
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 25 May 2020
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.00916
 Zewen Wen1,2†
Zewen Wen1,2† Yongpeng Shang1,2†
Yongpeng Shang1,2† Guangjian Xu1†
Guangjian Xu1† Zhangya Pu3
Zhangya Pu3 Zhiwei Lin1,2
Zhiwei Lin1,2 Bing Bai1
Bing Bai1 Zhong Chen1
Zhong Chen1 Jinxin Zheng1
Jinxin Zheng1 Qiwen Deng1*
Qiwen Deng1* Zhijian Yu1,2*
Zhijian Yu1,2*Opportunistic infections caused by multidrug-resistant Enterococcus faecalis strains are a significant clinical challenge. Eravacycline (Erava) is a synthetic fluorocycline structurally similar to tigecycline (Tige) that exhibits robust antimicrobial activity against Gram-positive bacteria. This study investigated the in vitro antimicrobial activity and heteroresistance risk of Eravacycline (Erava) in clinical E. faecalis isolates from China along with the mechanism of Erava resistance. A total of 276 non-duplicate E. faecalis isolates were retrospectively collected from a tertiary care hospital in China. Heteroresistance to Erava and the influence of tetracycline (Tet) resistance genes on Erava susceptibility were examined. To clarify the molecular basis for Erava resistance, E. faecalis variants exhibiting Erava-induced resistance were selected under Erava pressure. The relative transcript levels of six candidate genes linked to Erava susceptibility were determined by quantitative reverse-transcription PCR, and their role in Erava resistance and heteroresistance was evaluated by in vitro overexpression experiments. We found that Erava minimum inhibitory concentrations (MICs) against clinical E. faecalis isolates ranged from ≤0.015 to 0.25 mg/l even in strains harboring Tet resistance genes. The detection frequency of Erava heteroresistance in isolates with MICs ≤ 0.06, 0.125, and 0.25 mg/l were 0.43% (1/231), 7.5% (3/40), and 0 (0/5), respectively. No mutations were detected in the 30S ribosomal subunit gene in Erava heteroresistance-derived clones, although mutations in this subunit conferred cross resistance to Tige in Erava-induced resistant E. faecalis. Overexpressing RS00630 (encoding a bone morphogenetic protein family ATP-binding cassette transporter substrate-binding protein) in E. faecalis increased the frequency of Erava and Tige heteroresistance, whereas RS12140, RS06145, and RS06880 overexpression conferred heteroresistance to Tige only. These results indicate that Erava has potent in vitro antimicrobial activity against clinical E. faecalis isolates from China and that Erava heteroresistance can be induced by RS00630 overexpression.
Enterococcus faecalis is a Gram-positive facultative anaerobe that normally exists as a commensal microbe colonizing the gastrointestinal or urinary tract of humans and animals (Arias and Murray, 2012). E. faecalis is an important nosocomial pathogen that is linked to various infections, including urinary tract infection, sepsis, endocarditis, and peritonitis (Arias and Murray, 2012; Mercuro et al., 2018). E. faecalis exhibits both intrinsic and acquired resistance to several commonly used antibiotics, such as aminoglycosides and macrolides, and can serve as a model for the emergence of antibiotic resistance (Arias and Murray, 2012). Given the increasing number of reports of multidrug resistant E. faecalis strains, including those resistant to vancomycin (VAN) and linezolid (LZD), there is an urgent need to develop effective measures to treat infections caused by E. faecalis (Van Harten et al., 2017; Sparo et al., 2018; Fiore et al., 2019; García-Solache and Rice, 2019).
Eravacycline (Erava) is a novel fluorocycline antibacterial agent belonging to the tetracycline (Tet) drug class (Grossman et al., 2012). Erava has robust antimicrobial activity against a broad range of microorganisms, including Gram-positive, Gram-negative, anaerobic, and atypical bacteria (Sutcliffe et al., 2013; Livermore et al., 2016; Monogue et al., 2016; Zhanel et al., 2016). Intravenous Erava is newly approved in several countries for the treatment of adult patients with complicated intra-abdominal infections (cIAIs) caused by enteric microorganisms, including multidrug resistant, difficult-to-treat enterococci infection (Lee and Burton, 2019; Scott, 2019; Alosaimy et al., 2020). Against E. faecalis clinical isolates collected globally (Hackel et al., 2013; Sutcliffe et al., 2013; in 2013–2015; Olesky et al., 2017), in Canada (CANWARD surveillance study; 2014–2015; Zhanel et al., 2018), and the USA (2013–2016; Bouchillon et al., 2018), Erava exhibited potent in vitro activity, including against vancomycin-resistant E. faecalis isolates. However, the in vitro antimicrobial activity of Erava in clinical E. faecalis isolates from China has not been comprehensive investigated.
In our earlier work, we described Erava heteroresistance in Staphylococcus aureus and Klebsiella pneumonia isolates with comparatively low Erava minimum inhibitory concentrations (MICs), which suggested that the resistance emerged as a result of antibiotic treatment failure (Zhang F. et al., 2018; Zheng J. X. et al., 2018). However, it is not known whether Erava heteroresistance is also found in E. faecalis. Recent evidence has attributed the development of resistance to the new class of Tet antibiotics including tigecycline (Tige) to genetic mutations in the Tet binding site of the 30S ribosomal subunit, which includes 16S rRNA and 30S ribosome protein S10 (Nguyen et al., 2014; Grossman, 2016). Regulators of several efflux pumps or membrane proteins, such as SoxS, MarA, RamA, and Rob, have also been implicated in Tige resistance (Nguyen et al., 2014; Grossman, 2016), although the mechanistic details are not well-understood. In addition, tet(X) orthologs, which encode Tet destructases and inactivate Tet antibiotics, represent a unique enzymatic Tet resistance mechanism (Grossman, 2016; He et al., 2019; Sun et al., 2019). Moreover, it is unclear whether Erava susceptibility is influenced by Tet binding site mutations or overexpression of the efflux pump and membrane proteins.
The present study investigated the in vitro antimicrobial activity of and potential for the development of heteroresistance to Erava in clinical E. faecalis isolates collected at a tertiary care hospital in China. We also examined the influence of Tet resistance genes on Erava susceptibility and analyzed mutations in the 30S ribosomal subunit in Erava heteroresistance-derived clones and E. faecalis isolates exhibiting Erava-induced resistance. The genetic and molecular factors affecting Erava susceptibility were assessed by the efflux pump inhibition, qRT-PCR, and in vitro overexpression experiments.
Non-duplicate clinical isolates of E. faecalis (n = 276 strains) were collected from January 1, 2011, to December 31, 2015, at the clinical microbiology laboratory of Shenzhen Nanshan People’s Hospital, a tertiary care hospital in Shenzhen, China (Lin et al., 2019). The isolates were from urine, blood, bile, wound exudate, drainage liquid, sputum, and aseptic body fluids and were originally identified and tested for susceptibility to clinically relevant antibiotics using the VITEK 2 Compact automatic microbial analysis system (Biomérieux, Marcy l’Etoile, France). Species-appropriate quality control strains were used to ensure that the isolates met the standards recommended by Clinical and Laboratory Standards Institute (CLSI) guidelines. E. faecalis ATCC29212 was used as reference strains for quality control. Procedures involving human participants were performed in accordance with the ethical standards of Shenzhen Nanshan People’s Hospital and the Declaration of Helsinki 1964 and its later amendments. For this particular study, formal consent was not required.
