
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 31 October 2019
Sec. Evolutionary and Genomic Microbiology
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.02480
Parts of this article's content have been modified or rectified in:
Erratum: Corrigendum: Taxogenomics Resolves Conflict in the Genus Rhodobacter: A Two and Half Decades Pending Thought to Reclassify the Genus Rhodobacter
Corrigendum: Taxogenomics Resolves Conflict in the Genus Rhodobacter: A Two and Half Decades Pending Thought to Reclassify the Genus Rhodobacter
The genus Rhodobacter is taxonomically well studied, and some members are model organisms. However, this genus is comprised of a heterogeneous group of members. 16S rRNA gene-based phylogeny of the genus Rhodobacter indicates a motley assemblage of anoxygenic phototrophic bacteria (genus Rhodobacter) with interspersing members of other genera (chemotrophs) making the genus polyphyletic. Taxogenomics was performed to resolve the taxonomic conflicts of the genus Rhodobacter using twelve type strains. The phylogenomic analysis showed that Rhodobacter spp. can be grouped into four monophyletic clusters with interspersing chemotrophs. Genomic indices (ANI and dDDH) confirmed that all the current species are well defined, except Rhodobacter megalophilus. The average amino acid identity values between the monophyletic clusters of Rhodobacter members, as well as with the chemotrophic genera, are less than 80% whereas the percentage of conserved proteins values were below 70%, which has been observed among several genera related to Rhodobacter. The pan-genome analysis has shown that there are only 1239 core genes shared between the 12 species of the genus Rhodobacter. The polyphasic taxonomic analysis supports the phylogenomic and genomic studies in distinguishing the four Rhodobacter clusters. Each cluster is comprised of one to seven species according to the current Rhodobacter taxonomy. Therefore, to address this taxonomic discrepancy we propose to reclassify the members of the genus Rhodobacter into three new genera, Luteovulum gen. nov., Phaeovulum gen. nov. and Fuscovulum gen. nov., and provide an emended description of the genus Rhodobacter sensu stricto. Also, we propose reclassification of Rhodobacter megalophilus as a sub-species of Rhodobacter sphaeroides.
The genus Rhodobacter (Rba.) was proposed by Imhoff et al. (1984) to accommodate those species of Rhodopseudomonas (Rhodopseudomonas capsulata [formerly Rhodonostoc capsulatum (Molisch, 1907)], Rhodopseudomonas spheroides, Rhodopseudomonas sulfidophila and Rhodopseudomonas adriatica) with vesicular intracytoplasmic membrane (ICM) architecture as Rba. capsulatus (type species of the genus), Rba. sphaeroides, Rba. adriaticus and Rba. sulfidophilus. Subsequently, based on 16S rRNA gene sequence comparisons, requirement of NaCl for optimal growth, final oxidation product of sulfide, polar lipid composition and sulfide tolerance, Rba. sulfidophilus, Rba. euryhalinus, and Rba. adriaticus were reclassified as Rhodovulum sulfidophilum, Rhodovulum euryhulinum, and Rhodovulum adriaticum (Hiraishi and Ueda, 1994). Rhodopseudomonas blastica was also reclassified into the genus Rhodobacter, although it should be emphasized that Rba. blasticus is now the only member of the genus with a lamellar ICM architecture (Hiraishi and Ueda, 1994).
Rhodobacter is the type genus of the family ‘Rhodobacteraceae’ which presently contains more than 150 validly named genera1. However, it must be noted that the name ‘Rhodobacteraceae’ is illegitimate2 as the relationship between this suprageneric grouping and the family Hyphomonadaceae (Lee et al., 2005) has not yet been resolved. Lee et al. (2005) grouped members of the family ‘Rhodobacteraceae’ into five-well defined groups, based on phylogenetic analysis. These are the Rhodobacter, Rhodovulum, Amaricoccus, Roseobacter, and Paracoccus groups. The Rhodobacter group consists of 17 genera: Albirhodobacter, Cereibacter, Defluviimonas, Falsirhodobacter, Gemmobacter, Haematobacter, Paenirhodobacter, Pararhodobacter, Plastorhodobacter, Pseudorhodobacter, Rhodobaca, Rhodobacter, Roseicitreum, Roseibaca, Roseinatronobacter, Sinorhodobacter, and Thioclava.
