
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 07 April 2020
Sec. Systems Microbiology
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.00606
 Yongshuai Peng1,2†
Yongshuai Peng1,2† Shanshan Zhao1†
Shanshan Zhao1† Kunlun Wang1
Kunlun Wang1 Jinxing Song1
Jinxing Song1 Yaqun Yan1
Yaqun Yan1 Yongchun Zhou1
Yongchun Zhou1 Ke Shi1
Ke Shi1 Fuchun Jian1
Fuchun Jian1 Rongjun Wang1
Rongjun Wang1 Longxian Zhang1,3
Longxian Zhang1,3 Changshen Ning1,3*
Changshen Ning1,3*The genus Anaplasma (Rickettsiales: Anaplasmataceae), which includes the species Anaplasma capra, Anaplasma bovis, Anaplasma ovis, and Anaplasma phagocytophilum, is responsible for a wide variety of infections in both human and veterinary health worldwide. Multiple infections with these four Anaplasma pathogens have been reported in many cases. We introduce a novel multiplex PCR for the simultaneous detection of A. capra, A. bovis, A. ovis, and A. phagocytophilum, based on species-specific primers against the groEL (A. capra and A. bovis), msp4 (A. ovis), and 16S rRNA (A. phagocytophilum) genes. To verify the specificity of the PCR reactions, we evaluated four sets of primers to analyze samples containing different blood pathogens. The sensitivity of the multiplex PCR was evaluated by amplifying 10-fold dilutions of total genomic DNA extracted from sheep blood infected with A. capra, A. bovis, A. ovis, or A. phagocytophilum. The reproducibility of the assay was evaluated by testing 10-fold dilutions of total genomic DNA extracted from sheep blood infected with these pathogens from 100 to 10–3 ng/μL per reaction in triplicate on three different days. A total of 175 field blood DNA samples were used to evaluate the reproducibility of multiplex PCR compared with the simplex PCRs. PCR primers used in this study were confirmed to be 100% species-specific using blood pathogens previously identified by other methods. The lower limit of detection of the multiplex PCR with good repeatability enabled the detection of A. capra, A. bovis, A. ovis and A. phagocytophilum at concentrations of 3 × 10–5, 5 × 10–7, 2 × 10–5, and 7 × 10–7 ng/μL, respectively. There was no significant difference between conventional and multiplex PCR protocols used to detect the four Anaplasma species (P > 0.05). The results of the multiplex PCR revealed that the A. capra groEL gene, the A. bovis groEL gene, the A. ovis msp4 gene, and the A. phagocytophilum 16S rRNA gene were reliable target genes for species identification in clinical isolates, being specific for each of the four target Anaplasma species. Our study provides an effective, sensitive, specific, and accurate tool for the rapid differential clinical diagnosis and epidemiological surveillance of Anaplasma pathogens in sheep and goats.
The genus Anaplasma (Rickettsiales: Anaplasmataceae) comprises tick-transmitted obligate intracellular bacterial species including Anaplasma capra, Anaplasma bovis, Anaplasma ovis, and Anaplasma phagocytophilum, which are responsible for a wide variety of different infections in both human and veterinary health (Guo et al., 2018; Kundave et al., 2018; Guimarães et al., 2019). A. capra is a potential novel tick-borne Anaplasma species, which was identified in goats in China (Liu et al., 2012) and provisionally nominated it “Anaplasma capra” as the causative agent of human infections as reported by Li et al. (2015). This pathogen may be responsible for anaplasmosis cases and may be a substantial public health concern; it also appears to be widely distributed in China and South Korea (Li et al., 2015; Peng et al., 2018; Yang et al., 2018a). A. bovis infects monocytes and tissue macrophages of small mammals and ruminants (Dumler et al., 2001). The DNA of this pathogen has been detected in cattle, goats, sheep, dogs, and some small wild mammals (Fukui and Inokuma, 2019; Giglioti et al., 2019; Yang et al., 2019). It can cause a variety of clinical symptoms, including weight loss, fever, anemia, listlessness, and death in some cases (Aktas and zübek, 2015). A. ovis is an intra-erythrocytic rickettsial pathogen that mainly affects domestic sheep and goats, but has also been reported to be present in some wild ruminants and dogs with mild clinical symptoms (Zaid et al., 2019). However, symptoms may be exacerbated by stressors such as co-infection, elevated temperature, and animal movement disorders throughout Asia, Africa, Europe and the United States (Hornok et al., 2011; Cabezas-Cruz et al., 2019; Enkhtaivan et al., 2019; Liu et al., 2019). This pathogen also poses a potential threat to humans (Wei et al., 2017). The first human case of A. ovis infection was reported in Cyprus and was characterized by a fever, hepatosplenomegaly, and lymphadenopathy (Chochlakis et al., 2010). A. phagocytophilum mainly affects small ruminants in Hungary, Romania, Italy, Kenya, South Africa, China, Vietnam, Morocco, and Mexico (Ait Lbacha et al., 2017; Seo et al., 2018a; Han et al., 2019; Hornok et al., 2019), but can also cause human granulocytic anaplasmosis. The pathogen infects neutrophils and survives by inhibiting or delaying important antimicrobial mechanisms in host cells (Goel et al., 2018; Lee et al., 2018; Dehhaghi et al., 2019). Simultaneous infection of these four Anaplasma agents or mixed infections by different Anaplasma species has also been reported (Halajian et al., 2018; Wang et al., 2018; Seo et al., 2018b).
Traditionally, laboratory detection of Anaplasma species in animals or humans was dependent on the microscopic examination of blood samples (Giglioti et al., 2019). However, using this method, it was difficult to clearly distinguish between A. capra, A. bovis, A. ovis, and A. phagocytophilum, as they are highly similar in terms of their morphological characteristics (Liu et al., 2012; Li et al., 2015; Wei et al., 2017; Lee et al., 2018). It was also difficult to distinguish these agents from other blood pathogenic agents, such as members of the Theileria and Babesia genera (Al-Hosary et al., 2018; Pradeep et al., 2019). Especially when the pathogen load is low, it is difficult to distinguish between these pathogens by microscopic examination. In recent years, molecular methods have been increasingly used in microbiology laboratories, such as duplex PCR based assays and loop-mediated isothermal amplification assays, as well as other recently developed methods that could lead to the development of new diagnostic tests to identify different Anaplasma species (Cui et al., 2017; Yang et al., 2018a; Giglioti et al., 2019). The development of multiplex PCR can enable positive detection without the need for additional reagents or increased amounts of input DNA (Hao et al., 2019). However, primer design for multiplex PCR becomes more challenging when the number of PCR reactions increases. Hence, to the best of our knowledge, no multiplex PCR assay for the identification of clinically relevant Anaplasma species in field blood samples has previously been reported.
