
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 13 February 2020
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.00195
 Lang Yang1,2†
Lang Yang1,2† Yanfeng Lin1,2†
Yanfeng Lin1,2† Lanfen Lu3†
Lanfen Lu3† Mei Xue4†
Mei Xue4† Hui Ma5†
Hui Ma5† Xuguang Guo6
Xuguang Guo6 Kaiying Wang1,2
Kaiying Wang1,2 Peihan Li1,2
Peihan Li1,2 Xinying Du2
Xinying Du2 Kezong Qi4*
Kezong Qi4* Peng Li2*
Peng Li2* Hongbin Song2*
Hongbin Song2*The emergence of carbapenem-resistant Enterobacteriaceae poses a significant threat to public health worldwide. Here, we reported a multidrug-resistant Escherichia coli strain with two different blaNDM–5-carrying plasmids from China. Illumina short-read and MinION long-read whole genome sequencing were performed. Genomic analysis found that one blaNDM–5 gene together with mphA was located on a 55-kb IncX3 plasmid, while the other blaNDM–5 gene was on a novel 68-kb IncFII plasmid. Susceptibility testing and quantitative reverse transcription PCR results further indicated that the transconjugants with the IncX3 plasmid exhibited higher-level carbapenem resistance and expression of blaNDM–5 than those with both plasmids or the IncFII plasmid. Two other β-lactamase genes (blaCTX–M–15 and blaOXA–1) were also detected on another 160-kb IncF plasmid. This is the first report of coexistence of two blaNDM–5-carrying plasmids in a single bacterial isolate, highlighting the genetic complexity of NDM-5 carbapenemase circulation, and the urgent need for continued active surveillance.
Prevalence of carbapenemase-producing Enterobacteriaceae is a major concern around the world. NDM, one of the most important carbapenemases, represents a serious threat to patient care and public health. Since NDM-1 was first reported in 2009 (Yong et al., 2009), NDM-producing organisms has caused various types of clinical infections (Cornaglia et al., 2011). To date, 21 NDM variants have been discovered worldwide (Liu et al., 2018).
NDM-5, one of the most common variants encountered among Enterobacteriaceae, was first identified in an Escherichia coli isolate from a patient in the United Kingdom in 2011 with a recent history of hospitalization in India. It has two amino acid substitutions (Val88Leu and Met154Leu) in comparison with NDM-1, and confers increased resistance to extended-spectrum cephalosporins and carbapenems (Hornsey et al., 2011). Most strains usually harbored one single copy of the blaNDM gene. Coexistence of two copies of the blaNDM gene is rarely detected. Recent studies reported the presence of two tandem copies of blaNDM–1 in the chromosomes of an ST167 E. coli from China (Shen et al., 2017) and a Pseudomonas aeruginosa strain from Serbia (Jovcić et al., 2013). Duplication of the blaNDM–5 gene on an IncF plasmid was observed in an E. coli strain (Feng et al., 2018), which showed identical carbapenem resistance levels to those conferred by an IncX3 plasmid with a single copy. However, no studies have reported coexistence of two blaNDM–5 genes on different plasmids in the same strain. The effect of two blaNDM–5-carrying plasmids on carbapenem resistance still remained unclear.
In the present study, we report an E. coli strain harboring two blaNDM–5 genes, which were carried by IncX3 and IncFII plasmids, respectively. Susceptibility testing indicated that the transconjugant with the IncX3 plasmid alone showed the highest imipenem and meropenem resistance among all the transconjugants. Illumina short-read and MinION long-read whole genome sequencing was performed to gain an insight into the genomic and plasmid features.
The blaNDM–5-positive E. coli strain GZ04-0086 was recovered from the stool sample of a patient through routine surveillance in Guangzhou, China, in 2018. The species level identification was performed using Vitek 2 compact system (bioMérieux, France). Presence of common genes encoding carbapenemases and extended-spectrum β-lactamases was determined by PCR screening (Balm et al., 2012; Zong and Zhang, 2013; Elumalai et al., 2014). The complete blaNDM gene was amplified with previously described primers and confirmed by sequencing (Zou et al., 2015). Written informed consent was obtained from the patient. All experimental protocols were approved by the Institutional Ethics Committees of Academy of Military Medical Sciences.
Bacterial genomic DNA from strain GZ04-0086 was prepared in agarose plugs and digested with the S1 endonuclease (Takara, Dalian, China). DNA fragments were separated by pulsed-field gel electrophoresis (PFGE) through a CHEF-DR III system (Bio-Rad, Hercules, CA, United States) for 15 h at 6 V/cm, with initial and final pulse time of 0.22 s and 26.29 s, respectively. The plasmid DNA was transferred to a positively charged nylon membrane (Solabio) and hybridized with the digoxigenin-labeled probe specific to blaNDM–5.
Conjugation experiment was performed by filter mating using strain GZ04-0086 as donors and azide-resistant E. coli J53 as the recipient. The donor and recipient cultures were mixed at a ratio of 4:1 and collected on a membrane filter (pore size, 0.22 μm). The filter was placed on a BHI agar plate and incubated at 37°C overnight without shaking. The mixture was transferred into BHI agar plates containing 4 μg/ml meropenem and 200 μg/ml sodium azide. The transconjugants were selected after 72 h of incubation. The plasmid transfer frequency was calculated as the ratio of transconjugants to recipient cells. Presence of either one or both of the blaNDM–5-carrying plasmids in the transconjugants was further confirmed by S1-PFGE.
