- 1Laboratory C15, Division of Infections, Immunity and Microbes, School of Life Sciences, University of Nottingham, Nottingham, United Kingdom
The predatory bacterium B. bacteriovorus grows and divides inside the periplasm of Gram-negative bacteria, forming a structure known as a bdelloplast. Cell division of predators inside the dead prey cell is not by binary fission but instead by synchronous division of a single elongated filamentous cell into odd or even numbers of progeny cells. Bdellovibrio replication and cell division processes are dependent on the finite level of nutrients available from inside the prey bacterium. The filamentous growth and division process of the predator maximizes the number of progeny produced by the finite nutrients in a way that binary fission could not. To learn more about such an unusual growth profile, we studied the role of DivIVA in the growing Bdellovibrio cell. This protein is well known for its link to polar cell growth and spore formation in Gram-positive bacteria, but little is known about its function in a predatory growth context. We show that DivIVA is expressed in the growing B. bacteriovorus cell and controls cell morphology during filamentous cell division, but not the number of progeny produced. Bacterial Two Hybrid (BTH) analysis shows DivIVA may interact with proteins that respond to metabolic indicators of amino-acid biosynthesis or changes in redox state. Such changes may be relevant signals to the predator, indicating the consumption of prey nutrients within the sealed bdelloplast environment. ParA, a chromosome segregation protein, also contributes to bacterial septation in many species. The B. bacteriovorus genome contains three ParA homologs; we identify a canonical ParAB pair required for predatory cell division and show a BTH interaction between a gene product encoded from the same operon as DivIVA with the canonical ParA. The remaining ParA proteins are both expressed in Bdellovibrio but are not required for predator cell division. Instead, one of these ParA proteins coordinates gliding motility, changing the frequency at which the cells reverse direction. Our work will prime further studies into how one bacterium can co-ordinate its cell division with the destruction of another bacterium that it dwells within.
Introduction
B. bacteriovorus is a small predatory bacterium that invades and replicates within other Gram-negative bacteria, forming a rounded structure called a ‘bdelloplast’. Inside the dead prey bacterium, the Bdellovibrio cell elongates to form a multiploid filament, before synchronous septation liberates odd or even numbered progeny (Fenton et al., 2010a). Predatory mutants of B. bacteriovorus can be saved and cultured slowly by host-independent (HI) axenic growth, requiring an amino acid rich medium (Seidler and Starr, 1969; Cotter and Thomashow, 1992). HI growth also involves septation of a filamentous cell, akin to that in the bdelloplast, although HI cells are pleiomorphic, and division can occur either synchronously or asynchronously (Cotter and Thomashow, 1992; Hobley et al., 2012b). During Bdellovibrio predatory growth in the prey bdelloplast (and as HI cells), cellular components must be partitioned along the filament prior to division to ensure that they are faithfully segregated prior to synchronous septation. This contrasts the binary fission model of division, seen in most other bacteria, where conventional septation results in two daughter cells.
Pre-divisional partitioning is a process required for the organization of prokaryote cellular components, including chromosomes, plasmids, individual proteins [such as E. coli proteins UidR (transcriptional repressor), HisG (ATP phosphoribosyltransferase) and MalI (transcriptional repressor)], chemotaxis clusters and carboxysomes (Bignell and Thomas, 2001; Ringgaard et al., 2011, Roberts et al., 2012; Cho, 2015, Kuwada et al., 2015). The partitioning of DNA ensures that cell division does not occur across nucleoids, whilst the partitioning of proteins ensures that each daughter cell receives the prerequisite components for optimum fitness. Whilst some partitioning events may be stochastic, other events require active organization (Huh and Paulsson, 2011). A major checkpoint in the division cycle is the segregation of chromosomes, such that each progeny has a complete copy of the genome. In many bacterial species, such as Vibrio and Caulobacter, chromosome segregation is driven by a three component ParABS system which guides newly replicated chromosomes to bacterial cell poles, facilitating DNA segregation (Hui et al., 2010; Mierzejewska and Jagura-Burdzy, 2012, Espinosa et al., 2017). Here we identify a canonical ParAB system in B. bacteriovorus and show that it is required for efficient predatory growth.
In many bacteria, chromosomal segregation during cell division is controlled by the three element ParABS system via a ratchet diffusion model, as recently reviewed (Jindal and Emberly, 2019). ParB binds to centromere-like parS DNA sequences forming the ParBS nucleoprotein complex. ParA, an ATPase, will dimerize and bind DNA non-specifically in the presence of ATP. Chromosome segregation is facilitated by ParA-ATP interacting with ParBS complexes which activates the ATPase activity of ParA, causing it to dissociate from the chromosome, and for the ParBS complex to move. ParA-ADP can then be phosphorylated and will bind to DNA again after a delay. Therefore, as ParBS moves, it leaves an area of DNA behind it with no bound ParA-ATP, preventing the direction of ParBS movement from reversing. This allows the ParBS to move unidirectionally along a concentration gradient of bound ParA-ATP. This is a very attractive model but it is unclear how this system could maintain the position of multiple B. bacterivorous chromosomes along the growing filamentous cell prior to division.
In addition to parA and parB, there are several genes in the B. bacteriovorus genome that code for proteins that have been identified in other bacteria as key control elements for cell division. One of these is a divIVA homolog, hereafter referred to as divIVABd, which has been shown to encode a protein with a number of cell growth and septation related roles in Gram-positive bacteria, including septal site selection and chromosome segregation (Thomaides et al., 2001; Hammond et al., 2019). In Gram-negative bacteria, few studies have focused on the function of DivIVA homologs, which are typically limited to some Oligoflexia and Deltaproteobacteria (Akiyama et al., 2003), so we aimed to establish the function of DivIVABd in B. bacteriovorus HD100, a Gram-negative predatory bacterium which grows inside prey.
Other bacterial genomes containing divIVA homologs can have non-conventional methods for septation. Mycobacteria are reported to have both symmetrical septation, producing two identical daughter cells, and asymmetric septation, where one daughter cell is significantly larger than the other (Kieser and Rubin, 2014; Vijay et al., 2014). In contrast, Streptomyces growth and division is characterized by polar (apical) growth of branched hyphae and the dispersion of spores, much like fungi (Flardh et al., 2012). The background of other proteins in addition to DivIVA (such as Noc and MinCD which are involved in the co-ordination of division site), can vary between Gram-positive bacteria so there is clearly no “one size fits all” scenario (Monahan et al., 2014; Hammond et al., 2019). In addition, in Streptococci and other bacteria, which like B. bacteriovorus lack MinCD, it was noted that post translational modifications of the DivIVA protein can also occur, and in Streptococci that a conserved alanine residue A78 is important for mediating DivIVA protein-protein interactions, suggesting a different regulatory network for septation control (Fadda et al., 2007). In these and other Gram-positive bacterial systems, DivIVA has been at least partially characterized as having varied roles, from regulating septation to polar cell wall growth (Hempel et al., 2008; Kang et al., 2008, Flardh et al., 2012; Ginda et al., 2013, Hammond et al., 2019). As apical growth is seen for the B. bacteriovorus filament, we postulated that DivIVA may be involved in this growth mode (Fenton et al., 2010a).
In Bacillus subtilis, the coiled-coil DivIVA protein is localized to negatively curved membranes by an N-terminus membrane-binding domain (Lenarcic et al., 2009). This localization allows DivIVA to interact with partner proteins to facilitate septum formation at mid-cell and chromosome segregation (Edwards and Errington, 1997; Marston and Errington, 1999, Thomaides et al., 2001). In other species the DivIVA protein directs hyphal tip extension in Streptomyces coelicolor (Flärdh, 2003; Hempel et al., 2008) and regulates polar growth in Corynebacterium glutamicum and Mycobacterium smegmatis (Kang et al., 2008; Letek et al., 2008, Donovan and Bramkamp, 2014; Kieser and Rubin, 2014, Meniche et al., 2014). However, in the cyanobacterium Synechococcus elongatus the DivIVA homolog Cdv3 does not contain the conserved residues linked to negative curvature sensing (MacCready et al., 2017).
Previous work has shown DivIVA and ParA directly interact in M. smegmatis (Ginda et al., 2013; Ramirez et al., 2013), establishing a link between the functions of both proteins. In other bacteria, DivIVA interacts with MinD, a ParA-like ATPase that functions in division site selection, either directly, such as in Listeria monocytogenes (Kaval et al., 2014), or indirectly, such as in B. subtilis where the interaction occurs via MinJ (Patrick and Kearns, 2008; Van Baarle and Bramkamp, 2010, Eswaramoorthy et al., 2011). In the M. smegmatis model, DivIVA directs subpolar addition to the cell wall during division (Kang et al., 2008), suggesting there is coordination between cell elongation and chromosome segregation. Given these roles across bacteria, DivIVA is a strong candidate to provide this type of coordination in B. bacteriovorus where complex, multi-septa division occurs along a filamentous cell (Fenton et al., 2010a).
Bacterial chromosomes often also encode orphan ParA-like proteins, which are additional ParA homologs not encoded from a canonical parAB locus. These ParA-like proteins perform roles distinct from ParA, for example PldP determines division sites in Corynebacterium glutamicum (Donovan et al., 2010; Donovan and Bramkamp, 2014), a ParA/Soj-like protein in M. tuberculosis interacts with the MzF6 protein regulating cell growth (Ramirez et al., 2013), and PpfA is involved in partitioning cytoplasmic chemotaxis clusters in Rhodobacter sphaeroides (Thompson et al., 2006; Roberts et al., 2012). These orphan parA-like genes are often, but not exclusively, found within operons containing genes for the processes in which they are involved. For example, parC (partitioning of chemotaxis) genes have been identified in chemotaxis operons in numerous bacterial species (Ringgaard et al., 2011). In Vibrio cholerae, ParC is involved in partitioning chemotaxis proteins, thus playing a role in chemotaxis itself, and influencing swimming and swarming (Ringgaard et al., 2011). To date no ParA-like proteins have been reported to influence gliding motility, however, discrete cellular locations for gliding motor complexes, at different points along a cell axis, are required for gliding function and Par proteins could participate in their positioning.
Bacterial gliding is a process characterized by the non-flagellar movement of a single cell on a solid surface and has been previously observed in many bacterial species, including B. bacteriovorus (Spormann, 1999; Mendez et al., 2008, Lambert et al., 2011; Asada et al., 2012, Zhu and McBride, 2016). Gliding motility can be subdivided into two categories; social (S)-motility, surface movement using pilus retraction in Myxococcus xanthus; and adventurous (A)-gliding motility, characterized by the movement of individual cells (McBride and Zhu, 2013; Jakobczak et al., 2015, Zhu and McBride, 2016). B. bacteriovorus is known to exhibit a form of A-motility which uses gliding engines and is independent of pili (Lambert et al., 2011). Abolition of gliding (for example, in a diguanylyl cyclase (dgcA) mutant), renders the Bdellovibrio cells unable to glide out and exit a prey cell after lysis (Hobley et al., 2012a). Thus gliding may be particularly relevant when Bdellovibrio prey upon bacteria within biofilms.
The B. bacteriovorus HD100 genome contains two orphan parA-like genes (bd1326 and bd2331), in addition to the typical parAB locus (bd3906-5). Both of these parA-like genes in B. bacteriovorus are located at loci where neighboring genes encode putative proteins with unknown functions, rather than a ParB homolog. A study into the early prey invasion ‘predatosome’ of B. bacteriovorus revealed that both the parA-like genes were up-regulated during HI filamentous growth, but not at the 30 min stage of prey invasion by flagellate predators in liquid cultures (Lambert et al., 2010). However, this work focused upon early invasion events, and not those at later time points. If these ParA-like proteins function during division, as canonical ParA does, then it is reasonable to expect them to be up-regulated later in the cell cycle once the bdelloplast is established at a time when the B. bacteriovorus filamentous cell is rapidly growing. If relevant to gliding motility, we would not expect to see this up-regulation when the cell is flagellate or sessile. Hereafter, the three ParA homologs of B. bacteriovorus will be referred to as ParA1 (Bd1326), ParA2 (Bd2331) and ParA3 (Bd3906), the latter which is co-expressed with the gene for ParB (Bd3905).
In this study, we demonstrate that DivIVABd participates in septal positioning during predatory replication with resultant effects on progeny morphology. We postulate that its regulatory role may be linked to cell partitioning through interaction with a protein product of its co-transcribed neighboring gene and the canonical parA3 gene-product. DivIVA may also interact with proteins that could signal nutritional or oxygenic status within the bdelloplast prior to predator septation. We also show by fluorescent tagging that the canonical ParA3 is associated with the cell division process and that the tag affects protein function and perturbs B. bacteriovorus cell length. We further demonstrate that two additional ParA homologs, ParA1 and ParA2, are expressed in B. bacteriovorus, with gene deletion and fluorescent localization assays revealing that both are non-essential, but that ParA2 contributes to gliding motility behavior.
These results prime further biochemical studies on the changing nature of the bdelloplast environment as B. bacteriovorus grow within it. We show here that cell division, morphological and movement behaviors of intra-bacterial B. bacteriovorus are responding to DivIVABd or Par protein controls and that some of these may be sensing changes in bdelloplast biochemistry.
Materials and Methods
Bioinformatics
Gene and protein sequences for B. bacteriovorus HD100 were acquired from Xbase (Chaudhuri et al., 2008). Gene and protein homologs were found with the NCBI BLAST program suite (Altschul et al., 1990). Pairwise alignments were generated in EMBOSS Needle and multiple alignments in EMBOSS ClustalO (Rice et al., 2000; Sievers and Higgins, 2018). Alignments were visualized in ESPript 3.0 (Robert and Gouet, 2014). Statistics and graphs were processed in IBM SPSS Statistics for Windows, Version 25.0. Armonk, NY: IBM Corp. Protein trees were generated through MEGA-X version 10.0.05 using the Maximum Likelihood method, with 1000 bootstraps (Kumar et al., 2018) and visualized in FigTree version 1.4.41.
