
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 30 January 2020
Sec. Infectious Agents and Disease
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.00073
 Rundong Wang1,2†
Rundong Wang1,2† Yijia Deng1†
Yijia Deng1† Qi Deng1*
Qi Deng1* Dongfang Sun1
Dongfang Sun1 Zhijia Fang1
Zhijia Fang1 Lijun Sun1*
Lijun Sun1* Yaling Wang1*
Yaling Wang1* Ravi Gooneratne3
Ravi Gooneratne3Vibrio parahaemolyticus (Vp), a major food-borne pathogen, is responsible for severe infections such as gastroenteritis and septicemia, which may be accompanied by life-threatening complications. While studies have evaluated factors that affect the virulence of the pathogen, none have investigated the interaction of Vp with gut microbiota. To address this knowledge gap, we compared the effect of Vp on gut bacterial community structure, immunity, liver and kidney function, in pseudo germ-free (PGF) mice and normal (control) mice. Significant damage to the ileum was observed in normal mice compared with the PGF mice. The inflammatory factors IL-1β, IL-6, and TNF-α in normal mice were ∼2.5-fold higher than in the PGF mice, and liver (ALT, AST, ALP) and kidney (BUN) function indices were ∼1.6-fold higher. The Vp infection substantially reduced species composition and richness of the gut microbial communities. In particular, there was a shift in keystone taxa, from protective species of genera Bacteroides, Lactobacillus, Bifidobacterium, and Akkermansia in the gut of control mice to opportunistic pathogens Enterobacteriaceae, Proteus, Prevotella, and Sutterella in Vp-infected mice, thus affecting microbiota-related biological functions in the mice. Specifically, pathways involved in infectious diseases and ion channels were significantly augmented in infected mice, while the pathways involved in metabolism, digestion and cell growth declined. We propose that the normal mice are more prone to Vp infection because of the alteration in gut-microbe-mediated functions. All these effects reduce intestinal resistance, with marked damage to the gut lining and pathogen leakage into the blood culminating in liver and kidney damage. These findings greatly advance our understanding of the mechanisms underlying interactions between Vp, the gut microbiota and the infected host.
Vibrio parahaemolyticus (Vp) is a Gram-negative halophilic bacterium that is widely disseminated in estuarine environments. Vp has been found to be responsible for 20–30% of food poisoning cases in Japan and seafood borne infections in many Asian countries (Letchumanan et al., 2014). Vp also recognized as the leading cause of human foodborne infection from seafood consumption in the United States (Su and Liu, 2007; Shinoda, 2011). The characteristics of Vp infection include abdominal pain, acute gastroenteritis, and septicemia, which in its most severe form (10 to 15% of sufferers) can present with life-threatening complications such as hemolytic uremic syndrome and liver damage (Ryan, 1976; Mertens et al., 1979; Daniels et al., 2000). Most outbreaks of food poisoning are caused by strains of the serotype O3: K6. Vp infections occur mostly via the ingestion of contaminated raw, undercooked, or mishandled marine products (Matsumoto et al., 2000; Newton et al., 2012; Zarei et al., 2012). The main virulence factors that are thermostable direct hemolysin (TDH)-a pore forming protein that contributes to the cytotoxicity and enterotoxicity of Vp in host, and TDH related hemolysin, which plays a similar role as TDH in the disease pathogenesis (Broberg et al., 2011). In addition, Vp also encodes adhesion and type III secretion systems to ensure its survival in the host vivo environment (Dean, 2011; Raghunath, 2015; Li et al., 2019). However, these virulence factors alone cannot directly damage the liver and kidney (Wang et al., 2016). Hence, further research and explanation need to be sought for why the clinical outcomes vary following people’s exposure to Vp.
The gut microbiota performs a diversity of important functions in the host, including promoting the development of the immune system, stimulating metabolic function and repressing pathogenic bacteria (Hooper et al., 2012). There are two major phyla that dominate the human gut- Bacteroidetes and Firmicutes phyla. The Bacteroidetes phyla constitutes of Bacteroides and Prevotella species, while the Firmicutes is made up of Lactobacillus and Clostridium species (Torres et al., 2001). It has been indicated that there is a correlation between human diseases and bacterial communities in the gut (Turnbaugh et al., 2008). Vibrio cholerae can cause explosive diarrhea in which extensive disruption of the intestinal microbes promotes infection by this bacterium (Midani et al., 2018). Microbiota sampled in travelers showed a higher risk of Campylobacter infection for those with a specific microbial profile (Kampmann et al., 2016). Diarrheagenic Escherichia coli can induce changes in the gut microbiota and distinctive components of these can alter how virulence genes are expressed, thus promoting the infection process (Curtis et al., 2014; Singh et al., 2015). Thus, these compelling evidence suggested that bacterial communities in the host gut may contribute to the pathogenesis of enteric infections. Vp is a significant intestinal pathogen but whether specific gut microbiota in a host resist or promote infection is not known. To advance our understanding of Vp pathogenesis, further research is needed on interactions between gut bacterial communities and Vp.
The term “pseudo germ-free (PGF) mice” refers to mice demonstrably free from intestinal microbes as a result of treatment with mixed antibiotics (Liu et al., 2012) and such mice are an invaluable experimental tool for examining interactions between disease in a host and its gut microbiota (Gordon and Pesti, 1971; Lee et al., 2012; Grover and Kashyap, 2014). Due to recent advances in the application of next generation sequencing technology, it is now possible to gradually unveil the role of the gut microbiome in disease development (Fraher et al., 2012). Use of 16S rRNA gene sequencing to determine the abundance of microbes in different treated samples has also promoted the identification of known and potentially new microorganisms (Yarza et al., 2014).
The main aim of the study was to elucidate the relationship between host gut bacterial communities and Vp infection and advance our understanding of Vp pathogenesis and to develop novel interventions for disease prevention. To achieve this we constructed a PGF mouse as an animal model to evaluate the role of gut microbiota in the Vp pathogenicity in vivo. In addition we used 16S rRNA gene sequencing to help identify gut microbiota that could serve as biomarkers of Vp pathogenicity.
All experimental protocols and animal handling procedures were carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals (National Institutes of Health Publication No. 80–23, revised 1996). This study was approved by the Ethical Committee on Animal Experimentation of the College of Food Science and Technology, Guangdong Ocean University (No. SYXK2014-0053).
Two-month-old male specific-pathogen-free (SPF) C57BL/6J mice (20 ± 5 g) were obtained from the animal center of Guangdong Province. The mice were housed in stainless steel cages and allowed to acclimatize to environmental conditions for 5 days before experiments. The animals had free access to distilled water and sterilized food; the room temperature was maintained at 20 ± 2°C with a relative humidity of 60 ± 5% on a 12 h light–dark cycle. The mice were randomly allocated to four groups for treatment: control SPF mice, PGF mice, SPF mice infected with Vp, and PGF mice infected with Vp.
The procedure for preparing PGF mice was based on previous studies (Clarke et al., 2010; Wang et al., 2011; Ge et al., 2017) but modified to deliver antibiotics by gavage rather than in the animals’ drinking water. This was done to ensure consistency of exposure. The SPF C57BL/6J mice in the PGF group received a mixture of non-absorbable antibiotics – vancomycin (200 mg/kg), metronidazole (200 mg/kg), and neomycin (300 mg/kg) by orally gavage twice a day for 7 days. Each mouse was housed in a micro-isolator cage under germ-free conditions with free access to sterilized water and food.
Tests to evaluate the pseudo-germ-free model in comparison to normal SPF C57BL/6J mice (controls) were carried out on days 3 and 6 after the final oral gavage with of mixed antibiotics. The same procedure was employed for control mice as is described below for the model mice.
Five pseudo-germ-free mice were euthanized by exsanguination following ether vapor narcosis (via a funnel) after overnight fasting. Blood was sampled by percutaneous cardiac puncture. Half the volume of blood collected was centrifuged at 3,500 rpm for 10 min and the serum used to assess physiological function. Detection kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) were used to measure three liver-specific enzymes [aspartate aminotransferase (AST), alanine aminotransferase (ALT), and alkaline phosphatase (ALP)] and the kidney-specific index, blood urea nitrogen (BUN). The remaining blood was transferred into tubes containing EDTA for measurement of red blood cells (RBC), white blood cells (WBC), hemoglobin (Hb), and platelet concentrations, using an automatic hematology analyzer (Cell – DYN 3700, Abbott Diagnostics, Abbott Park, IL, United States).
The ileum and colon contents were collected following the blood collection. The mice were sacrificed by cervical dislocation and dissected under a biosafety hood. The ileum was examined for damage, flushed with ice-cold sterilized PBS, and the ileal contents collected aseptically. Samples were placed in 1.5-mL sterile tubes, snap-frozen on dry ice, and stored at −80°C. The numbers of microbiota were measured by a plate counting method as described below.
The samples were weighed, mixed with sterile 0.01 M phosphate-buffered saline solution (PBS, pH 7.2) at 1 g: 9 mL ratio and homogenized for 30 s in a Stomacher® in a sterile environment. The homogenates were diluted (10×) in sterile PBS solution, and a 100-μL volume of homogenate was plated onto selective media in triplicate and incubated at 37°C for 24 h (for aerobes) or 48 h (for anaerobes). Colony forming units (CFU) were counted manually to determine the density of viable cells in each sample (expressed as CFU/g). The triplicates were analyzed independently with the whole process completed within 20 min.