In this study, we developed an accurate, specific and sensitive multiplex PCR, using a combination of specially designed primers and previously reported primers, for the identification of Anaplasma species in field blood samples from animals or patients suspected of being infected with members of this genus.
Preserved DNA of Anaplasma spp. (A. capra, A. bovis, A. ovis, A. phagocytophilum) was used as positive controls in this study. DNA samples positive for Anaplasma marginale, Anaplasma platys, Theileria ovis, Theileria annulata, Theileria uilenbergi, Theileria luwenshuni, Babesia motasi, and Toxoplasma gondii were used as negative controls. T. ovis and T. uilenbergi positive samples were provided by Professor Mengqi from Tarim University and the others were maintained at −80°C at the Laboratory of Parasitology of Henan Agricultural University. All the control DNA samples were evaluated with a spectrophotometer (Nanodrop Onec; Thermo Fisher Scientific, Wilmington, DE, United States) prior to use to ensure that the concentration of DNA samples used in the test were >20 ng/μL and verified it by specific primers before use. Double-distilled (dd) H2O was used as a blank control.
DNA of field samples was extracted from 250 μL of blood using the Blood DNA Kit (OMEGA Bio-Tek, Norcross, GA, United States), in accordance with the manufacturer’s instructions. The DNA of each sample was eluted in 200 μL of elution buffer. All DNA samples were examined using the spectrophotometer to test their quality and quantity, and were stored at −20°C until use.
Primers were designed against regions of the A. bovis groEL gene and the A. phagocytophilum 16S rRNA gene using Premier 5 (Premier Biosoft International, Palo Alto, CA, United States). These regions were identified through alignments of nucleotide sequences obtained in this study and sequences available from the GenBank database (KX987399, KU585932, and KY425449 for A. bovis; KY242452, KR002115, LC060987, KF569915, and KC916737 for A. phagocytophilum). The specificity of each primer set was evaluated using the Basic Local Alignment Search Tool (BLAST) from the National Center for Biotechnology Information (NCBI) database1. For the detection of A. capra and A. ovis targeting the groEL and msp4 genes, respectively, previously described primers were used (Torina et al., 2012; Yang et al., 2016). The primers are described in Table 1. The primer sets were synthesized by Sangon Biotech Company (Shanghai, China).
To ensure optimal amplification conditions for the four target sequences in a multiplex PCR assay, a range of PCR-related parameters were evaluated such as the annealing temperatures of the primers, the dosage of primers, and the concentrations of La Taq DNA polymerase. To examine these parameters, a number of tests were performed in 25 μL reaction volumes.
PCR conditions were investigated including the variation of the annealing temperature from 57 to 64°C, the concentration of primer sets from 0.08 to 0.56 μM, the concentration of La Taq DNA polymerase (TaKaRa, Dalian, China) from 0.75 to 1.75 U, and the concentration of PCR buffer (10×) and the dNTPs (2.5 mM) from 1.5 to 3 μL and 2 to 6 μL, respectively. The optimization of the multiplex PCR conditions was based on a previous report by Henegariu et al. (1997).
For the construction of plasmids containing the A. capra groEL gene, the A. bovis groEL gene, the A. ovis msp4 gene, and the A. phagocytophilum 16S rRNA gene, the corresponding PCR products of 874, 529, 347 and 172 bp were cloned into pMD-18T (TaKaRa) and then propagated in Escherichia coli DH5α competent cells (Sangon Biotech). Plasmid DNA was purified from transformed cells using the SanPrep Column Plasmid Mini-Prep Kit (Sangon Biotech) and quantified using a spectrophotometer (Nanodrop Onec; Thermo Fisher Scientific).
To generate standard curves for quantitative determinations and to assess the amplification efficiency, plasmids were 10-fold-diluted in elution buffer (Sangon Biotech), representing 100–10–4 ng/μL DNA template. Aliquots of each dilution were frozen at −20°C until use. To minimize the potential for contamination, the standard plasmid DNA was stored in a separate laboratory.
To rule out the possibility of cross-reactions with other closely related pathogens of the blood, the specificity of the multiplex PCR was evaluated by testing positive control DNA from blood samples infected with A. capra, A. bovis, A. ovis, A. phagocytophilum, A. marginale, A. platys, Theileria ovis, T. annulata, T. uilenbergi, T. luwenshuni, B. motasi, and T. gondii. The negative control was ddH2O, and 29.4 ng/μL was the lowest DNA concentration used in the assay.
Ten-fold dilutions of total genomic DNA extracted from the blood of sheep infected with A. capra, A. bovis, A. ovis, and A. phagocytophilum were used to evaluate the sensitivity of the multiplex PCR. Each experiment consisted of triplicate tests on two replicates.
The reproducibility of the assay was evaluated by testing 10-fold dilutions of total genomic DNA extracted from sheep blood infected with A. capra, A. bovis, A. ovis, and A. phagocytophilum from 100 to 10–3 ng/μL per reaction in triplicate on three different days. Kendall’s coefficient of concordance (Kendall’s W) was calculated to evaluate the repeatability of the method. Its value can be between 0 and 1; the higher the value, the better the repeatability.
To quantify the pathogen DNA of A. capra, A. bovis, A. ovis, and A. phagocytophilum in clinical samples, the qTOWER3 G QPCR System (Analytikjena Technologies, Jena, Germany) was used for the real-time PCR assay performed in a final volume of 10 μL containing 5 μL of TB GreenTM Premix Ex TaqTM (TaKaRa, Dalian, China), 0.4 μM of each primer and 2 μL of the DNA template. The thermal profile of the PCR was as follows: 30 s at 95°C for denaturation, followed by 40 cycles of denaturation at 95°C for 5 s, annealing at 63°C for 30 s, and extension at 72°C for 45 s. The final extension step was 1 min at 72°C.
A total of 175 blood samples (sheep 99, goats 76) were collected at different time points from a simplex farm in Linyou County, Shanxi Province. The sheep and goats in this farm were grazed perennially and highly infested with ticks. DNA of these samples was used to evaluate the efficiency of the simplex and multiplex PCR protocols. Conventional PCR conditions as previously described were performed and compared in terms of efficiency and capability to the multiplex PCR. The used primer sets are shown in Table 1. Upon detecting Anaplasma in field samples by the multiplex PCR, all positive PCR products were sent to a sequencing company for sequencing, and the results were consistent with expectations. Infection rates were assessed using the Chi-square test with Yates’ correction, and a P-value ≤0.05 was considered to represent statistical significance.
