activity, and reduces the possibly that the broken chain can reanneal into the crystal surface (Kurasin and Valjamae, 2011). In virtually, all organisms employing GH7 cellulases to degrade lignocellulosic biomass possess multiple genes of GH7 cellulases. To date, the CAZy database lists nearly 5000 GH7 sequences, including EGs and CBHs, and these GH7 cellulases offer complicated evolutionary branches with each other (Lombard et al., 2014).
In addition, unlike some prevalent GH families, such as GH5, GH6, GH12, and GH45, GH7 enzymes mainly consist in fungi, but have not been found in bacteria or archaea (Payne et al., 2015). The cellulose depolymerization of GH7 could be employed by some biomass-degrading fungi, such as the white-rot basidiomycete Phanerochaete chrysosporium, for nutrition and to promote entry into plant tissue (Martinez et al., 2004; Vanden Wymelenberg et al., 2006). So far, there is little research on the GH7 enzymes of phytopathogenic pathogens. Here we have isolated and characterized a gene of oomycete P. sojae, named PsGH7a, encoding a cellobiohydrolases belonging to the glycoside hydrolase family 7. PsGH7a is up regulated during early infection, and the protein product promotes the invasion of Phytophthora pathogens. The deletion of PsGH7a had pronounced effects on P. sojae virulence, delaying the rot of hypocotyls and reducing the lesion size on soybean leaves.
Materials and Methods
Phylogenetic Analysis of PsGH7a Homologs
All sequences of PsGH7a Homologs from oomycetes and fungi were obtained from NCBI (National Center for Biotechnology Information) website. Sequence alignments were generated via the Clustal Omega program (Sievers et al., 2011). The phylogenetic tree was constructed using MEGA 6.0 program via a neighbor joining algorithm with 1,000 bootstrap replicates.
Plant and Phytophthora Cultivation
Nicotiana benthamiana, soybean (Glycine max), and tomato (Solanum lycopersicum L.) plants were grown in the chamber at 25°C with a cycle of 16 h of high light intensity and 8 h of darkness. P. sojae strain P6497 and Phytophthora capsici strain LT1534 and all transformants were grown on 10% V8 medium at 25°C in the dark. 1 × 1-mm hyphal plugs were cultured in V8 liquid medium. After 48 h, mycelia were collected for RNA and DNA extraction. For the expression pattern analysis, mycelia (MY), as well as infection stages (10 min, 30 min, 1, 3, 6, 12, and 24 h), were collected as described previously (Ye et al., 2011).
Nucleic Acid Manipulation and Quantitative PCR Assay
The DNAMAN software was used to analyze the genes and help to design primers. The signal peptides of the proteins were predicted at the SignalP4.0 Server2. Genomic DNA of the P. sojae strains for gDNA PCR or biomass assay was isolated using HP Plant DNA kit (OMEGA Bio-Tek, Norcross, GA, United States), respectively. Total RNA of P. sojae was extracted using EZNA Total RNA Kit I (OMEGA Bio-Tek, Norcross, GA, United States). The concentration of DNA or RNA was measured using a spectrophotometer, and the mass was determined by agarose gel electrophoresis. cDNA was synthesized with HiScript Reverse Transcriptase Kit (Vazyme, Nanjing, China). The PsGH7a gene was amplified using Phanta Super-Fidelity DNA Polymerase (Vazyme, Nanjing, China) from the cDNA. The mutagenesis of PsGH7aE236A was generated by using Mut Express II Fast Mutagenesis Kit V2 (Vazyme, Nanjing, China) following the previous protocol (Han et al., 2018).
Quantitative RT-PCR was performed with the PsGH7a forward primer TCAAGGAACCTACGGCATCAC and the reverse primer AGTTCACTCTCGACGTGGAC. The actin gene (PsActin = Ps108986) was used as an internal reference to detect PsGH7a transcription levels changes. Relative P. sojae biomass in infected plan tissue were quantified with qPCR as described previously (Wang et al., 2011). Primers used in this study are listed (Supplementary Table S2).
Heterologous Expression and Immunoblotting
After PCR progress, the purified PsGH7a PCR products and pPIC9K vector were incubated with EcoRI and NotI (Fermentas, Glen Burnie, MD, United States) and ligated using T4 DNA ligase (Fermentas, Glen Burnie, MD, United States). The recombinant plasmid pPIC9K/PsGH7a was linearized with the restriction enzyme SacI and then transformed into Pichia pastoris GS115. The transformants were seeded onto minimal dextrose (MD) plates and minimal methanol (MM) plates and were then screened on yeast peptone dextrose (YPD) agar medium supplemented with different concentrations of G418 (geneticin; Sangon, Shanghai, China) for the selection of multicopy integrants.
Methanol-induced enzyme expression was performed with shaking cultivation (200 rpm, 28°C) for 7 days based on the Pichia Expression Kit (Invitrogen, Carlsbad, CA, United States). Subsequently, the cell-free supernatant was harvested by centrifugation at 8,000 rpm for 20 min. The obtained supernatant liquor was adjusted with ammonium sulfate with 80% saturation at 4°C overnight (Han et al., 2018). Then, the precipitate was dissolved in phosphate buffer solution (PBS, 20 mM, pH 7.4). After dialysis with PBS, the crude extract was centrifuged at 8,000 rpm for 20 min, and the supernatant liquor was collected. Subsequently, the crude enzyme was purified using Ni2+ affinity chromatography through a HisTrap HP column (GE Healthcare, Waukesha, United States). The purified enzyme was preserved and used for subsequent assays. Protein immunoblots were performed as previously described (Wang et al., 2011). PsGH7a and the mutant were assessed using the anti-6xHis-tag primary antibody (Abclonal, College Park, MD, United States). Purified PsGH7a proteins are diluted into 100 nM for infiltration, as described by Ma (Ma Z. et al., 2015).
Detection of Enzyme Active
The 3,5-dinitrosalicylic acid (DNS) method was employed for evaluating the cellulase activity using barley β-D-glucan and Avicel (Sigma-Aldrich) as the substrates (Song et al., 2016), respectively. The reaction system contained 150 μL of 0.2% (w/v) β-D-glucan or 1% (w/v) Avicel and 15 μg of the purified enzyme in a 300 μL reaction mixture. The hydrolysis reaction was performed at optimal 60°C and pH 4.0 for 30 min, and then terminated by adding 300 μL of DNS solution in a boiling water bath for 10 min. After the sample was cooled down to the room temperature, the absorbance was measured at 540 nm, as described by Miller (Miller, 1959). The standard curve was quantified with D-glucose. One international unit (IU) of enzymatic activity is defined as the amount of enzyme capable of releasing 1 μmol of reducing sugars per minute (Hua et al., 2018). Each experiment was performed in triplicate.
Phytophthora Infection Assay on N. benthamiana
Nicotiana benthamiana leaves were harvested after infiltration and maintained on wet filter paper in Petri dishes. Infiltrated regions were inoculated with hyphal agar plugs (diameter 5 mm) of P. sojae and P. capsici, as previously described (Yang et al., 2017). The expanding lesions were photographed at 36 h after inoculation. Three independent biological replicates were included.
CRISPR/Cas9 Knockout PsCBH7a
The PsGH7a gene sequence was introduced into the website3, and at least two sgRNA sequences were obtained by screening the specificity of the sgRNA sequence and self-circulation. The sgRNA was constructed in the pYF515 vector using the double digestion method with Nhe I and Bsa I as restriction sites. The PsGH7a gene was located on the FungiDB website, and upstream 1000 bp sequence and downstream 1000 bp sequence of PsGH7a in the whole genome of strain P6497 were found. The upstream 1000 bp and downstream 1000 bp fragment of PsGH7a was connected to the pBluescript II KS + vector, and the mCherry gene was inserted between them. The plasmids pYF515 and pBluescript II SK + were co-transformed into protoplast of P. sojae via Polyethylene-Glycol (PEG)-mediated transformation (Hua et al., 2008). After G418 resistance screening, gDNA was extracted from the transformants. The PsGH7a gene was trying to be amplified from gDNA of the transformants using PsGH7a-specific primers, and those without detected amplicons were selected for subsequent assays. The truncated fragments were sequenced to identify positive transformants.