The choice of selective media was based on the composition of dominant microbiota in ileum and colon. In this test, we used Bifidobacterium, Lactobacillus, E. coli, and Enterococcus to characterize the gut flora (Hooper and Gordon, 2001; Qin et al., 2010). For aerobes, E. coli and Enterococcus were detected using eosin methylene blue and enterococcus chromogenic media. For anaerobes, Bifidobacterium and Lactobacillus were detected by TPY agar and MRS agar media and incubated in an anaerobic tank.
Vibrio parahaemolyticus ATCC 33847 is a typical pathogen, containing tdh and tlh genes, two kinds of type of III secretion systems (T3SS1 and T3SS2) and the T6SS2 that co-exists with T3SS2 system, was used in this study. The Vp was stored in 25% glycerol at −20°C before being grown at 37°C for 24 h in Luria Broth (LB; BLBT, Beijing, China) containing 3% NaCl. The inoculum was thrice passaged in LB-3% NaCl. The final density of inoculant was adjusted to ∼109 CFU/mL and used to infect mice.
Mice were fasted overnight then randomly divided into three groups of 5 mice each: control SPF mice, PGF model mice and SPF mice for inoculation. The PGF and SPF mice were gavaged with a suspension of Vp (as described above) at 0.2 mL/10 g body weight (bw). The control group was gavaged with fresh sterilized LB medium. The three groups of mice were observed for 12 h before sample collection. This in vivo experiment was performed in triplicate.
The mice were anesthetized with ether, as described in section “Evaluation of Antibiotic Treatment on Serum and Gut Flora,” and blood samples collected by cardiac puncture. The mice were sacrificed by cervical dislocation and dissected under a biosafety hood. The ileum was examined for any damage, flushed with ice-cold sterilized PBS, and the ileal contents collected aseptically and stored at −80°C for gut microbiota analyses.
The blood samples were centrifuged at 3,500 rpm for 10 min and serum collected. Three liver-specific enzymes (AST, ALT, and ALP) and the kidney-specific index BUN were detected using kits (Nanjing Jiancheng Bioengineering Institute, China). The pro-inflammatory markers [Interleukin-1β (IL-1β), Interleukin-6 (IL-6), and Tumor Necrosis Factor (TNF-α)] and an anti-inflammatory marker [(Interleukin-10 (IL-10)] were determined by enzyme-linked immunosorbent assay (ELISA) kits (Neobioscience Technology Co., Shenzhen, China) according to the manufacturer’s protocols.
To better understand the effect of Vp infection on the richness and species composition of the gut microbiota, high-throughput sequencing of the V3–V4 region of the 16S rRNA gene was used. The metagenomic DNA was extracted from ileal samples using the Power Max DNA isolation kit provided by MoBio Laboratories, Carlsbad, CA, following to the manufacturer’s recommendations, and quantified by UV spectroscopy (NanoDrop ND-1000 spectrophotometer, Thermo Fisher Scientific, Waltham, MA, United States). The V3–V4 region of the 16S rRNA gene was amplified from metagenomic DNA using a pair of universal primers (515F 5′-GTGCCAGCM GCCGC GGTAA-3′, 806R 5′-GGACTACHVGGGTWTCTAAT-3′) (Shang Q. et al., 2017). The PCR products of each sample were checked and purified using Agencourt AMPure XP Beads (Beckman Coulter, Indianapolis, IN, United States) and the PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA, United States). Equimolar amounts of PCR products were pooled and sequenced using the Illumina HiSeq4000 platform at GUHE Info Technology Co. (Hangzhou, China).
The QIIME software package (version 1.9.0) was employed to filter raw sequencing reads in terms of quality using the script, as previously described (Caporaso et al., 2010; Shang Q. et al., 2017).
Vsearch (v1.11.1) was applied to pick the operational taxonomic units (OTUs) at an identity of 97% (Rognes et al., 2016). A Venn diagram was constructed to compare the compositional OTUs of the bacterial communities in each group by using R packages (v3.2.0) as previously documented (Zaura et al., 2009).
Operational taxonomic unit-level alpha diversity indices, such as Chao1, Ace, Simpson index and Shannon index for evaluation of community richness and species composition, were calculated by Mothur software packages (v1.30.1) (Ling et al., 2014).
Principal coordinate analysis (Ramette, 2007) was performed to compare the structural differences in microbial communities. Principal components 1 (PC1) and 2 (PC2) were subjected to the Wilcoxon rank-sum test to determine the significance of differentiation of microbiota structure among control mice and Vp-infected mice.
Linear discriminant analysis Effect Size (LEfSe) was performed to identify and characterize gut microbiota differences between the control group and Vp-infected group, using the online version of Galaxy3 (Segata et al., 2011). Linear discriminant analysis (LDA) was performed using a one-against-all strategy; a significance value of less than 0.05 combined with an effect size threshold of 2 was selected.
The OTU table was normalized by the 16S rRNA copy number per chromosome, and the Kyoto Encyclopedia of Genes and Genomes (KEGG) classified functions by Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (Langille et al., 2013; Zhang et al., 2019) using the Galaxy platform1. This process was conducted to obtain the metabolic pathways of bacteria. The significant differences in KEGG pathways between Vp-infected mice and control mice were tested using the Statistical Analysis of Metagenomic Profiles software package (v2.1.3) (Parks et al., 2014), and results shown by heatmap.
All data were analyzed using the software SPSS 19.0 (SPSS Inc., Chicago, IL, United States). Differences between the means and changes in the diversity of gut bacterial community were compared by one-way analysis of variance, followed by Tukey’s post hoc test, with the level of significance set at p < 0.05.
The effect of mixed antibiotics on the gut microbiota of mice was evaluated by a plate counting method with media selective for aerobes and anaerobes. The dominant intestinal microbiota in the PGF mice were significantly reduced in density by the mixed antibiotic treatment compared with untreated mice (Table 1). On day 3 following the last oral gavage, there was no Bifidobacterium and Lactobacillus, and the numbers of E. coli (1.01 ± 0.02 log10 CFU/g) and Enterococcus (0.12 ± 0.01 log10 CFU/g) were very significantly lower (p < 0.01) than in the control group. In the PGF mice, the gut microbiota was almost completely eliminated and there was no sign of recovery on day 6 when sampling results were similar to day 3.
The effect of mixed antibiotics on physiological functions of mice was assessed by hemogram and serum biochemical indices. There were no significant differences in hematological parameters between normal untreated mice and PGF mice on days 3 or 6 following treatment with mixed antibiotics (Table 2). The concentration of WBC, RBC, Hb, and platelets of PGF mice were not significantly different (p > 0.05) to normal mice and neither were the serum biochemical parameters AST, ALT, ALP, and BUN. Therefore, it appears that exposure to mixed antibiotics only removed intestinal microbial flora and did not affect the monitored hematological parameters in mice over 6 days.
The ileal damage differences were compared by observing ileal fluid accumulation and ileal wall bleeding. When the two groups of mice were administered Vp, both the PGF group and normal (SPF) mice showed ileal bleeding, ileal fluid and intestinal damage compared with the untreated controls and these effects were most marked in normal SPF mice (with intact gut microbiota). The ileal damage caused by Vp in PGF mice and normal mice is shown in Figure 1.
