- Department of Biochemistry and Biomedical Sciences, Faculty of Health Sciences, McMaster University, Hamilton, ON, Canada
The family Caryophanaceae/Planococcaceae is a taxonomically heterogeneous assemblage of >100 species classified within 13 genera, many of which are polyphyletic. Exhibiting considerable phylogenetic overlap with other families, primarily Bacillaceae, the evolutionary history of this family, containing the potent mosquitocidal species Lysinibacillus sphaericus, remains incoherent. To develop a reliable phylogenetic and taxonomic framework for the family Caryophanaceae/Planococcaceae and its genera, we report comprehensive phylogenetic and comparative genomic analyses on 124 genome sequences from all available Caryophanaceae/Planococcaceae and representative Bacillaceae species. Phylogenetic trees were constructed based on multiple datasets of proteins including 819 core proteins for this group and 87 conserved Firmicutes proteins. Using the core proteins, pairwise average amino acid identity was also determined. In parallel, comparative analyses on protein sequences from these species have identified 92 unique molecular markers (synapomorphies) consisting of conserved signature indels that are specifically shared by either the entire family Caryophanaceae/Planococcaceae or different monophyletic clades present within this family, enabling their reliable demarcation in molecular terms. Based on multiple lines of investigations, 18 monophyletic clades can be reliably distinguished within the family Caryophanaceae/Planococcaceae based on their phylogenetic affinities and identified molecular signatures. Some of these clades are comprised of species from several polyphyletic genera within this family as well as other families. Based on our results, we are proposing the creation of three novel genera within the family Caryophanaceae/Planococcaceae, namely Metalysinibacillus gen. nov., Metasolibacillus gen. nov., and Metaplanococcus gen. nov., as well as the transfer of 25 misclassified species from the families Caryophanaceae/Planococcaceae and Bacillaceae into these three genera and in Planococcus, Solibacillus, Sporosarcina, and Ureibacillus genera. These amendments establish a coherent taxonomy and evolutionary history for the family Caryophanaceae/Planococcaceae, and the described molecular markers provide novel means for diagnostic, genetic, and biochemical studies. Lastly, we are also proposing a consolidation of the family Planococcaceae within the emended family Caryophanaceae.
Introduction
The family Planococcaceae is a diverse assemblage of bacteria within the order Bacillales comprising 14 validly published genera (viz. Planococcus, Bhargavaea, Chryseomicrobium, Filibacter, Indiicoccus, Jeotgalibacillus, Kurthia, Marinibacillus, Paenisporosarcina, Planomicrobium, Psychrobacillus, Savagea, Sporosarcina, and Ureibacillus) (based on updated information available from the Names for Life Server in September 20191). It contains >100 species with varying morphology which are Gram-variable, spore forming or non-spore forming, motile or non-motile that are usually aerobic (Ludwig et al., 2009; Shivaji et al., 2014; Yilmaz et al., 2014). Of these species, Lysinibacillus sphaericus is of particular importance as some strains of this species produce proteins/toxins that exhibit potent activity against mosquito larvae, and thus have been widely used as biocontrol agents for disease-transmitting mosquitoes (Baumann et al., 1991; Berry, 2012). However, the absence of any known characteristics exclusive to all members of the family Planococcaceae and a lack of other reliable means for classifying its members has made demarcation of this family very difficult (Ludwig et al., 2009; Shivaji et al., 2014). Although several phylogenetic studies have focused on specific genera within Planococcaceae, the evolutionary history of the family as a whole remains unclear as evidenced by the observation that different taxonomic databases/studies indicate different genera belonging to this family (Yoon et al., 2001a; Yarza et al., 2010; Parte, 2014; Shivaji et al., 2014; Mual et al., 2016; Maayer et al., 2019).
The current classification of species within this family relies on a limited number of phenotypic characteristics, 16S rRNA gene signature nucleotides, and the branching observed in phylogenetic trees based on 16S rRNA gene sequences (Dai et al., 2005; Ludwig et al., 2009; Shivaji et al., 2014). Utilizing these methods, several genera have been frequently added and removed from this family in recent years providing a better insight into their interrelationships than was previously attainable (Yoon et al., 2001a; Arora et al., 2011; Shivaji et al., 2014; Xu et al., 2015; Tetz and Tetz, 2018). However, the studies based on 16S rRNA gene sequences have low discriminatory power at the species and genus levels resulting in poorly resolved interrelationships of the members of the family Planococcaceae in phylogenetic trees based on 16S rRNA (Konstantinidis and Tiedje, 2005; Yarza et al., 2010; Maayer et al., 2019). For instance, the family remains polyphyletic as it exhibits considerable overlap with species from genera belonging to the families Bacillaceae and Incertae sedis 19 (Farrow et al., 1992; Seiler et al., 2013; Shivaji et al., 2014; Xu et al., 2015; Mual et al., 2016; Maayer et al., 2019). Furthermore, it has been well documented that several genera within the family Planococcaceae (viz. Planococcus, Planomicrobium, Filibacter, and Sporosarcina) do not form distinct clades and exhibit polyphyletic branching in phylogenetic trees (Shivaji et al., 2014; Xu et al., 2015; Maayer et al., 2019). The nomenclature of the family Planococcaceae also presents a taxonomic anomaly as highlighted recently by Tindall (2019). The taxonomic anomaly results from the fact that the family Planococcaceae was validly published in 1949 (Krasil’nikov, 1949), but it includes within it the family Caryophanaceae, which was validly published in 1939 (Peshkoff, 1939). Based on the International Code of Nomenclature of Prokaryotes (ICNP) (Parker et al., 2019), due to the earlier valid publication of the name Caryophanaceae, this name has priority over the family name Planococcaceae (Tindall, 2019). To rectify this anomaly, in the present work we are proposing a unification of the family Planococcaceae within the emended family Caryophanaceae. Therefore, hence forward, we will be referring to this family as either the Caryophanaceae or Caryophanaceae/Planococcaceae family. The present study was undertaken with the aim of gaining a robust understanding of the interrelationships among the Caryophanaceae species using multiple genomic sequences-based approaches to provide reliable means for demarcating this family and different genus level taxa within this family.
Due to significant advancements in genome sequencing technology, genome sequences are now available for the majority of the named Caryophanaceae species, providing an excellent representation of the overall genetic diversity that exists within this family. This genomic data has already been employed, to a limited extent, to study the phylogeny and taxonomy of a small number of Caryophanaceae/Planococcaceae species in recent years and continues to develop as a promising resource for clarifying the evolutionary history of Caryophanaceae species (Xu et al., 2015; Maayer et al., 2019). In addition, genome sequences are also available for >150 other species belonging to the family Bacillaceae, its closest phylogenetic relative (Ludwig et al., 2009; Yakoubou et al., 2010; Shivaji et al., 2014; Patel and Gupta, 2019). In the present study, we have used genomic information from Caryophanaceae and Bacillaceae species to comprehensively examine the interrelationships among species within these families using phylogenomic and comparative genomic approaches. Based on genome sequences, phylogenetic trees were constructed based on four large datasets of protein sequences. These trees not only confirm the polyphyletic nature of the family Caryophanaceae and the presence of polyphyletic genera within it, but they also consistently identified 18 distinct clades within the family, some of which consisted of genera that are not currently classified as belonging to the family Caryophanaceae/Planococcaceae. In parallel, detailed comparative analyses of protein sequences from these species have identified 92 novel molecular markers in the form of conserved signature indels (CSIs) which are either specific for the entire family Caryophanaceae or specific clades/genera within this family, which are reliably observed in all constructed phylogenomic trees. The identified CSIs provide novel and reliable means for the demarcation of the family Caryophanaceae as well as different observed species groups within this family in molecular terms (Gao and Gupta, 2012; Adeolu et al., 2016; Gupta, 2016; Dobritsa et al., 2017; Patel and Gupta, 2019). Based on the results from our analyses, we propose here the creation of three novel genera (viz. Metasolibacillus gen. nov., Metalysinibacillus gen. nov., and Metaplanococcus gen. nov.) within the family Caryophanaceae and also propose the transfer of 25 misclassified species from the families Caryophanaceae and Bacillaceae into the different reliably demarcated genera that are now part of the emended family Caryophanaceae.
Materials and Methods
Phylogenetic and Genomic Analysis
Phylogenetic trees were constructed for 124 species comprising all available Caryophanaceae/Planococcaceae and some representative Bacillaceae species whose complete genomes were available in the NCBI genome database along with Streptococcus pyogenes, S. mitis, Lactococcus piscium, and L. lactis, which were used to root the trees2. Phylogenetic analyses were carried out as in our earlier work (Patel and Gupta, 2018; Gupta et al., 2019) using an internally developed pipeline (Adeolu et al., 2016). Using CD-HIT program (Fu et al., 2012), protein families were identified that were present in at least 80% of the input genomes and shared >50% in sequence identity and sequence lengths. Clustal Omega program (Sievers et al., 2011) was used for the creation of multiple sequence alignments (MSAs) and after removal of poorly aligned regions with TrimAl (Capella-Gutierrez et al., 2009) sequences were concatenated. Maximum-likelihood trees based on the alignments were constructed using FastTree 2 (Price et al., 2010) based on the Whelan and Goldman (2001) model. Optimization of the trees was carried out using RAxML 8 (Stamatakis, 2014) based on Le and Gascuel (2008) model. RAxML 8 was also used to calcuate the SH-like statistical support values for different nodes and the trees were drawn using MEGA 6 (Tamura et al., 2013). The sequence alignments of the conserved core genome proteins were also used for calculation of the pairwise average amino acid sequence identity (AAI) between each pair of genomes (Thompson et al., 2013).
In addition to the protein-based trees, an unrooted 16S rRNA-based phylogenetic tree was also constructed using gene sequences for all available Planococcaceae and some representative Bacillaceae species (109 total species) retrieved from the All-Species Living Tree Project (Yilmaz et al., 2014). Type strains were obtained, if available, for all species which were then aligned using ClustalX 2.1 (Jeanmougin et al., 1998). Non-conserved regions and positions with gaps were removed from the alignment. A maximum-likelihood phylogenetic tree based on this alignment, which consisted of 1348 positions, was created using MEGA 6 (Tamura et al., 2013) using Kimura 2-parameter model (Kimura, 1980) based on 1000 bootstrap replicates as described in earlier work (Patel and Gupta, 2018). Similar results were obtained when MEGA X instead of MEGA6 was used for construction of the 16S rRNA tree.
Identification of Conserved Signature Indels
Conserved signature indels were identified using the method detailed by Gupta (2014). For these analyses, BLASTp searches on protein sequences from the genomes of Lysinibacillus boronitolerans, Lysinibacillus endophyticus, Planococcus citreus, and Sporosarcina ureae were carried out against the NCBI non-redundant database and sequences for 10–15 divergent Planococcaceae and Bacillaceae species and six to eight outgroup species were retrieved. Subsequent analyses on these protein sequences were carried out as previously described (Patel and Gupta, 2018; Gupta et al., 2019). “Briefly, MSAs of different proteins were created using ClustalX 2.1 (Jeanmougin et al., 1998) and inspected for sequence gaps of fixed lengths which were flanked by at least four to five conserved residues in the neighboring 40–50 amino acids and appeared to be shared by either most or all homologs belonging to a certain group (Gupta, 2014). Query sequences encompassing the gap and flanking regions (40–100 amino acids long) were subjected to a second BLASTp search and the resulting top 500–1000 hits were examined to assess the group specificity of the identified CSIs. Signature files reported here were created using the SIG_CREATE and SIG_STYLE programs that are available on Gleans.net (Gupta, 2014). Unless otherwise stated, the CSIs described here are exclusive to the indicated groups of species and absent in other homologs (in the top 500 BLASTp hits examined). Due to space constraints, sequence information is shown for only a limited number of species in the main figures; however, unless otherwise indicated, the described CSIs are also present in other members of the indicated groups.” For some of the proteins containing these CSIs, homologs were not present in all species from a given clade. Detailed information for all CSIs identified in this study is provided in the Supplementary Material.
Results
Phylogenetic Analysis of the Family Caryophanaceae/Planococcaceae
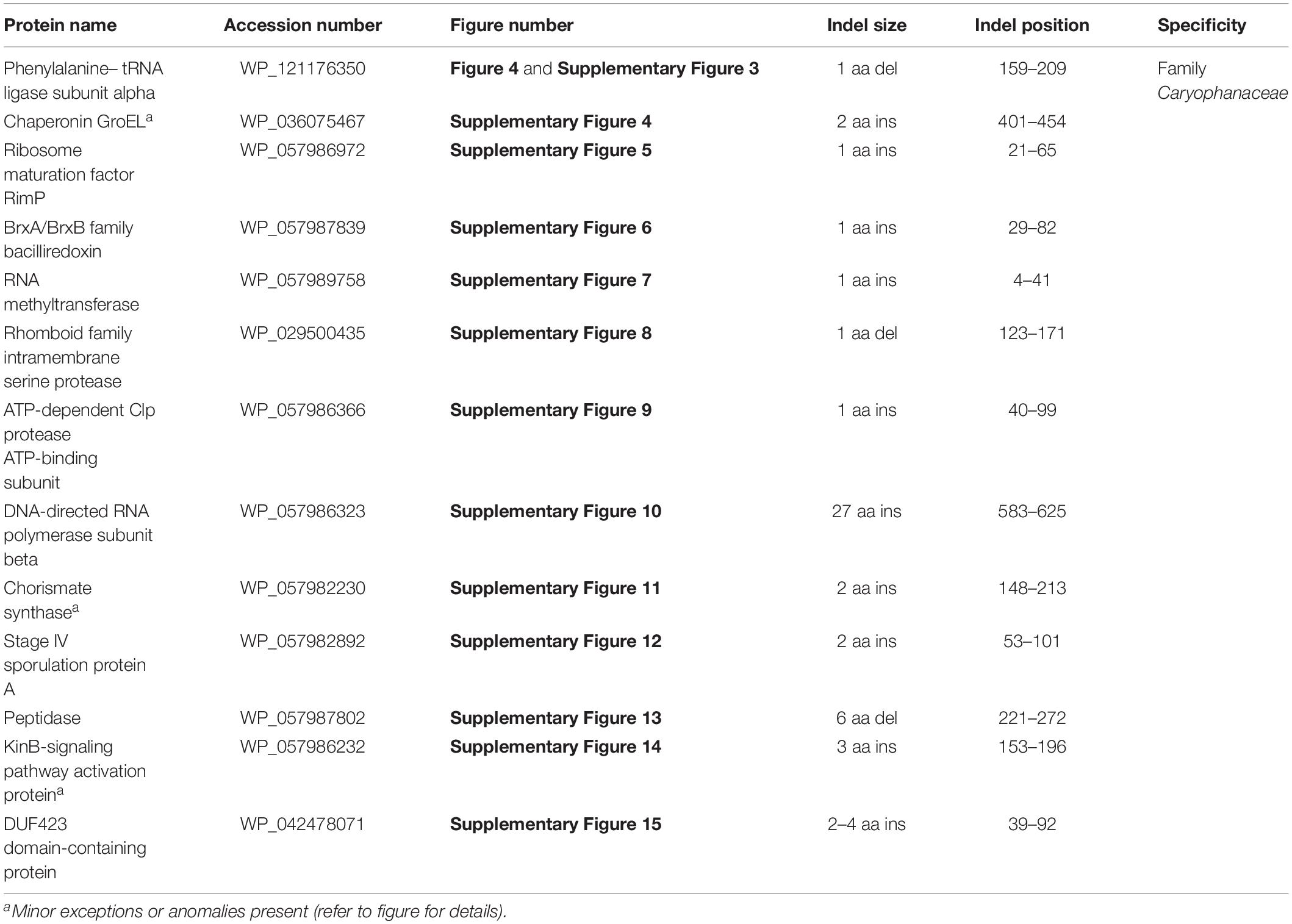
Using 124 genomes of all available genome-sequenced Caryophanaceae species and representative species from the family Bacillaceae, several phylogenetic trees were constructed based on different datasets of proteins. Each species in these trees is represented by a single genomic sequence generally of the type strain of the species when available. The trees were constructed based on concatenated sequences for (i) 819 core genome proteins for this selection of species consisting of 256,546 aligned amino acids (Figure 1A); (ii) 87 proteins commonly shared by species of the phylum Firmicutes (Wang and Wu, 2013) containing 26,445 aligned amino acids (Figure 1B); (iii) the DNA helicase II and DNA polymerase I proteins (Supplementary Figure 1A), and (iv) the two major subunits (RpoB and RpoC) of the RNA polymerase protein (Supplementary Figure 1B). All constructed trees exhibited nearly identical branching patterns and interrelationships among different Caryophanaceae and Bacillaceae species with high statistical support values at virtually every node. Hence, all trees reveal consistent, robust, and reliable species’ relationships among the family Caryophanaceae. In every tree, the family Caryophanaceae is comprised of 18 distinct clades of species from several different genera, some of which are currently classified as belonging to the family Bacillaceae (viz. Bacillus, Lysinibacillus, Viridibacillus, and “Edaphobacillus”) or the family “Incertae sedis 19” (viz. Rummeliibacillus and Solibacillus). In these trees, species from the genera Caryophanon, Chryseomicrobium, Kurthia, Rummeliibacillus, Viridibacillus, “Edaphobacillus,” and “Tetzosporium” all branched separately or formed independent monophyletic clades, while species from the remaining genera either exhibited polyphyletic branching or were not clearly separated from species of other closely related genera.
