- 1Department of Hospital Infection Control, The First Affiliated Hospital of Nanchang University, Nanchang, China
- 2Department of Respiratory and Critical Care, The First Affiliated Hospital of Nanchang University, Nanchang, China
- 3Department of Infectious Disease, The First Affiliated Hospital of Nanchang University, Nanchang, China
Objective: Regional dissemination is the major cause of the widespread prevalence of a plasmid-encoding NDM-1 enzyme. We investigated the drug resistance, joint efficiency, and gene environment of a Klebsiella pneumoniae strain carrying blaNDM–1 gene.
Materials and Methods: Carbapenem-non-susceptible strains were analyzed using the VITEK 2 Compact. Strains carrying blaNDM–1 were identified using polymerase chain reaction and sequencing. Antimicrobial susceptibility testing and plasmid conjugation experiments were then conducted. Strains carrying blaNDM–1 were subjected to Southern blot analysis. After the gene mapping of blaNDM–1, library construction, and sequencing, plasmids were subsequently spliced and genotyped using the software Glimmer 3.0, and then analyzed using Mauve software.
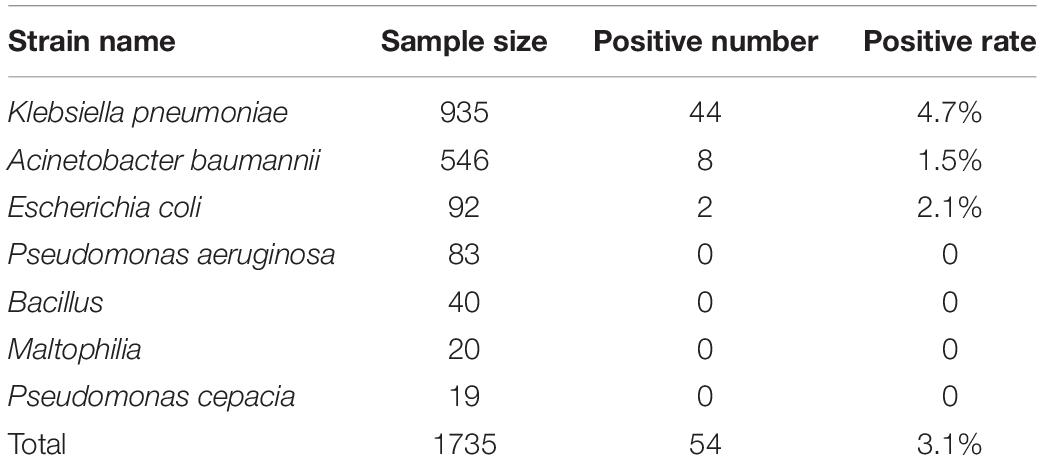
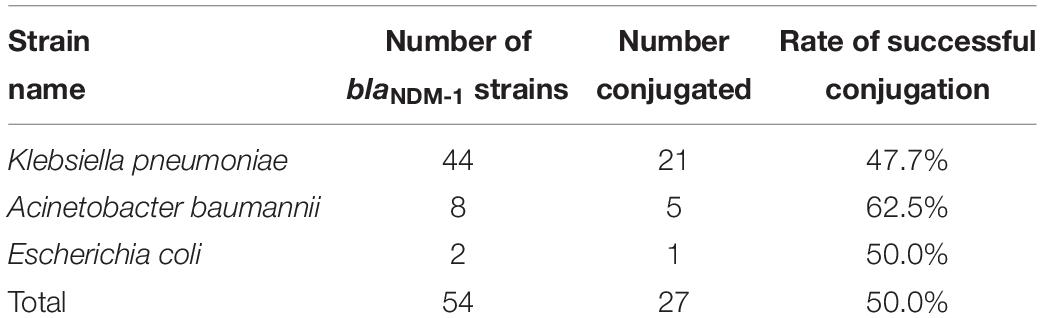
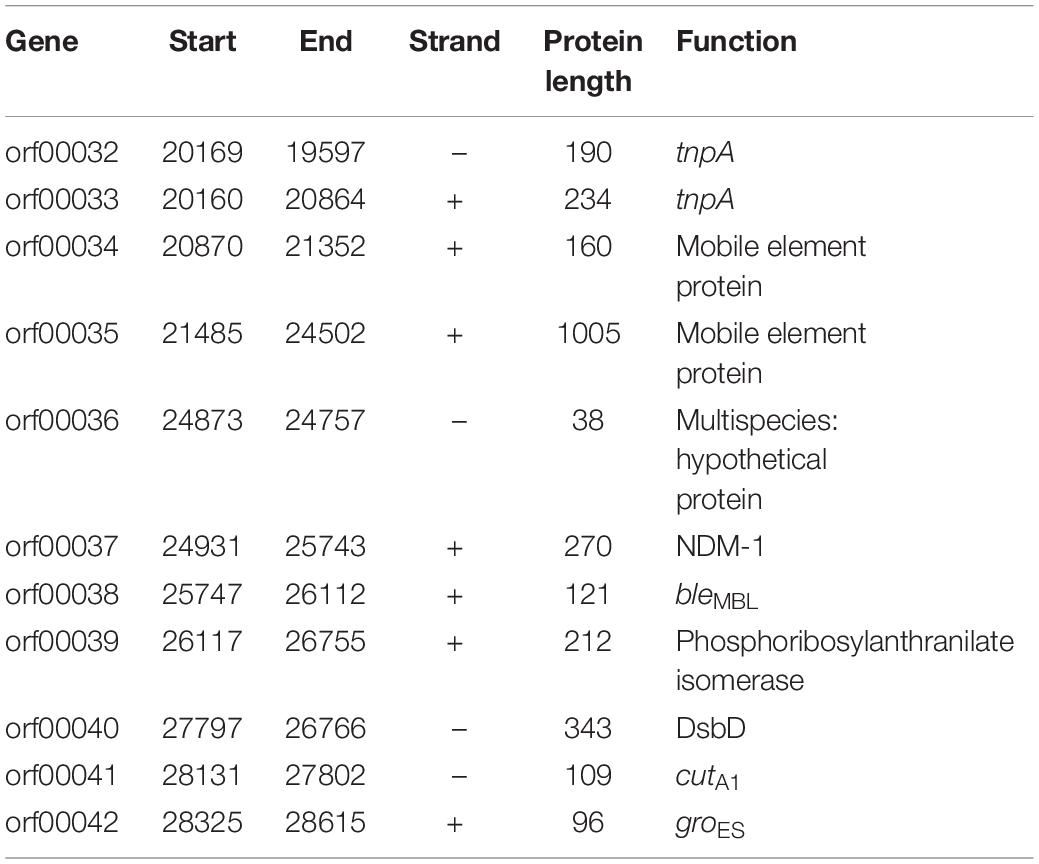
Results: Among 1735 carbapenem-non-susceptible strains, 54 strains of blaNDM–1-positive bacteria were identified, which consisted of 44 strains of K. pneumoniae, 8 strains of Acinetobacter baumannii and 2 strains of Escherichia coli. Strains carrying blaNDM–1 had a resistance rate of more than 50% in most antibiotics. Plasmid conjugation between strains carrying blaNDM–1 and E. coli strain J53 had a success rate of 50%. Southern blot analysis indicated that each strain had multiple plasmids containing blaNDM–1. Among the five plasmids containing blaNDM–1 in K. pneumoniae for sequencing, two plasmids with complete sequences were obtained. The findings were as follows: (i) The p11106 and p12 plasmids were highly similar to pNDM-BTR; (ii) the p11106 and p12 plasmids showed differences in the 20–30 kb region (orf00032–orf00043) from the other six plasmids; and (iii) blaNDM–1 was located at orf00037, while ble was found at orf00038. Two tnpA genes were located in the upstream region, and orf00052 (tnpA) in the 36 kb region was in the downstream sequence.
