
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol., 28 February 2020
Sec. Infectious Agents and Disease
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.00321
This article is part of the Research TopicRole Of Small Colony Variants (SCVs) And Persisters In Staphylococcus aureus Pathogenesis And Development Of Therapeutic ApproachesView all 9 articles
Staphylococcus aureus remains a great burden on the healthcare system. Despite prescribed treatments often seemingly to be successful, S. aureus can survive and cause a relapsing infection which cannot be cleared. These infections are in part due to quasi-dormant sub-population which is tolerant to antibiotics and able to evade the host immune response. These include Small Colony Variants (SCVs). Because SCVs readily revert to non-SCV cell types under laboratory conditions, the characterization of SCVs has been problematic. This mini-review covers the phenotypic and genetic changes in stable SCVs including the selection of SCVs by and interactions with other bacterial species.
Staphylococcus aureus is a Gram positive, facultative anaerobe and non-motile opportunistic bacterium. Approximately 30% of the human population are asymptomatic carriers and 60% are intermittent carriers. The anterior nares serve as the major reservoir and thereby becomes a source for systemic infections (Tong et al., 2015) as S. aureus is also an important human pathogen responsible for morbidity and mortality worldwide (Tong et al., 2015; Kahl et al., 2016). These infections can be difficult to clear due to a persistent reservoir of S. aureus which survives antibiotic treatment.
Even within a genetically clonal population, it is now recognized that there exists a variety of phenotypes which can be referred to as alternative lifestyles (Balaban et al., 2004; Wood et al., 2013). In contrast to classical resistance mechanisms, these phenotypes confer tolerance to antimicrobials, where there is limited or no growth, but not cell death (Keren et al., 2004). The alternative lifestyles arise through disruptions to cellular activities and not through the acquisition of new virulence genes (Balaban et al., 2004; Wood et al., 2013). These lifestyles involve forming quasi-dormant sub-populations during infection which have increased fitness in unfavorable conditions. While the mechanisms may differ, these sub-populations generally create a reservoir of S. aureus within an anatomical niche which are difficult to clear by the host immune response or therapeutic means and can revert to their parental, active cell type (Keren et al., 2004; Singh et al., 2009). These phenotypic switches in S. aureus include formation of biofilms, persister cells and Small Colony Variants (SCVs).
Staphylococcus aureus SCVs are characterized by small colony size, impeded growth, loss of carotenoid pigment (Proctor et al., 2006; Melter and Radojevič, 2010) non-hemolytic and coagulase negative (Thomas, 1955; Quie, 1969; Melter and Radojevič, 2010; Bui et al., 2015). SCVs exhibit changes in structural morphology and dysfunction in cell separation (Kahl et al., 2003a). While they have been extensively studied there remain unknown nuances to their development and reversion. It is known that they form through auxotrophy in elements of the electron transport chain and ATP production (Kahl et al., 2003a; Proctor et al., 2006). These are either single or a combination of auxotrophy in the biosynthesis of menadione, hemin or thymidine (Kahl et al., 2003a; Kohler et al., 2008; Melter and Radojevič, 2010; Maduka-Ezeh et al., 2012; Dean et al., 2014; Horiuchi et al., 2015) CO2 (Thomas, 1955) and fatty acids (Schleimer et al., 2018). Many SCV isolates have no defined auxotrophism (Edwards, 2012) and various mutations in the electron transport chain that result in SCV (Proctor, 2019) are not observed in clinical isolates.
Although complex, reduced ATP production is associated with variations in cell-wall biosynthesis and carotenoid pigment (Proctor et al., 2006, 2014). Global genetic change has been observed in the switch to SCVs, with increased biofilm formation (Singh et al., 2010), autolysis (Bui et al., 2015), expression of adhesive proteins (Mirani et al., 2015) and decreased expression of secreted virulence factors (Melter and Radojevič, 2010; Tuchscherr et al., 2010; Ou et al., 2016).
Small Colony Variants persist intracellularly, avoiding clearance by the host immune response (Melter and Radojevič, 2010; Kahl et al., 2016). Examples of SCV infections include the lungs in patients with cystic fibrosis (Kahl et al., 1998, 2003b; Schwerdt et al., 2018), bovine mastitis (Atalla et al., 2011), osteomyelitis (Proctor et al., 1995), and foreign body infections (Baddour and Christensen, 1987; Von Eiff et al., 1999) including prosthetic-periprosthetic joint infection (PJI) (Yang et al., 2018). SCVs have an innate tolerance of antibiotics which is not associated with resistance genes (Edwards, 2012). The lack of an electrochemical gradient in their cell wall prevents aminoglycosides from entering the cell and the growth dormancy reduces the effectiveness of antibiotics that target metabolic processes of actively growing cells (Proctor et al., 1998; Melter and Radojevič, 2010; Garcia et al., 2013). This allows selection of SCV and chronic infection in antibiotic treatment, such as the use of gentamicin in bone cement for prosthetic joint implants (Chang et al., 2013). SCVs also have greater intracellular uptake and survival within non-professional phagocytic cells (fibroblast, epithelial, endothelial, osteoblast, osteocytes, keratinocytes) and professional phagocytes (Garzoni and Kelley, 2009). In vitro cell culture models of non-professional phagocytic cells shows a dependence on fibronectin binding proteins (FbNPs) for adhesion and activating cytoskeletal remodeling (Sendi and Proctor, 2009). Many surface adhesion molecules are upregulated in SCVs (Gómez-González et al., 2010; Bui and Kidd, 2015). Down regulation of α-toxin is required to keep the host cell alive and allow persistence intracellularly (Garzoni and Kelley, 2009; Sendi and Proctor, 2009).
Clinically isolated SCVs are often not stable and revert to their non-SCV cell type when cultured in the laboratory (Edwards, 2012; Kriegeskorte et al., 2014a; Bui et al., 2015). Also, clinical isolates of SCVs from diseased tissues give no indication of the parental type, leaving questions as to the genetic and molecular mechanisms involved during the transition from the original cell to its SCV phenotype. While SCVs can be abundant in persistent infections (Bates et al., 2003), identifying and culturing clinical samples is indeed difficult. The use of genetically stable SCVs (sSCV) has greatly improved our understanding of SCVs (Kriegeskorte et al., 2014a; Bui et al., 2015). Models of sSCV S. aureus include mutants auxotrophic to menadione (Schaaff et al., 2003; Lannergård et al., 2008; Dean et al., 2014; Pader et al., 2014), hemin (Balwit et al., 1994; Von Eiff et al., 1997; Schaaff et al., 2003), thymidine (Balwit et al., 1994; Von Eiff et al., 1997; Besier et al., 2007; Chatterjee et al., 2008; Kriegeskorte et al., 2014a; Kittinger et al., 2019), fatty acids (Bazaid et al., 2018; Schleimer et al., 2018) CO2 (Thomas, 1955; Gómez-González et al., 2010), chorismite synthesis (precursor for aromatic amino acids and menaquinone biosynthesis) (Zhang et al., 2017), selection in gentamicin (Balwit et al., 1994) and serial passage in mice models immunized against capsular polysaccharide (Tuchscherr et al., 2008).