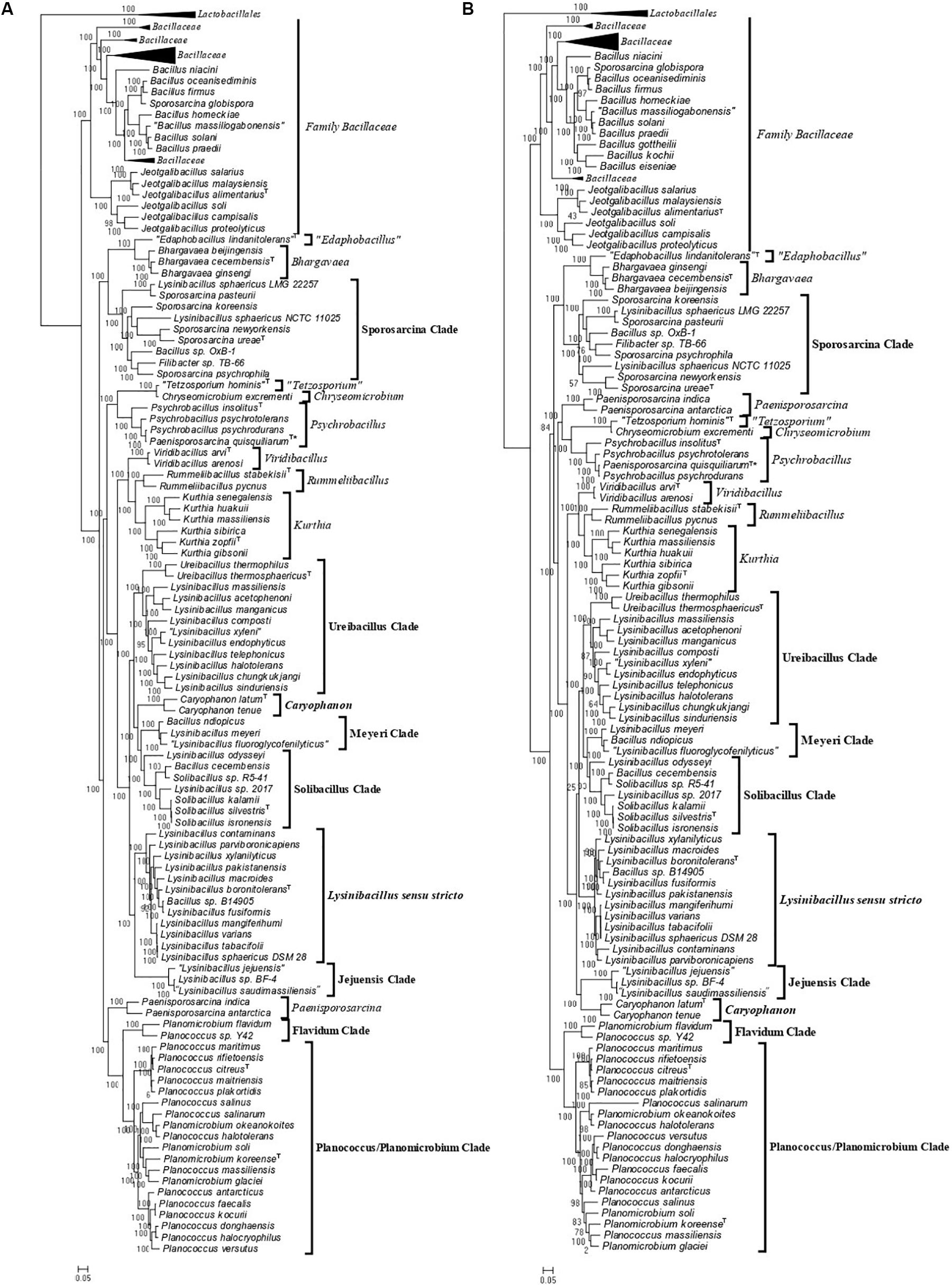
Figure 1. Maximum-likelihood phylogenetic trees for 124 genome sequenced members of the family Caryophanaceae/Planococcaceae and some representative members of the family “Bacillaceae” based on (A) 819 core proteins for this group of species, and (B) a set of 87 conserved proteins that are part of the phyloeco marker set for the phylum Firmicutes (Wang and Wu, 2013). Both trees were rooted using genome sequences of Streptococcus pyogenes, Streptococcus mitis, Lactococcus piscium, and Lactococcus lactis (labeled as Lactobacillales). SH-like statistical support values are indicated at each branch node. All clades observed in this study are labeled and presented with square brackets. The specific clades of interest are indicated in bold. A superscript “T” indicates the type species of a specific genus and the asterisk (∗) indicates the genome of Psychrobacillus quisquiliarum seems to be contaminated. The scale bars at the bottom represent 0.05 changes per amino acid position for each tree.
As an example of the polyphyletic and paraphyletic branching of species from genera that are part of the emended family Caryophanaceae, members of the genus Lysinibacillus were consistently found to form six different clusters, some of which were interspersed with species from other genera within the family Caryophanaceae. Eleven Lysinibacillus species branched together with the type species, L. boronitolerans, to form the Lysinibacillus sensu stricto clade; three other Lysinibacillus species formed a distant cluster, which we have marked as the “Jejuensis clade.” Ten other Lysinibacillus species are seen branching alongside Ureibacillus species with small branch separation to form the “Ureibacillus clade”; two Lysinibacillus species are seen branching with Bacillus ndiopicus to form the “Meyeri clade,” while two other Lysinibacillus species are observed to be interspersed between Solibacillus species forming the “Solibacillus clade.” Finally, two non-type strains of L. sphaericus are observed branching within the “Sporosarcina clade” which is primarily comprised of Sporosarcina species. Other genera which also displayed polyphyletic branching are Planococcus and Planomicrobium whose species are interspersed among one another within a larger clade of 19 species which we have called the “Planococcus/Planomicrobium clade” and a smaller clade of two species which we have called the “Flavidum clade.” Several Bacillus species are also seen branching within the family Caryophanaceae, further contributing to the polyphyly of the genera Solibacillus, Sporosarcina, and Lysinibacillus. In the trees based on genome sequences, Paenisporosarcina quisquiliarum, which is the type species of Paenisporosarcina, is also found to branch independently from other Paenisporosarcina species within a clade comprised of all genome-sequenced Psychrobacillus species making both Paenisporosarcina and Psychrobacillus polyphyletic. However, as clarified in the section “Discussion,” the anomalous branching of P. quisquiliarum within Psychrobacillus is very likely due to a mislabeling of this genome. Finally, the genus Jeotgalibacillus and Sporosarcina globispora, which are currently classified as belonging to the family Caryophanaceae/Planococcaceae, are seen branching with the representative species of the family Bacillaceae in all constructed phylogenetic trees. All 18 identified clades are depicted and labeled in the phylogenetic trees (Figure 1 and Supplementary Figure 1) with square brackets and the clades of interest, which were further investigated, are differentiated by bold labels.
As the genome scale and concatenated protein-based trees were limited to only those Caryophanaceae species for which genomic sequences were available, a 16S rRNA-based phylogenetic tree for 109 species of the family Caryophanaceae and some representative Bacillaceae species was also constructed to discern the relative branching of all named Caryophanaceae species whose 16S rRNA gene sequences were available in the All-Species Living Tree Project (Figure 2; Yilmaz et al., 2014). Although the different nodes in the 16S rRNA tree are not as strongly supported, the overall branching pattern observed is very similar to that seen in the protein trees. For instance, Lysinibacillus, Viridibacillus, Rummeliibacillus, and Solibacillus are all seen branching within the family Caryophanaceae/Planococcaceae and the genera Lysinibacillus, Sporosarcina, Planococcus, and Planomicrobium also exhibited polyphyletic branching in this tree as well. Furthermore, all species of the Lysinibacillus sensu stricto, Meyeri, “Planococcus/Planomicrobium,” “Sporosarcina,” “Ureibacillus,” Caryophanon, and Jeotgalibacillus clades are also seen generally clustering together in the 16S rRNA-based tree and are labeled accordingly in Figure 2 with square brackets. Apart from the genome-sequenced species of the observed clades indicated in Figure 1 and Supplementary Figure 1, the 16S rRNA tree shows additional non-genome sequenced species also branching within these clades which are highlighted in red in Figure 2. Jeotgalibacillus is also seen branching with the family Bacillaceae in this tree while S. globispora is seen branching alongside other Sporosarcina species in the family Caryophanaceae. Lastly, unlike the genome sequenced-based trees (Figure 1 and Supplementary Figure 1), Psychrobacillus species and Paeinsporosarcina species form independent monophyletic clades in the 16S rRNA tree and P. quisquiliarum branches with the other Paenisporosarcina species, as expected.
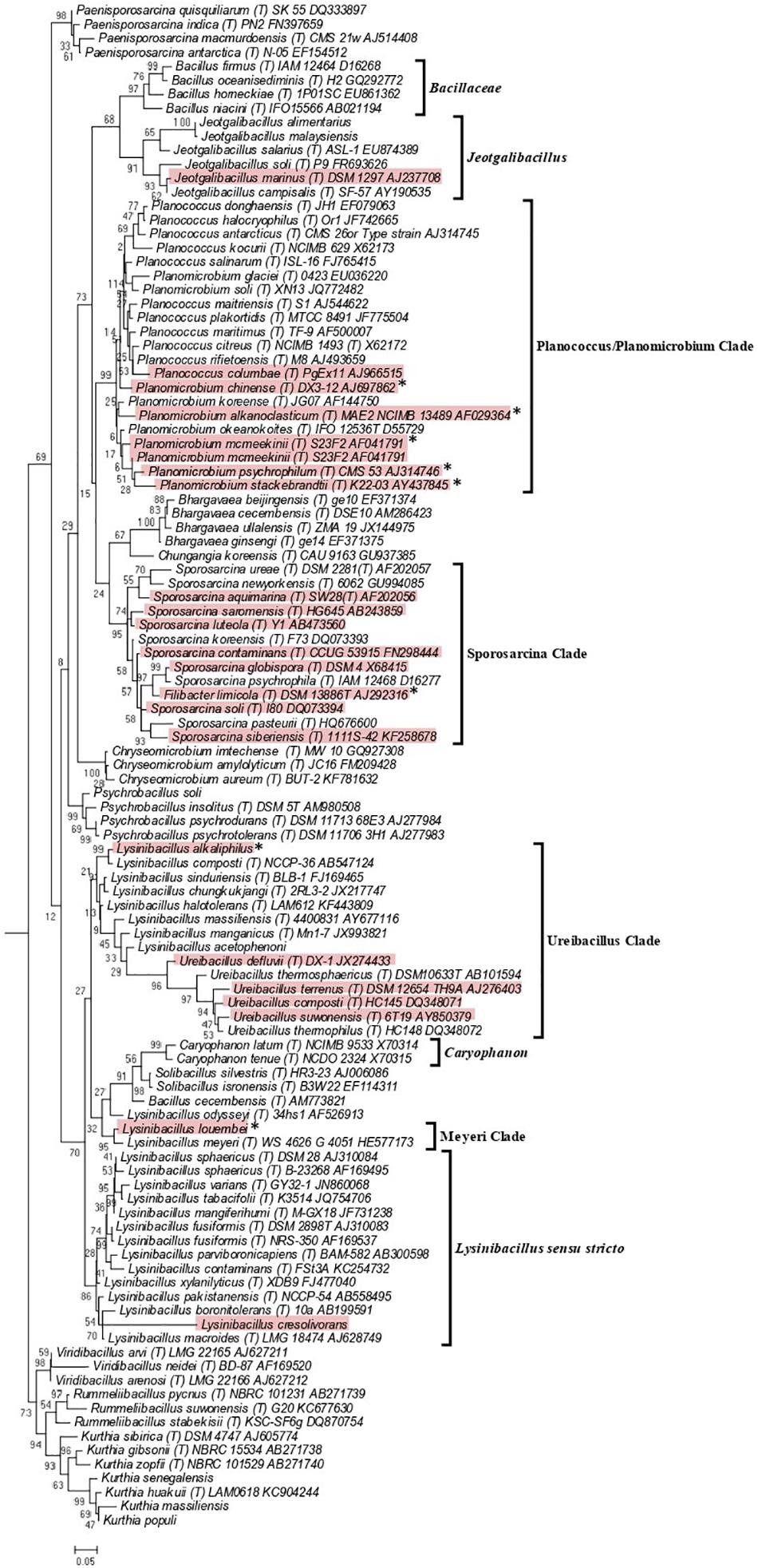
Figure 2. Maximum-likelihood phylogenetic tree for 109 Caryophanaceae/Planococcaceae and Bacillaceae species based on 16S rRNA gene sequences retrieved from the All-Species Living Tree Project (Yilmaz et al., 2014). The evolutionary history was inferred based on the Kimura 2-parameter model (Kimura, 1980). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The scale bar at the bottom represents 0.05 changes per nucleotide position. All species which had their 16S rRNA gene sequence of their type strains are indicated by (T) followed by the name of the strains. The proposed clades seen in this tree are labeled with square brackets and all non-genome-sequenced species which are part of these clades are highlighted in red with the species which we have proposed be transferred labeled with an asterisk (∗). This tree is unrooted.
Genome Similarity Among Caryophanaceae Species
The genetic relatedness across the family Caryophanaceae was measured by calculating the pairwise average amino acid identity (AAI) between species pairs using a concatenated sequence alignment of 819 core proteins shared by Caryophanaceae/Planococcaceae species and some representative Bacillaceae species (Konstantinidis and Tiedje, 2005; Konstantinidis and Stackebrandt, 2013; Thompson et al., 2013). The resulting AAI information is depicted in Figure 3 in the form of a matrix where genome pairs exhibiting a greater degree of similarity are represented with a darker shade of red. The highest AAI is observed between species that form the 18 identified clades in the phylogenetic trees mentioned above. Although comparison of AAI values provides a useful means for distinguishing prokaryotic taxa based on their genomic similarities (Konstantinidis and Stackebrandt, 2013; Sangal et al., 2016), there is no reliable threshold value that is generally applicable for the distinction of genus level taxa (Qin et al., 2014; Gupta, 2019). However, based on their AAI values, the nine clades of interest, which are boxed and labeled with black arrows in Figure 3, can be clearly distinguished from each other. The mean AAI values of species from these nine clades are all >70% and the deviation from mean in these values is 5% or less in most cases. The observed AAI values support the distinctness of the identified clades. A detailed AAI matrix with the AAI values for each pair of species is provided in Supplementary Figure 2.
Figure 3. A matrix indicating the percentage of average amino acid identities shared by members of the family Caryophanaceae/Planococcaceae analyzed in this study based on 819 core proteins for this set of species. Higher amino acid identity shared by a pair of species are colored more darkly (red). The specific clades observed based on higher interspecies similarity are boxed with blue borders and labeled with arrows. The mean and standard deviation in AAI values for different clades of interest are indicated. The “Caryophanon” and “Flavidum” clades have only 1 AAI similarity values and hence the mean and standard deviation values for these clades could not be determined. The family Bacillaceae and the genus Jeotgalibacillus are indicated with boxes with black borders. A detailed amino acid matrix with the numerical values underlying this amino acid matrix and the species names is provided in Supplementary Figure 2.
Comparative Genomic Analysis of Different Monophyletic Clades of the Family Caryophanaceae
The results of our comprehensive phylogenetic studies indicated the existence of several clades of species comprised of distinct genera within the family Caryophanaceae. However, the topology of phylogenetic trees can be influenced by many variables (Gupta, 1998, 2016; Klenk and Goker, 2010). Thus, it is important to confirm the presence of the observed clades and their genetic distinctness by other means. Molecular markers, such as conserved signature insertions and deletions (CSIs) within genes/proteins that are exclusively shared by members from a given group of species, have proven very useful in the reliable demarcation of different species clades and for clarifying their evolutionary relationships and taxonomy (Baldauf and Palmer, 1993; Rokas and Holland, 2000; Adeolu and Gupta, 2014; Bhandari and Gupta, 2014; Gupta, 2014, 2016; Naushad et al., 2014; Dobritsa et al., 2017; Hu et al., 2019). The most parsimonious explanation for the presence of these clade-specific CSIs is that the genetic changes leading to them occurred in a common ancestor of the group(s) and they were then vertically inherited by subsequent descendants (Baldauf and Palmer, 1993; Gupta, 1998, 2014, 2016; Rokas and Holland, 2000). Thus, in view of their unique shared ancestry, the CSIs represent synapomorphic characteristics that provide reliable evidence, independent of the topology observed in phylogenetic trees, of the evolutionary relatedness of a given group of species. Hence, in this study we carried out comprehensive comparative genomic analyses of protein sequences from genomes of Caryophanaceae and some representative Bacillaceae species to identify CSIs that are specific for different novel and distinct monophyletic clades of Caryophanaceae species. These analyses have identified 13 CSIs which are specific for the emended family Caryophanaceae, and 79 CSIs that are distinctive characteristics of the nine clades of interest (all of which are also observed in the phylogenetic trees) within this family. The results of these analyses are briefly described in the subsections below.
