
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 25 February 2020
Sec. Food Microbiology
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.00302
 Yameng Liu
Yameng Liu Mark A. Hayes*
Mark A. Hayes*Salmonella is an important pathogen and is a world-wide threat to food safety and public health. Surveillance of serotypes and fundamental biological and biochemical studies are supported by a wide variety of established and emerging bioanalytical techniques. These include classic serotyping based on the Kauffmann–White nomenclature and the emerging whole genome sequencing strategy. Another emerging strategy is native whole cell biophysical characterization which has yet to be applied to Salmonella. However, this technique has been shown to provide high resolution differentiation of serotypes with several other paired strains of other microbes and pathogens. To demonstrate that biophysical characterization might be useful for Salmonella serotyping, the closely related strains sv. Cubana and sv. Poona were chosen for study. These two serovars were subjected to biophysical measurements on a dielectrophoresis-based microfluidic device that generated full differentiation of the unlabeled and native cells. They were differentiated by the ratio of electrophoretic (EP) to dielectrophoretic (DEP) mobilities. This differentiation factor is 2.7 ± 0.3 × 1010 V/m2 for sv. Cubana, versus 2.2 ± 0.3 × 1010 V/m2 for sv. Poona. This work shows for the first time the differentiation, concentration, and characterization of the Salmonella serotypes by exploiting their biophysical properties. It may lead to a less expensive and more decentralized new tool and method for microbiologists, complimenting and working in parallel with other characterization methods.
There is an increasing array of methods to characterize microorganisms from whole genome sequencing to traditional culturing strategies (Chiou et al., 2015; Ibrahim and Morin, 2018). For Salmonella, a common foodborne pathogen that can cause disease in humans, the characterization must allow tracking of the contamination source by using appropriate subtyping tools (Tang et al., 2019). The “gold standard” classifying subtle differences between salmonella strains is based on the Kauffmann–White nomenclature (Grimont and Weill, 2007), representing a traditional phenotyping method that is logistically challenging, as it requires the use of more than 150 specific antisera and well-trained personnel to interpret the results (Diep et al., 2019). One emerging and unproven strategy is to directly assess the biophysical characteristics of the native and unlabeled cells toward correlating their properties with specific serotypes. In this study, two closely related serovars based on the similar antigens indicated in the Kauffmann–White categorization scheme are tested and were differentiated in their native state with simple electric field interactions.
The common microbe Salmonella is thought to be responsible for 450 deaths, 23,000 hospitalizations, and 1.4 million illnesses each year in the United States (Bishop et al., 2011). The typical symptom is abdominal pain and is diagnosed as gastroenteritis, with severe infections becoming life threatening. Food safety incidents and recalls continue in recent years, mostly associated with processed products (Pillai and Ricke, 2002; Maciorowski et al., 2004; Park et al., 2008; Hanning et al., 2009), and other food commodities (e.g., meat products, eggs, and vegetables) (Greig and Ravel, 2009; Wu et al., 2017; Ricke et al., 2018; Tang et al., 2019). These occurrences necessitate accurate and relatively rapid subtyping tools for identifying the original source of contamination (Olaimat and Holley, 2012; Barco et al., 2013; Shi et al., 2015). Salmonella is a diverse pathogen and there are over 2500 Salmonella serotypes (2007 data), which have been described (Grimont and Weill, 2007). Of these, 99% of human isolates belong to the subspecies Salmonella enterica subsp. enterica (also described equivalently as “subspecies I”).
The immunoreactivities to O and H antigens of each isolate define the serotype, where a substantial diversity exists within the antigens. A cell surface lipopolysaccharide structure makes up the O antigen and typically consists of four to six sugars. The various specific antigens can differ by the linkages between sugars, covalent bonds between the units, or differences in the sugars themselves. These are divided into “O group antigens” (specific sugar configuration of the O antigen structure) and “ancillary O antigens” (additional carbohydrates). On the other hand, a proteinaceous antigen, flagellin or H antigen, is located on the flagellum in a filamentous portion. The core structural elements of these proteins which provide the filamentous structure, C′ and N′ termini, are conserved. The middle region of flagellin is exposed on the surface and is antigenically variable. Like many taxonomic and categorization schemes, those associated with Salmonella are evolving and therefore include modern and systematic definitions along with archaic terms still in common usage.
The two serotypes characterized in this study are S. enterica serotype Cubana and S. enterica serotype Poona. These strains belong to a group equivalently referred to as Group O:13 and Group G, and commonly Group O:13 (G) (Table 1 and Figure 1; Grimont and Weill, 2007). Noting that there are more than 1500 serovars (sv.) in S. enterica (Figure 1B), these serotypes are considered to be closely related with respect to the total breadth of serovars. Both of these serovars have been characterized with whole genome sequencing studies (Hoffmann et al., 2014; USFDA, 2014). This will provide a framework for assigning the quantified differences in biophysical properties presented here to specific biochemical origins.
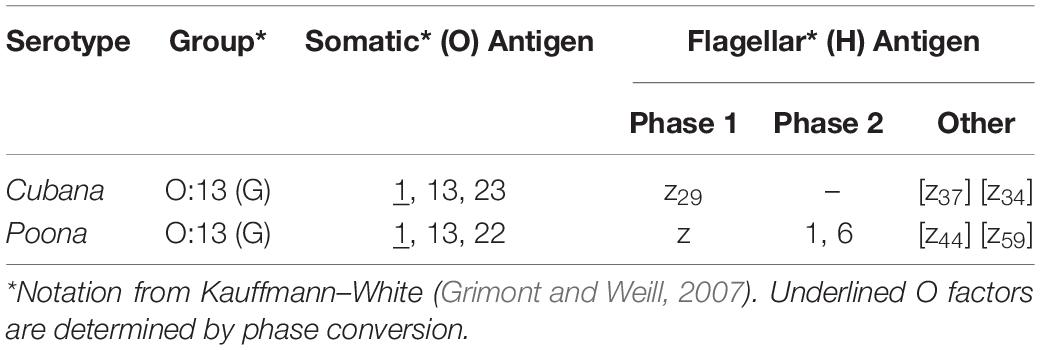
Table 1. Listing of specific known antigens for two Salmonella strains using Kauffmann–White nomenclature.