Antimicrobial susceptibility to Erava (AdooQ Bioscience, Irvine, CA, United States) was determined by the agar dilution method. MICs of doxycycline, minocycline (Aladdin, Shanghai, China), LZD, and VAN (Macklin, Shanghai, China) were determined by the broth microdilution method according to CLSI guidelines (CLSI-M100-S26). Susceptibility breakpoints of antibiotics, including ampicillin, erythromycin, ciprofloxacin, gentamycin, nitrofurantoin, trimethoprim-sulfamethoxazole, Tet, VAN, and LZD were defined according to CLSI guidelines. The MIC range of Erava for the quality control strain E. faecalis ATCC29212 was 0.015–0.06 μg/ml according to CLSI, and the Erava susceptibility breakpoint in E. faecalis was defined as ≤0.5 mg/l according to the EUCAST 2019 standard. To characterize antimicrobial activity, MICs of Erava in E. faecalis were categorized into four levels (≤0.0625, 0.125, 0.25, and 0.5 mg/l) (Lin et al., 2019).
Bacterial DNA was extracted using Lysis Buffer for Microorganism to Direct PCR (Takara Bio, Otsu, Japan) according to the manufacturer’s instructions. Tet resistance genes in E. faecalis isolates, including tet(K), tet(L), tet(M), tet(S), tet(O), tet(W), tet(U), and tet(X) variants, were amplified by PCR, as previously described (Zheng et al., 2017; Bai et al., 2018; He et al., 2019; Lin et al., 2019), using a commercial kit (Takara Bio) according to the manufacturer’s protocol. All primers used are listed in the Supplementary Table S2. Mutations in genes encoding the 30S ribosomal subunit, including four separate copies of the 16S rRNA gene, and the ribosomal protein S10 gene was detected by PCR and sequence alignment using previously described primer sets (Supplementary Table S3) (Lin et al., 2019).
PAP experiments were performed as previously described (Zhang F. et al., 2018; Zheng J. X. et al., 2018; Lin et al., 2019), with minor modifications. Briefly, 50 μl aliquots (∼108 CFU/ml) were spread onto Müller-Hinton (MH) agar plates containing various concentrations of Erava (0.25, 0.5, 1.0, and 2.0 mg/l). Colonies were counted after incubation at 37°C for 24 h. Erava heteroresistance was defined as an Erava-susceptible isolate (MIC ≤ 0.5 mg/l) with subpopulations growing in the presence of 0.5 mg/l Erava and with a detection limit of 5 CFU/plate. Three resistant clones were randomly selected from the plates for experiments. The MICs of Erava and Tige were determined as described above (Zhang F. et al., 2018; Zheng J. X. et al., 2018). The frequency of Erava heteroresistance was separately statistically analyzed for clinical isolates with Erava MICs of ≤0.125 and 0.25 mg/l, respectively.
The role of the efflux pump in Erava-heteroresistant E. fecalis isolates was evaluated using the efflux pump inhibitors (EPIs) Phe-Arg-β-naphthylamide (PAβN; Sigma-Aldrich, St. Louis, MO, United States) and carbonyl cyanide m-chlorophenylhydrazone (CCCP, Sigma-Aldrich). Erava MICs of Erava-resistant subpopulations of heteroresistant isolates were determined by the agar dilution method in the presence and absence of PAβN (50 mg/l) or CCCP (16 mg/l) (Zhang F. et al., 2018). The inhibition was considered significant if the MIC decreased at least fourfold in the presence of EPIs (Zhang F. et al., 2018; Zheng J. X. et al., 2018).
Three parental E. faecalis isolates, including two clinical isolates (FC1 and FC2), and the OG1RF strain were used to generate Erava-resistant E. faecalis. The isolates were serially subcultured in MH broth containing increasing concentrations of Erava, with MIC equivalents as the initial concentration (0.06 mg/l) and 2×, 4×, 8×, 16×, and 32× MICs (Zhang F. et al., 2018; Lin et al., 2019). Strains were serially cultured for four passages before exposure to the next concentration. Isolates from passages at each concentration were selected for three generations on MH plates without antibiotics for further experiments, including the detection of mutations at Tet binding sites in the 30S ribosomal subunit, MIC assays, and quantitative reverse transcription (qRT)-PCR analysis.
Erava is structurally similar to Omadacycline, another member of the Tet drug class. Our previous study showed that heteroresistance to Omadacycline in E. faecalis was potentially linked to 11 genes, i.e., RS11300, RS11485, RS10660, RS06880, RS12140, RS02205, RS05865, RS06145, RS09080, RS12590, and RS00630 (Lin et al., 2019). To determine whether these genes are also involved in Erava resistance in E. faecalis, we measured their relative transcript levels in clones exhibiting Erava-induced resistance by qRT-PCR. Total bacterial RNA was extracted using the RNeasy Mini kit (Qiagen GmbH, Hilden, Germany), and cDNA was synthesized using the PrimeScript RT reagent kit (Takara Bio). qRT-PCR was performed as previously described (Zheng J. X. et al., 2018). The recA gene was used as an internal control. All primers used for RT-PCR are listed in the Supplementary Table S4. Expression levels of target genes in clinical isolates were normalized to those in E. faecalis strain OG1RF.
Recombinant plasmids for overexpression of RS02205, RS12140, RS06145, RS06880, RS11485, and RS00630 were constructed as previously described (Lin et al., 2019) and separately transformed into one to three Erava-susceptible isolates with comparatively low transcript levels of the corresponding genes. All primers used for generating constructs in the present study are listed in the Supplementary Table S5. The MICs and heteroresistance of derivative isolates were evaluated after overexpression of the candidate genes were confirmed by qRT-PCR.
Continuous variables were analyzed with the Student’s t test and by one-way factorial analysis of variance using SPSS v17.0 software (SPSS Inc., Chicago, IL, United States). Differences were considered statistically significant at P < 0.05.
The distribution of Erava MICs against E. faecalis is shown in Table 1, with a focus on isolates resistant to the commonly used antibiotics LZD, VAN, ampicillin, gentamycin, erythromycin, ciprofloxacin, nitrofurantoin, and trimethoprim-sulfamethoxazole. Rates of antimicrobial susceptibility (MIC50/90) and resistance to these antibiotics and Erava are summarized in Supplementary Table S1. The range of Erava MIC values against the 276 clinical E. faecalis isolates analyzed in this study was 0.015–0.25 mg/l, and the MIC50/90 was 0.06/0.125 μg/ml. The MIC of Erava was lower than those of doxycycline and minocycline. Notably, Erava MIC was 0.03/0.125 mg/l against multidrug resistant E. faecalis, including 51 LZD-intermediate, 13 LZD-resistant, and two VAN-intermediate isolates. In addition, the MIC50/MIC90 of Erava was 0.06/0.06–0.125 mg/l against E. faecalis harboring one or more Tet resistance genes, including tet(M) and tet(L) (Table 2). Erava MIC for E. faecalis isolates harboring Tet resistance genes was ≤0.25 mg/l, indicating that Erava can overcome the antimicrobial resistance conferred by Tet resistance genes.
