- Key Laboratory of Prevention and Control Agents for Animal Bacteriosis, Institute of Animal Husbandry and Veterinary, Hubei Academy of Agricultural Sciences, Wuhan, China
Campylobacter jejuni and Campylobacter coli are major food-borne pathogens that cause bacterial gastroenteritis in humans, and poultry is considered as their most important reservoir. Macrolides, such as erythromycin, are the first-line choice for treatment of campylobacteriosis. In this study, of the 143 Campylobacter isolates recovered from poultry in central China during 2015–2017, 25.2% were erythromycin resistant. A2075G substitution in 23S ribosomal RNA (rRNA) and ribosomal methylase encoded by erm(B) were found in 4.2 and 4.9% isolates, respectively, and correlated with erythromycin resistance. The polymorphisms of CmeR-Box were also analyzed in our isolates. Among them, 9.1% isolates harbored a point deletion or insertion within the CmeR-Box, and we first showed that point deletion or insertion, but not substitution, in CmeR-Box led to high expression of cmeABC, which was significantly associated with erythromycin resistance (p < 0.05). These results suggest that point deletion or insertion in CmeR-Box, A2075G substitution in 23S rRNA, and presence of erm(B) are three main factors to erythromycin resistance in C. jejuni and C. coli.
Introduction
Campylobacter species, especially Campylobacter jejuni and Campylobacter coli, are major food-borne pathogens that cause bacterial gastroenteritis in humans (Huang et al., 2009; Kaakoush et al., 2015). Campylobacter is widespread in animals, with poultry as the most important reservoir (Kaakoush et al., 2015). Contaminated poultry products are recognized as the main source of human infection (Zhang T. et al., 2016). In view of the high incidence of fluoroquinolone resistance, including in China (Zhang et al., 2017b), macrolides, such as erythromycin, are the first-line choice for treatment of campylobacteriosis (Bolinger and Kathariou, 2017). Although erythromycin has been limited for use in animal production in China in 2000, the incidence of erythromycin resistance in Campylobacter continues to increase (Zhang A. et al., 2016; Du et al., 2018). Therefore, the surveillance of erythromycin-resistant Campylobacter is important not only for animal breeding but also for public health.
Macrolides are antibiotics that act by binding to the bacterial 50S ribosomal subunit to obstruct the ribosomal exit tunnel, resulting in inhibition of protein synthesis in bacteria (Dinos, 2017). The A2075G and A2074C/G substitutions in the 23S ribosomal RNA (rRNA) are the most common mechanism for erythromycin resistance in Campylobacter (Hao et al., 2009; Perez-Boto et al., 2010; Zhao et al., 2016). Recently, the erm(B) gene, which encodes a horizontally transferable ribosomal methylase, was identified in Campylobacter (Qin et al., 2014). erm(B) can dimethylate a single adenine in the 23S rRNA, leading to the inhibition of the binding of macrolides to the 50S subunit of bacterial ribosomes (Qin et al., 2014; Wang et al., 2014). In addition, various native efflux pumps are encoded in bacteria, which provide baseline resistance levels (Trastoy et al., 2018). cmeABC is an important efflux system in C. jejuni and C. coli (Lin et al., 2002), and the inactivation of cmeB results in a significant decrease in the minimum inhibitory concentrations (MICs) of various antibiotics (Ge et al., 2005). CmeR acts as transcriptional repressor by binding to the promoter of cmeABC operon to control its expression (Lin et al., 2005a). Mutations in the regulatory region of cmeABC promoter (CmeR-Box) have been reported to confer fluoroquinolone resistance in Campylobacter (Zhang et al., 2017a; Du et al., 2018), but the effect of these mutations on macrolide resistance has not been investigated.
In this study, to uncover the prevalence and the underlying molecular basis of erythromycin-resistant Campylobacter in central China, resistance analysis was conducted, and the mutations on macrolide targets and the present of erm(B) were screened. In addition, the polymorphisms of CmeR-Box in the promoter of the cmeABC efflux pump were also investigated.
Materials and Methods
Ethics Statement
All animal studies were conducted in strict accordance with the animal welfare guidelines of the World Organization for Animal Health. The protocols were approved by the Hubei Provincial Animal Care and Use Committee (approval number SCXK 2015-0021).
Bacterial Isolation
From 2015 to 2017, 143 Campylobacter isolates were collected from chickens or chicken meats in central China (three farms and four markets in Hubei, two farms and three markets in Henan, two farms and two markets in Jiangxi, one farm and two markets in Anhui, and one farm in Hunan), and all the chickens were from commercial broiler flocks. In brief, freshly collected anal and meat swabs were kept into Cary–Blair modified transport media (AMRESCO, United States) and transported to the laboratory for Campylobacter isolation. The samples were resuspended in phosphate-buffered saline (PBS) first, and then inoculated in Bolton broth containing Campylobacter growth supplement (Oxoid, United Kingdom) and Campylobacter Bolton broth selective supplement (Oxoid, United Kingdom) for 24 h at 42°C under microaerobic condition. After inoculation, 100 μl of the culture was spread onto a modified charcoal cefoperazone deoxycholate agar (mCCDA, Oxoid) plate containing Campylobacter CCDA-selective supplement (Zhang A. et al., 2016). The suspected Campylobacter colonies were identified by polymerase chain reaction (PCR) targeting 16S ribosomal DNA (rDNA) and sequencing as described (Weisburg et al., 1991), and the primers were as follows: 27F, 5′-AGAGTTTGATCMTGGCTCAG-3′; 1492R, 5′-TACGGYTACCTTGTTACGACTT-3′. C. jejuni and C. coli were differentiated by hippuric acid hydrolysis test and PCR test targeting C. jejuni-specific hipO gene and C. coli-specific asp gene (Persson and Olsen, 2005; Keller and Shriver, 2014). To generate a microaerobic environment, all the bacterial culture processes were carried out at 42°C in air tight jars containing the AnaeroPack (Mitsubishi, Japan).
