
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 03 December 2019
Sec. Microbial Physiology and Metabolism
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.02748
Lysobacter species are emerging as novel sources of antibiotics, but the regulation of their physiological metabolism is still poorly understood. In this work, we extracted AHL (acyl-homoserine lactone) autoinducers, identified the structures of AHLs and described the AHL quorum-sensing system in Lysobacter brunescens OH23. AHLs were isolated from the supernatant of L. brunescens OH23, and ESI-MS/MS (electrospray ionization mass spectrometry) analysis revealed biosynthesis of three different AHL chemical structures by L. brunescens OH23: N-(3-oxohexanoyl)- homoserine lactone (HSL), 3-OH-C10-HSL and C8-HSL. The growth rate of AHL quorum-sensing knockout mutants was dramatically increased compared to that of wildtype. Sucrose consumptions were also twice as high in AHL quorum-sensing knockout mutants than that in wildtype in early-log phase. Additionally, expression of key genes related to sucrose metabolism α-glucosidase was enhanced in AHL quorum-sensing knockout mutants, which indicated that AHL quorum sensing negatively regulates sucrose uptake and metabolism which further affects the growth rate of L. brunescens. Furthermore, autolysis was strongly induced in AHL quorum-sensing knockout mutants compared to wildtype, suggesting that AHL quorum sensing plays a negative regulatory role in cell autolysis. Moreover, compared to wildtype, XSAC (Xanthomonas-specific antibiotic compound) production was significantly increased in AHL knockout mutants in the early-log and late-log phases, and surface motility capabilities are also enhanced also in AHL knockout mutants; the normalized data of XSAC production and surface motility and expressions of key genes related to these two phenotypes reveal that growth rare and autolysis strongly affects XSAC biosynthesis and surface motility rather than AHL quorum-sensing system. Our results show that the AHL quorum-sensing system negatively regulates cell growth and autolysis, and further maintain nutrition homeostasis and population stability in L. brunescens.
Lysobacter, belonging to Xanthomonadaceae, is a type of gram-negative bacteria that inhibits the growth of a variety of plant pathogens by producing various active secondary metabolites (Folman et al., 2003). Lysobactin, originally isolated from Lysobacter sp. ATCC53042, inhibits a different pathogenic gram-positive and gram-negative bacteria (O’Sullivan et al., 1988). WAP-8294A, cyclic lipodepsipeptides, were isolated from Lysobacter sp. 8294 and inhibited clinical MRSA (methicillin-resistant Staphylococcus aureus) in mice better than did vancomycin (Kato et al., 1998; Zhang et al., 2011). In addition, Li et al. found that the highly thermostable antifungal factor HSAF (dihydromaltophilin) from L. enzygenes C3 inhibits various fungi (Yu et al., 2007).
Originally discovered in X. campestris pv. campestris (Xcc), the diffusible signal factor (DSF) quorum-sensing system is widely distributed among gram-negative bacteria, particularly Xanthomonas campestris and Xanthomonas oryzae (Barber et al., 1997; Wang et al., 2004). The DSF signaling molecule was identified as an unsaturated fatty acid with a chemical structure of cis-11-methyl-2-dodecenoic acid (Wang et al., 2004). Subsequently, it was discovered that Xcc can synthesize DSF molecules with at least three different structures, including DSF, cis-2-dodecenoic acid (BDSF) and [(2Z,5Z)-11-methyldodeca-2,5-dienoci acid] (CDSF) (He et al., 2010). Although the structures of DSF signaling molecules vary, their biosynthesis in Xanthomonas is dependent on the gene cluster rpf (regulation of pathogenicity factor) (He et al., 2006). Among the genes in this cluster, rpfF encodes an enoyl-CoA hydratase responsible for the biosynthesis of DSF signaling molecules (Almeida et al., 2012). In addition, the perception and conduction of DSF signals is dependent on the two-component system RpfC/RpfG in bacteria, namely, the histidine kinase (HK) RpfC and response regulator (RR) RpfG; RpfC is responsible for the perception of signaling molecules and RpfG for signal transduction (Slater et al., 2000).
The acyl-homoserine lactone (AHL)-mediated quorum-sensing system is also common in gram-negative bacteria, and the signaling molecules of the AHL-mediated quorum-sensing system are mainly synthesized by a class of genes homologous to luxI and luxR, which were the first quorum-sensing genes identified in Vibrio fischeri (Fuqua and Winans, 1994). The main function of the synthetic LuxI/LuxR quorum-sensing system in V. fischeri is regulation of luminescence according to the density of the population (Fuqua and Winans, 1994). In general, LuxI proteins are synthetases of AHLs and are responsible for the synthesis of related signaling molecules. LuxR family proteins are AHL molecular receptors, and the AHL autoinducers bind to LuxR proteins, which in turn regulate downstream gene expression and ultimately the relevant biological phenotype (Fuqua and Winans, 1994; Tsai and Winans, 2010). Typical LuxR-like receptor proteins require AHLs during protein folding or for evading hydrolysis by other hydrolases in the cell (Zhu and Winans, 1999, 2001). However, some special LuxR-like proteins do not depend on AHLs and, even in the presence of AHLs, inhibit the function of LuxR (Zhu and Winans, 1999, 2001). The AHL signaling molecules reported to date include N-(3-oxohexanoyl)-homoserine lactone (HSL) (V. fischeri), N-(3-oxooctanoyl)-HSL (A. tumefaciens), N-butanoyl-HSL (Aeromonas hydrophila), N-octanoyl-HSL (Burkholderia cepacia), N-(3-oxododecanoyl)-HSL (Pseudomonas aeruginosa), and N-hexanoyl-HSL (Rhizobium leguminosarum) (Miller and Bassler, 2001).
The quorum-sensing system is involved in cell-to-cell communication, regulates transcription factor activity and alters bacterial phenotypes, including biofilm, virulence, and symbiosis processes (Zheng et al., 2015; Papenfort and Bassler, 2016). During nodulation in Mesorhizobium tianshanense, the number of nodules per plant inoculated with AHL-QS mutants was significantly decreased compared to that inoculated with the wildtype strain, which indicated that the AHL quorum-sensing system was required during the symbiotic process between the bacterium and host plant Glycyrrhiza (licorice) (Cao et al., 2009). In Burkholderia cepacian, the AHL quorum-sensing system positively regulates swarming motility and biofilm formation, and complementation of the LuxR regulator cepR was able to restore swarming motility (Huber et al., 2001). In contrast, Pseudomonas syringae pv. Actinidiae (Psa) encodes three solo LuxR and negatively biosynthesize AHL autoinducers; Psa also senses the neighboring AHL-positive bacteria by responding to the AHLs signals, which might important for Psa fitness and virulence in the host kiwifruit (Cellini et al., 2019). In Lysobacter enzymogenes, overexpression of the solo LuxR type protein LesR led to a decrease in HSAF (Heat Stable Antifungal Factor) production, acceleration of cell aggregation and enhancement of melanin-like pigment production (Qian et al., 2014). In Burkholderia glumae, the AHLs quorum sensing system mutant grew more rapidly than wildtype strain and the glucose uptake was also increased in AHL QS mutant, which indicating that the AHL quorum sensing system mediated the homeostasis of nutrient metabolism of individual cells, especially under the nutrition limit condition (An et al., 2014).
In this study, we isolated AHL autoinducers and investigated the function of the AHL quorum-sensing system in L. brunescens. To this end, we disrupted the key genes lbsI and lbsR and assessed the growth, autolysis, motility and Xanthomonas-specific antibiotic compound (XSAC) production in L. brunescens. According to our results, the lbsI and lbsR mutants grew more rapidly than that of wildtype OH23 at the early-log phase, and the AHL quorum-sensing system altered the growth rate by modulating sucrose uptake. Moreover, autolysis in the lbsI and lbsR mutants occurred faster than that in wildtype, which indicated that the AHL quorum-sensing system negatively regulates autolysis in L. brunescens. In XSAC production and surface motility assays, the normalized data of XSAC production and surface motility showed no significant difference between wildtype OH23 and AHL knockout mutants. Moreover, expression of key genes related to XSAC production and surface motility did not differ, which indicated that the effects of growth and autolysis on these phenotypes are much greater than the direct regulation of AHL quorum sensing system. Taken together, AHL quorum sensing downregulates the growth rate and autolysis, indicating that AHL quorum sensing system is involved in regulation of nutrition consumption to maintain population stable and cooperative in L. brunescens OH23.
The bacterial strains and plasmids used in this study are listed in Table 1, and the primers used are listed in Table 2. Escherichia coli strains were grown on LB medium (Lysogeny Broth containing 10 g tryptone, 5 g yeast extract, and 10 g sodium chloride in 1 l of distilled water, pH 7.0-7.2) at 37°C (Sambrook and Russell, 2001). L. brunescens and its derivative mutants were grown on NB medium (Nutrient Broth containing 5 g tryptone, 1 g yeast extract, 3 g beef extract, and 10 g sucrose in 1 l of distilled water, pH 7.0–7.2) at 28°C (Atlas and Parks, 1997). The AHL detection strains were grown on AT medium [5 g glucose, 2 g (NH4)2SO4, 0.078 g MgSO4, 0.0076 g CaCl2, 0.005 g FeSO4⋅7H2O, 0.0022 g MnSO4⋅H2O, and 10.7 g KH2PO4 in 1 l of distilled water, pH 7.0–7.2] (Fuqua and Winans, 1994). All solid media contained 1.5% agar, and antibiotics were added at the following concentrations: rifamycin, 20 μg/ml; gentamicin, 8 μg/ml. Sucrose was added to a final concentration of 4% in media used for counterselection of the in-frame deletion strain and its derivatives. In-frame deletions of lbsR and lbsI were constructed by cloning the regions flanking lbsR and lbsI into the suicide vector pJQ200SK containing a sacB counterselectable marker. The resulting plasmids were introduced into L. brunescens by conjugation, and deletion mutants were selected for double homologous recombination events.