Conserved Signature Indels Specific for the Family Caryophanaceae
The emended family Caryophanaceae consists of >100 genome-sequenced species that consistently group together in different phylogenetic trees (Figure 1 and Supplementary Figure 1). A specific grouping of these species is also supported by our identification of 13 CSIs that are exclusively shared by all of the genome-sequenced species from this clade. One example of such a CSI consisting of a 1 aa deletion in a highly conserved region of the protein phenylalanine–tRNA ligase alpha subunit, which is specific for this family, is presented in Figure 4. Detailed sequence information for this CSI and the 12 other CSIs that are also specific for the family Caryophanaceae are provided in Supplementary Figures 3–15 and some of their characteristics are summarized in Table 1. The unique shared presence of these CSIs in the indicated groups of species serves to reliably demarcate the members of the family Caryophanaceae in molecular terms.
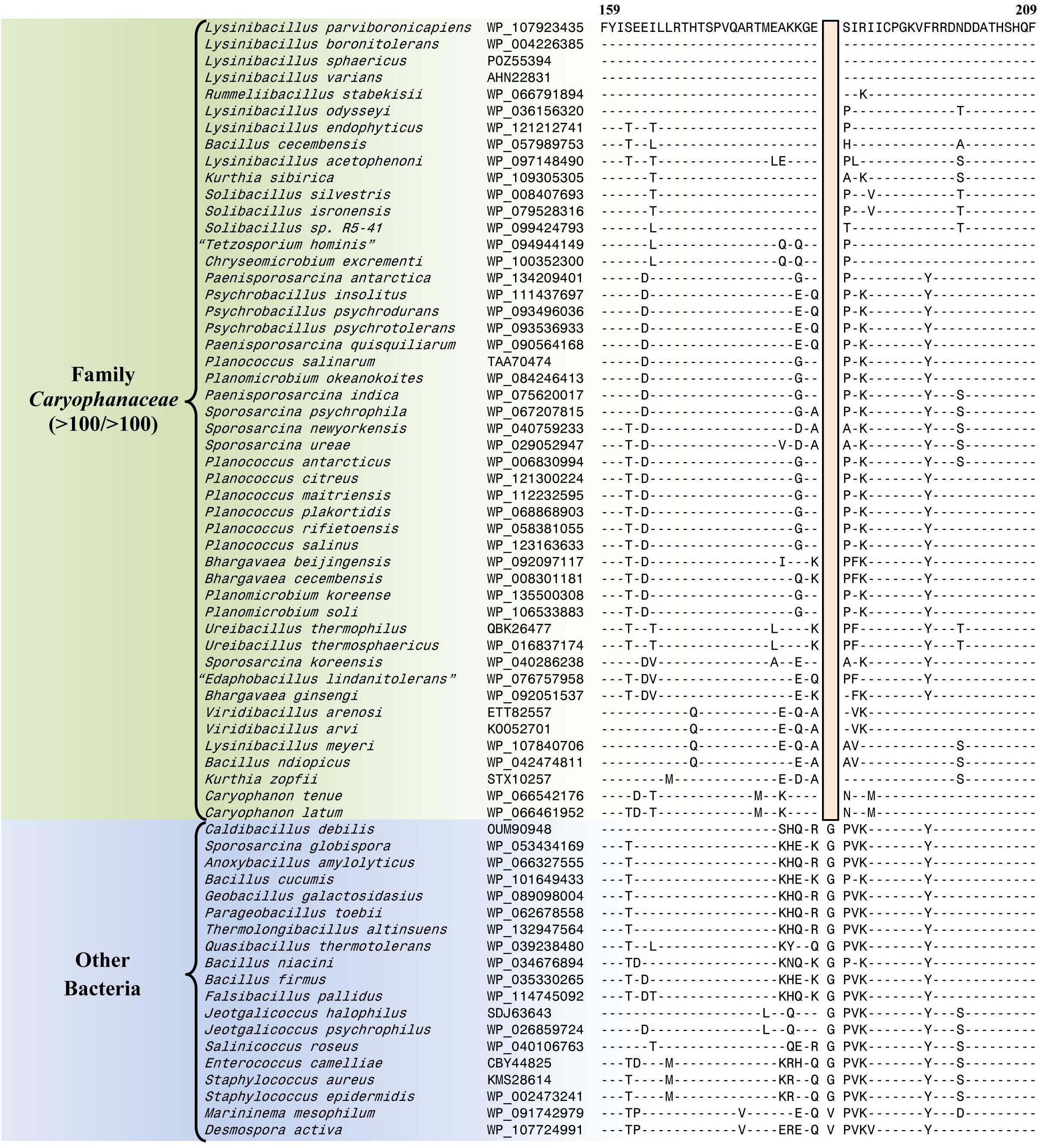
Figure 4. Partial sequence alignment of the phenylalanine–tRNA ligase subunit alpha protein showing a one amino acid deletion (boxed) that is exclusively shared by all members of the emended family Caryophanaceae. Sequence information for a limited number of Caryophanaceae species and other bacteria are shown here, but unless otherwise indicated, similar CSIs were detected in all members of the indicated group and not detected in any other species in the top 1000 BLASTp hits. The dashes (-) in this alignment and all other alignments presented in this paper indicate identity with the residue in their respective top sequences. Accession numbers for each sequence are indicated in the second column. Detailed sequence alignments for this CSI as well as additional CSIs specific for the family Caryophanaceae are presented in Supplementary Figures 3–15 and some of their characteristics are summarized in Table 1.
Conserved Signature Indels Specific for the Lysinibacillus sensu stricto Clade
The Lysinibacillus sensu stricto clade is a monophyletic clade consisting of 11 genome-sequenced Lysinibacillus species, including L. boronitolerans (the nomenclatural type of the genus Lysinibacillus), and Bacillus sp. B14905 which consistently group together in different phylogenetic trees (Figure 1 and Supplementary Figure 1). Dunlap (2019) has recently indicated that of the Lysinibacillus species which are part of this clade, L. mangiferihumi, L. tabacifolii, and L. varians are later heterotypic synonyms of L. sphaericus. Additionally, after this work was completed, a new Lysinibacillus species, L capsici, has been described, which based on its branching in a phylogenetic tree and close similarity to other members of this clade, is also indicated to be a part of this clade (Burkett-Cadena et al., 2019). Nonetheless, a specific grouping of all genome-sequenced species, which are indicated to be part of the Lysinibacillus sensu stricto clade, is strongly supported by our identification of six CSIs that are exclusively shared by all of these species. One example of such a CSI consisting of a 1 aa insertion in the bacillithiol biosynthesis deacetylase BshB2 protein is presented in Figure 5A. Detailed sequence information for this CSI and the five other CSIs that are specific for the Lysinibacillus sensu stricto clade are provided in Supplementary Figures 16–21 and some of their characteristics are summarized in Table 2. Lysinibacillus pakistanensis does not appear in the CSIs presented here because it is not available in the NCBI non-redundant protein database at the time of writing this paper.
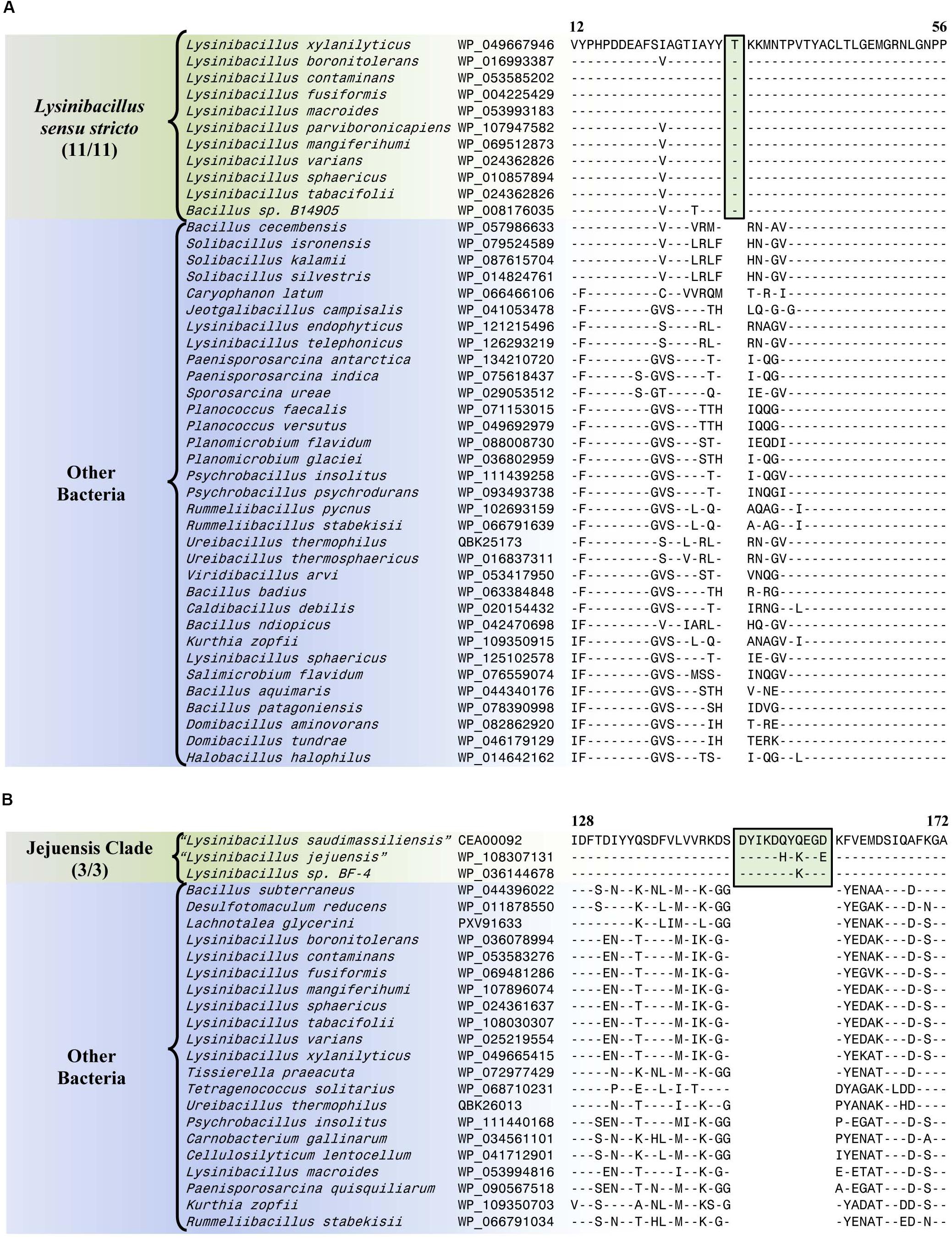
Figure 5. Partial sequence alignment of (A) the bacillithiol biosynthesis deacetylase BshB2 protein showing a one amino acid insertion (boxed) that is exclusively shared by all members of the Lysinibacillus sensu stricto clade, and (B) the arginine-binding extracellular protein ArtP precursor containing an 11 amino acid insertion that is exclusively shared by all members of the Jejuensis clade. Detailed sequence alignments for these CSIs as well as additional CSIs specific for these clades are presented in Supplementary Figures 16–21 for the Lysinibacillus sensu stricto clade and Supplementary Figures 22–38 for the Jejuensis clade and some of their characteristics are summarized in Table 2.
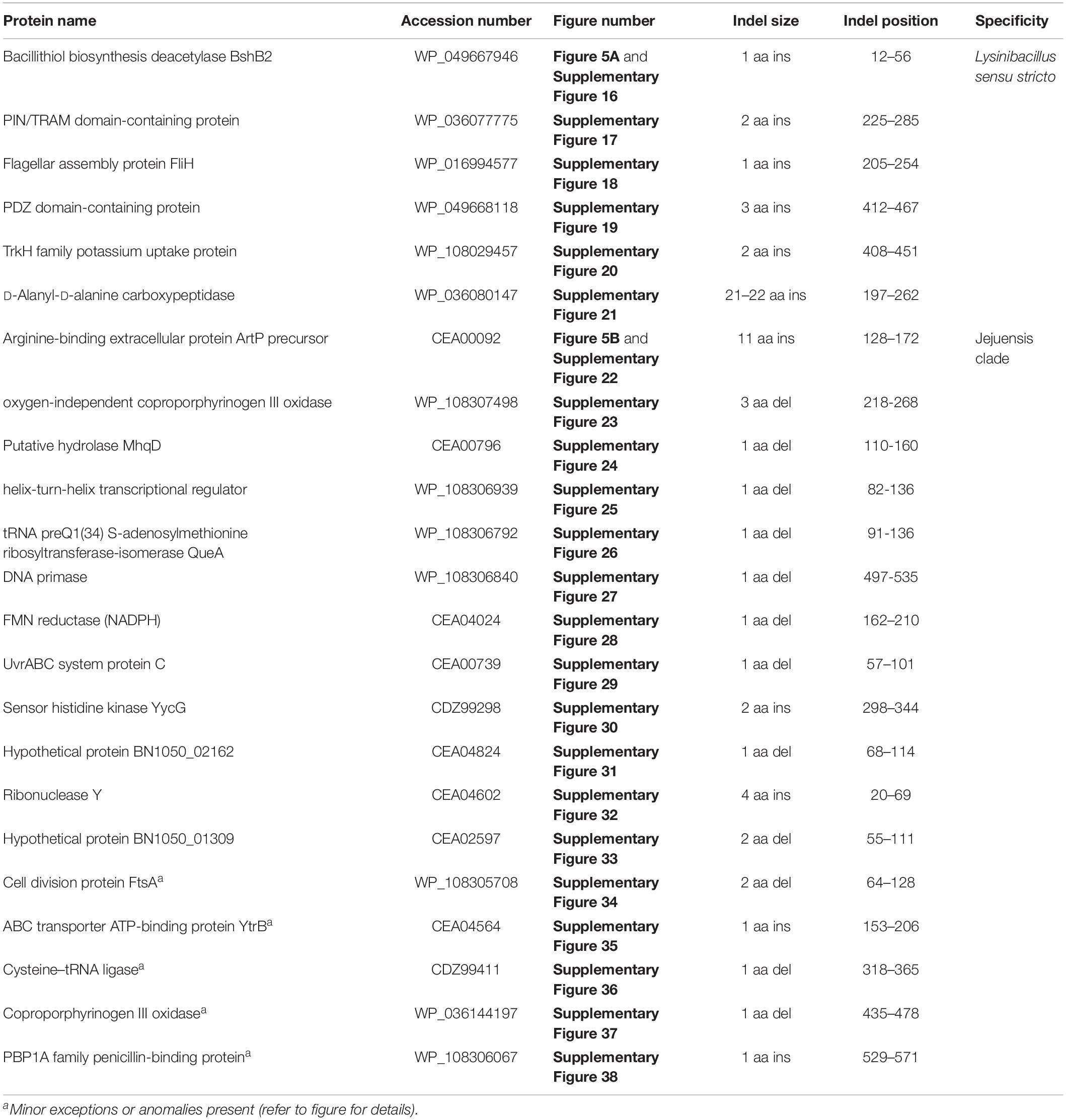
Table 2. Summary of conserved signature indels specific for the members of the Lysinibacillus sensu stricto clade and the Jejuensis clade.
Conserved Signature Indels Specific for the Jejuensis Clade
The “Jejuensis clade” is a monophyletic clade consisting of the genome-sequenced species “Lysinibacillus jejuensis,” “Lysinibacillus saudimassiliensis,” and Lysinibacillus sp. BF-4 which consistently group together in different phylogenetic trees (Figure 1 and Supplementary Figure 1). A specific grouping of these three species is also supported by our identification of 17 CSIs that are exclusively shared by them. One example of such a CSI consisting of an 11 aa insertion in the arginine-binding extracellular protein ArtP precursor is presented in Figure 5B. Detailed sequence information for this CSI and the 16 other CSIs that are specific for the “Jejuensis clade” are provided in Supplementary Figures 22–38 and some of their characteristics are summarized in Table 2.
Conserved Signature Indels Specific for the “Ureibacillus Clade”
The “Ureibacillus clade” is a monophyletic clade consisting of the genome-sequenced species Ureibacillus thermosphaericus, U. thermophilus, and 10 Lysinibacillus species which consistently group together in different phylogenetic trees (Figures 1, 2 and Supplementary Figure 1). A specific grouping of these 12 species is also supported by our identification of three CSIs that are exclusively shared by them. One example of such a CSI consisting of a 1 aa insertion in the MFS transporter protein is presented in Figure 6A. Detailed sequence information for this CSI and the two other CSIs that are specific for the “Ureibacillus clade” are provided in Supplementary Figures 39–41 and some of their characteristics are summarized in Table 3.

Figure 6. Partial sequence alignments of (A) the MFS transporter protein showing a one amino acid insertion (boxed) that is exclusively shared by all members of the Ureibacillus clade and (B) the DUF456 domain-containing protein containing a one amino acid deletion (boxed) that is exclusively shared by all members within the Meyeri clade. Detailed sequence alignments for these CSIs as well as additional CSIs specific for these clades are presented in Supplementary Figures 39–41 for the Ureibacillus clade and Supplementary Figures 42–53 for the Meyeri clade and some of their characteristics are summarized in Table 3.
