- 1Department of Microbiology, Zhongshan School of Medicine, Sun Yat-sen University, Guangzhou, China
- 2Key Laboratory of Tropical Diseases Control, Ministry of Education, Sun Yat-sen University, Guangzhou, China
- 3Clinical Laboratory, Fifth Affiliated Hospital, Sun Yat-sen University, Zhuhai, China
- 4Department of Microbiology and Immunology, Faculty of Pharmaceutical Sciences and Drug Manufacturing, Misr University for Science and Technology, Cairo, Egypt
- 5Department of Tropical Disease Biology, Liverpool School of Tropical Medicine, Liverpool, United Kingdom
- 6Centre for Drugs and Diagnostics, Liverpool School of Tropical Medicine, Liverpool, United Kingdom
- 7School of Pharmaceutical Sciences (Shenzhen), Sun Yat-sen University, Guangzhou, China
There is growing evidence that GreA aids adaptation to stressful environments in various bacteria. However, the functions of GreA among mycobacteria remain obscure. Here, we report on cellular consequences following deletion of greA gene in Mycobacterium spp. The greA mutant strain (ΔgreA) was generated in Mycobacterium smegmatis, Mycobacterium tuberculosis (MTB) H37Ra, and M. tuberculosis H37Rv. Deletion of greA results in growth retardation and poor survival in response to adverse stress, besides rendering M. tuberculosis more susceptible to vancomycin and rifampicin. By using RNA-seq, we observe that disrupting greA results in the differential regulation of 195 genes in M. smegmatis with 167 being negatively regulated. Among these, KEGG pathways significantly enriched for differentially regulated genes included tryptophan metabolism, starch and sucrose metabolism, and carotenoid biosynthesis, supporting a role of GreA in the metabolic regulation of mycobacteria. Moreover, like Escherichia coli GreA, M. smegmatis GreA exhibits a series of conservative features, and the anti-backtracking activity of C-terminal domain is indispensable for the expression of glgX, a gene was down-regulated in the RNA-seq data. Interestingly, the decrease in the expression of glgX by CRISPR interference, resulted in reduced growth. Finally, intracellular fitness significantly declines due to loss of greA. Our data indicates that GreA is an important factor for the survival and resistance establishment in Mycobacterium spp. This study provides new insight into GreA as a potential target in multi-drug resistant TB treatment.
Introduction
Mycobacterium tuberculosis (MTB), the causative agent of tuberculosis (TB) has influenced human populations since ancient times. In 2018, M. tuberculosis caused seven million TB cases and was responsible for 1.5 million deaths worldwide (Harding, 2019). The administration of anti-TB drugs has led to the emergence of drug-resistant strains of MTB (Kumar et al., 2017) with approximately 5% of infections being caused by multidrug-resistant (MDR) strains globally (Wright et al., 2009). China has a high prevalence of drug-resistant TB and is the region with second largest number of MDR cases worldwide (Yang et al., 2017). Therefore, exploring the cause of MDR-TB will be pivotal within the implementation of effective public health strategies to reduce MDR-TB.
Transcription (the first step of gene expression) is fundamentally important to all life. Once RNA polymerase (RNAP) initiates the process of transcription, it is vital for the enzyme to carry out elongation and termination to ensure full-length RNA synthesis. Several transcription factors can bind to RNAP, modifying its properties by affecting transcription processivity and fidelity through modulating pausing, arrest, and termination (Tehranchi et al., 2010; Li et al., 2015). The transcript cleavage factor, GreA, interacts with the RNAP secondary channel and stimulates the intrinsic transcript cleavage activity of RNAP for the removal of the aberrant RNA 3′ ends. Therefore, polymerization activity can be restarted from the end of a cleaved RNA allowing transcription to resume (Komissarova and Kashlev, 1997).
Gre factors are widely distributed in prokaryotes. There is growing evidence that Gre factors participate in each step during the transcription process, including initiation, elongation, and fidelity (Yuzenkova et al., 2014; Traverse and Ochman, 2018). In addition, it has been reported that GreA is essential for the survival of bacteria under stress (Volker et al., 1994; Nogales et al., 2002). Moreover, GreA, which can facilitate RecBCD-mediated resection and inhibits RecA, plays an important role in impeding DNA break repair in Escherichia coli (Sivaramakrishnan et al., 2017), indicating a role for GreA in adapting to stressful environments.
The genome of M. tuberculosis contains a Gre-factor encoded by rv1080c. Recent in vitro experiments demonstrated that Mycobacterium smegmatis GreA exhibits chaperone-like activity (Joseph et al., 2019). It has been shown that M. tuberculosis GreA failed to rescue E. coli RNAP stalled elongation complexes (China et al., 2011). This suggested that the function of M. tuberculosis GreA is sufficiently different from the other well-studied bacterial GreA systems to warrant further investigation. Gumber et al. (2009) revealed that greA was found to be upregulated in anaerobic and acidic conditions in Mycobacterium avium. Moreover, further studies showed that greA was upregulated in M. tuberculosis following treatment with ofloxacin, moxifloxacin, and streptomycin (Sharma et al., 2010; Lata et al., 2015), indicating an important role for GreA in the environmental adaptation.
Here, we aimed to dissect the role of GreA and determine any influence on the fitness of mycobacteria. We revealed that GreA serves as a transcriptional factor that coordinates the expression of genes involved in metabolism in M. smegmatis. Our data suggests that GreA may be a novel target for adjunctive therapy.
Materials and Methods
Ethics Statement
All animal experiments were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals, and the experimental procedures were approved by the Ethics Committee of Zhongshan School of Medicine on Laboratory Animal Care (reference number: 2016-159), Sun Yat-sen University.
Animal Studies in C57BL/6 Mice
Female C57BL/6 mice were purchased from Animal Supply Center, Sun Yat-sen University. Mice were bred and maintained under specific pathogen-free conditions at the animal facility of the Sun Yat-sen University. Adult mice between 6–8 weeks of age were used, and infected mice were maintained under biosafety two conditions. Mice were infected with 1 × 107 colony-forming units (CFUs) of H37Ra, ΔgreA and comp-ΔgreA, through the intraperitoneal injection. Four mice from each group were sacrificed after 14- and 21- days post-infection, and the lung and spleen homogenates (prepared by homogenizing aseptically removed organs in 1 ml of sterile saline) were plated on 7H10 agar supplemented with 10% OADC (BD Biosciences), in triplicates. CFUs of M. tuberculosis were counted after 3-4 weeks of incubation at 37°C.
Bacterial Strains and Generation of greA Mutants
A mutant (ΔgreA) strain of M. tuberculosis was constructed as described previously (Jain et al., 2014). For the deletion of greA in M. tuberculosis H37Rv, PCR was performed to synthesize fragments bearing the 1000 and 980 bps of flanking regions of endogenous greA of M. tuberculosis, resulting in a deletion of the gene (primer set Mtb greA-LL and Mtb greA-LR for the 5′ region and primer set Mtb greA-RL and Mtb greA-RR for the 3′ flanking region). Amplicons corresponding to upstream and downstream flanking regions were digested with Van91I and cloned into the Van91I digested p0004s plasmid that contains a hygromycin resistance cassette and the sacB gene to be able to select for sucrose sensitivity. This allelic exchange substrate was introduced into the PacI site of phasmid phAE159 and electroporated into M. smegmatis mc21 155 to obtain high titers of phage phAE159. Subsequently, the M. tuberculosis wild-type strain was incubated with high titers of the corresponding phage to create greA knockouts. Colonies that had deleted endogenous greA were selected on hygromycin-containing plates and verified for sucrose sensitivity. Since the H37Ra genome is highly similar to that of H37Rv with respect to gene content, similar experiments have been performed for generation of H37Ra mutant strain. For unmarking of the deletion mutations generated by the helper plasmid pYUB870 (SacB, tnpR, and KanR). After transformation by electroporation with pYUB870 and plating onto medium containing kanamycin, 5 kanamycin-resistance colonies were obtained and screened by steaking on 7H10 agar alone and on 7H10 agar with 50 μg/ml hygromycin. A hygromycin-sensitive clone was grown in liquid medium in the absence of antibiotic selection. Further confirmation of res-hyg-sacB-res cassette deletion was achieved by PCR analysis and sequencing.
For the construction of M. smegmatis ΔgreA strain, an in-frame deletion of the gene was enabled using Xer site-specific recombination as previously described (van Kessel and Hatfull, 2007). Briefly, two DNA fragments, approximately 500 bp, flanking greA of M. smegmatis were acquired through PCR and cloned at the borders of the hygromycin excisable cassette. The resulting DNA fragment was purified and electroporated into the competent cells of M. smegmatis harboring pJV53 plasmid. The hygromycin-resistant colonies were grown for ten generations in the absence of hygromycin and kanamycin to allow excision of the hygromycin cassette and the loss of pJV53. Finally, unmarked ΔgreA strain was confirmed by PCR and sequencing.
For the complementation strain of M. smegmatis (comp-ΔgreA) and M. tuberculosis H37Ra (comp-ΔgreA), greA from M. smegmatis strain and greA from M. tuberculosis H37Ra strain was cloned into a pMV306-Phsp60 plasmid, and then the recombinant plasmid pMV306-Phsp60-greA was transformed into M. smegmatis ΔgreA and M. tuberculosisΔgreA strains, resulting in M. smegmatis comp-ΔgreA and M. tuberculosis comp-ΔgreA, respectively. The comp-ΔgreA strains were selected on Middlebrook 7H10 agar medium (complemented with 10% OADC) containing 30 μg/ml kanamycin.
The open reading frame (ORF) of greA fused with 2 × FLAG was amplified on a template of M. smegmatis mc2 155 (WT) chromosomal DNA using primer pairs. The two single-point mutants of greA were generated using overlapping PCR. All of the greA variants were cloned into a pMV306-Phsp60 plasmid. The glgX (MSMEG_3186) gene included its native promoter were cloned into pSMT3-LxEGFP vector in order to perform flow cytometry experiment. As a result, all the plasmid constructs were confirmed via direct DNA sequencing and PCR assay.
Construction of CRISPR Interference (CRISPRi) Targeting Constructs in M. smegmatis
CRISPRi targeting constructs in M. smegmatis was performed as previously described (Singh et al., 2016). Briefly, we have designed oligos to target two sites in the glgX gene. The oligos were annealed to generate a double-stranded insert which was phosphorylated by T4 polynucleotide kinase and then ligated into the BbsI digested pRH2521 to obtain pRH2521-sgRNA. pRH2521 containing sgRNA-targeting the genes of interest was transformed into M. smegmatis competent cells into which the dcas9-containing vector pRH2502 was previously introduced. Transformants were selected for hygromycin and kanamycin resistance. Addition of anhydrotetracycline (ATc) was repeated every 48 h to maintain induction of dcas9 and sgRNA for experiments.
