- 1Department of Technical Biochemistry, Institute of Biochemistry and Technical Biochemistry, University of Stuttgart, Stuttgart, Germany
- 2LIEC UMR7360, Faculté des Sciences et Technologies, Université de Lorraine, Vandoeuvre-lès-Nancy, France
- 3CNRS, LIEC UMR7360, Faculté des Sciences et Technologies, Vandoeuvre-lès-Nancy, France
In the soil-dwelling organism Pseudomonas putida KT2440, the rare earth element (REE)-utilizing, and pyrroloquinoline quinone (PQQ)-dependent ethanol dehydrogenase PedH is part of a periplasmic oxidation system that is vital for growth on various alcoholic volatiles. Production of PedH and its Ca2+-dependent counterpart PedE is inversely regulated in response to lanthanide (Ln3+) bioavailability, a mechanism termed the REE-switch. In the present study, we demonstrate that copper, zinc, and in particular, iron availability influences this regulation in a pyoverdine-independent manner by increasing the minimal Ln3+ concentration required for the REE-switch to occur by several orders of magnitude. A combined genetic and physiological approach reveals that an ABC-type transporter system encoded by the gene cluster pedA1A2BC is essential for efficient growth on 2-phenylethanol with low (nanomolar) Ln3+ concentrations. In the absence of pedA1A2BC, a ∼100-fold higher La3+-concentration is needed for PedH-dependent growth but not for the ability to repress growth based on PedE activity. From these results, we conclude that cytoplasmic uptake of lanthanides through PedA1A2BC is essential to facilitate REE-dependent growth on 2-phenylethanol under environmental conditions with poor REE bioavailability. Our data further suggest that the La3+/Fe2+/3+ ratio impacts the REE-switch through the mismetallation of putative La3+-binding proteins, such as the sensor histidine kinase PedS2, in the presence of high iron concentrations. As such, this study provides an example for the complexity of bacteria-metal interactions and highlights the importance of medium compositions when studying physiological traits in vitro in particular in regard to REE-dependent phenomena.
Introduction
Metal ions are essential for all living organisms as they play important roles in stabilizing macromolecular cellular structures, by catalyzing biochemical reactions or acting as cofactors for enzymes (Gray, 2003; Merchant and Helmann, 2012). They can, however, also be toxic to cells at elevated levels through the generation of reactive oxygen species or by aspecific interactions such as mismetallation (Cornelis et al., 2011; Dixon and Stockwell, 2014; Foster et al., 2014). Bacteria have hence developed a sophisticated toolset to maintain cellular metal homeostasis (Andrews et al., 2003; Schalk and Cunrath, 2016; Chandrangsu et al., 2017; Semrau et al., 2018). Common mechanisms include release of metal-specific scavenger molecules, the activation of high-affinity transport systems, the production of metal storage proteins, and the expression of specific efflux pumps.
As it is the case for all strictly aerobic bacteria, the soil-dwelling organism Pseudomonas putida KT2440 has a high demand for iron. However, the bioavailability of this metal is very poor under most oxic environmental conditions due to the fast oxidation of Fe2+- and the low solubility of Fe3+-species (Andrews et al., 2003). One strategy of many bacteria to overcome this challenge is to excrete self-made peptide-based siderophores (such as pyoverdines) into the environment that bind Fe3+ with high affinity, and thereby increase its bioavailability (Salah El Din et al., 1997; Cornelis and Andrews, 2010; Baune et al., 2017). A second adaptation of P. putida cells to iron-limitation is a change in the proteomic inventory to limit the use of Fe-containing enzymes, exemplified by the switch from the Fe-dependent superoxide dismutase (SOD) to a Mn-dependent isoenzyme or by re-routing of entire metabolic pathways (Kim et al., 1999; Sasnow et al., 2016). In contrast, when Fe bioavailability is high, the production of the bacterioferritins Bfrα and Bfrβ is increased to enable intracellular storage and thereby improve cellular fitness under potential future conditions of iron starvation (Chen et al., 2010). The regulatory mechanisms for metal homeostasis of P. putida cells in response to other essential metal ions such as Co, Cu, Mg, Mo, Ni, and Zn are less well explored. Genes encoding for transport systems associated with the uptake and efflux of these metals can, however, be found in its genome (Nelson et al., 2002; Belda et al., 2016), and some of these have been studied in more detail (Miller et al., 2009; Ray et al., 2013).
We have recently reported that P. putida KT2440 is capable of using rare earth elements (REEs) of the lanthanide series (Ln3+) when growing on several alcoholic substrates (Wehrmann et al., 2017, 2019). Under these conditions, the cells use the pyrroloquinoline quinone (PQQ)-dependent ethanol dehydrogenase (EDH) PedH, to catalyze their initial oxidation within the periplasm. Like many other organism, P. putida harbors an additional, Ln3+-independent functional homolog of PedH termed PedE that depends on a Ca2+ ion as metal cofactor (Takeda et al., 2013; Wehrmann et al., 2017). Depending on the availability of REEs in the environment, P. putida tightly regulates PedE and PedH production (Wehrmann et al., 2017, 2018). In the absence of Ln3+, growth is solely dependent on PedE whereas PedH transcription is repressed. The situation immediately changes in the presence of small amounts of Ln3+ (low nM range) leading to a strong induction of the Ln3+-dependent enzyme PedH and repression of its Ca2+-dependent counterpart PedE. For P. putida KT2440 the PedS2/PedR2 two component system (TCS) is a central component of this inverse regulation (Wehrmann et al., 2018). Notably, the REE-switch in P. putida was also found to be influenced by environmental conditions, as the critical La3+ concentrations required to support PedH-dependent growth differ dramatically depending on the medium used, ranging from 5 nM up to 10 μM (Wehrmann et al., 2017).
Lanthanides are only poorly available in natural environments (often picomolar concentrations) due to the formation of low soluble hydroxide and/or phosphate complexes (Meloche and Vrátný, 1959; Firsching and Brune, 1991). The presence of active uptake systems to facilitate REE-dependent growth in bacteria has thus been favored by many researchers (Markert, 1987; Tyler, 2004; Aide and Aide, 2012; Gu et al., 2016; Gu and Semrau, 2017; Cotruvo et al., 2018; Picone and Op den Camp, 2019). A transcriptomic study of M. trichosporium OB3b cells observed that multiple genes encoding for different active transport systems were among the most regulated in the presence of cerium (Gu and Semrau, 2017). In addition, it has been found that a specific TonB-dependent receptor protein as well as a TonB-like transporter protein are highly conserved in bacteria that carry genes encoding for Ln3+-dependent MDHs (Keltjens et al., 2014; Wu et al., 2015). Only very recently, different studies indeed identified both an ABC-transporter and TonB-dependent receptor proteins that are needed for REE-dependent growth of methano- and methylotrophs on methanol, strongly suggesting the existence of an uptake system that specifically transports a Ln3+-chelator complex in these organisms (Groom et al., 2019; Ochsner et al., 2019; Roszczenko-Jasińska et al., 2019).
With the present study, we show that a homologous ABC-transporter system, encoded by the gene cluster pedA1A2BC, is essential for lanthanide-dependent growth on 2-phenylethanol in the non-methylotrophic organism P. putida KT2440 under low (nanomolar) concentrations of REEs. Notably, no homolog of the TonB-dependent receptor proteins found in methanotrophic or methylotrophic strains could be identified within the genome of P. putida KT2440 indicating either a lack or substantial differences in the chemical nature of such a Ln3+-specific chelator system. Finally, we show that the siderophore pyoverdine plays no essential role for growth on 2-phenylethanol under low REE concentrations but provide compelling evidence that in addition to Cu2+ and Zn2+ the Fe2+/3+ to Ln3+ ratio significantly alters the REE-switch most likely through mismetallation.
Materials and Methods
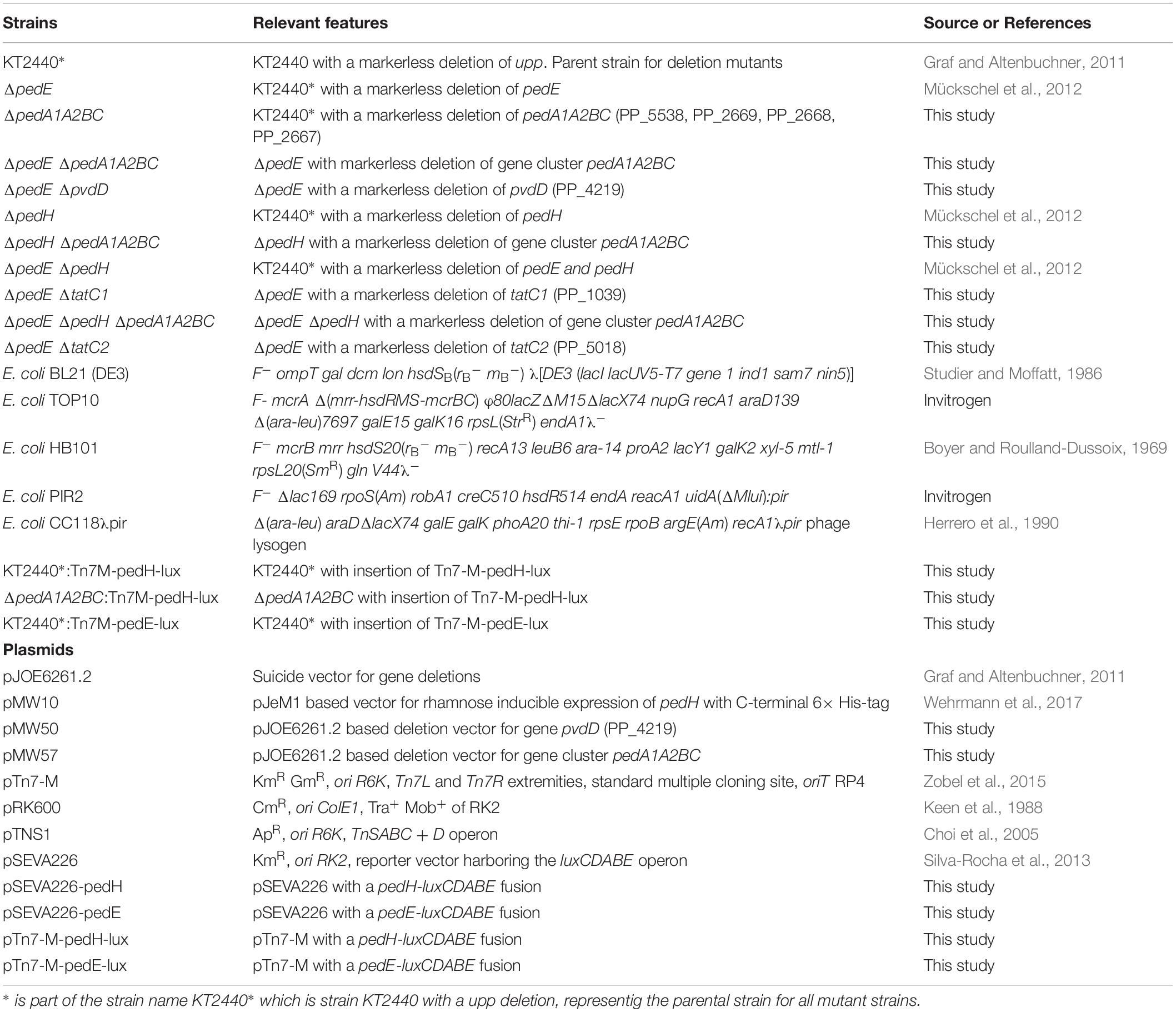
Bacterial Strains, Plasmids, and Culture Conditions
The Escherichia coli and Pseudomonas putida KT2440 strains and the plasmids used in this study are described in Table 1. Maintenance of strains was routinely performed on solidified (1.5% agar) LB medium (Maniatis et al., 1982). If not stated otherwise, strains were grown in liquid LB medium (Maniatis et al., 1982) or a modified M9 salt medium (Wehrmann et al., 2017) supplemented with 5 mM 2-phenylethanol, 5 mM 2-phenylacetaldehyde, 5 mM phenylacetic acid, or 25 mM succinate (pre-cultures) as carbon and energy source at 28°C to 30°C and shaking. 40 μg mL–1 kanamycin or 15 μg mL–1 gentamycin for E. coli and 40 μg mL–1 kanamycin, 20 μg mL–1 5-fluorouracil, or 30 μg mL–1 gentamycin for P. putida strains was added to the medium for maintenance and selection, if indicated.