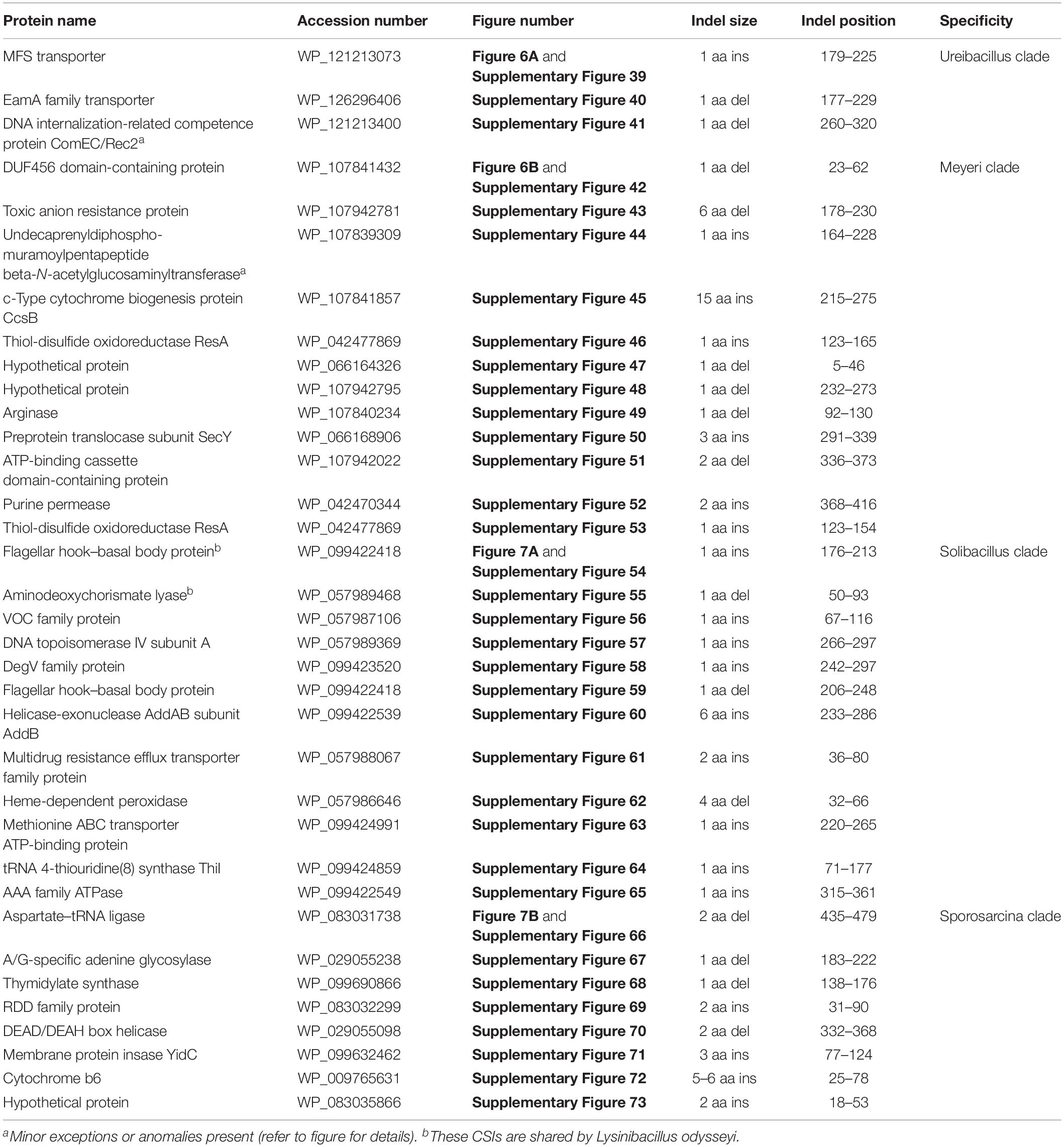
Table 3. Summary of conserved signature indels specific for the Ureibacillus clade, the Meyeri clade, the Solibacillus clade, and the Sporosarcina clade.
Conserved Signature Indels Specific for the “Meyeri Clade”
The “Meyeri clade” is a monophyletic clade consisting of the genome-sequenced species Lysinibacillus meyeri, “Lysinibacillus fluoroglycofenilyticus,” and B. ndiopicus which consistently group together in different phylogenetic trees (Figure 1 and Supplementary Figure 1). A specific grouping of these three species is also supported by our identification of 12 CSIs that are exclusively shared by them. One example of such a CSI consisting of a 1 aa deletion in the DUF456 domain-containing protein is presented in Figure 6B. Detailed sequence information for this CSI and the 11 other CSIs that are specific for the “Meyeri clade” are provided in Supplementary Figures 42–53 and some of their characteristics are summarized in Table 3.
Conserved Signature Indels Specific for the “Solibacillus Clade”
The “Solibacillus clade” is a monophyletic clade consisting of four genome-sequenced Solibacillus species including Solibacillus silvestris (the nomenclatural type of the genus Solibacillus), Lysinibacillus odysseyi, Lysinibacillus sp. 2017, and Bacillus cecembensis which consistently group together in different phylogenetic trees (Figure 1 and Supplementary Figure 1). A specific grouping of these seven species is also supported by our identification of 12 CSIs that are exclusively shared by them. One example of such a CSI consisting of a 1 aa insertion in the flagellar hook–basal body protein is presented in Figure 7A. Detailed sequence information for this CSI and the 11 other CSIs that are specific for the “Solibacillus clade” are provided in Supplementary Figures 54–65 and some of their characteristics are summarized in Table 3.
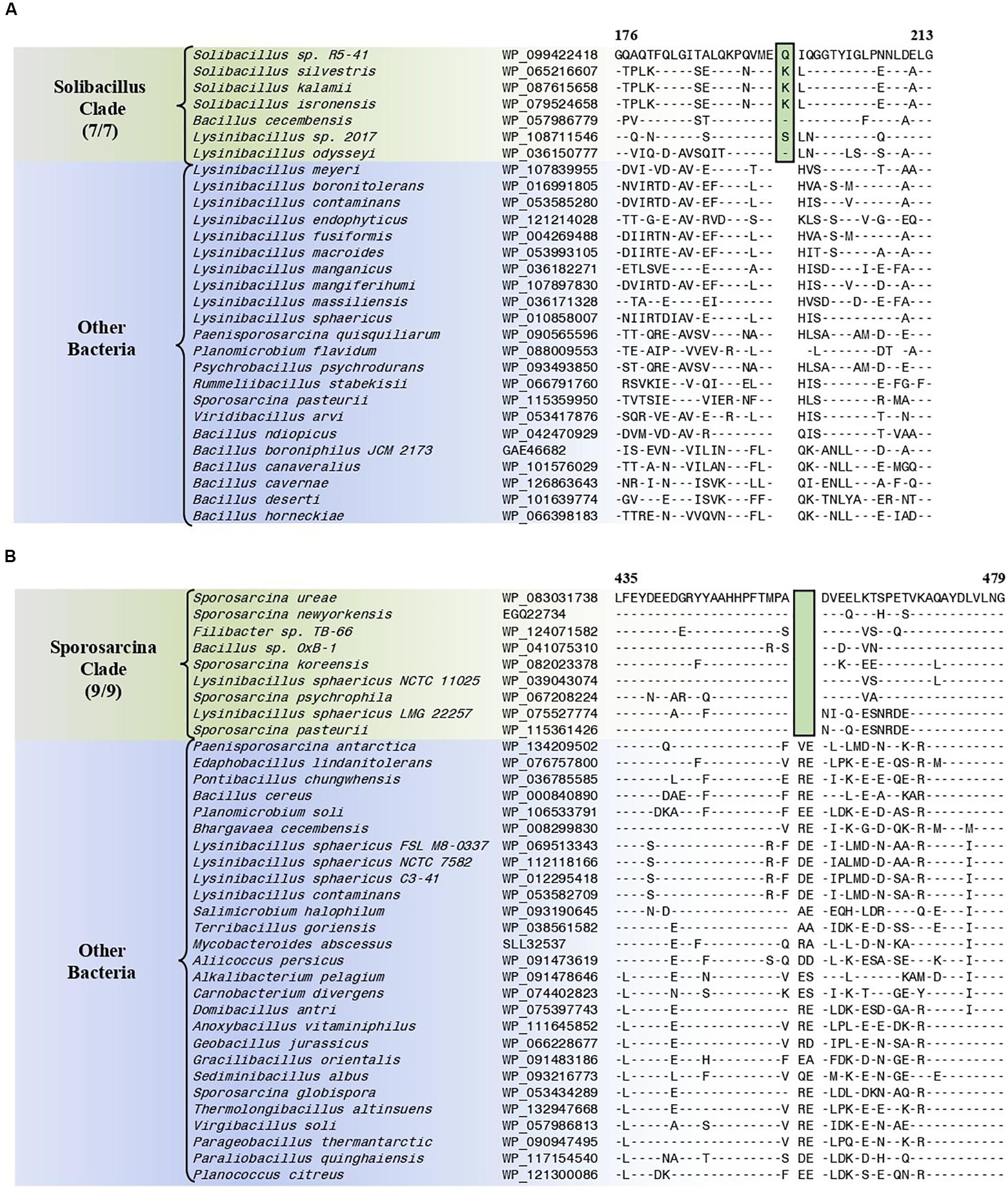
Figure 7. Partial sequence alignments of (A) the flagellar hook–basal body protein showing a one amino acid insertion (boxed) that is exclusively shared by all members of the Solibacillus clade, and (B) the aspartate–tRNA ligase protein containing a two amino acid deletion (boxed) that is exclusively shared by all members of the Sporosarcina clade. Detailed sequence alignments for these CSIs as well as additional CSIs specific for these clades are presented in Supplementary Figures 54–65 for the Solibacillus clade, and Supplementary Figures 66–73 for the Sporosarcina clade and some of their characteristics are summarized in Table 3.
Conserved Signature Indels Specific for the “Sporosarcina Clade”
The “Sporosarcina clade” is a monophyletic clade consisting of five genome-sequenced Sporosarcina species including S. ureae (the nomenclatural type of the genus Sporosarcina), L. sphaericus LMG 22257, L. sphaericus NCTC 11025, Bacillus OxB-1, and Filibacter sp. TB-66 which consistently group together in different phylogenetic trees (Figure 1 and Supplementary Figure 1). A specific grouping of these nine species is also supported by our identification of eight CSIs that are exclusively shared by them. One example of such a CSI consisting of a 2 aa deletion in the aspartate–tRNA ligase protein is presented in Figure 7B. Detailed sequence information for this CSI and the seven other CSIs that are specific for the “Sporosarcina clade” are provided in Supplementary Figures 66–73 and some of their characteristics are summarized in Table 3.
Conserved Signature Indels Specific for the “Planococcus/Planomicrobium Clade”
The “Planococcus/Planomicrobium” clade is a monophyletic clade consisting of 15 genome-sequenced Planococcus species including P. citreus, and 4 Planomicrobium species which consistently group together in different phylogenetic trees (Figure 1 and Supplementary Figure 1). A specific grouping of these 19 species is also supported by our identification of five CSIs that are exclusively shared by them. One example of such a CSI consisting of a 2 aa insertion in penicillin-binding protein 2 is presented in Figure 8A. Detailed sequence information for this CSI and the four other CSIs that are specific for the “Planococcus/Planomicrobium clade” are provided in Supplementary Figures 74–78 and some of their characteristics are summarized in Table 4.
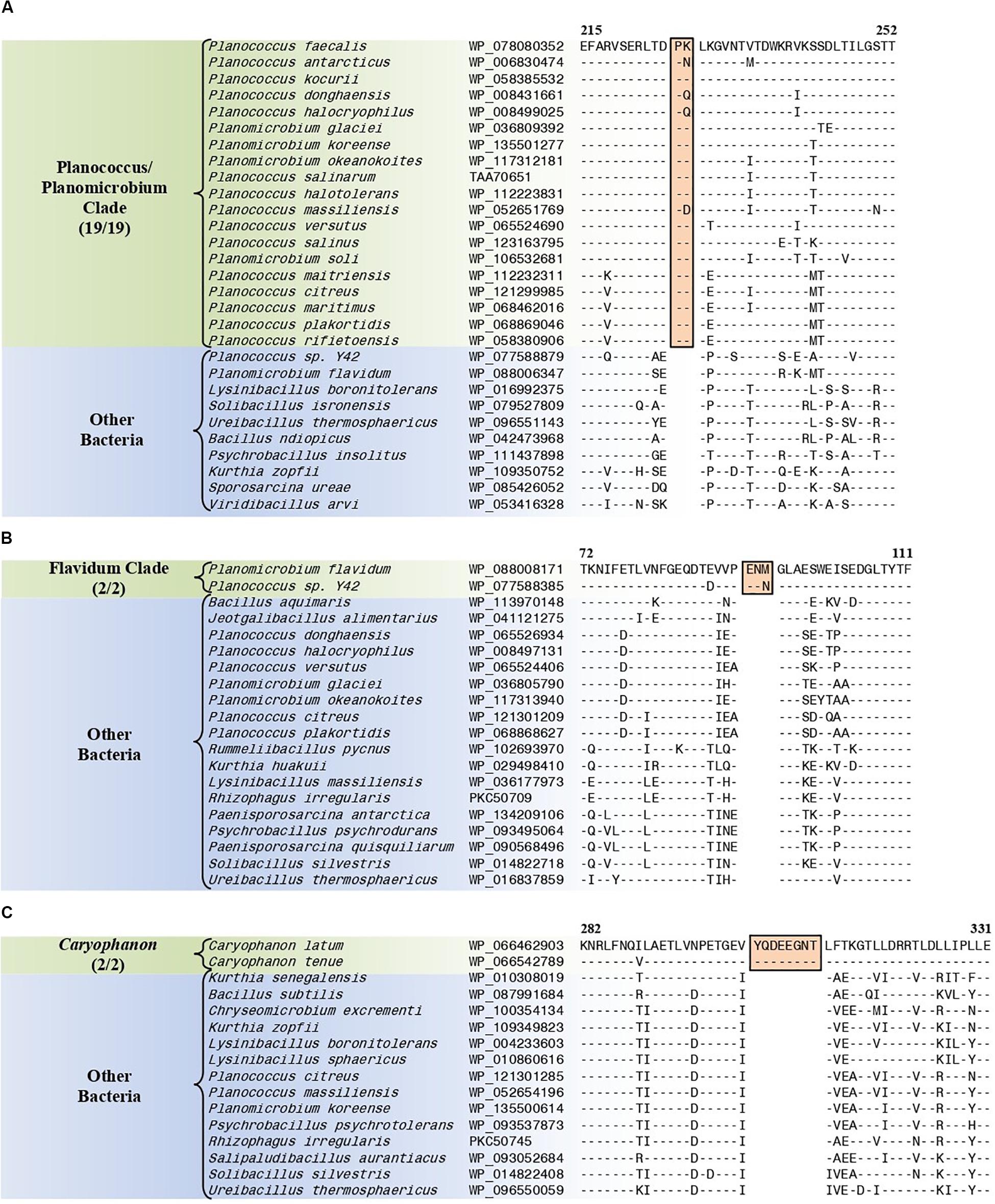
Figure 8. Partial sequence alignments of (A) the penicillin-binding protein 2 protein showing a two amino acid insertion (boxed) that is exclusively shared by all members of the Planococcus/Planomicrobium clade, (B) the ABC transporter substrate-binding protein containing a three amino acid insertion (boxed) that is exclusively shared by all members within the Flavidum clade, and (C) the DNA-directed RNA polymerase subunit beta protein containing an eight amino acid insertion that is exclusively shared by all members of the genus Caryophanon. Detailed sequence alignments for these CSIs as well as additional CSIs specific for these clades are presented in Supplementary Figures 74–78 for the Planococcus/Planomicrobium clade, Supplementary Figures 79–86 for the Flavidum clade, and Supplementary Figures 87–94 for the genus Caryophanon and some of their characteristics are summarized in Table 4.
Table 4. Summary of conserved signature indels specific for the Planococcus/Planomicrobium clade, the Flavidum clade, and the genus Caryophanon.
Conserved Signature Indels Specific for the “Flavidum Clade”
The “Flavidum clade” is a monophyletic clade consisting of the genome-sequenced species Planomicrobium flavidum, and Planococcus sp. Y42 which consistently group together in different phylogenetic trees (Figure 1 and Supplementary Figure 1). A specific grouping of these two species is also supported by our identification of eight CSIs that are exclusively shared by them. One example of such a CSI consisting of a 3 aa insertion in ABC transporter substrate-binding protein is presented in Figure 8B. Detailed sequence information for this CSI and the seven other CSIs that are specific for the “Flavidum clade” are provided in Supplementary Figures 79–86 and some of their characteristics are summarized in Table 4.
Conserved Signature Indels Specific for the Genus Caryophanon
The genus Caryophanon is a monophyletic clade consisting of the genome-sequenced species Caryophanon latum, and Caryophanon tenue which consistently group together in different phylogenetic trees (Figure 1 and Supplementary Figure 1). A specific grouping of these two species is also supported by our identification of eight CSIs that are exclusively shared by them. One example of such a CSI consisting of an 8 aa insertion in the DNA-directed RNA polymerase subunit beta protein is presented in Figure 8C. Detailed sequence information for this CSI and the seven other CSIs that are specific for the genus Caryophanon are provided in Supplementary Figures 87–94 and some of their characteristics are summarized in Table 4.