Antimicrobial Sensitivity Assays
Antimicrobial sensitivity assays for M. tuberculosis have been performed as previously described (Goodsmith et al., 2015). Mycobacteria strains were grown to early log phase and were diluted to an optical density of 0.02 in 7H9 media containing 0.05% Tween 80 and 10% OADC. Then the bacterial strains were exposed to two-fold dilutions of the following antimicrobials; rifampicin, vancomycin, bedaquiline, streptomycin, ofloxacin, isoniazid, Capreomycin, amikacin, and ethambutol (Sigma-Aldrich, United States). The minimum inhibitory concentrations (MICs) were recorded as the minimum concentrations at which the growth was inhibited by at least 90%, as compared to a control containing no antibiotic.
For M. smegmatis, the MICs at which no bacterial growth was observed were recorded after 3–5 days of incubation. For M. tuberculosis, MICs at which no bacterial growth was observed were recorded after approximately 2 weeks of incubation.
Growth Inhibition Test
Mycobacterium smegmatis had been grown to early log phase and was diluted to an optical density of 0.02 in 7H9 broth medium containing 10% OADC. Then, M. smegmatis culture was mixed with each antibiotic, at the final concentration to 1/100th the MIC, including streptomycin, and rifampicin. The OD600 value of the bacteria was monitored hourly.
Analysis of in vitro Response to Adverse Stress
Exponentially growing bacterial cultures (OD600 0.4–0.6) were pelleted and washed three times with 7H9/Tween-80.
In M. smegmatis, for the low pH stress, the strains were washed twice in 7H9 acidified to pH 4.5 with HCl, supernatants were adjusted to an OD600 = 0.5, and CFU was determined after 24 h. For heat shock stresses, M. smegmatis strains were incubated at 48°C in a water bath for 6 h, CFUs were counted after 3∼4 days of incubation. For other stresses, M. smegmatis strains prepared as above were adjusted to OD600 = 0.5, and SDS, H2O2, or antibiotics were added, then the CFUs were determined after 24 h.
For M. tuberculosis H37Rv strains, the cells were treated only in 7H9 broth medium with 0.5% SDS and were incubated for 24 h. Then bacterial cells were diluted 10-fold and spotted on Middlebrook 7H10 agar media. While for M. tuberculosis H37Ra strain, the cells were resuspended in Middlebrook 7H9 broth under 0.5% SDS and were incubated for 8 h. The CFUs were counted after 4 weeks of incubation.
Synthesis of cDNA and RT-PCR
Equal amounts of bacteria of M. smegmatis strains, which were grown to logarithmic phase, were collected. Total RNA and cDNA from M. smegmatis were extracted as previously described (Rustad et al., 2009). cDNA was prepared and amplified as previously described (Alland et al., 1998). Control reactions were performed using primers specific to sigA (Manganelli et al., 1999) (see Supplementary Table S1). PCR products were separated on 2% agarose gels. The mean intensity of each band was analyzed using ImageJ software. The relative expression was calculated by normalizing the intensity of the glgX band with that of the sigA band amplified with the same cDNA sample.
Cell Culture
Murine macrophage-like RAW264.7 cells (ATCC; TIB-71) were cultured on Dulbecco’ s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin (GIBCO, Invitrogen) with 5% CO2 at 37°C.
Macrophage Infection Study
RAW264.7 cells were seeded at 3 × 105 cells/well in a 24-well plate and incubated for 18 h. Homogenized bacterial suspension at a multiplicity of infection (MOI) of 10, was transformed into adherent RAW264.7 cells. The extracellular bacteria were removed by washing three times with phosphate buffer saline (PBS) after incubation for 2 h. Cells were further incubated for 48 h with 5% CO2. Then cell lysates were serially diluted 10-fold and spotted on 7H10-OADC agar. CFUs of M. smegmatis were counted after 3∼4 days of incubation at 37°C, while CFUs of M. tuberculosis were counted after 4 weeks days of incubation at 37°C.
Structural Modeling
The architecture of GreA in full length was modeled with Swiss-Model2. The GreA of E. coli [PDB: 1grj.1] has functioned as a structural template, and the ribbon structure was presented with the PyMol software.
RNA Preparation and RNA-Seq Analysis
Wild-type M. smegmatis mc2155 as well as the greA deletion strain were grown to the log-phase (OD600, 0.4) in Middlebrook 7H9-ADC. Total RNA was prepared from wild-type and mutant strains using TRIzol reagent (Invitrogen), followed by DNase I treatment. Approximately 1-μg total RNA samples were treated by the Ribo-Zero rRNA removal procedure (Illumina) to enrichment for mRNA. Approximately 1 μg of RNA was used for library preparation using a ScriptSeq (v2) RNA-seq kit and high-throughput sequencing on an Illumina NextSeq platform. All raw sequence reads by RNA-Seq were initially pre-processed by Trimmomatic (v0.36) to trim the adaptor sequences and remove low-quality sequences. The remaining clean reads were mapped to the M. smegmatis mc2 155 genomes using Tophat2. The alignment results were input into Cufflinks (v2.2.1). Unless otherwise stated, the unit of expression level in our analyses is FPKM. Cuffdiff (v2.2.1) was used to test for differential expression. We defined genes as differentially expressed using the following criteria: FPKM > 10 and FDR < 0.001. The data for visualization was generated by R (R Development Core Team, Vienna, Austria). The genome sequences and the annotations of M. smegmatis mc2 155 were obtained from the GenBank2. We analyzed differences in the enrichment of Gene Ontology (GO) categories and KEGG pathways for the DEGs using the DAVID (v6.8) functional annotation analysis tool3.
Chromatin Immunoprecipitation
ChIP was performed using log-phase (OD600, 0.4) liquid medium grown cultures of M. smegmatis strains producing GreA-FLAG. As a negative control, WT log-phase culture was used. The cells were fixed with 1% formaldehyde for 30 min, the reaction was quenched with 150 mM glycine for 15 min, and then the cells were washed three times with cold PBS (54,000 rpm, 5 min, 4°C) and frozen at −80°C. To prepare lysates, pellets were resuspended in FA-1 buffer (HEPES-KOH at 50 mM [pH 7.5], NaCl at 140 mM, EDTA at 1 mM, Triton X-100 at 1%, and protease inhibitor cocktail [Sigma-Aldrich]), disintegrated with silica beads (0.1 mm) for 45 min, and sonicated on ice using ten cycles of a 10-s pulse followed by a 50-s pause. The obtained lysates were centrifuged (5 min, 12,000 rpm, 4°C) and frozen at −80°C in 5% glycerol. For immunoprecipitation, 200 μg of total protein was incubated on a rotary shaker (Thermo Fisher Scientific, China Co., Ltd.) for 4 h at 4°C with a 15-μl packed-gel volume of anti-FLAG M2 magnetic beads (Sigma-Aldrich) and then washed twice with FA-1 buffer. Samples were processed in a final volume of 0.5 ml in two biological replicates, with input DNA controls (200 μg of total protein alone) included for each replicate. Samples were washed using a magnetic separator with sequential applications of FA-1 buffer, FA-2 buffer (HEPES-KOH at 50 mM [pH 7.5], NaCl at 500 mM, EDTA at 1 mM, Triton X-100 at 1%, and protease inhibitor cocktail), and Tris–EDTA buffer solution (TE) (Tris–HCl at 10 mM [pH 8.0], EDTA at 1 mM). Immunoprecipitated samples were de-cross-linked overnight in TE containing 1% SDS at 65°C and then digested with proteinase K (final concentration, 0.05 mg/ml) for 1.5 h at 55°C. The immunoprecipitated DNA was extracted using Cycle-Pure Kit (OMEGA Bio-tek, Inc.).
Library Construction and Illumina Sequencing
The library of DNA fragments was prepared using a QIAseq Ultralow Input library kit (Qiagen). Briefly, the protocol includes DNA end-repair, sequencing adapter ligation, cleanup, and PCR amplification. At the end of the procedure, quantification and quality evaluations were done using a NanoPhotometer ® spectrophotometer (IMPLEN), a Qubit® 2.0 Fluorometer (Life Technologies), and a 2100 Bioanalyzer (Agilent). Second-generation sequencing was performed using a HiSeq 1500 sequencing platform (Illumina).
Analysis of CHIP-Seq Data
The obtained FASTQ files were filtered according to read quality, and adapter sequences were trimmed using the Trimmomatic software (Usadel Lab; Aachen University, Aachen, Germany). The filtered FASTQ files were mapped to the genome of M. smegmatis strain mc2 155 (from GenBank) using the BWA v0.7.12 (Burrows-Wheeler aligner), and the Bwa mem algorithm was applied. The bam files were sorted and indexed. Then the bam files for the ChIP and input samples were subjected to MACS analysis (MACS2 software) for ChIP-Seq peak detection. Peak calling was performed without building a model using a shift size of 150 bp. The ChIP-Seq peaks were uploaded into the R environment as bed files, and the peaks were annotated ChIP-Seq data.
Statistical Analysis
Statistical analysis was performed using Prism (version 6.0c; GraphPad Software). Data were analyzed using the paired Student’s t-test, and in the comparisons of data from three or more conditions, analysis of variance (ANOVA) was used. A p-value of 0.05 or less was considered significant.
Results
Mutants for greA Caused a Delay in Mycobacterial Growth
To explore the function of greA in mycobacteria, we deleted the greA (MSMEG_5263) gene by allelic exchange facilitated via recombination in M. smegmatis mc2155 which is widely used as a model to study non-pathogenic aspects of mycobacterial physiology. We have detected colonies of small sizes in the greA mutant strain compared with the parental strain (Figure 1A), it was observed that ΔgreA strain grows slower than the wild-type M. smegmatis and the phenotypes that were complemented by expression of greA (comp-ΔgreA) from M. smegmatis on the chromosome (Figure 1B). To confirm the phenotype of GreA in M. tuberculosis strain, we knocked out greA (rv1080c) in M. tuberculosis H37Rv and H37Ra by specialized transducing mycobacteriophages (Supplementary Figure S1). Following the observation that a growth defect was caused by greA deficiency in M. smegmatis, we found that colonies of small sizes were also observed in the ΔgreA strain of H37Ra and H37Rv (Figure 1A) and a growth defect was detected in the ΔgreA strain of H37Ra (Figure 1C). Our observation was consistent with a previous study (China et al., 2011).