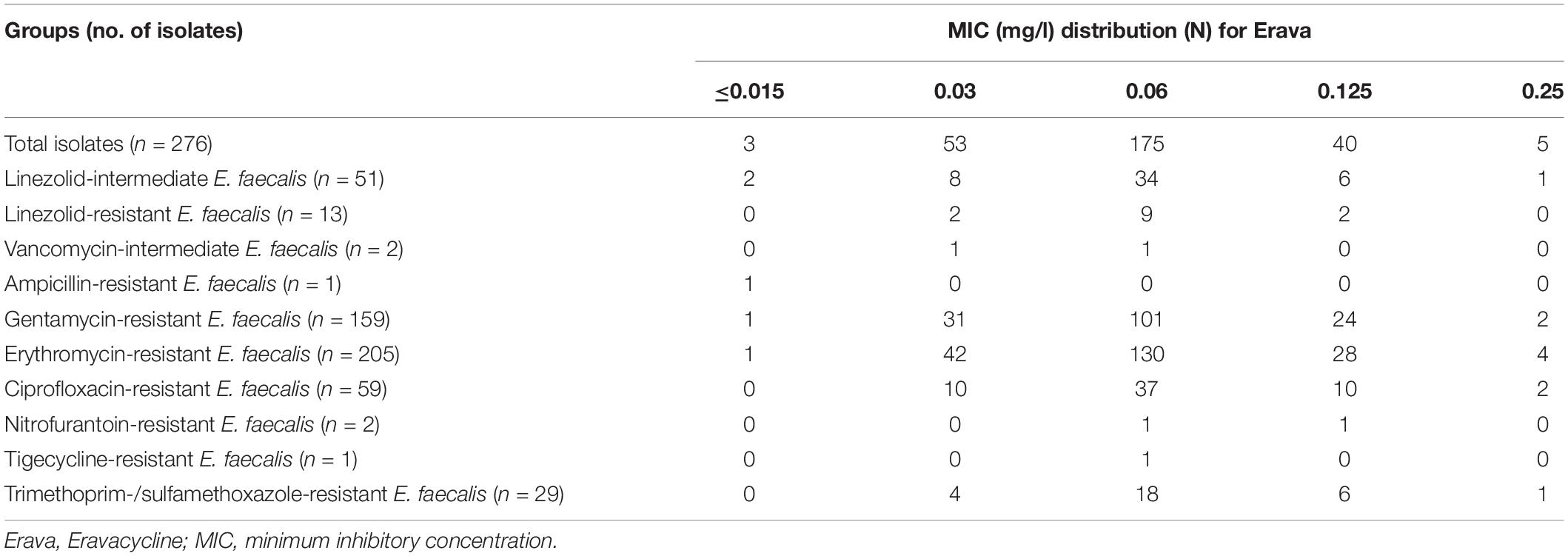
Table 1. MIC distributions for Eravacycline against E. faecalis resistant to commonly used antibiotics.
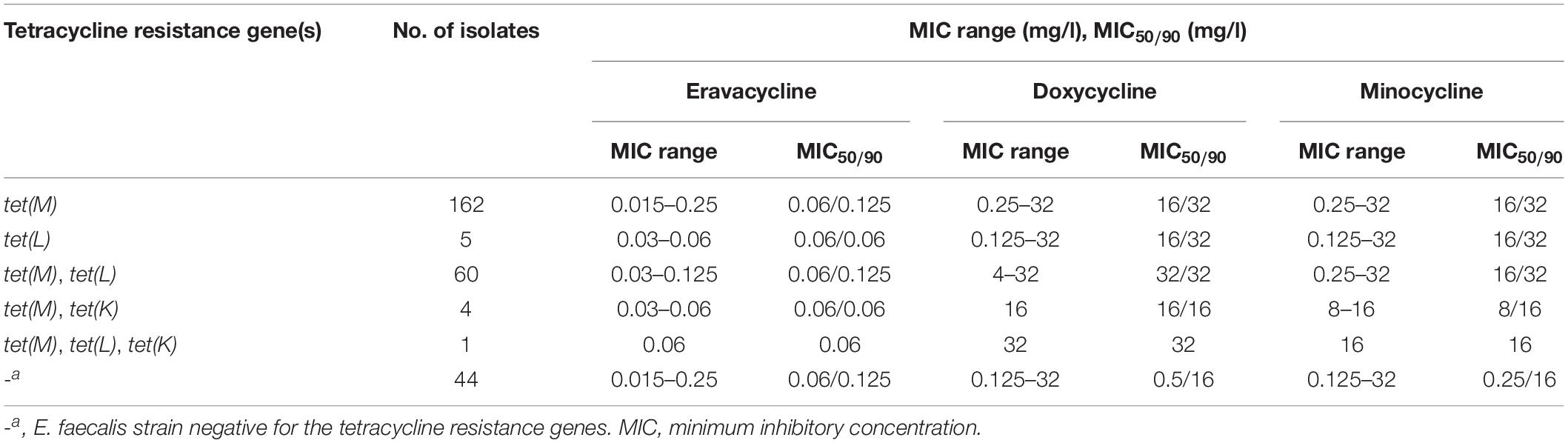
Table 2. Antimicrobial activity of Eravacycline against E. faecalis harboring tetracycline resistance genes.
Erava MICs were categorized into three levels – i.e., < 0.125, 0.125, and 0.25 mg/l – and the rate of Erava heteroresistance in E. faecalis with various MIC levels was evaluated by PAP. Erava heteroresistance was detected in 0.43% (1/231) and 7.5% (3/40) of isolates with Erava MICs of <0.125 and 0.125 mg/l, respectively. No Erava heteroresistance was observed among isolates with an MIC of 0.25 mg/l. Three heteroresistance-derived clones were selected for further analysis. MIC ranges of Erava and Tige were similarly elevated in these clones (0.5–4 mg/l for Erava and 0.5–8 mg/l for Tige) (Table 3).
To investigate the mechanism of Erava heteroresistance in E. faecalis, we screened for mutations in the Tet binding site of the 30S ribosomal subunit. However, there were no mutations in four copies of the 16S rRNA and 30 ribsomal protein S10 genes in heteroresistance-derived clones (Table 3). Moreover, Erava and Tige MICs were four times lower in clones treated with CCCP, although PAβN had no effect (Table 3), indicating that efflux pumps or membrane proteins contribute to the emergence of Erava heteroresistance.