Liquid Medium Growth Experiments
Liquid growth experiments were performed in biological triplicates by monitoring the optical density at 600 nm (OD600) during growth in modified M9 medium supplemented with the corresponding carbon and energy sources (see above). For all experiments, washed cells from overnight cultures grown with succinate at 30°C and 180 rpm shaking were utilized to inoculate fresh medium with an OD600 of 0.01 to 0.05. Depending on the culture vessel, the incubation was carried out in 1 ml medium per well for 96-well 2 ml deep-well plates (Carl Roth) at 350 rpm shaking and 30°C or 200 μL medium per well for 96-well microtiter plates (Sarstedt) at 180 rpm shaking and 28°C. If needed, different concentrations of LaCl3 (Sigma-Aldrich) were added to the medium.
Construction of Plasmids
The 600 bp regions upstream and downstream of gene pvdD, gene cluster pedA1A2BC and genes tatC1 and tatC2 were amplified from genomic DNA of P. putida KT2440 using primer pairs MWH56/MWH57 and MWH58/MWH59, MWH94/MWH95 and MWH96/MWH97, PBtatC1.1/PBtatC1.2 and PBtatC1.3/PBtaC1.4 and PBtatC2.1/PBtatC2.2 and PBtatC2.3/PBtatC2.4 to construct the deletion plasmids pMW50, pMW57, pJOE-tatC1 and pJOE-tatC2 (Table 2). The BamHI digested pJOE6261.2 as well as the two up- and downstream fragments were therefore joined together using one-step isothermal assembly (Gibson, 2011) and subsequently transformed into E. coli BL21(DE3) or TOP10 cells. Sanger sequencing confirmed the correctness of the plasmids.
For measuring promoter activity of pedE and pedH in vivo, plasmids pTn7-M-pedH-lux and pTn7-M-pedE-lux were constructed. The DNA regions encompassing the promoters from pedE and pedH genes were amplified by PCR using the primer pairs p2674-FSac/p2674-RPst and p2679-FSac/p2679-RPst (Wehrmann et al., 2017). The PCR products were digested with SacI and PstI and inserted upstream the luxCDABE operon hosted by plasmid pSEVA226. The cargo module bearing the pedE-lux or pedH-lux fusion was then passed from the resulting pSEVA226-based constructs to pTn7-M as PacI/SpeI fragments.
Strain Constructions
For the deletion of chromosomal genes a previously described method for markerless gene deletions in P. putida KT2440 was used (Graf and Altenbuchner, 2011). In short, after transformation of the integration vectors carrying the up- and downstream region of the target gene, clones that were kanamycin (Kan) resistant and 5-fluorouracil (5-FU) sensitive were selected and one clone was incubated in liquid LB medium for 24 h at 30°C and 180 rpm shaking. Upon selection for 5-FU resistance and Kan sensitivity on minimal medium agar plates, clones that carried the desired gene deletion were identified by colony PCR.
Integration of the pTn7-M based pedH-lux and pedE-lux fusions into the chromosome of P. putida KT2440 was performed by tetraparental mating using PIR2/pTn7-M-pedH-lux or PIR2/pTn7-M-pedE-lux as the donor, E. coli CC118 λpir/pTNS1 and E. coli HB101/pRK600 as helper strains and appropriate KT2440 strain as the recipient (Zobel et al., 2015). Briefly, cultures of the four strains grown under selective conditions were mixed, spotted on LB agar and incubated overnight at 28°C. Transconjugants were selected on cetrimide agar (Sigma-Aldrich) containing gentamicin. Correct chromosomal integration of mini-Tn7 was checked by colony PCR using Pput-glmSDN and PTn7R primers as described elsewhere (Choi et al., 2005).
Promoter Activity Assays
Pseudomonas putida harboring a Tn7-based pedH-lux or pedE-lux fusion were grown overnight in M9 medium with 25 mM succinate, washed three times in M9 medium with no added carbon source and suspended to an OD600 of 0.1 in the same medium with 1 mM 2-phenylethanol. For luminescence measurements, 198 μl of cell suspension was added to 2 μl of a 100-fold-concentrated metal salt solution in white 96-well plates with a clear bottom (μClear; Greiner Bio-One). Microtiter plates were incubated in a FLX-Xenius plate reader (SAFAS, Monaco) at 30°C with orbital shaking (600 rpm, amplitude 3 mm) and light emission and OD600 were recorded after the indicated time periods. Promoter activity was expressed as relative light units (RLU) normalized to the corresponding OD600. Experiments were performed in triplicates and data are presented as the mean value with error bars representing the standard deviation.
Results
The REE-Switch in P. putida KT2440 Is Influenced by the Presence of Iron, Copper, and Zinc
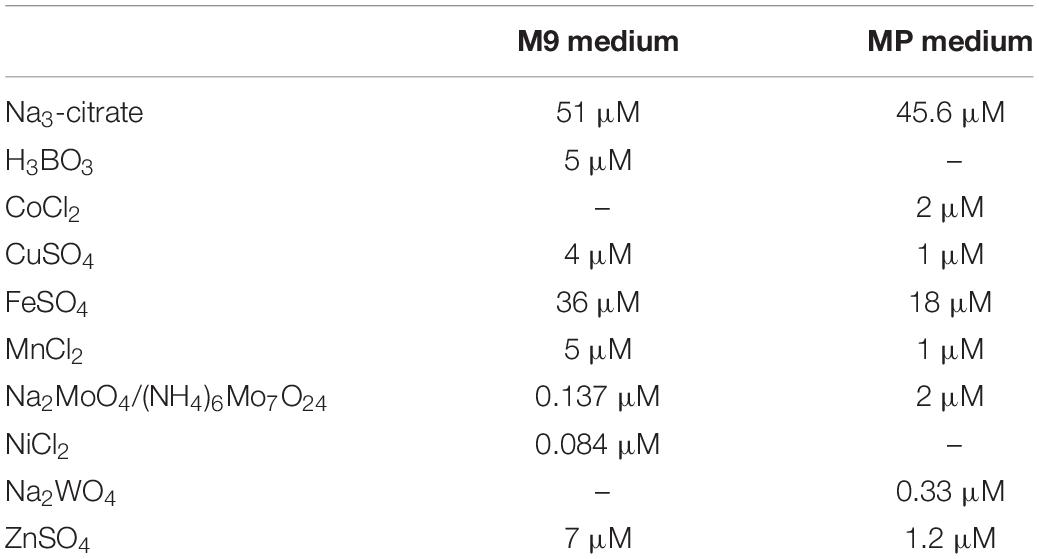
Pseudomonas putida KT2440 makes use of a periplasmic oxidation system to grow on a variety of alcoholic substrates. Crucial to this system are two PQQ-dependent ethanol dehydrogenases (PQQ-EDHs), which share a similar substrate scope but differ in their metal cofactor dependency (Wehrmann et al., 2017). PedE makes use of a Ca2+-ion whereas PedH relies on the bioavailability of different REE. During our studies, we found that the critical REE concentration that supports growth based on PedH activity differs dramatically depending on the minimal medium used. In a modified M9 medium, concentrations of about 10 μM of La3+ were necessary to observe PedH-dependent growth on 2-phenylethanol while only about 20–100 nM La3+ were required in MP medium (Wehrmann et al., 2017). One major difference between the two minimal media lies in their trace element composition and the respective metal ion concentrations (Table 3). The concentrations of copper, iron, manganese and zinc are between 2× and 7× higher in the modified M9 medium compared to MP medium, and other trace elements such as boron, cobalt, nickel or tungsten are only present in one out of the two media. To study the impact of the trace element solution (TES) on growth in the presence of La3+, we used the ΔpedE strain growing on 2-phenylethanol in M9 minimal medium in the presence and absence of TES.
While a critical La3+ concentration of 10 μM or higher was needed in the presence of TES to support PedH-dependent growth, this concentration dropped to as little as 10 nM La3+ in the absence of TES (Figure 1A). Similarly, inhibition of PedE-dependent growth by La3+ in strain ΔpedH differed dramatically depending on the presence of TES (Figure 1B). In the presence of TES, the addition of ≥100 μM of La3+ was required for growth inhibition in the ΔpedH strain within 48 h of incubation, whereas a minimum of only ≥1 μM La3+ was required in the absence of TES. From these experiments, we conclude that also a non-complemented minimal medium contains low, but sufficient, amounts of essential trace elements to allow growth even in the absence of TES. To find out whether the trace element mixture or a single trace element was causing the observed differences, we analyzed the growth of strain ΔpedE in more detail (Figure 2A). For concentrations of H3BO3, Na2MoO4, NiSO4, and MnCl2 similar to those found in complemented M9 medium, PedH-dependent growth on 2-phenylethanol in the presence of 10 nM La3+ was observed. In contrast, upon the individual supplementation with 4 μM CuSO4, 36 μM FeSO4 or 7 μM ZnSO4 PedH-dependent growth could not be observed with 10 nM La3+, as it was the case upon the supplementation with TES. Since citrate is used as a metal chelator in TES, we further tested the impact of citrate on growth inhibition of CuSO4, FeSO4 and ZnSO4 when used as additional supplement (Figure 2B). The addition of 50 μM of Na3-citrate restored growth of the ΔpedE strain in the presence of 4 μM CuSO4 and 7 μM ZnSO4, even though cell growth was still impaired for Zn-containing medium. However, in cultures containing 36 μM FeSO4 the addition of citrate had no effect, strongly indicating that FeSO4 is predominantly responsible for the inhibition of PedH-dependent growth of the ΔpedE strain under low La3+ concentrations in a TES complemented M9 medium.