Discussion
The family Caryophanaceae is a taxonomically heterogeneous assemblage of species from diverse genera and current methods of classification, such as 16S rRNA-based phylogenetic trees, have proven inadequate in clarifying the evolutionary history and composition of this family (Shivaji et al., 2014; Maayer et al., 2019). With no shared characteristics or reliable means for its demarcation, the family Caryophanaceae has become a polyphyletic assemblage of bacterial genera with conflicting classifications (Ludwig et al., 2009; Yarza et al., 2010; Parte, 2014; Shivaji et al., 2014; Maayer et al., 2019). Fortunately, recent improvements in genome sequencing technology have provided a plethora of genome sequence data for the majority of species from this family3, providing an exclusive and previously unavailable resource for resolving the interrelationships of different species forming the family Caryophanaceae via multiple independent approaches (Gupta, 2014, 2016; Dobritsa et al., 2017).
Using available genome sequences for 124 Caryophanaceae/Planococcaceae and some representative Bacillaceae species, in the present work, we have performed comprehensive phylogenomic analyses based on several large datasets of protein sequences. All of the constructed trees were observed to form extremely similar branching patterns and interrelationships and also consistently displayed a strongly supported monophyletic clade containing all Caryophanaceae/Planococcaceae genera (except Jeotgalibacillus). Within this clade, a number of genera (viz. Lysinibacillus, Viridibacillus, “Edaphobacillus,” Solibacillus, and Rummeliibacillus) belonging to other families within the order Bacillales were also interspersed. Strong independent evidence that the species from the above genera form a monophyletic grouping distinct from all other Bacillales families and genera is provided by our identification of 13 CSIs in 13 different proteins that are uniquely shared by species from all of the genera contained within this clade, but not by other Bacillales species. Thus, the identified CSIs in conjunction with the results from our comprehensive phylogenetic studies reliably demarcate a monophyletic grouping of species, which represent the emended family Caryophanaceae. The emended family Caryophanaceae now encompasses both the families Planococcaceae and Caryophanaceae and it rectifies the taxonomic anomaly that the family name Planococcaceae is a later heterotypic synonym of Caryophanaceae (Tindall, 2019).
Within the emended family Caryophanaceae, in all phylogenetic trees based on genome sequences, 18 identical and distinct clades were consistently observed (labeled with square brackets in Figure 1 and Supplementary Figure 1). The distinctness of these species clades was also strongly supported by the results from our AAI analysis. More importantly, our identification of multiple highly specific molecular markers (i.e., CSIs) in important proteins that are uniquely shared by the members of these clades clearly distinguish the members of these clades from each other as well as all other Bacillales species. Molecular markers of this kind provide independent evidence that the species from these clades are specifically related to each other and they shared common ancestors exclusive of all other species. Figure 9 presents a summary diagram showing the species compositions of these clades as well as the numbers of distinct CSIs that were identified in this work. In addition to the genome sequenced species (shown in bold), this summary diagram also includes some additional species (non-bolded), which based on their branching in the 16S rRNA tree (Figure 2) are also a part of the indicated clades. Of these 18 clades, 10 clades were monophyletic groupings comprising of different species from known genera (viz. Rummeliibacillus, Chryseomicrobium, Viridibacillus, Rummeliibacillus, Kurthia, Caryophanon, Psychrobacillus, Paenisporosarcina, “Edaphobacillus,” and “Tetzosporium”). In contrast to these clades, the remaining eight clades were either comprised of species from multiple different genera or constituted novel species clades identified in the present work. These species clades were investigated in greater detail in this work and the taxonomic implications of these results are discussed below.
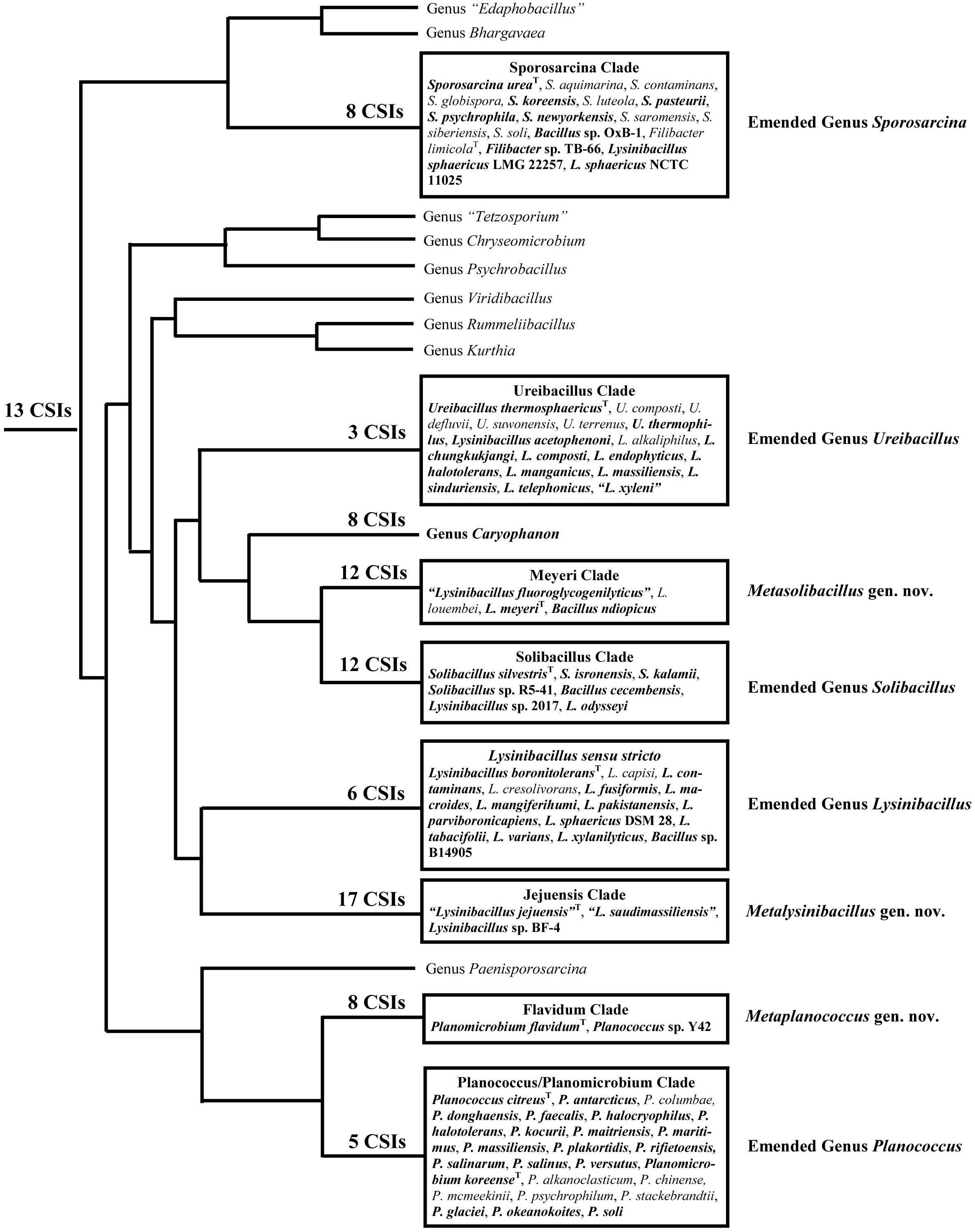
Figure 9. A conceptual diagram based on results obtained from phylogenetic studies, AAI similarity analysis, and several identified CSIs indicating the evolutionary relationships among the Caryophanaceae species. The total numbers of identified CSIs that are specifically shared by species from each of these clades are indicated at their respective nodes. All clades analyzed in this study are shown in boxes containing the species they comprise. Of these species, the ones in bold are genome-sequenced while the rest are placed within these clades based on our 16S rRNA analysis.
Before discussing the taxonomic implications of our results, it should be indicated that in our analyses of genome sequences, anomalous results were observed with genome sequences for two species viz. P. quisquiliarum (GenBank assembly accession: GCA_900109875.1) and S. globispora (GenBank assembly accession: GCA_001274725.1). In phylogenetic trees based on the deposited genome sequence for P. quisquiliarum, it branches separately from other Paenisporosarcina species, within a clade comprised of different genome-sequenced Psychrobacillus species, making both the genera Paenisporosarcina and Psychrobacillus polyphyletic. However, the 16S rRNA sequences from this genome exhibit only 94.24% sequence identity to the reference 16S rRNA sequence for the type strain of P. quisquiliarum (GenBank assembly accession: GCA_900109875.1), but >99.9% sequence similarity to the 16S rRNA sequence for the type strain of Psychrobacillus psychrodurans. Thus, we suspect that the deposited genome sequence for P. quisquiliarum (GenBank assembly accession: GCA_900109875.1) is closely related to P. psychrodurans, accounting for its anomalous branching in different protein trees (Figure 1 and Supplementary Figure 1). Similarly, in our protein trees, S. globispora branches separately from all other Sporosarcina species and deeply within the Bacillaceae species (Figure 1 and Supplementary Figure 1). The 16S rRNA sequences from this genome exhibit only 94.40% sequence identity to the reference 16S rRNA sequence for the type strain from S. globispora (GenBank assembly accession: GCA_001274725.1), indicating that the deposited genome sequence does not correspond to this species. As the sequences for both these species reliably group with the sequences of other species from these genera in the 16S rRNA trees, we have chosen to ignore the anomalous results obtained from their deposited genomes in our discussion. However, the genomes for the type strains of these two species should be sequenced again to confirm that the results obtained from the deposited genomes are anomalous.
As noted above, the work presented here allows reliable identification and demarcation of eight different species clades within the emended family Caryophanaceae that are comprised of either species from multiple different genera or novel species clades from the existing genera (Figure 9). Similar groupings of species from these genera have also been observed in earlier studies based on other types of analyses using a limited number of genome sequences as well as some chemotaxonomic characteristics and branching in 16S rRNA trees (Seiler et al., 2013; Shivaji et al., 2014; Xu et al., 2015; Maayer et al., 2019). The species from these clades are also observed to form distinct groupings in the Genome Taxonomy Database (GTDB) based on phylogenetic analyses of 120 ubiquitous single-copy proteins (Parks et al., 2018). Some characteristics of the species from these eight clades and their taxonomic implications are indicated below.
The genus Lysinibacillus is known to be highly polyphyletic (Kampfer et al., 2013; Xu et al., 2015; Gomez-Garzon et al., 2016; Mual et al., 2016), and in our work, species from this genus are seen forming six different clades. Of these clades, the clade marked Lysinibacillus sensu stricto, which is distinguished by six identified CSIs, contains the type species L. boronitolerans as well as 11 other Lysinibacillus species including the type strain for the important entomopathogenic bacteria L. sphaericus. One unnamed Bacillus sp. is also a part of this clade. As this clade is distinct from all other clades, we are proposing that the emended genus Lysinibacillus be restricted to only the species from this clade. Another clade, referred to here as the “Ureibacillus clade” is comprised of different species from the genus Ureibacillus as well 10 Lysinibacillus species. These species consistently group together in different phylogenetic trees including both protein sequences-based and 16S rRNA-based trees (Figures 1, 2 and Supplementary Figure 1). A specific grouping of different genome sequenced species from this clade is also supported by three specific CSIs that are exclusively shared by them. In contrast, no CSI was identified that is only shared by the Lysinibacillus species that are part of this clade. Hence, to bring taxonomic clarity to this clade of species, we are proposing the transfer of all 10 Lysinibacillus species which are part of this clade to the emended genus Ureibacillus. The name combinations proposing these transfers are listed in Table 5.
The “Meyeri” and “Jejuensis” clades are two new species clades identified in this work. These two clades are reliably distinguished from all other clades by different means including our identification of 12 and 17 novel CSIs, respectively, which are uniquely shared by the species from these clades. Of these two clades, the “Meyeri clade” is comprised of B. ndiopicus and two Lysinibacillus species, whereas the “Jejuensis clade” is comprised of two named and one unnamed Lysinibacillus species. As both these clades are novel clades, with no designated type species, we are proposing the transfer of species from the “Meyeri” and “Jejuensis” clades into two novel genera named Metalysinibacillus gen. nov. and Metasolibacillus gen. nov., respectively. The descriptions of these genera are provided below and the new name combinations proposing the transfer of species from these two clades into the two proposed genera are listed after the section “Discussion” and in Table 6, respectively. The “Solibacillus clade” demarcated by our studies not only contains all species from the genus Solibacillus, but also encompasses two Lysinibacillus species as well as B. cecembensis. This grouping is strongly supported by 12 CSIs that are uniquely shared by all seven species forming this clade. In light of the strong evidence supporting the distinctness of this clade, we are proposing the transfer of B. cecembensis and the Lysinibacillus species, which are observed to branch within this clade, to the emended genus Solibacillus forming a taxonomically homogeneous clade/genus. The name combinations proposing these transfers are listed in Table 6.
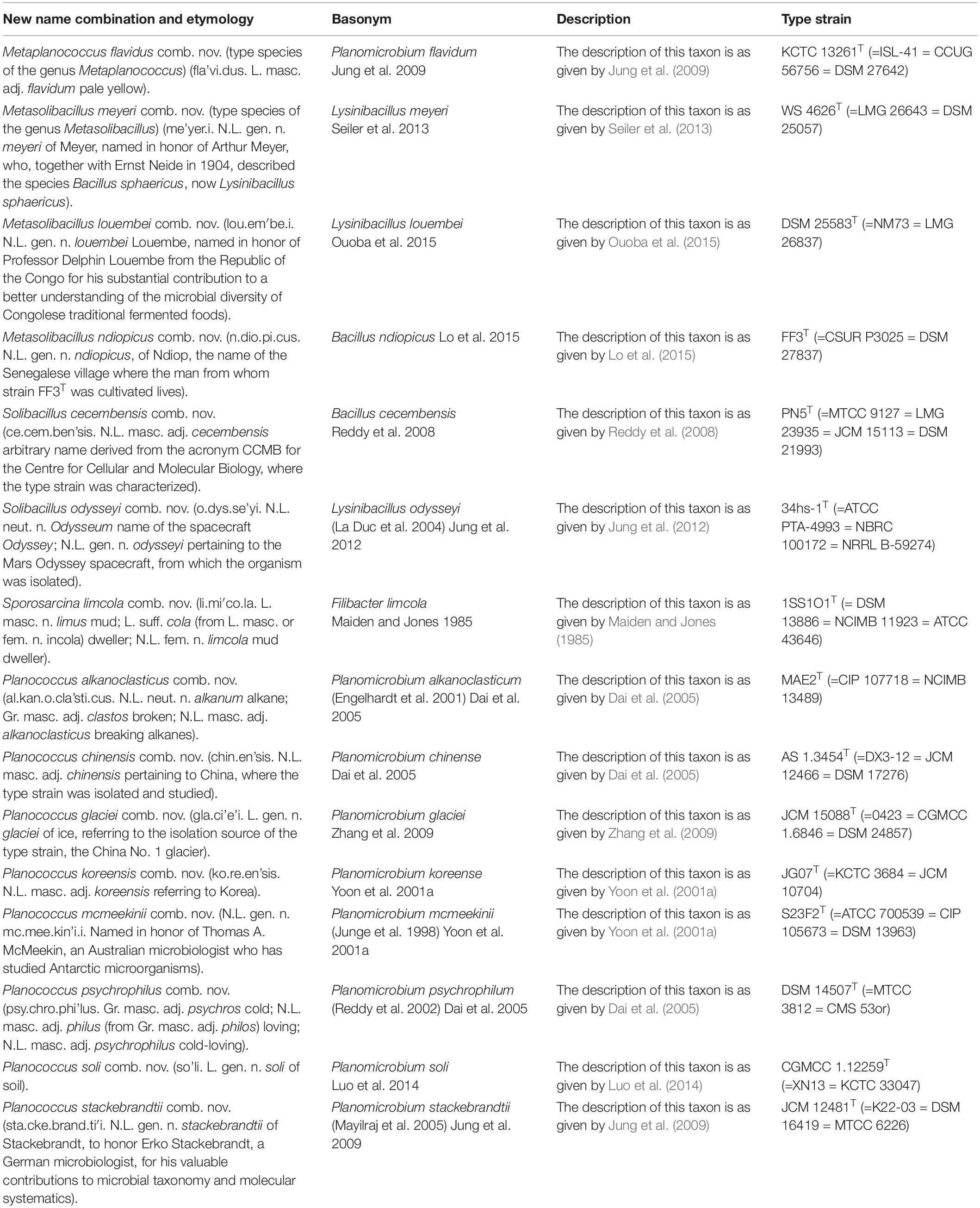
Table 6. Descriptions of the new combinations in Metaplanococcus gen. nov., Metasolibacillus gen. nov, emended genus Solibacillus, emended genus Sporosarcina, and emended genus Planococcus.
