- 1College of Food Science and Engineering, Northwest University, Xi’an, China
- 2College of Food Science and Engineering, Northwest A&F University, Yangling, China
The osmotolerant Zygosaccharomyces rouxii is known for its trait to survive in extreme high sugar environments. This ability determines its role in the fermentation process and leads to yeast spoilage in the food industry. However, our knowledge of the gene expression in response to high sugar stress remains limited. Here, we conducted RNA-sequencing (RNA-seq) under different sugar concentrations of the spoilage yeast, Z. rouxii, which exhibit extremely high tolerance to sugar stress. The obtained differentially expressed genes (DEGs) are significantly different to that of the Saccharomyces cerevisiae, which is sensitive to extreme high sugar stress. Most of the DEGs participated in the “glucan synthesis,” “transmembrane transport,” “ribosome,” etc. In this work, we also demonstrated that the gene ZYRO0B03476g (ZrKAR2) encoding Kar2p can significantly affect the growth of Z. rouxii under high sugar stress. In addition, we combined with a previous study on the genome sequence of Z. rouxii, indicating that several gene families contain significantly more gene copies in the Z. rouxii lineage, which involved in tolerance to sugar stress. Our results provide a gene insight for understanding the high sugar tolerance trait, which may impact food and biotechnological industries and improve the osmotolerance in other organisms.
Introduction
Osmotolerant yeast dominate much of the fermentation process and yeast spoilage in food industries (Watanabe et al., 2004; Dakal et al., 2014; Niu et al., 2016). Most of these yeast species rarely occur on extremely high sugar environments (such as concentrated apple juice, honey, etc.). Among all the osmotolerant yeast, members of the genus Zygosaccharomyces are famous not only because of their spoilage ability but also because of their high tolerance to sugar stress (Zuehlke et al., 2013; Alonso et al., 2015; Marvig et al., 2015; Wang et al., 2015; Xiang et al., 2018). Zygosaccharomyces rouxii species, which is native to concentrated apple juice, nougat, etc. (Martorell et al., 2007; Wang et al., 2015), is characterized by extraordinary adaptation to sugar stress. Notably, at high sugar stress, it maintains higher growth than other yeast species and can survive at glucose concentrations up to 75% w/v (Dakal et al., 2014). However, 60% w/v extremely high sugar stress was reported to inhibit the growth of Saccharomyces cerevisiae (Silva et al., 2005; Dakal et al., 2014). Dakal et al. (2014) have identified S. cerevisiae as moderately osmotolerant yeast and Z. rouxii as osmotolerant yeast.
Currently, S. cerevisiae appears as a cell model to unravel molecular mechanism in response to hyperosmotic stress (Hohmann, 2002; Klipp et al., 2005; De and Al, 2011; Dakal et al., 2014). In S. cerevisiae, Hog1 mitogen-activated protein kinase MAPK pathway responds to hyperosmotic stress and ultimately leads to increased synthesis and retention of glycerol (Jiménez-Martí et al., 2011; Dakal et al., 2014; Ene et al., 2015; Mitchell et al., 2015). Several reports have investigated the response of S. cerevisiae to mild sugar stress by conducting global or particular transcriptomic analyses. An upregulation of glycerol and trehalose biosynthetic genes was found in S. cerevisiae exposed to 20% w/v sugar stress. Erasmus et al. (2003) reported that a particular analysis of the response of wine yeast to 40% w/v sugar stress upregulated the genes of the glycolytic and the pentose phosphate pathway. Jiménez-Martí et al. (2011) also found that the ΔYHR087W mutant (S. cerevisiae) reduced the expression of heat shock proteins Hsp104 and Hsp78 in response to 20% w/v sugar stress.
Proteomic characterization of Z. rouxii response to 60% w/v extreme high sugar stress by our group (Guo et al., 2016) has revealed that most of the differential expressed proteins are involved in carbohydrate and energy metabolism, amino acid metabolism, response to stimulus (mainly heat shock proteins), etc. Among them, Kar2p (belongs to Hsp70 family) is the most prominently upregulated protein, increasing approximately 29-fold. Interestingly, we have unexpectedly discovered that Vaupotic et al. (2008) also reported that in extreme halotolerant black yeast Hortaea werneckii and in the adaptation to high amounts of sorbitol (55% w/v), the mitochondria preferentially accumulate Kar2p and Hsp60. However, Pham and Wright (2008) reported that the expression of heat shock proteins remains almost unchanged in S. cerevisiae during 30% w/v glucose stress (mild stress). Recently, Liu et al. (2020) found that antioxidant enzymes of Z. rouxii under the D-fructose stress were related to the resistance characteristics.
In this study, based on the previous genomics study (Genolévures et al., 2009), we studied the response mechanism of Z. rouxii under extreme high glucose stress by physiological response, gene knockout, and global transcriptional response. We further examine gene expression differences following sugar stress treatment in a comparison with S. cerevisiae, which is sensitive to extreme high sugar environments. Our study highlighted the genetic bases of sugar tolerance in the Z. rouxii.
Materials and Methods
Materials
Zygosaccharomyces rouxii (BW-WHX-12-54) was isolated from apple juice concentrated by our group (Wang et al., 2015), which was identified by sequencing of the D1/D2 domain of the 26S ribosomal gene and registered at the National Center for Biotechnology Information, United States (GenBank Accession Number KC544459). S. cerevisiae (ATCC 38531) was bought from the American Type Culture Collection (ATCC). Phosphate buffer saline (PBS, pH 7.0) and Geneticin (G418) were bought from Sigma-Aldrich (St. Louis, MO, United States). TRIzol was bought from Invitrogen (Carlsbad, United States). M-MuLV reverse transcriptase (RNase H–) was bought from Roche (Basel, Switzerland). NEBNext® UltraTM RNA Library Prep Kit bought from New England Biolabs (NEB, United States). For PCR reactions, Taq DNA polymerase from TaKaRa Biotechnology (Co., Ltd., Dalian, China) was used. Yeast extract, peptone, D-(+)-glucose, and peptide were purchased from local supplier.
Strain Construction
According to the protein Kar2p detected by Guo et al. (2016), we identified its encoding gene ZYRO0B03476g (ZrKAR2) in Z. rouxii. We designed the primers (ZrKAR2-F, ZrKAR2-R) to confirm ZrKAR2. Then, we designed the interrupt primers (ZrKAR2-LF, ZrKAR2-LR). All primers are shown in Supplementary Table S1. To delete the ZrKAR2, the PUG6 plasmid was used as a template, and ZrKAR2-LF and ZrKAR2-LR were used as interrupt primers to amplify KanMX gene. Then, this KanMX PCR product containing a ZrKAR2 homogeneous arm was obtained and sequenced. The correct PCR products were transferred to Z. rouxii according to LiAc methods (Simons et al., 1998). The transformed yeast was applied to a YPD plate containing G418. Positive colonies were confirmed by re-sequencing.
Strains and Growth Conditions
Zygosaccharomyces rouxii BW-WHX-12-54 and S. cerevisiae ATCC38531 were grown in YPD (10 g/L yeast extract, 20 g/L peptone, and 20 g/L glucose) broth at 30°C until the culture was in late exponential phase. YPD medium (2% w/v glucose) was used as basic and normal stress medium. YPD containing 40% w/v glucose (40% YPD) and 60% w/v glucose (60% YPD) were used as mild and extreme high sugar stress medium, respectively. For analyses, yeast cells were grown at 30°C. Then, late exponential phase yeast cells (about 2 × 109 CFU/ml) were exposed to 60% YPD, diluted to a density of 2 × 107 CFU/ml for 4 h.
For detecting the growth of Z. rouxii and its mutant strain, purified yeast colonies were inoculated to a 200-well plate pre-filled with 250 μl of YPD and 60% YPD medium. Individual growth curves were obtained using an automatic growth curve analyzer (Bioscreen, Finland) at 30°C, the absorbance at 600 nm was measured, and it was automatically detected for 98 h.
Transmission Electron Microscopy and Atomic Force Microscopy
The transmission electron microscopy (TEM) samples were performed as described previously (Simons et al., 1998), including cutting ultrathin sections by using EM UC7 (Leica, Germany). TEM images were obtained by using a TECNAI G2 SPIRIT TEM (FEI, United States) at an accelerating voltage of 120 kV.
Atomic force microscopy (AFM) images of yeast cells trapped in freshly stripped mica sheet were recorded with a NanoScope V America (Bruker, United States), in contact mode (Pillet et al., 2014), using SiO4 cantilevers. Images and cell diameter were analyzed by NanoScope Analysis 1.5.
