
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 31 January 2020
Sec. Microbial Physiology and Metabolism
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.00059
This article is part of the Research Topic Acetogens - From The Origin Of Life To Biotechnological Applications View all 17 articles
The hydrogen-dependent carbon dioxide reductase is a soluble enzyme complex that directly utilizes hydrogen (H2) for the reduction of carbon dioxide (CO2) to formate in the first step of the acetyl-coenzyme A- or Wood-Ljungdahl pathway (WLP). HDCR consists of 2 catalytic subunits, a hydrogenase and a formate dehydrogenase (FDH) and two small subunits carrying iron-sulfur clusters. The enzyme complex has been purified and characterized from two acetogenic bacteria, from the mesophile Acetobacterium woodii and, recently, from the thermophile Thermoanaerobacter kivui. Physiological studies toward the importance of the HDCR for growth and formate metabolism in acetogens have not been carried out yet, due to the lack of genetic tools. Here, we deleted the genes encoding HDCR in T. kivui taking advantage of the recently developed genetic system. As expected, the deletion mutant (strain TKV_MB013) did not grow with formate as single substrate or under autotrophic conditions with H2 + CO2. Surprisingly, the strain did also not grow on any other substrate (sugars, mannitol or pyruvate), except for when formate was added. Concentrated cell suspensions quickly consumed formate in the presence of glucose only. In conclusion, HDCR provides formate which was essential for growth of the T. kivui mutant. Alternatively, extracellularly added formate served as terminal electron acceptor in addition to CO2, complementing the growth deficiency. The results show a tight coupling of multi-carbon substrate oxidation to the WLP. The metabolism in the mutant can be viewed as a coupled formate + CO2 respiration, which may be an ancient metabolic trait.
Acetogenic bacteria thrive on the production of acetic acid from H2 + CO2. As such, they are abundant in the environment (Drake et al., 2008), since they link primary fermenters and aceticlastic methanogens in the anoxic food chain (Schink and Stams, 2006). The metabolism of acetogens can be separated into three parts: substrate oxidation (oxidative branch), disposal of reducing equivalents (reductive branch) and redox balancing by electron-bifurcating hydrogenase and/or ferredoxin (Fd)-oxidizing, partly energy-conserving enzyme complexes (Schuchmann and Müller, 2016). H2 as electron donor in chemolithotrophic metabolism is primarily oxidized by an electron-bifurcating hydrogenase (Schuchmann and Müller, 2012) providing NADH and reduced ferredoxin (Fdred) for CO2 reduction. The terminal electron accepting pathway is the presumably ancient Wood-Ljungdahl pathway (WLP; Ljungdahl, 1986; Wood et al., 1986), which is also important in anabolism, since a fraction of its product, acetyl-coenzyme A, is provided for growth. Energy conservation is tightly coupled to redox balancing, since a part of the Fdred is oxidized by energy-conserving membrane-bound enzyme complexes (Schuchmann and Müller, 2014), by the Rnf complex, as present in e.g., Acetobacterium woodii (Biegel and Müller, 2010), or by energy-converting hydrogenases Ech encoded in the genome of Moorella thermoacetica (Pierce et al., 2008) or as present in Thermoanaerobacter kivui (Hess et al., 2014; Schoelmerich and Müller, 2019).
The first step in the methyl-branch of the WLP is the reduction of CO2 to formate, which is catalyzed by a formate dehydrogenase (FDH). In some acetogenic microorganisms, FDH occurs in a complex with a hydrogenase and two small subunits to form a hydrogen-dependent carbon dioxide reductase (HDCR) (Schuchmann and Müller, 2013). A distinct property of the soluble enzyme complex is the direct use of H2 as electron donor. Therefore, HDCR is the second H2-oxidizing hydrogenase in acetogenic catabolism besides the electron-bifurcating hydrogenase. The enzyme complex has been purified from two acetogenic bacteria, the mesophile Acetobacterium woodii (Schuchmann and Müller, 2013) and the thermophile Thermoanaerobacter kivui (Schwarz et al., 2018). HDCR contained four subunits, a hydrogenase, a formate dehydrogenase and two small subunits bearing iron-sulfur clusters, which are likely to be involved in electron transfer from the hydrogenase to the formate dehydrogenase. HDCR catalyzes formate-dependent hydrogen formation, as determined by the concentrations of H2, CO2 and formate. The reduction of CO2 to formate with hydrogen as electron donor is close to the thermodynamic equilibrium (E0′ [CO2/formate] = –432 mV; E0′ [2 H+/H2] = –414 mV), and HDCR catalyzed formate oxidation to CO2 at comparable rates. Moreover, the reactions were catalyzed with high turnover frequencies (TOF) of up to 101,600 h–1 in A. woodii and 10,000,000 h–1 in T. kivui, making HDCRs promising candidate enzymes for biotechnological applications such as H2 storage or H2 release from stored formate (Pereira, 2013; Schuchmann and Müller, 2013; Müller, 2019).
Many acetogens are metabolically versatile, and able to utilize electron donors other than H2, such as sugars, products of primary fermentations such as alcohols or C1 compounds (methanol, formate, CO), or methylated nitrogen compounds such as glycine betaine, in addition to H2 (Diekert and Wohlfarth, 1994; Schuchmann and Müller, 2016). Here, we studied the function of the HDCR complex in vivo using genetic tools (Basen et al., 2018), with focus on its role in the catabolic conversion of multi-carbon substrates. Our hypotheses were that (i) HDCR is essential in formate oxidation during growth on formate as sole substrate, since it is the only FDH annotated in the genome (Hess et al., 2014), and (ii) in heterotrophic metabolism, HDCR, its product formate and the Wood-Ljungdahl pathway are essential unless electrons are disposed elsewhere, e.g., as H2 through the reaction of the electron-bifurcating hydrogenase (Figure 1). Interestingly, the generation of a mutant, strain TKV_MB013, that lacked the genes predicted to encode for the subunits of HDCR, was only possible if formate was supplied in addition to sugars, and the phenotype of the strain was characterized in detail.
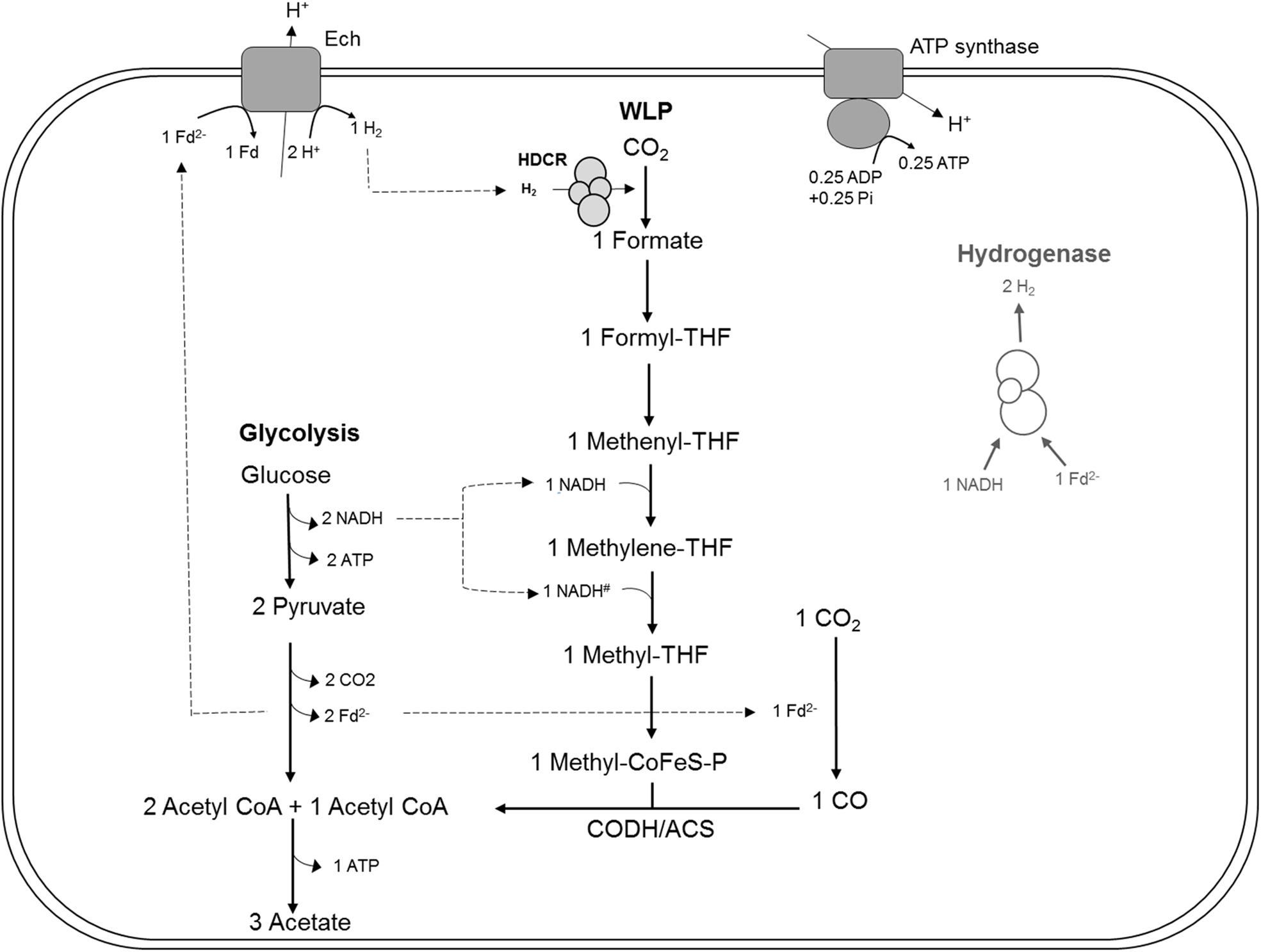
Figure 1. Bioenergetic model of glucose metabolism in T. kivui, modified after Hess et al. (2014), highlighting the assumed role of the hydrogen-dependent carbon dioxide reductase (HDCR) in heterotrophic growth. Re-oxidation of ferredoxin (Fdred) may be catalyzed by the electron-converting hydrogenase (Ech) complex, and evolved hydrogen (H2) may be re-oxidized by HDCR. Alternatively, it may be catalyzed in conjunction with NADH re-oxidation by electron-bifurcating hydrogenase HydABC (transparent, on the right), putatively making HDCR dispensable during heterotrophic growth of T. kivui. The stoichiometry of Ech and ATP synthase have been assumed to be 1 proton translocated per one H2 evolved in the methanogen Methanosarcina mazei (Welte et al., 2010), in the thermophilic archaeon Pyrococcus furiosus (Sapra et al., 2003) and in T. kivui (Schoelmerich and Müller, 2019). The ATP synthase of T. kivui is proton-dependent (Hess et al., 2014), and the ratio of 0.25 ATP per proton translocated is supported by thermodynamic calculations in acetogens (Schuchmann and Müller, 2014). #, The NADH-dependence of methylene-THF reductase has been assumed, based on the model of A. woodii and no biochemical evidence for electron-bifurcation. CoFeS-P, Corrinoid iron-sulfur protein.