To clarify the mechanism of Erava resistance in E. faecalis, resistance was induced in vitro under Erava selection pressure in strain OG1RF and two clinical isolates (FC1 and FC2). The MICs of Erava and Tige as well as mutations in the 30S ribosomal subunit were characterized in the resistant isolates. Erava and Tige MICs showed an increasing trend in E. faecalis isolates exhibiting Erava-induced resistance (Table 4); the increase in Erava MICs was accompanied by a higher frequency of mutations in the 16S rRNA gene. The mutation site varied across the four 16S rRNA gene copies, with high frequencies of the A1461G polymorphism in RR2 and of C1265T and C1191T in RR3. In addition, a K57Q amino acid substitution was a mutation hotspot in 30S ribosomal protein S10 of Erava-resistant E. faecalis.
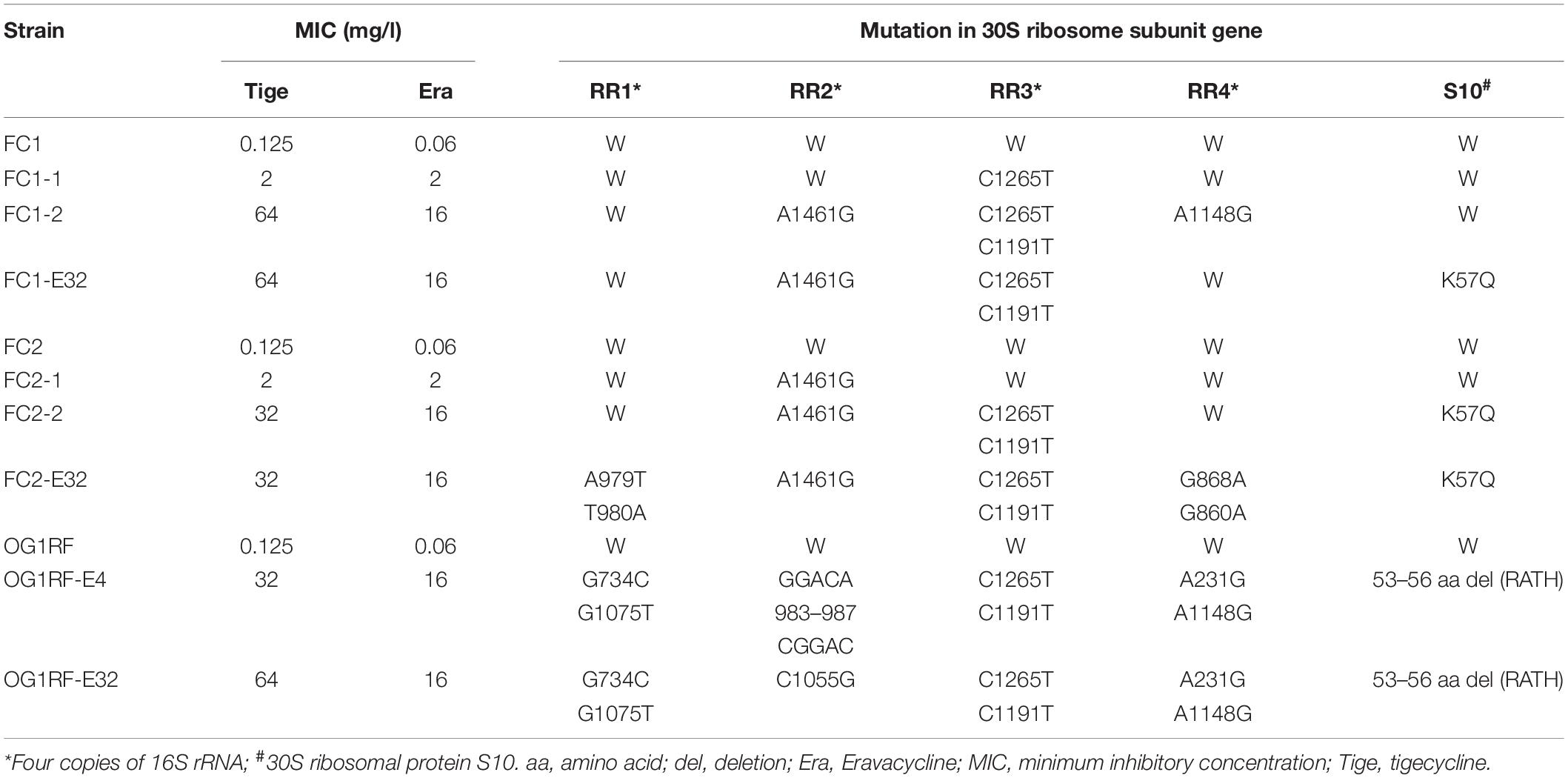
Table 4. MIC values and mutations in tetracycline binding sites in E. faecalis isolates exhibiting Erava-induced resistance.
Compared to the parent strains, the relative expression of six of the candidate genes (RS02205, RS12140, RS06145, RS06880, RS11485, and RS00630) was upregulated in the Erava-heteroresistant derivatives (Figure 1), suggesting that these genes might contribute to Erava resistance in E. faecalis.
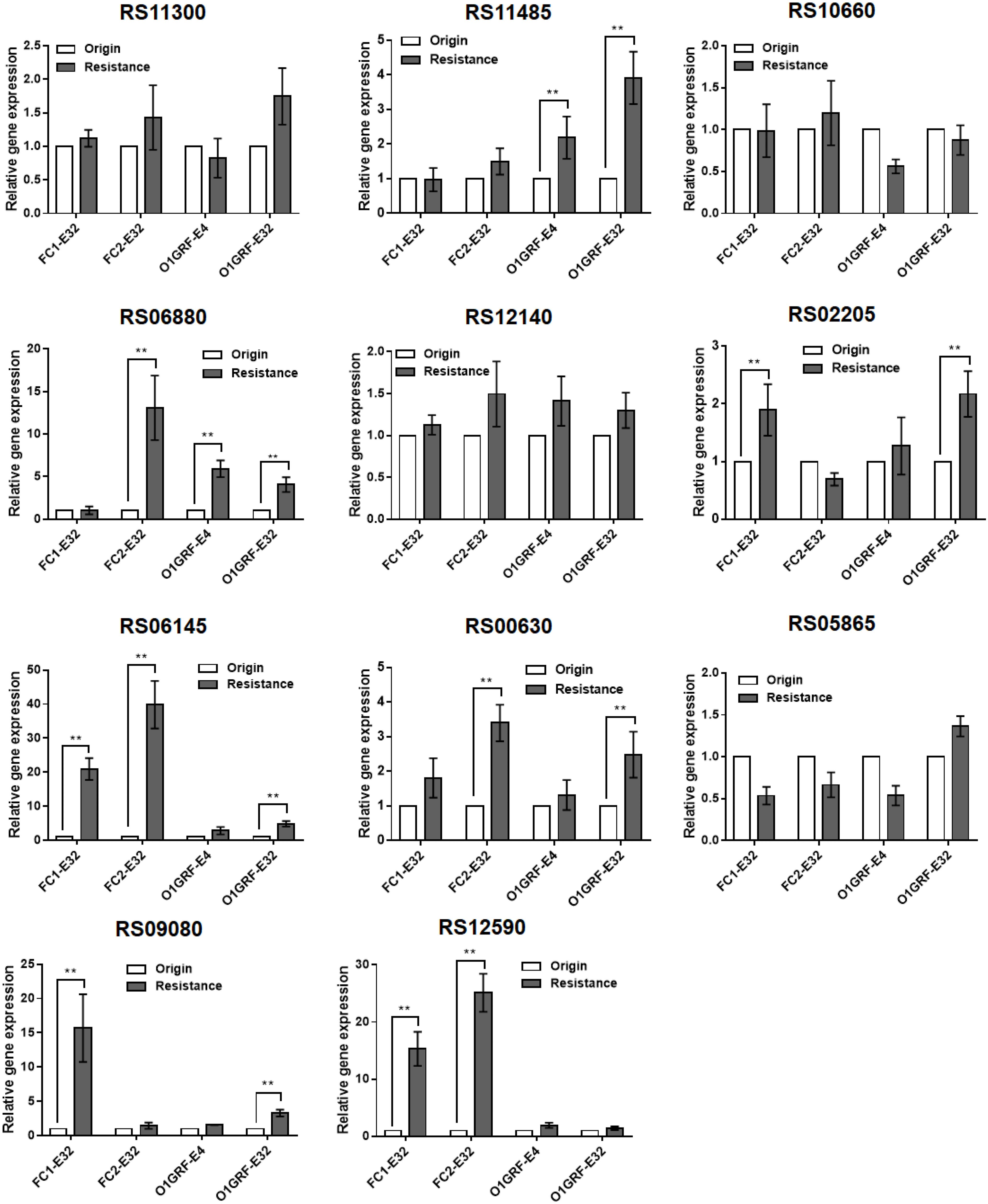
Figure 1. Correlation between transcript levels of 11 candidate genes and Erava-induced resistance in E. faecalis isolates. Transcript levels were determined by qRT-PCR, with the housekeeping gene recA serving as an internal control. The original (wild-type) strain