Antimicrobial susceptibility testing of strain GZ04-0086 and the transconjugants was performed using Vitek 2 compact system following the manufacturer’s instructions. The results were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI, 2017). E. coli ATCC 25922 was used as the control strain. Minimum inhibitory concentration (MIC) values of imipenem and meropenem were further determined using MIC test strips (MTS, Liofilchem, Italy).
Transcriptional expression of blaNDM–5 in strain GZ04-0086 and the transconjugants was determined by quantitative RT-PCR (qRT-PCR). Total RNA was isolated from bacterial cultures using RNApure Bacteria Kit (DNase I) (CWBio, China). RT-PCR was performed using UltraSYBR One Step RT-qPCR Kit (CWbio, China) in CFX96 Real-Time System (Bio-Rad, United States) with specific primers (NDM-RT-F 5′-GATTGCGACTTATGCCAATG-3′ and NDM-RT-R 5′- TCGATCCCAACGGTGATATT-3′) and 16S rRNA primers as previously described (Yu et al., 2019). Expression levels were normalized relative to the transcriptional level of the constitutive 16S rRNA and interpreted by the ΔΔCt method (Livak and Schmittgen, 2001). Statistical analysis was performed using a student t test to compare the log-transformed expression levels of blaNDM–5 among strain GZ04-0086 and the transconjugants.
Genomic DNA was extracted from cultured bacterium using High Pure PCR Template Preparation Kit (Roche). Whole genome sequencing was carried out using both the Illumina MiSeq platform and the Oxford Nanopore MinION platform. The de novo hybrid assembly of short Illumina reads and long MinION reads was performed using Unicycler (v0.4.8) (Wick et al., 2017). Genome sequences were annotated using RAST (Aziz et al., 2008). The sequence type was determined through the MLST web server (Larsen et al., 2012). Genome sequences of seven E. coli ST44 isolates provided by BacWGSTdb (Ruan and Feng, 2016) were used to construct a Maximum-Likelihood phylogenetic tree using RAxML (v8.2.4) (Stamatakis, 2014). Plasmid replicon types and the sequence types of IncF plasmids were identified using PlasmidFinder and pMLST (Carattoli et al., 2014).
