- 1Guangdong Province Key Laboratory of Microbial Signals and Disease Control, Integrative Microbiology Research Centre, South China Agricultural University, Guangzhou, China
- 2Guangdong Laboratory for Lingnan Modern Agriculture, South China Agricultural University, Guangzhou, China
The ubiquitous second messenger c-di-GMP is involved in regulation of multiple biological functions including the important extracellular matrix exopolysaccharides (EPS). But how c-di-GMP metabolic proteins influence EPS and their enzymatic properties are not fully understood. Here we showed that deletion of proE, which encodes a protein with GGDEF-EAL hybrid domains, significantly increased the transcriptional expression of the genes encoding EPS production in Pseudomonas aeruginosa PAO1 and changed the bacterial colony morphology. Our data showed that ProE is a very active phosphodiesterase (PDE), with a high enzyme activity in degradation of c-di-GMP. Interestingly, the optimal activity of ProE was found in the presence of Co2+, unlike other PDEs that commonly rely on Mg2+ or Mn2+ for best performance. Furthermore, we identified three widely conserved novel residues that are critical for the function of ProE through site-directed mutagenesis. Subsequent study showed that ProE, together with other three key PDEs, i.e., RbdA, BifA, and DipA regulate the EPS production in P. aeruginosa PAO1. Moreover, by using the GFP-fusion approach, we observed that these four EPS associated-PDEs showed a polar localization pattern in general. Taken together, our data unveil the molecular mechanisms of ProE in regulation of EPS production, and provide a new insight on its enzymatic properties in degradation of c-di-GMP.
Introduction
The cyclic dinucleotide (c-di-GMP) is a conserved second messenger in many bacteria species (Romling et al., 2013). It plays important roles in regulation of biofilm formation, motility, virulence, development, and cell cycle progression (Jenal and Malone, 2006; Hengge, 2009; Jenal et al., 2017). The synthesis and degradation of cyclic di-GMP is controlled by two classes of enzymes, i.e., the GGDEF domain containing diguanylate cyclase (DGC), EAL or HD-GYP domain containing phosphodiesterase (PDE). C-di-GMP acts by binding to specific effectors, including PilZ containing protein (Xu et al., 2016), inactive GGDEF or EAL domains (Navarro et al., 2009; Whitney et al., 2012), riboswitches (Sudarsan et al., 2008) and transcription factors (Tao et al., 2010; Baraquet et al., 2012), to regulate downstream genes associated with different biological functions. Among the products regulated by c-di-GMP, EPS are the key component of extracellular matrix involved in surface adhesion, cell-cell interactions and biofilm formation (Bazaka et al., 2011; Koo et al., 2013; Matthysse, 2014), and are regulated by c-di-GMP binding proteins such as Alg44, PelD, and BcsA (Lee et al., 2007; Merighi et al., 2007; Morgan et al., 2014).
The opportunistic human pathogen Pseudomonas aeruginosa can cause severe infections in cystic fibrosis patients and immunocompromised individuals (Lee and Zhang, 2015; Ahator and Zhang, 2019). The pathogen produces at least three polysaccharides, including alginate, Psl, and Pel (Ryder et al., 2007). Among them, alginate is the predominant extracellular polysaccharide of the extracellular matrix in mucoid strains (Hentzer et al., 2001), while Pel and Psl polysaccharides were primarily utilized for biofilm formation by non-mucoid strains (Wozniak et al., 2003). P. aeruginosa PAO1 has two operon which can synthesis Pel and Psl, respectively (Friedman and Kolter, 2004). The production of Pel and Psl are regulated by transcription factors such as FleQ and AmrZ (Hickman and Harwood, 2008; Jones et al., 2014), and DGCs or PDEs (Liang, 2015).
Pseudomonas aeruginosa PAO1 encodes a total of forty-one proteins which are involved in the metabolism of c-di-GMP (Kulasakara et al., 2006). During the last decade, more than half of c-di-GMP metabolic proteins have been functionally characterized (Hickman et al., 2005; Hoffman et al., 2005; Kulasekara et al., 2005; Merritt et al., 2007; Roy et al., 2012; Basu Roy and Sauer, 2014). Previous studies showed that Pel was regulated by DGCs WspR, YfiN, SadC, and RoeA (Guvener and Harwood, 2007; Merritt et al., 2007, 2010; Malone et al., 2010, 2012), and by PDEs BifA and RbdA (Kuchma et al., 2007; An et al., 2010). Interestingly, Psl can act as a signal to activate SiaD and SadC, two DGCs, thus increasing the production of Psl and other components of the biofilm in P. aeruginosa PAO1 (Irie et al., 2012). A comprehensive study showed that more than half of these c-di-GMP metabolic proteins can influence the EPS production in PA14 (Ha et al., 2014).
However, the enzymatic properties and underlying mechanism with which c-di-GMP metabolic proteins modulate EPS production remain to be further studied. In this study, we identified the gene PA5295, designated as proE for its role as an important phosphodiesterase regulator of EPS. The proE gene encodes a dual-domain protein consisting of GGDEF-EAL domains. Genetic and biochemical analyses revealed the role of proE as a highly active PDE, which negatively regulates EPS production in strain PAO1. Three novel conserved residues of ProE were identified to play key roles in c-di-GMP metabolism. Our in vitro analysis shown that purified ProE was more active in degradation of c-di-GMP than the previously characterized highly active PDE RocR (Kulasakara et al., 2006; Rao et al., 2008; Chen et al., 2012). Furthermore, we provided evidence that ProE with other three PDEs together controlled the EPS production in P. aeruginosa. Our subcellular localization analysis indicated that these EPS-associated proteins were more or less localized in cell poles.
Materials and Methods
Bacterial Strains, Plasmids, Media and Growth Conditions
Bacterial strains and plasmids used in this study are listed in Supplementary Table S1. Bacteria were routinely maintained at 37°C in Lysogenic Broth (LB). Antibiotics at the following concentrations were added when necessary: gentamicin, 50 μg/ml for P. aeruginosa; and gentamicin, 25 μg/ml; kanamycin, 50 μg/ml for Escherichia coli.
Construction of In-Frame Deletion Mutants and Complementation
The plasmids and primers used in this study are listed in Supplementary Tables S1, S2, in the Supplementary Material, respectively. To generate the proE deletion mutant of P. aeruginosa, two PCR fragments flanking proE were amplified. After purification with NucleoSpin Gel and PCR Clean-up kit (Macherey Nagel), the two flanking fragments were ligated with the linear vector pK18 (digested with EcoRI and BamHI) by One Step Cloning Kit (Vazyme Biotech, Nanjing, China). The resultant construct was transformed into E. coli DH5α competent cells by heat shock at 42°C and introduced into strain PAO1 through triparental mating. In frame-deletion was performed as described previously (An et al., 2010). The generated proE deletion mutant was confirmed by PCR and DNA sequencing. Single-deletion mutants ΔproE, ΔfleQ, ΔrbdA, ΔbifA, ΔdipA, ΔpelA, ΔpslA, and ΔPA5294, double-deletion mutants ΔproEΔpelA, ΔproEΔpslA, ΔfleQΔpelA, ΔfleQΔpslA, and ΔfleQΔproE, and the triple-deletion mutants ΔproEΔpelAΔpslA, ΔfleQΔpelAΔpslA were generated by the same procedure using corresponding primers (Supplementary Table S2).
For in trans complementation, the coding region with native promoter was amplified by the primers listed in Supplementary Table S2, the PCR products were cloned into the plasmid pBBR1-MCS5 digested by EcoRI and BamHI. The resultant construct was mobilized into E. coli DH5α and sequenced before introducing it into the corresponding mutants by tri-parental mating and then confirmed by PCR analysis.
Colony Morphology Assay
Overnight cultures were diluted 1/1000 in ddH2O. One microliter of cells were spotted onto T-agar plates (10 g/l tryptone, 1% agar) supplemented with 40 mg/ml Congo red and 15 mg/ml Coomassie brilliant blue R-250 (Sigma-Aldrich, United States) and incubated at 25°C for 2 days prior to observation and taking photographs.
RNA Extraction and Quantitative Real-Time PCR (qRT-PCR)
Bacteria were grown in LB medium and harvested at the mid-exponential phase (OD600 about 0.5), and RNA samples were prepared using the RNAprep Pure Cell/Bacteria Kit (TIANGEN, Beijing, China), following the manufacturer’s protocol. The integrity and purity of RNA was determined by agarose gel electrophoresis and the concentration was measured by NanoDrop 2000C (Thermo Fisher Scientific, Waltham, MA, United States). The first-strand cDNA was reversely transcribed by using the FastKing RT kit (with gDNase) (Tiangen Biotech, Co., Ltd., Beijing, China) with 1.5 μg RNA. Quantitative real-time PCR (qRT-PCR) was performed on a QuantStudio 6 Flex Real-Time PCR System using PowerUp SYBR green master mix (Applied Biosystems, United States) with the following PCR procedure: 50°C for 2 min, 95°C for 2 min, (95°C, 15 s; 60°C, 1 min) × 40 cycles. The experiment was repeated three times, each time with triplicates. All the primers are listed in Supplementary Table S2. The relative expression levels of the target genes were normalized to the housekeeping gene rplU and the gene expression level was calculated by using 2–ΔΔCT method (Kuchma et al., 2007).
Protein Cloning, Expression, and Purification
The DNA fragments encoding proE, rocR, and wspR were amplified with the primers listed in Supplementary Table S2 and cloned into the expression vector pET28b (+) (Novagen) between the BamHI and HindIII restriction sites. For protein expression, 10 ml overnight culture of the expression strains of ProE, RocR, and WspR were added to 1 L of LB medium, respectively. The bacterial culture was grown at 37°C until it reached an OD600 about 0.5 before addition of 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG) at 18°C overnight. The cell pellet was resuspended in 25 ml lysis buffer, which consists of 50 mM Tris–HCl (pH 8.0), 200 mM NaCl. The cells were then lysed by sonication, after centrifugation at 12,000 rpm for 1 h, the supernatant was filtered by 0.45 μm filter membranes (Pall Corporation, United States) and then incubated with 5 ml of Ni2+-nitrilotriacetic acid resin (Clontech, Japan) for 2 h on ice. The resin was washed with 50 ml of washing buffer (lysis buffer with 20 mM imidazole). The proteins were eluted using a stepped gradient method with the elution buffer containing 50 mM Tris–HCl (pH 8.0), 200 mM NaCl, and 50 mM, 100 mM, 200 mM, 300 mM, or 400 mM imidazole. After sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis, fractions with purity higher than 95% were pooled together and desalted using a HiPrep 26/10 Desalting column (GE Healthcare, United States). Proteins were concentrated using an Amicon concentrator (Merck Millipore, Germany) and flash-frozen in liquid nitrogen, then the protein samples were stored at -80°C prior to use.
Enzymatic Activity Assay
Enzyme activity analyses were performed following the methods described previously, with minor modifications (An et al., 2010). For PDE activity assay, 0.08 μM ProE or RocR were added to reaction buffer containing 50 μM c-di-GMP, 100 mM Tris–HCl (pH 8.0), 20 mM KCl, 5 mM MgCl2, in a final volume of 50 μl. The reaction mixture was incubated at 37°C for 20 min, then stopped by adding 1/10 volume of 1 M CaCl2 and heating at 95°C for 10 min. The PDE activity was assessed by monitoring the formation of the product 5′-pGpG from the hydrolysis of c-di-GMP. To test the effect of GTP, a final concentration of 50 μM GTP was added into the reaction mixture and incubated for 10 min at room temperature prior to reaction, then the enzyme reaction was performed as described above.
For DGC activity assay, 500 μM GTP and 5 μM ProE or WspR were added to reaction buffer containing 75 mM Tris–HCl (pH 7.8), 250 mM NaCl, 25 mM KCl, 10 mM MgCl2 in a final volume of 50 μl. The reaction mixture was incubated at 37°C for 120 min. High-performance liquid chromatography (HPLC) analyses were performed by running the samples through a reverse-phase C18 column (YMC-Pack ODS-A, 250 × 4.6 mm, 5 μm) on a HPLC chromatographic system (Agilent 1260 Infinity, United States) at an injection volume of 10 μl, using the solvents and elution gradient as previous described (Chua et al., 2015), at a flow rate of 1.0 ml/min and with a detection wavelength of 254 nm.
Site-Directed Mutagenesis
The constructs pBBR1-MCS5-proE and pET28b-proE were used as templates for site-directed mutagenesis. PCR was carried out by using Phanta Max Super-Fidelity DNA Polymerase (Vazyme Biotech, Nanjing, China) and relevant mutagenic PCR primers (Supplementary Table S2). The PCR products were treated with DpnI to digest the methylated and hemimethylated DNA, then ligated by One Step Cloning Kit (Vazyme Biotech, Nanjing, China) prior to transformation of E. coli strain DH5α. The resultant mutations were confirmed by PCR and DNA sequencing analysis.
Fluorescence Microscopy
For microscopy observation, the strains contain fluorescent fusion (s) were grown overnight on LB plates with gentamicin (50 μg/ml), then using sterile toothpicks to pick single colony into 10 μl PBS and 1 μl culture was spotted on a slide which was coated with 1% agarose. Phase contrast and fluorescence microscopy were performed by using Observer Z1 equipped with sCMOS camera (Zeiss, Germany).