Members of the genus Rhodobacter perform anoxygenic photosynthesis, fix dinitrogen and play a key role in bio-geochemical cycles. At present, the genus Rhodobacter comprises 16 validly named species: Rba. aestuarii, Rba. azollae, Rba. alkalitolerans, Rba. azotoformans, Rba. blasticus, Rba. capsulatus, Rba. johrii, Rba. lacus, Rba. maris, Rba. megalophilus, Rba. ovatus, Rba. sediminis, Rba. sphaeroides, Rba. veldkampii, Rba. Vinaykumarii, and Rba. viridis. Species of the genus Rhodobacter have ovoid to rod-shaped-cells, are Gram-stain negative, facultative photoheterotrophic and have vesicular/lamellar ICM architecture, bacteriochlorophyll-a, carotenoids of the spheroidene series and have Q10 as their major quinone. Some of the members are capable of growing photolithoautotrophically in the presence of sulfide/H2 as an electron donor (Imhoff, 2005; Girija et al., 2010; Suresh et al., 2017; Gandham et al., 2018).
Based on 16S rRNA gene sequence phylogenetic analysis we previously demonstrated the heterogeneity of the genus Rhodobacter, due to a large diversity of interspersing chemotrophs dividing the members into five monophyletic clusters (Suresh et al., 2017). Apart from being polyphyletic, the members of the genus Rhodobacter also have phenotypic differences which mainly include: heterogeneity in polar lipids (presence and absence glycolipids/diphosphatidylglycerol [DPG]), ICM architecture (vesicular/lamellar), sulfur metabolism and cell division (budding/binary fission). The phenotypic diversity among the taxa of the genus Rhodobacter was critically commented upon 21/2 decades back (Hiraishi and Ueda, 1994). However, due to a lack of strong evidence, reclassification of this genus has not yet been done. A taxogenomics (phylogenomics) helped in the reclassification of Mycobacterium (Gupta et al., 2018), Phaeobacter (Breider et al., 2014), Gammaproteobacterial methanotrophs (Orata et al., 2018), Geobacillus (Aliyu et al., 2016), Burkholderia (Lopes-Santos et al., 2017), Populibacter (Li et al., 2017), and Roseobacter group (Wirth and Whitman, 2018). In the present communication, based on phylogenetic, pan-genomic, phylogenomic/taxogenomic analysis supported by phenotypic and chemotaxonomic differences, we propose the reclassification of the genus Rhodobacter into three new genera and emended description of genus Rhodobacter sensu stricto.
All the 16 current type species of the genus Rhodobacter were cultured and maintained on the medium described by Lakshmi et al. (2011) in completely filled screw cap test tubes (10 × 100 mm) under photoheterotrophic conditions using pyruvate as a carbon source. Pure cultures were maintained on agar slants or as lyophilized cultures were preserved at 4°C.
Type strains of twelve species of the genus Rhodobacter and type species of different genera of the Rhodobacter group in the family ‘Rhodobacteraceae’ (Pujalte et al., 2014), whose genomes were available along with representatives from each of the remaining four groups (Lee et al., 2005) were considered in this study. Escherichia coli ATCC 11775T and Pseudomonas aeruginosa DSM 50071T genomes were included as out groups. The genome sequences of type strains of Rba. blasticus, Rba. veldkampii and Cereibacter changlensis were shared by Prof. Meyer Terrance, prior to their release to the NCBI database. All the other genome sequences used in the present study were downloaded from the NCBI database (Supplementary Table S1).
Phylogenetic analysis was performed with MEGA7 software (Kumar et al., 2016). For 16S rRNA gene-based phylogenetic analysis, sequences of all Rhodobacter spp. and related members were extracted from the NCBI database. From all 32 genomes selected in this study including the two outgroups, ninety-two core genes were retrieved using the Up-to-date Bacterial Core Gene (UBCG) tool (Na et al., 2018). A concatenated sequence was used to construct the phylogenetic tree. For both trees (16S rRNA gene and UBCG tree) distances were calculated by using the Kimura2-parameter (Kimura, 1980) in a pairwise deletion procedure and Poisson model (Zuckerkandl and Pauling, 1965) for the protein sequences of PufX, PufL,M and photosynthetic gene cluster (PGC) based phylogenetic trees. Neighbor-Joining (NJ), Maximum Likelihood (ML) and Minimum Evolution (ME) methods in the MEGA7 software were used to construct phylogenetic trees. Bootstrap analysis was carried out with 1000 resampling.