The complete sequences of the chromosome of strain GZ04-0086 and plasmids pCTXM-GZ04, pNDM5-GZ04_B, pNDM5-GZ04_A, and p13k-GZ04 have been deposited in GenBank under the accession numbers CP042336 to CP042340, respectively.
Escherichia coli strain GZ04-0086 was recovered from the stool sample of a patient in Guangzhou, China, in 2018. GZ04-0086 was resistant to most tested antibiotics including penicillins, cephalosporins, fluoroquinolones and carbapenems, but remained susceptible to amikacin, aztreonam, and trimethoprim-sulfamethoxazole (Table 1). PCR amplification and sequencing confirmed the presence of blaNDM–5. S1-PFGE indicated that GZ04-0086 contained four different plasmids (<20 kb, ∼55 kb, ∼65 kb, and ∼160 kb) (Figure 1A). Interestingly, southern blotting further revealed the simultaneous presence of the blaNDM–5 gene on the ∼55-kb and ∼65-kb plasmids (designated as pNDM5-GZ04_A and pNDM5-GZ04_B, respectively), which were both transferable to E. coli J53. The transfer frequency of the blaNDM–5 gene was 1.07 × 10–3 per donor cell. Transconjugants containing pNDM5-GZ04_A (J53-A), pNDM5-GZ04_B (J53-B) or both plasmids (J53-AB) were obtained and confirmed by S1-PFGE (Figure 1B).
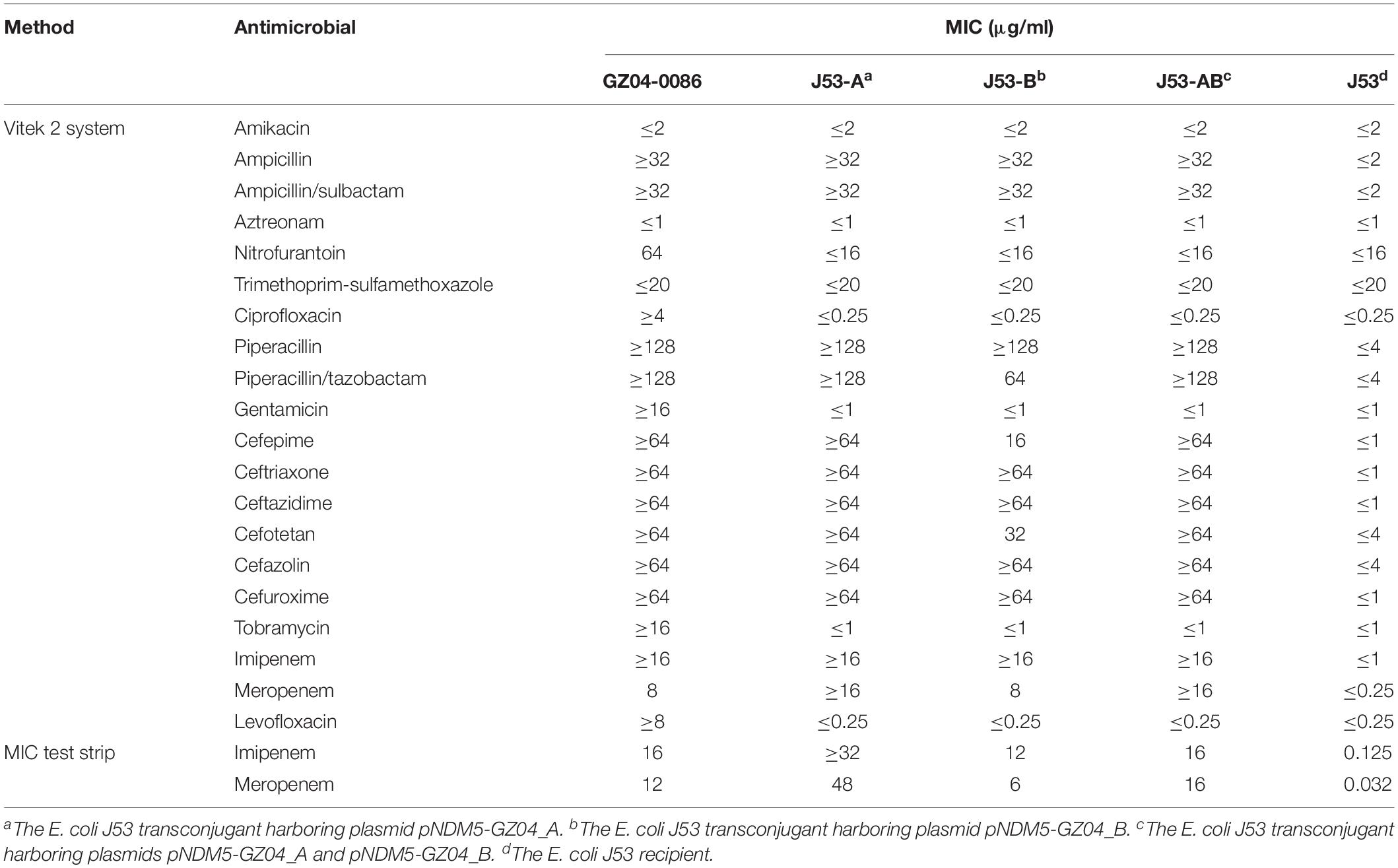
Table 1. Antibiotic susceptibilities of E. coli strain GZ04-0086, the E. coli J53 transconjugants and the E. coli J53 recipient.
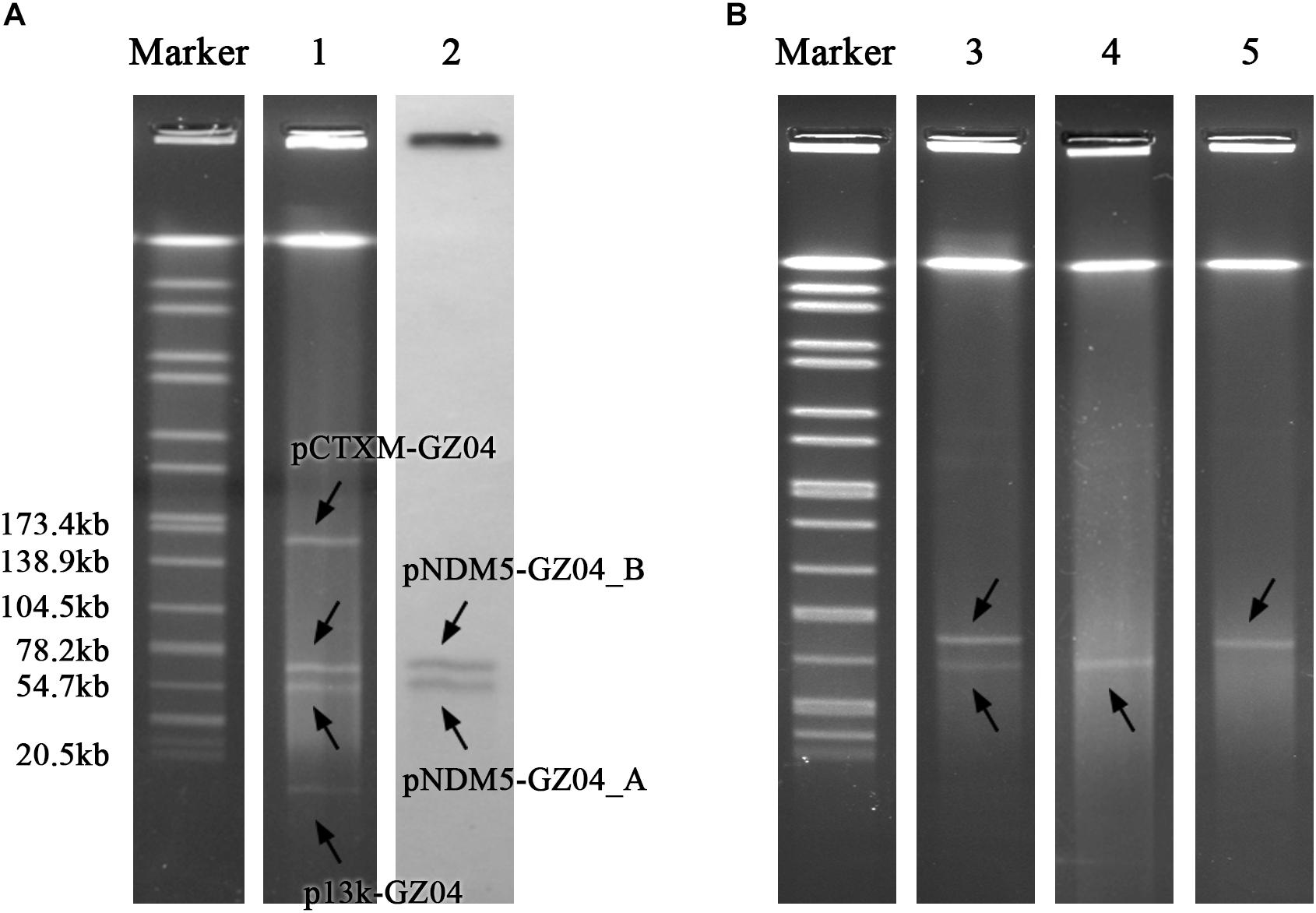
Figure 1. (A) S1-PFGE pattern for strain GZ04-0086 and southern blotting for the blaNDM–5 gene. (B) S1-PFGE pattern for E. coli J53 transconjugants. Lanes: Marker, Salmonella serotype Braenderup strain H9812 as a reference size standard; 1, PFGE result for S1-digested plasmid DNA of strain GZ04-0086; 2, southern blot hybridization with the probe specific to blaNDM–5; 3, 4, and 5, PFGE patterns for S1-digested plasmid DNA of E. coli transconjugants J53-AB, J53-A, and J53-B, respectively.