After optimization, the optimum multiplex PCR assay was performed in a final volume of 25 μL, containing 2.5 μL of 10× PCR La buffer, 4 μL of dNTPs at 2.5 mM, 1.25 U of La Taq DNA polymerase, 0.32 μM of each primer, and 2 μL of the DNA template. The thermal profile of the PCR was as follows: 5 min at 94°C for denaturation, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 63°C for 30 s, and extension at 72°C for 1 min. The final extension step was 10 min at 72°C. The PCR products (5 μL) were analyzed using a UV gel imaging system following electrophoresis in a 1.5% agarose gel and staining with DNA GREEN (Solarbio, Beijing, China) (Figures 1A–C).
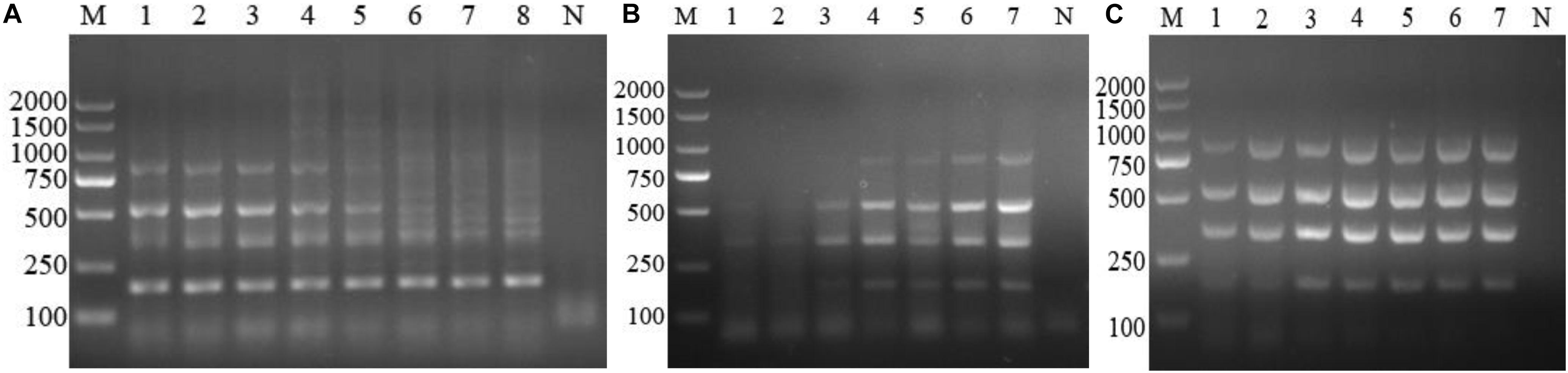
Figure 1. Optimization of the multiplex PCR components. (A) Annealing temperature gradients, M: DL2000 marker. Lanes 1–8: 64, 63, 62, 61, 60, 59, 58, and 57°C, respectively. (B) Dose of Anaplasma primers, lane M: DL2000 marker; Lanes 1–7: 0.08, 0.16, 0.24, 0.32, 0.4, 0.48, and 0.56 μM, respectively; Lane N: negative control. (C) La Taq DNA polymerase. M: DL2000 marker; Lanes 1–7: 0.75, 1.0, 1.25, 1.5, 1.75, 2, and 2.25 U, respectively; Lane N: negative control.
To characterize the specificity of the reactions, the four sets of primers were evaluated using samples containing A. capra, A. bovis, A. ovis, A. phagocytophilum, A. marginale, A. platys, Theileria ovis, T. annulata, T. uilenbergi, T. luwenshun, B. motasi, or T. gondii DNA, and ddH2O was used as a blank control. Fragments of the expected sizes were generated from the positive DNA templates. Using the mixed DNA of the four Anaplasma species as templates, four bands of expected sizes were observed, and the target bands for infection with a simplex Anaplasma agent appeared separately. No amplification signals were detected with the negative control (A. marginale, A. platys, Theileria ovis, T. annulata, T. uilenbergi, T. luwenshuni, B. motasi, and T. gondii) and blank control templates (Figure 2). The results showed that the reaction products could be distinguished by gel electrophoresis and the primer sets used in this study were species-specific.