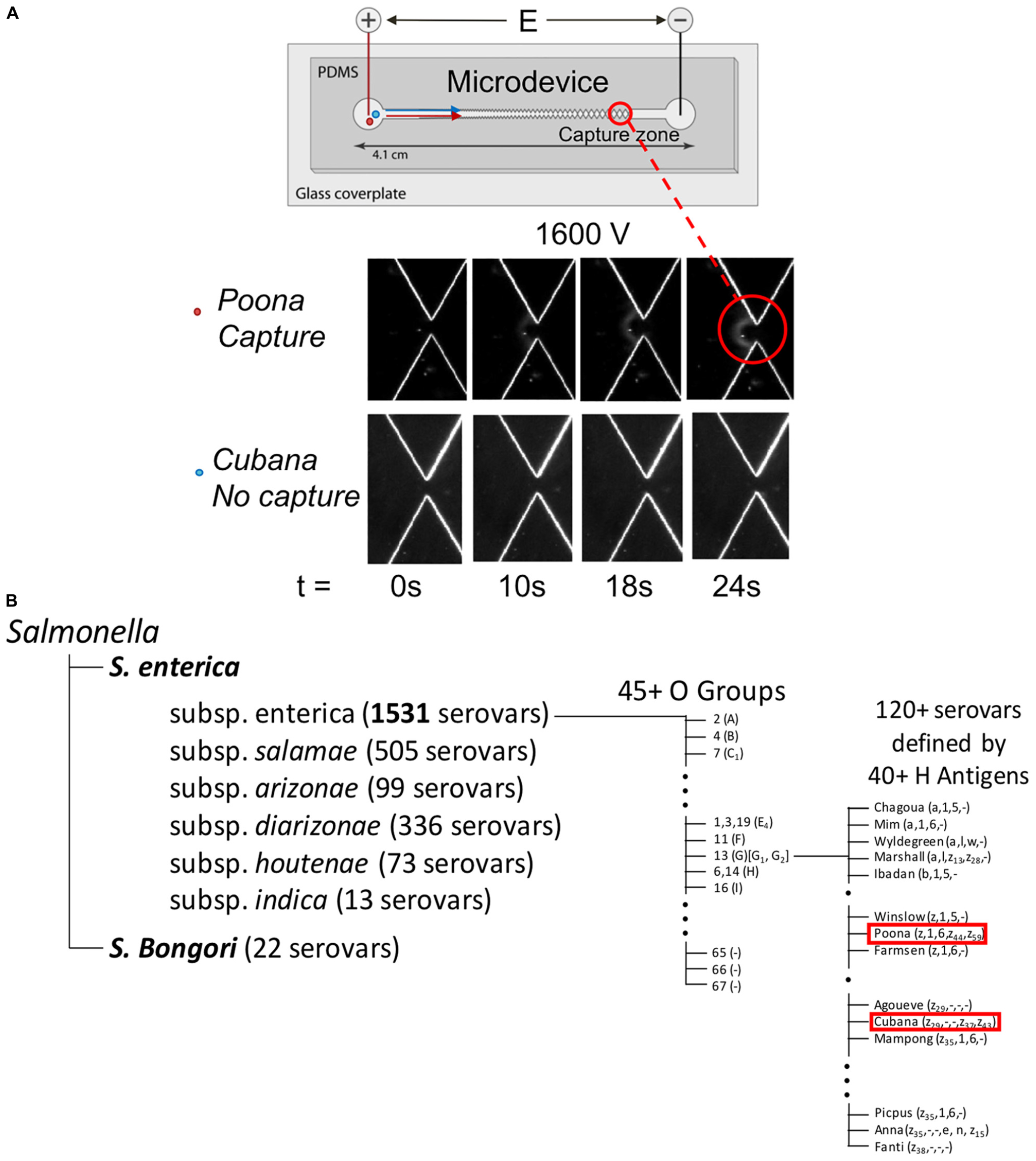
Figure 1. Graphic representation of microdevice (A) and Kauffmann–White nomenclature showing the relationship by phenotypic characterization between sv. Cubana and sv. Poona (B). In part (A), capture and concentration behavior of Salmonella serotypes in the DC-iDEP device. Insets images (bottom) of sv. Cubana and sv. Poona at 0, 10, 18, 24 s of 1600 V applied. The images were recorded at a 27-μm gate. With same potential (1600 V) applied, sv. Poona was captured but sv. Cubana was able to pass through. The graphic shown in part (B) indicates sv. Cubana and sv. Poona are closely associated with each other in comparison to the array of some 2500+ Salmonella serovars.
In this study, we demonstrate a rapid biophysical differentiation of two closely related strains of Salmonella, sv. Cubana and sv. Poona, using constant voltage gradient insulator-based dielectrophoresis (DC-iDEP) (Figure 1). The distinction is reflected by a different voltage at which each strain begins to capture, defining a specific characteristic and deterministic property for each strain. With some additional measurements, the specific forces regarding the electrokinetic (EK) and dielectrophoretic (DEP) mobilities are determined. These values allow some insights into the molecular and structural origins of the differentiation (Hilton and Hayes, 2019). This provides strong evidence that the simple measurement of the native and unlabeled cells may provide another valuable tool in the determination of serovars and basic science studies of Salmonella.
