
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 18 February 2020
Sec. Extreme Microbiology
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.00171
 Giuliana d’Ippolito*†
Giuliana d’Ippolito*† Simone Landi*†
Simone Landi*† Nunzia Esercizio
Nunzia Esercizio Mariamichella Lanzilli
Mariamichella Lanzilli Marco Vastano
Marco Vastano Laura Dipasquale
Laura Dipasquale Nirakar Pradhan
Nirakar Pradhan Angelo Fontana
Angelo Fontana
Capnophilic lactic fermentation (CLF) is a novel anaplerotic pathway able to convert sugars to lactic acid (LA) and hydrogen using CO2 as carbon enhancer in the hyperthermophilic bacterium Thermotoga neapolitana. In order to give further insights into CLF metabolic networks, we investigated the transcriptional modification induced by CO2 using a RNA-seq approach. Transcriptomic analysis revealed 1601 differentially expressed genes (DEGs) in an enriched CO2 atmosphere over a total of 1938 genes of the T. neapolitana genome. Transcription of PFOR and LDH genes belonging to the CLF pathway was up-regulated by CO2 together with 6-phosphogluconolactonase (6PGL) and 6-phosphogluconate dehydratase (EDD) of the Entner–Doudoroff (ED) pathway. The transcriptomic study also revealed up-regulation of genes coding for the flavin-based enzymes NADH-dependent reduced ferredoxin:NADP oxidoreductase (NFN) and NAD-ferredoxin oxidoreductase (RNF) that control supply of reduced ferredoxin and NADH and allow energy conservation-based sodium translocation through the cell membrane. These results support the hypothesis that CO2 induces rearrangement of the central carbon metabolism together with activation of mechanisms that increase availability of the reducing equivalents that are necessary to sustain CLF. In this view, this study reports a first rationale of the molecular basis of CLF in T. neapolitana and provides a list of target genes for the biotechnological implementation of this process.
Thermotoga neapolitana is a hyperthermophilic anaerobic bacterium of the order Thermotogales (Belkin et al., 1986). The taxonomic group shares a rod shape and complex outer envelope called toga that surrounds the bacterial cell and forms a periplasmatic space around the poles (Angel et al., 1993). T. neapolitana and other sister species are good candidates for the sustainable and efficient conversion of food and agriculture residues to hydrogen (H2) by Dark Fermentation (Conners et al., 2006; Manish and Banerjee, 2008; Hallenbeck and Ghosh, 2009; Guo et al., 2010; d’Ippolito et al., 2010; Elleuche et al., 2014; Pradhan et al., 2015, 2016a).
In the last years, we reported that T. neapolitana also operates a novel, anaplerotic process named capnophilic lactic fermentation (CLF) for the synthesis of almost enantiopure L-lactic acid (LA) without affecting H2 production (Dipasquale et al., 2014; d’Ippolito et al., 2014; Pradhan et al., 2016b, 2019; Nuzzo et al., 2019). The metabolic process is activated by CO2 (capnophilic means “requiring CO2”) and, nominally, is dependent on a Janus pathway including a catabolic branch leading to acetyl-CoA (AcCoA) from sugars by glycolysis, and an anabolic branch that combines AcCoA and CO2 to give LA through reduction of newly synthesized pyruvate (PYR) by a NADH-dependent lactic dehydrogenase (LDH) (Figure 1).
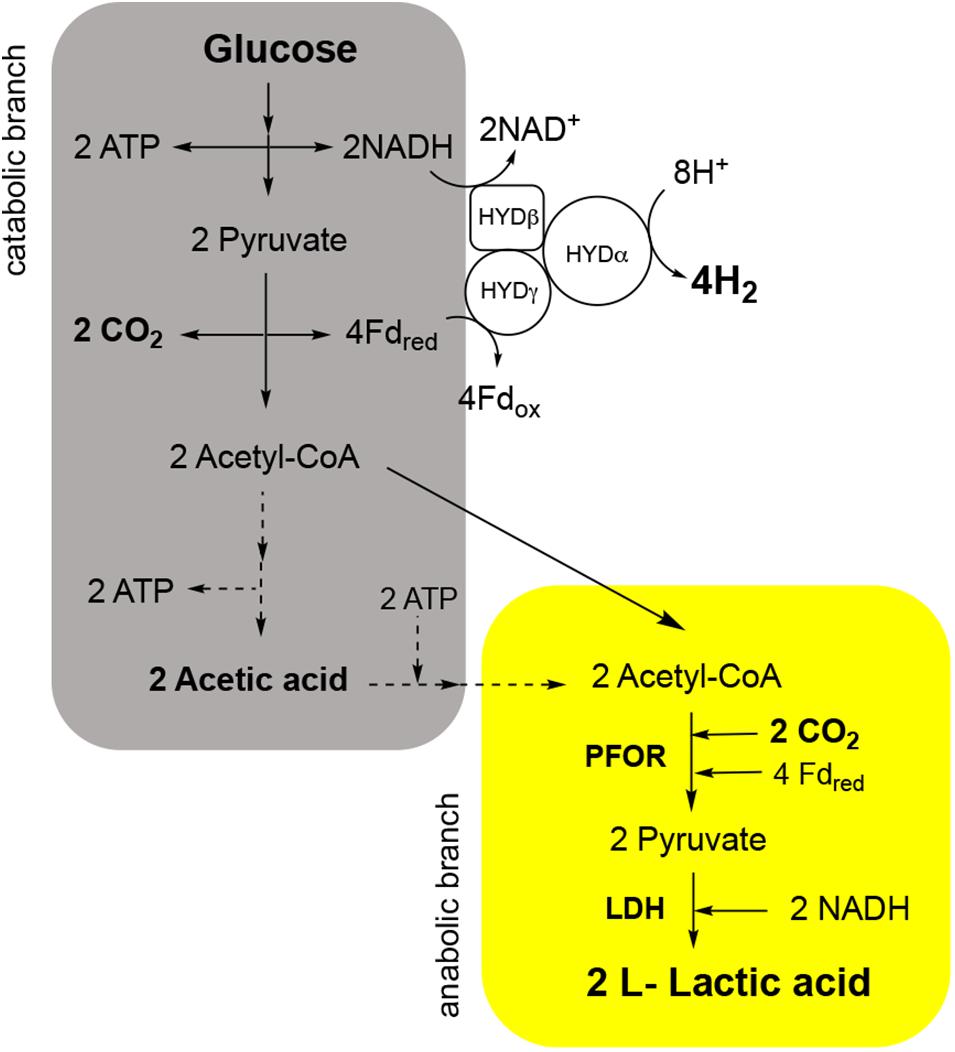
Figure 1. Capnophilic lactic fermentation (CLF) pathway in T. neapolitana with details of the enzymatic reactions in the catabolic (gray) and anabolic (yellow) branch in relation to production and use of CO2. PFOR, pyruvate:ferredoxin oxidoreductase; LDH, lactate dehydrogenase; HYD, hydrogenase.
This second part of the pathway requires an additional burden of reducing equivalents and determines an unconceivable deviation from Dark Fermentation model for carbon and hydrogen balance (Dipasquale et al., 2014). We showed that CLF is not equally active in all members of the order Thermotogales (Dipasquale et al., 2018) but the almost complete absence of molecular and biochemical studies on metabolism of this group of bacteria has been an insurmountable barrier to explore this process that presumably involves crosstalking of several pathways. The aim of the present work was to investigate the role of CO2 as biochemical trigger in T. neapolitana and to correlate metabolic change with or without CO2 to hydrogen and LA production. We based our analysis on a differential RNA-sequencing of the strain T. neapolitana subsp. capnolactica, a mutant that shows an incremented operation of CLF (Pradhan et al., 2017). Transcriptome studies were associated to the experimental response of the bacterium to CO2. Under our experimental conditions, CO2 sparging generates a complex equilibrium between carbon dioxide as a gas or dissolved in the aqueous phase, and its hydrated derivatives H2CO3, , . For simplicity we refer to all these chemical forms as CO2 throughout the manuscript.