To detect whether L. brunescens biosynthesizes AHL autoinducers, a highly efficient detection strain, A. tumefaciens KYC55 (pJZ372) (pJZ384) (pJZ410), was used to examine the biosynthetic ability of OH23. The autoinducer synthase gene traI in the highly efficient detection strain was disrupted, and a plasmid (PT7-traR) capable of overexpressing the A. tumefaciens TraR protein and a reporter plasmid containing the PtraI-lacZ promoter were introduced. The combination of TraR and AHL autoinducers convert it to an active form that binds to the promoter sequence of traI, initiating its transcription and expression of lacZ. Therefore, overexpression of TraR increases its binding to the autoinducer, enhances expression of the reporter lacZ gene and increases the sensitivity of the detection strain to the autoinducer (Zhu et al., 2003).
L. brunescens and AHL mutants were grown in NB liquid medium at 28°C at 180 rpm; the cultures were collected at the indicated time points and centrifuged at 12,000 rpm for 10 minutes, and the supernatant was extracted with the same volume of ethyl acetate, as previously described (Ling et al., 2019). Two liters of the organic phase of every treatment was evaporated at 47°C, and the crude extract was dissolved in 2 ml dimethyl sulfoxide (DMSO). Two microliters of the crude extract and approximately 107 AHL bioassay strain cells per ml were added to AT medium and incubated at 28°C at 180 rpm for 12 h followed by assays for β-galactosidase specific activity (Miller, 1972; Fuqua and Winans, 1994; Zhu and Mekalanos, 2003). A thin-layer chromatography (TLC) assay was used to analyze the AHLs biosynthesized by L. brunescens strains, as previously described (Zheng et al., 2015), using a sensitive AHL indicator strain. Briefly, concentrated crude extracts were applied to C18 reversed-phase TLC plates (Whatman) and incubated at 28°C for 12 h.
For the identification of AHLs, the supernatant was extracted with the same volume of chromatographically pure dichloromethane, and AHLs were identified by ESI-MS/MS. The structures of AHLs were calculated by the following formulas: for Cn ×2 + 4-HSL, n = (m/z-1-171)/28; for 3-hydroxyl-Cn×2+4-HSL, n = (m/z-1-187)/28; for 3-oxo-Cn×2+4-HSL, n = (m/z-1-185)/28 (Yang et al., 2009).
L. brunescens OH23 and the AHL knockout mutants were cultured in NB medium at 28°C with shaking at 180 rpm until the OD600 was approximately 1.0 [which corresponds to approximately 109 CFU/ml (Colony Forming Units/ml), about 18 to 22 h]. One milliliter of culture for each strain was transferred to 50 ml of new liquid NB medium, and the cultures were incubated at 28°C with shaking at 180 rpm. To measure growth, the OD600 value was determined every 12 h for each culture using a BioPhotometer Plus (Eppendorf, Germany) until each culture reached the stationary phase. Three replicates were performed for each treatment, and the experiment was repeated three times.
One milliliter (OD600 was approximately 1.0, about 18 to 22 h) of culture for test strain was transferred to 50 ml of new liquid NB medium and incubated at 28°C with shaking at 180 rpm; samples were collected every 12 h. The cells were removed by centrifugation, and the supernatants were used to analyze the concentrations of sucrose by using a sucrose detection kit (Solarbio, Beijing Solarbio Science & Technology Co., Ltd.).
Autolysis assays were performed as described by Mani et al. with modifications (Chitnis et al., 1990; Mani et al., 1993). Bacteria were grown in NB liquid medium at 28°C with shaking at 180 rpm to an OD600 of approximately 0.7 (about 16 to 18 h). The cells were washed twice with PBS, resuspended in the same volume of PBS and incubated at 28°C with shaking at 180 rpm. Optical density was measured at the indicated time points. The results were normalized to an OD600 at time zero, and percent lysis was calculated using the formula% autolysis = [OD600 (t0)-OD600 (t)]/OD600 (t0).
The ability of the wildtype and AHL mutants to produce XSAC was measured by an anti-Xanthomonas activity assay (diameter of inhibition zone), which was described previously (Ling et al., 2019). Briefly, the pathogenic strain X. oryzae pv. oryzae RS105 was incubated in NB liquid medium at 28°C with shaking at 180 rpm until the culture OD600 was approximately 1.0; L. brunescens OH23, ΔlbsR and ΔlbsI mutants were incubated in NB liquid medium at 28°C with shaking at 180 rpm until the indicated time points. For X. oryzae pv. oryzae RS105, 100 ml of liquefied NB solid medium was incubated at 45°C for 30 min, mixed with 108 cells, and then poured into plates for the anti-Xanthomonas activity assay. The cultures of L. brunescens OH23, ΔlbsR and ΔlbsI were centrifuged, and the supernatants were separately incubated at 85°C for 30 min; 30 μl of the treated supernatant from the cultures at the indicated time points was added to the hole in the selection plates. All plates were cultured at 28°C, and the zones of inhibition were photographed and compared after 2 days. XSAC production was evaluated, as represented by the diameter of the inhibition zone. Three replicates were performed for each treatment, and the experiment was repeated three times.
The surface motility assay of L. brunescens wildtype OH23, ΔlbsR and ΔlbsI was performed as previously described (Ling et al., 2019). Briefly, NB semi-solid medium containing 0.3% agar was used, and 2.5 μl of L. brunescens wildtype OH23, ΔlbsR or ΔlbsI (109 CFU/ml, OD600 was approximately 1.0 for all strains, about 18 to 22 h) was spotted onto the surface. The plates were incubated at 28°C for 4 days, and the surface motility of each strain was photographed, measured, and compared. Three replicates were performed for each treatment, and the experiment was repeated three times.
L. brunescens OH23, ΔlbsR and ΔlbsI mutants were each grown in 5 ml NB liquid medium until the OD600 was approximately 1.0 (about 18 to 22 h) or until to the indicated time points. Three milliliters of cells were transferred to a sterile centrifuge tube and centrifuged for 3 min at 12,000 rpm. RNA was extracted using TRIzol solution (TaKaRa Biocompany) following the manufacturer’s instructions and 250 ng RNA of every sample was used for further experiments. For DNA removal and reverse transcription PCR, HiScript III RT SuperMix for qPCR (+ gDNA wiper) (Vazyme Biotech Co., Ltd.) was used in this study. The real-time PCR assay was carried out as previously described (Ling et al., 2019). A QuantStudio 6 Flex Real-Time PCR System (Thermo Fisher Scientific) and HiScript II One Step qRT-PCR SYBR Green Kit (Vazyme Biotech Co., Ltd.) were used to detect gene expression. Gene expression was calculated by the 2−ΔΔCT method, and recA cDNA was used as an internal control in all reactions. The primers used for real-time PCR are listed in Table 2. Three replicates were performed for each treatment, and the experiment was repeated three times.
Statistical analyses were calculated using SPSS (Statistical Package, Version 21.0). The variables were subjected to Student’s t test and tested for significance at P < 0.05 (∗), P < 0.01 (∗∗), P < 0.001 (∗∗∗), and P < 0.0001 (****).
Wildtype OH23 was grown in 10 l NB liquid medium until the OD600 was approximately 2.0, which corresponds to approximately 2 × 109 CFU/ml. Extraction of the crude AHLs from the supernatant was performed using the same method previously described (Holden et al., 1999), and the extracted AHL signaling activities in L. brunescens OH23 were detected by an Agrobacterium-based AHL bioassay (Zhu et al., 2003). The AHL autoinducers from the wildtype strain OH23 had higher induction capabilities, and lacZ expression was dramatically induced in Agrobacterium. Furthermore, we detected autoinducer activity in different cell growth stages. As shown in Figure 1A, autoinducer production by wildtype was at a low level in the early-log phase; however, as the density of the cells increased, the concentration of autoinducer also increased, and AHL induction activities reached the highest value in the late-log phase. In Mesorhizobium tianshanense, the concentration of AHL signaling molecules increased with the growth of the strain and reached the highest concentration during the stationary phase (Zheng et al., 2006). The AHL autoinducers could be detected in L. brunescens supernatants, and AHL autoinducers show lower activity at low cell concentration and higher activity at high cell concentration which is a typical phenomenon of cell-density-dependent AHL quorum sensing system.