Bacterial Two-Hybrid Assays
Protein-protein interaction was detected by using the BacterioMatch II two-hybrid system (Stratagene) following the manufacturer’s protocol. Bait and prey plasmids harboring pBT-proE and pTRG-proE were co-transformed into the reporter strain XL1-Blue MRF’ Kan by electroporation. The co-transformed cells were grown on M9+His-deficient medium containing 5 mM 3-AT for 24–72 h at 30°C. Colonies that grew on these plates were selected as positive colonies. Then the positive colonies were subsequently picked and re-streaked on M9+ His-deficient plate containing 5 mM 3-AT and 12.5 mg/ml streptomycin. Normal growth on the selective screening medium indicates a positive protein-protein interaction.
3D Homology of ProE
The amino acid sequence for ProE (PA5295) was obtained from the P. aeruginosa database. Generation of the 3D homology for ProE was performed using the SWISS-MODEL workspace (Waterhouse et al., 2018). Three templates 4Y9M (PA3825-EAL-Apo), 4Y9P (PA3825-EAL-Ca2+, c-di-GMP), and 5M3C (PA0575-GGDEF-EAL-Ca2+,GTP) were selected to generate the 3D models of ProE. The molecular visualization, model analysis and image design of the ProE models, and the interactions between the wild type and mutant residues were performed using PyMOL (V. 2.3.5).
Results
Mutation of proE Results in Wrinkly Colony Morphology in P. aeruginosa PAO1
In the process of construction of Tn5 mutant library of P. aeruginosa strain PAO1, we found that mutation of PA5295, which encodes a hypothetical protein containing a GGDEF domain and an EAL domain (Figure 1A), resulted in wrinkly colonies. A previous study by Ha et al. (2014) showed that deletion of PA5295 resulted in increased EPS production and decreased motility in P. aeruginosa strain PA14, but how PA5295 influences these phenotype changes was not determined. In addition, strains PAO1 and PA14 differ in the genes encoding EPS production, with the former containing the psl and pel gene clusters and the later containing only the pel gene cluster (Kuchma et al., 2007), suggesting that the scope and impact of PA5295 in these two strains might not be the same.
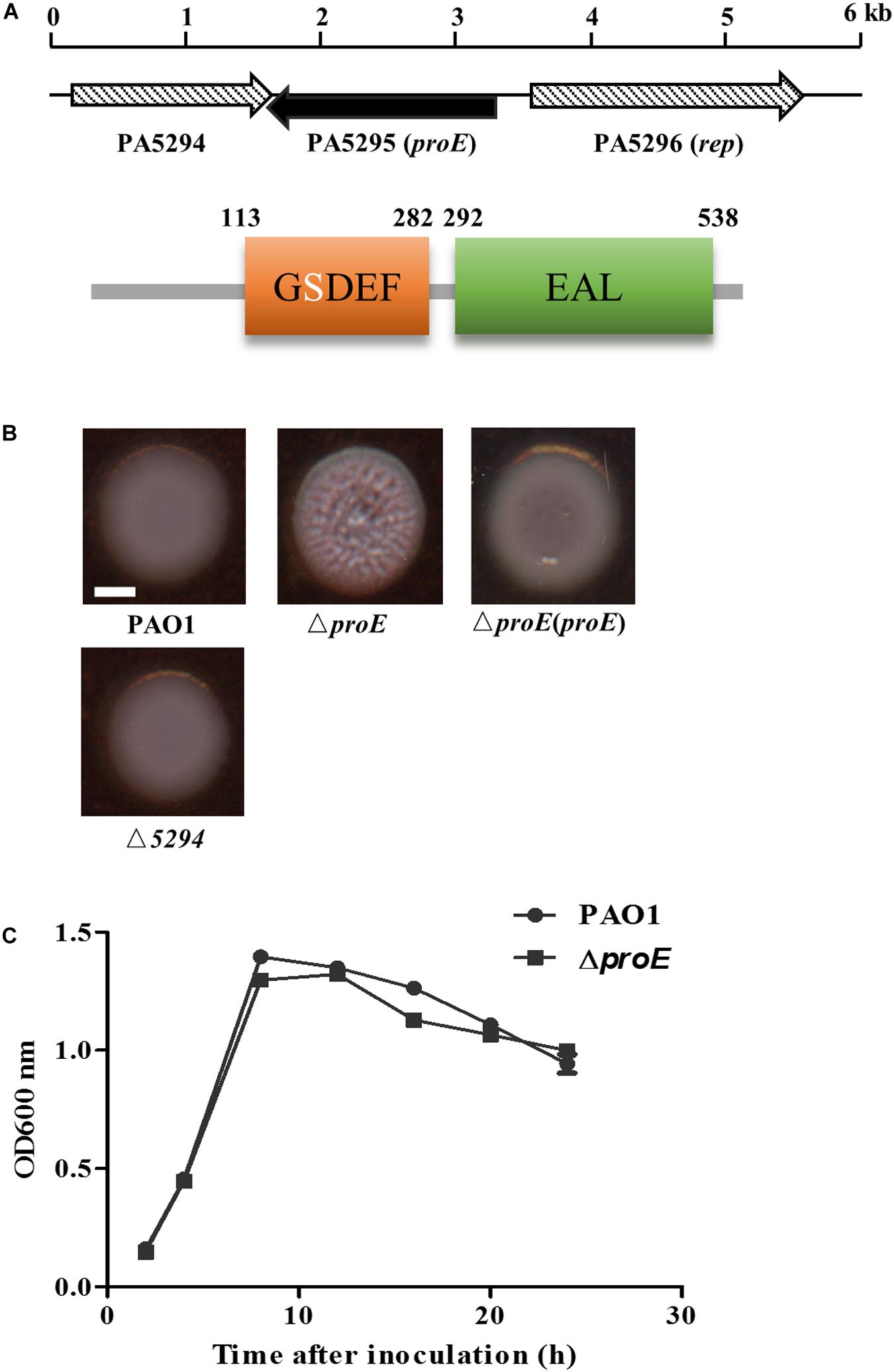
Figure 1. The proE mutant showed a wrinkly colony morphology. (A) Genetic organization and domain structures of ProE, and the active site of the GGDEF domain is degenerate. (B) Mutation of proE affected the colony morphology on Congo-Red plate after growth on 25°C for 2 days, nor does mutation of PA5294 have the similar phenotype. Scale bar = 2 mm. (C) Growth curves of PAO1 and ΔproE. The data are means of three replicates and error bars indicate standard deviation.
To explore the function of PA5295 in P. aeruginosa PAO1, we generated an in-frame deletion mutant of PA5295. The deletion mutant didn’t show growth defect compared with the wild type strain PAO1 (Figure 1B). Then we tested its phenotypes including motility, biofilm formation, and EPS production. We found that the PA5295 null mutant formed colonies showing wrinkled appearance, which was different to the colony morphology of the wild-type strain PAO1 on tryptone agar plates (Figure 1B). For verification, the wild type proE was cloned into the expression plasmid pBBR1-MCS5, the resulting construct was introduced into ΔproE, which fully restored the colony morphology of ΔproE to the level of wild type strain PAO1 (Figure 1B).
Deletion of proE did not affect biofilm formation (Supplementary Figure S1), which was consistent with the findings of the previous studies (Kulasakara et al., 2006; Ha et al., 2014). However, unlike in strain PA14 (Ha et al., 2014), the proE null mutation did not affect bacterial motility in strain PAO1 (Supplementary Figure S1), indicating the regulatory divergence in different P. aeruginosa strains. Given its role in regulation of the EPS gene expression in P. aeruginosa PAO1, as described below, the PA5295 was designated as proE.
Genome organization analysis showed that the upstream gene of proE is PA5296 known as rep, encoding an ATP-dependent DNA helicase, and its downstream gene is PA5294, which encodes a putative multidrug efflux and H + -coupled pump protein belonging to the MATE family (He et al., 2004). We noticed that the ORF of PA5294 has an 89 bp overlap with proE in opposite transcriptional directions. To determine whether proE and PA5294 are functional associated, we constructed an PA5294 null mutant and assayed its morphology on a Congo-Red plate, the result showed that mutation of PA5294 did not result in obvious morphology changes compared with the wild type PAO1 (Figure 1B).
ProE Negatively Regulates the Transcriptional Expression of pel and psl
Previous studies have shown that the wrinkly colony was mainly caused by increased production of Pel or Psl EPS, such as deletion of wspF increases the pel and psl transcription thus causing formation of wrinkly colonies (Hickman et al., 2005). In P. aeruginosa, the expression of pel and psl were transcriptionally regulated by the c-di-GMP binding protein FleQ, which is also the master regulator of flagella gene expression (Hickman and Harwood, 2008; Baraquet et al., 2012; Baraquet and Harwood, 2016). To use FleQ as a control, we constructed a fleQ null mutant, and compared the colony morphology of the mutants ΔfleQ and ΔproE. The results showed that similar to ΔproE, ΔfleQ also generated wrinkly colonies, agreeable with previous report (Hickman and Harwood, 2008). Then we generated deletions of pelA and pslA in the ΔproE, ΔfleQ and wild type background, the results showed that deletion of pelA in the ΔproE or ΔfleQ background significantly decreased its ability to bind to Congo-Red with relatively white colonies but wrinkly morphology remained (Figure 2A). In contrast, deletion of pslA in the ΔproE or ΔfleQ background did not affect Congo-Red binding but substantially reduced the wrinkly colony morphology, while it still remained partial wrinkly at edge of the colony in the ΔproE background (Figure 2A), and double deletion of pelA and pslA in ΔproE or ΔfleQ background fully restored the colony morphology similar to that of wild type PAO1 (Figure 2A), which clearly established the link between the wrinkly colony morphology and the increased expression of pel and psl genes in the proE deletion mutant. We also generated the proE and fleQ double mutant, curiously, which produced even smaller, rough and red colony appearance than the corresponding single deletion mutants (Figure 2A), suggesting a synogentics effect of two regulators on the colony morphology of P. aeruginosa. Consistent with this notion, we found that deletion of proE did not affect the transcriptional expression of fleQ and vice versa (Supplementary Figure S2).
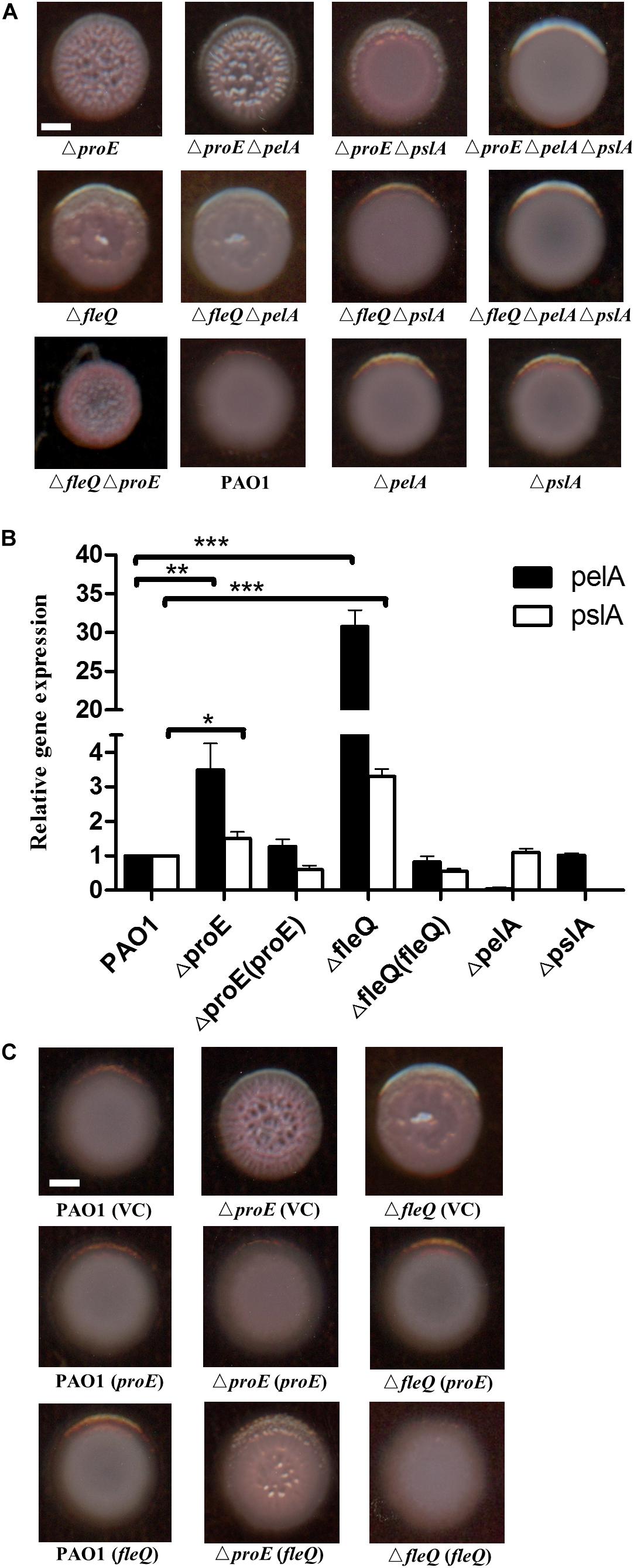
Figure 2. ProE regulated EPS production was dependent on pel and psl. (A) The wrinkly colony is only abolished when both pelA and pslA are mutated in the ΔfleQ background, while a pslA mutation is not sufficient to confer smooth colony morphology in the ΔproE background. All strains were growth on Congo-Red plates. (B) The relative gene expression of pelA and pslA in PAO1, ΔproE, ΔfleQ, Δ proE (proE),ΔfleQ (fleQ),ΔpelA, and ΔpslA by qRT-PCR analysis. The data are means of three replicates and error bars indicate standard deviation. *P < 0.05; **P < 0.01; ***P < 0.0001. (C) Complementation of proE can restore the wrinkly colony phenotype of ΔfleQ, and complementation of fleQ can also substantially alleviate the wrinkly colony phenotype of ΔproE. Scale bar = 2 mm.
