- 1CAS Key Laboratory of Synthetic Biology, Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China
- 2University of Chinese Academy of Sciences, Beijing, China
- 3Shanghai Tolo Biotechnology Company Limited, Shanghai, China
- 4Department of Microbiology and Li Ka Shing Institute of Health Sciences, The Chinese University of Hong Kong, Shatin, Hong Kong
- 5College of Life Sciences, Shanghai Normal University, Shanghai, China
Because of the remarkable efficacy in treating mycobacterial infections, rifamycin and its derivatives are still first-line antimycobacterial drugs. It has been intensely studied to increase rifamycin yield from Amycolatopsis mediterranei, and nitrate is found to provide a stable and remarkable stimulating effect on the rifamycin production, a phenomenon known as “nitrate-stimulating effect (NSE)”. Although the NSE has been widely used for the industrial production of rifamycin, its detailed molecular mechanism remains ill-defined. And our previous study has established that the global nitrogen regulator GlnR may participate in the NSE, but the underlying mechanism is still enigmatic. Here, we demonstrate that GlnR directly controls rifamycin biosynthesis in A. mediterranei and thus plays an essential role in the NSE. Firstly, GlnR specifically binds to the upstream region of rifZ, which leads us to uncover that rifZ has its own promoter. As RifZ is a pathway-specific activator for the whole rif cluster, GlnR indirectly upregulates the whole rif cluster transcription by directly activating the rifZ expression. Secondly, GlnR specifically binds to the upstream region of rifK, which is also characterized to have its own promoter. It is well-known that RifK is a 3-amino-5-hydroxybenzoic acid (AHBA, the starter unit of rifamycin) synthase, thus GlnR can promote the supply of the rifamycin precursor by directly activating the rifK transcription. Notably, GlnR and RifZ independently activate the rifK transcription through binding to different sites in rifK promoter region, which suggests that the cells have a sophisticated regulatory mechanism to control the AHBA biosynthesis. Collectively, this study reveals that GlnR activates the rif cluster transcription in both direct (for rifZ and rifK) and indirect (for the whole rif cluster) manners, which well interprets the phenomenon that the NSE doesn’t occur in the glnR null mutant. Furthermore, this study deepens our understanding about the molecular mechanism of the NSE.
Introduction
Being a broad-spectrum antibiotic with unique property of inhibiting the prokaryotic RNA polymerase activity (Wehrli et al., 1968; Campbell et al., 2001), rifamycin and its derivatives (e.g., rifampicin, rifabutin, rifapentine, and rifaximin) are still broadly used as the first-line antimycobacterial agents (Rothstein, 2016). To increase rifamycin yield, multiple strategies have been tested, including both engineering the producer Amycolatopsis mediterranei (a rare actinomycete) (Peano et al., 2014; Kumari et al., 2016) and optimizing fermentation conditions (Jiao et al., 1979; Lee et al., 1983; Mejia et al., 1998). Among those tested conditions, supplementation of nitrate was found to be able to remarkably stimulate rifamycin production, which has become known as the “nitrate-stimulating effect (NSE)” (Jiao et al., 1979). This phenomenon was first reported by our laboratory about 40 years ago, and has been widely exploited in the industrial production of rifamycin SV.
With the assistance of stable isotope tracing technology, it was found that the nitrogen atom of nitrate can be incorporated into the rifamycin at last, and both intracellular nitrogen metabolites and the biosynthesis of rifamycin precursors are greatly promoted upon nitrate supplementation (Jiao et al., 1984; Jin and Jiao, 1986). Later, to better understand rifamycin biosynthesis, the rifamycin biosynthetic gene cluster (rif cluster) and the complete genome of A. mediterranei were sequenced (August et al., 1998; Zhao et al., 2010). The rif cluster, consisting of 43 genes (i.e., from rifS to rifZ, Supplementary Figure S1) (Floss and Yu, 1999; Li et al., 2017), includes genes involved in the synthesis of the starter unit AHBA (3-amino-5-hydroxybenzoic acid), assembly and modification of the polyketide backbone, downstream processing and conversion of rifamycin, and resistance (Yu et al., 1999; Xu et al., 2003, 2005; Floss and Yu, 2005; Floss et al., 2011; Yuan et al., 2011). In addition, the genome annotation results of A. mediterranei U32 indicate that the rif cluster contains two canonical transcriptional regulators: the LuxR-family transcriptional regulator RifZ and the TetR-family transcriptional regulator RifQ (Zhao et al., 2010). Recently, our studies have revealed that RifZ is a pathway-specific regulator of the rif cluster and directly activates the transcription of all rif cluster genes (Li et al., 2017), and RifQ could reduce the intracellular toxicity of rifamycin via directly regulating the expression of the rifamycin exporter-encoding gene rifP (Lei et al., 2018).
About the molecular mechanism of NSE, our previously published results of the RNA-seq analysis in A. mediterranei U32 have provided more comprehensive insights: supplementation of nitrate is able to remarkably increase the transcripts of the rif cluster genes to accumulate more rifamycin-biosynthesis enzymes; furthermore, the transcriptions of some other genes (such as glnA and nas operon) are also profoundly activated by nitrate supplementation to furnish more precursors for rifamycin biosynthesis (Shao Z.H. et al., 2015). Accordingly, in that work we speculate that nitrate could stimulate the rifamycin production via increasing both the precursor supply and the biosynthetic enzyme accumulation (Shao Z.H. et al., 2015). In addition, our previous genetic analyses have established that GlnR may participate in the nitrate-mediated regulation on rifamycin biosynthesis, as the deletion of glnR from A. mediterranei resulted in the loss of NSE (Yu et al., 2007); however, the underlying molecular mechanism remains ill-defined.
In actinomycetes, GlnR is an OmpR-type transcriptional regulator, and those early studies show that most GlnR-regulated genes (e.g., amtB, nirB, nasA, ureA, glnA, and ald) are involved in the assimilation and utilization of a variety of nitrogen sources, such as ammonium, nitrate and urea (Fink et al., 2002; Tiffert et al., 2008; Wang and Zhao, 2009; Wang et al., 2012, 2013, 2014). Accordingly, the GlnR in actinomycetes is mainly considered as a global nitrogen metabolism regulator. Later, increasing evidences demonstrate that GlnR also regulates the transcription of other genes involved in carbon metabolism, antibiotic biosynthesis and so on (Liao et al., 2015; Qu et al., 2015; Shao Z. et al., 2015; Cen et al., 2016; He et al., 2016; You et al., 2017; Liu et al., 2019).
In this study, the in vivo analyses clearly reveal that GlnR positively regulates rif cluster transcription and rifamycin biosynthesis in A. mediterranei U32. Then the EMSAs and DNase I footprinting assays show that GlnR can specifically bind to the upstream regions of genes rifK and rifZ, respectively, which lead us to disclose that both genes in fact have their own promoters on the basis of primer extension assays. It is well-known that RifK is responsible for the rifamycin starter unit AHBA biosynthesis, and RifZ is the pathway-specific regulator for the whole rif cluster transcription. Therefore, one may easily conclude that GlnR both directly and indirectly regulates rif cluster genes transcription to control rifamycin biosynthesis, which well interprets our previous findings that the deletion of glnR resulted in the loss of NSE in A. mediterranei.
Results
GlnR Positively Regulates Rifamycin Biosynthesis Through Activating the Transcription of rif Cluster
To explore the role of GlnR in the NSE in A. mediterranei U32, we carefully compared the time course of bacterial growth and rifamycin yield between the glnR– and glnR+ strains. The glnR null mutant (ΔglnR) was constructed using double crossover recombination (Yu et al., 2007; Lin et al., 2014). To obtain the glnR complemented strain (glnR+), the glnR gene (with its native promoter) was integrated into the chromosome of ΔglnR, employing the integrative plasmid pDZL803 as the vector (Lin et al., 2014; Li et al., 2017). As a control, ΔglnR was integrated with the vector to obtain ΔglnR/803 (Lin et al., 2014), which was used to investigate the influence of the empty plasmid on both bacterial growth and rifamycin production.
In liquid Bennet medium supplemented with 80 mM nitrate, the above four strains showed similar growth rates before 48 h (Figure 1A). After 48 h, the wild type U32 and glnR+ strains started to produce a large amount of rifamycin, but ΔglnR and ΔglnR/803, both of which showed a higher growth rate, produced significantly less rifamycin (only ∼15% of the wild type) by 120 h (Figure 1). Moreover, although the ΔglnR showed a little higher growth rate than the wild type U32 when grown in liquid Bennet medium without nitrate, it had a significantly lower yield of rifamycin than the yield of U32 (Supplementary Figure S2), which was similar with the above results under nitrate condition. Based on these results, one may conclude that GlnR has weakly negative effect on bacterial growth but positively regulates the rifamycin biosynthesis in A. mediterranei.