The phylogenomic tree was also deduced from the genome translated amino acids using the automated pipeline of the Patric online server3. In brief, the Patric server begins with amino acid sequence files for each genome. Homologous proteins were identified in two rounds. BLAST was used in first round in which the genome of each distinct species is searched against other genomes and an MCL (Markov Cluster) algorithm was used to cluster the top scoring hits, which are initial seed sets for the homology groups. Alignment of seed sets was carried out by MUSCLE, and hmmbuild was used to build Hidden Markov Models (HMMs). The phylogenomic tree was generated with FastTree and RAxML from the concatenated alignment. Instead of bootstraps, trees are built from random samples of 50% of the homology groups used for the main tree, in a process referred to as gene-wise jackknifing. Hundred of these 50% gene-wise jackknife trees are made using FastTree/RAxML, and the support values shown indicate the number of times a particular branch was observed in the support trees.
The pan-genome analysis of genomes of type strains of 12 species of the genus Rhodobacter was carried out using the BPGA pipeline (Chaudhari et al., 2015), with default parameters to check the inter species variation and core genome. The pan-genome analysis was also carried out for proposed genera. In order to ensure the correct assignation at the species level of each analyzed genome, the ANI and the dDDH were calculated between the genomes. The OrthoANIu values were calculated using the OrthoANI tool (Yoon et al., 2017). The dDDH values were calculated with the GGDC software and values obtained with the formula 2 were considered (Auch et al., 2012). Average amino acid identity (AAI) and percentage of conserved protein (POCP) was calculated among the selected genomes as these are considered as important for genus classification. The AAI was calculated using the AAI calculator developed by Kostas lab (Rodriguez and Konstantinidis, 2014). The POCP was determined as described by Qin et al. (2014).
Polar lipids were extracted from 1g freeze-dried cells with methanol/chloroform/saline (2:1:0.8, by vol.) as described by Kates (1986). Lipids were separated using silica gel TLC (Kieselgel 60 F254; Merck) by two-dimensional chromatography using chloroform/methanol/water (65:25:4 by vol.) in the first dimension and chloroform/methanol/acetic acid/water (80:12:15:4 by vol.) in the second dimension (Tindall, 1990a, b). The total lipid profile was visualized by spraying with 5% ethanolic molybdophosphoric acid and was further characterized by spraying with ninhydrin (specific for amino groups), molybdenum blue (specific for phosphates), Dragendorff (quaternary nitrogen), α-naphthol (specific for sugars) (Oren et al., 1996; Kates, 1972).
Based on 16S rRNA gene sequence phylogenetic analysis, the sixteen species of the genus Rhodobacter form five distinct clades (Figure 1) with interspersing chemotrophs (members of other described genera in the family ‘Rhodobacteraceae’). Clade I, the Rba. sphaeroides clade, contain Rba. sphaeroides, Rba. johrii, Rba. megalophilus, Rba. azotoformans, Rba. ovatus and Rba. alkalitolerans. Clade II contain the Rba. capsulatus clade, which includes Rba. capsulatus, Rba. viridis, Rba. azollae, Rba. sediminis, Rba. aestuarii, Rba. maris, and Rba. lacus. Clade III, the Rba. blasticus, clade contains only a single species, Rba. blasticus. Clade IV is the Rba. veldkampii clade containing only Rba. veldkampii while clade V is represented by Rba. vinaykumarii. The 16S rRNA gene pairwise sequence similarities among Rhodobacter species were calculated using LALIGN tool4 and the results showed that some of the Rhodobacter species (between clade I and clade II) have 94.0% sequence similarity (Supplementary Table S2), which is less than the recommended value for genus delineation (Rosselló-Móra and Amann, 2015).