To confirm the findings described above, the transcript levels of pelA and pslA in different backgrounds were determined at an optical density at 600 nm (OD600) about 0.5. The quantitative real-time polymerase chain reaction (qRT-PCR) results showed that the transcriptional expression of pelA was increased by 3.5 and 30.8 fold in ΔproE and ΔfleQ, respectively, compare with the wild type (Figure 2B). The expression of pslA was also increased 1.5 and 3.3 fold in ΔproE and ΔfleQ, respectively (Figure 2B). The transcript levels of pelA and pslA were higher in ΔfleQ than in ΔproE, agreeable with the role of FleQ as the downstream regulator of EPS production in P. aeruginosa. We also determined the gene expression of pelA and pslA in the complemented strains ΔproE (proE) and ΔfleQ (fleQ), the result showed that transcriptional expression of pelA and pslA was decreased to the wild type level, respectively (Figure 2B).
To further explore whether proE and fleQ could functionally replace with each other, we introduced the expression constructs pBBR1-MCS5-proE and pBBR1-MCS5-fleQ into ΔfleQ, ΔproE and wild type PAO1, respectively, using empty vector as a control. The results showed that empty vector or overexpression constructs didn’t influence the colony morphology (Figure 2C). It appeared rational that in trans expression of fleQ, which encodes a c-di-GMP effector, could alleviate the wrinkly colony morphology of ΔproE (Figure 2C). Intriguingly, however, the wrinkly colony of ΔfleQ was fully rescued by complementation with proE (Figure 2C). Given the finding that EPS production was de-repressed after FleQ binds to c-di-GMP (Hickman and Harwood, 2008; Baraquet et al., 2012; Baraquet and Harwood, 2016), it seemed not logical that in trans expression of proE, which would decrease intracellular c-di-GMP level, could still result in reduced EPS production in ΔfleQ. One plausible explanation is that in addition to FleQ, there is another c-di-GMP-dependent repressor that could inhibit EPS production. Similar to FleQ, whose function in suppression of EPS production is also inactivated by c-di-GMP. Taken together, the above data showed that ProE negatively regulated EPS production by downregulating the transcriptional expression of pel and psl.
Sequence Alignment of the GGDEF Domain and EAL Domain of ProE
ProE is a hybrid protein with a GGDEF domain (113–282 aa) and an EAL domain (292–538 aa), which are known to be involved in c-di-GMP biosynthesis and degradation, respectively. To understand whether these two domains are well conserved, we firstly compared the amino acid sequence of GGDEF domain of ProE with the functionally characterized DGCs. We found that the ProE contains most conserved residues, however, three key residues, including c-di-GMP binding site (E192), GG(D/E)EF signature motif (S202), and GTP binding site (R276) are mutated (Supplementary Figure S3), suggesting ProE is likely to be a degenerated DGC. Then we compared the amino acid sequence of EAL domain of ProE with other functional characterized PDEs. Previous studies found that several key residues are critical for the EAL domain to perform its PDE activity (Rao et al., 2008, 2009; Tchigvintsev et al., 2010; Yang et al., 2017). We thus asked whether these key residues are also conserved in the EAL domain of ProE, we found that the most essential residues for PDE structure and activity are conserved, except that the conserved loop 6 [DDFG(A/T)GYSS] has a mutation with Y455 being changed to F455 (449DDFGTGFSS457) (Figure 3). Phylogenetic tree analysis showed that ProE was quite conserved within the Pseudomonas genus (Supplementary Figure S4). And based on our bioinformatic analysis by using the SMART program1 and Pseudomonas website2, ProE lacks N-terminal transmembrane or signal sensing domain, which suggests that ProE is likely to be a soluble cytoplasmic protein, and may not directly interact with signal(s).
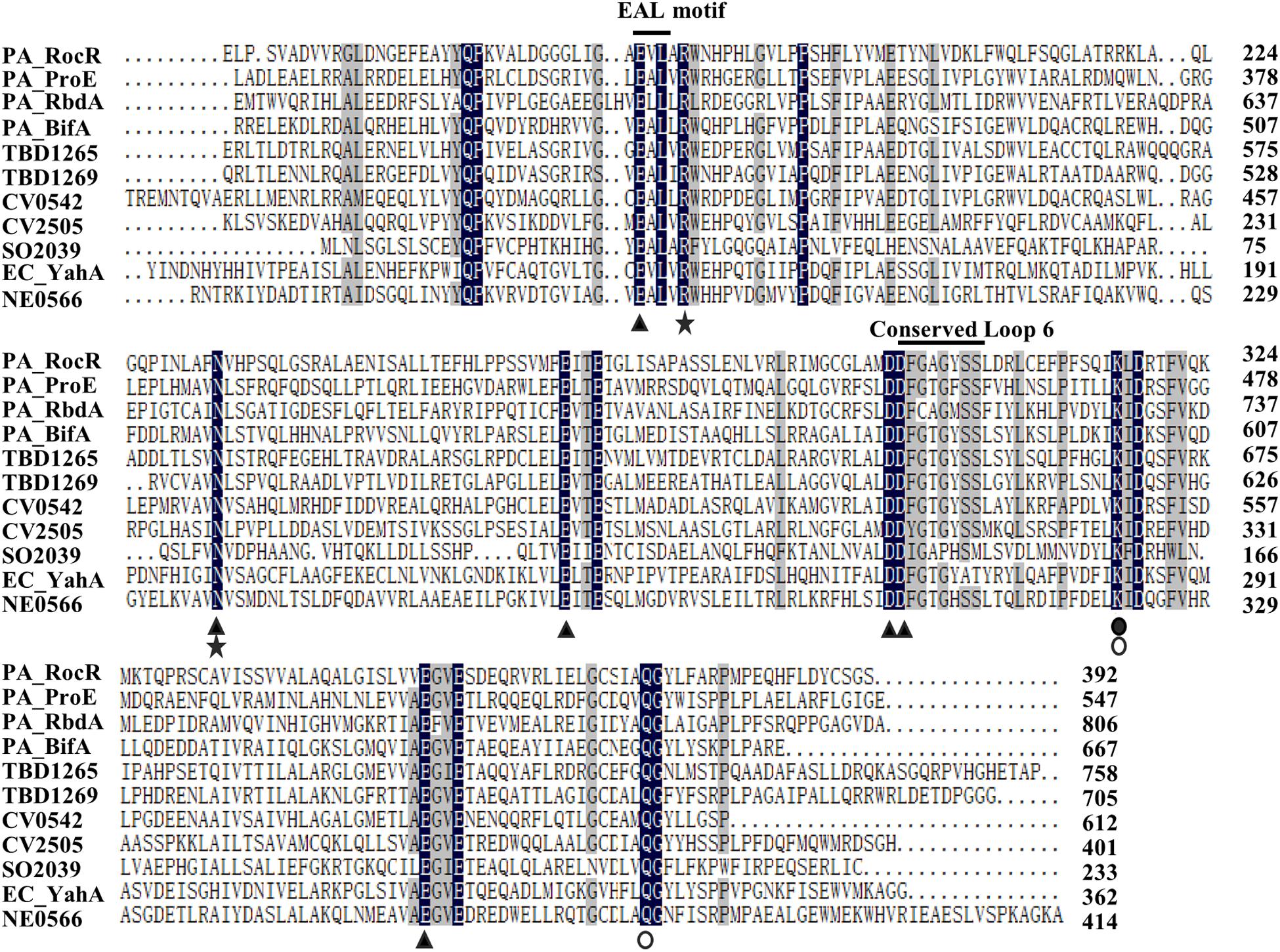
Figure 3. Sequence alignment of EAL-containing phosphodiesterases. The amino acid sequence of RocR (Q9HX69), ProE (Q9HTQ9), RbdA (Q9I580), BifA (Q9HW35) from P. aeruginosa, TBD1265 (Q3SJE6), TBD1269 (Q3SJE2) from Thiobacillus denitrificans, CV0542 (Q7P0M4), CV2505 (Q7NV41) from Chromobacterium violaceum, SO2039 (Q8EFE2) from Shewanella oneidensis, YahA (P21514) from Escherichia coli, NE0566 (Q82WU5) from Nitrosomonas europaea. The amino acids highlighted with black stands for 100% similarity, and gray indicates similarity level ≥75%. E328, N387, E419, D449, D450, E506 (filled triangle) are conserved residues which may bind to metal ion (Tamayo et al., 2005; Rao et al., 2008; Tchigvintsev et al., 2010; Yang et al., 2017). The conserved loop 6 (DFG(T/A)GYSS) is essential for the dimerization of EAL domain and Mg2+, c-di-GMP binding (Rao et al., 2009). R332, N387 are (asterisks) reported to bind with substrate (Rao et al., 2008; Tchigvintsev et al., 2010; Yang et al., 2017). K470 (filled circle) is essential for coordinating with water molecule for catalysis and interacting with residue E419 residue (Rao et al., 2008; Tchigvintsev et al., 2010; Yang et al., 2017). Q526 is reported to interact with E328 and K470 (open circle) (Tchigvintsev et al., 2010; Yang et al., 2017).
ProE Is an Active Phosphodiesterase
To determine the enzyme activity of ProE, we expressed and purified the recombinant ProE, we also purified the previously functionally characterized DGC WspR and PDE RocR as positive controls (Hickman et al., 2005; Rao et al., 2008; Supplementary Figure S5). To test DGC activity of ProE, we incubated ProE with GTP for 2 h, then analyzed the products in the reaction mixture by HPLC. The result showed that while the control protein WspR could convert most GTP into c-di-GMP, ProE could not synthesis c-di-GMP (Supplementary Figure S6), suggesting that in agreement with sequence alignment analysis, ProE is not a functional DGC.
Then we tested the PDE activity of ProE, after incubation with c-di-GMP at 37°C for 20 min, the reaction was stopped. HPLC analysis showed that the degradation product pGpG was produced with a retention time at 3.7 min, indicating that ProE is an active PDE (Figure 4D). Interestingly, we noticed that ProE was more active than RocR (Figures 4B,D). Quantitative analysis showed that the enzyme activity of ProE was about 4 folds higher than RocR (Figure 4G), which was shown to be a highly active PDE (Kulasakara et al., 2006).
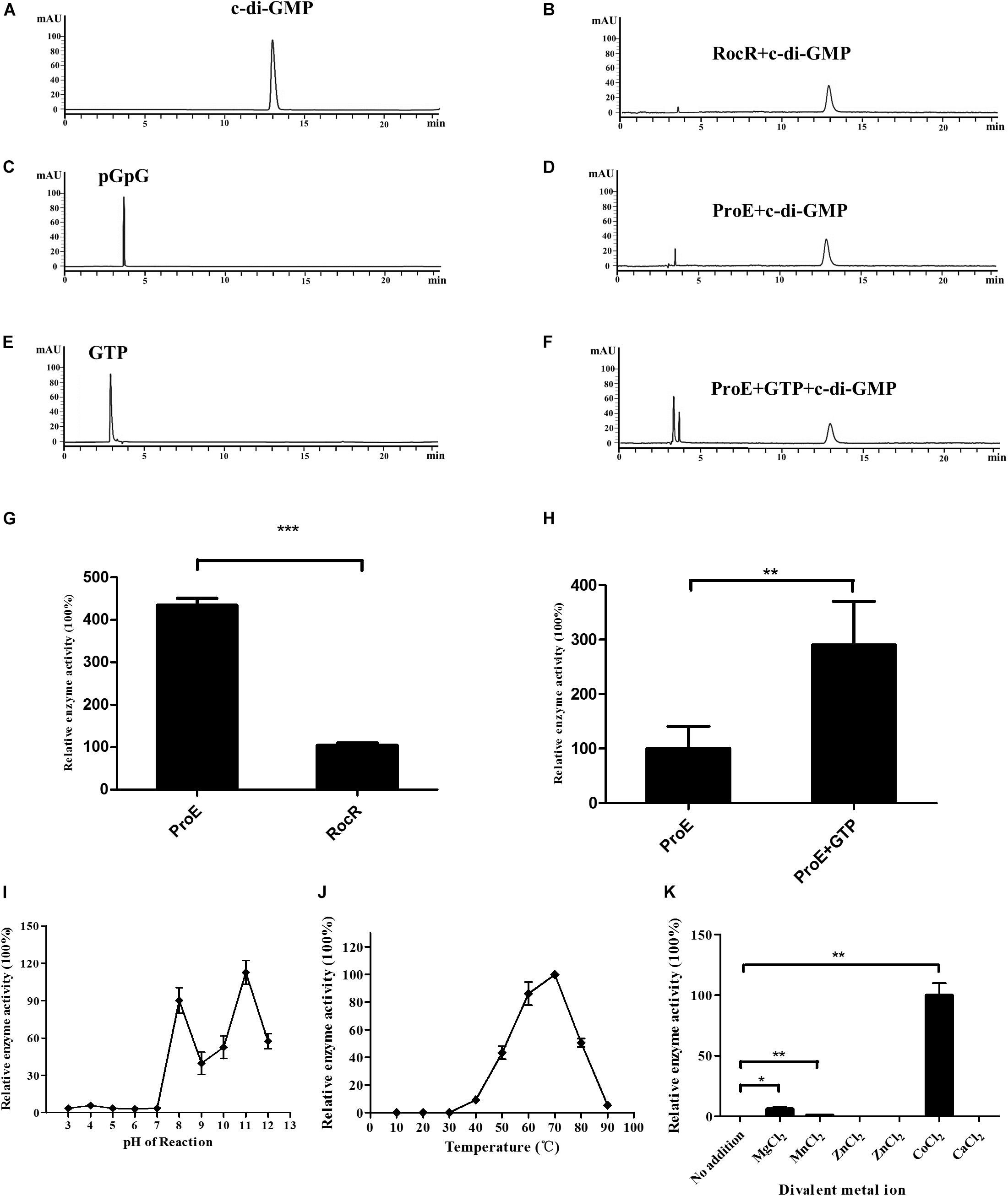
Figure 4. Purified ProE is an active phosphodiesterase. (A,C,E) The standard of c-di-GMP (A), pGpG (C), GTP (E) in a final concentration of 100 μM was prepared in the reaction buffer, and 10 μl was injected for HPLC analysis. (B) RocR and (D) ProE after incubation with c-di-GMP at a final concentration of 50 μM at 37°C for 20 min. (F) ProE incubated with 50 μM GTP for 5 min at room temperature prior to addition of c-di-GMP, then the enzyme activity was analysis by HPLC. (G) Quantification of the enzyme activity of ProE and RocR. (H) Quantification of the enzyme activity of ProE and ProE with GTP prior to addition substrate. (I) The influence of pH on enzyme activity. The pH of the phosphodiesterase reaction was adjusted to 3–12 prior to the addition of enzyme and substrate (37°C, 5 mM MgCl2). ProE activity was higher in alkaline condition. (J) The influence of temperature on enzyme activity. The ProE phosphodiesterase reaction was performed at a range of temperatures from 10°C to 90°C (pH 8.0, 5 mM MgCl2). The highest rate of c-di-GMP hydrolysis was achieved at 60°C and 70°C. At temperatures lower than 30°C, no activity was detectable. (K) Dependence of the phosphodiesterase activity of ProE on divalent metal cations. Reaction buffer (37°C, pH 8.0) The data are means of three replicates and error bars indicate standard deviation. *P < 0.05; **P < 0.01, ***P < 0.0001.