Pathogenicity Assay
Pathogenicity of the transformants was tested by hyphal inoculation. Hyphal agar plugs were inoculated on hypocotyls of potted soybean seedlings or leaves of soybean cultivar Williams, which is compatible with P. sojae strain P6497. Soybeans leaves from the second-leaf stage were used for leaf infection while the hypocotyls for hypocotyl infection. Then, the hyphal plugs were inoculated on hypocotyls or leaves and incubated at 25°C in the dark for 2 days before sampling. Place the leaves of the inoculated hypha pieces in a wet filter paper dish in the dark at 25°C for 3 days. Pictures were taken and relative virulence was measured by qRT-PCR. The ratios of P. sojae DNA to soybean DNA were quantified in the infected plants tissues. All these assays were repeated independently at least three times.
Results
Identification and Phylogenetic Analysis of PsGH7a
Seven candidate genes encoding GH7 cellulases (PsGH7a to PsGH7g) were identified in the P. sojae (strain P6497) genome. According to the predictions on SignalP Sever-5.0 (see footnote 2), four of these seven proteins (PsGH7a to PsGH7d) contain potential signal peptide, indicating those are secreted enzymes (Supplementary Table S1).
The PsGH7a gene is 1395 bp long with no introns (NCBI Gene ID: 20663650), encodes a 464-aa GH family 7 protein (NCBI Reference Sequence: XP_009531399.1). The expression pattern was investigated based on global digital gene expression profiling as reported (Ye et al., 2011). PsGH7a was highly expressed during the cyst germination and infection stages (Supplementary Table S1), and the expression pattern during infection was confirmed via quantitative reverse transcription (qRT)-PCR (Supplementary Figure S1). This indicates that PsGH7a may be required during P. sojae invasion and infection.
Based on Protein BLAST (Basic Local Alignment Search Tool)4 searches, the homologs of PsGH7a in some oomycetes and some other pathogen species were identified. The phylogenetic tree was constructed using PsGH7a protein sequence and the homologs from oomycetes and fungi (Figure 1). The result showed that PsGH7a-homologous proteins are widespread among plant pathogenic oomycetes and fungi. The NCBI-blast results revealed PsGH7a shares a high degree of identity (>85%) within Phytophthora genus (Supplementary Figure S2). Of these, PsGH7a shared 90.7% identity with the Phytophthora fragariae GH7 cellobiohydrolase (KAE9199421), 90.5% identity with the Phytophthora infestans GH7 family cellobiohydrolase (XP_002902727) and 90.0% identity with the Phytophthora parasitica GH7 family protein (ETL91310). Also, PsGH7a shared 86.97% identity with the GH7 family protein (RMX63503.1) of Peronospora effusa, which is an obligate downy mildew pathogen. That indicates PsGH7a may contribute to the biotrophic phase of pathogens.
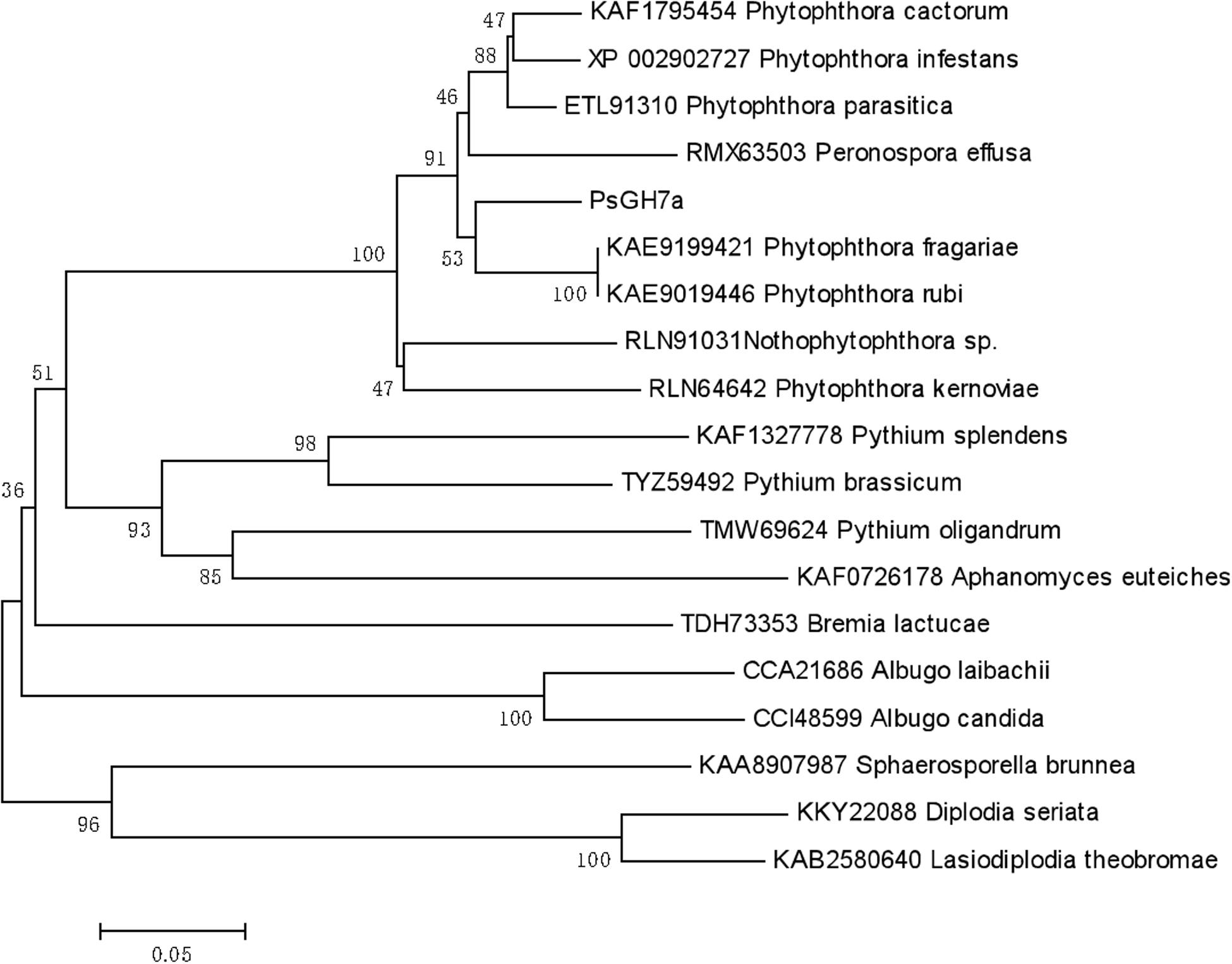
Figure 1. Phylogenetic tree of PsGH7a and its homologous enzymes from oomycetes and fungi. The phylogenetic tree was constructed through the MEGA 6.0 program with a neighbor-joining algorithm using 1,000 bootstrap replicates.
Biochemical Characterization of PsGH7a
To determine the biochemical properties, mature PsGH7a was heterologously expressed in Pichia pastoris and purified following a previously described protocol (Ma Y. A. et al., 2015). Western blotting exhibited that the purified PsGH7a protein appeared as a single band with an approximate molecular weight of 48 kDa (Supplementary Figure S3). The recombinant PsGH7a is able to efficiently hydrolyze the natural cellulose material of β-1,4-glucan (Figure 2). Evidently, PsGH7a is able to catalyze the hydrolysis of avicel, which is a typical characteristic of cellobiohydrolase (Figure 2). However, PsGH7a could not efficiently hydrolyze carboxymethyl cellulose (CMC) (Figure 2) and this phenomenon is also detected by other previous reports on cellobiohydrolases (Baramee et al., 2017; Han et al., 2018). Lichenin also could not be efficiently hydrolyzed (Figure 2), because it is composed of glucose units by the main β-1,3-1,4-glycosidic bonds which is not the preference for a β-1,4-cellobiohydrolase. Although previous reports have demonstrated that some glycoside hydrolases are bifunctional cellulase-xylanase enzymes (Hua et al., 2018; Lee et al., 2018; Chu et al., 2019), PsGH7a has no catalytic ability on the hydrolysis of xylan (Figure 2).