The properties of particles dispersed in a buffer or solvent in the presence of an external electric field exhibit behavior directly according to their physical makeup. In this study, biophysical behaviors are identified within a DC-iDEP device by the applied voltage and location of cell capture. Particles in the device experience DEP and EK forces. Higher order electrophysical effects are described by DEP mobility (μDEP). It can be expressed as (Jones, 1995; Nili and Green, 2014; Hilton and Hayes, 2019):
where εm is the permittivity of the medium, r is the radius of the particle, fCM is the Clausius–Mossotti factor, and η is the medium viscosity. EK force is the combination of electrophoretic (EP) force (first order effects, monopole moment) and electroosmotic flow (EOF). Reflecting these processes, EK mobility (μEK) is defined by
where μEP is EP mobility and μEOF is the EOF mobility. And they can be described by
where εm is the permittivity of the medium, ζp is the EK (zeta) potential of the particle, and ζm is the EK (zeta) potential of the medium/wall system. The value of μEK was determined for both strains by particle tracking velocimetry (Crowther et al., 2019; Hilton and Hayes, 2019) at various applied voltages based on:
where is the velocity of the particle in an open channel.
The capture of the particles can be observed when the flux of particles () is zero in the channel with the condition:
where is the gradient of the electric field and E and are the scalar and vector electric field, respectively. The ratio of EK to DEP mobilities (EKMr) which relates size, conductivity, surface charge, and other factors of the particle to the electric field properties are used to distinguish the subtle differences between the two strains of Salmonella. The specific cell features which are reflected in this term are under debate, but the magnitude of this measured property will not change (Pethig, 2019). The electric field and the gradient of the electric field combination are simulated to provide the EKMr for each strain.
sv. Cubana (ATCC 12007) and sv. Poona (ATCC BAA-1673) were obtained from ATCC. Each strain was grown on triple sugar iron agar for 4 days at ambient temperature. Ten milliliters of sterile 3% tryptic soy broth was inoculated, and the serotype solutions were incubated in a shaker/incubator at 250 rpm (37°C) for 19 h. The concentration of cells is about 109 CFU/mL. The cultures were stored at 4°C.
A volume of 100 μl of each culture was dissolved into 900 μl 5 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer (pH = 7.3) solution and centrifuged for 5 min at 2000 × g. The supernatant was removed, and the washing procedure was repeated three times with HEPES buffer solution. The sample was suspended in 1 ml 5 mM HEPES buffer solution prior to use. Microbial cultures are required at Biosafety Level 1 or 2 or 3. All the experiments were performed with Biosafety Level 2 space and procedures.
A microchannel described in a previous work (Staton et al., 2010) and used for other cellular studies (Jones et al., 2014, 2015; Crowther et al., 2019) was used for the biophysical behavior study of Salmonella strains. In brief, opposing pairs of triangles were designed to constitute the sawtooth shape of the channel (Figure 1A). The length between the inlet and outlet of the channel is 4.2 cm. The increasing size of the triangles in the channel restrict the narrowest width of the pathway of each gate from 945 to 27 μm with a depth of 16.9 ± 1 μm. Soft lithography was used to fabricate the microchannels using PDMS (Sylgard 184, Dow/Corning, Midland, MI, United States).
Finite element modeling (COMSOL, Inc., Burlington, MA, United States) of the distribution of the electric field in the microchannel was performed as previously detailed (Crowther and Hayes, 2017). The AC/DC module was used to interrogate the, and in an accurately scaled 2D model of the microchannel.
The microdevice channel was treated with 5% (w/v) bovine serum album in 2 mM phosphate buffer at pH 7.4 and rinsed with 5 mM HEPES buffer solution before introducing the prepared bacterial sample. Dielectrophoresis behaviors of the two strains were observed by an Olympus IX70 inverted microscope with a 4× or 10× objective. Images and videos were recorded by QICAM cooled CCD camera (QImaging, Inc., Surrey, BC) and Streampix III image capture software (Norpix, Inc., Montreal, QC). A voltage between 0 – 3000 V was applied to platinum electrodes (0.404-mm external diameter, 99.9% purity, Alfa Aesar, Ward Hill, MA) connected to the inlet (+) and outlet (ground) to capture and study behaviors of Salmonella strains. Analysis and error assessment were based on 4 individual trials for each strain.
The biophysical behavior of sv. Cubana and sv. Poona were investigated in the DC-iDEP device. Each strain was tested separately in various devices of the same design. Both strains produced a pattern of collected cells appearing as distinct arcs near a 27 μm gate at appropriate applied voltages (Figures 1A, 2; Jones et al., 2014, 2015; Crowther et al., 2019). With 1600 V applied, sv. Poona showed capture behavior whereas the same electric field conditions did not capture sv. Cubana. The intensity of concentrated sv. Poona increases with the time of 1600 V applied at 0, 10, 18, 24 s. The strain dispersed as expected when the electric field was removed, which indicated the effective removal of the EK and DEP forces on the particles. With higher voltages applied, capture of sv. Cubana was then observed.