It was reported that GTP can allosterically control PDE activity (Christen et al., 2005). We then measured the PDE activity of ProE in the presence or absence of GTP under the same reaction conditions. Similar to other GGDEF-EAL fusion domain proteins (Christen et al., 2005; An et al., 2010; Bellini et al., 2017; Mantoni et al., 2018), the PDE activity of ProE was greatly enhanced by about 2.9 folds after supplementation with 50 μM GTP in the reaction mix prior to addition of c-di-GMP (Figures 4D,F,H).
Previous studies showed that several PDEs are active homodimers (Barends et al., 2009; Minasov et al., 2009; Robert-Paganin et al., 2012; Sundriyal et al., 2014; Bellini et al., 2017; Mantoni et al., 2018), whereas RocR has a unusual tetrameric structure (Chen et al., 2012),the dimerization of EAL domain is critical for the function PDEs (Rao et al., 2009; Bellini et al., 2017; Mantoni et al., 2018). And the GGDEF domain also plays important role as a scaffold to ensure such dimerization (Mantoni et al., 2018). By using bacterial two-hybrid approach, we found that ProE could interact with ProE (Supplementary Figure S7), implying that ProE is also likely to exist as an oligomer.
To test the substrate specificity of ProE, we added other three nucleotide second messengers including cAMP, cGMP, and c-di-AMP into the reaction mix and incubated at 37°C for 20 min prior to HPLC analysis. The results showed that ProE could only degrade c-di-GMP, but not cAMP, cGMP, or c-di-AMP (Supplementary Figure S8), indicating that ProE is a c-di-GMP specific PDE.
Analysis of the Optimal ProE Reaction Conditions
To investigate the optimal reaction conditions for the PDE activity of ProE, we tested the effect of various environmental factors including pH, temperature and divalent cations on ProE activity. The results showed that ProE displayed a low enzyme activity from pH 3 to 7, and enzyme activity was substantially increased at pH ranging from 8 to 11 (Figure 4I). Enzyme activity of ProE was also determined at temperature ranging from 10°C to 90°C, and ProE showed a very low activity when the temperature was lower than 30°C with the maximum enzyme activity being at 70°C (Figure 4J). PDE enzymes commonly need divalent cation for their enzyme activity. Several cations, including Mg2+, Mn2+, and Co2+ could significantly boost the enzyme activity of PDEs, whereas Ca2+, Fe2+, Ni2+, Zn2+ don’t support the PDE activity (Bobrov et al., 2005; Schmidt et al., 2005; Tamayo et al., 2005; Barends et al., 2009; Tchigvintsev et al., 2010). We test the ProE activity by addition of above divalent cations to the reaction buffer, respectively. Similar to other known PDEs, the cations Ca2+, Fe2+, Ni2+, Zn2+ didn’t support the enzyme activity of ProE (Figure 4K). However, unlike other PDEs, which showed the highest enzyme activity in the presence of Mn2+ or Mg2+ (Bobrov et al., 2005; Schmidt et al., 2005; Tamayo et al., 2005; Barends et al., 2009; Tchigvintsev et al., 2010), Co2+ was shown to be the best metal ion for the enzyme activity of ProE (Figure 4K).
Identification Three Novel Residues Critical for ProE Activity
According to the amino acid sequence alignment, we identified a total of seventeen conserved residues in the EAL domains of the functional PDEs (Figure 3). To verify the roles of these conserved residues in the hydrolysis of c-di-GMP, we mutated these residues to alanine (Ala, A) and assayed the activity of purified mutant proteins. Among these proteins, four proteins showed a decreased enzyme activity (less than 55% of the wild type enzyme) (Q314, PA343, D472, E509) with unknown functions, nine proteins with mutations in the metal-binding residues showed an extremely low or abolished enzyme activity (E328, R332, N387, E419, E422, D449, D450, K470, E506, Q526), agreeable with the above observation that metal ions played a critical role in ProE catalysis (Figure 5A and Supplementary Table S3). Three proteins with mutations in the residues involved in substrate binding or coordination with water molecule (R332, N387, K470), as well as two proteins with mutations in the residues involved in interaction with other residues (K470, Q526) also showed compromised enzyme activity (Figure 5A and Supplementary Table S3). Significantly, this study unveiled three novel residues which were critical for ProE activity (P315, L330, G527) (Figure 5A and Supplementary Table S3). The proteins with mutation in these residues showed a very low enzyme activity (less than 10% of the wild type enzyme), indicating their indispensable roles in catalysis. Their functional roles in catalysis remain to be further characterized.
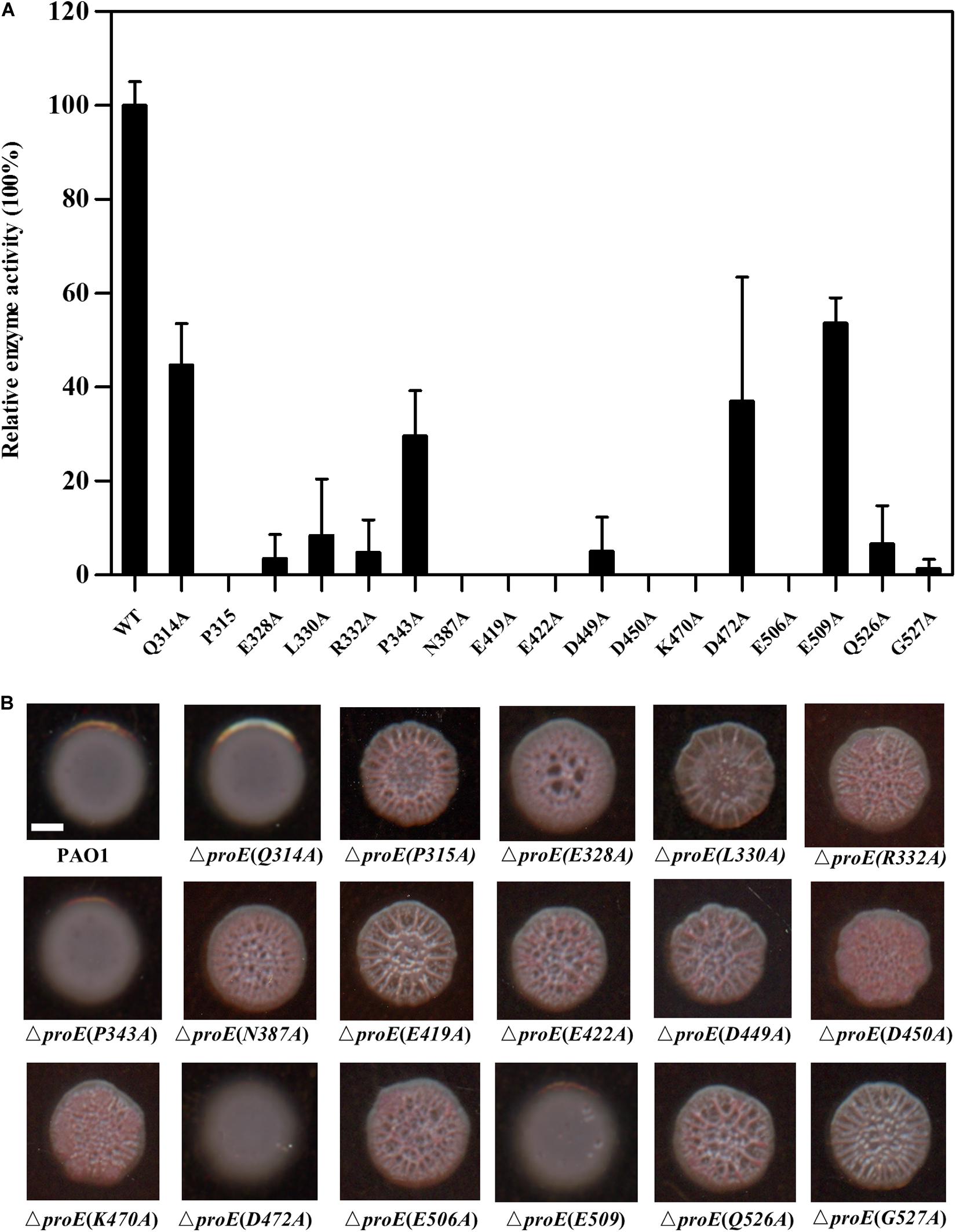
Figure 5. Functional analysis of the conserved residues of EAL domain of ProE. (A) After site-directed mutagenesis, the enzyme activity of mutant proteins were analyzed. The data are means of three replicates and error bars indicate standard deviation. (B) Effect of conserved residues on the colony morphology of P. aeruginosa strain PAO1 and its derivatives on Congo-Red plates. The native proE gene is deleted, and the proE alleles are according to complementation of mutated pBBR1-MCS5-proE into ΔproE. Scale bar = 2 mm.
We wanted to know whether the above key residues are also important for the ProE function in vivo. To address this question, we introduced the proE derivatives into ΔproE and assayed their colony morphology by Congo-Red plate. The results showed that substitution of thirteen residues with Ala failed to restore the colony morphology of ΔproE, including P315, L330, E328, R332, N387, E419, E422, D449, D450, K470, E506, Q526, and G527 (Figure 5B), which was consistent with our in vitro data, and confirmed the important roles of these residues for ProE function. The remaining four ProE derivatives (Q314, P343, D472, E509) could restore the ΔproE colony morphology to the wild type PAO1 level (Figure 5B). Our in vitro enzyme assay, showed that these ProE derivatives could still degrade c-di-GMP with activity about 29.6–53.7% of the wild type ProE. It is likely that these derivatives were overexpressed due to the multi-copy nature of the complementary expression vector, thus keeping the concentration of intracellular c-di-GMP to a level similar to wild type PAO1.
ProE EAL Domain 3D Model Comparison and Analysis
To further analysis the role of the identified conserved residues, especially the newly identified three residues, i.e., P315, L330, and G527. We use homology modeling approach to computationally generate structural models based on two previously characterized PDEs PA3825 and RmcA (Bellini et al., 2017; Mantoni et al., 2018). The Apo and the ligand-bound (Ca2+ and c-di-GMP) templates from PA3825 were used to generate two different structures, whereas the other models were obtained from the ligand-bound (Calcium and GTP) RmcA. The superposed model of the ProE with the x-ray structures of PA3825 and RmcA revealed very high similarities in the conformation of the EAL domain (RMSD < 1) (Figures 6A–C). The PA3825 Apo and PA3825 (Ca2+, c-di-GMP) models showed low structural differences when superposed on the ProE model with RMSD of 0.064 Å and 0.045 Å, respectively, for 214 atoms (Figures 6A,B). Superposition of the x-ray structure model of RmcA-GGDEF-EAL-Ca2+GTP with the ProE model also exhibited a high structural similarity (0.190 Å for 721 atoms) (Figure 6C). As expected, the superposed structures of the Apo-ProE and ligand-bound ProE models demonstrated a significant variation in conformation around the alpha 5 helix, which was reported to enable catalytic metal interaction and dimerization (D160 and D161 in PA3825: D449 and D450 in ProE) (Rao et al., 2009; Bellini et al., 2017; Figure 6D).
