
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 23 January 2020
Sec. Food Microbiology
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.03059
This article is part of the Research Topic Natural Bioactive Substances As An Alternative Solution Against Food Pathogenic Bacteria: Interaction With The QS System View all 9 articles
Staphylococcus aureus is an important pathogen that is associated with nosocomial infections, as well as food poisoning. This bacterium is resistant to antimicrobial agents and can survive in a wide range of environmental conditions. The aim of this study was to measure the uptake and release of amino acids by S. aureus at mid-exponential and stationary phases of growth following exposure to a combination of conditions including variations in temperature, pH and NaCl. Bacterial cells were grown up to mid-exponential and stationary phases in tryptic soy broth (TSB), where the supernatants were collected for analyses of amino acids to determine the uptake and release characteristics. The uptake/release of amino acids was estimated by subtracting the initial levels of the free amino acids in the media from those measured at mid-exponential and stationary phases of growth. When cells were grown at ideal conditions, the analyses revealed that significant uptake of amino acids had occurred by stationary phase compared with the mid-exponential phase. A substantial release of valine and tyrosine into the external media was observed by cells at stationary phase. At both phases, the uptake and release patterns were significantly different between cells grown under ideal control conditions, when compared with those grown under various combinations of sub-optimal environmental conditions. The analyses of the supernatants harvested from controls and treatment groups at exponential phase indicated that the total uptake of amino acids was reduced approximately five times by cells grown with addition of 2.5% NaCl or with pH6 at 35°C, and 2-fold by cells grown at pH8 at 35°C. However, the final quantities of amino acids taken up by cells grown to stationary phase did not significantly alter between control and treated samples. Valine was found to be the most abundant amino acid that was significantly released into the media at stationary phase by both control and treated samples. It was evident that diverse environmental conditions resulted in differential patterns of amino acid uptake and release during adaptation to designated conditions.
Staphylococcus aureus is an important pathogen capable of causing a wide range of infections from mild skin and food-borne intoxications to life-threatening sepsis and endocarditis (Josse et al., 2017). Staphylococci have been shown to alter their cytoplasmic composition in response to alterations in the environment such as variation in temperatures, pH and osmolality (Alreshidi et al., 2013; Onyango and Alreshidi, 2018). One of the key areas of interest in food processing industries is the study of bacterial adaptation in harsh environmental conditions, as the understanding of bacterial adaptation and survival mechanisms would pave ways to reduce food-borne illnesses (Zeaki et al., 2014). Survival of bacteria relies on their ability to induce the optimum metabolic homeostasis required for growth in the altered environments (Wolf et al., 2008; Solis et al., 2014; Alreshidi et al., 2015; Onyango and Alreshidi, 2018). Adjustments to environmental influences can lead to significant changes in morphological structure, including cell/colony sizes, and induce the production of biofilms (Zhu et al., 2007; Onyango et al., 2012; Onyango et al., 2013; Crompton et al., 2014).
Small colony variants (SCVs) can be induced as a response to altered environmental conditions including variations in pH, osmotic pressure, temperatures and exposures to antimicrobial agents, etc. (von Eiff, 2008; Onyango et al., 2012; Onyango et al., 2013; Proctor et al., 2014). The occurrence of SCVs have been observed in medical samples, animals (Tkadlec et al., 2015) and even food (Li et al., 2016). SCVs are a slow-growing sub-population of bacteria that are characterized with decreased metabolic activity and increased resistance to antimicrobial agents (von Eiff et al., 2000). SCVs of S. aureus were found to exhibit enhanced biofilm formation in comparison to their corresponding wild-type strains (Singh et al., 2010). The exposure of S. aureus to variations in NaCl concentrations led to increased production of biofilm, with concomitant increase in the expression of genes associated with biofilm such as the icaA gene. Thus, the use of NaCl in processed and canned foods may increase biofilm production of S. aureus (Lee et al., 2014). Further, biofilm formation can occur in response to other non-ideal growth conditions such as exposure to alkaline environments (Jones et al., 2015; Jaishankar and Srivastava, 2017). Both SCV phenotypes and biofilm greatly contribute to the profound resistance and stable adhesion, which eventually assist the bacteria to survive under changed environmental conditions (Yao et al., 2005; Li et al., 2016).
A prior investigation has shown that selective amino acids were taken up by the biofilm cultures of S. aureus when compared with their corresponding planktonic counterparts (Ammons et al., 2014). Hence, it was hypothesized that amino acid uptake is essential for staphylococcal biofilm development and adjustment to changes in pH (Beenken et al., 2004; Zhu et al., 2007). Another study showed that limited proline in the media impaired staphylococcal growth and survival (Schwan et al., 2004). likewise, combinations of subtle environmental parameters led to substantial modifications in amino acid uptake profiles associated with significant patterns of cytoplasmic amino acids (Murphy et al., 2018). Similarly, significant changes in cytoplasmic amino acid compositions of S. aureus in response to variation in pH, temperature and osmotic pressure were observed (Alreshidi et al., 2016). Exometabolome analysis of different strains of S. aureus exhibited substantial differences in the uptake of extracellular metabolites (Dorries and Lalk, 2013) and cells grown in culture with limited supplies of glucose also altered their uptake of metabolites (Liebeke et al., 2011). This was proposed as an adaptive mechanism by the bacteria to utilize amino acids for energy and alternative resources to support growth based on nutrient availabilities from the surrounding environment (Zhu et al., 2007; Stipetic et al., 2016). To date, the studies have demonstrated that the exposure of S. aureus to subtle variations in temperature, pH and NaCl resulted in significant alterations in cytoplasmic amino acids, proteins and external morphological structures compared with bacterial cells grown under normal conditions (Crompton et al., 2014; Wehrli et al., 2014; Alreshidi et al., 2015; Alreshidi et al., 2016; Alreshidi et al., 2019). It has also been shown that S. aureus consumed a significant amount of glycine when incubated in minimal media supplemented with 5% NaCl compared to the cells grown in normal conditions (Alreshidi et al., 2019). The present study investigated whether S. aureus could adjust the amino acid uptake profiles from growth media in response to varied environmental parameters (pH, temperature and NaCl) when grown to mid-exponential and stationary phases. It was hypothesized that S. aureus would require a rapid adaptation to changes in the temperature, pH and osmolality that require simultaneous rapid alterations in the demand of biosynthesis of metabolites and proteins essential for bacterial survival and growth. Therefore, to do so, the bacterium would alter its amino acid uptake profiles to obtain the optimum metabolism required for adaptation and survival following exposure to changes in the environmental conditions.
The strain of S. aureus used in current study was isolated from a patient who hospitalized due to prolonged muscle pain (Butt et al., 1998). This bacterial strain used in the following studies to investigate metabolic and proteomic changes to environmental stresses (Alreshidi et al., 2015, 2016, 2019). The bacterium has been maintained as culture stock on horse blood agar (HBA) and preserved appropriately on sterile glass beads at −80°C with a consistent sub-culturing to preserve cells viability. The identity of the bacteria strain was confirmed regularly using APITM Staph biochemistry and through PCR (Brown et al., 2001).
Staphylococcus aureus was cultured in a sub-optimal of environmental conditions for analyses of amino acid uptake by S. aureus following growth in tryptic soy broth media (TSB) (Oxoid Ltd-Australia). The reference control included cells grown under ideal conditions of pH7 at 37°C with no added NaCl in tryptic soy broth medium (TSB) and three sets of experimental conditions were applied with (1) pH7 at 37°C with NaCl added (2.5%) in TSB; (2) 35°C and pH6 with no added NaCl in TSB; and (3) 35°C and pH8 with no added NaCl in TSB.
An overnight starter culture (50 ml) of S. aureus was grown for 16 h in Tryptic Soy Broth (TSB) at 37°C with constant agitation (120 rpm) to be used as an inoculum for the growth experiments. Replicates of each condition containing 95 ml TSB culture media were inoculated with 5 ml of overnight culture (OD600 = 0.1 ≈ 108 CFU/ml)in 500 ml conical flasks and incubated at 37°C with constant agitation (120 rpm). Growth was monitored by aseptically monitoring the absorbance at 600 nm of the cultures to estimate cell numbers, so that the cultures could be harvested at either mid-exponential or stationary phases. Culture supernatants were generated by centrifugation at 6,000 × g for 25 min and the supernatants removed for subsequent analyses.