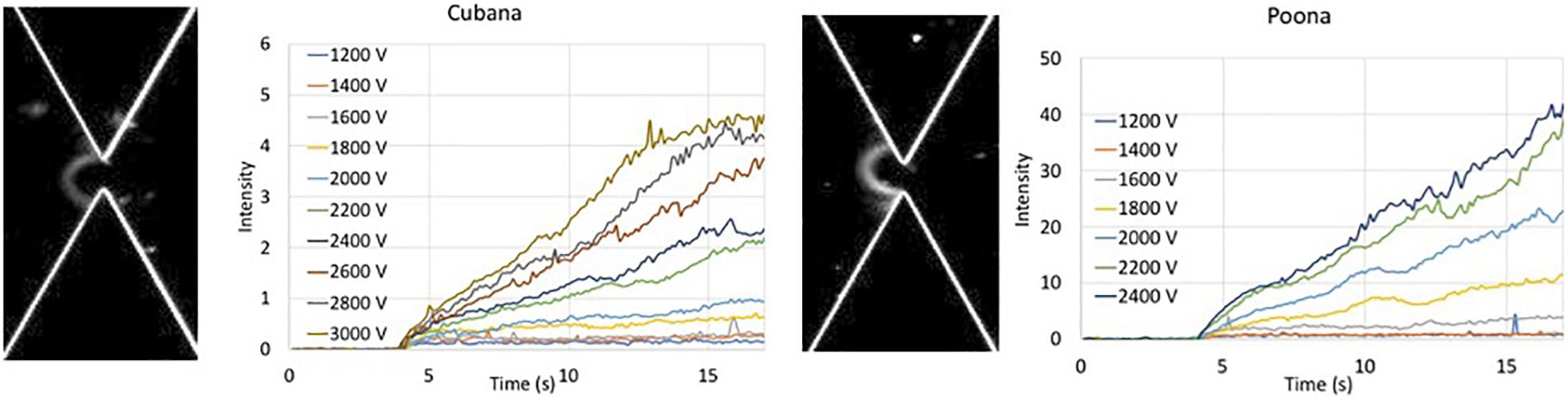
Figure 2. Intensity of collected bolus of Salmonella versus time at various applied voltages. Intensity of sv. Cubana was recorded at voltages from 1200 to 3000 V. Intensity of sv. Poona was recorded at voltages from 1200 to 2400 V. Images are recorded at 20 s of 2400 V for sv. Cubana (Left) and sv. Poona (Right).
The intensity within the capture area was recorded where the increased intensity reflected the collection of the cells. The intensity curves of sv. Cubana from 1200 to 3000 V and sv. Poona from 1200 to 2400 V in 200 V increments were plotted (Figure 2). The intensities increase with higher applied voltages for both sv. Cubana and sv. Poona.
Data were analyzed at a constant time (10 s after voltage applied) for sv. Cubana and sv. Poona (Figure 3). For the blue data points at lower voltages, where no capture occurred, no significant change of the intensity is observed and is comparable to the background. The orange data points at higher voltages, from 2000 V for sv. Cubana and from 1600 V for sv. Poona, were used for plotting the linear regression line of the increased intensity reflective of cell accumulation. Error bars reflect standard error of the mean (SEM).
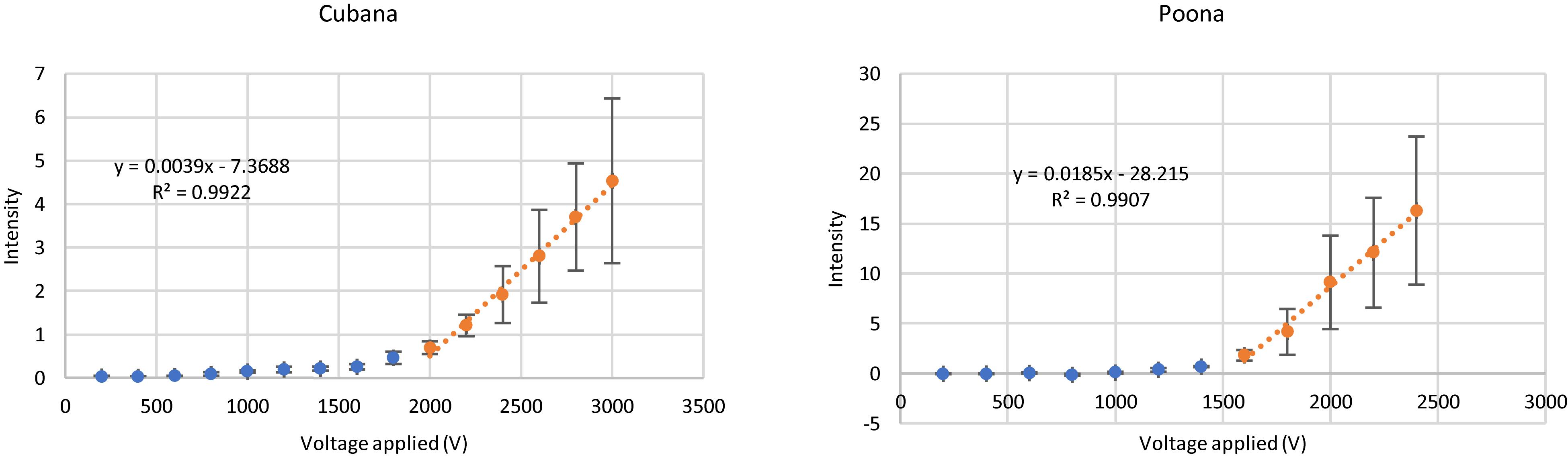
Figure 3. Capture behaviors of sv. Cubana and sv. Poona at various applied potentials. Intensity was recorded at a 27-μm gate when the potential was applied for 10 s. Error bars represent SEM.
The slope and intercept of the linear fits were used to determine the rate of particle accumulation and onset voltage for capture (Hilton and Hayes, 2019). In this way, initial capture voltages were determined to be 1889 ± 228 V for sv. Cubana and 1525 ± 196 V for sv. Poona. Using the multiphysics calculations to determine the field and gradient values, the EKMr was determined to be 2.7 ± 0.3 × 1010 V/m2 for sv. Cubana and 2.2 ± 0.3 × 1010 V/m2 for sv. Poona. These are well differentiated and sufficiently different to be considered statistically significant.