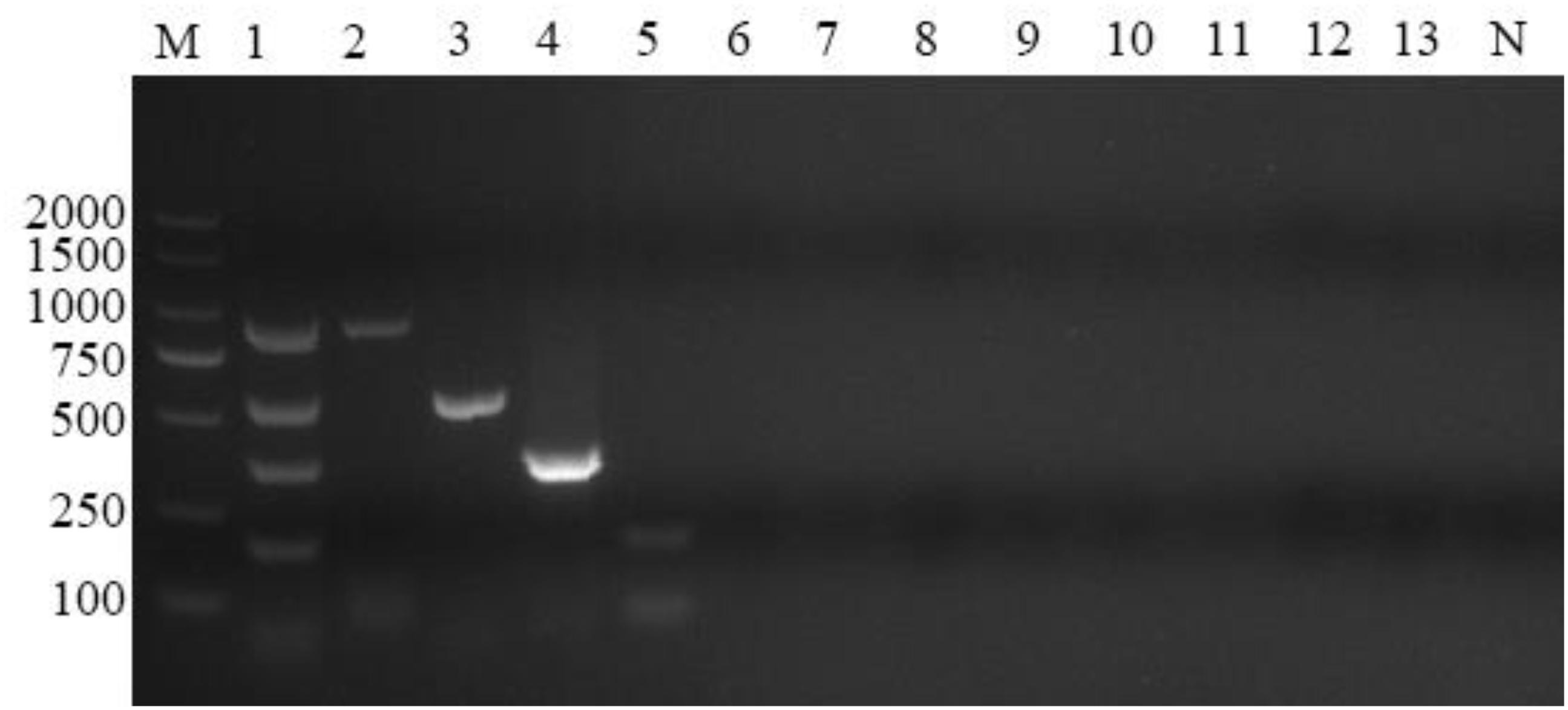
Figure 2. Multiplex PCR specificity test. Four primer sets could amplify genus-specific bands of 874, 529, 347, and 172 bp only in samples positive for Anaplasma capra, Anaplasma bovis, Anaplasma ovis, and Anaplasma phagocytophilum, respectively. M: DL2000 marker; Lane 1: mixed DNA samples from the four Anaplasma species; Lanes 2–13: samples positive for A. capra, A. bovis, A. ovis, A. phagocytophilum, A. marginale, A. platys, Theileria ovis, Theileria annulata, Theileria uilenbergi, Theileria luwenshuni, Babesia motasi, and Toxoplasma gondii, respectively; Lane N: negative control.
The optimized multiplex PCR was evaluated by amplifying 10-fold serial dilutions (100–10–4 ng/μL) of the mixture DNA infected with these pathogens extracted from sheep blood. The lower limit of detection of the multiplex PCR could detect A. capra, A. bovis, A. ovis, and A. phagocytophilum at concentrations of 3 × 10–5, 5 × 10–7, 2 × 10–5, and 7 × 10–7 ng/μL, respectively (Figure 3).
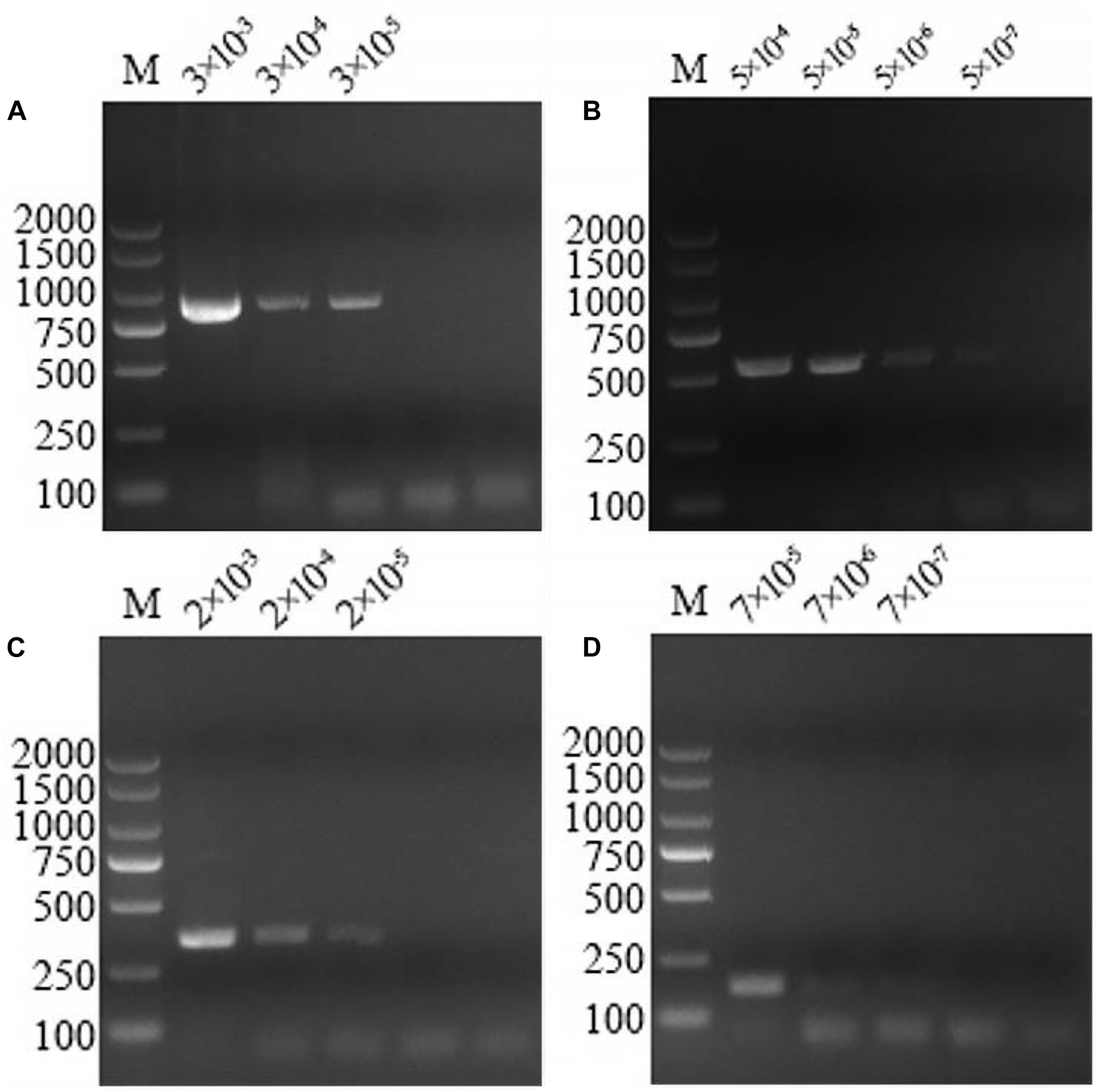
Figure 3. Multiplex PCR sensitivity tests on serially diluted positive DNA sample templates. At the top of the figure is dilution factor, with a unit of ng/μL. The left and right sides of the figure show the molecular weight of the DNA. (A) The sensitivity of the multiplex PCR assay for Anaplasma capra was 3 × 10–5 ng/μL. (B) The sensitivity of the multiplex PCR assay for Anaplasma bovis was 5 × 10–7 ng/μL. (C) The sensitivity of the multiplex PCR assay for Anaplasma ovis was 2 × 10–5 ng/μL. (D) The sensitivity of the multiplex PCR assay for Anaplasma phagocytophilum was 7 × 10–7 ng/μL.