Preliminary analyses of the tryptic soy broth media (TSB) were performed to determine that it contained 23 amino acids (listed in Table 1) and their derivatives, which were available for direct uptake by the bacterial cells. Utilization of these amino acids by the bacteria for growth was assessed by collecting five ml of S. aureus cultures grown in TSB media at both mid-exponential and stationary phases. The collected supernatants were filtered through a Millex® membrane filter (0.22 mμ) and stored at −20°C for further analysis. 100 μl of sterile TSB media and the filtered supernatants harvested at mid-exponential phase were diluted with 300 μl of sterile Milli-Q water, and the 100 μl harvested at stationary phase was diluted with 100 μl with Milli-Q water. The dilution was done to avoid overwhelming the analytical capacity of the gas chromatography column. An aliquot of 100 μl of diluted supernatants was then processed and analyzed using a commercial analytical kit (Phenomenex® EZ: faastTM). The technique was processed according to the manufacturer’s instructions. The derivatized amino acids were then analyzed using an Agilent gas chromatograph (Hewlett Packard HP 6890 series) coupled with flame ionization detector (GC-FID) which was calibrated to measure more than 40 amino acid metabolites as previously described (Onyango et al., 2012). The injection volume was 2 μl with splitless mode and flow rate of the carrier gas (Helium) was 0.5 ml/min. Nor-valine was used as an internal standard to calculate the concentrations of amino acids present in the sample as nmol/100 μl. The results were normalized by multiplied by dilution factors.
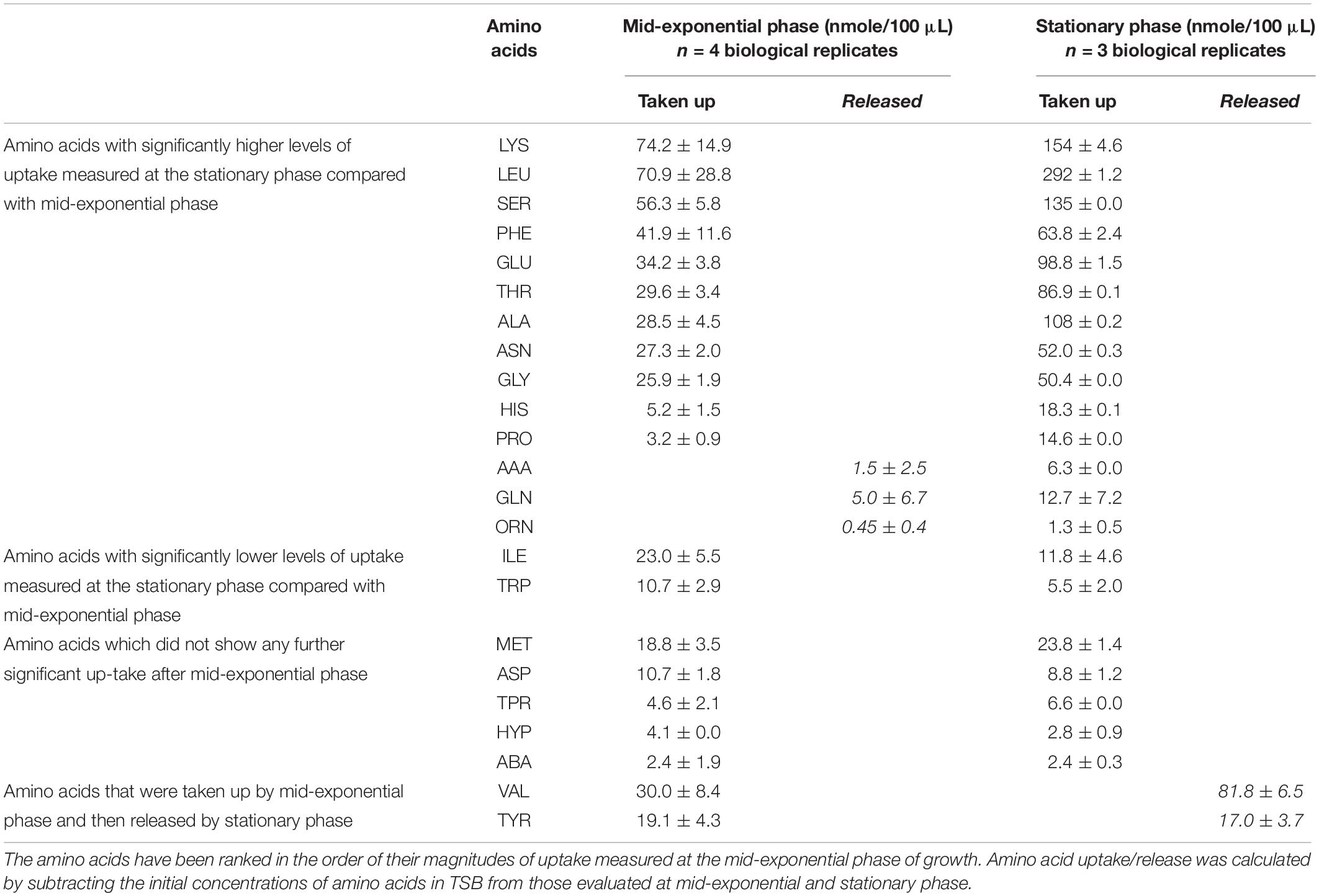
Table 1. Comparisons of the quantities of amino acids taken up or released by S. aureus at mid-exponential and stationary phases, during growth under optimal conditions (mean ± SD, P < 0.05).
The data generated from gas chromatography and flame ionization (GC−FID) were evaluated via ANOVA to detect the uptake of amino acids that were significantly altered following growth in variations in temperature, pH and NaCl (Statistica, TIBCO Software Inc. [2017], data analysis software system, version 13)1. Principal component analyses (PCA) were also performed. The data were subjected to mean centering and unit variance scaling before PCA calculations. The model complexity and validity were assessed by cross validation as applied in the software.
Cultures of S. aureus were grown in TSB to mid-exponential and stationary phases of growth under optimal conditions (pH7, 37°C and no added NaCl). The analyses of the amino acid compositions in cultures supernatants revealed the presence of 23 amino acids and amino acid derivatives (Table 1). The analysis indicated that lysine, leucine and serine were taken up in the greatest quantities at both phases, and it was noted that the uptake of leucine was 4 times higher at stationary phase compared with the exponential phase, whereas serine and lysine were 2.4 and 2-times higher respectively. Valine and tyrosine were taken up during growth to mid-exponential phase of growth, but it was evident that by stationary phase, these amino acids had been released back into the external medium to yield a final concentration higher than present in the original TSB medium. The uptake of five amino acids did not significantly change between phases, including aspartic acid, tryptophan and methionine, despite their high quantity in the initial growth media. Small quantities of glutamine were released into the medium by mid-exponential phase, but some uptake was evident by stationary phase. Isoleucine and tryptophan were taken up by mid exponential phase, but then appeared to be released back into the medium by the stationary phase.
The differences in the uptake of amino acids from the culture supernatants were additionally investigated through principal component analysis (PCA). As per the PCA plot, the biological replicates for each phase were tightly grouped, but well resolved by the uptake/release of amino acids (Figure 1A). The samples of amino acid uptake/release estimated at the mid-exponential phase were spread along with the negative component 1 while the replicates of amino acid uptake from culture supernatants measured at the stationary phase of growth were distributed at the positive side of component 1 (Figure 1A). The PCA loading scatterplot analysis indicated that distinct amino acid uptake/release contributed substantially to the discrimination of growth phases (Figure 1B).
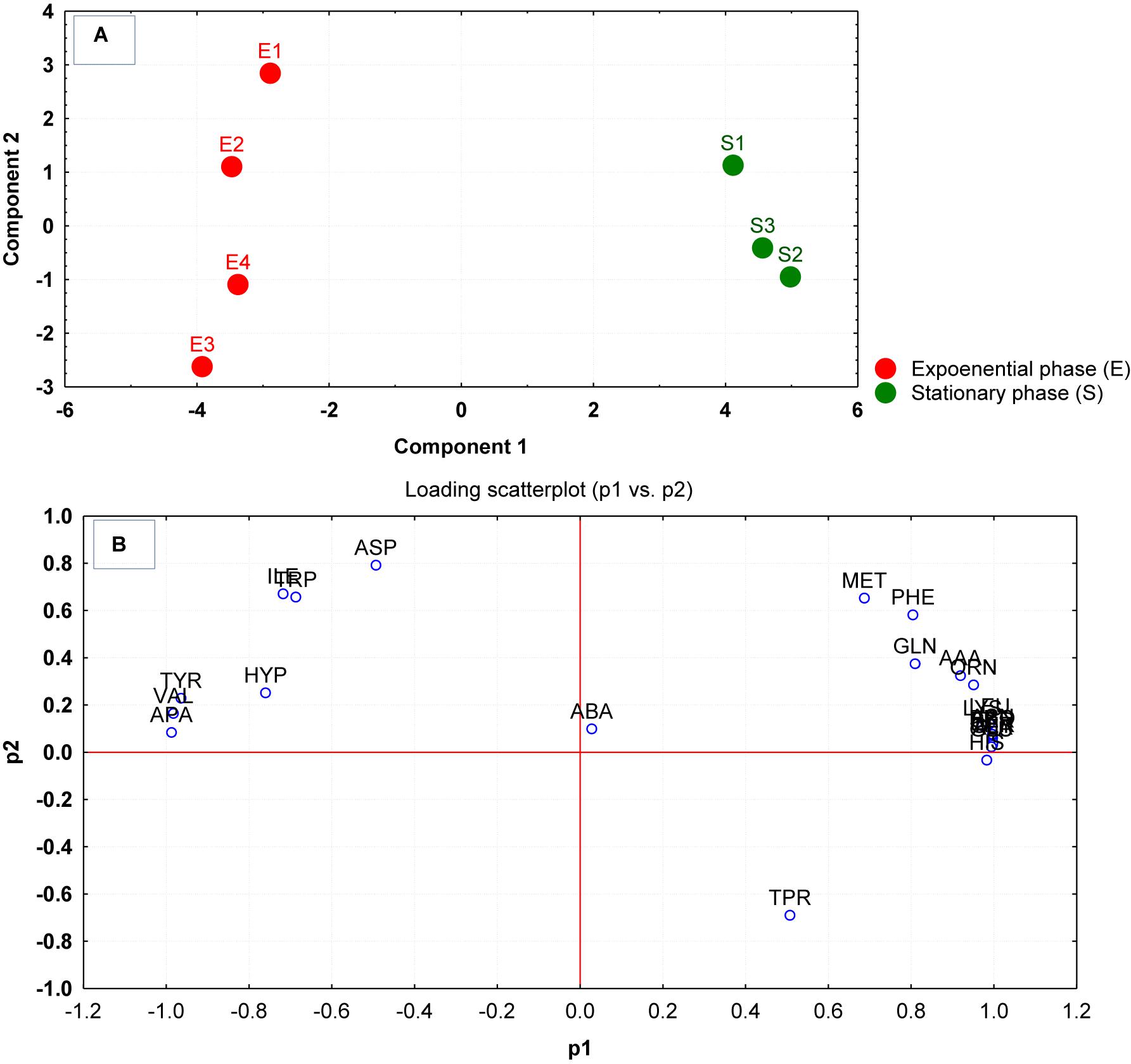
Figure 1. (A) Principal component analysis (PCA) scores generated from the amino acid compositions of the culture supernatants after growth were plotted for each replicate measured at the mid-exponential (E1–E4) and stationary phase (S1–S4). (B) The corresponding loading scatter plot of the amino acids (abbreviations from Table 1) indicates the influence of amino acid uptake/release on the clustering and separation on the PCA plot.