As HDCR likely fulfills an essential function during growth on formate or on H2 + CO2, but potentially not during growth on sugars (Figure 1), we aimed to delete the genes encoding the four subunits forming the active enzyme, fdhF (TKV_c19990), hycB3 (TKV_c19980), hycB4 (TKV_c19970), and hydA2 (TKV_c19960) in T. kivui (Schwarz et al., 2018). These consecutive HDCR genes are part of a gene cluster that also contains a fifth gene, fdhD (TKV_c19950), presumably encoding a formate dehydrogenase maturation protein (Schwarz et al., 2018). FdhD was not deleted, since it was not identified as part of the enzyme complex in T. kivui. For ease of understanding, we refer to the four targeted genes fdhF, hycB3, hycB4, and hydA2 as the HDCR genes in the following. In order to create the HDCR genes deletion in T. kivui, plasmid pMBTkv012 was designed (Supplementary Figure S1), carrying approximately 1000 bp regions flanking the HDCR genes. Apart from these upstream (5′) and downstream (3′) flanking regions (UFR and DFR, respectively), the plasmid also contained the pyrE cassette as selectable marker, to be introduced into the pyrE-deficient uracil-auxotrophic strain TKV_MB002. The genetic system has been described in detail recently (Basen et al., 2018). In brief, we selected for uracil-prototrophs in the first selection round, and for the loss of the plasmid including pyrE with 5-fluoroorotic acid (5-FOA) in the second round of selection (Figure 2A), as described previously (Basen et al., 2018). Initially, we used glucose as only substrate, but after screening >50 colonies, we did not obtain any mutant lacking the HDCR gene cluster. In a second approach, we added formate (50 mM) in addition to glucose during the selection as it is the product of HDCR (Figure 1). We then obtained five genotypically “clean” HDCR deletion mutants out of 6 screened colonies/isolates as verified by PCR analysis after the second round of selection (Figure 2B), while the gene locus in the 6th picked colony likely reverted to the wild type gene locus. The markerless deletion of the genes encoding HDCR in mutant 5 was verified by sequencing and immunoblotting (Figure 2C), and the mutant was designated T. kivui strain TKV_MB013 (ΔpyrE, ΔfdhF hycB3 hycB4 hydA2). Unlike cell-free extracts of the wild type, cell-free extracts of strain TKV_MB013 neither carried out H2-dependent formate production nor formate-dependent H2 production (Figure 2D), an activity specific to the HDCR from T. kivui or A. woodii (Schuchmann and Müller, 2013; Schwarz et al., 2018). Taken together, all genetic and biochemical evidence suggest that T. kivui strain TKV_MB013 was devoid of the genes encoding HDCR.
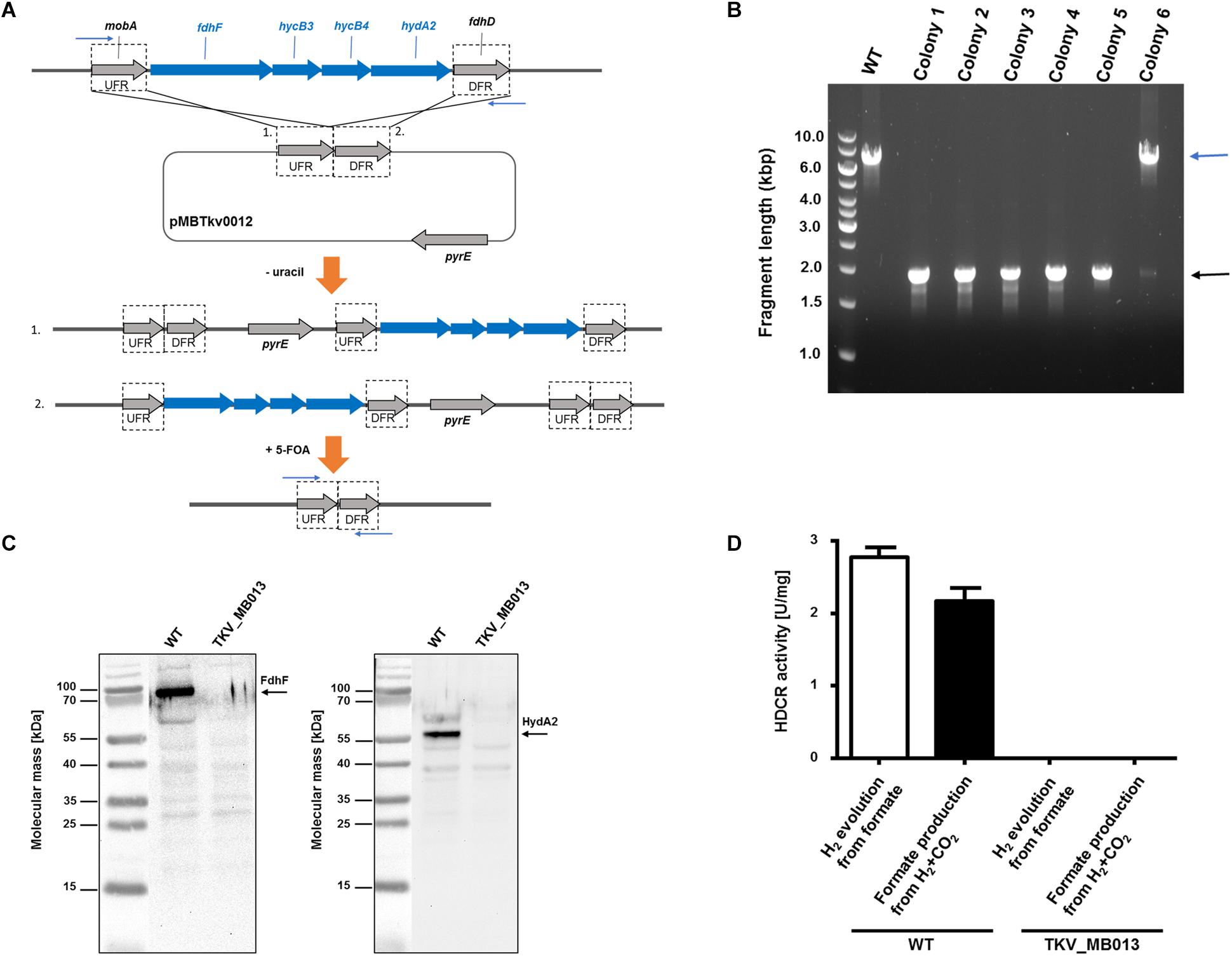
Figure 2. Deletion of the genes encoding HDCR, fdhF (TKV_c19990), hycB3 (TKV_c19980), hycB4 (TKV_c19970) and hydA2 (TKV_c19960). (A) Strategy for deletion using plasmid pMBTkv0012. 1. and 2. refer to insertion of the plasmid in the first round of selection on uracil prototrophy at the upstream flanking region (UFR) or at the downstream flanking region (DFR), respectively. (B) DNA fragments separated by agarose gel electrophoresis after PCR amplification of the HDCR gene locus, using primers outside the flanking regions (indicated as blue arrows). wild type, WT; colonies 1–6). (C) Detection of the HDCR subunits FdhF (left side) and HydA2 (right side) in cell free extracts of T. kivui DSM2030 (wild type, WT) or of T. kivui TKV_MB013. Cells were grown in complex medium (Leigh et al., 1981) with 28 mM glucose and 50 mM formate. 40 μg of cytoplasmic fractions were separated via denaturing gel electrophoresis, and then transferred to a nitrocellulose membrane. The presence of FdhF and HydA2 was determined immunologically with antibodies raised against corresponding His-tagged proteins, purified by affinity chromatography. For comparison of the molecular masses, the PageRuler® Prestained Protein Ladder (Thermo Scientific, Dreieich, Germany; left side of both images) was loaded onto the same gel, a picture of the membrane with and without chemiluminescence was taken (ChemoStar, INTAS, Göttingen, Germany), and both images were assembled using the ChemoStar TS software (INTAS, Göttingen, Germany). (D) Hydrogen evolution from formate (white bars) and formate formation from H2 + CO2 (black bars) of cytoplasmic fractions from T. kivui wild type (WT) and the HDCR deletion strain (T. kivui TKV_MB013). Cells were grown in complex medium with 28 mM glucose and 50 mM formate, and harvested in the late exponential growth phase. 0.3 mg of cytoplasmic protein was incubated in the reaction buffer (100 mM HEPES, 20 mM MgSO4, 0.0001% resazurin, 0.5 mM DTE, pH 7.0) at 64°C, and formate (150 mM) or H2 + CO2 (80:20 [v:v], 1.1 × 105 Pa) were added as substrate. H2 production from formate was measured in the gas phase, and formate production from H2 + CO2 was measured in the liquid phase (n = 3).