Thermotoga neapolitana subsp. capnolactica (DSM 33003) derives from the DSMZ 4359T strain that was stimulated in our laboratory under saturating concentration of CO2 (Pradhan et al., 2017). Bacterial cells were grown in a modified ATCC 1977 culture medium containing 10 ml/L of filter-sterilized vitamins and trace element solution (DSM medium 141) together with 10 g/L NaCl, 0.1 g/L KCl, 0.2 g/L MgCl2.6H2O, 1 g/L NH4Cl, 0.3 g/L K2HPO4, 0.3 g/L KH2PO4, 0.1 g/L CaCl2.2H2O, 1 g/L cysteine–HCl, 2 g/L yeast extract, 2 g/L tryptone, 5 g/L glucose, and 0.001 g/L resazurin (d’Ippolito et al., 2010).
Bacterial precultures (30 mL) were incubated overnight at 80°C without shaking and used to inoculate (6% v/v) cultures in 120 ml serum bottles with a final culture volume of 30 mL. Oxygen was removed by heating until solution was colorless. Cultures were sparged with CO2 gas (CLF condition, three bottles) or N2 gas (control, three bottles) for 5 min at 30 mL/min. pH was monitored and adjusted to approximately 7.5 by 1 M NaOH. Sparging followed by pH adjustment was repeated every 24 h. Inoculated bottles were maintained in a heater (Binder ED720) at 80°C. Cell growth was determined by optical density (OD) at 540 nm (UV/Vis Spectrophotometer DU 730, Beckman Coulter). Samples (2 ml of medium) were collected from each bottle after 0, 24, and 48 h. After centrifugation at 16,000 × g for 15 min (Hermle Z3236K), residues and supernatants were kept at −20°C until analysis. Cell morphology was monitored by microscope observation (Axio VertA1, Carl Zeiss, magnification of 100×).
Gas (H2 and CO2) measurements were performed by gas chromatography (GC) on an instrument (Focus GC, Thermo Scientific) equipped with a thermoconductivity detector (TCD) and fitted with a 3 m molecular sieve column (Hayesep Q). N2 was used as carrier gas. Gas sampling was carried out at 24 and 48 h.
Glucose concentration was determined by the dinitrosalicylic acid method calibrated on a standard solution of 2 g/L glucose (Bernfeld, 1995). Organic acids were measured by ERETIC 1H NMR as described by Nuzzo et al. (2019). All experiments were performed on a Bruker DRX 600 spectrometer equipped with an inverse TCI CryoProbe. Peak integration, ERETIC measurements, and spectrum calibration were obtained by the specific subroutines of Bruker Top-Spin 3.1 program. Spectra were acquired with the following parameters: flip angle = 90°, recycle delay = 20 s, SW = 3000 Hz, SI = 16K, NS = 16, RG = 1. An exponential multiplication (EM) function was applied to the FID for line broadening of 1 Hz. No baseline correction was used.
RNA (three replicates for CLF and three replicates for control) was extracted by standard method with TRIzol (Invitrogen, Carlsbad, CA, United States). Concentration in each sample was determined by a ND-1000 Spectrophotometer (NanoDrop) and quality assessed by Agilent 2100 Bioanalyzer and Agilent RNA 6000 nano kit (Agilent Technologies, Santa Clara, CA, United States). Hi-quality RNA samples were used for high-throughput sequencing. Indexed libraries were prepared from 4 μg/ea purified RNA with TruSeq Stranded mRNA Sample Prep Kit (Illumina) according to the manufacturer’s instructions. Libraries were quantified using the Agilent 2100 Bioanalyzer (Agilent Technologies) and pooled such that each index-tagged sample was present in equimolar amounts, with final concentration of the pooled samples of 2 nM. The pooled samples were subject to cluster generation and sequencing using an Illumina HiSeq 2500 System (Illumina) in a 2 × 100 paired-end format at a final concentration of 8 pmol. RNA extraction and the sequencing service was provided by Genomix4life SRL (Baronissi, Salerno, Italy).
Quality check of the sequenced reads was performed using the FAST QC software1. The obtained high-quality reads were used for mapping by the Bowtie software (Langmead and Salzberg, 2012). Genome of T. neapolitana DSM_4359 at GenBank database (assembly accession: GCA_000018945.1) was used as reference. The quality statistics of the mapping process is showed in the Supplementary Table 1. The counting of the mapped reads was performed by the software HTScount. In order to define the set of expressed genes, raw read counts were normalized using the TMM method (Trimmed mean). Differentially expressed genes (DEGs) were obtained by the DESeq2 package of R language at a false discovery rate (FDR) ≤ 0.05 (Love et al., 2014). Expression values were reported as “Fold Change” (ratio of the normalized expression value in sample over control). Values <1 are shown as (−1/FC) to display negative regulation. Cluster analysis was performed using the MeV software based on the respective normalized reads count values (Howe et al., 2011). The number of clusters was determined by Figure of Merit (FOM) analysis. Clusters were generated by employing k-means clustering with Euclidian distances. In gene ontology enrichment analysis (GOEA), categories with a number of entries lower than 4 were excluded in the final output.
RNAseq results were validated by real-time PCR. Triplicate quantitative assays were performed using a Platinum SYBR Green qPCR SuperMix (Life Technologies, Carlsbad, CA, United States). Cells from control cultures (N2 sparging) were used as calibrators and RNA 16S served as endogenous reference gene (Okonkwo et al., 2017). Calculation of gene expression was carried out using the 2–ΔΔCt method as in Livak and Schmittgen (2001). For each sample, mRNA amount was calculated relatively to the calibrator sample for the corresponding genes. Primers used for genes expression analysis are listed in Supplementary Table 2.
Bacterial cells were grown under CLF condition and control as described above. Samples were collected after 12, 15, 36, 39, 42, and 45 h. Proteins extraction was performed on 1.5 ml of culture. After centrifugation at 10,000 × g for 20 min at 4°C, the pellets were suspended in cracking buffer (100 mM Tris–HCl pH 7.5, 30% glycerol, 2% SDS, 4 mM EDTA, 28 mM β-mercaptoethanol) and incubated at 100°C for 10 min. After centrifugation at 10,000 × g at 4°C for 1 h, the supernatants were recovered and protein content was measured by Bradford reagent (Bio-Rad, Hercules, CA, United States). Protein fractionation was performed on SDS polyacrylamide gel using a precast 4–15% Mini-PROTEAN TGX Stain Free (Bio-Rad, Hercules, CA, United States) and electro-transferred on polyvinylidene difluoride membrane (PVDF) using the Trans-Blot Turbo Transfer System (Bio-Rad, Hercules, CA, United States) following the manufacturer’s instructions. The detection of protein was performed by primary antibody raised in rabbit (1:1000; PRIMM Srl, Milan, Italy) against peptide 149–359 of T. neapolitana PYR synthase subunit A (PFOR-A) (NCBI database protein accession number YP_002534223.1) and against peptide 1-192 of T. neapolitana Fe–Fe hydrogenase subunit β (β-HYD) (NCBI database protein accession number AAC02685.1). Horseradish peroxidase (HRP)-conjugated anti-rabbit secondary antibody was used for staining (Sigma; 1:7000). Western blot filters were visualized by ClarityTM Western ECL substrate (Bio-Rad, Hercules, CA, United States) and subjected to analysis using Image Lab 6.0 Software (Bio-Rad, Hercules, CA, United States). Analysis included the determination of intensity of total Stain-Free fluorescence and intensity of PFOR-A and β-HYD blots for each lane by the “Lane and Bands” tool. Results were expressed as relative percentage of antibody blot intensity respect to intensity of total proteins.
The raw sequencing data from this study are stored in the NCBI SRA database2 and are retrievable under the accession code PRJNA574556.
Each experiment was performed at least in triplicate. Values were expressed as mean ± standard deviation (SD). The statistical significance of OD540, qRT-PCR, sugar consumption, organic acid, and H2 yields was evaluated through Student’s t-test (p ≤ 0.05). The differentially expressed data from the RNA-seq approach were filtered using an FDR ≤ 0.05.