Kendall’s W values for inter-assay reproducibility were found to range from 0.80 to 0.90 (Supplementary Appendix). This shows that the optimized multiplex PCR has good repeatability.
Out of a total of 175 samples, multiplex PCR revealed 41 (23.4%), 68 (38.9%), 33 (18.9%), and 105 (60.0%) blood samples positive for A. capra, A. bovis, A. ovis, and A. phagocytophilum, respectively. By comparison, the positive detection rates of these four Anaplasma species using simplex PCR and conventional PCR as previously described were 25.1, 47.4, 20.0, and 60.6, and 24.0, 44.6, 20.6, and 58.3%, respectively (Table 2). There was no significant difference among the three methods used to detect the four Anaplasma species (P > 0.05). An example of the multiplex PCR results for the detection of Anaplasma spp. in field samples is provided in Figure 4.
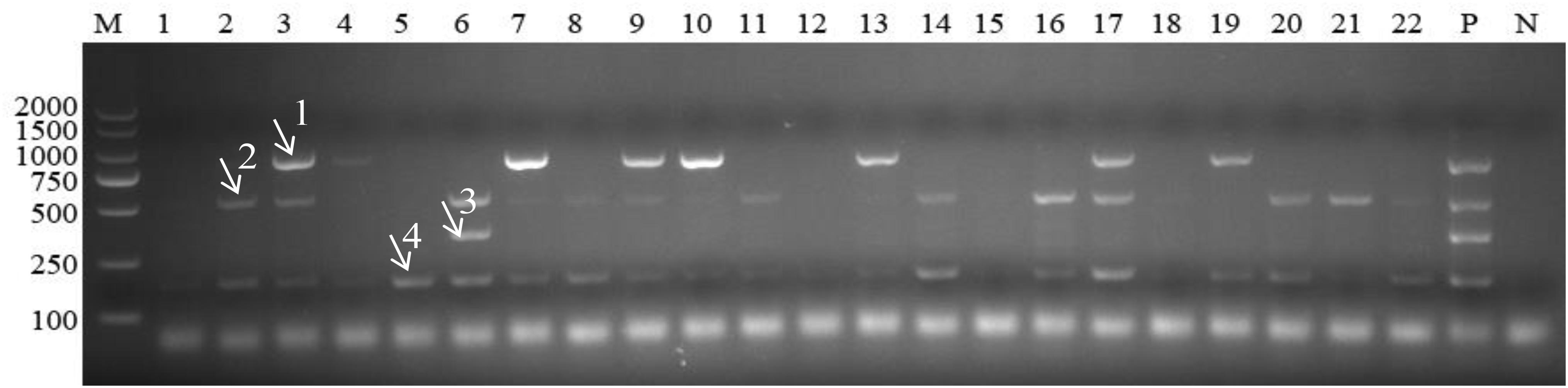
Figure 4. Detection of Anaplasma in field samples using multiplex PCR. M: DL2000 marker; Lanes 1–22: field samples; P: positive control; N: negative control. Arrows with numbers (1–4) indicate the amplicons of Anaplasma capra 874 bp, Anaplasma bovis 529 bp, Anaplasma ovis 347 bp, and Anaplasma phagocytophilum 172 bp.
Anaplasma species are some of the most widespread microbes with potential human and animal pathogenicity that are transmitted by ticks (Kim et al., 2014; Sun et al., 2015; Ben Said et al., 2018). Among these, A. capra, A. phagocytophilum, and A. ovis are tick-borne zoonotic pathogens that are significant for public health (Chochlakis et al., 2010; Li et al., 2015; Lee et al., 2018), they have been detected in many domestic and wild animals across the world (Kawahara et al., 2006; Seo et al., 2018b; Yang et al., 2018b; Cabezas-Cruz et al., 2019). Additionally, recent reports have presented evidence of co-infection with two or more Anaplasma species in cattle, sheep, goats, and ixodid ticks worldwide (Rjeibi et al., 2018; Seo et al., 2018b; Wang et al., 2018; Zhou et al., 2018; Han et al., 2019). It is thus important to detect several Anaplasma species in a single DNA sample by using a single PCR reaction. This would also reduce the cost of the analysis, which is important particular in developing countries.
PCR assays have been used to sensitively and specifically detect Anaplasma species in many diagnostic laboratories. In fact, numerous reports have been published on the PCR detection of simplex Anaplasma species in infected animals and humans (Torina et al., 2012; Chi et al., 2013; Lee et al., 2018). Duplex PCR assays for the detection of co-infection with A. marginale (targeting the msp4 gene) and A. phagocytophilum (targeting the msp2 gene) in cattle (M’ghirbi et al., 2016) and duplex real-time PCR assays for the detection of co-infection with A. marginale (targeting the msp1β gene) and Anaplasma centrale (targeting the groEL gene) in sheep (Decaro et al., 2008) were previously reported. However, no previous study has developed an efficient and convenient PCR assay that would be more convenient and efficient for field diagnosis than the simplex or duplex PCR assays to detect multiple Anaplasma agents. Early diagnosis and treatment are required to reduce the morbidity and mortality associated with these diseases worldwide (Zhuo et al., 2019). Hence the present multiplex PCR assay could support the need for epidemiological assessments of the global distribution of A. capra, A. bovis, A. ovis, and A. phagocytophilum in animals and humans.