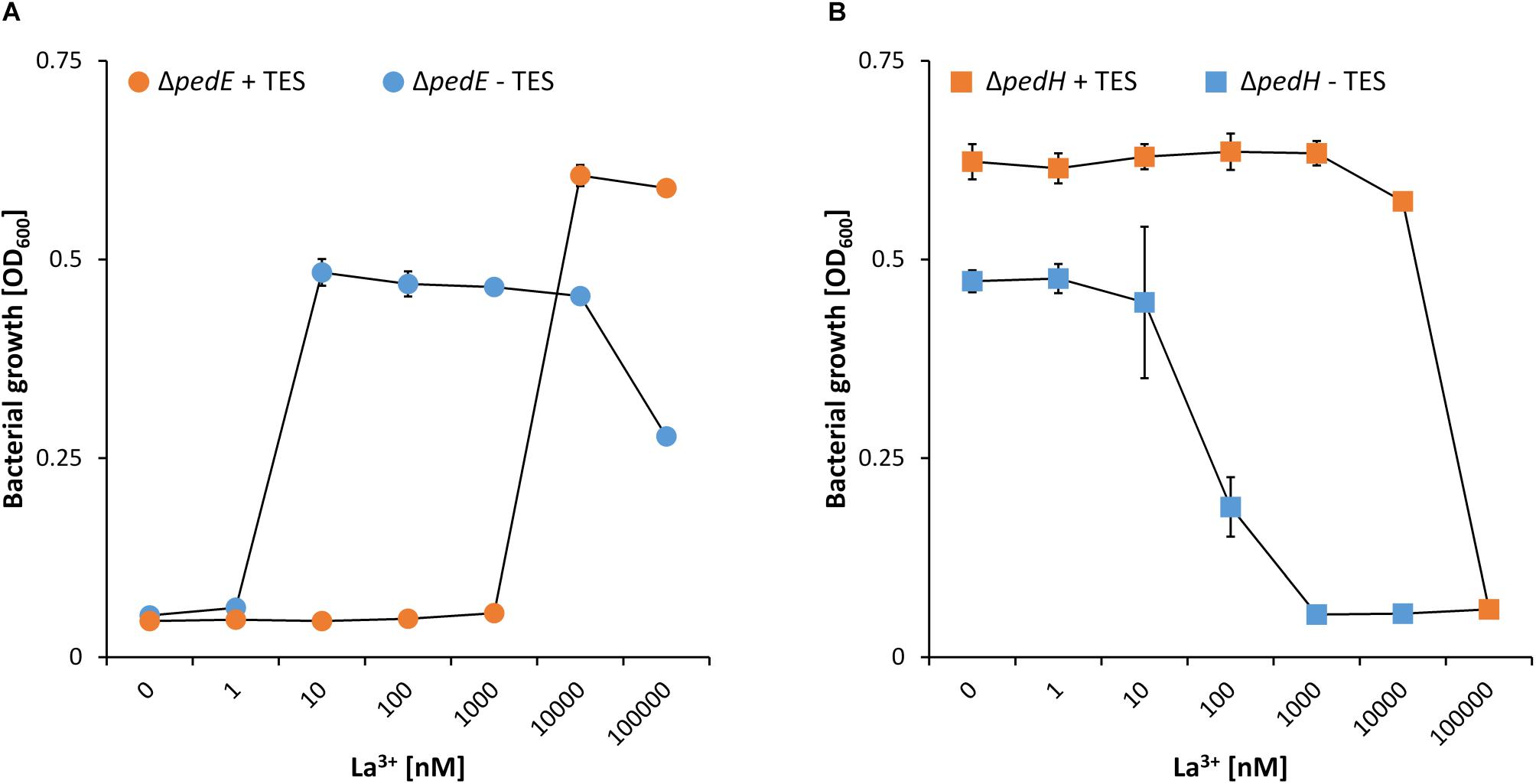
Figure 1. Growth of strain ΔpedE (A, dots) and ΔpedH (B, squares) in 1 mL liquid M9 medium in 96-well deep-well plates on 5 mM 2-phenylethanol and various concentrations of La3+ in the presence (orange) or absence (blue) of trace element solution (TES). OD600 was determined upon 48 h of incubation at 30°C and 350 rpm. Data are presented as the mean values of biological triplicates and error bars represent the corresponding standard deviations.
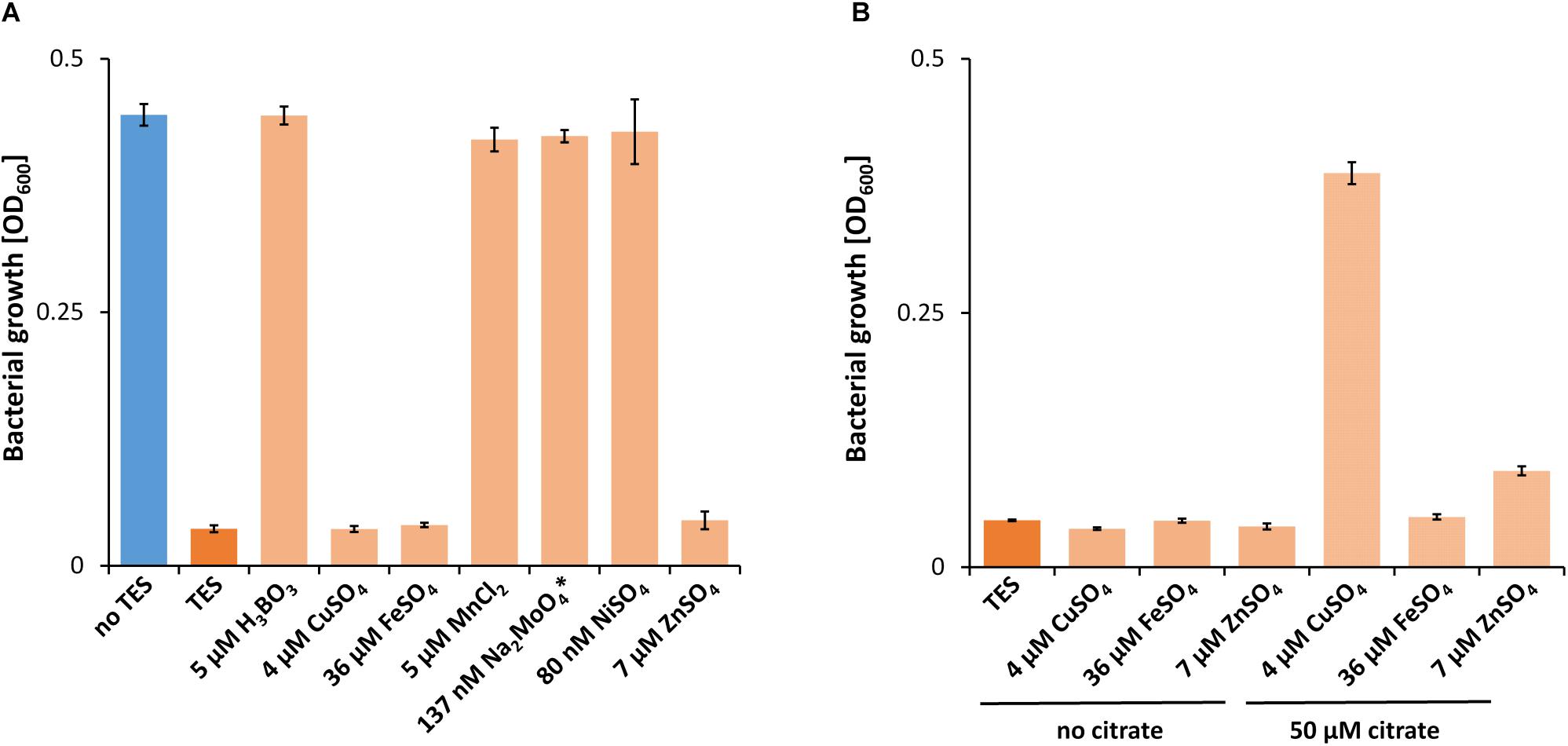
Figure 2. (A) Growth of ΔpedE in 1 mL liquid M9 medium in 96-well deep-well plates on 5 mM 2-phenylethanol and 10 nM La3+ in the presence of TES or individual components thereof. (B) Growth of ΔpedE as described in (A) with and without the additional supplementation of 50 μM Na3-citrate. OD600 was determined upon 48 h of incubation at 30°C and 350 rpm. Data are presented as the mean values of biological triplicates and error bars represent the corresponding standard deviations. ∗Tested condition with 137 nM NaMoO4 also contains 5 μM H3BO3 and 84 nM NiSO4.
La3+-Dependent Growth on 2-Phenylethanol Under Low La3+ Conditions Does Not Depend on Pyoverdine Production
To acquire iron under restricted conditions, P. putida KT2440 can excrete two variants of the siderophore pyoverdine (Salah El Din et al., 1997). Beside their great specificity toward Fe3+, different pyoverdines can also chelate other ions including Al3+, Cu2+, Eu3+ or Tb3+, although with lower affinity (Braud et al., 2009a, b). To test whether pyoverdine production in response to low iron conditions facilitates growth on 2-phenylethanol under low La3+ conditions, the mutant strain ΔpedEΔpvdD was constructed. This strain is no longer able to produce the two pyoverdines due to the loss of the non-ribosomal peptide synthetase pvdD (PP_4219; formerly known as ppsD) (Matilla et al., 2007), which was confirmed upon growth on agar plates (Figure 3C). In experiments with varying FeSO4 supplementation, we found that PedH-dependent growth on 2-phenylethanol of strain ΔpedE was only observed for FeSO4 concentrations ≤10 μM under low (10 nM) La3+ conditions (Figure 3A). With ≥20 μM FeSO4 in the medium, no growth was observed. Strain ΔpedEΔpvdD exhibited the same FeSO4-dependent growth phenotype as the parental strain under low La3+ concentrations. Under high (10 μM) La3+ conditions, strain ΔpedE exhibited PedH-dependent growth under any FeSO4 concentration tested (Figure 3B). Notably, strain ΔpedEΔpvdD showed nearly the same growth pattern as ΔpedE under high La3+ concentrations, with the exception of the condition where no FeSO4 was added to the medium. Under this condition, no growth was observed.