Conclusion: blaNDM–1-containing bacteria exhibit multidrug resistance, which rapidly spreads and is transferred through efficient plasmid conjugation; the multidrug resistance of these bacteria may be determined by analyzing their drug-resistant plasmids. The presence of ble and tnpA genes suggests a possible hypothesis that blaNDM–1 originates from A. baumannii, which is retained in K. pneumoniae over a long period by transposition of mobile elements.
Introduction
The clinical application of sulfa drugs can be traced back to the 1930s, which marked a new era of antimicrobial therapy. Once exposed to antibacterial drugs, bacteria spontaneously change their metabolic pathways or produce corresponding inactivating substances to resist antibiotics, exhibiting drug resistance. Notably, the abuse of antibiotics poses selective pressure of bacteria, conferring a survival advantage on drug-resistant bacteria. Consequently, numerous drug-resistant bacteria are spread in different pathogens.
New Delhi metallo-β-lactamase1 (NDM-1), also known as metallo-β-lactamase or metal-β-lactamase, was first isolated from a highly infectious and pathogenic multidrug-resistant strain of Klebsiella pneumoniae in 2009 (Yong et al., 2009). Cases of infection with the blaNDM–1 gene have subsequently been reported in more than 20 countries and regions worldwide, including the United Kingdom and India (Kumarasamy et al., 2010). The therapeutic efficacy of multiple antibiotic treatments for bacteria carrying blaNDM–1 is usually unsatisfactory. Therefore, bacterial strains carrying blaNDM–1 are also called superbugs. With a prolonged length of stay, bacteria-carrying blaNDM–1 have a higher probability of being isolated from stool samples. However, infection with blaNDM–1 cannot be determined from clinical symptoms and signs (Bush and Fisher, 2011). Superbugs pose a serious challenge to antibiotic therapy.
The blaNDM–1 gene is mainly distributed in plasmids and occasionally in the chromosomes of Escherichia coli, Pseudomonas aeruginosa, and Proteus mirabilis (Girlich et al., 2015; Rahman et al., 2015; Shen et al., 2017). In clinical practice, blaNDM–1 plasmids from different bacterial species isolated from the same patient typically have a similar structure, suggesting the significance of plasmids in the spread of blaNDM–1. Plasmids containing blaNDM–1 vary in size from 30–300 kb, and exist in different types, such as IncA/C, IncL/M, and IncR (Carattoli et al., 2015; Gamal et al., 2016). Specifically, IncA/C has a wide range of hosts and can exist in multiple strains, such as Enterobacteriaceae, Pseudomonas, Acinetobacter, and Vibrio cholerae, providing convenience for blaNDM–1 in different species of bacterial hosts (Wailan and Paterson, 2014).
The sequences and genetic modes of blaNDM–1 have been identified in previous studies. However, the blaNDM–1 gene environment is yet to be determined. In addition, studies mostly focus on blaNDM–1 in Acinetobacter (Bontron et al., 2016; Wang et al., 2017) and rarely on blaNDM–1 carried by K. pneumoniae. In the present study, 1735 carbapenem-non-susceptible bacteria were collected from The First Affiliated Hospital of Nanchang University. These strains were sequenced, conjugated, and compared with the corresponding plasmids. This study aimed to analyze the differences in the blaNDM–1 gene environment and provide directions in clarifying the origin and propagation of the blaNDM–1 gene.
Materials and Methods
Sample Collection and Identification
Approval was obtained from the Medical Ethics Committee of The First Affiliated Hospital of Nanchang University and informed consent was obtained from each subject. Carbapenem-non-susceptible bacteria were then collected from The First Affiliated Hospital of Nanchang University from January 2013 to December 2016. A total of 1735 carbapenem-non-susceptible bacteria were isolated and then identified using VITEK 2 Compact (Pioneering Diagnostics, France) at the Microbiology Laboratory at The First Affiliated Hospital of Nanchang University.
Polymerase Chain Reaction and Sequencing
Polymerase chain reaction (PCR) templates were prepared by boiling. Fresh bacteria were harvested and diluted in 500 μL ddH2O for 10 min in a boiling water bath. The supernatant was collected as PCR templates. Subsequently, a PCR system with a total volume of 50 μL was prepared, consisting of 25 μL of Taq Mix (Takara, Dalian), 2 μL of forward primer, 2 μL of reverse primer, 2 μL of template, and 19 μL of ddH2O. Subsequently, 29 cycles of PCR were conducted. The blaNDM–1 primer sequences were forward 5’-GGCGGAAGGCTCATCACGA-3’ and reverse 5’-CGCAACACAGCCTGACTTTC-3’. The amplified product was 287 bp.
The PCR products were analyzed using electrophoresis with 1% agarose gel and 1 × TAE at 120 V for 25 min. Strains verified by sequencing to contain blaNDM–1 were used as markers. Electrophoresis results were obtained using an ultraviolet (UV) transilluminator. Positive PCR products were sequenced (Synbio Technology, Suzhou, China), and the sequencing results were compared using the software BLAST. Strains that were positive for PCR and matched the sequencing results were identified as those containing blaNDM–1.
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing was conducted using the zone of inhibition test, and the comprehensive drug resistance of each antibiotic was determined using the E-test. The antibiotics screened in this study included imipenem, meropenem, ertapenem, amikacin, amoxicillin/clavulanicacid, aztreonam, ceftriaxone, ceftazidime, cephalosporins, cefoxitin, cefazolin, ciprofloxacin, gentamicin, levofloxacin, piperacillin/tazobactam, trimethoprim/sulfamethoxazole, tetracycline, ticarcillin/clavulanicacid, and tobramycin.
Plasmid Conjugation
The receptor strain was sodium azide-resistant E. coli strain J53. The donor and receptor strains were implanted in the Mueller-Hinton plate and cultured at 37°C for 16–18 h. Strains in appropriate amounts were inoculated in a glass tube containing 5 mL of LB medium and then cultured at 37°C for 16–18 h. Subsequently, 400 μL of the donor strain and 200 μL of the receptor strain were added to a glass tube containing 800 μL of LB broth medium and then cultured at 37°C for 16–18 h. Meanwhile, the donor strain, screened in 180 μg/mL sodium azide, and the receptor strain, screened in 0.5 μg/mL of imipenem, were used as blank controls. Exactly 100 μL of the aforementioned mixture was added to the Mueller-Hinton plate and cultured at 37°C for 16–18 h. Conjugation strains in good condition were ultimately identified using the VITEK 2 compact automatic microbial identification instrument.
Southern Blot Analysis
Southern blot analysis was conducted using 1% agarose gel with 1 × TAE and run on 120 V electrophoresis for 40 min. The gel was incubated in 0.25 mol/L HCl for 15 min, 0.5 mol/L NaOH for 20 min twice, and 0.1 mol/L phosphate buffer for 15 min twice. Membrane transfer was conducted in a 20 × saline-sodium citrate buffer. The membrane was washed in a 2 × saline–sodium citrate buffer and then dried in a baking oven at 80°C for 2 h. PCR products carrying blaNDM–1 were labeled using the DIG High Prime DNA Labeling and Detection Starter Kit I (Roche, United States). DIG-labeled DNA products were prepared and examined using Southern blot analysis, and images were obtained using a UV transilluminator.