Antimicrobial Susceptibility Test
According to the European Committee on Antimicrobial Susceptibility Testing guidelines (EUCAST, 2018), erythromycin resistance was first determined by the disk diffusion method on Mueller–Hinton agar (Oxoid, United Kingdom) using erythromycin disks with 15 μg. After incubation for 40 h at 42°C, the diameters (in mm) of the inhibition zones were measured, and <20 mm (C. jejuni) or <24 mm (C. coli) was determined to be resistant. Then, the MICs of erythromycin-resistant strains were measured by the broth dilution method. In brief, twofold serial dilutions of erythromycin was used at the concentrations of 2–2,048 μg/ml, and 5 × 105 CFU/ml of each isolate was incubated in Mueller–Hinton broth containing serial dilutions of erythromycin under microaerobic condition at 42°C for 24 h. The MICs were determined as the lowest concentration of the agent that completely inhibits visible growth. The antibiotic disks and powder were purchased from Oxoid, United Kingdom. The Escherichia coli ATCC 25922 was used as a quality control strain.
MLST
Multilocus sequence typing (MLST) was carried out in all erythromycin-resistant isolates. In brief, genomic DNA of the Campylobacter isolates was extracted using MiniBEST Universal Genomic DNA Extraction Kit (TaKaRa, Dalian, China) according to the manufacturer’s instructions. MLST analysis was conducted by sequencing seven Campylobacter housekeeping genes (aspA, glnA, gltA, glyA, pgm, tkt, and uncA) as previously described (Dingle et al., 2001). Allele numbers, sequence types (STs) and clonal complexes (CCs) were assigned using the Campylobacter MLST database1. The calculated tree of the erythromycin-resistant isolates was constructed using the SliptsTree 4 version 1.2 based on the ST types.
Detection of erm(B) and Mutations in 23S rRNA and CmeR-Box
The presence of erm(B) was screened by PCR as previously reported (Wang et al., 2014), and the primers used were as follows: ermB-F, 5′-GGGCATTTAACGACGAAACTGG-3′; ermB-R, 5′-CTGTGGTATGGCGGGTAAGT-3′. Polymorphisms present on the amplified fragment of the 23S rRNA and on the promoter of cmeABC operon (CmeR-Box) were investigated using PCR and double-stranded DNA sequencing as previously described (Corcoran et al., 2005; Perez-Boto et al., 2010). The primers for amplifying the fragment of the 23S rRNA and CmeR-Box were as follows: 23S-F, 5′-GCTCGAAGGTTAATTGATG-3′ and 23S-R, 5′-GCTCTTGGCAGAACAAC-3′; Cbox1-F, 5′-GG TTGTTACAGGTTGAGGC-3′ and Cbox1-R, 5′-AGCTTAC GCAAAGGATAATG-3′ for C. jejuni; and Cbox2-F, 5′-GGTT GTTACAGGTTGAGGC-3′ and Cbox2-R, 5′-AGCTTACGCAAAGGATAATG-3′ for C. coli.
Electrophoresis Mobility Shift Assay
Binding of recombinant CmeR protein to the promoters of the cmeABC operon, which contained different CmeR-box sequences, was performed by electrophoresis mobility shift assay (EMSAs) as previously described (Zhang et al., 2012; Grinnage-Pulley and Zhang, 2015). Briefly, the DNA fragments containing CmeR-Box sequences were amplified from genomic DNA of Campylobacter of the Campylobacter isolates. To obtain the CmeR protein, the coding sequence of cmeR was amplified and cloned into vector pET-28a (Novagen, Shanghai, China). Escherichia coli BL21(DE3) was transformed with the recombinant plasmid pET28a-cmeR, and then, the expression was induced by the addition of 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at 18°C for 12 h. The recombinant CmeR protein was purified with Ni–NTA agarose (Bio-Rad, Shanghai, China) under native conditions, according to the manufacturer’s instructions. Binding reactions were carried out in a 20 μl volume containing 0.1 μg promoter DNA and different amounts of purified recombinant CmeR protein (0, 2, 4, and 8 μg), and incubated at room temperature for 30 min. Electrophoresis was carried out with 5% native polyacrylamide gels at 100 V for 1 h. The gels were stained with 1 μg/ml of ethidium bromide. To compare the bonding abilities, the optical densities of bound DNA and free DNA were measured using Clinx Image Analysis software (Clinx Science Instruments Co., Ltd., Shanghai, China). The proportions of bound DNA were calculated according to the optical densities values (0% was “−”; >0 to ≤50% was “+”; >50 to ≤95% was “++”; >95% was “+++”).
Real-Time RT-PCR
To analyze the effect of CmeR-Box polymorphism on the expression of cmeABC, five C. jejuni isolates (QCJ3, JSJ27, XZJ48, WHJ54, and XTJ10) were chosen. The QCJ3 strain contained a wild-type CmeR-Box, the JSJ27 strain contained a point substitution in CmeR-Box, the XZJ48 (ST-7510) and WHJ54 (ST-7512) strains contained the same point deletion in CmeR-Box, and the XTJ10 (ST-7508) strain contained a point insertion in CmeR-Box. None of the isolates contain the erm(B) and/or A2075G substitution in the 23S rRNA. Total RNAs of the selected Campylobacter isolates were isolated as follows: overnight cultured bacteria were diluted 1:100 in fresh Mueller–Hinton broth and then incubated to mid-log phase (OD600 = 0.5) at 42°C under microaerobic condition. Eight micrograms per milliliter (two times of MIC breakpoint by EUCAST, and the breakpoint of erythromycin is 4 μg/ml) of erythromycin was added, and the bacteria were collected at 0, 5, and 120 min post-treatment. Total RNA was isolated and purified using the RNeasy Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. Then, the isolated RNA was reverse transcribed to complementary DNA (cDNA), and the expression levels of the cmeA gene were assessed by real-time reverse transcription PCR (RT-PCR) using SYBRGreen detection (TAKARA BIO INC., Dalian, China) in an ABI7500 system (Thermo Fisher Scientific, CA, United States). The primers for cmeA were as follows: qcmeA-F, 5′-CTGACAAGTTTAGCAGGGTA-3′, qcmeA-R, 5′-GCAGCAAAGAAGAAGCACCA-3′. The 16S rRNA and gapdh genes were used as the internal control. The primers were as follows: q16S-F, 5′-TACCTGGGCTTGATATCCTA-3′, q16S-R, 5′-GGACTTAACCCAACATCTCA-3′; qgapdh-F, 5′-AGGCAGTGTTGATAGTGAAGG-3′, qgapdh-R, 5′-CAATTTGTGCGCCGTGTT-3′. The expression level of cmeA before treatment of the strain harboring wild-type CmeR-Box (C. jejuni strain QCJ3) was used as control condition. Each assay was carried out with at least three biological replicates. Differences in relative transcript abundance level were calculated using the 2–ΔΔCT method (Pfaffl, 2001), and ≥2-fold changes were considered as differentially expressed.