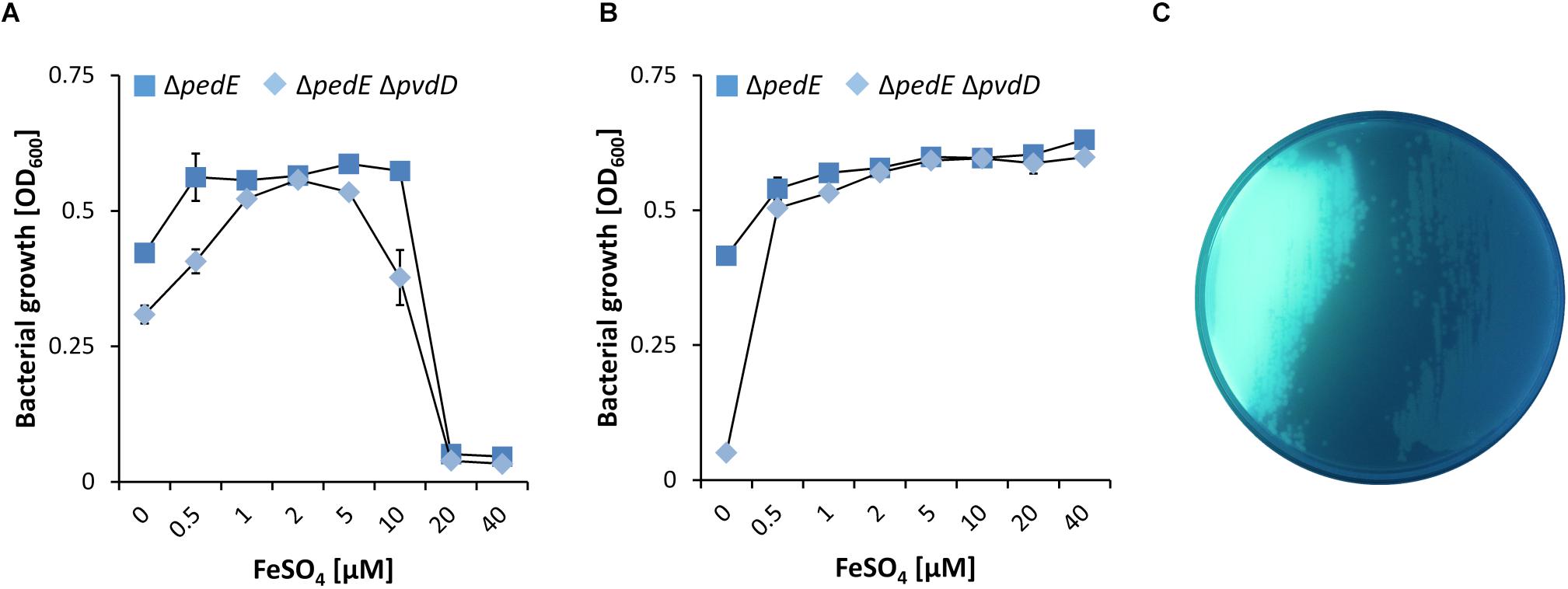
Figure 3. (A,B) Growth of ΔpedE (blue squares) and ΔpedEΔpvdD (light blue diamonds) in 1 mL liquid M9 medium in 96-well deep-well plates without TES on 5 mM 2-phenylethanol and various concentrations of FeSO4 in the presence of 10 nM La3+ (A) or 10 μM La3+ (B). OD600 was determined upon 48 h of incubation at 30°C and 350 rpm. Data are presented as the mean values of biological triplicates and error bars represent the corresponding standard deviations. (C) Pyoverdine production by strains ΔpedE (left) and ΔpedEΔpvdD (right) grown on cetrimide agar plates examined under blue light.
The REE-Switch Is Influenced by the Fe2+/3+ to La3+ Ratio
From the data described above, it can be speculated that beside the PedH-dependent growth also the inhibition of PedE-dependent growth on 2-phenylethanol is dependent on the Fe2+/3+ to La3+ ratio. In the presence of 10 nM La3+, pedE promoter activity was comparably high and increased with increasing FeSO4 concentrations. In addition, strain ΔpedH grew readily on 2-phenylethanol under all these conditions even with no FeSO4 supplementation (Figure 4A). When 10 μM La3+ was available, no growth of the ΔpedH mutant was observed in presence of ≤20 μM FeSO4 and the pedE promoter activities were low (Figure 4B). However, when 40 μM FeSO4 were present in the medium, representing a fourfold excess compared to La3+, PedE-dependent growth and an increased pedE promoter activity was detected.
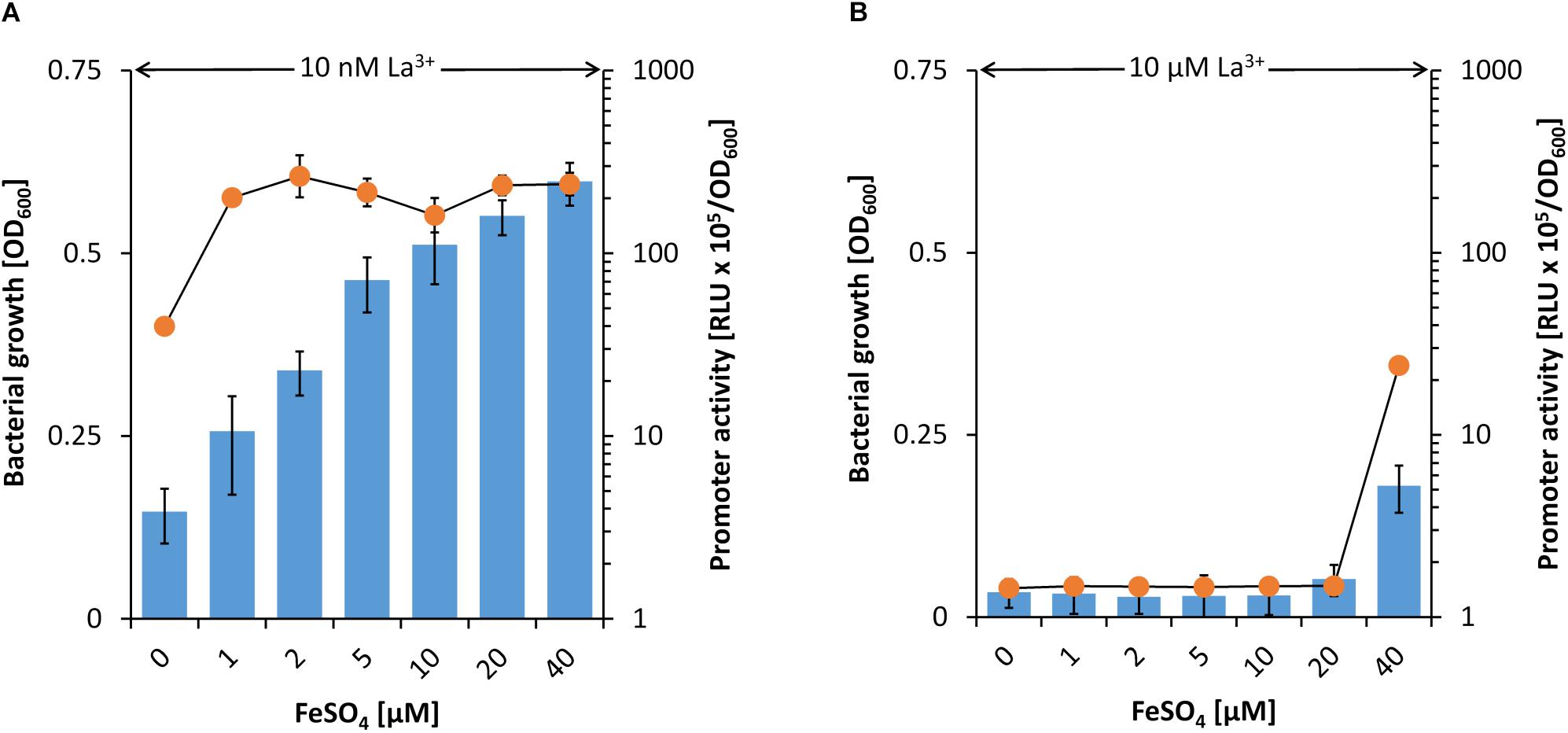
Figure 4. Activities of the pedE promoter (blue bars) in strain KT2440∗ during incubation in M9 medium with 2-phenylethanol, no TES and 10 nM (A) or 10 μM La3+ (B) as well as different FeSO4 concentrations. Promoter activities were determined upon 8 h of incubation at 600 rpm and 30°C. Growth of strain ΔpedH (orange dots) in M9 medium on 2-phenylethanol, no TES and 10 nM (A) or 10 μM (B) La3+ as well as different FeSO4 concentrations. Cells were incubated for 48 h in 96 deep-well plates at 30°C and 350 rpm prior to OD600 measurements. Data are presented as the mean values of biological triplicates and error bars represent the corresponding standard deviations.
PedA1A2BC Is Required for La3+-Dependent Growth on 2-Phenylethanol in Presence of Low La3+ Concentrations
Due to the very low concentrations of REEs (nM range) required for REE-dependent growth, it is commonly speculated that specific REE uptake systems must exist. From our previous results, we can conclude that the produced pyoverdines do not contribute to such a system. A search of the genomic context of the ped gene cluster identified a putative ABC transporter system located nearby the two PQQ-EDHs encoding genes pedE and pedH (Figure 5A). The ABC-transporter is predicted to be encoded as a single transcript by the online tool “Operon-mapper” (Taboada et al., 2018). It consists of four genes encoding a putative permease [pedC (PP_2667)], an ATP-binding protein [pedB (PP_2668)], a YVTN beta-propeller repeat protein of unknown function [pedA2 (PP_2669)], and a periplasmic substrate-binding protein [pedA1 (PP_5538)]. While efflux systems are usually composed of the transmembrane domains and nucleotide binding domains, ABC-dependent import system additionally require a substrate binding protein for functional transport (Biemans-Oldehinkel et al., 2006). As the gene pedA1 is predicted to be such a substrate binding protein, it is very likely that this transporter represents an import system.