Plasmid Sequencing
Five qualified plasmids containing blaNDM–1 were used to construct a sequencing library. These 5 vectors were from 4 strains, which showed high resistance to the antibiotics and successfully conjugated with E. coli J53. The data are provided as Supplementary Material and Supplementary Table S2. Briefly, 1 μg of plasmid was placed in a Covaris tube, and the DNA was separated into 400 bp fragments using Covaris S2 (Covaris, United States). Small DNA fragments were generated for library construction using the NEXTflex DNA Sequencing Kit compatible with Biomek FXp (Bio Scientific, United States). The library fragments were subjected to paired-end sequencing (2 × 150 bp) on the HiSeq2500 Sequencing System (Illumina, United States).
Clean reads after pre-processing were assembled using Velvetver.1.2.03 software. Gene prediction and annotation analyses were performed using Glimmer 3.0 software. The p11106 and p12 plasmids were compared with the plasmid sequences without blaNDM–1 of the seven species. Similarities in the plasmid sequences were depicted using Mauve software. Gene functions in different regions were annotated and analyzed.
Results
Screening and Identification of Strains Carrying blaNDM–1
Among the 1735 carbapenem-non-susceptible strains harvested in this experiment, 54 strains (3.1%) were blaNDM–1-positive. These strains consisted of 44 strains of K. pneumoniae, 8 strains of A. baumannii, and 2 strains of E. coli. The blaNDM–1 gene was not found in P. aeruginosa, Enterobacter cloacae, Bacillus, Maltophilia, or Pseudomonas cepacia. All sequencing results of the 54 strains were consistent with the NCBI database1. The positive rates of each strain are listed in Table 1.
These 54 multidrug-resistant strains were obtained from 43 patients at The First Affiliated Hospital of Nanchang University from January 2013 to December 2016. The patients were from different cities and provinces. Temporal and regional differences suggested that the same strains could not be causing the outbreak. Patient records are shown in Supplementary Table S1.
Determination of Drug Resistance
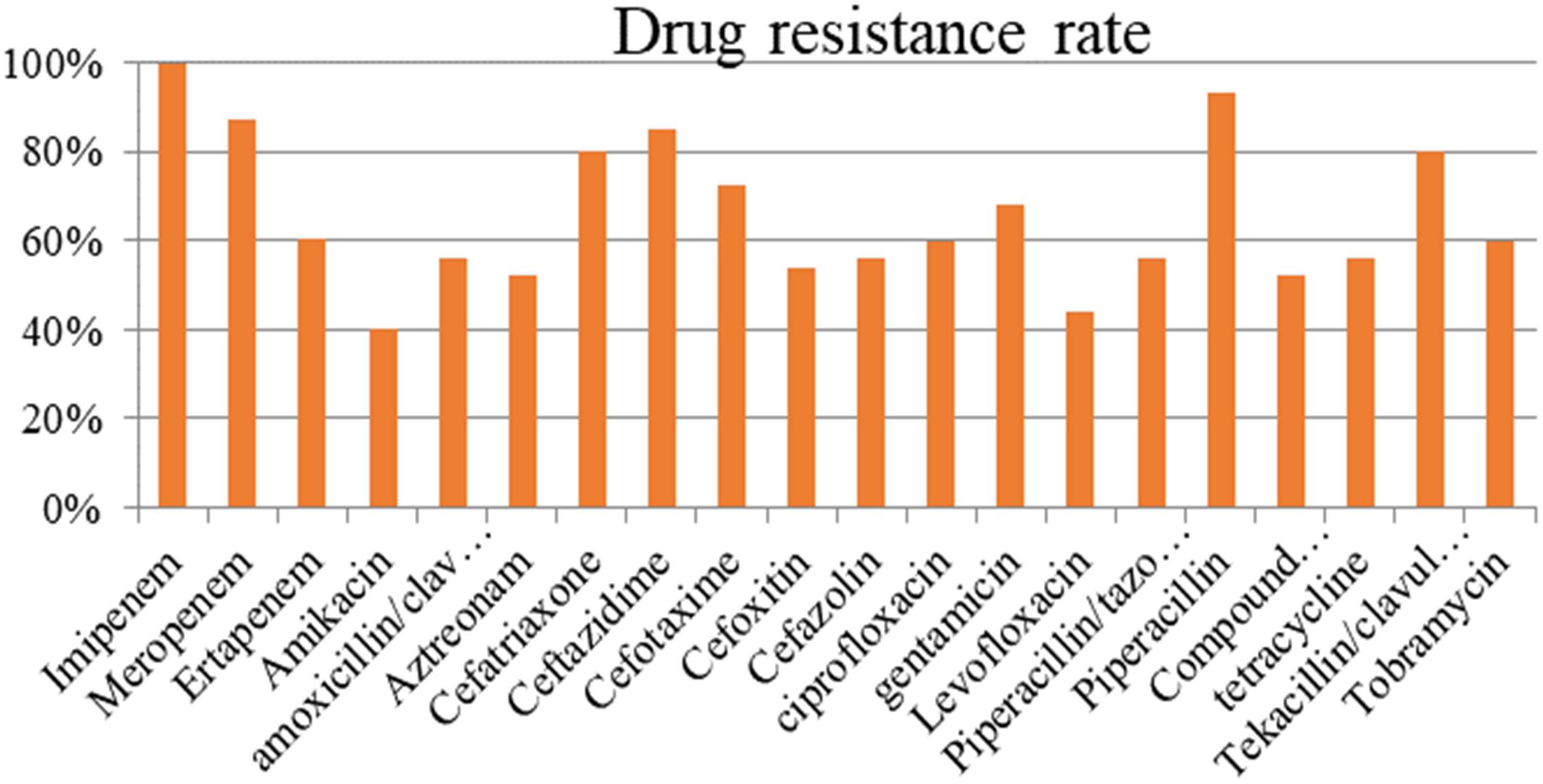
The resistance rate of the strains carrying blaNDM–1 exceeded 50% in most antibiotics (Figure 1). The tested antibiotics exhibited nearly 100% resistance to imipenem and more than 90% resistance to meropenem and piperacillin. Meanwhile, the blaNDM–1-positive strains showed the lowest resistance (40%) to amikacin.
Plasmid Conjugation
The plasmid conjugation experiment was conducted on all blaNDM–1-positive strains and J53. A total of 27 strains (21, K. pneumoniae strains; 5, A. baumannii strains; and 1, E. coli strain) successfully conjugated their plasmids containing the blaNDM–1 gene (Table 2). The success rate of plasmid conjugation was 50%.
Location of the blaNDM–1 Gene
BlaNDM–1-positive strains that were successfully conjugated were subjected to Southern blot analysis to detect the location of blaNDM–1. The results further demonstrated the conjugation of blaNDM–1 in E. coli strain J53 (Figure 2).