Bacterial Strain Growth
B. bacteriovorus HD100 was used throughout and maintained in Ca/HEPES buffer preying upon E. coli S17-1 prey as previously described (Supplementary Tables S1A,B) (Lambert et al., 2006, 2016). Host independent B. bacteriovorus strains were grown as previously described (Lambert and Sockett, 2013). Prey were grown in YT broth for 16 h at 37 °C with shaking at 200 rpm (Supplementary Table S1C).
Reverse Transcription PCR
RNA extraction and Reverse Transcriptase PCR (RT-PCR) assays were performed as previously described (Lambert et al., 2006, 2016). Using a Promega SV total RNA isolation kit, total RNA was extracted from samples taken throughout the time course. RT-PCR assays were performed using the Qiagen One-step RT-PCR kit. Reaction conditions are as follows: One cycle of 50 °C for 30 min then 95 °C for 15 min, followed by 25-30 cycles of 94 °C for 1 min, 48 °C for 1 min, 72 °C for 2 min, and then a 10 min extension at 72 °C after the 25-30 cycles, and ultimately held at 4 °C prior to gel analysis of products. Primers used for amplification are listed in Supplementary Table S2A. Primers used specifically for operon walking are listed in Supplementary Table S2B.
Cloning Fluorescently Tagged and Deletion Constructs
Fluorescent constructs were made using previously described methods (Fenton et al., 2010b). Genes were cloned into a vector by removal of the stop codon such that the C-terminus of the gene product fused with the mCherry, or mTFP gene. This was then subcloned into the vector pK18mobsacB and then introduced into the B. bacteriovorus via a single crossover event at the 5’ end of the gene, such that the ORF-tag is transcribed from the endogenous promoter in a merodiploid strain. To illuminate the cytoplasm of the B. bacteriovorus cells within prey, we used a previously published strain with full gene replacement of cytoplasmic marker protein Bd0064 with Bd0064mCerulean (Raghunathan et al., 2019). Genomic deletions of specific B. bacteriovorus genes were made using methods previously described (Capeness et al., 2013; Lambert and Sockett, 2013, Lambert et al., 2016). Primers used for amplification of genes are listed in Supplementary Tables S2C,D.
Fluorescent and Phase Contrast Microscopy
Microscopy images were acquired using a Nikon Eclipse Ti-E widefield inverted microscope equipped with an Andor Neo sCMOS camera, as previously described (Kuru et al., 2017). Semi synchronous predatory prey lysate cultures for time course microscopy through the predatory cycle were prepared as previously described (Lambert and Sockett, 2013). Images were processed in either SimplePCI software (HCImage.com) or FIJI (ImageJ) (Schneider et al., 2012).
Gliding Motility Assays
Timelapse video microscopy was used to take images of B. bacteriovorus HD100 strains immobilized on 1% agarose/CaHEPES (Lambert et al., 2011). Single cells were selected by generating random regions of interest (ROIs) containing approximately 5–10 cells in each field of view. Following the cells through each frame allowed the time at which each cell started gliding to be observed. After an individual cell had been gliding for one hour (allowing an establishment period), the number of direction changes (reversals) was counted manually and recorded. To determine the gliding status and position of any fluorescent foci, cells were immobilized as above. After 400 min of incubation (a time chosen to allow prolonged incubation on a surface, and for gliding motility to commence), images were acquired every 15 min for an hour and analyzed in SimplePCI.
Predation Efficiency Assays
Predatorily grown (prey/host dependent HD) B. bacteriovorus strains containing fluorescent tags were assayed against a fliC1 merodiploid which served as an antibiotic marked “wild type” equivalent control. 50 ml predatory cultures were grown and filtered through a 0.45 μm filter to remove any remaining prey cells, these were matched by protein concentration using a Lowry assay and subsequently enumerated to confirm the number of B. bacteriovorus added. To serve as bacterial prey, a 50 ml culture of luminescent S17-1 pCL100, which encodes luxCDABE, was also grown under standard conditions. A 1.5 ml sample of each B. bacteriovorus strain was heat-treated at 105°C for 5 min and allowed to cool to room temperature, generating the ‘heat killed’ control cells. Equivalent amounts of each Lowry-matched B. bacteriovorus strain was made up to 64 μl with the heat-killed B. bacteriovorus preparation (64, 32, 16 and 8 μl live B. bacteriovorus plus 0, 32, 48 or 56 μl heat-killed cells). Predatory cells were mixed with 200 μl of E. coli S17-1 pCL100 and diluted to an OD600 of 1.0 in CaHEPES, in a 96-well microtiter plate. Control wells had 64 μl heat-killed B. bacteriovorus only for each strain. The plates were then covered with Breathe-Easy membrane. The reduction in luminescence due to the killing of the E. coli S17-1 pCL100 prey cells by B. bacteriovorus was measured over time using a BMG FluoStar microplate reader.
Cell Morphology Measurements Using MicrobeJ and Detection of Fluorescent Foci of DivIVA
After image acquisition, cell measurements were generated using MicrobeJ, a plugin for the FIJI software, as described previously in Kuru et al., 2017 (Schindelin et al., 2012; Ducret et al., 2016, Kuru et al., 2017). To detect fluorescent foci of DivIVA-mCherry; the rounded, invaded, E. coli prey cells (bdelloplasts) were detected in the phase channel by defining circularity as 0.96-max and length as 1-max, with all other parameters as default. B. bacteriovorus cells (with cytoplasmic Bd0064-mCerulean) were detected by the medial axis method in the mCerulean channel as maxima 1, by defining area as 0.15-max, with all other parameters as default. They were associated with the bdelloplasts with a tolerance of 0.1. Fluorescent foci of DivIVA-mCherry were detected as maxima 2 in the red channel by the “fit shape to circle” method by defining area as 0-0.25, with all other parameters as default. These were associated to maxima 1 with a tolerance of 0.1. Manual inspection of the analyzed images confirmed that the vast majority of cells were correctly assigned. In cases where bdelloplasts appeared to be infected by two Bdellovibrio, these were manually removed from the analysis. The shape measurements including the angularity, area, aspect ratio, circularity, curvature, length, roundness, sinuosity, solidity and width were measured for each type of cell.
Pairwise Bacterial Two Hybrid and Liquid β-Galactosidase Assays
Pairwise Bacterial Two Hybrid (BTH) assays were performed to test for interactions between DivIVABd, ParA3 and other B. bacteriovorus proteins using protocols previously described (Battesti and Bouveret, 2012). Liquid β-galactosidase assays were performed using the single-step protocol (Schaefer et al., 2016). Primers used for gene amplification are listed in Supplementary Table S2E.
B. bacteriovorus HD100 Bacterial Two Hybrid Library Construction and Assay
A B. bacteriovorus HD100 BTH library was constructed following methods, adapted from those previously described (Handford et al., 2009; Houot et al., 2012). Briefly, the HD100 genome was extracted, using the Sigma-Aldrich GenElute Bacterial Genomic DNA Kit, and restriction-digested into fragments between 500 bp and 2000 bp which were extracted via the Sigma-Aldrich GenElute Gel Extraction Kit, according to manufacturer’s instructions. This was then ligated into four plasmids; pUT18, pUT18C and pUT18 + 1, where + 1 denotes an additional nucleotide added in the linker region to account for the frame of the genome fragments. The ligations were transformed into E. coli DH5α, and the resulting transformants were pooled. Plasmids were extracted using the Sigma-Aldrich GenElute Plasmid Miniprep Kit, resulting in a library of mixed genomic fragments for each of the plasmids.
To perform the assay, a pKT25 plasmid containing the bait gene was transformed into the E. coli BTH101 strain, which was made chemically competent. These were transformed with 1μl of library plasmids. Incubation was for 48 h at 29oC on MacConkey agar supplemented with 50 mg/ml Ampicillin, 25 mg/ml Kanamycin, 40 mg/ml X-gal and 20 mg/ml IPTG. Blue colonies were grown in high salt Mu media and the plasmids, bait and library were extracted via miniprep. Competent E. coli DH5α were then transformed with both plasmids and cured for the pKT25 plasmid. The library plasmid was then extracted. This was retested with the BTH pairwise protocol against the bait plasmid to confirm transformants remain blue on the supplemented MacConkey agar. These were then sequenced to determine the interacting protein fragments encoded in the library fragments. Once the genes in the fragments were identified, full length gene copies were cloned into pUT18C and pKT25 and used again in the pairwise assay above.
Results
B. bacteriovorus DivIVABd Is Encoded on a Four Gene Operon
DivIVA proteins are usually found in Gram-positive genomes. Despite this, the Gram-negative B. bacteriovorus genome contains a DivIVA homolog encoded by bd0464 and referred to as DivIVABd (Figure 1A). Comparing this protein to features of DivIVA in Bacillus subtilis (detailed in Supplementary Information S1) revealed that DivIVABd can likely dimerize, but substitutions in R18 suggest that it cannot inherently sense negative curvature, likely requiring other protein interactions to localize to curved bacterial cell poles.
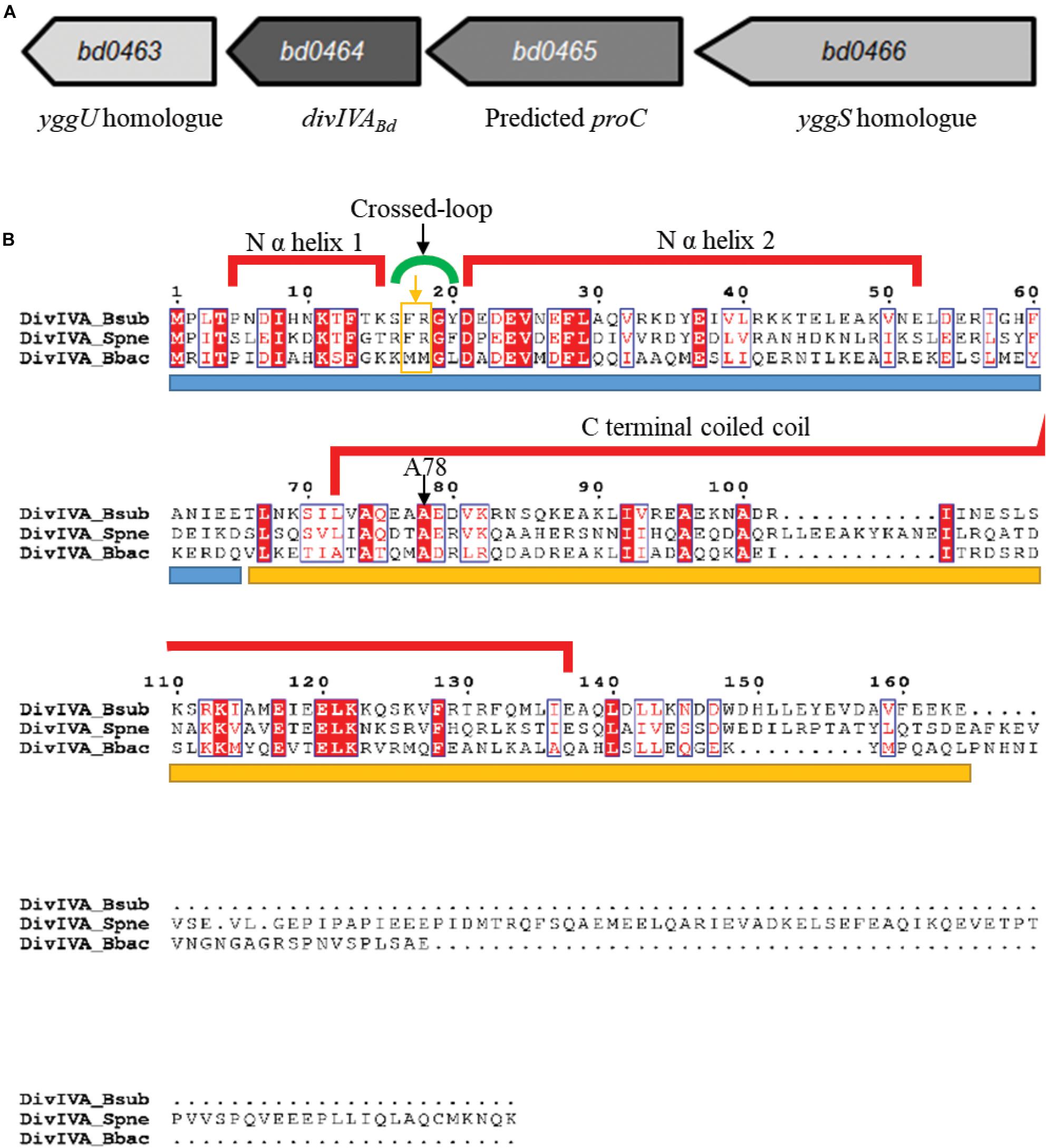
Figure 1. Genomic context of the B. bacteriovorus divIVABd gene and protein alignment of DivIVABd with Gram-positive DivIVA homologs. (A) Map of the B. bacteriovorus divIVA gene and neighboring region. The divIVABd gene is downstream of genes encoding a predicted yggS homolog (Bd0466 accession: NP_967454.1) and pyrroline-5-carboxylate reductase (Bd0465 accession: NP_967453.1) and upstream of a gene encoding an YggU homolog (Bd0463 accession: NP_967451.1). (B) Alignment of DivIVABb with DivIVA homologs in Streptococcus pneumoniae (DivIVASpn; accession: AAC95445.1) and Bacillus subtilis (DivIVABsub; accession: P71021.1). Sequences are colored and annotated based on the DivIVABsub crystal structure. The blue bar under the sequences indicates the N terminal domain and the yellow bar, the C terminal domain. The A78 residue, conserved in DivIVABd, was shown to be important for DivIVA function in B. subtilis, and an A78T substitution in S. pneumoniae DivIVA disturbed protein localization and perturbed interactions with FtsK and late-stage divisome components. Residues F17 and R18 are highlighted by an orange box and arrow underneath the green cross-link indicator.