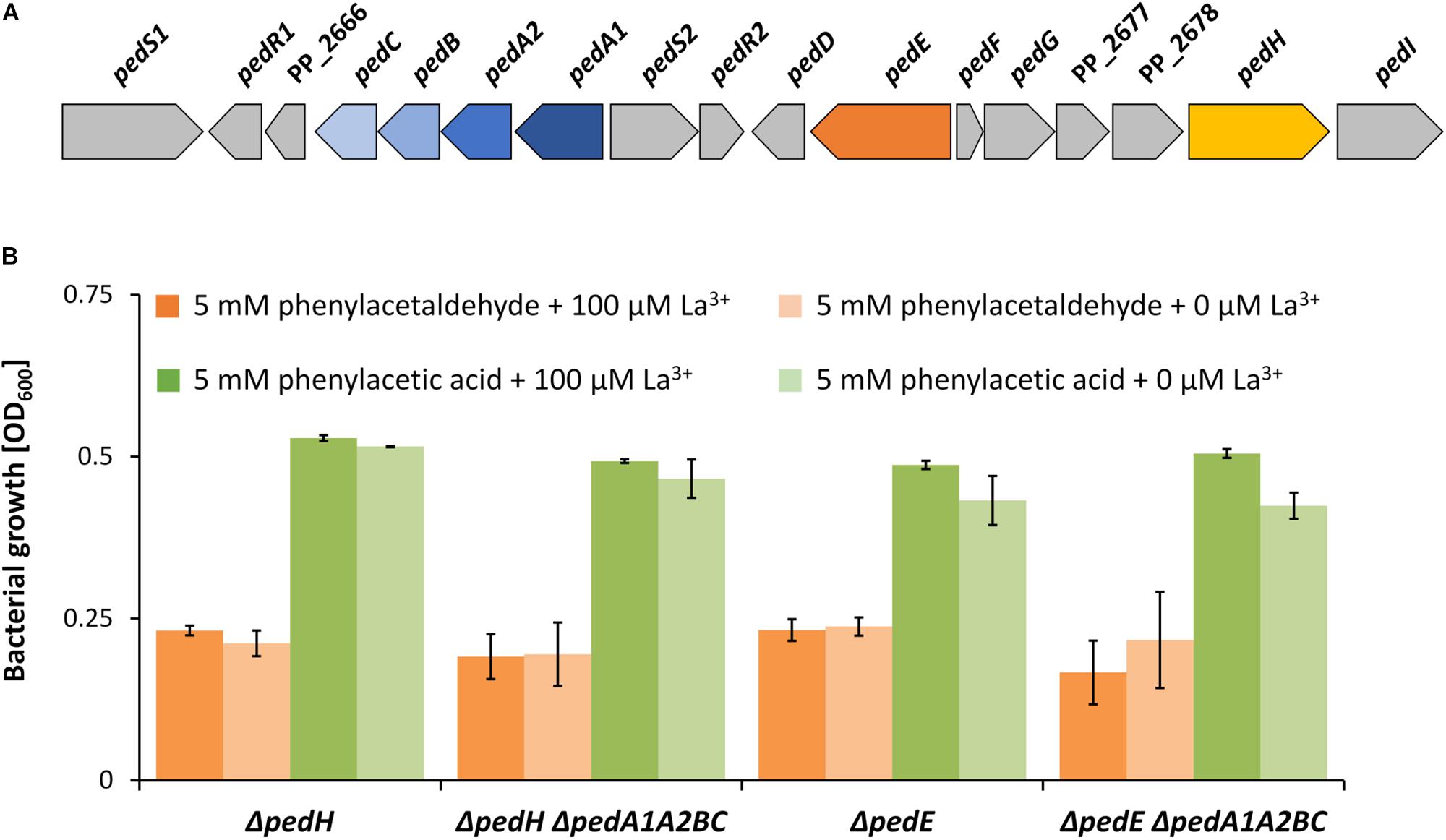
Figure 5. (A) Genomic organization of the ped cluster in Pseudomonas putida KT2440. Nomenclature in analogy to P. putida U as suggested by Arias et al. (2008). (B) Growth of ΔpedH, ΔpedHΔpedA1A2BC, ΔpedE, and ΔpedEΔpedA1A2BC strains in liquid M9 medium with TES on 5 mM phenylacetaldehyde (orange bars) or 5 mM phenylacetic acid (green bars) and either 0 μM La3+ (dark green and dark orange bars) or 100 μM La3+ (light green and light orange bars). OD600 was determined upon 48 h of incubation at 30°C and 180 rpm. Data are presented as the mean values of biological triplicates and error bars represent the corresponding standard deviations.
ABC-dependent importers can be specific for carbon substrates or metal ions. Growth experiments with ΔpedE, ΔpedH, ΔpedEΔpedA1A2BC and ΔpedHΔpedA1A2BC demon- strated that independent of La3+ (100 μM) availability, all strains were capable of growing on the oxidized degradation intermediates of 2-phenylethanol, namely 2-phenylacetaldehyde and phenylacetic acid, within 48 h of incubation (Figure 5B). This indicates, that the transport system is not involved in carbon substrate uptake.
When subsequently different La3+ concentrations were tested, we found that PedH-dependent growth on 2-phenylethanol of strain ΔpedEΔpedA1A2BC was inhibited for the first 48 h of incubation, irrespectively of the presence or absence of TES (Figure 6A). This was in contrast to the ΔpedE deletion strain, which grew in the presence of ≥10 nM La3+ or ≥10 μM La3+ depending on TES availability (Figures 1A, 6A pale symbols and lines). Upon an increased incubation time of 120 h, however, strain ΔpedEΔpedA1A2BC eventually did grow with 1 and 10 μM La3+ or 100 μM La3+ depending on TES addition (Figure 6B). Notably, also the critical REE concentration for PedH-dependent growth of ΔpedEΔpedA1A2BC was increased by 100-fold compared to the ΔpedE strain under all conditions tested. Assuming that PedA1A2BC is specific for REE uptake, growth with the Ca2+-dependent enzyme PedE should not be influenced by a loss of the transporter function. When we tested the ΔpedH and ΔpedHΔpedA1A2BC strain, we indeed could not find any difference in growth as both strains exhibited a similar inhibition pattern for concentrations ≥1 μM La3+ or ≥100 μM La3+ depending on the absence or presence of TES in the medium (Figure 6C).
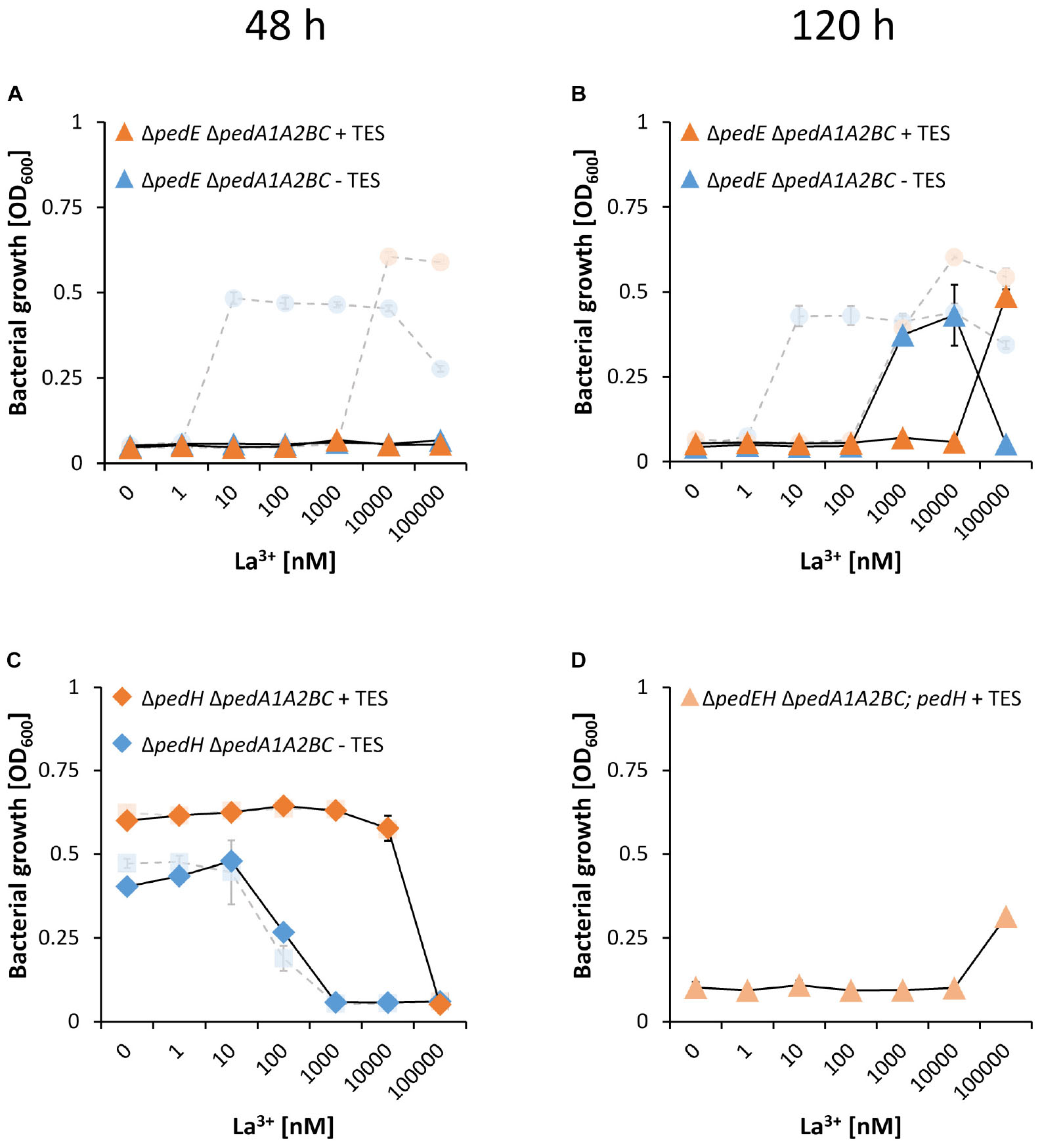
Figure 6. Growth of strains ΔpedEΔpedA1A2BC (A,B, triangles) and ΔpedHΔpedA1A2BC (C, diamonds) in liquid M9 medium on 5 mM 2-phenylethanol with (orange) or without (blue) TES and various concentrations of La3+. Pale circles and squares represent the growth of ΔpedE and ΔpedH parental strains. Growth of strains ΔpedE and ΔpedH in (A,C) represents the restated data from Figure 1 for better comparability. (D) Growth of strain ΔpedEΔpedHΔpedA1A2BC harboring plasmid pMW10 (light orange triangles) in liquid M9 medium on 5 mM 2-phenylethanol, 20 μg/ml kanamycin, TES and various La3+ concentrations. OD600 was determined upon 48 h (A,C) or 120 h (B,D) of incubation at 30°C and 350 rpm. Data are presented as the mean values of biological triplicates, and error bars represent the corresponding standard deviations.
PedA1A2BC Does Not Influence La3+-Dependent Expression of pedH
ABC-transporter systems, or the transported compounds, can be involved in transcriptional regulation of specific target genes (Biemans-Oldehinkel et al., 2006). Thus, the impaired growth on 2-phenylethanol under low La3+ concentrations of the ΔpedEΔpedA1A2BC strain might be caused by the lack of transcriptional activation of the pedH gene. To test this hypothesis, strain ΔpedEΔpedHΔpedA1A2BC was complemented with a pedH gene independent of its natural promoter. Growth analysis of this strain on 2-phenylethanol in the presence of TES and varying La3+ concentrations revealed no difference in the growth pattern when compared to strain ΔpedEΔpedA1A2BC (Figure 6D). This indicated that the impaired growth phenotype of the ABC-transporter mutant is not due to a lack of transcriptional activation of pedH. To further validate this conclusion, pedH promoter activities were measured during incubation with 2-phenylethanol in strain ΔpedA1A2BC and its parental strain in the absence and presence of 10 μM La3+ (Figure 7A). Both strains showed a very similar induction of the pedH promoter in response to La3+ supplementation over time, which was about 20-fold higher at the end of the experiment (3 h) compared to the incubations in the absence of La3+ (26-fold for KT2440∗:Tn7-pedH-lux and 23-fold in ΔpedA1A2BC:Tn7-pedH-lux).