Cultures of S. aureus (4 replicates each) were grown to mid-exponential phase of growth under various combinations of sub-optimal environmental conditions with temperature ranging from 35 to 37°C, pH 6–8 and higher NaCl (0–2.5%). The supernatants were collected at mid-exponential phase of growth to analyze the uptake/release of amino acids. The amino acid uptake and release determined in each of the replicates were reproducible within each treatment group with specific changes in the uptake/release of amino acids (Table 2). On analyzing the uptake of the amino acids in controls, lysine, leucine, and serine were found to be the major amino acids extracted from the culture media, accounting for 14.5, 13.9, and 11% of total amino acids consumed, respectively (Table 2). However, when bacterial cells were grown under the alternative environmental conditions designated B–D in Table 2, the uptake of lysine was greatly diminished and leucine was actually released into the external medium in treatments B and C. Serine and glycine became the major amino acids taken up from the media in treatments B–D. Various specific alterations in amino acid uptake and release associated with the different environmental conditions were evident in Table 2. At pH8 and 35°C with no added NaCl, glutamic acid was the second most consumed amino acid after serine. The total quantities of amino acids taken up was 3.5 times lower when bacterial cells were grown in the presence of 2.5% NaCl in treatment B. Reduction of temperature to 35°C under more acidic (C) or alkali (D) conditions resulted in a 3.3 and 1.8-fold decline in the total uptake of amino acids, respectively.
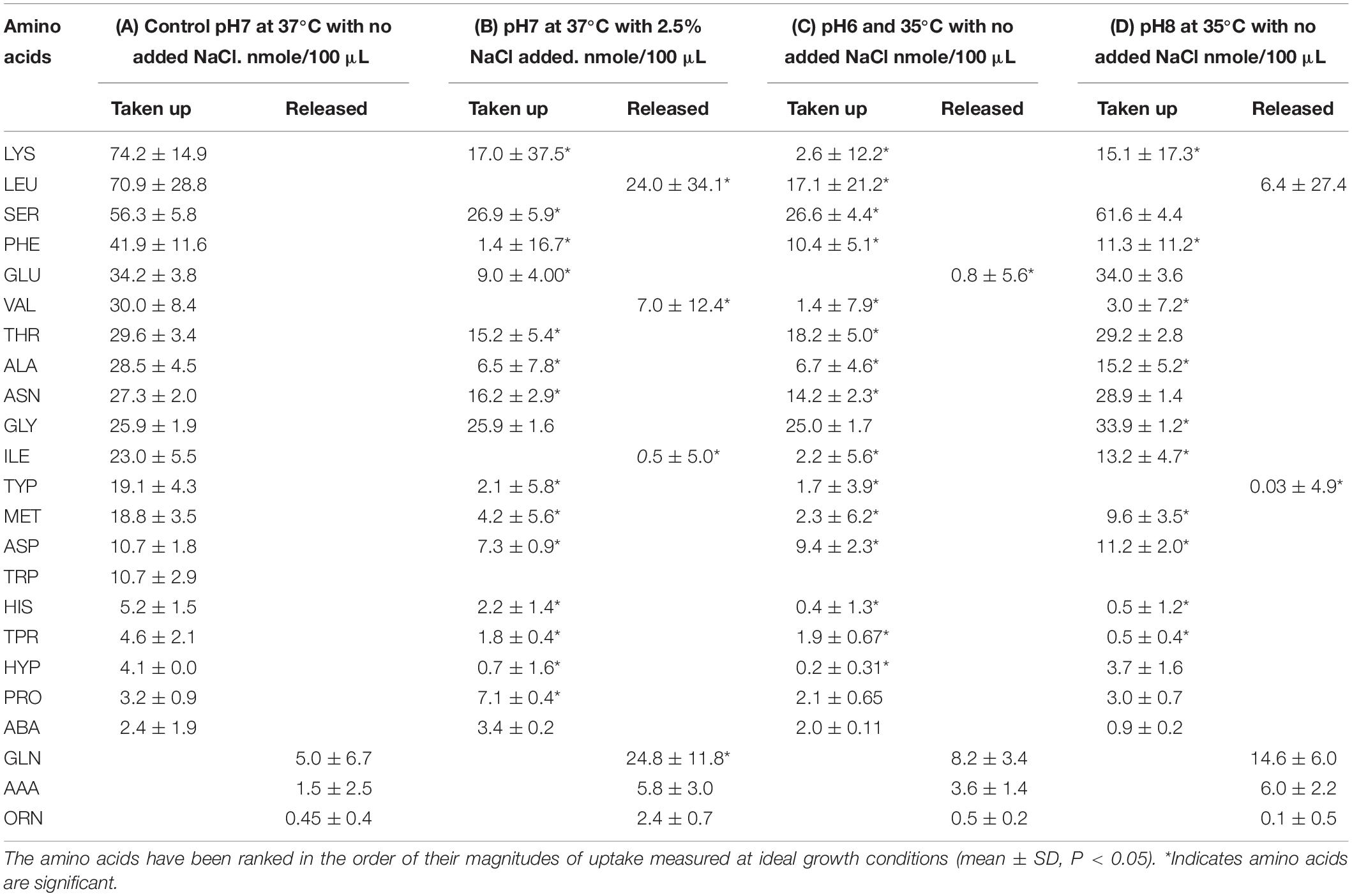
Table 2. Comparisons of the quantities of amino acids taken up or released by S. aureus at the mid-exponential phase of growth under optimal conditions (A) compared with growth under sub-optimal environmental conditions (B–D).
The total quantities of amino acids released was almost six times higher when bacterial cells were exposed to 2.5% NaCl whereas 4.4 and 2.9 times higher when cells were grown at 35°C under more acidic (C) or alkali (D) conditions, respectively.
Multivariate analyses using PCA revealed significant variations between the amino acid compositions of the culture supernatants from the different treatment regimens (Figure 2A). The result of the PCA indicated that the cells grown at pH 8 and 35°C with no added NaCl (D) were the most distinct group from the control and other treatment groups. Groups B and C had more similar but separable amino acid uptake profiles, and groups B–D were very well distinguished from those grown under ideal conditions (A). The loading scatterplot indicates the significance of the variables (amino acids) on PCA plot separation and clustering of the experimental groups (Figure 2B).