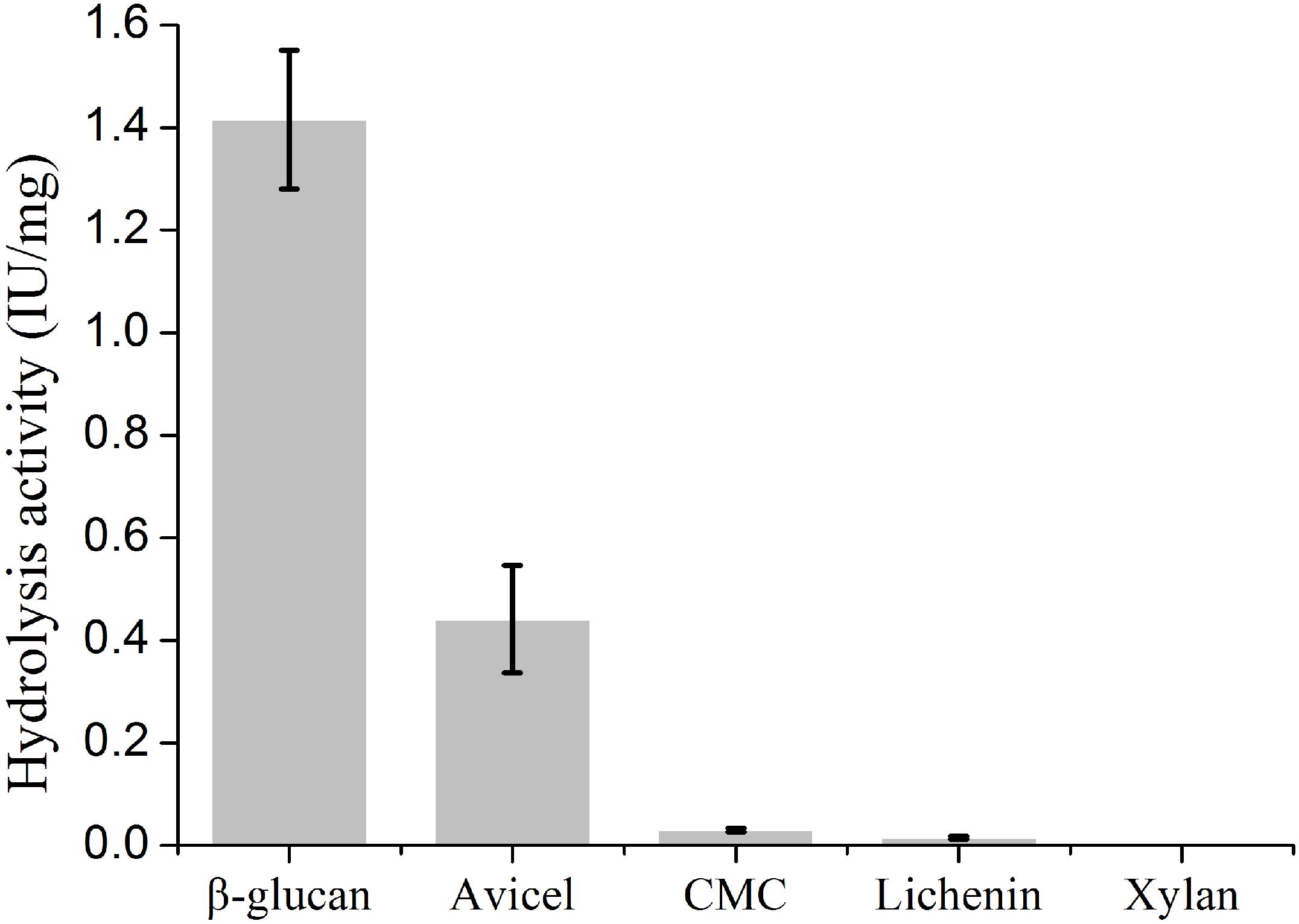
Figure 2. Substrate specific activity of cellobiohydrolase PsGH7a. Values are means ± SD of three replications.
PsGH7a Is an Elicitor and Induces Hypersensitive Responses in Various Plants
The recombinant PsGH7a protein was infiltrated into expanded leaves of N. benthamiana. 5 days later, trypan-blue staining indicates the areas of cell death, which were enlarged with increasing concentrations of PsGH7a protein from 20 to 100 nM (Figure 3A). Compared to the other reported fungal and oomycete cellulase elicitors, the response in N. benthamiana is weakened and restricted to the infiltration site (Ma Y. A. et al., 2015; Ma Z. et al., 2015; Gui et al., 2017). To examine the host specificity, purified PsGH7a (100 nM) was infiltrated into expanded leaves of soybean (Glycine max) and obvious symptoms appeared at 5 days after infiltration (Figure 3B). Thus, PsGH7a protein can elicits hypersensitive response in the host, soybean.
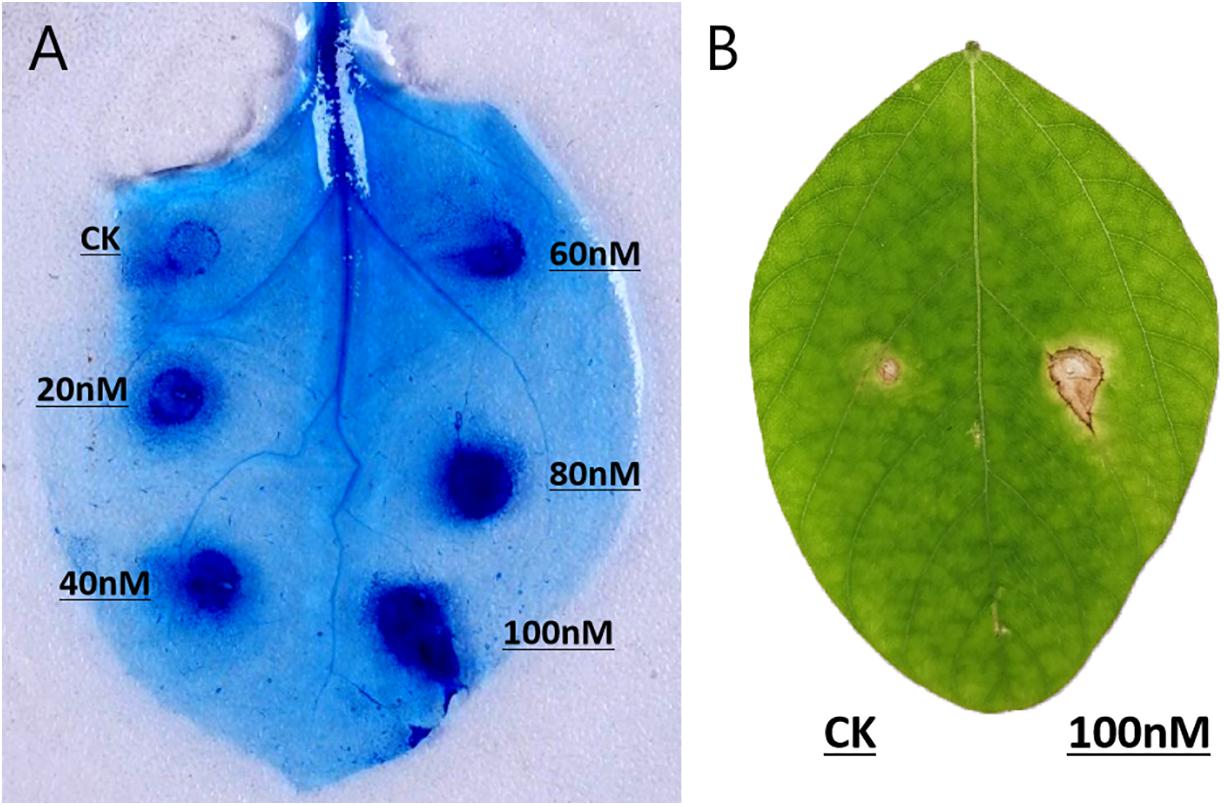
Figure 3. PsGH7a induces hypersensitive responses in various plants. (A) Protein solutions (20, 40, 60, 80, and 100 nM) and use buffer as control (CK) were infiltrated in N. benthamiana leaves, and then stained with trypan blue at 5 days post infiltration. (B) Soybean leaves were infiltrated with 100 nM protein solution and control (CK), and pictures were taken after 5 days.
PsGH7a Promotes the Invasion of Phytophthora
To investigate the function of PsGH7a, a malfunction mutation was constructed. According to the structure determination of homologous GH7 cellobiohydrolases, the highly conserved residue Glu236 served as a catalytic acid in the glycosyl group hydrolysis and the conserved residue Glu241 acted as the acid/base for increasing the nucleophilicity of the catalytic water (Figure 4A and Supplementary Figure S2; Momeni et al., 2013). When the catalytic residue Glu236 was substituted with Ala, the intrinsic functional hydrogen bond in carboxyl of Glu236 was eliminated and could not effectively induce the nucleophilic attack on the anomeric carbon atom to cleave the glucosidic bond through a covalent glycosyl-enzyme intermediate (Davies et al., 1993; Vocadlo and Davies, 2008; Supplementary Figure S4). Besides, in mutant PsGH7aE236A, the extended distance between Ala236 and Glu241 is adverse to the substrate binding, and this single mutation was not sufficient to induce comformational change of the active site architecture. Therefore, the mutant on the Glu236 (PsGH7AE236A) did not affect the stability and molecular mass of PsGH7a, and can not induce cell death at 5 dpi (Supplementary Figure S3).