was used as the reference (expression = 1.0). The Y axis represents relative fold change compared to the reference. Resistance, the expression level of corresponding gene in the resistant strain; origin, the expression level of corresponding gene in the origin strain. Resistant isolates and the reference strain are shown in Table 4. Data represent mean ± SD of three technical repeats. **P < 0.01 (Student’s t-test).
To explore the functional significance of the upregulated candidate genes in Erava heteroresistance, we transformed clinical E. faecalis isolates with low endogenous expression of the target genes with recombinant overexpression plasmids and evaluated their susceptibility to Erava and Tige. Target gene overexpression was confirmed by qRT-PCR. Introduction of the exogenous genes did not increase Erava or Tige MIC in the absence of antibiotic (Table 5). However, PAP results showed that overexpression of OG1RF_RS00630 encoding a bone morphogenetic protein (BMP) family ATP-binding cassette (ABC) transporter substrate-binding protein conferred Erava and Tige heteroresistance compared to the parent isolate. In addition, overexpression of OG1RF_RS06145 encoding a molybdate ABC transporter substrate-binding protein, OG1RF_RS12140 encoding an ABC transporter ATP-binding protein, and OG1RF_RS06880 encoding a coenzyme A-binding protein conferred heteroresistance to Tige but not Erava.
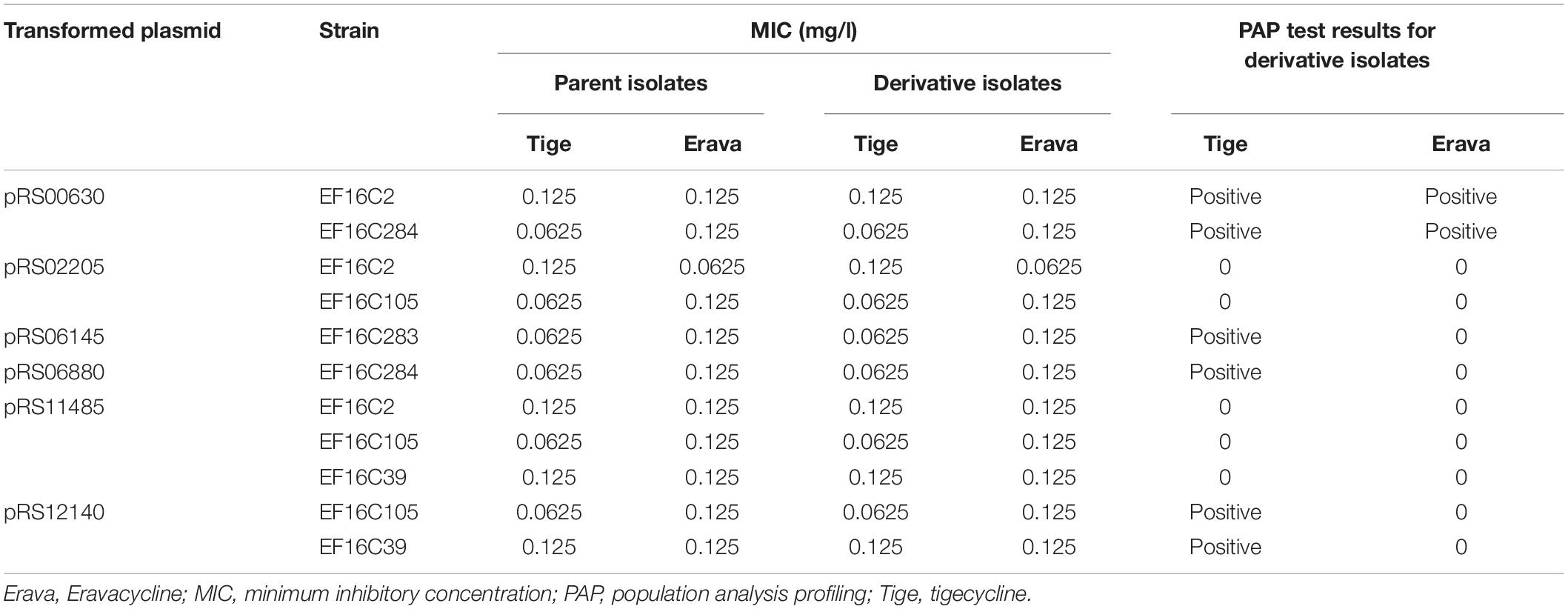
Table 5. Tige and Erava MICs in E. faecalis derivatives overexpressing candidate genes and their influence on PAP.