Two of the clades shown in Figures 1, 9 are comprised of species from the genera Planococcus and Planomicrobium. Of these two genera, the genus Planomicrobium was created in 2001 by the transfer of three Planococcus species on the basis of cell morphology and 16S rRNA-based phylogenetic analysis to this new genus (Yoon et al., 2001a). However, our analyses reveal that both these genera are polyphyletic, and their species are interspersed among one another within a larger clade consisting of all of the species from these two genera (see Figure 1 and Supplementary Figure 1). Within this large clade, the species Planococcus flavidum together with an unnamed Planococcus species form a deeper branching lineage (designated as the “Flavidum clade”) relative to the rest of the species from these two genera (designated as the “Planococcus/Planomicrobium clade”). In our work, while we have identified multiple CSIs that are specific for the species from the “Flavidum” or “Planococcus/Planomicrobium” clade, no CSI was identified that was specifically shared by the species from only the genus Planococcus or the genus Planomicrobium. To account for these results and to clarify the taxonomy of species from these two genera, we are proposing that all of the species from these two genera, which comprise the “Planococcus/Planomicrobium clade” be united within the emended genus Planococcus, which has priority [Rule 24b (1)] over the genus Planomicrobium (Migula, 1894; Skerman et al., 1980; Nakagawa et al., 1996; Yoon et al., 2001a, 2010; Parker et al., 2019). Further, due to the phylogenetic and molecular distinctness of the “Flavidum clade,” we are proposing the transfer of species from this clade into a new genus named Metaplanococcus gen. nov. The name combinations proposing these taxonomic changes are listed in Table 6. After this work was completed, a new genus Indiicoccus containing the sole species I. explosivorum, which branches peripherally to the Planococcus clade has also been described (Pal et al., 2019). It is unclear at present whether this new species shares any of the CSIs that are specific for members of the genera Planococcus or Metaplanococcus.
Lastly, the clade marked as the “Sporosarcina clade” includes within it all of the species from the genus Sporosarcina along with Bacillus sp. OxB-1, two strains of L. sphaericus and Filibacter sp. TB-66. In the 16S rRNA tree (Figure 2), the species Filibacter limicola, which is the type species of the genus Filibacter, also reliably branches with the other Sporosarcina species. To clarify the taxonomy of this clade, we are proposing that all of the species which are part of the “Sporosarcina clade” should be transferred to the emended genus Sporosarcina, which has priority over the genus Filibacter (Kluyver and Van Niel, 1936; Skerman et al., 1980; Maiden and Jones, 1985; Yoon et al., 2001b). As the type strain of the species L. sphaericus is a part of the genus Lysinibacillus, the two L. sphaericus strains which branch within the “Sporosarcina clade” are described as two new species viz. Sporosarcina sphaericus sp. nov., and Sporosarcina urealyticus sp. nov.
Based on the results presented here, we have developed a reliable and coherent phylogenetic framework for understanding the evolutionary relationships as well as a classification scheme for the members of the family Caryophanaceae. In the proposed classification scheme, the family Caryophanaceae and the different monophyletic clades (genera) that form this family are reliably delineated both by means of extensive phylogenetic analyses as well as by our identification of large numbers of highly specific molecular markers (CSIs) that are specifically shared by the members of these clades. It is important to note that the CSIs described in this study for the family Caryophanaceae and for its different genera have several important applications. Earlier work on CSIs has shown that they exhibit a high degree of predictive ability to be found in other members of the group they represent (whose genome sequences are not yet available or in species which are not yet discovered) (Gao and Gupta, 2012; Adeolu and Gupta, 2014; Bhandari and Gupta, 2014; Sawana et al., 2014; Adeolu et al., 2016; Gupta et al., 2016, 2018; Dobritsa et al., 2017; Patel and Gupta, 2018; Gupta, 2019). Hence, the presence or absence of these CSIs in the genome sequences of other species (including unnamed species) can be used to determine if they belong to the family Caryophanaceae or any of the other genera for which CSIs are described in this study. Additionally, earlier studies on CSIs provide evidence that these molecular characteristics are functionally important for the group of organisms for which they are specific (Singh and Gupta, 2009; Khadka and Gupta, 2017). Hence, genetic and biochemical studies on understanding the functional significance of these CSIs are expected to lead to the identification of novel biochemical and/or other characteristics that are distinctive properties of the described groups of bacteria.
The emended descriptions of the family Caryophanaceae as well as the descriptions of various novel and emended species and genera are given below. The new name combinations for species that results from the proposed taxonomic changes are listed in Tables 5, 6.
Emended Description of the Family Caryophanaceae Peshkoff 1939 (Approved Lists 1980)
(Ca.ry.o.pha.na.ce’ae. N.L. neut. n. Caryophanon, type genus of the family: suff. -aceae, ending to denote a family; N.L. fem. pl. n. Caryophanaceae, the Caryophanon family).
The family Caryophanaceae is circumscribed here based on the monophyletic grouping of different taxa from this family in phylogenetic trees based on multiple large datasets of protein sequences and conserved signatures indels in multiple proteins listed below that are specifically shared by the members of this family. The emended family Caryophanaceae presently contains the following genera: Caryophanon, Bhargavaea, “Chryseomicrobium,” “Edaphobacillus,” Indii- coccus, Kurthia, Lysinibacillus, Metalysinibacillus, Metasoli- bacillus, Metaplanococcus, Paenisporosarcina, Planococcus, Psychrobacillus, Rummelibacillus, Solibacillus, Sporosarcina, “Tetzosporium,” Ureibacillus, and Virdibacillus. As both Caryophanon and Planococcus are part of the same family and, according to the Rule 24b (1) of the Code (Parker et al., 2019), the name Caryophanaceae (Peshkoff, 1939) has priority over the name Planococcaceae (Krasil’nikov, 1949), the family name Caryophanaceae is used for the description of this family. As a result, the name Planococcaceae is now a later heterotypic synonym for the Caryophanaceae. Cells from members of the family Caryophanaceae can be cocci or rods, sometimes forming filaments or trichomes. Most species are strictly aerobic heterotrophs, although some are also facultatively aerobes. Cells are generally motile by flagella or gliding and they may or may not form endospores. Most species are catalase-positive and oxidase positive or negative. Members of this family can be reliably distinguished from all other Firmicutes genera based on the shared presence of CSIs described in this work in all or most of the following proteins: phenylalanine–tRNA ligase subunit alpha, chaperonin GroEL, ribosome maturation factor RimP, BrxA/BrxB family bacilliredoxin, RNA methyltransferase, Rhomboid family intramembrane serine protease, ATP-dependent Clp protease ATP-binding subunit, DNA-directed RNA polymerase subunit beta, Chorismate synthase, Stage IV sporulation protein A, peptidase, KinB-signaling pathway activation protein, and DUF423 domain-containing protein.
Type genus: Caryophanon Peshkoff 1939 (Approved Lists 1980).
Emended Description of the Genus Caryophanon Peshkoff 1939 (Approved Lists 1980)
Caryophanon. (Ca.ry.o’pha.non. Gr. neut. n. karyon, nut, kernel, nucleus; Gr. masc.adj. phaneros, bright, conspicuous; N.L. neut. n. Caryophanon, that which has a conspicuous nucleus).
The emended genus Caryophanon contains the type species Caryophanon latum. Cells are Gram-positive, aerobic, and motile by means of peritrichous flagella. The members of this genus form a monophyletic clade in 16S rRNA gene tree and phylogenetic trees based on multiple large datasets of protein sequences. Members of this genus can be reliably distinguished from other genera within the family Caryophanaceae based on the shared presence of CSIs described in this work in all or most of the following proteins: DNA-directed RNA polymerase subunit beta, peroxide-responsive transcriptional repressor PerR, ADP-forming succinate-CoA ligase subunit beta, tRNA (N(6)-L-threonylcarbamoyl adenosine (37)-C(2)-methylthiotransferase MtaB, magnesium transporter, dephospho-CoA kinase, ATP synthase subunit I, and bifunctional DNA-formamidopyrimidine glycosylase/DNA-(apurinic or apyrimidinic site) lyase.
Type species: Caryophanon latum Peshkoff 1939 (Approved Lists 1980).
Emended Description of the Genus Lysinibacillus Ahmed et al. 2007 emend. Jung et al. 2012
Lysinibacillus (Ly.si.ni.ba.cil’lus. N.L. neut. n. lysinum lysine; L. masc. n. bacillus a small staff or rod; N.L. masc. n. Lysinibacillus lysine bacillus, referring to the presence of the Lys–Asp type of peptidoglycan in the cell wall).
The emended genus Lysinibacillus contains the type species L. boronitolerans. The members of this genus are generally motile, rod-shaped cells that produce ellipsoidal or spherical endospores which lie terminally in a swollen sporangium. Cell-wall peptidoglycan of the studied species has been reported to contain lysine and aspartic acid (Ahmed et al., 2007). Members of this emended genus form a monophyletic clade in a 16S rRNA gene tree and trees based on multiple large datasets of protein sequences. Further, members of this genus can be reliably distinguished from all other genera within the family Caryophanaceae based on the shared presence of CSIs described in this work in all or most of the following proteins: bacillithiol biosynthesis deacetylase BshB2, PIN/TRAM domain-containing protein, flagellar assembly protein FliH, PDZ domain-containing protein, TrkH family potassium uptake protein, and D-alanyl-D-alanine carboxypeptidase.
Type species: Lysinibacillus boronitolerans Ahmed et al. 2007.
Emended Description of the Genus Ureibacillus (Andersson et al. 1996) Fortina et al. 2001
Ureibacillus (Ur.e.i. ba.cil’lus. N. L. fem. n. urea urea; N. L. masc. n. Bacillus a bacterial genus; N.L. masc. n. Ureibacillus a ureolytic Bacillus-like organism).
The emended genus Ureibacillus contains the type species U. thermosphaericus, which was originally described as Bacillus thermosphaericus (Andersson et al., 1996). The members of this genus are generally motile, rod-shaped cells, and some species are known to produce ellipsoidal or spherical endospores which lie terminally in a swollen sporangium (Fortina et al., 2001). The genus includes some thermophilic bacteria. The members of this genus form a monophyletic clade in a 16S rRNA gene tree, and trees based on multiple large datasets of protein sequences. Members of this genus can be reliably distinguished from other genera within the family Caryophanaceae based on the shared presence of CSIs described in this work in all or most of the following proteins: MFS transporter, EamA family transporter, and DNA internalization-related competence protein ComEC/Rec2. Description of a new species that is part of this genus is provided below; new name combinations for some other species that are part of this emended genus are described in Table 5.
Type species: Ureibacillus thermosphaericus Fortina et al. 2001.
Description of Ureibacillus xyleni sp. nov.
(xy.le’.ni. N.L. neut. n. xylenum, xylene; N.L. gen. n. xyleni: of/from xylene).
The description of this taxon is as given by Begum et al. (2016) for “Lysinibacillus xyleni.” The type strain is NBRC 105753T (=DSM 23555 = KCTC 13604 = CCUG 57912 = JC22).
Emended Description of the Genus Solibacillus Krishnamurthi et al., 2009 emend. Mual et al., 2016
Solibacillus (So.li.ba.cil’lus. L. neut. n. solum soil; N.L. masc. n. Bacillus a bacterial genus; N.L. masc. n. Solibacillus a Bacillus-like organism isolated from soil).
The emended genus Solibacillus contains the type species S. silvestris. The members of this genus are rod-shaped cells staining generally Gram-positive. Some species are reported to form round endospores terminally in swollen sporangia (Krishnamurthi et al., 2009). The members of this genus form a monophyletic clade in phylogenetic trees based on multiple large datasets of protein sequences. Members of this genus can be reliably distinguished from other genera within the family Caryophanaceae based on the shared presence of CSIs described in this work in all or most of the following proteins: Flagellar hook–basal body protein (2 indels), aminodeoxychorismate lyase, VOC family protein, DNA topoisomerase IV subunit A, DegV family protein, helicase–exonuclease AddAB subunit AddB, multidrug resistance efflux transporter family protein, heme-dependent peroxidase, methionine ABC transporter ATP-binding protein, tRNA 4-thiouridine(8) synthase ThiI, and AAA family ATPase. New name combinations for some species that are part of the emended genus are described in Table 6.
Type species: Solibacillus silvestris (Rheims et al., 1999) Krishnamurthi et al., 2009.
Emended Description of the Genus Sporosarcina Kluyver and Van Niel 1936 (Approved Lists 1980) emend. Yoon et al. 2001b
Sporosarcina. (Spo.ro.sar.ci’na. Gr. fem. n. spora a seed, a spore; N.L. fem. n. Sarcina generic name; N.L. fem. n. Sporosarcina spore-forming Sarcina).
The emended genus Sporosarcina contains the type species S. ureae. Cells exhibit Gram-positive or Gram-variable staining. Studied species form round endospores and are generally motile. Facultatively anaerobic or strictly aerobic. The members of this genus form a monophyletic clade in 16S rRNA gene tree and phylogenetic trees based on multiple large datasets of protein sequences. Members of this genus can be reliably distinguished from other genera within the family Caryophanaceae based on the shared presence of CSIs described in this work in all or most of the following proteins: aspartate–tRNA ligase, A/G-specific adenine glycosylase, thymidylate synthase, RDD family protein, DEAD/DEAH box helicase, membrane protein insase YidC, cytochrome b6, and a hypothetical protein (accession no. WP_083035866).
Type species: Sporosarcina ureae (Beijerinck, 1901) Kluyver and Van Niel 1936 (Approved Lists 1980).
Description of Sporosarcina sphaerica sp. nov.
(sphae’ri.ca. L. fem. adj. sphaerica spherical).
The type strain for this species was isolated from human lung by A.M.R. Mackenzie (Southampton General Hospital) in 1975 and characterized as Lysinibacillus sphaericus NCTC 11025. This strain consistently branches within the genus Sporosarcina, distinctly from all other Lysinibacillus species. This strain also shares all of the conserved indels that are specific for the genus Sporosarcina, leading to its assignment as a novel species within this genus. The type strain for this species is NCTC 11025.
Description of Sporosarcina ureilytica sp. nov.
(u.re.i.ly’ti.ca. N.L. fem. n. urea, urea; N.L. masc. adj. lyticus from Gr. masc. adj. lytikos, able to loosen, able to dissolve; N.L. fem. adj. ureilytica, urea dissolving).
The type strain for this Gram-positive species was isolated by Dick (2004) and originally described as a Bacillus sphaericus strain. This strain was transferred along with other B. sphaericus strains to the genus Lysinibacillus when this genus was created (Ahmed et al., 2007). This strain branches reliably within the genus Sporosarcina and shares all conserved indels specific for this genus, leading to its assignment as a novel Sporosarcina species. The type strain for this species is LMG 22257.
Emended Description of the Genus Planococcus Migula 1894 (Approved Lists 1980) emend. Yoon et al. 2010
Planococcus. (Plan.o.coc’cus. Gr. masc. n. planes, a wanderer; N.L. masc. n. coccus (from Gr. masc. n. kokkos, grain, seed), coccus; N.L. masc. n. Planococcus, motile coccus).
The emended genus Planococcus contains the type species P. citreus and it encompasses most of the species from the genus Planomicrobium. Cells exhibit Gram-positive or Gram-variable staining and they are cocci or short rods and generally motile. The members of this genus form a monophyletic clade in a 16S rRNA gene tree and phylogenetic trees based on multiple large datasets of protein sequences. Members of this genus can be reliably distinguished from other genera within the family Caryophanaceae based on the shared presence of conserved signatures indels described in this work in all or most of the following proteins: penicillin-binding protein 2, hypothetical protein (WP_065528121), NADPH-dependent 7-cyano-7-deazaguanine reductase QueF, ACT domain-containing protein, and methylmalonyl-CoA mutase.
Type species: Planococcus citreus Migula 1894 (Approved Lists 1980).
Description of Metasolibacillus gen. nov.
Metasolibacillus (Me.ta.so.li.ba.cil’lus. Gr. adv. meta besides; N.L. masc. n. Solibacillus a bacterial genus name; N.L. masc. n. Metasolibacillus a genus besides Solibacillus).
Motile, rod-shaped, endospores forming cells exhibit Gram-positive staining. Grow aerobically in the range of 10–45°C, with optimal growth at 30–37°C. Studied species are reported to be positive for catalase and Voges–Proskauer tests. Species from this genus form a monophyletic clade in phylogenetic trees based on multiple large datasets of protein sequences. Members of this genus can be reliably distinguished from other genera within the family Caryophanaceae based on the shared presence of CSIs described in this work in all or most of the following proteins: DUF456 domain-containing protein, toxic anion resistance protein, undecaprenyldiphospho-muramoylpentapeptide beta-N-acetylglucosaminyltransferase, c-type cytochrome biogenesis protein CcsB, thiol-disulfide oxidoreductase ResA, hypothetical protein (accession no. WP_066164326), hypothetical protein (accession no. WP_107942795), Arginase, preprotein translocase subunit SecY, ATP-binding cassette domain-containing protein, purine permease, and thiol-disulfide oxidoreductase ResA.
Type species: Metasolibacillus meyeri comb. nov.
Description of Metasolibacillus fluoroglycofenilyticus sp. nov.
(flu.o.ro.gly.co.fe.ni.ly’ti.cus. N.L. neut. n. fluoroglycofenum, fluoroglycofen; N.L. masc. adj. lyticus (from Gr. masc. adj. lutikos) able to dissolve; N.L. masc. adj. fluoroglycofenilyticus, fluoroglycofen degrading).