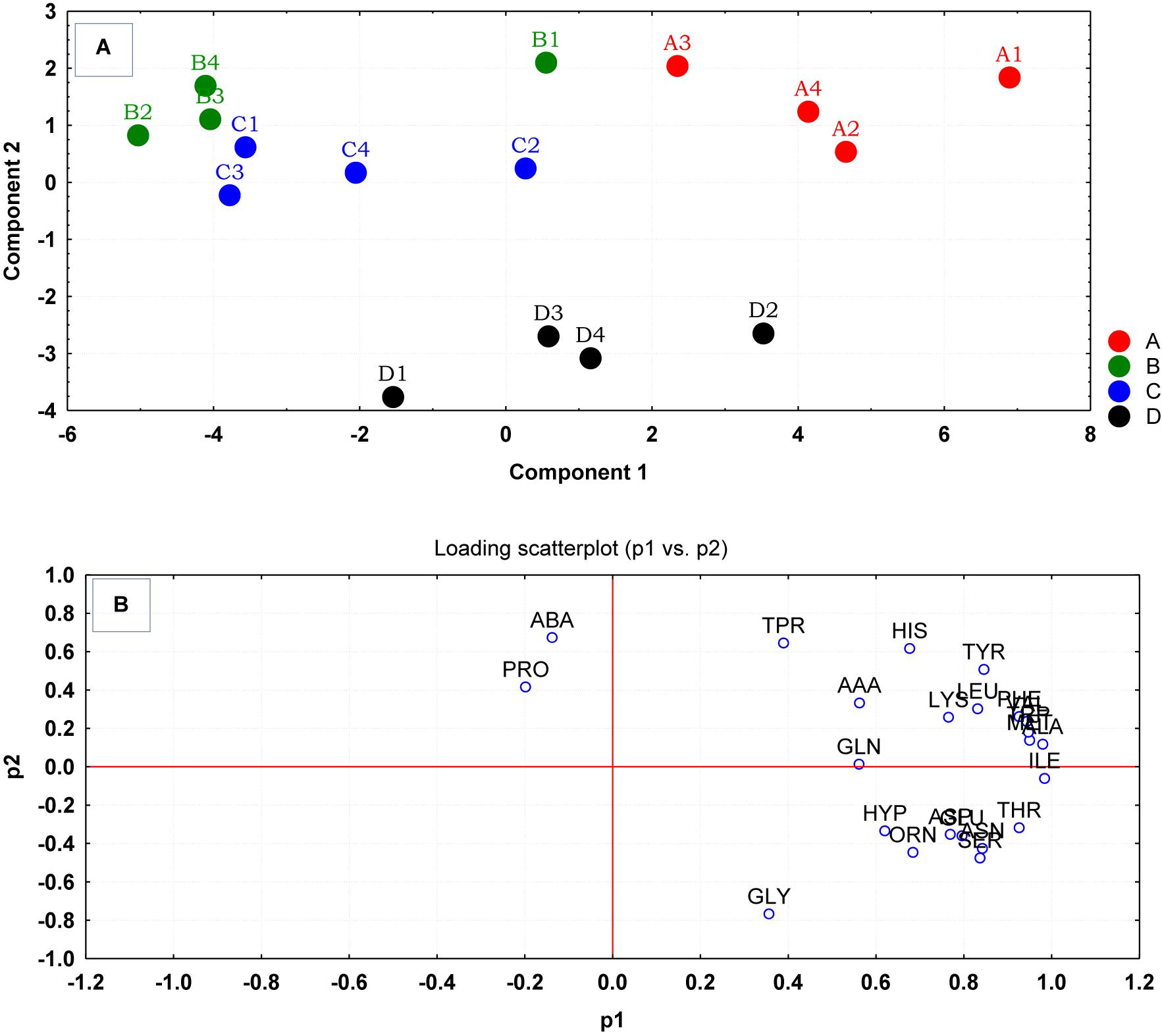
Figure 2. (A) Principal component analysis (PCA) scores generated from the amino acid uptake and release data, were plotted for each replicate culture of S. aureus grown under ideal conditions (A) and those grown under sub-optimal conditions (B–D) evaluated at the mid-exponential phase of growth. (B) The loading scatterplot of the amino acids (abbreviations from Table 1) indicated the influence of variables in PCA plot. The cells were grown under the following conditions: (A1–A4) Control cultures were grown under ideal conditions at pH7 and 37°C with no added NaCl; (B1–B4) Cultures were grown at pH 7 and 37°C with 2.5% NaCl; (C1–C4) Cultures were grown at 35°C and pH6 with no added NaCl; and (D1–D4) Cultures were grown at 35°C and pH8 with no added NaCl.
Cultures of S. aureus grown under the same environmental conditions A–D, were harvested at the stationary phase of growth to assess the impact on the total uptake and release of amino acids during culture growth. Under ideal growth conditions (A), lysine, leucine, and serine continued to be the most utilized amino acids from the medium (Table 3). This trend was similar for treatments B and D, but treatment C under the alkaline conditions had taken up much lower levels of lysine. The profile of amino acid uptake and release was differential between the treatments with numerous significant differences observed between utilization rates of the amino acids. It was interesting to note that the uptake of alanine and phenylalanine, as the fourth and seventh most utilized amino acids from the medium, was the same across all treatments.
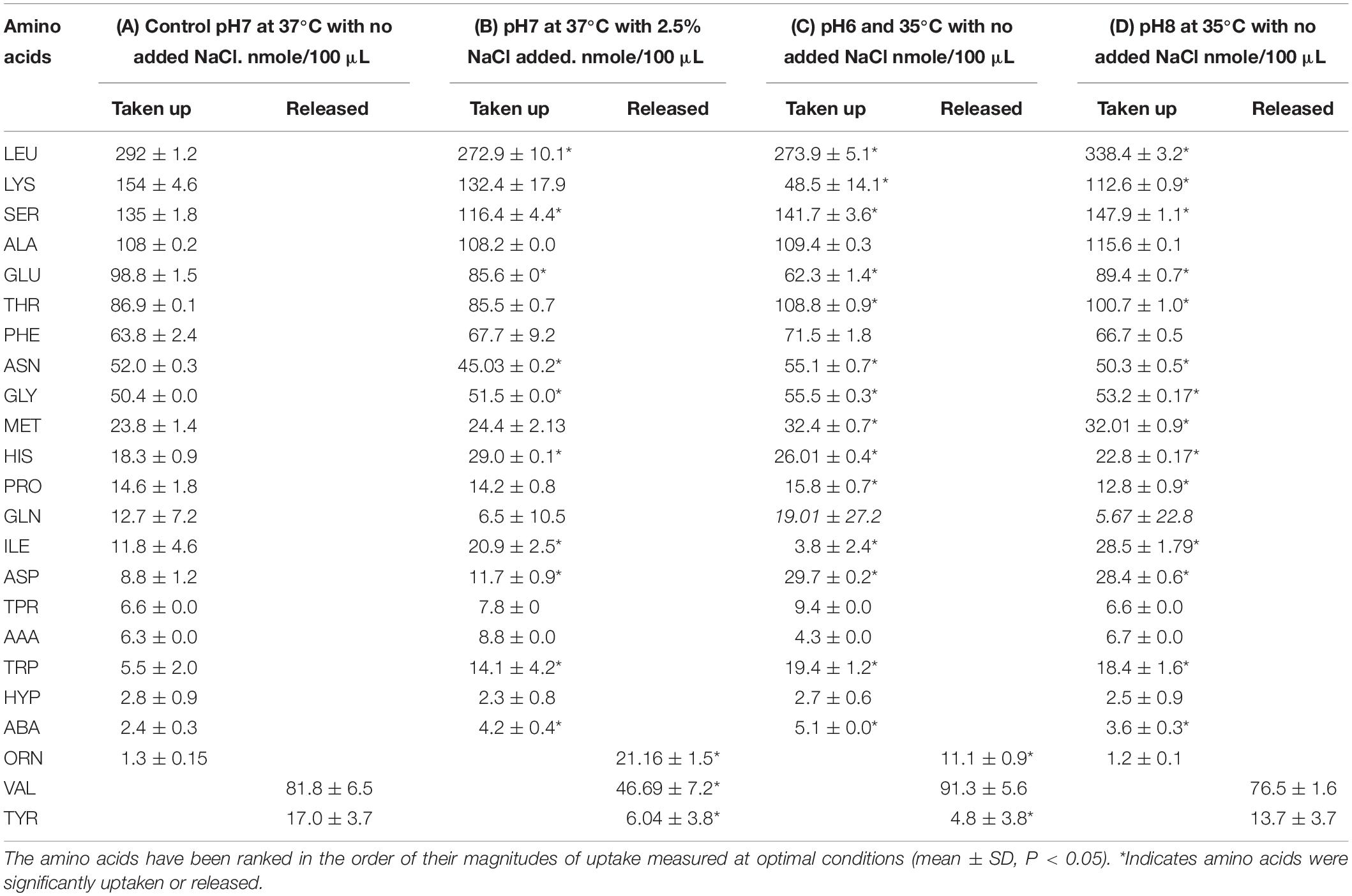
Table 3. Comparisons of the quantities of amino acids taken up or released by S. aureus at the stationary phase of growth under optimal conditions (A) compared with growth under sub-optimal environmental conditions (B–D).