DivIVA homologs of Gram-positive bacteria are typically encoded downstream of a cell division and cell wall (dcw) cluster and the ftsZ gene (Ayala et al., 1994; Cha and Stewart, 1997, Massidda et al., 1998; Ramirez-Arcos et al., 2005). Although B. bacteriovorus does have a dcw cluster, divIVABd (bd0464, accession number: NP_967452.1) is not found near this region (Figure 1A), rather it is downstream of a yggS homolog, bd0466, which encodes a protein similar to the N-terminal barrel domain of an alanine racemase (LeMagueres et al., 2005; Ito et al., 2013). In E. coli, this domain binds the coenzyme Pyridoxal Phosphate (PLP), with the required residues conserved in Bd0466 (Ito et al., 2013). Recently, the function of YggS has been studied in other bacteria, implicating it in the regulation of PLP and biosynthesis of amino acids isoleucine and valine (Ito et al., 2013, 2016, Prunetti et al., 2016; Ito et al., 2019). In species such as Bacillus subtilis, Staphylococcus aureus, and Streptococcus pneumoniae, a yggS protein is also found upstream of the divIVA homolog.
Between bd0466 and divIVABd is the gene bd0465, which encodes a pyrroline-5-carboxylate reductase, ProC, homolog (Figure 1A). ProC proteins catalyze the final step in the biosynthesis pathway that converts glutamate to proline (Nocek et al., 2005).
Alignment of the B. bacteriovorus DivIVABd with DivIVA homologs of Streptococcus pneumoniae and Bacillus subtilis (Figure 1B) showed that the N-terminal domain and coiled-coil domains [confirmed using Multicoil prediction (Wolf et al., 1997)] are conserved between the proteins. A NEEDLE global alignment showed that DivIVABd and DivIVABsub share 24.6% protein identity and 47.5% protein similarity, with differences predominantly at the C-terminus. This is typical of DivIVA proteins, as the C-terminus of different DivIVA homologs tends to show greater variability (Tavares et al., 2008).
DivIVABd-mCherry Is Localized at the Poles of B. bacteriovorus Attack Phase Cells and Growing Filaments
As DivIVABd is lacking residues shown to be essential for negative curvature sensing, F17 and R18 (highlighted in Figure 1B), we wanted to assess its localization in host-dependent B. bacteriovorus cells. The protein was labeled with C-terminal mCherry fluorescent protein and attack-phase B. bacteriovorus cells were analyzed by fluorescent microscopy. This revealed DivIVABd-mCherry localizes to both poles of host-dependent B. bacteriovorus cells (Figure 2). This is consistent with the localization pattern of DivIVA homologs in Bacillus and relevant to the monopolar localization in Streptococci and Mycobacterium. (Marston et al., 1998; Fadda et al., 2007, Nguyen et al., 2007).
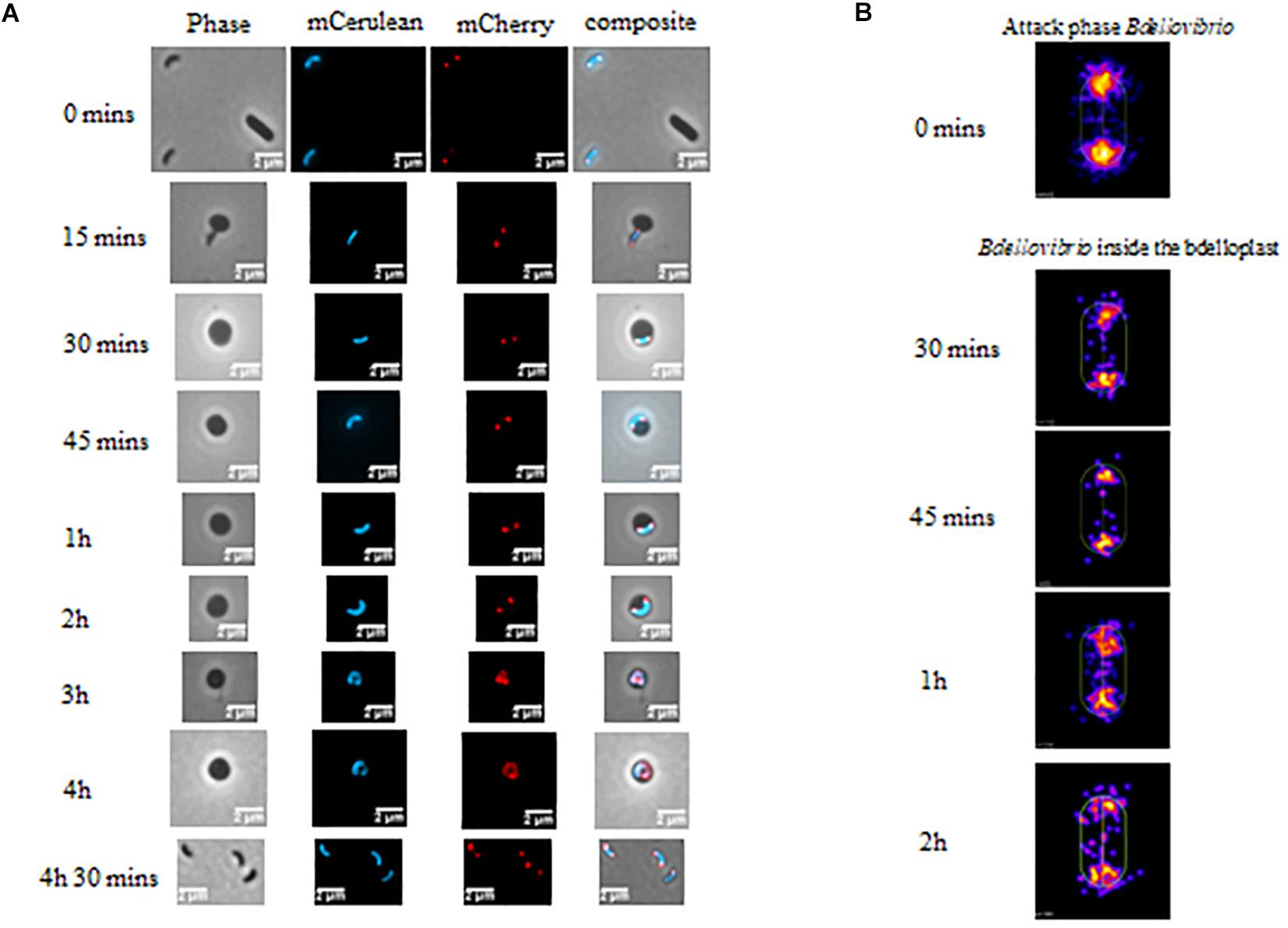
Figure 2. (A) Phase and epifluorescence microscopy displaying the location of DivIVABd tagged with mCherry. Localization was observed during invasion and growth within E. coli prey cells. The Bdellovibrio cytoplasm is constitutively fluorescent with Bd0064-mCerulean to visualize the cell within the bdelloplast. DivIVA-mCherry localizes to the poles of the Bdellovibrio, except at the 3–4 h timepoints, where it is less clear due to the growing filament extending and twisting in three dimensions as it elongates (see Supplementary Figure S1). Images are representative of three biological repeats. Scale bars are 2 μm. (B) Cellular position maps of fluorescent foci of DivIVA-mCherry detected by MicrobeJ. All of the cells detected (including free swimming attack phase cells) were measured at time 0, but only cells within bdelloplasts were measured at the other timepoints. Data are pooled from three independent experiments (N values of cells at each timepoint: T0- 845, T30- 110, T45–65, T1h- 226, and T2h- 120).
Fluorescent localization of DivIVABd-mCherry was then assessed at stages of intracellular growth. This showed that the fluorescently tagged protein was found at both poles of the growing B. bacteriovorus filament during predation for up to 2 h in the prey bdelloplast (Figure 2). At later timepoints (3–4 h post-invasion), the situation was less clear as the growing filament twists in three dimensions as it grows and divides in the bdelloplast. A mixture of some single or double foci and more diffuse DivIVABd-mCherry fluorescence was seen. This likely represents the DivIVABd-mCherry migrating to the newly forming poles of the B. bacteriovorus progeny (Figure 2 and further examples in Supplementary Figure S1).
To determine whether expression of DivIVABd-mCherry affected progeny number and the general predation rate, we conducted an assay of predation on luminescent prey (Lambert et al., 2003). This clearly showed that the predation rate of the DivIVABd-mCherry strain was not different to that of the FliC1 merodiploid (“wild-type” equivalent) control (Supplementary Figure S2A; Mann-Whitney test p = 0.995; n = 35). In addition, measuring intraperiplasmic growth area of the B. bacteriovorus (as total area of mCerulean fluorescence) showed no significant difference for this strain compared to wild-type (Supplementary Figure S2B), suggesting that the C-terminal mCherry tagging of DivIVABd has no detrimental effect on growth.
Cells Lacking DivIVABd Show Morphological Changes in Attack-Phase B. bacteriovorus Progeny Cells
DivIVA is essential in Streptomyces coelicolor, Enterococcus faecalis, and Mycobacterium smegmatis, so we wanted to determine if this is also the case in B. bacteriovorus (Flärdh, 2003; Ramirez-Arcos et al., 2005, Kang et al., 2008). Deletion of divIVABd was possible in predatory B. bacteriovorus, demonstrating that DivIVABd is not essential for B. bacteriovorus predatory growth and division.
Transmission electron microscopy of progeny (attack phase) cells recently emerged from bdelloplasts revealed that the B. bacteriovorus ΔdivIVABd strain had a morphological defect resulting in shorter, wider cells (Figure 3). Wild-type B. bacteriovorus HD100 pSUP404.2 (empty vector) cells had a mean length of 1.40 ± 0.04 μm and mean width of 0.36 ± 0.01 μm (n = 75). The ΔdivIVABd mutant had a shorter mean length of 1.01 ± 0.03 μm (p < 0.001; n = 75) and larger mean width of 0.42 ± 0.01 μm (p < 0.01; n = 75) (Figure 4). Additionally, rare doublet cells were observed in the attack phase population (at less than 1%) (Figure 3C compared to D). These appeared to have an incompletely divided septum, which pinched in between the non-divided cells, resulting in a single longer cell. These morphological defects were also apparent through analysis of attack phase cell images by phase contrast microscopy and analyzed using the MicrobeJ plugin (Ducret et al., 2016). This analysis showed wild type cells to have a mean length of 1.34 ± 0.06 μm and width of 0.47 ± 0.004 μm (n = 2,302). In comparison, the ΔdivIVABd strain was shorter, 1.01 ± 0.004 μm (p < 0.001, n = 2796), and marginally wider, 0.50 ± 0.004 μm (p < 0.001, n = 2796).
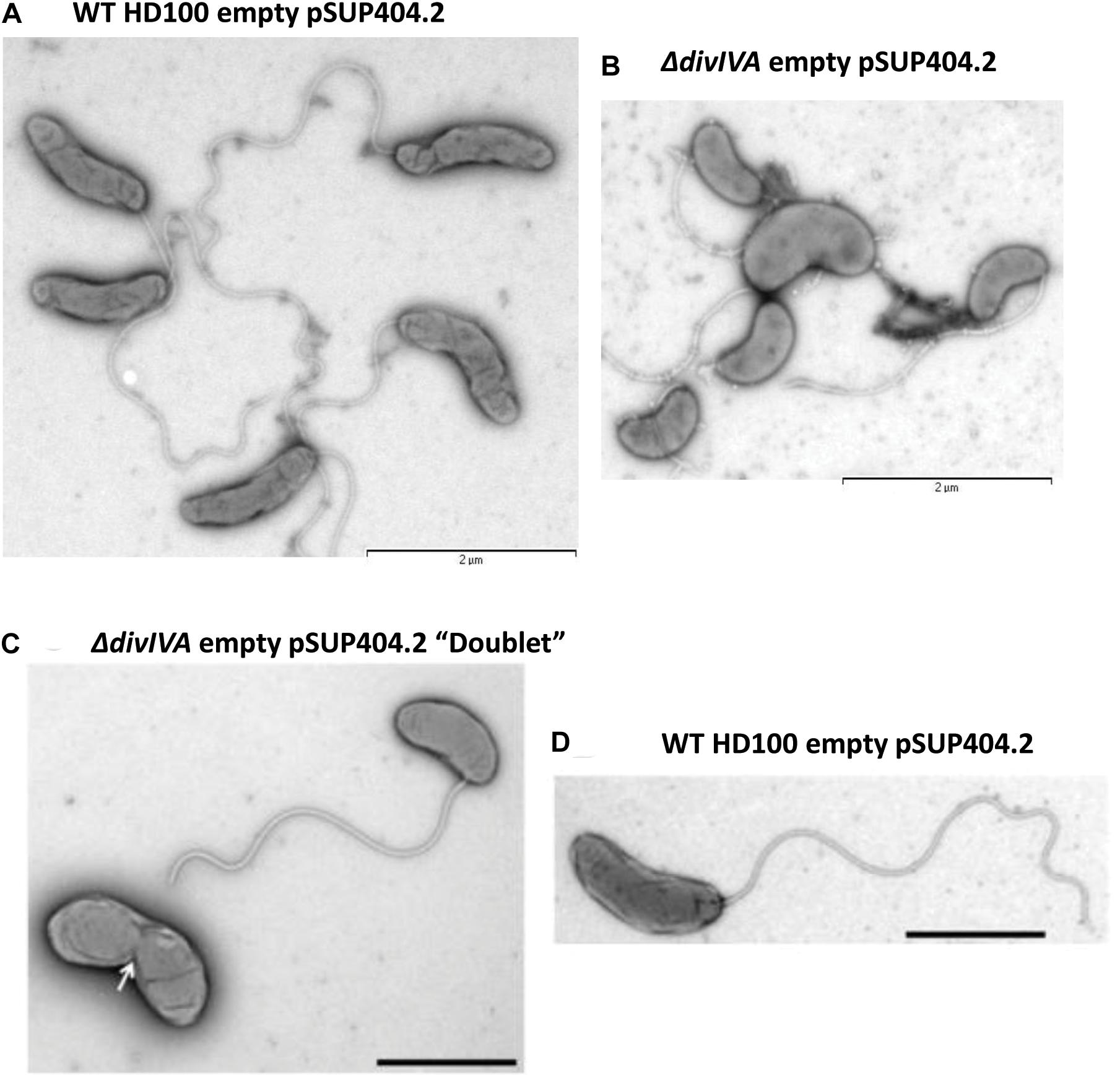
Figure 3. Morphological changes in the attack phase cells in B. bacteriovorusΔdivIVABd deletion strain grown with E. coli S17-1. TEM images of (A) Wild-type HD100 (pSUP404.2) cells are long and slender, whilst ΔdivIVABd (pSUP404.2) cells (B) are shorter and wider. (C) A rare a doublet cell of ΔdivIVABd with pinched septum. (D) B. bacteriovorus HD100 wild-type attack phase cell. Cells were stained with 0.5% uranyl acetate. Scale bars are 2 μm.