The EK behaviors of the strains were determined according to Eq. 6 by particle tracking to monitor the velocity while varying electric field strength (Figure 4). The slopes of the linear fits determine the sv. Cubana μEK to be 5.0 ± 0.5 × 10–8 m2/Vs and sv. Poona μEK to be 6.7 ± 0.3 × 10–8 m2/Vs. With EKMr and μEK values, μDEP of sv. Cubana was calculated to be 1.8 ± 0.3 × 10–18 m4/V2s and for sv. Poona it was determined to be 3.0 ± 1.3 × 10–18 m4/V2s.
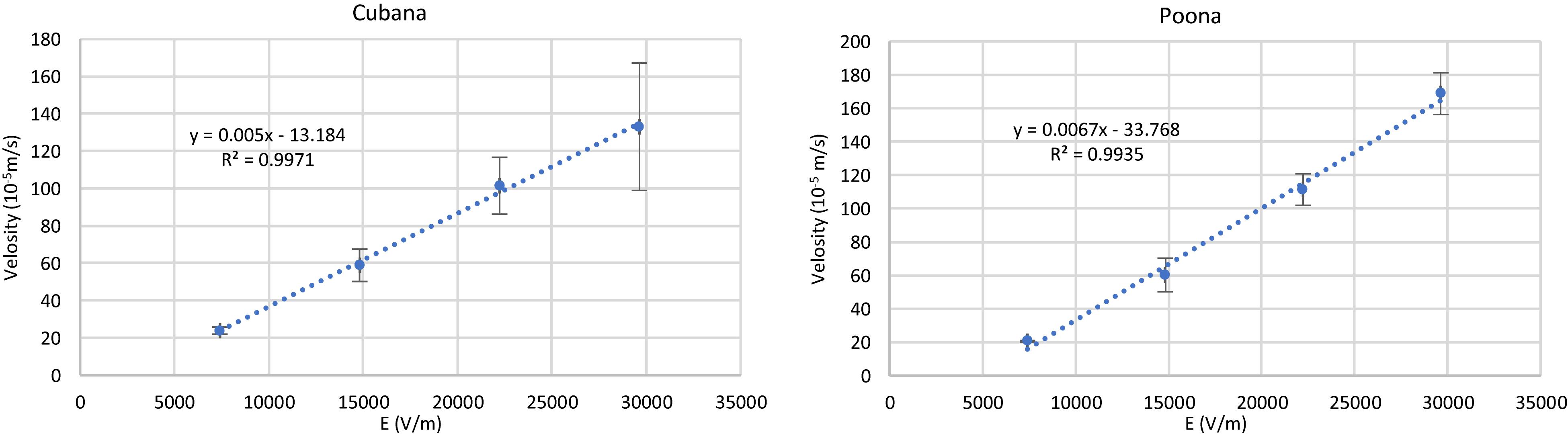
Figure 4. Plots of velocity of sv. Cubana and sv. Poona with varying electric field to determine μEK. (). Error bars represent SEM.
The two closely related Salmonella strains were differentiated by mobility comparisons (Figure 5). They can be both distinguished by EKMr/EK and EKMr/DEP mobilities. The two strains are demonstrated to have different biophysical behaviors distinguished by the DC-iDEP device.
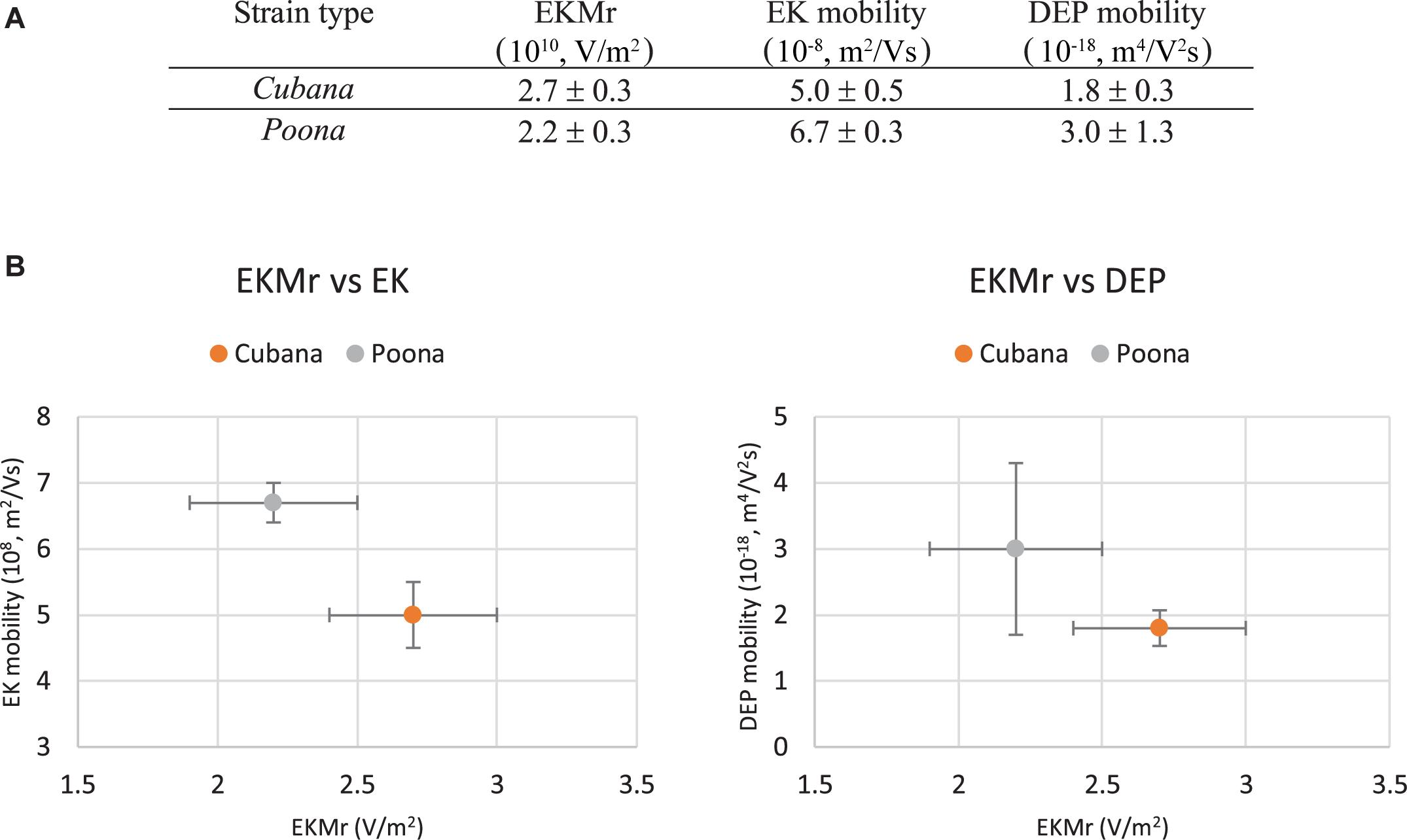
Figure 5. Mobilities of sv. Cubana and sv. Poona (A). Electrokinetic mobility is determined from velocimetry measurements in the more open parts of the device channel and was used with EKMr to calculate the DEP mobility. The EKMr is plotted versus the EK and DEP mobilities separately (B). The strains are well differentiated by all metrics used. Error bars represent SEM.
