
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 08 November 2019
Sec. Food Microbiology
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.02644
 Zhenyu Liu1
Zhenyu Liu1 Lingying Zhao2
Lingying Zhao2 Qin Zhang3
Qin Zhang3 Nan Huo3
Nan Huo3 Xiaojing Shi3
Xiaojing Shi3 Linwei Li1
Linwei Li1 Liyan Jia4
Liyan Jia4 Yuanyuan Lu3
Yuanyuan Lu3 Yong Peng5
Yong Peng5 Yanbo Song3*
Yanbo Song3*The pulsed electric field (PEF) technology has been widely applied to inactivate pathogenic bacteria in food products. Though irreversible pore formation and membrane disruption is considered to be the main contributing factor to PEF’s sterilizing effects, the exact molecular mechanisms remain poorly understood. In this study, by using mass spectrometry (MS)-based label-free quantitative proteomic analysis, we compared the protein profiles of PEF-treated and untreated Escherichia coli. We identified a total of 175 differentially expressed proteins, including 52 candidates that were only detected in at least two of the three samples in one experiment group but not in the other group. Functional analysis revealed that the differential proteins were primarily involved in the regulation of cell membrane composition and integrity, stress response, as well as various metabolic processes. Quantitative reverse-transcription polymerase chain reaction (qRT-PCR) analysis was conducted on the genes of selected differential proteins at varying PEF intensities, which were known to result in different cell killing levels. The qRT-PCR data confirmed that the proteomic results could be reliably used for further data interpretation, and that the changes in the expression levels of the differential candidates were, to a large extent, caused directly by the PEF treatment. The findings of the current study offered valuable insight into PEF-induced cell inactivation.
The pulsed electric field (PEF) technology is a mild, non-thermal method that processes various biological materials with intermittent high-intensity electric energy (Clark, 2006). Compared to other thermal processing techniques, PEF generates substantially less heat, which ensures better preservation of flavor, color and nutrition in food products. PEF also offers obvious advantages over both enzyme- and chemical-based cold processing methods in that it does not introduce any potentially quality-degrading additives. One of the main applications of PEF in food and beverage industry is the inactivation of microbial pathogens. For example, liquid whole egg processed by combination of PEF and mild thermal treatment at 55°C exhibited significantly longer shelf-life at 4°C than those treated with heat alone (Hermawan et al., 2004). In another study, Listeria monocytogenes populations in different liquid milk products were reduced by a factor of 104 with a 0.6-ms PEF treatment at 50°C (Reina et al., 1998). Recently, PEF has also been evaluated for the preservation of solid food products, such as fruits (Sotelo et al., 2015; Yu et al., 2017), and for facilitating the extraction of valuable chemicals from plants and fungi (Goettel et al., 2013; Xue and Farid, 2015).
Currently, studies that aim to investigate the mechanism of PEF-induced cell inactivation have focused predominantly on its deleterious effects on membrane integrity (Unal et al., 2002; Wu, 2008). It is generally accepted that cell membranes exposed to PEF are locally distorted by electromechanical compression, culminating in pore formation and even membrane rupture (Weaver and Chizmadzhev, 1996). In comparison, there have been very few studies on whether PEF also affects other cellular components or processes, particularly proteins (Li et al., 2011; Rivas et al., 2013). In this regard, it has been demonstrated that PEF could induce significant structural changes in solid-state egg white proteins (Qian et al., 2016). Meanwhile, PEF-processed malting barley seeds showed reduced α-amylase expression compared to the untreated controls (Dymek et al., 2012). In recent years, the rapid development of proteomic technologies has allowed researchers to systematically analyze proteome-wide changes in response to external stimuli. For example, Rivas et al. (2013) examined the proteomic changes in PEF-damaged Escherichia coli DH5α cells with or without a 1-h recovery in Luria-Bertani (LB) medium after the treatment. The recovery and non-recovery groups showed different sets of differentially expressed proteins, which were mainly associated with cell metabolism, membrane structure, aerobic respiration, and protein folding (Rivas et al., 2013). There have also been a number of other studies regarding the effects of PEF treatment or electroporation on the mRNA and/or protein expression in various prokaryotic and eukaryotic cells (Mlakar et al., 2009; Heller et al., 2010; Zhao et al., 2018). However, further research is needed in order to provide a more comprehensive and detailed elucidation of how PEF disrupts various aspects of cellular structures and functions to achieve its sterilizing effects.
The E. coli strain CGMCC44102 was obtained from the China General Microbiological Culture Collection Center (CGMCCC), and cultured in LB medium at 150 rpm and 37°C to a final optical density (OD) of 1.0 at 600 nm. Subsequently, 1 mL of the culture broth was collected and centrifuged at 8000 rpm, 4°C for 2 min. The cell pellet was re-suspended in 2 mL of ice-cold 0.1% (w/v) bacteriological peptone solution (Solarbio Life Science, China) and divided into 70 μL aliquots. For PEF treatment, each aliquot was pipetted into a new, sterile 1.0-mm electroporation cuvette and pulsed using a BTX ECM830 Square Wave Electroporation System (BTX, United States) based on a protocol described previously (Liu Z.Y. et al., 2016). The cell density in the aliquot was approximately 108 CFU/mL and electric conductivity was 0.19 mS/cm. The general setup of the device and the wave shape of the pulse were illustrated in Figure 1. After treatment, the cells were serially diluted and counted based on a previously described protocol (Aronsson et al., 2005) to calculate the inhibition rate. To achieve different cell killing levels, we employed different sets of pulse conditions as previously described. For a cell killing extent of 95 ± 2.0%, pulse intensity, number and duration were set to 14.5 kV⋅cm–1, 26 and 67 μs, respectively. The killing extent was 51 ± 1.3% with the treatment parameters of 6.10 kV⋅cm–1, pulse number of 54, pulse duration of 77 μs. And when the pulse intensity was 2.88 kV⋅cm–1, pulse number was 62, pulse duration was 82 μs, the killing extent was 29 ± 0.7%. Based on measurement on an RC05 Infrared Thermal Imaging Camera (Rinch Industrial, China), the culture temperature showed an average increase of less than 3°C following the PEF treatment, which would not significantly reduce the viability of the cells.
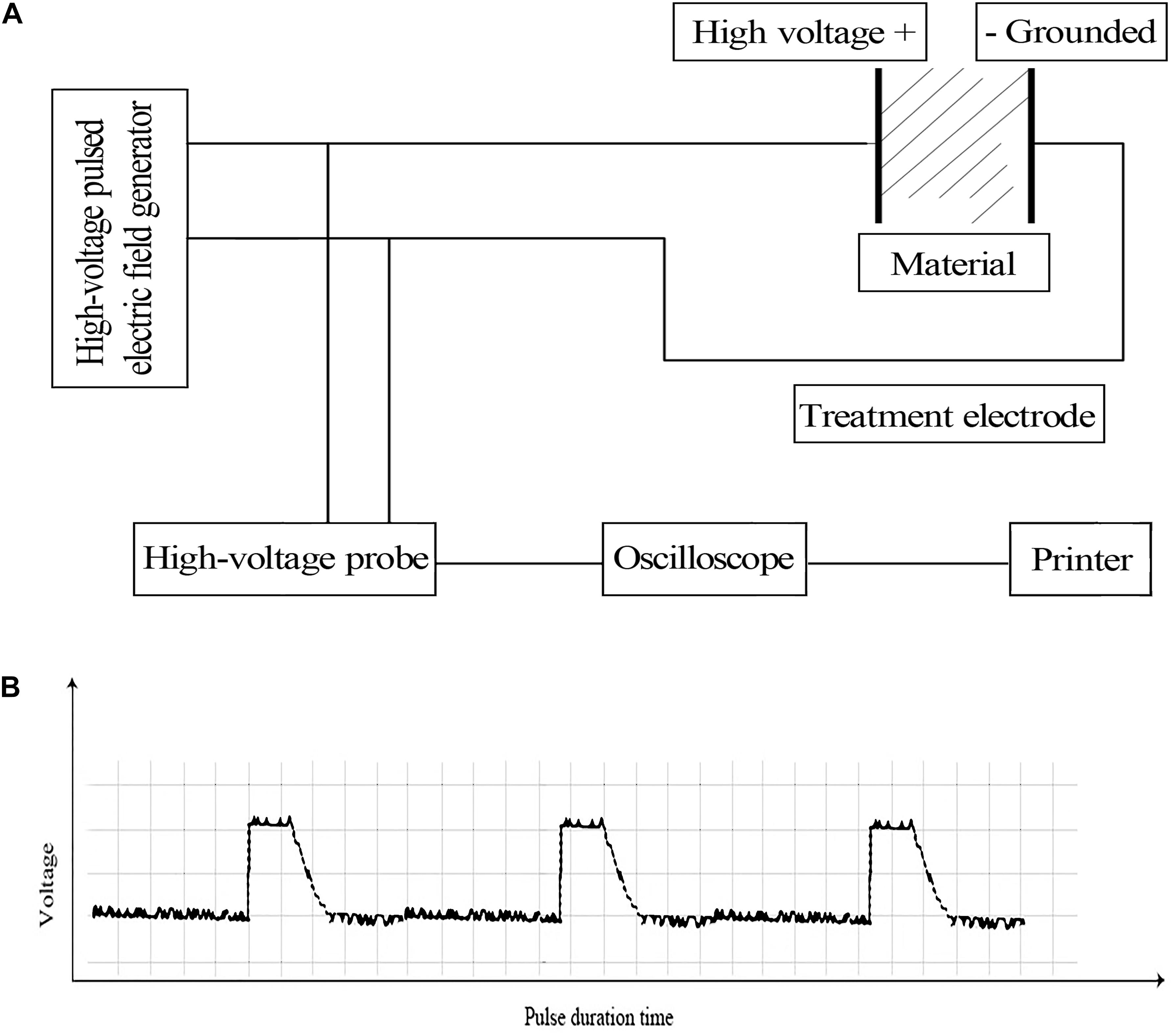
Figure 1. (A) Schematic diagram of the PEF device setup (Liu Z.Y. et al., 2016) and (B) wave shape of the pulse.