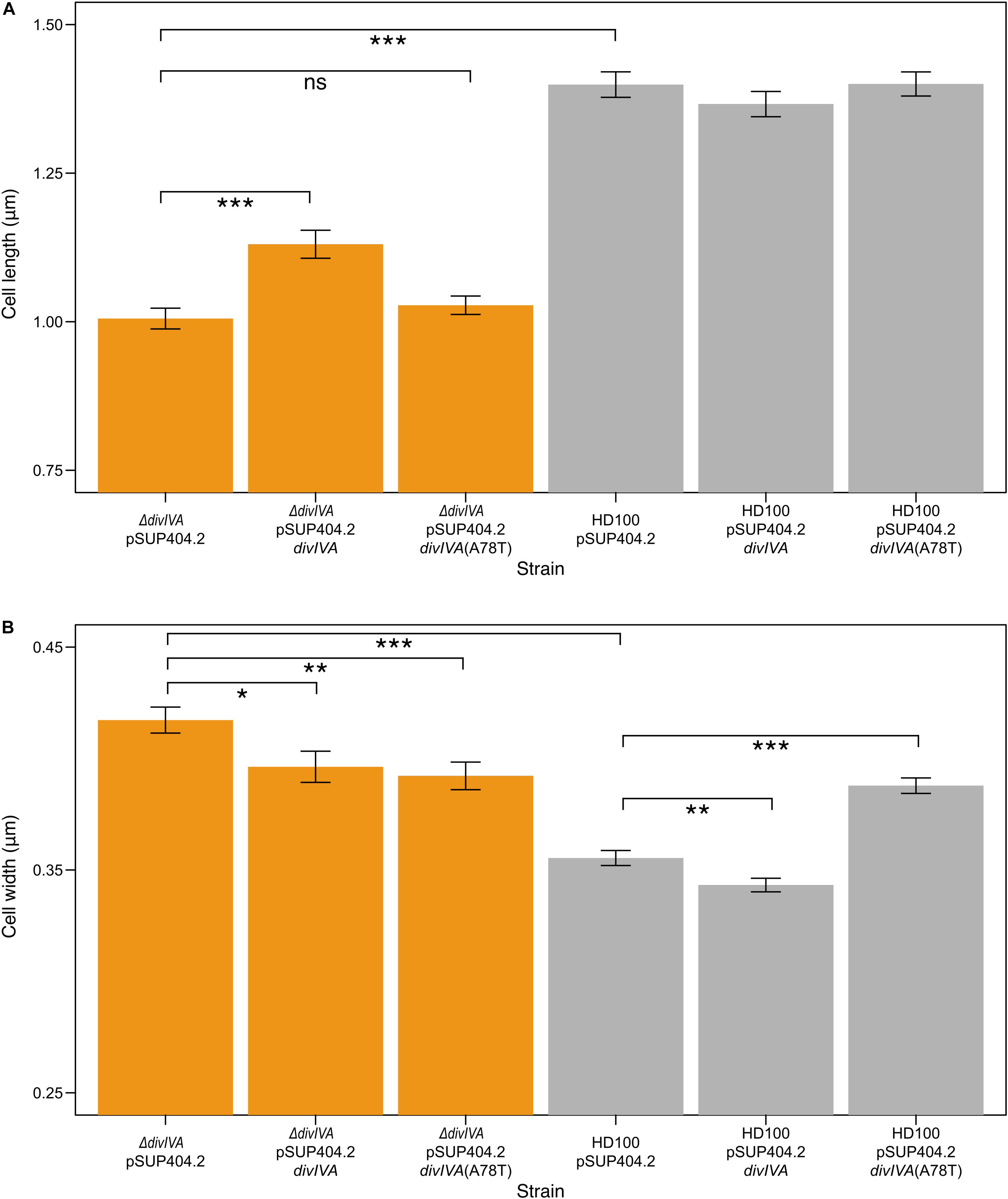
Figure 4. ΔdivIVABd strains are shorter and wider than wild type, and are partially restored through complementation. Bar charts showing the mean lengths (A) and widths (B) of B. bacteriovorus strains containing a pSUP404.2 plasmid either empty, or encoding DivIVABd, or a mutant DivIVABd(A78T). ΔdivIVABd strains are significantly shorter (P < 0.001) than wild type, with partial restoration of length when divIVABd is introduced on plasmid pSUP404.2 (P < 0.001). Average width of ΔdivIVABd strains is significantly greater than wild type (P < 0.001) with partial restoration when complemented with divIVABd (P < 0.05) or divIVABd(A78T) (P < 0.01). Width measurements show that wild type HD100 containing a plasmid with divIVABd are thinner (P < 0.01), and wider with divIVABd(A78T) (P < 0.001), suggesting that in trans expression levels may perturb DivIVA function. n = 75 for each population and data are from three biological repeats (all significance calculated as *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, using a t-test). Images were acquired through transmission electron microscopy and analyzed in SimplePCI.
To confirm these effects on morphology were due to the lack of DivIVABd, we introduced a wild type divIVABd gene expressed in trans on plasmid pSUP404.2 into the ΔdivIVABd strain. When compared to ΔdivIVABd containing an empty control vector this partially restored cell length and width of attack phase cells when imaged by TEM (p < 0.001, n = 75; Figure 4). This confirms that the deletion strain phenotype is attributable to the loss of a functional divIVABd locus. Further, there were no observed doublet cells in the complemented strains. To investigate this link between cell division and divVIAbd further, we complemented these strains with a divIVABd-A78T mutated copy of the gene (Figure 4). In Streptococcus pneumoniae, divIVA-A78T mutants had impaired cell division but normal chromosome segregation (Fadda et al., 2007). The ΔdivIVABd strain complemented with divIVA-A78T had a mean length of 1.03 ± 0.13 μm, which was not significantly different to ΔdivIVABd with an empty pSUP404.2 plasmid (n = 75). However, its width, 0.39 ± 0.05 μm, was partially restored and significantly lower than ΔdivIVABd (p < 0.01, n = 75).
B. bacteriovorus divIVA Is Expressed Throughout Growth and Is Co-transcribed With bd0465 and bd0466
Using a semi-quantitative Reverse Transcription PCR (RT-PCR) approach, transcription of the divIVABd gene and the neighboring genes bd0465 and bd0466 (Figure 1) were assessed across the B. bacteriovorus predatory cycle (Figure 5). We hypothesized that transcription of the gene cluster containing divIVABd might be up-regulated when the gene products were required for function. This approach showed that all three genes were constitutively expressed throughout the host dependent cycle (Figure 5).
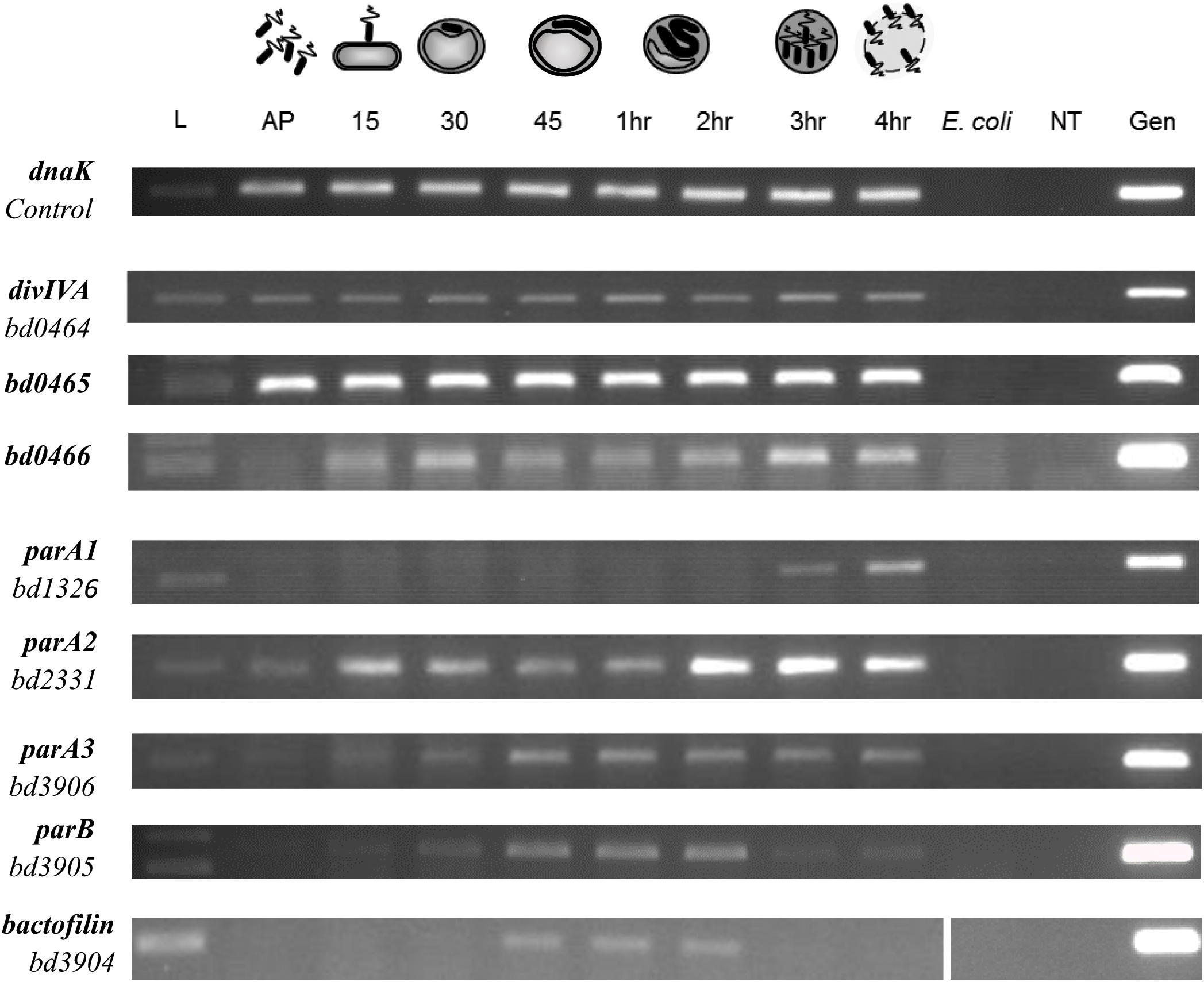
Figure 5. Expression pattern of B. bacteriovorus HD100 parA-like genes, parB, bactofilin, divIVABd, and its upstream genomic neighbors bd0465 and bd0466. Expression was assessed alongside control gene dnaK throughout the predatory life cycle using RT-PCR with transcript specific primers. RNA was prepared from identical volumes of infection cultures with B. bacteriovorus HD100 predator and E. coli S17-1 prey at time-points throughout the predatory life cycle. AP: RNA from attack-phase B. bacteriovorus, 15–45: RNA from 15–45 min time-points through the predatory cycle, 1–4 h: RNA from 1–4 h through the time-course, E. coli: E. coli S17-1 only template RNA, -ve: no template negative control, Gen: B. bacteriovorus HD100 genomic DNA template as positive control. Images are representative of two biological repeats.
To validate the hypothesis that divIVABd is expressed in an operon with its neighboring genes bd0465 and bd0466 (Figure 1), we wanted to determine whether these genes were co-transcribed. RNA-seq analysis suggested that transcription occurs across this cluster of genes, but that divIVABd has its own promoter, as it had a higher transcription level than bd0465 and bd0466 (Reads Per Kilobase Million for bd0466 = 9.56, bd0465 = 121.04, divIVABd = 1554.641) (Capeness et al., 2013). Promoter walking confirmed that bd0465 and divIVABd were co-transcribed, as well as showing that divIVABd has its own promoter. An RT-PCR assay confirmed that bd0466 and bd0465 were also co-transcribed (Supplementary Figure S3).