Multivariate analyses using PCA revealed significant variations between the amino acid compositions of the culture supernatants from the different treatment regimens (Figure 3A). Measured amino acids were subjected to multivariate analysis using PCA. The PCA analysis revealed an apparent separation of reference control and treatment samples due to their amino acid uptake and release profiles analyzed at stationary phase of growth. It was also evident from PCA plot that cells gown with lower temperature of 35°C and pH6 (C) had very different amino acid uptake and release profiles compared to equivalent cells grown at pH8 (D). The loading scatterplot corresponding to PCA scores indicates that amino acid uptake/release that contributed to the significance of the clustering/separation observed on the PCA plot, and was unique to the control and treatment regimens (Figure 3B).
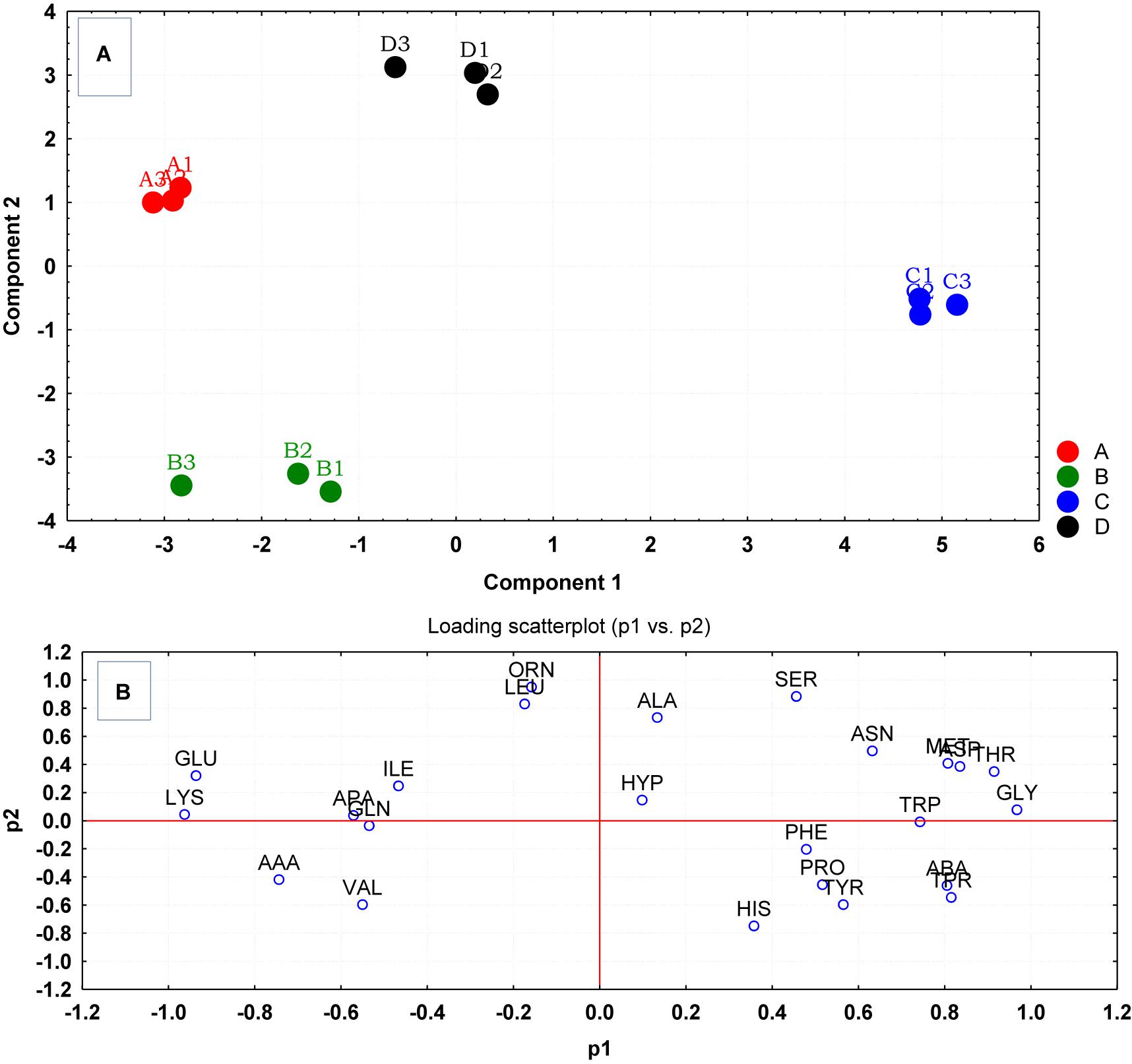
Figure 3. (A) Principal component analysis (PCA) scores generated from the amino acid uptake and release data, were plotted each replicate culture of S. aureus grown under ideal conditions (A) and those grown under sub-optimal conditions (B–D) evaluated at stationary phase. (B) The loading scatterplot of the amino acids (abbreviations from Table 1) indicates the significance of amino acid variables on PCA plot. The cells were grown under the following conditions: (A1–A4) Control cultures were grown under ideal conditions at pH7 and 37°C with no added NaCl; (B1–B4) Cultures were grown at pH 7 and 37°C with 2.5% NaCl; (C1–C4) Cultures were grown at 35°C and pH6 with no added NaCl; and (D1–D4) Cultures were grown at 35°C and pH8 with no added NaCl.
The amino acid compositions of S. aureus culture supernatants from the four environmental treatment regimens (A–D) were compared between the mid-exponential and stationary phases of growth using PCA. The PCA analysis rendered a two-component model as validated by cross-validation (CV). The PCA scores for mid-exponential and stationary phases displayed two clear clusters greatly separated by component 1 scores where the mid-exponential phase had a negative component 1 score and the stationary phase had positive component 1 scores (Figure 4). Explanation of 95% of the data was achieved (i.e., R2 = 0.70 and Q2 = 0.64) and the eigenvalue was 16.3 as compared to 2.3 for PC2. The loading scatterplot showed the influence of key variables on the clustering and separation. The stationary phase was characterized via a generally greater level of amino acids taken up from the medium, whilst the mid-exponential phase control cells (A) were characterized by having taken up valine and tyrosine in high quantities before releasing these amino acids back into the medium by the time of assessment at stationary phase.
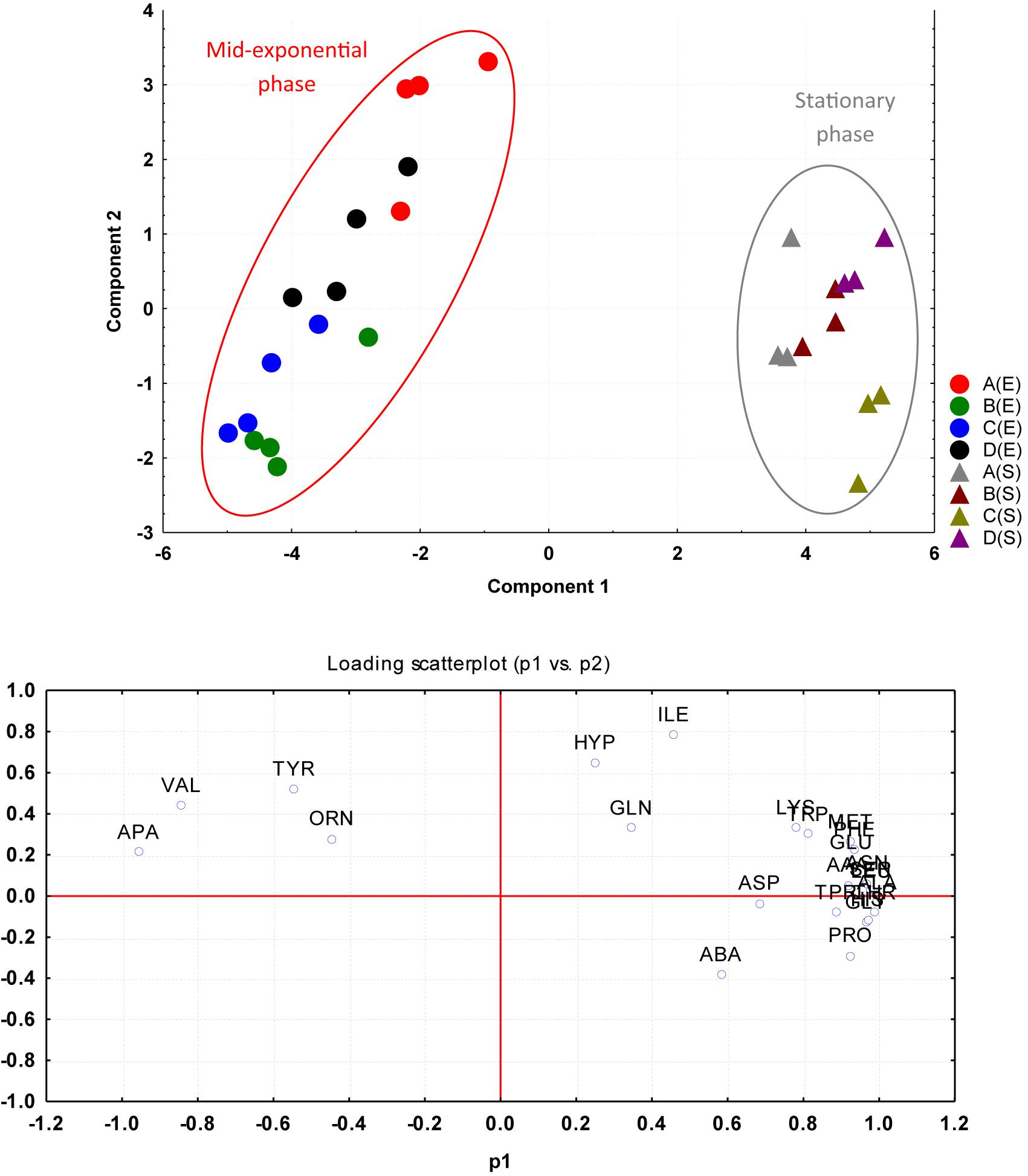
Figure 4. PCA plotted from the four environmental treatment regimens (A–D) were compared between mid-exponential (E series) and stationary (S series) phases of growth based on the amino acid uptake and release data. The cells were grown under the following conditions: (A) Control cultures were grown under ideal conditions at pH7 and 37°C with no added NaCl; (B) Cultures were grown at pH 7 and 37°C with 2.5% NaCl; (C) Cultures were grown at 35°C and pH6 with no added NaCl; and (D) Cultures were grown at 35°C and pH8 with no added NaCl.