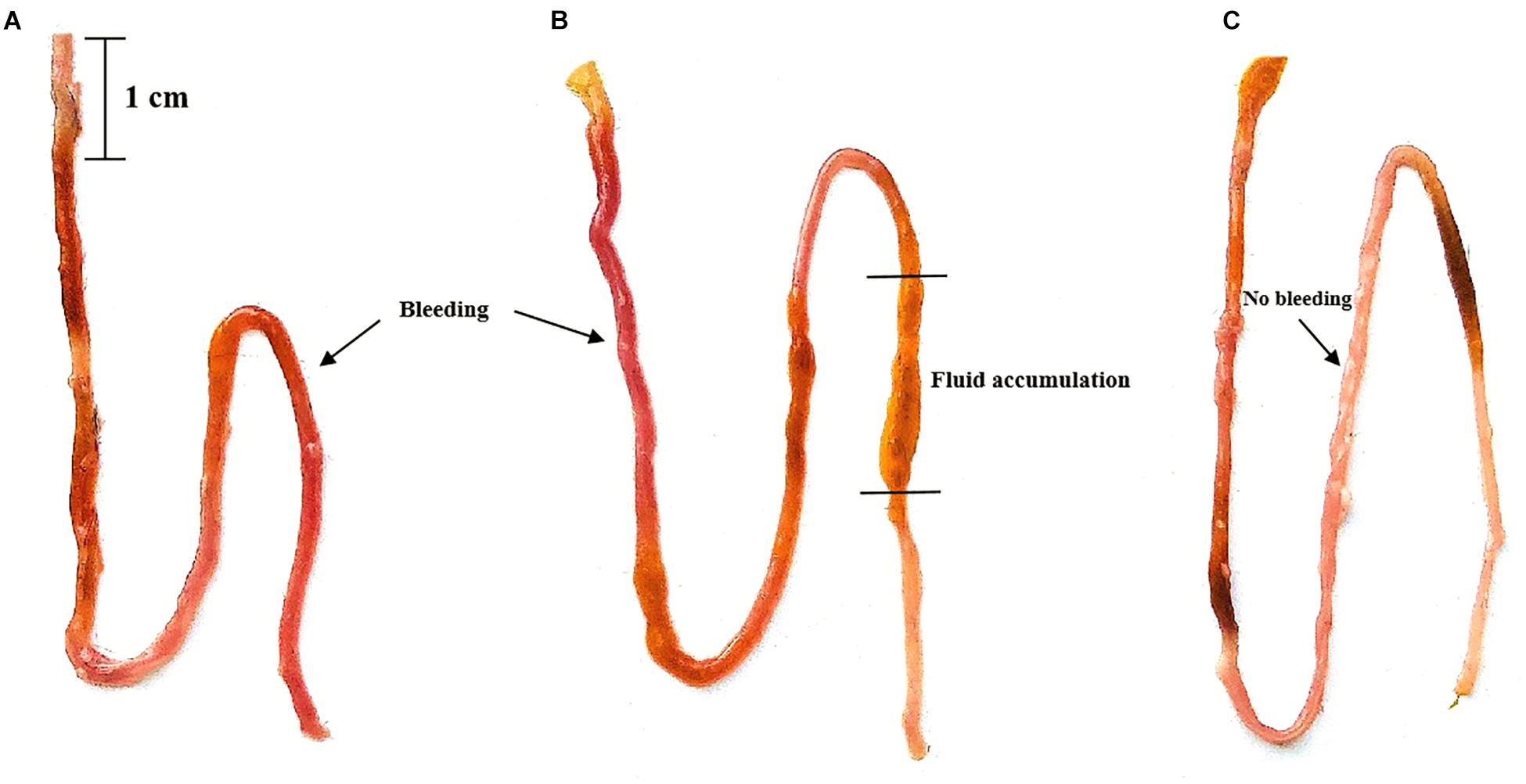
Figure 1. Effects of oral gavage with Vibrio parahaemolyticus (Vp) on the ileum of pseudo germ-free and normal (SPF) mice. (A) Pseudo germ-free mouse, (B) SPF mouse, (C) control mouse. The control mice were given fresh sterilized LB medium instead of Vp.
Damage to the metabolism of mice caused by Vp may be revealed by liver and kidney function tests. Therefore, serum biochemical parameters indicative of liver and kidney function were measured at 12 h in normal SPF and PGF mice gavaged with Vp (Figure 2). AST, ALT, ALP, and BUN were significantly elevated (p < 0.05) in infected SPF and PGF groups compared with the control group. Moreover, AST, ALT, ALP, and BUN activities in the SPF group were 1.5, 1.7, 1.9, and 1.4 times higher, respectively, than in the PGF group.
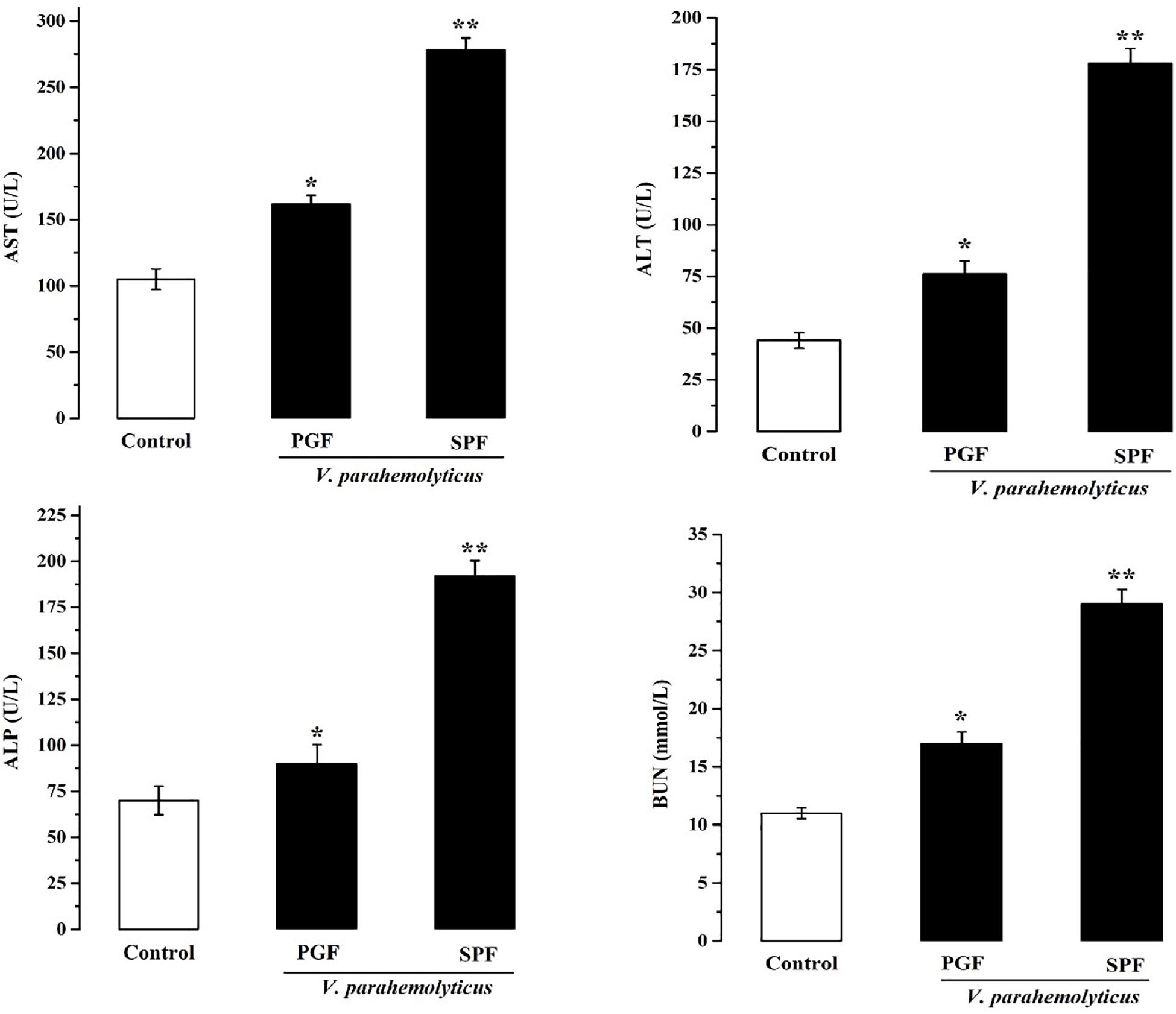
Figure 2. Effects of Vibrio parahaemolyticus on serum AST, ALT, ALP, and BUN indices of pseudo germ-free (PGF) and normal (SPF) mice. Data are expressed as the mean ± SEM (n = 5) compared with the control group gavaged with fresh sterilized LB medium. *p < 0.05; **p < 0.01.
Inflammation is the body’s response to infection. The pro-inflammatory and anti-inflammatory marker studies showed that IL-1β, IL-6, and TNF-α were significantly increased (p < 0.05) and that IL-10 was significantly reduced (p < 0.05) in the Vp-infected SPF and PGF groups compared with the control group (Figure 3). In the SPF group, the pro-inflammatory markers IL-1β, IL-6, and TNF-α were 1.6, 4.4, and 1.4 times that of the PGF group, and the anti-inflammatory marker IL-10 was approximately half that in the PGF mice.