Alternative methods of inducing the transition to SCV and formation of sSCV have been reported. Bui et al., grew the clinical blood isolate, WCH-SK2 for a prolonged time-period by continuous culture under nutrient limiting conditions and a low growth rate using an in vitro system allows single parameters, outside the complexity of in vivo conditions (Bui et al., 2015). Introduced oxidative stress and growth over 209 generations (60 days) produced a sSCV which dominated the population. Accumulated oxidative stress causes damage to DNA, and the SOS response creates mismatch repairs and increased rate of mutation. Previous studies have shown the frequency of SCV formation increases with the rate of mutation (Schaaff et al., 2003; Vestergaard et al., 2015; Lacoma et al., 2019).
This methodology has been replicated to select for sSCV in other S. aureus strains; such as MW2, community acquired MRSA blood isolate (Lee et al., unpublished). Unlike sSCV created in the laboratory (by site directed mutations of specific genes), continuous culture considers the phenotypic and genetic responses in a time-dependent manner within nutrient limiting conditions. This enables one strain to be observed in transition from a parental population to one with a diversity of cell types and then dominated by SCVs.
Whole genome sequencing of WCH-SK2 and WCH-SK2-SCV revealed 24 genetic events; single nucleotide polymorphisms (SNPs) or insertions-deletions (indels) which could be implicated with the switch to a sSCV while under nutrient starved, stressed conditions (Bui et al., 2015; Table 1). The instability of some clinical SCV isolates suggests their phenotype is transcriptionally controlled (and/or post-transcription) rather than by stable SNPs. The in vivo environment with combined stressors likely pressures S. aureus to become intracellular and remain as a SCV.
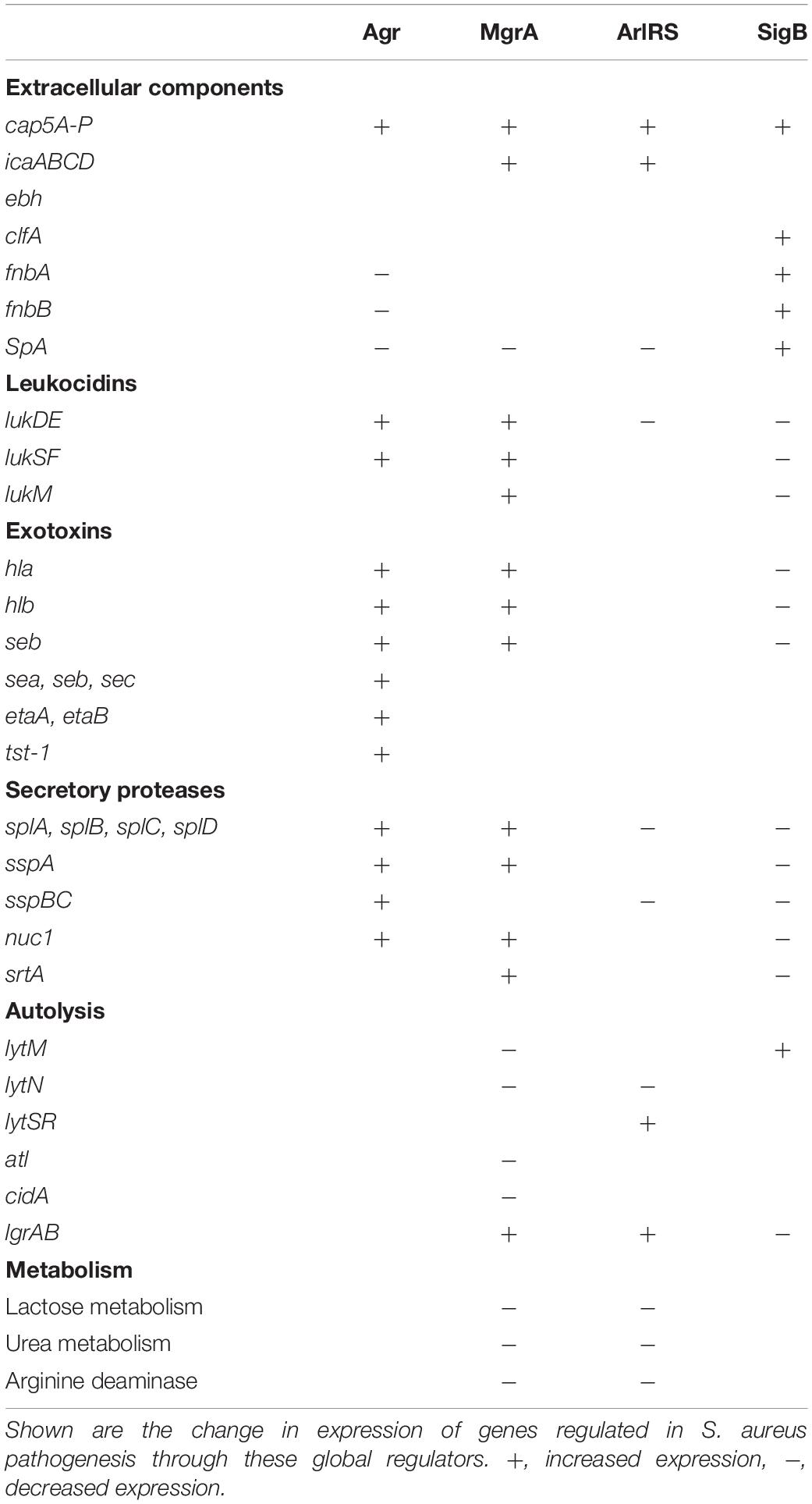
Table 1. The switch to a Small Colony Variant (SCV) by WCH-SK2 through continuous culture was associated with genetic events (SNPs) in global regulators of virulence.
The transition of WCH-SK2 to an sSCV was suggested to be associated with a SNP in the DNA binding domain of mgrA (R92C change in MgrA). This mutation could impact the binding kinetics (protein-DNA interaction); previous mutations studied in MgrA suggests mutations at this point would make MgrA non-functional. The loss of mgrA function has not been previously reported in any SCV clinical isolates, but present a new perspective for sSCVs. The global regulator MgrA is known to function downstream to the two-component ArlRS (Crosby et al., 2016; Kwiecinski et al., 2019) and controls genes including the upregulation of capsular polysaccharide, α-toxin, leukocidins, coagulase and protein A (Luong et al., 2006; Lei et al., 2019), all of which are virulence factors downregulated in SCVs (Proctor et al., 2014; Bui and Kidd, 2015). The loss of mgrA enhances autolysis (Ingavale et al., 2003, 2005), invasion of HeLa cells (Lei et al., 2019), increases biofilm formation (Trotonda et al., 2008; Crosby et al., 2016) and increases expression of microbial surface components recognizing adhesive matrix molecules (MSCRAMM) such as Ebh, a large 1.1-MDa protein (Crosby et al., 2016) and indeed other surface proteins (Kwiecinski et al., 2019). While the loss of function of MgrA has not been characterized in the context of formation or stability of SCVs, the downregulation of other homologous SarA family proteins have been associated with the formation of SCV (Kahl et al., 2005; Kriegeskorte et al., 2014b; Mirani et al., 2015).
Another major determinant for the production of toxins and extracellular enzymes is the quorum sensing two component system (TCS), Agr. Quorum sensing is controlled by secretion of the agr inducing peptide (AIP) to upregulate agr expression in surrounding S. aureus. The downregulation or loss of agr has previously been reported in clinical SCVs (Vaudaux et al., 2002; Kahl et al., 2005; Moisan et al., 2006; Kriegeskorte et al., 2014b). RNAIII is the effector molecule of the Agr TCS and controls the upregulation of secreted proteins and toxins and downregulation of cell surface proteins (Rescei et al., 1986; Abdelnour et al., 1993; Arvidson and Tegmark, 2001; Novick, 2003; Cheung et al., 2004). SCVs can display changes in RNAIII in persistent infections through the RNA degrasome, production of small RNAs and toxin-antitoxins (Proctor et al., 2014).