The sv. Cubana and sv. Poona. are closely related with regards to the Kauffmann–White nomenclature (Figure 1B). According to this classification scheme, there is one known difference in the O Group (Cubana – O: 23; Poona – O: 22) and several differences in the H Antigens, sv. Cubana having z29, [z37] and [z49] and sv. Poona expressing z, 1, 6, [z44] and [z59] (notation details retained from nomenclature guide) (Grimont and Weill, 2007). Even though these serovars are considered “close” with the Kauffman–White classification system, there are still clear and identifiable differences in the chemical structure of the surface of the cell which may influence how they interact with an electric field.
Dielectrophoresis and cellular impedance spectroscopy have demonstrated a capability to differentiate cells based upon changes in the biochemical makeup of the cellular structure with labels (Labeed et al., 2003, 2011; Chin et al., 2006; Coley et al., 2007; Flanagan et al., 2008; Jones et al., 2015; Su et al., 2016; Fernandez et al., 2017; Rohani et al., 2018; Crowther et al., 2019; Liu et al., 2019; Hilton et al., 2020). When fully developed and vetted, this approach of serotyping will require less expense and expertise compared to producing antisera in agglutination test utilizing O and H antiserum and will not require expert genomic information interpretation skills in whole genome sequencing for identifying Salmonella (Ng and Kirkness, 2010; Ashton et al., 2016).
The relationship between biophysical behaviors, zeta potential, and the mobilities has been described (Hilton and Hayes, 2019). Zeta potential has a linear relationship with EK mobility and results in the difference in the onset voltage and the concentration slope. The differences in the conductivity of the serotypes could also affect the capture onset potential because the change of the DEP mobility. However, the conductivity and permittivity of the medium contribute little to the capture onset potentials for the behaviors of two serotypes but has a significant effect to the accumulation slope.
The EK mobility is significantly different between these strains, supporting the conclusion that the surface charge is changed (Figure 4; Hilton and Hayes, 2019). This a reasonable result since the surface antigens are known to be different between sv. Cubana and sv. Poona. In addition, the DEP mobility differs between the strains, showing that both surface and interior electrophysical properties differ, although it is impossible to assign a specific ratio to the relative effect from each (Hilton and Hayes, 2019). The biophysical differences between the two strains are reasonable with regards to the biological and biochemical alterations noted in the nomenclature alone, without considering other undocumented effects. The eventual impact of these results is yet to be understood in the serotyping laboratory; it is not known what other techniques will be enhanced by using this as a pre-screening or concentrating tool or if it will eventually develop into a standalone serotyping mechanism for well-known and vetted samples. An interesting question which remains to be answered is whether the magnitude of the differences in biophysical properties have any correlation with the total known and identified differences in the strains.
Previous DC-iDEP work successfully distinguished closely related strains of microbes (Jones et al., 2014, 2015; Crowther et al., 2019; Hilton, 2019; Hilton et al., 2020). The connectivity between biophysical properties and current notions of “relatedness” of two strains is unknown. The biophysical properties relate in a non-linear fashion with traditional cataloging systems [genetics, transcriptome, proteomics, molecular recognition (immune-, selex), and/or metabolic assessments]. In the author’s laboratory, all previously attempted paired biophysical differentiation were successful and include: Staphylococcus epidermidis (gentamicin resistant/susceptible) (Jones et al., 2015), various strains of Escherichia coli (Jones et al., 2014), Listeria monocytogenes strains (Crowther et al., 2019), Staphylococcus aureus (methicillin resistance/susceptible) (Hilton et al., 2020), and Klebsiella pneumoniae (Hilton, 2019). Within the species Salmonella, some interesting future works would include the differentiation paired serotypes Salmonella typhimurium compared to Salmonella typimurium monophasic variant (Salmonella 1,4,[5],12:i:-) and Salmonella Indiana (O4,12;z;1,7) compared to Salmonella Loubomo (O4,12;z;1,6).
In developing and discussing this technique over many venues, some themes emerged which have proven instructive. First is that the effects of biologically important changes may not induce a measurable change in the cells using the electric field effects. The current study undermines this concept, along with many previous quantitative assessments (Jones et al., 2014, 2015; Crowther et al., 2019; Hilton and Hayes, 2019; Liu et al., 2019). The second theme is that the variation in the biological entities will be too great to decipher. Interestingly enough, this high-resolution capability provides a tool to investigate the origins and structure of that variability. Within any population of cells some will have a biologically significant change and others will have routine property variances which are not biological differentiators. This technique can allow for quantitative determination of biophysical-to-biological action connectivity.