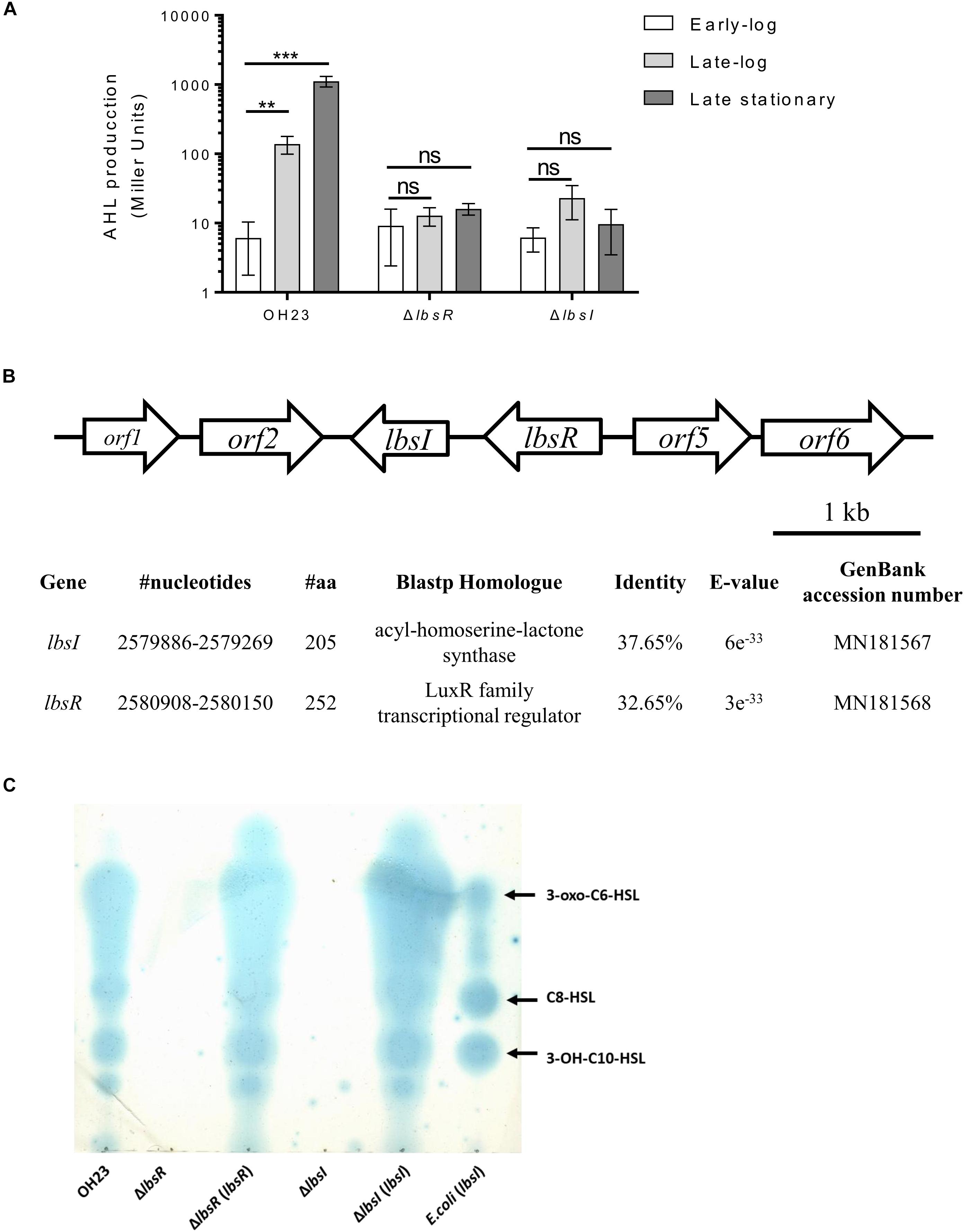
Figure 1. The location of the AHL lbsI/lbsR quorum-sensing system in L. brunescens OH23 and the AHL structures and AHLs of L. brunescens OH23 and its mutants. (A) AHL production in L. brunescens and its QS mutants in different growth phases. Statistical analyses of AHL production were performed using Student’s t-test compared with wildtype OH23 at the early-log phase. ∗∗P < 0.01; ∗∗∗P < 0.001; and ns (ns: no significance), P > 0.05. (B) Physical map of the LbsR/LbsI quorum sensing system gene cluster in L. brunescens. The identity results were represented by BLASTp comparison between the proteins encoded by lbsI and lbsR of L. brunescens and P. aeruginosa (taxid: 287). The protein similarity was compared using BioEdit 7.0.9.0. (C) TLC analysis of AHL contents. The supernatants of wildtype OH23, ΔlbsI, ΔlbsR, ΔlbsI (lbsI), ΔlbsR (lbsR) and E. coli (lbsI) were extracted and loaded onto a C18 reverse-phase TLC plate, followed by an overlay of the agar medium seeded with JZA1 bioassay strains. Each lane was loaded with 2 μl of concentrated extract (Miller Unit ≈ 500).
To confirm the L. brunescens AHL structures, the culture supernatant of the wildtype OH23 strain was extracted with dichloromethane, and concentrated AHLs were subjected to ESI-MS/MS analysis. Analysis of the mass spectrum revealed that the wildtype strain of L. brunescens mainly biosynthesized three types of AHL molecules, 3-OH-C10-HSL (MW271), C8-HSL (MW 227) and 3-oxo-C6-HSL (MW 213), with distinct peak times (Supplementary Figure S1).
As mentioned above, AHL autoinducer signals have been reported in different bacteria, and three AHL autoinducers were isolated from L. brunescens. To identify genes related to AHL biosynthesis, random transposon insertional mutagenesis was used to screen AHL-deficient mutants (Supplementary Table S1). All transconjugants were grown in NB liquid medium until the OD600 was approximately 1.0, and supernatants were concentrated and used for liquid AHL bioassays. From approximately 3000 random transconjugants, 2 mutants exhibited deficiency in inducing the β-galactosidase activity of the A. tumefaciens indicator strain (Supplementary Figure S2A), indicating that the transposon-inserted gene might be responsible for AHL biosynthesis. The transposon flanking region was sequenced to identify genes potentially related to the AHL biosynthesis pathway (Supplementary Figures S2B,C). Based on BLAST searches of the transposon flanking region1, we found that the AHL-related proteins from L. brunescens have homology to those of P. aeruginosa (taxid: 287), with levels of protein identity of 32.62% (LbsR) and 37.65% (LbsI). LbsR is a LuxR family transcriptional regulator involved in AHL biosynthesis modulation, and LbsI is a GNAT family N-acetyltransferase responsible for the biosynthesis of AHLs (Figure 1B).
In Mesorhizobium tianshanense and Pseudomonas syringae, the disruption of LuxR-type regulator and LuxI-type synthase leaded to the AHL biosynthesis capabilities abolished (Quinones et al., 2004; Zheng et al., 2006). Meanwhile, the expression of MrtI requires the intact MrtR and AHL signals in M. tianshanense (Zheng et al., 2006). To demonstrate the role of the LbsR/LbsI system in the biosynthesis of AHL signaling molecules in L. brunescens, in-frame knockout experiments were performed on lbsI and lbsR, and the concentrated supernatant AHL activity of the mutants was evaluated. As shown in Figure 1A, mutation of lbsI and lbsR abolished the ability to produce AHLs during the entire bacterial life cycle, which suggested that lbsI and lbsR are essential for AHL biosynthesis in L. brunescens. Further confirmation by a TLC assay showed that the lbsI and lbsR mutants completely lacked the ability to produce any AHL molecules (Figure 1C). Two possible reasons that the lbsI and lbsR mutants abolished AHL biosynthesis capabilities under testing conditions. One is that the LbsI is responsible for the biosynthesis of AHL, which means that lbsI is indispensable in AHL biosynthesis. The second possible reason is that the regulation of LbsR protein is required in the synthesis of AHL signaling molecules, just like MrtR-MrtI regulation system in M. tianshanense (Zheng et al., 2006). lbsI and lbsR gene overexpression plasmids were transferred into the lbsI and lbsR knockout mutants, respectively, by conjugation. As shown in Figure 1C, the complemented strains ΔlbsR (lbsR) and ΔlbsI (lbsI) displayed strong activities in inducing A. tumefaciens indicator strain β-galactosidase expression compared to that treated by concentrated supernatants from ΔlbsR and ΔlbsI. The AHL TLC bioassay results indicated restoration of the biosynthesis activity of the AHL autoinducer in the deletion mutants.
To further understand the function of the AHL synthase gene lbsI, the gene was overexpressed under the Plac promoter in the broad-host vector pBBR1-MCS5 and electrotransformed into E. coli DH5α to obtain E. coli (lbsI). The culture supernatants were concentrated and tested for AHL activities with the A. tumefaciens indicator strain. As illustrated in Figure 1C, TLC assays revealed three blue halos for the culture supernatant of E. coli (lbsI), which indicated that lbsI is responsible for the synthesis of three AHL molecules in E. coli (lbsI), and this result was consistent with the ESI-MS/MS results.
To investigate whether AHL quorum sensing regulates the growth rate in L. brunescens, we measured the growth of wildtype OH23, ΔlbsR, ΔlbsI, and the corresponding complementation strains in NB medium (Figure 2A, Supplementary Figure S4A, and Supplementary Table S1). The ΔlbsR and ΔlbsI knockout mutants grew faster in the early-log and late-log phase than that in wild-type OH23; the cell density of the mutant strain significantly decreased during the stationary phase compared to that of the wild type strain (Figure 2A). In the early exponential growth phase, the growth rates of ΔlbsR (pBBR) and ΔlbsI (pBBR) increased faster than that of OH23 (pBBR), which is consistent with the results observed for the ΔlbsR and ΔlbsI mutants (Supplementary Figure S4A). Moreover, no significant difference in growth rate was found between the complementation strains ΔlbsR (lbsR) and ΔlbsI (lbsI) and OH23 (pBBR) (Supplementary Figure S4A) and ΔlbsI mutant could show the same growth rate with wildtype OH23 in the condition of adding 2 μM C8-HSL (Supplementary Figure S4B).