All the transconjugants acquired resistance to ampicillin, ampicillin/sulbactam, piperacillin, imipenem, meropenem, and cephalosporins. However, decreased MICs of piperacillin/tazobactam, cefepime, cefotetan and meropenem were observed in transconjugant J53-B compared with those of J53-A and J53-AB. Interestingly, susceptibility testing using MIC test strips revealed that J53-A had the highest MICs of imipenem (≥32 μg/ml) and meropenem (48 μg/ml) among all the transconjugants. J53-AB had MICs of 16 μg/ml for imipenem and meropenem, while J53-B had lower MICs (imipenem, 12 μg/ml; meropenem, 6 μg/ml). The results of qRT-PCR also indicated that the expression of blaNDM–5 in J53-A was 1.37-fold higher than that in J53-AB and 1.56-fold higher than that in J53-B (Figure 2). These slightly varied expression levels of blaNDM–5 were consistent with the imipenem and meropenem MICs in all the transconjugants. Raw sequence reads mapped to regions specific to plasmids pNDM5-GZ04_A and pNDM5-GZ04_B revealed an average read depth of 251× and 198×, respectively, which implied corresponding copy numbers of blaNDM–5-carrying plasmids in E. coli GZ04-0086 (Supplementary Figure S1). However, strain GZ04-0086 exhibited a relatively low transcriptional level of blaNDM–5 compared with all the transconjugants but still had an imipenem MIC of 16 μg/ml and a meropenem MIC of 12 μg/ml.
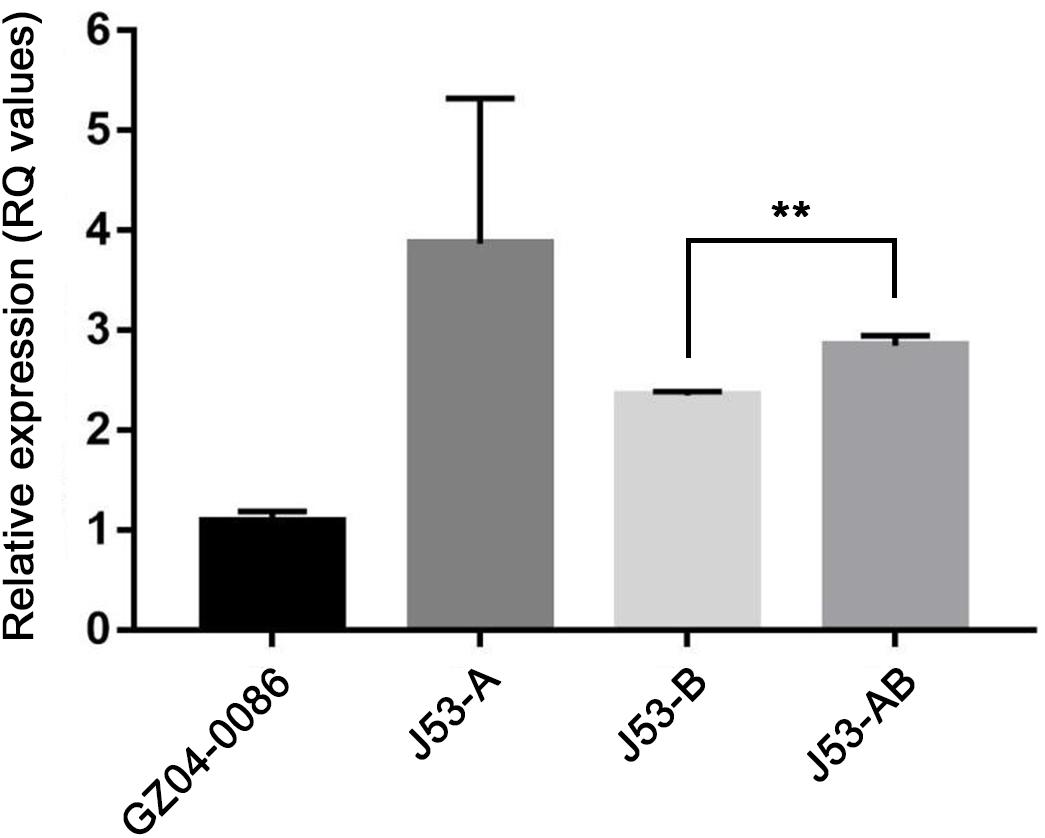
Figure 2. Relative expression of the blaNDM–5 gene among strain GZ04-0086 and transconjugants J53-A, J53-B and J53-AB. Strain GZ04-0086 was used as the reference. Asterisks above lines indicate a statistically significant difference (** P < 0.01) as determined by Student t test.