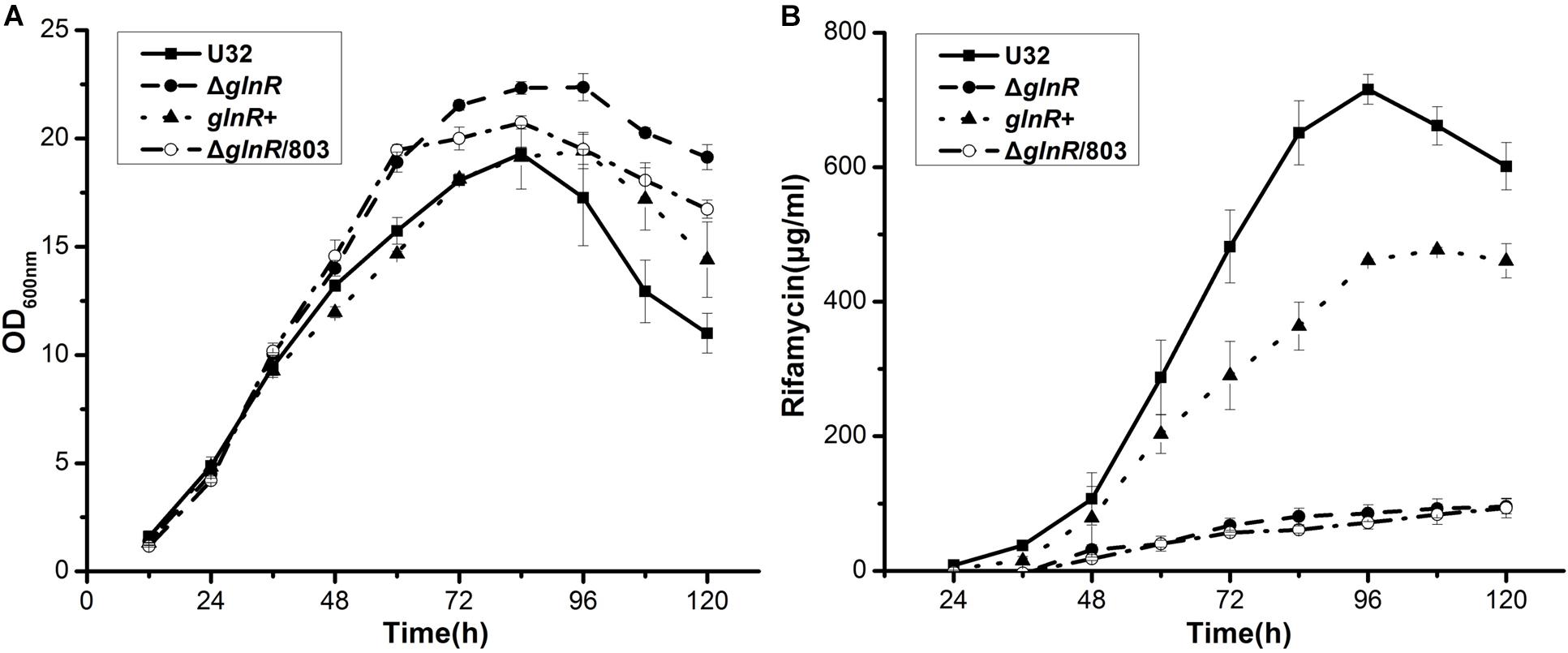
Figure 1. Effects of the glnR deletion on bacterial growth and rifamycin production in A. mediterranei when cultured in liquid Bennet medium containing 80 mM KNO3. (A) Growth curves of four A. mediterranei strains were determined by measuring the OD600 values of the cultures. (B) The rifamycin yield was measured with the differential spectrophotometric method. Error bars represented the SD from three independent biological replicates. U32, the wild type; ΔglnR, the glnR null mutant; glnR+, ΔglnR complemented with an intact glnR; ΔglnR/803, glnR null mutant integrated with the vector pDZL803.
Then we used quantitative real-time PCR (qRT-PCR) to determine the potential regulatory influence of GlnR on the transcription of rif cluster. The same four strains were cultured in liquid Bennet medium supplemented with 80 mM nitrate, and samples were taken for RNA extraction at 24, 48, and 72 h, which respectively represented the early mid-logarithmic phase, mid-logarithmic phase, and early stationary phase (ref to the Figure 1A). Our previous work has established that the rif cluster consists of 10 polycistronic operons (Supplementary Figure S1) (Li et al., 2017), and here we chose at least one gene from each of these operons to perform transcription analysis. It was found that the transcriptional levels of nearly all of the tested genes in glnR– strains (i.e., ΔglnR and ΔglnR/803) were remarkably reduced compared to those in the wild type, especially at 24 h, when their ratio was only ∼1% (Figure 2). When the mutant ΔglnR was complemented with an intact glnR, the transcriptional levels of most rif genes in glnR+ were restored to levels comparable with the wild type (Figure 2). In addition, the transcriptional levels of rif genes were compared between the wild type U32 and ΔglnR when grown in liquid Bennet medium without nitrate supplementation, and it was found that the tested genes in the ΔglnR again showed profoundly reduced transcriptional levels at both 24 h and 48 h (Supplementary Figure S3). These results clearly demonstrate that GlnR is a transcriptional activator for the whole rif cluster.
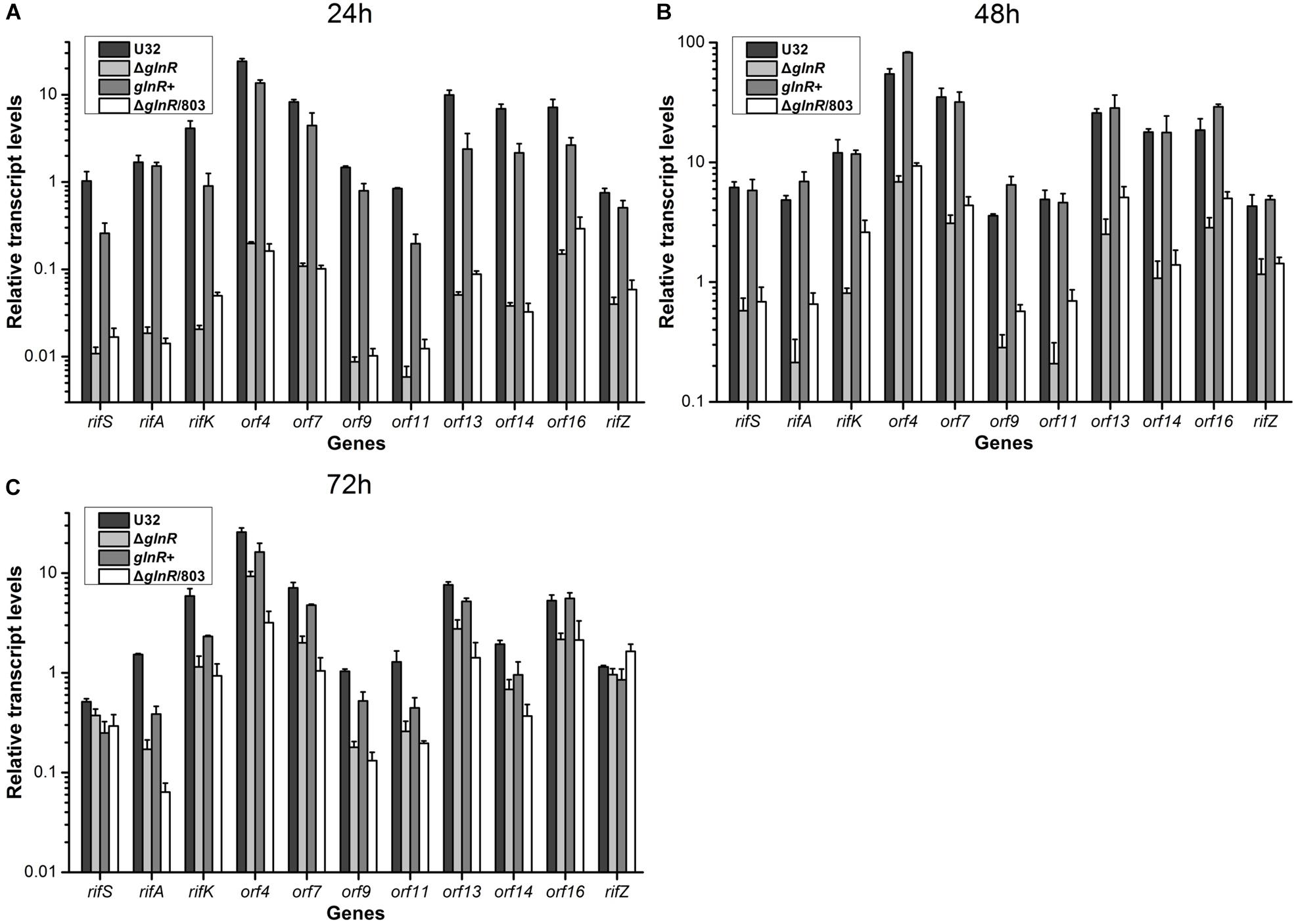
Figure 2. Transcriptional analyses of the rif cluster genes in A. mediterranei strains when cultured in liquid Bennet medium containing 80 mM KNO3. The relative transcriptional levels of 11 representative genes in the rif cluster were measured using qRT-PCR at 24 h (A), 48 h (B), and 72 h (C), respectively. The rpoB gene was used as the internal control, and the transcriptional level of the rifS gene in U32 at 24 h was used as the reference (i.e., its value was adjusted to 1). Error bars represented the SD from at least three independent biological replicates. U32, the wild type; ΔglnR, the glnR null mutant; glnR+, ΔglnR complemented with an intact glnR; ΔglnR/803, ΔglnR integrated with the empty plasmid pDZL803.
GlnR Indirectly Activates the Whole rif Cluster Transcription Through Regulating the Expression of the Pathway-Specific Regulator RifZ
Given our findings that GlnR regulates the transcription of all the rif genes we tested, and considering the known fact that RifZ is a pathway-specific activator for the whole rif cluster, we wondered whether GlnR may indirectly regulate the transcription of all rif genes by activating the transcription of the rifZ gene. Note that the rifZ co-transcribes with the upstream gene rifJ and these two genes together form an operon (Li et al., 2017), we then performed EMSA and DNase I footprinting assay to test whether GlnR directly bound to the promoter region of this operon but both obtained negative results (Figure 3A and Supplementary Figure S4), which indicated that GlnR cannot directly bind to the promoter of the rifJ-rifZ operon.