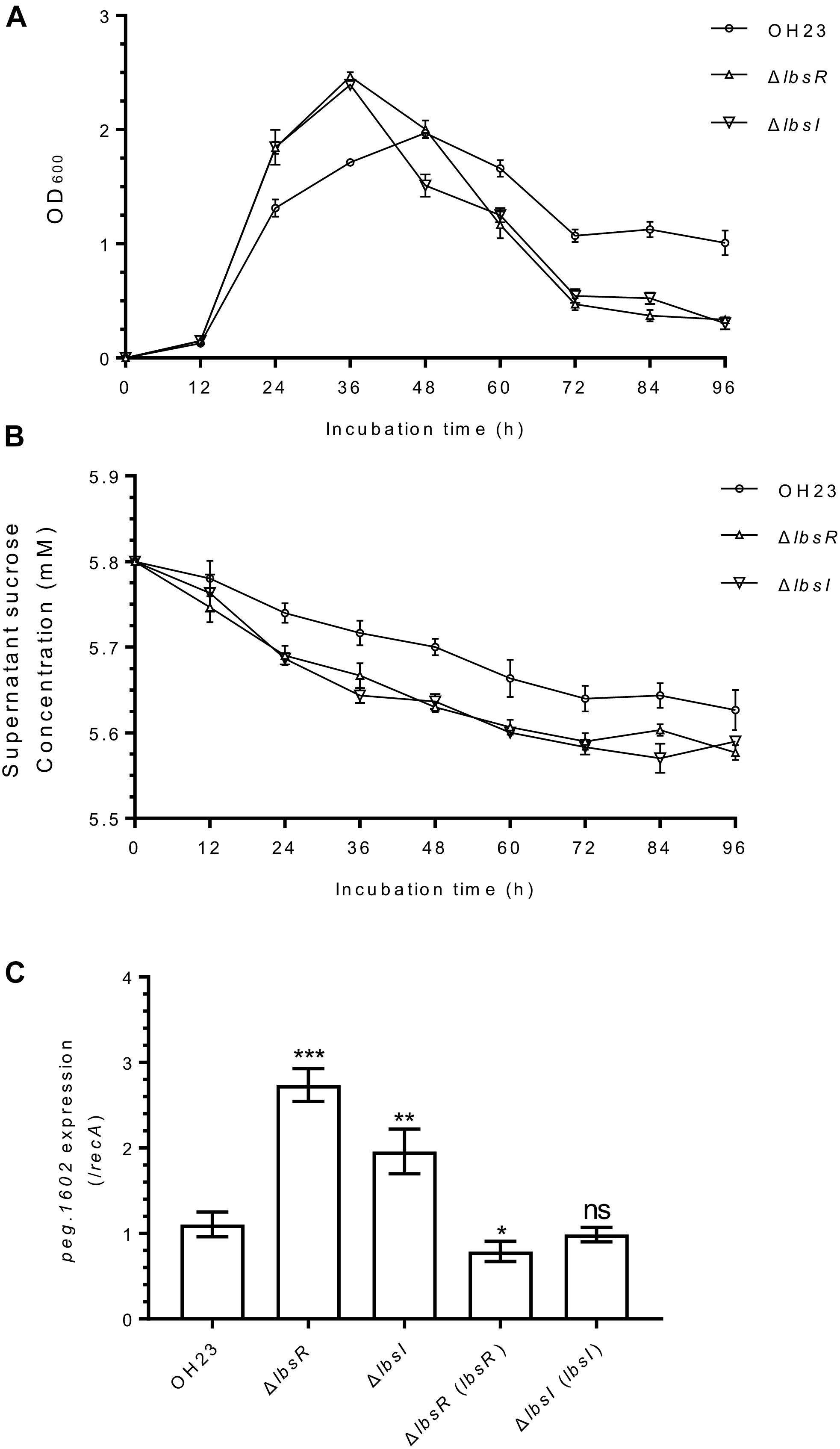
Figure 2. The growth and sucrose consumption-related gene expression of L. brunescens OH23 and its AHL mutants. (A) The growth of wildtype OH23 and its AHL mutants. Wildtype OH23 and AHL mutants (OD600 of approximately 1.0) were grown in NA liquid medium at 28°C, and the OD600 was measured at the time points indicated. The data are the combination of three individual experiments. (B) Sucrose consumptions of wildtype OH23 and its AHL mutants. The samples from growth measurement assay were centrifuged and the supernatants were used for sucrose concentration detection by using sucrose detection kit. (C) α-glucosidase gene peg.1106 expression in wildtype OH23 and AHL mutants. The growth of mutants was compared with that of strain OH23 at the same time points. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; and ns (ns: no significance), P > 0.05 (Student’s t-test).
AHL quorum-sensing knockout mutants of Burkholderia glumae grow more rapidly than do wildtype at the early exponential stage, and the AHL quorum-sensing system modulates the growth rate by invoking a phosphoenolpyruvate-dependent sugar phosphotransferase system (PTS) to affect the glucose uptake rate (An et al., 2014). In X. axonopodis pv. glycines, a unique sucrose hydrolase was identified that is responsible for sucrose metabolism to glucose and fructose (Kim et al., 2004). To determine whether the AHL-quorum sensing system uses sucrose metabolism to regulate the growth rate, we first detected the sucrose consumption in wildtype OH23 and AHL knockout mutants. As the strain grows, the sucrose content in the supernatant gradually decreases (Figure 2B). At 48 h, the sucrose consumption in the AHL knockout mutant strain was approximately twice than that of the wildtype OH23. In the later stage of growth, as the growth of AHL knockout mutants gradually slowed down in AHL knockout mutants, and the difference in sucrose consumption between AHL knockout mutants and wildtype OH23 gradually decreased. We also found a α-glucosidase responsible for sucrose metabolism (Supplementary Figure S3), and homology analysis revealed that peg.1602 (GenBank number: MN557391), encoding 540 amino acids, is homologous to α-glucosidase from Lysobacter sp. TY2-98 (85%), with an E-value of 0.0. The enzyme encoded by peg.1602 showed strong activity in digesting sucrose to glucose and fructose (Supplementary Figure S3B and Supplementary Table S1). Therefore, we detected expression of peg.1602 to evaluate the sucrose metabolism activity in L. brunescens. Expression of peg.1602 in OH23, ΔlbsR, ΔlbsI and complementation strains was detected using real-time PCR. As shown in Figure 2C and Supplementary Figure S4C, peg.1602 expression was significantly higher in ΔlbsR and ΔlbsI than in wildtype OH23, whereas peg.1602 expression did not differ significantly between wildtype OH23, the complementation strains and chemical complementation of ΔlbsI, which indicated that the AHL quorum-sensing system is involved in the regulation of peg.1602 expression. The classical AHL QS system positively regulates the growth in M. tianshanense (Cao et al., 2009). Meanwhile, some AHL QS system also negatively regulates the growth in B. glumae and the AHL QS system might involve in the regulation of consumption of nutrition (An et al., 2014). Based on AHL quorum sensing system, the cells could sense other bacteria by the AHLs from intercellular and control the population size and nutrition consumption. Taken together, the AHL quorum-sensing system regulates the growth rate and restricts the sucrose consumption in L. brunescens, indicating that AHL quorum sensing system plays important roles in coordinating nutrition distribution in the population group to achieve stability and cooperation of the population, especially in the nutritional limit conditions.
Because the OD600 value of the AHL quorum-sensing knockout mutants was significantly lower than that of wildtype OH23 in the late stationary phase (Figure 3A), we speculated that AHL quorum sensing may be involved in the autolysis process in L. brunescens. During autolysis, peptidoglycan hydrolase digests the cell wall, which leads to a decrease in turbidity (OD600); thus, a turbidimetric assay can be used to calculate the degree of autolysis. As presented in Figure 3A, autolysis processes began at 1 h in wildtype and the ΔlbsR and ΔlbsI mutants, with percentages of 4.73, 10.84, and 12.79%, respectively. In addition, autolysis percentages in the ΔlbsR and ΔlbsI mutants reached over 30% after 4 h of incubation, whereas the wildtype strain required over 7 h to achieve 30% autolysis. In S. aureus, upregulation of the peptidoglycan hydrolase gene lytM further promotes autolysis (Michel et al., 2006). Homology analysis revealed that peg.977 (GenBank number: MN557390), encoding 367 amino acids, is homologous to peptidoglycan hydrolase gene lytM from S. aureus (55%), with an E-value of 2 × e–014. To further confirm that AHL quorum sensing regulates autolysis in L. brunescens, we examined peg.977 expression in wildtype OH23, the ΔlbsR, ΔlbsI mutants and chemical complementation of ΔlbsI strain. The results showed the peg.977 expression increases ∼3-fold in the ΔlbsR and ΔlbsI mutants compared to wildtype OH23; the peg.977 expression is downregulated in ΔlbsR (lbsR) and ΔlbsI (lbsI); the peg.977 expression shows no significant difference between wildtype OH23 and chemical complementation of ΔlbsI strain (Figure 3B and Supplementary Figure S5). Taken together, AHL quorum sensing negatively regulates autolysis to control and maintain the population in L. brunescens.