Several molecular techniques have been proposed for detecting and characterizing species belonging to the Anaplasma genus. Previous studies indicated that the 16S rRNA, 18S rRNA, groEL, ankA, citrate synthase (gltA), and major surface protein (msps) sequences are highly conserved in Anaplasma spp.; thus, they have been used widely to classify the Anaplasma genus (Lew et al., 2003; Lee et al., 2018; Song et al., 2018; Cabezas-Cruz et al., 2019). Currently, epidemiological investigation of A. capra uses an assay based on the 16S rRNA, gltA and msp4 genes (Yang et al., 2018a). Molecular detection of A. capra, A. bovis, A. ovis, and A. phagocytophilum in infected ixodid ticks was based on the 16S rRNA, gltA, msp4, and 16S rRNA genes, respectively (Han et al., 2019). Additionally, molecular evidence of coinfection of Anaplasma species in small ruminants was obtained by targeting the 16S rRNA and msp4 genes of A. bovis, A. ovis, and A. phagocytophilum (Yang et al., 2019). Here, we developed a multiplex PCR assay, which was able to specifically detect A. capra, A. bovis, A. ovis, and A. phagocytophilum from field samples. These detection limits of A. bovis and A. phagocytophilum were higher than those reported for the detection of Anaplasma spp. in a duplex PCR assay by Cui et al. (2017).
This method had one limitation. A. capra and A. ovis have slightly lower detection limits. The primers used to amplify A. capra and A. ovis thus need further improvement. Considering the convenience and high efficiency of this method, it may still be suitable for the detection and epidemiological investigation of Anaplasma in humans, animals and vector ticks.
The results of the multiplex PCR revealed that the A. capra groEL gene, the A. bovis groEL gene, the A. ovis msp4 gene, and the A. phagocytophilum 16S rRNA gene are reliable target genes for species identification in clinical isolates, as they are specific for each of the four target Anaplasma species. Other, closely related pathogens of blood tested negative in this PCR assay. In conclusion, our study provides an effective, sensitive, specific, and accurate tool for the rapid differential clinical diagnosis and epidemiological surveillance of Anaplasma pathogens (A. capra, A. bovis, A. ovis, and A. phagocytophilum) in sheep and goats.
All datasets generated for this study are included in the article/Supplementary Material.
This research was conducted in accordance with the Chinese Laboratory Animal Administration Act (1988) after review and approval of its protocol by the Research Ethics Committee of Henan Agricultural University. Appropriate permission was obtained from the farm owners before the collection of blood specimens from their sheep and goats.
YP analyzed the samples, interpreted data, and wrote the manuscript. SZ conducted the laboratory examination of samples and sequences analysis. SZ, KW, JS, YY, YZ, and KS collected the samples. FJ, RW, LZ, and CN revised the manuscript, designed and supervised the study. All authors read and approved the final version of the manuscript.
This work was supported by the Earmarked Fund for China Modern Agro-industry Technology Research System (nycytx-38) and the National Key R&D Program of China (2018YFD0502100).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00606/full#supplementary-material
Ait Lbacha, H., Alali, S., Zouagui, Z., El Mamoun, L., Rhalem, A., Petit, E., et al. (2017). High prevalence of Anaplasma spp. in small ruminants in morocco. Transbound Emerg. Dis. 64, 250–263. doi: 10.1111/tbed.12366
Aktas, M., and Zübek, S. (2015). Bovine anaplasmosis in Turkey: first laboratory confirmed clinical cases caused by Anaplasma phagocytophilum. Vet. Microbiol. 178, 246–251. doi: 10.1016/j.vetmic.2015.05.021
Al-Hosary, A., Ahmed, L., Ahmed, J., Nijhof, A., and Clausen, P. H. (2018). Epidemiological study on tropical theileriosis (Theileria annulata infection) in the Egyptian Oases with special reference to the molecular characterization of Theileria spp. Ticks Tick Borne Dis. 9, 1489–1493. doi: 10.1016/j.ttbdis.2018.07.008
Ben Said, M., Belkahia, H., and Messadi, L. (2018). Anaplasma spp. in North Africa: a review on molecular epidemiology, associated risk factors and genetic characteristics. Ticks Tick Borne Dis. 9, 543–555. doi: 10.1016/j.ttbdis.2018.01.003
Cabezas-Cruz, A., Gallois, M., Fontugne, M., Allain, E., Denoual, M., Moutailler, S., et al. (2019). Epidemiology and genetic diversity of Anaplasma ovis in goats in Corsica. France. Parasit Vectors. 12:3. doi: 10.1186/s13071-018-3269-7
Chi, Q., Liu, Z., Li, Y., Yang, J., Chen, Z., Yue, C., et al. (2013). Development of a real-time PCR assay for detection and quantification of Anaplasma ovis infection. Transbound Emerg. Dis. 60, (Suppl. 2), 119–124. doi: 10.1111/tbed.12139
Chochlakis, D., Ioannou, I., Tselentis, Y., and Psaroulaki, A. (2010). Human anaplasmosis and Anaplasma ovis variant. Emerging Infect Dis. 16, 1031–1032. doi: 10.3201/eid1606.090175
Cui, Y., Zhang, Y., Jian, F., Zhang, L., Wang, R., Cao, S., et al. (2017). Development of duplex PCR for simultaneous detection of Theileria spp. and Anaplasma spp. in sheep and goats. Exp. Parasitol. 176, 1–7. doi: 10.1016/j.exppara.2017.01.011
de la Fuente, J., Atkinson, M. W., Naranjo, V., Fernandez de Mera, I. G., Mangold, A. J., Keating, K. A., et al. (2007). Sequence analysis of the msp4 gene of Anaplasma ovis strains. Vet. Microbiol. 119, 375–381. doi: 10.1016/j.vetmic.2006.09.011