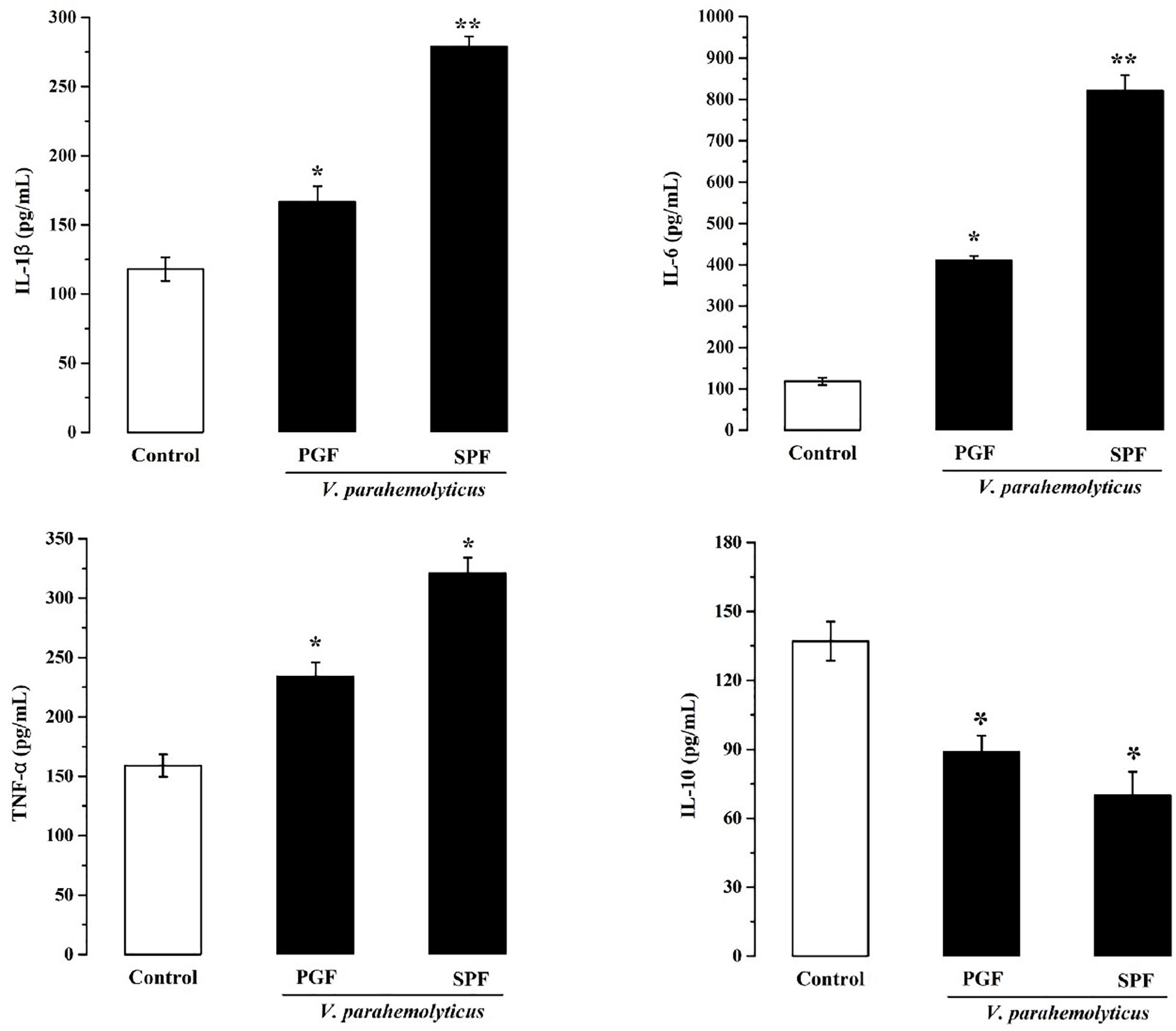
Figure 3. Effects of Vibrio parahaemolyticus on the serum IL-1β, IL-6, TNF-α, and IL-10 indices of pseudo germ-free mice and normal mice. Data are expressed as the mean ± SEM (n = 5) compared with the control group gavaged with fresh sterilized LB medium. *p < 0.05; **p < 0.01.
To better understand the effect of Vp infection on the richness and species composition of the gut microbiota, high-throughput sequencing of the V3–V4 region of the 16S rRNA gene was used. The pyrosequencing coverage of more than 99.1% indicates an adequate sequencing depth for all samples (Table 3). To estimate bacterial richness and composition of the gut microbiota, the Chao1, Ace, Shannon and Simpson indices were calculated (Table 3). Both richness and composition of the gut microbiota were reduced following Vp infection and these differences were statistically significant (p < 0.05). These preliminary results suggest that Vp infection could alter the community structure of gut microbiota.

Table 3. Evaluation of community richness and species composition (Chao1, Ace, Shannon and Simpson indices) of mouse gut microbiota after V. parahaemolyticus (Vp) infection+.
To analyze the microbiota in ileal contents, sequences with at least 97% similarity were combined into OTUs. As shown in a Venn diagram, 780 OTUs were identified in ileal contents of the control group. In the Vp-infected groups 255 of these OTUs decreased and 100 were different from the controls (Figure 4A). Overall, 32.7% of ileal OTUs changed following Vp infection. At the phylum level (Figure 4B), Bacteroidetes, Firmicutes, Fusobacteria, Actinobacteria, Proteobacteria (which includes Vp), and Verrucomicrobia were most abundant in the ileum, making up 62.8, 25.0, 1.3, 5.1, 1.1, and 4.7% of all identified OTUs, respectively. The proportions of Bacteroidetes and Verrucomicrobia were decreased in the ileum following Vp infection whereas Firmicutes, Fusobacteria, Actinobacteria, and Proteobacteria were increased. At the genus level (or family level where microbe could not be identified to genus) (Figure 4C), the abundance of Enterobacteriaceae, Clostridiales, Prevotella, and Vibrio increased while S24_7, Bacteroides, Lactobacillus, Bifidobacterium, and Akkermansia decreased following Vp infection. In addition, at the genus level, abundance of Rikenellaceae, Ruminococcus, and Faecalibacterium were also significantly different from the control group (data not shown). Moreover, according to the UniFrac principal coordinates analysis, the ileal content samples of the control group clustered in the lower left quadrant, while those of the Vp-infected groups clustered in the upper right quadrant, indicating that gut microbiota changed significantly following exposure to Vp (Figure 4D).
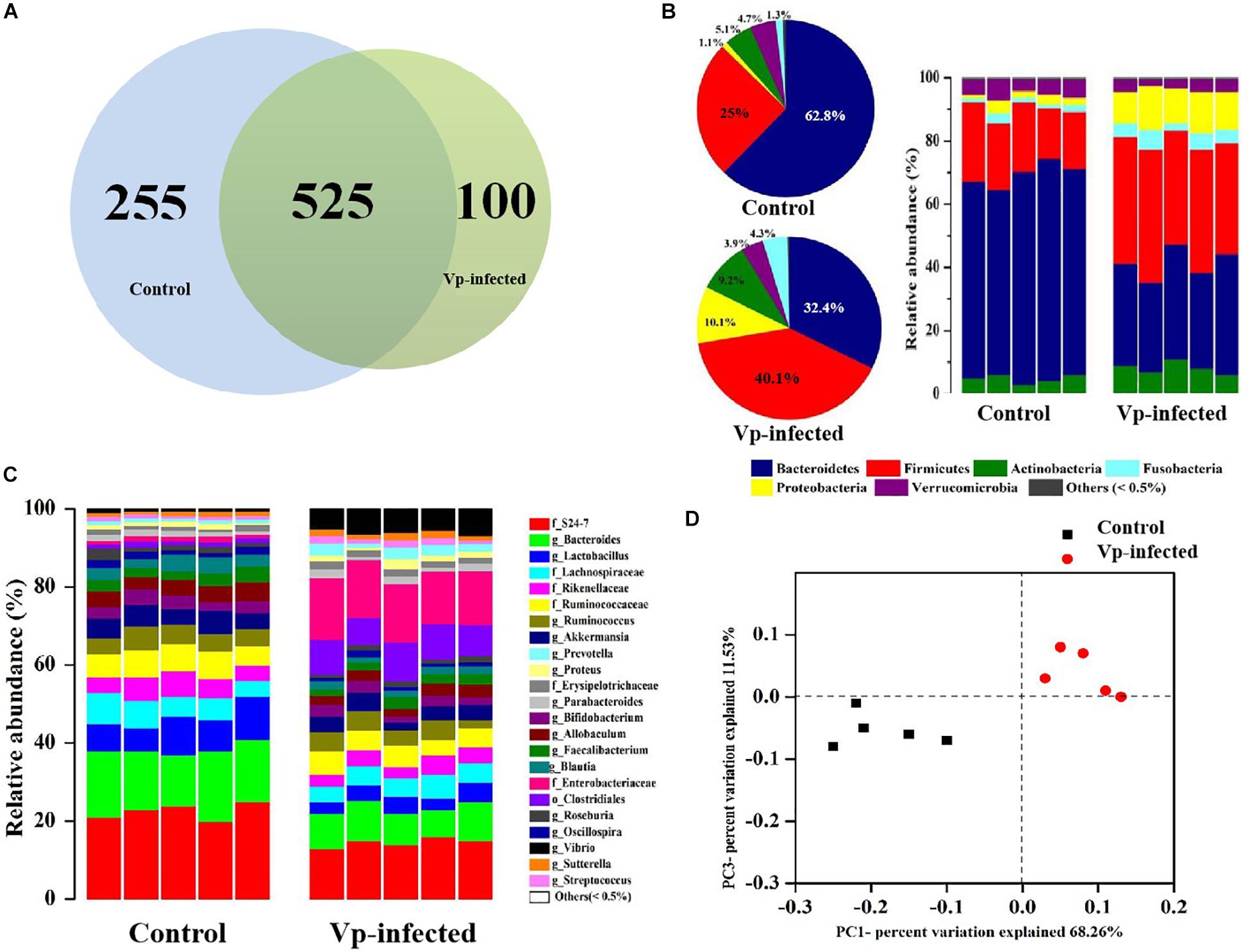
Figure 4. Vibrio parahaemolyticus-induced gut microbiota dysbiosis. (A) Venn diagram analysis of unique/shared OTUs in the gut microbiota of experimental mice following V. parahaemolyticus (Vp) infection. (B,C) The ileum microbiome composition profiles at the phylum and genus level in the control group and Vp-infected groups (each color represents one bacterial taxon). (D) UniFrac principal co-ordinates analysis estimates of ileum microbiota in the control and Vp-infected groups.