The electroporated aliquots were pooled together to a final volume of 3 mL (∼43 aliquots) and divided in a clean 1.5 mL microcentrifuge tube placed on ice. This was used as an experimental replicate. The pooled E. coli suspension was then centrifuged at 4°C, 5,000 × g for 5 min, washed twice with and then re-suspended in ice-cold phosphate-buffered saline (PBS), and then quickly flash-frozen in liquid nitrogen for storage at −80°C. For controls, a mock treatment without the application of PEF was performed, with identical downstream procedures. The whole experiment described above was conducted in triplicate.
The cell pellet of strain was suspended in 200 μL SDT lysis buffer consisting of 4% (w/v) sodium dodecyl sulfate (SDS), 0.15 M Tris–HCl at pH 7.6 and 100 mM dithiothreitol (DTT), and transferred to a 2-mL tube pre-filled with 1/3 vol of quartz sand and ten steel grinding beads. The resultant mixture was subsequently homogenized twice on a Fastprep-24 homogenizer (MP Biochemicals, United States) at 6.0 m/s, followed by ten cycles of 10-s sonication at 80 W with 15-s intervals. The homogenate was boiled for 15 min and centrifuged at 14,000 × g for 40 min, after which the supernatant was sterilized with a 0.22-μm filter. Total protein in the filtrate was quantified with a BCA Protein Assay Kit (Bio-Rad, United States). All filtered protein extracts were stored at −80°C until use.
Based on the BCA assay results, 200 μg of the extracted protein were denatured with 30 μL of SDT buffer and incubated at 56°C for 30 min. Low-molecular-weight components, including SDS and DTT, were removed by repeated ultrafiltration through a Microcon 10-kDa Centrifugal Filter Unit (Millipore, United States) with UA buffer, which consisted of 8 M urea in 150 mM Tris–HCl at pH 8.0, as the wash buffer. The sample was mixed with 100 μL of 100 mM iodoacetamide in UA, followed by a 30-min incubation in darkness. The filter unit was subsequently washed with 100 μL of UA twice and 100 μL of 25 mM NH4HCO3 twice. The resultant protein sample was digested by adding 4 μg of trypsin (Promega, United States) in 40 μL of 25 mM NH4HCO3 to the filter unit and then incubating at 37°C for 18 h. The filter unit was centrifuged at 13,400 rpm for 30 min and the peptide products were collected as a flow-through. The filter unit was next washed with 40 μL of 25 mM NH4HCO3 and centrifuged as above. The two flow-through were combined, desalted on an Empore C-18 Standard Density Solid-Phase Extraction Cartridge (bed I.D. 7 mm, volume 3 mL; Sigma-Aldrich, United States) and concentrated by vacuum centrifugation, followed by reconstituting the peptides in 40 μL of 0.1% (v/v) formic acid. Peptide quantification was performed by measuring the absorbance at 280 nm on an ultraviolet-visible spectrophotometer.
Proteomic analysis was performed on an Easy-nLC 1,000 System (Thermo Fisher Scientific, United States) coupled to a Q Exactive HF Quadrupole-Orbitrap Mass Spectrometer (Thermo Fisher Scientific, United States). Each peptide sample was loaded onto an Acclaim PepMap100 C18 reverse-phase trap column (100 μm × 2 cm, nanoViper fitting; Thermo Fisher Scientific, United States) connected to an Easy C18 reverse-phase analytical column (75 μm × 10 cm, 3 μm resin; Thermo Fisher Scientific, United States) pre-equilibrated in buffer A [0.1% (v/v) formic acid]. The peptides were separated with a linear gradient of buffer B [84% (v/v) acetonitrile and 0.1% (v/v) formic acid] at a constant flow rate of 300 nL/min. The gradient of B was first increased from 0 to 55% over 110 min, then to 100% over 5 min, before being maintained at 100% for another 5 min.
Following the separation, the eluted peptides were immediately analyzed under positive-ion mode with peptide recognition enabled. The ions were first subjected to one survey scan in the m/z range of 300–1,800 at a mass resolution of 70,000 at m/z 200. Automatic gain control target, maximum inject time and dynamic exclusion were set to 3 × 106, 10 ms and 40.0 s, respectively. MS2 spectra were obtained by using a data-dependent top 10 method to dynamically select the most abundant precursor ions from the survey scans for high-energy collisional dissociation (HCD) fragmentation. The resolution of the MS2 scans was set to 17,500 at m/z 200. Isolation width, normalized collision energy and underfill ratio were set to 2 m/z, 30 eV and 0.1%, respectively.
The raw mass spectrometry (MS) data for each sample were combined and imported into MaxQuant (version 1.5.3.17, Max Planck Institute of Biochemistry, Germany) (Cox and Mann, 2008). For protein identification, the MS data were searched against the Uniprot E. coli protein database represented by the file uniprot_Escherichia_coli_1124415_20180910.fasta. Software settings were adjusted as follows: enzyme-trypsin; max missed cleavage-2; fixed modification-carbamidomethyl (C); variable modification-oxidation (M), acetyl (Protein N-term); main search ppm-6; MS/MS tolerance ppm-20; database pattern-reverse; include contaminants-true; false discovery rate (FDR) threshold-0.01; peptides for quantification-unique + razor; match between runs-2 min; protein quantification-label-free quantification (LFQ) (Cox et al., 2014); minimum ratio count-1.
Protein quantification was performed based on the LFQ algorithm as previously described (Cox et al., 2014; Liu et al., 2018). Intensity-based absolute quantification (iBAQ) in MaxQuant was performed on the identified peptides to quantify protein abundance. The significance of the differentially expressed proteins between the samples was examined with the cutoff values fold change >1.5 and p ≤ 0.05 by t-test.
For gene ontology (GO1) analysis, the sequences of differentially expressed proteins were batch-searched against a local SwissProt E. coli protein database using the NCBI BLAST + client software (version 2.2.28) to identify any functionally annotated homologs. These annotations were then transferred and combined with those of the corresponding query E. coli proteins. To reduce the workload, only the top 10 blast hits of each query sequence with an E-value below 1 × 10–3 were selected. These candidates were annotated by Blast2GO (version 3.3.5; Stefan et al., 2008) with an E-value filter of 1 × 10–6, GO weight of 5, annotation cutoff of 75, and default gradual EC weights. Sequences that could not be annotated under these settings were then subjected a new round of annotation using more permissive parameters. Subsequently, the sequences that could not be annotated and those without BLAST hits were then searched against database by using InterProScan (Sun et al., 2016) to retrieve functional annotations of their conserved motifs. The combined GO annotation results were plotted by R scripts. For Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotation, the sequences of differentially expressed proteins were searched via BLAST against the KEGG Database at https://www.genome.jp/kegg/kaas (Moriya et al., 2007).
Next, enrichment analyses were conducted using Fisher’s exact test in order to unearth the cellular and physiological roles of the differentially expressed proteins. The GO terms were categorized into three subcategories, including biological process, molecular function and cellular component. The Benjamini–Hochberg method was used to calculate the adjusted p-values. p < 0.05 was considered statistically significant.
Hierarchical clustering was performed to reveal global differences in protein expression profiles between the PEF treatment group and the control group. To this end, the data from protein relative expression were analyzed by Cluster 3.0 (Lu et al., 2012) using the average-linkage method for clustering, with similarity between genes expressed in Euclidean distance. The obtained dendrogram was visualized in Java Treeview (version 3.0) (Zhu et al., 2014). In addition, a heat map was also generated as a visual aid.
The differentially expressed proteins were imported into STRING to analyze their potential interactions (version 10.5) (Zhou et al., 2012). The results were downloaded in XGMML format and visualized in the form of an interaction network by importing into Cytoscape (version 3.2.1) (Zhu et al., 2014). The degree value of each protein node was calculated to evaluate its importance in the PPI network.
Parallel reaction monitoring analysis of selected proteins were conducted as previously described (Qu et al., 2018). Briefly, digested peptides were prepared as elucidated above. Then, 2 μg of the peptides were spiked with 20 fmol standard (PRTC: GISNEGQNASIK) and separated on an Easy nLC 1,200 System (Thermo Fisher Scientific, United States) with the same buffer A, B and flow rate as above. The gradient of B was set as follows: 5–10% over 2 min, 10–30% over 43 min, 30–100% over 10 min and then 100% for another 5 min. The eluted peptides were immediately analyzed under positive-ion mode on a Q-Exactive HF mass spectrometer (Thermo Fisher Scientific, United States). The ions were first subjected to one survey scan in the m/z range of 300 to 1,800 at a mass resolution of 60,000 at m/z 200. Automatic gain control target and maximum inject time were set to 3 × 106 and 200 ms, respectively. The full MS1 scan were followed by 20 MS2 scans at a resolution of 17,500 at m/z 200 (Liu et al., 2019). Precursor ions were fragmented via the HCD method at normalized collision energy of 27 eV. Isolation window and maximum injection time were set to 1.6 Th and 120 ms, respectively. The obtained raw PRM data were imported into Skyline (version 3.5.0) (MacLean et al., 2010) for analysis.
Following the PEF treatment, total RNA was immediately extracted using anRNAiso Plus Kit (TaKaRa, Japan), followed by reverse transcription with a PrimeScript Reverse Transcriptase Kit (TaKaRa, Japan). All gene-specific primers were designed using Primer Premier 5.0 software (Premier Biosoft International, United States) and synthesized by Sangon Biotech, China. Gene expression levels were analyzed by qRT-PCR using SYBR Green qPCR Mix (TaKaRa, United States) on a CFX96 Real-Time PCR System (Bio-Rad, United States) according to the manufacturers’ instructions. Glucan biosynthesis protein G (mdoG) was used as a control (Heng et al., 2011). All reactions were performed with at least three replicates. Fold changes were calculated based on the 2–ΔΔCt method (Pfaffl, 2001).
All statistical analyses were performed using the SPSS software (version 13.0; IBM, United States). Data were expressed as mean ± standard error of mean (SEM). Differences between two groups were analyzed using Student’s t-test. p < 0.05 was considered statistically significant.