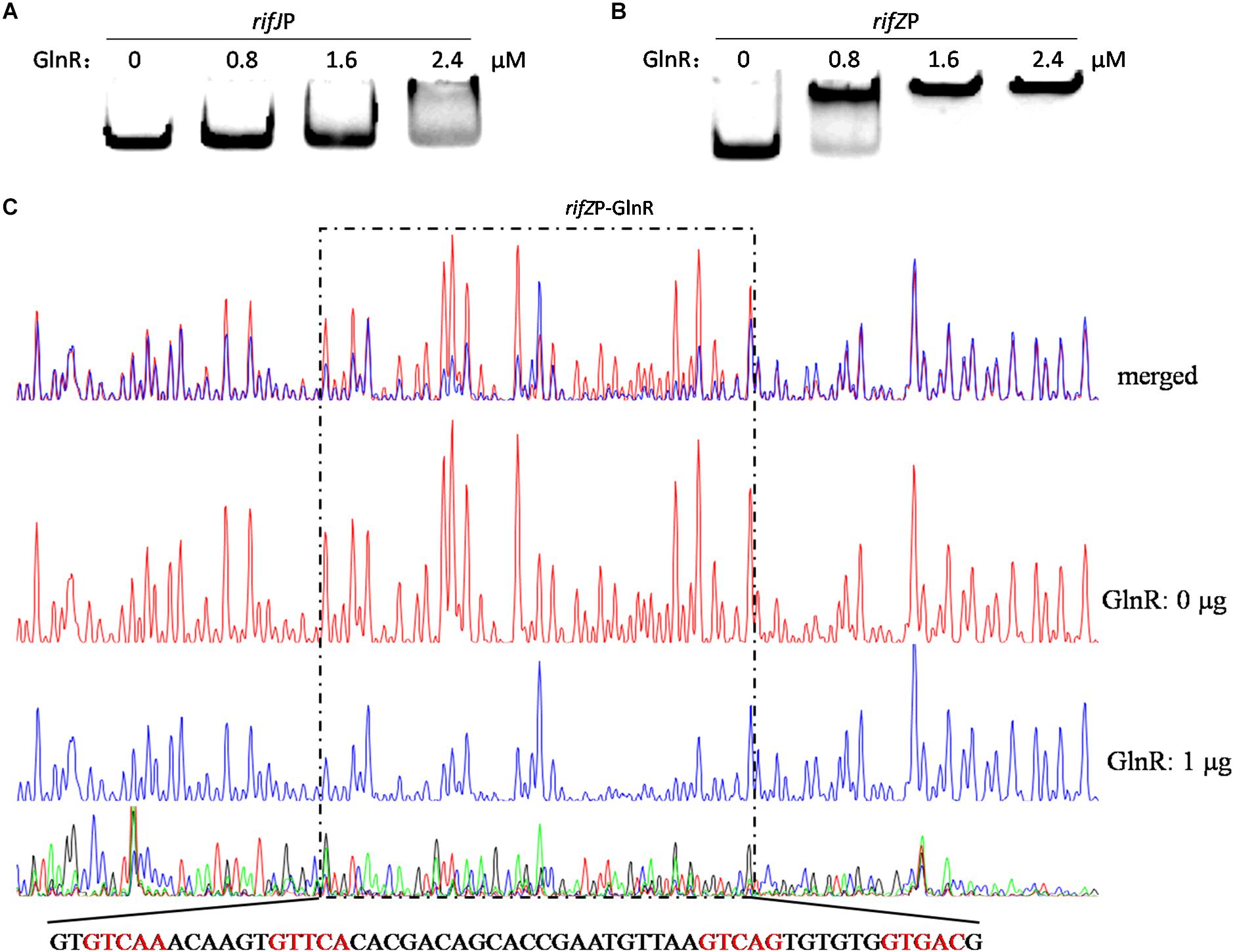
Figure 3. Analyses of the GlnR binding affinity to the rifZ promoter region. (A) Analysis of the GlnR binding affinity to the promoter region of the rifJ-rifZ operon using EMSA. (B) Analysis of the GlnR binding affinity to the rifZ promoter region using EMSA. (C) Characterization of the GlnR-protected sequences in the rifZ promoter via DNase I footprinting assay. Reactions were performed either without (the control reaction, red line) or with (the experimental reaction, blue line) the addition of the GlnR protein, and the electrophoretograms were merged to ease the identification of the GlnR-protected region, which was indicated by a dashed box. The precise DNA sequence was shown at the bottom, and the four predicted typical GlnR-binding sites were highlighted in red.
Noticeably, the length of the intergenic region between rifJ and rifZ is more than 600 bps. More importantly, from the previously published RNA-seq data set (Shao Z.H. et al., 2015), we found that the transcription of rifZ had a more than 10-fold increase relative to the transcription of rifJ (Supplementary Table S1). Thus, we speculate that in addition to sharing the promoter of the operon, rifZ probably also has its own promoter to be operated by transcriptional regulators. To test this hypothesis, we firstly performed EMSA and found GlnR was able to specifically bind to the DNA sequence immediately preceding the gene rifZ (i.e., the rifZ promoter) (Figure 3B). Subsequently, we employed the DNase I footprinting assay to precisely determine the GlnR-protected DNA sequence in the rifZ promoter, and identified a 56-bp region (Figure 3C). In this region, we found two DNA sequences (GTCAC-n6-CTGAC, TGAAC-n6-TTGAC), which well meet the criteria of the GlnR binding sites (i.e., the fourth base should be ‘A’) (Wang et al., 2015). Here both the EMSA and DNase I footprinting results indicate that GlnR can specifically bind to the rifZ promoter and thereby directly operate the transcription of the pathway-specific regulator-encoding gene rifZ.
Transcription Initiation Site Analysis Reveals rifZ Has Its Own Promoter
To fully understand this GlnR-mediated transcriptional regulation mechanism, we characterized the transcription initiation site (TIS) of the gene rifZ using the primer extension assay. In detail, two specific primers were designed to be complementary to either the rifZ coding DNA sequence (CDS) or the rifZ promoter region (Figure 4B and Supplementary Figure S5A), and both the primer extension results showed that the transcription of rifZ began from an adenine (A) site, which was located at the −328th nucleotide (nt) position relative to the translational start site of rifZ (Figure 4 and Supplementary Figures S5B,C). In addition, we also verified the rifZ TIS via qRT-PCR using two pairs of primers located upstream and downstream of the TIS, respectively. And results showed that the transcriptional level of the TIS downstream region was much higher than that of the upstream region (Supplementary Figure S5D), which further proves this TIS characterized by the primer extension assay.
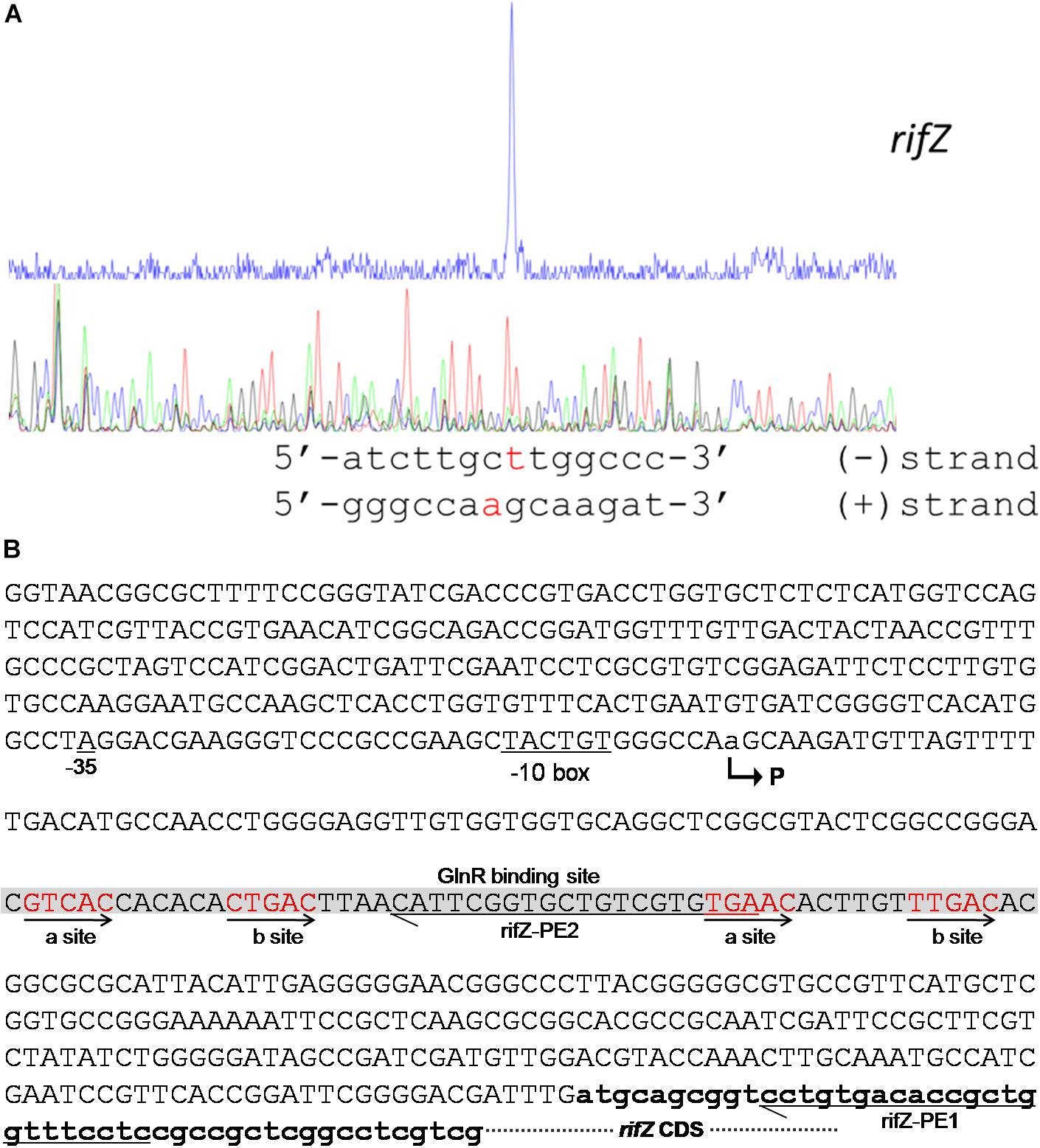
Figure 4. Characterization of the rifZ promoter. (A) Characterization of the rifZ TIS via primer extension assay. During the assays two different primers were used, and the primer extension result using the primer rifZ-PE2 was shown here. Locations of both primers as well as the primer extension result using the primer rifZ-PE1 could be found in Supplementary Figure S5. After the primer extension reaction, the samples were analyzed by the electrophoresis, whose results were shown in the upper panel. Sequencing results of the promoter region were shown in the bottom panel, and the identified TIS of rifZ was marked in red. (B) Schematic of the cis-elements in the rifZ promoter. The TIS of rifZ was indicated by a bent arrow and labeled as “P”. The GlnR-protected sequences were shown in gray background and the GlnR-binding sites (a site and b site) were shown in red and indicated by arrows. Those two primers (rifZ-PE1 and rifZ-PE2) used for primer extension assays in Supplementary Figure S5 were also labeled.