In stressful environments, S. aureus employs alternative sigma factor B (SigB) to sense changes in the environment and alter its gene expression profile accordingly. The SigB expression profile acts in opposition to the Agr TCS where there is an increased expression of cell surface proteins for colonization and decreased expression of secreted proteins and toxins (Kullik et al., 1998; Bischoff et al., 2004; Jonsson et al., 2004). RsbU positively regulates sigB in a growth phase dependent manner, with SigB expression highest during late exponential phase (Senn et al., 2005). SigB can regulate the switch to dormancy in S. aureus in opposition to agr and its regulation of virulence. High levels of SigB have been found in clinical SCVs isolated from cystic fibrosis (Moisan et al., 2006; Mitchell et al., 2008, 2013), osteomyelitis (Tuchscherr et al., 2017) and bovine mastitis (Mitchell et al., 2010a). SigB activity is associated with downregulation of the agr system (Yarwood and Schlievert, 2003) and immunogenic virulence factors (enterotoxins, hemolysins, secreted proteases) and shown to allow SCVs to persist intracellularly within human endothelium (Tuchscherr et al., 2015). This also is associated with the increase in expression of FnBPs such as FnbA which then contributes to biofilm formation, adhesion and intracellular persistence. Furthermore, sigB expression is required for intracellular replication of SCVs, and was shown to confer greater fitness in a pulmonary mouse model (Mitchell et al., 2013). SigB and subsequent agr repression is required for formation of SCVs in response to aminoglycoside stress (Mitchell et al., 2010a).
The diverse population of bacteria within the human microbiome means S. aureus is rarely in isolation during infection or commensal carriage. The specific nature of the microbial population within a niche creates a vast array of complex interactions between S. aureus and other bacterial species and this affects its ability to colonize, acquire nutrients and proliferate. The local bacteria are known to impact on S. aureus cell types. One such interaction is a co-culture of S. aureus and P. aeruginosa which has been found to select for SCV or persisters of S. aureus. Various S. aureus infections are frequently isolated alongside P. aeruginosa such as in soft tissue infections, diabetic foot infections, osteomyelitis and within cystic fibrosis airways (Kahl et al., 2016). These two species can act competitively or cooperatively, such as P. aeruginosa secretions of LasB; an elastase which removes lung surfactant and prevents uptake by macrophages to allow effective colonization and persistence in the lung (Hotterbeekx et al., 2017).
Conversely, the secretion of an antistaphyloccocal metabolite, 4-hydroxy-2-heptylquinoline-N-oxide (HQNQ), by P. aeruginosa inhibits S. aureus growth through interrupting its electron transport chain and ATP production (Machan et al., 1992; Proctor, 2019). Long term exposure to physiological concentrations of HQNQ in combination with aminoglycosides in vitro resulted in high proportions of menadione SCVs (Hoffman et al., 2006). Indeed, in the co-existence of these bacteria, S. aureus becomes less susceptible to vancomycin and other antibiotics (Orazi and O’Toole, 2017; Radlinski et al., 2017). This has also been recognized through clinically relevant analyses (Mitchell et al., 2010b; Fugère et al., 2014).
No other interaction between S. aureus and other bacterial species has been found to directly induce SCV formation. However, if we consider the selective pressures which allow SCV to survive and dominate a population of cells through increased fitness over their parental cell types; including a reduced nutrient availability, antibiotics, phagocytosis, extreme pH; we can deduce that the interactions between S. aureus and other bacterial species which negatively affect S. aureus pathogenesis and survival may also be selecting for SCV (or indeed, other quasi-dormant cell types). Corynebacterium spp. and other Staphylococci are known to inhibit S. aureus virulence (in particular, through blocking agr function), nutrient acquisition and adhesion (Iwase et al., 2010; Wollenberg et al., 2014; Ramsey et al., 2016). Streptococcus spp. and Staphylococcus lugdunensis produce exoproducts which actively kill S. aureus (Zipperer et al., 2016; Wu et al., 2019). Many of these species we describe later are carried in the nares (Lina et al., 2003; Huttenhower et al., 2012), a potentially ideal anatomical niche for selecting and forming SCVs. Thymidine auxotroph SCVs have been isolated from the nares in a patient with AIDS (von Eiff et al., 2004) and pulmonary fibrosis (Cleeve et al., 2006). There are cases of S. aureus progressing from nasal colonization to bacteremia without the acquisition of additional virulence genes and SNPs in arlS and agrA (Benoit et al., 2018); genetic profiles previously reported in clinical SCVs (Kahl et al., 2005; Kohler et al., 2008; Kriegeskorte et al., 2014b). However, in contrast, S. aureus nasal colonization has been shown to favor dispersed cell-types rather than biofilm formation (Krismer and Peschel, 2011) which may imply SCV are less fit in the nares. The interactions between S. aureus and other bacterial species has not been researched in detail, and so we review the interactions in the context of selection of SCV.
Staphylococcus epidermidis is commonly found to out-compete S. aureus in the nares (Lina et al., 2003; Frank et al., 2010; Lee et al., 2019). S. epidermidis can block agr quorum sensing in S. aureus (Otto et al., 1999) and production of the serine protease, Esp, which inhibits S. aureus colonization through inhibiting biofilm formation and synergistically increasing the ability of Human-beta defensin 2 to clear S. aureus (Iwase et al., 2010). In a similar fashion, Staphylococcus caprae, a skin commensal, also interferes with S. aureus colonization and produces an AIP which blocks S. aureus agr sensing (Paharik et al., 2017).
Staphylococcus lugdunensis produces a peptide antibiotic, lugdunin, which has bactericidal effects against S. aureus (Zipperer et al., 2016). The mechanism of action of lugdunin against S. aureus has not been determined, and so whether this bactericidal effect selects for SCV is unclear. However, it has been shown to act synergistically with the innate immune response, where lugdunin increases recruitment of monocytes and neutrophils to keratinocytes (Bitschar et al., 2019).
Propionibacterium spp. produces an exoproduct coproporphyrin III (CIII) which induces aggregation and biofilm formation in S. aureus within acidic conditions (pH 4–6) (Wollenberg et al., 2014). SarA was shown to be involved in CIII mediated biofilm, however, the role of other biofilm regulators was not tested in this study.
Streptococcus pneumoniae production of hydrogen peroxide is able to kill S. aureus within in vitro conditions (Uehara et al., 2001; Regev-Yochay et al., 2006; Wu et al., 2019). Hydrogen peroxide can oxidize iron groups to damage proteins, or generate OH– to cause DNA damage (Keyer and Imlay, 1996). However, a study using the nasal cavities of neonatal rats showed this hydrogen peroxide is not enough to affect S. aureus colonization (Margolis, 2009). It is known that over prolonged periods of time S. aureus in its SCV state can tolerate hydrogen peroxide (Painter et al., 2015) and so S. aureus may switch to a SCV when assaulted by S. pneumoniae generated hydrogen peroxide.