Bd0465-mCherry and Bd0466-mTFP Are Localized in the Cytoplasm and Pairwise BTH Testing Shows Bd0465 Interacts With DivIVABd, Bd0466, and ParA3
Given that DivIVABd fluorescence was seen at polar foci (Figure 2), we assessed the localization of Bd0465 and Bd0466 through fluorescent microscopy. To do so Bd0465-mCherry and Bd0466-mTFP constructs were conjugated into wild type B. bacteriovorus HD100. In contrast to the polar foci of DivIVABd-mCherry, both tagged proteins were ubiquitously expressed in the cytoplasm in attack phase cells. Images were taken throughout the predatory life cycle, but the fluorescence was too faint to accurately determine localization when the B. bacteriovorus was within the bdelloplast (Supplementary Figure S4).
To determine if DivIVABd interacts with its co-transcribed neighboring proteins we used the Bacterial Two Hybrid assay. In addition, we also tested a selection of known B. bacteriovorus cell division related proteins. Bacterial Two Hybrid (BTH) assays were performed in a pairwise manner between candidate proteins. For confirmation and quantification, the positive interactions were subjected to β-galactosidase assays. These assays showed significant results (p < 0.001) for interactions between Bd0465-DivIVABd, Bd0465-Bd0466 and Bd0465-ParA3 (Figure 6 and Supplementary Figure S5). No interaction was detected between DivIVABd-Bd0463, and for DivIVABd with ParA1, ParA2 or ParA3. We later turned to analyze Par protein function (see below) as there is a potential three-way interaction between Bd0465-DivIVABd, and Bd0465-ParA3. Further potential DivVIAbd interacting partners were sought by an unbiased BTH library screening method (Supplementary Information S2).
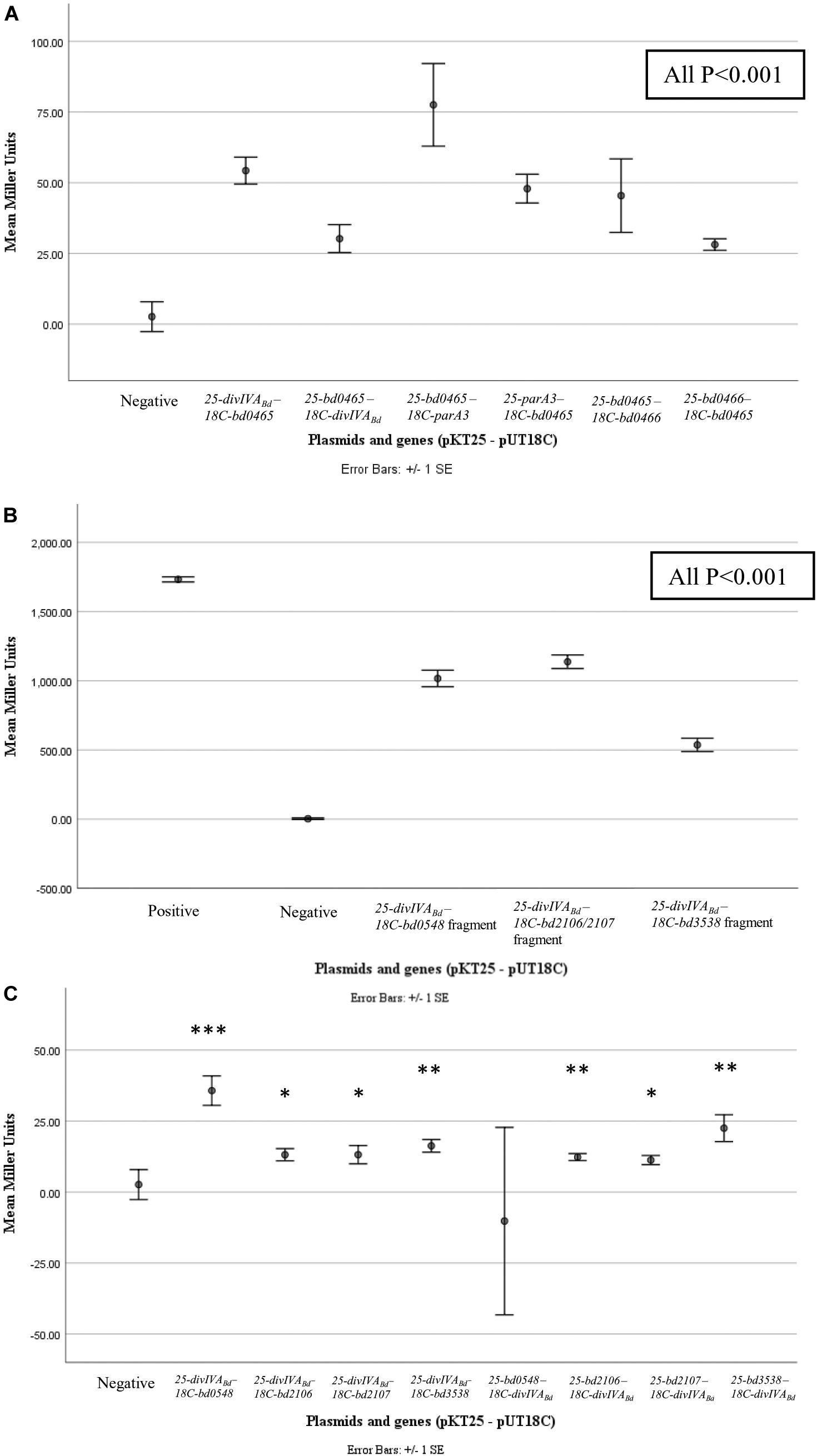
Figure 6. β-galactosidase assay results for pairwise and library screening bacterial two hybrid interactions. Points represent average Miller Units of cotransformants (n = 16), error bar represent standard error. (A) shows significant interactions between DivIVABd (Bd0464) and Bd0465, Bd0465 and ParA3 (Bd3906), and Bd0465 and Bd0466 (all P < 0.001) when compared to the negative control. (B) shows significant interactions between protein fragments encoded from BTH library plasmids, specifically partial proteins of Bd0548, Bd2106, Bd2107, and Bd3538, with DivIVABd plasmids (all P < 0.001). (C) shows results for interactions between DivIVABd and full length proteins Bd0548, Bd2106, Bd2107, and Bd3538 (***P < 0.001, **P < 0.01, and *P < 0.05). Data are from two biological repeats. Individual data points are presented in Supplementary Figure S5.
B. bacteriovorus Has Three Genes With Homology to Bacillus subtilis soj (parA)
Our cell morphology phenotype and fluorescence-localization experiments suggest that DivIVABd may have an ancillary role in co-ordinating division with cell growth (Figures 2–4). Its link to canonical ParA3 (bd3906) through three-way (Bd0465-DivIVABd, and Bd0465-ParA3) BTH protein interactions (Figure 6) then led us to investigate the role of chromosome-partitioning genes. Three genes in the B. bacteriovorus HD100 genome encode proteins with similarity to Soj, the ParA ortholog in B. subtilis. The product of bd3906 shares 57.71% protein identity with Soj, whilst the other predicted proteins of ParA1 and ParA2 share 27.94% and 35.48% identity, respectively (Supplementary Figure S6). Gene parA3 is located upstream of a parB homolog, in the arrangement typically seen at the parAB locus. Attempts at deleting parA3 were unsuccessful, despite screening many host dependent and host independent exconjugants (Chang et al., 2011), suggesting that ParA3 may be essential in B. bacteriovorus. The products of the genes neighboring parA1 and parA2 (bd1327 and bd2329) do not show any strong homology to ParB, or to any other proteins of known function, and show only weak homology to each other (12% sequence identity). Adjacent to parA1 is a bolA-like gene (bd1328). BolA is a transcription factor involved in the regulation of penicillin-binding proteins PBP5 and PBP6, and of MreB (Guinote et al., 2011; Singh and Montgomery, 2014).
Expression of parA-like Genes Peaks Toward the End of the B. bacteriovorus Predatory Cycle, and all Three Genes Have Differing Transcription Patterns
RT-PCR was carried out on all three parA-like genes in B. bacteriovorus (Figure 5). This demonstrated that expression of parA1 peaks at 3–4 h post infection at the very late stage of B. bacteriovorus filamentous cell division and prey exit. Expression of parA2 peaks at 2–4 h (with a peak at 15 min post-infection), whilst the canonical parA3 shows an increase in expression up to 45 min post-infection, then exhibiting consistent expression until the end of the predatory cycle. These later parA2 and parA3 profiles are consistent with stable expression across the B. bacteriovorus filamentous growth phase (Figure 5).
RT-PCR analysis also suggested that all three parA genes are encoded within operons (Supplementary Figure S7). Co-expression of parA1, bd1327, and bd1328 (encoding a BolA homolog) was observed at the 3 h time point, whilst parA2 was found to be co-transcribed with bd2329 at 3 h post-infection. Co-transcription of parA3, bd3905 (parB) and bd3904 (encoding a bactofilin homolog) was observed at 1 h post-infection.
Fluorescent Tagging of the Canonical ParA3 in B. bacteriovorus Creates Some Longer Attack-Phase Cells and Slows the Overall Rate of Predation
When parA3(bd3906) was originally tagged with mTFP, the mean length of attack phase cells increased compared to wild type (16.7% increase) suggesting that the tagged protein was only partly functional (Figure 7). Quantification and analysis of electron microscopy images showed the distribution of lengths, with a higher number of very long cells observed in the parA3-mTFP population. This was repeated with ParA3-mCherry (to be comparable with the other two ParA1 and 2 strains tagged with mCherry) and a similar small excess of very long cells was detected in the population. Next we measured the predation rate of B. bacteriovorus strains containing tagged parA1, parA2, and parA3 (Figure 8). These data showed only the strains containing the parA3-mTFP had a significantly slower overall predatory growth (Figure 8). To see if tagging parA3 affected nucleoid position and copy number, attack phase cells were stained with DAPI. Through fluorescent microscopy and image analysis in SimplePCI software, lengths of nucleoid and cell were measured and the ratios compared. This ratio was significantly lower for bd3906-mTFP (mean nucleoid length/cell length ratio of 0.55 ± 0.15 μm) than wild type (0.63 ± 0.1 μm. P < 0.05) suggesting a constant nucleoid length which is independent of cell length (Supplementary Figure S8). However, there was no significant difference between the proportion of anucleate cells in wild type (0/804 cells) and Bd3906-mTFP cells (1/702 cells) (Fisher’s exact test p = 0.466).
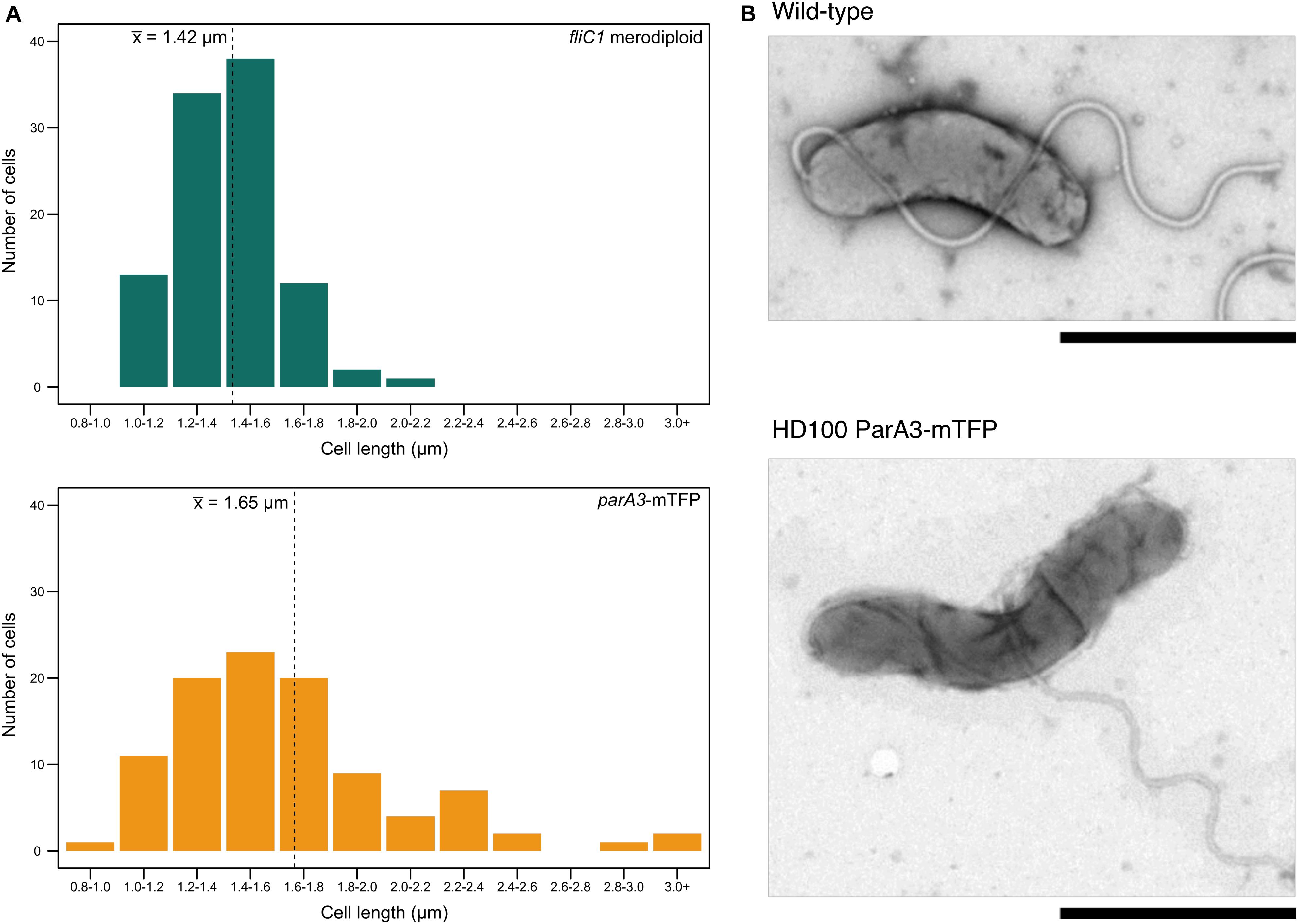
Figure 7. Cell length distributions for attack phase cells of wild type and ParA3-mTFP strains. (A) Distribution of cell lengths for B. bacteriovorus fliC1 merodiploid strain (“wild type” equivalent, green) and HD100 ParA3-mTFP (yellow) populations (n = 100). The mean is denoted by the dotted line. Representative TEM images of both populations are shown in (B). Scale bars are 1 μm and data are from three biological repeats.