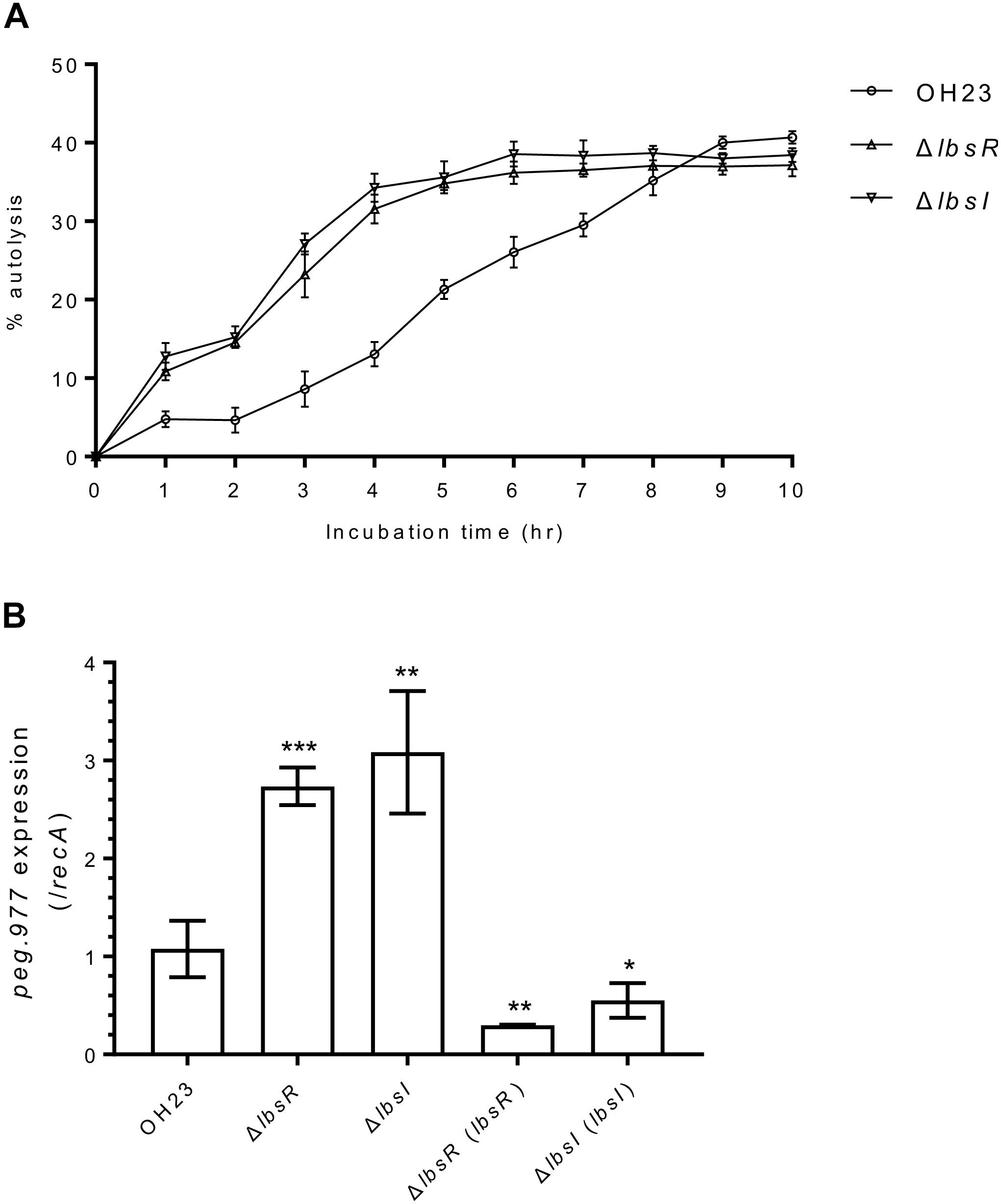
Figure 3. The autolysis and peptidoglycan hydrolase-related gene expression of L. brunescens OH23 and its AHL mutants. (A) Autolysis of wildtype OH23 and AHL mutant whole cells in PBS buffer (pH 7.2). 5 × 108 cells of wildtype OH23 and its AHL mutants were washed twice with PBS (pH 7.2) and resuspended in equal volume PBS. The cultures were incubated at 28°C with shaking at 180 rpm. OD600 was measured at the indicated time points. The data are the combination of three individual experiments. (B) Peptidoglycan hydrolase peg.977 expression in wildtype OH23 and AHL mutants. The data are the combination of three individual experiments. Statistical analyses of peg.977 expression were performed using Student’s t-test compared with wildtype OH23. ∗P < 0.05; ∗∗P < 0.01; and ∗∗∗P < 0.001 (Student’s t-test).
In a previous study, the AHL-type quorum-sensing system was found to affect the growth rate, swimming motility and infection virulence of Acidovorax avenae (Fan et al., 2011). Moreover, the AHL-type quorum-sensing LuxR family-type regulator is required for rice virulence in Xanthomonas oryzae pv. oryzae and negatively regulates heat-stable antifungal factor (HSAF) biosynthesis in Lysobacter enzymogenes (Ferluga et al., 2007; Qian et al., 2014). To investigate whether the AHL quorum-sensing system modulates XSAC biosynthesis in L. brunescens, we detected the anti-Xanthomonas abilities of wildtype OH23, ΔlbsR and ΔlbsI to evaluate the production of XSAC biosynthesis. As depicted in Figures 4A,B, anti-Xanthomonas abilities increased with the growth of OH23, and the diameters of the inhibition zone were 0.23 ± 0.08 cm, 0.85 ± 0.05 cm, and 0.83 ± 0.24 cm from the early-log phase to the late log phase to the stationary phase, respectively. Compared to wildtype OH23, the diameters of the inhibition zones of ΔlbsR and ΔlbsI increased 278.57 and 271.43% in the early-log phase and 37.25 and 56.86% in the late-log phase, respectively. In the late stationary phase, XSAC production was dramatically decreased in the ΔlbsR and ΔlbsI mutants, and the diameters of the inhibition zones were 0.07 ± 0.02 cm and 0.05 ± 0.01 cm, respectively.
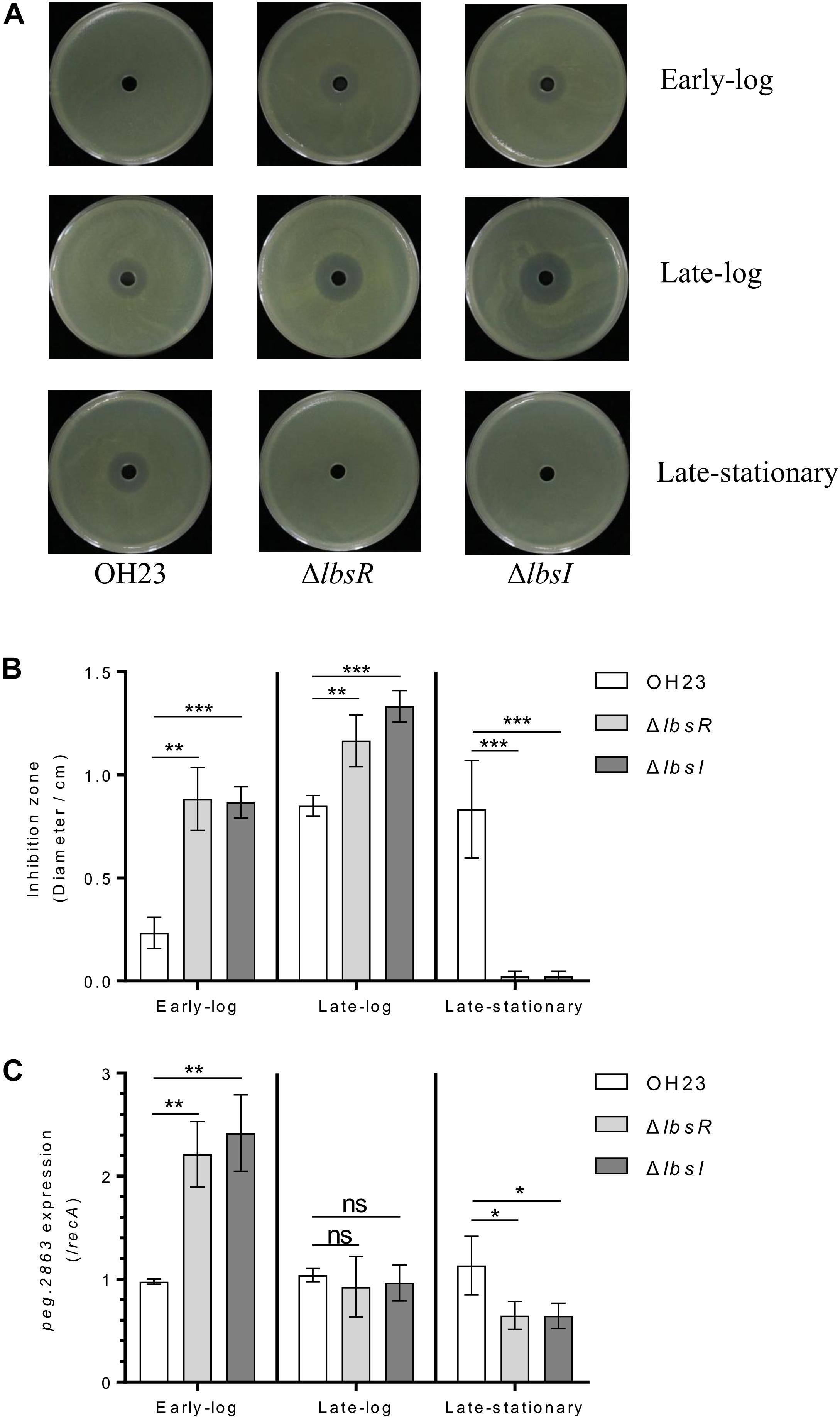
Figure 4. XSAC production in L. brunescens OH23 and its AHL mutants. (A) Cultures of L. brunescens OH23 or AHL mutants were tested for antimicrobial activity against X. oryzae pv. oryzae RS105 at the indicated time points. 105 CFU cells of X. oryzae pv. oryzae RS105 were spread onto the surface of each NB plate, and 30 μl of test supernatants of L. brunescens and AHL mutants (samples collected from indicated time points) was added into the central holes of NB plates. The plates were incubated at 28°C for 4 days and the size of inhibition zones were using to calculate for the antimicrobial activity. (B) Analysis of the images of X. oryzae pv. oryzae RS105 growth inhibition zones shown in A. (C) peg.2863 (key gene involved in XSAC biosynthesis) expression in wildtype OH23, ΔlbsI and ΔlbsR. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; and ns, P > 0.05 (Student’s t-test).