To fully ascertain the effects of Vp on gut microbiota and identify biomarker microbiota that differ between the Vp-infected groups and the control group, we further analyzed the gut microbiota at different taxonomic levels using the LEfSe algorithm with significance set at p < 0.05. As shown in the taxonomic cladograms, LEfSe analysis visualized and confirmed the modulatory effects of Vp on the gut microbiota at different taxonomic levels in C57BL/6J mice (Figure 5). Specifically, Vp decreased the Bacteroidetes. At genus level, Bacteroides, Lactobacillus, Bifidobacterium, Blautia, Acidaminococcus, Allobacullum, and Akkermansia decreased. Among the intestinal microbes that were enriched by Vp – Enterobacteriaceae, Streptococcaceae, Proteus, Vibrio, Prevotella, Sutterella, Dietzia, Adhaeribacter, Desulfovibrio, Paraprevotella, Sphingobacteriaceae, and Kocuria – most belonged to the phyla Firmicutes, Actinobacteria, and Proteobacteria. Thus, the presence of these sensitive taxa may serve to indicate existence of Vp infection.
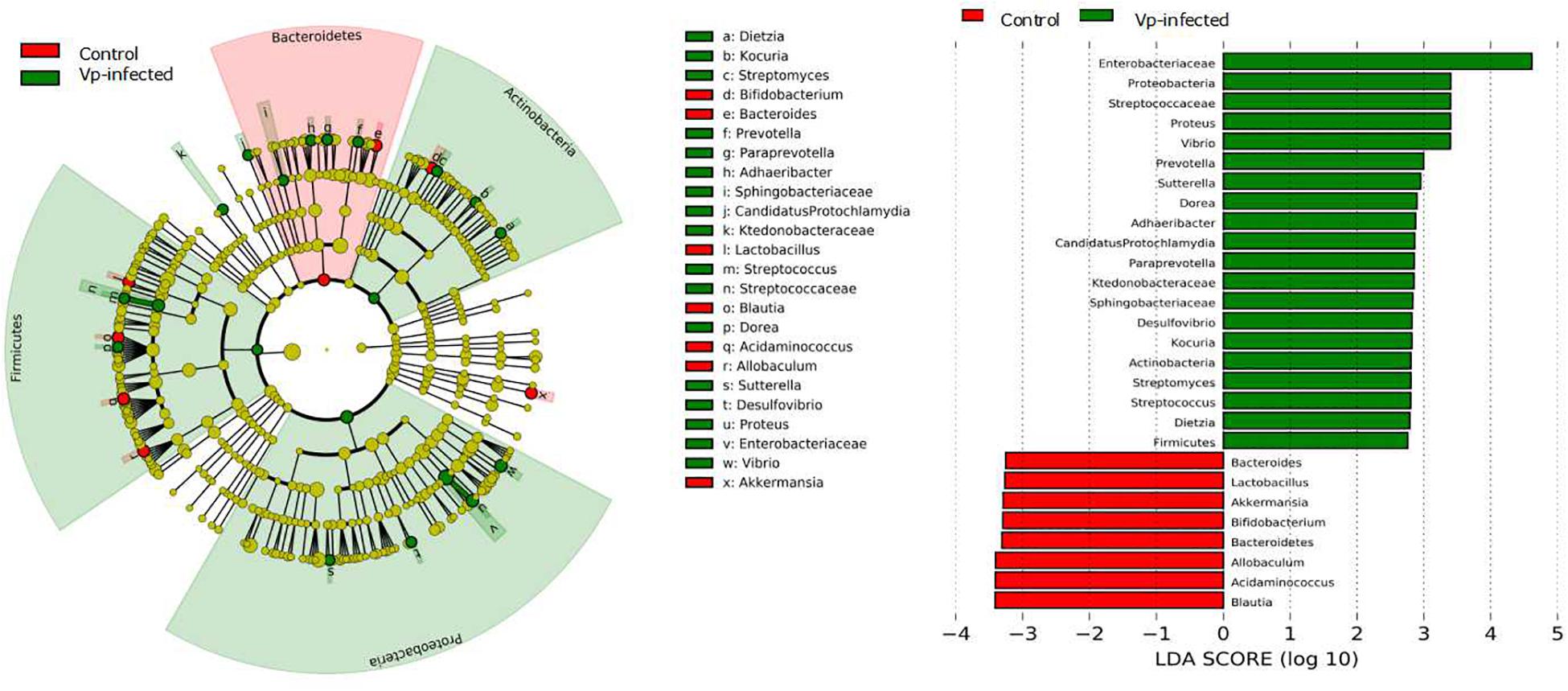
Figure 5. Identification of significant differences in bacterial taxa between Vp-infected and control groups. Cladogram shows the differences and phylogenic location. In each section, the diameter of the circle is proportional to the abundance of the taxon. Ileum microbial communities from Vp-infected and control mice were compared using LEfSe (red = taxon significantly enriched in control; green = taxon significantly enriched in Vp-infected; yellow = not-significant). Histogram of the LDA scores computed for features differentially abundant between the Vp-infected and control groups. LEfSe scores represent the degree of consistent difference in relative abundance between features in two groups of analyzed microbial communities. The clades of the histogram identify statistical and biological differences between the communities. Taxonomic groups showing LDA scores >2.0 with p < 0.05.
To evaluate whether alterations in gut-bacterial-mediated functions were caused by infection, we compared predicted bacterial KEGG pathways between the control and Vp-infected mice. The results showed that KEGG pathways were significantly influenced by Vp infection (Figure 6). In particular, KEGG pathways known to be involved in pathogen infection, such as infectious diseases (e.g., V. cholerae infection, V. cholerae pathogenic cycle, bacterial invasion of epithelial cells and epithelial cell signaling in Helicobacter pylori infection), ion channel pores, bacterial toxins and the cAMP signaling pathway were significantly induced following Vp infection. In contrast, metabolic functional processes (e.g., drug metabolism-other enzymes, carbohydrate/energy/biotin/butanoate/fatty acid metabolism), digestive activities (e.g., bile secretion, carbohydrate digestion and absorption), cell motility, cell growth and death and genetic information processing, exhibited an opposite pattern.

Figure 6. Predicted KEGG pathways of mice gut microbiota. Changes in the abundance of KEGG metabolic pathways in the Vp-infected group relative to the control group at 12 h. The red color intensity indicates the richness of metabolic pathways.
The Vp pathogen has become more widespread in recent years with more than 50 million people in estuarine areas at risk of Vp infection from unsafe food (Xu et al., 1994; Shinoda, 2011; Romilio et al., 2017; Lee et al., 2018). Although the relationship between virulence factors and pathogenicity is recognized (Vongxay et al., 2008; Hiyoshi et al., 2010; Broberg et al., 2011), we are far from delineating the pathogenesis of Vp in the host intestine. More recent research has reported that the gut microbiota is altered in response to enteric pathogen infections and diseases (Lofgren et al., 2011; Zhang et al., 2012; Tuomisto et al., 2014). Hence, an improved understanding of how the host gut microbiota affects Vp pathogenicity could support the design of better infection prevention methods.
In this study, differences in damage caused by Vp in PGF mice and normal mice were investigated to better understand the role of gut microbiota during Vp infection. In normal mice, inflammation (Figure 3) and damage to the intestines (Figure 1), liver and kidney (Figure 2) were more marked than in PGF mice. Other studies have shown that exogenous enteric pathogens can induce dysbiosis of the gut microbiota resulting in reduced intestinal resistance to harmful bacteria, and increased intestinal inflammation and permeability, providing an opportunity for a systemic infection by these pathogens (Wang et al., 2011; Kamada et al., 2012). Clostridium difficile infected patients usually show decreased diversity of gut microbiota and increased facultative anaerobes in the intestines, which together may promote peptic ulcers, gastroenteritis and colitis (Zalig and Rupnik, 2014). It is suggested (Anonymous, 2012) that Salmonella could interact with gut microbiota, resulting in dysbiosis and intestinal inflammation, thus providing a growth advantage for Salmonella. The resulting damage to the intestinal barrier, could, allow bacterial toxins and pathogens to enter the body, leading to a systemic infection. Thus, we hypothesize that the normal steady-state network interactions between intestinal flora is destabilized by the increased number of Vp, resulting in marked changes to gut microbiota diversity and abundance. This together with the pathogenic Vp itself leads to the induction of markers of both inflammation and liver and kidney damage in our infected mice.