We used an untargeted LC-MS/MS approach to systematically investigate the effects of PEF on E. coli proteome. To this end, we applied PEF to freshly grown E. coli cells under abovementioned conditions that resulted in a 95 ± 2.0% reduction of bacterial population. In the meantime, we set up a control group in which cell aliquots from the same culture were similarly processed, but without the application of PEF. In total, we detected 16,829 peptides from our MS data and identified 2,252 proteins by sequence comparison with the Uniprot E. coli protein database. We then calculated the relative abundances of all identified proteins in the two experimental groups. Comparison of the expression data revealed 123 differentially expressed proteins based on the criteria of fold-change >1.5 or <0.67 and p < 0.05. Among them, 99 proteins exhibited significantly increased expression levels in PEF-processed E. coli, whereas 24 were found to be down-regulated (Table 1). The annotations, accession numbers and fold-change values of all differential proteins were summarized in Table 1. Notably, 33 proteins were confidently detected in at least two PEF-treated E. coli cultures but not in any of the controls, whereas another 19 were identified in at least two control samples but showed no apparent expression in the PEF treatment group. These candidates were therefore also considered as differentially expressed between the two groups and subsequently merged with the 123 proteins mentioned above. It is worth emphasizing that the reason that some proteins were only detected in two of the three samples in an experimental group could be caused by a combination of biochemical, analytical and statistical factors, such as miscleavage, ionization competition, ion suppression, peptide misidentification, ambiguous matching, etc. (Lazar et al., 2016).
To better compare the global proteomic profiles of the PEF-treated E. coli samples and the controls, we performed hierarchical clustering analysis of the 123 differentially expressed proteins and illustrated the results in an expression heat map with dendrogram (Figure 2). As depicted, the plot indicated clear differences in the number of detected proteins between the two experimental groups.
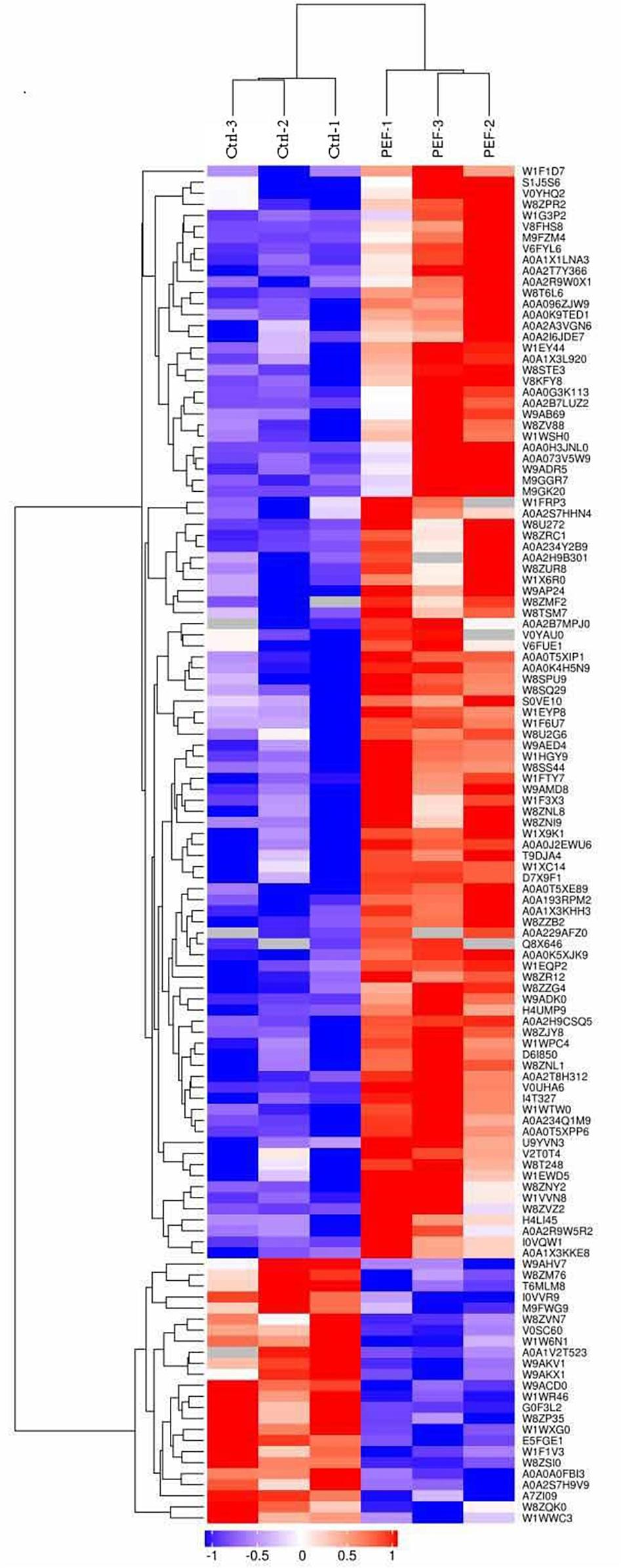
Figure 2. Hierarchical clustering of 123 differentially expressed proteins (confidently detected in all six samples; fold-change >1.5 or <0.67, and p < 0.05; see Table 1). Results are illustrated using a heat map with a dendrogram. The Uniprot ID of each protein is listed in the column to the right of the heat map. The color bar located below the heat map denotes the extent of change in expression level, with red indicating up-regulation and blue down-regulation.
We next performed functional analysis of all 175 differentially expressed proteins (including the ones that could not be confidently detected in all samples as explained above) to shed light on their potential molecular and cellular roles. GO annotation by Blast2GO and InterProScan indicated that the differential proteins were predominantly involved in catalytic activity (47.43%) and binding (38.86%) under the subcategory of molecular function (Figure 3A). For biological process, the overwhelming majority of the candidates could be assigned to GO terms of cellular process (36%) and metabolic process (38.29%) (Figure 3A). Subsequent GO enrichment analysis demonstrated that PEF-treated E. coli underwent a wide range of metabolic alterations, such as those associated with sulfate, sulfide, propionate, 2-methylcitrate and short-chain fatty acids. In addition, we also identified significant changes in the transmembrane transport of inorganic anions, particularly phosphate ions. Other notably enriched GO terms included enzyme binding and enzyme inhibitor activity, including ribonuclease inhibitor activity, endoribonuclease inhibitor activity and methyl isocitrate lyase activity (Figure 3B).
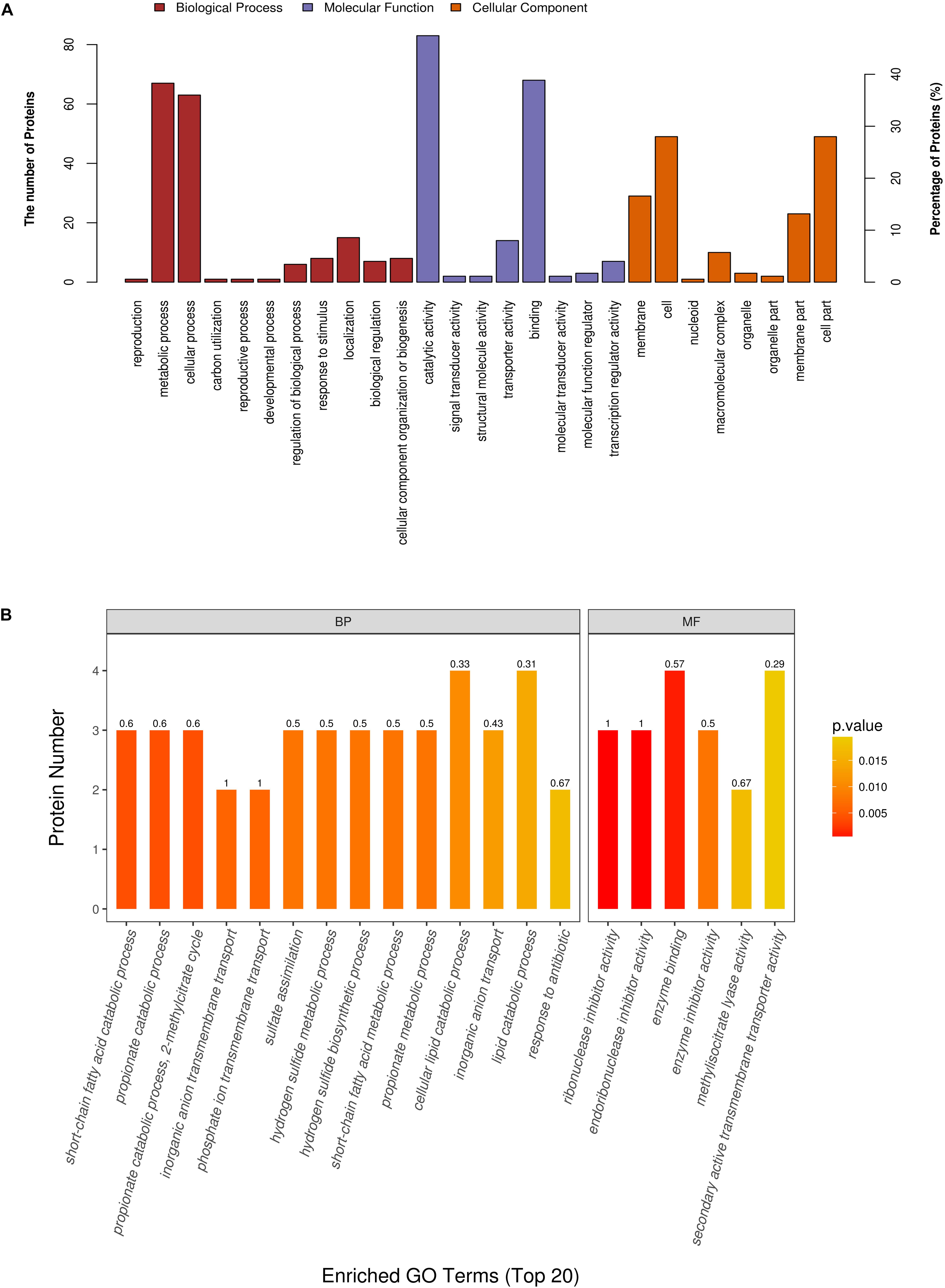
Figure 3. Gene ontology (GO) annotation and enrichment analysis of all 175 differentially expressed proteins. (A) The primary Y axis denotes the number of annotated proteins categorized to each GO term. The secondary Y axis represents the percentage of annotated proteins belonging to each GO term in all differential proteins. GO terms are classified into three subcategories, including biological process (BP, red), molecular function (MF, purple) and, cellular compartment (CC, orange). (B) The color gradient from orange to red represents the p-value; the closer the color to red, the lower the p-value and the higher the significance level corresponding to the enrichment. The small number above each column is the rich factor, which denotes the ratio of the number of differential proteins enriched to a given GO term to the number of all annotated proteins categorized to the same GO term.