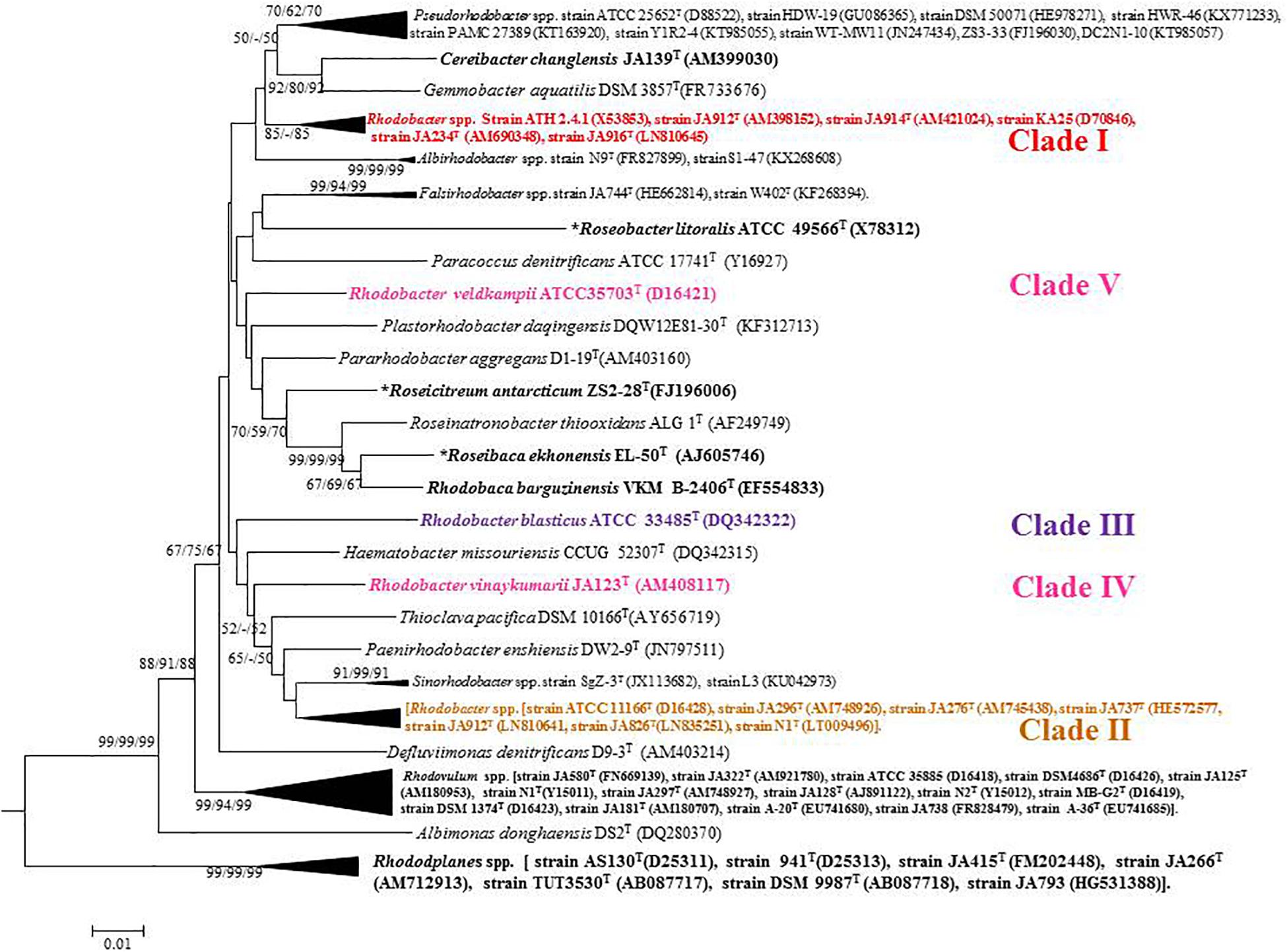
Figure 1. Phylogenetic tree based on 16S rRNA gene sequences showing the phylogenetic relationships of the genus Rhodobacter of the family ‘Rhodobacteraceae.’ The tree was computed with MEGA 7 software and rooted by using Escherichia coli and Pseudomonas aeruginosa as out-group. The GenBank accession numbers for 16S rRNA gene sequences are shown in parentheses. Bootstrap percentages refer to NJ/ML/ME analysis. Bar, 0.01 nucleotide substitution per position. Phototrophic bacteria are indicated by bold letters and aerobic anoxygenic phototrophic bacteria with bold letters and star.