The outcomes from the current study indicated that the patterns of amino acid uptake and release activities observed in S. aureus at the mid-exponential phase of growth were different to those observed at stationary growth phases under optimal growth conditions. It was clear that the demands for amino acids altered throughout the various stages of growth and this varied in relation to variations in the initial environmental conditions involving pH, temperature and osmolality. Amino acids represent an essential resource for sustaining cell integrity and metabolic homeostasis since in addition to their utilization in protein synthesis; they are utilized as precursors for the biosynthesis of primary and secondary metabolites nucleotides, complex lipids and cell wall components. The presence of free amino acids in the complex growth medium facilitates optimal growth capacity under ideal environmental conditions.
The shift from mid-exponential phase to stationary phase is known to involve changes in the environmental conditions such as diminishing levels of nutrients, accumulation of toxic waste products due to metabolism, and changes in pH. Significant uptakes of lysine, leucine and serine at the stationary phase were noted in this study. S. aureus possibly consumes these amino acids to balance pH in the cytoplasm that could occur as a result of proton influx (Liebeke et al., 2011). In the stationary phase cultures, the release of large quantities of valine into the external medium was noted. Valine release was observed from E. coli biofilm cultures (Valle et al., 2008) and this amino acid was secreted by S. aureus COL when it was grown in the eukaryotic cell culture medium (Dorries and Lalk, 2013). Bacteria have been previously reported to produce secondary metabolites and small molecules in the stationary phase of growth (Martin and Liras, 1989; Lopez et al., 2010). These function as signaling molecules to start the process of biofilm induction or inhibiting the growth of other organisms sharing the same environments (Lopez and Kolter, 2010; Santos et al., 2019). At stationary phase, the population density increases significantly, and the production of extracellular molecules that participate in cell-cell communications (quorum sensing) show a high correlation with cell density (Beenken et al., 2004; Resch et al., 2006). Thus the release of some amino acids in the current study could potentially function as a quorum-sense which is involved in the development and maintenance of biofilm. This study showed, as expected, that cells grown to stationary phase progressively consumed additional amounts of amino acids compared after growth to the mid-exponential phase. However, the amino acids were consumed at varying rates, which presumably reflected differential metabolic demands. It is not yet clear why valine and tyrosine would be initially taken up during growth to the mid-exponential phase and then released at high quantities by stationary phase. The levels of both of these amino acids in the external medium at stationary phase indicated that significant quantities of these amino acids were synthesized de novo. It has been shown that biofilm cultures consumed and secreted significantly higher amount of metabolites compared to the planktonic cells, suggesting that the increased uptake is a result of high bacterial requirement to synthesize cell wall components and extracellular matrix, which represent a protective mechanism against undesirable conditions present in the stationary phase (Ammons et al., 2014). Lysine uptake increased significantly at the stationary phase, indicating an intermediate proton force in the enhanced uptake of lysine (Liebeke et al., 2011; Stipetic et al., 2016). Interestingly, the uptake of some amino acids did not change significantly between growth phases, possibly pointing out that these amino acids may not be as important as other amino acids, and that these are not part of the adaptive mechanism in the stationary phase.
Exposing S. aureus to subtle changes in temperature, pH and osmolality resulted in significant differences in the uptake of amino acids harvested at mid-exponential phase. This indicated that the bacterium had to consume different quantities of amino acids to adapt to changes in the environmental conditions. A previous study exposing the bacterium to the same environmental conditions resulted in different cytoplasmic amino acid profiles (Alreshidi et al., 2016). The reduction in cytoplasmic amino acids is greatly correlated with the reduction of amino acid uptake in this study. This decreased uptake of amino acids is concomitant with reduced amino acids in the cytoplasm, suggesting that the bacterium had undergone a reduced metabolic activity to adapt to the changes in temperature, pH and osmolality (Vandecasteele et al., 2004; Wolf et al., 2008; Tremaroli et al., 2009; Zhu et al., 2009).
It has been shown that the staphylococcal biofilms consumed more arginine and accumulated ornithine in the culture media (Zhu et al., 2007), as was observed for ornithine in the current study. A high production of polysaccharide intercellular adhesin has been associated with reduced tricarboxylic acid activity (Vuong et al., 2005). Hence, this may explain the reduction in amino acid uptake, as it possibly associates with changes in external morphology such as cell size, phenotypic shift and biofilm development (Onyango et al., 2012, 2013). It has been demonstrated that SVCs have a reduced metabolic activity with a decrease of toxins productions. The activity of tricarboxylic acid cycle (TCA) was highly associated with polysaccharide intercellular adhesin (PIA) production in biofilm (Vuong et al., 2005; Sadykov et al., 2008, 2010; Zhu et al., 2009), it showed that active TCA led to repression of PIA production and hence reduced the synthesis of virulence factors (Somerville and Proctor, 2009). Therefore, it has been suggested that the uptake of amino acids would be critical for TCA activation and therefore, pathogenicity. Metabolomic studies have demonstrated that amino acid catabolism is important for the synthesis of intermediates oxaloacetate, oxoglutarate, phosphoenolpyruvate, and pyruvate for TCA and gluconeogenesis (Vuong et al., 2005; Chatterjee et al., 2006; Fleury et al., 2009). On the basis of these results, it has been suggested that the bacterium regularly detecting and adapting to the changes in the external environmental conditions by inducing the best operative and competent phenotypes for survival in stressed environments (de Jonge et al., 1992; Casadevall, 2008; Onyango et al., 2012, 2013).
The assessment of uptake and release patterns of amino acids by S. aureus at the stationary phase under various stress-induced conditions revealed remarkable amino acid uptake and release profiles, suggesting that the bacterium alters its amino acid uptake to achieve ideal metabolism related to the external environmental conditions. Surprisingly, the total quantities of amino acids consumed at the stationary phase measurements were very similar under all conditions, yet the individual uptake profiles of amino acids were significantly different. Prior research showed that Escherichia coli continued synthesizing proteins at constant rates at stationary phase for several days (Gefen et al., 2014). This may illustrate the higher uptake of amino acids at stationary phase in the present study. The productions of proteins at stationary phase may be linked with the need of the bacterium to combat complex environments present in the stationary phase including altered pH and osmolality. It has been reported that the production of proteins at stationary phase increased by 121% as compared with exponential phase (Ou et al., 2004; Jaishankar and Srivastava, 2017). The release of amino acids in this study may relate to peptidoglycan synthesis and quorum-sensing system (De Kievit et al., 2001; Aliashkevich et al., 2018). Many bacteria release D-amino acids at a stationary stage into the growth medium, which influences the regulation of peptidoglycan components, thickness, and quantity (Aliashkevich et al., 2018). Therefore, the release of these amino acids might be an acclimation mechanism for bacteria to the altered environment (Lam et al., 2009).
PCA analysis of the whole data sets of mid-exponential and stationary phases resulted in two different clusters, one representative of amino acid uptake data analyzed at mid-exponential phase and the second cluster representative of amino acids uptake obtained at stationary phase. This clearly indicated that regardless of alterations in environmental factors applied in this study, the different phases of growth inferred the most influence on uptake characteristics. Stationary phase is considered as a complex environment, and physiologically is comparable to biofilm (Jaishankar and Srivastava, 2017). In stationary phase, extraordinary adjustments take place including morphological, physiological changes and DNA/protein ratio was found to be higher (Nystrom, 2004; Jaishankar and Srivastava, 2017).