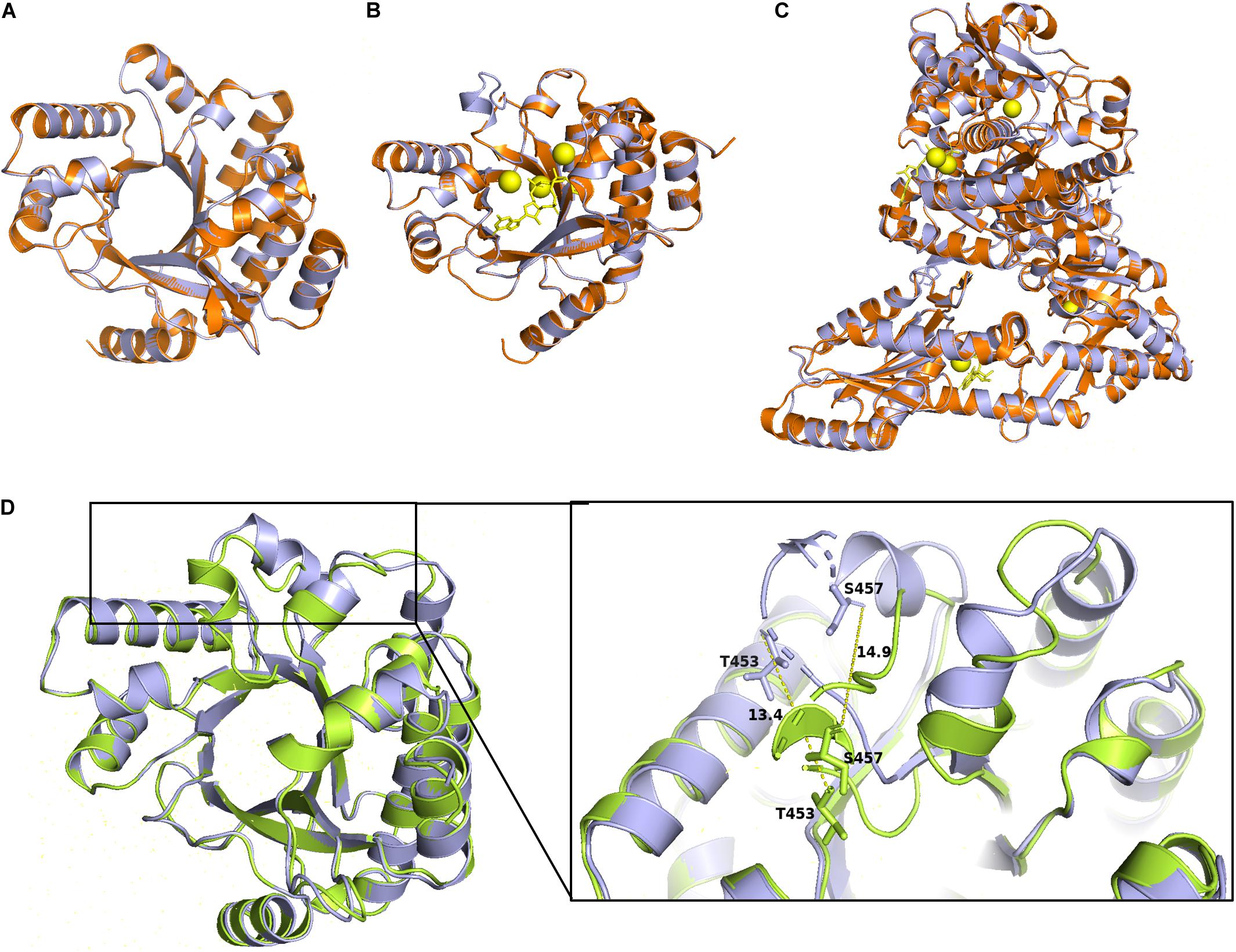
Figure 6. Homology models of the ProE with PA3825 and RmcA. Superimposition of the wild type ProE models with templates of 4y9m (PA3825-Apo), RMSD = 0.064 (214 to 214 atoms) (A), and 4y9p (PA3825-Ca2+ and c-di-GMP), RMSD = 0.04 (213 to 213 atoms) (B), and 5m3C (RmcA-Ca2+-GTP), RMSD = 0.190 (721 to 721 atoms) (C). ProE and templates are shown in lightblue and orange cartoons, respectively. C-di-GMP and GTP are shown as yellow sticks, calcium ions are shown in yellow spheres. (D) Superposition of Apo-ProE and ligand-bound ProE models reveals conformational changes due to ligand interaction. Apo-ProE and ligand-bound ProE are shown in lightblue and lime cartoons, respectively. The number represents the distance between residues (Å).
Then we map the identified conserved residues in ProE with the PA3825-Apo, PA3825(Ca2+, c-di-GMP), RmcA-GGDEF-EAL-Ca2+GTP templates, the result showed that the most residues are distributed in the ligand binding region (Figure 7A). Superposition of the conserved residues of ProE model with PA3825(Ca2+, c-di-GMP), showed that the residues E328, N387, E419, D449, D450, D472, and E506 enable catalytic metal ion (Ca2+) interaction (Figure 7B). The residues R332, N387, E509 interact with the c-di-GMP molecule (Figure 7C). Additionally, from the c-di-GMP bound ProE model, E509 is located in the dimerization interface and enables the formation of dimers between the two monomers (Figure 7D). The residue E422 forms polar interaction with L461 and G452, whiles K470 forms a polar interaction with Q526 and a water-mediated hydrogen bond with the calcium ion (Figure 7E). Polar interactions of E422 with L461 and G452, respectively, were affected by the E422A mutation (Figure 7F). Likewise, the water-mediated hydrogen bonds between K470, calcium ion and c-di-GMP were affected by the K470A mutation (Figure 7F).
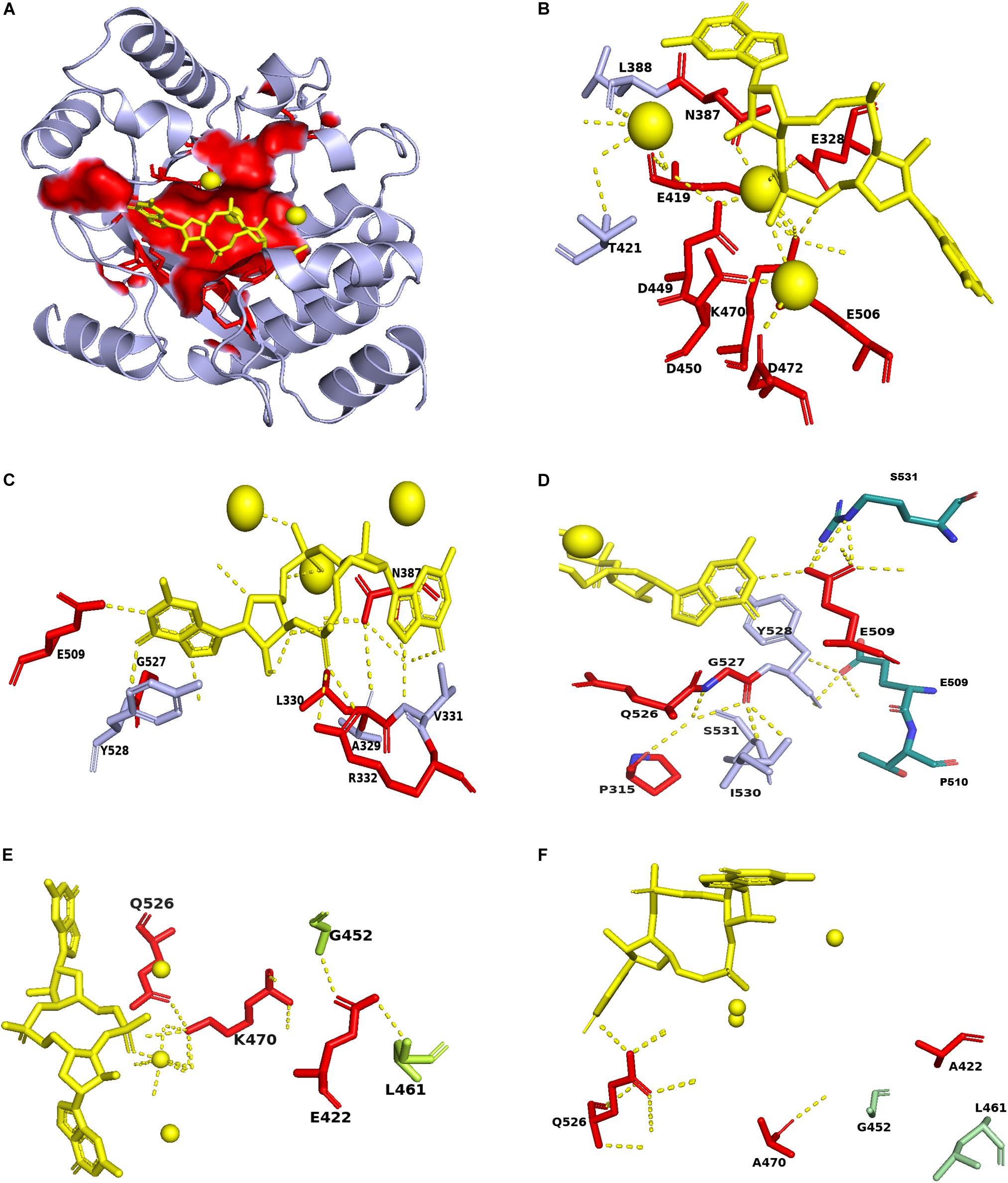
Figure 7. Mapping of conserved residues identified in ProE with homolog model. (A) The surface structure showing the localization of the conserved residues in the ProE model around the calcium ions and c-di-GMP. (B) The labeled residues indicate those interact with the calcium ions in ProE. (C) The labeled residues indicate those form direct or indirect polar contacts with c-di-GMP. (D) The residues E509 and Y528 interact with S531 and E509, respectively, from another monomer to allow dimer formation. (E) Interaction of conserved residues K470 and E422 with their adjacent residues. (F) Interaction of K470A and E422A with their adjacent residues. Calcium ions and c-di-GMP molecules are shown as yellow spheres and sticks, respectively. Polar interaction are shown as yellow dashes.
Next, we investigated the potential roles in catalysis of the three newly identified residues P315, L330, and G527 using the model generated from PA3825(Ca2+, c-di-GMP). We observed a water-mediated hydrogen bond interaction between the residue P315 and G527 (Supplementary Figure S9A), however, this interaction was not abolished in both P315A and G527A mutants (Supplementary Figures S9B,C). Substitution of P315 with A315 did not affect the interaction of the neighboring residues, i.e., Q314 with S531, and Q526 with E328 (Supplementary Figures S9D,E). No conformational changes were observed when the wild type ProE and P315A-ProE mutant models were superposed with the two ligand-bound PA3825 templates (RMSD = 0, 212 atoms) (Supplementary Figure S9F).
From the ProE model generated with the PA3825 (Ca2+c-di-GMP) template, L330A mutation did not seem to affect the polar interaction between L330 and H312, nor did it affect the neighboring residues E328, A329, V331, and R332 (Supplementary Figures S10A,B). Also, the mutation L330A did not result in a conformational change when superposed with the wild type ProE model generated with 4Y9P (RMSD = 0, 227 atoms) (Supplementary Figure S10C).
The dipeptide bond between G527 and Y528 enables interaction with the c-di-GMP molecule. It is unclear how this G527 influences binding of c-di-GMP as G527A mutation did not seem to affect c-di-GMP polar interaction as well as the interaction mediated by adjacent residues (Supplementary Figures S10D,E). Substitution of the adjacent residue Q526 with A526, however, resulted in loss of polar interaction between A505 and K470 (Supplementary Figure S10E).
According to above analysis we re-confirm the role of residues involved in substrate binding (R332 and N387), metal ion binding (E328, N387, E419, D449, D450, and E506), and residue-residue interaction (K470 and Q526) (Figure 7 and Supplementary Figures S9, S10, Supplementary Table S3). We found that the residues in which mutation resulted in decreased enzyme activity in our study and previous study with unknown functions (Supplementary Table S3), such as E422, D472, and E509, may play role in residue-residue interaction, metal ion binding and substrate binding, respectively (Figure 7 and Supplementary Figure S9). As for the newly identified three residues P315, L330, and G527, homolog model analysis seem to preclude their involvement in substrate or metal-ion binding (Figure 7 and Supplementary Figures S9, S10). How these residues affect the enzyme activity of ProE remain to be further investigated.
EPS Production Was Synergistically Regulated by Several PDEs in P. aeruginosa PAO1
In previous work, rbdA was shown to modulate biofilm dispersal by negatively regulating EPS production and positively regulating bacterial motility in our lab and other groups (An et al., 2010; Liu et al., 2018). Roy et al. (2012) reported that mutation of dipA (another name pch) can cause increased Congo-Red binding, suggesting its role in EPS production. In PA14 strain, bifA was demonstrated to inversely regulate biofilm formation and swarming motility, and the regulation on biofilm formation was dependent on pel (Kuchma et al., 2007). To assess the roles and compare the impact of these PDEs in regulating EPS production in P. aeruginosa PAO1, we generated the deletion mutants of rbdA, bifA, and dipA, respectively. Then we tested the effect of mutation on EPS production by Congo-Red plate assay. The results indicated that mutation of rbdA, bifA, and dipA, respectively, resulted in enhanced Congo-red binding, with colonies showing different extents of wrinkles. Among them, ΔrbdA and ΔbifA showed a rough appearance, and ΔdipA produced wrinkly colonies like ΔproE (Figure 8A). The results suggest that although they are all involved c-di-GMP metabolism and EPS production, the extent of their regulation may not be exactly the same. We were curious whether these PDEs could functionally replace each other. The results showed that proE, rbdA, bifA, and dipA could functionally restore the colony morphology of each other to the wild type PAO1 level (Figure 8A). These data support the notion that P. aeruginosa might use different PDEs for subtle control of c-di-GMP homeostasis and EPS production.