Transcriptome Analysis
Late-exponential-phase Z. rouxii cells grown in YPD served as the baseline of control (Z_C). Late-exponential-phase Z. rouxii (about 2 × 109 CFU) cells were exposed to 60% YPD for 4 h (final density was about 2 × 107 CFU/ml) as stressed-Z. rouxii (Z_stress). Late-exponential-phase S. cerevisiae cells grown in YPD served as the baseline of control (S_C). Late-exponential-phase S. cerevisiae (about 2 × 109 CFU) cells were exposed to 60% YPD for 4 h (final density was about 2 × 107 CFU/ml) as stressed-S. cerevisiae (S_stress). Z. rouxii cells and S. cerevisiae cells were sampled at 4 h after 60% w/v high sugar stress (Supplementary Figure S1). Each sample had three biological replicates. All these 12 samples were harvested and subsequently flash-frozen with liquid nitrogen and stored at −80°C for RNA extraction.
Total RNA was extracted using TRIzol regent (Invitrogen, United States) per the manufacturer’s protocol and treated with RNase-free DNase I. A NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE, United States), a Qubit Fluorometer 2.0 (Life Technologies, Carlsbad, CA, United States), the agarose gel electrophoresis, and an Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA, United States) were used to determine concentration, purity, and integrity of RNA samples (Lin et al., 2013; Wang et al., 2018).
The cDNA libraries were constructed according to earlier report (Lai et al., 2017; Wang et al., 2018). A total amount of 1 μg of RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, United States) following the manufacturer’s recommendations and index codes were added to attribute sequences to each sample. Briefly, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperature in NEB Next First Strand Synthesis Reaction Buffer (5×). First-strand cDNA was synthesized using random hexamer primer and M-MuLV Reverse Transcriptase (RNase H–). Second-strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. The cDNA libraries were then sequenced on the Illumina Hiseq2000 platform (San Diego, CA, United States, 2010). FastQC (version 0.11.2) was used for evaluating the quality of sequenced data. The raw reads were generated from control and treatment samples. After filtering the adaptor sequences, the clean reads of Z. rouxii and S. cerevisiae were mapped onto the reference genome of Z. rouxii CBS732 and the reference genome of S. cerevisiae S288C by HISAT2 (version 2.0). Gene expression values were quantified by HTSeq v0.6.1. DESeq v1.10.1 was used to determine differentially expressed genes (DEGs) between control and treatment samples. Genes were considered as significant differentially expressed if |log2(fold change)| > 0 and p-adj < 0.05. Subsequent enrichment analysis of DEGs by GO enrichment analysis (GOSeq Release2.12) was performed based on a previous report (Wang et al., 2016). The RNA-seq data have been deposited in the National Center for Biotechnology Information (NCBI), with accession code PRJNA437612.
Validation of RNA-Seq Data by q-PCR
To validate the RNA-seq data, the expression level of five interest genes related to sugar adaptation (2% w/v and 60% w/v) was analyzed by real-time quantitative PCR (q-PCR). The q-PCR experiments were performed according to the method of Guo et al. (2016). Primers were designed using Primer 5, and they are listed in Supplementary Table S1. Total RNA was extracted and purified with Yeast RNAiso kit (TaKaRa Biotechnology Co., Ltd., Dalian, China) following the user protocol. Reverse transcription was performed using PrimeScriptTM RT reagent Kit with gDNA Eraser (TaKaRa Biotechnology Co., Ltd., Dalian, China). Real-time quantitative PCR reactions were performed in 96-Well Optical Reaction Plates (Bio-Rad) in trice using the SYBR® Premix Ex TaqTM II (Tli RNaseH Plus) (Takara Biotechnology Co., Ltd., Dalian, China) and analyzed on the iCycler iQ5 2.0 Standard Edition Optical System (Bio-Rad). ZYRO0F02772g (ZrACT1) was used as the internal control.
Results
Overall Description of the Transcriptomic Response to 60% w/v Sugar Stress
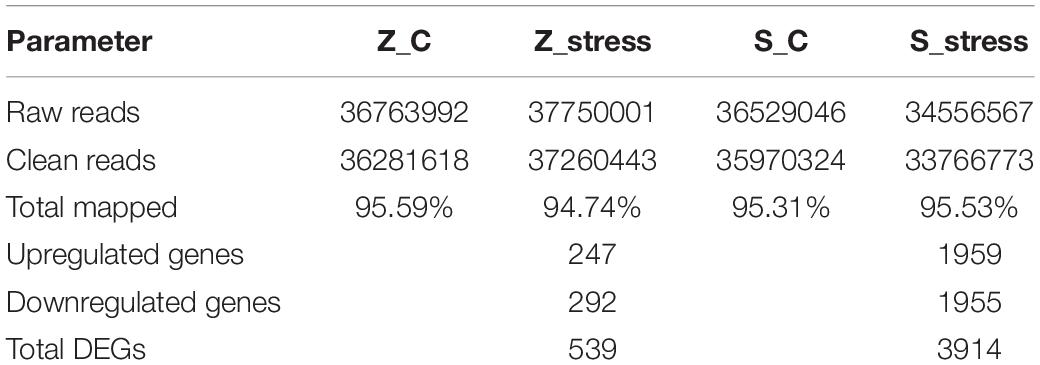
To examine the genome-wide responses to sugar stress of this extreme osmotolerant Z. rouxii, transcriptome analysis was carried out to assess the specific response at mRNA levels. The details in assembly and annotation information are shown in Table 1. Via RNA-seq, clean reads were obtained. Therein, 95.59 and 94.74% of total clean reads from the Z_C and Z_stress group were aligned to reference sequences (Z. rouxii CBS732). 95.31 and 95.53% of total clean reads from S_C and S_stress group were aligned to reference sequences (S. cerevisiae S288C). A total of 539 genes in Z. rouxii and 3914 genes in S. cerevisiae were considered as significant changes in abundance under 60% w/v sugar stress.
Furthermore, to better analyze the functions and interactions of the DEGs in Z. rouxii, GO enrichment analyses were performed. Figure 1 shows the top 30 enriched functional categories and the bottom 30 enriched functional categories of these DEGs. We found these DEGs of Z. rouxii enriched in “glucan biosynthesis” (GO:0051274), “transmembrane transport” (GO:0055085), “ribosome” (GO:0003735), etc. Those DEGs enriched in sensitive pathways will be further analyzed.
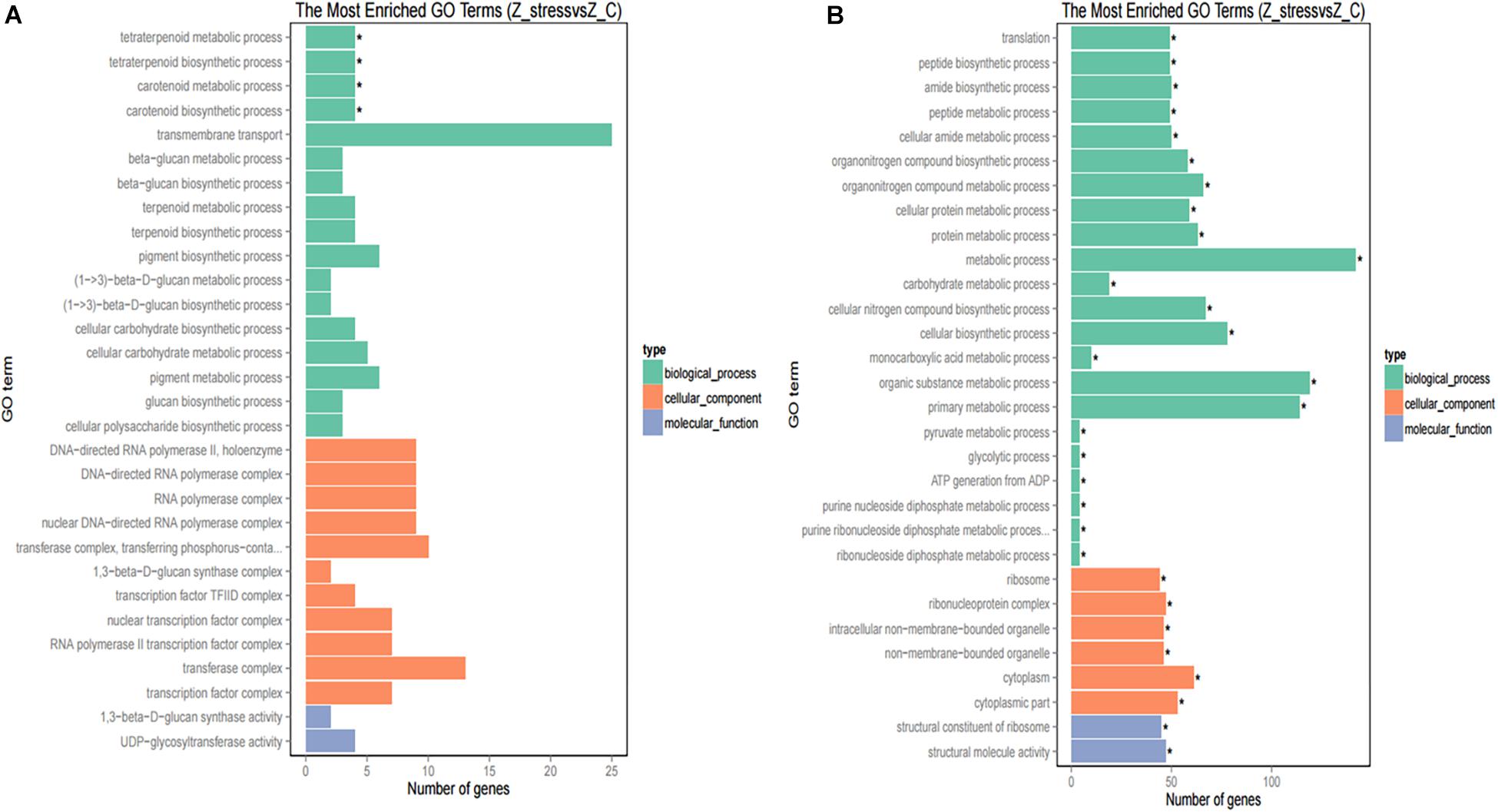
Figure 1. GO functional classification of upregulated DEGs (A) and downregulated DEGs (B). The ordinate means GO term, the abscissa means the number of DEGs of each GO term. “*” means significant enrichment.