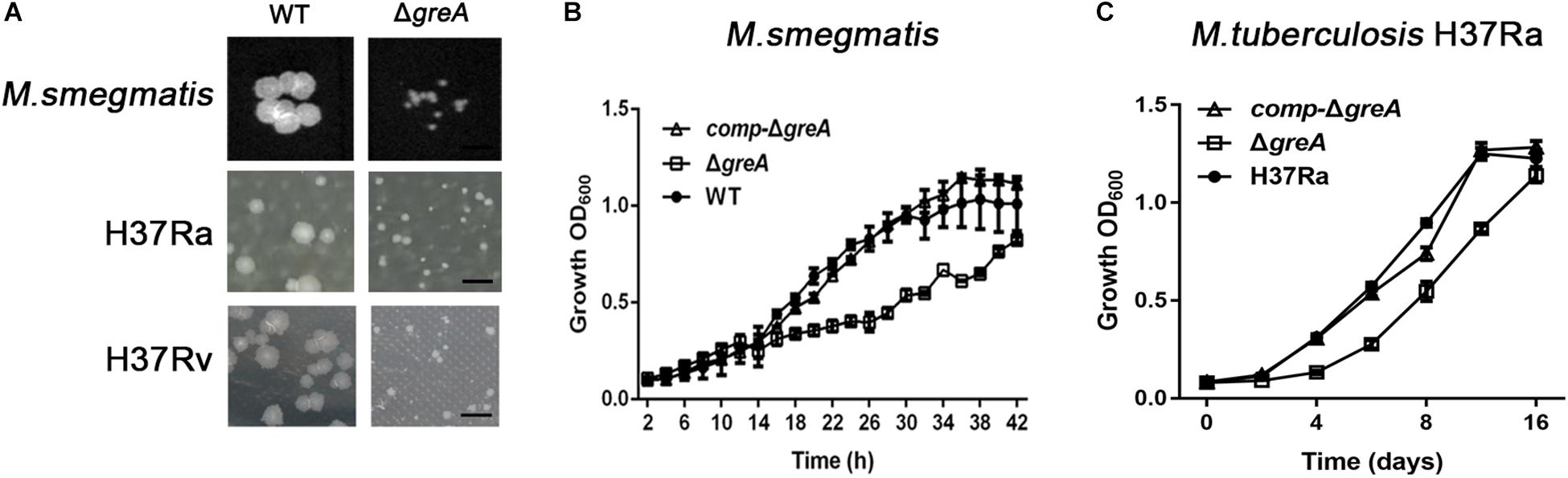
Figure 1. Mutants for greA on growth, colonial morphology in mycobacteria. (A) The colony size of WT and ΔgreA strains from M. smegmatis, H37Ra and H37Rv. The bar corresponds to 0.5 cm. The growth of WT, ΔgreA, and comp-ΔgreA strains from (B) M. smegmatis and (C) M. tuberculosis H37Ra in 7H9 broth. Bacteria were diluted to an initial OD600 of 0.02 and the OD600 was monitored at the designated time points. Data are shown as mean ± SEM of three independent experiments.
Deletion of greA Increased the Sensitivity to Adverse Stress
It has been reported that GreA is essential for the survival of E. coli under stress conditions (Sharma and Chatterji, 2010). Herein, we investigated the role of GreA in the environmental adaptation of mycobacteria. The WT, ΔgreA, and comp-ΔgreA strains were treated with adverse stresses. When compared to parental bacteria and complement strain, the ΔgreA strain was more sensitive to SDS, hydrogen peroxide, and starvation (Figures 2A–C). However, there were no differences in the survival rate of bacteria which were treated with low pH and heat shock (Figures 2D,E). In addition, we also found that the survivability of M. tuberculosis ΔgreA mutant strain was significantly reduced compared to H37Rv strain upon the treatment with 0.5% SDS (Figure 2F) and a similar phenotype was also observed in M. tuberculosis H37Ra strain (Figure 2G). Moreover, the greA gene also complemented the phenotype of ΔgreA in M. tuberculosis H37Ra (Figure 2G). These results indicated that mycobacteria GreA is required for adaptation to these stressful conditions.
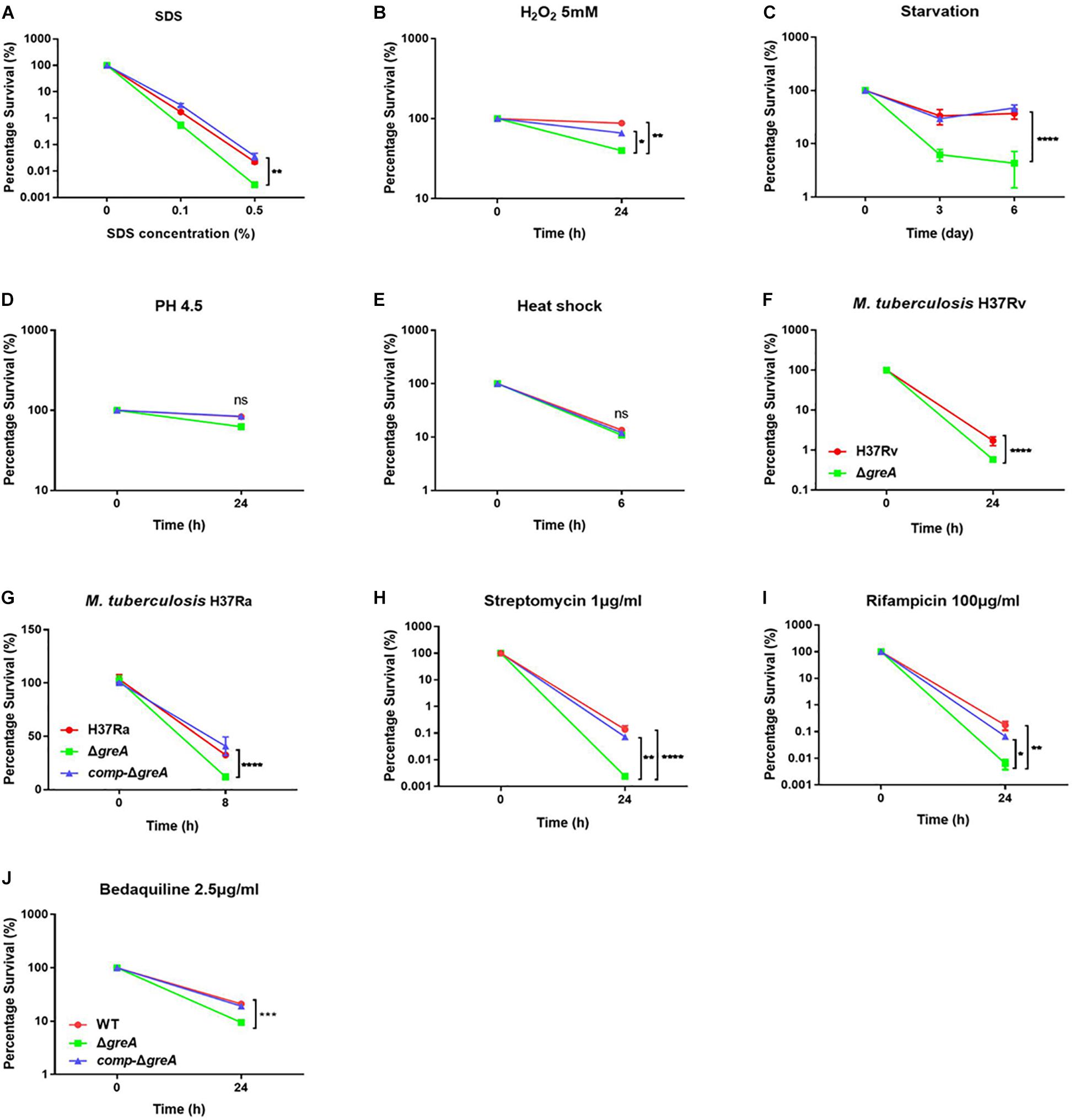
Figure 2. GreA is required for resistance to adverse stress. (A–E, H–J) Survival of M. smegmatis strains during adverse stress. The strain symbols listed in the (J). WT, ΔgreA and comp-ΔgreA strains were treated for at the indicated time with varying concentrations of SDS (A), 5mM H2O2 (B), PBS + 0.05% Tween 80 (C), low pH (pH4.5) (D), heat shock (48°C) (E), 1 μg/ml of streptomycin (H), 100 μg/ml of rifampicin (I), or 2.5 μg/ml of bedaquiline (J). After treatment, dilutions were plated on 7H10 and survival is expressed as a ratio of CFUs compared to untreated cultures. (F) M. tuberculosis H37Rv strains were spotted on the 7H10 plates after they were treated under 0.5% SDS for 24 h. (G) WT, ΔgreA, and comp-ΔgreA strains from M. tuberculosis H37Ra were subjected to 0.5% SDS for 8 h. CFU enumeration was performed after incubation for the indicated time. These experiments were performed three times with similar results. Data are shown as mean ± SEM of three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns, p ≥ 0.05.
Of note, the greA gene was found upregulated under stress with antibiotics in M. tuberculosis (Sharma et al., 2010; Lata et al., 2015). We speculated that ΔgreA strain might be more susceptible to antibiotics. To test this hypothesis, we performed antibiotic susceptibility tests where, M. smegmatis WT, ΔgreA, and comp-ΔgreA strains were challenged with streptomycin, rifampicin, and bedaquiline. We found that ΔgreA strain was more susceptible to the tested antibiotics compared with both parental and complemented bacteria (Figures 2H–J). Moreover, we also evaluated the susceptibility to antibiotics through the MIC test in M. tuberculosis, and we found that M. tuberculosisΔgreA strain showed increased susceptibility to rifampicin, bedaquiline, and vancomycin. Surprisingly, the MIC of the ΔgreA strain decreased by 4-fold with rifampicin and by 8-fold with vancomycin compared with MTB H37Rv (Table 1), suggesting that greA plays an important role in the antibiotic susceptibility in mycobacteria.
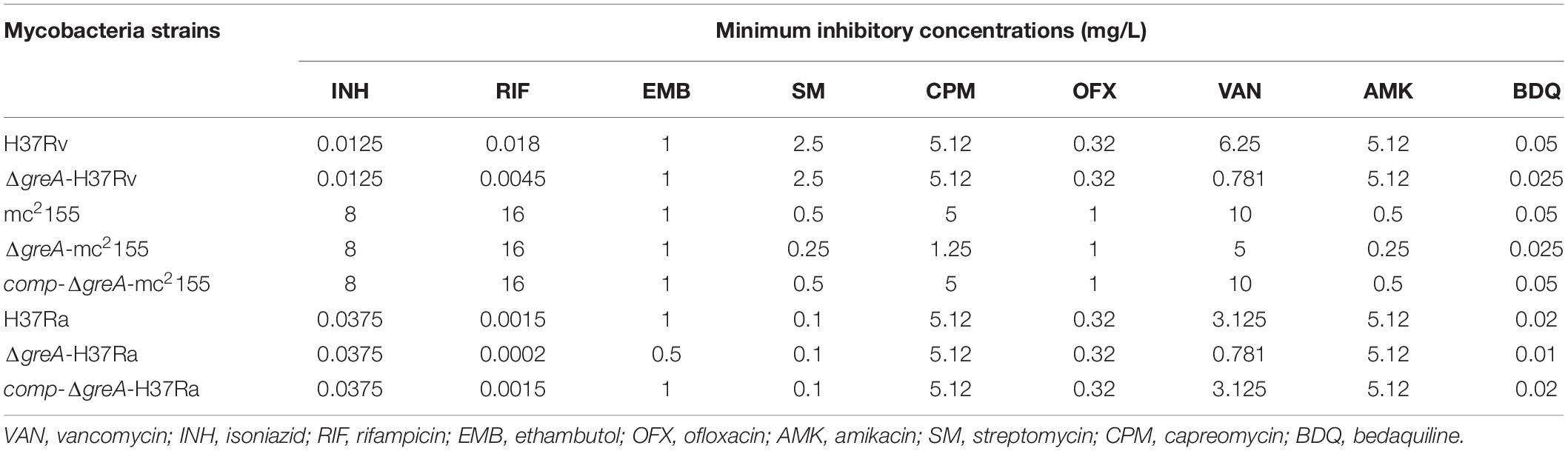
Table 1. Changes of MICs of wild-type mycobacteria and ΔgreA strains against different antimicrobial agents.