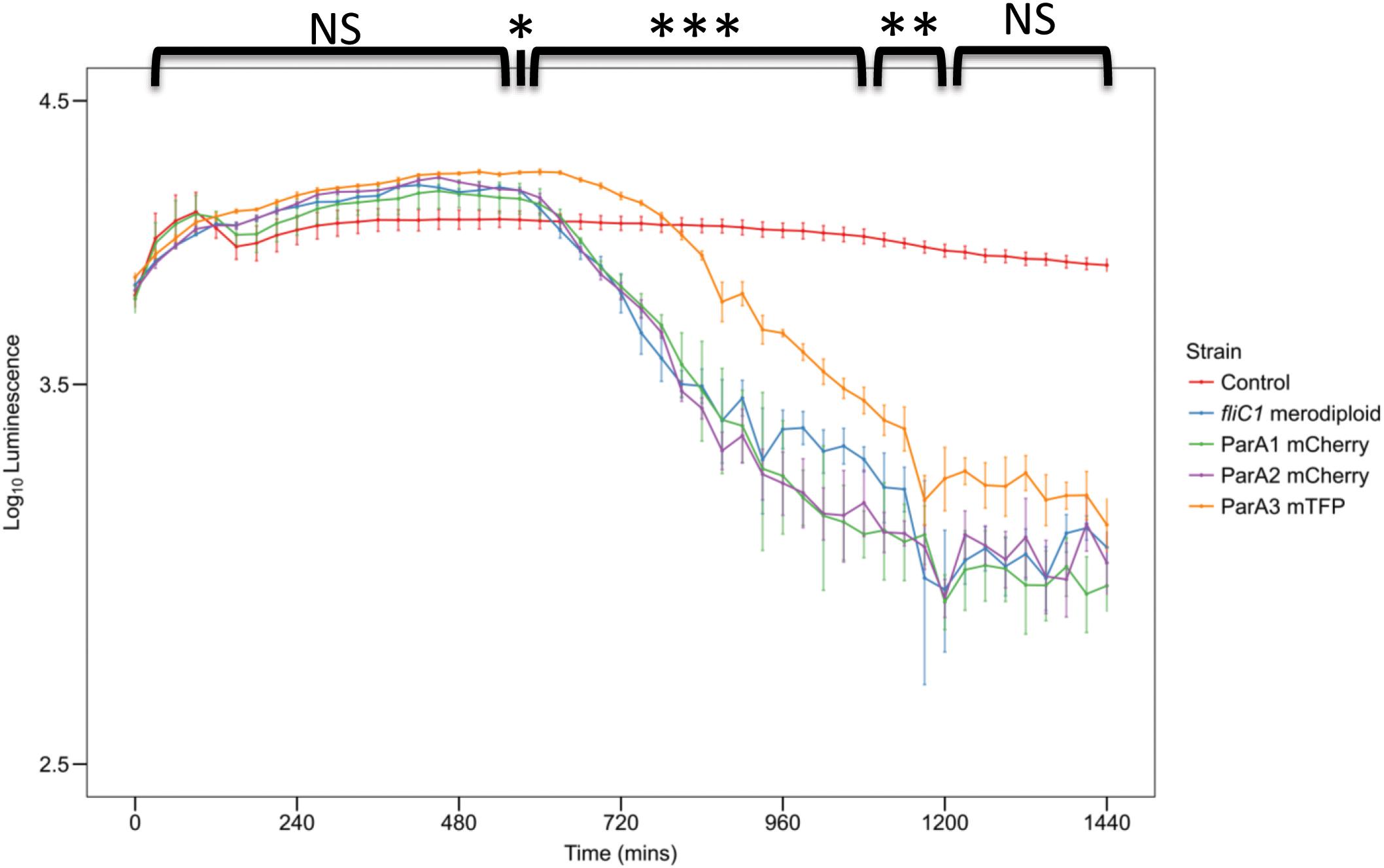
Figure 8. Plot of prey luminescence decrease over time comparing the predation rates of B. bacteriovorus cells expressing C terminally, fluorescently tagged ParA1, ParA2, or ParA3. Predation rate is measured as the drop in luminescence from the prey as they are preyed upon by Bdellovibrio, compared to wild type equivalent strain B. bacteriovorus FliC1 merodiploid (blue) (n = 12 technical replicates per strain for three biological repeats). Only the tagged ParA3 strain (orange) is delayed in causing a logarithmic drop in luminescence (which represents killing of the luminescent prey) compared to the other 3 strains. Control: E. coli prey only. ***p < 0.001, **p < 0.01, *p < 0.05, and NS not significant by Mann-Whitney U test.
ParA2 May Have a Role in Gliding Motility
Initial experiments on deletions of parA1 and parA2 implicate a role for ParA2 in gliding motility behavior, including the number of reversals of gliding direction after the first hour on a solid surface and the number of cells initiating productive (non-rapidly reversing) gliding behavior after that hour-long adaptation period (Supplementary Information S3 and Supplementary Figures S9–S11). However, as both strains could be constructed in predatory cultures, neither was required for predatory growth and division. Using the top homologous sequences from pBLAST, protein trees were constructed to show the relationship between the three ParA homologs in B. bacteriovorus and proteins of other prokaryotes (Supplementary Figure S12 and Supplementary Information S4).
Discussion
The constitutive expression of DivIVABd and its bi-polar localization throughout 2 h of predatory growth until diffusion around the time point of septation and new cell pole specification, suggested that DivIVABd coordinates important processes from the cell poles during predatory growth.
Morphological changes in the divIVA deletion strain may be a consequence of aberrant cell wall regulation and changes in septal positioning. Consequent adaptation to conserve cell volume could cause the short-wider morphology of B. bacteriovorus progeny that had a divIVABd deletion. To determine whether this is the case, septal and sub-polar peptidoglycan labeling would be required, which is beyond the scope of this study. We note that in M. smegmatis incorporation of new sub-polar peptidoglycan is coordinated by a DivIVA homolog (Kang et al., 2008; Ginda et al., 2013).
Interestingly, DivIVABd-mCherry was present at both poles in newly formed B. bacteriovorus progeny, suggesting possible inheritance of at least one focus of the protein rather than de novo synthesis of both foci in progeny cells. Due to the constraints of B. bacteriovorus growing in 3D inside another cell, as a long twisting filament, prior to and during septation it is hard to tell when the polar DivIVABd migrates to the septa. However, some diffusion, but still some single foci, of DivIVABd-mCherry were observed at the three hour time point of the predatory cycle (Figure 2 and Supplementary Figure S1). This contrasts with the bipolar fluorescence seen at two hours, suggesting a transition in DivIVA position from filament growth to septation, facilitating progeny inheritance of the protein. In B. subtilis, DivIVA has been shown to migrate between the two poles of a dividing cell (Bach et al., 2014). Although MinC and MinD are not present in B. bacteriovorus, as they are in B. subtilis, DivIVABd may still direct septal selection through other as-yet unidentified processes, resulting in a similar function.
Although preliminary, a number of potential interacting partners with DivIVABd have been identified, and have been highlighted (Figure 6 and Supplementary Figure S5) for future investigation. Probing a B. bacteriovorus Bacterial Two Hybrid library identified four putative DivIVABd-interacting proteins beyond those encoded by the divIVABd operon (see discussion below). Although diverse, all these proteins would be expected to change in levels in a B. bacteriovorus filament that has been growing for a long period in a bdelloplast, consuming oxygen and nutrients. Bd0548 MenE is a cytoplasmic protein associated with synthesis of a menaquinone to allow electron transport in more anaerobic conditions, such as those generated by continued enclosed growth of B. bacteriovorus inside the bdelloplast (Sharma et al., 1996). A pair of proteins Bd2106/7, associated with haem biosynthesis and disulfide bond formation in other bacteria (Zapun et al., 1993; Chan et al., 2006; Heras et al., 2009), were also found to interact with DivIVABd. Again, changes in synthesis of electron transport components such as Fe-S proteins and cytochromes could occur to remodel electron transport as the occupied bdelloplast was depleted for oxygen when the B. bacteriovorus filament reaches a greater biomass. The final DivIVABd interactor identified by BTH was Bd3538, a TrmJ homolog. This is a potential oxidative stress responsive protein in Gram-negative bacteria, which is intriguing as deletion of divIVA in Streptococci leads to an oxidative stress phenotype (Jaroensuk et al., 2016; Ni et al., 2018). Again, this could be associated with oxidative changes in the bdelloplast toward the end of filamentous growth of the B. bacteriovorus and could signal the need to divide and exit the dead prey cell. While the β-galactosidase assays for interaction of the library fragments were strongly positive, whole gene interactions were less substantial, prompting future work to determine the strength and extent of these protein interactions.
The pairwise protein-protein interactions with products encoded from the divIVABd operon, shown by BTH analysis, suggest that Bd0465 binds ParA3 (from the canonical ParAB pair) and that Bd0465 binds DivIVA (Bd0464) and Bd0466. This may coordinate the functions of DivIVABd and ParA3 of the canonical ParAB complex. ParA3, but neither of the other two other ParAs, gave aberrant B. bacteriovorus cell length distributions when fluorescently tagged, with ParA2 only affecting gliding motility reversals when deleted and ParA1 having no measureable phenotype. Therefore the Bd0465- DivIVABd -ParA3 interaction could act to synchronize the actions of DivIVABd with chromosome segregation, as is thought to be the case in M. smegmatis, albeit through an indirect interaction (Kang et al., 2008; Ginda et al., 2013, Vijay et al., 2014). It may possibly relate to Corynebacterium glutamicum, where DivIVA was shown by protein-protein studies to interact with ParB (Hammond et al., 2019). We also note (Figure 5) that the bd3904 bactofilin gene of B. bacteriovorus is cotranscribed with bd3905 parB. In Myxococcus, ParB interaction with bactofilin scaffolds emanating from each cell pole restrains the chromosome segregation machinery near poles until needed; something similar may operate in B. bacteriovorus (Lin et al., 2017).
The interaction between Bd0465, a ProC homolog, and Bd0466, an YggS homolog, has not been recorded in any other bacteria. It is possible that this interaction is a part of a signaling pathway that continues from Bd0466 to both ParA3 and DivIVABd. YggS proteins bind pyridoxal phosphate (PLP), and a change in the level of this molecule may signal to the division machinery to coordinate growth and septation. Recent work has suggested that changes in PLP levels are associated with changes in flux through the biosynthetic pathways for amino acids (Ito et al., 2013; Prunetti et al., 2016, Ito et al., 2019). One can see the necessity for such a signal due to B. bacteriovorus producing progeny at a timepoint when prey cell resources have been depleted. At a critical point in the predatory life cycle, the invading B. bacteriovorus cell must detect when remaining nutrients from the inside of the single prey cell are insufficient to produce another progeny cell. At this time septation must occur and the B. bacteriovorus cell must switch from growth and biosynthesis to the less metabolically active, prey-hunting attack phase progeny. The Bd0466 interaction with Bd0465 may represent a pivotal part of this signaling pathway. This is further supported by the constitutive expression of both bd0465 and bd0466, as is the case with DivIVABd, and the diffuse, cytoplasmic localization of the Bd0465/0466 proteins in the predator cells. This may lead to a specific association between Bd0466 and Bd0465 occurring when PLP levels change; then in turn this could affect the Bd0465 association with ParA3, possibly releasing ParA3 for partitioning which may lead to Bd0465 association with DivIVABd. Demonstrating the validity of this idea and the dynamics of such a catch and release type of mechanism and its effects on cell division inside the bdelloplast, is beyond this study, but we hope that our results will prime such work.
Conclusion
In summary we report that Gram-negative B. bacteriovorus HD100 uses DivIVA to define cell proportions during synchronous division from a long filamentous cell inside a dead prey bacterium. This process may involve an interaction with a canonical ParA protein and proteins that bind indicators of the nutritional contents and oxidative status of the dead prey cell.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material or can be obtained by contacting the corresponding author.
Author Contributions
DM carried out the DivIVA mutagenesis, phenotyping, some phylogenetic analyses, ParA protein tagging, gene deletion, and phenotyping work jointly with ES. LR carried out BTH library construction and analysis, further ParA3 protein tagging and phylogenetic analyses. RT carried out gene cloning for BTH analyses. CL carried out transcriptional analyses and contributed to luminsecent prey assays. AF carried out some fluorescent tagging, transcriptional analysis of bactofilin gene, and initial phylogenetic analyses. RS, DM, and AF devised the initial projects and supervised and helped to interpret the work with inputs from the other authors, drafted the manuscript with LR and edited it on receipt of comments from our co-authors.