Sparging of CO2 did not affect cell growth (OD540) in comparison to control under N2 but increased significantly glucose consumption rate. In agreement with previous reports (Dipasquale et al., 2014; Pradhan et al., 2017), the overall effect of CO2 on the fermentation process was the increase of LA production without changing H2 yield (except for a slight increase at 24 h) in comparison to N2-treated cells. This outcome was more evident at 48 h when lactic/acetic acid ratio was 0.23 under N2 and 0.39 under CO2 (Table 1).
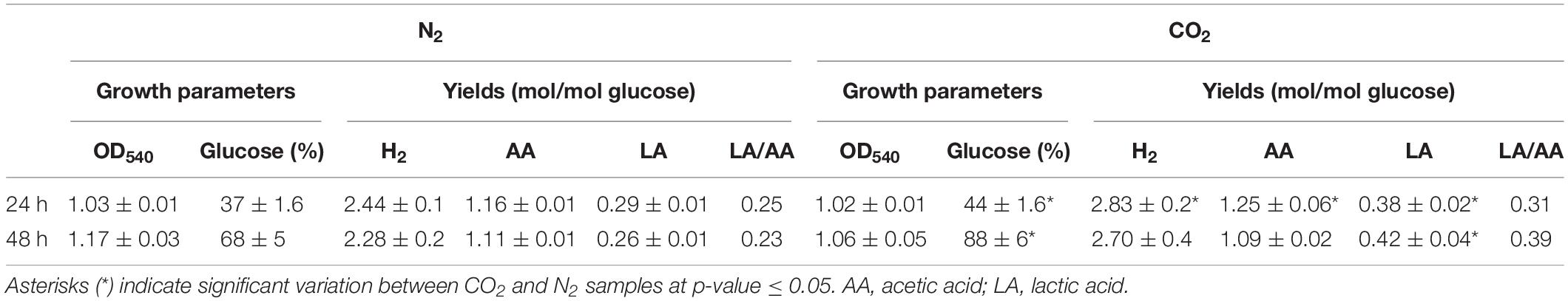
Table 1. Growth parameters (OD540 and percentage of glucose consumption) and product yields (H2, acetic acid, and lactic acid – LA) in T. neapolitana cultures sparged with N2 and CO2 after 24 and 48 h.
In order to clarify the molecular effects of CO2, an RNA-sequencing approach was performed by using N2-treated cells as control. The transcriptomic analysis gave from 19,390,095 to 31,800,340 reads with a percentage of alignment rate >98.59% (Supplementary Table 1). We identified 1601 DEGs between experiments under CO2 (CLF samples) and N2 (control) (see Supplementary Information). In particular, 612 DEGs showed a significant fold change up to ≥1.5 whereas 593 DEGs were down-regulated (fold change below −1.5) (Figure 2). Considering the whole genome of T. neapolitana, CO2-dependent DEGs accounted for 83% of the bacterial genes. GOEA of DEGs highlighted change of 45 metabolic networks under CLF conditions, with 26 related to up-regulation and 19 to down-regulation (Supplementary Figure 1). The analysis indicated increased expression of genes related to RNA translation (GO:0006412), ribosome organization (GO:0015934, GO:0015935), transcription factor (TF) (GO:0003700), and amino acids biosynthesis (GO:0009089, GO:0009088, GO:0006526, GO:0009097, GO:0009098, GO:0009073, GO:0006541). Expression and regulation of proteins related to active transport, including major facilitator proteins, and various membrane channels and antiporters, put forward a general down-regulation of sugar transport and vice versa an increased mobilization of phosphate, polyamine, and amino acids. The GOEA also suggested a significant impact of CO2 on organization of the cell-wall (GO:0071555) with a marked down-regulation of the degradation of polysaccharides of the outer membrane (xylan catabolic process, GO:0045493; endoglucanase activity, GO:0004519). Interestingly, two of the three genes encoding for the outer membrane proteins that form the structure of toga (Petrus et al., 2012), namely CTN_RS01400 and CTN_RS01395, showed a similar decrease of expression. A number of TFs were also regulated by CO2 treatment, including a number of TFs involved in the regulation of sugar catabolism (AraC and LacI). As expected by the simultaneous production of LA and H2, we found the enrichment of metabolic categories related to redox reactions (Cell redox homeostasis – GO:0045454; Electron carrier activity – GO:0009055; and Oxidoreductase activity – GO:0016491) and carbon metabolism.
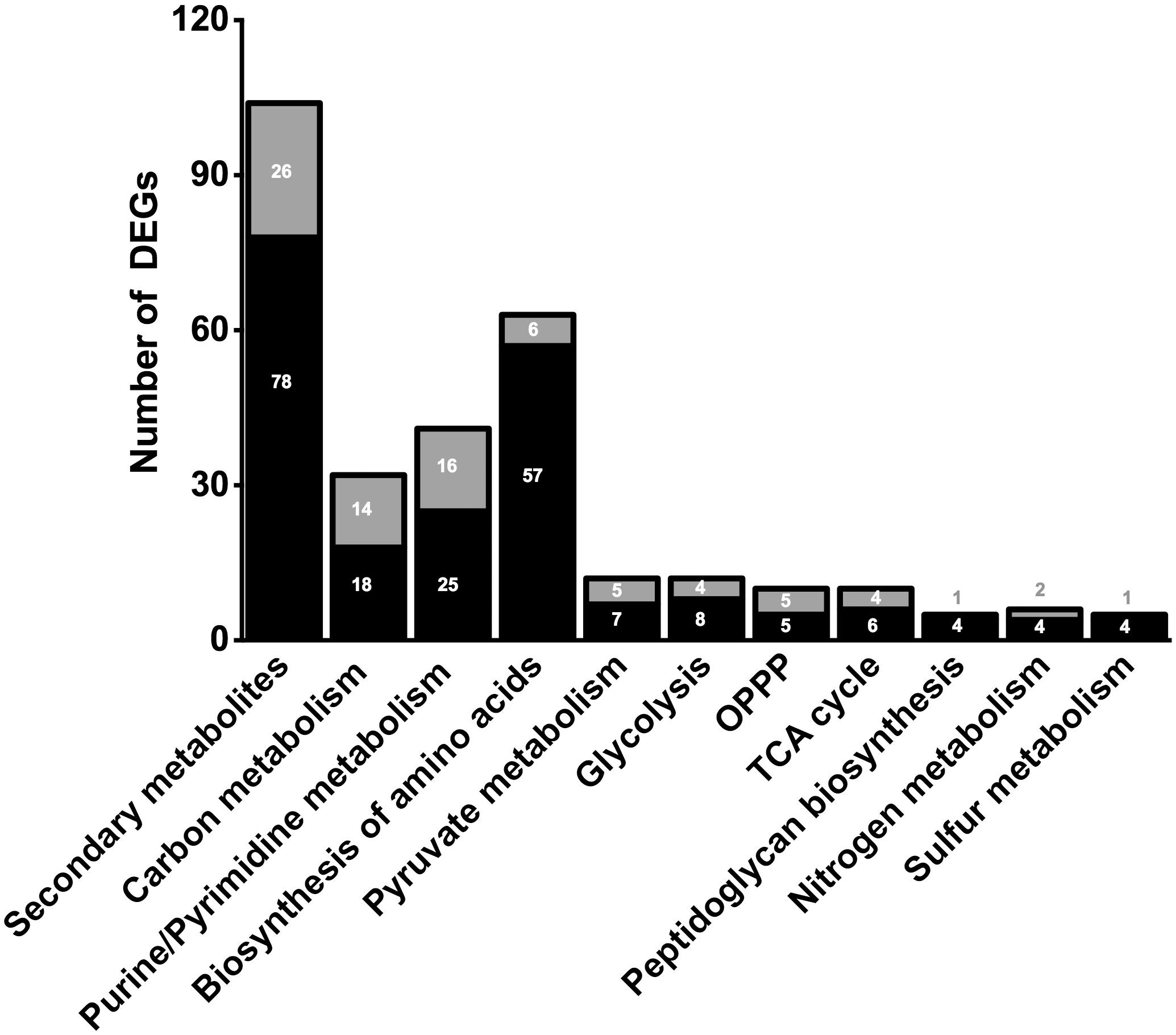
Figure 2. Number of significant (≤0.05 FDR) up-regulated (black) and down-regulated (gray) DEGs of selected metabolic pathways. Number of genes in correlated pathways were identified using KEGG categories.