Both S. aureus and Cornyebacterium spp. are common nasal colonizers (Frank et al., 2010) and in vitro co-colonization of S. aureus with Corynebacterium spp. results in a shift of S. aureus from a virulent to a commensal state, with a strongly inhibited agr (Ramsey et al., 2016). Co-cultures with Corynebacterium spp. (including nasal strains C. striatum, C. amycolatum, C. accolens, C. pseudodiptheriticum and a soil strain C. glutamicum) interferes with AIP-1 (agr activator molecule) and increases clearance of S. aureus within a murine model of infection (Ramsey et al., 2016). This co-culture is associated with a global change in gene regulation, with a notably increased expression of spa (260-fold) and down-regulation of agr. The loss of agr function has been associated with resistance to the bacteriocidal effects of C. pseudodiptheriticum (Hardy et al., 2019), however, the bacteriocidal mechanism by C. pseudodiptheriticum has not been established. The loss of agr activity and increased expression of adhesive surface proteins are common characteristics of SCV and this is an indicator that S. aureus may resist clearance by Corynebacterium spp. by switching to SCV.
To advance both diagnosis and treatment protocols for S. aureus infections, it is vital to understand the molecular mechanisms that select for or induce the formation of SCVs. Current therapeutic treatments are becoming less effective against persistent S. aureus infections. Given the tendency for SCVs to revert, and the difficulty in their culturing, means research into S. aureus SCVs still remains a challenge. The use of continuous culture to select for SCVs amongst a population of cells has a great potential in discovering the molecular mechanisms involved in the transition into a SCV state which other sSCV models cannot clearly define.
Another aspect of SCV formation to consider is the influence of the microbiota co-existing with S. aureus. Studies with S. aureus and P. aeruginosa have shown this interaction selects for quasi-dormant cell types. The conditions in the nares, interactions with other bacterial species and their ability to persist intracellularly may favor the formation of SCVs. These can then transit around the human body and evade the host immune response. This provides a new perspective on the nasal carriage of S. aureus and the increased risk of endogenous S. aureus infection (Von Eiff et al., 2001; Kluytmans and Wertheim, 2005; Stanaway et al., 2007; Haleem et al., 2014; Dunyach-Remy et al., 2017). This includes immunocompromised sites such as the diabetic foot (Lavery et al., 2006) or a foreign body surface for attachment and biofilm production (Kahl et al., 2016; Yang et al., 2018).
JL, PZ, and SK each contributed to the design, construction, and writing of this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abdelnour, A., Arvidson, S., Bremell, T., Ryden, C., and Tarkowski, A. (1993). The accessory gene regulator (agr) controls Staphylococcus aureus virulence in a murine arthritis model. Infect. Immun. 61, 3879–3885. doi: 10.1128/iai.61.9.3879-3885.1993
Arvidson, S., and Tegmark, K. (2001). Regulation of virulence determinants in Staphylococcus aureus. Int. J. Med. Microbiol. 291, 159–170. doi: 10.1078/1438-4221-00112
Atalla, H., Gyles, C., and Mallard, B. (2011). Staphylococcus aureus small colony variants (SCVs) and their role in disease. Anim. Health Res. Rev. 12, 33–45. doi: 10.1017/S1466252311000065
Baddour, L. M., and Christensen, G. D. (1987). Prosthetic valve endocarditis due to small-colony staphylococcal variants. Rev. Infect. Dis. 9, 1168–1174. doi: 10.1093/clinids/9.6.1168
Balaban, N. Q., Merrin, J., Chait, R., Kowalik, L., and Leibler, S. (2004). Bacterial persistence as a phenotypic switch. Science 305, 1622–1625. doi: 10.1126/science.1099390
Balwit, J. M., van Langevelde, P., Vann, J. M., and Proctor, R. (1994). Gentamicin-resistant menadione and hemin auxotrophic Staphylococcus aureus persist within cultured endothelial cells. J. Infect Dis. 170, 1033–1037. doi: 10.1093/infdis/170.4.1033
Bates, D. M., Von Eiff, C., McNamara, P. J., Peters, G., Yeaman, M. R., and Bayer, A. S. (2003). Staphylococcus aureus menD and hemB mutants are as infective as the parent strains, but the menadione biosynthetic mutant persists within the kidney. J. Infect. Dis. 187, 1654–1661. doi: 10.1086/374642
Bazaid, A. S., Forbes, S., Humphreys, G. J., Ledder, R. G., OCualain, R., and McBain, A. J. (2018). Fatty acid supplementation reverses the small colony variant phenotype in triclosan-adapted Staphylococcus aureus: genetic, proteomic and phenotypic analyses. Sci. Rep. 8:3876. doi: 10.1038/s41598-018-21925-6
Benoit, J. B., Frank, D. N., and Bessesen, M. T. (2018). Genomic evolution of Staphylococcus aureus isolates colonizing the nares and progressing to bacteremia. PLoS One 13:e0195860. doi: 10.1371/journal.pone.0195860
Besier, S., Ludwig, A., Ohlsen, K., Brade, V., and Wichelhaus, T. A. (2007). Molecular analysis of the thymidine-auxotrophic small colony variant phenotype of Staphylococcus aureus. Int. J. Med. Microbiol. 297, 217–225. doi: 10.1016/j.ijmm.2007.02.003
Bischoff, M., Dunman, P., Kormanec, J., Macapagal, D., Murphy, E., and Mounts, W. (2004). Microarray-based analysis of the Staphylococcus aureus σB regulon. J. Bacteriol. 186, 4085–4099. doi: 10.1128/jb.186.13.4085-4099.2004
Bitschar, K., Sauer, B., Focken, J., Dehmer, H., Moos, S., and Konnerth, M. (2019). Lugdunin amplifies innate immune responses in the skin in synergy with host- and microbiota-derived factors. Nat. Commun. 10:2730. doi: 10.1038/s41467-019-10646-7
Bui, L. M. G., Hoffmann, P., Turnidge, J. D., Zilm, P. S., and Kidd, S. P. (2015). Prolonged growth of a clinical Staphylococcus aureus strain selects for a stable small-colony-variant cell type. Infect. Immun. 83, 470–481. doi: 10.1128/IAI.02702-14
Bui, L. M. G., and Kidd, S. P. (2015). A full genomic characterization of the development of a stable small colony variant cell-type by a clinical Staphylococcus aureus strain. Infect. Genet. Evol. 36, 345–355. doi: 10.1016/j.meegid.2015.10.011
Chang, Y., Tai, C. L., Hsieh, P. H., and Ueng, S. W. (2013). Gentamicin in bone cement: a potentially more effective prophylactic measure of infectionin joint arthroplasty. Bone Joint Res. 2, 220–226. doi: 10.1302/2046-3758.210.2000188
Chatterjee, I., Kriegeskorte, A., Fischer, A., Deiwick, S., Theimann, N., Proctor, R. A., et al. (2008). In vivo mutations of thymidylate synthase (Encoded by thyA) are responsible for thymidine dependency in clinical small-colony variants of Staphylococcus aureus. J. Bacteriol. 190, 834–842. doi: 10.1128/jb.00912-07
Cheung, A. L., Bayer, A. S., Zhang, G., Gresham, H., and Xiong, Y. Q. (2004). Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immun. Med. Microbiol. 40, 1–9. doi: 10.1016/s0928-8244(03)00309-2