Statistical Analysis
The Fisher test was used to identify the correlation between resistance and mutations, and the Student’s t-test was used to compare the expression levels of target genes of different strains. The analysis was carried out by SPSS 19.0. A probability (p) value of <0.05 was considered statistically significant.
Results
Prevalence of Erythromycin-Resistant Campylobacter
A total of 143 Campylobacter strains, including 83 C. jejuni and 60 C. coli isolates, were isolated and tested for erythromycin susceptibility. Among them, 36 (25.2%) Campylobacter isolates were erythromycin resistant, including 25 (30.1%, MIC > 4 mg/l) C. jejuni and 11 (18.3%, MIC > 8 mg/l) C. coli (Figure 1). These results showed high erythromycin resistance rates of C. jejuni and C. coli isolates in our study.

Figure 1. Dendrogram of multilocus sequence typing (MLST) profiles among the 36 erythromycin-resistant Campylobacter isolates. Corresponding minimum inhibitory concentrations (MICs) of erythromycin, erythromycin-resistant genes, mutations found, and the CmeR-Box sequence are listed for each isolate. aUA means unallocated in campylobacters. bThe underline indicates that two of the three copies were mutated. cUnderline means point substitution, “(A)” means point insertion, “–” means point deletion.
Mutations in 23S rRNA of Campylobacter Isolates
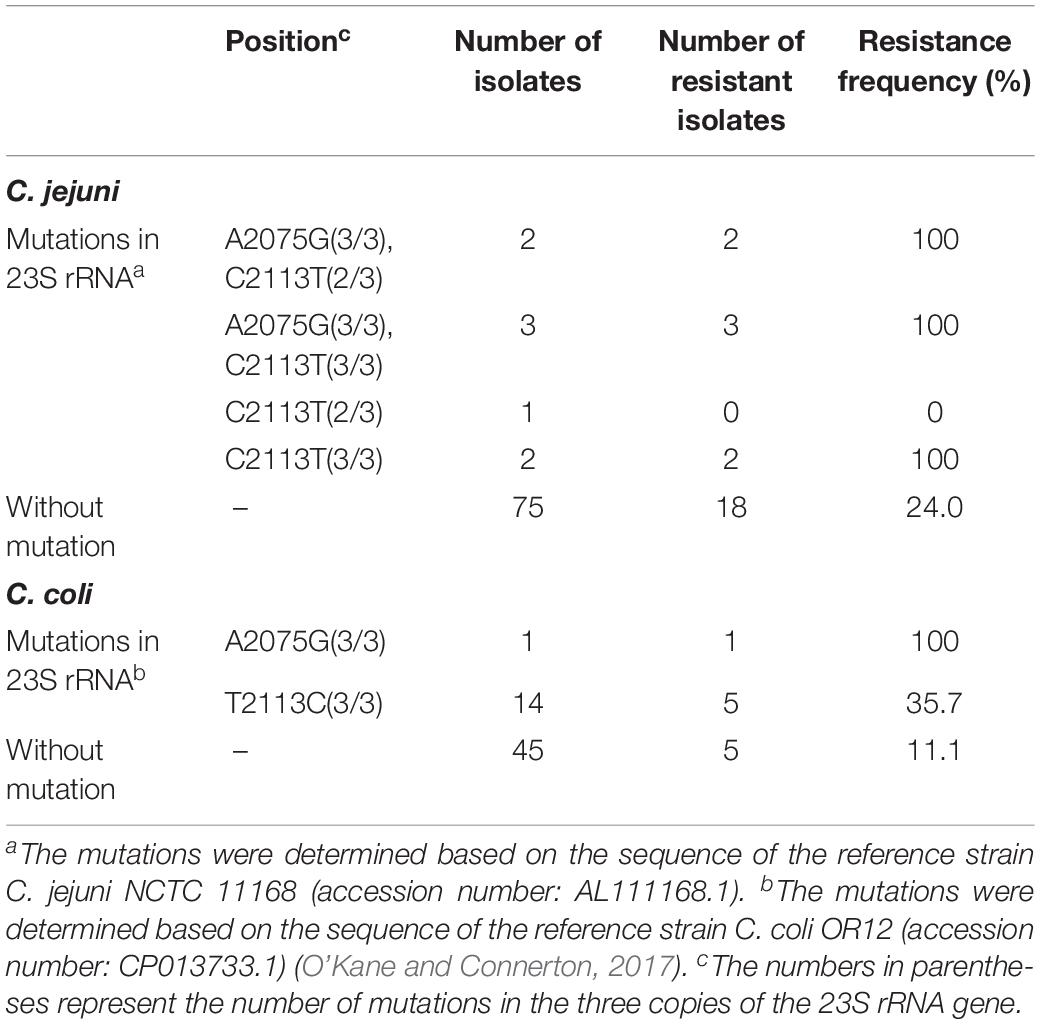
Polymorphisms on the 23S rRNA were analyzed in all Campylobacter isolates. As shown in Table 1, Figure 1, and Supplementary Material, point substitutions on 23S rRNA were found in 23 isolates (8 C. jejuni and 15 C. coli isolates). According to published sequences, the Campylobacter genome encodes three copies of 23S rRNA (Sheppard and Maiden, 2015). Except for the isolates that contained A2075G substitution, 16 isolates contained the C2113T (C. jejuni) or T2113C (C. coli) substitution in all three copies of 23S rRNA, while one C. jejuni contained this substitution in two of three copies (double peak and the peak for guanine was two times higher than the peak for adenine), and among them, 7 of the 17 isolates were erythromycin resistant; thus, the correlation was not significant (p > 0.05). The A2075G substitution was observed in 6 (5 C. jejuni and 1 C. coli isolates) of the 23 isolates, and all of them were erythromycin resistant. These results suggested that the A2075G, but not the C2113T or T2113C, substitution in 23S rRNA was responsible for erythromycin resistance in Campylobacter.