Although large numbers of microbes reside normally in the host intestinal tract, many species of which are beneficial to the host body, there is potential for some undesirable microbiota to affect the intestinal immune system (Furusawa et al., 2013) and cause inflammatory reactions in the host (Garrett et al., 2010). It was apparent that the diversity (richness and composition) of gut microbiota in the ileum declined during Vp infection (Table 2), indicating that Vp can displace some lineages such as Bacteroidetes and Verrucomicrobia (Figure 4B). According to the diversity resistance hypothesis, a less diverse community has a reduced chance of harboring species with antagonistic traits toward pathogens and vice versa (Kennedy et al., 2002). Also, where gut bacterial diversity is reduced, the vacant niches may be taken up by invading pathogens (Dillon et al., 2005; Mallon et al., 2015; Nie et al., 2017). Similarly, Chen et al. (2011) reported that a reduced abundance of Bacteroidetes in parallel with an increase in Proteobacteria and Fusobacteria species in samples of gut microbiota taken from a diseased host were attended with a reduced gut bacterial diversity compared with healthy controls. 16S rRNA gene sequencing revealed a change in 32.7% of ileal OTUs following Vp infection (Figure 4A), which was significantly different from the control group (Figure 4D). Thus, Vp infections can substantially alter the richness and composition of the bacterial community in the mouse gut, which may create niches for colonization by pathogens because of reduced competition, which, in turn, could possibly induce more serious inflammation, gut epithelial damage, impediment to the growth of the good resident species and proliferation of indigenous opportunistic pathogenic bacteria (Thiennimitr et al., 2011; Ayres et al., 2012; Kamada et al., 2013). Besides, dysbiosis of microbiota composition has been shown to play a contributory role in the pathogenesis and progression of liver and kidney diseases (Chen et al., 2011). This assertion is supported by the present finding that following Vp gavage, normal mice showed higher levels of markers for liver and kidney damage than the PGF mice, possibly due to altered metabolic pathways in the normal gut microbiota resulting in variation in the composition and richness of gut microbiota species.
Given that microbiota in the host gut are sensitive to Vp infection, we identified some microbiota that could potentially be used as biomarkers indicative of infection. Notably, seven normally present OTUs were absent in the Vp-infected group (Figure 5). Among them, affiliated with Lactobacillus, Bifidobacterium, and Akkermansia, are probiotics, which have been shown to protect the host from mucosal inflammation by stimulating the anti-inflammatory cytokine IL-10 or down-regulating inflammatory cytokines (Shang Q.S. et al., 2017). The relative abundance of Lactobacillus was significantly decreased compared with Bifidobacterium and Akkermansia following Vp infection (Figure 4C). Lactobacillus can generate reactive oxygen species and produce bacteriocin molecules capable of killing pathogenic microbes (Jones et al., 2012). Other studies have shown this potential to restore gut function by stimulating mucosal and humoral immune responses to eliminate enteropathogens (Wardwell et al., 2011; Berrilli et al., 2012). Lactobacillus species also play an important protective role in Salmonella enterica infection (Burkholder and Bhunia, 2009). We therefore anticipate that Lactobacillus might be used as the biomarker indicating degree of protection against Vp infection. Other protective species may be Bacteroides, Blautia, and Allobacullum, which are involved in the production of short-chain fatty acids, antibacterial peptides and bile acids, all of which can support intestinal resistance to pathogenic bacteria (Kang et al., 2013; Zhang et al., 2016). In contrast, 17 OTUs were increased in response to Vp infection. Among them, Enterobacteriaceae including pathogenic species such as E. coli, Shigella and Salmonella (Lupp et al., 2007), Proteus, Prevotella and Sutterella have been linked to inflammation and disease (He et al., 2013; Zackular et al., 2013; Zhang et al., 2015), and in particular Prevotella correlates with the severity of liver and kidney diseases. Prevotella was significantly enriched during Vp infection in the mice in our study (Figure 5). This is consistent with previous studies that have suggested the Enterobacteriaceae family are more likely to increase as a result of colonization by enteric pathogens (Stecher et al., 2010; David et al., 2015). Besides, when the gut microbiota is perturbed so that it displays an abundance of the pathobiont Prevotella, this, along with induction of inflammation, is considered a cardinal marker of Entamoeba histolytica infection (Burgess and Petri, 2016). We hypothesize that Prevotella increased susceptibility to and colonization by Vp and therefore it has a strong correlation with Vp infection. In summary, both emergence or absence of intestinal bacterial communities can be associated with disease (Vigne et al., 2013; Xiong et al., 2015) and our microbiome profiling offered some feasible biomarkers for the diagnosis of Vp infection. If humans are infected with Vp, apart from using bacteriophage therapy to control and inhibit the virulence of Vibrio species (Letchumanan et al., 2016), adjusting the biomarker proportions of the species could also be used as a treatment.
There is evidence that changes in intestinal microbiota can significantly alter the biological functioning of the gut bacterial community in the host (Zhu et al., 2016). Consistent with this supposition, we found that many bacterial KEGG pathways were significantly different between the controls and Vp-infected mice (Figure 6). Infectious disease pathways, such as those used in V. cholerae and H. pylori infection, bacterial invasion of epithelial cells and epithelial cell signaling, were significantly enriched in Vp-infected mice compared to the controls. In contrast, the pathways involved in metabolism, digestion and cell growth decreased after Vp infection. Similarly, responses between infectious diseases and energy metabolism pathways were divergent in the gut microbiota of Vibrio-infected shrimp (Zhu et al., 2016). It has been proposed that changes in metabolic pathways are correlated with variations in their functions (Letchumanan et al., 2017). Therefore, Vp colonization markedly altered gut microbial function in the mice resulting in the effects on the progression of infection.
In conclusion, this is the first attempt to explore the pathogenicity of Vp and its role in modulating gut microbiota. This study suggests that shifts in the make-up and quantity of gut microbiota populations play a potentially pathogenic role in Vp infection. These changes in the gut bacterial community lead to a change in gut microbe functions. This provides new insight for targeting interventions in response to changes in gut microbiota and improved understanding and prognosis of Vp infection. From a microbial ecological point of view, the findings reported here significantly improve our knowledge of how gut microbiota respond to Vp infection. However, the data in this study are derived from just a single sampling 12 h after infection, whereas mice intestinal communities are dynamic and change over time during the different stages of infection. Thus, a time-series design with multiple samplings is required to verify these ecological patterns in a real infection process.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
The animal study was reviewed and approved by the College of Food Science and Technology, Guangdong Ocean University.
RW, LS, and YW designed the study. RW and YD performed the gut microbiota experiments and wrote the manuscript. QD and DS performed the pseudo germ-free mouse model. ZF and YW performed the statistical analyses. RG provided advice and revised the manuscript.
This research was supported by grants from the National Science Foundation (No. 31371746) and the Guangdong Provincial Special Fund for Technological Innovation of Modern Agricultural Industry (No. 2019KJ151).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Anonymous (2012). Gut microbiota: Salmonella has a growth advantage in the inflamed gut. Nat. Rev. Gastroenterol. Hepatol. 9:5. doi: 10.1038/nrgastro.2011.227
Ayres, J. S., Trinidad, N. J., and Vance, R. E. (2012). Lethal inflammasome activation by a multidrug-resistant pathobiont upon antibiotic disruption of the microbiota. Nat. Med. 18, 799–806. doi: 10.1038/nm.2729