In this study, we investigated the in vitro antimicrobial activity of Erava against 276 non-duplicate clinical E. faecalis isolates from China. Consistent with the in vitro antimicrobial activity analyses results of Erava against clinical isolates collected globally (Hackel et al., 2013; Sutcliffe et al., 2013; Olesky et al., 2017), in Canada (Zhanel et al., 2018), and the USA (Bouchillon et al., 2018), the overall Erava MIC was ≤0.25 μg/ml against multidrug-resistant E. faecalis with intermediate or complete resistance to LZD and intermediate resistance to Vancomycin. Previous studies have seldom reported Tet-resistant E. faecalis or E. faecium with Erava MIC90 ≤0.125 and 0.06 μg/ml, respectively (Sutcliffe et al., 2013; Bai et al., 2018; Lan et al., 2019; McCarthy, 2019; Morrissey et al., 2019; Wang et al., 2019). Two primary mechanisms known to confer acquired resistance to Tet, the presence of Tet resistance genes encoding efflux pumps tet(K) and tet(L) and ribosomal protection proteins tet(M), had minimal or no effect on the in vitro antibacterial activity of Erava against clinical E. faecalis isolates. This likely stems from the fact that Erava have the potential to overcome these mechanisms (Grossman et al., 2012). Our results further confirm that Erava could be effective in the treatment of infectious diseases caused by E. faecalis, including isolates harboring one or more Tet resistance genes, including tet(M) and tet(L). Nevertheless, other Tet resistance genes, such as tet(U) and tet(X), involved in two other Mechanism of Tet resistance were not detected in our study and need to be further investigated in E. faecalis.
Our previous studies on the rate of Erava heteroresistance in K. pneumonia and S. aureus provided a basis for evaluating the risk of Erava resistance development (Zhang F. et al., 2018; Zheng J. X. et al., 2018). Our present findings further demonstrate that increasing Erava MICs in clinical E. faecalis isolates are associated with enhanced risk of heteroresistance. The concomitant increases in Erava and Tige MICs in clones exhibiting Erava-induced heteroresistance imply that Erava exposure leads to cross resistance to Tige. Thus, although Erava has relatively low MICs against clinical E. faecalis isolates, surveillance and early detection of Erava resistance is important; moreover, screening for Erava heteroresistance in clinical E. faecalis isolates should be considered as a routine diagnostic procedure.
Mutations affecting Tet binding to the 30S ribosomal subunit is a major factor responsible for both Tet and Tige resistance in several bacterial species (Nguyen et al., 2014; Grossman, 2016). As no mutations were detected in the Tet binding site of the 30S ribosomal subunit in Erava heteroresistant E. faecalis in the present work, it is likely that resistance to Tet and Erava involve distinct molecular mechanisms. However, mutations in the 16S rRNA gene and 30S ribosomal protein S10 are frequently detected in E. faecalis isolates exhibiting Erava-induced resistance, suggesting that they are contributing factors. Especially the mutation of C1191T in 16S rRNA RR3, which also occurred frequently in Omadacycline-induced resistance strains with MIC ≥ 8 mg/l (Lin et al., 2019), was only present in isolates with Erava MIC of 16 mg/l, suggesting that this site might play a major role in the occurrence of Erava resistance as well as Omadacycline resistance. It is intriguing to examine the impact of direct base mutagenesis on Erava or Omadacycline MIC to confirm the possibility. It has been demonstrated, however, that mutation of rpsJ encoding ribosomal protein S10 was sufficient to confer Tet resistance. It is worth noting that the mutation sites of Erava-induced resistant E. faecalis isolates were only present in residues 53–57. Of particular interest, a modification (R53Q-Δ54–57ATHK) that occurred in Tige-resistant E. faecalis strain selected in vitro resulted in a 4-fold increase in Tige resistance (Beabout et al., 2015). Most recently, Dabul et al. (2019) reported that a deletion of amino acids at position 56–59 (HKYK) was present at three Tige-resistant clinical E. faecalis isolates. Furthermore, our previous study also proposed that two amino acid substitutions H56Y and K57R appeared in E. faecalis OG1RF derivative isolates resistant to Omadacycline, another Tet class of antimicrobial drugs (Lin et al., 2019). Besides, plenty of studies have established that changes or deletions in residues 53–60 in the 30S ribosomal subunit protein S10 were linked to Tet resistance in Gram-positive bacteria Bacillus subtilis, Enterococcus faecium, and S. aureus and in Gram-negative bacteria E. coli, Acinetobacter baumannii, Neisseria gonorrhoeae, and K. pneumoniae (Williams and Smith, 1979; Wei and Bechhofer, 2002; Hu et al., 2005; Villa et al., 2014; Cattoir et al., 2015; Niebel et al., 2015). It has been proposed that this region of the S10 protein near the Tet binding pocket and its mutation may alter the interaction of Tets and 16S rRNA (Brodersen et al., 2000; Hu et al., 2005). Here, mutations, including a K57Q substitution and deletion of 53–56 (RATH), at the same region of the S10 protein were observed in Erava-induced resistant E. faecalis isolates, highlighting the relevance of alteration in this specific region of S10 protein for Tets including Erava resistance in E. faecalis.
Moreover, the increases in Erava and Tige MICs were reversed in the presence of CCCP (Table 3), indicating that efflux pumps or membrane proteins participate in the early phase of heteroresistance emergence. We previously showed that efflux pumps AcrAB, OqxAB, and MacAB contribute to the Erava resistant phenotype in K. pneumonia (Zheng J. X. et al., 2018). The RND-type efflux pump AcrEF was observed overexpression in two Erava-resistant K. pneumonia isolates (Zheng J. X. et al., 2018). Besides, adeB is involved in the regulation of efflux pump adeABC, and the Erava MIC in adeB-hyperexpressing A. baumannii was shown to be reduced by eightfold by disrupting the gene adeB (Abdallah et al., 2015). Similar to that of Omadacycline, our current results provide further evidence that ATP-binding cassette family efflux protein RS00630 overexpression contributes to heteroresistance to both Erava and Tige in E. faecalis. However, additional studies are needed to clarify the Erava resistance mechanism of RS00630. Although several other efflux pump coding genes were overexpressed in parts of Erava-induced resistance strains, the transgenic overexpression of these genes did not confer Erava resistance or heteroresistance. It turns out that these candidate genes do not correlate to Erava resistance. In addition, the clinical isolates tested in our study were limited by area and country, as these isolates were exclusively collected in one hospital. Therefore, it is important that other isolates are utilized to verify our work to understand the efficacy and underlying the resistance mechanism of these new antibiotics.
Erava showed excellent in vitro antimicrobial effects against clinical isolates of E. faecalis in China, including LZD non-susceptible strains with significantly lower MICs than doxycycline or minocycline. However, Erava heteroresistance may occur in E. faecalis isolates with MIC ≥ 0.125 mg/l. Overexpression of RS00630 encoding a BMP family ABC transporter substrate-binding protein in E. faecalis enhanced heteroresistance to both Erava and Tige, whereas RS12140, RS06145, and RS06880 were associated with heteroresistance to Tige only. Additional studies are needed to determine how RS00630 mediates antibiotic heteroresistance in E. faecalis. Our findings suggest that the emergence of Erava heteroresistance in E. faecalis clinical isolates – especially those with high Erava MICs – should be closely monitored.
All datasets generated for this study are included in the article/Supplementary Material.
Procedures involving human participants were performed in accordance with the ethical standards of Shenzhen Nanshan People’s Hospital and the Declaration of Helsinki 1964 and its later amendments. For this particular study, formal consent was not required.
ZY and QD conceived and designed the project. ZW, YS, and GX performed the molecular biology experiments and PAP tests. ZP, ZL, and BB performed the efflux inhibition experiment. ZC and JZ performed the MIC test. ZY, QD, and ZW wrote the manuscript with input from all other authors. All authors participated in data analysis.