Out of 16 type strains of Rhodobacter, genome sequences of 12 type strains are available. Genome sequences of Rba. alkalitolerans (Gandham et al., 2018), Rba. azollae, Rba. lacus (Suresh et al., 2017) and Rba. sediminis (Subhash and Lee, 2016) are not available. A genomic summary of the twelve species (and some related strains) is presented in Figure 2 and Supplementary Table S3. Clade I members typically have large genomes (4.3–4.7 Mb) and high G+C content (68.2–69.1 mol%). An exception in Clade I is Rba. ovatus, which has an 3.8 Mb genome and genomic GC content of 66.5 mol%. Clade II members have 3.6–3.9 Mb genomes and genomic GC content of 61.0–66.6 mol%. Clade III members have a genome size of 3.6–3.7 Mb, 66.4–66.5 GC mol%, while clade IV and V have 3.3, 3.5 Mb and 65.0, 68.2 GC mol%, respectively. Phylogenetic analysis based on 92 bacterial core genes (UBCG) also showed (Figure 3) that members of Rhodobacter are not monophyletic, as observed in the 16S rRNA gene-based phylogenetic tree (Figure 1), with the exception of Rba. veldkampii and Rba. vinaykumarii forming one cluster unlike in the 16S rRNA gene-based phylogenetic tree.
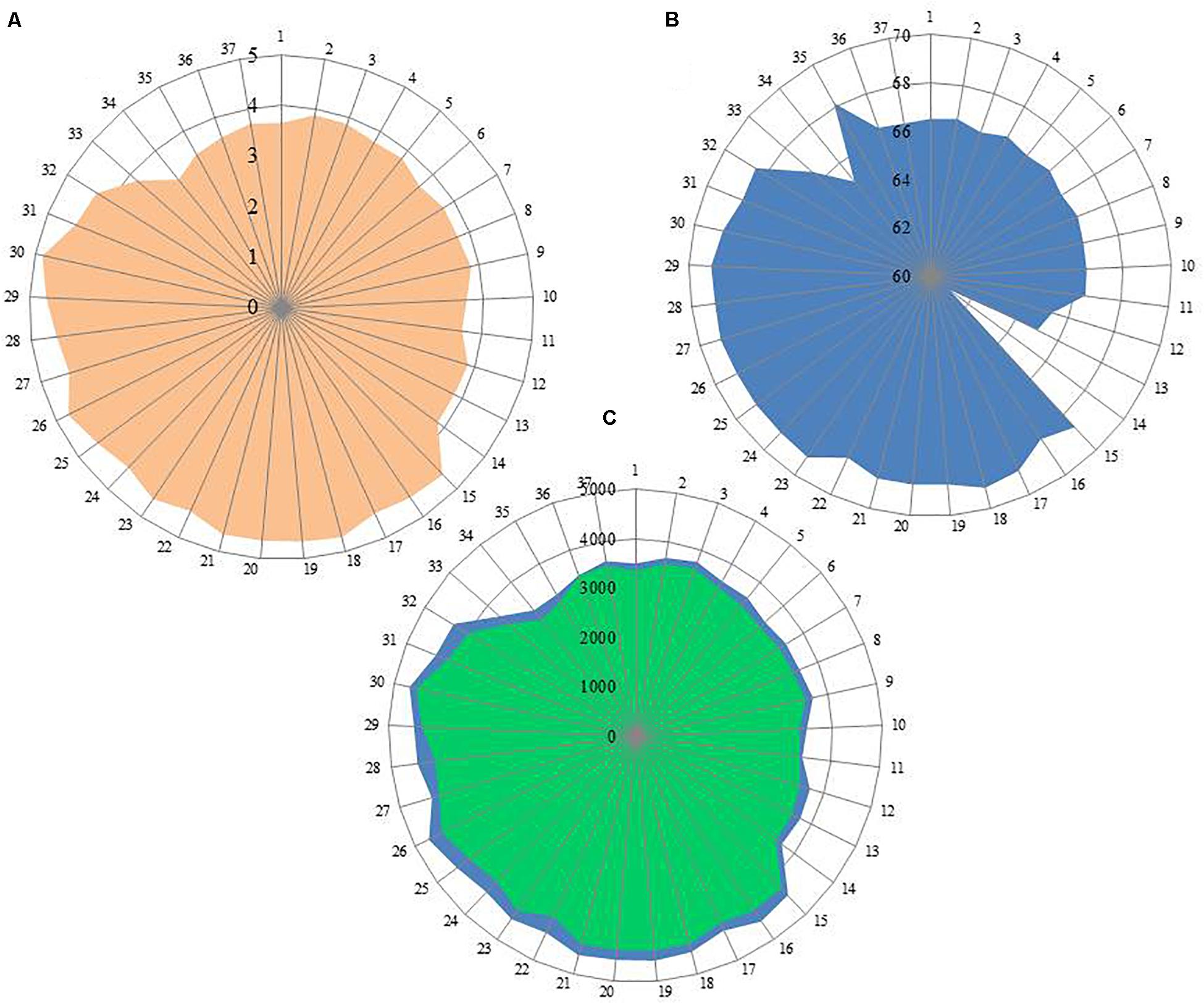
Figure 2. Genomic information of genus Rhodobacter members. (A) Genome size (Mb); (B) G+C content calculated from Genome in mol%; (C) No of genes in blue color and no of proteins in green color. Each spoke in the circle represent one strain and the numbering same as in Supplementary Table S3.
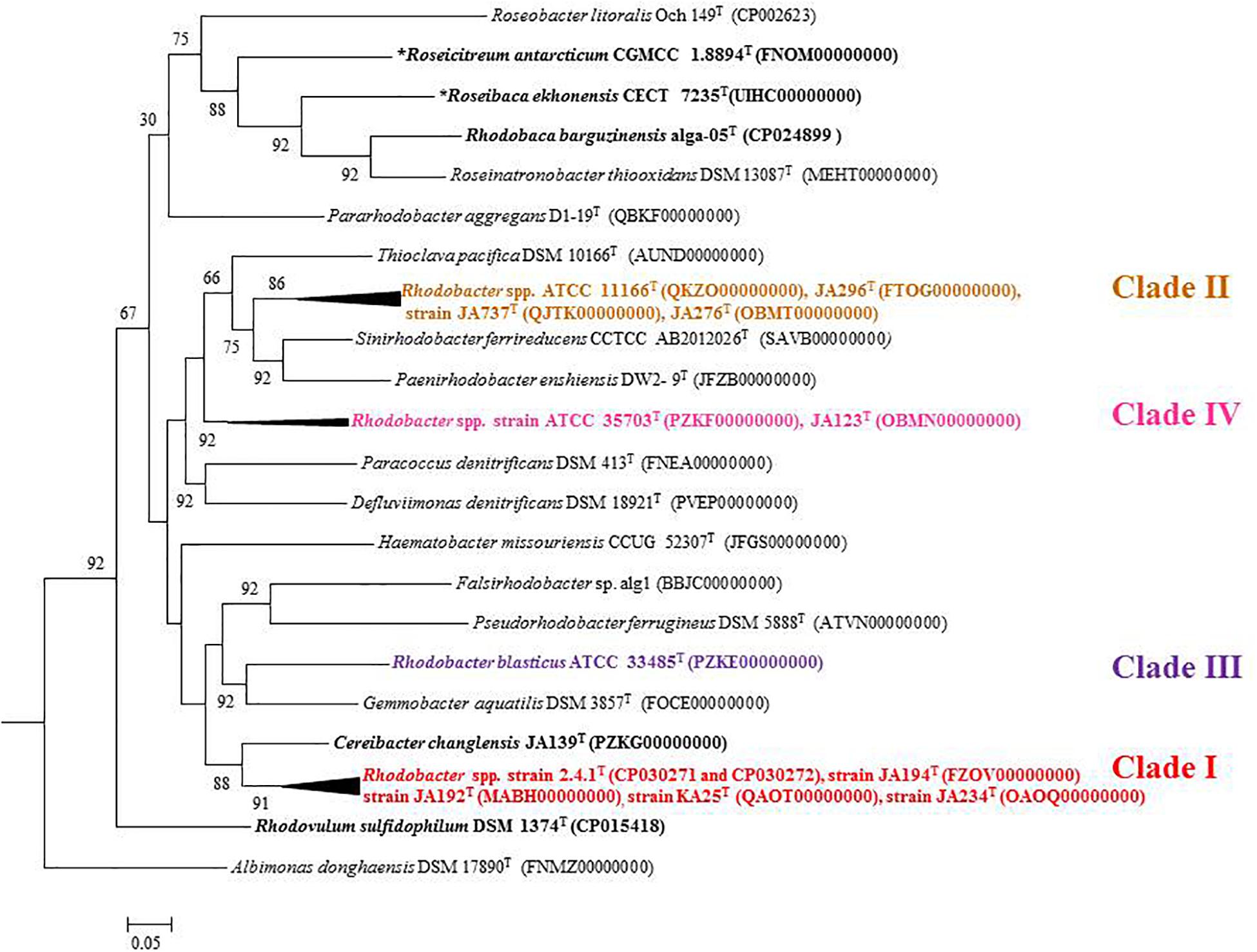
Figure 3. Phylogenetic tree constructed using the 92 bacterial core gene sequences. The tree was constructed using MEGA 7 software. Bootstrap percentages refer to NJ/ML/ME analysis. Phototrophic bacteria are indicated by bold letters and aerobic anoxygenic phototrophic bacteria with bold letters and star.