Presence of erm(B) in Campylobacter Isolates
The ribosomal methylase erm(B) gene was found in seven Campylobacter isolates (4.9%), including three C. jejuni and four C. coli. These seven isolates were also erythromycin resistant, and none of them contained an A2075G substitution in 23S rRNA (Figure 1; Wang et al., 2014). Although the incidence of erm(B) was not high, the seven erm(B)-harboring strains were isolated from five different regions.
Polymorphism Analysis of CmeR-Box in Campylobacter Isolates
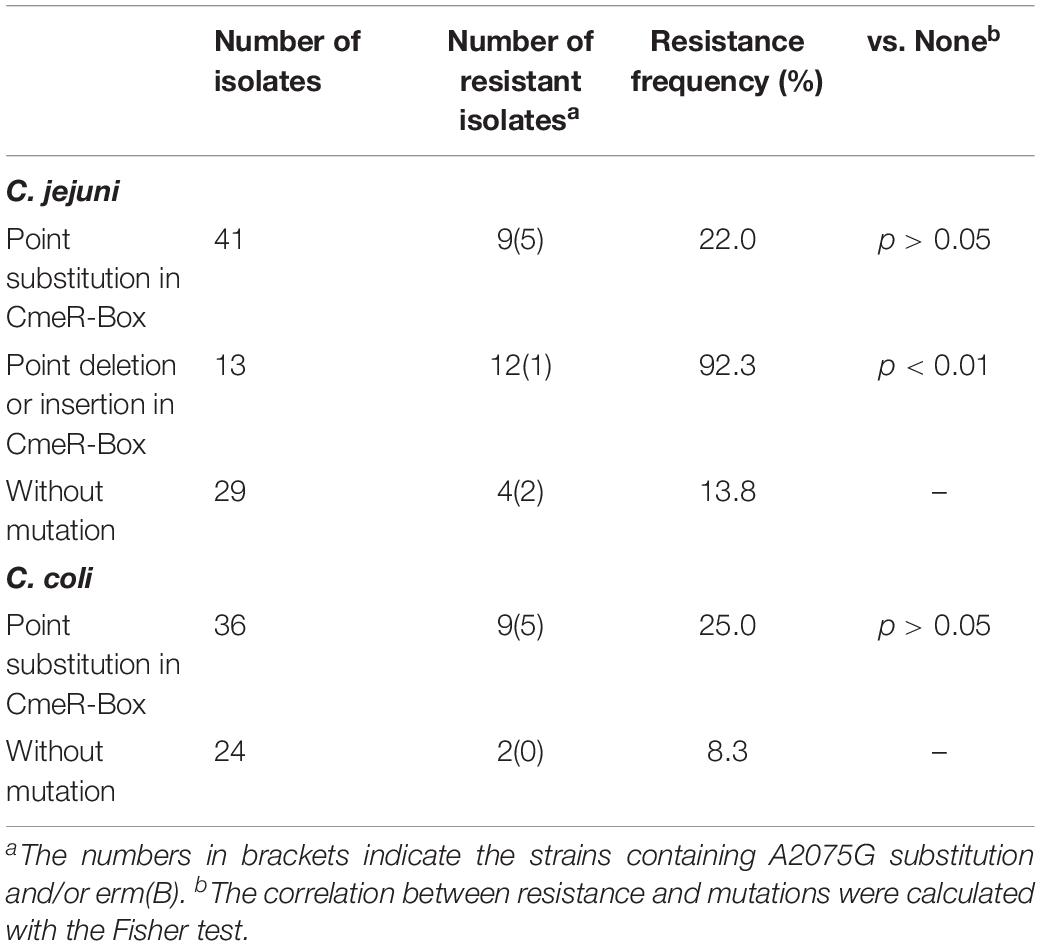
Polymorphisms of CmeR-Box were analyzed in all isolates, and seven CmeR-Box variants were identified (Figure 1 and Table 2). Among them, 13 (9.1%) isolates contained a point deletion/insertion within the inverted sequences (Figure 2A), and 12 out of 13 were resistant to erythromycin. Among the 12 resistant isolates, 11 did not contain erm(B) or a mutation on 23S rRNA. Statistical analysis showed that the occurrence of point deletion/insertions significantly correlated with erythromycin resistance (p < 0.05), suggesting an important role in the erythromycin resistance phenotype. In contrast, point substitutions in CmeR-Box were found in 41 (49.4%) C. jejuni isolates and 36 (60.0%) C. coli isolates, but among them, only 9 (22.0%) C. jejuni and 9 (25.0%) C. coli isolates were erythromycin resistant, and among the 18 resistant isolates, most of them also harbor mutations in 23S rRNA and/or the erm(B) gene (Figure 1). Statistical analysis showed that nucleotide substitution was not correlated with erythromycin resistance (p > 0.05). Our results suggested that point deletions or insertions, but not nucleotide substitution, in the CmeR-Box was significantly associated with erythromycin resistance.