Funding
BBSRC Ph.D. studentships supported DM, LR, AF, and ES was supported via a joint BBSRC Rothamsted-Reading Nottingham Food Security Studentship and also was an undergraduate at Nottingham University when some of the research was done. CL was funded by BBSRC BB/M010325/1 when the work was done. RT was funded partially by BBSRC and by University of Nottingham.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Grant Buchanan from Tracy Palmer’s lab at the Centre for Bacterial Cell Biology at Newcastle University for protocols and advice for the BTH library construction. DM and AF were supported by BBSRC Ph.D. studentships, LR by a Ph.D. studentship from the Nottingham BBSRC Doctoral Training Partnership and ES was initially an undergraduate project student, supervised by DM and RS at University of Nottingham, and then was a Ph.D. student supported by a University of Nottingham- Rothamsted-University of Reading BBSRC Food Security Studentship. CL was funded by BBSRC grant BB/M010325/1. We thank Marilyn Whitworth, Julie Beavon, Julie Rodgers, and Paul Knowles from the team of Central Support Technicians for general lab maintenance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00542/full#supplementary-material
Footnotes
References
Akiyama, T., Inouye, S., and Komano, T. (2003). Novel developmental genes, frucd, of Myxococcus xanthus: involvement of a cell division protein in multicellular development. J. Bacteriol. 185, 3317–3324. doi: 10.1128/jb.185.11.3317-3324.2003
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410.
Asada, M., Goto, Y., Yahata, K., Yokoyama, N., Kawai, S., Inoue, N., et al. (2012). Gliding motility of Babesia bovis merozoites visualized by time-lapse video microscopy. PLoS One 7:e0035227 doi: 10.1371/journal.pone.0035227
Ayala, J. A., Garrido, T., De Pedro, M. A., and Vicente, M. (1994). Molecular Biology of Bacterial Septation. Bacterial Cell Wall. Amsterdam: Elsevier Science.
Bach, J. N., Albrecht, N., and Bramkamp, M. (2014). Imaging Diviva dynamics using photo-convertible and activatable fluorophores in Bacillus subtilis. Front. Microbiol. 5:53. doi: 10.3389/fmicb.2014.00059
Battesti, A., and Bouveret, E. (2012). The bacterial two-hybrid system based on adenylate cyclase reconstitution in Escherichia coli. Methods 58, 325–334. doi: 10.1016/j.ymeth.2012.07.018
Bignell, C., and Thomas, C. M. (2001). The bacterial Para-Parb partitioning proteins. J. Biotechnol. 91, 1–34. doi: 10.1016/s0168-1656(01)00293-0
Capeness, M. J., Lambert, C., Lovering, A. L., Till, R., Uchida, K., Chaudhuri, R., et al. (2013). Activity of Bdellovibrio hit locus proteins, Bd0108 and Bd0109, links type iva pilus extrusion/retraction status to prey-independent growth signalling. PLoS One 8:e0079759. doi: 10.1371/journal.pone.0079759
Cha, J. H., and Stewart, G. C. (1997). The diviva minicell locus of Bacillus subtilis. J. Bacteriol. 179, 1671–1683. doi: 10.1128/jb.179.5.1671-1683.1997
Chan, A. C. K., Lelj-Garolla, B., Rosell, F. I., Pedersen, K. A., Mauk, A. G., and Murphy, M. E. P. (2006). Cofacial heme binding is linked to dimerization by a bacterial heme transport protein. J. Mol. Biol. 362, 1108–1119. doi: 10.1016/j.jmb.2006.08.001
Chang, C. Y., Hobley, L., Till, R., Capeness, M., Kanna, M., Burtt, W., et al. (2011). The Bdellovibrio bacteriovorus twin-arginine transport system has roles in predatory and prey-independent growth. Microbiol. Sgm 157, 3079–3093. doi: 10.1099/mic.0.052449-0
Chaudhuri, R. R., Loman, N. J., Snyder, L. A. S., Bailey, C. M., Stekel, D. J., and Pallen, M. J. (2008). xbase2: a comprehensive resource for comparative bacterial genomics. Nucleic Acids Res. 36, D543–D546.
Cho, H. (2015). The role of cytoskeletal elements in shaping bacterial cells. J. Microbiol. Biotechnol. 25, 307–316. doi: 10.4014/jmb.1409.09047
Cotter, T. W., and Thomashow, M. F. (1992). Identification of a Bdellovibrio-Bacteriovorus genetic-locus, hit, associated with the host-independent phenotype. J. Bacteriol. 174, 6018–6024. doi: 10.1128/jb.174.19.6018-6024.1992
Donovan, C., and Bramkamp, M. (2014). Cell division in corynebacterineae. Front. Microbiol. 5:132. doi: 10.3389/fmicb.2014.00132
Donovan, C., Schwaiger, A., Kramer, R., and Bramkamp, M. (2010). Subcellular localization and characterization of the parab system from Corynebacterium glutamicum. J. Bacteriol. 192, 3441–3451. doi: 10.1128/JB.00214-10
Ducret, A., Quardokus, E. M., and Brun, Y. V. (2016). Microbej, a tool for high throughput bacterial cell detection and quantitative analysis. Nat. Microbiol. 1:16077. doi: 10.1038/nmicrobiol.2016.77
Edwards, D. H., and Errington, J. (1997). The Bacillus subtilis Diviva protein targets to the division septum and controls the site specificity of cell division. Mol. Microbiol. 24, 905–915. doi: 10.1046/j.1365-2958.1997.3811764.x
Espinosa, E., Barre, F. X., and Galli, E. (2017). Coordination between replication, segregation and cell division in multi-chromosomal bacteria: lessons from Vibrio cholerae. Int. Microbiol. 20, 121–129. doi: 10.2436/20.1501.01.293
Eswaramoorthy, P., Erb, M. L., Gregory, J. A., Silverman, J., Pogliano, K., Pogliano, J., et al. (2011). Cellular architecture mediates Diviva ultrastructure and regulates min activity in Bacillus subtilis. mBio 2:e00257-11. doi: 10.1128/mBio.00257-11
Fadda, D., Santona, A., D’ulisse, V., Ghelardini, P., Ennas, M. G., Whalen, M. B., et al. (2007). Streptococcus pneumoniae Diviva: localization and interactions in a Mincd-free context. J. Bacteriol. 189, 1288–1298. doi: 10.1128/jb.01168-06
Fenton, A. K., Kanna, M., Woods, R. D., Aizawa, S. I., and Sockett, R. E. (2010a). Shadowing the actions of a predator: backlit fluorescent microscopy reveals synchronous non-binary septation of predatory Bdellovibrio inside prey and exit through discrete bdelloplast pores. J. Bacteriol. 192, 6329–6335. doi: 10.1128/JB.00914-10
Fenton, A. K., Lambert, C., Wagstaff, P. C., and Sockett, R. E. (2010b). Manipulating each Mreb of Bdellovibrio bacteriovorus gives diverse morphological and predatory phenotypes. J. Bacteriol. 192, 1299–1311. doi: 10.1128/JB.01157-09
Flärdh, K. (2003). Essential role of Diviva in polar growth and morphogenesis in Streptomyces coelicolor A3(2). Mol. Microbiol. 49, 1523–1536. doi: 10.1046/j.1365-2958.2003.03660.x
Flardh, K., Richards, D. M., Hempel, A. M., Howard, M., and Buttner, M. J. (2012). Regulation of apical growth and hyphal branching in Streptomyces. Curr. Opin. Microbiol. 15, 737–743. doi: 10.1016/j.mib.2012.10.012
Ginda, K., Bezulska, M., Ziolkiewicz, M., Dziadek, J., Zakrzewska-Czerwinska, J., and Jakimowicz, D. (2013). Para of Mycobacterium smegmatis co-ordinates chromosome segregation with the cell cycle and interacts with the polar growth determinant Diviva. Mol. Microbiol. 87, 998–1012. doi: 10.1111/mmi.12146
Guinote, I. B., Matos, R. G., Freire, P., and Arraiano, C. M. (2011). Bola affects cell growth, and binds to the promoters of penicillin-binding proteins 5 and 6 and regulates their expression. J. Microbiol. Biotechnol. 21, 243–251.
Hammond, L. R., White, M. L., and Eswara, P. J. (2019). viva la Diviva!. J. Bacteriol. 201:e00245-19. doi: 10.1128/JB.00245-19
Handford, J. I., Ize, B., Buchanan, G., Butland, G. P., Greenblatt, J., Emili, A., et al. (2009). Conserved network of proteins essential for bacterial viability. J. Bacteriol. 191, 4732–4749. doi: 10.1128/JB.00136-09
Hempel, A. M., Wang, S.-B., Letek, M., Gil, J. A., and Flardh, K. (2008). Assemblies of Diviva mark sites for hyphal branching and can establish new zones of cell wall growth in Streptomyces coelicolor. J. Bacteriol. 190, 7579–7583. doi: 10.1128/JB.00839-08
Heras, B., Shouldice, S. R., Totsika, M., Scanlon, M. J., Schembri, M. A., and Martin, J. L. (2009). Dsb proteins and bacterial pathogenicity. Nat. Rev. Microbiol. 7, 215–225. doi: 10.1038/nrmicro2087
Hobley, L., Fung, R. K. Y., Lambert, C., Harris, M., Dabhi, J. M., King, S. S., et al. (2012a). Discrete Cyclic di-Gmp-dependent control of bacterial Predation versus axenic growth in Bdellovibrio bacteriovorus. PLoS Pathog. 8:e1002493. doi: 10.1371/journal.ppat.1002493
Hobley, L., Lerner, T. R., Williams, L. E., Lambert, C., Till, R., Milner, D. S., et al. (2012b). Genome analysis of a simultaneously predatory and prey-independent, novel Bdellovibrio bacteriovorus from the River Tiber, supports in silico predictions of both ancient and recent lateral gene transfer from diverse bacteria. BMC Genomics 13:670. doi: 10.1186/1471-2164-13-670
Houot, L., Fanni, A., De Bentzmann, S., and Bordi, C. (2012). A bacterial two-hybrid genome fragment library for deciphering regulatory networks of the opportunistic pathogen Pseudomonas aeruginosa. Microbiol. Sgm 158, 1964–1971. doi: 10.1099/mic.0.057059-0
Huh, D., and Paulsson, J. (2011). Random partitioning of molecules at cell division. Proc. Natl. Acad. Sci. U.S.A. 108, 15004–15009. doi: 10.1073/pnas.1013171108
Hui, M. P., Galkin, V. E., Yu, X., Stasiak, A. Z., Stasiak, A., Waldor, M. K., et al. (2010). Para2, a Vibrio cholerae chromosome partitioning protein, forms left-handed helical filaments on Dna. Proc. Natl. Acad. Sci. U.S.A. 107, 4590–4595. doi: 10.1073/pnas.0913060107
Ito, T., Iimori, J., Takayama, S., Moriyama, A., Yamauchi, A., Hemmi, H., et al. (2013). Conserved pyridoxal protein that regulates Ile and Val metabolism. J. Bacteriol. 195, 5439–5449. doi: 10.1128/JB.00593-13
Ito, T., Yamamoto, K., Hori, R., Yamauchi, A., Downs, D. M., Hemmi, H., et al. (2019). Conserved Pyridoxal 5 ’-Phosphate-binding protein yggs impacts amino acid metabolism through Pyridoxine 5 ’-Phosphate in Escherichia coli. Appl. Environ. Microbiol. 85:e00430-19. doi: 10.1128/AEM.00430-19
Ito, T., Yamauchi, A., Hemmi, H., and Yoshimura, T. (2016). Ophthalmic acid accumulation in an Escherichia coli mutant lacking the conserved pyridoxal 5 ’-phosphate-binding protein Yggs. J. Biosci. Bioeng. 122, 689–693. doi: 10.1016/j.jbiosc.2016.06.010
Jakobczak, B., Keilberg, D., Wuichet, K., and Sogaard-Andersen, L. (2015). Contact- and protein transfer-dependent stimulation of assembly of the gliding motility machinery in Myxococcus xanthus. PLoS Genet. 11:e1005341. doi: 10.1371/journal.pgen.1005341
Jaroensuk, J., Atichartpongkul, S., Chionh, Y. H., Wong, Y. H., Liew, C. W., Mcbee, M. E., et al. (2016). Methylation at position 32 of trna catalyzed by Trmj alters oxidative stress response in Pseudomonas aeruginosa. Nucleic Acids Res. 44, 10834–10848. doi: 10.1093/nar/gkw870
Jindal, L., and Emberly, E. (2019). Dna segregation under Par protein control. PLoS One 14:e0218520. doi: 10.1371/journal.pone.0218520
Kang, C. M., Nyayapathy, S., Lee, J. Y., Suh, J. W., and Husson, R. N. (2008). Wag31, a homologue of the cell division protein Diviva, regulates growth, morphology and polar cell wall synthesis in mycobacteria. Microbiol. Sgm 154, 725–735. doi: 10.1099/mic.0.2007/014076-0
Kaval, K. G., Rismondo, J., and Halbedel, S. (2014). A function of Diviva in Listeria monocytogenes division site selection. Mol. Microbiol. 94, 637–654. doi: 10.1111/mmi.12784
Kieser, K. J., and Rubin, E. J. (2014). How sisters grow apart: mycobacterial growth and division. Nat. Rev. Microbiol. 12, 550–562. doi: 10.1038/nrmicro3299
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). Mega X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kuru, E., Lambert, C., Rittichier, J., Till, R., Ducret, A., Derouaux, A., et al. (2017). Fluorescent D-amino-acids reveal bi-cellular cell wall modifications important for Bdellovibrio bacteriovorus predation. Nat. Microbiol. 2, 1648–1657. doi: 10.1038/s41564-017-0029-y