Strain GZ04-0086 was subjected to whole genome sequencing using both the MiSeq and the MinION platforms. Complete sequences of a 4.81-Mb chromosome and four plasmids were obtained through a de novo hybrid assembly. GZ04-0086 belonged to phylogenetic group A and sequence type ST44. E. coli ST44 has been frequently isolated from clinical settings (Kanamori et al., 2017; Runcharoen et al., 2017; Sonda et al., 2018). A phylogenetic analysis of E. coli ST44 revealed that GZ04-0086 was closest to isolates DO40 from canine and RDK06_431D from a patient in Tanzania, with 51 and 54 SNPs, respectively (Supplementary Figure S2). The blaCTX–M–15, blaOXA–1, tet(B), tet(D), and aac(6′)-Ib-cr genes were found on a 159,610-bp IncF [F36:F31:A4:B1] plasmid designated as pCTXM-GZ04. Another plasmid (designated as p13k-GZ04) had a length of 12,602 bp and was typed as Col440I (97% coverage and 93% identity).
The blaNDM–5-carrying plasmid pNDM5-GZ04_A had 55,253 bp in length. The plasmid belonged to the incompatibility type IncX3. A BLAST search against NCBI revealed that pNDM5-GZ04_A presented 84% query coverage and 99% identity with plasmid pNDM_MGR194 in K. pneumoniae from India (Krishnaraju et al., 2015; Figure 3). Both plasmids shared the same backbone sequence containing genes for plasmid replication (repB), conjugation/T4SS (virB genes), maintenance (topB and hns) and partition (parA), and the genetic structure of the blaNDM–5 gene, which was organized as IS3000-ISAba125-IS5-blaNDM–5-ble-trpF-tat-ΔctuA1-IS26. The accessory region between mpr and umuD genes of pNDM_MGR194 was composed of a Tn2 and the blaNDM–5 genetic structure, while that of pNDM-GZ04_A consisted of a Tn2 and an 18-kb novel composite transposon Tn6638. Tn6638 was flanked by two IS26 elements in the same orientation. In addition to the blaNDM–5 genetic structure, a 5.7-kb fragment of a Tn3 family transposon and a 3.2-kb remnant of an IS26-mphA-mrx-mphR-IS6100 region were also carried by Tn6638. The 5.7-kb fragment contained Tn3 tnpA, tnpR and an ABC-transporter-ATPase-encoding gene, which was previously present in a Tn3 family transposon in plasmid pH226B from E. coli (Zurfluh et al., 2016). The 3.2-kb remnant had undergone the deletion of the IS6100 and the truncation of the mphR gene compared with the prototype mphA-carrying region in plasmid pRSB101 (Szczepanowski et al., 2004).
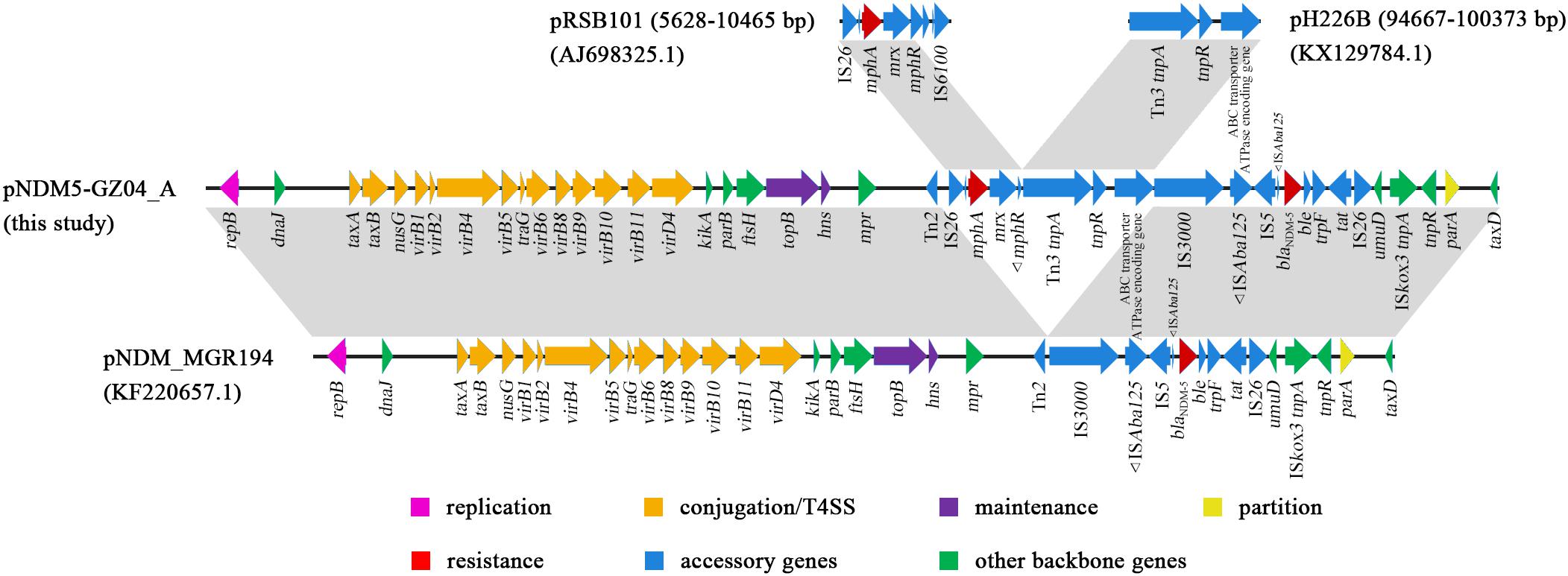
Figure 3. The comparative schematic diagram of plasmids pNDM5-GZ04_A, pNDM_MGR194, and fragments of pRSB101 and pH226B. The open reading frames are denoted by arrows. Regions with >99% identity among different plasmids are indicated by light gray.