To further confirm whether the AHL-type quorum-sensing system controls XSAC biosynthesis, we collected wildtype OH23, ΔlbsR and ΔlbsI cells at different cell growth phased and extracted RNA to detect expression of peg.2863 (a key gene related to the biosynthesis of XSAC) (Ling et al., 2019). In the early-log phase, the expression of peg.2863 in AHL knockout mutants were significantly enhanced comparted to wildtype Oh23; in the mid-log phase, expression of peg.2863 showed no significant difference between wildtype OH23 and ΔlbsR or ΔlbsI.; in the late-stationary, the peg.2863 expression was downregulated in AHL knockout mutants (Figure 4C). Combining the different growth rates presented in Figure 2A, the diameter of inhibition zone was normalized by the OD600, and the inhibition activities in late-log phase were shown no significant difference and between wildtype and AHL knockout mutants (Supplementary Figure S6A), which indicating that cell densities decrease dramatically in the AHL mutants due to autolysis in the late stationary phase and cause the AHL mutants to completely lack activity against Xanthomonas oryzae pv. oryzae RS105. In L. enzymogenes, the yield of HSAF accumulates with growth and reaches a maximum concentration at the stationary phase (Tang et al., 2018). In addition, the biosynthesis of HSAF is regulated by the DSF quorum sensing system in L. enzymogenes, and the loss of the key DSF quorum sensing system genes leads to a significant decrease in HSAF production (Han et al., 2015). Similarly, in L. brunescens, the capability of XSAC biosynthesis is completed lost in DSF quorum sensing knockout mutants (Ling et al., 2019). The inhibition zones of AHL knockout mutants was significantly bigger than that in wildtype OH23 in early-log and late-log phase; the differences of antagonistic capabilities between AHL knockout mutants and wildtype OH23 are not significant when the yield of all XSAC was normalized by OD600 value. The AHL quorum-sensing system is involved in the growth and autolysis of L. brunescens, and the growth rate and autolysis negatively affect XSAC biosynthesis. Taken together, the different XSAC production capacities between wildtype OH23 and the AHL mutants might be due to the different growth rates in the early-log phase and late-log phase.
In Pseudomonas syringae, the AHL-mediated quorum-sensing system negatively regulates swarming motility (Quinones et al., 2005). To investigate whether the AHL-type quorum-sensing system participates in surface motility in L. brunescens, we tested the surface motility of wildtype OH23, ΔlbsR and ΔlbsI on NB semi-solid (0.3% agar) motility medium plates for 4 days at 28°Ñ. As illustrated in Figures 5A,B, the surface motility diameter of wildtype OH23 was 4.27 ± 0.17 cm over four days. However, the average surface motility diameters of ΔlbsR and ΔlbsI reached 6.51 ± 0.25 cm and 6.28 ± 0.13 cm, respectively. In order to eliminate side-effects of growth rates, the diameter of surface motility was normalized by the OD600. And the diameters of surface motility were shown no significant difference and between wildtype and AHL knockout mutants (Supplementary Figure S6B). Moreover, pilA1 gene expression showed no significant difference between wildtype OH23 and the AHL-type QS mutants, indicating that surface motility was indirectly regulated by the AHL-type quorum-sensing system and might be affected by the growth rate in L. brunescens (Figrue 5C). In Psa, when, the motility of the bacteria and the expression of some genes related to motility are significantly enhanced while adding 1 μM of AHL signaling molecules. In addition, the expression level of pilA gene was significantly decreased in the three luxR knockout mutants compared to wildtype, indicating that solo LuxRs can mediate Psa response to environmental AHL signaling molecules and regulate motility, biofilm formation and virulence (Cellini et al., 2019). In L. brunescens, the DSF quorum sensing system positively regulates the XSAC biosynthesis and surface motility, and the DSF quorum sensing system also positively regulates the expression of peg.2863 and pilA1 (Ling et al., 2019). Taken together, the DSF quorum sensing system positively regulates the synthesis and motility of XSAC, while the AHL quorum sensing system affects cell growth and autolysis which further affecting the XSAC biosynthesis and surface motility.
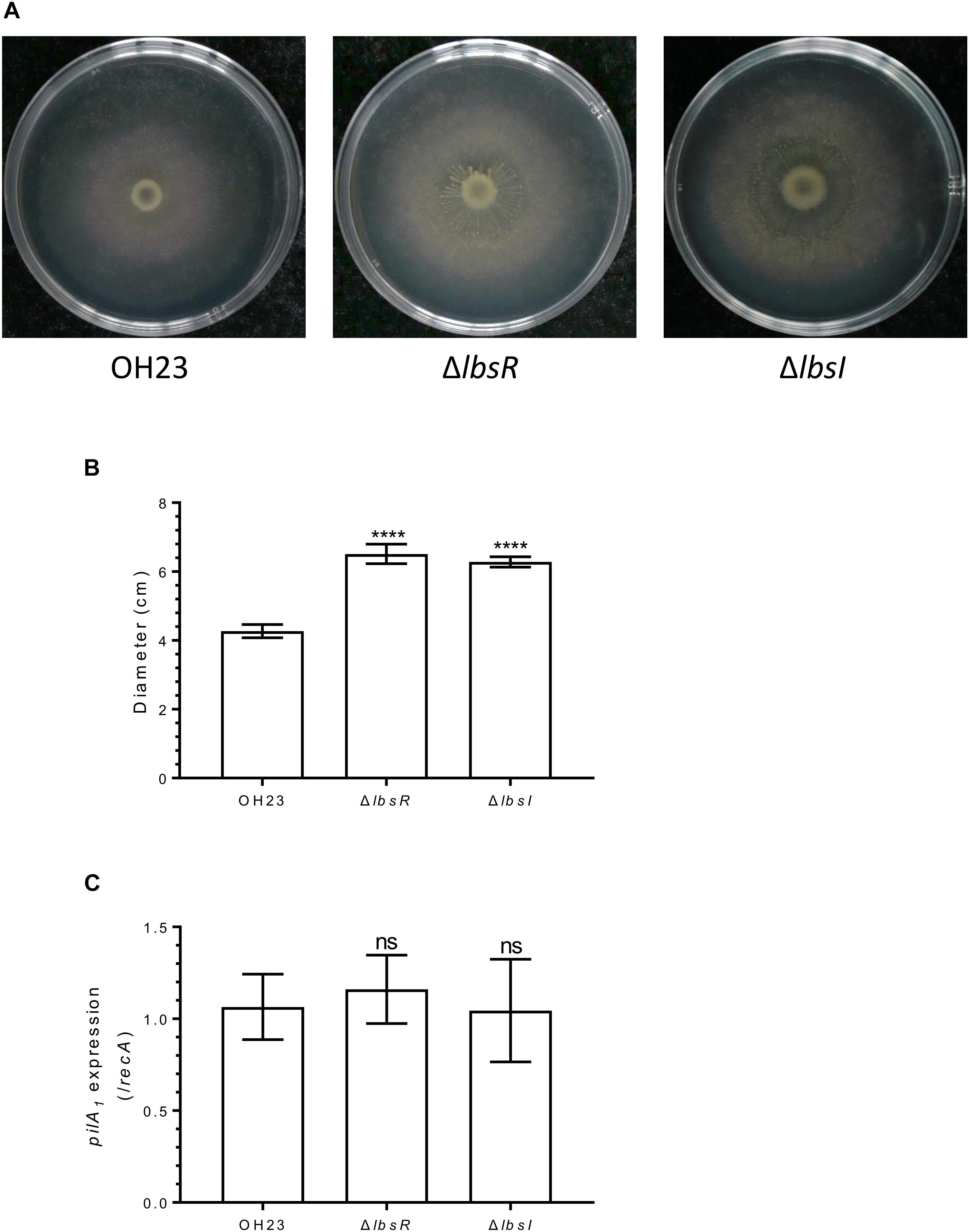
Figure 5. Surface motility in L. brunescens OH23 and its AHL mutants. (A) Surface motility of wildtype OH23 and AHL mutants. Each strain was grown in liquid NB medium until the culture reached an OD600 of approximately 1.0, after which 3 μl was inoculated onto the surface of 0.3% Agra NB plates. The plates were inoculated at 28°C for 4 days. (B) Analysis of the images of surface motility shown in A. (C) pilA1 expression in wildtype OH23, ΔlbsI, and ΔlbsR. ****P < 0.0001; and ns, P > 0.05 (Student’s t-test).
In this study, we report the role of the AHL quorum-sensing system in the regulation of cell growth and autolysis, and our data revealed the sucrose consumption and autolysis system to be negatively regulated by the AHL quorum-sensing system. First, we characterized three different AHLs in L. brunescens, including 3-OH-C10-HSL, C8-HSL and 3-oxo-C6-HSL. Second, we observed that the AHL mutants grew more rapidly than did wildtype OH23; the sucrose consumption rates were high, as was expression of the α-glucosidase gene in AHL mutants. These findings indicate that the AHL quorum-sensing system negatively regulates sucrose consumption and that the growth rate is increased in AHL QS mutants compared to wildtype OH23. Third, the autolysis rate of the AHL QS mutants was higher than that of wildtype, and expression of the key gene peg.977 was increased in the AHL QS mutants, suggesting that the autolysis process was strongly influenced by the AHL quorum-sensing system. Moreover, XSAC production and surface motility might be affected by growth and autolysis and that the AHL quorum-sensing system indirectly regulates XSAC biosynthesis and surface motility. Altogether, the AHL quorum sensing system is involved in growth and autolysis regulation, indicating the AHL quorum sensing system regulates nutrition consumption to maintain population stable and cooperative in L. brunescens.