The identification of amino acid uptake profiles that distinguish between control and treatment cultures will have great implications in the translational research for manipulating adaptation and survival of S. aureus in response to subtle changes in the environmental conditions. Indeed, the analysis of amino acid uptake/release presented in this study indicated that even within small changes in the environmental conditions, a significant difference in amino acid uptake patterns can be observed. In addition, specific changes in amino acid uptake would provide the basis for adaptation strategies that could lead to evolutionary existence and ideal acclimatization of this bacterium to allow it to capitalize on infective opportunities. It is obvious that certain systematic mechanisms were initiated to survive during the alterations in temperature, pH, and osmolality to acquire optimal metabolism status by consuming specific amino acids. It is thus concluded that these changes in the uptake and release of amino acids are critical for the adaptation processes in response to changes in subtle-environmental conditions.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
All authors conceived and designed the experiments. MA and RD performed the experiments, analyzed the data, and wrote the manuscript with support from all authors.
This work was supported by the University of Ha’il (grant no. 160967). This work was also supported by the University of Newcastle (grant no. G0189306), Australia funds as well as the Harold Stannet Williams and Judith Mason Research Foundation and the Gideon Lang Research Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Aliashkevich, A., Alvarez, L., and Cava, F. (2018). New insights into the mechanisms and biological roles of D-amino acids in complex eco-systems. Front. Microbiol. 9:683. doi: 10.3389/fmicb.2018.00683
Alreshidi, M. M., Dunstan, R. H., Gottfries, J., Macdonald, M. M., Crompton, M. J., Ang, C. S., et al. (2016). Changes in the cytoplasmic composition of amino acids and proteins observed in Staphylococcus aureus during growth under variable growth conditions representative of the human wound site. PLoS One 11:e0159662. doi: 10.1371/journal.pone.0159662
Alreshidi, M. M., Dunstan, R. H., Macdonald, M. M., Smith, N. D., Gottfries, J., and Roberts, T. K. (2015). Metabolomic and proteomic responses of Staphylococcus aureus to prolonged cold stress. J. Prot. 121, 44–55. doi: 10.1016/j.jprot.2015.03.010
Alreshidi, M. M., Dunstan, R. H., Macdonald, M. M., Smith, N. D., Gottfries, J., and Roberts, T. K. (2019). Amino acids and proteomic acclimation of Staphylococcus aureus when incubated in a defined minimal medium supplemented with 5% sodium chloride. Microbiologyopen 8:e00772. doi: 10.1002/mbo3.772
Alreshidi, M. M., Dunstan, R. H., Onyango, L. A., and Roberts, T. K. (2013). “Staphylococcal phenomics: metabolomic and proteomic responses to environmental stessors,” in Microbial Pathogens and Strategies for Combating Them: Science, Technology and Education, ed. A. Mendez-Vilas (Spain: Formatex Research Center).
Ammons, M. C., Tripet, B. P., Carlson, R. P., Kirker, K. R., Gross, M. A., Stanisich, J. J., et al. (2014). Quantitative Nmr metabolite profiling of methicillin-resistant and methicillin-susceptible Staphylococcus aureus discriminates between biofilm and planktonic phenotypes. J. Prot. Res. 13, 2973–2985. doi: 10.1021/pr500120c
Beenken, K. E., Dunman, P. M., Mcaleese, F., Macapagal, D., Murphy, E., Projan, S. J., et al. (2004). Global gene expression in Staphylococcus aureus biofilms. J. Bacteriol. 186, 4665–4684.
Brown, G. K., Martin, A. R., Roberts, T. K., and Aitken, R. J. (2001). Detection of ehrlichia platys in dogs in Australia. Aust. Vet. J. 79, 554–558. doi: 10.1111/j.1751-0813.2001.tb10747.x
Butt, H. L., Dunstan, R. H., Mcgregor, N. R., Roberts, T. K., Zerbes, M., and Klineberg, I. J. (1998). An association of membrane-damaging toxins from coagulase-negative staphylococci and chronic orofacial muscle pain. J. Med. Microbiol. 47, 577–584. doi: 10.1099/00222615-47-7-577
Casadevall, A. (2008). Evolution of intracellular pathogens. Annu. Rev. Microbiol. 62, 19–33. doi: 10.1146/annurev.micro.61.080706.093305
Chatterjee, I., Somerville, G. A., Heilmann, C., Sahl, H. G., Maurer, H. H., and Herrmann, M. (2006). Very low ethanol concentrations affect the viability and growth recovery in post-stationary-phase Staphylococcus aureus populations. Appl. Environ. Microbiol. 72, 2627–2636. doi: 10.1128/aem.72.4.2627-2636.2006
Crompton, M. J., Dunstan, R. H., Macdonald, M. M., Gottfries, J., Von Eiff, C., and Roberts, T. K. (2014). Small changes in environmental parameters lead to alterations in antibiotic resistance, cell morphology and membrane fatty acid composition in Staphylococcus lugdunensis. PLoS One 9:e92296. doi: 10.1371/journal.pone.0092296
de Jonge, B. L., Chang, Y. S., Gage, D., and Tomasz, A. (1992). Peptidoglycan composition of a highly methicillin-resistant Staphylococcus aureus strain. the role of penicillin binding protein 2A. J. Biol. Chem. 267, 11248–11254.
De Kievit, T. R., Gillis, R., Marx, S., Brown, C., and Iglewski, B. H. (2001). Quorum-sensing genes in Pseudomonas aeruginosa biofilms: their role and expression patterns. Appl. Environ. Microbiol. 67, 1865–1873. doi: 10.1128/aem.67.4.1865-1873.2001
Dorries, K., and Lalk, M. (2013). Metabolic footprint analysis uncovers strain specific overflow metabolism and D-isoleucine production of Staphylococcus aureus Col and Hg001. PLoS One 8:e81500. doi: 10.1371/journal.pone.0081500
Fleury, B., Kelley, W. L., Lew, D., Gotz, F., Proctor, R. A., and Vaudaux, P. (2009). Transcriptomic and metabolic responses of Staphylococcus aureus exposed to supra-physiological temperatures. BMC Microbiol. 9:76. doi: 10.1186/1471-2180-9-76
Gefen, O., Fridman, O., Ronin, I., and Balaban, N. Q. (2014). Direct observation of single stationary-phase bacteria reveals a surprisingly long period of constant protein production activity. Proc. Natl. Acad. Sci. U.S.A. 111, 556–561. doi: 10.1073/pnas.1314114111
Jaishankar, J., and Srivastava, P. (2017). Molecular basis of stationary phase survival and applications. Front. Microbiol. 8:2000. doi: 10.3389/fmicb.2017.02000
Jones, E. M., Cochrane, C. A., and Percival, S. L. (2015). The effect of pH on the extracellular matrix and biofilms. Adv. Wound Care 4, 431–439. doi: 10.1089/wound.2014.0538
Josse, J., Laurent, F., and Diot, A. (2017). Staphylococcal adhesion and host cell invasion: fibronectin-binding and other mechanisms. Front. Microbiol. 8:2433. doi: 10.3389/fmicb.2017.02433
Lam, H., Oh, D. C., Cava, F., Takacs, C. N., Clardy, J., De Pedro, M. A., et al. (2009). D-amino acids govern stationary phase cell wall remodeling in bacteria. Science 325, 1552–1555. doi: 10.1126/science.1178123
Lee, S., Choi, K. H., and Yoon, Y. (2014). Effect of NaCl on biofilm formation of the isolate from Staphylococcus aureus outbreak linked to ham. Korean J. Food Sci. Anim. Resour. 34, 257–261. doi: 10.5851/kosfa.2014.34.2.257
Li, W., Li, Y., Wu, Y., Cui, Y., Liu, Y., Shi, X., et al. (2016). Phenotypic and genetic changes in the life cycle of small colony variants of Salmonella enterica serotype Typhimurium induced by streptomycin. Ann. Clin. Microbiol. Antimicrob. 15:37. doi: 10.1186/s12941-016-0151-3
Liebeke, M., Dorries, K., Zuhlke, D., Bernhardt, J., Fuchs, S., Pane-Farre, J., et al. (2011). A metabolomics and proteomics study of the adaptation of Staphylococcus aureus to glucose starvation. Mol. Biosyst. 7, 1241–1253. doi: 10.1039/c0mb00315h
Lopez, D., and Kolter, R. (2010). Extracellular signals that define distinct and coexisting cell fates in Bacillus subtilis. Fems Microbiol. Rev. 34, 134–149. doi: 10.1111/j.1574-6976.2009.00199.x
Lopez, D., Vlamakis, H., and Kolter, R. (2010). Biofilms. Cold Spring Harb. Perspect. Biol. 2:a000398. doi: 10.1101/cshperspect.a000398