After deletion of the HDCR gene cluster and the absence of the protein in strain TKV_MB013 was confirmed, we analyzed the growth phenotype of the strain on all different substrates with and without formate as electron acceptor in addition to CO2. Initially, we tested its ability to utilize formate as sole electron donor. As expected, the strain did not grow when 300 mM formate was supplied as sole electron donor (Table 1) due to the absence of HDCR, the sole formate oxidizing enzyme encoded in the genome. The wild type grew to an optical density (OD600) of 0.22 ± 0.017. Second, we tested the ability of strain TKV_MB013 to grow chemolithoautotrophically with H2 + CO2, and this was also not observed (Figure 3A), due to the absence of HDCR as essential formate providing enzyme. In contrast, the wild type grew to an OD600 of 0.57 ± 0.02. To indisputably assign this growth deficiency to the loss of the HDCR genes, an ectopic insertion of the wild type genes fdhF, hycB3, hycB4, and hydA2 into the genome of the mutant strain TKV_MB013 was performed, resulting in strain TKV_MB019. The genes were inserted in between the convergent genes TKV_c24500 and TKV_c24520. The addition of the HDCR genes, controlled by the presumably strong promoter of the S-layer protein of T. kivui, complemented the growth effect, therefore we conclude that formate, provided by HDCR activity, is essential for chemolithoautotrophic growth of T. kivui on H2 + CO2. This was expected since formate is an intermediate in the methyl branch of the WLP. To prove this hypothesis, we added formate in addition to H2 + CO2. In that experiment, formate represented the electron acceptor in the methyl branch of the WLP, and indeed, it supported growth of strain TKV_MB013 on H2 + CO2 (Figure 3B), with a similar growth rate and final OD600 as the wild type.
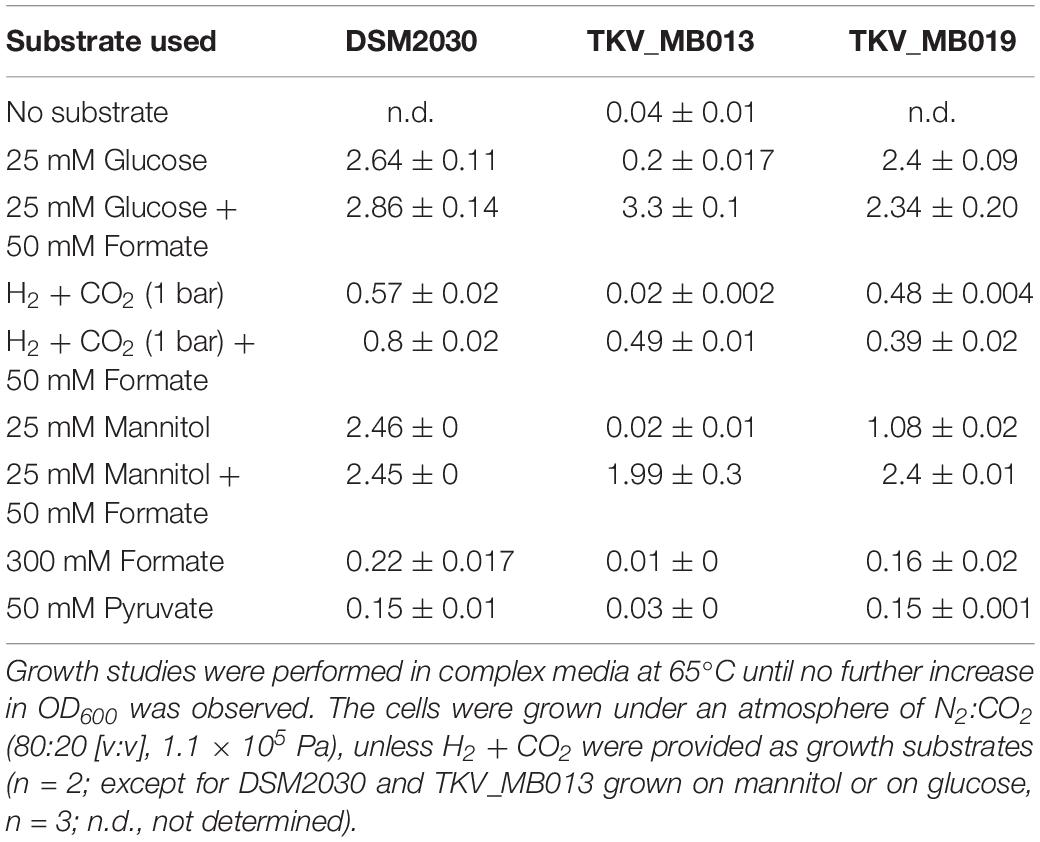
Table 1. Average maximal optical densities (OD600) from stationary phase cultures of T. kivui DSM2030, the mutant T. kivui TKV_MB013 lacking the genes encoding HDCR, and its daughter strain T. kivui TKV_MB019, with the HDCR encoding genes re-introduced into the genome.
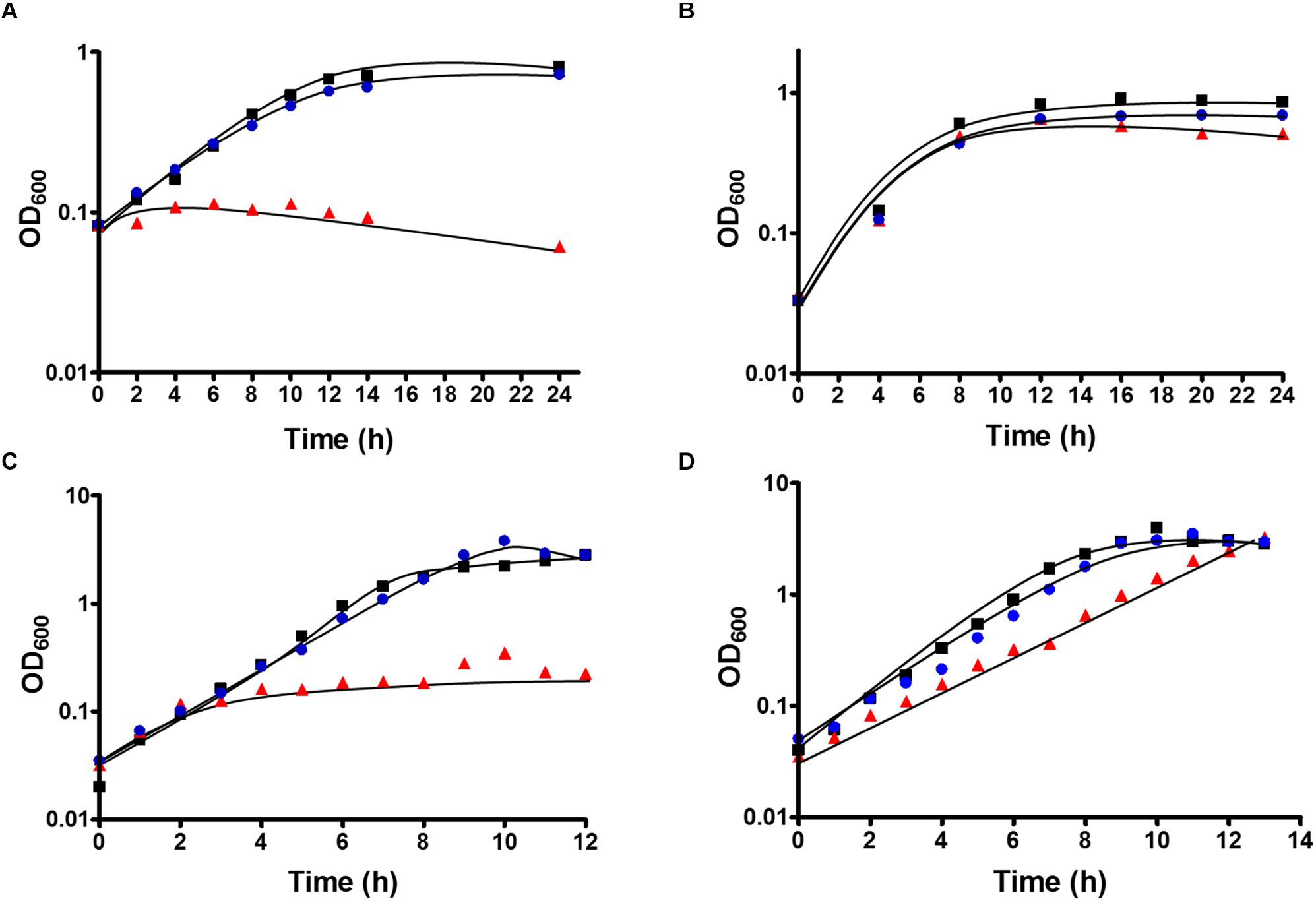
Figure 3. Growth of T. kivui strain TKV_MB013 on (A) H2 + CO2 (80:20 [v:v], 2 × 105 Pa), (B) H2 + CO2 (80:20 [v:v], 2 × 105 Pa) + 50 mM formate (C) 25 mM glucose or (D) 25 mM glucose + 50 mM formate. The cells were grown at 65°C in 100 ml serum bottles containing 25 ml of complex media under an atmosphere of N2:CO2 (80:20 [v:v], 1.1 × 105 Pa), unless H2 + CO2 were provided as growth substrates. TKV_MB013, red triangles; TKV_MB013 plus re-introduced HDCR genes in a different genome location, blue circles; or wild type, black squares. The growth experiments were performed in biological triplicates, and a representative growth curve is shown.