On the other hand, the differentially expressed proteins could be mapped to 81 KEGG pathways. The top 5 KEGG pathways with the greatest numbers of protein candidates included those associated with ABC transporters, two-component system, propanoate metabolism, phenylalanine, tyrosine and tryptophan biosynthesis, as well as sulfur metabolism (Figure 4A). Further examination suggested that sulfur metabolism was significantly enriched (Figure 4B), which comprised four differentially expressed proteins W8ZVZ2 (sulfate adenylyltransferase subunit 1, EC:2.7.7.4, CysND), W8ZV88 (Sulfite reductase [NADPH] hemoprotein beta-component, EC:1.8.1.2, CysJI), I0VXI2 (Sulfate-binding protein, CysPUWA) and A0A1V2T430 [Adenylyl-sulfate kinase (Fragment), EC:2.7.1.25, CysC]. Among them, W8ZVZ2 and W8ZV88 were up-regulated in PEF-treated E. coli cells by 2.0-fold and 1.8-fold, respectively, compared to the untreated cells, whereas I0VXI2 and A0A1V2T430 were only detected in PEF-treated cells. The metabolic roles of these proteins were illustrated in Figure 4C.
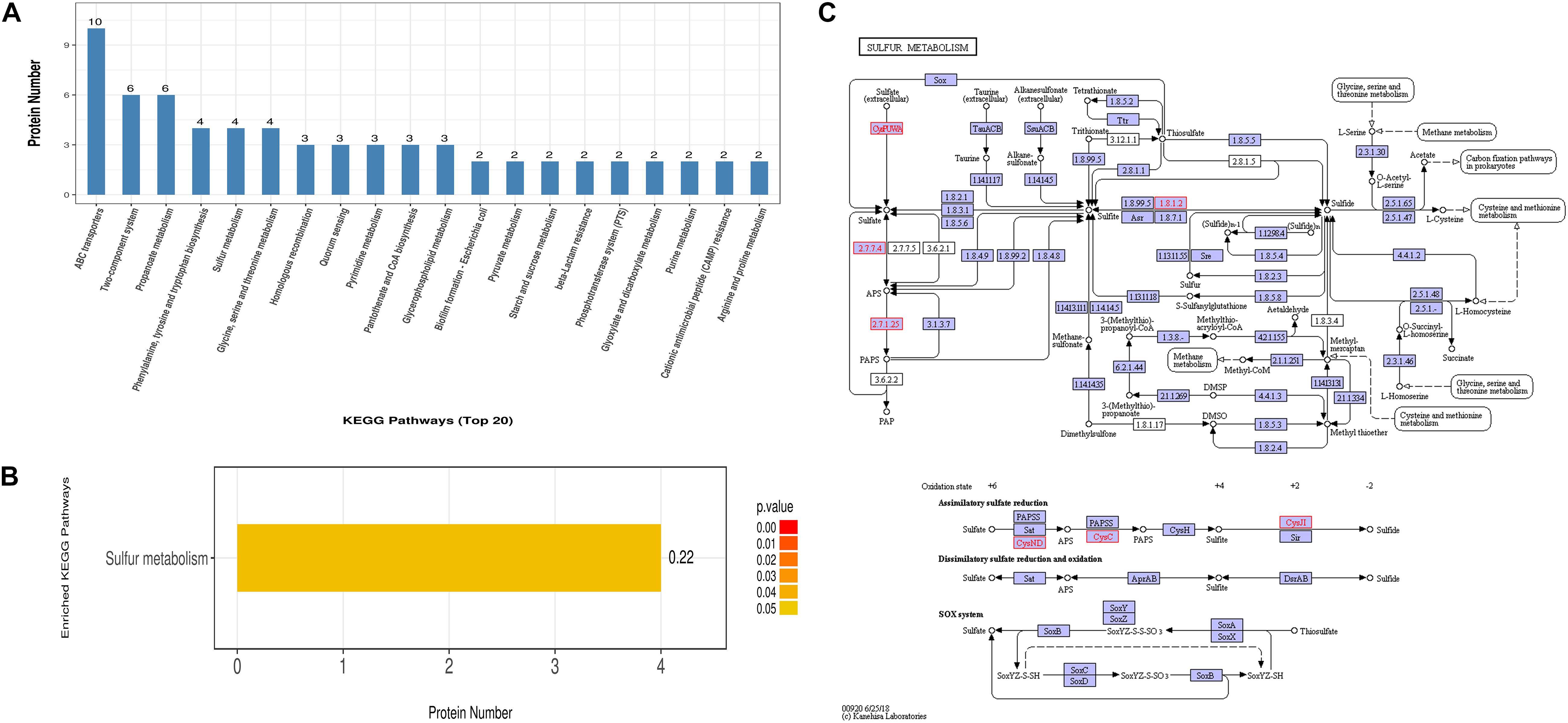
Figure 4. KEGG pathway and enrichment analysis of all 175 differentially expressed proteins. (A) The Y axis indicates the number of annotated proteins that are categorized into each KEGG pathway. Only the top 20 pathways are shown. (B) Sulfur metabolism is the only KEGG pathway found to be significantly enriched. (C) An illustration of the cellular processes associated with sulfur metabolism pathways. All differential proteins captured in this study are highlighted in red. Small circles (o) represent small-molecule metabolites (Source: Kanehisa et al., 2019).
It is a well-established concept that cellular processes are often the results of specific interactions between two or more proteins rather than the actions of a single protein. Based on these considerations, we constructed a PPI network of all the differentially expressed proteins that we identified in order to gain a deeper understanding of how the various PEF-altered biological functions in E. coli cells were interconnected with each other. As shown in Figure 5, the network map comprised 123 nodes and 208 interactions. W8ZUR8 (uncharacterized protein elaB), predicted by GO annotation to participate in ribosome binding, represented the largest node with ten putative associations. Other differential proteins predicted to interact with seven or more partners included W9ADR5 (uncharacterized protein yccJ), W9ADK0 (uncharacterized protein cbJ), A0A1 × 3L920 (beta-lactamase induction protein AmpE), W8SS44 (hydrolase ycaC), W8ZR12 (tryptophan synthase alpha chain, trpA), and V6FUM8 (universal stress protein B UspB). Notably, all these proteins were up-regulated in the PEF treatment group, with V6FUM8 detected only in PEF-processed samples.
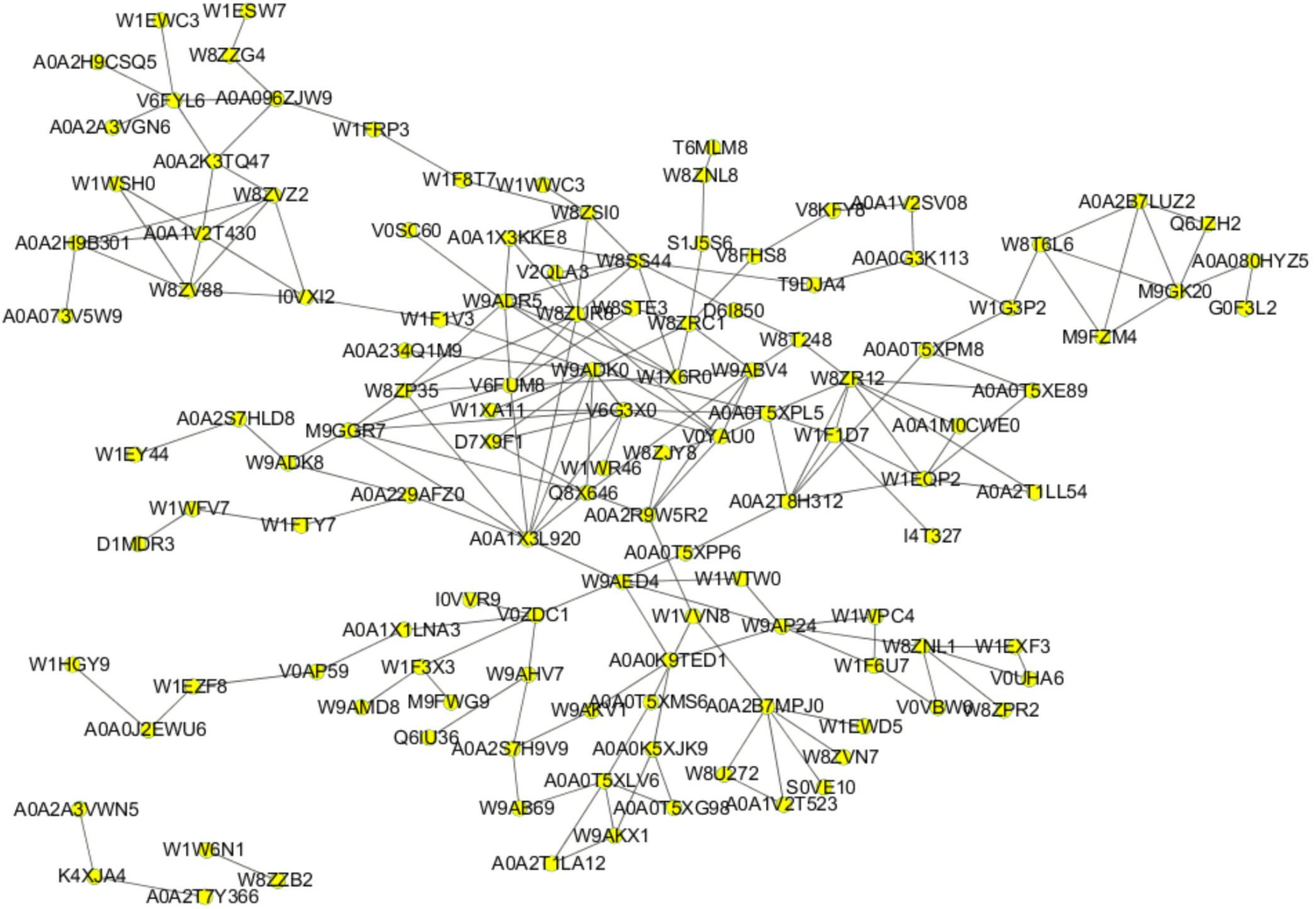
Figure 5. PPI network based on 124 proteins from all 175 differentially expressed proteins. Nodes and lines represent the protein and their interactions, respectively.