Therefore, the primer extension results further demonstrate that the gene rifZ undoubtedly has its own promoter. Although the DNase I footprinting-confirmed GlnR binding sequences are positioned downstream of the rifZ TIS, we have already established that GlnR positively regulates the transcription of rifZ on the basis of the genetic experimental results above (Figures 1, 2). Therefore, we speculate that here the GlnR-mediated regulation is unlikely to accord with a typical activation model, wherein an activator binds to the sequence preceding a gene’s TIS and recruits the RNA polymerase complex to initiate the transcription.
GlnR Directly Activates the Transcription of the AHBA Synthase-Encoding Gene rifK
Besides rifZ, we also tested whether GlnR bound to the promoter regions of other genes in the rif cluster. Firstly, our previous work has established that the rif cluster consists of 10 polycistronic operons, and these operons in fact shares only six promoter regions as four pairs of them are divergently transcribed (refer to the Supplementary Figure S1) (Li et al., 2017). Then, we re-analyzed the transcription profiles of all the rif genes in the previously published RNA-seq data set (Shao Z.H. et al., 2015), and found a fascinating phenomenon in their transcription levels: besides the gene rifZ mentioned above, there are still six intra-operonic genes had a ≥2-fold transcription increase relative to the transcription of their immediately preceding gene (Supplementary Table S1). Thus, it appears that in addition to sharing the promoter of the given operon, these six genes probably also have their own promoter. Finally, coupled with another two experimentally verified intra-operonic promoters of rifP (Lei et al., 2018) and rifZ (just characterized in the Figure 4), there are collectively at least 14 promoters within the rif cluster.
Since the two promoters of rifZ and rifJ-rifZ operon had been respectively tested above, here we only analyzed the GlnR binding affinity to the other 12 promoter regions using EMSA. The EMSA results showed that GlnR was able to significantly bind to the rifK promoter with a relatively higher binding affinity (Figure 5A). Furthermore, from the Figure 5A, it can be found that GlnR might also bind to another four promoters (rifSP, orf35P, rifPP and orf6P), but the binding affinity to them was much reduced relative to the promoters of both rifK and rifZ.
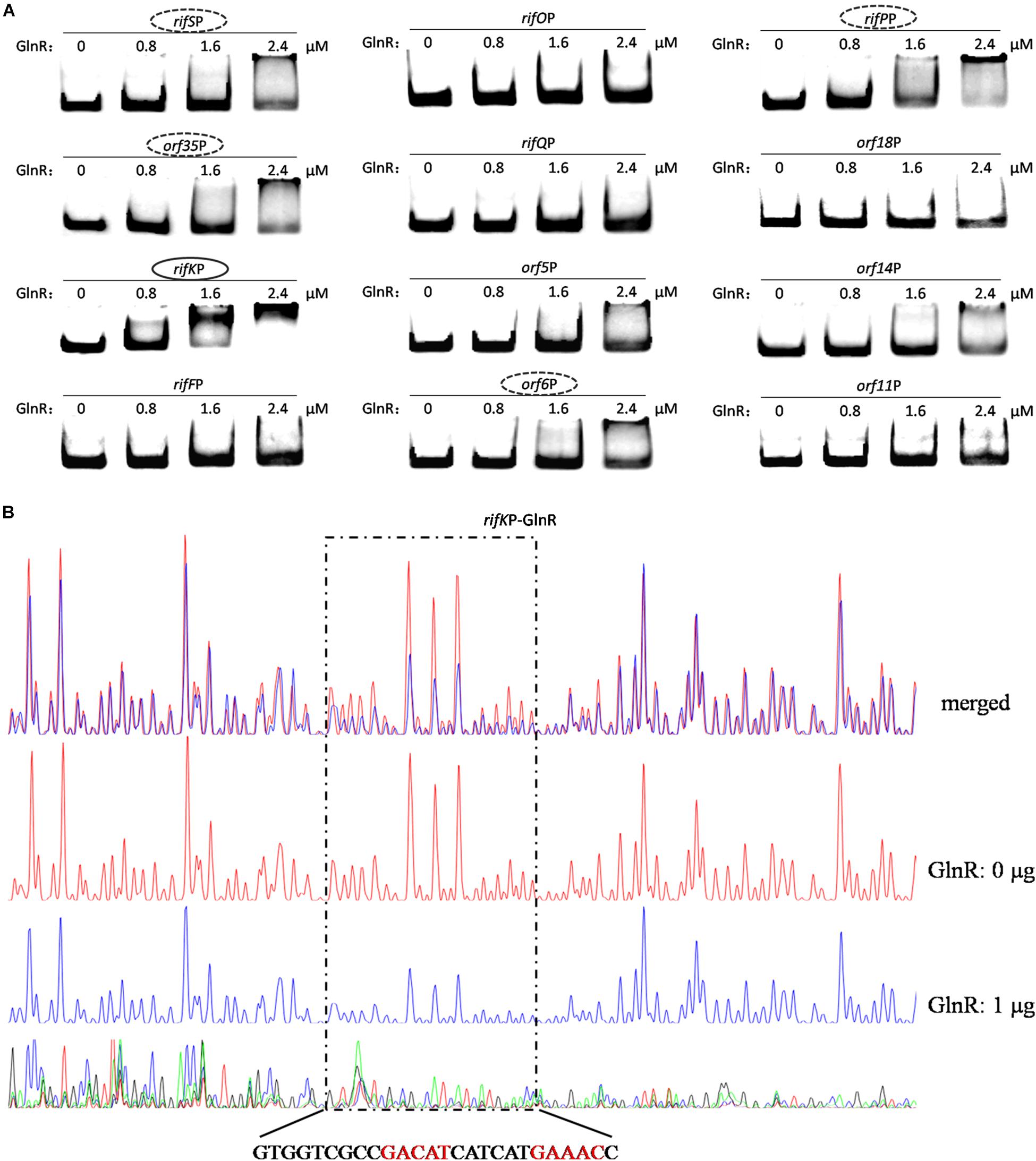
Figure 5. Analyses of the GlnR binding affinity to the rifK promoter region. (A) Analyses of the GlnR binding affinity to 12 promoter regions within the rif cluster using EMSA. To prevent the non-specific binding between the GlnR and probes, 2 μg sheared salmon sperm DNA was added into each reaction. Based on the amount of band shift, putative GlnR-targeted promoters were indicated by circles (with strong binding affinity) and dashed circles (with weak binding affinity). (B) Characterization of the GlnR-protected sequences in the rifK promoter via DNase I footprinting assay. The reactions were performed either without (the control reaction, red line) or with (the experimental reaction, blue line) the addition of the GlnR protein. The precise DNA sequences protected by GlnR were shown at the bottom, and the two predicted typical GlnR-binding sites were highlighted in red. Results of the DNase I footprinting assays of GlnR to other four putative targeted promoters (i.e., the dashed circles in Figure 5A) could be found in Supplementary Figure S6.
To verify these (possibly) positive EMSA results in Figure 5A and further understand the binding behavior, we next performed the DNase I footprinting assay to precisely determine the GlnR binding sequences in the five promoter regions, and found that GlnR only specifically protected a region in the rifK promoter (Figure 5B) but protected no region in the other four promoters (where the GlnR binding affinity was much reduced during the EMSAs) (Supplementary Figure S6). Therefore, these DNase I footprinting results lead us to accurately identify the true positive results of EMSAs, where too much regulatory protein easily generate false positive results.
In the rifK promoter, GlnR protected a region from the −280th nt to the −255th nt relative to the rifK translational start site (Figure 6B), which did not overlap with the protected region by RifZ, another direct activator for rifK transcription (Li et al., 2017). And in the GlnR-protected region, a sequence of “GACAT-n6-GAAAC” was found, which well meets the criteria of the GlnR binding sites (Wang et al., 2015). As both GlnR and RifZ activate rifK transcription and their binding sequences are not overlapped, we speculate that these two regulators might be cooperative in regulating the transcription of rifK. Thereafter, to clarify the relative importance of GlnR and RifZ for the expression of rifK, we measured the transcriptional profiles of rifK in three strains (U32, ΔglnR and ΔrifZ) when cultured in liquid Bennet medium with 80 mM nitrate. Interestingly, it was found that the rifK’s transcriptional level in ΔrifZ was a little higher than in ΔglnR at 24 h, while its level in ΔrifZ was much lower than in ΔglnR at 48 and 72 h (Supplementary Figure S7). These results indicate that GlnR may be relatively more important for the rifK transcription comparing with RifZ during the early phase, and then the RifZ gradually surpasses the GlnR and plays a dominant role in the rifK transcription.