Interestingly, heterotrophic growth on glucose also depended on the presence of the HDCR genes. T. kivui strain TKV_MB013 only grew to an OD600 of 0.2 (Figure 3C), which was only slightly higher than growth without any substrate added (0.04 ± 0.01). In contrast, the wild type grew to an OD600 of 2.64 ± 0.01, which is comparable to what has been observed before (Basen et al., 2018). Again, the ectopic insertion of the HDCR genes into the genome of strain TKV_MB013 complemented the growth deficiency. While the addition of formate did not influence the wild type, formate again stimulated growth of T. kivui strain TKV_MB013 (Figure 3D), with a similar OD600 of 3.2 reached after 13 h. Interestingly, the addition of formate did not completely restore the growth behavior in strain TKV_MB013, since only a lower growth rate of 0.38 h–1 was reached (vs. 0.56 h–1 in the wild type). We then tested whether formate addition was essential for growth with all known electron donors for strain TKV_MB013, and this was indeed the case (Table 1). Unlike the wild type, T. kivui strain TKV_MB013 neither grew on fructose, mannose (data not shown), pyruvate nor on the recently identified novel substrate mannitol (Moon et al., 2019), to higher optical densities than 0.06, unless formate was added as electron acceptor (Table 1). This unambiguously showed that formate, produced by HDCR, and likely the complete WLP as terminal electron accepting pathway, is essential to growth of the acetogen T. kivui.
To study substrate consumption and product formation of T. kivui strain TKV_MB013, experiments with concentrated suspensions of resting cells were performed. The cells were concentrated to 10× in defined medium and subjected to a short-termed incubation at 65°C. As expected, when glucose was omitted from the medium and when formate was the sole electron donor, the latter was not consumed, due to the absence of HDCR, and no acetate was produced. When formate was provided as electron acceptor in addition to CO2, glucose was completely consumed (22.7 ± 1.9 mM), while the formate concentration decreased from 40.1 ± 2.9 to 19 ± 0.8 mM. Acetate (76.4 ± 3.2 mM) was the sole product (Figure 4A), while only a low concentration of H2 was detected in the headspace, corresponding to 0.3 mM, if all H2 was dissolved in the medium. Therefore, the results of the cell suspension experiments with the HDCR mutant strain TKV_MB013 reflected the ones from growth experiments. Formate strongly stimulated glucose consumption in the HDCR deficient T. kivui strain TKV_MB013, and formate consumption was strictly coupled to glucose consumption. The theoretically assumed stoichiometry based on glucose oxidation to acetate and CO2 and concomitant formate and CO2 reduction to acetate in the mutant strain, TKV_MB013 is depicted in eq. 1.
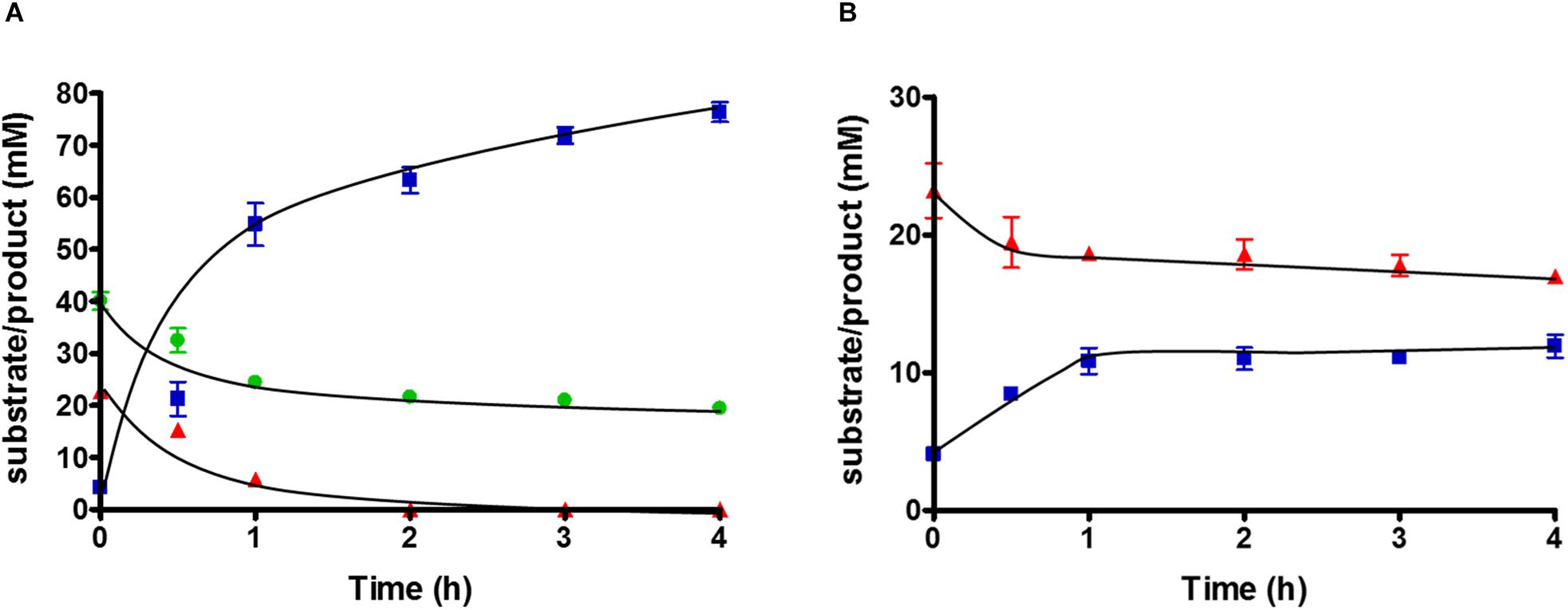
Figure 4. Substrate conversion by 10 ml of 10-fold concentrated cell suspensions of T. kivui strain TKV_MB013 (1 mg ml–1 protein), pre-grown with glucose and formate, with (A) glucose + formate or (B) glucose only. Glucose, red triangles; acetate, blue squares; formate, green circles. Experiments were performed in triplicate at 65°C in defined medium, under an atmosphere of N2:CO2 (80:20 [v:v], 1.1 × 105 Pa).
Carbon from glucose and formate was stoichiometrically recovered in acetate carbon, under the assumption that one CO2 was consumed for each formate consumed (109 ± 14%, n = 3). The electron balance, based on oxidation of glucose and reduction of formate and CO2 to acetate was nearly closed as well (99 ± 13%, n = 3). Therefore, we assume that no other products were present in high concentrations. The measured average acetate to glucose ratio was slightly higher than three (3.2 ± 0.3; n = 3), as expected. In conclusion, the observed conversion of glucose and formate to acetate by the HDCR mutant, T. kivui strain TKV_MB013 is nearly reflected by theoretically assumed stoichiometry (eq. 1), which reveals that formate served as electron acceptor for the oxidation of H2 and organic electron donors (Figure 5).