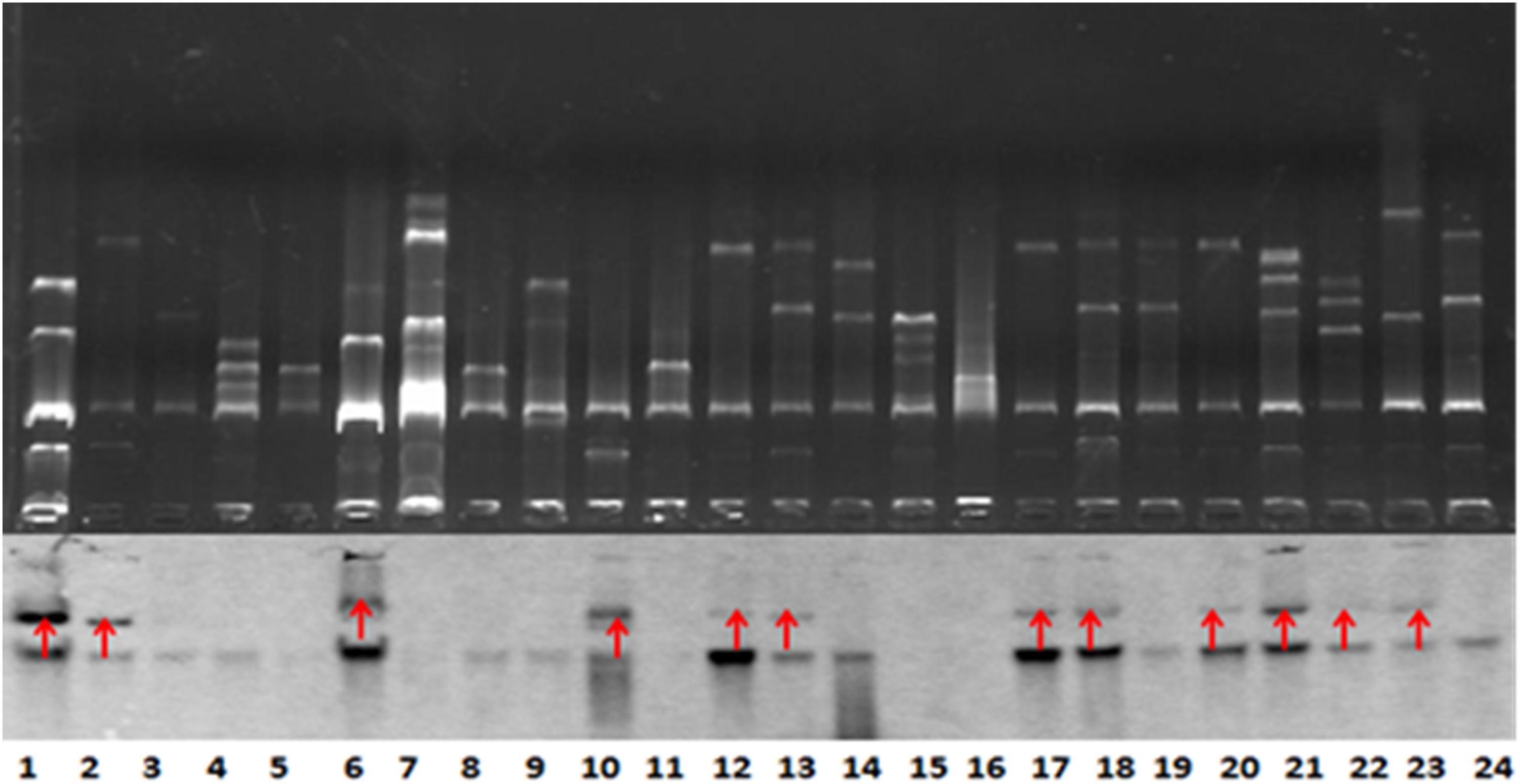
Figure 2. Southern blot analysis for blaNDM–1 localization. The conjugation of blaNDM–1-positive strain J53 was visualized. The 12 strains are denoted by red arrows. Lanes 12, 13, 17, 18, 20, 21, 22, and 23 are visualized bands that indicate Klebsiella pneumoniae strains carrying blaNDM–1. Lanes 6 and 10 indicate Acinetobacter baumannii strains carrying blaNDM–1. Lanes 1 and 2 indicate Escherichia coli strains carrying blaNDM–1. The lower part of this figure is mirrored with the top part.
Sequencing of Plasmids
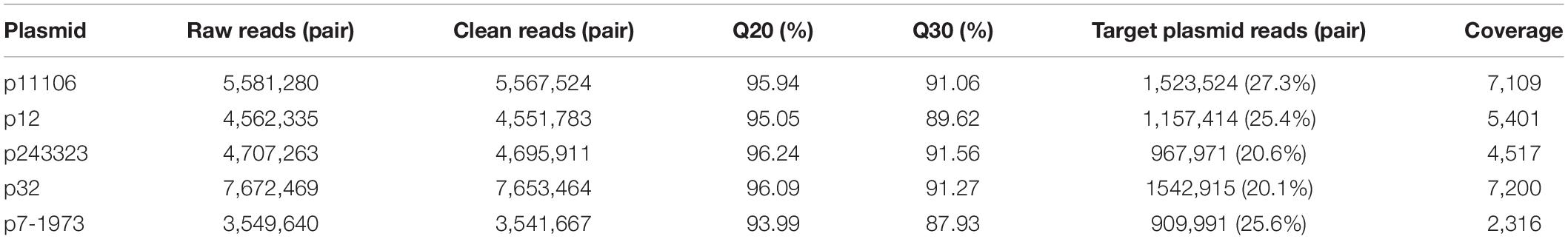
Plasmid libraries were constructed from genomic DNA. Qualified plasmid DNAs were sequenced, and the results suggested excellent qualifications for library construction and sequencing (Table 3). After removal of DNA from host genes and other plasmid DNAs, approximately 20% of the reads were extracted from target plasmid DNAs with over 2000× coverage.
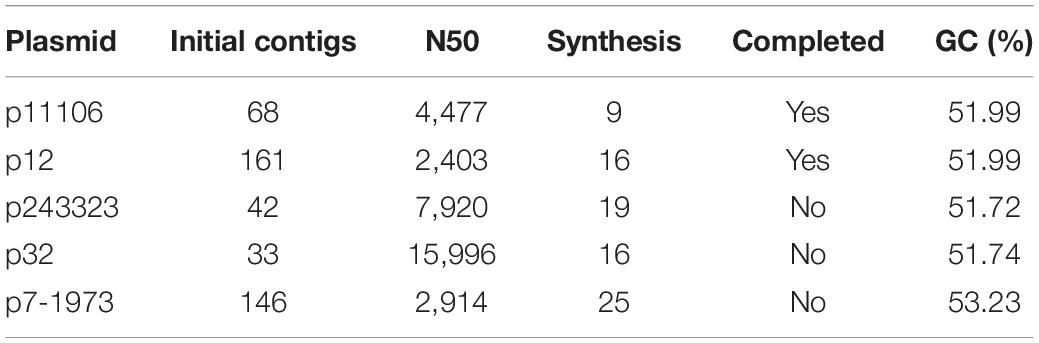
Subsequently, the clean reads of each plasmid were synthesized using Velvet. Large-fragment sequence assembly is presented in Table 4. Although coverage was sufficiently high (>2000×), the synthesis was not satisfactory, with the contigs between 33 and 161. Thus, artificial gap closing with the KU862632.1 strain was conducted using the Cytoscape platform, and two complete plasmid (p12 and p11106) sequences were generated. Synthesis of p243323, p32, and p7-1973 failed (Table 4).