In accordance with the observation that the greA mutant strain has an increase in the sensitivity to antibiotics, we found that the growth of ΔgreA strain was more retarded despite at low antibiotic concentrations used when compared to WT and complemented strains (Supplementary Figure S2).
In vivo Transcription Analysis of GreA-Responsive Genes
Given our observations that inactivation of greA in M. smegmatis results in reduced survivability under stress, we next sought to identify the proteins being regulated by GreA. To understand how GreA may affect gene expression at the transcriptional level, we performed whole-transcriptome RNA-seq analyses. We found that disrupting greA resulted in the differential regulation of 195 genes in M. smegmatis during the exponential growth. Among these, 28 were positively regulated and 167 were negatively regulated (Figure 3A and Supplementary Table S2). Moreover, our findings for enriched KEGG pathways are summarized in Figure 3B and Supplementary Table S3. Three pathways were significantly overrepresented. Three genes are involved in tryptophan metabolism, and four genes are involved in starch and sucrose metabolism. Interestingly, the glgX gene, which encodes the debranching enzyme of the catabolic enzymes, was involved with starch and sucrose metabolism (Schneider et al., 2000). Chromosomal inactivation of glgX in Corynebacterium glutamicum led to slower growth (Seibold and Eikmanns, 2007), it is suggested that GreA mediated regulation of the growth might be partly associated with glgX. This outcome demonstrated that GreA has a profound effect on cellular gene regulation, especially on metabolic associated genes. Collectively, these transcriptional changes are consistent with the scenario that greA gene expression is required for optimal growth of mycobacteria, whereas loss of greA results in slower growth and reduced tolerance to adverse environments.
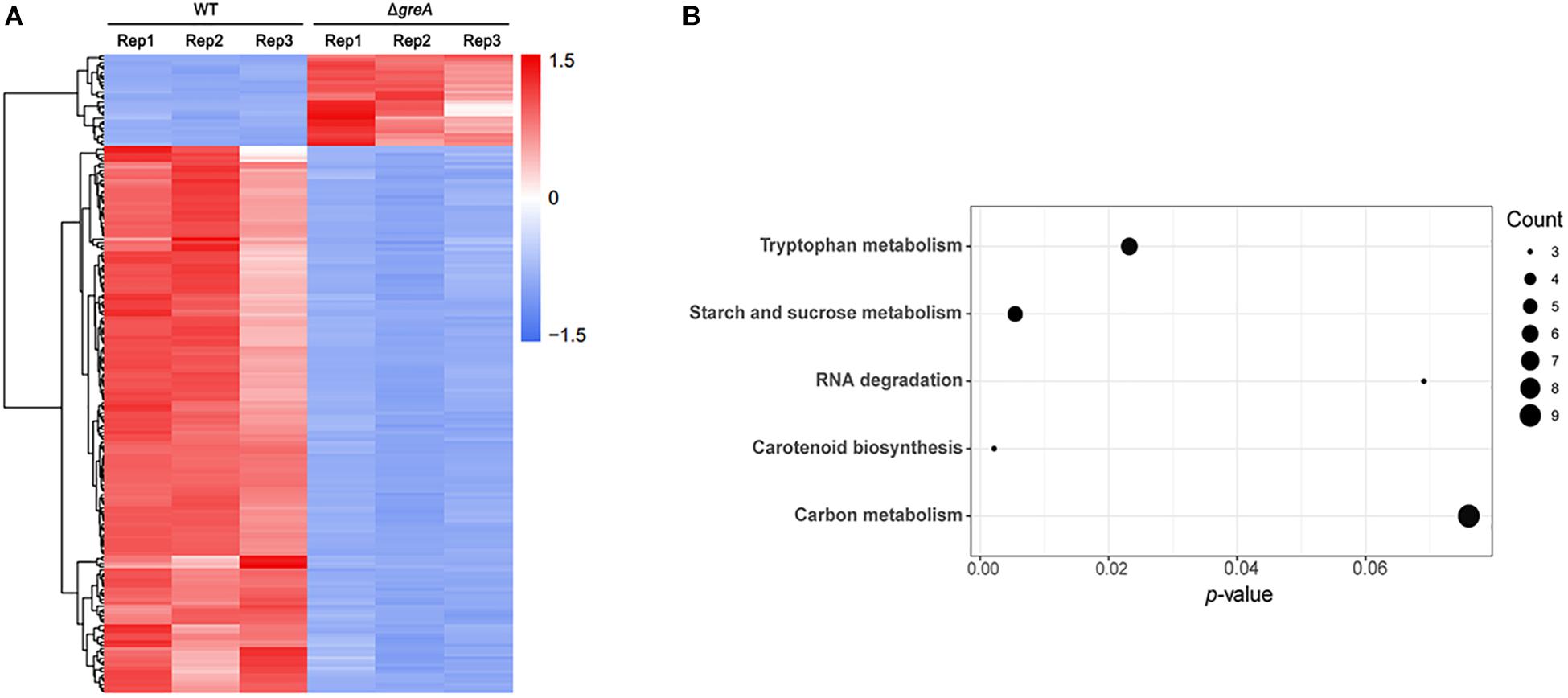
Figure 3. Differentially regulated genes were identified between ΔgreA strain and WT strain by RNA-seq. (A) Heat maps showing the expression profiles of genes upregulated or downregulated by deletion of the greA gene in M smegmatis. (B) Summary of the KEGG reference pathway results. Three biological replicates were tested.
Identification of GreA-Binding Sites Using ChIP-Seq
To examine whether GreA binds to specific chromosomal regions, we performed chromatin immunoprecipitation followed by deep sequencing (CHIP-seq) and then sought to verify the binding mode of GreA. CHIP-seq experiments were performed by using M. smegmatis strains producing GreA proteins fused with two repeats of the FLAG epitope (FLAG2). GreA-FLAG2-DNA nucleoprotein complexes fixed with formaldehyde in the log phase of growth were immunoprecipitated using magnetic beads. To exclude unspecific interactions with magnetic beads, we used a WT strain lacking the FLAG epitope as a negative control for our CHIP-seq experiments. Enriched regions were determined by comparison to the background noise level, which was estimated versus the input DNA of each ChIP-Seq replicate. Using the data obtained from this analysis, we established chromosomal binding maps for the analyzed proteins. The GreA-FLAG2 binding sites were distributed evenly along the M. smegmatis (Figure 4). The identified GreA-FLAG2 binding sites included 1005 CHIP-seq peaks that were confirmed in two biological replicates and absent in WT strain (Supplementary Table S4). Among the 1005 CHIP-seq peaks identified for GreA-FLAG2, we observed that several binding sites were located downstream of the gene (∼0.7% of all ChIP-Seq peaks). However, GreA binding sites seemed to be more frequent within promoters (∼84.8% of all ChIP-Seq peaks), and ∼14.5% of all ChIP-Seq peaks were identified in gene bodies (Supplementary Figure S3). Notably, from the CHIP-seq dataset, we found that one of the peaks was located at the coding sequence of glgX, suggesting that GreA might regulate the transcription of glgX.

Figure 4. Heat maps of the distributions of GreA-FLAG binding sites in two biological replicates. Two biological replicates were tested.
The Full Activity of the GreA Is Crucial in M. smegmatis Growth
It is known that E. coli GreA consists of two domains: an N-terminal extended coiled-coil domain (NTD, residues 1–75) and a C-terminal globular domain (CTD, residues 76–158) (Stebbins et al., 1995; Koulich et al., 1997). The NTD carries the structural determinants conferring the cleavage activity and, presumably, the anti-arrest and readthrough activities. The CTD participates in binding to RNAP (Koulich et al., 1998). Alignment of the mycobacterial GreA with E. coli GreA revealed the similar conserved features (Figure 5A). In addition, GreA from M. smegmatis is almost identical to that of M. tuberculosis GreA, indicating its similar function in mycobacteria. To gain further structural insight into the conserved domain of mycobacterial GreA, structural modeling by the Swiss-Model program was performed using E. coli GreA (PDB accession no. 1grj.1) as a structural template (Figure 5B). The ribbon structure of M. smegmatis GreA, together with E. coli GreA were generated with the PyMol software. Of note, structural comparison of the two GreA proteins illustrated that the coiled-coil domain and RNAP interaction of CTD constitute almost identical motifs (Figure 5C). To further explore their physiological role in the growth phenotype of GreA, we applied overlapping PCR to generate the following two single point mutations of GreA (D43N, S127E). As expected, the D43N and S127E mutations completely disrupted the optimal growth of M. smegmatis GreA (Figure 5D), the observation is in line with a previous in vitro study (China et al., 2011). Collectively, these observations strongly support the conclusion that the full activity of the GreA is required for M. smegmatis growth.