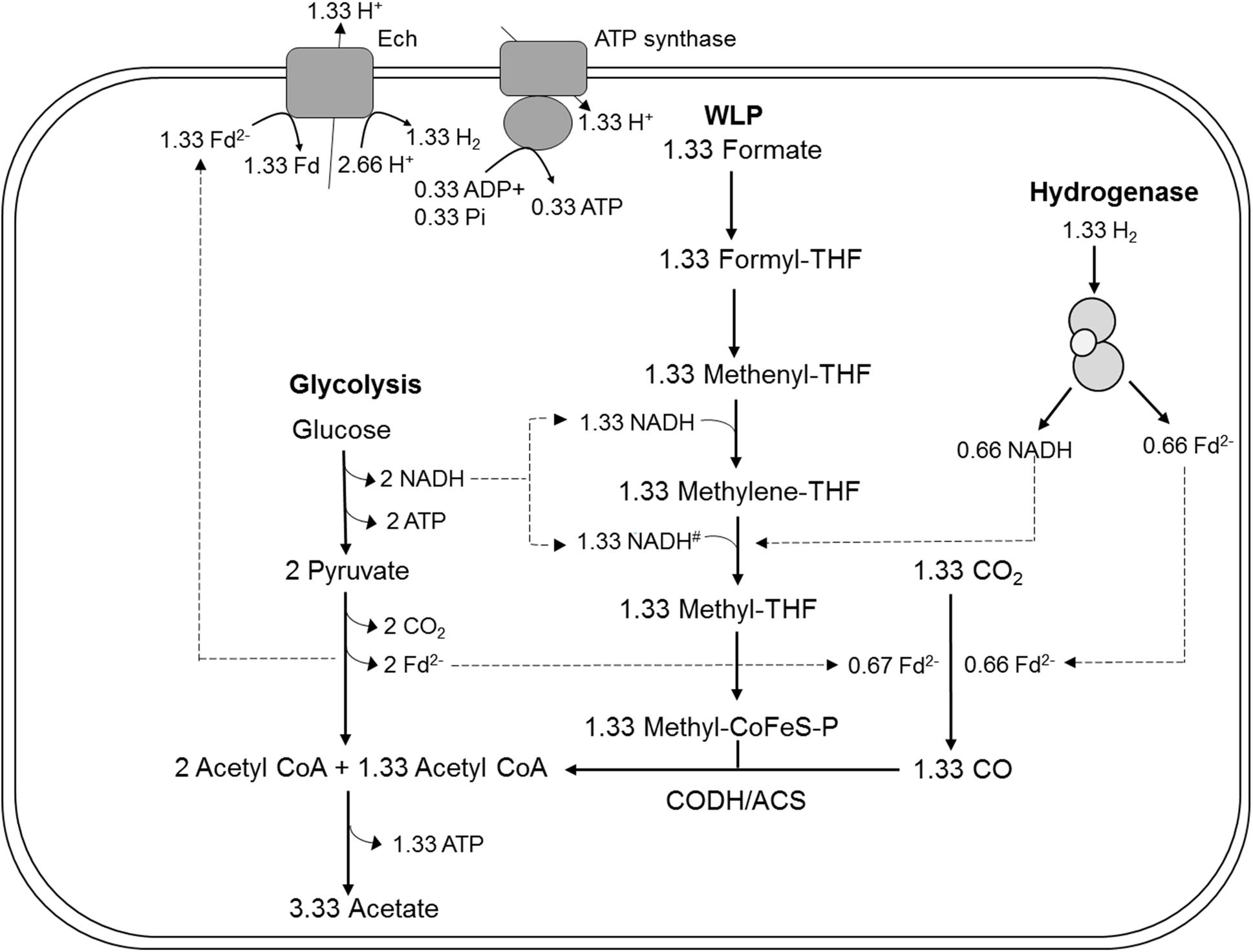
Figure 5. Model for acetogenesis from glucose with formate + CO2 as electron acceptors in T. kivui TKV_MB013 lacking the genes encoding HDCR. WLP, Wood-Ljungdahl-pathway; HDCR, hydrogen-dependent carbon dioxide reductase; Fd2–, reduced ferredoxin; Ech, electron-converting hydrogenase; CoFeS-P, corronoid iron-sulfur protein. For explanations the redox cofactor specificities and of the proposed stoichiometries, please see Figure 1.
Since acetogens may dispose electrons from glucose oxidation onto H+ to form H2, we tested the effect of omitting formate from the cell suspension experiments with T. kivui strain TKV_MB013. Glucose was consumed, but only from 23.2 ± 3.4 to 17 ± 0.8 mM, and acetate production stopped at a concentration of 11.9 ± 1.45 mM (Figure 4B). More H2 was produced (7.8 ± 0.3 mM, if all hydrogen was dissolved) than in the corresponding experiments with formate. This indicates some reducing equivalents (10.5%) from glucose oxidation were indeed channeled toward H2 when formate was omitted. Growth to higher OD600 than 0.2 (Figure 3C), however, was not observed.
Acetogens utilize C1-compounds of intermediate redox state such as formate or the methyl groups of methanol, methylamines or methoxylated compounds via the WLP (Kerby et al., 1983; Schuchmann and Müller, 2016). Formate-H2 interconversion, catalyzed by formate:H2 lyase (FHL), has been studied well in the facultative anaerobe Escherichia coli (Mcdowall et al., 2014; Trchounian and Sawers, 2014; Pinske and Sargent, 2016). In contrast to E. coli, acetogens utilize formate as sole source of energy and carbon. In principle, acetogens may utilize formate as electron acceptor and electron donor at the same time. Three mols of formate must be oxidized to provide sufficient (6) mols of reductant for the reduction of formate and of CO2 to one mol of acetate through the Wood-Ljungdahl pathway (Bertsch and Müller, 2015), according to:
Formate oxidation to CO2 is catalyzed by formate dehydrogenase (FDH). The T. kivui genome only contains one fdh copy, fdhF (TKV_c19990). Therefore, the deletion of the HDCR gene cluster including fdhF completely abolished its ability to thrive on formate as sole substrate and electron donor (Table 1), which may be different in other acetogens such as A. woodii or Treponema primitia (Matson et al., 2010) that contain several fdh copies, or genes encoding FHLs in addition to fdh genes (Poehlein et al., 2015).
Acetogens catalyze formate reduction in the methyl branch of the WLP. Formate is provided by FDH, which is bound to hydrogenase in the HDCR complex in at least two acetogens, A. woodii and T. kivui. So, in contrast to the membrane-bound FHL that primarily oxidizes formate to CO2 in mixed acid fermentation (Pinske and Sargent, 2016), the metabolic function of HDCR is not only formate oxidation but also catabolic CO2 reduction to formate in the WLP. Therefore, the ability to use CO2 as sole electron acceptor through the WLP was abolished by the HDCR genes deletion. The T. kivui mutant lacking the HDCR genes, strain TKV_MB013, depended on the complementation with formate as additional electron acceptor for growth (Figure 3C) and for complete and efficient glucose oxidation (Figure 4A). While our observation has been made - somewhat artificially - with a mutant strain, a similar kind of formate metabolism in native acetogens has been described before, e.g., in Butyribacterium methylotrophicum. This bacterium has been reported to utilize CO or H2 + CO2 and formate simultaneously (Kerby et al., 1983; Kerby and Zeikus, 1987). The acetogen Acetobacterium woodii has also been reported to co-utilize formate with CO (Bertsch and Müller, 2015), however, the study had a different focus, since the authors report that CO is only used in the presence of formate. The determined stoichiometries of 1:1.8:1 (formate:CO:acetate) suggest that formate was mainly used as electron acceptor, while CO was used as electron donor (Figure 5). In that scenario, HDCR may be dispensable in the metabolism of A. woodii.
The utilization of formate as electron acceptor may indeed be an ancient metabolic trait. Acetogenesis itself supposedly is one of the oldest types of metabolism (Weiss et al., 2016). The first organisms may have thrived on the oxidation of molecular H2 with CO2, and there are only two groups of organisms that thrive on the conversion of these compounds – methanogenic archaea and acetogenic bacteria. Interestingly, many genes of the WLP essential for CO2 fixation in both groups, and the genes essential to acetogenesis, were described as part of the genome of the Last Universal Common Ancestor (Weiss et al., 2016). The role of formate in Early Life, however, is not clear. Strikingly, in acetogens the genes of the WLP are clustered (Poehlein et al., 2015), also in T. kivui (Hess et al., 2014), however, e.g., in A. woodii (Poehlein et al., 2012) and Clostridium aceticum the genes encoding the formate dehydrogenase/HDCR are separate from this cluster. One may speculate that the WLP may have evolved as three independent parts - CO2 reduction to formate, formate reduction to a methyl group and the CODH/ACS reaction. In the present study, one of the parts, CO2 reduction to formate was removed, which could be complemented by the addition of formate. In an Early Earth environment, organic acids have been reported as prevalent forms of carbon (Amend et al., 2013). Formate has been reported to be thermodynamically more stable than CO2 under alkaline conditions, and significant concentrations of formate were found at the alkaline Lost City hydrothermal field (Lang et al., 2010), an environment discussed to have supported the Evolution of Life (Weiss et al., 2016). Thus, formate may have been present as electron acceptor in Early Earth, and a coupled formate + CO2 respiration, as described in here, may have allowed the conservation of energy in a primordial environment.
The results also indicate a tight coupling of CO2 reduction in the WLP to the oxidation of multi-carbon substrates during heterotrophic growth in T. kivui. We initially considered that the HDCR reaction may not be essential for growth on sugars, despite many acetogens have been described as “homoacetogens” such as Moorella thermoacetica (Fontaine et al., 1942). The term “homoacetogenesis” refers to the conversion of 1 mol of glucose to 3 mols of acetate (eq. 3), in analogy to homolactate fermentation (Drake et al., 2008). The oxidative part of homoacetate fermentation yields only two mols of acetate, 2 mols of CO2 and 8 reducing equivalents (eq. 4), while in the reductive part, catalyzed by the WLP, two mols of CO2 are reduced to the third mol of acetate (eq. 5), depleting the reducing equivalents produced in glucose oxidation. This metabolism has been observed during heterotrophic growth of wild type T. kivui, with 2.3–3 mols of acetate formed from 1 mol of glucose (Leigh et al., 1981).