The other blaNDM–5-carrying plasmid pNDM5-GZ04_B had a size of 68,545 bp and belonged to the incompatibility type IncFII [F2:A-:B-]. A BLAST search revealed that pNDM5-GZ04_B showed 84% query coverage and 99% identity to another blaNDM–5-carrying plasmid pCC1410-2 in K. pneumoniae from Korea (Shin et al., 2017) but differed significantly from pNDM5-GZ04_A (Figure 4). However, pNDM5-GZ04_B shared the same blaNDM–5 genetic context (IS3000-ISAba125-IS5-blaNDM–5-ble-trpF-tat-ΔctuA1-IS26) with pNDM5-GZ04_A, while pCC1410-2 had a different context organized as IS26-blaNDM–5-ble-trpF-tat. The 52-kb backbone of pNDM5-GZ04_B contained a set of core genes for plasmid replication (repA), conjugation/T4SS (tra and trb genes), stability (stdB) and segregation (parM). This backbone was highly identical to those of F2:A-:B- plasmids (Pu et al., 2016; Nahid et al., 2017; Shin et al., 2017) including pCC1410-2 but lacked a 5-kb region carrying the psiA, psiB, spo0J and ssbF genes associated with plasmid SOS inhibition, partition and recombination.
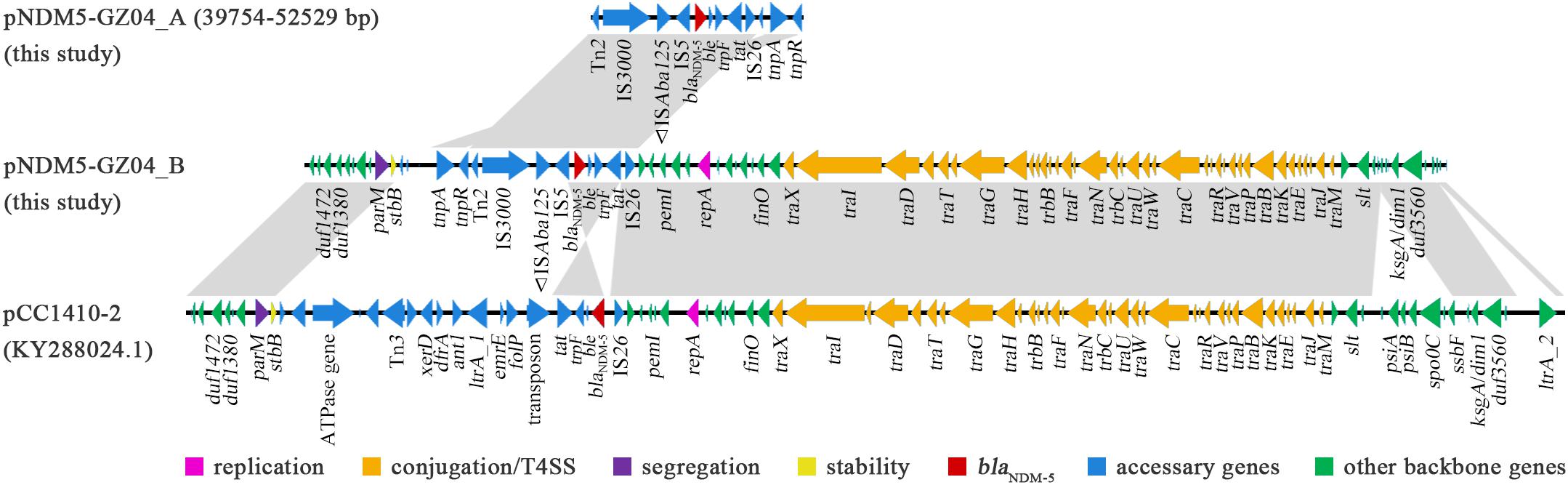
Figure 4. The comparative structures of plasmids pNDM5-GZ04_B, pCC1410-2, and fragment of pNDM5-GZ04_A. The open reading frames are shown by arrows. Regions with > 99% identity among different plasmids are denoted by light gray.
Emergence of NDM-producing Enterobacteriaceae represents an additional challenge for clinical management and infection control. A variety of plasmids have contributed to the dissemination of the blaNDM gene among diverse Enterobacteriaceae species throughout the world. pNDM_MGR194-like IncX3 plasmids have been found to be a major vehicle for blaNDM–5 (Wu et al., 2019). It was likely that pNDM5-GZ04_A had evolved from these pNDM_MGR194-like plasmids and undergone horizontal gene transfer which resulted in the additional insertion. Plasmids pNDM5-GZ04_A and pNDM5-GZ04_B had different backbones but the same blaNDM–5 genetic structure, suggesting possible occurrence of similar transposition events. It was noteworthy that a second blaNDM–5 gene could not be detected based on the Illumina short reads alone due to the repeated appearance of the same blaNDM–5 genetic context. MinION sequencing played a crucial role in characterizing the coexistence of two blaNDM–5 genes.