Although DEGs of S. cerevisiae (3914) were about 7.2 times more than that of Z. rouxii (539) (Table 1), Z. rouxii had more percent genes enriched in “beta glucan synthesis,” “transmembrane transport,” and “structural constituent of ribosome” involved in sugar stress (Figure 2A). Clustering analysis suggested that many of the genes showed different regulatory patterns in response to sugar between these two species (Figure 2B).
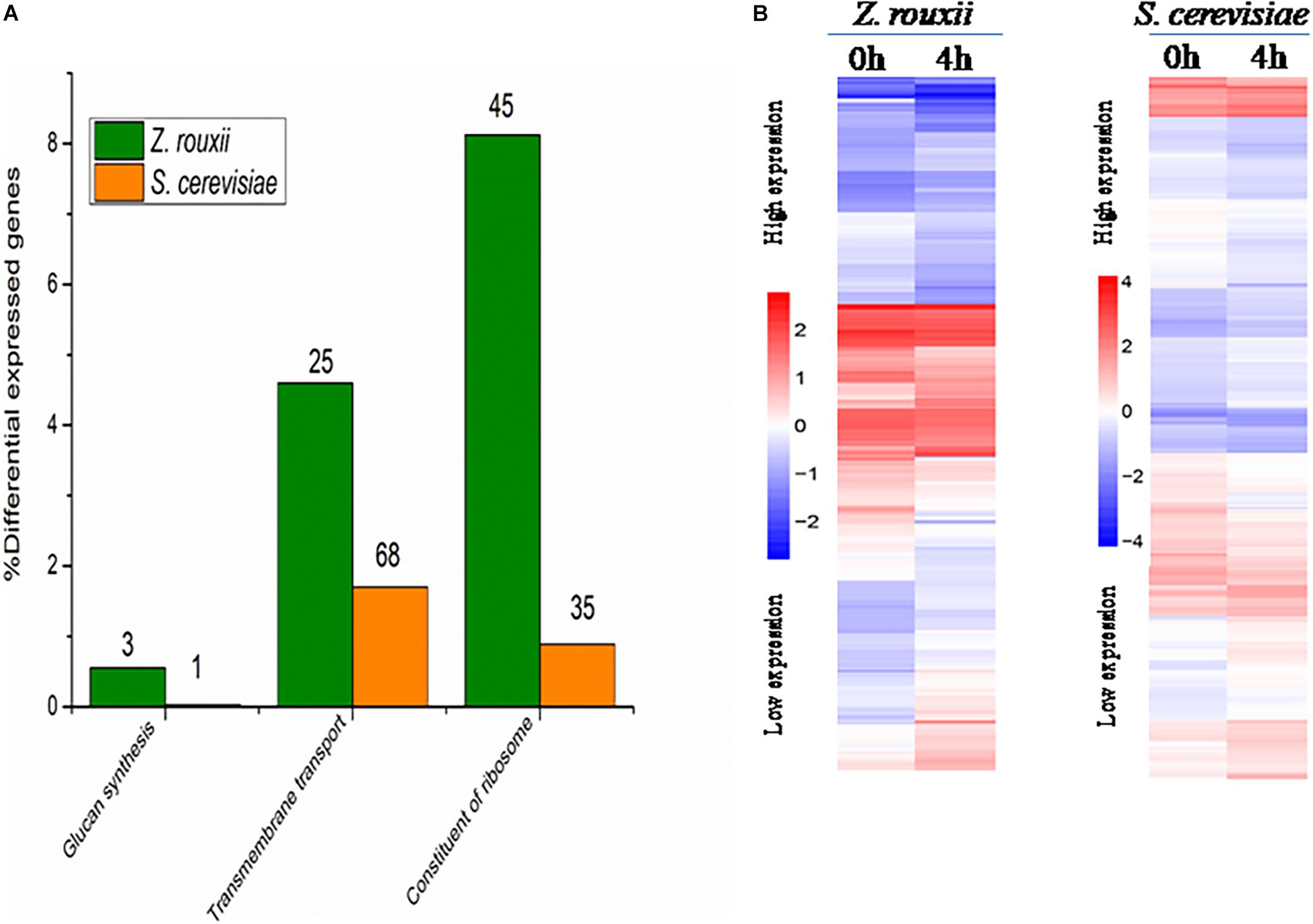
Figure 2. (A) DEGs proportions in Z. rouxii and S. cerevisiae. The number of DEGs of each class is indicated above each bar. (B) Expression of the genes identified in Z. rouxii and S. cerevisiae. The heatmap was generated from hierarchical cluster analysis of genes. The transcript levels were determined by fragments per kilobase of exon per million fragments mapped (FPKM).
In addition, we combined DEGs of Z. rouxii with the gene family analysis of Z. rouxii reported by Genolévures et al. (2009). We found that several DEGs we measured belong to Z. rouxii specific expansion gene families (Supplementary Table S2). Although these expansion gene families listed in Supplementary Table S2 are come from the genome of Z. rouxii CBS 732 (measured by Genolévures et al.), the strain we used in this study is the same species as the one Genolévures et al. (2009) used. Moreover, the reference genome we used is also Z. rouxii CBS732 and the alignment rate of the two strains was approximately 95% (Table 1). These DEGs have not been reported in the previous sugar tolerance responses studies (Erasmus et al., 2003; Jiménez-Martí et al., 2011; Dakal et al., 2014). These findings will have important significance for revealing the osmotic stress mechanism of extremophile microorganisms.
Copy number within gene families has been reported to vary greatly between closely related, divergent species (Genolévures et al., 2009; Ma et al., 2013). These gene families related to sugar stress were substantially expanded in Z. rouxii compared with other yeast species (Supplementary Table S2). For example, the GL3C0007 gene family, encoding proteins similar to the FLR1 plasma membrane multidrug transporter, expanded from one member in Kluyveromyces lactis genome to four in the Z. rouxii genome. Four of these genes occurred as tandem gene array (TGA) in Z. rouxii (Genolévures et al., 2009). In this study, we found that ZYRO0E09966g in the GL3C0007 gene family was upregulated 2.1-fold in response to extreme high sugar stress. The gene family (GL3C0055) encoding NADPH-dependent oxidoreductase also had more copies (10 copies compared to 0 to 4 for other species) of genes in the Z. rouxii genome. Catalase has been reported in affecting D-fructose tolerance of Z. rouxii (Liu et al., 2020). The gene family encoding this enzyme was also expanded in the Z. rouxii. These findings are similar to the reports of Ma et al. (2013) that several gene families related to high salt stress were substantially expanded in extreme plant desert poplar compared with other plant species.
Furthermore, we verified five interest genes related to sugar adaptation in the RNA-seq data by q-PCR. As shown in Supplementary Figure S2, these results were agreement with the data of transcriptomic analysis. Therefore, it suggested that the RNA-seq data were reliable.