Bacterial pan-genome analysis was carried out between the type strains of the genus Rhodobacter. In the 12 analyzed species of the genus Rhodobacter, 1239 genes were identified as core genes. The species of the Rba. sphaeroides clade have 2294 genes as a core genome with 6256 genes as accessory (Supplementary Figure S1), whereas species of the Rba. capsulatus clade share 2225 genes as a core genome with 2588 genes as the accessory genome (Supplementary Figure S2). The distribution of core genome, accessory genome and unique genes is given in Supplementary Table S4.
dDDH values were calculated between 12 species of genus Rhodobacter and other closely related genera. dDDH values ranged from 18.6 to 59.0% between type strains of the genus Rhodobacter (Supplementary Table S5) except between the pair of Rba. sphaeroides 2.4.1T and Rba. megalophilus JA194T (81.6%). The results of dDDH also matched with the data obtained using the Type (Strain) Genome Server (TYGS) (Meier-Kolthoff and Göker, 2019). Orthologous Average Nucleotide Identity (OrthoANI) varied between 68.25 and 94.93% between type strains of the genus Rhodobacter. The only exception was again the pair, Rba. sphaeroides 2.4.1T and Rba. megalophilus JA194T, where it was 97.96% (Supplementary Table S5).
A POCPs threshold below 50% was proposed to determine if a species belongs to the two different genera (Qin et al., 2014). The POCP values within the genus Rhodobacter ranged from 54.2 to 88.2%. The lowest POCP value was between Rba. megalophilus JA194T and Rba. aestuarii JA296T and highest POCP value was between Rba. megalophilus JA194T and Rba. sphaeroides 2.4.1T. The POCP values among the clade I species ranged from 66.5 to 88.2%, whereas for clade II values were between 72.1 to 79.9%, and was 71.1% between Rba. veldkampii and Rba. vinaykumarii. The POCP value between Rba. capsulatus ATCC 11166T and Rba. sphaeroides 2.4.1T was 59%. POCP values among Rhodobacter clades and related genera are above 50% (Supplementary Table S6).
The AAI among the different clades of Rhodobacter along with chemotrophs were below 80% (Supplementary Table S6). The AAI values within the genus Rhodobacter ranged from 63.04 to 98.56%. The lowest AAI value was between Rba. megalophilus JA194T and Rba. aestuarii JA296T and highest AAI value was between Rba. megalophilus JA194T and Rba. sphaeroides 2.4.1T. The AAI values in clade I range from 98.56 to 80.37%, whereas clade II values range from 77.86% to 89.21%, and 72.85% between Rba. veldkampii and Rba. vinaykumarii. The AAI value between the Rba. capsulatus ATCC 11166T and Rba. sphaeroides 2.4.1T was 64.36%. AAI values between the type species of all the genera under study were below the recommended cut-off value of 80% (Luo et al., 2014; Supplementary Table S6). The average AAI value between the members of clade I and clade II was 64.1%, between clade I and clade III was 66.64%, between clade I and clade IV was 67.18%, between clade II and clade III was 63.72%, between clade II and IV was 68.53% and between the clade III and IV was 65.14%. These values are well below the recommended cut-off of 80% used for the genus delineation (Luo et al., 2014).