Cleeve, V. J., Perry, J. D., Cresswell, G., and Orr, K. E. (2006). Thymidine-dependent meticillin-resistant Staphylococcus aureus: a potentially unrecognized reservoir of MRSA in hospital patients? J. Hosp. Infect. 63, 228–229. doi: 10.1016/j.jhin.2005.11.009
Crosby, H. A., Schlievert, P. M., Merriman, J. A., King, J. M., Salgado-Pabon, W., and Horswill, A. R. (2016). The Staphylococcus aureus global regulator mgra modulates clumping and virulence by controlling surface protein expression. PLoS Pathog. 12:e1005604. doi: 10.1371/journal.ppat.1005604
Dean, M. A., Olsen, R. J., Wesley Long, S., Rosato, A. E., and Musser, J. M. (2014). Identification of point mutations in clinical Staphylococcus aureus strains that produce small-colony variants auxotrophic for menadione. Infect. Immun. 82, 1600–1605. doi: 10.1128/IAI.01487-13
Dunyach-Remy, C., Courtais-Coulon, C., DeMattei, C., Jourdan, N., Schuldiner, S., and Sultan, A. (2017). Link between nasal carriage of Staphylococcus aureus and infected diabetic foot ulcers. Diabetes Metab. 43, 167–171. doi: 10.1016/j.diabet.2016.09.003
Edwards, A. M. (2012). Phenotype switching is a natural consequence of Staphylococcus aureus replication. J. Bacteriol. 194, 5404–5412. doi: 10.1128/JB.00948-12
Frank, D. N., Feazel, L. M., Bessesen, M. T., Price, C. S., Janoff, E. N., and Pace, N. R. (2010). The human nasal microbiota and Staphylococcus aureus. PLoS One 5:e10598. doi: 10.1371/journal.pone.0010598
Fugère, A., Lalonde Séguin, D., Mitchell, G., Déziel, E., Dekimpe, V., and Cantin André, M. (2014). Interspecific small molecule interactions between clinical isolates of Pseudomonas aeruginosa and Staphylococcus aureus from adult cystic fibrosis patients. PLoS One 9:e86705. doi: 10.1371/journal.pone.0086705
Garcia, L. G., Lemaire, S., Kahl, B. C., Becker, K., Proctor, R. A., and Denis, O. (2013). Antibiotic activity against small-colony variants of Staphylococcus aureus: review of in vitro, animal and clinical data. J. Antimicrob. Chemother. 68, 1455–1464. doi: 10.1093/jac/dkt072
Garzoni, C., and Kelley, W. L. (2009). Staphylococcus aureus: new evidence for intracellular persistence. Trends Microbiol. 17, 59–65. doi: 10.1016/j.tim.2008.11.005
Gómez-González, C., Acosta, J., Villa, J., Barrado, L., Sanz, F., Orellana, M. A., et al. (2010). Clinical and molecular characteristics of infections with CO2-dependent small-colony variants of Staphylococcus aureus. J. Clin. Microbiol. 48, 2878–2884. doi: 10.1128/JCM.00520-10
Haleem, A., Schultz, J. S., Heilmann, K. P., Dohrn, C. L., Diekema, D. J., and Gardner, S. E. (2014). Concordance of nasal and diabetic foot ulcer staphylococcal colonization. Diagn. Microbiol. Infect. Dis. 79, 85–89. doi: 10.1016/j.diagmicrobio.2014.01.014
Hardy, B. L., Dickey, S. W., Plaut, R. D., Riggins, D. P., Stibitz, S., and Otto, M. (2019). Corynebacterium pseudodiphtheriticum exploits Staphylococcus aureus virulence components in a novel polymicrobial defense strategy. mBio 10:e02491-18. doi: 10.1128/mBio.02491-18
Hoffman, L. R., Déziel, E., DArgenio, D. A., Lépine, F., Emerson, J., and McNamara, S. (2006). Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 103, 19890–19895. doi: 10.1073/pnas.0606756104
Horiuchi, K., Matsumoto, T., Ota, Y., Kasuga, E., Negishi, T., and Yaguchi, T. (2015). Addition of thymidine to culture media for accurate examination of thymidine-dependent small-colony variants of methicillin-resistant Staphylococcus aureus: a pilot study. J. Microbiol. Methods 110, 40–44. doi: 10.1016/j.mimet.2015.01.007
Hotterbeekx, A., Kumar-Singh, S., Goossens, H., and Malhotra-Kumar, S. (2017). In vivo and In vitro interactions between Pseudomonas aeruginosa and Staphylococcus spp. Front. Cell. Infect. Microbiol. 7:106. doi: 10.3389/fcimb.2017.00106
Huttenhower, C., Gevers, D., Knight, R., Abubucker, S., Badger, J. H., and Chinwalla, S. T. (2012). Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. doi: 10.1038/nature11234
Ingavale, S. S., Van Wamel, W., and Cheung, A. L. (2003). Characterization of RAT, an autolysis regulator in Staphylococcus aureus. Mol. Microbiol. 48, 1451–1466. doi: 10.1046/j.1365-2958.2003.03503.x
Ingavale, S. S., van Wamel, W., Luong, T. T., Lee, C. Y., and Cheung, A. L. (2005). Rat/MgrA, a regulator of autolysis, is a regulator of virulence genes in Staphylococcus aureus. Infect. Immun. 73, 1423–1431. doi: 10.1128/iai.73.3.1423-1431.2005
Iwase, T., Uehara, Y., Shinji, H., Tajima, A., Seo, H., and Takada, K. (2010). Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 465, 346–349. doi: 10.1038/nature09074
Jonsson, I. M., Arvidson, S., Foster, S., and Tarkowski, A. (2004). Sigma factor B and RsbU are required for virulence in Staphylococcus aureus-induced arthritis and sepsis. Infect. Immun. 72, 6106–6111. doi: 10.1128/iai.72.10.6106-6111.2004
Kahl, B. C., Becker, K., and Löffler, B. (2016). Clinical significance and pathogenesis of staphylococcal small colony variants in persistent infections. Clin. Microbiol. Rev. 29, 401–427. doi: 10.1128/CMR.00069-15
Kahl, B. C., Belling, G., Becker, P., Chatterjee, I., Wardecki, K., and Hilgert, K. (2005). Thymidine-dependent Staphylococcus aureus small-colony variants are associated with extensive alterations in regulator and virulence gene expression profiles. Infect. Immun. 73, 4119–4126. doi: 10.1128/iai.73.7.4119-4126.2005
Kahl, B. C., Belling, G., Reichelt, R., Herrmann, M., Proctor, R. A., and Peters, G. (2003a). Thymidine-dependent small-colony variants of Staphylococcus aureus exhibit gross morphological and ultrastructural changes consistent with impaired cell separation. J. Clin. Microbiol. 41, 410–413. doi: 10.1128/jcm.41.1.410-413.2003
Kahl, B. C., Duebbers, A., Lubritz, G., Haeberle, J., Koch, H. G., and Ritzerfeld, B. (2003b). Population dynamics of persistent Staphylococcus aureus isolated from the airways of cystic fibrosis patients during a 6-year prospective study. J. Clin. Microbiol. 41, 4424–4427. doi: 10.1128/jcm.41.9.4424-4427.2003
Kahl, B. C., Herrmann, M., Everding, A. S., Koch, H. G., Becker, K., and Harms, E. (1998). Persistent infection with small colony variant strains of Staphylococcus aureus in patients with cystic fibrosis. J. Infect. Dis. 177, 1023–1029. doi: 10.1086/515238
Keren, I., Kaldalu, N., Spoering, A., Wang, Y., and Lewis, K. (2004). Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 230, 13–18. doi: 10.1016/s0378-1097(03)00856-5
Keyer, K., and Imlay, J. A. (1996). Superoxide accelerates DNA damage by elevating free-iron levels. Proc. Natl. Acad. Sci. U.S.A. 93, 13635. doi: 10.1073/pnas.93.24.13635
Kittinger, C., Toplitsch, D., Folli, B., Masoud, L. L., and Zarfel, G. (2019). Phenotypic Stability of Staphylococcus aureus small colony variants (SCV) isolates from cystic fibrosis (CF) Patients. Int. J. Env. Res. Public Health 16:1940. doi: 10.3390/ijerph16111940