This work was supported by grants from the National Natural Science Foundation of China (Nos. 81170370 and 81601797), Science, Technology, and Innovation Commission of Shenzhen Municipality of Key Funds (No. JCYJ20180508162403996) and Basic Research Funds (Nos. JCYJ20180302144340004, JCYJ20180302144721183, JCYJ20180302144345028, JCYJ20180302144431923, and JCYJ20180302144403714), Shenzhen Health and Family Planning Commission (Nos. SZXJ2018027 and SZXJ2017032), Sanming Project of Medicine in Shenzhen, Shenzhen Nanshan District Scientific Research Program of the People’s Republic of China (Nos. 2018010, 2018011, and 2018021), and Provincial Medical Funds of Guangdong (No. A2018163).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to thank the reviewer(s) for their valuable comments and suggestions, which have greatly improved on the quality of the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00916/full#supplementary-material
Abdallah, M., Olafisoye, O., Cortes, C., Urban, C., Landman, D., and Quale, J. (2015). Activity of eravacycline against Enterobacteriaceae and Acinetobacter baumannii, including multidrug-resistant isolates, from New York City. Antimicrob. Agents Chemother. 59, 1802–1805. doi: 10.1128/AAC.04809-14
Alosaimy, S., Abdul-Mutakabbir, J. C., Kebriaei, R., Jorgensen, S. C. J., and Rybak, M. J. (2020). Evaluation of eravacycline: a novel fluorocycline. Pharmacotherapy 40, 221–238. doi: 10.1002/phar.2366
Arias, C. A., and Murray, B. E. (2012). The rise of the Enterococcus: beyond vancomycin resistance. Nat. Rev. Microbiol. 10, 266–278. doi: 10.1038/nrmicro2761
Bai, B., Hu, K., Li, H., Yao, W., Li, D., Chen, Z., et al. (2018). Effect of tedizolid on clinical Enterococcus isolates: in vitro activity, distribution of virulence factor, resistance genes and multilocus sequence typing. FEMS Microbiol Lett. 365:fnx284. doi: 10.1093/femsle/fnx284
Beabout, K., Hammerstrom, T. G., Perez, A. M., Magalhães, B. F., Prater, A. G., Clements, T. P., et al. (2015). The ribosomal S10 protein is a general target for decreased tigecycline susceptibility. Antimicrob. Agents Chemother. 59, 5561–5566. doi: 10.1128/AAC.00547-15
Bouchillon, S., Hawser, S., Monti, F., et al. (2018). “Surveillance of the in vitro activity of eravacycline and comparators against clinical isolates from the US from 2013-2016. [abstract no. Global-MO11],” in Proceedings of the Surgical Infection Society Meeting, Madrid.
Brodersen, D. E., Clemons, W. M. Jr., Carter, A. P., Morgan-Warren, R. J., Wimberly, B. T., et al. (2000). The structural basis for the action of the antibiotics Tetracycline, Pactamycin, and Hygromycin B on the 30S Ribosomal Subunit. Cell 103, 1143–1154. doi: 10.1016/S0092-8674(00)00216-6
Cattoir, V., Isnard, C., Cosquer, T., Odhiambo, A., Bucquet, F., Guérin, F., et al. (2015). Genomic analysis of reduced susceptibility to tigecycline in Enterococcus faecium. Antimicrob. Agents Chemother. 59, 239–244. doi: 10.1128/AAC.04174-14
Dabul, A. N. G., Avaca-Crusca, J. S., Navais, R. B., Merlo, T. P., Van Tyne, D., Gilmore, M. S., et al. (2019). Molecular basis for the emergence of a new hospital endemic tigecycline-resistant Enterococcus faecalis ST103 lineage. Infect. Genet. Evol. 67, 23–32. doi: 10.1016/j.meegid.2018.10.018
Fiore, E., Van Tyne, D., and Gilmore, M. S. (2019). Pathogenicity of Enterococci. Microbiol. Spectr. 7:10.1128/microbiolspec.GPP3-0053-2018. doi: 10.1128/microbiolspec.GPP3-0053-2018
García-Solache, M., and Rice, L. B. (2019). The Enterococcus: a model of adaptability to its environment. Clin. Microbiol. Rev. 32:e00058-18. doi: 10.1128/CMR.00058-18
Grossman, T. H. (2016). Tetracycline antibiotics and resistance. Cold Spring Harb. Perspect. Med. 6:a025387. doi: 10.1101/cshperspect.a025387
Grossman, T. H., Starosta, A. L., Fyfe, C., O’Brien, W., Rothstein, D. M., Mikolajka, A., et al. (2012). Target- and resistance-based mechanistic studies with TP-434, a novel fluorocycline antibiotic. Antimicrob. Agents Chemother. 56, 2559–2564. doi: 10.1128/aac.06187-11
Hackel, M., Bouchillon, S., Biedenbach, D., and Sutcliffe, J. (2013). “Comparative analysis of eravacycline (TP-434) by broth microdilution, and disk diffusion [abstract no. E-. (1180)],” in Proceedings of the 53rd Annual Interscience Conferrence on Antimicrobial Agents and Chemotherapy, Denver, CO.