Sparging by CO2 induced up-regulation of glucokinase (GCK – CTN_RS05065) and glucose 6-phosphate isomerase (PGI – CTN_RS01845) that catalyze the starting steps of Embden–Meyerhof–Parnas glycolysis (EMP) (Table 2). On the other hand, we found that other enzymes of EMP were significantly down-regulated. Inspection of other genes related to central carbon metabolism revealed a significant transcription of glucose-6-phosphate dehydrogenase (G6PDH – CTN_RS07110) and 6-phosphogluconolactonase (6PGL – CTN_RS07115). These enzymes preside over the synthesis of 6-phospho gluconate that is the first metabolite of the main alternative pathways of glucose catabolism, namely ED and oxidative pentose phosphate (OPP) pathway. On the whole, these data suggest a diversion of carbon flux from EMP to ED or OPP pathways (Figure 3). This evidence was further validated by RT-PCR that showed up-regulation of 6-phosphogluconate dehydratase (EDD – CTN_RS00550) that control the key step of ED and down-regulation of 6-phosphofructokinase (PFK – CTN_RS02345) that is the regulator enzyme of EMP (Supplementary Figure 2). Analysis of the genes of Krebs cycle reveals that T. neapolitana apparently lacks the whole pathway. However, we found a significant upregulation of the expression of citrate synthase (CTN_RS01940) and isocitrate dehydrogenase (CTN_RS07145) (Table 2).
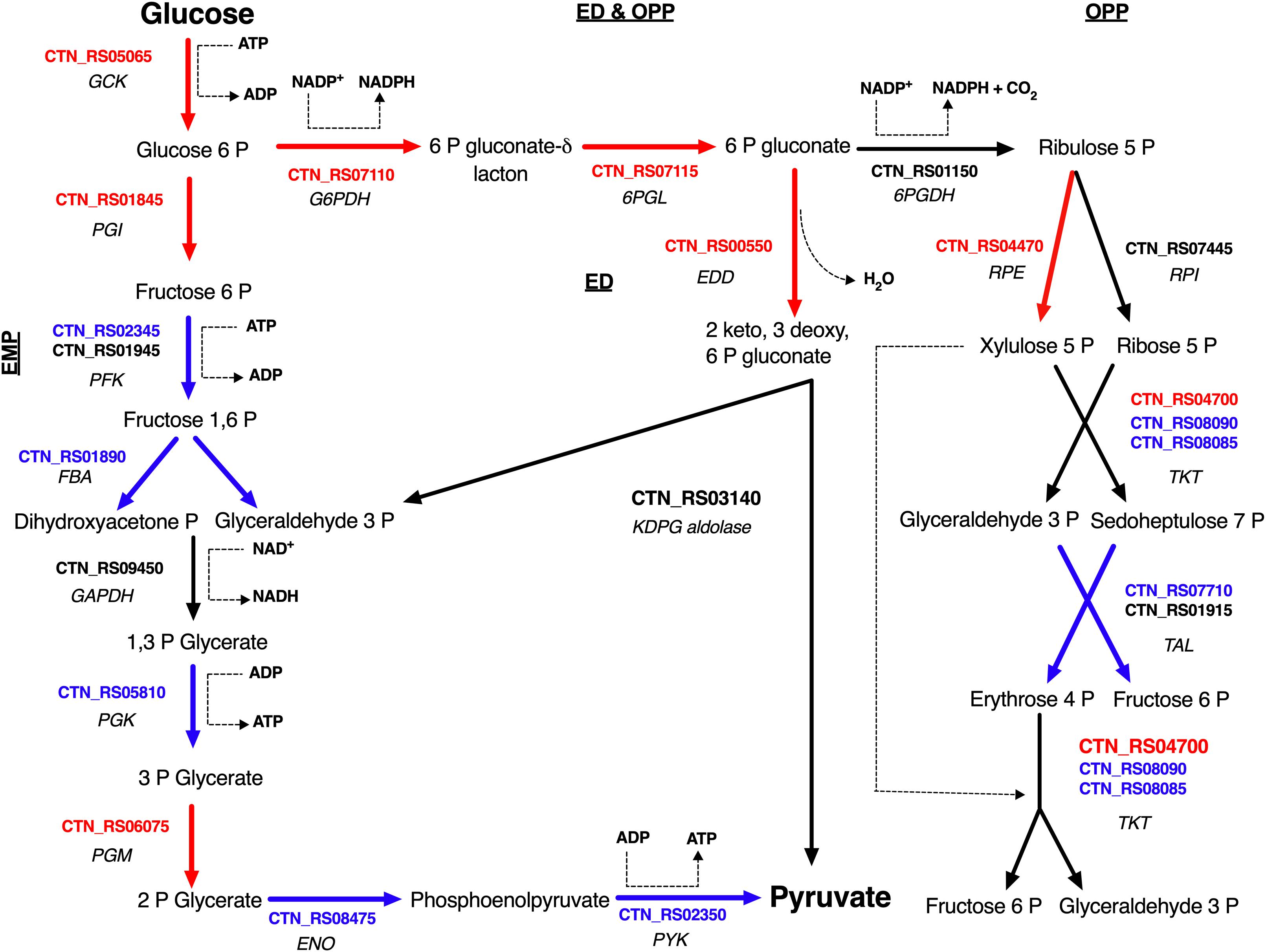
Figure 3. Glucose utilization model in T. neapolitana cells under CO2. Genes up- and down-regulated are highlighted in red and blue, respectively; NDE genes with 1.5 < fc < −1.5 are in black. GCK, glucokinase; PGI, glucose-6-phosphate isomerase; PFK, 6-phosphofructokinase; FBA, fructose-bisphosphate aldolase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PGK, glycerate kinase; PGM, phosphoglycerate mutase; ENO, enolase (Aka phosphoenolate hydratase); PYK, pyruvate kinase; G6PDH, glucose-6-phosphate dehydrogenase; 6PGL, 6-phosphogluconolactonase; EDD, 6-phophoglucono dehydratase; KDPG aldolase, 2-dehydro-3-deoxyphosphogluconate aldolase; 6PGDH, 6-phosphogluconate dehydrogenase; RPE, ribulose 5 phosphate epimerase; RPI, ribulose 5 phosphate isomerase; TKT, transketolase; TAL, transaldolase.
Genes encoding key enzymes of the anabolic branch of CLF pathway were differential expressed by CO2 (Table 3). Pyruvate:ferredoxin oxidoreductase (PFOR, E.C. 1.2.7.1) is a heterotetramer enzyme that operates the reversible coupling of AcCoA and CO2 to PYR under CLF conditions (d’Ippolito et al., 2014). Each subunit of this enzyme (CTN_RS03380, CTN_RS03385, CTN_RS03390, and CTN_RS03395) showed a significant increase of expression. In consideration of the double function of PFOR as catabolic (from PYR to AcCoA) and anabolic (from AcCoA and CO2 to PYR) enzyme, the up-regulation is in good agreement with both the general acceleration of metabolism induced by CO2 and the enhanced demand of PYR to feed the downfield synthesis of LA. According to this view, LDH (E.C. 1.1.1.27 – CTN_RS03950) was also up-regulated under the experimental conditions. A specific biochemical character of CLF is the increase of the rate of H2 synthesis during the fermentation process (Dipasquale et al., 2014). The molecular analysis revealed a significant up-regulation of the expression of the α-subunit of the bacterial hydrogenase (CTN_RS05285) together with two of the three proteins (CTN_RS06525, HYD E; CTN_RS06535, HYD G) that are necessary to the complex process of maturation of this enzyme. The α-subunit (HYDα) contains the catalytic H-cluster (Schut and Adams, 2009) that is committed to reduction of H+ to hydrogen gas. In their model of the hydrogenase of Thermotoga maritima, Schut and Adams (2009) have discussed a trimeric bifurcating enzyme that catalyzes multiple electron transfer events simultaneously. HYDα is the final collector of this transfer of electrons from NADH and reduced ferredoxin; thus, it is plausible that the experimentally observed boost of the H2 production rate requires an accelerated turnover of this protein under CLF conditions. The CO2-induced upregulation of PFOR and HYD was further confirmed by western blotting analysis carried out on PFORα and the β-subunit of hydrogenase (HYDβ) (Figure 4).