Despite the presence of blaNDM–5, the transconjugant J53-B exhibited significantly lower carbapenem resistance than that of J53-A. This decrease might be associated with the lower copy number of pNDM5-GZ04_B. Our analysis also revealed a lower copy number of pNDM5-GZ04_B, which might be the key reason for the reduced expression of blaNDM–5 in J53-B. Previous studies demonstrated that two copies of blaNDM–5 in the same plasmid would not lead to stronger carbapenem resistances (Feng et al., 2018), while coexistence of two plasmids harboring blaNDM–1 and blaKPC–2, respectively, conferred higher resistances (Wu et al., 2016, 1). However, GZ04-0086 and J53-AB also showed lower carbapenem resistance than that of J53-A. The coexistence of these two blaNDM-5-carrying plasmids showed no additive effect on imipenem and meropenem resistance levels. The varied copy numbers and inter-plasmids competition seem to offset each other on carbapenem MICs and expression level of blaNDM–5, which still need further investigation.
In summary, our study characterized a multidrug-resistant ST44 E. coli harboring two blaNDM–5 genes, which were carried by different plasmids. However, coexistence of the two blaNDM–5-carrying plasmids did not lead to stronger carbapenem resistance. Both plasmids were transferable and might serve as new reservoirs for further dissemination of blaNDM–5. Our findings highlighted the threat of NDM-5 carbapenemase circulation and the urgency of stringent surveillance and control measures.
The datasets generated for this study can be found in the NCBI GenBank, CP042336, CP042337, CP042338, CP042339, and CP042340.
The studies involving human participants were reviewed and approved by the Institutional Ethics Committees of Academy of Military Medical Sciences. The patients/participants provided their written informed consent to participate in this study.
LY, YL, and LL performed the experiments. LY, MX, and HM analyzed the data. XG collected the samples. KW and PHL performed library construction and genome sequencing. HM and XD prepared the tables and figures. LY and PL prepared the manuscript. KQ, PL, and HS designed the study and revised the manuscript. All authors read and approved the final manuscript.
This study was supported by grants from the National Science and Technology Major Project (No. 2018ZX10201001-003), Beijing Municipal Natural Science Foundation (No. 5172029), and Beijing Nova Program (No. Z181100006218110).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00195/full#supplementary-material
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genom. 9:75. doi: 10.1186/1471-2164-9-75
Balm, M. N. D., Ngan, G., Jureen, R., Lin, R. T. P., and Teo, J. (2012). Molecular characterization of newly emerged blaKPC-2-producing Klebsiella pneumoniae in Singapore. J. Clin. Microbiol. 50, 475–476. doi: 10.1128/JCM.05914-11
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
CLSI (2017). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Seventh Informational Supplement. M100-S27. Wayne, PA: CLSI.
Cornaglia, G., Giamarellou, H., and Rossolini, G. M. (2011). Metallo-β-lactamases: a last frontier for β-lactams? Lancet Infect. Dis. 11, 381–393. doi: 10.1016/S1473-3099(11)70056-1
Elumalai, S., Muthu, G., Selvam, R. E. M., and Ramesh, S. (2014). Detection of TEM-, SHV- and CTX-M-type β-lactamase production among clinical isolates of Salmonella species. J. Med. Microbiol. 63, 962–967. doi: 10.1099/jmm.0.068486-0
Feng, Y., Liu, L., McNally, A., and Zong, Z. (2018). Coexistence of Two blaNDM–5 genes on an IncF plasmid as revealed by nanopore sequencing. Antimicrob. Agents Chemother. 62:e0110-18. doi: 10.1128/AAC.00110-18
Hornsey, M., Phee, L., and Wareham, D. W. (2011). A novel variant, NDM-5, of the New Delhi metallo-β-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob. Agents Chemother. 55, 5952–5954. doi: 10.1128/AAC.05108-11
Jovcić, B., Lepsanović, Z., Begović, J., Rakonjac, B., Perovanović, J., Topisirović, L., et al. (2013). The clinical isolate Pseudomonas aeruginosa MMA83 carries two copies of the blaNDM-1 gene in a novel genetic context. Antimicrob. Agents Chemother. 57, 3405–3407. doi: 10.1128/AAC.02312-12
Kanamori, H., Parobek, C. M., Juliano, J. J., Johnson, J. R., Johnston, B. D., Johnson, T. J., et al. (2017). Genomic analysis of multidrug-resistant Escherichia coli from north carolina community hospitals: ongoing circulation of CTX-M-producing ST131-H30Rx and ST131-H30R1 strains. Antimicrob. Agents Chemother. 61:e00912-17. doi: 10.1128/AAC.00912-17
Krishnaraju, M., Kamatchi, C., Jha, A. K., Devasena, N., Vennila, R., Sumathi, G., et al. (2015). Complete sequencing of an IncX3 plasmid carrying blaNDM-5 allele reveals an early stage in the dissemination of the blaNDM gene. Indian J. Med. Microbiol. 33, 30–38. doi: 10.4103/0255-0857.148373
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/JCM.06094-11