He, T., Wang, R., Liu, D., Walsh, T. R., Zhang, R., Lv, Y., et al. (2019). Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 4, 1450–1456. doi: 10.1038/s41564-019-0445-2
Hu, M., Nandi, S., Davies, C., and Nicholas, R. A. (2005). High-level chromosomally mediated tetracycline resistance in Neisseria gonorrhoeae results from a point mutation in the rpsJ gene encoding ribosomal protein S10 in combination with the mtrR and penB resistance determinants. Antimicrob. Agents Chemother. 49, 4327–4334. doi: 10.1128/AAC.49.10.4327-4334.2005
Lan, S. H., Chang, S. P., Lai, C. C., Lu, L. C., and Chao, C. M. (2019). The efficacy and safety of Eravacycline in the treatment of complicated intra-abdominal infections: a systemic review and meta-analysis of randomized controlled trials. J Clin Med. 8:E866. doi: 10.3390/jcm8060866
Lee, Y. R., and Burton, C. E. (2019). Eravacycline, a newly approved fluorocycline. Eur. J. Clin. Microbiol. Infect. Dis. 38, 1787–1794. doi: 10.1007/s10096-019-03590-3
Lin, Z., Pu, Z., Xu, G., Bai, B., Chen, Z., Sun, X., et al. (2019). Omadacycline efficacy against Enterococcus faecalis isolated in China: activity, heteroresistance, and resistance mechanisms. Antimicrob Agents Chemother. 23:aac.02097-19. doi: 10.1128/aac.02097-19
Livermore, D. M., Mushtaq, S., Warner, M., and Woodford, N. (2016). In vitro activity of Eravacycline against carbapenem-resistant Enterobacteriaceae and Acinetobacter baumannii. Antimicrob. Agents Chemother. 60, 3840–3844. doi: 10.1128/aac.00436-16
McCarthy, M. W. (2019). Clinical pharmacokinetics and pharmacodynamics of Eravacycline. Clin. Pharmacokinet. 58, 1149–1153. doi: 10.1007/s40262-019-00767-z
Mercuro, N. J., Davis, S. L., Zervos, M. J., and Herc, E. S. (2018). Combatting resistant enterococcal infections: a pharmacotherapy review. Expert Opin. Pharmacother. 19, 979–992. doi: 10.1080/14656566.2018.1479397
Monogue, M. L., Thabit, A. K., Hamada, Y., and Nicolau, D. P. (2016). Antibacterial efficacy of Eravacycline in vivo against Gram-positive and Gram-negative organisms. Antimicrob. Agents Chemother. 60, 5001–5005. doi: 10.1128/aac.00366-16
Morrissey, I., Olesky, M., Hawser, S., Lob, S. H., Karlowsky, J. A., Corey, G. R., et al. (2019). In vitro activity of Eravacycline against Gram-negative Bacilli isolated in clinical laboratories worldwide from 2013 to 2017. Antimicrob. Agents Chemother. 16:AAC.01699-19. doi: 10.1128/aac.01699-19
Nguyen, F., Starosta, A. L., Arenz, S., Sohmen, D., Donhofer, A., and Wilson, D. N. (2014). Tetracycline antibiotics and resistance mechanisms. Biol. Chem. 395, 559–575. doi: 10.1515/hsz-2013-0292
Niebel, M., Quick, J., Prieto, A. M., Hill, R. L., Pike, R., Huber, D., et al. (2015). Deletions in a ribosomal protein-coding gene are associated with tigecycline resistance in Enterococcus faecium. Int. J. Antimicrob. Agents 46, 572–575. doi: 10.1016/j.ijantimicag.2015.07.009
Olesky, M., Bassetti, M., Corey, R., et al. (2017). “In vitro global surveillance of eravacycline and comparators against Enterobacteriaceae, Acinetobacter baumannii, Stenotrophomonas maltophilia, including multidrug-resistant (MDR) isolates, over a three-year period (2013–15). [abstract no. 20],” in Proceedings of the ASM/ESCMID Conference on Drug Development to Meet the Challenges of Antimicrobial Resistance, Boston, MA.
Scott, L. J. (2019). Eravacycline: a review in complicated intra-abdominal infections. Drugs 79, 315–324. doi: 10.1007/s40265-019-01067-3
Sparo, M., Delpech, G., and García Allende, N. (2018). Impact on public health of the spread of high-level resistance to gentamicin and vancomycin in Enterococci. Front. Microbiol. 9:3073. doi: 10.3389/fmicb.2018.03073
Sun, J., Chen, C., Cui, C. Y., Zhang, Y., Liu, X., Cui, Z. H., et al. (2019). Plasmid-encoded tet(X) genes that confer high-level tigecycline resistance in Escherichia coli. Nat. Microbiol. 4, 1457–1464. doi: 10.1038/s41564-019-0496-4
Sutcliffe, J. A., O’Brien, W., Fyfe, C., and Grossman, T. H. (2013). Antibacterial activity of Eravacycline (TP-434), a novel fluorocycline, against hospital and community pathogens. Antimicrob. Agents Chemother. 57, 5548–5558. doi: 10.1128/aac.01288-13
Van Harten, R. M., Willems, R. J. L., Martin, N. I., and Hendrickx, A. P. A. (2017). Multidrug-resistant Enterococcal infections: new compounds, novel antimicrobial Therapies? Trends Microbiol. 25, 467–479. doi: 10.1016/j.tim.2017.01.004
Villa, L., Feudi, C., Fortini, D., García-Fernández, A., and Carattoli, A. (2014). Genomics of KPC-producing Klebsiella pneumoniae sequence type. 512 clone highlights the role of RamR and ribosomal S10 protein mutations in conferring tigecycline resistance. Antimicrob. Agents Chemother. 58, 1707–1712. doi: 10.1128/AAC.01803-13
Wang, L., Liu, D., Lv, Y., Cui, L., Li, Y., Li, T., et al. (2019). Novel plasmid-mediated tet(X5) gene conferring resistance to tigecycline, Eravacycline, and Omadacycline in a clinical Acinetobacter baumannii isolate. Antimicrob. Agents Chemother. 64:e01326-19. doi: 10.1128/aac.01326-19
Wei, Y., and Bechhofer, D. H. (2002). Tetracycline induces stabilization of mRNA in Bacillus subtilis. J. Bacteriol. 184, 889–894. doi: 10.1128/jb.184.4.889-894.2002
Williams, G., and Smith, I. (1979). Chromosomal mutations causing resistance to tetracycline in Bacillus subtilis. Mol. Gen. Genet. 177, 23–29. doi: 10.1007/bf00267249
Zhanel, G. G., Baxter, M. R., Adam, H. J., Sutcliffe, J., and Karlowsky, J. A. (2018). In vitro activity of eravacycline against 2213 Gram-negative and 2424 Gram-positive bacterial pathogens isolated in Canadian hospital laboratories: CANWARD surveillance study 2014–2015. Diagn. Microbiol. Infect. Dis. 91, 55–62. doi: 10.1016/j.diagmicrobio.2017.12.013
Zhanel, G. G., Cheung, D., Adam, H., Zelenitsky, S., Golden, A., Schweizer, F., et al. (2016). Review of Eravacycline, a novel fluorocycline antibacterial agent. Drugs 76, 567–588. doi: 10.1007/s40265-016-0545-8
Zhang, F., Bai, B., Xu, G. J., Lin, Z. W., Li, G. Q., Chen, Z., et al. (2018). Eravacycline activity against clinical S. aureus isolates from China: in vitro activity. MLST profiles and heteroresistance. BMC Microbiol. 18:211. doi: 10.1186/s12866-018-1349-7
Zheng, J. X., Lin, Z. W., Sun, X., Lin, W. H., Chen, Z., Wu, Y., et al. (2018). Overexpression of OqxAB and MacAB efflux pumps contributes to Eravacycline resistance and heteroresistance in clinical isolates of Klebsiella pneumoniae. Emerg. Microbes Infect. 7:139. doi: 10.1038/s41426-018-0141-y
Keywords: Eravacycline, Enterococcus faecalis, heteroresistance, tetracycline resistance genes, resistance mechanism
Citation: Wen Z, Shang Y, Xu G, Pu Z, Lin Z, Bai B, Chen Z, Zheng J, Deng Q and Yu Z (2020) Mechanism of Eravacycline Resistance in Clinical Enterococcus faecalis Isolates From China. Front. Microbiol. 11:916. doi: 10.3389/fmicb.2020.00916
Received: 19 January 2020; Accepted: 17 April 2020;
Published: 25 May 2020.
Edited by:
Rustam Aminov, University of Aberdeen, United KingdomReviewed by:
Ana R. Freitas, University of Porto, PortugalCopyright © 2020 Wen, Shang, Xu, Pu, Lin, Bai, Chen, Zheng, Deng and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiwen Deng, cWl3ZW5kZW5nQGhvdG1haWwuY29t; Zhijian Yu, eXV6aGlqaWFuc211QDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.