Martin, M. F., and Liras, P. (1989). Organization and expression of genes involved in the biosynthesis of antibiotics and other secondary metabolites. Annu. Rev. Microbiol. 43, 173–206. doi: 10.1146/annurev.micro.43.1.173
Murphy, G. R., Dunstan, R. H., Macdonald, M. M., Gottfries, J., and Roberts, T. K. (2018). Alterations in amino acid metabolism during growth by Staphylococcus aureus following exposure to H2O2 - a multifactorial approach. Heliyon 4:e00620. doi: 10.1016/j.heliyon.2018.e00620
Nystrom, T. (2004). Stationary-phase physiology. Annu. Rev. Microbiol. 58, 161–181. doi: 10.1146/annurev.micro.58.030603.123818
Onyango, L. A., and Alreshidi, M. M. (2018). Adaptive metabolism in Staphylococci: survival and persistence in environmental and clinical settings. J. Pathog. 2018:1092632. doi: 10.1155/2018/1092632
Onyango, L. A., Dunstan, R. H., Gottfries, J., Von Eiff, C., and Roberts, T. K. (2012). Effect of low temperature on growth and ultra-structure of Staphylococcus spp. PLoS One 7:e29031. doi: 10.1371/journal.pone.0029031
Onyango, L. A., Hugh Dunstan, R., Roberts, T. K., Macdonald, M. M., and Gottfries, J. (2013). Phenotypic variants of staphylococci and their underlying population distributions following exposure to stress. PLoS One 8:e77614. doi: 10.1371/journal.pone.0077614
Ou, J., Wang, L., Ding, X., Du, J., Zhang, Y., Chen, H., et al. (2004). Stationary phase protein overproduction is a fundamental capability of Escherichia coli. Biochem. Biophys. Res. Commun. 314, 174–180. doi: 10.1016/j.bbrc.2003.12.077
Proctor, R. A., Kriegeskorte, A., Kahl, B. C., Becker, K., Loffler, B., and Peters, G. (2014). Staphylococcus aureus small colony variants (Scvs): a road map for the metabolic pathways involved in persistent infections. Front. Cell Infect. Microbiol. 4:99. doi: 10.3389/fcimb.2014.00099
Resch, A., Leicht, S., Saric, M., Pasztor, L., Jakob, A., Gotz, F., et al. (2006). Comparative proteome analysis of Staphylococcus aureus biofilm and planktonic cells and correlation with transcriptome profiling. Proteomics 6, 1867–1877. doi: 10.1002/pmic.200500531
Sadykov, M. R., Olson, M. E., Halouska, S., Zhu, Y., Fey, P. D., Powers, R., et al. (2008). Tricarboxylic acid cycle-dependent regulation of Staphylococcus epidermidis polysaccharide intercellular adhesin synthesis. J. Bacteriol. 190, 7621–7632. doi: 10.1128/JB.00806-08
Sadykov, M. R., Zhang, B., Halouska, S., Nelson, J. L., Kreimer, L. W., Zhu, Y., et al. (2010). Using Nmr metabolomics to investigate tricarboxylic acid cycle-dependent signal transduction in Staphylococcus epidermidis. J. Biol. Chem. 285, 36616–36624. doi: 10.1074/jbc.M110.152843
Santos, T., Viala, D., Chambon, C., Esbelin, J., and Hebraud, M. (2019). Listeria monocytogenes biofilm adaptation to different temperatures seen through shotgun proteomics. Front. Nutr. 6:89. doi: 10.3389/fnut.2019.00089
Schwan, W. R., Wetzel, K. J., Gomez, T. S., Stiles, M. A., Beitlich, B. D., and Grunwald, S. (2004). Low-proline environments impair growth, proline transport and in vivo survival of Staphylococcus aureus strain-specific putP mutants. Microbiology 150, 1055–1061. doi: 10.1099/mic.0.26710-0
Singh, R., Ray, P., Das, A., and Sharma, M. (2010). Enhanced production of exopolysaccharide matrix and biofilm by a menadione-auxotrophic Staphylococcus aureus small-colony variant. J. Med. Microbiol. 59, 521–527. doi: 10.1099/jmm.0.017046-0
Solis, N., Parker, B. L., Kwong, S. M., Robinson, G., Firth, N., and Cordwell, S. J. (2014). Staphylococcus aureus surface proteins involved in adaptation to oxacillin identified using a novel cell shaving approach. J. Prot. Res. 13, 2954–2972. doi: 10.1021/pr500107p
Somerville, G. A., and Proctor, R. A. (2009). At the crossroads of bacterial metabolism and virulence factor synthesis in Staphylococci. Microbiol. Mol. Biol. Rev. 73, 233–248. doi: 10.1128/MMBR.00005-09
Stipetic, L. H., Dalby, M. J., Davies, R. L., Morton, F. R., Ramage, G., and Burgess, K. E. (2016). A novel metabolomic approach used for the comparison of Staphylococcus aureus planktonic cells and biofilm samples. Metabolomics 12:75.
Tkadlec, J., Varekova, E., Pantucek, R., Doskar, J., Ruzickova, V., Botka, T., et al. (2015). Characterization of Staphylococcus aureus strains isolated from czech cystic fibrosis patients: high rate of ribosomal mutation conferring resistance to mls antibiotics as a result of long-term and low-dose azithromycin treatment. Microb. Drug Resist. 21, 416–423. doi: 10.1089/mdr.2014.0276
Tremaroli, V., Workentine, M. L., Weljie, A. M., Vogel, H. J., Ceri, H., Viti, C., et al. (2009). Metabolomic investigation of the bacterial response to a metal challenge. Appl. Environ. Microbiol. 75, 719–728. doi: 10.1128/AEM.01771-08
Valle, J., Da Re, S., Schmid, S., Skurnik, D., D’ari, R., and Ghigo, J. M. (2008). The amino acid valine is secreted in continuous-flow bacterial biofilms. J. Bacteriol. 190, 264–274. doi: 10.1128/jb.01405-07
Vandecasteele, S. J., Peetermans, W. E., Carbonez, A., and Van Eldere, J. (2004). Metabolic activity of Staphylococcus epidermidis is high during initial and low during late experimental foreign-body infection. J. Bacteriol. 186, 2236–2239. doi: 10.1128/jb.186.8.2236-2239.2004
von Eiff, C. (2008). Staphylococcus aureus small colony variants: a challenge to microbiologists and clinicians. Int. J. Antimicrob. Agents 31, 507–510. doi: 10.1016/j.ijantimicag.2007.10.026
von Eiff, C., Proctor, R. A., and Peters, G. (2000). Small colony variants of Staphylococci: a link to persistent infections. Berl Munch Tierarztl Wochenschr 113, 321–325.
Vuong, C., Kidder, J. B., Jacobson, E. R., Otto, M., Proctor, R. A., and Somerville, G. A. (2005). Staphylococcus epidermidis polysaccharide intercellular adhesin production significantly increases during tricarboxylic acid cycle stress. J. Bacteriol. 187, 2967–2973. doi: 10.1128/jb.187.9.2967-2973.2005
Wehrli, P. M., Lindberg, E., Svensson, O., Sparén, A., Josefson, M., Dunstan, R. H., et al. (2014). Exploring bacterial phenotypic diversity using factorial design and Ftir multivariate fingerprinting. Chemometrics 30, 283–289.
Wolf, C., Hochgrafe, F., Kusch, H., Albrecht, D., Hecker, M., and Engelmann, S. (2008). Proteomic analysis of antioxidant strategies of Staphylococcus aureus: diverse responses to different oxidants. Proteomics 8, 3139–3153. doi: 10.1002/pmic.200701062
Yao, Y. F., Sturdevant, D. E., and Otto, M. (2005). Genomewide analysis of gene expression in Staphylococcus epidermidis biofilms: insights into the pathophysiology of S-epidermidis biofilms and the role of phenol-soluble modulins in formation of biofilms. J. Infect. Dis. 191, 289–298. doi: 10.1086/426945
Zeaki, N., Cao, R., Skandamis, P. N., Radstrom, P., and Schelin, J. (2014). Assessment of high and low enterotoxin A producing Staphylococcus aureus strains on pork sausage. Int. J. Food Microbiol. 18, 44–50. doi: 10.1016/j.ijfoodmicro.2014.05.010
Zhu, Y., Weiss, E. C., Otto, M., Fey, P. D., Smeltzer, M. S., and Somerville, G. A. (2007). Staphylococcus aureus biofilm metabolism and the influence of arginine on polysaccharide intercellular adhesin synthesis, biofilm formation, and pathogenesis. Infect. Immun. 75, 4219–4226. doi: 10.1128/iai.00509-07
Keywords: amino acid uptake, S. aureus, valine, stress response and adaptation, phenotypic shift
Citation: Alreshidi MM, Dunstan RH, Macdonald MM, Gottfries J and Roberts TK (2020) The Uptake and Release of Amino Acids by Staphylococcus aureus at Mid-Exponential and Stationary Phases and Their Corresponding Responses to Changes in Temperature, pH and Osmolality. Front. Microbiol. 10:3059. doi: 10.3389/fmicb.2019.03059
Received: 17 November 2019; Accepted: 18 December 2019;
Published: 23 January 2020.
Edited by:
Filomena Nazzaro, Italian National Research Council (CNR), ItalyReviewed by:
Abderrahmen Merghni, Tunis El Manar University, TunisiaCopyright © 2020 Alreshidi, Dunstan, Macdonald, Gottfries and Roberts. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mousa M. Alreshidi, TW8uYWxyZXNoaWRpQHVvaC5lZHUuc2E=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.