In theory, reducing equivalents may take an alternative route, especially, in case that the electron-accepting WLP is impaired. For example, the Fdred and NADH produced in T. kivui sugar oxidation may be oxidized by an electron-confurcating hydrogenase (Schut and Adams, 2009). In result, 4 H2 were produced per glucose oxidized, in addition to two acetate and two CO2 (Figure 1A). That type of metabolism was originally described for the thermophilic bacterium Thermotoga maritima (Schut and Adams, 2009), but it is widespread especially among other thermophilic microorganisms. Indeed, in cell suspension experiments with the T. kivui HDCR mutant, 7–8 mM of H2 (if all headspace H2 was dissolved) formed from glucose in the absence of formate (Figure 4B). However, the reducing equivalents present in H2 represented only a minor fraction of the total reducing equivalents (10.5%), and the rate of glucose oxidation was much lower than in the experiments with formate present. In growth experiments, cells only reached a low cell density (Figure 3C), even after prolonged incubation times of 1 week (data not shown). Cultures did also not reach higher cell densities when grown with a proportionally larger gaseous headspace (data not shown), indicating that the headspace hydrogen concentration itself was not inhibitory. Therefore, we assume that, while electron-channeling toward H2 production was possible in principle, it may not be fast enough to support growth to high cell densities. For example, the electron-bifurcating hydrogenase HydABC of T. kivui may be prone to H2 oxidation rather than to H2 formation, as described for membrane-bound hydrogenases of the hyperthermophilic archaeon Pyrococcus furiosus (Mcternan et al., 2014). Alternatively, growth on glucose without a functional WLP or HDCR may be impaired by a non-functional C1-metabolism (since the WLP provides C1-units for anabolic reactions) or by a modified ratio of reduced redox carriers. The latter was observed for A. woodii, where a functional Rnf complex was essential for providing Fdred during growth on low-energy heterotrophic substrates such as lactate or ethanol (Westphal et al., 2018).
Thermoanaerobacter kivui strain LKT-1 (DSM2030), referred to as wild type, strain TKV_MB002 (ΔpyrE, previous name strain TKV002) and strain TKV_MB013 (ΔpyrE ΔTKV_c19960-TKV_c19990) were cultivated under strict anoxic condition at 65°C in complex or defined media as described previously (Weghoff and Müller, 2016; Basen et al., 2018). Complex media contained Na2HPO4 × 2H2O, 50 mM; NaH2PO4 × 2H2O, 50 mM; K2HPO4, 1.2 mM; KH2PO4, 1.2 mM; NH4Cl, 4.7 mM; (NH4)2SO4, 1.7 mM; NaCl, 7.5 mM; MgSO4 × 7 H2O, 0.37 mM; CaCl2 × 2 H2O, 42 μM; Fe(II)SO4 × 7 H2O, 7.2 μM; KHCO3, 54 mM; cysteine-HCl × H2O, 3 mM; resazurin, 4.4 μM; 0.2% (w/v) yeast extract, 10 ml/l trace element solution DSM141 and 10 ml/l vitamin solution DSM141. Defined media was prepared similarly, as complex media without addition of yeast extract. The medium was flushed with N2:CO2 (80:20 [v:v], 1.1 × 105 Pa) before autoclaving. The pH of the medium was 7.5 after flushing. All gases were purchased from Praxair Deutschland GmbH (Düsseldorf, Germany).
Growth experiments were carried out in 20 ml Hungate glass tubes or serum bottles sealed with butyl rubber stoppers under an atmosphere of N2:CO2 (80:20 [v:v], 1.1 × 105 Pa), unless denoted otherwise (Basen et al., 2018). Usually, a concentration of 25 mM of different organic electron donors such as glucose or mannitol was chosen, and 50 mM formate as electron acceptor. Non-gaseous substrates were added from sterile anoxic solutions. If H2 + CO2 were used as substrates, tubes were only filled with medium to 1/4 of the volume, and the remaining headspace was replaced with H2:CO2 (80:20 [v:v], 2 × 105 Pa). To determine the growth behavior, all cultures were inoculated to an optical density of 0.03–0.08 from a pre-culture grown to the exponential growth phase with the same substrate, and then incubated at 65°C under slow shaking. The determination of the cell density was carried out in three biological replicates. Growth in liquid medium was monitored by measuring the optical density at 600 nm. Plating and cultivation on solid media was carried out according to Basen et al. (2018).
Plasmid pMBTkv0012 (Supplementary Figure S1) was used for the deletion of HDCR gene cluster consisting of fdhF, hycB3, hycB4, and hydA2 (TKV_c19960-TKV_c19990). The plasmid was generated by inserting 899 and 1001 bp regions adjacent to the four genes of the cluster (upstream flanking region, UFR, and downstream flanking region, DFR, respectively) into the plasmid pMBTkv005 (Basen et al., 2018). The UFR and DFR were amplified by using the primers NP001 (5′- GCTCG GTACC CGGGG ATCCT AAAGT TTAGT GCATT ACCCC TAAAA TAATG G) and NP002 (5′- CCACT ACCAA CAAAA TTTAA CAAAA CCTCC TCTTA TAACA AAGCA GAAAG G) for UFR, and NP003 (5′- GGAGG TTTTG TTAAA TTTTG TTGGT AGTGG GTTGT AAACA ATCC) and NP004 (5′- GCCGC ATGCC TGCAG GTCGA CTCTA GAGTT ATGTT TAATT TTCTT CCAAC CTCAA CGG) for DFR, followed by the fusion of the PCR products, restriction digest with XbaI and BamHI, and by ligation into plasmid pMBTkv005.
Thermoanaerobacter kivui ΔpyrE was transformed with the plasmid pMBTkv0012, taking advantage of its natural competence for DNA uptake (Basen et al., 2018). The first round of selection was performed in defined media without uracil in the presence of 25 mM glucose + 50mM formate, to select for transformants with the plasmid integrated into the genome. To verify the integration of plasmid pMBTkv012, genomic DNA was extracted and the HDCR gene region was amplified by PCR with the oligonucleotides, NP005 (5′- GATAG GTGAT ACAAT TGAAG TGC) and NP006 (5′- CGCCT CTTGC AAAAC CCG), both binding outside the HDCR gene cluster. Mutants containing the plasmid and growing in the absence of uracil were subjected to a second round of selection as described previously (Basen et al., 2018). Cells were plated on agar with a defined medium containing 50 μM uracil and 5 mM 5-fluoroorotic acid (5-FOA), selecting against the pyrE gene. The substrates used were 25 mM glucose + 50mM formate. The genotype of the cells was again checked by using primer pairs NP005/NP006 binding outside and amplifying the complete HDCR gene locus as well as the primer pairs NP001/SJ003 (5′- AGC CGC ATG CCT GCA GGT CGA CTC TAG ATT CAT ATT GAG GCA ATA GTT CAA TAG CC), P9fw (5′- AAA GAT GGT AAA CAG GAA AAG G)/NP007 (5′- CAG GTG TTA AAT CTC CCA AAT), and PBseq10 (5′- GCT CCG GCT ATT AGA GTT TC)/P18brev (5′- GCG TTA TGC CTA CCT ATA TCT TC) each pair leading to the amplification of part of the HDCR gene cluster. The loss of the HDCR gene cluster in the selected T. kivui Δhdcr mutant, strain TKV_MB013, was additionally verified by sequencing.
Plasmid pSJ002 (Supplementary Figure S2) was constructed to reintroduce the HDCR gene cluster back into the TKV_MB013 genome, between the convergent genes TKV_c24500 (annoted as AAA family ATPase) and TKV_c24520 (annoted as hydroxylamine reductase), therefore likely not causing polar effects (Basen et al., 2018).
Plasmid pJM006 was used as backbone. Plasmid pJM006 was derived from plasmid pMBTkv007 (Basen et al., 2018), with pyrE under control of the promoter controlling gyrase from Thermoanaerobacter sp. strain X514, and directly adjacent to the 3′-end, gene Teth514_0627 from Thermoanaerobacter sp. strain X514 under control of the promoter of the S-layer protein from T. kivui. pJM006 except for adhE from Thermoanaerobacter sp. strain X514 was amplified by PCR using primers SJ0012 (5′- GAG AAA AAA AGT ATA AAA TTT AAT TTA AAA ATT TCA CAG CAA) and SJ0013 (5′- TTT ACC ATC TTT CAT ACA GTC AAT CCT CCT CCT TG). The HDCR gene cluster of T. kivui was amplified by using SJ0010 (5″>- GAG GAG GAT TGA CTG TAT GAA AGA TGG TAA ACA GGA AAA) and SJ0011 (5′- TTT TAA ATT AAA TTT TAT ACT TTT TTT CTC GGT GTA TAT TTA G). The PCR products were then fused to generate the plasmid pSJ002, using Gibson Assembly Mastermix (NEB, Frankfurt/Main, Germany). TKV_MB013 was transformed with plasmid pSJ002. Selection for the transformants was performed by using defined media without uracil in the presence of the substrate 25 mM glucose and 50 mM formate.