Cell Wall
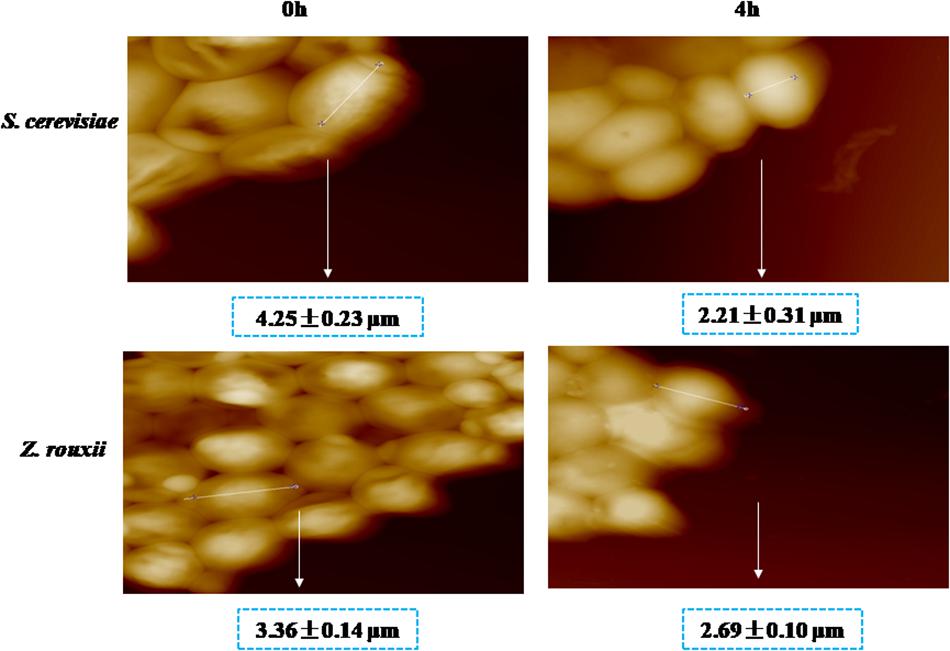
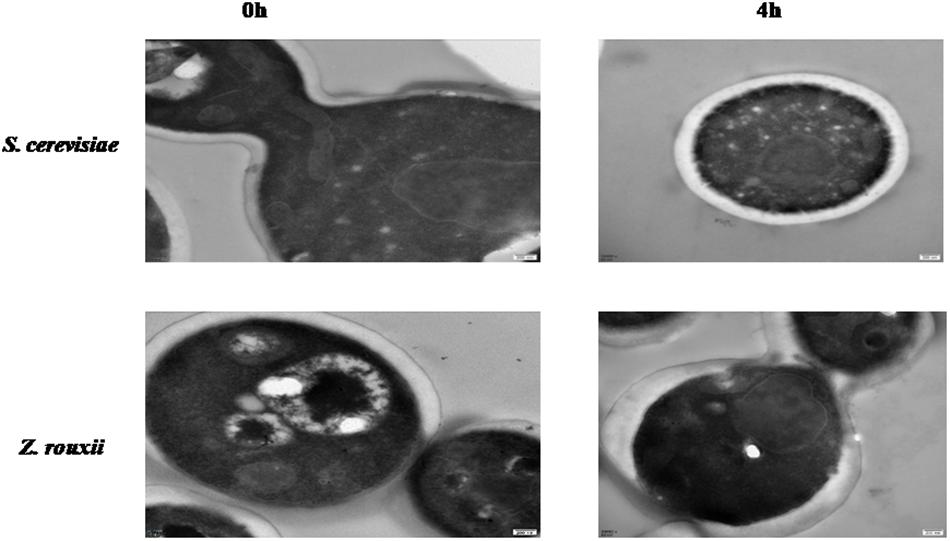
Previous reports suggested that the reduction in cell volume during the hyperosmotic stress is accompanied by a thicker β-glucan-chitin layer of the wall (Ene et al., 2015). These phenomena are mediated by cell wall remodeling enzymes (included chitin synthase CHS1, β-1,3-glucan synthase FKS, cell wall transglycosylases UTR2, etc.). In this study, we tested this by comparing the cell size and cell walls of Z. rouxii and S. cerevisiae cells before sugar addition and after osmo-adaptation. Similar to previous results (Formosa et al., 2013; Ene et al., 2015), there was a significant decrease in cell mean size following sugar stress exposure for both Z. rouxii and S. cerevisiae cells (Figure 3 and Supplementary Figure S3). Concomitant with the loss in cell volume, the cell wall became thicker (Figure 4).
The upregulated “glucan synthesis” (Figure 1A and Table 2) supported this phenomenon. We found that genes encoding cell wall remodeling enzymes that influence cross-linking in the β-glucan-chitin network exhibited similar and different regulatory patterns in response to 60% w/v sugar stress between Z. rouxii and S. cerevisiae (Table 2). On the one hand, several genes were both upregulated after 4 h of sugar stress in Z. rouxii and high-sugar-sensitive S. cerevisiae. For example, FKS1, encoding β-1,3-glucan synthase involved in glucan synthesis, affects cell wall integrity (Qiu et al., 2018) and is the target of antifungal agent (Xie et al., 2017). UTR2, encoding the putative cell wall transglycosylases, was involved in linking chitin to β-glucan (Brennan et al., 2013; Ene et al., 2015). It has been reported that overexpression of UTR2 increased the osmotic stress resistance (Ene et al., 2015). In our experiments, FKS1 and UTR2 increased similarly in response to sugar stress in Z. rouxii and S. cerevisiae. Although UTR2 is not overexpressed in Z. rouxii, Z. rouxii still has extremely high resistance to osmotic stress. We also found that cell wall remodeling enzymes exhibited the different regulatory patterns in response to 60% w/v sugar stress between Z. rouxii and S. cerevisiae. For example, the alternative subunit β-1,3-glucan synthase ZYRO0D06974g (FKS3) and 1,6-glucansynthase encoding gene ZYRO0G18898g (KRE9) were upregulated after 4 h of sugar stress in Z. rouxii but were maintained at control levels in S. cerevisiae. In addition, CHS1, a gene encoding chitin synthase, is essential for cell wall integrity. The transcript of this gene was upregulated after 4 h of sugar stress in Z. rouxii, but was downregulated at control levels in S. cerevisiae.
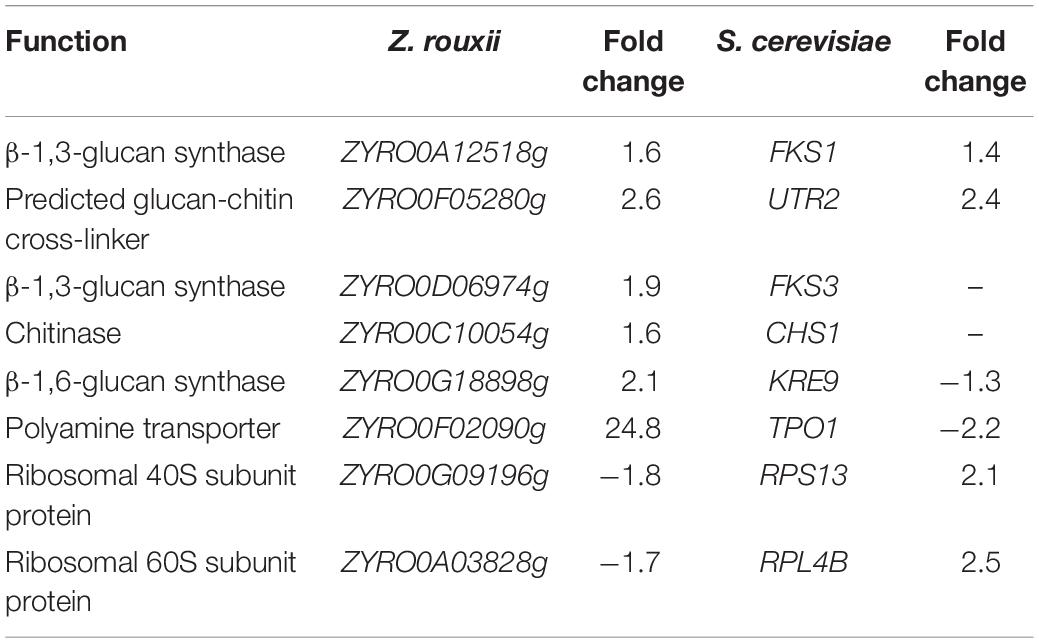
Table 2. Transcript levels of the genes showing different expression patterns between Z. rouxii and S. cerevisiae after 60% w/v glucose treatment.