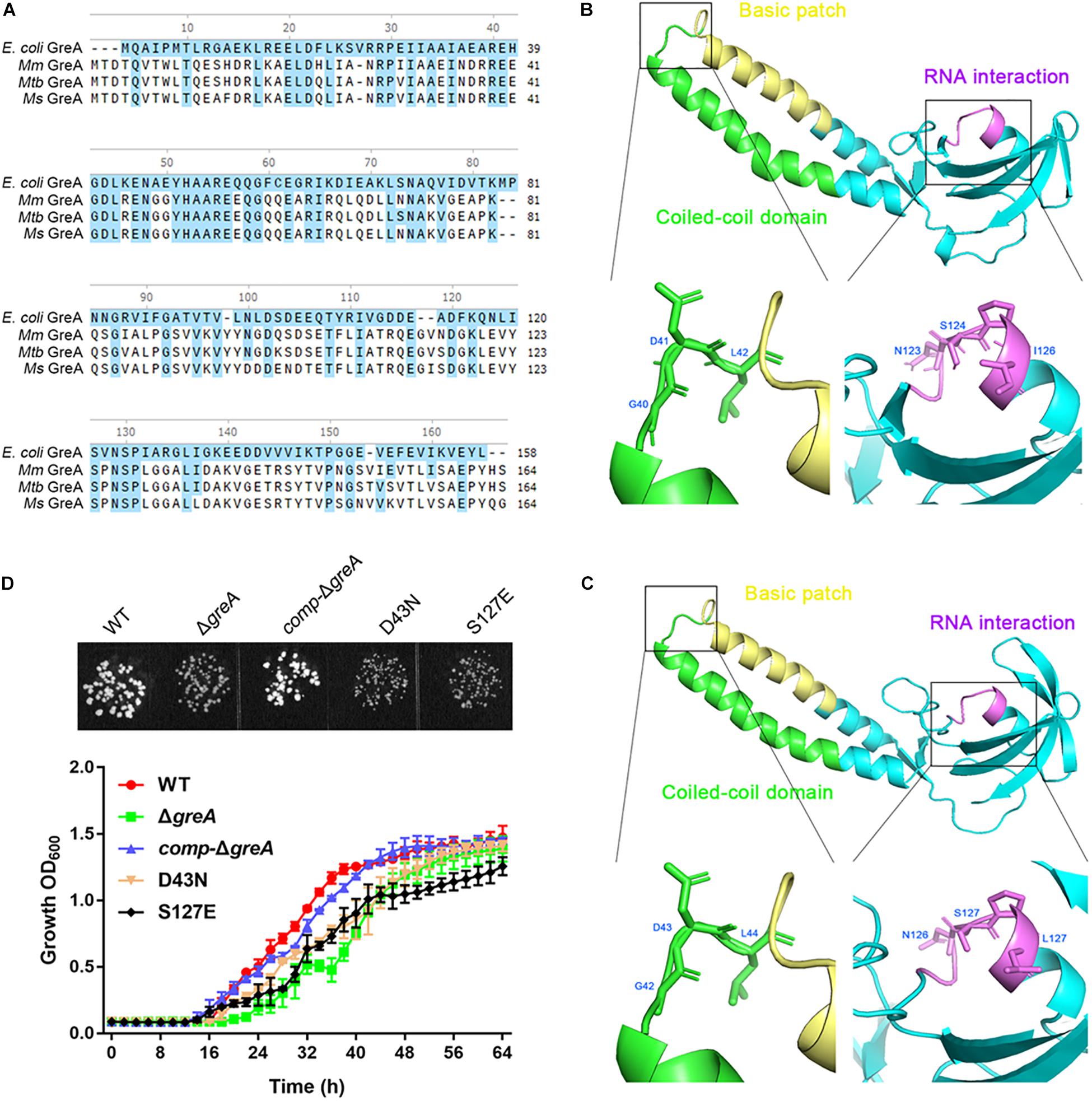
Figure 5. Optimal growth of M. smegmatis is required for the full activity of the GreA. (A) Multiple sequence alignment of E. coli GreA (158 aa) with M. smegmatis GreA (164 aa), M. tuberculosis GreA (164 aa), and M. marinum (165 aa). The conservative residues of E. coli GreA (B) and M. smegmatis GreA (C). PyMol is applied to generate the photographs of coiled-coil domain, basic patch, and RNAP interaction. (D) The growth of M. smegmatis strains harboring greA and/or its point mutants. Three biological replicates were tested. Data are shown as mean ± SEM of three independent experiments.
The glgX Gene Regulated by GreA Is Involved in the Optimal Growth of M. smegmatis
To extend our in silico finding, we performed genetic experiments to confirm the relationship between GreA and a candidate gene. We selected the candidate gene based on their down-regulation in the RNA-seq data, the enrichment in CHIP-seq data, and the potential role of their gene product in the growth phenotype. Taking these into consideration, we focused on the function of the glgX gene. To determine whether the reduced growth of M. smegmatis greA mutant strain was attributed to the decreased expression of glgX, we applied the mycobacterial CRISPRi for rapid validation. Initially, we introduced a plasmid encoding the gene of green fluorescent protein (GFP) fused with the glgX gene, including its native promoter, into both wild type and mutant M. smegmatis strains. Then we determined the mean fluorescence intensity of GFP by the flow cytometry. The mean fluorescence intensity of GFP was significantly decreased in ΔgreA strain compared to WT strain (Figure 6A), confirming that disruption of greA was responsible for the reduced expression of glgX. Moreover, the silencing efficiency was measured by semi-quantitative RT-PCR that showed a ∼80% decrease in the expression of the glgX gene (Figure 6B). As expected, the decrease in the expression of glgX, resulted in reduced growth compared to the strain without induction of ATc (Figures 6C,D), indicating that loss of greA has led to a partial defect of growth by down-regulating glgX.
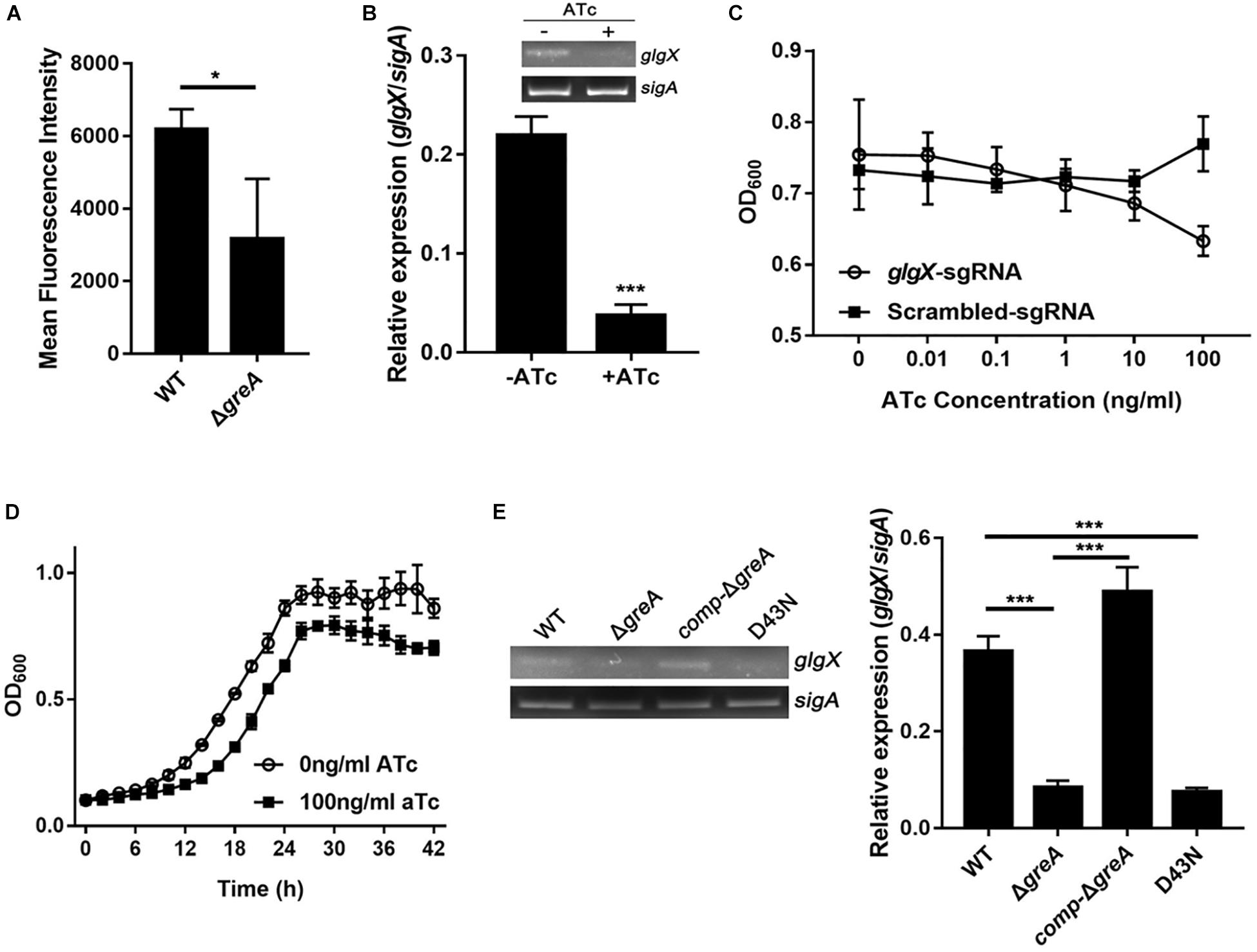
Figure 6. The glgX gene regulated by GreA is involved in the optimal growth of M. smegmatis. (A) Bacteria was collected at logarithmic phase. The mean fluorescence intensity of GFP in the M. smegmatis strains harboring a plasmid encoding gfp fusion with the glgX gene including its native promoter. (B) Expression of glgX was determined by semi-quantitative RT-PCR with or without induction of ATc. (C,D) In vitro growth of culture with or without induction of ATc. (E) Results of semi-quantitative RT-PCR of glgX in M. smegmatis strains harboring greA and/or its point mutants. (B,E) The gel images shown are representative of three repeat experiments. The PCRs of sigA served as controls for normalization of the cDNAs. The relative expression of glgX versus that of sigA was quantitated using ImageJ. The bar graphs show the means of two independent experiments. The error bars indicate the standard errors of the means (SEM). Three biological replicates were tested. *p ¡ 0.05, ***p < 0.001.
In E. coli, the residues D41 of the GreA protein are required to prevent the backtracking of paused complexes, thereby suppressing the transcriptional pauses (Vinella et al., 2012). Since the GreA protein of E. coli and M. smegmatis shares the residues mentioned above, we determined if the anti-backtracking activity of GreA was associated with the regulation glgX. To achieve this, the transcriptional expression of glgX was detected in M. smegmatis strains. Interestingly, transcriptional expression of glgX was fully restored in comp-ΔgreA strain. However, this has not been observed in the D43N strain and greA mutant strain (Figure 6E), indicating that the anti-backtracking activity of GreA is required for the transcriptional expression of the glgX gene.
Deletion of greA Attenuated the Intracellular Survivability
The persistence inside a host macrophage is a vital characteristic that distinguishes the pathogenic mycobacteria from non-pathogenic strains (Singh et al., 2008; Yaseen et al., 2015). RAW264.7 cells were infected with WT, ΔgreA or complemented strains of M. smegmatis. The numbers of the surviving intracellular bacteria were assessed after 24 and 48 h post-infection. A significant reduction in the number of viable intracellular mycobacteria was observed after 24 and 48 h in the ΔgreA strain (Figure 7A). Moreover, the survivability of H37Rv and ΔgreA strain was assessed by the detection of the bacterial CFUs inside the macrophage. We found that the survivability of greA mutant strain was significantly decreased in macrophages after 48 h when compared to the H37Rv strain (Figure 7C). In addition, the RAW264.7 cells were infected with WT, ΔgreA, or complemented strains of M. tuberculosis H37Ra. In this case, no significant difference between H37Ra and ΔgreA strain for intracellular survivability was observed at 48 h post-infection (Figure 7B). The findings lead us to suppose that this survival phenotype may be the result of the growth defect of M. tuberculosis lacking greA. It has been demonstrated that strain H37Ra is avirulent, and does not multiply in mice (Larson and Wicht, 1964). Thus, we believe that in vivo experiments may help to clarify this ambiguous phenomenon. To do this, mice were infected with WT, ΔgreA, or complemented strains of M. tuberculosis H37Ra. After 14-days of infection, we observed a decrease of bacterial burden in mice infected with the H37Ra greA mutant strain (Figure 7D).