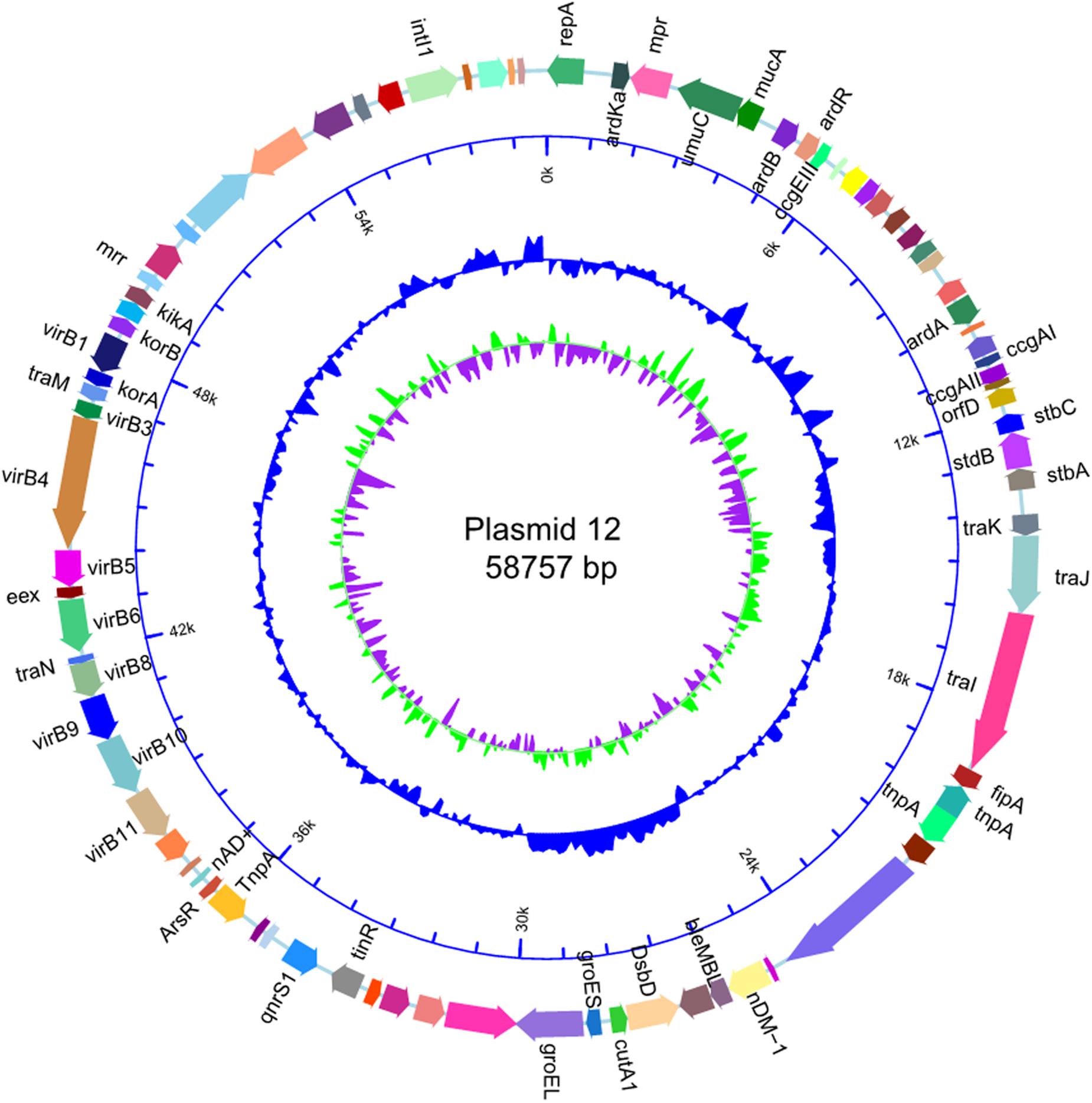
The size of p12 was the same as that of p11106 (58757 bp). Cytosine was located on 50396 bp of p11106, thymine was located in p12, and the remaining regions were the same. In the following gene prediction and annotation analysis, p12 was used as an example. As depicted in Figure 3, 46 genes were reverse transcribed, and 39 were forward transcribed without chain specificity (p = 0.627). We uploaded the sequencing data to the NCBI database under the SRA (Sequence Read Archive) accession number PRJNA596354.
Comparative Analysis of the blaNDM–1 Gene Environment
Plasmid sequences from seven species were subjected to similarity analysis using Mauve software. The baseline characteristics of the selected plasmids are listed in Table 5. p11106 and p12 were compared with the aforementioned plasmids, and their similarity information is depicted in Figure 4 and Table 6.
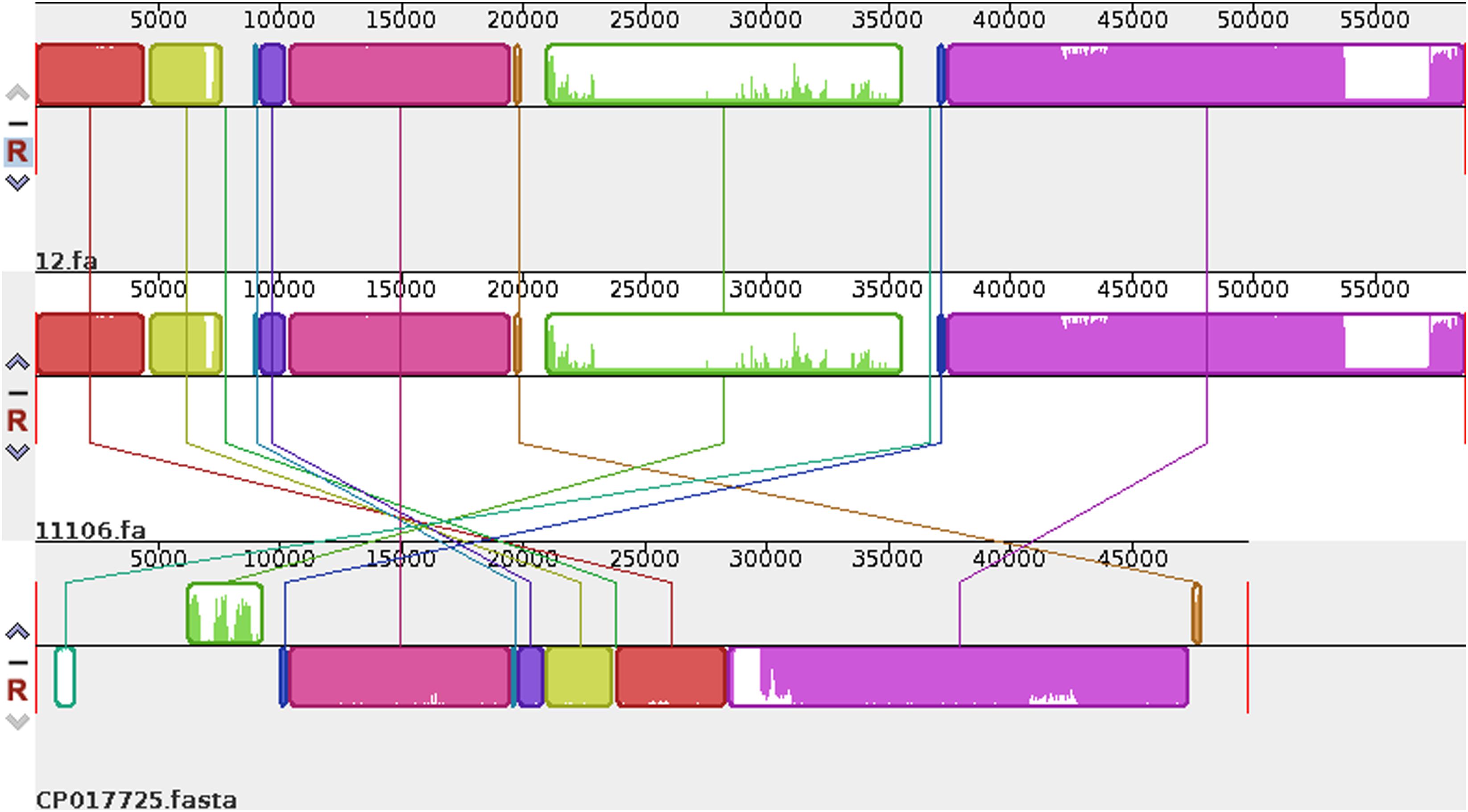
Figure 4. Comparison between p12 and p11106 with pSARB26_02. Differences between p12 and p11106 with pSARB26_02 (CP017725) were mainly enriched in the 20–37 kb region (green region), with orf00032–orf00052 in p11106 and p12. Two TnpA transposases are located in this region (orf00032 mapped 20169–19597 bp, negative strand; orf00033 mapped 20160–20864 bp, positive strand), indicating that gene insertion is highly possible in this region of p11106 and p12. Similarly, additional 3.5 kb sequences present in the 3’ UTR of p11106 and p12 were mapped orf00078–orf00082. This may also be explained by gene transposition or insertion.