The proteomic data were verified by selecting five differentially expressed proteins for PRM analysis, including M9GK20 (Citrate synthase), W8ZVZ2 (Sulfate adenylyltransferase subunit 1), V6FYL6 (Bifunctional polymyxin resistance protein ArnA), M9FZM4 (2-methylcitrate dehydratase) and W9AMD8 (Phosphate-binding protein PstS). We quantified the levels of 1–3 unique peptides for each selected protein in the PEF-processed E. coli and untreated controls. As shown in (Table 2), all five proteins were significantly up-regulated as a result of the PEF treatment, which was consistent with the results of the untargeted proteomic analysis. This suggested that the proteomic data that we obtained were sufficiently reliable for subsequent interpretation.
In the present study, we systematically analyzed PEF-induced changes in the protein expression profile of E. coli through MS-based label-free quantitative proteomics. Our experimental data echoed previous findings (Rivas et al., 2013; Yun et al., 2016; Zhao et al., 2018), that exposure to PEF exerted significant detrimental effects on cell membrane integrity, triggered a host of cellular stress response mechanisms, and altered important metabolic pathways. The results of our current study could increase our understanding of the mechanisms responsible for PEF-induced inactivation of bacterial cells in food products.
We selected a list of significantly up-regulated proteins and measured their mRNA levels at different PEF intensities (which resulted in different cell killing levels). The detailed information and the selection rationale for each protein were summarized in (Table 3). As illustrated in Figure 6, all candidates demonstrated increased transcription at all PEF intensities that we tested, confirming the validity of our proteomic results. However, the genes showed different expression trends with increasing PEF intensities. In most cases, we did not observe a positive correlation between the mRNA level of the gene and the level of cell death. These results suggested that PEF treatment could directly stimulate the expression of the E. coli proteins, though indirect contribution via cell lysis could not be completely eliminated. In addition, at high intensities, PEF exerted a clear inhibitory effect on the transcription of most of the genes.
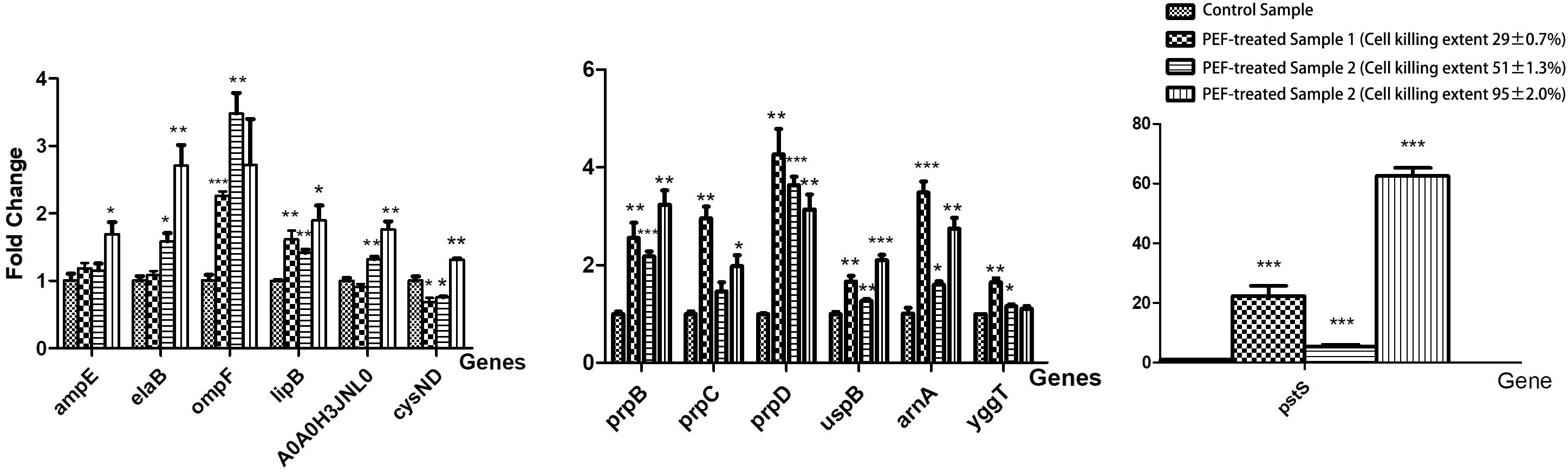
Figure 6. The effect of PEF treatment on the expression of selected E. coli genes under different pulse conditions (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001). The gene expression data are arranged in separate column charts due the fold-change values of pstS being significantly greater than those of other genes. The pulse conditions and cell killing extent for each sample are as follows: control sample: untreated; PEF-treated sample 1: pulse intensity – 2.88 kV cm–1, pulse number – 62, pulse duration – 82 μs, cell killing extent – 29 ± 0.7%; PEF-treated sample 2: pulse intensity – 6.10 kV cm–1, pulse number – 54, pulse duration – 77 μs, cell killing extent – 51 ± 1.3%; PEF-treated sample 3: pulse intensity – 14.5 kV cm–1, pulse number – 26, pulse duration – 67 μs, cell killing extent – 95 ± 2.0%.
Notable among the most up-regulated proteins in the PEF treatment group are several enzymes related to the methylcitrate cycle and the TCA cycle, including 2-methylisocitrate lyase (PrpB; W8T6L6), the dual-functional aconitate hydratase B (alsoaconitase, AcnB; A0A2B7LUZ2), 2-methylcitrate dehydratase (PrpD; M9FZM4), and the rate-limiting citrate synthase (prpC; M9GK20). The methylcitrate cycle functions in a similar manner as the TCA cycle, and is responsible for converting propionyl-CoA to pyruvate for further catabolism (Luo et al., 2016; Serafini et al., 2019). As a result, both the TCA cycle and the methylcitrate cycle are functionally connected to and downstream of the β-oxidation of fatty acids. Since increased lipid oxidation is a well-established phenomenon under oxidative stress (Esterbauer et al., 1991) or PEF treatment (La et al., 2015; Yun et al., 2016), we speculate that the augmented expression of TCA or methylcitrate cycle-associated enzymes could reflect a pre-emptive metabolic defense mechanism of the stressed E. coli cells. This is supported by the results discussed in several studies with regard to the anti-oxidative roles of the TCA cycle (Tretter and Adam, 2000; Mailloux et al., 2010). It is worth mentioning that AcnB contains a critical Fe-S cluster that is particularly susceptible to oxidative stress. In fact, there is evidence that excessive accumulation of reactive oxygen species (ROS) could dramatically reduce the protein level of AcnB in superoxide dismutase-deficient Salmonella enterica (Thorgersen and Downs, 2009). Therefore, the effects of PEF treatment and oxidative stress on the expression of AcnB and other enzymes mentioned above could be both stimulatory and inhibitory. Another top-ranking up-regulated enzyme is bifunctional proline utilization A (PutA; A0A0G3K113), which catalyzes the conversion of proline to glutamate via two consecutive steps of dehydrogenation (Menzel and Roth, 1981). PutA is known to be involved in redox homeostasis and its deletion in E. coli has been shown to substantially increased cell susceptibility to oxidative injuries (Zhang et al., 2015). It has been postulated that the protective role of PutA against oxidative stress could be attributed to the generation of a low level of hydrogen peroxide by proline oxidative metabolism, which builds resistance in cells in a pre-adaptive manner (Zhang et al., 2015). Taken together, these results suggested that PEF treatment and the possibly associated oxidative stress could activate the stress response mechanism in the E. coli cells, leading to induced expression of various oxidation-combating enzymes.
Cell membrane functions as the main barrier against harmful substances and regulates the exchange of essential materials between the cytoplasm and the environment. When exposed to a strong electric field, a cell becomes polarized due to the migration of ions in its cytoplasmic fluid. The realignment of opposite charges on both sides of the cell creates electrostatic attraction that squeezes the cell and its membrane (Zimmermann et al., 1974). Meanwhile, the various membrane constituents of the cell are also perturbed by the electromechanical forces. The polar lipids, including phospholipids and cholesterol, represent obvious targets. As the electric field intensifies, both electromechanical compression and polar lipid rearrangement will reach a breaking point that leads to pore formation (Zimmermann et al., 1974; Serpersu et al., 1985; Weaver and Chizmadzhev, 1996; Unal et al., 2002; Liu Z.W. et al., 2016). One of the direct consequences of this is increased membrane permeability, which not only allows entry of deleterious substances, but also generates an osmotic imbalance (Aronsson et al., 2005). Obviously, the electric induction of a few small pores is usually non-lethal, as the fluidity of the biolipid layer allows rapid restoration of a stable membrane structure (Aronsson et al., 2005). However, a sufficiently strong electric field is capable of creating an increased number of large pores on the membrane, leading to irreversible cell damage, disruption, and eventually death (Locke et al., 2006; Lin et al., 2012). Numerous studies have been performed to determine the boundary between PEF-induced temporary electroporation and permanent sterilization. Based on these results, it is generally believed that irreversible cell injury occurs when the field strength reaches 5–15 kV⋅cm–1 (Guerrero and Welti, 2016). This is consistent with observations that PEF at an intensity above 15 kV⋅cm–1 is necessary for membrane breakdown (Lampe, 1999; Lebovka et al., 2001). However, some researchers have noticed microbial resistance to field strengths as high as 19 kV⋅cm–1 (Ulmer et al., 2002). As a result, it has been argued that PEF with a field strength of 25 kV⋅cm–1 or above should be used to ensure pathogen inactivation in food processing (Toepfl et al., 2007).