Immunological detection of the presence or absence of HDCR subunits in cell-free extracts of T. kivui strains was performed using antisera containing antibodies specific for the formate dehydrogenase (FdhF, encoded by TKV_c19990) and the hydrogenase subunit HydA2 of HDCR (encoded by TKV_c19960). First, genes encoding both subunits were cloned into plasmids pRT001 (fdhF) and pRT002 (hydA2). For pRT001, primers PRT1d (5′- TTT GTT TAA CTT TAA GAA GGA GAT ATA CAT ATG AAA GAT GGT AAA CAG G) and PRT2b (5′- CAA GCT TGT CGA CTC AAT GGT GAT GGT GAT GGT GTT TTC CTC CCT TTT CCT TTG C) were used to amplify the fdhF fragment, followed by digestion with restriction enzymes NdeI and SalI. For pRT002, hydA2 fragment was amplified using primers PRT3 (5′- TTT GTT TAA CTT TAA GAA GGA GAT ATA CAT ATG TCT GCA AAT AAA GCT ATA ATT AAT ATA G) and PRT4 (5′- GTG GTG GTG CTC GAG TGC GGC CGC AAG CTT GTC GAC TTA ATG GTG ATG GTG ATG GTG TAC TTT TTT TCT CGG TGT ATA TTT AG), again followed by digestion with NdeI and SalI. Fragments were cloned into vector pET21a, which was digested using the same restriction enzymes according to manufacturer’s guidelines (NEB, Frankfurt/Main, Germany). The recombinant, His-tagged versions of FDH or HydA2 were produced in E. coli BL21(DE3), purified by affinity chromatography according to standard procedures (Sambrook and Russell, 2001), and sent for rabbit immunization (Davids Biotechnologie, Regensburg, Germany). For Western Blot analysis, 40 μg of T. kivui wild type (WT) or TKV_MB013 cell extract was separated via denaturing polyacrylamide gel electrophoresis (12%), and immunoblotting onto a nitrocellulose membrane (Protran BA 83; GE Healthcare, United Kingdom) was performed according to standard procedures (Sambrook and Russell, 2001) with goat-anti-rabbit IG, conjugated to horseradish peroxidase (dilution of 1:10,000; Bio-Rad, München, Germany). Rabbit antisera were diluted 1:15,000 (FdhF) and 1:10,000 (HydA2), respectively. The chemiluminescence signal was detected using a chemiluminescence detector (ChemoStar, INTAS, Göttingen, Germany). For comparison of the molecular masses of the detected proteins, two images were recorded of the same membrane, one with and one without chemiluminescence detection; and both images were assembled using the ChemoStar TS software (INTAS, Göttingen, Germany).
Specific HDCR activity in the cell-free extract of T. kivui was determined as formate-dependent H2 production or as H2- dependent formate production from CO2 as reported in Schwarz et al. (2018), but at 64°C. Cells for cytoplasmic fraction preparations were harvested in late exponential growth phase and 0.3 mg of cytoplasmic fraction was used for measuring enzymatic activity, respectively. Experiments were conducted in serum bottles and samples for H2 or formate measurement were taken about every 2 min. H2 evolution from formate was measured in 950 μl reaction buffer (100 mM HEPES, 20 mM MgSO4, 0.0001% Resazurin, 0.5 mM DTE, pH 7.0, N2 atmosphere) with 150 mM formate as a substrate. Formate production from H2/CO2 (80:20 [v:v] 1.1 × 105 Pa) was measured in 5 ml reaction buffer (100 mM HEPES, 20 mM MgSO4, 0.0001% Resazurin, 0.5 mM DTE, pH 7.0) in a two-step enzyme assay. After starting the assay, samples were taken from the liquid phase every 2 min and stored on ice. Determination of formate concentration was then performed using a commercially available formic acid-kit (Boehringer Mannheim/R-Biopharm AG, Mannheim/Darmstadt, Germany). Concentrations of purified proteins or proteins in the cell-free extract were determined as described previously (Bradford, 1976).
The stoichiometry of metabolite conversion was determined using concentrated suspensions of resting T. kivui cells. Initially, 500 ml cultures of T. kivui TKV_MB013 (ΔpyrE, ΔfdhF hycB3 hycB4 hydA2; HDCR deletion mutant) were grown in defined media, in the presence of formate, to the mid exponential phase (OD600 of 0.97 to 1.01), and then harvested by centrifugation (AvantiTMJ-25 and JA-10 Fixed-Angle Rotor; Beckman Coulter, Brea, CA, United States) at 12,700 × g, 4°C for 10 min. The supernatant was discarded and cells were re-suspended in 50 ml of defined media. The centrifugation step was repeated, and then, cells were re-suspended again in 50 ml of defined media, and distributed to 10 ml into Hungate tubes. All steps were performed in an anoxic glove box (Coy Laboratory Products, Grass Lake, United States) with an atmosphere of N2:CO2 (80:20 [v:v], 1.1 × 105 Pa) plus approximately 2% H2. The Hungate tubes were closed with butyl rubber stoppers inside the chamber, taken out, and then H2 was removed by exchange of the gaseous headspace against N2:CO2 (80:20 [v:v], 1.1 × 105 Pa). As substrates, 25 mM glucose + 50 mM formate, 25 mM glucose or 50 mM formate were added to the Hungate tubes. The experiment was started by incubation of the concentrated resting cells in a water bath set to 65°C, under slow shaking. 1 ml of subsamples were taken for protein, substrate and product measurements. The protein concentration was determined according to Schmidt et al. (1963).
Organic acid and H2 production were measured by gas chromatography, in accordance with Weghoff and Müller (2016). Consumption of the substrates glucose and formate was determined by high performance liquid chromatography (HPLC, P680 HPLC Pump, ASI-100 Automated Sample Injector and thermostatted Column Compartment TCC-100, Dionex, Sunnyvale, CA, United States). For the sample preparation, cells were spun down by centrifugation at 13,000 rpm for 5 min and 200 μl of supernatant was filled into 2 ml vials containing 400 μl flat bottom glass insert (Agilent Technologies). A HyperREZ XP Carbohydrate H+ ion exchange column (Thermo Fisher Scientific, Waltham, MA, United States) was used for separation. For elution, degassed 5 mM sulfuric acid was used at a flow rate of 0.6 ml/min. The temperature of the oven was set at 65°C. 10 μl of sample was injected by auto-sampler and analyzed with a refractive index detector (RefractoMax 520; Dionex, Sunnyvale, CA, United States) set at 55°C.
The datasets generated for this study are available on request to the corresponding author.
VM and MB designed the study. SJ and HD performed the experiments and prepared the figures. All authors analyzed the data and wrote the manuscript.
VM and MB are grateful to Deutsche Forschungsgemeinschaft (DFG) for funding. VM was also supported by an Advanced Grant of the European Research Council under the European Union’s Horizon 2020 Research and Innovation Program (grant agreement no. 741791). SJ was funded by a fellowship from Deutscher Akademischer Austauschdienst (DAAD). HD was supported by the Deutsche Bundesstiftung Umwelt (DBU) (Ph.D. grant no. 20016/446). We acknowledge financial support by the Deutsche Forschungsgemeinschaft and Universität Rostock/Universitätsmedizin Rostock within the funding programme Open Access Publishing.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors gratefully acknowledge Nils Peiter for his help to obtain plasmid pMBTkv012.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00059/full#supplementary-material
Amend, J. P., Larowe, D. E., Mccollom, T. M., and Shock, E. L. (2013). The energetics of organic synthesis inside and outside the cell. Philos. Trans. R. Soc. B 368:20120255. doi: 10.1098/rstb.2012.0255
Basen, M., Geiger, I., Henke, L., and Müller, V. (2018). A genetic system for the thermophilic acetogenic bacterium Thermoanaerobacter kivui. Appl. Environ. Microbiol. 84:e02210-17. doi: 10.1128/AEM.02210-17
Bertsch, J., and Müller, V. (2015). CO metabolism in the acetogen Acetobacterium woodii. Appl. Environ. Microbiol. 81, 5949–5956. doi: 10.1128/AEM.01772-15
Biegel, E., and Müller, V. (2010). Bacterial Na+-translocating ferredoxin: NAD+ oxidoreductase. Proc. Natl. Acad. Sci. U.S.A. 107, 18138–18142. doi: 10.1073/pnas.1010318107
Bradford, M. M. (1976). Rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Diekert, G., and Wohlfarth, G. (1994). Metabolism of homoacetogens. Antonie Van Leeuwenhoek 66, 209–221. doi: 10.1007/bf00871640
Drake, H. L., Gössner, A. S., and Daniel, S. L. (2008). Old acetogens, new light. Ann. N. Y. Acad. Sci. 1125, 100–128. doi: 10.1196/annals.1419.016
Fontaine, F. E., Peterson, W. H., Mccoy, E., Johnson, M. J., and Ritter, G. J. (1942). A new type of glucose fermentation by Clostridium thermoaceticum. J. Bacteriol. 43, 701–715. doi: 10.1128/jb.43.6.701-715.1942
Hess, V., Poehlein, A., Weghoff, M. C., Daniel, R., and Müller, V. (2014). A genome-guided analysis of energy conservation in the thermophilic, cytochrome-free acetogenic bacterium Thermoanaerobacter kivui. BMC Genomics 15:1139. doi: 10.1186/1471-2164-15-1139
Kerby, R., Niemczura, W., and Zeikus, J. G. (1983). Single-carbon catabolism in acetogens: analysis of carbon flow in Acetobacterium woodii and Butyribacterium methylotrophicum by fermentation and 13C nuclear magnetic resonance measurement. J. Bacteriol. 155, 1208–1218. doi: 10.1128/jb.155.3.1208-1218.1983
Kerby, R., and Zeikus, J. G. (1987). Anaerobic catabolism of formate to acetate and CO2 by Butyribacterium methylotrophicum. J. Bacteriol. 169, 2063–2068. doi: 10.1128/jb.169.5.2063-2068.1987
Lang, S. Q., Butterfield, D. A., Schulte, M., Kelley, D. S., and Lilley, M. D. (2010). Elevated concentrations of formate, acetate and dissolved organic carbon found at the Lost City hydrothermal field. Geochim. Cosmochim. Acta 74, 941–952. doi: 10.1016/j.gca.2009.10.045
Leigh, J. A., Mayer, F., and Wolfe, R. S. (1981). Acetogenium kivui, a new thermophilic hydrogen-oxidizing, acetogenic bacterium. Arch. Microbiol. 129, 275–280. doi: 10.1007/bf00414697
Ljungdahl, L. G. (1986). The autotrophic pathway of acetate synthesis in acetogenic bacteria. Annu. Rev. Microbiol. 40, 415–450. doi: 10.1146/annurev.mi.40.100186.002215
Matson, E. G., Zhang, X. N., and Leadbetter, J. R. (2010). Selenium controls transcription of paralogous formate dehydrogenase genes in the termite gut acetogen, Treponema primitia. Environ. Microbiol. 12, 2245–2258. doi: 10.1111/j.1462-2920.2010.02188.x
Mcdowall, J. S., Murphy, B. J., Haumann, M., Palmer, T., Armstrong, F. A., and Sargent, F. (2014). Bacterial formate hydrogenlyase complex. Proc. Natl. Acad. Sci. U.S.A. 111, E3948–E3956.