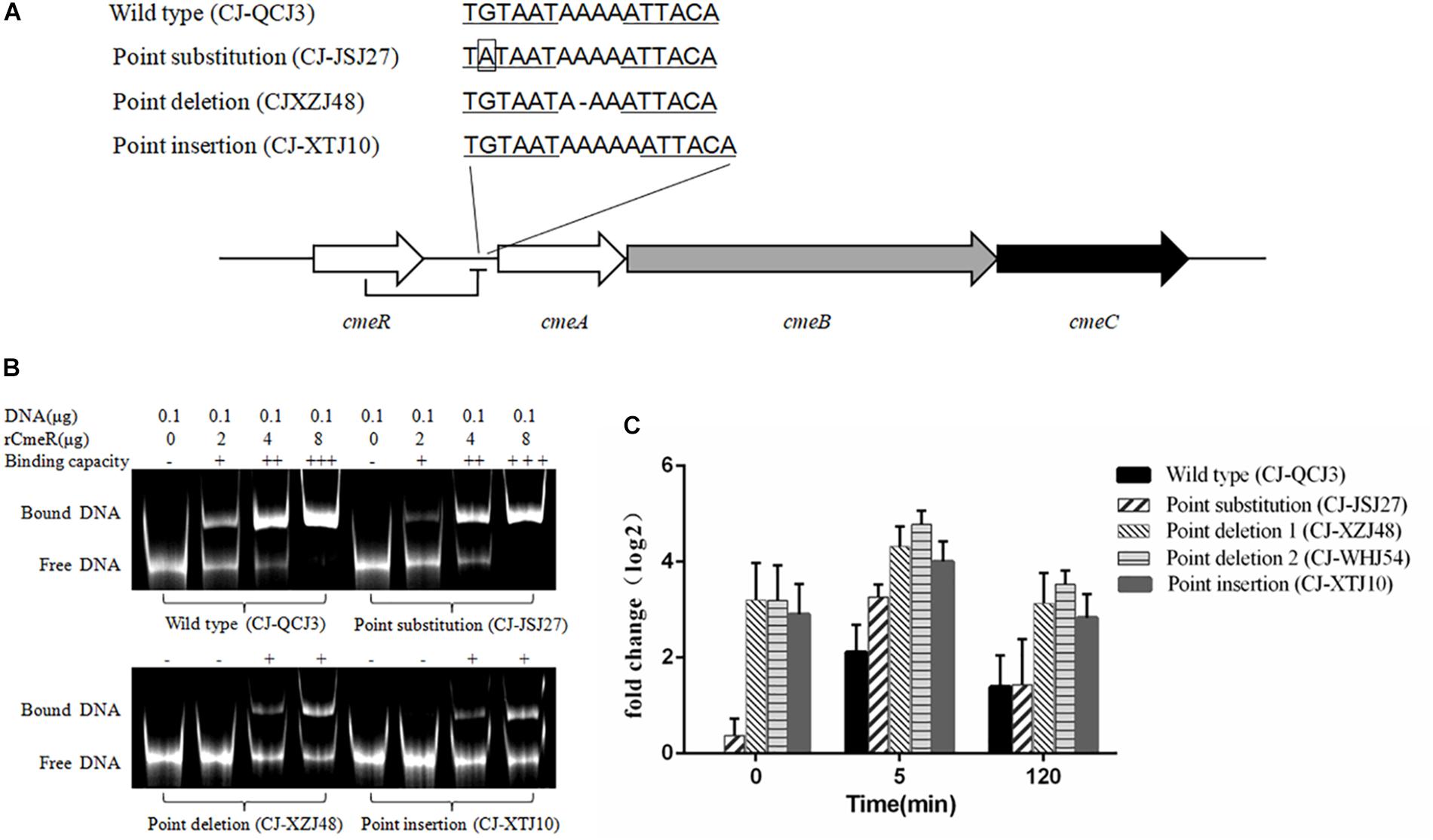
Figure 2. cmeA expression levels in Campylobacter isolates with different CmeR-Box sequences. (A) CmeR-Box sequences in the Campylobacter isolates. (B) Binding of CmeR to the variant cmeABC promoter DNA. (C) The expression levels of cmeA gene at 5- and 120-min post-erythromycin treatment in the Campylobacter isolates harboring different CmeR-Box sequences. Each assay was carried out at least in three biological replicates, and the bars mean the standard deviation in each assay.
EMSAs and Expression Analysis of cmeABC
To further understand the effect of point deletion/insertion in the CmeR-Box on erythromycin resistance, the binding of CmeR to the CmeR-Box with a point deletion/insertion was confirmed with the EMSAs. As shown in Figure 2B, the binding of CmeR to CmeR-Box with a point deletion/insertion was significantly decreased, suggesting that the cmeABC operon in these isolates was highly derepressed and associated with erythromycin resistance. Analysis of the expression levels of cmeA at the time point 0 min further confirmed these results (Figure 2C). Although cmeA expression could be induced by the addition of erythromycin in all the isolates, as compared with the isolate without a mutation in the CmeR-Box, higher expression was observed in the isolates harboring a point deletion/insertion before and after erythromycin treatment (p < 0.05). It is worth noting that, in the isolates harboring a point deletion/insertion, cmeA expression was originally high before erythromycin treatment and only increased by approximately twofold at 5 min post-treatment and recovered at 2 h post-treatment. In contrast, in the wild-type isolate and isolate with a point substitution, although cmeA expression was upregulated by more than fourfold at 5 min post-treatment, their expression levels were still lower than in isolates with a point deletion/insertion.
Genetic Diversity Analysis
The genetic diversity of erythromycin-resistant Campylobacter isolates was analyzed with MLST, as previously described (Dingle et al., 2001). As shown in Figure 1, the 36 erythromycin-resistant Campylobacter isolates were distributed along 23 STs and 8 CCs. The dominant CC was CC353, and seven isolates could not be assigned to a CC. Five C. jejuni strains with an A2075G substitution in the 23S rRNA belonged to three CCs (CC607, CC464, and CC52), and one could not be assigned to a CC. Three C. jejuni erm(B)-harboring strains belonged to two different CCs (CC353 and CC21), and one was not assigned. These results suggest a diverse distribution of genotypes in C. jejuni resistant to erythromycin. In contrast, 10 out of 11 erythromycin-resistant C. jejuni with a point deletion in the CmeR-Box were distributed between two STs both belonging to the dominant CC353, suggesting a common ancestor. In C. coli isolates, three erm(B)-harboring isolates shared the same CC (CC828) clustered with an unallocated (UA) strain, suggesting a common ancestor.
Discussion
Campylobacter jejuni and Campylobacter coli are major food-borne pathogens worldwide, and poultry is recognized as the most important reservoir of these pathogens (Crawshaw, 2019; Thomas et al., 2019). Usually, erythromycin is the first-line choice for treatment of campylobacteriosis (Bolinger and Kathariou, 2017). Therefore, to investigate the prevalence and erythromycin resistance mechanisms of C. jejuni and C. coli is not only important for the poultry industry but also for public health.
In our study, 30.1% C. jejuni and 18.3% C. coli isolates were erythromycin resistant. As previously reported, the prevalence of macrolide resistance is common in C. coli but remains low in C. jejuni. For example, data from the National Antimicrobial Resistance Monitoring System (NARMS) report of the U.S. Food and Drug Administration on human clinical isolates and food-producing animal isolates indicated that the resistance rate in C. jejuni was under 4% in 2014 (FDA, 2014). In a previous investigation in China, the global resistance rate of Campylobacter to erythromycin was 18.4%, and among the resistant strains, 97.5% were C. coli and 2.5% were C. jejuni (Zhang A. et al., 2016). However, in this study, high erythromycin resistance rates were found in both C. jejuni and C. coli isolated from central China, which pressed us to further understand the molecular mechanisms of erythromycin resistance in these isolates.