Overall, 29 of the 175 differentially expressed proteins that we identified in our current proteomic study are localized on the cell membrane and/or potentially involved in membrane functions. Particularly, 16 of these proteins participate in the transmembrane transport of various small metabolites and macromolecules, including phosphate ions, zinc, glycine betaine, trehalose, as well as short peptides, carbohydrates and lipids in general. There is ample evidence that sugar molecules, especially trehalose, exert a protective effect on the physical structure and integrity of cell membrane (Crowe, 2002). It has been hypothesized that carbohydrates can increase the internal cohesion of cell membrane by forming stabilizing hydrogen bonds with its various components (Locke et al., 2006). Another theory argues that sucrose and trehalose can mitigate mechanical disruption of cell membrane by forming an amorphous glass structure (Crowe et al., 1998). In support of these roles, Pereira et al. demonstrated that trehalose could effectively reduce the physical stress that stretching induced on cell membrane by forming hydrogen bonds with the polar lipid components (Pereira and Hünenberger, 2008). Glycine betaine is another well-known cryoprotectant and osmoprotectant that is often accumulated in cells in response to external abiotic stress stimuli. Several studies have shown that transport of glycine betaine across bacterial cell membrane can be activated by osmotic shock and low temperature (Gerhardt et al., 2000; Guillot et al., 2000). Therefore, it seems more plausible that the altered expression of membrane transporters could be a cellular defense mechanism against PEF-induced structural disruptions, though it is also possible that the electromechanical forces might directly affect the expression or stability of these proteins.
It is worth mentioning that some of the differential membrane proteins were only detectable in the PEF treatment group. A careful survey of their functions implied that these proteins might have been activated by PEF for different reasons. The outer membrane protein F (ompF; W9ADK8) directly contributes to pore formation and has been shown to help E. coli cells maintain their structural stability (Nogami and Mizushima, 1983). However, the effects of ompF seem to be mixed, as Tan et al. (2017) reported that its deletion led to substantial enhancement of membrane integrity. Based on these findings, we speculated that the augmented expression of ompF could either be a sign of increased membrane damage or reflect the cellular response to PEF-induced perturbations of intracellular compounds. Universal stress protein B (uspB; V6FUM8) and other members of the USP family can be induced by a wide range of abiotic stress stimuli (Nachin et al., 2005) and participate in a variety of cytoprotective activities such as DNA protection (Gustavsson et al., 2002), arrest of cell growth (Neidhardt and Nyström, 2010) and re-adaptation of cell metabolism to nutrient shortage (Tkaczuk et al., 2013). The E. coli sensor histidine kinase (A0A2R9W207) belongs to the two-component signal-transducing system implicated in stimuli perception (Mascher et al., 2006). Thus, its stimulated expression could be the result of increased PEF-dependent stresses. The prenyltransferase family protein (ubiA; A0A080HYZ5) is responsible for lipidating a wide range of suitable acceptor compounds. Recent studies pointed out that many lipophilic products of prenyltransferase serve as important components of bacterial cell membrane (Li, 2016), raising the possibility that its up-regulation by PEF could be a cellular repair mechanism. We are especially intrigued to discover that a macrolide-specific efflux protein (MacA; H4L9U9) exhibited detectable expression only in PEF-treated cells. Mostly known for their antibiotic activities, macrolides are recently speculated to play an additional role in membrane organization. For example, Tyteca et al. (2003) examined the mechanism of azithromycin-based inhibition of endocytosis and found that the macrolide compound could diminish membrane fluidity by binding to the polar heads of phospholipids. This is further corroborated by findings that macrolide efflux pumps can be up-regulated by stress signals unrelated to drug resistance (Bolla, 2014).
In addition to Rivas et al.’s (2013) protein profiling study based on 2D gel electrophoresis, there have been several other proteomics or transcriptomics investigations of PEF-treated E. coli. Chueca and coworkers treated E. coli MG1655 cells, buffered at pH 4.0, with 50 exponential waveform electric pulses with a frequency of 0.08 Hz and pulse duration of 2 μs, followed by DNA microarray-based transcriptomic analysis (Chueca et al., 2015). Based on their data, a total of 47 genes showed differential expression as a result of the treatment. One of their major findings was the significantly elevated mRNA expression of several genes associated with the TCA cycle, including (sdhA, sdhB, sdhC, sdhD). Although these genes were different from those that encoded the differential proteins identified in this study, their finding of a link between PEF treatment and alterations in TCA-associated cell metabolism mirrored what our results unearthed in this study. In addition, Chueca et al.’s (2015) study also indicated the up-regulation of genes involved in cell respiration and membrane function, as well as the down-regulation of several mRNAs related to acid shock response, possibly as a result of the mild acidic environment of the cell suspension during the treatment. On the other hand, it has come to our attention that a recent PEF study by Guionet et al. (2014) did not result in as extensive changes in cells as what we have observed in our current study. Specifically, they reported that treating E. coli cells with 500 consecutive 60-ns pulses at 107 V m–1, followed by a 1-h proteomic recovery in LB at room temperature, only led to the differential expression of one protein (Guionet et al., 2014). We speculated that the discrepancies could have stemmed from the fact that they used considerably shorter pulses and detected proteins by two-dimensional gel electrophoresis, which, in theory, tends to afford a lower diversity of protein candidates.
The development of resistance in pathogenic microbes against PEF treatment poses a significant problem to food preservation. In some cases, such resistance seems to be temperature- and/or pH-dependent (Somolinos et al., 2008), suggesting the involvement of enzymes and/or other bioactive molecules that confer damage mitigation. Liu and coworkers pointed out that the composition of membrane fatty acids plays a key role in the PEF resistance of E. coli by regulating membrane fluidity (Liu et al., 2017). A similar conclusion has also been obtained by Cebrián (Cebrián et al., 2016). On the other hand, microbial cells can activate an oxidative stress response upon PEF treatment, which has been shown to produce excessive ROS (Pakhomova et al., 2012). As evidence, Tanino et al. (2012) reported the up-regulation of several genes associated with oxidation stress, including those encoding glutathione synthase and superoxide dismutase, in PEF-stressed yeasts. Furthermore, the authors confirmed that the attenuation of such oxidative injuries strongly correlated with the PEF resistance of the cells (Tanino et al., 2012). García et al. (2010) suggested that enzymes involved in lipid biosynthesis and energy production are required for post-PEF membrane repair. In support of these findings, we demonstrated that PEF treatment significantly augmented the expression of several proteins involved in sulfate transport and reduction. This would in turn result in increased levels of sulfide and other reductive sulfur-containing species, presumably exerting a cytoprotective effect against PEF-induced oxidative stress (Shatalin et al., 2011; Ono et al., 2014; Wu and Gao, 2017). Taken together, these studies offered clear evidence that microorganisms can adopt a number of strategies to combat the detrimental effects of PEF.
In summary, we reported the first systematic proteomic profiling study on PEF-treated E. coli. The results that we obtained suggested that the application of PEF could significantly impact the membrane integrity of the cells and could potentially stimulate a wide range of cellular defense mechanisms. These results can help researchers better understand PEF-induced microbial inactivation and develop more effective PEF-based food processing methods.
The datasets generated for this study can be found in the ProteomeXchange Consortium using accession number PXD014365 and also via the iProX partner repository (Ma et al., 2019).
ZL, LZ, and YS completed the experimental design and modification of the manuscript. ZL, QZ, NH, and XS E. coli completed the experimental part. LL, LJ, and YL completed the writing and translation of the manuscript. YP, XS, and YS completed the data analysis and interpretation of the manuscript.
This work was supported by the National Natural Science Foundation of China (Grant No. 31772651).
YP was employed by company the Shanghai Applied Protein Technology Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Aronsson, K., Rönner, U., and Borch, E. (2005). Inactivation of Escherichia coli, listeria innocua and saccharomyces cerevisiae in relation to membrane permeabilization and subsequent leakage of intracellular compounds due to pulsed electric field processing. Int. J. Food Microbiol. 99, 19–32. doi: 10.1016/j.ijfoodmicro.2004.07.012
Bolla, J. R. (2014). Bacterial Multidrug Efflux Pumps: Structure, Function and Regulation. Ph.D. thesis, Iowa State University, Ames, IA.