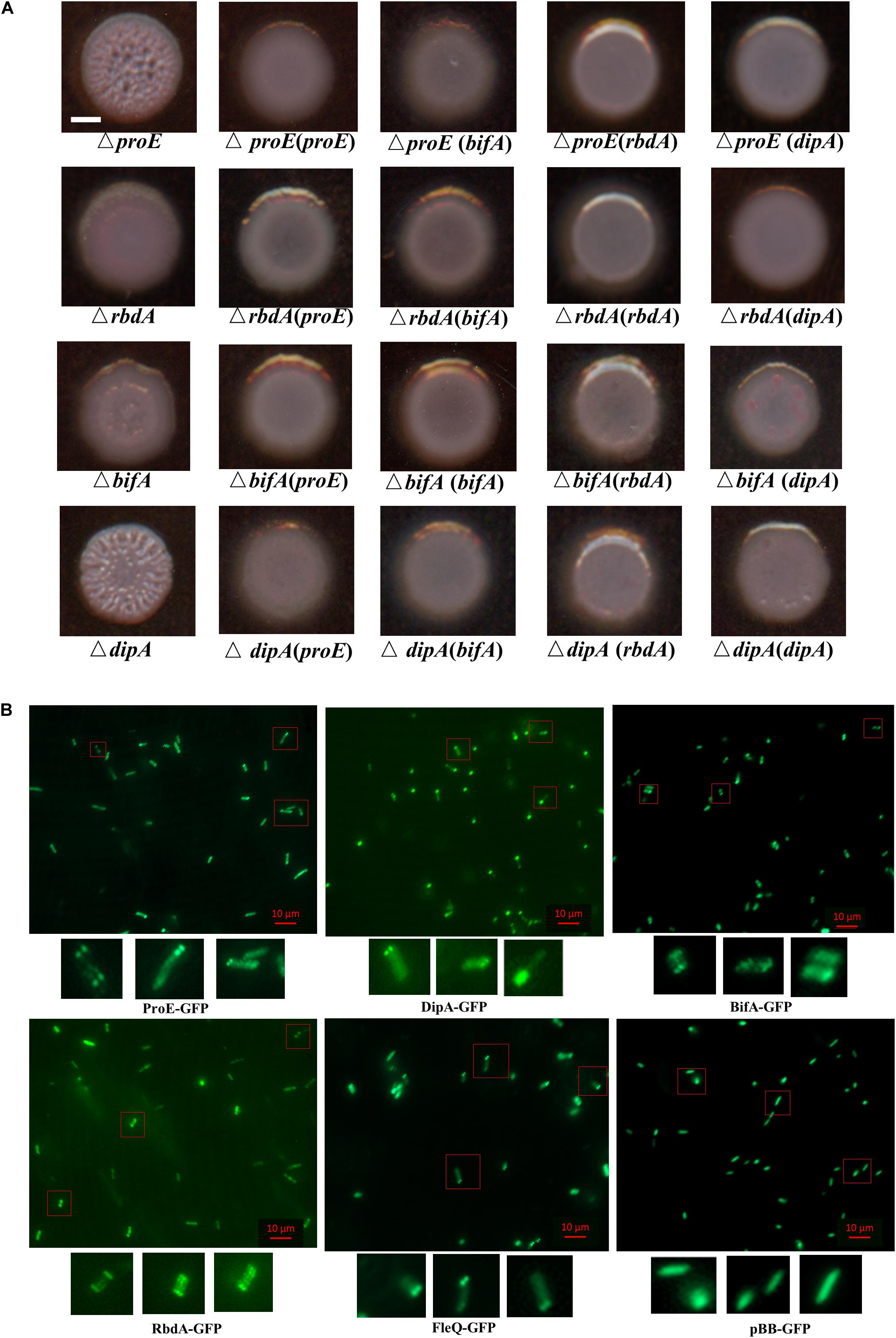
Figure 8. Functional and subcellular localization analysis of EPS-associated phosphodiesterases. (A) ProE and other three phosphodiesterases synergically regulated the colony morphology in P. aeruginosa. Deletion of either proE, rbdA, bifA or dipA can cause wrinkly morphology at different extent, and the functional defect can be restored by each other. Scale bar = 2 mm. (B) Subcellular localization of GFP fusion proteins. Bacterial colonies expressing indicated GFP fusion proteins were picked from overnight LB agar plates, resuspended in PBS and 2 μl culture was spotted onto a glass slide coated with 1% agarose. Images were taken by using epifluorescent microscopy. Scale bar = 10 μM.
Subcellular Localization of EPS Associated PDEs and Master Regulator FleQ
Previous studies showed that a set of proteins involved in c-di-GMP metabolism share a distinct subcellular localization in P. aeruginosa and other bacterial species (Guvener and Harwood, 2007; Yang et al., 2012; Kulasekara et al., 2013; Valentini et al., 2016; Jain et al., 2017; Xue et al., 2018; Li et al., 2019). We wanted to know whether the EPS associated proteins ProE, RbdA, BifA, DipA, and FleQ share a link between their function and subcellular localization. To address this question, we constructed C-terminal GFP fusions of ProE, RbdA, BifA, DipA, and FleQ, respectively, then their subcellular localizations were observed in P. aeruginosa PAO1 cells under fluorescence microscopy (Figure 8B). As shown in Figure 8B, DipA showed polar localization in most cells (more than 50%), which was consistent with previous study (Kulasekara et al., 2013). ProE, BifA, and FleQ showed mostly polar localization, including unipolar localization, bipolar localization. RbdA showed less polar localization, and a fraction of cells showed multiple-point distributions. As control, we also introduced the empty GFP into PAO1, the GFP proteins are uniformly distributed in the cells (Figure 8B). The above findings seem to suggest a general pattern of polar localization of these EPS-associated regulators, which may be important for precise regulation of EPS production in bacteria cells.
Discussion
In this study, we showed that ProE played a key role in regulating EPS production in P. aeruginosa strain PAO1 through modulation of the transcriptional expression of the pel and psl gene clusters (Figures 1, 2). We further demonstrated that ProE was a highly active PDE, whose maximum activity required the cation Co2+ (Figure 4). Furthermore, by scanning mutagenesis, we identified three novel key residues for the function of ProE in vitro and in vivo (Figure 5 and Supplementary Table S3), and these three residues are conserved in other functional PDEs. Moreover, we showed that several PDEs, including RbdA, BifA, DipA, and ProE, could functionally replace each other in regulation of the EPS-related colony morphology, and these enzymes, together with the EPS regulator FleQ, shared a general pattern of polar localization, which may suggest a link between localization and functionality in regulation of EPS production (Figure 8). These findings expand our understandings on the enzymatic properties of c-di-GMP metabolic enzymes, and provide a new insight on the regulatory mechanisms of c-di-GMP metabolic proteins on EPS production and colony morphology.
The proE gene was identified as its mutation led to formation of wrinkly colonies. Previous studies showed that wrinkly colony morphology is caused by enhanced EPS production (Hickman et al., 2005; Hickman and Harwood, 2008). Unlike other functionally characterized PDEs that commonly influence biofilm formation and motility (Kuchma et al., 2007; An et al., 2010; Yi et al., 2010; Deng et al., 2012; Roy et al., 2012; Chao et al., 2013; Rossello et al., 2017). The function of ProE seems to be specific to EPS production. Deletion of proE resulted in significantly elevated transcriptional expression of the pel and psl genes responsible for EPS production, but did not influence other phenotypes like biofilm formation or motility (Figure 1 and Supplementary Figure S1). And interestingly, we noticed that ProE showed some similarities with PA3177, which controls the metabolism of intracellular c-di-GMP without influences biofilm formation. Given that ProE plays a key role in regulating EPS production, which are critical for antibiotics resistance (Murakami et al., 2017), it is possible that ProE may function like PA3177 by regulating the biofilm-mediated antibiotics resistance (Poudyal and Sauer, 2018). The phylogenetic analysis showed that ProE is conserved in other Pseudomonas species as well, including Pseudomonas syringae pv. tomato DC3000, Pseudomonas savastanoi pv. phaseolicola 1448A, Pseudomonas putida F1 and Pseudomonas fluorescens SBW25 (Supplementary Figure S4), suggesting that ProE is a widely conserved c-di-GMP metabolic enzyme. It would be interesting to determine whether ProE may also play a role in regulating EPS production and colony morphology in these bacterial species. Prior to this study, the biological functions of ProE has only been investigated in P. aeruginosa strain PA14, in which ProE was shown to play a role in regulation of EPS biosynthesis and bacterial motility (Ha et al., 2014). In contrast, the ProE of strain PAO1, which shares the same amino acid sequences with its counterpart in strain PA14, only regulated EPS biosynthesis without affecting bacterial motility. These findings may suggest that the functional divergence of ProE occurred in the process of bacterial evolution. The underpinning regulatory mechanisms of functional divergence await further investigations.
In previous studies, several groups used proE for heterogenous complementation, which could rescue the c-di-GMP dependent phenotypes (Malone et al., 2010, 2012; Davis et al., 2013; Skotnicka et al., 2016a, b; Schmid et al., 2017; Skotnicka and Sogaard-Andersen, 2017; Berne et al., 2018; Laventie et al., 2019), suggesting its PDE activity in vivo. However, the enzyme properties of ProE have not yet been characterized in detail. Our data showed that ProE was a very active PDE with catalytic activity much higher than RocR (Figures 4B,D,G), which was shown to be one of the most active enzymes among the P. aeruginosa PDEs (Kulasakara et al., 2006). It is not clear yet what features account for the high catalytic activity of ProE, but several clues may be worthy of further consideration. Firstly, compared with another highly active PDE RocR, these two highly active enzymes differs in domain structures with RocR has an extra REC domain at N-terminal, whereas ProE contains only the GGDEF and EAL domains. It was believed that under certain conditions, the REC domain of RocR could be phosphorylated, which might induce the enzyme structural changes and present the EAL domain at an active status (Chen et al., 2012). In contrast, ProE lacks the N-terminal REC domain and thus its EAL domain is likely keeping at ready and active status, and ProE has a GTP binding domain that may further increase its enzyme activity. These may explain the observed ultra high activity of ProE in comparison with RocR. Second, ProE showed the maximum enzyme activity in the presence of Co2+ (Figure 4K), rather than Mg2+ or Mn2+ commonly used by others PDEs (Bobrov et al., 2005; Schmidt et al., 2005; Tamayo et al., 2005; Barends et al., 2009; Tchigvintsev et al., 2010). It has been well documented that different cations may generate variable impact on enzyme structure and activity. Similarly, it was shown recently that another two PDEs, CnpB from Mycobacterium tuberculosis and VcEAL (VC0395_A1247) from Vibrio cholerae also showed the best performance with Co2+ (Yang et al., 2014; Yadav et al., 2019). It is likely that ProE may share a similar metal-ion-dependent catalysis mechanism with these enzymes. Nevertheless, cation ion seems only affect catalysis but doesn’t influence the enzyme substrate specificity. ProE degraded only c-di-GMP (Supplementary Figure S8), whereas CnpB and VcEAL could degrade not only c-di-GMP, but also c-di-AMP or cGAMP, respectively (Yang et al., 2014; Yadav et al., 2019).
By using bioinformatics analysis and site-directed mutagenesis, we found a total of seventeen residues important for the function of ProE (Figure 5). While most of these residues are previously identified and known to be involved in metal-ion and substrate binding, water molecule coordination, residue-residue interaction or dimerization (Supplementary Table S3), we also identified three novel residues, i.e., P315, L330, and G527, which are also critical for the function of ProE. These three residues are well conserved in other functional PDEs, suggesting their potential roles in these PDEs (Figure 3), but the role of these residues remains unknown, it seems that they may not play a role in substrate or metal-ion binding by our homolog model analysis (Figures 6, 7 and Supplementary Figures S9, S10). Further structural and biochemical studies will gain more insights into the detailed mechanisms of these residues in ProE.
Deletion of the four PDEs known for c-di-GMP degradation, including ProE, RbdA, DipA, and BifA, respectively, caused colony morphological changes and the mutant phenotypes could be rescued by any of these four PDEs (Figure 8A), suggesting that they are all involved in regulation of EPS production in strain PAO1. This can be interpreted that over-expression of these PDEs dramatically decreased the concentration of intracellular c-di-GMP, which frees and enables FleQ to interact with the promoter of pel and psl, thus decreasing the expression of pel and psl (Baraquet et al., 2012; Baraquet and Harwood, 2016), and rescued the EPS over-production phenotype. In previous study, several DGCs and PDEs have been found to show polar localization including SadC (Merritt et al., 2010; Zhu et al., 2016), DipA (Kulasekara et al., 2013), WspR (Guvener and Harwood, 2007), HsbD (Valentini et al., 2016). By using C-terminal tagged fusion proteins to explore their subcellular localization, we found that DipA showed polar localization (Figure 8B), which agreed with the previous study (Kulasekara et al., 2013). Intriguingly, ProE, RbdA, and BifA, as well as then EPS regulator FleQ, also showed polar localization with varied extent or percentages (Figure 8B). In particular, ProE and DipA, whose null mutants showing more obvious wrinkly phenotype that the other two PDEs (Figure 8A), showed higher degree of polar localization (Figure 8B). These evidences seem to suggest that there is a link between EPS production and cell pole localization. It is possible that somehow EPS production was regulated at cell pole when the local c-di-GMP concentration reach a threshold. The underlying mechanism remains to be further studied by in situ exploration of the local c-di-GMP concentration and EPS production.