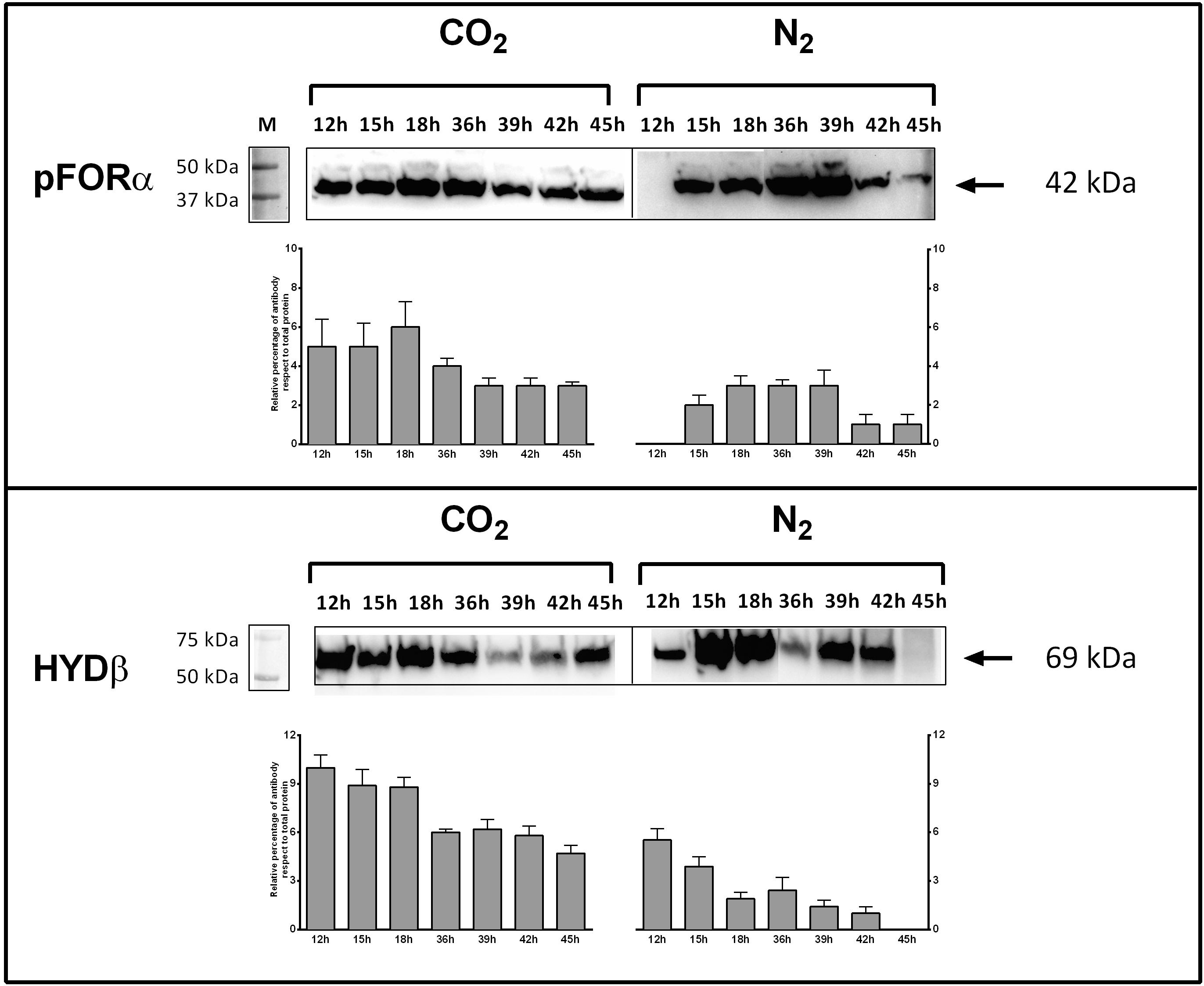
Figure 4. Expression of pyruvate:ferredoxin oxidoreductase (PFOR) and Fe–Fe hydrogenase (HYD) in T. neapolitana subsp. capnolactica. Bacterial cultures were grown under N2 or CO2, and collected at 12, 15, 18, 36, 39, 42, and 45 h. PFOR levels were detected by using a polyclonal antibody raised in rabbits against peptide 149–359 of subunit α. HYD levels were detected by using a polyclonal antibody raised in rabbits against peptide 1–192 of subunit β. Western blot filters were subjected to analysis using Image Lab 6.0 Software (Bio-Rad, Hercules, CA, United States). Analysis included the intensity determination of total Stain-Free fluorescence and intensity of PFOR-A and β-HYD blots for each lane. Results were expressed as relative percentage of antibody blot intensity respect intensity of total proteins.
Enhancement of cell reductants, e.g., NAD(P)H or ferredoxin (Fd), represents a major requirement to sustain the simultaneous production of H2 and LA during CLF (Dipasquale et al., 2014; d’Ippolito et al., 2014). RNA-seq approach revealed up-regulation of several genes involved in redox reactions and electron transport. Intriguingly, among up-regulated DEGs we found a significant expression of genes coding for enzymes related to the flavin-based oxidoreductase enzymes NADH-dependent reduced ferredoxin:NADP oxidoreductase (NFN) (CTN_RS04025 and CTN_RS04020) and NAD-Ferredoxin oxidoreductase (RNF) (CTN_RS02165, CTN_RS02160, and CTN_RS02155). Both these proteins are energy conservation systems. NFN controls supply of reduced ferredoxin and NADH by oxidation of NADPH while RNF couples reversible consumption of NADH with synthesis of reduced Fd and trans-membrane transport of Na+ or H+ (Hess et al., 2013). K-means cluster analysis of DEGs identified five different groups of genes with similar expression profile (not shown). One of the up-regulated cluster (Supplementary Figure 3) included genes coding for PFOR (CTN_RS03385, CTN_RS03390, and CTN_RS03395), flavin-based protein complexes (RNF and NFN – CTN_RS02160, CTN_RS02165, CTN_RS04020, and CTN_RS04025), thioredoxin (CTN_RS05860 and CTN_RS08515), ferredoxin (CTN_RS08225), Na/P cotransporter family protein (CTN_RS07190), and G6PDH (CTN_RS07110). The clustering was coherent with the fermentation results and suggested a CO2-stimulated rearrangement of the bacterial metabolism that links NAD(P)H from ED/OPP, PYR synthesis by PFOR, and production of NADH by flavin-based energy conservation systems. According to the experimental results, up-regulation of these pathways could be consistent with increase of the availability of redox potential for the synthesis of LA and H2 during CLF.
Thermophilic bacteria, such as Thermotogales, have been acquiring an emerging attention in biotechnology for hydrogen production by digestion of organic residues (Conners et al., 2006; Elleuche et al., 2014; Pradhan et al., 2015). Among thermophilic bacteria, T. neapolitana shows an interesting potential for the simultaneous production of hydrogen and LA by CLF (Dipasquale et al., 2014; d’Ippolito et al., 2014; Pradhan et al., 2017). In the present study, a selected strain of this bacterium, namely T. neapolitana subsp. capnolactica (Pradhan et al., 2017) was characterized by comparative analysis of gene expression profiling under CO2 (CLF conditions) and N2 (control). The study showed transcriptional changes that well correlate with the promoting effect of CO2 on both the synthesis of LA and the rate of hydrogen production and glucose consumption. The appearance of this bacterial phenotype was accompanied by variation in the expression of 1601 genes related to central biochemical pathways including glycolysis, TCA, cell wall construction, and protein synthesis. Under light microscopy, the cells showed a typical rod shape with a more homogeneous length than the control bacteria under N2 that vice versa were round-shaped (Supplementary Figure 4). In agreement with the acceleration of the fermentation rate, CO2 also triggered modification of the expression of 105 genes related to control of extracellular transport. Latif et al. (2015) have recently underlined that adaptation of Thermotoga maritima to achieve improved growth fitness requires plasticity of the ABC transporters. We found that expression of several ABC transporters of T. neapolitana was differently regulated (Supplementary Information), which is consistent with the modulation of these proteins in response to the different environmental conditions.