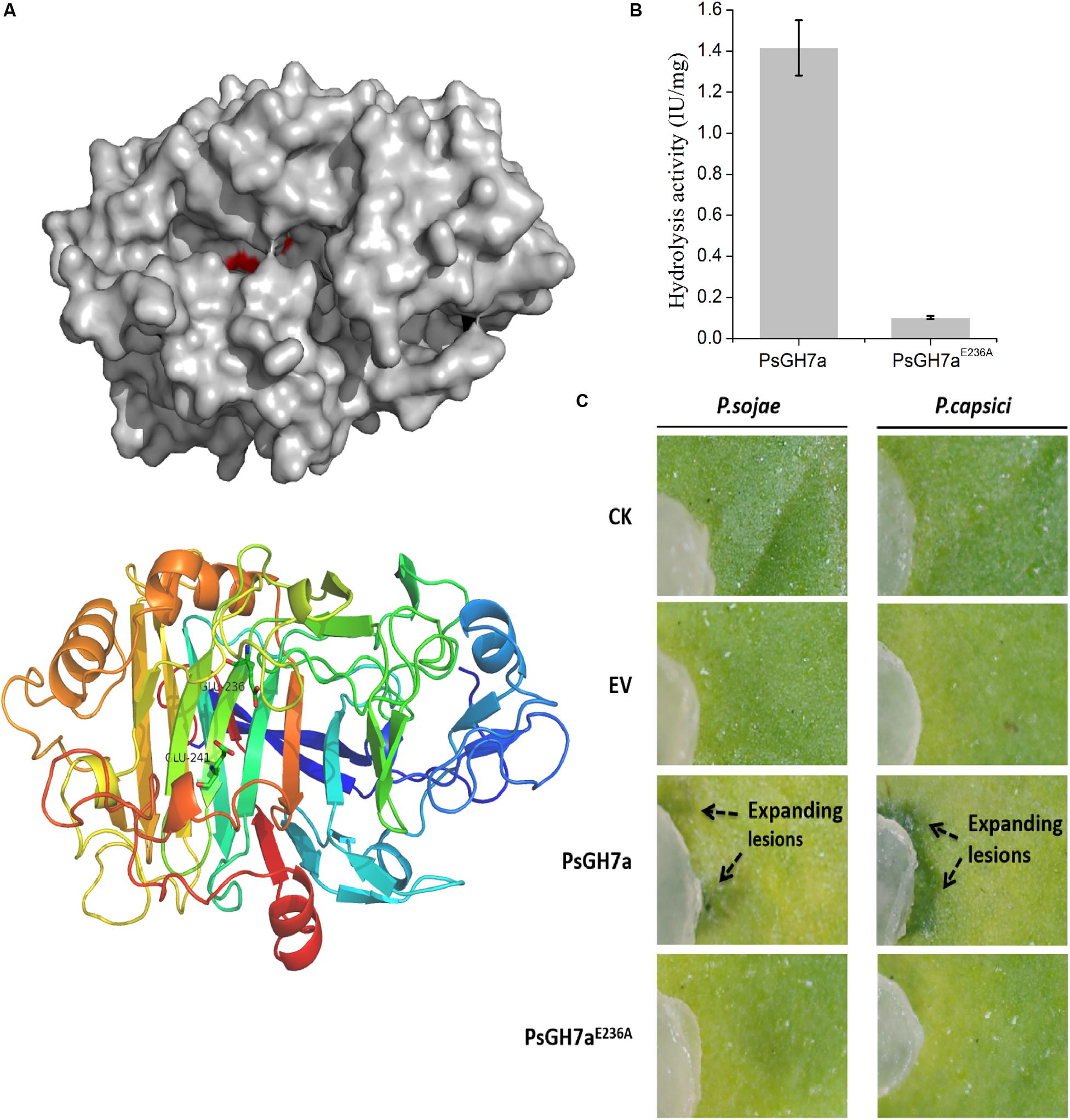
Figure 4. The hydrolysis of PsGH7a promotes Phytophthora invasion. (A) Predicted three dimensional structure of the PsGH7a protein obtained using the homologous Phanerochaete chrysosporium cellobiohydrolase Cel7D as the template (PDB: 1z3t). The color gradient shows the sequence from the N terminus (blue) to the C terminus (red). In particular, the putative catalytic residues Glu236 and Glu241 are presented as green sticks. (B) Cellulase activity (IU) of the wild-type and the mutant form PsGH7aE236A. One unit (U) of cellulase activity was defined as the amount of cellulase that catalyzed the liberation of reducing sugar equivalent to 1.0 μg glucose/min under assay conditions. Three independent biological replicates were used for each protein. (C) After the PsGH7a and the mutant form PsGH7aE236A proteins were infiltrated into N. benthamiana leaves, P. sojae and P. capsici hyphal plugs were put on the infiltration sites. The buffer was infiltrated as a control (CK); the cell-free supernatant from Pichia pastoris GS115 which contains a empty vector (EV) was also infiltrated as another control. Arrow indicates enlarged lesion area. The pictures were taken at 3 days after inoculation.
The catalytic activity assay indicates that PsGH7aE236A showed much lower activity than wild type (Figure 4B). The purified PsGH7a and PsGH7aE236A was infiltrated into N. benthamiana leaves, and hyphal plugs of P. sojae and P. capsici were put on the infiltration sites. Three days after inoculation, the lesions on the area with PsGH7a were significantly larger than others on the leaves (Figure 4C). The results indicated that the infiltration of PsGH7a in leaves increased their susceptibility to P. sojae and P. capsici, and its enzymic activity is also required.
CRISPR/Cas9 Genome Editing for PsGH7a Knockout
The contribution of PsGH7a to P. sojae virulence was investigated through CRISPR/Cas9 genome editing. The PsGH7a-knockout mutants of P. sojae (strain P6497) via CRISPR/Cas9 system were generated following a previously described protocol (Fang and Tyler, 2016). Two single guide RNAs (sgRNAs) with independent targeting were designed to disrupt the PsGH7a coding region (Figure 5A). The plasmid pBS KS + containing a homologous donor DNA (the mCherry gene with PsGH7a flanking sequences are used as a template for fragment homologous substitution, Figure 5A) and pYF515 vectors carrying each sgRNA information and hSpCas9 sequence were introduced into P. sojae via protoplast transformation as described (Hua et al., 2008). Six independent transformants were identified by gDNA PCR (Supplementary Figure S5) and sequencing screening, which all showed normal filamentous growth. The sequencing result showed that the PsGH7a gene was replaced by inserted mCherry in the mutants TG1 and TG6 (Figure 5B). The transformant TG3 is failed to acquire PsGH7a deletion and be used as the assay control (CK) (Supplementary Figure S5).