The description of this taxon is as given by Cheng et al. (2015) for “Lysinibacillus fluoroglycofenilyticus.” The type strain is cmg86T (=KCTC 33183 = CCTCC AB 2013247).
Description of Metalysinibacillus gen. nov.
Metalysinibacillus (Me.ta.ly.si.ni.ba.cil’lus. Gr. adv. meta besides; N.L. masc. n. Lysinibacillus a bacterial genus name; N.L. masc. n. Metalysinibacillus a genus besides Lysinibacillus).
Gram-positive, rod-shaped cells able form endospores. Some species exhibit positive catalase and Voges–Proskauer tests. Grow optimally under aerobic condition in the temperature range of 30–37°C. Species from this genus form a monophyletic clade in phylogenetic trees based on multiple large datasets of protein sequences. Members of this genus can be reliably distinguished from other genera within the family Caryophanaceae based on the shared presence of CSIs described in this work in all or most of the following proteins: arginine-binding extracellular protein ArtP precursor, oxygen-independent coproporphyrinogen III oxidase, putative hydrolase MhqD, helix-turn-helix transcriptional regulator, tRNA preQ1(34) S-adenosylmethionine ribosyltransferase-isomerase QueA, DNA primase, FMN reductase (NADPH), UvrABC system protein C, sensor histidine kinase YycG, hypothetical protein BN1050_02162, ribonuclease Y, hypothetical protein BN1050_01309, cell division protein FtsA, ABC transporter ATP-binding protein YtrB, cysteine–tRNA ligase, coproporphyrinogen III oxidase, and PBP1A family penicillin-binding protein.
Type species: Metalysinibacillus jejuensis.
Description of Metalysinibacillus jejuensis sp. nov.
(je.ju.en’sis. N.L. masc. adj. jejuensis, referring to Jeju Island in the Republic of Korea, where the type strain was isolated).
The description of this taxon is as given by Kim et al. (2013b) for “Lysinibacillus jejuensis.” The type strain is DSM 28310T (=N2-5 = KCTC 13837).
Description of Metalysinibacillus saudimassiliensis sp. nov.
(sau.di.mas.si.li.en’sis. Arab. n. as-Sa’udiyah, name of Saudi Arabia: L. masc. adj. massiliensis pertaining to Marseille; N.L. fem. adj. saudimassiliensis, referring to Saudi Arabia and Marseille).
The description of this taxon is as given by Papadioti et al. (2017) for “Lysinibacillus saudimassiliensis.” The type strain is 13S34_airT (=CSUR P1222).
Description of Metaplanococcus gen. nov.
Metaplanococcus (Me.ta. pla.no.coc’cus. Gr. adv. meta besides; N.L. masc. n. Planococcus, a bacterial genus name; N.L. masc. n. Metaplanococcus, a genus besides Planococcus).
Cells are Gram-positive to Gram-variable and cocci or short rods (0.4–0.8 × 0.4–1.6 μm); motile by means of a single polar flagellum. Other phenotypic characteristics of this genus are as described by Jung et al. (2009) for P. flavidum. The members of this genus form a monophyletic clade adjoining to the genus Planococcus in phylogenetic trees constructed based on multiple large datasets of protein sequences. Members of this genus can be reliably distinguished from the members of genus Planococcus and other Caryophanaceae genera based on the shared presence of CSIs described in this work in all or most of the following proteins: ABC transporter substrate-binding protein, methionine–tRNA ligase, MetQ/NlpA family ABC transporter substrate-binding protein, ABC transporter permease spore protease YyaC, N-acetyl-alpha-D-glucosaminyl L-malate synthase BshA, orotidine-5′-phosphate decarboxylase, and phospho-N-acetylmuramoyl-pentapeptide-transferase.
Type species: Metaplanococcus flavidus comb. nov.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
RG obtained funding for this work, planned and supervised the entire study, carried out the construction of phlylogenetic trees, helped in the creation of sequence alignments and identification of CSIs, and wrote large sections of the manuscript and finalized it. SP was primarily responsible for the identification of CSIs from sequence alignments, determining the specificity of the CSIs, formatting of different figures and tables for publications, and in the writing of a draft manuscript.
Funding
This work was supported by the research Grant No. RGPIN-2019-206397 from the Natural Science and Engineering Research Council of Canada awarded to RG.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We express our sincere thanks and appreciation to Professor Aharon Oren for editing/correcting the etymology and protologs for the names of newly proposed taxa and new name combinations.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02821/full#supplementary-material
Footnotes
- ^ https://www.namesforlife.com/
- ^ http://www.ncbi.nlm.nih.gov/genome
- ^ http://www.ncbi.nlm.nih.gov/genome
References
Adeolu, M., Alnajar, S., Naushad, S., and Gupta, R. S. (2016). Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int. J. Syst. Evol. Microbiol. 66, 5575–5599. doi: 10.1099/ijsem.0.001485
Adeolu, M., and Gupta, R. S. (2014). A phylogenomic and molecular marker based proposal for the division of the genus Borrelia into two genera: the emended genus Borrelia containing only the members of the relapsing fever Borrelia, and the genus Borreliella gen. nov. containing the members of the Lyme disease Borrelia (Borrelia burgdorferi sensu lato complex). Antonie Van Leeuwenhoek 105, 1049–1072. doi: 10.1007/s10482-014-0164-x
Ahmed, I., Yokota, A., Yamazoe, A., and Fujiwara, T. (2007). Proposal of Lysinibacillus boronitolerans gen. nov. sp. nov., and transfer of Bacillus fusiformis to Lysinibacillus fusiformis comb. nov. and Bacillus sphaericus to Lysinibacillus sphaericus comb. nov. Int. J. Syst. Evol. Microbiol. 57, 1117–1125.
Andersson, M., Laukkanen, M., Nurmiaho-Lassila, E. L., Rainey, F. A., Niemela, S. I., and Salkinoja-Salonen, M. (1996). Bacillus thermosphaericus sp. nov. In list no. 56, validation of the publication of new names and new combinations previously effectively published outside the IJSEB. Int. J. Syst. Bacteriol. 46, 362–363.
Arora, P. K., Chauhan, A., Pant, B., Korpole, S., Mayilraj, S., and Jain, R. K. (2011). Chryseomicrobium imtechense gen. nov., sp. nov., a new member of the family Planococcaceae. Int. J. Syst. Evol. Microbiol. 61, 1859–1864. doi: 10.1099/ijs.0.023184-0
Azmatunnisa, M., Rahul, K., Lakshmi, K. V., Sasikala, C., and Ramana, C. (2015). Lysinibacillus acetophenoni sp. nov., a solvent-tolerant bacterium isolated from acetophenone. Int. J. Syst. Evol. Microbiol. 65, 1741–1748. doi: 10.1099/ijs.0.000170
Baldauf, S. L., and Palmer, J. D. (1993). Animals and fungi are each other’s closest relatives: congruent evidence from multiple proteins. Proc. Natl. Acad. Sci. U.S.A. 90, 11558–11562.
Baumann, P., Clark, M. A., Baumann, L., and Broadwell, A. H. (1991). Bacillus sphaericus as a mosquito pathogen: properties of the organism and its toxins. Microbiol. Rev. 55, 425–436.
Begum, M. A., Rahul, K., Sasikala, C., and Ramana, C. V. (2016). Lysinibacillus xyleni sp. nov., isolated from a bottle of xylene. Arch. Microbiol. 198, 325–332. doi: 10.1007/s00203-016-1194-8
Beijerinck, M. W. (1901). Anhäufungsversuche mit ureumbakterien. Ureumspaltung durch urease und katabolismus. Zentralblatt fur bakteriologie parasitenkunde infektionskrankheiten und hygiene. Abteilung II 7, 33–61.
Berry, C. (2012). The bacterium, Lysinibacillus sphaericus, as an insect pathogen. J. Invertebr. Pathol. 109, 1–10. doi: 10.1016/j.jip.2011.11.008
Bhandari, V., and Gupta, R. S. (2014). Molecular signatures for the phylum (class) Thermotogae and a proposal for its division into three orders (Thermotogales, Kosmotogales ord. nov. and Petrotogales ord. nov.) containing four families (Thermotogaceae, Fervidobacteriaceae fam. nov., Kosmotogaceae fam. nov. and Petrotogaceae fam. nov.) and a new genus Pseudothermotoga gen. nov. with five new combinations. Antonie Van Leeuwenhoek 105, 143–168.
Burkett-Cadena, M., Sastoque, L., Cadena, J., and Dunlap, C. A. (2019). Lysinibacillus capsici sp. nov, isolated from the rhizosphere of a pepper plant. Antonie Van Leeuwenhoek 112, 1161–1167. doi: 10.1007/s10482-019-01248-w
Capella-Gutierrez, S., Silla-Martinez, J. M., and Gabaldon, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Cheng, M., Zhang, H., Zhang, J., Hu, G., Zhang, J., He, J., et al. (2015). Lysinibacillus fluoroglycofenilyticus sp. nov., a bacterium isolated from fluoroglycofen contaminated soil. Antonie Van Leeuwenhoek 107, 157–164. doi: 10.1007/s10482-014-0313-2
Dai, X., Wang, Y. N., Wang, B. J., Liu, S. J., and Zhou, Y. G. (2005). Planomicrobium chinense sp. nov., isolated from coastal sediment, and transfer of Planococcus psychrophilus and Planococcus alkanoclasticus to Planomicrobium as Planomicrobium psychrophilum comb. nov. and Planomicrobium alkanoclasticum comb. nov. Int. J. Syst. Evol. Microbiol. 55, 699–702.
Dick, J. (2004). UGent - Fac. Bio-Ingenieurswetenschappen, Vakgroep Biochemische en Microbiële Technologie - Laboratorium voor Microbiële Ecologie en Technologie (LabMET) (Bacillus Sphaericus). Brussels: Belgian Co-ordinated Collections of Micro-organisms.
Dobritsa, A. P., Linardopoulou, E. V., and Samadpour, M. (2017). Transfer of 13 species of the genus Burkholderia to the genus Caballeronia and reclassification of Burkholderia jirisanensis as Paraburkholderia jirisanensis comb. nov. Int. J. Syst. Evol. Microbiol. 67, 3846–3853. doi: 10.1099/ijsem.0.002202
Dunlap, C. A. (2019). Lysinibacillus mangiferihumi, Lysinibacillus tabacifolii and Lysinibacillus varians are later heterotypic synonyms of Lysinibacillus sphaericus. Int. J. Syst. Evol. Microbiol. 69, 2958–2962. doi: 10.1099/ijsem.0.003577
Engelhardt, M. A., Daly, K., Swannell, R. P., and Head, I. M. (2001). Isolation and characterization of a novel hydrocarbon-degrading, Gram-positive bacterium, isolated from intertidal beach sediment, and description of Planococcus alkanoclasticus sp. nov. J. Appl. Microbiol. 90, 237–247.
Farrow, J. A., Ash, C., Wallbanks, S., and Collins, M. D. (1992). Phylogenetic analysis of the genera Planococcus, Marinococcus and Sporosarcina and their relationships to members of the genus Bacillus. FEMS Microbiol. Lett. 72, 167–172.
Fortina, M. G., Pukall, R., Schumann, P., Mora, D., Parini, C., Manachini, P. L., et al. (2001). Ureibacillus gen. nov., a new genus to accommodate Bacillus thermosphaericus (Andersson et al. 1995), emendation of Ureibacillus thermosphaericus and description of Ureibacillus terrenus sp. nov. Int. J. Syst. Evol. Microbiol. 51, 447–455.
Fu, L., Niu, B., Zhu, Z., Wu, S., and Li, W. (2012). CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152. doi: 10.1093/bioinformatics/bts565
Gao, B., and Gupta, R. S. (2012). Phylogenetic framework and molecular signatures for the main clades of the phylum Actinobacteria. Microbiol. Mol. Biol. Rev. 76, 66–112. doi: 10.1128/MMBR.05011-11
Glazunova, O. O., Raoult, D., and Roux, V. (2006). Bacillus massiliensis sp. nov., isolated from cerebrospinal fluid. Int. J. Syst. Evol. Microbiol. 56, 1485–1488.
Gomez-Garzon, C., Hernandez-Santana, A., and Dussan, J. (2016). Comparative genomics reveals Lysinibacillus sphaericus group comprises a novel species. BMC Genom. 17:709. doi: 10.1186/s12864-016-3056-9
Gupta, R. S. (1998). Protein phylogenies and signature sequences: a reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiol. Mol. Biol. Rev. 62, 1435–1491.
Gupta, R. S. (2014). “Identification of conserved indels that are useful for classification and evolutionary studies,” in Methods in Microbiology, eds M. Goodfellow, I. Sutcliffe, and J. Chun, (Oxford: Academic Press), 153–182.
Gupta, R. S. (2016). Impact of genomics on the understanding of microbial evolution and classification: the importance of Darwin’s views on classification. FEMS Microbiol. Rev. 40, 520–553. doi: 10.1093/femsre/fuw011
Gupta, R. S. (2019). Distinction between Borrelia and Borreliella is more robustly supported by molecular and phenotypic characteristics than all other neighbouring prokaryotic genera: response to Margos’ et al. “The genus Borrelia reloaded” (PLoS ONE 13(12): e0208432). PLoS One 14:e0221397. doi: 10.1371/journal.pone.0221397
Gupta, R. S., Lo, B., and Son, J. (2018). Phylogenomics and comparative genomic studies robustly support division of the genus Mycobacterium into an emended genus Mycobacterium and four novel genera. Front. Microbiol. 9:67. doi: 10.3389/fmicb.2018.00067
Gupta, R. S., Naushad, S., Fabros, R., and Adeolu, M. (2016). A phylogenomic reappraisal of family-level divisions within the class Halobacteria: proposal to divide the order Halobacteriales into the families Halobacteriaceae, Haloarculaceae fam. nov., and Halococcaceae fam. nov., and the order Haloferacales into the families, Haloferacaceae and Halorubraceae fam nov. Antonie Van Leeuwenhoek 109, 565–587. doi: 10.1007/s10482-016-0660-2
Gupta, R. S., Son, J., and Oren, A. (2019). A phylogenomic and molecular markers based taxonomic framework for members of the order Entomoplasmatales: proposal for an emended order Mycoplasmatales containing the family Spiroplasmataceae and emended family Mycoplasmataceae comprised of six genera. Antonie Van Leeuwenhoek 112, 561–588. doi: 10.1007/s10482-018-1188-4
Hayat, R., Ahmed, I., Paek, J., Sin, J., Ehsan, M., Iqbal, M., et al. (2014). Lysinibacillus composti sp. nov., isolated from compost. Ann. Microbiol. 64, 1081–1088.
Hu, D., Zang, Y., Mao, Y., and Gao, B. (2019). Identification of molecular markers that are specific to the class Thermoleophilia. Front. Microbiol. 10:1185. doi: 10.3389/fmicb.2019.01185
Jeanmougin, F., Thompson, J. D., Gouy, M., Higgins, D. G., and Gibson, T. J. (1998). Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23, 403–405.
Jung, M. Y., Kim, J. S., Paek, W. K., Styrak, I., Park, I. S., Sin, Y., et al. (2012). Description of Lysinibacillus sinduriensis sp. nov., and transfer of Bacillus massiliensis and Bacillus odysseyi to the genus Lysinibacillus as Lysinibacillus massiliensis comb. nov. and Lysinibacillus odysseyi comb. nov. with emended description of the genus Lysinibacillus. Int. J. Syst. Evol. Microbiol. 62, 2347–2355. doi: 10.1099/ijs.0.033837-0
Jung, Y. T., Kang, S. J., Oh, T. K., Yoon, J. H., and Kim, B. H. (2009). Planomicrobium flavidum sp. nov., isolated from a marine solar saltern, and transfer of Planococcus stackebrandtii Mayilraj et al., 2005 to the genus Planomicrobium as Planomicrobium stackebrandtii comb. nov. Int. J. Syst. Evol. Microbiol. 59, 2929–2933. doi: 10.1099/ijs.0.009191-0
Junge, K., Gosink, J. J., Hoppe, H. G., and Staley, J. T. (1998). Arthrobacter, Brachybacterium and Planococcus isolates identified from antarctic sea ice brine. description of Planococcus mcmeekinii, sp. nov. Syst. Appl. Microbiol. 21, 306–314.
Kampfer, P., Martin, K., and Glaeser, S. P. (2013). Lysinibacillus contaminans sp. nov., isolated from surface water. Int. J. Syst. Evol. Microbiol. 63, 3148–3153. doi: 10.1099/ijs.0.049593-0
Khadka, B., and Gupta, R. S. (2017). Identification of a conserved 8 aa insert in the PIP5K protein in the Saccharomycetaceae family of fungi and the molecular dynamics simulations and structural analysis to investigate its potential functional role. Proteins 85, 1454–1467. doi: 10.1002/prot.25306
Kim, S.-J., Jang, Y.-H., Hamada, M., Ahn, J.-H., Weon, H.-Y., Suzuki, K., et al. (2013a). Lysinibacillus chungkukjangi sp. nov., isolated from Chungkukjang, Korean fermented soybean food. J. Microbiol. 51, 400–404. doi: 10.1007/s12275-013-2664-1
Kim, J.-Y., Park, S.-H., Oh, D.-C., and Kim, Y.-J. (2013b). Lysinibacillus jejuensis sp. nov., isolated from swinery waste. J. Microbiol. 51, 872–876. doi: 10.1007/s12275-013-2500-7
Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120.