Previous study showed that the degree of cross-linking in the β-glucan-chitin network (a stiffer cell wall) is likely to increase the osmotic resistance of cells and constrains the rate of change in cell size, thereby decreasing compromise cell integrity (Ene et al., 2015). Our results supported this view that the high sugar resistance yeast Z. rouxii seems to have a stiffer cell wall (high degree of cross-linking in the β-glucan-chitin network) during the 60% w/v sugar stress and the change amplitude of cell size in Z. rouxii was smaller than that of S. cerevisiae during the 60% w/v sugar stress.
Transmembrane Transport
Under 60% w/v sugar stress, 25 genes in Z. rouxii involved in transmembrane transport were upregulated, including ZYRO0E09966g (FLR1), ZYRO0F02090g (TPO1), ZYRO0E10054g (TPO1), ZYRO0G14256g (NAB6), ZYRO0F17446g (GNP1), ZYRO0B03784g (ORF), ZYRO0C08140g (FCY2), ZYRO0G21252g (ZSP1), ZYRO0F14652g (DTR1), ZYRO0C04598 (THI7), ZYRO0F12606g (ZRC1), ZYRO0B06688g (ITR2), ZYRO0A00902g (PHO84), ZYRO0F14630g (SSU1), ZYRO0A00308g (AGP3), ZYRO0A04312g (ATP16), ZYRO0G20614g (YHC3), ZYRO0E02772g, ZYRO0B16896g, ZYRO0B03784g, ZYRO0G04796g, ZYRO0D17732g, ZYRO0C06424g, ZYRO0E09306g, and ZYRO0A11396g. Among them, polyamine transporter ZYRO0F02090g (TPO1) was extensively upregulated 24.8-fold after 4 h of sugar stress in Z. rouxii, but was downregulated 2.2-fold in S. cerevisiae. A previous study indicated that transcript levels of this gene are maintained at control levels in S. cerevisiae under 20% w/v and 40% w/v sugar stress (Erasmus et al., 2003; Jiménez-Martí et al., 2011). Krüger et al. (2013) have reported on the function of TPO1 in response to hydrogen peroxide (H2O2) stress. They found that TPO1 controls S. cerevisiae cell cycle delay and mediates the induction of antioxidant proteins such as Hsp70 and Hsp90. Furthermore, in overexpressing TPO1 cells, the induction of Hsps was delayed. Similar to this, in our experiments, ZYRO0F02090g (TPO1) was upregulated 24.8-fold and Z. rouxii had a cell cycle delay (Table 2 and Figure 5). ZrKAR2, encoding Kar2p (Hsp70), was downregulated 5.7-fold after 4 h of sugar stress but was increased about 2.9- and 8.2-fold after 8 and 20 h of sugar stress in Z. rouxii (Supplementary Figure S4). Then, Kar2p in Z. rouxii was significantly increased about 29-fold after about 27 h of 60% w/v sugar concentrations (Guo et al., 2016).
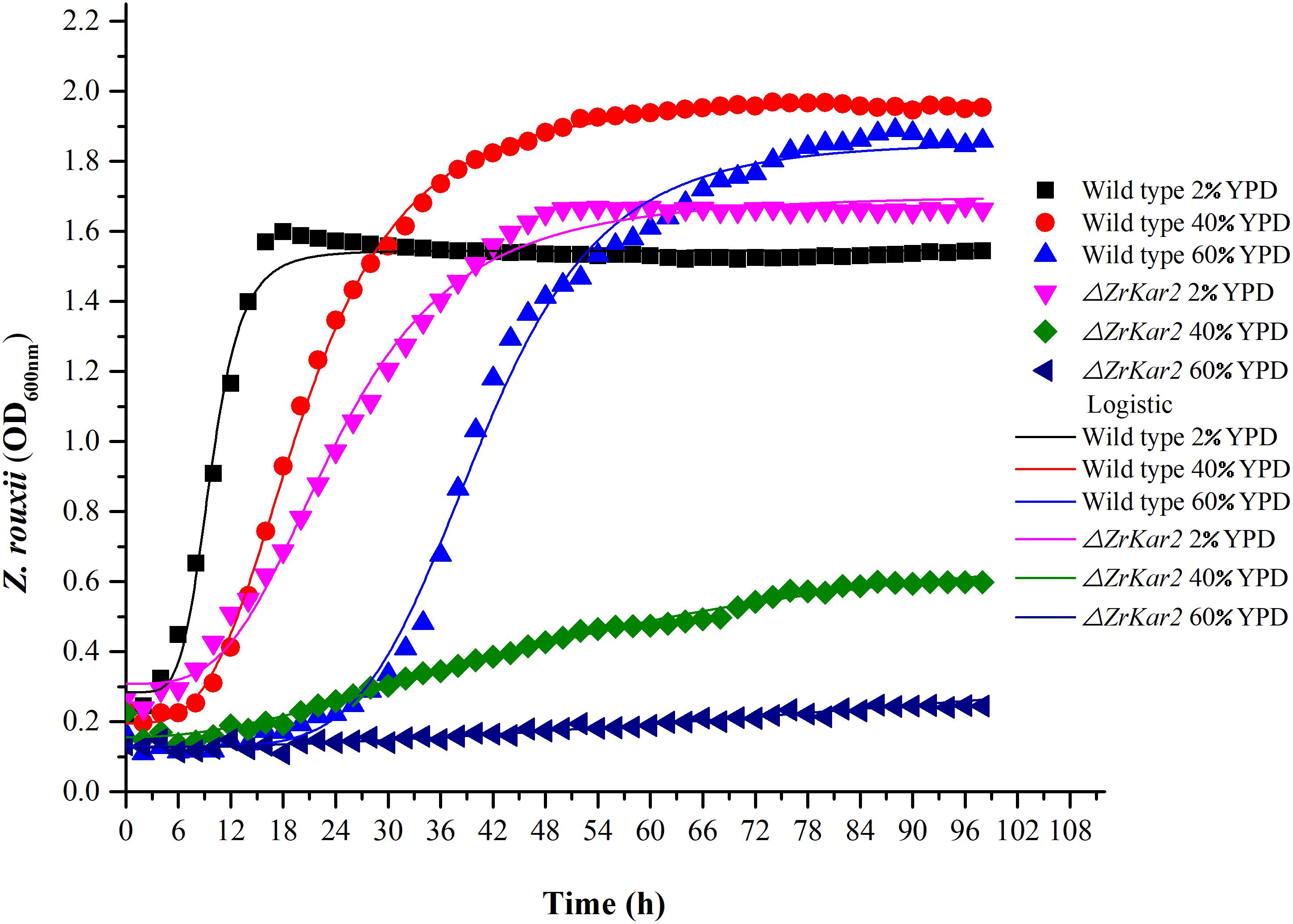
Figure 5. The growth of wild-type strain and mutant strain under 2, 40, and 60% w/v sugar concentrations.
Sugar Stress Resistance
To further uncover the role of ZrKAR2, we deleted this gene. As shown in Figure 5, wild-type strain of Z. rouxii grows well at normal sugar concentration (2% w/v), mild high sugar stress (40% w/v), and extremely high sugar stress (60% w/v). Moreover, as the sugar concentration increases, the adaptation time of the wild type to the exponential phase increases (cell cycle delay). However, the growth of ΔZrKar2 strain grows well at 2% w/v sugar concentration but was seriously inhibited under both 40% w/v sugar concentration and 60% w/v sugar stress. That is, the deletion of ZrKar2 did not affect its growth under low sugar conditions but causes the Z. rouxii to lose its high sugar tolerance. These results further demonstrated that the gene ZrKAR2 plays key important roles in the osmotic resistance of Z. rouxii. Finally, the logistic equation was applied to fit the growth curve of wild-type and ΔZrKar2 mutant strains. Except for the R2 of the fitting equation of mutant strain during 60% w/v sugar stress (0.95), the R2 values of other fitting equations are all greater than 0.99, which indicates that the equation can well fit the growth of Z. rouxii under different sugar stresses.
In addition, as shown in Figure 5, we speculated that the cell cycle delay in wild-type strains and ΔZrKar2 mutant strains was caused by the overexpression of TPO1 in cells. Due to the limitations of transcriptomics, overexpressed genes often do not necessarily coincide with genes that confer resistance to a particular stressor. Whether overexpression of TPO1 had the same function as reported in S. cerevisiae (Krüger et al., 2013) that controls the cell cycle delay and induces the expression of Kar2p in Z. rouxii still requires further verification.