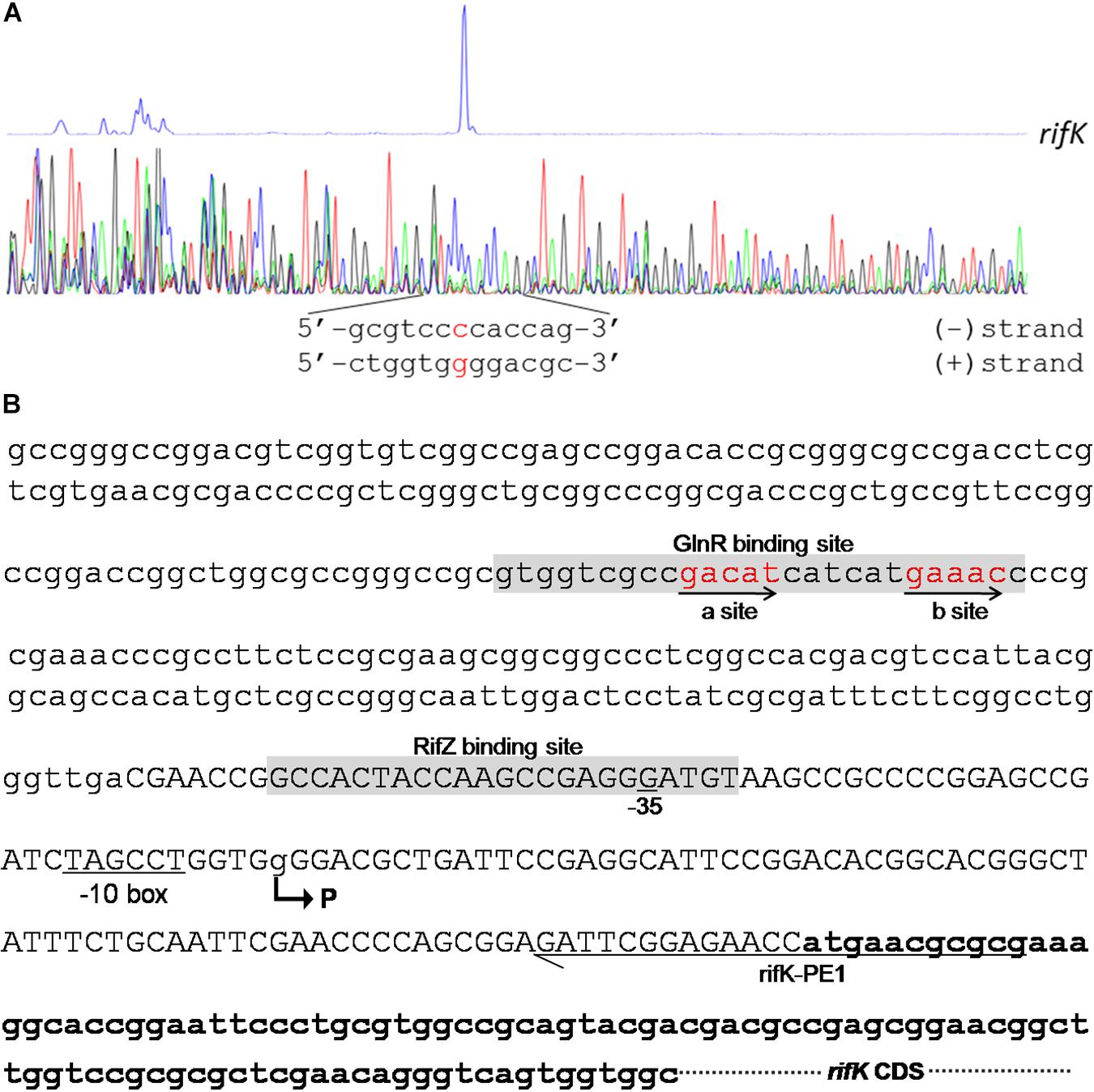
Figure 6. Characterization of the rifK promoter. (A) Characterization of the TIS of rifK via primer extension assay. (B) Schematic of the cis-elements in the rifK promoter. The TIS of rifK was indicated by a bent arrow and labeled as “P”. Both the GlnR-protected and RifZ-protected sequences were shown in gray background, and the typical GlnR-binding sites (a site and b site) were shown in red and indicated by arrows. The primer (rifK-PE1) used for the primer extension assay in Figure 6A was also labeled.
Transcription Initiation Site Analysis Reveals rifK Has Its Own Promoter
To further investigate the molecular mechanism of GlnR-mediated activation of the rifK transcription, we performed primer extension assay to identify the TIS of the gene rifK using a primer complementary to the −13th nt to the 11th nt of the rifK CDS, and found rifK transcribed from a guanine (G) site which is located at the −79th nt position relative to the rifK translational start site (Figure 6). Noticeably, the -10 box (TAGCCT) of the rifK promoter was very similar to the previously characterized −10 box of promoters in S. coelicolor and A. mediterranei (Yu et al., 2006; Wang et al., 2013). In addition, as the GlnR-protected region is located upstream of the −35 box in the rifK promoter (Figure 6B), we speculate that GlnR might activate the rifK transcription through recruiting the RNA polymerase complex using a mode just similar to other typical transcriptional activators.
Discussion
The NSE, i.e., the phenomenon that nitrate-stimulated remarkable increase of rifamycin biosynthesis in A. mediterranei, was initially demonstrated by our laboratory about 40 years ago (Jiao et al., 1979). Then it was noticed that the NSE did not occur in the glnR null mutant, however, the underlying molecular mechanism is still unknown (Yu et al., 2007). In the present study, we uncover the mechanism of GlnR controlling the rifamycin biosynthesis in A. mediterranei U32 via multifarious experiments both in vivo and in vitro.
The phenotypic analyses (Figure 1 and Supplementary Figure S2) showed that, unlike the previous work in Streptomyces (He et al., 2016), the cellular growth under our cultivation conditions was not impaired by glnR deletion at all, which ruled out the impact of the growth factor on gene transcription and on antibiotic production, thus making our conclusions from this study (especially the Figures 1, 2 and Supplementary Figures S2, S3) more reliable. The Supplementary Figure S2B showed that ΔglnR had a significantly lower yield of rifamycin comparing with the wild type U32 when grown in liquid Bennet medium without nitrate, whereas in our previous work, the rifamycin yield of ΔglnR was a little bit higher than that of U32 when grown on Bennet agar plates without nitrate supplementation (Yu et al., 2007). Here we speculated this difference might be caused by the cultivation conditions (i.e., liquid medium versus solid medium).
During the transcriptional assays, we noticed that the transcriptional levels of rif genes in the wild type U32 stayed high till 72 h under nitrate condition (Figure 2) but had dropped dramatically by the time point 48 h without nitrate supplementation (Supplementary Figure S3). These results were quite consistent with our previous RNA-seq results, where the nitrate supplementation well maintained the high-level transcription of the rif cluster (a key character/mechanism of NSE) (Shao Z.H. et al., 2015). And from the Figure 2, we found that the high-level transcription of the rif cluster under nitrate condition is severely impaired by the glnR deletion (especially at 24 and 48 h), which demonstrates that one reason for GlnR being necessary for NSE is its remarkable up-regulation on the rif cluster transcription. Interestingly, the Supplementary Figure S3 shows that GlnR can also up-regulate the rif cluster transcription under no nitrate condition (i.e., NSE cannot occur), indicating that NSE needs other factors besides the GlnR-mediated regulation on the rif cluster transcription. Furthermore, the magnitude of the differences in transcriptional levels between the glnR– and glnR+ strains decreased significantly over time (Figure 2 and Supplementary Figure S3), indicating that the rif cluster (especially rifZ gene) may be controlled by other unknown mechanisms as well, which will be the focus of our future research.
All of the in vivo evidence in this study have clearly demonstrated that GlnR is an activator for the transcription of rifZ and rif cluster. However, the in vitro experimental results show that the GlnR binding regions are located downstream of the rifZ TIS, which is different from the binding locations of the typical transcriptional activators. In fact, some previous reports have shown that transcriptional regulators can have binding sites downstream of the TIS but still function as activators, e.g., Rns (Munson and Scott, 2000), MetR (Cowan et al., 1993), PhoP (Liu et al., 1998; Qi and Hulett, 1998), DnaA (Szalewska-Palasz et al., 1998), and FleQ (Baraquet and Harwood, 2015), with multiple distinct proposed regulatory mechanisms. Therefore, here we speculate that GlnR probably also activates rifZ transcription via an atypical activating model, which is worthy of detailed investigation in the future.
In addition, based on the results of both EMSA and DNase I footprinting assay (Figures 3, 5), we noticed that GlnR had lower binding affinity for the rifK promoter than for the rifZ promoter. For a regulator, an increased number of binding sites in its target sequences usually indicates higher binding affinity. Quite consistent with the different GlnR binding affinities against the two promoters, there are four typical GlnR-binding sites in the rifZ promoter, but only two typical GlnR-binding sites in the rifK promoter.