This work demonstrates the differentiation of the two close strains of Salmonella, sv. Cubana and sv. Poona, by DC-iDEP device. The variable capture conditions are accomplished without labels or otherwise altering the cells, the effects occur due to the native condition of the organisms with the setting of electric field properties. These results support the concept that biophysical separation and concentration will potentially become a useful tool in the microbiology laboratory to aid in serotyping of Salmonella.
All datasets generated for this study are included in the article/supplementary material.
YL was responsible for data collection, experimental condition testing, and manuscript preparation. MH worked on the overall instruction on the research and manuscript preparation.
The study was supported by the National Institutes of Health, grant number 1R21AI130855-01.
MH is an investor in and a collaborator with Charlot Biosciences, a company which has licensed the underlying technology presented in this paper.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
LaKeta Kemp is acknowledged for discussions associated with the formation of the overall project.
Ashton, P. M., Nair, S., Peters, T. M., Bale, J. A., Powell, D. G., Painset, A., et al. (2016). Identification of Salmonella for public health surveillance using whole genome sequencing. PeerJ 4:e1752. doi: 10.7717/peerj.1752
Barco, L., Barrucci, F., Olsen, J. E., and Ricci, A. (2013). Salmonella source attribution based on microbial subtyping. Int. J. Food Microbiol. 163, 193–203. doi: 10.1016/j.ijfoodmicro.2013.03.005
Bishop, R., Erdman, M. M., Fields, P., Fullerton, K., Jackson, K., and Mahon, B. (2011). CDC. National Salmonella Surveillance Overview. Atlanta: US Department of Health and Human Services.
Chin, S., Hughes, M. P., Coley, H. M., and Labeed, F. H. (2006). Rapid assesment of early biophysical changes in K562 cells during apoptosis determined using dielectrophoresis. Int. J. Nanomed. 1, 333–337.
Chiou, C. S., Torpdahl, M., Liao, Y. S., Liao, C. H., Tsao, C. S., Liang, S. Y., et al. (2015). Usefulness of pulsed-field gel electrophoresis profiles for the determination of Salmonella serovars. Int. J. Food Microbiol. 214, 1–3. doi: 10.1016/j.ijfoodmicro.2015.07.016
Coley, H. M., Labeed, F. H., Thomas, H., and Hughes, M. P. (2007). Biophysical characterization of MDR breast cancer cell lines reveals the cytoplasm is critical in determining drug sensitivity. Biochim. Biophys. Acta Gen. Subj. 1770, 601–608. doi: 10.1016/j.bbagen.2006.12.002
Crowther, C. V., and Hayes, M. A. (2017). Refinement of insulator-based dielectrophoresis. Analyst 142, 1608–1618. doi: 10.1039/c6an02509a
Crowther, C. V., Hilton, S. H., Kemp, L., and Hayes, M. A. (2019). Isolation and identification of Listeria monocytogenes utilizing DC insulator-based dielectrophoresis. Anal. Chim. Acta 1068, 41–51. doi: 10.1016/j.aca.2019.03.019
Diep, B., Barretto, C., Portmann, A.-C., Fournier, C., Karczmarek, A., Voets, G., et al. (2019). Salmonella serotyping; comparison of the traditional method to a microarray-based method and an in silico platform using whole genome sequencing data. Front. Microbiol. 10:2554. doi: 10.3389/fmicb.2019.02554
Fernandez, R. E., Rohani, A., Farmehini, V., and Swami, N. S. (2017). Review: microbial analysis in dielectrophoretic microfluidic systems. Anal. Chim. Acta 966, 11–33. doi: 10.1016/j.aca.2017.02.024
Flanagan, L. A., Lu, J., Wang, L., Marchenko, S. A., Jeon, N. L., Lee, A. P., et al. (2008). Unique dielectric properties distinguish stem cells and their differentiated progeny. Stem Cells 26, 656–665. doi: 10.1634/stemcells.2007-0810
Greig, J. D., and Ravel, A. (2009). Analysis of foodborne outbreak data reported internationally for source attribution. Int. J. Food Microbiol. 130, 77–87. doi: 10.1016/j.ijfoodmicro.2008.12.031
Grimont, P. A. D., and Weill, F.-X. (2007). Antigenic Formulae of the Salmonella Serovars. Paris, FR: Pasteur Institute.
Hanning, I. B., Nutt, J. D., and Ricke, S. C. (2009). Salmonellosis outbreaks in the united states due to fresh produce: sources and potential intervention measures. Foodborne Pathog. Dis. 6, 635–648. doi: 10.1089/fpd.2008.0232
Hilton, S. H., Crowther, C., McLaren, A., and Hayes, M. A. (2020). Biophysical differentiation of susceptibility and chemical differences in Staphylococcus aureus. Analyst 140, 5152–5161.
Hilton, S. H., and Hayes, M. A. (2019). A mathematical model of dielectrophoretic data to connect measurements with cell properties. Anal. Bioanal. Chem. 411, 2223–2237. doi: 10.1007/s00216-019-01757-7
Hilton, S. L. (2019). Analyzing Cellular Properties with Dielectrophoresis. Ph.D. Thesis, Arizona State University, Tempe, AZ.