Decaro, N., Carelli, G., Lorusso, E., Lucente, M. S., Greco, G., Lorusso, A., et al. (2008). Duplex real-time polymerase chain reaction for simultaneous detection and quantification of Anaplasma marginale and Anaplasma centrale. J. Vet. Diagn Invest. 20, 606–611. doi: 10.1177/104063870802000511
Dehhaghi, M., Kazemi Shariat Panahi, H., Holmes, E. C., Hudson, B. J., Schloeffel, R., and Guillemin, G. J. (2019). Human tick-borne diseases in Australia. Front. Cell Infect. Microbiol. 9:3. doi: 10.3389/fcimb.2019.00003
Dumler, J. S., Barbet, A. F., Bekker, C. P., Dasch, G. A., Palmer, G. H., Ray, S. C., et al. (2001). Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. Evol. Microbiol. 51, 2145–2165. doi: 10.1099/00207713-51-6-2145
Enkhtaivan, B., Narantsatsral, S., Davaasuren, B., Otgonsuren, D., Amgalanbaatar, T., Uuganbayar, E., et al. (2019). Molecular detection of Anaplasma ovis in small ruminants and ixodid ticks from Mongolia. Parasitol. Int. 69, 47–53. doi: 10.1016/j.parint.2018.11.004
Fukui, Y., and Inokuma, H. (2019). Subclinical infections of Anaplasma phagocytophilum and Anaplasma bovis in dogs in Ibaraki. Japan.Jpn. J. Infect. Dis. 72, 168–172. doi: 10.7883/yoken.JJID.2018.470
Giglioti, R., Bassetto, C. C., Okino, C. H., de Oliveira, H. N., and de Sena Oliveira, M. C. (2019). Development of a loop-mediated isothermal amplification (LAMP) assay for the detection of Anaplasma marginale. Exp. Appl. Acarol. 77, 65–72. doi: 10.1007/s10493-018-0327-y
Goel, R., Westblade, L. F., Kessler, D. A., Sfeir, M., Slavinski, S., Backenson, B., et al. (2018). Death from transfusion-transmitted Anaplasmosis. New York, USA, 2017. Emerging Infect. Dis. 24, 1548–1550. doi: 10.3201/eid2408.172048
Guimarães, A., Raimundo, J. M., Peixoto, M. P., da Silva, C. B., Pires, M. S., Santos, H. A., et al. (2019). Molecular detection, characterization of Anaplasma spp. in domestic cats from Rio de Janeiro state. Acta Trop. 191, 239–242. doi: 10.1016/j.actatropica.2019.01.003
Guo, W. P., Huang, B., Zhao, Q., Xu, G., Liu, B., Wang, Y. H., et al. (2018). Human-pathogenic Anaplasma spp., and Rickettsia spp. in animals in Xi’an. China. PLoS Negl. Trop. Dis. 12:e0006916. doi: 10.1371/journal.pntd.0006916
Halajian, A., Palomar, A. M., Portillo, A., Heyne, H., Romero, L., and Oteo, J. A. (2018). Detection of zoonotic agents and a new Rickettsia strain in ticks from donkeys from South Africa: implications for travel medicine. Travel. Med. Infect. Dis. 26, 43–50. doi: 10.1016/j.tmaid.2018.10.007
Han, R., Yang, J. F., Mukhtar, M. U., Chen, Z., Niu, Q. L., Lin, Y. Q., et al. (2019). Molecular detection of Anaplasma infections in ixodid ticks from the qinghai-tibet plateau. Infect. Dis Poverty. 8:12. doi: 10.1186/s40249-019-0522-z
Hao, X., Liu, R., He, Y., Xiao, X., Xiao, W., Zheng, Q., et al. (2019). Multiplex PCR methods for detection of several viruses associated with canine respiratory and enteric diseases. PLoS One. 14:e0213295. doi: 10.1371/journal.pone.0213295
Henegariu, O., Heerema, N. A., Dlouhy, S. R., Vance, G. H., and Vogt, P. H. (1997). Multiplex PCR: critical parameters and step-by-step protocol. Biotechniques 23, 504–511. doi: 10.2144/97233rr01
Hornok, S., De, L. F. J., Biró, N., Ig, F. D. M., Meli, M. L., Elek, V., et al. (2011). First molecular evidence of Anaplasma ovis and Rickettsia spp. in keds (Diptera: Hippoboscidae) of sheep and wild ruminants. Vector Borne Zoonotic Dis. 11, 1319–1321. doi: 10.1089/vbz.2011.0649
Hornok, S., Szoke, K., Meli, M. L., Sandor, A. D., Gorfol, T., Estok, P., et al. (2019). Molecular detection of vector-borne bacteria in bat ticks (Acari: Ixodidae, Argasidae) from eight countries of the Old and New Worlds. Parasit. Vectors 12:50. doi: 10.1186/s13071-019-3303-4
Kawahara, M., Rikihisa, Y., Lin, Q., Isogai, E., Tahara, K., Itagaki, A., et al. (2006). Novel genetic variants of Anaplasma phagocytophilum, Anaplasma bovis, Anaplasma centrale, and a novel Ehrlichia sp. in wild deer and ticks on two major islands in Japan. Appl. Environ. Microbiol. 72, 1102–1109. doi: 10.1128/AEM.72.2.1102-1109.2006
Kim, K., Yi, J., Oh, W., Kim, N., Choi, S., Choe, P., et al. (2014). Human granulocytic anaplasmosis, South Korea, 2013. Emerging Infect Dis. 20, 1708–1711. doi: 10.3201/eid2010.131680
Kundave, V. R., Ram, H., Banerjee, P. S., Garg, R., Mahendran, K., Ravikumar, G., et al. (2018). Development of multiplex PCR assay for concurrent detection of tick borne haemoparasitic infections in bovines. Acta Parasitol. 63, 759–765. doi: 10.1515/ap-2018-0090
Lee, S. H., Park, S., Lee, Y. S., Lee, H. K., and Hwang, S. D. (2018). Diagnosis and molecular characteristics of human infections caused by Anaplasma phagocytophilum in South Korea. J. Microbiol. 56, 847–853. doi: 10.1007/s12275-018-8385-8
Lew, A. E., Gale, K. R., Minchin, C. M., Shkap, V., and Waal, D. T. D. (2003). Phylogenetic analysis of the erythrocytic Anaplasma species based on 16S rDNA and GroEL (HSP60) sequences of A. marginale, A. centrale, and A. ovis and the specific detection of A. centrale vaccine strain. Vet. Microbiol. 92, 145–160. doi: 10.1016/S0378-1135(02)00352-8
Li, H., Zheng, Y. C., Ma, L., Jia, N., Jiang, B. G., Jiang, R. R., et al. (2015). Human infection with a novel tick-borne Anaplasma species in China: a surveillance study. Lancet Infect. Dis. 15, 663–670. doi: 10.1016/S1473-3099(15)70051-4