Cebrián, G., Condón, S., and Mañas, P. (2016). Influence of growth and treatment temperature on staphylococcus aureus resistance to pulsed electric fields: relationship with membrane fluidity. Innov. Food Sci. Emerg. Technol. 37, 161–169. doi: 10.1016/j.ifset.2016.08.011
Chueca, B., Pagán, R., and García-Gonzalo, D. (2015). Transcriptomic analysis of Escherichia coli MG1655 cells exposed to pulsed electric fields. Innov. Food Sci. Emerg. Technol. 29, 78–86. doi: 10.1016/j.ifset.2014.09.003
Cox, M., Hein, C., Luber, I., Paron, N., Nagaraj, N., and Mann, M. (2014). Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed maxlfq. Mol. Cell. Proteomics 13, 2513–2526. doi: 10.1074/mcp.M113.031591
Cox, M., and Mann, M. (2008). Maxquant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372. doi: 10.1038/nbt.1511
Crowe, J. H., Carpenter, J. F., and Crowe, L. M. (1998). The role of vitrification in anhydrobiosis. Annu. Rev. Physiol. 60, 73–103. doi: 10.1146/annurev.physiol.60.1.73
Crowe, L. M. (2002). Lessons from nature: the role of sugars in anhydrobiosis. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 131, 505–513. doi: 10.1016/s1095-6433(01)00503-7
Dymek, K., Dejmek, P., Panarese, V., António, A. V., Wadsö, L., Finnie, C., et al. (2012). Effect of pulsed electric field on the germination of barley seeds. LWT Food Sci. Technol. 47, 161–166. doi: 10.1016/j.lwt.2011.12.019
Esterbauer, H., Schaur, R. J., and Zollner, H. (1991). Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and elated aldehydes. Free Rad. Biol. Med. 11, 81–128. doi: 10.1016/0891-5849(91)90192-6
García, D., Mañas, P., Gómez, N., Raso, J., and Pagán, R. (2010). Biosynthetic requirements for the repair of sublethal membrane damage in Escherichia coli cells after pulsed electric fields. J. Appl. Microbiol. 100, 428–435. doi: 10.1111/j.1365-2672.2005.02795.x
Gerhardt, P. N. M., Tombras, S. L., and Smith, G. M. (2000). Osmotic and chill activation of glycine betaine porter ii in listeria monocytogenes membrane vesicles. J. Bacteriol. 182, 2544–2550. doi: 10.1128/JB.182.9.2544-2550.2000
Goettel, M., Eing, C., Gusbeth, C., Straessner, R., and Frey, W. (2013). Pulsed electric field assisted extraction of intracellular valuables from microalgae. Algal Res. 2, 401–408. doi: 10.1016/j.algal.2013.07.004
Guerrero, B. J. Á, and Welti, C. J. (2016). “Pulsed electric fields,” in Encyclopedia of Food and Health, eds B. Caballero, P. M. Finglas, and F. Toldrá, (Cambridge, MA: Academic Press), 561–565. doi: 10.1016/B978-0-12-384947-2.00579-1
Guillot, A., Obis, D., and Mistou, M. Y. (2000). Fatty acid membrane composition and activation of glycine-betaine transport in lactococcuslactis, subjected to osmotic stress. Int. J. Food Microbiol. 55, 47–51. doi: 10.1016/S0168-1605(00)00193-8
Guionet, A., Joubert-Durigneux, V., Packan, D., Cheype, C., Garnier, J. P., David, F., et al. (2014). Effect of nanosecond pulsed electric field on Escherichia coli in water: inactivation and impact on protein changes. J. Appl. Microbiol. 117, 721–728. doi: 10.1111/jam.12558
Gustavsson, N., Diez, A., and Nyström, T. (2002). The universal stress protein paralogues of Escherichia coli are co-ordinately regulated and co-operate in the defence against dna damage. Mol. Micobiol. 43, 107–117. doi: 10.1046/j.1365-2958.2002.02720.x
Heller, L. C., Cruz, Y. L., Ferraro, B., Yang, H., and Heller, R. (2010). Plasmid injection and application of electric pulses alter endogenous mRNA and protein expression in B16. F10 mouse melanomas. Cancer Gene Ther. 17, 864–871. doi: 10.1038/cgt.2010.43
Heng, S. S. J., Chan, O. Y. W., Keng, B. M. H., and Ling, M. H. (2011). Glucan biosynthesis protein G is a suitable reference gene in Escherichia coli K-12. ISRN Microbiol. 2011:469053. doi: 10.5402/2011/469053
Hermawan, N., Evrendilek, G. A., Dantzer, W. R., Zhang, Q. H., and Richter, E. R. (2004). Pulsed electric field treatment of liquid whole egg inoculated with Salmonella enteritidis. J. Food Saf. 24, 71–85. doi: 10.1111/j.1745-4565.2004.tb00376.x
Kanehisa, M., Sato, Y., Furumichi, M., Morishima, K., and Tanabe, M. (2019). New approach for understanding genome variations in KEGG. Nucleic Acids Res. 47, D590–D595. doi: 10.1093/nar/gky962
La, H. J., Choi, G. G., Cho, C., Seo, S. H., Srivastava, A., Jo, B. H., et al. (2015). Increased lipid productivity of Acutodesmus dimorphus using optimized pulsed electric field. J. Appl. Phycol. 28, 931–938. doi: 10.1007/s10811-015-0674-6
Lampe, J. W. (1999). Health effects of vegetables and fruits: assessing mechanisms of action in human experimental studies. Am. J. Clin. Nutr. 70, 475–490. doi: 10.1016/j.molstruc.2008.02.018
Lazar, C., Gatto, L., Ferro, M., Bruley, C., and Burger, T. (2016). Accounting for the multiple natures of missing values in label-free quantitative proteomics data sets to compare imputation strategies. J. Proteome Res. 15, 1116–1125. doi: 10.1021/acs.jproteome.5b00981
Lebovka, N. I., Bazhal, M. I., and Vorobiev, E. I. (2001). Pulsed electric field breakage of cellular tissues: visualisation of percolative properties. Innov. Food Sci. Emerg. Technol. 2, 113–125. doi: 10.1016/S1466-8564(01)00024-8
Li, J., Liao, X., Zhong, K., and Zhang, Y. (2011). Inactivation mechanism of pulsed electric fields on Saccharomyces cerevisiae. Trans. Chin. Soc. Agric. Eng. 27, 355–360. doi: 10.3969/j.issn.1002-6819.2011.04.062
Li, W. (2016). Bringing bioactive compounds into membranes: the ubia superfamily of intramembrane aromatic prenyltransferases. Trends Biochem. Sci. 41, 356–370. doi: 10.1016/j.tibs.2016.01.007
Lin, S., Guo, Y., You, Q., Yin, Y., and Liu, J. (2012). Preparation of antioxidant peptide from egg white protein and improvement of its activities assisted by high-intensity pulsed electric field. J. Sci. Food Agric. 92, 1554–1561. doi: 10.1002/jsfa.4742
Liu, J. J., Kirti, S., Luca, Z., Chongguang, C., Humphrey, S. J., Yi-Ting, C., et al. (2018). In vivo brain gpcr signaling elucidated by phosphoproteomics. Science 360:eaao4927. doi: 10.1126/science.aao4927
Liu, Z., Lv, J., Zhang, Z., Li, H., Yang, B., Chen, W., et al. (2019). Integrative transcriptome and proteome analysis identifies major metabolic pathways involved in pepper fruit development. J. Proteome Res. 18, 982–994. doi: 10.1021/acs.jproteome.8b00673
Liu, Z. Y., Song, Y. B., Guo, Y. M., Wang, H. T., and Liu, J. T. (2016). Optimization of pulsed electric field pretreatment parameters for preserving the quality of Raphanus sativus. Drying Technol. 34, 692–702. doi: 10.1080/07373937.2015.1070859
Liu, Z. W., Han, Z., Zeng, X. A., Sun, D. W., and Aadil, R. M. (2016). Effects of vesicle components on the electro-permeability of lipid bilayers of vesicles induced by pulsed electric fields (pef) treatment. J. Food Eng. 179, 88–97. doi: 10.1016/j.jfoodeng.2016.02.003
Liu, Z. W., Zeng, X. A., Ngadi, M., and Han, Z. (2017). Effect of cell membrane fatty acid composition of Escherichia coli on the resistance to pulsed electric field (pef) treatment. LWT Food Sci. Technol. 76, 18–25. doi: 10.1016/j.lwt.2016.10.019
Locke, B. R., Sato, M., Sunka, P., Hoffmann, M. R., and Chang, J. S. (2006). Electrohydraulic discharge and nonthermal plasma for water treatment. Ind. Eng. Chem. Res. 45, 882–905. doi: 10.1021/ie050981u
Lu, Y., Liu, X., Shi, S., Su, H., Bai, X., Cai, G., et al. (2012). Bioinformatics analysis of proteomic profiles during the process of anti-thy1 nephritis. Mol. Cell. Proteomics 11:M111.008755. doi: 10.1074/mcp.M111.008755
Luo, H., Zhou, D., Liu, X., Nie, Z., Quiroga-Sánchez, D. L., and Chang, Y. (2016). Production of 3-Hydroxypropionic acid via the propionyl-CoA pathway using recombinant Escherichia coli strains. PLoS One 11:e0156286. doi: 10.1371/journal.pone.0156286
Ma, J., Chen, T., Wu, S., Yang, C., Bai, M., Shu, K., et al. (2019). iProX: an integrated proteome resource. Nucleic Acids Res. 47, D1211–D1217. doi: 10.1093/nar/gky869
MacLean, B., Tomazela, D. M., Shulman, N., Chambers, M., Finney, G. L., Frewen, B., et al. (2010). Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968. doi: 10.1093/bioinformatics/btq054
Mailloux, R. J., Lemire, J., and Appanna, V. D. (2010). Metabolic networks to combat oxidative stress in Pseudomonas fluorescens. Antonie Van Leeuwenhoek 99, 433–442. doi: 10.1007/s10482-010-9538-x
Mascher, T., Helmann, J. D., and Unden, G. (2006). Perception in bacterial signal-transducing histidine kinases. Microbiol. Mol. Biol. Rev. 70, 910–938. doi: 10.1128/MMBR.00020-06
Menzel, R., and Roth, J. (1981). Purification of the putA gene product. A bifunctional membrane-bound protein from Salmonella typhimurium responsible for the two-step oxidation of proline to glutamate. J. Biol. Chem. 256, 9755–9761.