These molecular changes were driven by a number of key TFs that were differentially expressed under CO2. In particular, we observed different regulation of genes of the LacI family, such as LacI, AraC, and XylR, that are tightly linked to distinct regulons of carbohydrate catabolism (Ravcheev et al., 2014). These proteins have been also identified as regulators of sugar metabolism in Thermotogales, including T. neapolitana and T. maritima (Rodionov et al., 2013). Thus, control of their expression seems to corroborate the observation that the metabolic acceleration triggered by CO2 is sustained by a change of sugar catabolism in association with a more effective import/export of substrates.
According to this view, under CLF conditions, the bacterial cells showed an interesting shift of glucose utilization through down-regulation of EMP and activation of the alternative ED and/or OPP (Figure 3). EMP is energetically more effective than other glycolytic pathways because of the higher ATP yields per glucose unit. However, the energetic cost for the synthesis of the proteins carrying out the 10 reactions of this pathway may represent a growth-limiting factor (Molenaar et al., 2009; Flamholz et al., 2013). Recently, Singh et al. (2018) have discussed the role of OPP in providing additional reductants for H2 production in T. maritima. ED and OPP show a number of energetic advantages per glucose unit [reduced protein cost or increased amount of NAD(P)H, respectively] in comparison to EMP, thus switch from EMP to ED or OPP can be functional to the increased demand of reducing equivalents during CLF.
The analysis of the transcripts suggested that another pool of NADH could derive from the flavin-based oxido-reductase enzymes NFN and RNF that are both up-regulated by CO2. In T. maritima, NFN is a bifurcating enzyme that couples the oxidation of NADPH to the exergonic reduction of NAD+ and the endergonic reduction of Fd (Demmer et al., 2015). Electron bifurcation is one the mechanisms of energy conservation that biological organisms use to minimize free-energy loss in redox reactions. Bifurcating enzymes catalyze endoergonic and exoergonic electron transfer reactions to circumvent thermodynamic barriers and to carry out reactions that have unfavorable free energy variations (Buckel and Thauer, 2018). Deletion of the NFN complex (Tsac_2085 NfnA and Tsac_2086 NfnB) genes led to the total loss of NADPH-dependent activity and reduces ethanol fermentation in the thermophilic anaerobe Thermoanaerobacterium saccharolyticum (Lo et al., 2015). On the other hand, the RNF complex couples the reversible flow of electrons from reduced ferredoxin to NAD+. The process is associated with the transport of Na+ across the cell membrane and the direction of the electron flow is dependent on the sodium gradient (Biegel et al., 2011; Buckel and Thauer, 2013; Hess et al., 2013). The high production of hydrogen in T. neapolitana is associated to consumption of hydrogen ions (H+). RNF activity could be related to Na+ or H+ transport to balance the loss of H+ and to maintain the ionic homeostasis in the cell. As suggested in Figure 5, the combination of RNF with NFN may generate a cyclic process leading to production of NADH. In this view, synthesis of LA could be another mechanism to get rid of the excess of reducing equivalents. A similar cycle has been postulated between RNF and ferredoxin-dependent NADH oxidoreductase (FNOR) in T. saccharolyticum in order to operate a net transfer of protons across the cell membrane and eliminate the proton gradient that could be lethal for the cells (Tian et al., 2016).
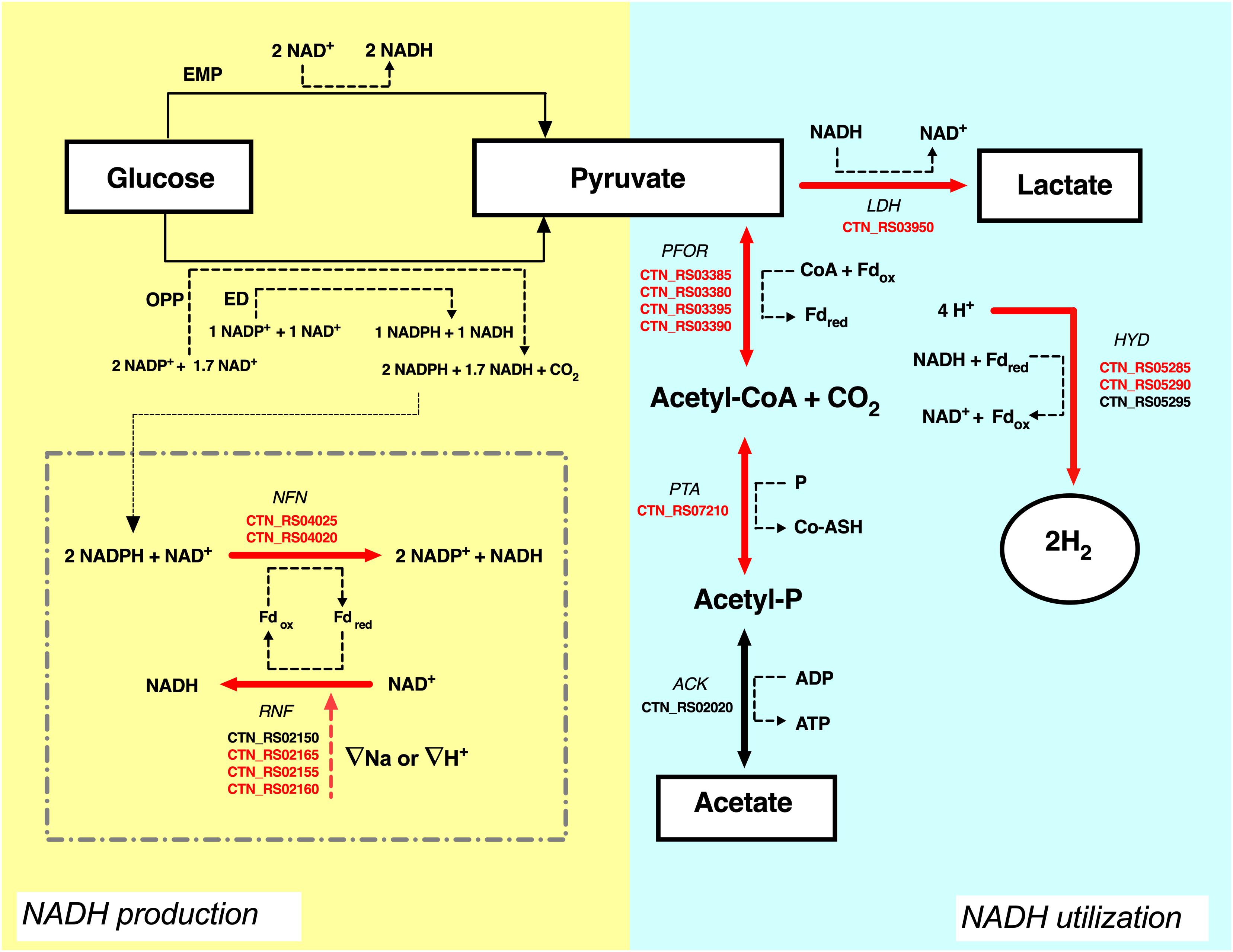
Figure 5. Proposal of carbon and NADH flow in T. neapolitana under CO2. Electron transfer form glucose (donor) to lactate and hydrogen (final acceptors) is shown. Suggested role of NFN-RNF cycle is shown in the dotted square. Up-regulated genes (≥1.5-fold change) are highlighted in red, NDE genes with 1.5 < fc < −1.5 are in black. ATP is omitted. For explanations see text. ACK, acetate kinase; PTA, phospshotransacetylase; PFOR, pyruvate:ferredoxin oxidoreductase; LDH, lactate dehydrogenase; HYD, hydrogenase; RNF, NAD-ferredoxin oxidoreductase; NFN, NADH-dependent reduced ferredoxin:NADP oxidoreductase; Fd, ferredoxin; EMP, Embden–Meyerhof–Parnas pathway; ED, Entner–Doudoroff pathway; OPP, oxidative pentose phosphate pathway.