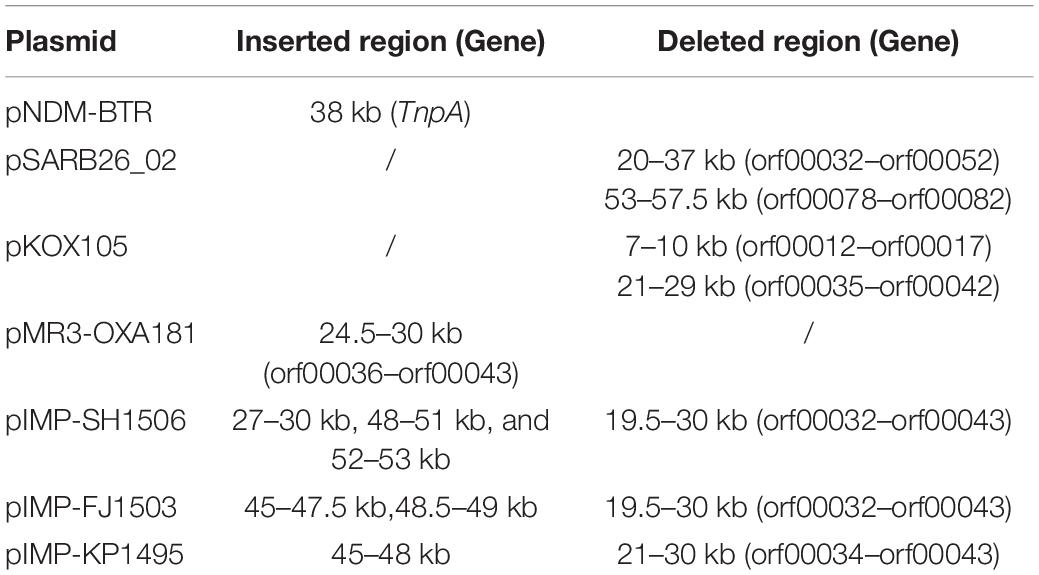
In addition to the plasmid pNDM-BTR, differences between p11106 and p12, and the remaining seven plasmids were mainly enriched in the 20–30 kb region. Although the plasmid pMR3-OXA181 was not absent in this region, it exhibited a considerably low homology to p11106 and p12. The corresponding genes mapping this 10 kb region were orf00032–orf00043. Moreover, gene annotation showed that two genes (TnpA and a mobile element protein) with similar functions were located in the upstream sequences of this region. BlaNDM–1 was also located in these sequences (orf00037). IMP-4 and β-lactamase were absent in this region. The gene annotations of orf00032–orf00043 are presented in Table 7.
As shown in Table 7, (i) the p11106 and p12 plasmids were highly similar to pNDM-BTR; (ii) the p11106 and p12 plasmids showed differences in the 20–30 kb region (orf00032–orf00043) from the other six plasmids; and (iii) blaNDM–1 was located in the middle of the orf00032–orf00043 region, whereas ble was found nearby. Two tnpA genes were on the top, and orf00052 (tnpA) in the 36 kb region was in the downstream sequence.
Discussion
Among the 1735 carbapenem-non-susceptible strains, 54 (3.1%) of blaNDM–1-positive bacteria were identified, consisting of 44 strains (4.7%) of K. pneumoniae, 8 strains (1.5%) of A. baumannii, and 2 strains (2.1%) of E. coli. Our findings are consistent with previous studies showing that blaNDM–1 is mainly carried by K. pneumoniae, E. coli, A. baumannii, and E. cloacae (Deshpande et al., 2010; Rolain et al., 2010; Zarfel et al., 2011; Dongmei et al., 2017). Among the collected strains carrying blaNDM–1 in our hospital, K. pneumoniae (4.7%) had the highest positive rate, which was inconsistent with other reports that identified Acinetobacter and Klebsiella as the major blaNDM–1-positive bacteria (Chen et al., 2014; Zhang et al., 2014; Shengshu et al., 2015). Both Acinetobacter and Klebsiella are significant in the regional dissemination of blaNDM–1-positive bacteria. K. pneumoniae could be the vital preservation host of blaNDM–1.
Antimicrobial susceptibility testing indicated that NDM-1-resistant bacteria exhibited strong resistance to most antibiotics (except for amikacin), with a total resistance rate above 50%. The drug resistance of NDM-1 to aminoglycosides, β-lactam antibiotics/β-lactamase inhibitors, and carbapenems are relatively high (Shengshu et al., 2015). NDM-1 has a drug resistance of up to 100% for dolipenem, ampicillin, furadantin, cefazolin, cefuroxime, cefotaxime, ceftriaxone sodium, ceftazidime, and cefoxitin. Its drug resistance rates to some carbapenems are as follows: meropenem, 97.30%; imipenem, 97.30%; and ertapenem, 92.00%. However, the drug resistance rates of NDM-1 to polymyxin B (37.93%), amikacin (32.65%), tigecycline (7.69%), and polymyxin E (2.33%) are significantly lower, compared with other antibiotics. NDM-1-resistant bacteria might be mostly multidrug-resistant, and plasmids containing blaNDM–1 can perform multiple drug-resistant gene transfers (Barguigua et al., 2015; Sarkar et al., 2015; Villa et al., 2015; Liu et al., 2016).
Our experiment obtained 27 successfully conjugated J53 strains resistant to sodium azide, which is consistent with previous studies (Wailan et al., 2015; Kocsis et al., 2016). Conjugation with K. pneumoniae, A. baumannii, and E. coli as donor plasmids containing blaNDM–1 achieved a success rate of approximately 50%. Southern blot analysis confirmed that blaNDM–1 was mainly expressed in plasmids, and each drug-resistant strain carrying blaNDM–1 could contain multiple blaNDM–1-positive plasmids. The blaNDM–1-positive plasmids exhibited a relatively strong ability for conjugation transfer. Specifically, IncA/C showed adaptation to a wide range of hosts. It can be found in Enterobacteriaceae, Pseudomonas, Aeromonas, Vibrio cholera, and other bacterial genera, and can be transmitted within or between strains through conjugation (Wailan and Paterson, 2014; Carattoli et al., 2015; Gamal et al., 2016).
Sequencing of five plasmids obtained from the collected K. pneumoniae strains was highly qualified (coverage >2000×). Regardless, the synthesis of p243323, p32, and p7-1973 failed; only p11106 and p12 were successfully synthesized. This result could be explained by the complexity of the plasmid structure and interruption of the host genome and other plasmids during plasmid extraction. Among the sequencing reads, only 20–30% of the plasmids were identified to carry theblaNDM–1 gene, significantly increasing the difficulty of synthesis. The only difference between p11106 and p12 was the 50396 bp (orf00074 was mapped) region, where c.587T > C occurred in p11106 and thus resulted in p.196Ile > Thr. A total of 85 genes were contained in the p11106 sequence, and most of them had functions that could be identified using BLAST.
The structures of plasmids p11106 and p12 were similar, and their strains Kp.11 and Kp.12 were also similar in resistance (Supplementary Table S2). This finding suggests that resistant plasmids are a key factor in determining the drug resistance of strains.