The datasets generated for this study can be found in the GenBank: MN181567, MN181568, MN557391, and MN557390.
JL, LZ, GW, YZ, and TJ conducted the experiments. FL designed and conducted the experiments and revised the manuscript. JL, LZ, GW, and YZ contributed to the writing of the manuscript.
This study was supported by the National Natural Science Foundation of China (31872018), the Earmarked Fund for China Agriculture Research System (CARS-28-16), China Postdoctoral Science Foundation (2017M610310), Jiangsu Agricultural Science and Technology Innovation Funds (CX(16)1049), and Jiangsu Provincial Key Technology Support Program (BE2015354).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02748/full#supplementary-material
FIGURE S1 | ESI-MS/MS analysis of AHLs and AHL production in different time pints in L. brunescens. (A) According to the ESI-MS results, the structures of AHLs were calculated by using the following formulas: n = [(m/z-1-171)/28]Cn ×2+4-HSL; n = [(m/z-1-187)/28] 3-hydroxyl-Cn×2+4-HSL; n = [(m/z-1-185)/28] 3-oxo-Cn×2+4-HSL (Bainton et al., 1992). (B) The supernatants of wildtype OH23 and its AHL mutants were collected at indicated time points, and AHL activities were measured by detection strain A. tumefaciens KYC55 (pJZ372) (pJZ384) (pJZ410).
FIGURE S2 | Identification of genes involved in AHL quorum sensing system in L. brunescens. (A) Screening the genes related to AHL quorum sensing system and AHL productions of mutants were detected by using the detection strain A. tumefaciens KYC55 (pJZ372) (pJZ384) (pJZ410). ∗∗∗P < 0.001 (Student’s t test). (B) Results of Arbitrary PCR of mutants. (C) Identification of transposon insertion position and genetic map of lbsR and lbsI. The triangles represent the transposon insertion positions.
FIGURE S3 | Sequence alignment and function identification of α-glucosidase in L. brunescens. (A) Amino acid sequence alignment of peg.1602. L. brunescens, MN557391; Xanthomonadaceae bacterium, TXH70081.1; Lysobacter sp. TY2-98, WP_115647852.1; Thermomonas haemolytica, WP_114958989.1. (B) SDS-PAGE of the different fractions in the purification of α-glucosidase from L. brunescens. (C) Enzyme activity detection of α-glucosidase from OH23.
FIGURE S4 | Growth of L. brunescens AHL-complemented strains. (A) Growth of gentamicin-marked complemented strains. Gentamicin-marked wildtype OH23 [OH23 (pBBR)], AHL-complemented strains [ΔlbsI (lbsI) and ΔlbsR (lbsR)] and AHL control strains [ΔlbsI (pBBR) and ΔlbsR (pBBR)] (OD600 of approximately 1.0) were grown in NA liquid medium with gentamicin at 28°C, and the OD600 value was measured at the time points indicated. The data are the combination of three individual experiments. (B) Growth of chemical complemented strains. 2 μM (final concentration) C8-HSL was then added to the cultures of ΔlbsI treatments and the OD600 value was measured at the time points indicated. (C) α-glucosidase gene peg.1602 expression in wildtype OH23, ΔlbsI and chemical complemented strain. The cells were collected at the OD600≈1.0 and RNA were extracted by using TRIzol solution (TaKaRa Biocompany). The real-time PCR assay was carried out by using QuantStudio 6 Flex Real-Time PCR System (Thermo Fisher Scientific) and HiScript II One Step qRT-PCR SYBR Green Kit (Vazyme Biotech Co., Ltd.). The data are the combination of three individual experiments. Statistical analyses were performed using Student’s t-test compared with wildtype OH23. ∗∗∗P < 0.001 and ns, P > 0.05 (Student’s t-test).
FIGURE S5 | Peptidoglycan hydrolase gene peg.977 expression in wildtype OH23, ΔlbsI and chemical complemented strain. The data are the combination of three individual experiments. Statistical analyses of peg.1602 expression were performed using Student’s t-test compared with wildtype OH23. ∗P < 0.05 and ∗∗P < 0.01 (Student’s t-test).
FIGURE S6 | Diameter of inhibition zone and surface motility ormalized by OD600. (A) Diameter of inhibition zone normalized by OD600. (B) Diameter of surface motility zone normalized by OD600. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; and ns, P > 0.05 (Student’s t-test).
Almeida, R. P., Killiny, N., Newman, K. L., Chatterjee, S., Ionescu, M., and Lindow, S. E. (2012). Contribution of rpfB to cell-to-cell signal synthesis, virulence, and vector transmission of Xylella fastidiosa. Mol. Plant Microbe Interact. 25, 453–462. doi: 10.1094/MPMI-03-11-0074
An, J. H., Goo, E., Kim, H., Seo, Y. S., and Hwang, I. (2014). Bacterial quorum sensing and metabolic slowing in a cooperative population. Proc. Natl. Acad. Sci. U.S.A. 111, 14912–14917. doi: 10.1073/pnas.1412431111
Atlas, R. M., and Parks, L. C. (1997). Handbook of Microbiological Media. Boca Raton, FL: CRC Press.
Bainton, N. J., Stead, P., Chhabra, S. R., Bycroft, B. W., Salmond, G. P., Stewart, G. S., et al. (1992). N-(3-oxohexanoyl)-L-homoserine lactone regulates carbapenem antibiotic production in Erwinia carotovora. Biochem. J. 288(Pt 3), 997–1004. doi: 10.1042/bj2880997
Barber, C. E., Tang, J. L., Feng, J. X., Pan, M. Q., Wilson, T. J., Slater, H., et al. (1997). A novel regulatory system required for pathogenicity of Xanthomonas campestris is mediated by a small diffusible signal molecule. Mol. Microbiol. 24, 555–566. doi: 10.1046/j.1365-2958.1997.3721736.x
Cao, H., Yang, M., Zheng, H., Zhang, J., Zhong, Z., and Zhu, J. (2009). Complex quorum-sensing regulatory systems regulate bacterial growth and symbiotic nodulation in Mesorhizobium tianshanense. Arch. Microbiol. 191, 283–289. doi: 10.1007/s00203-008-0454-7
Cellini, A., Donati, I., Fiorentini, L., Vandelle, E., Polverari, A., Venturi, V., et al. (2019). N-Acyl homoserine lactones and lux solos regulate social behaviour and virulence of Pseudomonas syringae pv. actinidiae. Microb. Ecol. doi: 10.1371/journal.pone.0087862 [Epub ahead of print].
Chitnis, S. N., Prasad, K. S. N., and Bhargava, P. M. (1990). Isolation and characterization of autolysis-defective mutants of Escherichia-Coli that are resistant to the lytic activity of seminalplasmin. J. Gen. Microbiol. 136, 463–469. doi: 10.1099/00221287-136-3-463
Fan, J. Q., Qian, G. L., Chen, T., Zhao, Y. Q., Liu, F. Q., Walcott, R. R., et al. (2011). The acyl-homoserine lactone (AHL)-type quorum sensing system affects growth rate, swimming motility and virulence in Acidovorax avenae subsp citrulli. World J. Microbiol. Biotechnol. 27, 1155–1166. doi: 10.1007/s11274-010-0562-9
Ferluga, S., Bigirimana, J., Hofte, M., and Venturi, V. (2007). A LuxR homologue of Xanthomonas oryzae pv. oryzae is required for optimal rice virulence. Mol. Plant Pathol. 8, 529–538. doi: 10.1111/j.1364-3703.2007.00415.x
Folman, L. B., Postma, J., and Van Veen, J. A. (2003). Characterisation of Lysobacter enzymogenes (Christensen and Cook 1978) strain 3.1T8, a powerful antagonist of fungal diseases of cucumber. Microbiol. Res. 158, 107–115. doi: 10.1078/0944-5013-00185
Fuqua, W. C., and Winans, S. C. (1994). A Luxr-Luxi type regulatory system activates agrobacterium ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J. Bacteriol. 176, 2796–2806. doi: 10.1128/jb.176.10.2796-2806.1994
Han, Y., Wang, Y., Tombosa, S., Wright, S., Huffman, J., Yuen, G., et al. (2015). Identification of a small molecule signaling factor that regulates the biosynthesis of the antifungal polycyclic tetramate macrolactam HSAF in Lysobacter enzymogenes. Appl. Microbiol. Biotechnol. 99, 801–811. doi: 10.1007/s00253-014-6120-x
He, Y. W., Wang, C., Zhou, L., Song, H., Dow, J. M., and Zhang, L. H. (2006). Dual signaling functions of the hybrid sensor kinase RpfC of Xanthomonas campestris involve either phosphorelay or receiver domain-protein interaction. J. Biol. Chem. 281, 33414–33421. doi: 10.1074/jbc.m606571200
He, Y. W., Wu, J., Cha, J. S., and Zhang, L. H. (2010). Rice bacterial blight pathogen Xanthomonas oryzae pv. oryzae produces multiple DSF-family signals in regulation of virulence factor production. BMC Microbiol. 10:187. doi: 10.1186/1471-2180-10-187
Holden, M. T., Ram Chhabra, S., De Nys, R., Stead, P., Bainton, N. J., Hill, P. J., et al. (1999). Quorum-sensing cross talk: isolation and chemical characterization of cyclic dipeptides from Pseudomonas aeruginosa and other gram-negative bacteria. Mol. Microbiol. 33, 1254–1266. doi: 10.1046/j.1365-2958.1999.01577.x
Huber, B., Riedel, K., Hentzer, M., Heydorn, A., Gotschlich, A., Givskov, M., et al. (2001). The cep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology 147, 2517–2528. doi: 10.1099/00221287-147-9-2517
Kato, A., Nakaya, S., Kokubo, N., Aiba, Y., Ohashi, Y., Hirata, H., et al. (1998). A new anti-MRSA antibiotic complex, WAP-8294A - I. Taxonomy, isolation and biological activities. J. Antibiot. 51, 929–935. doi: 10.7164/antibiotics.51.929
Kim, H. S., Park, H. J., Heu, S., and Jung, J. (2004). Molecular and functional characterization of a unique sucrose hydrolase from Xanthomonas axonopodis pv. glycines. J. Bacteriol. 186, 411–418. doi: 10.1128/jb.186.2.411-418.2004
Kolter, R., Inuzuka, M., and Helinski, D. R. (1978). Trans-complementation-dependent replication of a low molecular weight origin fragment from plasmid R6K. Cell 15, 1199–1208. doi: 10.1016/0092-8674(78)90046-6
Kovach, M. E., Elzer, P. H., Hill, D. S., Robertson, G. T., Farris, M. A., and Roop, R. M. II, et al. (1995). Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176. doi: 10.1016/0378-1119(95)00584-1
Ling, J., Zhu, R., Laborda, P., Jiang, T., Jia, Y., Zhao, Y., et al. (2019). LbDSF, the lysobacter brunescens quorum-sensing system diffusible signaling factor, regulates anti- xanthomonas XSAC biosynthesis, colony morphology, and surface motility. Front. Microbiol. 10:1230. doi: 10.3389/fmicb.2019.01230
Mani, N., Tobin, P., and Jayaswal, R. K. (1993). Isolation and characterization of autolysis-defective mutants of Staphylococcus-aureus created by Tn917-Lacz mutagenesis. J. Bacteriol. 175, 1493–1499. doi: 10.1128/jb.175.5.1493-1499.1993
Michel, A., Agerer, F., Hauck, C. R., Herrmann, M., Ullrich, J., Hacker, J., et al. (2006). Global regulatory impact of ClpP protease of Staphylococcus aureus on regulons involved in virulence, oxidative stress response, autolysis, and DNA repair. J. Bacteriol. 188, 5783–5796. doi: 10.1128/jb.00074-06
Miller, J. H. (1972). Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
Miller, M. B., and Bassler, B. L. (2001). Quorum sensing in bacteria. Annu. Rev. Microbiol. 55, 165–199.