Mutations in 23S rRNA, and single adenine methylation in the 23S rRNA by ribosomal methylase erm(B), are two well-known mechanisms for erythromycin resistance in Campylobacter (Zhao et al., 2016). In this study, A2075G substitution in 23S rRNA and ribosomal methylase encoded by erm(B) were found in 4.2 and 4.9% isolates, respectively, and all of them were erythromycin resistant. As previously reported, the A2075G substitution is one of the most prevalent genetic events conferring high-level resistance to erythromycin (Zhao et al., 2016), as further evidenced by our results. Substitutions at 2113 site were found in our isolates. Based on previous reports and the sequenced reference strains, including C. jejuni NCTC 11168 and C. coli OR12 (Parkhill et al., 2000; Perez-Boto et al., 2010; O’Kane and Connerton, 2017), the dominant nucleotide at 2113 site is “C” in C. jejuni and “T” in C. coli, which were defined as wild type in both species in this study. Correlation analysis showed that this substitution was not correlated with erythromycin resistance (p > 0.05). We infer that this substitution exists extensively in Campylobacter and does not affect erythromycin resistance. Some of the resistant isolates with this substitution could be explained by other resistance factors, such as erm(B) in strains QCJ11, XTJ11, XTJ12, and XTJ13. However, the causes of resistance on the part of isolates are still unknown and require further study.
erm(B)-harboring Campylobacter isolates were first reported in 2014 in China (Qin et al., 2014) and then detected in Spain and the United States (Florez-Cuadrado et al., 2016). In this study, although the incidence of erm(B) was not high, seven erm(B)-harboring strains were isolated from five different regions. Our results suggested that erm(B) might be widespread at the regional distribution in central China.
Drug efflux is another important resistance mechanism conferred by native efflux systems in bacteria. The cmeABC operon, which encodes a drug efflux pump and plays an important role in drug resistance in Campylobacter (Lin et al., 2005a), is negatively regulated by CmeR by binding to a 16-base inverted repeat sequence [CmeR-Box, TGTAATA(or T)TTTATTACA] in the promoter region (Yan et al., 2006). In our isolates, a couple of CmeR-Box variants were identified (Figure 2A and Table 2), including point substitutions and deletion/insertion within the inverted sequences. It is interesting that point deletions/insertions were found in 13 (9.1%) isolates, and 12 out of 13 isolates were resistant, which was correlated to erythromycin resistance (p < 0.05). However, Campylobacter harboring point deletions in CmeR-Box were seldom reported and not well characterized (Abd El-Tawab et al., 2019). In contrast, point substitutions were not correlated to erythromycin resistance (p > 0.05). Substitutions in CmeR-Box have been reported to cause overexpression of cmeABC although not affecting the susceptibility of C. jejuni to most tested antimicrobials including erythromycin (Grinnage-Pulley and Zhang, 2015). It is worth noting that all of the mutations identified in CmeR-Box were point substitutions in that report, so their result was consistent with our finding that there was no correlation between point substitutions in CmeR-Box and erythromycin resistance. In our study, point deletion/insertion in CmeR-Box was identified in C. jejuni isolates, and our results suggested that point deletion or insertion, but not substitution, in CmeR-Box was significantly associated with erythromycin resistance.
To further investigate the effect of point deletion/insertion in CmeR-Box on resistance, the binding of CmeR to the various CmeR-Box and the expressional levels of cmeABC were detected. It is well-known that CmeR can interact with bile salts and transduce this signaling, but little is known about the capacity of antibiotics to induce cmeABC expression (Lin et al., 2005b). In this study, the expression of cmeA was induced after erythromycin treatment. We found that the expression of cmeR, which encodes the repressor of cmeABC, was slightly reduced 5 min post-treatment, and there were no significant difference between these tested isolates (data not shown). We inferred that the upregulated expression of cmeABC after erythromycin treatment was due to the reduced expression of its repressor CmeR. Comparing with the wild-type CmeR-Box and CmeR-Box with point substitution, an obvious decrease in binding ability of CmeR to CmeR-Box with a point deletion/insertion, and subsequent higher overexpression of cmeABC, was showed. These results support why point deletion or insertion, but not substitutions, in CmeR-Box was significantly associated with erythromycin resistance. Functional CmeR is a dimer, and each monomer binds to one half of the inverted repeat of the CmeR-Box (Gu et al., 2007; Lei et al., 2011). We inferred that, although point substitutions reduced the bonding strength of CmeR to CmeR-Box, the bonding strength was still moderate (Figure 2B). Therefore, the expression of cmeABC was only slightly increased (Figure 2B) in these isolates harboring CmeR-Box with point substitutions. The slightly increased expression of cmeABC might result in increased MICs, but the MICs did not reach the breakpoint in these isolates. In contrast, a point deletion/insertion changed the distance between two consecutive binding sequences, leading to an obvious decrease in binding ability and subsequent overexpression of cmeABC (Figure 2), which conferred high level of erythromycin resistance. With regard to that one susceptible isolate carrying point deletion in CmeR-Box, we infer that there might be some unknown mutations, such as the mutations in the cmeABC proteins, which affect its resistant ability. CC-JMJ1 is a specific highly resistant isolate, which does not contain any mutations in CmeR-Box and 23S rRNA, and for erm(B), we infer that it may contain a previously reported resistance-enhancing variant of the efflux pump cmeABC, or due to other unknown mechanisms (Yao et al., 2016).
Conclusion
In summary, 25.2% of Campylobacter isolates in central China were erythromycin resistant. The A2075G substitution in 23S rRNA and the presence of erm(B) were identified as two important factors that lead to erythromycin resistance. Furthermore, this is the first study to report that a point deletion/insertion, but not substitution, in CmeR-Box could significantly increase the expression of cmeABC, which plays important roles in erythromycin resistance. These findings will help us to further understand the mechanism of erythromycin resistance in Campylobacter.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Ethics Statement
All animal studies were conducted in strict accordance with the animal welfare guidelines of the World Organization for Animal Health. The protocols were approved by the Hubei Provincial Animal Care and Use Committee (approval number SCXK 2015-0021).