Hoffmann, M., Muruvanda, T., Pirone, C., Korlach, J., Timme, R., Payne, J., et al. (2014). First Fully closed genome sequence of Salmonella enterica subsp. enterica Serovar cubana associated with a food-borne outbreak. Genom. Announc. 2:e01112-14. doi: 10.1128/genomeA.01112-14
Ibrahim, G. M., and Morin, P. M. (2018). Salmonella serotyping using whole genome sequencing. Front. Microbiol. 9:2993. doi: 10.3389/fmicb.2018.02993
Jones, P. V., DeMichele, A. F., Kemp, L., and Hayes, M. A. (2014). Differentiation of Escherichia coli serotypes using dc gradient insulator dielectrophoresis. Anal. Bioanal. Chem. 406, 183–192. doi: 10.1007/s00216-013-7437-5
Jones, P. V., Huey, S., Davis, P., McLemore, R., McLaren, A., and Hayes, M. A. (2015). Biophysical separation of Staphylococcus epidermidis strains based on antibiotic resistance. Analyst 140, 5152–5161. doi: 10.1039/c5an00906e
Labeed, F. H., Coley, H. M., Thomas, H., and Hughes, M. P. (2003). Assessment of multidrug resistance reversal using dielectrophoresis and flow cytometry. Biophys. J. 85, 2028–2034. doi: 10.1016/s0006-3495(03)74630-x
Labeed, F. H., Lu, J. T., Mulhall, H. J., Marchenko, S. A., Hoettges, K. F., Estrada, L. C., et al. (2011). Biophysical characteristics reveal neural stem cell differentiation potential. PLoS One 6:e25458. doi: 10.1371/journal.pone.0025458
Liu, Y. M., Jiang, A. A., Kim, E., Ro, C., Adams, T., Flanagan, L. A., et al. (2019). Identification of neural stem and progenitor cell subpopulations using DC insulator-based dielectrophoresis. Analyst 144, 4066–4072. doi: 10.1039/c9an00456d
Maciorowski, K. G., Jones, F. T., Pillai, S. D., and Ricke, S. C. (2004). Incidence, sources, and control of food-borne Salmonella spp. in poultry feeds. Worlds Poult. Sci. J. 60, 446–457. doi: 10.1079/wps200428
Ng, P. C., and Kirkness, E. F. (2010). “Whole genome sequencing,” in Genetic Variation, (Totowa, NJ: Humana Press), 215–226.
Nili, H., and Green, N. G. (2014). Higher-order dielectrophoresis of nonspherical particles. Phys. Rev. E 89:063302.
Olaimat, A. N., and Holley, R. A. (2012). Factors influencing the microbial safety of fresh produce: a review. Food Microbiol. 32, 1–19. doi: 10.1016/j.fm.2012.04.016
Park, S. Y., Woodward, C. L., Kubena, L. F., Nisbet, D. J., Birkhold, S. G., and Ricke, S. C. (2008). Environmental dissemination of foodborne Salmonella in preharvest poultry production: reservoirs, critical factors, and research strategies. Crit. Rev. Environ. Sci. Technol. 38, 73–111. doi: 10.1080/10643380701598227
Pethig, R. (2019). Limitations of the Clausius-Mossotti function used in dielectrophoresis and electrical impedance studies of biomacromolecules. Electrophoresis 40, 2575–2583. doi: 10.1002/elps.201900057
Pillai, S. D., and Ricke, S. C. (2002). Bioaerosols from municipal and animal wastes: background and contemporary issues. Can. J. Microbiol. 48, 681–696. doi: 10.1139/w02-070
Ricke, S. C., Kim, S. A., Shi, Z., and Park, S. H. (2018). Molecular-based identification and detection of Salmonella in food production systems: current perspectives. J. Appl. Microbiol. 125, 313–327. doi: 10.1111/jam.13888
Rohani, A., Moore, J. H., Su, Y. H., Stagnaro, V., Warren, C., and Swami, N. S. (2018). Single-cell electro-phenotyping for rapid assessment of clostridium difficile heterogeneity under vancomycin treatment at sub-MIC (minimum inhibitory concentration) levels. Sens. Actuat. B Chem. 276, 472–480. doi: 10.1016/j.snb.2018.08.137
Shi, C. L., Singh, P., Ranieri, M. L., Wiedmann, M., and Switt, A. I. M. (2015). Molecular methods for serovar determination of Salmonella. Crit. Rev. Microbiol 41, 309–325. doi: 10.3109/1040841X.2013.837862
Staton, S. J. R., Chen, K. P., Taylor, T. J., Pacheco, J. R., and Hayes, M. A. (2010). Characterization of particle capture in a sawtooth patterned insulating electrokinetic microfluidic device. Electrophoresis 31, 3634–3641. doi: 10.1002/elps.201000438
Su, Y. H., Rohani, A., Warren, C. A., and Swami, N. S. (2016). Tracking inhibitory alterations during interstrain clostridium difficile interactions by monitoring cell envelope capacitance. ACS Infect. Dis. 2, 544–551. doi: 10.1021/acsinfecdis.6b00050
Tang, S., Orsi, R. H., Luo, H., Ge, C., Zhang, G., Baker, R. C., et al. (2019). Assessment and comparison of molecular subtyping and characterization methods for Salmonella. Front. Microbiol. 10:1591. doi: 10.3389/fmicb.2019.01591
USFDA (2014). Whole Genome Sequence S. Enterica sv. Poona. ATCC BAA-1673 GCA_000493295.1. Available at: https://img.jgi.doe.gov/cgi-bin/m/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=2558860441 (accessed October 20, 2019).
Keywords: dielectrophoresis, Salmonella, electrokinetics, label-free, microfluidics, serotype, Cubana, Poona
Citation: Liu Y and Hayes MA (2020) Differential Biophysical Behaviors of Closely Related Strains of Salmonella. Front. Microbiol. 11:302. doi: 10.3389/fmicb.2020.00302
Received: 04 January 2020; Accepted: 11 February 2020;
Published: 25 February 2020.
Edited by:
Lin Lin, Jiangsu University, ChinaCopyright © 2020 Liu and Hayes. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mark A. Hayes, bWhheWVzQGFzdS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.