Mlakar, V., Todorovic, V., Cemazar, M., Glavac, D., and Sersa, G. (2009). Electric pulses used in electrochemotherapy and electrogene therapy do not significantly change the expression profile of genes involved in the development of cancer in malignant melanoma cells. BMC Cancer 9:299. doi: 10.1186/1471-2407-9-299
Moriya, Y., Itoh, M., Okuda, S., Yoshizawa, A. C., and Kanehisa, M. (2007). Kaas: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35, W182–W185. doi: 10.1093/nar/gkm321
Nachin, L., Nannmark, U., and Nystrom, T. (2005). Differential roles of the universal stress proteins of Escherichia coli in oxidative stress resistance, adhesion, and motility. J. Bacteriol. 187, 6265–6272. doi: 10.1128/JB.187.18.6265-6272.2005
Neidhardt, F. C., and Nyström, T. (2010). Expression and role of the universal stress protein, uspa, of Escherichia coli during growth arrest. Mol. Microbiol. 11, 537–544. doi: 10.1111/j.1365-2958.1994.tb00334.x
Nogami, T., and Mizushima, S. (1983). Outer membrane porins are important in maintenance of the surface structure of Escherichia coli cells. J. Bacteriol. 156, 402–408. doi: 10.1111/j.1365-2672.1983.tb02640.x
Ono, K., Akaike, T., Sawa, T., Kumagai, Y., Wink, D. A., Tantillo, D. J., et al. (2014). Redox chemistry and chemical biology of h2s, hydropersulfides, and derived species: implications of their possible biological activity and utility. Free Rad. Biol. Med. 77, 82–94. doi: 10.1016/j.freeradbiomed.2014.09.007
Pakhomova, O. N., Khorokhorina, V. A., Bowman, A. M., Rodaitė-Riševičienė, R., Saulis, G., Xiao, S., et al. (2012). Oxidative effects of nanosecond pulsed electric field exposure in cells and cell-free media. Arch. Biochem. Biophys. 527, 55–64. doi: 10.1016/j.abb.2012.08.004
Pereira, C. S., and Hünenberger, P. H. (2008). Effect of trehalose on a phospholipid membrane under mechanical stress. Biophys. J. 95, 3525–3534. doi: 10.1529/biophysj.108.131656
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Qian, J. Y., Ma, L. J., Wang, L. J., and Jiang, W. (2016). Effect of pulsed electric field on structural properties of protein in solid state. LWT Food Sci. Technol. 74, 331–337. doi: 10.1016/j.lwt.2016.07.068
Qu, H., Yu, H., Gu, R., Chen, D., Chen, X., Huang, Y., et al. (2018). Proteomics for studying the effects of L. rhamnosus LV108 against non-alcoholic fatty liver disease in rats. RSC Adv. 8, 38517–38528. doi: 10.1039/c8ra06771f
Reina, L. D., Tony, J. Z., Howard, Z. Q., and Yousef, A. E. (1998). Inactivation of listeria monocytogenes in milk by pulsed electric field. J. Food Prot. 61, 1203–1206. doi: 10.4315/0362-028X-61.9.1203
Rivas, A., Pina-Pérez, M. C., Rodriguez, V. S., Zuñiga, M., Martinez, A., and Rodrigo, D. (2013). Sublethally damaged cells of Escherichia coli by pulsed electric fields: the chance of transformation and proteomic assays. Food Res. Int. 54, 1120–1127. doi: 10.1016/j.foodres.2013.01.014
Serafini, A., Tan, L., Horswell, S., Howell, S., Greenwood, D. J., Hunt, D. M., et al. (2019). Mycobacterium tuberculosis requires glyoxylate shunt and reverse methylcitrate cycle for lactate and pyruvate metabolism. Mol. Microbiol 112, 1284–1307. doi: 10.1111/mmi.14362
Serpersu, E. H., Kinosita, K., and Tsong, T. Y. (1985). Reversible and irreversible modification of erythrocyte membrane permeability by electric field. Biochim. Biophys. Acta 812, 779–785. doi: 10.1016/0005-2736(85)90272-X
Shatalin, K., Shatalina, E., Mironov, A., and Nudler, E. (2011). H2S: a universal defense against antibiotics in bacteria. Science 334, 986–990. doi: 10.1126/science.1209855
Somolinos, M., García, D., Mañas, P., Condón, S., and Pagán, R. (2008). Effect of environmental factors and cell physiological state on pulsed electric fields resistance and repair capacity of various strains of Escherichia coli. Int. J. Food Microbiol. 124, 260–267. doi: 10.1016/j.ijfoodmicro.2008.03.021
Sotelo, G. K. A., Hamid, N., Oey, I., Noemi, G. M., Ma, Q., and Leong, S. Y. (2015). Effect of pulsed electric fields on the flavour profile of, red-fleshed sweet cherries (prunusavium var. stella). Molecules 20, 5223–5238. doi: 10.3390/molecules20035223
Stefan, G., Juan, M. G., Terol, J., Williams, T. D., Nagaraj, S. H., María, J. N., et al. (2008). High-throughput functional annotation and data mining with the blast2go suite. Nucleic Acids Res. 36, 3420–3435. doi: 10.1093/nar/gkn176
Sun, Q., Zhang, N., Wang, J., Cao, Y., Li, X., Zhang, H., et al. (2016). A label-free differential proteomics analysis reveals the effect of melatonin on promoting fruit ripening and anthocyanin accumulation upon postharvest in tomato. J. Pineal Res. 61, 138–153. doi: 10.1111/jpi.12315
Tan, Z., Black, W., Yoon, J. M., Shanks, J. V., and Jarboe, L. R. (2017). Improving Escherichia coli membrane integrity and fatty acid production by expression tuning of fadl and OmpF. Microb. Cell Factor. 16:38. doi: 10.1186/s12934-017-0650-8
Tanino, T., Sato, S., Oshige, M., and Ohshima, T. (2012). Analysis of the stress response of yeast saccharomyces cerevisiae toward pulsed electric field. J. Electrostat. 70, 212–216. doi: 10.1016/j.elstat.2012.01.003
Thorgersen, M. P., and Downs, D. M. (2009). Oxidative stress and disruption of labile iron generate specific auxotrophic requirements in Salmonella enterica. Microbiology 155, 295–304. doi: 10.1099/mic.0.020727-0
Tkaczuk, K. L., A Shumilin, I., Chruszcz, M., Evdokimova, E., Savchenko, A., and Minor, W. (2013). Structural and functional insight into the universal stress protein family. Evol. Appl. 6, 434–449. doi: 10.1111/eva.12057
Toepfl, S., Heinz, V., and Knorr, D. (2007). High intensity pulsed electric fields applied for food preservation. Chem. Eng. Process. 46, 537–546. doi: 10.1016/j.cep.2006.07.011
Tretter, L., and Adam, V. V. (2000). Inhibition of Krebs cycle enzymes by hydrogen peroxide: a key role of α-ketoglutarate dehydrogenase in limiting NADH production under oxidative stress. J. Neurosci. 20, 8972–8979. doi: 10.1523/JNEUROSCI.20-24-08972.2000
Tyteca, D., Schanck, A. Y., Dufrêne, F., Deleu, M., Courtoy, P. J., Tulkens, P. M., et al. (2003). The macrolide antibiotic azithromycin interacts with lipids and affects membrane organization and fluidity: studies on langmuir-blodgett monolayers, liposomes and j774 macrophages. J. Membr. Biol. 192, 203–215. doi: 10.1007/s00232-002-1076-7
Ulmer, H. M., Heinz, V., Gänzle, M. G., Knorr, D., and Vogel, R. F. (2002). Effects of pulsed electric fields on inactivation and metabolic activity of Lactobacillus plantarum in model beer. J. Appl. Microbiol. 93, 326–335. doi: 10.1046/j.1365-2672.2002.01699.x
Unal, R., Yousef, A. E., and Dunne, C. P. (2002). Spectrofluometric assessment of bacterial cell damage by pulsed electric field. Innov. Food Sci. Emerg. Technol. 3, 247–254. doi: 10.1016/S1466-8564(02)00033-4
Weaver, J. C., and Chizmadzhev, Y. A. (1996). Theory of electroporation: a review. Bioelectrochem. Bioenerg. 41, 135–160. doi: 10.1016/s0302-4598(96)05062-3
Wu, G., and Gao, H. (2017). Endogenous production and physiological functions of hydrogen sulfide in facultative anaerobic bacteria. Acta Microbiol. Sin. 57, 170–178.
Wu, V. C. H. (2008). A review of microbial injury and recovery methods in food. Food Microbiol. 25, 735–744. doi: 10.1016/j.fm.2008.04.011
Xue, D., and Farid, M. M. (2015). Pulsed electric field extraction of valuable compounds from white button mushroom (Agaricus bisporus). Innov. Food Sci. Emerg. Technol. 29, 178–186. doi: 10.1016/j.ifset.2015.03.012
Yu, Y., Jin, T. Z., and Xiao, G. (2017). Effects of pulsed electric fields pretreatment and drying method on drying characteristics and nutritive quality of blueberries. J. Food Process. Preserv. 41:e13303. doi: 10.1111/jfpp.13303
Yun, O., Zeng, X. A., Brennan, C. S., Han, Z., Yun, O., Zeng, X. A., et al. (2016). Effect of pulsed electric field on membrane lipids and oxidative injury of Salmonella typhimurium. Int. J. Mol. Sci. 17:1374. doi: 10.3390/ijms17081374
Zhang, L., Alfano, J. R., and Becker, D. F. (2015). Proline metabolism increases katG expression and oxidative stress resistance in Escherichia coli. J. Bacteriol. 197, 431–440. doi: 10.1128/JB.02282-14
Zhao, M., Zhao, D., Ma, Y., Hu, Z., and Wei, Z. (2018). Quantitative proteomic analysis of cell responses to electroporation, a classical gene delivery approach. Proteomics 18:1800127. doi: 10.1002/pmic.201800127
Zhou, S., Yi, T., Liu, R., Bian, C., Qi, X., He, X., et al. (2012). Proteomics identification of annexin a2 as a key mediator in the metastasis and proangiogenesis of endometrial cells in human adenomyosis. Mol. Cell. Proteomics 11, 3929–3936. doi: 10.1074/mcp.M112.017988
Zhu, G., Cai, G., Liu, Y., Tan, H., Yu, C., Huang, M., et al. (2014). Quantitative itraqlc-ms/ms proteomics reveals transcription factor crosstalk and regulatory networks in hypopharyngeal squamous cell carcinoma. J. Cancer 5, 525–536. doi: 10.7150/jca.9207
Keywords: pulsed electric field, E. coli, proteomics, cell inactivation, molecular mechanisms
Citation: Liu Z, Zhao L, Zhang Q, Huo N, Shi X, Li L, Jia L, Lu Y, Peng Y and Song Y (2019) Proteomics-Based Mechanistic Investigation of Escherichia coli Inactivation by Pulsed Electric Field. Front. Microbiol. 10:2644. doi: 10.3389/fmicb.2019.02644
Received: 02 July 2019; Accepted: 30 October 2019;
Published: 08 November 2019.
Edited by:
Eugenia Bezirtzoglou, Democritus University of Thrace, GreeceReviewed by:
Peter Setlow, University of Connecticut Health Center, United StatesCopyright © 2019 Liu, Zhao, Zhang, Huo, Shi, Li, Jia, Lu, Peng and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanbo Song, MTE1MzYyNTAzOUBxcS5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.