Ribosome
Most of the genes involved in “ribosome” in the Z. rouxii response to 60% w/v sugar concentration were downregulated. This response seems to be consistent with other yeasts under different stresses, for example, Zygosaccharomyces parabilii under lactic acid stress (Ortizmerino et al., 2018), Pichia pastoris under methanol stress, and S. cerevisiae response to silicon encapsulation stress (Fazal et al., 2017). For example, ribosomal protein encoding genes, RPS13 and RPL4B, were downregulated after 4 h (Table 2) of extreme high sugar stress in Z. rouxii. Transcript levels of these genes are also strongly downregulated by pH stress in P. pastoris (Sauer et al., 2004). It is reported that cell synthesis ribosome requires energy, reducing gene expression associated with ribosome biogenesis and minimizing energy consumption (Fazal et al., 2017). Decreasing “ribosome” seems to be a general stress response in yeast (Fazal et al., 2017).
Discussion
On the one hand, extreme high sugar tolerance ability makes Z. rouxii an increasing threat to high sugary food industries (Dakal et al., 2014; Vermeulen et al., 2015). On the other hand, Z. rouxii also played a central role in the production of traditional fermented foods (such as soy sauce) (Watanabe et al., 2013). Extreme yeasts produce and accumulate large amounts of osmo-protective metabolites. Although this feature has been widely exploited in industrial bioprocesses, its molecular mechanism has been poorly investigated (Dakal et al., 2014). In addition, researchers explore how cell adaptation to hyperosmotic stress can help reveal pathological features associated with hyperosmotic stress such as pustular shock, diabetes, etc. (Silva et al., 2005). Combining with existing reports and our group’s research, we considered that Z. rouxii is an excellent candidate for analyzing extreme glucose tolerance in cells.
Our protein results (Guo et al., 2016) and gene knockout results (Figure 5) have suggested that ZrKAR2 protects Z. rouxii from the extreme high sugar stress. Simons et al. (1998) demonstrated that cell wall 1,6-β-glucan synthesis in S. cerevisiae depends on Kar2p (Hsp70 family). Although this study did not verify the direct relationship between Kar2p and glucan synthesis reported by previous researchers, in our results, both Kar2p and glucan synthesis play an important role in the extreme high glucose tolerance of Z. rouxii.
Hsp70 family has been reported to interact with virtually unfolded or misfolded proteins to regulate protein stability and activity (Wang et al., 2004; Hsieh et al., 2013). Hsp90 acts downstream of Hsp70 (Moran Luengo et al., 2018) and its contribution to protein folding is unclear. Moran Luengo et al. (2018) demonstrated that Hsp90 takes a key role in protein folding by breaking the folding barrier caused by Hsp70, empowering protein clients to fold on their own. Hsieh et al. (2013) found that Hsp90 in Z. rouxii responded to environmental stress. In our extreme high sugar stress, Hsp90-encoding gene, ZYRO0E07986g, was downregulated 5.3-fold. Whether Hsp90 and Kar2p work together to regulate the activity of target protein in Z. rouxii during 60% w/v sugar stress still requires further study.
Hao et al. (2018) reported that Hsp70 may modulate stress-activated MAPK signaling by inhibition of p38 (the mammalian homolog of Hog1 from yeast) to protect against heat stress-induced injury in rat small intestine. Glycerol production and retention under hyperosmotic stress are mediated by the Hog1 MAPK signaling cascade, which enhances glycerol synthesis. Recent reports have further confirmed that the Hog1 MAPK pathway is involved in both salt stress and sugar stress (Jiménez-Martí et al., 2011; Guo et al., 2016; Solieri et al., 2016; Wang et al., 2019). Combining recent reports with our results, we found the overlapping subset of salt- and sugar-responsive genes. For example, Iwaki et al. (2001) demonstrated that transcripts of ZrGCY1 and ZrGCY2 genes increased in salt-stressed (12% w/v) ATCC42981 cells compared to unstressed ones, suggesting that this salt concentration elicits the Gcy-Dak pathway (Iwaki et al., 2001), which includes the oxidation of glycerol to dihydroxyacetone (DHA). But in our experiments, ZYRO0F10032g (GCY1) decreased. These results highlighted that yeasts exploit different strategies to adapt in osmotic and salt stress. Solieri et al. (2014) also suggested that the differences in stress response could imply different adaptation mechanisms to sugar stress and salt stress. We speculated that yeast cell downregulated GCY1 in a high glucose environment in order to retain glycerol.
In addition, we also found the overlapping subset of salt- and sugar-responsive genes related to “structural constituent of ribosome.” We found that in our experiment condition, ribosome protein encoding genes, ZYRO0G11000g (RPL38) and ZYRO0A12606g (RPL26), were upregulated in Z. rouxii exposed to extreme high sugar stress but were downregulated in Z. rouxii under 12% w/v salt stress. These results also indicated that yeast have different strategies in response to sugar and salt stress.
In this study, we found that several genes involved in extreme high sugar stress tolerance, such as “transmembrane transport,” were selectively expanded in the Z. rouxii genome (Supplementary Table S2). In addition, we demonstrated by gene knockout that ZrKAR2 encoding Kar2p (Hsp70) significantly affected the growth of Z. rouxii at high sugar concentrations. According to previous reports and our results, we found that Kar2p in Z. rouxii seems to be associated with “glucan synthesis” and “transmembrane transport” and contributes to sugar stress resistance (Figure 6). Our analyses taken together suggest that Z. rouxii may have increased its sugar tolerance through duplication and/or mediates multiple genes involved in transmembrane transport, cell wall remodeling enzymes, ribosome, and Hsp defense response. These findings are important for an improved understanding of yeast adaptation to sugar stress and engineering the osmotolerance in other organisms.
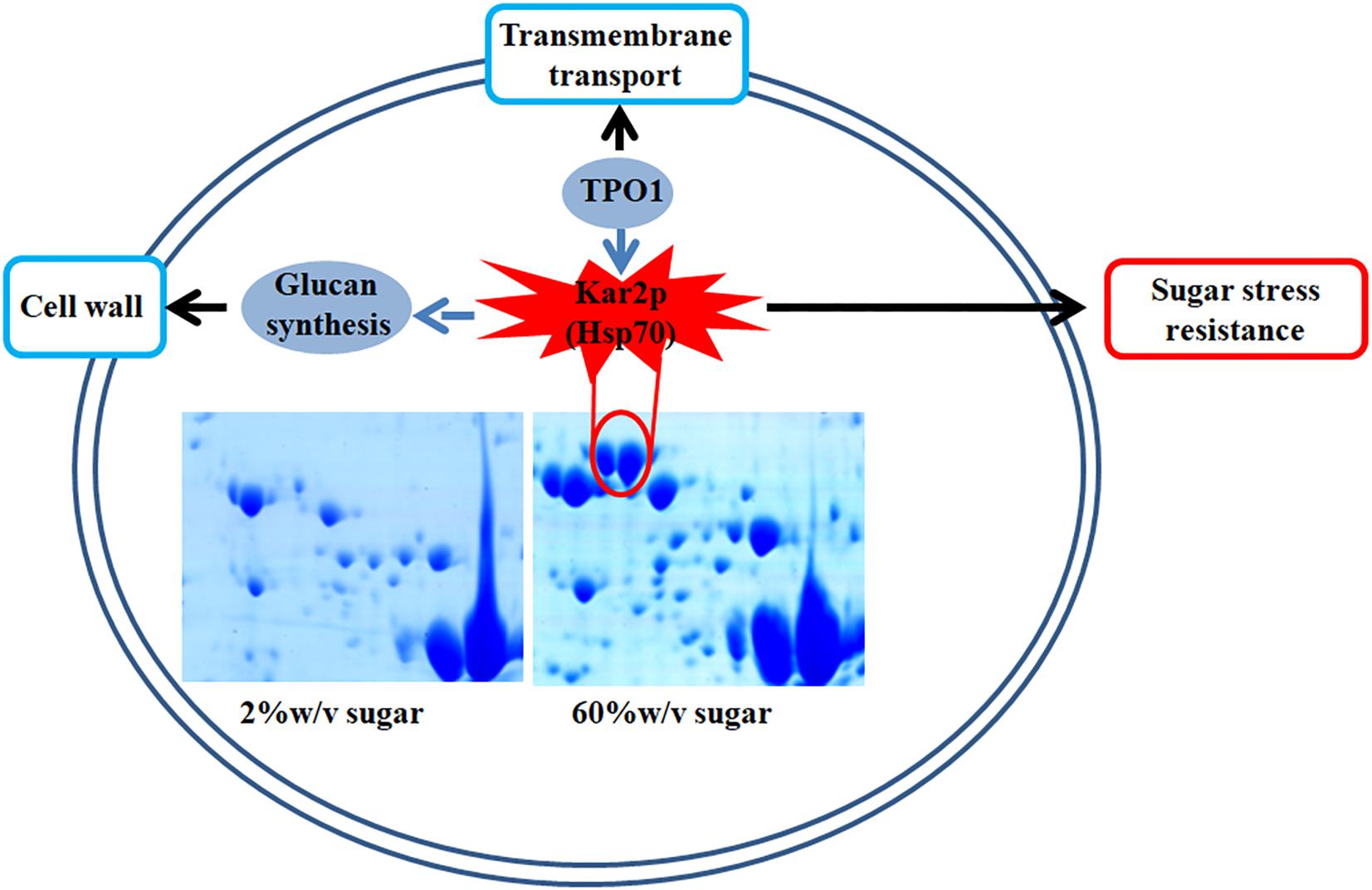
Figure 6. A schematic diagram illustrating the potential relationship of Kar2p (Hsp70 family), glucan synthesis, transmembrane transport, and sugar stress resistance.