Kluytmans, J. A., and Wertheim, H. F. (2005). Nasal carriage of Staphylococcus aureus and prevention of nosocomial infections. Infection 33, 3–8. doi: 10.1007/s15010-005-4012-9
Kohler, C., von Eiff, C., Liebeke, M., McNamara, P. J., Lalk, M., and Proctor, R. A. (2008). A defect in menadione biosynthesis induces global changes in gene expression in Staphylococcus aureus. J. Bacteriol. 190, 6351–6364. doi: 10.1128/JB.00505-08
Kriegeskorte, A., Block, D., Drescher, M., Windmüller, N., Mellmann, A., Baum, C., et al. (2014b). Inactivation of thyA in Staphylococcus aureus attenuates virulence and has a strong impact on metabolism and virulence gene expression. mBio 5:e01447-14. doi: 10.1128/mBio.01447-14
Kriegeskorte, A., Grubmüller, S., Huber, C., Kahl, B. C., von Eiff, C., and Proctor, R. A. (2014a). Staphylococcus aureus small colony variants show common metabolic features in central metabolism irrespective of the underlying auxotrophism. Front. Cell. Infect. Microbiol. 4:141. doi: 10.3389/fcimb.2014.00141
Krismer, B., and Peschel, A. (2011). Does Staphylococcus aureus nasal colonization involve biofilm formation? Future Microbiol. 6, 489–493. doi: 10.2217/fmb.11.37
Kullik, I., Giachino, P., and Fuchs, T. (1998). Deletion of the alternative sigma factor σ(B) in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J. Bacteriol. 180, 4814–4820. doi: 10.1128/jb.180.18.4814-4820.1998
Kwiecinski, J. M., Crosby, H. A., Valotteau, C., Hippensteel, J. A., Nayak, M. K., and Chauhan, A. K. (2019). Staphylococcus aureus adhesion in endovascular infections is controlled by the arlRS–MgrA signaling cascade. PLoS Pathog. 15:e1007800. doi: 10.1371/journal.ppat.1007800
Lacoma, A., Edwards, A. M., Young, B. C., Domínguez, J., Prat, C., and Laabei, M. (2019). Cigarette smoke exposure redirects Staphylococcus aureus to a virulence profile associated with persistent infection. Sci. Rep. 9:10798. doi: 10.1038/s41598-019-47258-6
Lannergård, J., Von Eiff, C., Sander, G., Cordes, T., Seggewiß, J., and Peters, G. (2008). Identification of the genetic basis for clinical menadione-auxotrophic small-colony variant isolates of Staphylococcus aureus. Antimicrob. Agents Chemother. 52, 4017–4022. doi: 10.1128/AAC.00668-08
Lavery, L. A., Armstrong, D. G., Wunderlich, R. P., Mohler, M. J., Wendel, C. S., and Lipsky, B. A. (2006). Risk factors for foot infections in individuals with diabetes. Diabetes Care 29, 1288–1293. doi: 10.2337/dc05-2425
Lee, D. C., Kananurak, A., Tran, M. T., Connolly, P. A., Polage, C. R., and Iwase, T. (2019). Bacterial colonization of the hospitalized newborn: competition between Staphylococcus aureus and Staphylococcus epidermidis. Paediatr. Inf. Dis. J. 38, 682–686. doi: 10.1097/INF.0000000000002285
Lei, M. G., Gudeta, D. D., Luong, T. T., and Lee, C. Y. (2019). MgrA negatively impacts Staphylococcus aureus invasion by regulating capsule and FnbA. Infect. Immun. 87:e00590-19. doi: 10.1128/IAI.00590-19
Lina, G., Boutite, F., Tristan, A., Bes, M., Etienne, J., and Vandenesch, F. (2003). Bacterial competition for human nasal cavity colonization: role of staphylococcal agr alleles. Appl. Environ. Microbiol. 69, 18–23. doi: 10.1128/aem.69.1.18-23.2003
Luong, T. T., Dunman, P. M., Murphy, E., Projan, S. J., and Lee, C. Y. (2006). Transcription profiling of the mgrA regulon in Staphylococcus aureus. J. Bacteriol. 188, 1899–1910. doi: 10.1128/jb.188.5.1899-1910.2006
Machan, Z. A., Graham, W. T., Pitt, T. L., Cole, P. J., and Wilson, R. (1992). 2-heptyl-4-hydroxyquinoline N-oxide, an antistaphylococcal produced by Pseudomonas aeruginosa. J. Antimicrob. Chemother. 30, 615–623. doi: 10.1093/jac/30.5.615
Maduka-Ezeh, A., Seville, M. T., Kusne, S., Vikram, H. R., Blair, J. E., and Greenwood-Quaintance, K. (2012). Thymidine auxotrophic Staphylococcus aureus small-colony variant endocarditis and left ventricular assist device infection. J. Clin. Microbiol. 50, 1102–1105. doi: 10.1128/JCM.01170-11
Margolis, E. (2009). Hydrogen peroxide-mediated interference competition by Streptococcus pneumoniae has no significant effect on Staphylococcus aureus nasal colonization of neonatal rats. J. Bacteriol. 191, 571–575. doi: 10.1128/JB.00950-08
Melter, O., and Radojevič, B. (2010). Small colony variants of Staphylococcus aureus—review. Folia Microbiol. 55, 548–558. doi: 10.1007/s12223-010-0089-3
Mirani, Z. A., Aziz, M., and Khan, S. I. (2015). Small colony variants have a major role in stability and persistence of Staphylococcus aureus biofilms. J. Antibiot. 68, 98–105. doi: 10.1038/ja.2014.115
Mitchell, G., Brouillette, E., Séguin, D. L., Asselin, A. E., Jacob, C. L., and Malouin, F. (2010a). A role for sigma factor B in the emergence of Staphylococcus aureus small-colony variants and elevated biofilm production resulting from an exposure to aminoglycosides. Microb. Pathog. 48, 18–27. doi: 10.1016/j.micpath.2009.10.003
Mitchell, G., Fugère, A., Gaudreau, K., Brouillette, E., Frost, E. H., and Cantin, A. M. (2013). SigB is a dominant regulator of virulence in Staphylococcus aureus small-colony variants. PLoS One 8:e65018. doi: 10.1371/journal.pone.0065018
Mitchell, G., Lamontagne, C. A., Brouillette, E., Grondin, G., Talbot, B. G., and Grandbois, M. (2008). Staphylococcus aureus SigB activity promotes a strong fibronectin-bacterium interaction which may sustain host tissue colonization by small-colony variants isolated from cystic fibrosis patients. Mol. Microbiol. 70, 1540–1555. doi: 10.1111/j.1365-2958.2008.06511.x
Mitchell, G., Séguin, D. L., Asselin, A. E., Déziel, E., Cantin, A. M., and Frost, E. H. (2010b). Staphylococcus aureus sigma B-dependent emergence of small-colony variants and biofilm production following exposure to Pseudomonas aeruginosa 4-hydroxy-2-heptylquinoline-N-oxide. BMC Microbiol. 10:33. doi: 10.1186/1471-2180-10-33
Moisan, H., Brouillette, E., Jacob, C. L., Langlois-Bégin, P., Michaud, S., and Malouin, F. (2006). Transcription of virulence factors in Staphylococcus aureus small-colony variants isolated from cystic fibrosis patients is influenced by SigB. J. Bacteriol. 188, 64–76. doi: 10.1128/jb.188.1.64-76.2006
Novick, R. P. (2003). Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48, 1429–1449. doi: 10.1046/j.1365-2958.2003.03526.x
Orazi, G., and O’Toole, G. A. (2017). Pseudomonas aeruginosa alters Staphylococcus aureus sensitivity to vancomycin in a biofilm model of cystic fibrosis infection. mBio 8:e00873-17. doi: 10.1128/mBio.00873-17
Otto, M., Süßmuth, R., Vuong, C., Jung, G., and Götz, F. (1999). Inhibition of virulence factor expression in Staphylococcus aureus by the Staphylococcus epidermidis agr pheromone and derivatives. FEBS Lett. 450, 257–262.