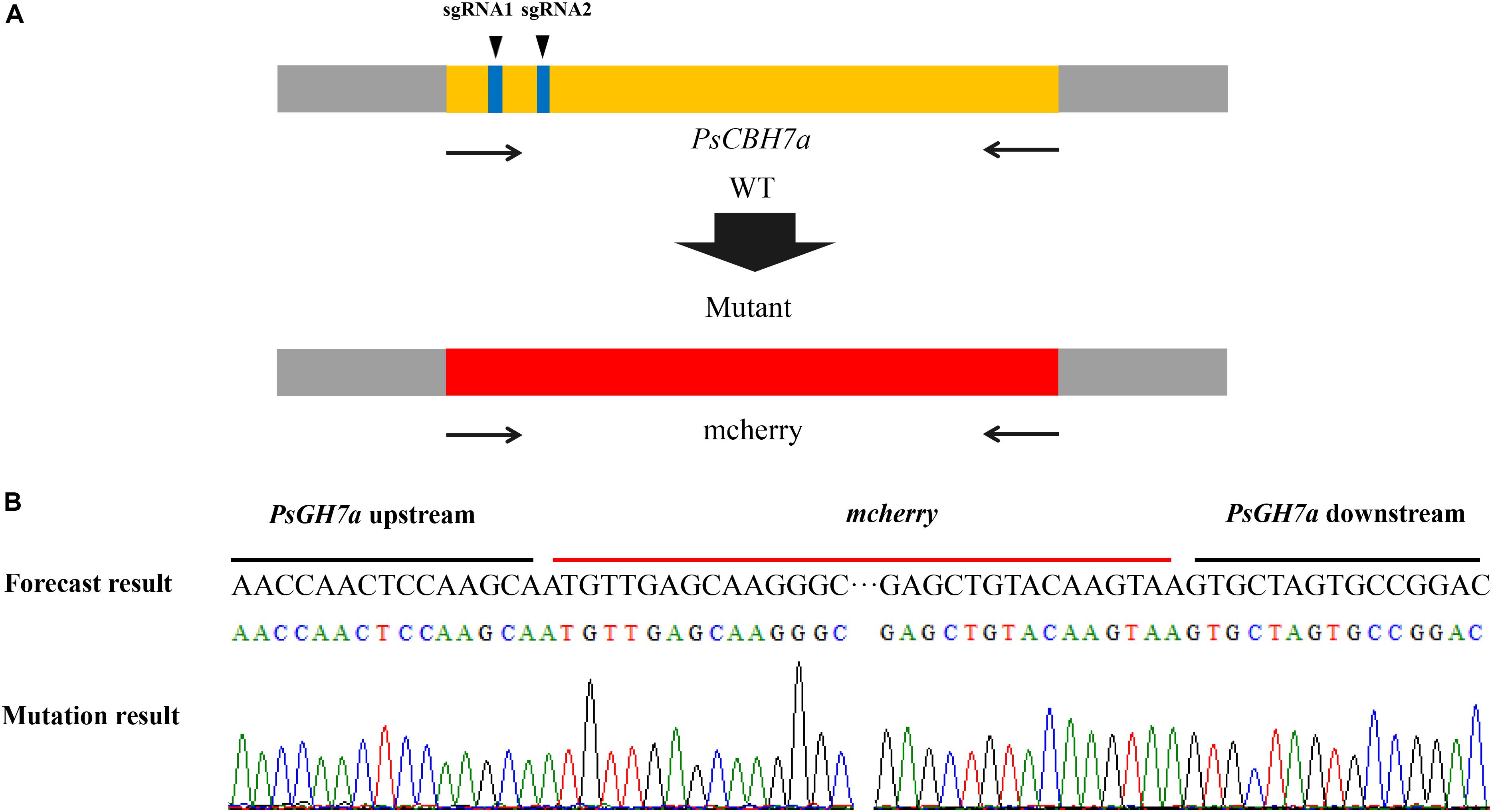
Figure 5. CRISPR/Cas9 Genome Editing for PsGH7a Knockout. (A) sgRNA1, sgRNA2 are designed to target PsGH7a, and the mCherry gene with PsGH7a flanking sequences are used as template for fragment homologous substitution. (B) The sequencing results consist with the forecast result, indicates the PsGH7a gene have been successfully replaced as mcherry. The DNA used for sequencing are obtained from transformants, and the sequencing peaks are attached.
PsGH7a Is Required for Full Virulence of P. sojae
Potted seedlings of the soybean susceptible cultivar (Williams, without any known resistance genes) were inoculated with hyphal plugs on the wounded hypocotyls, respectively. After 4 days, the recipient strain P6497 killed soybean seedlings quickly while the transformants did not (Figure 6A). Consistent with these results, the virulence of these transformants was damnified on soybean leaves also. Hyphal agar plugs were inoculated on the opposite leaves of Williams, and moisturized in petri dishes for 2 days. The lesion regions caused by TG1 and TG6 were much smaller than WT and CK (Figure 6B). The biomass quantification showed the relative amount of P. sojae DNA of the mutants were significantly reduced in the inoculated soybean hypocotyls and leaves compared to WT and CK (Figures 6C,D). These results suggest that PsGH7a is required for full virulence of P. sojae in soybean infection.
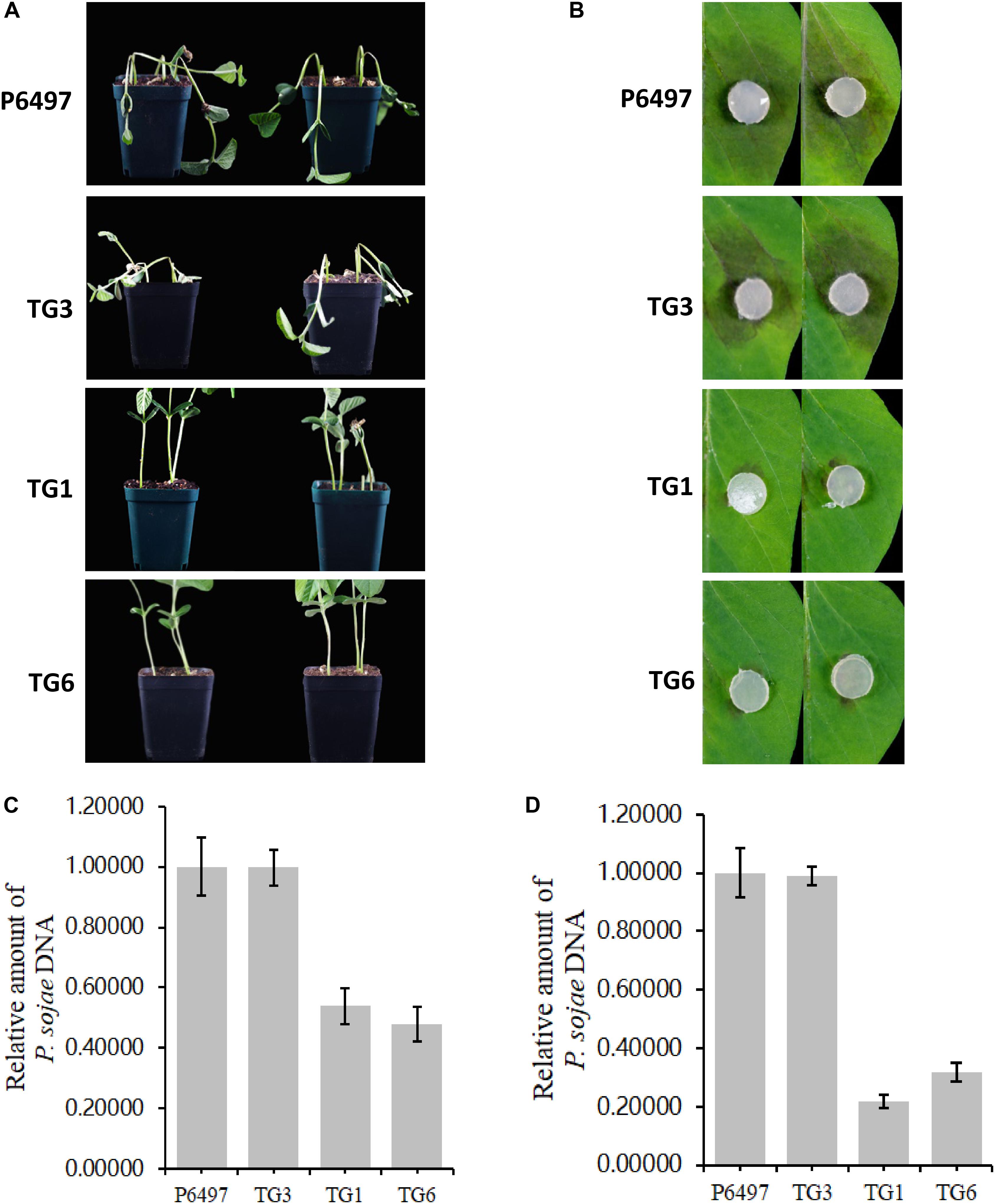
Figure 6. PsGH7a is required for full virulence of P. sojae. (A) P6497 wild-type and transformants hyphae plugs infected wounded hypocotyls, the pictures were taken at 3 days later. (B) P6497 and transformants hyphae plugs infected soybean leaves, the pictures were taken at 3 days later. (C) Determination of P. sojae DNA content in infected hypocotyls. Hypocotyls were collected at 3 days after infection with P6497 and transformants, and DNA was extracted for real-time PCR. (D) Determination of P. sojae DNA content in leaves. Leaves were collected 3 days after infection with P6497 and transformants, and DNA was extracted for real-time PCR. Three independent replicates of each real-time PCR.