Liu, Z., Ma, M., Wang, Z., Wang, J., Peng, Y., Li, Y., et al. (2012). Molecular survey and genetic identification of Anaplasma species in goats from central and southern China. Appl. Environ. Microbiol. 78, 464–470. doi: 10.1128/AEM.06848-11
Liu, Z., Peasley, A. M., Yang, J., Li, Y., Guan, G., Luo, J., et al. (2019). The Anaplasma ovis genome reveals a high proportion of pseudogenes. BMC Genomics 20:69. doi: 10.1186/s12864-018-5374-6
M’ghirbi, Y., Beji, M., Oporto, B., Khrouf, F., Hurtado, A., and Bouattour, A. (2016). Anaplasma marginale and A. phagocytophilum in cattle in Tunisia. Parasit. Vectors. 9:556. doi: 10.1186/s13071-016-1840-7
Peng, Y., Wang, K., Zhao, S., Yan, Y., Wang, H., Jing, J., et al. (2018). Detection and Phylogenetic Characterization of Anaplasma capra: an emerging pathogen in sheep and Goats in China. Front. Cell Infect. Microbiol. 8:283. doi: 10.3389/fcimb.2018.00283
Pradeep, R. K., Nimisha, M., Sruthi, M. K., Vidya, P., Amrutha, B. M., Kurbet, P. S., et al. (2019). Molecular characterization of South Indian field isolates of bovine Babesia spp. and Anaplasma spp. Parasitol. Res. 118, 617–630. doi: 10.1007/s00436-018-6172-4
Rjeibi, M. R., Ayadi, O., Rekik, M., and Gharbi, M. (2018). Molecular survey and genetic characterization of Anaplasma centrale. A. marginale and A. bovis in cattle from Algeria. Transbound Emerg Dis. 65, 456–464. doi: 10.1111/tbed.12725
Seo, M. G., Ouh, I. O., Kwon, O. D., and Kwak, D. (2018a). Molecular detection of Anaplasma phagocytophilum-like Anaplasma spp. and pathogenic A. phagocytophilum in cattle from South Korea. Mol. Phylogenet Evol. 126, 23–30. doi: 10.1016/j.ympev.2018.04.012
Seo, M. G., Ouh, I. O., Lee, H., Geraldino, P. J. L., Rhee, M. H., Kwon, O. D., et al. (2018b). Differential identification of Anaplasma in cattle and potential of cattle to serve as reservoirs of Anaplasma capra, an emerging tick-borne zoonotic pathogen. Vet. Microbiol. 226, 15–22. doi: 10.1016/j.vetmic.2018.10.008
Song, R., Wang, Q., Guo, F., Liu, X., Song, S., Chen, C., et al. (2018). Detection of Babesia spp., Theileria spp. and Anaplasma ovis in Border Regions, northwestern China. Transbound Emerg Dis. 65, 1537–1544. doi: 10.1111/tbed.12894
Sun, X. F., Zhao, L., Wen, H. L., Luo, L. M., and Yu, X. J. (2015). Anaplasma species in China. Lancet Infect. Dis. 15, 1263–1264. doi: 10.1016/S1473-3099(15)00377-1
Torina, A., Agnone, A., Blanda, V., Alongi, A., D’Agostino, R., Caracappa, S., et al. (2012). Development and validation of two PCR tests for the detection of and differentiation between Anaplasma ovis and Anaplasma marginale. Ticks Tick Borne Dis. 3, 283–287. doi: 10.1016/j.ttbdis.2012.10.033
Wang, J., Zhang, Y., Cui, Y., Yan, Y., Wang, X., and Wang, R. (2018). A rapid, simple and sensitive loop-mediated isothermal amplification method to detect Anaplasma bovis in sheep and goats samples. Parasitol. Int. 67, 70–73. doi: 10.1016/j.parint.2017.03.005
Wei, R., Liu, H. B., Jongejan, F., Jiang, B. G., Chang, Q. C., Fu, X., et al. (2017). Cultivation of Anaplasma ovis in the HL-60 human promyelocytic leukemia cell line. Emerg. Microbes Infect. 6:e83. doi: 10.1038/emi.2017.70
Yang, B., Sun, E., Wen, Y., Ye, C., Liu, F., Jiang, P., et al. (2019). Molecular evidence of coinfection of Anaplasma species in small ruminants from Anhui Province. China. Parasitol. Int. 71, 143–146. doi: 10.1016/j.parint.2019.04.004
Yang, J., Han, R., Niu, Q., Liu, Z., Guan, G., Liu, G., et al. (2018a). Occurrence of four Anaplasma species with veterinary and public health significance in sheep, northwestern China. Ticks Tick Borne Dis. 9, 82–85. doi: 10.1016/j.ttbdis.2017.10.005
Yang, J., Liu, Z., Niu, Q., Mukhtar, M. U., Guan, G., Liu, G., et al. (2018b). A novel genotype of “Anaplasma capra” in wildlife and its phylogenetic relationship with the human genotypes. Emerg. Microbes Infect. 7:210. doi: 10.1038/s41426-018-0212-0
Yang, J., Liu, Z., Niu, Q., Liu, J., Han, R., Liu, G., et al. (2016). Molecular survey and characterization of a novel Anaplasma species closely related to Anaplasma capra in ticks, northwestern China. Parasit Vectors. 9:603. doi: 10.1186/s13071-016-1886-6
Zaid, T., Ereqat, S., Nasereddin, A., Al-Jawabreh, A., Abdelkader, A., and Abdeen, Z. (2019). Molecular characterization of Anaplasma and Ehrlichia in ixodid ticks and reservoir hosts from Palestine: a pilot survey. Vet. Med. Sci. 5, 230–242. doi: 10.1002/vms3.150
Zhou, Z., Wu, Y., Chen, Y., Wang, Z., Hu, S., Zhou, R., et al. (2018). Molecular and serological prevalence of Toxoplasma gondii and Anaplasma spp. infection in goats from Chongqing Municipality. China. Parasite. 25, 20. doi: 10.1051/parasite/2018024
Keywords: Anaplasma capra, Anaplasma bovis, Anaplasma ovis, Anaplasma phagocytophilum, multiplex PCR
Citation: Peng Y, Zhao S, Wang K, Song J, Yan Y, Zhou Y, Shi K, Jian F, Wang R, Zhang L and Ning C (2020) A Multiplex PCR Detection Assay for the Identification of Clinically Relevant Anaplasma Species in Field Blood Samples. Front. Microbiol. 11:606. doi: 10.3389/fmicb.2020.00606
Received: 12 November 2019; Accepted: 18 March 2020;
Published: 07 April 2020.
Edited by:
Spyridon Ntougias, Democritus University of Thrace, GreeceCopyright © 2020 Peng, Zhao, Wang, Song, Yan, Zhou, Shi, Jian, Wang, Zhang and Ning. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Changshen Ning, bm5sMTk4NkAxNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.