The genetic environment of a certain gene helps reveal the origin and genetic characteristics of the gene. Poirel et al. (2011) uncovered the full-length or truncated ISAba125 sequences in the upstream region of blaNDM–1 and conserved bleomycin (ble) resistance genes in the downstream region. They hypothesized that ISAba125 sequences are carried by blaNDM–1 when it transfers from the original host, and the blaNDM–1 and ble genes come from the same original host strain (Poirel et al., 2011). In 2012, Toleman analyzed all available blaNDM–1-associated sequences and found that ISAba125 sequences are always present in the 100 bp upstream region of blaNDM–1, and demonstrated that blaNDM–1 is a chimera. The chimeric process occurs in A. baumannii and is mediated by ISAba125 (Jones et al., 2014, 2015; Toleman et al., 2015). Our sequencing results identified certain differences in the 20–30 kb region between p11106, p12, and the compared plasmids, except for pNDM-BTR. On the basis of the aforementioned findings, we propose the following: (a) p11106 and p12 are structurally similar to pNDM-BTR, indicating that p11106 and p12 may present characteristics of pNDM-BTR (McGann et al., 2015); (b) the differences in the 20–30 kb region between p11106, p12, and the compared plasmids support the theory of gene polymorphism (Potron et al., 2011; Mata et al., 2012; Datta et al., 2017); (c) the existence of ble indicates that blaNDM–1 may originate from A. baumannii; and (d) the presence of two TnpA in the upstream region of blaNDM–1 and orf00052 (TnpA) in the 36 kb downstream region suggests that blaNDM–1 is acquired from plasmid transposition and long-term preservation (Campos et al., 2015; An et al., 2016; Khong et al., 2016).
This experiment screened drug-resistant strains carrying blaNDM–1 gene from carbapenem-non-susceptible bacteria collected from The First Affiliated Hospital of Nanchang University between January 2013 and December 2016. We assessed their drug resistance and plasmid conjugation transfer ability. Two complete plasmid sequences were obtained after sequencing analysis of five blaNDM–1-positive plasmids isolated from K. pneumoniae and compared with blaNDM–1-negative plasmids with known sequences. However, data on abundance were limited owing to the small sample size of drug-resistant strains as well as complexity and interferences during plasmid synthesis. In addition, the conclusion drawn from sequencing in this study is yet to be verified by molecular experiments. Gene deletion (IMP-4 and β-lactamase) occurred in p11106 and p12. The presence of a potential relationship between gene deletion and blaNDM–1 requires further study.
Conclusion
In conclusion, NDM-1-resistant bacteria exhibit multidrug resistance and spread to a certain extent in the hospital via efficient plasmid conjugation transfer. K. pneumoniae may be an important intermediate retention host in the dissemination process. Nosocomial blaNDM–1-carrying bacteria are of concern.
Drug-resistant plasmids may be a key factor in determining drug resistance. The p11106 and p12 plasmids containing the blaNDM–1 gene isolated from K. pneumoniae exhibit the characteristics of the pNDM-BTR plasmid and have a polymorphic gene environment. The existence of ble in the surrounding environment of blaNDM–1 suggests that blaNDM–1 is derived from A. baumannii. Meanwhile, the presence of TnpA in both the upstream and downstream regions of blaNDM–1 indicates that blaNDM–1 may be acquired from gene transposition and persists over a long period in K. pneumoniae strains.
Data Availability Statement
The datasets generated for this study are available from the corresponding author upon reasonable request.
Ethics Statement
Written informed consent was obtained from all participants, and the purpose and procedures of the study were explained to them. Ethical approval for this study was obtained from the Institutional Review Board of The First Affiliated Hospital of Nanchang University.
Author Contributions
TX and CC designed the study, collected and analyzed the data, and drafted the manuscript. TX, CC, JW, YL, and QZ contributed to the performance of the experiment and data collection. NC and XW reviewed the study design. WZ contributed to the review of data analysis and data interpretation. All authors have approved the final version of the manuscript.
Funding
This work was supported by the Fund Project of Jiangxi Health and Family Planning Commission (20181058) and the Natural Science Foundation of Jiangxi Province (20181BAB215033).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00700/full#supplementary-material
Footnotes
References
An, J., Guo, L., Zhou, L., Ma, Y., Luo, Y., Tao, C., et al. (2016). NDM-producing Enterobacteriaceae in a Chinese hospital, 2014-2015: identification of NDM-producing Citrobacterwerkmanii and acquisition of blaNDM-1-carrying plasmid in vivo in a clinical Escherichia coli isolate. J. Med. Microbiol. 65, 1253–1259. doi: 10.1099/jmm.0.000357
Barguigua, A., Zerouali, K., Katfy, K., El Otmani, F., Timinouni, M., and Elmdaghri, N. (2015). Occurrence of OXA-48 and NDM-1 carbapenemase-producing Klebsiella pneumoniae in a Moroccan university hospital in Casablanca, Morocco. Infect. Genet. Evol. 31, 142–148. doi: 10.1016/j.meegid.2015.01.010
Bontron, S., Nordmann, P., and Poirel, L. (2016). Transposition of Tn125 Encoding the NDM-1 Carbapenemase in Acinetobacter baumannii. Antimicrob. Agents Chemother. 60, 7245–7251. doi: 10.1128/AAC.01755-16
Bush, K., and Fisher, J. F. (2011). Epidemiological expansion, structural studies, and clinical challenges of new beta-lactamases from gram-negative bacteria. Annu. Rev. Microbiol. 65, 455–478. doi: 10.1146/annurev-micro-090110-102911
Campos, J. C., da Silva, M. J., dos Santos, P. R., Barros, E. M., Pereira Mde, O., Seco, B. M., et al. (2015). Characterization of Tn3000, a Transposon Responsible for blaNDM-1 Dissemination among Enterobacteriaceae in Brazil, Nepal, Morocco, and India. Antimicrob. Agents Chemother. 59, 7387–7395. doi: 10.1128/aac.01458-15
Carattoli, A., Seiffert, S. N., Schwendener, S., Perreten, V., and Endimiani, A. (2015). Differentiation of IncL and IncM Plasmids associated with the spread of clinically relevant antimicrobial resistance. PLoS One 10:e0123063. doi: 10.1371/journal.pone.0123063
Chen, C. J., Wu, T. L., Lu, P. L., Chen, Y. T., Fung, C. P., Chuang, Y. C., et al. (2014). Closely related NDM-1-encoding plasmids from Escherichia coli and Klebsiella pneumoniae in Taiwan. PLoS One 9:e104899. doi: 10.1371/journal.pone.0104899
Datta, S., Mitra, S., Chattopadhyay, P., Som, T., Mukherjee, S., and Basu, S. (2017). Spread and exchange of bla NDM-1 in hospitalized neonates: role of mobilizable genetic elements. Eur. J. Clin. Microbiol. Infect. Dis. 36, 255–265. doi: 10.1007/s10096-016-2794-6
Deshpande, P., Rodrigues, C., Shetty, A., Kapadia, F., Hedge, A., and Soman, R. (2010). New Delhi Metallo-beta lactamase (NDM-1) in Enterobacteriaceae: treatment options with carbapenems compromised. J. Assoc. Physicians India 58, 147–149.