Berrilli, F., Di, C. D., Cavallero, S. D., and Amelio, S. (2012). Interactions between parasites and microbial communities in the human gut. Front. Cell Infect. Microbiol. 2:141. doi: 10.3389/fcimb.2012.00141
Broberg, C. A., Calder, T. J., and Orth, K. (2011). Vibrio parahaemolyticus cell biology and pathogenicity determinants. Microb. Infect. 13, 992–1001. doi: 10.1016/j.micinf.2011.06.013
Burgess, S. L., and Petri, W. A. (2016). The intestinal bacterial microbiome and E. histolytica infection. Curr. Trop. Med. Rep. 3, 71–74. doi: 10.1007/s40475-016-0083-1
Burkholder, K. M., and Bhunia, A. K. (2009). Salmonella enterica serovar Typhimurium adhesion and cytotoxicity during epithelial cell stress is reduced by Lactobacillus rhamnosus GG. Gut Pathog. 1:14. doi: 10.1186/1757-4749-1-14
Caporaso, J. G., Kuczynski, J., Stombaugh, K., Bittinger, F. D., Bushman, E. K., Costello, N., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Met. 7, 335–336. doi: 10.1038/NMETH.F.303
Chen, Y., Yang, F., Lu, H., Wang, B. H., Chen, Y. B., Lei, D. J., et al. (2011). Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology 54, 562–572. doi: 10.1002/hep.24423
Clarke, T. B., Davis, K. M., Lysenko, E. S., Zhou, A. Y., Yu, Y. M., and Weiser, J. N. (2010). Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat. Med. 16, 228–231. doi: 10.1038/nm.2087
Curtis, M. M., Hu, Z., Klimko, C., Narayanan, S., Deberardinis, R., and Sperandio, V. (2014). The gut commensal Bacteroides thetaiotaomicron exacerbates enteric infection through modification of the metabolic landscape. Cell Host Microb. 16, 759–769. doi: 10.1016/j.chom.2014.11.005
Daniels, N. A., MacKinnon, L., Bishop, R., Altekruse, S., Ray, B., Hammond, M., et al. (2000). Vibrio parahaemolyticus infections in the United States, 1973-1998. J. Infect. Dis. 181, 1661–1666. doi: 10.1086/315459
David, L. A., Weil, A., Ryan, E. T., Calderwood, S. B., and Harris, J. B. (2015). Gut microbial succession follows acute secretory diarrhea in humans. mBio 6, 1–14. doi: 10.1128/mBio.00381-15
Dean, P. (2011). Functional domains and motifs of bacterial type III effector proteins and their roles in infection. FEMS Microbiol. Rev. 35, 1100–1125. doi: 10.1111/j.1574-6976.2011.00271.x
Dillon, R. J., Vennard, C. T., Buckling, A., and Charnley, A. K. (2005). Diversity of locust gut bacteria protects against pathogen invasion. Ecol. Lett. 8, 1291–1298. doi: 10.1111/j.1461-0248.2005.00828.x
Fraher, M. H., O’Toole, P. W., and Quigley, E. M. M. (2012). Techniques used to characterize the gut microbiota: a guide for the clinician. Nat. Rev. Gastroenterol. Hepatol. 9, 312–322. doi: 10.1038/nrgastro.2012.44
Furusawa, Y., Obata, Y., Fukuda, S., Endo, T. A., Nakato, G., Takahashi, D., et al. (2013). Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450. doi: 10.1038/nature12721
Garrett, W. S., Gordon, J. I., and Glimcher, L. H. (2010). Homeostasis and inflammation in the intestine. Cell 140, 859–870. doi: 10.1016/j.cell.2010.01.023
Ge, X., Zhao, W., Ding, C., Tian, H., Xu, L., Wang, H., et al. (2017). Potential role of fecal microbiota from patients with slow transit constipation in the regulation of gastrointestinal motility. Sci. Rep. 7:441. doi: 10.1038/s41598-017-00612-y
Gordon, H. A., and Pesti, L. (1971). The gnotobiotic animal as a tool in the study of host microbial relationships. Bacteriol. Rev. 35, 390–429. doi: 10.1128/mmbr.35.4.390-429.1971
Grover, M., and Kashyap, P. C. (2014). Germ-free mice as a model to study effect of gut microbiota on host physiology. Neurogastroenterol. Motil. 26, 745–748. doi: 10.1111/nmo.12366
He, X., Marco, M. L., and Slupsky, C. M. (2013). Emerging aspects of food and nutrition on gut microbiota. J. Agric. Food Chem. 61, 9559–9574. doi: 10.1021/jf4029046
Hiyoshi, H., Kodama, T., Iida, T., and Honda, T. (2010). Contribution of Vibrio parahaemolyticus virulence factors to cytotoxicity, enterotoxicity and lethality in mice. Infect. Immun. 78, 1772–1780. doi: 10.1128/IAI.01051-09
Hooper, L. V., and Gordon, J. I. (2001). Commensal host-bacterial relationships in the gut. Science 292, 1115–1118. doi: 10.1126/science.1058709
Hooper, L. V., Littman, D. R., and Macpherson, A. J. (2012). Interactions between the microbiota and the immune system. Science 336, 1268–1273. doi: 10.1126/science.1223490
Jones, R. M., Mercante, J. W., and Neish, A. S. (2012). Reactive oxygen production induced by the gut microbiota: pharmacotherapeutic implications. Curr. Med. Chem. 19, 1519–1529. doi: 10.2174/092986712799828283
Kamada, N., Kim, Y. G., Sham, H. P., Vallance, B. A., Puente, J. L., Martens, E. C., et al. (2012). Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science 336, 1325–1329. doi: 10.1126/science.1222195
Kamada, N., Chen, G. Y., Inohara, N., and Núnez, G. (2013). Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 14, 685–690. doi: 10.1038/ni.2608
Kampmann, C., Dicksved, J., Engstrand, L., and Rautelin, H. (2016). Composition of human faecal microbiota in resistance to Campylobacter infection. Clin. Microbiol. Infect. 22, 61.e1–61.e8. doi: 10.1016/j.cmi.2015.09.004
Kang, C. S., Ban, M., Choi, E. J., Moon, H. G., Jeon, J. S., Kim, D. K., et al. (2013). Extracellular vesicles derived from gut microbiota, especially Akkermansia muciniphila, protect the progression of dextran sulfate sodium-induced colitis. PLoS One 8:e76520. doi: 10.1371/journal.pone.0076520
Kennedy, T. A., Naeem, S., Howe, K. M., Knops, J. M. H., Tilman, D., and Reich, P. (2002). Biodiversity as a barrier to ecological invasion. Nature 417, 636–638. doi: 10.1038/nature00776
Langille, M. G., Zaneveld, G., Caporaso, J. G., McDonald, D., Knights, D., Reyes, J. A., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821. doi: 10.1038/nbt.2676
Lee, L. H., Mutalib, A. B., Law, J. W., Wong, S. H., and Letchumanan, V. (2018). Discovery on antibiotic resistance patterns of Vibrio parahaemolyticus in selangor reveals carbapenemase producing Vibrio parahaemolyticus in marine and freshwater fish. Front. Microbiol. 9:2513. doi: 10.3389/fmicb.2018.02513
Lee, S. H., An, J. H., Lee, H. J., and Jung, B. H. (2012). Evaluation of pharmacokinetic differences of acetaminophen in pseudo germ-free rats. Biopharm. Drug Dispos. 33, 292–303. doi: 10.1002/bdd.1799
Letchumanan, V., Chan, K., and Lee, L. (2014). Vibrio parahaemolyticus: a review on the pathogenesis, prevalence, and advance molecular identification techniques. Front. Microbiol. 5:705. doi: 10.3389/fmicb.2014.00705
Letchumanan, V., Chan, K. G., Pusparajah, P., Saokaew, S., Duangjai, A., Goh, B. H., et al. (2016). Insights into bacteriophage application in controlling Vibrio species. Front. Microbiol. 7:1114. doi: 10.3389/fmicb.2016.01114
Letchumanan, V., Chan, K. G., Khan, T. M., Bukhari, S. I., Ab Mutalib, N. S., Goh, B. H., et al. (2017). Bile sensing: the activation of Vibrio parahaemolyticus virulence. Front. Microbiol. 8:728. doi: 10.3389/fmicb.2017.00728
Ling, Z., Liu, X., Jia, X., Cheng, Y., Luo, Y., Yuan, L., et al. (2014). Impacts of infection with different toxigenic Clostridium difficile strains on faecal microbiota in children. Sci. Rep. 4:7485. doi: 10.1038/srep07485
Liu, H., Wu, B., Pan, G., He, L., Li, Z., Fan, M., et al. (2012). Metabolism and pharmacokinetics of mangiferin in conventional rats, pseudo-germ-free rats, and streptozotocin-induced diabetic rats. Drug Metab. Dispos. 40, 2109–2118. doi: 10.1124/dmd.112.045849
Li, L. Z., Meng, H. M., Gu, D., Li, Y., and Jia, M. D. (2019). Molecular mechanisms of Vibrio parahaemolyticus pathogenesis. Microbiol. Res. 222, 43–51. doi: 10.1016/j.micres.2019.03.003
Lofgren, J. L., Whary, M. T., Ge, Z., Muthupalani, S., Taylor, N. S., Mobley, M., et al. (2011). Lack of commensal flora in Helicobacter pylori-infected INS-GAS mice reduces gastritis and delays intraepithelial neoplasia. Gastroenterology 140, 210–220. doi: 10.1053/gastro.2010.09.048
Lupp, C., Robertson, M. L., Wickham, M. E., Sekirov, I., Champion, O. L., Gaynor, E. C., et al. (2007). Host-mediated inflammation disrupts the intestinal microbiota and the overgrowth of Enterobacteriaceae. Cell Host Microb. 2, 119–129. doi: 10.1016/j.chom.2007.06.010
Mallon, C. A., Elsas, J. D., and Salles, J. F. (2015). Microbial invasions: the process, patterns, and mechanisms. Trends Microbiol. 23, 719–729. doi: 10.1016/j.tim.2015.07.013
Matsumoto, C., Okuda, J., Ishibashi, M., Iwanaga, M., Garg, P., Ramamurthy, T., et al. (2000). Pandemic spread of an O3:K6 clone of Vibrio parahaemolyticus & emergence of related strains evidenced by Arbitrarily Primed PCR & toxRS sequence analysis. J. Clin. Microbiol. 38, 578–585. doi: 10.1128/jcm.38.2.578-585.2000
Mertens, A., Nagler, J., Hansen, W., and Gepts-Friedenreich, E. (1979). Halophilic, lactose-positive Vibrio in a case of fatal septicemia. J. Clin. Microbiol. 9, 233–235.