In summary, the results from this study demonstrated that ProE is an highly active PDE, which plays a key role in modulation of transcriptional expression of the pel and psl genes encoding EPS production in P. aeruginosa. The work also presents useful clues or questions for further investigations. For example, what are the precise roles of the three newly identified key residues, i.e., P315, L330, and G527, which are highly conserved in other PDEs. It is highly intriguing why ProE affects only EPS production, whereas other PDEs such as RbdA could affect multiple phenotypes. We reported previously that RbdA plays a key role in regulation of EPS production, bacterial motility, and biofilm formation (An et al., 2010). Given that EPS production is not only regulated by c-di-GMP but also controlled by quorum sensing (He et al., 2006), it is equally thought-provoking how P. aeruginosa could tap to multiple signal inputs or regulatory mechanisms in modulation of EPS production, and whether and if yes, how different regulation systems could interact or interplay with each other? The findings from this study lay down a footstone for probing these challenges.
Data Availability Statement
The datasets presented in this study can be found in online repositories, the amino acid sequences of ProE and its homolog in phylogenetic analysis are accessible in NCBI under accession numbers NP_253982.1, WP_034025707.1, WP_033966831.1, WP_033998527.1, WP_033969843.1, WP_073636980, PAZ 22036.1, WP_142901090.1, WP_031673977.1, WP_096084263.1, YP_793769.1, YP_002443268, WP_012077917.1, NP_789973.1, WP_015886513.1, WP_011064201.1, WP_003253539.1, WP_01 1167318.1, WP_011911732.1, WP_003206833.1, WP_0286836 56.1, and AAY49330.1.
Author Contributions
L-HZ and QF conceived and designed the experiments. QF, SA, TZ, ZL, QL, YL, and JH performed the experiments. QF, SA, and JZ analyzed the data. QF and L-HZ wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 31330002), the Key Projects of Guangzhou Science and Technology Plan (Grant No. 201804020066), and Guangdong Technological Innovation Strategy of Special Funds (Grant No. 2018B020205003).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01226/full#supplementary-material
Footnotes
References
Ahator, S. D., and Zhang, L. (2019). Small is mighty-chemical communication systems in Pseudomonas aeruginosa. Annu. Rev. Microbiol. 73, 559–578. doi: 10.1146/annurev-micro-020518-120044
An, S., Wu, J., and Zhang, L. H. (2010). Modulation of Pseudomonas aeruginosa biofilm dispersal by a cyclic-Di-GMP phosphodiesterase with a putative hypoxia-sensing domain. Appl. Environ. Microbiol. 76, 8160–8173. doi: 10.1128/AEM.01233-10
Baraquet, C., and Harwood, C. S. (2016). FleQ DNA binding consensus sequence revealed by studies of FleQ-dependent regulation of biofilm gene expression in Pseudomonas aeruginosa. J. Bacteriol. 198, 178–186. doi: 10.1128/JB.00539-15
Baraquet, C., Murakami, K., Parsek, M. R., and Harwood, C. S. (2012). The FleQ protein from Pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the pel operon promoter in response to c-di-GMP. Nucleic Acids Res. 40, 7207–7218. doi: 10.1093/nar/gks384
Barends, T. R., Hartmann, E., Griese, J. J., Beitlich, T., Kirienko, N. V., Ryjenkov, D. A., et al. (2009). Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature 459, 1015–1018. doi: 10.1038/nature07966
Basu Roy, A., and Sauer, K. (2014). Diguanylate cyclase NicD-based signalling mechanism of nutrient-induced dispersion by Pseudomonas aeruginosa. Mol. Microbiol. 94, 771–793. doi: 10.1111/mmi.12802
Bazaka, K., Crawford, R. J., Nazarenko, E. L., and Ivanova, E. P. (2011). Bacterial extracellular polysaccharides. Adv. Exp. Med. Biol. 715, 213–226. doi: 10.1007/978-94-007-0940-9_13
Bellini, D., Horrell, S., Hutchin, A., Phippen, C. W., Strange, R. W., Cai, Y., et al. (2017). Dimerisation induced formation of the active site and the identification of three metal sites in EAL-phosphodiesterases. Sci. Rep. 7:42166. doi: 10.1038/srep42166
Berne, C., Ellison, C. K., Agarwal, R., Severin, G. B., Fiebig, A., and Morton, R. I. III, et al. (2018). Feedback regulation of Caulobacter crescentus holdfast synthesis by flagellum assembly via the holdfast inhibitor HfiA. Mol. Microbiol. 110, 219–238. doi: 10.1111/mmi.14099
Bobrov, A. G., Kirillina, O., and Perry, R. D. (2005). The phosphodiesterase activity of the HmsP EAL domain is required for negative regulation of biofilm formation in Yersinia pestis. FEMS Microbiol. Lett. 247, 123–130. doi: 10.1016/j.femsle.2005.04.036
Chao, L., Rakshe, S., Leff, M., and Spormann, A. M. (2013). PdeB, a cyclic Di-GMP-specific phosphodiesterase that regulates Shewanella oneidensis MR-1 motility and biofilm formation. J. Bacteriol. 195, 3827–3833. doi: 10.1128/JB.00498-13
Chen, M. W., Kotaka, M., Vonrhein, C., Bricogne, G., Rao, F., Chuah, M. L., et al. (2012). Structural insights into the regulatory mechanism of the response regulator RocR from Pseudomonas aeruginosa in cyclic Di-GMP signaling. J. Bacteriol. 194, 4837–4846. doi: 10.1128/JB.00560-12
Christen, M., Christen, B., Folcher, M., Schauerte, A., and Jenal, U. (2005). Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J. Biol. Chem. 280, 30829–30837. doi: 10.1074/jbc.m504429200
Chua, S. L., Hultqvist, L. D., Yuan, M., Rybtke, M., Nielsen, T. E., Givskov, M., et al. (2015). In vitro and in vivo generation and characterization of Pseudomonas aeruginosa biofilm-dispersed cells via c-di-GMP manipulation. Nat. Protoc. 10, 1165–1180. doi: 10.1038/nprot.2015.067
Davis, N. J., Cohen, Y., Sanselicio, S., Fumeaux, C., Ozaki, S., Luciano, J., et al. (2013). De- and repolarization mechanism of flagellar morphogenesis during a bacterial cell cycle. Genes Dev. 27, 2049–2062. doi: 10.1101/gad.222679.113
Deng, Y., Schmid, N., Wang, C., Wang, J., Pessi, G., Wu, D., et al. (2012). Cis-2-dodecenoic acid receptor RpfR links quorum-sensing signal perception with regulation of virulence through cyclic dimeric guanosine monophosphate turnover. Proc. Natl. Acad. Sci. U.S.A. 109, 15479–15484. doi: 10.1073/pnas.1205037109
Friedman, L., and Kolter, R. (2004). Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J. Bacteriol. 186, 4457–4465. doi: 10.1128/jb.186.14.4457-4465.2004
Guvener, Z. T., and Harwood, C. S. (2007). Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol. Microbiol. 66, 1459–1473.
Ha, D. G., Richman, M. E., and O’toole, G. A. (2014). Deletion mutant library for investigation of functional outputs of cyclic diguanylate metabolism in Pseudomonas aeruginosa PA14. Appl. Environ. Microbiol. 80, 3384–3393. doi: 10.1128/AEM.00299-14
He, G. X., Kuroda, T., Mima, T., Morita, Y., Mizushima, T., and Tsuchiya, T. (2004). An H(+)-coupled multidrug efflux pump, PmpM, a member of the MATE family of transporters, from Pseudomonas aeruginosa. J. Bacteriol. 186, 262–265. doi: 10.1128/jb.186.1.262-265.2004
He, Y. W., Wang, C., Zhou, L., Song, H., Dow, J. M., and Zhang, L. H. (2006). Dual signaling functions of the hybrid sensor kinase RpfC of Xanthomonas campestris involve either phosphorelay or receiver domain-protein interaction. J. Biol. Chem. 281, 33414–33421. doi: 10.1074/jbc.m606571200
Hengge, R. (2009). Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7, 263–273. doi: 10.1038/nrmicro2109
Hentzer, M., Teitzel, G. M., Balzer, G. J., Heydorn, A., Molin, S., Givskov, M., et al. (2001). Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J. Bacteriol. 183, 5395–5401. doi: 10.1128/jb.183.18.5395-5401.2001
Hickman, J. W., and Harwood, C. S. (2008). Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol. Microbiol. 69, 376–389. doi: 10.1111/j.1365-2958.2008.06281.x
Hickman, J. W., Tifrea, D. F., and Harwood, C. S. (2005). A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. U.S.A. 102, 14422–14427. doi: 10.1073/pnas.0507170102
Hoffman, L. R., D’argenio, D. A., Maccoss, M. J., Zhang, Z., Jones, R. A., and Miller, S. I. (2005). Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 436, 1171–1175. doi: 10.1038/nature03912
Irie, Y., Borlee, B. R., O’connor, J. R., Hill, P. J., Harwood, C. S., Wozniak, D. J., et al. (2012). Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 109, 20632–20636. doi: 10.1073/pnas.1217993109
Jain, R., Sliusarenko, O., and Kazmierczak, B. I. (2017). Interaction of the cyclic-di-GMP binding protein FimX and the Type 4 pilus assembly ATPase promotes pilus assembly. PLoS Pathog. 13:e1006594. doi: 10.1371/journal.ppat.1006594
Jenal, U., and Malone, J. (2006). Mechanisms of cyclic-di-GMP signaling in bacteria. Annu. Rev. Genet. 40, 385–407.