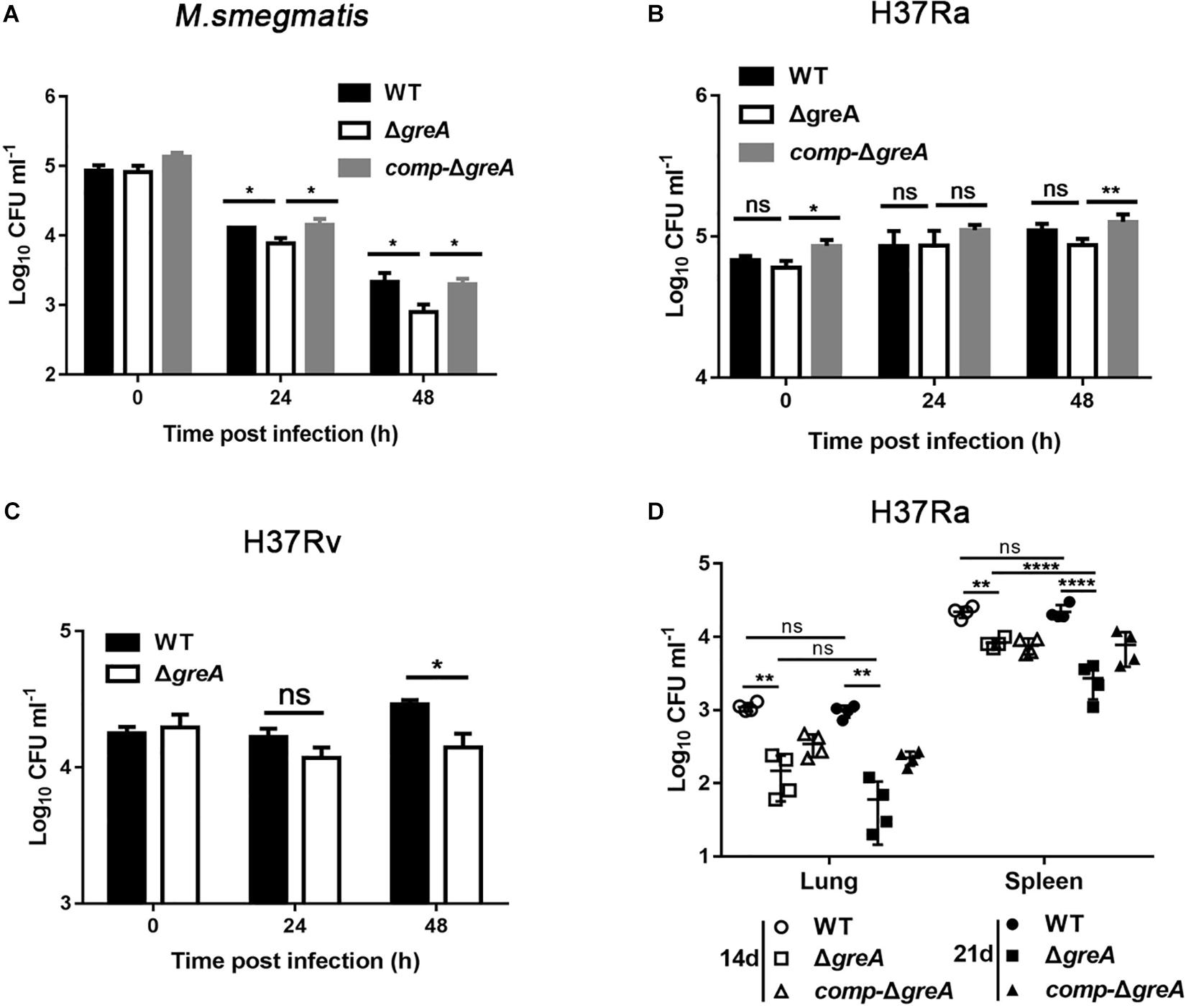
Figure 7. Intracellular fitness of ΔgreA mutant strain was evaluated in vivo and in vitro. RAW264.7 cells were seeded at 3 × 105 cells per well in 24-well culture plates. After adhesion, cells were infected with bacteria at MOI of 10. The extracellular bacteria were removed 2 h post infection by washing with PBS. The cells of M. smegmatis (A), M. tuberculosis H37Ra (B), and M. tuberculosis H37Rv (C) were further incubated for 24 and 48 h at 37°C. Then serially diluted (10-fold) cells lysates were spotted on a 7H10-OADC agar and cultured in 37°C. CFUs were counted after incubation for the indicated time for M. smegmatis and M. tuberculosis, respectively. (D) Mice were infected i.p. (1 × 107) with WT, ΔgreA, or comp-ΔgreA strains of M. tuberculosis H37Ra. The number of surviving bacilli by CFU counting at the indicated time. Data are shown as mean ± SEM of two independent experiments. *p < 0.05, **p < 0.01, ****p < 0.0001, ns, p ≥ 0.05.
Overall, these results emphasize that GreA facilitates the intracellular survival of mycobacteria, and thus revealed its critical role in both an in vitro and an in vivo infection model.
Discussion
GreA Is Essential for Sustaining Mycobacterial Growth and Environmental Adaptation
Few studies are available about the functional characteristics of GreA in Mycobacterium spp. A previous study showed that knocking down of greA resulted in growth retardation in M. tuberculosis H37Ra (China et al., 2011). For example, the absence of greA in S. pneumoniae severely perturbed its growth (Yuzenkova et al., 2014). Regarding the universal role of Gre factors in prokaryotes, it is not surprising that deletion of greA in M. tuberculosis resulted in growth retardation.
It has been convincingly shown in several studies that GreA is required for the survival of bacteria under stress. For instance, Wei and colleagues found that the deletion of greA in Sinorhizobium meliloti results in sensitivity to salt stress (Wei et al., 2004). Another study revealed that overexpression of GreA provides the host cells with enhanced resistance to heat shock and oxidative stress in E. coli (Li et al., 2012). A very recent study has shown that similar with E. coli GreA, M. smegmatis GreA exhibits chaperone-like activity in vitro, which prevents heat-induced aggregation of substrate protein (Joseph et al., 2019). Intriguingly, it has been established that double-deletion of greA and greB, encoding a transcript cleavage factors function-similar to that of GreA, causes heat sensitivity, indicating that another Gre factor may partially complement the phenotype of greA mutant. Our data shows that GreA is dispensable for the survival of M. tuberculosis under heat shock stress. It seems to be that the phenotype, in part, may be complemented by another copy of greA, since there are two genes (MSMEG_5263 and MSMEG_6292) encoding transcription elongation factor GreA in M. smegmatis (Mohan et al., 2015). Further experiments are required to address the role of MSMEG_6292 in M. smegmatis.
Moreover, the deletion of greA in M. smegmatis increased the susceptibility to bedaquiline, capreomycin, streptomycin, amikacin, and vancomycin (Table 1). Notably, our data has established that the growth defect of ΔgreA strain was more obvious despite the low antibiotic concentrations used. This finding is crucial, as it provides the evidence to support that a GreA inhibitor might be able to enhance the efficacy of the current anti-TB drugs.
Transcription Regulation by the M. smegmatis Transcription Elongation Factor GreA
Here, we showed that 167 genes were down-regulated in the M. smegmatis lacking the greA, while only 28 genes were positively regulated. Most of the down-regulated genes were associated with metabolism, supporting the view that GreA optimizes mycobacterial growth and environmental adaptation by the regulation of metabolism. For example, glgX, a gene involved in starch and sucrose metabolism, was significantly down-regulated in greA mutant strain and the inactivation of glgX in C. glutamicum led to slower growth (Seibold and Eikmanns, 2007). It is suggested that GreA mediated regulation of the growth might be partly associated with glgX. In addition, katG, a gene encoded catalase-peroxidase-peroxynitritase, plays a role in the intracellular survival of the mycobacteria within macrophages and in protection against reactive oxygen and nitrogen intermediates produced by phagocytic cells (Milano et al., 2001; Pym et al., 2002; Sassetti et al., 2003). Given the down-regulation of katG in the greA mutant strain, this could explain the role of GreA in mediating the regulation of oxidation resistance.
GreA has an impact on the regulation of gene expression in E. coli (Roberts et al., 2008) and S. pneumoniae (Yuzenkova et al., 2014) by affecting the transcription elongation. In addition to the effect of GreA on transcription elongation, competition between GreA and DksA has been reported. DksA, an RNAP-binding transcription factor, binds to the secondary channel of RNAP and plays a vital role in controlling polyP activation, which is required for stress survival and virulence in diverse pathogenic microbes (Gray, 2019).
Moreover, GreA and DksA, have an opposite regulation of a subset of genes in E. coli (Vinella et al., 2012). These data suggest that GreA might have a global role in the regulation of genes expressed in a prokaryotic organism. From our CHIP-seq dataset, it was revealed that GreA is symmetrically distributed on the M. smegmatis chromosome. However, we were unable to identify discrete binding sites for GreA. Of note, there is no evidence that the Gre factors can bind with nucleic acids. Indeed, the Gre factors are proteins interacting with the RNAPs, this might suggest that GreA indirectly regulates the gene expression by interaction with the RNAP complex.
GreA consists of two domains; a C-terminal globular domain and an extended N-terminal coiled-coil domain. The NTD is responsible for the GreA induction of nucleolytic activity, while the CTD determines the binding of GreA to RNAP. Both domains are required for full functional activity of GreA (Koulich et al., 1998). We also confirmed that the full activity of the GreA is crucial in M. smegmatis growth. Of note, it has been reported that, another Gre homolog, Rv3788 binds near the secondary channel of RNAP to inhibit transcription. However, the factor did not show any transcript cleavage stimulatory activity (China et al., 2012). Furthermore, our data demonstrated that glgX, a gene involved in starch and sucrose metabolism, regulated by the anti-backtracking activity of GreA is required for the optimal growth of M. smegmatis. Recently, studies in Salmonella enterica serovar Typhimurium demonstrated that GreA-mediated rescue of backtracked paused complexes during transcription is crucial in promoting hilD expression through its 3′-UTR (Gaviria-Cantin et al., 2017). In addition, GreA enhanced expression of several E. coli RNAP promoters in vitro (Erie et al., 1993; Feng et al., 1994; Hsu et al., 1995; Maddalena et al., 2016). Given that GreA has a multitude of effects on transcription, initiation, and elongation (Stepanova et al., 2007; Vinella et al., 2012), this could explain our failure to identify the binding motif of GreA. Based on the present study, we suppose that the anti-backtracking activity GreA is involved in the regulation of gene expression in mycobacteria. It would be interesting in future studies to examine the regulation of GreA on gene expression in vitro.