Midani, F. S., Weil, A. A., Chowdhury, F., Begum, Y. A., Khan, A. I., Debela, M. D., et al. (2018). Human gut microbiota predicts susceptibility to Vibrio cholerae infection. J. Infect. Dis. 218, 645–653. doi: 10.1093/infdis/jiy192
Newton, A., Kendall, M., Vugia, D. J., Henao, O. L., and Mahon, B. E. (2012). Increasing rates of vibriosis in the United States, 1996-2010: review of surveillance data from 2 systems. Clin. Infect. Dis. 54, S391–S395. doi: 10.1093/cid/cis243
Nie, L., Zhou, Q. J., Qiao, Y., and Chen, J. (2017). Interplay between the gut microbiota and immune responses of ayu (Plecoglossus altivelis) during Vibrio anguillarum infection. Fish Shellfish Immunol. 68, 479–487. doi: 10.1016/j.fsi.2017.07.054
Parks, D. H., Tyson, G. W., Hugenholtz, P., and Beiko, R. G. (2014). STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30, 3123–3124. doi: 10.1093/bioinformatics/btu494
Qin, J. J., Li, R. Q., Raes, J., Arumugam, M., and Burgdorf, K. S. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65. doi: 10.1038/nature08821
Ramette, A. (2007). Multivariate analyses in microbial ecology. FEMS Microbiol. Ecol. 62, 142–160. doi: 10.1111/j.1574-6941.2007.00375.x
Raghunath, P. (2015). Roles of thermostable direct hemolysin (TDH) and TDH-related hemolysin (TRH) in Vibrio parahaemolyticus. Front. Microbiol. 5:805. doi: 10.3389/fmicb.2014.00805
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahé, C. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584
Romilio, T. E., Katherine, G., and Nicolas, P. (2017). Insight into the origin and evolution of the Vibrio parahaemolyticus pandemic strain. Front. Microbiol. 8:1397. doi: 10.3389/fmicb.2017.01397
Ryan, W. J. (1976). Marine Vibrios associated with superficial septic lesions. J. Clin. Pathol. 29, 1014–1015. doi: 10.1136/jcp.29.11.1014
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genom. Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60
Shang, Q., Song, G., Zhang, M., Shi, J., Xu, C., Hao, J., et al. (2017). Dietary fucoidan improves metabolic syndrome in association with increased Akkermansia population in the gut microbiota of high-fat diet-fed mice. J. Funct. Foods 28, 138–146. doi: 10.1016/j.jff.2016.11.002
Shang, Q. S., Sun, W. X., Shan, X. D., Jiang, H., Cai, C., Hao, J. J., et al. (2017). Carrageenan-induced colitis is associated with decreased population of anti-inflammatory bacterium, Akkermansia muciniphila, in the gut microbiota of C57BL/6J mice. Toxicol. Lett. 279, 87–95. doi: 10.1016/j.toxlet.2017.07.904
Shinoda, S. (2011). Sixty years from the discovery of Vibrio parahaemolyticus and some recollections. Biocontrol. Sci. 16, 129–137. doi: 10.4265/bio.16.129
Singh, P., Teal, T. K., Marsh, T. L., Tiedje, J. M., Mosci, R., Jernigan, K., et al. (2015). Intestinal microbial communities associated with acute enteric infections and disease recovery. Microbiome 3:45. doi: 10.1186/s40168-015-0109-2
Stecher, B., Chaffron, S., Käppeli, R., Hapfelmeier, S., and Freedrich, S. (2010). Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS Pathog. 6:e1000711. doi: 10.1371/journal.ppat.1000711
Su, Y. C., and Liu, C. (2007). Vibrio parahaemolyticus: a concern of seafood safety. Food Microbiol. 24, 549–558. doi: 10.1016/j.fm.2007.01.005
Torres, M. E., Pirez, M. C., Schelotto, F., Varela, G., Parodi, V., Allende, F., et al. (2001). Etiology of children’s diarrhea in montevideo, uruguay: associated pathogens and unusual isolates. J. Clin. Microbiol. 39, 2134–2139. doi: 10.1128/JCM.39.6.2134-2139.2001
Turnbaugh, P. J., Backhed, F., Fulton, L., and Gordon, J. (2008). Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microb. 3, 213–223. doi: 10.1016/j.chom.2008.02.015
Thiennimitr, P., Winter, S. E., Winter, M. G., Xavier, M. N., Tolstikov, V., Huseby, D. L., et al. (2011). Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. PNAS 108, 17480–17485. doi: 10.1073/pnas.1107857108/-/DCSupplemental
Tuomisto, S., Pessi, T., Collin, P., Vuento, R., Aittonoemi, J., and Karhunen, P. J. (2014). Changes in gut bacterial populations and their translocation into liver and ascites in alcoholic liver cirrhotics. BMC Gastroenterol. 14:40. doi: 10.1186/1471-230X-14-40
Vigne, J. D., Lefèvre, C., and Patoumathis, M. (2013). Longitudinal analyses of gut mucosal microbiotas in ulcerative colitis in relation to patient age and disease severity and duration. J. Clin. Microbiol. 51, 849–856. doi: 10.1128/JCM.02574-12
Vongxay, K., Wang, S., Zhang, X., Wu, B., Hu, H., Pan, Z., et al. (2008). Pathogenetic characterization of Vibrio parahaemolyticus isolates from clinical and seafood sources. Int. J. Food Microbiol. 126, 71–75. doi: 10.1016/j.ijfoodmicro.2008.04.032
Wang, R. D., Sun, L. J., Wang, Y. L., Deng, Y. J., Liu, Y., Xu, D. F., et al. (2016). Hemolytic activity of the metabolites produced by Vibrio parahaemolyticus ATCC33847 in five food matrices and their toxicity toward mice organs. Mod. Food Sci. Technol. 32, 48–53.
Wang, Z., Klipfell, E., Bennett, B. J., Koeth, R., Levison, B. S., DuGar, B., et al. (2011). Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57–63. doi: 10.1038/nature09922
Wardwell, L. H., Huttenhower, C., and Garrett, W. S. (2011). Current concepts of the intestinal microbiota and the pathogenesis of infection. Curr. Infect. Dis. Rep. 13, 28–34. doi: 10.1007/s11908-010-0147-7
Xiong, J., Wang, K., Wu, J., Qiuqian, L., Yang, K., Qian, Y., et al. (2015). Changes in intestinal bacterial communities are closely associated with shrimp disease severity. Appl. Microbiol. Biotech. 99, 6911–6919. doi: 10.1007/s00253-015-6632-z
Xu, M., Yamamoto, K., Honda, T., and Xu, M. (1994). Construction and characterization of an isogenic mutant of Vibrio parahaemolyticus having a deletion in the thermostable direct hemolysin-related hemolysin gene (trh). J. Bacteriol. 176, 4757–4760. doi: 10.1128/jb.176.15.4757-4760.1994
Yarza, P., Yilmaz, P., Pruesse, E., Glöckner, F. O., Ludwig, W., Schleifer, K. H., et al. (2014). Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 12, 635–645. doi: 10.1038/nrmicro3330
Zackular, J. P., Baxter, N. T., Iverson, K. D., Sadler, W. D., Petrosino, J. F., Chen, G., et al. (2013). The gut microbiome modulates colon tumorigenesis. mBio 4:e00692-13. doi: 10.1128/mBio.00692-13
Zalig, S., and Rupnik, M. (2014). Clostridium difficile infection and gut microbiota. Semin. Colon Rectal Surg. 25, 124–127. doi: 10.1053/j.scrs.2014.05.005
Zaura, E., Keijser, B. J. F., Huse, S. M., and Crielaard, W. (2009). Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 9:259. doi: 10.1186/1471-2180-9-259
Zarei, M., Borujeni, M. P., Jamnejad, A., and Khezrzadeh, M. (2012). Seasonal prevalence of Vibrio species in retail shrimps with an emphasis on Vibrio parahaemolyticus. Food Control 25, 107–109. doi: 10.1016/j.foodcont.2011.10.024
Zhang, H. L., Yu, L. X., Yang, W., Tang, L., Lin, Y., Wu, H., et al. (2012). Profound impact of gut homeostasis on chemically-induced pro-tumorigenic inflammation and hepatocarcinogenesis in rats. J. Hepatol. 57, 803–812. doi: 10.1016/j.jhep.2012.06.011
Zhang, J., Guo, Z., Xue, Z., Sun, Z., Zhang, M., Wang, L., et al. (2015). A phylo-functional core of gut microbiota in healthy young Chinese cohorts across lifestyles, geography and ethnicities. ISME J. 9, 1979–1990. doi: 10.1038/ismej.2015.11
Zhang, Z., Wu, X., Cao, S., Wang, L., Wang, D., Yang, H., et al. (2016). Caffeic acid ameliorates colitis in association with increased Akkermansia population in the gut microbiota of mice. Oncotarget 7:31790. doi: 10.18632/oncotarget.9306
Zhang, Z., Zhang, W. W., Hu, Z. G., Li, C. H., Shao, Y., Zhao, X. L., et al. (2019). Environmental factors promote pathogen-induced skin ulceration syndrome outbreak by readjusting the hindgut microbiome of Apostichopus japonicus. Aquaculture 507, 155–163. doi: 10.1016/j.aquaculture.2019.03.054
Keywords: pathogenesis, Vibrio parahaemolyticus infection, gut microbiota, 16S rRNA gene, gene sequencing
Citation: Wang R, Deng Y, Deng Q, Sun D, Fang Z, Sun L, Wang Y and Gooneratne R (2020) Vibrio parahaemolyticus Infection in Mice Reduces Protective Gut Microbiota, Augmenting Disease Pathways. Front. Microbiol. 11:73. doi: 10.3389/fmicb.2020.00073
Received: 05 August 2019; Accepted: 14 January 2020;
Published: 30 January 2020.
Edited by:
Leonard Peruski, Centers for Disease Control and Prevention (CDC), United StatesReviewed by:
Learn-Han Lee, Monash University Malaysia, MalaysiaCopyright © 2020 Wang, Deng, Deng, Sun, Fang, Sun, Wang and Gooneratne. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qi Deng, NDY1MDExOTQ3QHFxLmNvbQ==; Lijun Sun, c3VuY2FtdEAxMjYuY29t; Yaling Wang, d2FuZ3lsY2hpbmFAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.