Here our study fully illustrates that the well-known global nitrogen regulator GlnR governs the rifamycin biosynthesis via activating the rif cluster transcription in both direct and indirect manners in A. mediterranei U32 (Figure 7). And combining the results of this study and some previous studies, nowadays we can have a more comprehensive insight that GlnR plays a significant role in NSE by acting as a global regulator to coordinate both primary nitrogen metabolism and secondary metabolism. In detail, when the U32 is cultured in Bennet medium with nitrate, which is a designated nitrogen-limited condition specially for U32 (Wang et al., 2014), GlnR activates the transcription of the primary nitrogen metabolism related genes (e.g., glnA and nas operon) to enhance the glutamine biosynthesis (Yu et al., 2006; Wang et al., 2013). And it also has been established that the glutamine provides the sole nitrogen atom for rifamycin biosynthesis (Jin and Jiao, 1986), therefore GlnR promotes the precursor supply for rifamycin biosynthesis by regulating the primary nitrogen metabolism. In addition to glutamine, the biosynthesis of AHBA is regulated in a very subtle way: on the one hand, GlnR directly activates the transcription of AHBA-synthase-encoding gene rifK via binding to its promoter; on the other hand, GlnR indirectly controls the rifK transcription via activating the expression of the pathway-specific regulator RifZ (Figure 7). As AHBA serves as the starter unit of rifamycin, such a subtle regulation on AHBA synthesis may further facilitate the rifamycin biosynthesis in cells. Besides the gene rifK, the GlnR also upregulates the whole rif cluster transcription indirectly via activating the rifZ expression (Figure 7). And the high-level transcription of the whole rif cluster could ensure the accumulation of rifamycin biosynthesis related enzymes in cells. In our opinion, it is quite compelling that the GlnR unexpectedly exerts such sophisticated regulation on rifamycin biosynthesis in A. mediterranei, which well interprets our previous findings that the NSE does not occur in the glnR null mutant.
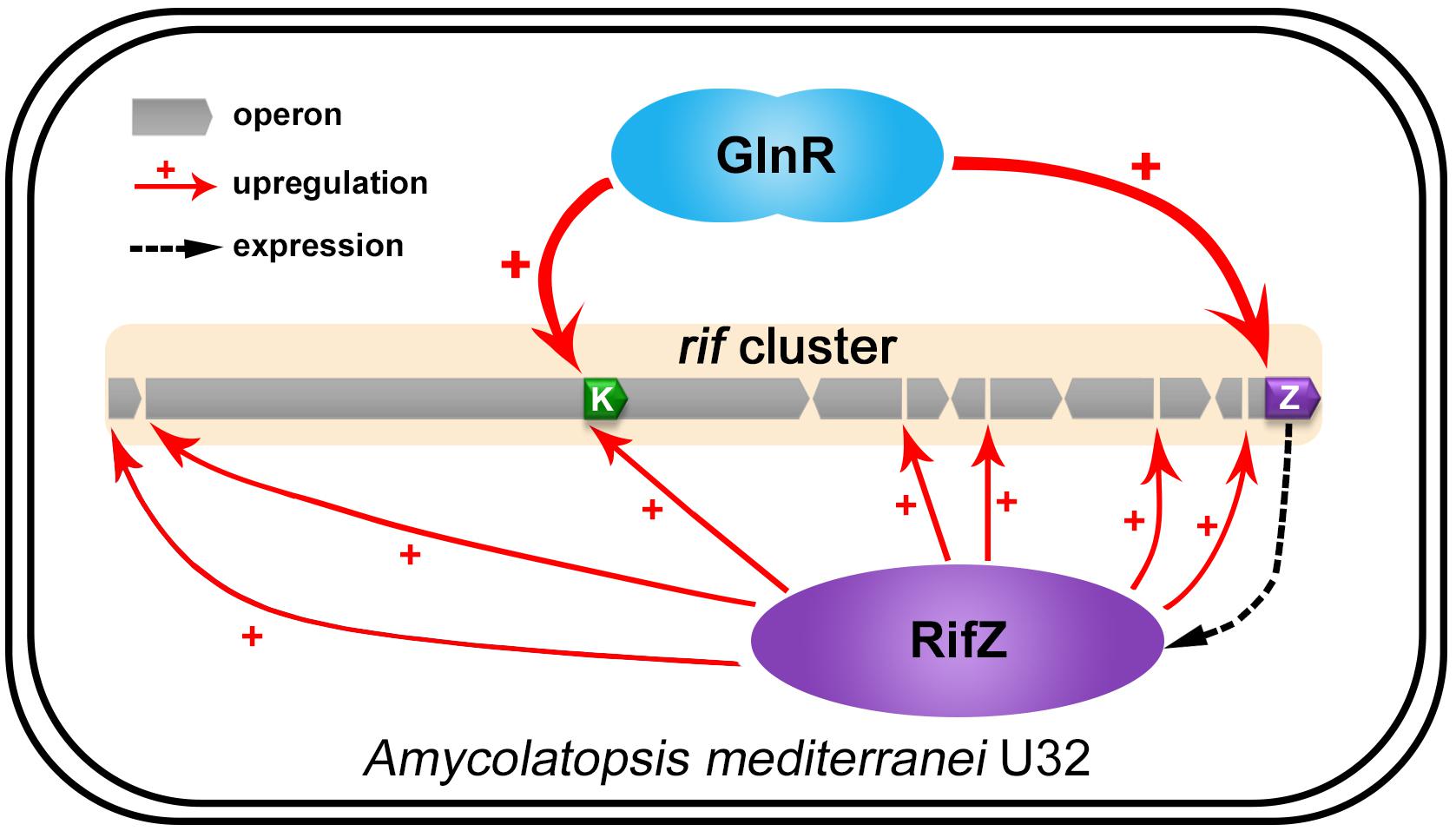
Figure 7. The model of GlnR-mediated activation for the rif cluster genes transcription in A. mediterranei. It has been established that RifZ functions as the pathway-specific activator for the whole rif cluster, therefore GlnR indirectly upregulates the transcription of all the rif cluster genes by activating the rifZ expression. Besides, the gene rifK encodes the AHBA synthase, GlnR therefore directly promotes the biosynthesis of the rifamycin starter unit AHBA by activating the rifK transcription. Taken together, GlnR both directly and indirectly activates the rif cluster genes transcription to enhance the rifamycin biosynthesis in cells, which further interprets the previous findings that the NSE did not occur in the glnR null mutant.
In actinomycetes, GlnR was typically viewed as a global nitrogen metabolism regulator before, governing many nitrogen metabolism related processes (Fink et al., 2002; Tiffert et al., 2008; Wang and Zhao, 2009; Wang et al., 2012, 2013, 2014). Recently, several studies have reported that the GlnR homologs in Streptomyces regulate the biosynthesis of some antibiotics, e.g., actinorhodin, avermectin and validamycin (Qu et al., 2015; He et al., 2016). Here our study also shows that the GlnR in A. mediterranei directly controls the rifamycin biosynthesis. Therefore, taking all these findings together, one may easily conclude that GlnR can function as a global regulator instead of merely a nitrogen metabolism regulator in actinomycetes, coordinating both primary metabolism and secondary metabolism in cells.
Materials and Methods
Bacterial Strains, Media, and Primers
All the primers used in this study are listed in Table 1. And all the bacterial strains and plasmids used here are listed in Table 2. The glnR null mutant (ΔglnR) was obtained by double crossover recombination (Yu et al., 2007; Lin et al., 2014). The glnR complemented strain (glnR+) was constructed by integration of the glnR gene with its native promoter into ΔglnR, using the integrative plasmid pDZL803 (Li et al., 2017), and the construction procedure was described by Lin et al. (2014). Escherichia coli DH10B was used for construction of the recombinant plasmids, and was grown in Luria-Bertani (LB) medium (Sambrook and Russell, 2001). A. mediterranei strains were grown aerobically in Bennet medium (10 g/L glycerol, 10 g/L glucose, 2 g/L tryptone, 1 g/L yeast extract, 1 g/L beef extract, pH 7.0) at 30°C (Yang et al., 2001; Shao Z.H. et al., 2015). If necessary, antibiotics were added at the following concentrations (μg/ml): ampicillin, 100; and kanamycin, 50.
Measurement of the Growth Curve and Rifamycin Yield
After streak cultivation on Bennet plates for 4–5 days, A. mediterranei strains were transferred into 50 ml liquid Bennet medium containing ceramic beads (diameter: 2.5 mm) to avoid mycelia aggregation. After incubation in a shaker at 220 rpm for about 36 h, they were inoculated into 200 ml fresh liquid Bennet medium containing ceramic beads and 0/80 mM KNO3 with the final OD600 = 0.2. The growth curves of different strains were determined by measuring the OD600 values with NanoDrop 2000C (Thermo Fisher Scientific) every 12 h.
After the strains being inoculated into 200 ml liquid Bennet medium with 0 or 80 mM KNO3 for 24 h, 1 ml bacterial suspensions were taken from the fermentation broth at 12 h intervals. For determining the rifamycin yield, the samples were centrifuged and the supernatant was collected. Then the rifamycin yield was measured using the spectrophotometric method as previously described (Pasqualucci et al., 1970). Briefly, 20 μl supernatant was diluted in 180 μl acetate buffer containing 0.1% NaNO2 or 0.1% vitamin C, respectively. Then, the optical density was read at the wavelength of 447 nm, with the NaNO2 group taken as a blank control. Finally, the yield of rifamycin was calculated based on a standard curve prepared by using pure rifamycin SV as standards.
RNA Extraction and Quantitative Real-Time PCR (qRT-PCR)
When A. mediterranei strains were grown in liquid Bennet medium with 0/80 mM KNO3 for 24, 48, and 72 h, 5–10 ml bacterial suspensions were collected and the mycelia were harvested at 4°C by centrifugation at 12,000 rpm for 5 min. Total RNA was then extracted from the mycelia using TRIzol reagent (Thermo Fisher Scientific) and SV Total RNA Isolation System (Promega), following the manufacturers’ instruction. Reverse transcription was performed with random primers using 1 μg total RNA as the template in a volume of 20 μl by the PrimeScript 1st Strand cDNA synthesis kit (TaKaRa), and the qRT-PCR was carried out with SYBR Premix Ex Taq II (Tli RNaseH Plus) (TaKaRa). At least three independent biological samples were tested in this experiment, and the relative transcriptional level of the tested genes was presented as means ± standard deviations (SD), using the rpoB gene as the internal control.