Author Contributions
YC, TZ, and HS conceived and designed the experiments and wrote the manuscript. YC, QL, QLo, and ZZ performed the experiments. GW and WZ analyzed the data.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from the National Key Research and Development Plan of China (2016YFD0501305) and the China Agriculture Research System (CARS-41-G13).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00203/full#supplementary-material
Footnotes
References
Abd El-Tawab, A. A., Ammar, A. M., Ahmed, H. A., and Hefny, A. A. (2019). Efflux pump inhibitors, Alpha-Tocopherol and aspirin: role in Campylobacter jejuni and Campylobacter coli fluoroquinolone resistance. Microb. Drug Resist. 25, 203–211. doi: 10.1089/mdr.2018.0086
Bolinger, H., and Kathariou, S. (2017). The current state of macrolide resistance in Campylobacter spp.: trends and impacts of resistance mechanisms. Appl. Environ. Microbiol. 83, e416–e417. doi: 10.1128/AEM.00416-17
Corcoran, D., Quinn, T., Cotter, L., O’halloran, F., and Fanning, S. (2005). Characterization of a cmeABC operon in a quinolone-resistant Campylobacter coli isolate of Irish origin. Microb. Drug Resist. 11, 303–308. doi: 10.1089/mdr.2005.11.303
Crawshaw, T. (2019). A review of the novel thermophilic Campylobacter, Campylobacter hepaticus, a pathogen of poultry. Transbound. Emerg. Dis. 66, 1481–1492. doi: 10.1111/tbed.13229
Dingle, K. E., Colles, F. M., Wareing, D. R., Ure, R., Fox, A. J., Bolton, F. E., et al. (2001). Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39, 14–23.
Dinos, G. P. (2017). The macrolide antibiotic renaissance. Br. J. Pharmacol. 174, 2967–2983. doi: 10.1111/bph.13936
Du, Y., Wang, C., Ye, Y., Liu, Y., Wang, A., Li, Y., et al. (2018). Molecular identification of multidrug-resistant Campylobacter species from Diarrheal patients and poultry meat in Shanghai China. Front. Microbiol. 9:1642. doi: 10.3389/fmicb.2018.01642
EUCAST (2018). Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 8.1, 2018. Available online at: http://www.eucast.org
FDA (2014). “The national antimicrobial resistance monitoring system (NARMS): enteric bacteria,” in NARMS Integrated Report: 2014, (Silver Spring, MA: US Food and Drug Administration).
Florez-Cuadrado, D., Ugarte-Ruiz, M., Quesada, A., Palomo, G., Domínguez, L., and Porrero, M. C. (2016). Description of an erm(B)-carrying Campylobacter coli isolate in Europe. J. Antimicrob. Chemother. 71, 841–843. doi: 10.1093/jac/dkv383
Ge, B., Mcdermott, P. F., White, D. G., and Meng, J. (2005). Role of efflux pumps and topoisomerase mutations in fluoroquinolone resistance in Campylobacter jejuni and Campylobacter coli. Antimicrob. Agents Chemother. 49, 3347–3354. doi: 10.1128/aac.49.8.3347-3354.2005
Grinnage-Pulley, T., and Zhang, Q. (2015). Genetic basis and functional consequences of differential expression of the CmeABC efflux pump in Campylobacter jejuni isolates. PLoS One 10:e0131534. doi: 10.1371/journal.pone.0131534
Gu, R., Su, C. C., Shi, F., Li, M., Mcdermott, G., Zhang, Q., et al. (2007). Crystal structure of the transcriptional regulator CmeR from Campylobacter jejuni. J. Mol. Biol. 372, 583–593. doi: 10.1016/j.jmb.2007.06.072
Hao, H., Dai, M., Wang, Y., Peng, D., Liu, Z., and Yuan, Z. (2009). 23S rRNA mutation A2074C conferring high-level macrolide resistance and fitness cost in Campylobacter jejuni. Microb. Drug Resist. 15, 239–244. doi: 10.1089/mdr.2009.0008
Huang, J. L., Xu, H. Y., Bao, G. Y., Zhou, X. H., Ji, D. J., Zhang, G., et al. (2009). Epidemiological surveillance of Campylobacter jejuni in chicken, dairy cattle and diarrhoea patients. Epidemiol. Infect. 137, 1111–1120. doi: 10.1017/S0950268809002039
Kaakoush, N. O., Castaño-Rodríguez, N., Mitchell, H. M., and Si, M. M. (2015). Global epidemiology of Campylobacter infection. Clin. Microbiol. Rev 28, 687–720. doi: 10.1128/CMR.00006-15
Keller, J. I., and Shriver, W. G. (2014). Prevalence of three campylobacter species, C. jejuni, C. coli, and C. lari, using multilocus sequence typing in wild birds of the Mid-Atlantic region, USA. J. Wildl. Dis. 50, 31–41. doi: 10.7589/2013-06-136
Lei, H. T., Shen, Z., Surana, P., Routh, M. D., Su, C. C., Zhang, Q., et al. (2011). Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni. Protein Sci. 20, 712–723. doi: 10.1002/pro.602
Lin, J., Akiba, M., Sahin, O., and Zhang, Q. (2005a). CmeR functions as a transcriptional repressor for the multidrug efflux pump CmeABC in Campylobacter jejuni. Antimicrob. Agents Chemother. 49, 1067–1075. doi: 10.1128/aac.49.3.1067-1075.2005
Lin, J., Cagliero, C., Guo, B., Barton, Y. W., Maurel, M. C., Payot, S., et al. (2005b). Bile salts modulate expression of the CmeABC multidrug efflux pump in Campylobacter jejuni. J. Bacteriol. 187, 7417–7424. doi: 10.1128/jb.187.21.7417-7424.2005
Lin, J., Michel, L. O., and Zhang, Q. (2002). CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob. Agents Chemother. 46, 2124–2131. doi: 10.1128/aac.46.7.2124-2131.2002
O’Kane, P. M., and Connerton, I. F. (2017). Characterisation of aerotolerant forms of a robust chicken colonizing Campylobacter coli. Front. Microbiol. 8:513. doi: 10.3389/fmicb.2017.00513
Parkhill, J., Wren, B. W., Mungall, K., Ketley, J. M., Churcher, C., Basham, D., et al. (2000). The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403, 665–668. doi: 10.1038/35001088
Perez-Boto, D., Lopez-Portoles, J. A., Simon, C., Valdezate, S., and Echeita, M. A. (2010). Study of the molecular mechanisms involved in high-level macrolide resistance of Spanish Campylobacter jejuni and Campylobacter coli strains. J. Antimicrob. Chemother. 65, 2083–2088. doi: 10.1093/jac/dkq268
Persson, S., and Olsen, K. E. (2005). Multiplex PCR for identification of Campylobacter coli and Campylobacter jejuni from pure cultures and directly on stool samples. J. Med. Microbiol. 54, 1043–1047. doi: 10.1099/jmm.0.46203-0
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45.