Discussion
Plants and pathogens are involved in a dynamic co-evolutionary struggle for survival. To cope with pathogen infection, plants rely on the physical barriers and layered innate immunity to program defenses (Jones and Dangl, 2006; Wang et al., 2019). Plant cell walls provide the primary physical defense, and may be dynamic strengthened for added defense during interaction with pathogens (Vorwerk et al., 2004). To penetrate plant cell walls and colonization of living host tissue, phytopathogens deploy secreted CWDEs as part of their arsenals (Hématy et al., 2009). The enzymes include glucanases, cellulases, polygalacturonases, pectinases and xyloglucanases (Vorwerk et al., 2004). For example, Ustilago maydis, the causal agents of corn smut, contains 33 such CWDE-encoding genes, (Kamper et al., 2006). However, there is little detailed information available about oomycete apoplastic CWDEs to date (Ma et al., 2017).
The CWDEs target multiple plant polysaccharides. Here we identified a P. sojae GH7 family protein, PsGH7a, which is a typical CBH GH7 family has been mainly found in fungi, our results showed that GH7 proteins are conserved in oomycetes. Both phytopathogenic fungi and oomycete are able to break the intact physical surfaces of host plants, which indicates the hydrolytic activity provided by GH7 enzymes maybe employed by these pathogens. Our results showed that the PsGH7a is highly conserved in both oomycetes and fungi (Figure 1). The PsGH7a shares a high degree of identity (>85%) within Phytophthora genus (Supplementary Figure S2), and shared 86.97% identity with the Peronospora effusa GH7 family protein (RMX63503.1). That indicates GH7a of Phytophthora may contribute to the biotrophic phase. The homologs from saprophytic oomycetes, including Aphanomyces euteiches, Pythium oligandrum, and Pythium brassicum, and Pythium splendens, share 60–65% identity with PsGH7a, are clustered to adjacent branch. The homologs from other obligate oomycetes, such as Bremia lactucae, Albugo candida and Albugo laibachii, are clustered to sub-groups. The homologs from fungi, including Sphaerosporella brunnea, Diplodia seriata, and Lasiodiplodia theobromae, are clustered into sub-population, that means they are evolved independently. These differences suggest that independent evolutionary events may have occurred in target sequences of oomycetes and fungi.
In the hydrolysis activity detection assay, PsGH7a is able to hydrolyze β-1,4-glucan and avicel. The main load-bearing component of plant cell wall is cellulose, which is the β-1,4-linked homopolymer of glucosides (Vorwerk et al., 2004). Thus, β-1,4-glucan represents the major component of cellulose in plant cell wall (Glass et al., 2013). In addition, the highly ordered arrangement of cellulose fibers connected by regular hydrogen bonds is termed as the crystalliferous region of cellulose, which is recognized as the critical traffic jam for reducing hydrolytic efficiency of cellulases on cellulose surface (Igarashi et al., 2011). The crystallinity of cellulosic avicel plays a major role in determining the rate of hydrolysis by cellulases, especially for CBH (Hall et al., 2010). As CBHs attach to cellulose chain ends, immediately triggering the internal bond cleavage by EGs, CBH can immediately capture the newly exposed reducing chain ends (Kurasin and Valjamae, 2011). So PsGH7a is identified as a cellobiohydrolase, provides the majority of hydrolytic turnover.
Usually, the hydrolysis activities of CWDE cocktails are required for phytopathogen virulence, which help to macerate plant tissues during infection (King et al., 2011). Silencing of P. sojae XEG1, which encodes a GH12-family xyloglucanase, severely reduced virulence (Ma Z. et al., 2015). Consistent with these results, pathogenicity assays showed the P. sojae lose its virulence when PsGH7a was knocked out. These results suggest that the PsGH7a plays key roles during P. sojae invasion. Furthermore, infiltration of PsGH7a even promote invasion of P. sojae and P. capsici on the non-host N. benthamiana leaves. So far, little is known about the differences in components of cell wall between hosts and non-hosts, and also the chemical composition is a factor in the outcome of the non-host disease resistance (Somerville et al., 2004). Usually, pathogenic CWDEs have obvious selectivity for hosts and good adaptability to the most preferred host (King et al., 2011), so it is possible to control fungus and oomycete disease by fine-tuned dynamic enhancement of plant cell walls. Our progress in defining P. sojae GH7a has provided new insights into the possible role of cell wall composition in controlling disease interactions.
In the co-evolution of plants and pathogens, some conserved CWDEs secreted by phytopathogens are recognized as pathogen-associated molecular patterns (PAMPs) by plant cell surface pattern recognition receptors (PRRs), and trigger plant immunity (PTI) (Wang and Wang, 2018). For example, an endocellulase from Rhizoctonia solani is an elicitor (Ma Y. A. et al., 2015). P. sojae XEG1 act as virulence factors and PAMPs in oomycetes (Ma Z. et al., 2015). Two GH12 proteins VdEG1 and VdEG3 produced by the fungus Verticillium dahliae Vd991 acted as PAMPs to trigger cell death (Gui et al., 2017). Plant cells also have molecular mechanisms for sensing and responding to cell wall derangement. Some of the cell wall degradation fragments produced by pathogen CWEDs, which are termed damage-associated molecular patterns (DAMPs), can elicit defensive responses by plant (Hématy et al., 2009). For example, oligogalacturonic acid, which is produced when degradation of pectins by polygalacturonase generates, can trigger plant defenses (D’Ovidio et al., 2004). We found that PsGH7a protein can elicits hypersensitive response in tobacco and soybean. The necrotic lesions usually appear at 3 to 5 days post protein infiltration, it is hard to say the response is due to PAMP or DAMP. For the protein mutant PsGH7a E236A showed much lower activity than wild type, and cannot induce HR on soybean leaves, that indicate the degradation product but not the protein triggers PTI. We thought maybe the degradation product but not the protein triggers cell death, and for some reason, the cell death spread out slowly from the wound, while control or the protein mutant PsGH7a E236A does not. Maybe that is a kind of DAMP-triggered immunity. Also, we noticed that the cell death was increased with increasing concentrations of enzyme, so we guess it is probably because more damage was created by PsGH7a, which had been proved as a cellobiohydrolase.
It is well reported a novel decoy strategy is used in Phytophthora pathosystems (Ma et al., 2017). P. sojae manipulates plant immunity by protecting the GH12-family xyloglucanase XEG1 through its truncated paralogous PsXLP1 as a decoy. It is worth to notice that the CAZy database lists nearly 5000 GH7 sequences, and the GH7 cellulases in Phytophthora offer complicated evolutionary branches with each other. The decoy pattern provides an explanation for the multiple, similar sequences from the same family. The cross fire at the front line of plant defense is always the most intense. Thus, the researches on Phytophthora CWDEs provide more novel viewing angles of evolutionary struggle between plants and pathogens.
Data Availability Statement
The datasets generated for this study can be found in the NCBI XP_009531399, NCBI KAE9199421.1, NCBI KAE9019446.1, NCBI ETL91310.1, NCBI KAF1795454.1, NCBI XP_002902727.1, NCBI RLN91031.1, NCBI RMX63503.1, NCBI RLN64642.1, NCBI KAF1327778.1, NCBI TYZ59492.1, NCBI TDH73353.1, NCBI CCA21686.1, NCBI KAF0726178.1, NCBI CCI48599.1, NCBI KAA8907987.1, NCBI KKY22088.1, and NCBI KAB2580640.1.