It is known that GreA influences not only cellular invasion but also replication in Francisella tularensis subsp. Novicida (Cui et al., 2018) and Salmonella typhimurium (Gaviria-Cantin et al., 2017). Our data also revealed that greA deletion caused a slight reduction of intracellular fitness in M. tuberculosis H37Rv, suggesting that GreA is required for the maintenance of intracellular survival of M. tuberculosis. The use of a non-pathogenic species, M. smegmatis, may have been a limitation of the present study. Although there is some evidence that M. smegmatis has been used as a model to study the function of interesting gene of M. tuberculosis (Wang et al., 2015, 2017; Yaseen et al., 2015, 2018), M. smegmatis isn’t closely related to M. tuberculosis in genetically. Very recently, based on the comprehensive phylogenomic analyses and comparative genomic studies, M. smegmatis categorized as a novel classification of Mycobacterium is distinguished as a different genus comparing to M. tuberculosis. (Gupta et al., 2019). It suggests that certain functions of GreA in pathogenic M. tuberculosis may be different from in M. smegmatis, especially with the virulence, since most experiments have been performed in M. smegmatis, instead of M. tuberculosis H37Rv. In a further study, we will provide evidence to elucidate the regulation role of GreA in pathogenic M. tuberculosis.
In conclusion, our study revealed that GreA plays an important role in mycobacterial biology where, the deletion of greA resulted in bacterial growth retardation, reduction of intracellular fitness, and more importantly from a clinical treatment point of view, improvement of antibiotic susceptibility. Overall, our results strongly suggest that GreA warrants being studied as a potential target for the development of MDR-TB inhibitor therapy.
Data Availability Statement
The datasets for this study can be found in GEO, under accession number GSE143764: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE143764.
Ethics Statement
All animal experiments were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals, and the experimental procedures were approved by the Ethics Committee of Zhongshan School of Medicine on Laboratory Animal Care (reference number: 2016-159), Sun Yat-sen University.
Author Contributions
GT conceived the study and supervised global data analysis. HC and AR Provided advice in design and co-edited manuscript. SF and YL Designed and performed the experiments, analyzed the data, and co-wrote the manuscript. WL and ME-S performed the construction of deletion of Gre in mycobacteria. ZhZ and CS performed the intracellular fitness experiment. ZiZ and ZG designed and performed the RNAseq experiment. LuL and LiL performed the ChIPseq experiment. YY and XW contributed to data interpretation and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work has been supported by grants from National Natural Science Foundation of China (Grant Numbers 81830103 and 81722030), the Guangdong Natural Science Foundation (Grant Number 2017A030306012), National Science and Technology Key Projects for Major Infectious Diseases (2017ZX10302301), and the Science and Technology Planning Project of Guangdong (Grant Number 2016A020219002).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Yaxin Chen of Sun Yat-sen University for data analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00413/full#supplementary-material
FIGURE S1 | Generation of greA gene replacement mutant. Schematic representation of the strategy used for the generation of greA gene replacement mutant and confirmation of greA disruption in M. smegmatis (A), M. tuberculosis H37Rv (B), and M. tuberculosis H37Ra (C), respectively.
FIGURE S2 | Deletion of greA altered susceptibility of antibiotics in M. smegmatis. WT, ΔgreA and comp-ΔgreA strains were exposed to 7H9 medium in the absent (control) or present of different antibiotics at 1/100th the MIC. The initial OD600 was 0.02. The growth curves were monitored for 42 h. (A) Streptomycin (MIC = 0.005 mg/L) and (B) Rifampicin (MIC = 0.16 mg/L).
FIGURE S3 | Analysis of binding sites identified from the ChIP-seq experiments. (A) Histogram showing the frequency distribution of distances between GreA ChIP-seq peak centers and TSSs. (B) Chromosomal binding map of HupB-FLAG.
TABLE S1 | All primers and strains used in this study.
TABLE S2 | Transcriptome of the M. smegmatis wild type and the ΔgreA mutant.
TABLE S3 | ChIP-seq of GreA: ChIP-seq binding sites.
TABLE S4 | Results of gene ontology analyses (DAVID; FDR < 0.05).
Footnotes
- ^ https://www.ncbi.nlm.nih.gov/genbank/
- ^ https://swissmodel.expasy.org/repository/uniprot/A0R2X1
- ^ https://david.ncifcrf.gov
References
Alland, D., Kramnik, I., Weisbrod, T. R., Otsubo, L., Cerny, R., Miller, L. P., et al. (1998). Identification of differentially expressed mRNA in prokaryotic organisms by customized amplification libraries (DECAL): the effect of isoniazid on gene expression in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U S A. 95, 13227–13232. doi: 10.1073/pnas.95.22.13227
China, A., Mishra, S., and Nagaraja, V. (2011). A transcript cleavage factor of Mycobacterium tuberculosis important for its survival. PLoS One 6:e21941. doi: 10.1371/journal.pone.0021941
China, A., Mishra, S., Tare, P., and Nagaraja, V. (2012). Inhibition of Mycobacterium tuberculosis RNA polymerase by binding of a Gre factor homolog to the secondary channel. J. Bacteriol. 194, 1009–1017. doi: 10.1128/JB.06128-11
Cui, G. L., Wang, J., Qi, X. Y., and Su, J. L. (2018). Transcription elongation factor GreA plays a key role in cellular invasion and virulence of Francisella tularensis subsp novicida. Sci. Rep. 8:6895. doi: 10.1038/s41598-018-25271-5
Erie, D. A., Hajiseyedjavadi, O., Young, M. C., and von Hippel, P. H. (1993). Multiple RNA polymerase conformations and GreA: control of the fidelity of transcription. Science 262, 867–873. doi: 10.1126/science.8235608
Feng, G. H., Lee, D. N., Wang, D., Chan, C. L., and Landick, R. (1994). GreA-induced transcript cleavage in transcription complexes containing Escherichia coli RNA polymerase is controlled by multiple factors, including nascent transcript location and structure. J. Biol. Chem. 269, 22282–22294.
Gaviria-Cantin, T., El Mouali, Y., Le Guyon, S., Romling, U., and Balsalobre, C. (2017). Gre factors-mediated control of hilD transcription is essential for the invasion of epithelial cells by Salmonella enterica serovar Typhimurium. PLoS Pathog. 13:e1006312. doi: 10.1371/journal.ppat.1006312
Goodsmith, N., Guo, X. V., Vandal, O. H., Vaubourgeix, J., Wang, R., Botella, H., et al. (2015). Disruption of an M. tuberculosis membrane protein causes a magnesium-dependent cell division defect and failure to persist in mice. PLoS Pathog. 11:e1004645. doi: 10.1371/journal.ppat.1004645
Gray, M. J. (2019). Inorganic polyphosphate accumulation in Escherichia coli is regulated by DksA but not by (p)ppGpp. J. Bacteriol. 201, e664–18. doi: 10.1128/JB.00664-18
Gumber, S., Taylor, D. L., Marsh, I. B., and Whittington, R. J. (2009). Growth pattern and partial proteome of Mycobacterium avium subsp. paratuberculosis during the stress response to hypoxia and nutrient starvation. Vet. Microbiol. 133, 344–357. doi: 10.1016/j.vetmic.2008.07.021
Gupta, R. S., Lo, B., and Son, J. (2019). Phylogenomics and comparative genomic studies robustly support division of the genus Mycobacterium into an emended genus Mycobacterium and four novel genera. Front. Microbiol. 13:67. doi: 10.3389/fmicb.2018.00067
Harding, E. (2019). WHO global progress report on tuberculosis elimination. Lancet Respir. Med. 8:19. doi: 10.1016/s2213-2600(19)30418-7
Hsu, L. M., Vo, N. V., and Chamberlin, M. J. (1995). Escherichia coli transcript cleavage factors GreA and GreB stimulate promoter escape and gene expression in vivo and in vitro. Proc. Natl. Acad. Sci. U S A. 92, 11588–11592. doi: 10.1073/pnas.92.25.11588
Jain, P., Hsu, T., Arai, M., Biermann, K., Thaler, D. S., Nguyen, A., et al. (2014). Specialized transduction designed for precise high-throughput unmarked deletions in Mycobacterium tuberculosis. MBio 5, e1245–14. doi: 10.1128/mBio.01245-14
Joseph, A., Nagaraja, V., and Natesh, R. (2019). Mycobacterial transcript cleavage factor Gre, exhibits chaperone-like activity. Biochim. Biophys. Acta. Proteins. Proteom. 1867, 757–764. doi: 10.1016/j.bbapap.2019.05.008
Komissarova, N., and Kashlev, M. (1997). Transcriptional arrest: Escherichia coli RNA polymerase translocates backward, leaving the 3′ end of the RNA intact and extruded. Proc. Natl. Acad. Sci. U S A. 94, 1755–1760. doi: 10.1073/pnas.94.5.1755
Koulich, D., Nikiforov, V., and Borukhov, S. (1998). Distinct functions of N and C-terminal domains of GreA, an Escherichia coli transcript cleavage factor. J. Mol. Biol. 276, 379–389. doi: 10.1006/jmbi.1997.1545
Koulich, D., Orlova, M., Malhotra, A., Sali, A., Darst, S. A., and Borukhov, S. (1997). Domain organization of Escherichia coli transcript cleavage factors GreA and GreB. J. Biol. Chem. 272, 7201–7210. doi: 10.1074/jbc.272.11.7201
Kumar, A., Saini, V., Kumar, A., Kaur, J., and Kaur, J. (2017). Modulation of trehalose dimycolate and immune system by Rv0774c protein enhanced the intracellular survival of Mycobacterium smegmatis in human macrophages cell line. Front. Cell Infect. Microbiol. 7:289. doi: 10.3389/fcimb.2017.00289
Larson, C. L., and Wicht, W. C. (1964). Infection of mice with Mycobacterium tuberculosis, strain H37Ra. Am. Rev. Respir. Dis. 90, 742–748.