Preparation of FAM-Labeled Probes
Promoter regions in rif cluster were PCR amplified with Phanta Max super-fidelity DNA polymerase (Vazyme), employing primers listed in Table 1. Then these PCR amplicons were separately cloned into the HincII site of pUC18H (Tolo Biotech.). After verification through DNA sequencing, these recombinant plasmids were used as the PCR templates to prepare FAM-labeled probes using primers of HincII-F and HincII-R (6-carboxyfluorescein [FAM] labeled). Finally, the FAM-labeled probes were purified by the Wizard SV gel and the PCR Clean-Up system (Promega), and quantified with NanoDrop 2000C (Thermo Fisher Scientific).
Electrophoretic Mobility Shift Assay (EMSA) and DNase I Footprinting Assay
The heterogenous expression and purification of the His-tagged GlnR of A. mediterranei U32 were performed as previously described (Wang et al., 2013). EMSA was carried out using the purified His-tagged GlnR and FAM-labeled probes, following the same procedures as described in our previous work (Li et al., 2017). DNase I footprinting assay was performed by Tolo Biotech. according to the method described by Wang et al. (2012). In brief, each FAM-labeled probe of about 300 ng was incubated with different amounts of His-tagged GlnR for 30 min at 25°C. After DNase I digestion, the stop solution was added to stop the reaction. The promoter region was sequenced using specific primer with FAM labeled at the 5′-ends, following the procedures described before (Wang et al., 2012). The samples together with the sequencing products were then purified and loaded into an ABI 3130 sequencer for electrophoresis analysis. The electropherograms were analyzed with PeakScanner v1.0 software (Applied Biosystems) to characterize the precise DNA sequences protected by GlnR.
Primer Extension Assay
Primer extension assay was carried out by Tolo Biotech (Lei et al., 2018). To prepare total RNA for primer extension assay, A. mediterranei U32 was inoculated into liquid Bennet medium with 80 mM KNO3 and incubated in a shaker at 30°C for about 36 h. Then, the mycelia were harvested by centrifugation, followed by extraction of the total RNA using a protocol the same as described above. For each assay, about 60 μg total RNA was used to characterize the TIS, employing FAM-labeled primers listed in Table 1. Sequencing analysis of the promoter region with the FAM-labeled primer was the same as that described before (Lei et al., 2018). The subsequent electrophoresis and analysis were the same as the procedures in the DNase I footprinting assay described above.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
XL performed most of the experiments and prepared the draft. YL performed the primer extension assay. YL and XL performed the DNase I footprinting experiment. CL prepared the FAM-labeled probes. JW and GZ designed the study. JW revised the manuscript and supervised the whole project.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31430004, 31670058, 31770057, and 81671988). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Conflict of Interest
YL was employed by the company Shanghai Tolo Biotechnology Company Limited.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Xuan Zheng for her assistance in drawing the model diagram of GlnR-mediated transcriptional activation of the rif cluster in Figure 7. We also thank Hua Yuan for his careful revision of this manuscript. We would like to dedicate this manuscript to the late Professor Ruishen Jiao in commemoration of the 10th anniversary of his passing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00319/full#supplementary-material
Abbreviations
AHBA, 3-amino-5-hydroxybenzoic acid; CDS, coding DNA sequences; EMSA, electrophoretic mobility shift assay; NSE, nitrate-stimulating effect; qRT-PCR, quantitative real-time PCR; TIS, transcription initiation site.
References
August, P. R., Tang, L., Yoon, Y. J., Ning, S., Muller, R., Yu, T. W., et al. (1998). Biosynthesis of the ansamycin antibiotic rifamycin: deductions from the molecular analysis of the rif biosynthetic gene cluster of Amycolatopsis mediterranei S699. Chem. Biol. 5, 69–79. doi: 10.1016/s1074-5521(98)90141-7
Baraquet, C., and Harwood, C. S. (2015). FleQ DNA binding consensus sequence revealed by studies of FleQ-dependent regulation of biofilm gene expression in Pseudomonas aeruginosa. J. Bacteriol. 198, 178–186. doi: 10.1128/JB.00539-515
Campbell, E. A., Korzheva, N., Mustaev, A., Murakami, K., Nair, S., Goldfarb, A., et al. (2001). Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 104, 901–912. doi: 10.1016/S0092-8674(01)00286-280
Cen, X. F., Wang, J. Z., Zhao, G. P., Wang, Y., and Wang, J. (2016). Molecular evidence for the coordination of nitrogen and carbon metabolisms, revealed by a study on the transcriptional regulation of the agl3EFG operon that encodes a putative carbohydrate transporter in Streptomyces coelicolor. Biochem. Biophys. Res. Commun. 471, 510–514. doi: 10.1016/j.bbrc.2016.02.044
Cowan, J. M., Urbanowski, M. L., Talmi, M., and Stauffer, G. V. (1993). Regulation of the Salmonella typhimurium metF gene by the MetR protein. J. Bacteriol. 175, 5862–5866. doi: 10.1128/jb.175.18.5862-5866.1993
Fink, D., Weissschuh, N., Reuther, J., Wohlleben, W., and Engels, A. (2002). Two transcriptional regulators GlnR and GlnRII are involved in regulation of nitrogen metabolism in Streptomyces coelicolor A3(2). Mol. Microbiol. 46, 331–347. doi: 10.1046/j.1365-2958.2002.03150.x
Floss, H. G., and Yu, T. W. (1999). Lessons from the rifamycin biosynthetic gene cluster. Curr. Opin. Chem. Biol. 3, 592–597. doi: 10.1016/s1367-5931(99)00014-9
Floss, H. G., and Yu, T. W. (2005). Rifamycin-mode of action, resistance, and biosynthesis. Chem. Rev. 105, 621–632. doi: 10.1021/cr030112j
Floss, H. G., Yu, T. W., and Arakawa, K. (2011). The biosynthesis of 3-amino-5-hydroxybenzoic acid (AHBA), the precursor of mC7N units in ansamycin and mitomycin antibiotics: a review. J. Antibiot. 64, 35–44. doi: 10.1038/ja.2010.139
He, J. M., Zhu, H., Zheng, G. S., Liu, P. P., Wang, J., Zhao, G. P., et al. (2016). Direct involvement of the master nitrogen metabolism regulator GlnR in antibiotic biosynthesis in Streptomyces. J. Biol. Chem. 291, 26443–26454. doi: 10.1074/jbc.M116.762476
Jiao, R. S., Chen, Y. M., Wu, M. G., and Gu, W. L. (1979). Studies on the metabolic regulation of biosynthesis of rifamycin by Norcadia (Amycolatopsis) mediterranei I. The stimulative effect of nitrate on biosynthesis of rifamycin SV by Nocardia (Amycolatopsis) mediterranei. Acta Phytophysiol. Sin. 05, 395–402.
Jiao, R. S., Liu, C. J., Jin, Z. K., Zhang, X. C., Ni, L. Y., and Lu, Z. M. (1984). The route of incorporation of nitrogen atom into rifamycin during its biosynthesis. Sci. Sin. B 27, 380–390.
Jin, Z. K., and Jiao, R. S. (1986). Further studies on the route of incorporation of nitrogen atom into rifamycin. Acta Microbiol. Sin. 26, 265–270.
Kumari, R., Singh, P., and Lal, R. (2016). Genetics and genomics of the genus Amycolatopsis. Indian J. Microbiol. 56, 233–246. doi: 10.1007/s12088-016-0590-598
Lee, J. G., Choi, C. Y., Seong, B. L., and Han, M. H. (1983). Optimal Ph profile in rifamycin-B fermentation. J. Ferment. Technol. 61, 49–53.