Kuwada, N. J., Traxler, B., and Wiggins, P. A. (2015). Genome-scale quantitative characterization of bacterial protein localization dynamics throughout the cell cycle. Mol. Microbiol. 95, 64–79. doi: 10.1111/mmi.12841
Lambert, C., Chang, C. Y., Capeness, M. J., and Sockett, R. E. (2010). The first bite- profiling the predatosome in the bacterial pathogen Bdellovibrio. PLoS One 5:e0008599. doi: 10.1371/journal.pone.0008599
Lambert, C., Evans, K. J., Till, R., Hobley, L., Capeness, M., Rendulic, S., et al. (2006). Characterizing the flagellar filament and the role of motility in bacterial prey-penetration by Bdellovibrio bacteriovorus. Mol. Microbiol. 60, 274–286. doi: 10.1111/j.1365-2958.2006.05081.x
Lambert, C., Fenton, A. K., Hobley, L., and Sockett, R. E. (2011). Predatory Bdellovibrio bacteria use gliding motility to scout for prey on surfaces. J. Bacteriol. 193, 3139–3141. doi: 10.1128/JB.00224-11
Lambert, C., Lerner, T. R., Bui, N. K., Somers, H., Aizawa, S. I., Liddell, S., et al. (2016). Interrupting peptidoglycan deacetylation during Bdellovibrio predator-prey interaction prevents ultimate destruction of prey wall, liberating bacterial-ghosts. Sci. Rep. 6:26010. doi: 10.1038/srep26010
Lambert, C., Smith, M. C. M., and Sockett, R. E. (2003). A novel assay to monitor predator-prey interactions for Bdellovibrio bacteriovorus 109 J reveals a role for methyl-accepting chemotaxis proteins in predation. Environ. Microbiol. 5, 127–132. doi: 10.1046/j.1462-2920.2003.00385.x
Lambert, C., and Sockett, R. E. (2013). Nucleases in Bdellovibrio bacteriovorus contribute towards efficient self-biofilm formation and eradication of preformed prey biofilms. FEMS Microbiol. Lett. 340, 109–116. doi: 10.1111/1574-6968.12075
LeMagueres, P., Im, H., Ebalunode, J., Strych, U., Benedik, M. J., Briggs, J. M., et al. (2005). The 1.9 angstrom crystal structure of alanine racemase from Mycobacterium tuberculosis contains a conserved entryway into the active site. Biochemistry 44, 1471–1481. doi: 10.1021/bi0486583
Lenarcic, R., Halbedel, S., Visser, L., Shaw, M., Wu, L. J., Errington, J., et al. (2009). Localisation of Diviva by targeting to negatively curved membranes. EMBO J. 28, 2272–2282. doi: 10.1038/emboj.2009.129
Letek, M., Ordonez, E., Vaquera, J., Margolin, W., Flardh, K., Mateos, L. M., et al. (2008). Diviva is required for polar growth in the Mreb-lacking rod-shaped actinomycete Corynebacterium glutamicum. J. Bacteriol. 190, 3283–3292. doi: 10.1128/JB.01934-07
Lin, L., Valeriano, M. O., Harms, A., Sogaard-Andersen, L., and Thanbichler, M. (2017). Bactofilin-mediated organization of the Parabs chromosome segregation system in Myxococcus xanthus. Nat. Commun. 8:1817. doi: 10.1038/s41467-017-02015-z
MacCready, J. S., Schossau, J., Osteryoung, K. W., and Ducat, D. C. (2017). Robust Min-system oscillation in the presence of internal photosynthetic membranes in cyanobacteria. Mol. Microbiol. 103, 483–503. doi: 10.1111/mmi.13571
Marston, A. L., and Errington, J. (1999). Selection of the midcell division site in Bacillus subtilis through Mind-dependent polar localization and activation of Minc. Mol. Microbiol. 33, 84–96. doi: 10.1046/j.1365-2958.1999.01450.x
Marston, A. L., Thomaides, H. B., Edwards, D. H., Sharpe, M. E., and Errington, J. (1998). Polar localization of the Mind protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev. 12, 3419–3430. doi: 10.1101/gad.12.21.3419
Massidda, O., Anderluzzi, D., Friedli, L., and Feger, G. (1998). Unconventional organization of the division and cell wall gene cluster of Streptococcus pneumoniae. Microbiology 144(Pt 11), 3069–3078. doi: 10.1099/00221287-144-11-3069
McBride, M. J., and Zhu, Y. T. (2013). Gliding motility and por secretion system genes are widespread among members of the Phylum bacteroidetes. J. Bacteriol. 195, 270–278. doi: 10.1128/JB.01962-12
Mendez, M., Huang, I. H., Ohtani, K., Grau, R., Shimizu, T., and Sarker, M. R. (2008). Carbon catabolite repression of type Iv pilus-dependent gliding motility in the anaerobic pathogen Clostridium perfringens. J. Bacteriol. 190, 48–60. doi: 10.1128/jb.01407-07
Meniche, X., Otten, R., Siegrist, M. S., Baer, C. E., Murphy, K. C., Bertozzi, C. R., et al. (2014). Subpolar addition of new cell wall is directed by Diviva in mycobacteria. Proc. Natl. Acad. Sci. U.S.A. 111, E3243–E3251. doi: 10.1073/pnas.1402158111
Mierzejewska, J., and Jagura-Burdzy, G. (2012). Prokaryotic Para-Parb-pars system links bacterial chromosome segregation with the cell cycle. Plasmid 67, 1–14. doi: 10.1016/j.plasmid.2011.08.003
Monahan, L. G., Liew, A. T. F., Bottomley, A. L., and Harry, E. J. (2014). Division site positioning in bacteria: one size does not fit all. Front. Microbiol. 5:7. doi: 10.3389/fmicb.2014.00019
Nguyen, L., Scherr, N., Gatfield, J., Walburger, A., Pieters, J., and Thompson, C. J. (2007). Antigen 84, an effector of pleiomorphism in Mycobacterium smegmatis. J. Bacteriol. 189, 7896–7910. doi: 10.1128/jb.00726-07
Ni, H., Fan, W. W., Li, C. L., Wu, Q. Q., Hongfen, H. F., Hui, D., et al. (2018). Streptococcus suis Diviva protein is a substrate of Ser/Thr Kinase Stk and involved in cell division regulation. Front. Cell. Infect. Microbiol. 8:85. doi: 10.3389/fcimb.2018.00085
Nocek, B., Chang, C., Li, H., Lezondra, L., Holzle, D., Collart, F., et al. (2005). Crystal structures of Delta(1)-pyrroline-5-carboxylate reductase from human pathogens Neisseria meningitides and Streptococcus pyogenes. J. Mol. Biol. 354, 91–106. doi: 10.1016/j.jmb.2005.08.036
Patrick, J. E., and Kearns, D. B. (2008). Minj (Yvjd) is a topological determinant of cell division in Bacillus subtilis. Mol. Microbiol. 70, 1166–1179. doi: 10.1111/j.1365-2958.2008.06469.x
Prunetti, L., El Yacoubi, B., Schiavon, C. R., Kirkpatrick, E., Huang, L. L., Bailly, M., et al. (2016). Evidence that Cog0325 proteins are involved in Plp homeostasis. Microbiol. Sgm 162, 694–706. doi: 10.1099/mic.0.000255
Raghunathan, D., Radford, P. M., Gell, C., Negus, D., Moore, C., Till, R., et al. (2019). Engulfment, persistence and fate of Bdellovibrio bacteriovorus predators inside human phagocytic cells informs their future therapeutic potential. Sci. Rep. 9:4293. doi: 10.1038/s41598-019-40223-3
Ramirez, M. V., Dawson, C. C., Crew, R., England, K., and Slayden, R. A. (2013). Mazf6 toxin of Mycobacterium tuberculosis demonstrates antitoxin specificity and is coupled to regulation of cell growth by a Soj-like protein. BMC Microbiol. 13:240. doi: 10.1186/1471-2180-13-240
Ramirez-Arcos, S., Liao, M., Marthaler, S., Rigden, M., and Dillon, J. A. (2005). Enterococcus faecalis diviva: an essential gene involved in cell division, cell growth and chromosome segregation. Microbiology 151, 1381–1393. doi: 10.1099/mic.0.27718-0
Rice, P., Longden, I., and Bleasby, A. (2000). Emboss: the European molecular biology open software suite. Trends Genet. 16, 276–277. doi: 10.1016/s0168-9525(00)02024-2
Ringgaard, S., Schirner, K., Davis, B. M., and Waldor, M. K. (2011). A family of Para-like Atpases promotes cell pole maturation by facilitating polar localization of chemotaxis proteins. Genes Dev. 25, 1544–1555. doi: 10.1101/gad.2061811
Robert, X., and Gouet, P. (2014). Deciphering key features in protein structures with the new Endscript server. Nucleic Acids Res. 42, W320–W324. doi: 10.1093/nar/gku316
Roberts, M. A. J., Wadhams, G. H., Hadfield, K. A., Tickner, S., and Armitage, J. P. (2012). Para-like protein uses non-specific chromosomal Dna binding to partition protein complexes. Proc. Natl. Acad. Sci. U.S.A. 109, 6698–6703. doi: 10.1073/pnas.1114000109
Schaefer, J., Jovanovic, G., Kotta-Loizou, L., and Buck, M. (2016). Single-step method for beta-galactosidase assays in Escherichia coli using a 96-well microplate reader. Anal. Biochem. 503, 56–57. doi: 10.1016/j.ab.2016.03.017
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. doi: 10.1038/nmeth.2019
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). Nih image to Imagej: 25 years of image analysis. Nat. Methods 9, 671–675. doi: 10.1038/nmeth.2089
Seidler, R. J., and Starr, M. P. (1969). Isolation and characterization of host-independent Bdellovibrios. J. Bacteriol. 100:769. doi: 10.1128/jb.100.2.769-785.1969
Sharma, V., Hudspeth, M. E. S., and Meganathan, R. (1996). Menaquinone (vitamin K-2) biosynthesis: localization and characterization of the mene gene from Escherichia coli. Gene 168, 43–48. doi: 10.1016/0378-1119(95)00721-0
Sievers, F., and Higgins, D. G. (2018). Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 27, 135–145. doi: 10.1002/pro.3290
Singh, S. P., and Montgomery, B. L. (2014). Morphogenes bola and mreb mediate the photoregulation of cellular morphology during complementary chromatic acclimation in Fremyella diplosiphon. Mol. Microbiol. 93, 167–182. doi: 10.1111/mmi.12649
Spormann, A. M. (1999). Gliding motility in bacteria: insights from studies of Myxococcus xanthus. Microbiol. Mol. Biol. Rev. 63, 621–641. doi: 10.1128/mmbr.63.3.621-641.1999
Tavares, J. R., De Souza, R. F., Meira, G. L., and Gueiros-Filho, F. J. (2008). Cytological characterization of Ypsb, a novel component of the Bacillus subtilis divisome. J. Bacteriol. 190, 7096–7107. doi: 10.1128/JB.00064-08
Thomaides, H. B., Freeman, M., El Karoui, M., and Errington, J. (2001). Division site selection protein Diviva of Bacillus subtilis has a second distinct function in chromosome segregation during sporulation. Genes Dev. 15, 1662–1673. doi: 10.1101/gad.197501
Thompson, S. R., Wadhams, G. H., and Armitage, J. P. (2006). The positioning of cytoplasmic protein clusters in bacteria. Proc. Natl. Acad. Sci. U.S.A. 103, 8209–8214. doi: 10.1073/pnas.0600919103
Van Baarle, S., and Bramkamp, M. (2010). The mincdj system in bacillus subtilis prevents minicell formation by promoting divisome disassembly. PLoS One 5:e0009850. doi: 10.1371/journal.pone.0009850
Vijay, S., Nagaraja, M., Sebastian, J., and Ajitkumar, P. (2014). Asymmetric cell division in Mycobacterium tuberculosis and its unique features. Arch. Microbiol. 196, 157–168. doi: 10.1007/s00203-014-0953-7
Wolf, E., Kim, P. S., and Berger, B. (1997). Multicoil: a program for predicting two- and three-stranded coiled coils. Protein Sci. 6, 1179–1189. doi: 10.1002/pro.5560060606
Zapun, A., Bardwell, J. C. A., and Creighton, T. E. (1993). The reactive and destabilizing disulfide bond of DsBA, a protein required for protein disulfide bond formation invivo. Biochemistry 32, 5083–5092. doi: 10.1021/bi00070a016
Keywords: predatory bacteria, Bdellovibrio, ParAB, DivIVA, septation, cell-morphology
Citation: Milner DS, Ray LJ, Saxon EB, Lambert C, Till R, Fenton AK and Sockett RE (2020) DivIVA Controls Progeny Morphology and Diverse ParA Proteins Regulate Cell Division or Gliding Motility in Bdellovibrio bacteriovorus. Front. Microbiol. 11:542. doi: 10.3389/fmicb.2020.00542
Received: 25 November 2019; Accepted: 12 March 2020;
Published: 21 April 2020.
Edited by:
Hari S. Misra, Bhabha Atomic Research Centre (BARC), IndiaReviewed by:
Dagmara Jakimowicz, University of Wrocław, PolandMarc Bramkamp, Ludwig Maximilian University of Munich, Germany
Copyright © 2020 Milner, Ray, Saxon, Lambert, Till, Fenton and Sockett. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Renee Elizabeth Sockett, bGl6LnNvY2tldHRAbm90dGluZ2hhbS5hYy51aw==
†These authors have contributed equally to this work
‡Present address: David S. Milner, Department of Zoology, University of Oxford, Oxford, United Kingdom; Andrew K. Fenton, Florey Institute, Department of Molecular Biology and Biotechnology, University of Sheffield, Sheffield, United Kingdom
 David S. Milner
David S. Milner Luke J. Ray†
Luke J. Ray† Renee Elizabeth Sockett
Renee Elizabeth Sockett