Lata, M., Sharma, D., Kumar, B., Deo, N., Tiwari, P. K., Bisht, D., et al. (2015). Proteome analysis of ofloxacin and moxifloxacin induced mycobacterium tuberculosis isolates by proteomic approach. Protein. Pept. Lett. 22, 362–371. doi: 10.2174/0929866522666150209113708
Li, J., Overall, C. C., Johnson, R. C., Jones, M. B., McDermott, J. E., Heffron, F., et al. (2015). ChIP-Seq analysis of the sigmaE regulon of Salmonella enterica Serovar Typhimurium reveals new genes implicated in heat shock and oxidative stress response. PLoS One 10:e0138466. doi: 10.1371/journal.pone.0138466
Li, K., Jiang, T., Yu, B., Wang, L., Gao, C., Ma, C., et al. (2012). Transcription elongation factor GreA has functional chaperone activity. PLoS One 7:e47521. doi: 10.1371/journal.pone.0047521
Maddalena, L. L., Niederholtmeyer, H., Turtola, M., Swank, Z. N., Belogurov, G. A., and Maerkl, S. J. (2016). GreA and GreB enhance expression of Escherichia coli RNA Polymerase promoters in a reconstituted transcription-translation system. ACS Synth. Biol. 5, 929–935. doi: 10.1021/acssynbio.6b00017
Manganelli, R., Dubnau, E., Tyagi, S., Kramer, F. R., and Smith, I. (1999). Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol. Microbiol. 31, 715–724. doi: 10.1046/j.1365-2958.1999.01212.x
Milano, A., Forti, F., Sala, C., Riccardi, G., and Ghisotti, D. (2001). Transcriptional regulation of furA and katG upon oxidative stress in Mycobacterium smegmatis. J. Bacteriol. 183, 6801–6806. doi: 10.1128/jb.183.23.6801-6806.2001
Mohan, A., Padiadpu, J., Baloni, P., and Chandra, N. (2015). Complete genome sequences of a Mycobacterium smegmatis laboratory strain (MC2 155) and isoniazid-resistant (4XR1/R2) mutant strains. Genome Announc. 3, e01520–14. doi: 10.1128/genomeA.01520-14
Nogales, J., Campos, R., BenAbdelkhalek, H., Olivares, J., Lluch, C., and Sanjuan, J. (2002). Rhizobium tropici genes involved in free-living salt tolerance are required for the establishment of efficient nitrogen-fixing symbiosis with Phaseolus vulgaris. Mol. Plant Microbe. Interact. 15, 225–232. doi: 10.1094/mpmi.2002.15.3.225
Pym, A. S., Saint-Joanis, B., and Cole, S. T. (2002). Effect of katG mutations on the virulence of Mycobacterium tuberculosis and the implication for transmission in humans. Infect. Immun. 70, 4955–4960. doi: 10.1128/iai.70.9.4955-4960.2002
Roberts, J. W., Shankar, S., and Filter, J. J. (2008). RNA polymerase elongation factors. Annu. Rev. Microbiol. 62, 211–233. doi: 10.1146/annurev.micro.61.080706.093422
Rustad, T. R., Roberts, D. M., Liao, R. P., and Sherman, D. R. (2009). Isolation of mycobacterial RNA. Methods Mol. Biol. 465, 13–21. doi: 10.1007/978-1-59745-207-6_2
Sassetti, C. M., Boyd, D. H., and Rubin, E. J. (2003). Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48, 77–84. doi: 10.1046/j.1365-2958.2003.03425.x
Schneider, D., Bruton, C. J., and Chater, K. F. (2000). Duplicated gene clusters suggest an interplay of glycogen and trehalose metabolism during sequential stages of aerial mycelium development in Streptomyces coelicolor A3(2). Mol. Gen. Genet. 263, 543–553. doi: 10.1007/s004380051200
Seibold, G. M., and Eikmanns, B. J. (2007). The glgX gene product of Corynebacterium glutamicum is required for glycogen degradation and for fast adaptation to hyperosmotic stress. Microbiology 153, 2212–2220. doi: 10.1099/mic.0.2006/005181-0
Sharma, P., Kumar, B., Singhal, N., Katoch, V. M., Venkatesan, K., Chauhan, D. S., et al. (2010). Streptomycin induced protein expression analysis in Mycobacterium tuberculosis by two-dimensional gel electrophoresis & mass spectrometry. Indian J. Med. Res. 132, 400–408.
Sharma, U. K., and Chatterji, D. (2010). Transcriptional switching in Escherichia coli during stress and starvation by modulation of sigma activity. FEMS Microbiol. Rev. 34, 646–657. doi: 10.1111/j.1574-6976.2010.00223.x
Singh, A. K., Carette, X., Potluri, L. P., Sharp, J. D., Xu, R., Prisic, S., et al. (2016). Investigating essential gene function in Mycobacterium tuberculosis using an efficient CRISPR interference system. Nucl. Acid. Res. 44:e143. doi: 10.1093/nar/gkw625
Singh, P. P., Parra, M., Cadieux, N., and Brennan, M. J. (2008). A comparative study of host response to three Mycobacterium tuberculosis PE_PGRS proteins. Microbiology 154(Pt 11), 3469–3479. doi: 10.1099/mic.0.2008/019968-0
Sivaramakrishnan, P., Sepulveda, L. A., Halliday, J. A., Liu, J., Nunez, M. A. B., Golding, I., et al. (2017). The transcription fidelity factor GreA impedes DNA break repair. Nature 550, 214–218. doi: 10.1038/nature23907
Stebbins, C. E., Borukhov, S., Orlova, M., Polyakov, A., Goldfarb, A., and Darst, S. A. (1995). Crystal structure of the GreA transcript cleavage factor from Escherichia coli. Nature 373, 636–640. doi: 10.1038/373636a0
Stepanova, E., Lee, J., Ozerova, M., Semenova, E., Datsenko, K., Wanner, B. L., et al. (2007). Analysis of promoter targets for Escherichia coli transcription elongation factor GreA in vivo and in vitro. J. Bacteriol. 189, 8772–8785. doi: 10.1128/jb.00911-07
Tehranchi, A. K., Blankschien, M. D., Zhang, Y., Halliday, J. A., Srivatsan, A., Peng, J., et al. (2010). The transcription factor DksA prevents conflicts between DNA replication and transcription machinery. Cell 141, 595–605. doi: 10.1016/j.cell.2010.03.036
Traverse, C. C., and Ochman, H. (2018). A genome-wide assay specifies only GreA as a transcription fidelity factor in Escherichia coli. G3 (Bethesda) 8, 2257–2264. doi: 10.1534/g3.118.200209
van Kessel, J. C., and Hatfull, G. F. (2007). Recombineering in Mycobacterium tuberculosis. Nat. Methods 4, 147–152. doi: 10.1038/nmeth996
Vinella, D., Potrykus, K., Murphy, H., and Cashel, M. (2012). Effects on growth by changes of the balance between GreA, GreB, and DksA suggest mutual competition and functional redundancy in Escherichia coli. J. Bacteriol. 194, 261–273. doi: 10.1128/JB.06238-11
Volker, U., Engelmann, S., Maul, B., Riethdorf, S., Volker, A., Schmid, R., et al. (1994). Analysis of the induction of general stress proteins of Bacillus subtilis. Microbiology 140(Pt 4), 741–752. doi: 10.1099/00221287-140-4-741
Wang, J., Ge, P. P., Qiang, L. H., Tian, F., Zhao, D. D., Chai, Q. Y., et al. (2017). The mycobacterial phosphatase PtpA regulates the expression of host genes and promotes cell proliferation. Nat. Commun. 8:244. doi: 10.1038/s41467-017-00279-z
Wang, J., Li, B. X., Ge, P. P., Li, J., Wang, Q., Gao, G. F., et al. (2015). Mycobacterium tuberculosis suppresses innate immunity by coopting the host ubiquitin system. Nat. Immunol. 16, 237–45. doi: 10.1038/ni.3096
Wei, W., Jiang, J., Li, X., Wang, L., and Yang, S. S. (2004). Isolation of salt-sensitive mutants from Sinorhizobium meliloti and characterization of genes involved in salt tolerance. Lett. Appl. Microbiol. 39, 278–283. doi: 10.1111/j.1472-765x.2004.01577.x
Wright, A., Zignol, M., Van Deun, A., Falzon, D., Gerdes, S. R., Feldman, K., et al. (2009). Epidemiology of antituberculosis drug resistance 2002-07: an updated analysis of the Global Project on Anti-Tuberculosis Drug Resistance Surveillance. Lancet 373, 1861–1873. doi: 10.1016/S0140-6736(09)60331-7
Yang, C., Luo, T., Shen, X., Wu, J., Gan, M., Xu, P., et al. (2017). Transmission of multidrug-resistant Mycobacterium tuberculosis in Shanghai, China: a retrospective observational study using whole-genome sequencing and epidemiological investigation. Lancet Infect. Dis. 17, 275–284. doi: 10.1016/S1473-3099(16)30418-2
Yaseen, I., Choudhury, M., Sritharan, M., and Khosla, S. (2018). Histone methyltransferase SUV39H1 participates in host defense by methylating mycobacterial histone-like protein HupB. EMBO J. 37, 183–200. doi: 10.15252/embj.201796918
Yaseen, I., Kaur, P., Nandicoori, V. K., and Khosla, S. (2015). Mycobacteria modulate host epigenetic machinery by Rv1988 methylation of a non-tail arginine of histone H3. Nat. Commun. 6:8922. doi: 10.1038/ncomms9922
Keywords: greA, Mycobacterium tuberculosis, Mycobacterium smegmatis, bacterial fitness, antibiotic susceptibility
Citation: Feng S, Liu Y, Liang W, El-Sayed Ahmed MAE-G, Zhao Z, Shen C, Roberts AP, Liang L, Liao L, Zhong Z, Guo Z, Yang Y, Wen X, Chen H and Tian G (2020) Involvement of Transcription Elongation Factor GreA in Mycobacterium Viability, Antibiotic Susceptibility, and Intracellular Fitness. Front. Microbiol. 11:413. doi: 10.3389/fmicb.2020.00413
Received: 02 January 2020; Accepted: 27 February 2020;
Published: 23 March 2020.
Edited by:
Yossef Av-Gay, The University of British Columbia, CanadaReviewed by:
Jianping Xie, Southwest University, ChinaHiroyuki Yamada, Japan Anti-tuberculosis Association, Japan
Copyright © 2020 Feng, Liu, Liang, El-Sayed Ahmed, Zhao, Shen, Roberts, Liang, Liao, Zhong, Guo, Yang, Wen, Chen and Tian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongtao Chen, MTM5MjY5MjU5NDFAMTM5LmNvbQ==; Guo-Bao Tian, dGlhbmdiQG1haWwuc3lzdS5lZHUuY24=
†These authors have contributed equally to this work
‡These authors share senior authorship
 Siyuan Feng
Siyuan Feng Yan Liu
Yan Liu Wanfei Liang
Wanfei Liang Mohamed Abd El-Gawad El-Sayed Ahmed
Mohamed Abd El-Gawad El-Sayed Ahmed Zihan Zhao
Zihan Zhao Cong Shen
Cong Shen Adam P. Roberts
Adam P. Roberts Lujie Liang
Lujie Liang Liya Liao3
Liya Liao3 Zhijuan Zhong
Zhijuan Zhong Zhaowang Guo
Zhaowang Guo Yongqiang Yang
Yongqiang Yang Xin Wen
Xin Wen Hongtao Chen
Hongtao Chen Guo-bao Tian
Guo-bao Tian