Author Contributions
QW and CH designed the experiments. YH, XT, and CH wrote the manuscript, and performed the experiments and data analysis. YH and CH expressed the proteins and test the catalytic activity. YH tested the virulence of Phytophthora. XT constructed the P. sojae mutants. QX, YJ, and XH participated in manuscript revision or experiment. QW revised the manuscript and provided the funding for this research. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31671985 and 31972249), Key R&D projects in Shandong Province (2019GNC106060 and 2019JZZY020608) and Natural Science Foundation of Shandong Province (ZR2018BC014).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01285/full#supplementary-material
Footnotes
- ^ http://www.cazy.org
- ^ http://www.cbs.dtu.dk/services/SignalP/
- ^ http://grna.ctegd.uga.edu/
- ^ https://blast.ncbi.nlm.nih.gov/Blast.cgi
References
Baker, P., Hill, P. J., Snarr, B. D., Alnabelseya, N., Pestrak, M. J., Lee, M. J., et al. (2016). Exopolysaccharide biosynthetic glycoside hydrolases can be utilized to disrupt and prevent Pseudomonas aeruginosa biofilms. Sci. Adv. 2:e1501632. doi: 10.1126/sciadv.1501632
Bakunina, I., Nedashkovskaya, O., Balabanova, L., Zvyagintseva, T., Rasskasov, V., and Mikhailov, V. (2013). Comparative analysis of glycoside hydrolases activities from phylogenetically diverse marine bacteria of the genus Arenibacter. Mar. Drugs. 11, 1977–1998. doi: 10.3390/md11061977
Baldauf, S. L. (2003). The deep roots of eukaryotes. Science 300, 1703–1706. doi: 10.1126/science.1085544
Baramee, S., Teeravivattanakit, T., Phitsuwan, P., Waeonukul, R., Pason, P., Tachaapaikoon, C., et al. (2017). A novel GH6 cellobiohydrolase from Paenibacillus curdlanolyticus B-6 and its synergistic action on cellulose degradation. Appl. Microbiol. Biotechnol. 101, 1175–1188. doi: 10.1007/s00253-016-7895-8
Baxter, L., Tripathy, S., Ishaque, N., Boot, N., Cabral, A., Kemen, E., et al. (2010). Signatures of adaptation to obligate biotrophy in the Hyaloperonospora arabidopsidis genome. Science 330, 1549–1551. doi: 10.1126/science.1195203
Ben-Daniel, B. H., Bar-Zvi, D., and Tsror Lahkim, L. (2012). Pectate lyase affects pathogenicity in natural isolates of Colletotrichum coccodes and in pelA gene-disrupted and gene-overexpressing mutant lines. Mol. Plant. Pathol. 13, 187–197. doi: 10.1111/j.1364-3703.2011.00740.x
Brito, N., Espino, J. J., and Gonzalez, C. (2006). The endo-beta-1,4-xylanase xyn11A is required for virulence in Botrytis cinerea. Mol. Plant. Microbe. Interact. 19, 25–32. doi: 10.1094/MPMI-19-0025
Chu, Y., Hao, Z., Wang, K., Tu, T., Huang, H., Wang, Y., et al. (2019). The GH10 and GH48 dual-functional catalytic domains from a multimodular glycoside hydrolase synergize in hydrolyzing both cellulose and xylan. Biotechnol. Biofuels. 12:279. doi: 10.1186/s13068-019-1617-2
Davies, G. J., Dodson, G. G., Hubbard, R. E., Tolley, S. P., Dauter, Z., Wilson, K. S., et al. (1993). Structure and function of endoglucanase V. Nature 365, 362–364. doi: 10.1038/365362a0
D’Ovidio, R., Mattei, B., Roberti, S., and Bellincampi, D. (2004). Polygalacturonases, polygalacturonase-inhibiting proteins and pectic oligomers in plant-pathogen interactions. Biochim. Biophys. Acta. 1696, 237–244. doi: 10.1016/j.bbapap.2003.08.012
Fang, Y., and Tyler, B. M. (2016). Efficient disruption and replacement of an effector gene in the oomycete Phytophthora sojae using CRISPR/Cas9. Mol. Plant. Pathol. 17, 127–139. doi: 10.1111/mpp.12318
Faure, D. (2002). The family-3 glycoside hydrolases: from housekeeping functions to host-microbe interactions. Appl. Environ. Microbiol. 68, 1485–1490. doi: 10.1128/aem.68.4.1485-1490.2002
Fleming, D., Chahin, L., and Rumbaugh, K. (2017). Glycoside hydrolases degrade polymicrobial bacterial biofilms in wounds. Antimicrob. Agents 61:e01998-16. doi: 10.1128/AAC.01998-16
Glass, N. L., Schmoll, M., Cate, J. H., and Coradetti, S. (2013). Plant cell wall deconstruction by ascomycete fungi. Annu. Rev. Microbiol. 67, 477–498. doi: 10.1146/annurev-micro-092611-150044
Gui, Y. J., Chen, J. Y., Zhang, D. D., Li, N. Y., Li, T. G., Zhang, W. Q., et al. (2017). Verticillium dahliae manipulates plant immunity by glycoside hydrolase 12 proteins in conjunction with carbohydrate-binding module 1. Environ. Microbiol. 19, 1914–1932. doi: 10.1111/1462-2920.13695
Haas, B. J., Kamoun, S., Zody, M. C., Jiang, R. H., Handsaker, R. E., Cano, L. M., et al. (2009). Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461, 393–398. doi: 10.1038/nature08358
Hall, M., Bansal, P., Lee, J. H., Realff, M. J., and Bommarius, A. S. (2010). Cellulose crystallinity-a key predictor of the enzymatic hydrolysis rate. FEBS. J. 277, 1571–1582. doi: 10.1111/j.1742-4658.2010.07585.x
Han, C., Li, W., Hua, C., Sun, F., Bi, P., and Wang, Q. (2018). Enhancement of catalytic activity and thermostability of a thermostable cellobiohydrolase from Chaetomium thermophilum by site-directed mutagenesis. Int. J. Biol. Macromol. 116, 691–697. doi: 10.1016/j.ijbiomac.2018.05.088
Hashimoto, W., Itoh, T., Maruyama, Y., Mikami, B., and Murata, K. (2007). Hydration of vinyl ether groups by unsaturated glycoside hydrolases and their role in bacterial pathogenesis. Int. Microbiol. 10, 233–243. doi: 10.2436/20.1501.01.32
Hématy, K., Cherk, C., and Somerville, S. (2009). Host-pathogen warfare at the plant cell wall. Curr. Opin. Plant Biol. 12:406. doi: 10.1016/j.pbi.2009.06.007
Henrissat, B., and Davies, G. J. (2000). Glycoside hydrolases and glycosyltransferases. Families, modules, and implications for genomics. Plant Physiol. 124, 1515–1519. doi: 10.1104/pp.124.4.1515
Hua, C., Li, W., Han, W., Wang, Q., Bi, P., Han, C., et al. (2018). Characterization of a novel thermostable GH7 endoglucanase from Chaetomium thermophilum capable of xylan hydrolysis. Int. J. Biol. Macromol. 117, 342–349. doi: 10.1016/j.ijbiomac.2018.05.189
Hua, C., Wang, Y., Zheng, X., Dou, D., Zhang, Z., Govers, F., et al. (2008). A Phytophthora sojae G-protein alpha subunit is involved in chemotaxis to soybean isoflavones. Eukaryot. Cell. 7, 2133–2140. doi: 10.1128/EC.00286-08
Igarashi, K., Uchihashi, T., Koivula, A., Wada, M., Kimura, S., Okamoto, T., et al. (2011). Traffic jams reduce hydrolytic efficiency of cellulase on cellulose surface. Science 333, 1279–1282. doi: 10.1126/science.1208386
Jones, J., and Dangl, J. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kamper, J., Kahmann, R., Bolker, M., Ma, L. J., Brefort, T., Saville, B. J., et al. (2006). Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 444, 97–101. doi: 10.1038/nature05248
King, B. C., Waxman, K. D., Nenni, N. V., Walker, L. P., Bergstrom, G. C., and Gibson, D. M. (2011). Arsenal of plant cell wall degrading enzymes reflects host preference among plant pathogenic fungi. Biotechnol. Biofuels 4:4. doi: 10.1186/1754-6834-4-4
Kurasin, M., and Valjamae, P. (2011). Processivity of Cellobiohydrolases is limited by the substrate. J. Biol. Chem. 286, 169–177. doi: 10.1074/jbc.M110.161059
Le Mauff, F., Bamford, N. C., Alnabelseya, N., Zhang, Y., Baker, P., Robinson, H., et al. (2019). Molecular mechanism of Aspergillus fumigatus biofilm disruption by fungal and bacterial glycoside hydrolases. J. Biol. Chem. 294, 10760–10772. doi: 10.1074/jbc.RA119.008511