As already reported by Krebs (1941), CO2 affects carboxylation reactions also in heterotrophs. In T. neapolitana, in addition to the indirect effect on the bacterial metabolism by gene regulation, CO2 plays a direct role as chemical reagent in the reaction to give PYR from acetate by PFOR. Thus, increase of the concentration of CO2 is very likely the driven force to reverse the oxidative decarboxylation of PYR and to yield carbon fixation. The occurrence of this reaction could explain the experimentally observed presence of PFOR even after the end of sugar consumption by glycolysis (d’Ippolito et al., 2014). At the moment we do not know whether this occurs for inversion of the functionality of PFOR or it is due to a newly synthesized pool of the enzyme (Figure 1). Synthetic PFOR is the key enzyme of the autotrophic fixation by the Wood–Ljungdahl pathway in acetogens (Furdui and Ragsdale, 2000). After our publication of the synthetic activity of PFOR in T. neapolitana (d’Ippolito et al., 2014), Xiong et al. (2016) have reported a similar mechanism in Clostridium thermocellum. In this work, the authors report assimilation of CO2 during cellobiose metabolism with fixation of over 40% of labeled carbon in PYR that derives from PFOR-dependent synthesis. This process is very similar to that of T. neapolitana thus suggesting that the activation of the anaplerotic reductive C1 pathway dependent on PFOR can be more common than it is generally believed. It is obvious that the energy requirements of this process need to be considered within a comprehensive organization of the cellular redox pathways.
In conclusion, the present work showed a correlation between the gene expression, the metabolic adaptation and the biochemical phenotype of the bacterium T. neapolitana under conditions that promote CLF. In according with acceleration of H2 synthesis, the CO2-induced effects clearly indicated an increased energetic metabolism of T. neapolitana that is sustained by a concerted reorganization of the cell metabolism including at least variation of central carbon metabolism, activation of anaplerotic oxidoreductase processes and increase of the exchange across the membrane. Similar effects on enhancement of the energetic metabolism or cell growth upon exposure to CO2 have been reported in other anaerobic bacteria, such as Streptococcus thermophilus (Arioli et al., 2009) and C. thermocellum (Xiong et al., 2016). The most significant trait of the molecular adaptation induced by CO2 in T. neapolitana is related to additional production of reductants by the EMP/ED-OPP switch and the bifurcating mechanisms based on the overexpression of the flavin-based complexes, such as NFN and RFN.
The raw sequencing data from this study are stored in the NCBI SRA database (http://www.ncbi.nlm.nih.gov) and are retrievable under the accession code PRJNA574556.
Gd’I conceived the whole research project, and wrote and amended the manuscript. SL conceived the idea of writing the manuscript, bioinformatic analysis, and wrote and amended the manuscript. NE performed the microbiology and western blot analysis. ML and MV performed the western blot. LD and NP performed the microbiology and sequencing analysis. AF planned and coordinated the whole research project and the experiments along with Gd’I, and wrote and amended the manuscript.
This research was funded by the BioRECO2VER Project, through the European Union’s Horizon 2020 Research and Innovation Programme under Grant Agreement No. 760431.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to thank Dr. Lucio Caso (CNR-ICB) for the technical support in microbiological activities.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00171/full#supplementary-material
Angel, A. M., Brunene, M., and Baumeister, W. (1993). The functional properties of Ompβ, the regularly arrayed porin of the hyperthermophilic bacterium Thermotoga neapolitana. FEMS Microbiol. 109, 231–236. doi: 10.1016/0378-1097(93)90025-w
Arioli, S., Roncada, P., Salzano, A. M., Deriu, F., Corona, S., Guglielmetti, S., et al. (2009). The relevance of carbon dioxide metabolism in Streptococcus thermophilus. Microbiol 155, 1953–1965. doi: 10.1099/mic.0.024737-24730
Belkin, S., Wirsen, C. O., and Jannasch, H. W. (1986). A new sulfur-reducing, extremely thermophilic eubacterium from a submarine thermal vent. Appl. Environ. Microbiol. 51, 1180–1185. doi: 10.1128/aem.51.6.1180-1185.1986
Biegel, E., Schmidt, S., González, J. M., and Müller, V. (2011). Biochemistry, evolution and physiological function of the Rnf complex, a novel ion-motive electron transport complex in prokaryotes. Cell. Mol. Life Sci. 68, 613–634. doi: 10.1007/s00018-010-0555-558
Buckel, W., and Thauer, R. K. (2013). Energy conservation via electron bifurcating ferredoxin reduction and proton/Na+ translocating ferredoxin oxidation. Biochim. Biophys. Acta 1827, 94–113. doi: 10.1016/j.bbabio.2012.07.002
Buckel, W., and Thauer, R. K. (2018). Flavin-based electron bifurcation, ferredoxin, flavodoxin, and anaerobic respiration with protons (Ech) or NAD+(Rnf) as electron acceptors: a historical review. Front. Microbiol. 14:401. doi: 10.3389/fmicb.2018.00401
Conners, S. B., Mongodin, E. F., Johnson, M. R., Montero, C. I., Nelson, K. E., and Kelly, R. M. (2006). Microbial biochemistry, physiology, and biotechnology of hyperthermophilic Thermotoga species. FEMS Microbiol. Rev. 30, 872–905. doi: 10.1111/j.1574-6976.2006.00039.x
Demmer, J. K., Huang, H., Wang, S., Demmer, U., Thauer, R. K., and Ermler, U. (2015). Insights into flavin-based electron bifurcation via the NADH-dependent reduced ferredoxin:NADP oxidoreductase structure. J. Biol. Chem. 290, 21985–21995. doi: 10.1074/jbc.M115.656520
Dipasquale, L., d’Ippolito, G., and Fontana, A. (2014). Capnophilic lactic fermentation and hydrogen synthesis by Thermotoga neapolitana: an unexpected deviation from the dark fermentation model. Int. J. Hydrogen Energ. 39, 4857–4862. doi: 10.1016/j.ijhydene.2013.12.183
Dipasquale, L., Pradhan, N., d’Ippolito, G., and Fontana, A. (2018). “Potential of hydrogen fermentative pathways in marine thermophilic bacteria: dark fermentation and capnophilic lactic fermentation in thermotoga and pseudothermotoga species,” in Grand Challenges in Marine Biotechnology, eds P. H. Rampelotto, and A. Trincone, (Berlin: Springer), 217–235. doi: 10.1007/978-3-319-69075-9_6
d’Ippolito, G., Dipasquale, L., and Fontana, A. (2014). Recycling of carbon dioxide and acetate as lactic acid by the hydrogen-producing bacterium Thermotoga neapolitana. ChemSusChem. 7, 2678–2683. doi: 10.1002/cssc.201402155
d’Ippolito, G., Dipasquale, L., Vella, F. M., Romano, I., Gambacorta, A., Cutignano, A., et al. (2010). Hydrogen metabolism in the extreme thermophile Thermotoga neapolitana. Int. J. Hydrogen Energ. 35, 2290–2295. doi: 10.1016/j.ijhydene.2009.12.044