Qin, S., Wang, Y., Zhang, Q., Zhang, M., Deng, F., Shen, Z., et al. (2014). Report of ribosomal RNA methylase gene erm(B) in multidrug-resistant Campylobacter coli. J. Antimicrob. Chemother. 69, 964–968. doi: 10.1093/jac/dkt492
Sheppard, S. K., and Maiden, M. C. (2015). The evolution of Campylobacter jejuni and Campylobacter coli. Cold Spring Harb. Perspect. Biol. 7:a018119. doi: 10.1101/cshperspect.a018119
Thomas, K. M., de Glanville, W. A., Barker, G. C., Benschop, J., Buza, J. J., and Cleaveland, S. (2019). Prevalence of Campylobacter and Salmonella in African food animals and meat: a systematic review and meta-analysis. Int. J. Food Microbiol. 315:108382. doi: 10.1016/j.ijfoodmicro.2019.108382
Trastoy, R., Manso, T., Fernandez-Garcia, L., Blasco, L., Ambroa, A., Perez Del Molino, M. L., et al. (2018). Mechanisms of bacterial tolerance and persistence in the gastrointestinal and respiratory environments. Clin. Microbiol. Rev. 31, e23–e18. doi: 10.1128/CMR.00023-18
Wang, Y., Zhang, M., Deng, F., Shen, Z., Wu, C., Zhang, J., et al. (2014). Emergence of multidrug-resistant Campylobacter species isolates with a horizontally acquired rRNAmethylase. Antimicrob. Agents Chemother. 58, 5405–5412. doi: 10.1128/AAC.03039-14
Weisburg, W. G., Barns, S. M., Pelletier, D. A., and Lane, D. J. (1991). 0.16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703. doi: 10.1128/jb.173.2.697-703.1991
Yan, M., Sahin, O., Lin, J., and Zhang, Q. (2006). Role of the CmeABC efflux pump in the emergence of fluoroquinolone-resistant Campylobacter under selection pressure. J. Antimicrob. Chemother. 58, 1154–1159. doi: 10.1093/jac/dkl412
Yao, H., Shen, Z., Wang, Y., Deng, F., Liu, D., Naren, G., et al. (2016). Emergence of a potent multidrug efflux pump variant that enhances Campylobacter resistance to multiple antibiotics. mBio 7:e001543-16. doi: 10.1128/mBio.01543-16
Zhang, A., Song, L., Liang, H., Gu, Y., Zhang, C., Liu, X., et al. (2016). Molecular subtyping and erythromycin resistance of Campylobacter in China. J. Appl. Microbiol. 121, 287–293. doi: 10.1111/jam.13135
Zhang, T., Cheng, Y., Luo, Q., Lu, Q., Dong, J., Zhang, R., et al. (2017a). Correlation between gyrA and CmeR box polymorphism and fluoroquinolone resistance in Campylobacter jejuni isolates in China. Antimicrob. Agents Chemother. 61:e00422-17. doi: 10.1128/AAC.00422-17
Zhang, T., Ding, Y., Li, T., Wan, Y., Li, W., Chen, H., et al. (2012). A Fur-like protein PerR regulates two oxidative stress response related operons dpr and metQIN in Streptococcus suis. BMC Microbiol. 12:85. doi: 10.1186/1471-2180-12-85
Zhang, T., Dong, J., Cheng, Y., Lu, Q., Luo, Q., Wen, G., et al. (2017b). Genotypic diversity, antimicrobial resistance and biofilm-forming abilities of Campylobacter isolated from chicken in Central China. Gut Pathog. 9, 62. doi: 10.1186/s13099-017-0209-6
Zhang, T., Luo, Q., Chen, Y., Li, T., Wen, G., Zhang, R., et al. (2016). Molecular epidemiology, virulence determinants and antimicrobial resistance of Campylobacter spreading in retail chicken meat in Central China. Gut Pathog. 8:48.
Keywords: Campylobacter, erythromycin resistance, 23S rRNA, erm(B), CmeR-Box
Citation: Cheng Y, Zhang W, Lu Q, Wen G, Zhao Z, Luo Q, Shao H and Zhang T (2020) Point Deletion or Insertion in CmeR-Box, A2075G Substitution in 23S rRNA, and Presence of erm(B) Are Key Factors of Erythromycin Resistance in Campylobacter jejuni and Campylobacter coli Isolated From Central China. Front. Microbiol. 11:203. doi: 10.3389/fmicb.2020.00203
Received: 13 March 2019; Accepted: 29 January 2020;
Published: 03 March 2020.
Edited by:
Ziad Daoud, University of Balamand, LebanonReviewed by:
Ana Belén Flórez, Spanish National Research Council, SpainLuis Collado, Austral University of Chile, Chile
Diana Machado, New University of Lisbon, Portugal
Sofia Santos Costa, New University of Lisbon, Portugal
Copyright © 2020 Cheng, Zhang, Lu, Wen, Zhao, Luo, Shao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huabin Shao, c2hoYjE5NjFAMTYzLmNvbQ==; Tengfei Zhang, dGZ6aGFuZzIzQDE2My5jb20=
†These authors have contributed equally to this work
 Yiluo Cheng†
Yiluo Cheng† Huabin Shao
Huabin Shao Tengfei Zhang
Tengfei Zhang