Data Availability Statement
The datasets generated for this study can be found in the RNA-seq data have been deposited in the National Center for Biotechnology Information (NCBI), with accession code PRJNA437612.
Author Contributions
HG and TY conceived and designed the experiments. HG performed the experiments. HG, YQ, YY, and CN analyzed the data. HG, JW, and YZ drafted the manuscript. All authors read and approved the final version of the manuscript.
Funding
This research was supported by the Scientific and Technology Cooperation Project in Hong Kong, Macao, and Taiwan of China (2015DFT30130), the National Basic Research Program of China (2013FY113400), the National Natural Science Foundation of China (31671866), and the National Agricultural Products Quality and Safety Risk Assessment Program (GJFP201701302).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.03157/full#supplementary-material
References
Alonso, A., Belda, I., Santos, A., Navascués, E., and Marquina, D. (2015). Advances in the control of the spoilage caused by Zygosaccharomyces species on sweet wines and concentrated grape musts. Food Control 51, 129–134. doi: 10.1016/j.foodcont.2014.11.019
Brennan, T. C., Krömer, J. O., and Nielsen, L. K. (2013). Physiological and transcriptional responses of Saccharomyces cerevisiae to d-limonene show changes to the cell wall but not to the plasma membrane. Appl. Environ. Microbiol. 79, 3590–3600. doi: 10.1128/AEM.00463-13
Dakal, T. C., Solieri, L., and Giudici, P. (2014). Adaptive response and tolerance to sugar and salt stress in the food yeast Zygosaccharomyces rouxii. Int. J. Food Microbiol. 185, 140–157. doi: 10.1016/j.ijfoodmicro.2014.05.015
De, N. E., and Al, E. (2011). Controlling gene expression in response to stress. Nat. Rev. Genet. 12, 833–845. doi: 10.1038/nrg3055
Ene, I. V., Walker, L. A., Schiavone, M., Lee, K. K., Martin-Yken, H., Dague, E., et al. (2015). Cell wall remodeling enzymes modulate fungal cell wall elasticity and osmotic stress resistance. mBio 6:e00986-15. doi: 10.1128/mBio.00986-15
Erasmus, D. J., Van der Merwe, G. K., and Van Vuuren, H. J. J. (2003). Genome-wide expression analyses: metabolic adaptation of Saccharomyces cerevisiae to high sugar stress. FEMS Yeast Res. 3, 375–399. doi: 10.1016/S1567-1356(02)00203-9
Fazal, Z., Pelowitz, J., Johnson, P. E., Harper, J. C., Brinker, C. J., and Jakobsson, E. (2017). Three-dimensional encapsulation of Saccharomyces cerevisiae in silicate matrices creates distinct metabolic states as revealed by gene chip analysis. ACS Nano 11, 3560–3575. doi: 10.1021/acsnano.6b06385
Formosa, C., Schiavone, M., Martin-Yken, H., Francois, J. M., Duval, R. E., and Dague, E. (2013). Nanoscale effects of caspofungin against two yeast species, Saccharomyces cerevisiae and Candida albicans. Antimicrob. Agents Chemother. 57, 3498–3506. doi: 10.1128/AAC.00105-13
Genolévures, C., Souciet, J. L., Dujon, B., Gaillardin, C., Johnston, M., Baret, P. V., et al. (2009). Comparative genomics of protoploid Saccharomycetaceae. Genome Res. 19, 1696–1709. doi: 10.1101/gr.091546.109
Guo, H., Niu, C., Liu, B., Wei, J. P., Wang, H. X., Yuan, Y. H., et al. (2016). Protein abundance changes of Zygosaccharomyces rouxii in different sugar concentrations. Int. J. Food Microbiol. 233, 44–51. doi: 10.1016/j.ijfoodmicro.2016.05.003
Hao, Y., Feng, Y., Li, J., and Gu, X. (2018). Role of MAPKs in HSP70’s protection against heat stress-induced injury in rat small intestine. Biomed Res. Int. 2018:1571406. doi: 10.1155/2018/1571406
Hohmann, S. (2002). Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 66, 300–372. doi: 10.1128/MMBR.66.2.300-372.2002
Hsieh, Y. Y., Hung, P. H., and Leu, J. Y. (2013). Hsp90 regulates nongenetic variation in response to environmental stress. Mol. Cell 50, 82–92. doi: 10.1016/j.molcel.2013.01.026
Iwaki, T., Kurono, S., Yokose, Y., Kubota, K., Tamai, Y., and Watanabe, Y. (2001). Cloning of glycerol-3-phosphate dehydrogenase genes (ZrGPD1 and ZrGPD2) and glycerol dehydrogenase genes (ZrGCY1 and ZrGCY2) from the salt-tolerant yeast Zygosaccharomyces rouxii. Yeast 18, 737–744. doi: 10.1002/yea.722
Jiménez-Martí, E., Zuzuarregui, A., Gomar-Alba, M., Gutiérrez, D., Gil, C., and del Olmo, M. (2011). Molecular response of Saccharomyces cerevisiae wine and laboratory strains to high sugar stress conditions. Int. J. Food Microbiol. 145, 211–220. doi: 10.1016/j.ijfoodmicro.2010.12.023
Klipp, E., Nordlander, B., Kruger, R., Gennemark, P., and Hohmann, S. (2005). Integrative model of the response of yeast to osmotic shock. Nat. Biotechnol. 23, 975–982. doi: 10.1038/embor.2013.165
Krüger, A., Vowinckel, J., Mülleder, M., Grote, P., Capuano, F., Bluemlein, K., et al. (2013). Tpo1-mediated spermine and spermidine export controls cell cycle delay and times antioxidant protein expression during the oxidative stress response. EMBO Rep. 14, 1113–1119. doi: 10.1038/embor.2013.165
Lai, T., Wang, Y., Fan, Y., Zhou, Y., Bao, Y., and Zhou, T. (2017). The response of growth and patulin production of postharvest pathogen Penicillium expansum to exogenous potassium phosphite treatment. Int. J. Food Microbiol. 244, 1–10. doi: 10.1038/embor.2013.165
Lin, J. Q., Zhao, X. X., Zhi, Q. Q., Zhao, M., and He, Z. M. (2013). Transcriptomic profiling of Aspergillus flavus in response to 5-azacytidine. Fungal Genet. Biol. 56, 78–86. doi: 10.1016/j.fgb.2013.04.007
Liu, H., Dai, L., Wang, F., Li, X., Liu, W., Pan, B., et al. (2020). A new understanding: gene expression, cell characteristic and antioxidant enzymes of Zygosaccharomyces rouxii under the D-fructose regulation. Enzyme Microb. Technol. 132:109409. doi: 10.1016/j.enzmictec.2019.109409
Ma, T., Wang, J., Zhou, G., Yue, Z., Hu, Q., Chen, Y., et al. (2013). Genomic insights into salt adaptation in a desert poplar. Nat. Commun. 4:2797. doi: 10.1038/ncomms3797
Martorell, P., Stratford, M., Steels, H., Fernándezespinar, M. T., and Querol, A. (2007). Physiological characterization of spoilage strains of Zygosaccharomyces bailii and Zygosaccharomyces rouxii isolated from high sugar environments. Int. J. Food Microbiol. 114, 234–242. doi: 10.1016/j.ijfoodmicro.2006.09.014
Marvig, C. L., Kristiansen, R. M., Madsen, M. G., and Nielsen, D. S. (2015). Identification and characterisation of organisms associated with chocolate pralines and sugar syrups used for their production. Int. J. Food Microbiol. 185, 167–176. doi: 10.1016/j.ijfoodmicro.2014.05.017
Mitchell, A., Wei, P., and Lim, W. A. (2015). Oscillatory stress stimulation uncovers an Achilles’ heel of the yeast MAPK signaling network. Science 350, 1379–1383. doi: 10.1126/science.aab0892