Jenal, U., Reinders, A., and Lori, C. (2017). Cyclic di-GMP: second messenger extraordinaire. Nat. Rev. Microbiol. 15, 271–284. doi: 10.1038/nrmicro.2016.190
Jones, C. J., Newsom, D., Kelly, B., Irie, Y., Jennings, L. K., Xu, B., et al. (2014). ChIP-Seq and RNA-Seq reveal an AmrZ-mediated mechanism for cyclic di-GMP synthesis and biofilm development by Pseudomonas aeruginosa. PLoS Pathog. 10:e1003984. doi: 10.1371/journal.ppat.1003984
Koo, H., Falsetta, M. L., and Klein, M. I. (2013). The exopolysaccharide matrix: a virulence determinant of cariogenic biofilm. J Dent. Res. 92, 1065–1073. doi: 10.1177/0022034513504218
Kuchma, S. L., Brothers, K. M., Merritt, J. H., Liberati, N. T., Ausubel, F. M., and O’toole, G. A. (2007). BifA, a cyclic-Di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 189, 8165–8178. doi: 10.1128/jb.00586-07
Kulasakara, H., Lee, V., Brencic, A., Liberati, N., Urbach, J., Miyata, S., et al. (2006). Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3’-5’)-cyclic-GMP in virulence. Proc. Natl. Acad. Sci. U.S.A. 103, 2839–2844. doi: 10.1073/pnas.0511090103
Kulasekara, B. R., Kamischke, C., Kulasekara, H. D., Christen, M., Wiggins, P. A., and Miller, S. I. (2013). c-di-GMP heterogeneity is generated by the chemotaxis machinery to regulate flagellar motility. eLife 2:e01402. doi: 10.7554/eLife.01402
Kulasekara, H. D., Ventre, I., Kulasekara, B. R., Lazdunski, A., Filloux, A., and Lory, S. (2005). A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol. Microbiol. 55, 368–380. doi: 10.1111/j.1365-2958.2004.04402.x
Laventie, B. J., Sangermani, M., Estermann, F., Manfredi, P., Planes, R., Hug, I., et al. (2019). A surface-induced asymmetric program promotes tissue colonization by Pseudomonas aeruginosa. Cell Host Microbe 25:e146. doi: 10.1016/j.chom.2018.11.008
Lee, J., and Zhang, L. (2015). The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 6, 26–41. doi: 10.1007/s13238-014-0100-x
Lee, V. T., Matewish, J. M., Kessler, J. L., Hyodo, M., Hayakawa, Y., and Lory, S. (2007). A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol. Microbiol. 65, 1474–1484. doi: 10.1111/j.1365-2958.2007.05879.x
Li, H., Xue, D., Tian, F., Yuan, X., Yang, F., Chen, H., et al. (2019). Xanthomonas oryzae pv. oryzae response regulator TriP regulates virulence and exopolysaccharide production via interacting with c-di-GMP phosphodiesterase PdeR. Mol. Plant Microbe Interact. 32, 729–739. doi: 10.1094/MPMI-09-18-0260-R
Liang, Z. X. (2015). The expanding roles of c-di-GMP in the biosynthesis of exopolysaccharides and secondary metabolites. Nat. Prod. Rep. 32, 663–683. doi: 10.1039/c4np00086b
Liu, C., Liew, C. W., Wong, Y. H., Tan, S. T., Poh, W. H., Manimekalai, M. S. S., et al. (2018). Insights into biofilm dispersal regulation from the crystal structure of the PAS-GGDEF-EAL region of RbdA from Pseudomonas aeruginosa. J. Bacteriol. 200:e00515-17. doi: 10.1128/JB.00515-17
Malone, J. G., Jaeger, T., Manfredi, P., Dotsch, A., Blanka, A., Bos, R., et al. (2012). The YfiBNR signal transduction mechanism reveals novel targets for the evolution of persistent Pseudomonas aeruginosa in cystic fibrosis airways. PLoS Pathog. 8:e1002760. doi: 10.1371/journal.ppat.1002760
Malone, J. G., Jaeger, T., Spangler, C., Ritz, D., Spang, A., Arrieumerlou, C., et al. (2010). YfiBNR mediates cyclic di-GMP dependent small colony variant formation and persistence in Pseudomonas aeruginosa. PLoS Pathog. 6:e1000804. doi: 10.1371/journal.ppat.1000804
Mantoni, F., Paiardini, A., Brunotti, P., D’angelo, C., Cervoni, L., Paone, A., et al. (2018). Insights into the GTP-dependent allosteric control of c-di-GMP hydrolysis from the crystal structure of PA0575 protein from Pseudomonas aeruginosa. FEBS J. 285, 3815–3834. doi: 10.1111/febs.14634
Matthysse, A. G. (2014). Attachment of agrobacterium to plant surfaces. Front. Plant Sci. 5:252. doi: 10.3389/fpls.2014.00252
Merighi, M., Lee, V. T., Hyodo, M., Hayakawa, Y., and Lory, S. (2007). The second messenger bis-(3’-5’)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol. Microbiol. 65, 876–895. doi: 10.1111/j.1365-2958.2007.05817.x
Merritt, J. H., Brothers, K. M., Kuchma, S. L., and O’toole, G. A. (2007). SadC reciprocally influences biofilm formation and swarming motility via modulation of exopolysaccharide production and flagellar function. J. Bacteriol. 189, 8154–8164. doi: 10.1128/jb.00585-07
Merritt, J. H., Ha, D. G., Cowles, K. N., Lu, W., Morales, D. K., Rabinowitz, J., et al. (2010). Specific control of Pseudomonas aeruginosa surface-associated behaviors by two c-di-GMP diguanylate cyclases. mBio 1:e00183-10. doi: 10.1128/mBio.00183-10
Minasov, G., Padavattan, S., Shuvalova, L., Brunzelle, J. S., Miller, D. J., Basle, A., et al. (2009). Crystal structures of YkuI and its complex with second messenger cyclic Di-GMP suggest catalytic mechanism of phosphodiester bond cleavage by EAL domains. J. Biol. Chem. 284, 13174–13184. doi: 10.1074/jbc.M808221200
Morgan, J. L., Mcnamara, J. T., and Zimmer, J. (2014). Mechanism of activation of bacterial cellulose synthase by cyclic di-GMP. Nat. Struct. Mol. Biol. 21, 489–496. doi: 10.1038/nsmb.2803
Murakami, K., Ono, T., Viducic, D., Somiya, Y., Kariyama, R., Hori, K., et al. (2017). Role of psl genes in antibiotic tolerance of adherent Pseudomonas aeruginosa. Antimicrob Agents Chemother. 61:e02587-16. doi: 10.1128/AAC.02587-16
Navarro, M. V., De, N., Bae, N., Wang, Q., and Sondermann, H. (2009). Structural analysis of the GGDEF-EAL domain-containing c-di-GMP receptor FimX. Structure 17, 1104–1116. doi: 10.1016/j.str.2009.06.010
Poudyal, B., and Sauer, K. (2018). The ABC of biofilm drug tolerance: the MerR-like regulator BrlR is an activator of ABC transport systems, with PA1874-77 contributing to the tolerance of Pseudomonas aeruginosa biofilms to tobramycin. Antimicrob Agents Chemother. 62:e01981-17. doi: 10.1128/AAC.01981-17
Rao, F., Qi, Y., Chong, H. S., Kotaka, M., Li, B., Li, J., et al. (2009). The functional role of a conserved loop in EAL domain-based cyclic di-GMP-specific phosphodiesterase. J. Bacteriol. 191, 4722–4731. doi: 10.1128/JB.00327-09
Rao, F., Yang, Y., Qi, Y., and Liang, Z. X. (2008). Catalytic mechanism of cyclic di-GMP-specific phosphodiesterase: a study of the EAL domain-containing RocR from Pseudomonas aeruginosa. J. Bacteriol. 190, 3622–3631. doi: 10.1128/JB.00165-08
Robert-Paganin, J., Nonin-Lecomte, S., and Rety, S. (2012). Crystal structure of an EAL domain in complex with reaction product 5’-pGpG. PLoS One 7:e52424. doi: 10.1371/journal.pone.0052424
Romling, U., Galperin, M. Y., and Gomelsky, M. (2013). Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 77, 1–52. doi: 10.1128/MMBR.00043-12
Rossello, J., Lima, A., Gil, M., Rodriguez Duarte, J., Correa, A., Carvalho, P. C., et al. (2017). The EAL-domain protein FcsR regulates flagella, chemotaxis and type III secretion system in Pseudomonas aeruginosa by a phosphodiesterase independent mechanism. Sci. Rep. 7:10281. doi: 10.1038/s41598-017-09926-3
Roy, A. B., Petrova, O. E., and Sauer, K. (2012). The phosphodiesterase DipA (PA5017) is essential for Pseudomonas aeruginosa biofilm dispersion. J. Bacteriol. 194, 2904–2915. doi: 10.1128/JB.05346-11
Ryder, C., Byrd, M., and Wozniak, D. J. (2007). Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr. Opin. Microbiol. 10, 644–648. doi: 10.1016/j.mib.2007.09.010
Schmid, N., Suppiger, A., Steiner, E., Pessi, G., Kaever, V., Fazli, M., et al. (2017). High intracellular c-di-GMP levels antagonize quorum sensing and virulence gene expression in Burkholderia cenocepacia H111. Microbiology 163, 754–764. doi: 10.1099/mic.0.000452
Schmidt, A. J., Ryjenkov, D. A., and Gomelsky, M. (2005). The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J. Bacteriol. 187, 4774–4781. doi: 10.1128/jb.187.14.4774-4781.2005
Skotnicka, D., Petters, T., Heering, J., Hoppert, M., Kaever, V., and Sogaard-Andersen, L. (2016a). Cyclic Di-GMP regulates type IV pilus-dependent motility in Myxococcus xanthus. J. Bacteriol. 198, 77–90. doi: 10.1128/JB.00281-15
Skotnicka, D., Smaldone, G. T., Petters, T., Trampari, E., Liang, J., Kaever, V., et al. (2016b). A minimal threshold of c-di-GMP is essential for fruiting body formation and sporulation in Myxococcus xanthus. PLoS Genet. 12:e1006080. doi: 10.1371/journal.pgen.1006080
Skotnicka, D., and Sogaard-Andersen, L. (2017). Type IV pili-dependent motility as a tool to determine the activity of c-di-GMP modulating enzymes in Myxococcus xanthus. Methods Mol. Biol. 1657, 157–165. doi: 10.1007/978-1-4939-7240-1_13
Sudarsan, N., Lee, E. R., Weinberg, Z., Moy, R. H., Kim, J. N., Link, K. H., et al. (2008). Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science 321, 411–413. doi: 10.1126/science.1159519
Sundriyal, A., Massa, C., Samoray, D., Zehender, F., Sharpe, T., Jenal, U., et al. (2014). Inherent regulation of EAL domain-catalyzed hydrolysis of second messenger cyclic di-GMP. J. Biol. Chem. 289, 6978–6990. doi: 10.1074/jbc.M113.516195
Tamayo, R., Tischler, A. D., and Camilli, A. (2005). The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. J. Biol. Chem. 280, 33324–33330. doi: 10.1074/jbc.m506500200
Tao, F., He, Y. W., Wu, D. H., Swarup, S., and Zhang, L. H. (2010). The cyclic nucleotide monophosphate domain of Xanthomonas campestris global regulator Clp defines a new class of cyclic di-GMP effectors. J. Bacteriol. 192, 1020–1029. doi: 10.1128/JB.01253-09
Tchigvintsev, A., Xu, X., Singer, A., Chang, C., Brown, G., Proudfoot, M., et al. (2010). Structural insight into the mechanism of c-di-GMP hydrolysis by EAL domain phosphodiesterases. J. Mol. Biol. 402, 524–538. doi: 10.1016/j.jmb.2010.07.050
Valentini, M., Laventie, B. J., Moscoso, J., Jenal, U., and Filloux, A. (2016). The diguanylate cyclase HsbD intersects with the HptB regulatory cascade to control Pseudomonas aeruginosa biofilm and motility. PLoS Genet. 12:e1006354. doi: 10.1371/journal.pgen.1006354
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. doi: 10.1093/nar/gky427
Whitney, J. C., Colvin, K. M., Marmont, L. S., Robinson, H., Parsek, M. R., and Howell, P. L. (2012). Structure of the cytoplasmic region of PelD, a degenerate diguanylate cyclase receptor that regulates exopolysaccharide production in Pseudomonas aeruginosa. J. Biol. Chem. 287, 23582–23593. doi: 10.1074/jbc.M112.375378
Wozniak, D. J., Wyckoff, T. J., Starkey, M., Keyser, R., Azadi, P., O’toole, G. A., et al. (2003). Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc. Natl. Acad. Sci. U.S.A. 100, 7907–7912. doi: 10.1073/pnas.1231792100
Xu, L., Venkataramani, P., Ding, Y., Liu, Y., Deng, Y., Yong, G. L., et al. (2016). A cyclic di-GMP-binding adaptor protein interacts with histidine kinase to regulate two-component signaling. J. Biol. Chem. 291, 16112–16123. doi: 10.1074/jbc.M116.730887
Xue, D., Tian, F., Yang, F., Chen, H., Yuan, X., Yang, C. H., et al. (2018). Phosphodiesterase EdpX1 promotes Xanthomonas oryzae pv. oryzae virulence, exopolysaccharide production, and biofilm formation. Appl. Environ. Microbiol. 84:e01717-18. doi: 10.1128/AEM.01717-18
Yadav, M., Pal, K., and Sen, U. (2019). Structures of c-di-GMP/cGAMP degrading phosphodiesterase VcEAL: identification of a novel conformational switch and its implication. Biochem. J. 476, 3333–3353. doi: 10.1042/BCJ20190399
Yang, C., Cui, C., Ye, Q., Kan, J., Fu, S., Song, S., et al. (2017). Burkholderia cenocepacia integrates cis-2-dodecenoic acid and cyclic dimeric guanosine monophosphate signals to control virulence. Proc. Natl. Acad. Sci. U.S.A. 114, 13006–13011. doi: 10.1073/pnas.1709048114
Yang, F., Tian, F., Sun, L., Chen, H., Wu, M., Yang, C. H., et al. (2012). A novel two-component system PdeK/PdeR regulates c-di-GMP turnover and virulence of Xanthomonas oryzae pv. oryzae. Mol. Plant Microbe Interact. 25, 1361–1369. doi: 10.1094/MPMI-01-12-0014-R
Yang, J., Bai, Y., Zhang, Y., Gabrielle, V. D., Jin, L., and Bai, G. (2014). Deletion of the cyclic di-AMP phosphodiesterase gene (cnpB) in Mycobacterium tuberculosis leads to reduced virulence in a mouse model of infection. Mol. Microbiol. 93, 65–79. doi: 10.1111/mmi.12641
Yi, X., Yamazaki, A., Biddle, E., Zeng, Q., and Yang, C. H. (2010). Genetic analysis of two phosphodiesterases reveals cyclic diguanylate regulation of virulence factors in Dickeya dadantii. Mol. Microbiol. 77, 787–800. doi: 10.1111/j.1365-2958.2010.07246.x
Keywords: Pseudomonas aeruginosa, c-di-GMP, phosphodiesterase, enzymatic properties, exopolysaccharide, Pel and Psl
Citation: Feng Q, Ahator SD, Zhou T, Liu Z, Lin Q, Liu Y, Huang J, Zhou J and Zhang L-H (2020) Regulation of Exopolysaccharide Production by ProE, a Cyclic-Di-GMP Phosphodiesterase in Pseudomonas aeruginosa PAO1. Front. Microbiol. 11:1226. doi: 10.3389/fmicb.2020.01226
Received: 15 March 2020; Accepted: 14 May 2020;
Published: 05 June 2020.
Edited by:
Hari S. Misra, Bhabha Atomic Research Centre (BARC), IndiaReviewed by:
Jacob Malone, John Innes Centre, United KingdomSerena Rinaldo, Sapienza University of Rome, Italy
Giorgio Giardina, Sapienza University of Rome, Italy
Copyright © 2020 Feng, Ahator, Zhou, Liu, Lin, Liu, Huang, Zhou and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lian-Hui Zhang, bGh6aGFuZzAxQHNjYXUuZWR1LmNu
 Qishun Feng
Qishun Feng Stephen Dela Ahator
Stephen Dela Ahator Tian Zhou
Tian Zhou Zhiqing Liu
Zhiqing Liu Qiqi Lin
Qiqi Lin Yang Liu1,2
Yang Liu1,2 Jianuan Zhou
Jianuan Zhou