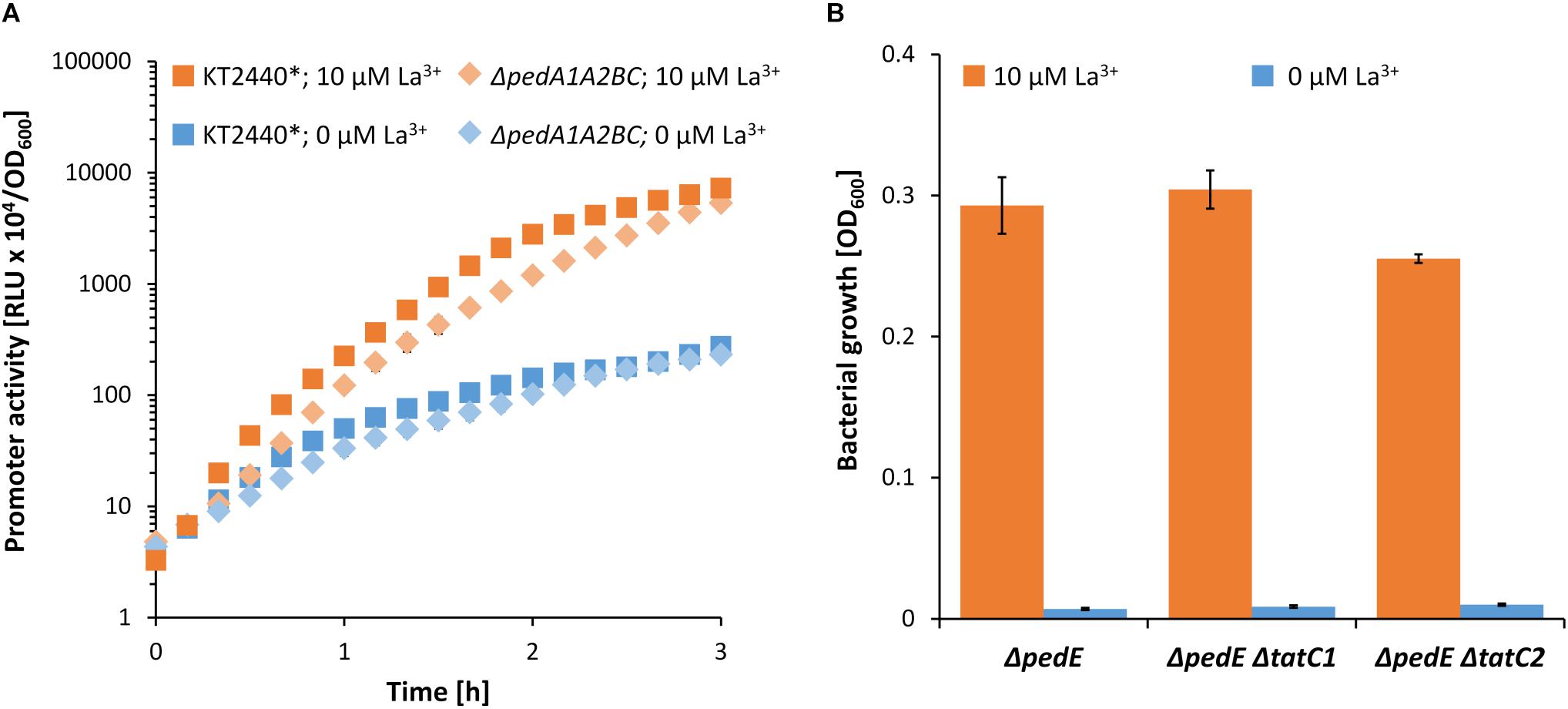
Figure 7. (A) Promoter activities of the pedH promoter in the KT2440∗ and ΔpedA1A2BC background during incubation on 2-phenylethanol in presence (orange) or absence (blue) of 10 μM La3+. Promoter activities were determined during 3 h of incubations at 600 rpm and 30°C. (B) Growth of strain ΔpedE ΔpedA1A2BC on 2-phenylethanol in presence (orange) or absence (blue) of 10 μM La3+. OD600 was determined upon 48 h of incubation at 30°C and 180 rpm. Experiments were conducted in presence of TES. Data are presented as the mean values of biological triplicates and error bars represent the corresponding standard deviations.
La3+-Dependent Growth on 2-Phenylethanol of Strain ΔpedE Is Not Impaired Upon Interference With the Twin-Arginine Translocation (Tat) System
In contrast to PedE, the signal peptide of PedH contains two adjacent arginine residues, which is an indication that it might be transported to the periplasm in a folded state via the Tat (twin-arginine translocation) protein translocation system (Berks, 2015). Therefore, one could argue that the transport of lanthanides into the cytoplasm might be beneficial as the incorporation into the active site of PedH could be more efficient during protein folding compared to the complementation of the apoenzyme in the periplasm. An initial analysis of the PedH signal peptide using different online software tools (TatP, PRED-TAT, SignalP 5.0, TatFind) could neither confirm nor refute this hypothesis (Rose et al., 2002; Bendtsen et al., 2005; Bagos et al., 2010; Almagro Armenteros et al., 2019). Therefore, we generated strains ΔpedE ΔtatC1 and ΔpedE ΔtatC2 in which the two individual TatC proteins [TatC1 (PP_1039) and TatC2 (PP_5018)] encoded in the genome of KT2440 are deleted. These strains should be restricted in the translocation of folded proteins into the periplasm, and if PedH would represent a Tat substrate, impaired growth on 2-phenylethanol in the presence of La3+ should be observable. However, neither a tatC1 nor a tatC2 mutation affected La3+-dependent growth on 2-phenylethanol (Figure 7B). Notably, various attempts to generate the double tatC1/C2 mutant strain were unsuccessful.
Discussion
In the present study, we reveal that iron availability severely affects the REE-switch in P. putida KT2440. This is evidenced by the reduction of the critical concentration of La3+ that is required both to promote PedH-dependent growth and for the repression of growth on 2-phenylethanol based on PedE activity. By using a ΔpvdD deletion strain, we demonstrate that the production of the iron chelating siderophore pyoverdine is not required for PedH-dependent growth on 2-phenylethanol under low La3+ conditions. Our data suggest that the observed effects during high Fe2+/3+/La3+ ratios are caused by mismetallation. In this scenario, the La3+-binding sites of proteins could be occupied by Fe2+/3+ ions that are in excess in the medium, and can also be present in the same 3+ oxidation state (Webb, 1970; Tripathi and Srivastava, 2006; Tottey et al., 2008; Foster et al., 2014). Transcriptional data show that pedE repression can be influenced by iron in a concentration dependent manner. Further, the impact of iron is not identical for PedE and PedH-dependent growth (100 fold vs. 1000 fold). Since PedE regulation is solely dependent on PedS2 (Wehrmann et al., 2017; 2018), these data are thus supportive of such a mismetallation hypothesis, assuming that the sensor histidine kinase PedS2 and PedH have different binding affinities to La3+ and/or Fe2+/3+.
The same hypothesis might similarly explain why under high La3+ concentrations and in the absence of Fe2+/3+ supplementation, a pyoverdine-deficient strain is strongly impaired in growth. In this scenario the Fe2+/3+ binding sites of pyoverdine-independent Fe transporters, such as the ferrichrome, ferrioxamine and ferric citrate uptake systems, might be occupied by La3+ and prevent binding of Fe2+/3+ ions (Jurkevitch et al., 1992; Cornelis, 2010). Consequently, a pyoverdine deficient strain would be unable to take up enough of this essential element that is, most likely, present at trace levels in the medium even without additional supplementation.
It is further interesting to point out that also micromolar Cu2+ and Zn2+ inhibited growth on 2-phenylethanol in presence of La3+ in the nanomolar range, although these metals do not exist in the same 3+ oxidation state under natural conditions. They are, however, the divalent transition metals that form the most stable complexes irrespective of the nature of the ligand, and as such also competitively bind non-cognate metal binding sites with high strength (Irving and Williams, 1953; Foster et al., 2014). Notably, Cu2+ has also been reported to interfere with REE-dependent regulation of PQQ-dependent methanol dehydrogenases in M. trichosporium OB3b (Gu et al., 2016; Gu and Semrau, 2017), and it is tempting to speculate that mismetallation might be involved in this process, too.
We provide compelling evidence that the predicted ABC-transporter PedA1A2BC is essential for PedH-dependent growth on 2-phenylethanol under low concentrations of La3+. Based on the PedE-dependent growth phenotype, we can further show that PedA1A2BC is not involved in transcriptional repression of pedE under low La3+ conditions. The fact that a ΔpedE ΔpedA1A2BC mutant strain can only grow with a 100-fold higher concentration of La3+ compared to the ΔpedE single mutant strongly indicates that PedA1A2BC functions as a La3+-specific importer into the cytoplasm. Recently, it was demonstrated that in several Methylobacterium extorquens strains a similar ABC-transporter system is required for Ln3+-dependent growth (Ochsner et al., 2019; Roszczenko-Jasińska et al., 2019). A BLAST analysis revealed that these ABC transporters show high similarities to all four genes of the pedA1A2BC operon (>43% sequence identity for pedA2 and >50% for pedA1, pedB and pedC) and that all bacterial strains that have been reported to produce Ln3+-dependent PQQ-ADHs thus far, carry homologs of this transporter system in their genome. Using a protein-based fluorescent sensor with picomolar affinity for REEs, Mattocks and collegues were able to demonstrate that M. extorquens indeed selectively takes up light REEs into its cytoplasm (Mattocks et al., 2019) and it was later shown that cytoplasmic REE-uptake depends on the presence of the previously identified ABC-transporter system (Roszczenko-Jasińska et al., 2019).
Since the PedH enzyme, like all currently known Ln3+-dependent enzymes, resides in the periplasm and since the purified apoenzyme of PedH can be converted into the catalytically active holoenzyme by Ln3+ supplementation in vitro, the question arises what the potential advantage of the postulated cytoplasmic Ln3+ uptake for P. putida would be. From our point of view, two different reasons can be imagined, namely that (i) the REE-dependent PedH protein is folded within the cytoplasm and the incorporation of the La3+-cofactor is only possible or more efficient during the folding process; or (ii) La3+ binds to a cytoplasmic protein that either represents a so-far uncharacterized transcriptional regulator or another REE-dependent enzyme.
It has been demonstrated that the location of protein folding can regulate metal binding (Tottey et al., 2008). As such, Ln3+ insertion during folding in the cytoplasm, where metal concentrations are tightly regulated, could provide a means of preventing the Ln3+ binding site of PedH from mismetallation with potentially competitive binders such as Cu2+, Zn2+, or Fe2+/3+ in the periplasm. However, we could not find evidence that PedH is a Tat substrate and consequently transported into the periplasm as a folded protein (Berks, 2015), as the individual tatC1 or tatC2 mutants both still showed PedH-dependent growth. However, it cannot be excluded that the two Tat systems are functionally redundant since attempts to generate a tatC1/C2 double mutant strain proved unsuccessful.
We can further conclude that the putative La3+ transport into the cytoplasm is not required to activate pedH transcription. It is, however, possible that additional genes/proteins required for PedH-dependent growth rely on, or are regulated by, the cytoplasmic presence of REEs. In this context, it is interesting to note that in a recent proteomic approach, we found that besides PedE and PedH, additional proteins of unknown function show differential abundance in response to La3+ availability (Wehrmann et al., 2019). It will hence be interesting to find out, whether any of these proteins is required for PedH function.