Liu, L., Feng, Y., McNally, A., and Zong, Z. (2018). blaNDM-21, a new variant of blaNDM in an Escherichia coli clinical isolate carrying blaCTX-M-55 and rmtB. J. Antimicrob. Chemother. 73, 2336–2339. doi: 10.1093/jac/dky226
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods San Diego Calif. 25, 402–408. doi: 10.1006/meth.2001.1262
Nahid, F., Zahra, R., and Sandegren, L. (2017). A blaOXA-181-harbouring multi-resistant ST147 Klebsiella pneumoniae isolate from Pakistan that represent an intermediate stage towards pan-drug resistance. PLoS One 12:e0189438. doi: 10.1371/journal.pone.0189438
Pu, X.-Y., Pan, J.-C., Gu, Y.-M., Zheng, W., Li, J., and Yu, H. (2016). Complete sequences and characterization of two novel plasmids carrying aac(6’)-Ib-cr and qnrS gene in Shigella flexneri. Microb. Drug Resist. Larchmt. N 22, 115–122. doi: 10.1089/mdr.2015.0082
Ruan, Z., and Feng, Y. (2016). BacWGSTdb, a database for genotyping and source tracking bacterial pathogens. Nucleic Acids Res. 44, D682–D687. doi: 10.1093/nar/gkv1004
Runcharoen, C., Raven, K. E., Reuter, S., Kallonen, T., Paksanont, S., Thammachote, J., et al. (2017). Whole genome sequencing of ESBL-producing Escherichia coli isolated from patients, farm waste and canals in Thailand. Genome Med. 9:81. doi: 10.1186/s13073-017-0471-8
Shen, P., Yi, M., Fu, Y., Ruan, Z., Du, X., Yu, Y., et al. (2017). Detection of an Escherichia coli sequence type 167 strain with two tandem copies of blaNDM-1 in the chromosome. J. Clin. Microbiol. 55, 199–205. doi: 10.1128/JCM.01581-16
Shin, J., Baek, J. Y., Chung, D. R., and Ko, K. S. (2017). Instability of the IncFII-type plasmid carrying blaNDM-5 in a Klebsiella pneumoniae isolate. J. Microbiol. Biotechnol. 27, 1711–1715. doi: 10.4014/jmb.1706.06030
Sonda, T., Kumburu, H., van Zwetselaar, M., Alifrangis, M., Mmbaga, B. T., Aarestrup, F. M., et al. (2018). Whole genome sequencing reveals high clonal diversity of Escherichia coli isolated from patients in a tertiary care hospital in Moshi, Tanzania. Antimicrob. Resist. Infect. Control 7:72. doi: 10.1186/s13756-018-0361-x
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinforma. Oxf. Engl. 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Szczepanowski, R., Krahn, I., Linke, B., Goesmann, A., Pühler, A., and Schlüter, A. (2004). Antibiotic multiresistance plasmid pRSB101 isolated from a wastewater treatment plant is related to plasmids residing in phytopathogenic bacteria and carries eight different resistance determinants including a multidrug transport system. Microbiol. Read. Engl. 150, 3613–3630. doi: 10.1099/mic.0.27317-0
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Wu, W., Espedido, B., Feng, Y., and Zong, Z. (2016). Citrobacter freundii carrying blaKPC-2 and blaNDM-1: characterization by whole genome sequencing. Sci. Rep. 6:30670. doi: 10.1038/srep30670
Wu, W., Feng, Y., Tang, G., Qiao, F., McNally, A., and Zong, Z. (2019). NDM metallo-β-Lactamases and their bacterial producers in health care settings. Clin. Microbiol. Rev. 32, 11–12. doi: 10.1128/CMR.00115-18
Yong, D., Toleman, M. A., Giske, C. G., Cho, H. S., Sundman, K., Lee, K., et al. (2009). Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53, 5046–5054. doi: 10.1128/AAC.00774-09
Yu, L., Li, W., Qi, K., Wang, S., Chen, X., Ni, J., et al. (2019). McbR is involved in biofilm formation and H2O2 stress response in avian pathogenic Escherichia coli X40. Poult. Sci. 98, 4094–4103. doi: 10.3382/ps/pez205
Zong, Z., and Zhang, X. (2013). blaNDM-1-carrying Acinetobacter johnsonii detected in hospital sewage. J. Antimicrob. Chemother. 68, 1007–1010. doi: 10.1093/jac/dks505
Zou, D., Huang, Y., Zhao, X., Liu, W., Dong, D., Li, H., et al. (2015). A novel New Delhi metallo-β-lactamase variant, NDM-14, isolated in a Chinese hospital possesses increased enzymatic activity against carbapenems. Antimicrob. Agents Chemother. 59, 2450–2453. doi: 10.1128/AAC.05168-14
Zurfluh, K., Klumpp, J., Nüesch-Inderbinen, M., and Stephan, R. (2016). Full-length nucleotide sequences of mcr-1-harboring plasmids isolated from extended-spectrum-β-Lactamase-producing Escherichia coli isolates of different origins. Antimicrob. Agents Chemother. 60, 5589–5591. doi: 10.1128/AAC.00935-16
Keywords: coexistence of two blaNDM–5 genes, Escherichia coli, transferability, Illumina short-read sequencing, MinION long-read sequencing
Citation: Yang L, Lin Y, Lu L, Xue M, Ma H, Guo X, Wang K, Li P, Du X, Qi K, Li P and Song H (2020) Coexistence of Two blaNDM–5 Genes Carried on IncX3 and IncFII Plasmids in an Escherichia coli Isolate Revealed by Illumina and Nanopore Sequencing. Front. Microbiol. 11:195. doi: 10.3389/fmicb.2020.00195
Received: 01 November 2019; Accepted: 28 January 2020;
Published: 13 February 2020.
Edited by:
Shaolin Wang, China Agricultural University (CAU), ChinaCopyright © 2020 Yang, Lin, Lu, Xue, Ma, Guo, Wang, Li, Du, Qi, Li and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kezong Qi, cWt6QGFoYXUuZWR1LmNu; Peng Li, amlla2VubGVlQDEyNi5jb20=; Hongbin Song, aG9uZ2JpbnNvbmdAMjYzLm5ldA==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.