Lei, C., Wang, J., Liu, Y., Liu, X., Zhao, G., and Wang, J. (2018). A feedback regulatory model for RifQ-mediated repression of rifamycin export in Amycolatopsis mediterranei. Microb. Cell Fact. 17:14. doi: 10.1186/s12934-018-0863-865
Li, C., Liu, X., Lei, C., Yan, H., Shao, Z., Wang, Y., et al. (2017). RifZ (AMED_0655) is a pathway-specific regulator for rifamycin biosynthesis in Amycolatopsis mediterranei. Appl. Environ. Microbiol. 83, e3201–e3216. doi: 10.1128/AEM.03201-3216
Liao, C. H., Yao, L., Xu, Y., Liu, W. B., Zhou, Y., and Ye, B. C. (2015). Nitrogen regulator GlnR controls uptake and utilization of non-phosphotransferase-system carbon sources in actinomycetes. Proc. Natl. Acad. Sci. U.S.A. 112, 15630–15635. doi: 10.1073/pnas.1508465112
Lin, W., Wang, Y., Han, X., Zhang, Z., Wang, C., Wang, J., et al. (2014). Atypical OmpR/PhoB subfamily response regulator GlnR of actinomycetes functions as a homodimer, stabilized by the unphosphorylated conserved Asp-focused charge interactions. J. Biol. Chem. 289, 15413–15425. doi: 10.1074/jbc.M113.543504
Liu, W., Qi, Y., and Hulett, F. M. (1998). Sites internal to the coding regions of phoA and pstS bind PhoP and are required for full promoter activity. Mol. Microbiol. 28, 119–130. doi: 10.1046/j.1365-2958.1998.00779.x
Liu, W. B., Liu, X. X., Shen, M. J., She, G. L., and Ye, B. C. (2019). The nitrogen regulator GlnR directly controls transcription of the prpDBC operon involved in methylcitrate cycle in Mycobacterium smegmatis. J. Bacteriol. 201:e00099-19. doi: 10.1128/JB.00099-19
Mejia, A., Barrios-Gonzalez, J., and Viniegra-Gonzalez, G. (1998). Overproduction of rifamycin B by Amycolatopsis mediterranei and its relationship with the toxic effect of barbital on growth. J. Antibiot. 51, 58–63. doi: 10.7164/antibiotics.51.58
Munson, G. P., and Scott, J. R. (2000). Rns, a virulence regulator within the AraC family, requires binding sites upstream and downstream of its own promoter to function as an activator. Mol. Microbiol. 36, 1391–1402. doi: 10.1046/j.1365-2958.2000.01957.x
Pasqualucci, C. R., Vigevani, A., Radaelli, P., and Gallo, G. G. (1970). Improved differential spectrophotometric determination of rifamycins. J. Pharm. Sci. 59, 685–687. doi: 10.1002/jps.2600590522
Peano, C., Damiano, F., Forcato, M., Pietrelli, A., Palumbo, C., Corti, G., et al. (2014). Comparative genomics revealed key molecular targets to rapidly convert a reference rifamycin-producing bacterial strain into an overproducer by genetic engineering. Metab. Eng. 26, 1–16. doi: 10.1016/j.ymben.2014.08.001
Qi, Y., and Hulett, F. M. (1998). PhoP-P and RNA polymerase sigmaA holoenzyme are sufficient for transcription of Pho regulon promoters in Bacillus subtilis: PhoP-P activator sites within the coding region stimulate transcription in vitro. Mol. Microbiol. 28, 1187–1197. doi: 10.1046/j.1365-2958.1998.00882.x
Qu, S., Kang, Q., Wu, H., Wang, L., and Bai, L. (2015). Positive and negative regulation of GlnR in validamycin A biosynthesis by binding to different loci in promoter region. Appl. Microbiol. Biotechnol. 99, 4771–4783. doi: 10.1007/s00253-015-6437-0
Rothstein, D. M. (2016). Rifamycins, alone and in combination. Cold Spring Harb. Perspect. Med. 6:a027011. doi: 10.1101/cshperspect.a027011
Sambrook, J., and Russell, D. W. (2001). Molecular Cloning : A Laboratory Manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press.
Shao, Z., Deng, W., Li, S., He, J., Ren, S., Huang, W., et al. (2015). GlnR-mediated regulation of ectABCD transcription expands the role of the GlnR regulon to osmotic stress management. J. Bacteriol. 197, 3041–3047. doi: 10.1128/JB.00185-115
Shao, Z. H., Ren, S. X., Liu, X. Q., Xu, J., Yan, H., Zhao, G. P., et al. (2015). A preliminary study of the mechanism of nitrate-stimulated remarkable increase of rifamycin production in Amycolatopsis mediterranei U32 by RNA-seq. Microb. Cell Fact. 14:75. doi: 10.1186/s12934-015-0264-y
Szalewska-Palasz, A., Wegrzyn, A., Blaszczak, A., Taylor, K., and Wegrzyn, G. (1998). DnaA-stimulated transcriptional activation of orilambda: Escherichia coli RNA polymerase beta subunit as a transcriptional activator contact site. Proc. Natl. Acad. Sci. U.S.A. 95, 4241–4246. doi: 10.1073/pnas.95.8.4241
Tiffert, Y., Supra, P., Wurm, R., Wohlleben, W., Wagner, R., and Reuther, J. (2008). The Streptomyces coelicolor GlnR regulon: identification of new GlnR targets and evidence for a central role of GlnR in nitrogen metabolism in actinomycetes. Mol. Microbiol. 67, 861–880. doi: 10.1111/j.1365-2958.2007.06092.x
Wang, J., Wang, Y., and Zhao, G. P. (2015). Precise characterization of GlnR Box in actinomycetes. Biochem. Biophys. Res. Commun. 458, 605–607. doi: 10.1016/j.bbrc.2015.02.010
Wang, J., and Zhao, G. P. (2009). GlnR positively regulates nasA transcription in Streptomyces coelicolor. Biochem. Biophys. Res. Commun. 386, 77–81. doi: 10.1016/j.bbrc.2009.05.147
Wang, Y., Cen, X. F., Zhao, G. P., and Wang, J. (2012). Characterization of a new GlnR binding box in the promoter of amtB in Streptomyces coelicolor inferred a PhoP/GlnR competitive binding mechanism for transcriptional regulation of amtB. J. Bacteriol. 194, 5237–5244. doi: 10.1128/JB.00989-912
Wang, Y., Li, C., Duan, N., Li, B., Ding, X. M., Yao, Y. F., et al. (2014). GlnR negatively regulates the transcription of the alanine dehydrogenase encoding gene ald in Amycolatopsis mediterranei U32 under nitrogen limited conditions via specific binding to its major transcription initiation site. PLoS One 9:e104811. doi: 10.1371/journal.pone.0104811
Wang, Y., Wang, J. Z., Shao, Z. H., Yuan, H., Lu, Y. H., Jiang, W. H., et al. (2013). Three of Four GlnR binding sites are essential for GlnR-mediated activation of transcription of the Amycolatopsis mediterranei nas operon. J. Bacteriol. 195, 2595–2602. doi: 10.1128/Jb.00182-113
Wehrli, W., Knusel, F., Schmid, K., and Staehelin, M. (1968). Interaction of rifamycin with bacterial RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 61, 667–673. doi: 10.1073/pnas.61.2.667
Xu, J., Mahmud, T., and Floss, H. G. (2003). Isolation and characterization of 27-O-demethylrifamycin SV methyltransferase provides new insights into the post-PKS modification steps during the biosynthesis of the antitubercular drug rifamycin B by Amycolatopsis mediterranei S699. Arch. Biochem. Biophys. 411, 277–288. doi: 10.1016/S0003-9861(03)0004-3
Xu, J., Wan, E., Kim, C. J., Floss, H. G., and Mahmud, T. (2005). Identification of tailoring genes involved in the modification of the polyketide backbone of rifamycin B by Amycolatopsis mediterranei S699. Microbiology 151(Pt 8), 2515–2528. doi: 10.1099/mic.0.28138-28130
Yang, L., Zhang, W., Chiao, J., Zhao, G., and Jiang, W. (2001). An eukaryotic-type serine/threonine protein kinase involved in the carbon source-dependent pigment biosynthesis in Amycolatopsis mediterranei U32. Biochem. Biophys. Res. Commun. 284, 357–362. doi: 10.1006/bbrc.2001.4980
You, D., Wang, M. M., and Ye, B. C. (2017). Acetyl-CoA synthetases of Saccharopolyspora erythraea are regulated by the nitrogen response regulator GlnR at both transcriptional and post-translational levels. Mol. Microbiol. 103, 845–859. doi: 10.1111/mmi.13595
Yu, H., Peng, W. T., Liu, Y., Wu, T., Yao, Y. F., Cui, M. X., et al. (2006). Identification and characterization of glnA promoter and its corresponding trans-regulatory protein GlnR in the rifamycin SV producing actinomycete, Amycolatopsis mediterranei U32. Acta Biochim. Biophys. Sin. 38, 831–843. doi: 10.1111/j.1745-7270.2006.00238.x
Yu, H., Yao, Y., Liu, Y., Jiao, R., Jiang, W., and Zhao, G. P. (2007). A complex role of Amycolatopsis mediterranei GlnR in nitrogen metabolism and related antibiotics production. Arch. Microbiol. 188, 89–96. doi: 10.1007/s00203-007-0228-227
Yu, T. W., Shen, Y. M., Doi-Katayama, Y., Tang, L., Park, C., Moore, B. S., et al. (1999). Direct evidence that the rifamycin polyketide synthase assembles polyketide chains processively. Proc. Natl. Acad. Sci. U.S.A. 96, 9051–9056. doi: 10.1073/pnas.96.16.9051
Yuan, H., Zhao, W., Zhong, Y., Wang, J., Qin, Z., Ding, X., et al. (2011). Two genes, rif15 and rif16, of the rifamycin biosynthetic gene cluster in Amycolatopsis mediterranei likely encode a transketolase and a P450 monooxygenase, respectively, both essential for the conversion of rifamycin SV into B. Acta Biochim. Biophys. Sin. 43, 948–956. doi: 10.1093/abbs/gmr091
Keywords: Amycolatopsis mediterranei, rifamycin biosynthesis, GlnR, nitrate-stimulating effect, rifZ, rifK
Citation: Liu X, Liu Y, Lei C, Zhao G and Wang J (2020) GlnR Dominates Rifamycin Biosynthesis by Activating the rif Cluster Genes Transcription Both Directly and Indirectly in Amycolatopsis mediterranei. Front. Microbiol. 11:319. doi: 10.3389/fmicb.2020.00319
Received: 10 October 2019; Accepted: 13 February 2020;
Published: 03 March 2020.
Edited by:
Harold J. Schreier, University of Maryland, Baltimore County, United StatesReviewed by:
Juan F. Martin, Universidad de León, SpainLinquan Bai, Shanghai Jiao Tong University, China
Copyright © 2020 Liu, Liu, Lei, Zhao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jin Wang, d2FuZ2owMUBob3RtYWlsLmNvbQ==
†These authors have contributed equally to this work
 Xinqiang Liu
Xinqiang Liu Yuanyuan Liu3†
Yuanyuan Liu3† Chao Lei
Chao Lei Guoping Zhao
Guoping Zhao Jin Wang
Jin Wang