In Methylobacterium extorquens PA1 and Methylobacterium extorquens AM1, almost identical TonB-dependent receptor proteins (>99% sequence identity) were found to be crucial for REE-dependent growth suggesting a specific Ln3+-binding chelator system in these organisms (Ochsner et al., 2019; Roszczenko-Jasińska et al., 2019). Interestingly, also in Methylotuvimicrobium buryatense 5GB1C a TonB-dependent receptor was identified that is crucial for the REE-switch to occur (Groom et al., 2019). The latter receptor only shows <20% sequence identity to those of the M. extorquens strains and thus could not have been identified by homology searches.
In Pseudomonas putida KT2440, no close homolog to any of the aforementioned TonB-dependent receptors can be identified (<30% sequence identity). P. putida preferentially resides in the rhizosphere whereas M. buryatense and M. extorquens PA1 were isolated from the sediment of a soda lake (pH 9 – 9.5) and the phyllosphere of Arabidopsis thaliana, respectively (Kaluzhnaya et al., 2001; Knief et al., 2010). One could speculate that a specific Ln3+-chelator system is perhaps not relevant in the rhizosphere due to the large reservoir of REEs within the soil and the usually acidic environment near the plant roots caused by the secretion of organic acids (Raghothama and Karthikeyan, 2005; Ramos et al., 2016). The lack of a homologous TonB-dependent receptor could also be explained by structural differences in the REE-specific chelator system that might be employed by P. putida compared to that of the methylotrophic bacteria. Lastly, it is also possible that in P. putida the REE uptake across the outer membrane proceeds via the same chelator systems that are used for pyoverdin independent Fe-acquisition. This could further provide another explanation for the impact of the Fe2+/3+ to La3+ ratio on the REE-switch.
Overall, the present study expands the crucial role of a conserved ABC-transporter system, which was very recently identified as Ln3+-specific inner membrane transport system in methano- and methylotrophs, to non-methylotrophic organisms. It further provides new insight into the complexity of bacterial-metal interactions and demonstrates that Cu, Zn and in particular Fe ions can strongly interfere with the REE-switch in P. putida, most likely through mismetallation. The body of knowledge how REEs impact protein function, gene regulation and consequently physiology of different microorganisms is rapidly increasing. As such, it will be very interesting to see when some of the most interesting questions, such as the cytoplasmic function of REEs or the nature and potential structural diversity of specific REE-chelator systems, will be resolved by future studies.
Data Availability Statement
All datasets generated for this study are included in the article/supplementary material.
Author Contributions
MW, PB, and JK designed the research. MW, CB, and PB performed the experiments. All authors analyzed the data. MW and JK wrote the manuscript with contributions of PB.
Funding
MW and JK was supported by an individual research grant from the Deutsche Forschungsgemeinschaft (DFG, KL 2340/2-1). CB and PB was supported by the French National Research Agency through the National Program “Investissements d’Avenir” with the reference ANR-10-LABX-21-01/LABEX RESSOURCES21.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
JK and MW would like to thank Prof. Bernhard Hauer for his continuous support.
References
Aide, M. T., and Aide, C. (2012). Rare earth elements: their importance in understanding soil genesis. ISRN Soil Sci. 2012, 1–11. doi: 10.5402/2012/783876
Almagro Armenteros, J. J., Tsirigos, K. D., Sønderby, C. K., Petersen, T. N., Winther, O., Brunak, S., et al. (2019). SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420–423. doi: 10.1038/s41587-019-0036-z
Andrews, S. C., Robinson, A. K., and Rodríguez-Quiñones, F. (2003). Bacterial iron homeostasis. FEMS Microbiol. Rev. 27, 215–237. doi: 10.1016/S0168-6445(03)00055-X
Arias, S., Olivera, E. R., Arcos, M., Naharro, G., and Luengo, J. M. (2008). Genetic analyses and molecular characterization of the pathways involved in the conversion of 2-phenylethylamine and 2-phenylethanol into phenylacetic acid in Pseudomonas putida U. Environ. Microbiol. 10, 413–432. doi: 10.1111/j.1462-2920.2007.01464.x
Bagos, P. G., Nikolaou, E. P., Liakopoulos, T. D., and Tsirigos, K. D. (2010). Combined prediction of tat and sec signal peptides with hidden markov models. Bioinformatics 26, 2811–2817. doi: 10.1093/bioinformatics/btq530
Baune, M., Qi, Y., Scholz, K., Volmer, D. A., and Hayen, H. (2017). Structural characterization of pyoverdines produced by Pseudomonas putida KT2440 and Pseudomonas taiwanensis VLB120. BioMetals 30, 589–597. doi: 10.1007/s10534-017-0029-27
Belda, E., van Heck, R. G. A., José Lopez-Sanchez, M., Cruveiller, S., Barbe, V., Fraser, C., et al. (2016). The revisited genome of Pseudomonas putida KT2440 enlightens its value as a robust metabolic chassis. Environ. Microbiol. 18, 3403–3424. doi: 10.1111/1462-2920.13230
Bendtsen, J. D., Nielsen, H., Widdick, D., Palmer, T., and Brunak, S. (2005). Prediction of twin-arginine signal peptides. BMC Bioinformatics 6:167. doi: 10.1186/1471-2105-6-167
Berks, B. C. (2015). The twin-arginine protein translocation pathway. Annu. Rev. Biochem. 84, 843–864. doi: 10.1146/annurev-biochem-060614-34251
Biemans-Oldehinkel, E., Doeven, M. K., and Poolman, B. (2006). ABC transporter architecture and regulatory roles of accessory domains. FEBS Lett. 580, 1023–1035. doi: 10.1016/j.febslet.2005.11.079
Boyer, H. W., and Roulland-Dussoix, D. (1969). A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41, 459–472. doi: 10.1038/s41598-018-31191-31191
Braud, A., Hannauer, M., Mislin, G. L. A., and Schalk, I. J. (2009a). The Pseudomonas aeruginosa pyochelin-iron uptake pathway and its metal specificity. J. Bacteriol. 191, 3517–3525. doi: 10.1128/JB.00010-19
Braud, A., Hoegy, F., Jezequel, K., Lebeau, T., and Schalk, I. J. (2009b). New insights into the metal specificity of the Pseudomonas aeruginosa pyoverdine-iron uptake pathway. Environ. Microbiol. 11, 1079–1091. doi: 10.1111/j.1462-2920.2008.01838.x
Chandrangsu, P., Rensing, C., and Helmann, J. D. (2017). Metal homeostasis and resistance in bacteria. Nat. Rev. Microbiol. 15, 338–350. doi: 10.1038/nrmicro.2017.15
Chen, S., Bleam, W. F., and Hickey, W. J. (2010). Molecular analysis of two bacterioferritin genes, bfralpha and bfrbeta, in the model rhizobacterium Pseudomonas putida KT2440. Appl. Environ. Microbiol. 76, 5335–5343. doi: 10.1128/AEM.00215-210
Choi, K.-H., Gaynor, J. B., White, K. G., Lopez, C., Bosio, C. M., Karkhoff-Schweizer, R. R., et al. (2005). A Tn7-based broad-range bacterial cloning and expression system. Nat. Methods 2, 443–448. doi: 10.1038/nmeth765
Cornelis, P. (2010). Iron uptake and metabolism in pseudomonads. Appl. Microbiol. Biotechnol. 86, 1637–1645. doi: 10.1007/s00253-010-2550-2552
Cornelis, P., and Andrews, S. C. (2010). Iron Uptake and Homeostasis in Microorganisms. Poole: Caister Academic Press.
Cornelis, P., Wei, Q., Andrews, S. C., and Vinckx, T. (2011). Iron homeostasis and management of oxidative stress response in bacteria. Metallomics 3, 540–549. doi: 10.1039/c1mt00022e
Cotruvo, J. A., Featherston, E. R., Mattocks, J. A., Ho, J. V., and Laremore, T. N. (2018). Lanmodulin: a highly selective lanthanide-binding protein from a lanthanide-utilizing bacterium. J. Am. Chem. Soc. 140, 15056–15061. doi: 10.1021/jacs.8b09842
Dixon, S. J., and Stockwell, B. R. (2014). The role of iron and reactive oxygen species in cell death. Nat. Chem. Biol. 10, 9–17. doi: 10.1038/nchembio.1416
Firsching, F. H., and Brune, S. N. (1991). Solubility products of the trivalent rare-earth phosphates. J. Chem. Eng. Data 36, 93–95. doi: 10.1021/je00001a028
Foster, A. W., Osman, D., and Robinson, N. J. (2014). Metal preferences and metallation. J. Biol. Chem. 289, 28095–28103. doi: 10.1074/jbc.R114.588145
Gibson, D. G. (2011). Enzymatic assembly of overlapping DNA fragments. Methods Enzymol. 498, 349–361. doi: 10.1016/B978-0-12-385120-8.00015-12
Graf, N., and Altenbuchner, J. (2011). Development of a method for markerless gene deletion in Pseudomonas putida. Appl. Environ. Microbiol. 77, 5549–5552. doi: 10.1128/AEM.05055-5011
Gray, H. B. (2003). Biological inorganic chemistry at the beginning of the 21st century. Proc. Natl. Acad. Sci. U.S.A. 100, 3563–3568. doi: 10.1073/pnas.0730378100
Groom, J., Ford, S. M., Pesesky, M. W., and Lidstrom, M. E. (2019). A mutagenic screen identifies a TonB-dependent receptor required for the lanthanide metal switch in the type I methanotroph Methylotuvimicrobium buryatense 5GB1C. J. Bacteriol. 201:e120-19. doi: 10.1128/JB.00120-19
Gu, W., Farhan Ul Haque, M., DiSpirito, A. A., and Semrau, J. D. (2016). Uptake and effect of rare earth elements on gene expression in Methylosinus trichosporium OB3b. FEMS Microbiol. Lett. 363:fnw129. doi: 10.1093/femsle/fnw129
Gu, W., and Semrau, J. D. (2017). Copper and cerium-regulated gene expression in Methylosinus trichosporium OB3b. Appl. Microbiol. Biotechnol. 101, 8499–8516. doi: 10.1007/s00253-017-8572-8572
Herrero, M., de Lorenzo, V., and Timmis, K. N. (1990). Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172, 6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990
Irving, H., and Williams, R. J. P. (1953). The stability of transition-metal complexes. J. Chem. Soc. 1953, 3192–3210. doi: 10.1039/jr9530003192
Jurkevitch, E., Hadar, Y., Chen, Y., Libman, J., and Shanzer, A. (1992). Iron uptake and molecular recognition in Pseudomonas putida: receptor mapping with ferrichrome and its biomimetic analogs. J. Bacteriol. 174, 78–83. doi: 10.1099/13500872-140-7-1697
Kaluzhnaya, M., Khmelenina, V., Eshinimaev, B., Suzina, N., Nikitin, D., Solonin, A., et al. (2001). Taxonomic characterization of new alkaliphilic and alkalitolerant methanotrophs from soda lakes of the southeastern transbaikal region and description of Methylomicrobium buryatense sp.nov. Syst. Appl. Microbiol. 24, 166–176. doi: 10.1078/0723-2020-2028
Keen, N. T., Tamaki, S., Kobayashi, D., and Trollinger, D. (1988). Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70, 191–197. doi: 10.1016/0378-1119(88)90117-90115
Keltjens, J. T., Pol, A., Reimann, J., and Op den Camp, H. J. M. (2014). PQQ-dependent methanol dehydrogenases: rare-earth elements make a difference. Appl. Microbiol. Biotechnol. 98, 6163–6183. doi: 10.1007/s00253-014-5766-5768
Kim, Y. C., Miller, C. D., and Anderson, A. J. (1999). Transcriptional regulation by iron of genes encoding iron- and manganese-superoxide dismutases from Pseudomonas putida. Gene 239, 129–135. doi: 10.1016/S0378-1119(99)00369-368
Knief, C., Frances, L., and Vorholt, J. A. (2010). Competitiveness of diverse Methylobacterium strains in the phyllosphere of Arabidopsis thaliana and identification of representative models, including M. extorquens PA1. Microb. Ecol. 60, 440–452. doi: 10.1007/s00248-010-9725-9723
Maniatis, T., Fritsch, E., Sambrook, J., and Laboratory, C. S. H. (1982). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory.