Elleuche, S., Schröder, C., Sahm, K., and Antranikian, G. (2014). Extremozymes-biocatalysts with unique properties from extremophilic microorganisms. Curr. Opin. Biotechnol. 29, 116–123. doi: 10.1016/j.copbio.2014.04.003
Flamholz, A., Noor, E., Bar-Even, A., Liebermeister, W., and Milo, R. (2013). Glycolytic strategy as a tradeoff between energy yield and protein cost. PNAS 110, 10039–10044. doi: 10.1073/pnas.1215283110
Furdui, C., and Ragsdale, S. W. (2000). The role of pyruvate ferredoxin oxidoreductase in pyruvate synthesis during autothrophic growth by the Wood-Ljungdalh pathway. J. Biol. Chem. 275, 28494–28499. doi: 10.1074/jbc.m003291200
Guo, X. M., Trably, E., Latrille, E., Carrère, H., and Steyer, J. P. (2010). Hydrogen production from agricultural waste by dark fermentation: a review. Int. J. Hydrogen Energ. 35, 10660–10673. doi: 10.1016/J.IJHYDENE.2010.03.008
Hallenbeck, P. C., and Ghosh, D. (2009). Advances in fermentative biohydrogen production: the way forward? Trends Biotechnol. 27, 287–297. doi: 10.1016/j.tibtech.2009.02.004
Hess, V., Schuchmann, K., and Müller, V. (2013). The ferredoxin:NAD+ oxidoreductase (Rnf) from the acetogen Acetobacterium woodii requires Na+ and is reversibly coupled to the membrane potential. J. Biol. Chem 288, 31496–31502. doi: 10.1074/jbc.M113.510255
Howe, E. A., Sinha, R., Schlauch, D., and Quackenbush, J. (2011). RNA-Seq analysis in MeV. Bioinformatics. 27, 3209–3210. doi: 10.1093/bioinformatics/btr490
Krebs, H. A. (1941). Carbon dioxide assimilation in heterotrophic organisms. Nature 147, 560–563. doi: 10.1038/147560a0
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923.Fast
Latif, H., Sahin, M., Tarasova, J., Tarasova, Y., Portnoy, V. A., Nogales, J., et al. (2015). Adaptive evolution of Thermotoga maritima reveals plasticity of the ABC transporter network. Appl. Environ. Microbiol. 81, 5477–5485. doi: 10.1128/AEM.01365-1315
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ÄÄCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lo, J., Zheng, T., Olson, D. G., Ruppertsberger, N., Tripathi, S. A., Tian, L., et al. (2015). Deletion of nfnAB in Thermoanaerobacterium saccharolyticum and its effect on metabolism. J. Bacteriol. 197, 2920–2929. doi: 10.1128/JB.00347-15
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21. doi: 10.1186/s13059-014-0550-8
Manish, S., and Banerjee, R. (2008). Comparison of biohydrogen production processes. Int. J. Hydrogen Ener. 33, 279–286. doi: 10.1016/j.ijhydene.2007.07.026
Molenaar, D., Van Berlo, R., De Ridder, D., and Teusink, B. (2009). Shifts in growth strategies reflect tradeoffs in cellular economics. Mol. Syst. Biol. 5, 1–10. doi: 10.1038/msb.2009.82
Nuzzo, G., Landi, S., Esercizio, N., Manzo, E., Fontana, A., and d’Ippolito, G. (2019). Capnophilic lactic fermentation from Thermotoga neapolitana: a resourceful pathway to obtain almost enantiopure l-lactic acid. Fermentation 5:34. doi: 10.3390/fermentation5020034
Okonkwo, O., Lakaniemi, A. M., Santala, V., Karp, M., and Mangayil, R. (2017). Quantitative real-time PCR monitoring dynamics of Thermotoga neapolitana in synthetic co-culture for biohydrogen production. Int. J. Hydrogen Energ. 43, 3133–3141. doi: 10.1016/j.ijhydene.2017.12.002
Petrus, A. K., Swithers, K. S., Ranjit, C., Wu, S., Brewer, H. M., Gogarten, J. P., et al. (2012). Genes for the major structural components of Thermotogales species’ togas revealed by proteomic and evolutionary analyses of OmpA and OmpB homologs. PLoS One 7:e40236. doi: 10.1371/journal.pone.0040236
Pradhan, N., Dipasquale, L., d’Ippolito, G., Fontana, A., Panico, A., Lens, P. N. L., et al. (2016a). Kinetic modeling of fermentative hydrogen production by Thermotoga neapolitana. Int. J. Hydrogen Energ. 41, 4931–4940. doi: 10.1016/j.ijhydene.2016.01.107
Pradhan, N., Dipasquale, L., d’Ippolito, G., Fontana, A., Panico, A., Pirozzi, F., et al. (2016b). Model development and experimental validation of capnophilic lactic fermentation and hydrogen synthesis by Thermotoga neapolitana. Water Res. 99, 225–234. doi: 10.1016/j.watres.2016.04.063
Pradhan, N., Dipasquale, L., d’Ippolito, G., Panico, A., Lens, P. N. L., Esposito, G., et al. (2015). Hydrogen production by the thermophilic bacterium Thermotoga neapolitana. Int. J. Hydrogen Energ. 16, 12578–12600. doi: 10.3390/ijms160612578
Pradhan, N., Dipasquale, L., d’Ippolito, G., Panico, A., Lens, P. N. L., Esposito, G., et al. (2017). Hydrogen and lactic acid synthesis by the wild-type and a laboratory strain of the hyperthermophilic bacterium Thermotoga neapolitana DSMZ 4359T under capnophilic lactic fermentation conditions. Int. J. Hydrogen Energ. 42, 16023–16030. doi: 10.1016/j.ijhydene.2017.05.052
Pradhan, N., d’Ippolito, G., Dipasquale, L., Esposito, G., Panico, A., Lens, P. N. L., et al. (2019). Simultaneous synthesis of lactic acid and hydrogen from sugars via capnophilic lactic fermentation by Thermotoga neapolitana cf capnolactica. Biomass Bioenerg 125, 17–22. doi: 10.1016/j.biombioe.2019.04.007
Ravcheev, D. A., Khoroshkin, M. S., Laikova, O. N., Tsoy, O. V., Sernova, N. V., Petrova, S. A., et al. (2014). Comparative genomics and evolution of regulons of the LacI-family transcription factors. Front. Microbiol. 5:294. doi: 10.3389/fmicb.2014.00294
Rodionov, D. A., Rodionova, I. A., Li, X., Ravcheev, D. A., Tarasova, Y., Portnoy, V. A., et al. (2013). Transcriptional regulation of the carbohydrate utilization network in Thermotoga maritima. Front. Microbiol. 4:244. doi: 10.3389/fmicb.2013.00244
Schut, G. J., and Adams, M. W. (2009). The iron-hydrogenase of Thermotoga maritima utilizes ferredoxin and NADH synergistically: a new perspective on anaerobic hydrogen production. J. Bacteriol. 191, 4451–4457. doi: 10.1128/JB.01582-08
Singh, R., White, D., Demirel, Y., Kelly, R., Noll, K., and Blum, P. (2018). Uncoupling Fermentative Synthesis of Molecular Hydrogen from Biomass Formation in Thermotoga maritime. Appl. Environ. Microbiol. 84:e00557-15..
Tian, L., Lo, J., Shao, X., Zheng, T., Olson, D. G., and Lynd, L. R. (2016). Ferredoxin:NAD oxidoreductase of Thermoanaerobacterium saccharolyticum and its role in ethanol formation. Appl. Environ. Microbiol. 82, 7134–7141. doi: 10.1128/aem.02130-16
Keywords: lactic acid, pyruvate, glycolysis, hydrogenase, thermophilic, RNA-seq
Citation: d’Ippolito G, Landi S, Esercizio N, Lanzilli M, Vastano M, Dipasquale L, Pradhan N and Fontana A (2020) CO2-Induced Transcriptional Reorganization: Molecular Basis of Capnophillic Lactic Fermentation in Thermotoga neapolitana. Front. Microbiol. 11:171. doi: 10.3389/fmicb.2020.00171
Received: 31 July 2019; Accepted: 24 January 2020;
Published: 18 February 2020.
Edited by:
Masahiro Ito, Toyo University, JapanCopyright © 2020 d’Ippolito, Landi, Esercizio, Lanzilli, Vastano, Dipasquale, Pradhan and Fontana. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Giuliana d’Ippolito, Z2RpcHBvbGl0b0BpY2IuY25yLml0; Simone Landi, cy5sYW5kaUBpY2IuY25yLml0; cy5sYW5kaTg3QGdtYWlsLmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.