O’Sullivan, J., Mccullough, J. E., Tymiak, A. A., Kirsch, D. R., Trejo, W. H., and Principe, P. A. (1988). Lysobactin, a novel antibacterial agent produced by Lysobacter sp. I. Taxonomy, isolation and partial characterization. J. Antibiot. 41, 1740–1744. doi: 10.7164/antibiotics.41.1740
Papenfort, K., and Bassler, B. L. (2016). Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 14, 576–588. doi: 10.1038/nrmicro.2016.89
Penfold, R. J., and Pemberton, J. M. (1992). An improved suicide vector for construction of chromosomal insertion mutations in bacteria. Gene 118, 145–146. doi: 10.1016/0378-1119(92)90263-o
Qian, G., Xu, F., Venturi, V., Du, L., and Liu, F. (2014). Roles of a solo LuxR in the biological control agent Lysobacter enzymogenes strain OH11. Phytopathology 104, 224–231. doi: 10.1094/PHYTO-07-13-0188-R
Quandt, J., and Hynes, M. F. (1993). Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127, 15–21. doi: 10.1016/0378-1119(93)90611-6
Quinones, B., Dulla, G., and Lindow, S. E. (2005). Quorum sensing regulates exopolysaccharide production, motility, and virulence in Pseudomonas syringae. Mol. Plant Microbe Interact. 18, 682–693. doi: 10.1094/mpmi-18-0682
Quinones, B., Pujol, C. J., and Lindow, S. E. (2004). Regulation of AHL production and its contribution to epiphytic fitness in Pseudomonas syringae. Mol. Plant Microbe Interact. 17, 521–531. doi: 10.1094/mpmi.2004.17.5.521
Sambrook, J., and Russell, D. W. (2001). Molecular Cloning : A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Slater, H., Alvarez-Morales, A., Barber, C. E., Daniels, M. J., and Dow, J. M. (2000). A two-component system involving an HD-GYP domain protein links cell-cell signalling to pathogenicity gene expression in Xanthomonas campestris. Mol. Microbiol. 38, 986–1003. doi: 10.1046/j.1365-2958.2000.02196.x
Song, Z. W., Zhao, Y. C., Zhou, X. Y., Wu, G. C., Zhang, Y. Q., Qian, G. L., et al. (2015). Identification and characterization of two novel DSF-controlled virulence-associated genes within the nodB-rhgB locus of Xanthomonas oryzae pv. oryzicola Rs105. Phytopathology 105, 588–596. doi: 10.1094/PHYTO-07-14-0190-R
Tang, B., Zhao, Y. C., Shi, X. M., Xu, H. Y., Zhao, Y. Y., Dai, C. C., et al. (2018). Enhanced heat stable antifungal factor production by Lysobacter enzymogenes OH11 with cheap feedstocks: medium optimization and quantitative determination. Lett. Appl. Microbiol. 66, 439–446. doi: 10.1111/lam.12870
Tsai, C. S., and Winans, S. C. (2010). LuxR-type quorum-sensing regulators that are detached from common scents. Mol. Microbiol. 77, 1072–1082. doi: 10.1111/j.1365-2958.2010.07279.x
Wang, L. H., He, Y., Gao, Y., Wu, J. E., Dong, Y. H., He, C., et al. (2004). A bacterial cell-cell communication signal with cross-kingdom structural analogues. Mol. Microbiol. 51, 903–912. doi: 10.1046/j.1365-2958.2003.03883.x
Yang, M., Sun, K., Zhou, L., Yang, R., Zhong, Z., and Zhu, J. (2009). Functional analysis of three AHL autoinducer synthase genes in Mesorhizobium loti reveals the important role of quorum sensing in symbiotic nodulation. Can. J. Microbiol. 55, 210–214. doi: 10.1139/w08-128
Yu, F. G., Zaleta-Rivera, K., Zhu, X. C., Huffman, J., Millet, J. C., Harris, S. D., et al. (2007). Structure and biosynthesis of heat-stable antifungal factor (HSAF), a broad-spectrum antimycotic with a novel mode of action. Antimicrob. Agents Chemother. 51, 64–72. doi: 10.1128/aac.00931-06
Zhang, W., Li, Y., Qian, G., Wang, Y., Chen, H., Li, Y. Z., et al. (2011). Identification and characterization of the anti-methicillin-resistant Staphylococcus aureus WAP-8294A2 biosynthetic gene cluster from Lysobacter enzymogenes OH11. Antimicrob. Agents Chemother. 55, 5581–5589. doi: 10.1128/AAC.05370-11
Zheng, H., Mao, Y., Zhu, Q., Ling, J., Zhang, N., Naseer, N., et al. (2015). The quorum sensing regulator CinR hierarchically regulates two other quorum sensing pathways in ligand-dependent and -independent fashions in Rhizobium etli. J. Bacteriol. 197, 1573–1581. doi: 10.1128/JB.00003-15
Zheng, H. M., Zhong, Z. T., Lai, X., Chen, W. X., Li, S. P., and Zhu, J. (2006). A LuxR/LuxI-type quorum-sensing system in a plant bacterium, Mesorhizobium tianshanense, controls symbiotic nodulation. J. Bacteriol. 188, 1943–1949. doi: 10.1128/jb.188.5.1943-1949.2006
Zhu, J., Chai, Y., Zhong, Z., Li, S., and Winans, S. C. (2003). Agrobacterium bioassay strain for ultrasensitive detection of N-acylhomoserine lactone-type quorum-sensing molecules: detection of autoinducers in Mesorhizobium huakuii. Appl. Environ. Microbiol. 69, 6949–6953. doi: 10.1128/aem.69.11.6949-6953.2003
Zhu, J., and Mekalanos, J. J. (2003). Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev. Cell 5, 647–656. doi: 10.1016/s1534-5807(03)00295-8
Zhu, J., and Winans, S. C. (1999). Autoinducer binding by the quorum-sensing regulator TraR increases affinity for target promoters in vitro and decreases TraR turnover rates in whole cells. Proc. Natl. Acad. Sci. U.S.A. 96, 4832–4837. doi: 10.1073/pnas.96.9.4832
Keywords: acyl-homoserine lactone, AHL quorum sensing, regulation, growth rate, autolysis, Lysobacter brunescens
Citation: Ling J, Zhou L, Wu G, Zhao Y, Jiang T and Liu F (2019) The AHL Quorum-Sensing System Negatively Regulates Growth and Autolysis in Lysobacter brunescens. Front. Microbiol. 10:2748. doi: 10.3389/fmicb.2019.02748
Received: 22 July 2019; Accepted: 12 November 2019;
Published: 03 December 2019.
Edited by:
Marie-Joelle Virolle, Centre National de la Recherche Scientifique (CNRS), FranceReviewed by:
Tomohiro Tobino, The University of Tokyo, JapanCopyright © 2019 Ling, Zhou, Wu, Zhao, Jiang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fengquan Liu, ZnFsaXUyMDAxMUBzaW5hLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.