Lee, K. T., Toushik, S. H., Baek, J. Y., Kim, J. E., Lee, J. S., and Kim, K. S. (2018). Metagenomic Mining and Functional Characterization of a Novel KG51 Bifunctional Cellulase/Hemicellulase from Black Goat Rumen. J. Agric. Food. Chem. 66, 9034–9041. doi: 10.1021/acs.jafc.8b01449
Lombard, V., Golaconda Ramulu, H., Drula, E., Coutinho, P. M., and Henrissat, B. (2014). The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic. Acids. Res. 42, D490–D495. doi: 10.1093/nar/gkt1178
Ma, Y. A., Han, C., Chen, J. Y., Li, H. Y., He, K., Liu, A. X., et al. (2015). Fungal cellulase is an elicitor but its enzymatic activity is not required for its elicitor activity. Mol. Plant. Pathol. 16, 14–26. doi: 10.1111/mpp.12156
Ma, Z., Song, T., Zhu, L., Ye, W., Wang, Y., Shao, Y., et al. (2015). A Phytophthora sojae Glycoside Hydrolase 12 protein is a major virulence factor during soybean infection and is recognized as a PAMP. Plant Cell 27, 2057–2072. doi: 10.1105/tpc.15.00390
Ma, Z., Zhu, L., Song, T., Wang, Y., Zhang, Q., Xia, Y., et al. (2017). A paralogous decoy protects Phytophthora sojae apoplastic effector PsXEG1 from a host inhibitor. Science 355, 710–714. doi: 10.1126/science.aai7919
Martinez, D., Berka, R. M., Henrissat, B., Saloheimo, M., Arvas, M., Baker, S. E., et al. (2008). Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina). Nat. Biotechnol. 26, 553–560. doi: 10.1038/nbt1403
Martinez, D., Larrondo, L. F., Putnam, N., Gelpke, M. D., Huang, K., Chapman, J., et al. (2004). Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat. Biotechnol. 22, 695–700. doi: 10.1038/nbt967
Miller, G. L. (1959). Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31, 426–428. doi: 10.1021/ac60147a030
Momeni, M. H., Payne, C. M., Hansson, H., Mikkelsen, N. E., Svedberg, J., Engström, Å., et al. (2013). Structural, biochemical, and computational characterization of the glycoside hydrolase family 7 cellobiohydrolase of the tree-killing fungus Heterobasidion Irregulare. J. Biol. Chem. 288, 5861–5872. doi: 10.1074/jbc.M112.440891
Payne, C. M., Knott, B. C., Mayes, H. B., Hansson, H., Himmel, M. E., Sandgren, M., et al. (2015). Fungal cellulases. Chem. Rev. 115, 1308–1448. doi: 10.1021/cr500351c
Pluvinage, B., Massel, P. M., Burak, K., and Boraston, A. B. (2019). Structural and functional analysis of four family 84 glycoside hydrolases from the opportunistic pathogen Clostridium perfringens. Glycobiology 30, 49–57. doi: 10.1093/glycob/cwz069
Shen, D., Wang, J., Dong, Y., Zhang, M., Tang, Z., Xia, Q., et al. (2020). The glycoside hydrolase 18 family chitinases are associated with development and virulence in the mosquito pathogen Pythium guiyangense. Fungal. Genet. Biol. 135:103290. doi: 10.1016/j.fgb.2019.103290
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
Snarr, B. D., Baker, P., Bamford, N. C., Sato, Y., Liu, H., Lehoux, M., et al. (2017). Microbial glycoside hydrolases as antibiofilm agents with cross-kingdom activity. Proc. Natl. Acad. Sci. U.S.A. 114, 7124–7129. doi: 10.1073/pnas.1702798114
Soanes, D. M., Richards, T. A., and Talbot, N. J. (2007). Insights from sequencing fungal and oomycete genomes: what can we learn about plant disease and the evolution of pathogenicity? Plant Cell 19, 3318–3326. doi: 10.1105/tpc.107.056663
Somerville, C., Bauer, S., Brininstool, G., Facette, M., Hamann, T., Milne, J., et al. (2004). Toward a systems approach to understanding plant cell walls. Science 306, 2206–2211. doi: 10.1126/science.1102765
Song, H. T., Gao, Y., Yang, Y. M., Xiao, W. J., Liu, S. H., Xia, W. C., et al. (2016). Synergistic effect of cellulase and xylanase during hydrolysis of natural lignocellulosic substrates. Bioresour. Technol. 219, 710–715. doi: 10.1016/j.biortech.2016.08.035
Turbe-Doan, A., Record, E., Lombard, V., Kumar, R., Levasseur, A., Henrissat, B., et al. (2019). Trichoderma reesei Dehydrogenase, a Pyrroloquinoline Quinone-Dependent Member of Auxiliary Activity Family 12 of the carbohydrate-active enzymes database: functional and structural characterization. Appl. Environ. Microbiol. 85:e00964-19. doi: 10.1128/AEM.00964-19
Tyler, B., Tripathy, S., Zhang, X., Dehal, P., Jiang, R., Aerts, A., et al. (2006). Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 313, 1261–1266. doi: 10.1126/science.1128796
van Wyk, N., Drancourt, M., Henrissat, B., and Kremer, L. (2017). Current perspectives on the families of glycoside hydrolases of Mycobacterium tuberculosis: their importance and prospects for assigning function to unknowns. Glycobiology 27, 112–122. doi: 10.1093/glycob/cww099
Vanden Wymelenberg, A., Minges, P., Sabat, G., Martinez, D., Aerts, A., Salamov, A., et al. (2006). Computational analysis of the Phanerochaete chrysosporium v2.0 genome database and mass spectrometry identification of peptides in ligninolytic cultures reveal complex mixtures of secreted proteins. Fungal. Genet. Biol. 43, 343–356. doi: 10.1016/j.fgb.2006.01.003
Vocadlo, D. J., and Davies, G. J. (2008). Mechanistic insights into glycosidase chemistry. Curr. Opin. Chem. Biol. 12, 539–555. doi: 10.1016/j.cbpa.2008.05.010
Vorwerk, S., Somerville, S., and Somerville, C. (2004). The role of plant cell wall polysaccharide composition in disease resistance. Trends Plant. Sci. 9, 203–209. doi: 10.1016/j.tplants.2004.02.005
Wang, Q., Han, C., Ferreira, A. O., Yu, X., Ye, W., Tripathy, S., et al. (2011). Transcriptional programming and functional interactions within the Phytophthora sojae RXLR effector repertoire. Plant Cell 23, 2064–2086. doi: 10.1105/tpc.111.086082
Wang, Y., Tyler, B. M., and Wang, Y. (2019). Defense and counterdefense during plant-pathogenic oomycete infection. Annu. Rev. of Microbiol. 73, 667–696. doi: 10.1146/annurev-micro-020518-120022
Wang, Y., and Wang, Y. (2018). Trick or treat: microbial pathogens evolved apoplastic effectors modulating plant susceptibility to infection. Mol. Plant. Microbe. Interact. 31, 6–12. doi: 10.1094/MPMI-07-17-0177-FI
Wood, T. M. (1985). Properties of cellulolytic enzyme systems. Biochem. Soc. Trans. 13, 407–410. doi: 10.1042/bst0130407
Yang, B., Wang, Q., Jing, M., Guo, B., Wu, J., Wang, H., et al. (2017). Distinct regions of the Phytophthora essential effector Avh238 determine its function in cell death activation and plant immunity suppression. New. Phytol. 214, 361–375. doi: 10.1111/nph.14430
Ye, W., Wang, X., Tao, K., Lu, Y., Dai, T., Dong, S., et al. (2011). Digital gene expression profiling of the Phytophthora sojae transcriptome. Mol. Plant. Microbe. Interact. 24, 1530–1539. doi: 10.1094/MPMI-05-11-0106
Keywords: Phytophthora sojae, glycoside hydrolase 7, virulence, soybean, cellobiohydrolase
Citation: Tan X, Hu Y, Jia Y, Hou X, Xu Q, Han C and Wang Q (2020) A Conserved Glycoside Hydrolase Family 7 Cellobiohydrolase PsGH7a of Phytophthora sojae Is Required for Full Virulence on Soybean. Front. Microbiol. 11:1285. doi: 10.3389/fmicb.2020.01285
Received: 27 February 2020; Accepted: 20 May 2020;
Published: 02 July 2020.
Edited by:
Meixiang Zhang, Nanjing Agricultural University, ChinaReviewed by:
Jianqiang Miao, China Agricultural University, ChinaYuling Meng, Northwest A&F University, China
Copyright © 2020 Tan, Hu, Jia, Hou, Xu, Han and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chao Han, aGFuY2g4N0AxNjMuY29t; Qunqing Wang, d2FuZ3F1bnFpbmdAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Xinwei Tan
Xinwei Tan Yuyao Hu
Yuyao Hu Yuli Jia
Yuli Jia Xiaoyuan Hou
Xiaoyuan Hou Qian Xu
Qian Xu Chao Han
Chao Han Qunqing Wang
Qunqing Wang