Moran Luengo, T., Kityk, R., Mayer, M. P., and Rudiger, S. G. D. (2018). Hsp90 breaks the deadlock of the Hsp70 chaperone system. Mol. Cell 70, 545–552. doi: 10.1016/j.molcel.2018.03.028
Niu, C., Yuan, Y. H., Hu, Z. Q., Wang, Z. L., Liu, B., Wang, H. X., et al. (2016). Accessing spoilage features of osmotolerant yeasts identified from kiwifruit plantation and processing environment in Shaanxi, China. Int. J. Food Microbiol. 232, 126–133. doi: 10.1016/j.ijfoodmicro.2016.03.012
Ortizmerino, R. A., Kuanyshev, N., Byrne, K. P., Varela, J. A., Morrissey, J. P., and Porro, D. (2018). Transcriptional response to lactic acid stress in the hybrid yeast Zygosaccharomyces parabailii. Appl. Environ. Microbiol. 84:e02294-17. doi: 10.1128/AEM.02294-17
Pham, T. K., and Wright, P. C. (2008). The proteomic response of Saccharomyces cerevisiae in very high glucose conditions with amino acid supplementation. J. Proteome Res. 7, 4766–4774. doi: 10.1021/pr800331s
Pillet, F., Lemonier, S., Schiavone, M., Formosa, C., Martinyken, H., Francois, J. M., et al. (2014). Uncovering by Atomic Force Microscopy of an original circular structure at the yeast cell surface in response to heat shock. BMC Biol. 12:6. doi: 10.1186/1741-7007-12-6
Qiu, Z., Wu, X., Gao, W., Zhang, J., and Huang, C. (2018). High temperature induced disruption of the cell wall integrity and structure in Pleurotus ostreatus mycelia. Appl. Microbiol. Biotechnol. 102, 6627–6636. doi: 10.1007/s00253-018-9090-6
Sauer, M., Branduardi, P., Gasser, B., Valli, M., Maurer, M., Porro, D., et al. (2004). Differential gene expression in recombinant Pichia pastoris analysed by heterologous DNA microarray hybridisation. Microb. Cell Fact. 3:17. doi: 10.1186/1475-2859-3-17
Silva, R. D., Sotoca, R., Johansson, B., Ludovico, P., Sansonetty, F., Silva, M. T., et al. (2005). Hyperosmotic stress induces metacaspase- and mitochondria-dependent apoptosis in Saccharomyces cerevisiae. Mol. Microbiol. 58, 824–834. doi: 10.1111/j.1365-2958.2005.04868.x
Simons, J. F., Ebersold, M., and Helenius, A. (1998). Cell wall 1,6-beta-glucan synthesis in Saccharomyces cerevisiae depends on ER glucosidases I and II, and the molecular chaperone BiP/Kar2p. EMBO J. 17, 396–405. doi: 10.1093/emboj/17.2.396
Solieri, L., Dakal, T. C., and Bicciato, S. (2014). Quantitative phenotypic analysis of multistress response in Zygosaccharomyces rouxii complex. FEMS Yeast Res. 14, 586–600. doi: 10.1111/1567-1364.12146
Solieri, L., Vezzani, V., Cassanelli, S., Dakal, T. C., Pazzini, J., and Giudici, P. (2016). Differential hypersaline stress response in Zygosaccharomyces rouxii complex yeasts: a physiological and transcriptional study. FEMS Yeast Res. 16:fow063. doi: 10.1093/femsyr/fow063
Vaupotic, T., Veranic, P., Jenoe, P., and Plemenitas, A. (2008). Mitochondrial mediation of environmental osmolytes discrimination during osmoadaptation in the extremely halotolerant black yeast Hortaea werneckii. Fungal Genet. Biol. 45, 994–1007. doi: 10.1016/j.fgb.2008.01.006
Vermeulen, A., Marvig, C. L., Daelman, J., Xhaferi, R., Nielsen, D. S., and Devlieghere, F. (2015). Strategies to increase the stability of intermediate moisture foods towards Zygosaccharomyces rouxii: the effect of temperature, ethanol, pH and water activity, with or without the influence of organic acids. Food Microbiol. 45, 119–125. doi: 10.1016/j.fm.2014.01.003
Wang, D., Hao, Z., Zhao, J., Jin, Y., Huang, J., Zhou, R., et al. (2019). Comparative physiological and transcriptomic analyses reveal salt tolerance mechanisms of Zygosaccharomyces rouxii. Process Biochem. 82, 59–67. doi: 10.1016/j.procbio.2019.04.009
Wang, H., Hu, Z., Long, F., Niu, C., Yuan, Y., and Yue, T. (2015). Characterization of osmotolerant yeasts and yeast−like molds from apple orchards and apple juice processing plants in China and investigation of their spoilage potential. J. Food Sci. 80, M1850–M1860. doi: 10.1111/1750-3841.12946
Wang, H., Lei, Y., Yan, L., Wan, L., Ren, X., Chen, S., et al. (2016). Functional genomic analysis of Aspergillus flavus interacting with resistant and susceptible peanut. Toxins (Basel) 8:46. doi: 10.3390/toxins8020046
Wang, W., Vinocur, B., Shoseyov, O., and Altman, A. (2004). Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 9, 244–252. doi: 10.1016/j.tplants.2004.03.006
Wang, Y., Feng, K., Yang, H., Zhang, Z., Yuan, Y., and Yue, T. (2018). Effect of cinnamaldehyde and citral combination on transcriptional profile, growth, oxidative damage and patulin biosynthesis of Penicillium expansum. Front. Microbiol. 9:597. doi: 10.3389/fmicb.2018.00597
Watanabe, J., Uehara, K., and Mogi, Y. (2013). Adaptation of the osmotolerant yeast Zygosaccharomyces rouxii to an osmotic environment through copy number amplification of FLO11D. Genetics 195, 393–405. doi: 10.1534/genetics.113.154690/-/DC1
Watanabe, Y., Tsuchimoto, S., and Tamai, Y. (2004). Heterologous expression of Zygosaccharomyces rouxii glycerol 3-phosphate dehydrogenase gene (ZrGPD1) and glycerol dehydrogenase gene (ZrGCY1) in Saccharomyces cerevisiae. FEMS Yeast Res. 4, 505–510. doi: 10.1016/S1567
Xiang, Q., Liu, X., Li, J., Liu, S., Zhang, H., and Bai, Y. (2018). Effects of dielectric barrier discharge plasma on the inactivation of Zygosaccharomyces rouxii and quality of apple juice. Food Chem. 254, 201–207. doi: 10.1016/j.foodchem.2018.02.008
Xie, J. L., Qin, L., Miao, Z., Grys, B. T., Diaz, J. D. L. C., Ting, K., et al. (2017). The Candida albicans transcription factor Cas5 couples stress responses, drug resistance and cell cycle regulation. Nat. Commun. 8:499. doi: 10.1038/s41467-017-00547-y
Keywords: Z. rouxii, S. cerevisiae, high sugar stress, osmotic resistance, RNA-sequencing
Citation: Guo H, Qiu Y, Wei J, Niu C, Zhang Y, Yuan Y and Yue T (2020) Genomic Insights Into Sugar Adaptation in an Extremophile Yeast Zygosaccharomyces rouxii. Front. Microbiol. 10:3157. doi: 10.3389/fmicb.2019.03157
Received: 09 August 2019; Accepted: 30 December 2019;
Published: 11 February 2020.
Edited by:
Rosanna Tofalo, University of Teramo, ItalyReviewed by:
Lisa Solieri, University of Modena and Reggio Emilia, ItalyNuno Pereira Mira, University of Lisbon, Portugal
Tikam Chand Dakal, Manipal University Jaipur, India
Copyright © 2020 Guo, Qiu, Wei, Niu, Zhang, Yuan and Yue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tianli Yue, eXVldGw0MjFAbnd1LmVkdS5jbg==
 Hong Guo
Hong Guo Yue Qiu2
Yue Qiu2 Jianping Wei
Jianping Wei Chen Niu
Chen Niu Tianli Yue
Tianli Yue