Markert, B. (1987). The pattern of distribution of lanthanide elements in soils and plants. Phytochemistry 26, 3167–3170. doi: 10.1016/S0031-9422(00)82463-82462
Matilla, M. A., Ramos, J. L., Duque, E., de Dios Alché, J., Espinosa-Urgel, M., and Ramos-González, M. I. (2007). Temperature and pyoverdine-mediated iron acquisition control surface motility of Pseudomonas putida. Environ. Microbiol. 9, 1842–1850. doi: 10.1111/j.1462-2920.2007.01286.x
Mattocks, J. A., Ho, J. V., and Cotruvo, J. A. (2019). A selective, protein-based fluorescent sensor with picomolar affinity for rare earth elements. J. Am. Chem. Soc. 141, 2857–2861. doi: 10.1021/jacs.8b12155
Meloche, C. C., and Vrátný, F. (1959). Solubility product relations in the rare earth hydrous hydroxides. Anal. Chim. Acta 20, 415–418. doi: 10.1016/0003-2670(59)80090-80098
Merchant, S. S., and Helmann, J. D. (2012). Elemental economy: microbial strategies for optimizing growth in the face of nutrient limitation. Adv. Microb. Physiol. 60, 91–210. doi: 10.1016/B978-0-12-398264-3.00002-4
Miller, C. D., Pettee, B., Zhang, C., Pabst, M., McLean, J. E., and Anderson, A. J. (2009). Copper and cadmium: responses in Pseudomonas putida KT2440. Lett. Appl. Microbiol. 49, 775–783. doi: 10.1111/j.1472-765X.2009.02741.x
Mückschel, B., Simon, O., Klebensberger, J., Graf, N., Rosche, B., Altenbuchner, J., et al. (2012). Ethylene glycol metabolism by Pseudomonas putida. Appl. Environ. Microbiol. 78, 8531–8539. doi: 10.1128/AEM.02062-2012
Nelson, K. E., Weinel, C., Paulsen, I. T., Dodson, R. J., Hilbert, H., Martins dos Santos, V. A. P., et al. (2002). Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4, 799–808. doi: 10.1046/j.1462-2920.2002.00366.x
Ochsner, A. M., Hemmerle, L., Vonderach, T., Nüssli, R., Bortfeld-Miller, M., Hattendorf, B., et al. (2019). Use of rare-earth elements in the phyllosphere colonizer Methylobacterium extorquens PA1. Mol. Microbiol. 111, 1152–1166. doi: 10.1111/mmi.14208
Picone, N., and Op den Camp, H. J. M. (2019). Role of rare earth elements in methanol oxidation. Curr. Opin. Chem. Biol. 49, 39–44. doi: 10.1016/j.cbpa.2018.09.019
Raghothama, K. G., and Karthikeyan, A. S. (2005). Phosphate acquisition. Plant Soil 274, 37–49. doi: 10.1007/s11104-004-2005-2006
Ramos, S. J., Dinali, G. S., Oliveira, C., Martins, G. C., Moreira, C. G., Siqueira, J. O., et al. (2016). Rare earth elements in the soil environment. Curr. Pollut. Reports 2, 28–50. doi: 10.1007/s40726-016-0026-24
Ray, P., Girard, V., Gault, M., Job, C., Bonneu, M., Mandrand-Berthelot, M.-A., et al. (2013). Pseudomonas putida KT2440 response to nickel or cobalt induced stress by quantitative proteomics. Metallomics 5, 68–79. doi: 10.1039/C2MT20147J
Rose, R. W., Brüser, T., Kissinger, J. C., and Pohlschröder, M. (2002). Adaptation of protein secretion to extremely high-salt conditions by extensive use of the twin-arginine translocation pathway. Mol. Microbiol. 45, 943–950. doi: 10.1046/j.1365-2958.2002.03090.x
Roszczenko-Jasińska, P., Vu, H. N., Subuyuj, G. A., Crisostomo, R. V., Cai, J., Raghuraman, C., et al. (2019). Lanthanide transport, storage, and beyond: genes and processes contributing to XoxF function in Methylorubrum extorquens AM1. bioRxiv doi: 10.1101/647677 [Preprint].
Salah El Din, A. L. M., Kyslík, P., Stephan, D., and Abdallah, M. A. (1997). Bacterial iron transport: structure elucidation by FAB-MS and by 2D NMR (1H, 13C, 15N) of pyoverdin G4R, a peptidic siderophore produced by a nitrogen-fixing strain of Pseudomonas putida. Tetrahedron 53, 12539–12552. doi: 10.1016/S0040-4020(97)00773-774
Sasnow, S. S., Wei, H., and Aristilde, L. (2016). Bypasses in intracellular glucose metabolism in iron-limited Pseudomonas putida. Microbiologyopen 5, 3–20. doi: 10.1002/mbo3.287
Schalk, I. J., and Cunrath, O. (2016). An overview of the biological metal uptake pathways in Pseudomonas aeruginosa. Environ. Microbiol. 18, 3227–3246. doi: 10.1111/1462-2920.13525
Semrau, J. D., DiSpirito, A. A., Gu, W., and Yoon, S. (2018). Metals and methanotrophy. Appl. Environ. Microbiol. 84:e2289-17. doi: 10.1128/AEM.02289-2217
Silva-Rocha, R., Martínez-García, E., Calles, B., Chavarría, M., Arce-Rodríguez, A., de las Heras, A., et al. (2013). The standard European vector architecture (SEVA): a coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Res. 41, D666–D675. doi: 10.1093/nar/gks1119
Studier, F. W., and Moffatt, B. A. (1986). Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189, 113–130. doi: 10.1016/0022-2836(86)90385-90382
Taboada, B., Estrada, K., Ciria, R., and Merino, E. (2018). Operon-mapper: a web server for precise operon identification in bacterial and archaeal genomes. Bioinformatics 34, 4118–4120. doi: 10.1093/bioinformatics/bty496
Takeda, K., Matsumura, H., Ishida, T., Samejima, M., Igarashi, K., Nakamura, N., et al. (2013). The two-step electrochemical oxidation of alcohols using a novel recombinant PQQ alcohol dehydrogenase as a catalyst for a bioanode. Bioelectrochemistry 94, 75–78. doi: 10.1016/j.bioelechem.2013.08.001
Tottey, S., Waldron, K. J., Firbank, S. J., Reale, B., Bessant, C., Sato, K., et al. (2008). Protein-folding location can regulate manganese-binding versus copper- or zinc-binding. Nature 455, 1138–1142. doi: 10.1038/nature07340
Tripathi, V. N., and Srivastava, S. (2006). Ni2 +-uptake in Pseudomonas putida strain S4: a possible role of Mg2 +-uptake pump. J. Biosci. 31, 61–67. doi: 10.1007/BF02705236
Tyler, G. (2004). Rare earth elements in soil and plant systems - a review. Plant Soil 267, 191–206. doi: 10.1007/s11104-005-4888-4882
Webb, M. (1970). The mechanism of acquired resistance to Co2 + and Ni2 + in gram-positive and gram-negative bacteria. Biochim. Biophys. Acta 222, 440–446. doi: 10.1016/0304-4165(70)90134-90130
Wehrmann, M., Berthelot, C., Billard, P., and Klebensberger, J. (2018). The PedS2/PedR2 two-component system is crucial for the rare earth element switch in Pseudomonas putida KT2440. mSphere 3, 1–12. doi: 10.1128/mSphere.00376-318
Wehrmann, M., Billard, P., Martin-Meriadec, A., Zegeye, A., and Klebensberger, J. (2017). Functional role of lanthanides in enzymatic activity and transcriptional regulation of pyrroloquinoline quinone-dependent alcohol dehydrogenases in Pseudomonas putida KT2440. mBio 8, e570–e517. doi: 10.1128/mBio.00570-517
Wehrmann, M., Toussaint, M., Pfannstiel, J., Billard, P., and Klebensberger, J. (2019). The cellular response towards lanthanum is substrate specific and reveals a novel route for glycerol metabolism in Pseudomonas putida KT2440. bioRxiv doi: 10.1101/567529 [Preprint],
Wu, M. L., Wessels, J. C. T., Pol, A., Op den Camp, H. J. M., Jetten, M. S. M., and van Niftrik, L. (2015). XoxF-type methanol dehydrogenase from the anaerobic methanotroph Candidatus Methylomirabilis oxyfera. Appl. Environ. Microbiol. 81, 1442–1451. doi: 10.1128/AEM.03292-3214
Keywords: lanthanides, rare earth elements, ABC-transporter, Pseudomonas putida, pyrroloquinoline quinone, PedH, mismetallation, metal homeostasis
Citation: Wehrmann M, Berthelot C, Billard P and Klebensberger J (2019) Rare Earth Element (REE)-Dependent Growth of Pseudomonas putida KT2440 Relies on the ABC-Transporter PedA1A2BC and Is Influenced by Iron Availability. Front. Microbiol. 10:2494. doi: 10.3389/fmicb.2019.02494
Received: 24 June 2019; Accepted: 16 October 2019;
Published: 31 October 2019.
Edited by:
Hari S. Misra, Bhabha Atomic Research Centre (BARC), IndiaReviewed by:
Rafael Silva-Rocha, University of São Paulo, BrazilPablo Ivan Nikel, Novo Nordisk Foundation Center for Biosustainability (DTU Biosustain), Denmark
Copyright © 2019 Wehrmann, Berthelot, Billard and Klebensberger. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Patrick Billard, cGF0cmljay5iaWxsYXJkQHVuaXYtbG9ycmFpbmUuZnI=; Janosch Klebensberger, amFub3NjaC5rbGViZW5zYmVyZ2VyQGl0Yi51bmktc3R1dHRnYXJ0LmRl
 Matthias Wehrmann
Matthias Wehrmann Charlotte Berthelot
Charlotte Berthelot Patrick Billard
Patrick Billard Janosch Klebensberger
Janosch Klebensberger