Ou, J. J., Drilling, A. J., Cooksley, C., Bassiouni, A., Kidd, S. P., and Psaltis, A. J. (2016). Reduced innate immune response to a Staphylococcus aureus small colony variant compared to its wild-type parent strain. Front. Cell. Infect. Microbiol. 6:187. doi: 10.3389/fcimb.2016.00187
Pader, V., James, E. H., Painter, K. L., Wigneshweraraj, S., and Edwards, A. M. (2014). The agr quorum-sensing system regulates fibronectin binding but not hemolysis in the absence of a functional electron transport chain. Inf. Immun. 82, 4337–4347. doi: 10.1128/IAI.02254-14
Paharik, A. E., Parlet, C. P., Chung, N., Todd, D. A., Rodriguez, I., and Van Dyke, M. J. (2017). Coagulase-negative staphylococcal strain prevents Staphylococcus aureus colonization and skin infection by blocking quorum sensing. Cell Host Microbe 22, 746.e5–756.e5. doi: 10.1016/j.chom.2017.11.001
Painter, K. L., Strange, E., Parkhill, J., Bamford, K. B., Armstrong-James, D., and Edwards, A. M. (2015). Staphylococcus aureus adapts to oxidative stress by producing H2O2 resistant small-colony variants via the SOS response. Inf. Immun. 83, 1830–1844. doi: 10.1128/IAI.03016-14
Proctor, R. A. (2019). Respiration and small colony variants of Staphylococcus aureus. Microbiol. Spectr. 7, 1–15. doi: 10.1128/microbiolspec.GPP3-0069-2019
Proctor, R. A., Kahl, B., Von Eiff, C., Vaudaux, P. E., Lew, D. P., and Peters, G. (1998). Staphylococcal small colony variants have novel mechanisms for antibiotic resistance. Clin. Infect. Dis. 27(Suppl. 1), S68–S74.
Proctor, R. A., Kriegeskorte, A., Kahl, B. C., Becker, K., Löffler, B., and Peters, G. (2014). Staphylococcus aureus small colony variants (SCVs): a road map for the metabolic pathways involved in persistent infections. Front. Cell. Infect. Microbiol. 4:99. doi: 10.3389/fcimb.2014.00099
Proctor, R. A., Van Langevelde, P., Kristjansson, M., Maslow, J. N., and Arbeit, R. D. (1995). Persistent and relapsing infections associated with small-colony variants of Staphylococcus aureus. Clin. Infect. Dis. 20, 95–102. doi: 10.1093/clinids/20.1.95
Proctor, R. A., von Eiff, C., Kahl, B. C., Becker, K., McNamara, P., and Herrmann, M. (2006). Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Microbiol. 4, 295–305. doi: 10.1038/nrmicro1384
Quie, P. G. (1969). Microcolonies (G-Variants) of Staphylococcus aureus. Yale J. Bio. Med. 41, 394–403.
Radlinski, L., Rowe Sarah, E., Kartchner Laurel, B., Maile, R., Cairns Bruce, A., and Vitko, P. (2017). Pseudomonas aeruginosa exoproducts determine antibiotic efficacy against Staphylococcus aureus. PLoS Biol. 15:e2003981. doi: 10.1371/journal.pbio.2003981
Ramsey, M. M., Freire, M. O., Gabrilska, R. A., Rumbaugh, K. P., and Lemon, K. P. (2016). Staphylococcus aureus shifts toward commensalism in response to Corynebacterium species. Front. Microbiol. 7:1230. doi: 10.3389/fmicb.2016.01230
Regev-Yochay, G., Trzciński, K., Thompson, C. M., Malley, R., and Lipsitch, M. (2006). Interference between Streptococcus pneumoniae and Staphylococcus aureus: In Vitro hydrogen peroxide-mediated killing by Streptococcus pneumoniae. J. Bacteriol. 188, 4996–5001. doi: 10.1128/jb.00317-06
Rescei, R., Kreiswirth, B., Reilly, M. O., Schlievert, P., Gruss, A., and Novick, R. P. (1986). Regulation of exoprotein gene expression is Staphylococcus aureus by agr. Mol. Gen. Genet. 202, 58–61. doi: 10.1007/bf00330517
Schaaff, F., Bierbaum, G., Baumert, N., Bartmann, P., and Sahl, H. G. (2003). Mutations are involved in emergence of aminoglycoside-induced small colony variants of Staphylococcus aureus. Int. J. Med Microbiol. 293, 427–435. doi: 10.1078/1438-4221-00282
Schleimer, N., Kaspar, U., Drescher, M., Seggewiß, J., von Eiff, C., and Proctor, R. A. (2018). The energy-coupling factor transporter module EcfAAT, a novel candidate for the genetic basis of fatty acid-auxotrophic small-colony variants of Staphylococcus aureus. Front. Microbiol. 9:1863. doi: 10.3389/fmicb.2018.01863
Schwerdt, M., Neumann, C., Schwartbeck, B., Kampmeier, S., Herzog, S., and Görlich, D. (2018). Staphylococcus aureus in the airways of cystic fibrosis patients – A retrospective long-term study. Int. J. Med. Microbiol. 308, 631–639. doi: 10.1016/j.ijmm.2018.02.003
Sendi, P., and Proctor, R. A. (2009). Staphylococcus aureus as an intracellular pathogen: the role of small colony variants. Trends Microbiol. 17, 54–58. doi: 10.1016/j.tim.2008.11.004
Senn, M. M., Giachino, P., Homerova, D., Steinhuber, A., Strassner, J., and Kormanec, J. (2005). Molecular analysis and organization of the σB operon in Staphylococcus aureus. J. Bacteriol. 187, 8006–8019. doi: 10.1128/jb.187.23.8006-8019.2005
Singh, R., Ray, P., Das, A., and Sharma, M. (2009). Role of persisters and small-colony variants in antibiotic resistance of planktonic and biofilm-associated Staphylococcus aureus: an in vitro study. J. Med. Microbiol. 58(Pt 8), 1067–1073. doi: 10.1099/jmm.0.009720-0
Singh, R., Ray, P., Das, A., and Sharma, M. (2010). Enhanced production of exopolysaccharide matrix and biofilm by a menadione-auxotrophic Staphylococcus aureus small-colony variant. J. Med. Microbiol. 59(Pt 5), 521–527. doi: 10.1099/jmm.0.017046-0
Stanaway, S., Johnson, D., Moulik, P., and Gill, G. (2007). Methicillin-resistant Staphyloccocus aureus (MRSA) isolation from diabetic foot ulcers correlates with nasal MRSA carriage. Diabetes Res. Clin. Pract. 75, 47–50. doi: 10.1016/j.diabres.2006.05.021