Mcternan, P. M., Chandrayan, S. K., Wu, C. H., Vaccaro, B. J., Lancaster, W. A., Yang, Q. Y., et al. (2014). Intact functional fourteen-subunit respiratory membrane-bound [NiFe]-hydrogenase complex of the hyperthermophilic archaeon Pyrococcus furiosus. J. Biol. Chem. 289, 19364–19372. doi: 10.1074/jbc.M114.567255
Moon, J., Henke, L., Merz, N., and Basen, M. (2019). A thermostable mannitol-1-phosphate dehydrogenase is required in mannitol metabolism of the thermophilic acetogenic bacterium Thermoanaerobacter kivui. Environ. Microbiol. 21, 3728–3736. doi: 10.1111/1462-2920.14720
Müller, V. (2019). New horizons in acetogenic conversion of one-carbon substrates and biological hydrogen storage. Trends Biotechnol. 37, 1344–1354. doi: 10.1016/j.tibtech.2019.05.008
Pereira, I. A. C. (2013). An enzymatic route to H2 storage. Science 342, 1329–1330. doi: 10.1126/science.1247698
Pierce, E., Xie, G., Barabote, R. D., Saunders, E., Han, C. S., Detter, J. C., et al. (2008). The complete genome sequence of Moorella thermoacetica (f. Clostridium thermoaceticum). Environ. Microbiol. 10, 2550–2573. doi: 10.1111/j.1462-2920.2008.01679.x
Pinske, C., and Sargent, F. (2016). Exploring the directionality of Escherichia coli formate hydrogenlyase: a membrane-bound enzyme capable of fixing carbon dioxide to organic acid. Microbiologyopen 5, 721–737. doi: 10.1002/mbo3.365
Poehlein, A., Cebulla, M., Ilg, M. M., Bengelsdorf, F. R., Schiel-Bengelsdorf, B., Whited, G., et al. (2015). The complete genome sequence of Clostridium aceticum: a missing link between Rnf- and cytochrome-containing autotrophic acetogens. mBio 6:e1168-15. doi: 10.1128/mBio.01168-15
Poehlein, A., Schmidt, S., Kaster, A. K., Goenrich, M., Vollmers, J., Thurmer, A., et al. (2012). An ancient pathway combining carbon dioxide fixation with the generation and utilization of a sodium ion gradient for ATP synthesis. PLoS One 7:e33439. doi: 10.1371/journal.pone.0033439
Sambrook, J., and Russell, D. W. (eds) (2001). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Sapra, R., Bagramyan, K., and Adams, M. W. W. (2003). A simple energy-conserving system: proton reduction coupled to proton translocation. Proc. Natl. Acad. Sci. U.S.A. 100, 7545–7550. doi: 10.1073/pnas.1331436100
Schink, B., and Stams, A. (2006). “Syntrophism among prokaryotes,” in The Prokaryotes – A Handbook on the Biology of Bacteria, 3rd Edn, eds M. Dworkin, S. Falkow, E. Rosenberg, K. H. Schleifer, and E. Stackebrandt, (New York, NY: Springer Science+Business Media, LLC), 309–336.
Schmidt, K., Jensen, S. L., and Schlegel, H. (1963). Die Carotinoide der Thiorhodaceae. Arch. Mikrobiol. 46, 117–126. doi: 10.1007/bf00408204
Schoelmerich, M. C., and Müller, V. (2019). Energy conservation by a hydrogenase-dependent chemiosmotic mechanism in an ancient metabolic pathway. Proc. Natl. Acad. Sci. U.S.A. 116, 6329–6334. doi: 10.1073/pnas.1818580116
Schuchmann, K., and Müller, V. (2012). A bacterial electron-bifurcating hydrogenase. J. Biol. Chem. 287, 31165–31171. doi: 10.1074/jbc.M112.395038
Schuchmann, K., and Müller, V. (2013). Direct and reversible hydrogenation of CO2 to formate by a bacterial carbon dioxide reductase. Science 342, 1382–1385. doi: 10.1126/science.1244758
Schuchmann, K., and Müller, V. (2014). Autotrophy at the thermodynamic limit of life: a model for energy conservation in acetogenic bacteria. Nat. Rev. Microbiol. 12, 809–821. doi: 10.1038/nrmicro3365
Schuchmann, K., and Müller, V. (2016). Energetics and application of heterotrophy in acetogenic bacteria. Appl. Environ. Microbiol. 82, 4056–4069. doi: 10.1128/AEM.00882-16
Schut, G. J., and Adams, M. W. W. (2009). The iron-hydrogenase of Thermotoga maritima utilizes ferredoxin and NADH synergistically: a new perspective on anaerobic hydrogen production. J. Bacteriol. 191, 4451–4457. doi: 10.1128/JB.01582-08
Schwarz, F. M., Schuchmann, K., and Müller, V. (2018). Hydrogenation of CO2 at ambient pressure catalyzed by a highly active thermostable biocatalyst. Biotechnol. Biofuels 11:237. doi: 10.1186/s13068-018-1236-3
Trchounian, A., and Sawers, G. R. (2014). Novel insights into the bioenergetics of mixed-acid fermentation: can hydrogen and proton cycles combine to help maintain a proton motive force? IUBMB Life 66, 1–7. doi: 10.1002/iub.1236
Weghoff, M. C., and Müller, V. (2016). CO metabolism in the thermophilic acetogen Thermoanaerobacter kivui. Appl. Environ. Microbiol. 82, 2312–2319. doi: 10.1128/AEM.00122-16
Weiss, M. C., Sousa, F. L., Mrnjavac, N., Neukirchen, S., Roettger, M., Nelson-Sathi, S., et al. (2016). The physiology and habitat of the last universal common ancestor. Nat. Microbiol. 1:16116. doi: 10.1038/nmicrobiol.2016.116
Welte, C., Kallnik, V., Grapp, M., Bender, G., Ragsdale, S., and Deppenmeier, U. (2010). Function of Ech hydrogenase in ferredoxin-dependent, membrane-bound electron transport in Methanosarcina mazei. J. Bacteriol. 192, 674–678. doi: 10.1128/JB.01307-09
Westphal, L., Wiechmann, A., Baker, J., Minton, N. P., and Müller, V. (2018). The Rnf complex is an energy-coupled transhydrogenase essential to reversibly link cellular NADH and ferredoxin pools in the acetogen Acetobacterium woodii. J. Bacteriol. 200:e00357-18. doi: 10.1128/JB.00357-18
Keywords: hydrogen oxidation, carbon dioxide reduction, hydrogen-dependent carbon dioxide reductase, acetogens, thermophiles, Thermoanaerobacter kivui, Wood-Ljungdahl pathway
Citation: Jain S, Dietrich HM, Müller V and Basen M (2020) Formate Is Required for Growth of the Thermophilic Acetogenic Bacterium Thermoanaerobacter kivui Lacking Hydrogen-Dependent Carbon Dioxide Reductase (HDCR). Front. Microbiol. 11:59. doi: 10.3389/fmicb.2020.00059
Received: 23 August 2019; Accepted: 13 January 2020;
Published: 31 January 2020.
Edited by:
Inês A. Cardoso Pereira, New University of Lisbon, PortugalReviewed by:
Constanze Pinske, Martin Luther University of Halle-Wittenberg, GermanyCopyright © 2020 Jain, Dietrich, Müller and Basen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mirko Basen, bWlya28uYmFzZW5AdW5pLXJvc3RvY2suZGU=
†Present address: Mirko Basen, Institute of Biological Science, University of Rostock, Rostock, Germany
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.