Klenk, H. P., and Goker, M. (2010). En route to a genome-based classification of Archaea and bacteria? Syst. Appl. Microbiol. 33, 175–182. doi: 10.1016/j.syapm.2010.03.003
Kluyver, A. J., and Van Niel, C. B. (1936). Prospects for a natural system of classification of bacteria. Zentralblatt fur Bakteriologie Parasitenkunde Infektionskrankheiten und Hygiene. Abteilung II 94, 369–403.
Kong, D., Wang, Y., Zhao, B., Li, Y., Song, J., Zhai, Y., et al. (2014). Lysinibacillus halotolerans sp. nov., isolated from saline-alkaline soil. Int. J. Syst. Evol. Microbiol. 64, 2593–2598. doi: 10.1099/ijs.0.061465-0
Konstantinidis, K. T., and Stackebrandt, E. (2013). “Defining taxonomic ranks,” in The Prokaryotes: Prokaryotic Biology and Symbiotic Associations, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson, (Berlin: Springer), 229–254.
Konstantinidis, K. T., and Tiedje, J. M. (2005). Towards a genome-based taxonomy for prokaryotes. J. Bacteriol. 187, 6258–6264.
Krasil’nikov, N. A. (1949). Guide to the Bacteria and Actinomycetes [Opredelitelv Bakterii i Actinomicetov] (Moscow: Akad. Nauk SSSR), 348.
Krishnamurthi, S., Chakrabarti, T., and Stackebrandt, E. (2009). Re-examination of the taxonomic position of Bacillus silvestris Rheims et al., 1999 and proposal to transfer it to Solibacillus gen. nov. as Solibacillus silvestris comb. nov. Int. J. Syst. Evol. Microbiol. 59, 1054–1058. doi: 10.1099/ijs.0.65742-0
La Duc, M. T., Satomi, M., and Venkateswaran, K. (2004). Bacillus odysseyi sp. nov., a round-spore-forming Bacillus isolated from the mars odyssey spacecraft. Int. J. Syst. Evol. Microbiol. 54, 195–201.
Le, S. Q., and Gascuel, O. (2008). An improved general amino acid replacement matrix. Mol. Biol. Evol. 25, 1307–1320. doi: 10.1093/molbev/msn067
Liu, H., Song, Y., Chen, F., Zheng, S., and Wang, G. (2013). Lysinibacillus manganicus sp. nov., isolated from manganese mining soil. Int. J. Syst. Evol. Microbiol. 63, 3568–3573. doi: 10.1099/ijs.0.050492-0
Lo, C. I., Padhmanabhan, R., Mediannikov, O., Caputo, A., Michelle, C., Faye, N., et al. (2015). High-quality genome sequence and description of Bacillus ndiopicus strain FF3(T) sp. nov. New Microbes New Infect. 8, 154–163. doi: 10.1016/j.nmni.2015.10.009
Ludwig, W., Schleifer, K. H., and Whitman, W. B. (2009). “Family VI. Planococcaceae Krasil’nikov, 1949,” in Bergey’s Manual of Systematic Bacteriology, eds P. De Vos, M. Garrity, D. Jones, R. Krieg, W. Ludwig, F. A. Rainey, et al. (New York: Springer), 348–385.
Luo, X., Zhang, J., Li, D., Xin, Y., Xin, D., and Fan, L. (2014). Planomicrobium soli sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 64, 2700–2705. doi: 10.1099/ijs.0.055426-0
Maayer, P., Aliyu, H., and Cowan, D. A. (2019). Reorganising the order Bacillales through phylogenomics. Syst. Appl. Microbiol. 42, 178–189. doi: 10.1016/j.syapm.2018.10.007
Maiden, M. F. J., and Jones, J. G. (1985). A new filamentous, gliding bacterium, Filibacter limicola gen. nov. sp. nov., from Lake Sediment. J. Gen. Microbiol. 130, 2943–2959.
Mayilraj, S., Prasad, G. S., Suresh, K., Saini, H. S., Shivaji, S., and Chakrabarti, T. (2005). Planococcus stackebrandtii sp. nov., isolated from a cold desert of the Himalayas, India. Int. J. Syst. Evol. Microbiol. 55, 91–94.
Migula, W. (1894). Über ein neues system der bakterien. Arbeiteen aus dem Bakteriol. Instit. der Tech. Hachschule zu Karlsruhe 1, 235–238.
Mual, P., Singh, N. K., Verma, A., Schumann, P., Krishnamurthi, S., Dastager, S., et al. (2016). Reclassification of Bacillus isronensis Shivaji et al. 2009 as Solibacillus isronensis comb. nov. and emended description of genus Solibacillus Krishnamurthi et al., 2009. Int. J. Syst. Evol. Microbiol. 66, 2113–2120.
Nakagawa, Y., Sakane, T., and Yokota, A. (1996). Emendation of the genus Planococcus and transfer of Flavobacterium okeanokoites Zobell and Upham 1944 to the genus Planococcus as Planococcus okeanokoites comb. nov. Int. J. Syst. Evol. Microbiol. 46, 866–870.
Naushad, S., Lee, B., and Gupta, R. S. (2014). Conserved signature indels and signature proteins as novel tools for understanding microbial phylogeny and systematics: identification of molecular signatures that are specific for the phytopathogenic genera Dickeya, Pectobacterium and Brenneria. Int. J. Syst. Evol. Microbiol. 64, 366–383. doi: 10.1099/ijs.0.054213-0
Ouoba, L. I. I., Vouidibio Mbozo, A. B., Thorsen, L., Anyogu, A., Nielsen, D. S., Kobawila, S. C., et al. (2015). Lysinibacillus louembei sp. nov., a spore-forming bacterium isolated from Ntoba Mbodi, alkaline fermented leaves of cassava from the Republic of the Congo. Int. J. Syst. Evol. Microbiol. 65, 4256–4262. doi: 10.1099/ijsem.0.000570
Pal, Y., Mayilraj, S., Paul, M., Schumann, P., and Krishnamurthi, S. (2019). Indiicoccus explosivorum gen. nov., sp. nov., isolated from an explosives waste contaminated site. Int. J. Syst. Evol. Microbiol. 69, 2555–2564. doi: 10.1099/ijsem.0.003541
Papadioti, A., Azhar, E. I., Bibi, F., Jiman-Fatani, A., Aboushoushah, S. M., Yasir, M., et al. (2017). ‘Lysinibacillus saudimassiliensis’ sp. nov., a new bacterial species isolated from air samples in the urban environment of Makkah, Saudi Arabia. New Microb. New Infect. 16, 25–27. doi: 10.1016/j.nmni.2016.12.011
Parker, C. T., Tindall, B. J., and Garrity, G. M. (2019). International code of nomenclature of prokaryotes. Int. J. Syst. Evol. Microbiol. 69, S1–S111. doi: 10.1099/ijsem.0.000778
Parks, D. H., Chuvochina, M., Waite, D. W., Rinke, C., Skarshewski, A., Chaumeil, P. A., et al. (2018). A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 36, 996–1004. doi: 10.1038/nbt.4229
Parte, A. C. (2014). LPSN–list of prokaryotic names with standing in nomenclature. Nucleic Acids Res. 42, D613–D616. doi: 10.1093/nar/gkt1111
Patel, S., and Gupta, R. S. (2018). Robust demarcation of fourteen different species groups within the genus Streptococcus based on genome-based phylogenies and molecular signatures. Infect. Genet. Evol. 66, 130–151. doi: 10.1016/j.meegid.2018.09.020
Patel, S., and Gupta, R. S. (2019). A Phylogenomics and comparative genomic framework for resolving the polyphyly of the genus Bacillus: proposal for six new genera of Bacillus species, Peribacillus gen. nov., Cytobacillus gen. nov., Mesobacillus gen. nov., Neobacillus gen. nov., Metabacillus gen. nov. and Alkalihalobacillus gen. nov. Int. J. Syst. Evol. Microbiol. doi: 10.1099/ijsem.0.003775 [Epub ahead of print].
Peshkoff, M. A. (1939). Cytology, karyology and cycle of development of new microbes, Caryophanon latum and Caryophanon tenue. Dokl. Akad. Nauk SSSR 25, 239–242.
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490
Qin, Q. L., Xie, B. B., Zhang, X. Y., Chen, X. L., Zhou, B. C., Zhou, J., et al. (2014). A proposed genus boundary for the prokaryotes based on genomic insights. J. Bacteriol. 196, 2210–2215. doi: 10.1128/JB.01688-14
Rahi, P., Kurli, R., Khairnar, M., Jagtap, S., Pansare, A. N., Dastager, S. G., et al. (2017). Description of Lysinibacillus telephonicus sp. nov., isolated from the screen of a cellular phone. Int. J. Syst. Evol. Microbiol. 67, 2289–2295. doi: 10.1099/ijsem.0.001943
Reddy, G. S., Prakash, J. S., Vairamani, M., Prabhakar, S., Matsumoto, G. I., and Shivaji, S. (2002). Planococcus antarcticus and Planococcus psychrophilus spp. nov. isolated from cyanobacterial mat samples collected from ponds in Antarctica. Extremophiles 6, 253–261.
Reddy, G. S., Uttam, A., and Shivaji, S. (2008). Bacillus cecembensis sp. nov., isolated from the Pindari glacier of the Indian Himalayas. Int. J. Syst. Evol. Microbiol. 58, 2330–2335. doi: 10.1099/ijs.0.65515-0
Rheims, H., Fruhling, A., Schumann, P., Rohde, M., and Stackebrandt, E. (1999). Bacillus silvestris sp. nov., a new member of the genus Bacillus that contains lysine in its cell wall. Int. J. Syst. Bacteriol. 49(Pt 2), 795–802.
Rokas, A., and Holland, P. W. (2000). Rare genomic changes as a tool for phylogenetics. Trends Ecol. Evol. 15, 454–459.
Sangal, V., Goodfellow, M., Jones, A. L., Schwalbe, E. C., Blom, J., Hoskisson, P. A., et al. (2016). Next-generation systematics: an innovative approach to resolve the structure of complex prokaryotic taxa. Sci. Rep. 6:38392. doi: 10.1038/srep38392
Sawana, A., Adeolu, M., and Gupta, R. S. (2014). Molecular signatures and phylogenomic analysis of the genus Burkholderia: proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harboring environmental species. Front. Genet. 5:429. doi: 10.3389/fgene.2014.00429
Seiler, H., Scherer, S., and Wenning, M. (2013). Lysinibacillus meyeri sp. nov., isolated from a medical practice. Int. J. Syst. Evol. Microbiol. 63, 1512–1518. doi: 10.1099/ijs.0.039420-0
Shivaji, S., Srinivas, T. N. R., and Reddy, G. S. N. (2014). “The family Planococcaceae,” in The Prokaryotes, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson, (Berlin: Springer), 303–345.
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
Singh, B., and Gupta, R. S. (2009). Conserved inserts in the Hsp60 (GroEL) and Hsp70 (DnaK) proteins are essential for cellular growth. Mol. Genet. Genom. 281, 361–373. doi: 10.1007/s00438-008-0417-3
Skerman, V. B. D., McGowan, V., and Sneath, P. H. A. (1980). Approved lists of bacterial names. Int. J. Syst. Bacteriol. 30, 225–420.
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tetz, G., and Tetz, V. (2018). Draft Genome Sequence of Tetzosporium hominis VT-49 gen. nov., sp. nov., isolated from the dental decay plaque of a patient with Periodontitis. Genom. Announc. 6:e01541-17. doi: 10.1128/genomeA.01541-17
Thompson, C. C., Chimetto, L., Edwards, R. A., Swings, J., Stackebrandt, E., and Thompson, F. L. (2013). Microbial genomic taxonomy. BMC Genom. 14:913. doi: 10.1186/1471-2164-14-913
Tindall, B. J. (2019). When treated as heterotypic synonyms the names Caryophanaceae Peshkoff, 1939 (Approved Lists 1980) and Caryophanales Peshkoff, 1939 (Approved Lists 1980) have priority over the names Planococcaceae Krasil’nikov, 1949 (Approved Lists 1980) and Bacillales Prevot 1953 (Approved Lists 1980) and Bacillales Prevot 1953 (Approved Lists 1980), respectively. Int. J. Syst. Evol. Microbiol. 69, 2187–2195. doi: 10.1099/ijsem.0.003354
Wang, Z., and Wu, M. (2013). A phylum-level bacterial phylogenetic marker database. Mol. Biol. Evol. 30, 1258–1262. doi: 10.1093/molbev/mst059
Whelan, S., and Goldman, N. (2001). A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18, 691–699.
Xu, K., Yuan, Z., Rayner, S., and Hu, X. (2015). Genome comparison provides molecular insights into the phylogeny of the reassigned new genus Lysinibacillus. BMC Genom. 16:140. doi: 10.1186/s12864-015-1359-x
Yakoubou, S., Xu, D., and Cote, J. (2010). Phylogeny of the order Bacillales inferred from 3′ 16S rDNA and 5′ 16S-23S ITS nucleotide sequences. Nat. Sci. 2, 990–997.
Yarza, P., Ludwig, W., Euzeby, J., Amann, R., Schleifer, K. H., Glockner, F. O., et al. (2010). Update of the All-Species Living Tree Project based on 16S and 23S rRNA sequence analyses. Syst. Appl. Microbiol. 33, 291–299. doi: 10.1016/j.syapm.2010.08.001
Yilmaz, P., Parfrey, L. W., Yarza, P., Gerken, J., Pruesse, E., Quast, C., et al. (2014). The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 42, D643–D648. doi: 10.1093/nar/gkt1209
Yoon, J. H., Kang, S. J., Lee, S. Y., Oh, K. H., and Oh, T. K. (2010). Planococcus salinarum sp. nov., isolated from a marine solar saltern, and emended description of the genus Planococcus. Int. J. Syst. Evol. Microbiol. 60, 754–758. doi: 10.1099/ijs.0.013136-0
Yoon, J. H., Kang, S. S., Lee, K. C., Lee, E. S., Kho, Y. H., Kang, K. H., et al. (2001a). Planomicrobium koreense gen. nov., sp. nov., a bacterium isolated from the Korean traditional fermented seafood jeotgal, and transfer of Planococcus okeanokoites (Nakagawa et al., 1996) and Planococcus mcmeekinii (Junge et al., 1998) to the genus Planomicrobium. Int. J. Syst. Evol. Microbiol. 51, 1511–1520.
Yoon, J. H., Lee, K. C., Weiss, N., Kho, Y. H., Kang, K. H., and Park, Y. H. (2001b). Sporosarcina aquimarina sp. nov., a bacterium isolated from seawater in Korea, and transfer of Bacillus globisporus (Larkin and Stokes 1967), Bacillus psychrophilus (Nakamura 1984) and Bacillus pasteurii (Chester 1898) to the genus Sporosarcina as Sporosarcina globispora comb. nov., Sporosarcina psychrophila comb. nov. and Sporosarcina pasteurii comb. nov., and emended description of the genus Sporosarcina. Int. J. Syst. Evol. Microbiol. 51, 1079–1086.
Yu, J., Guan, X., Liu, C., Xiang, W., Yu, Z., Liu, X., et al. (2017). Erratum to: Lysinibacillus endophyticus sp. nov., an indole-3-acetic acid producing endophytic bacterium isolated from corn root (Zea mays cv. Xinken-5). Antonie Van Leeuwenhoek 110:173.
Zhang, D. C., Liu, H. C., Xin, Y. H., Yu, Y., Zhou, P. J., and Zhou, Y. G. (2009). Planomicrobium glaciei sp. nov., a psychrotolerant bacterium isolated from a glacier. Int. J. Syst. Evol. Microbiol. 59, 1387–1390. doi: 10.1099/ijs.0.002592-0
Keywords: Planococcaceae and Caryophanaceae families, phylogenomic and comparative genomic analyses, conserved signature indels for different clades, Lysinibacillus, Metasolibacillus gen. nov., Metalysinibacillus gen. nov., Metaplanococcus gen. nov., emended descriptions of the Caryophanaceae/Planococcaceae genera
Citation: Gupta RS and Patel S (2020) Robust Demarcation of the Family Caryophanaceae (Planococcaceae) and Its Different Genera Including Three Novel Genera Based on Phylogenomics and Highly Specific Molecular Signatures. Front. Microbiol. 10:2821. doi: 10.3389/fmicb.2019.02821
Received: 27 September 2019; Accepted: 20 November 2019;
Published: 14 January 2020.
Edited by:
Iain Sutcliffe, Northumbria University, United KingdomReviewed by:
Martín Espariz, CONICET Instituto de Biología Molecular y Celular de Rosario (IBR), ArgentinaDaniel R. Zeigler, The Ohio State University, United States
Bernhard Schink, Universität Konstanz, Germany
Copyright © 2020 Gupta and Patel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Radhey S. Gupta, Z3VwdGFAbWNtYXN0ZXIuY2E=
 Radhey S. Gupta
Radhey S. Gupta Sudip Patel
Sudip Patel