Thomas, M. E. (1955). Studies on a CO2-dependent staphylococcus. J. Clin. Pathol. 8, 288–291. doi: 10.1136/jcp.8.4.288
Tong, S. Y., Davis, J. S., Eichenberger, E., Holland, T. L., and Fowler, V. G. Jr. (2015). Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 28, 603–661. doi: 10.1128/cmr.00134-14
Trotonda, M. P., Tamber, S., Memmi, G., and Cheung, A. L. (2008). MgrA represses biofilm formation in Staphylococcus aureus. Inf. Immun. 76, 5645–5654. doi: 10.1128/IAI.00735-08
Tuchscherr, L., Bischoff, M., Lattar, S. M., Noto Llana, M., Pförtner, H., and Niemann, S. (2015). Sigma factor SigB is crucial to mediate Staphylococcus aureus adaptation during chronic infections. PLoS Pathog. 11:e1004870. doi: 10.1371/journal.ppat.1004870
Tuchscherr, L., Buzzola, F. R., Alvarez, L. P., Lee, J. C., and Sordelli, D. O. (2008). Antibodies to capsular polysaccharide and clumping factor a prevent mastitis and the emergence of unencapsulated and small-colony variants of Staphylococcus aureus in mice. Infect. Immun. 76, 5738–5744. doi: 10.1128/IAI.00874-08
Tuchscherr, L., Geraci, J., and Löffler, B. (2017). Staphylococcus aureus regulator sigma B is important to develop chronic infections in hematogenous murine osteomyelitis model. Pathogens 6:31. doi: 10.3390/pathogens6030031
Tuchscherr, L., Heitmann, V., Hussain, M., Viemann, D., Roth, J., and Von Eiff, C. (2010). Staphylococcus aureus small-colony variants are adapted phenotypes for intracellular persistence. J. Infect. Dis. 202, 1031–1040. doi: 10.1086/656047
Uehara, Y., Kikuchi, K., Nakamura, T., Nakama, H., Agematsu, K., Kawakami, Y., et al. (2001). H2O2 Produced by viridans group streptococci may contribute to inhibition of methicillin-resistant Staphylococcus aureus colonization of oral cavities in newborns. Clin. Infect. Dis. 32, 1408–1413. doi: 10.1086/320179
Vaudaux, P., Francois, P., Bisognano, C., Kelley, W. L., Lew, D. P., and Schrenzel, J. (2002). Increased expression of clumping factor and fibronectin-binding proteins by hemB mutants of Staphylococcus aureus expressing small colony variant phenotypes. Inf. Immun. 70, 5428–5437. doi: 10.1128/iai.70.10.5428-5437.2002
Vestergaard, M., Paulander, W., and Ingmer, H. (2015). Activation of the SOS response increases the frequency of small colony variants. BMC Res. Notes 8:749. doi: 10.1186/s13104-015-1735-2
Von Eiff, C., Becker, K., Machka, K., Stammer, H., and Peters, G. (2001). Nasal carriage as a source of Staphylococcus aureus bacteremia. N. Engl. J. Med. 344, 11–16.
Von Eiff, C., Heilmann, C., Proctor, R., Woltz, C., Peters, G., and Götz, F. (1997). A site-directed Staphylococcus aureus hemB mutant is a small colony variant which persists intracellularly. J. Bacteriol. 179, 4706–4712. doi: 10.1128/jb.179.15.4706-4712.1997
von Eiff, C., Lubritz, G., Heese, C., Peters, G., and Becker, K. (2004). Effect of trimethoprim-sulfamethoxazole prophylaxis in AIDS patients on the formation of the small colony variant phenotype of Staphylococcus aureus. Diagn. Microbiol. Infect. Dis. 48, 191–194. doi: 10.1016/j.diagmicrobio.2003.10.010
Von Eiff, C., Vaudaux, P., Kahl, B., Lew, D., Emler, S., and Schmidt, A. (1999). Bloodstream infections caused by small-colony variants of coagulase-negative staphylococci following pacemaker implantation. Clin. Infect. Dis. 29, 932–934. doi: 10.1086/520462
Wollenberg, M. S., Claesen, J., Escapa, I. F., Aldridge, K. L., Fischbach, M. A., and Lemon, K. P. (2014). Propionibacterium-produced coproporphyrin III induces Staphylococcus aureus aggregation and biofilm formation. mBio 5:e01286-14. doi: 10.1128/mBio.01286-14
Wood, T. K., Knabel, S. J., and Kwan, B. W. (2013). Bacterial persister cell formation and dormancy. Appl. Environ. Microbiol. 79, 7116–7121. doi: 10.1128/AEM.02636-13
Wu, X., Gordon, O., Jiang, W., Antezana, B. S., Angulo-Samudio, U. A., del Rio, C., et al. (2019). Interaction between Streptococcus pneumoniae and Staphylococcus aureus generates OH radicals that rapidly kill Staphylococcus aureus strains. J. Bacteriol. 201:e00474-19. doi: 10.1128/JB.00474-19
Yang, D., Wijenayaka, A. R., Solomon, L. B., Pederson, S. M., Findlay, D. M., and Kidd, S. P. (2018). Novel insights into Staphylococcus aureus deep bone infections: the involvement of osteocytes. mBio 9:e00415-18. doi: 10.1128/mBio.00415-18
Yarwood, J. M., and Schlievert, P. M. (2003). Quorum sensing in Staphylococcus infections. J. Clin. Invest. 112, 1620–1625. doi: 10.1172/jci200320442
Zhang, P., Wright, J. A., Osman, A. A., and Nair, S. P. (2017). An aroD Ochre mutation results in a Staphylococcus aureus small colony variant that can undergo phenotypic switching via two alternative mechanisms. Front. Microbiol. 8:1001. doi: 10.3389/fmicb.2017.01001
Keywords: Staphylococcus aureus, small colony variants, continuous culture, prolonged growth, co-colonization
Citation: Lee J, Zilm PS and Kidd SP (2020) Novel Research Models for Staphylococcus aureus Small Colony Variants (SCV) Development: Co-pathogenesis and Growth Rate. Front. Microbiol. 11:321. doi: 10.3389/fmicb.2020.00321
Received: 10 December 2019; Accepted: 13 February 2020;
Published: 28 February 2020.
Edited by:
Lorena Tuchscherr, Jena University Hospital, GermanyReviewed by:
Daniel O. Sordelli, University of Buenos Aires, ArgentinaCopyright © 2020 Lee, Zilm and Kidd. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stephen P. Kidd, c3RlcGhlbi5raWRkQGFkZWxhaWRlLmVkdS5hdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.