Dongmei, N., Wanqing, Z., and Zhang, Z. (2017). Distribution of NDM-1 gene in strains of Acinetobacter sp. J. Clin. Transfus Lab. Med. 19, 364–367. doi: 10.1093/jac/dkr174
Gamal, D., Fernandez-Martinez, M., Salem, D., El-Defrawy, I., Montes, L. A., Ocampo-Sosa, A. A., et al. (2016). Carbapenem-resistant Klebsiella pneumoniae isolates from Egypt containing blaNDM-1 on IncR plasmids and its association with rmtF. Int. J. Infect. Dis. 43, 17–20. doi: 10.1016/j.ijid.2015.12.003
Girlich, D., Dortet, L., Poirel, L., and Nordmann, P. (2015). Integration of the blaNDM-1 carbapenemase gene into Proteus genomic island 1 (PGI1-PmPEL) in a Proteus mirabilis clinical isolate. J. Antimicrob. Chemother. 70, 98–102. doi: 10.1093/jac/dku371
Jones, L. S., Carvalho, M. J., Toleman, M. A., White, P. L., Connor, T. R., Mushtaq, A., et al. (2015). Characterization of plasmids in extensively drug-resistant acinetobacter strains isolated in India and Pakistan. Antimicrob. Agents Chemother. 59, 923–929. doi: 10.1128/aac.03242-14
Jones, L. S., Toleman, M. A., Weeks, J. L., Howe, R. A., Walsh, T. R., and Kumarasamy, K. K. (2014). Plasmid carriage of bla NDM-1 in clinical Acinetobacter baumannii isolates from India. Antimicrob. Agents Chemother. 58, 4211–4213. doi: 10.1128/aac.02500-14
Khong, W. X., Xia, E., Marimuthu, K., Xu, W., Teo, Y. Y., Tan, E. L., et al. (2016). Local transmission and global dissemination of New Delhi Metallo-Beta-Lactamase (NDM): a whole genome analysis. BMC Genomics 17:452. doi: 10.1186/s12864-016-2740-0
Kocsis, E., Guzvinec, M., Butic, I., Kresic, S., Crnek, S. S., Tambic, A., et al. (2016). blaNDM-1 carriage on IncR plasmid in Enterobacteriaceae strains. Microb. Drug Resist. 22, 123–128. doi: 10.1089/mdr.2015.0083
Kumarasamy, K. K., Toleman, M. A., Walsh, T. R., Bagaria, J., Butt, F., Balakrishnan, R., et al. (2010). Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10, 597–602. doi: 10.1016/s1473-3099(10)70143-2
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/s1473-3099(15)00424-7
Mata, C., Miro, E., Alvarado, A., Garcillan-Barcia, M. P., Toleman, M., Walsh, T. R., et al. (2012). Plasmid typing and genetic context of AmpC beta-lactamases in Enterobacteriaceae lacking inducible chromosomal ampC genes: findings from a Spanish hospital 1999-2007. J. Antimicrob. Chemother. 67, 115–122. doi: 10.1093/jac/dkr412
McGann, P., Snesrud, E., Ong, A. C., Appalla, L., Koren, M., Kwak, Y. I., et al. (2015). War wound treatment complications due to transfer of an IncN plasmid harboring bla(OXA-181) from Morganella morganii to CTX-M-27-producing sequence type 131 Escherichia coli. Antimicrob. Agents Chemother. 59, 3556–3562. doi: 10.1128/aac.04442-14
Poirel, L., Dortet, L., Bernabeu, S., and Nordmann, P. (2011). Genetic features of blaNDM-1-positive Enterobacteriaceae. Antimicrob. Agents Chemother. 55, 5403–5407. doi: 10.1128/aac.00585-11
Potron, A., Poirel, L., and Nordmann, P. (2011). Plasmid-mediated transfer of the bla(NDM-1) gene in gram-negative rods. FEMS Microbiol. Lett. 324, 111–116. doi: 10.1111/j.1574-6968.2011.02392.x
Rahman, M., Prasad, K. N., Pathak, A., Pati, B. K., Singh, A., Ovejero, C. M., et al. (2015). RmtC and RmtF 16S rRNA methyltransferase in NDM-1-Producing Pseudomonas aeruginosa. Emerg. Infect. Dis. 21, 2059–2062. doi: 10.3201/eid2111.150271
Rolain, J. M., Parola, P., and Cornaglia, G. (2010). New Delhi metallo-beta-lactamase (NDM-1): towards a new pandemia? Clin. Microbiol. Infect. 16, 1699–1701. doi: 10.1111/j.1469-0691.2010.03385.x
Sarkar, A., Pazhani, G. P., Chowdhury, G., Ghosh, A., and Ramamurthy, T. (2015). Attributes of carbapenemase encoding conjugative plasmid pNDM-SAL from an extensively drug-resistant Salmonella enterica serovar senftenberg. Front. Microbiol. 6:969. doi: 10.3389/fmicb.2015.00969
Shen, P., Yi, M., Fu, Y., Ruan, Z., Du, X., Yu, Y., et al. (2017). Detection of an Escherichia coli sequence type 167 strain with two tandem copies of blaNDM-1 in the chromosome. J. Clin. Microbiol. 55, 199–205. doi: 10.1128/jcm.01581-16
Shengshu, W., Jinzhu, S., Wenli, S., Zhi, H., Jianpeng, Y., and Wang, Y. (2015). Epidemiological analysis of NDM-1-positive bacteria in China. Mil. Med. Sci. 39, 825–830. doi: 10.1128/AAC.00165-11
Toleman, M. A., Bugert, J. J., and Nizam, S. A. (2015). Extensively drug-resistant New Delhi metallo-beta-lactamase-encoding bacteria in the environment, Dhaka, Bangladesh, 2012. Emerg. Infect. Dis. 21, 1027–1030. doi: 10.3201/eid2106.141578
Villa, L., Guerra, B., Schmoger, S., Fischer, J., Helmuth, R., Zong, Z., et al. (2015). IncA/C Plasmid Carrying bla(NDM-1), bla(CMY-16), and fosA3 in a Salmonella enterica Serovar Corvallis Strain Isolated from a Migratory Wild Bird in Germany. Antimicrob. Agents Chemother. 59, 6597–6600. doi: 10.1128/aac.00944-15
Wailan, A. M., and Paterson, D. L. (2014). The spread and acquisition of NDM-1: a multifactorial problem. Expert Rev. Anti Infect. Ther. 12, 91–115. doi: 10.1586/14787210.2014.856756
Wailan, A. M., Sartor, A. L., Zowawi, H. M., Perry, J. D., Paterson, D. L., and Sidjabat, H. E. (2015). Genetic contexts of blaNDM-1 in patients carrying multiple NDM-Producing strains. Antimicrob. Agents Chemother. 59, 7405–7410. doi: 10.1128/aac.01319-15
Wang, Y., Zhang, R., Li, J., Wu, Z., Yin, W., Schwarz, S., et al. (2017). Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat. Microbiol. 2:16260. doi: 10.1038/nmicrobiol.2016.260
Yong, D., Toleman, M. A., Giske, C. G., Cho, H. S., Sundman, K., Lee, K., et al. (2009). Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53, 5046–5054. doi: 10.1128/aac.00774-09
Zarfel, G., Hoenigl, M., Leitner, E., Salzer, H. J., Feierl, G., Masoud, L., et al. (2011). Emergence of New Delhi metallo-beta-lactamase, Austria. Emerg. Infect. Dis. 17, 129–130. doi: 10.3201/eid1701.101331
Keywords: carbapenem resistance, NDM-1, Klebsiella pneumoniae, plasmid, genetic characteristics
Citation: Xiang T, Chen C, Wen J, Liu Y, Zhang Q, Cheng N, Wu X and Zhang W (2020) Resistance of Klebsiella pneumoniae Strains Carrying blaNDM–1 Gene and the Genetic Environment of blaNDM–1. Front. Microbiol. 11:700. doi: 10.3389/fmicb.2020.00700
Received: 26 September 2019; Accepted: 25 March 2020;
Published: 30 April 2020.
Edited by:
Ilana L. B. C. Camargo, University of São Paulo, BrazilReviewed by:
Zhi Ruan, Zhejiang University, ChinaAndres Felipe Opazo-Capurro, University of Concepción, Chile
Copyright © 2020 Xiang, Chen, Wen, Liu, Zhang, Cheng, Wu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Zhang, bmR6aGFuZ3dlaTIwMTZAMTI2LmNvbQ==
†These authors have contributed equally to this work
 Tianxin Xiang1†
Tianxin Xiang1† Yang Liu
Yang Liu Wei Zhang
Wei Zhang