PufLM proteins are essential components of bacteria which perform photosynthesis with the help of photosystem II type of photosynthetic apparatus, and the phylogeny of both anaerobic and aerobic photosynthetic bacteria can be studied by PufLM protein analysis (Imhoff et al., 2018). Full length amino acid sequences of PufLM of 12 Rhodobacter type strains were extracted from their genomes and a phylogenetic tree was (Supplementary Figure S3) constructed using R. sulfidophilum as the out-group. All the Rhodobacter members were recovered in four different clades, similar to the 16S rRNA gene based phylogenetic tree although Rba. veldkampii and Rba. vinaykumarii formed a single cluster, as in the phylogenomic tree (Figure 3).
Crouch and Jones (2012) showed that some of the current members of the genus Rhodobacter (Rba. sphaeroides, Rba. azotoformans, Rba. blasticus) and C. changlensis (formerly Rba. changlensis) form RC-LH1 (photosynthesis reaction center-light-harvesting 1) complex dimers instead of monomers, as in Rba. capsulatus, Rba. veldkampii and Rba. vinaykumarii. The dimer formation is facilitated by the PufX protein, by interacting with two monomers of RC–LH1 complex, without disturbing their structure (Crouch and Jones, 2012). Deletion of the pufX gene in Rba. sphaeroides resulted in the formation of only RC-LH1 monomers and loss of the ability to grow phototrophically. However, in contrast, Rba. veldkampii in spite of having the PufX coding gene, forms only monomers and can grow phototrophically. This result indicated that RC-LH1 dimer formation is not only dependent on the presence or absence of PufX protein but also on some unidentified factors (Crouch and Jones, 2012). Mutational studies on PufX protein C-terminal sequences of Rba. sphaeroides reveal the importance of R49, G52 or R53 amino acids and alteration of these sequences prevents RC-LH1dimer formation (Qian et al., 2017).
To analyze members of Rhodobacter spp. for the presence of the sequence pattern required for RC-LH1 dimer formation, we extracted the PufX protein sequences of Rhodobacter members from NCBI and aligned them using CLUSTALW. Interestingly, all the members of Rba. sphaeroides clade have R49, G52 or R53 amino acids (Figure 4), while none of the other members of Rhodobacter outside of the Rba. sphaeroides clade has such a pattern. A phylogenetic tree (Supplementary Figure S4) constructed using the PufX protein sequences of Rhodobacter members and some phylogenetically related chemotrophic bacteria shows that Rhodobacter spp. are polyphyletic.

Figure 4. PufX protein sequence alignment using clustalW, showing the R49, G52 or R53 sequence pattern, which is important for RC-LH1 dimer formation.
All essential genes required for photosynthesis are organized in a continuous stretch called the photosynthetic gene cluster (PGC). Phylogenetic analysis of one or two photosynthetic genes (Imhoff et al., 2018) is not sufficient to study the photosynthetic gene phylogeny among the members of the ‘Rhodobacteraceae’ (Brinkmann et al., 2018). Thus, in addition to the PufLM or PufX protein based phylogenetic analysis, we studied the photosynthetic gene arrangement in the PGC to differentiate Rhodobacter spp. Comparative analysis of the PGC of Rhodobacter spp. (Figure 5) revealed that members of the Rba. sphaeroides clade lack the pufB gene sequence encoding for light-harvesting antenna LH1-beta subunit. In contrast the acsF gene encoding for the Mg-protoporphyrin IX oxidative cyclase, aerobic form, was not found in the genomes of members of the Rba. capsulatus clade (Figure 5). In the Rba. sphaeroides clade, urea metabolism genes were seen after the cycA gene, whereas in the remaining clades these genes are scattered elsewhere in their genomes but not adjacent to the PGC. The PGC gene encoded protein sequences were concatenated and used to construct a phylogenetic tree (Supplementary Figure S5), which was congruent with the PufLM protein, PufX protein and 16S rRNA gene phylogenetic trees. In the PGC based phylogenetic tree Rhodobacter spp. formed four monophyletic clades (Supplementary Figure S5).
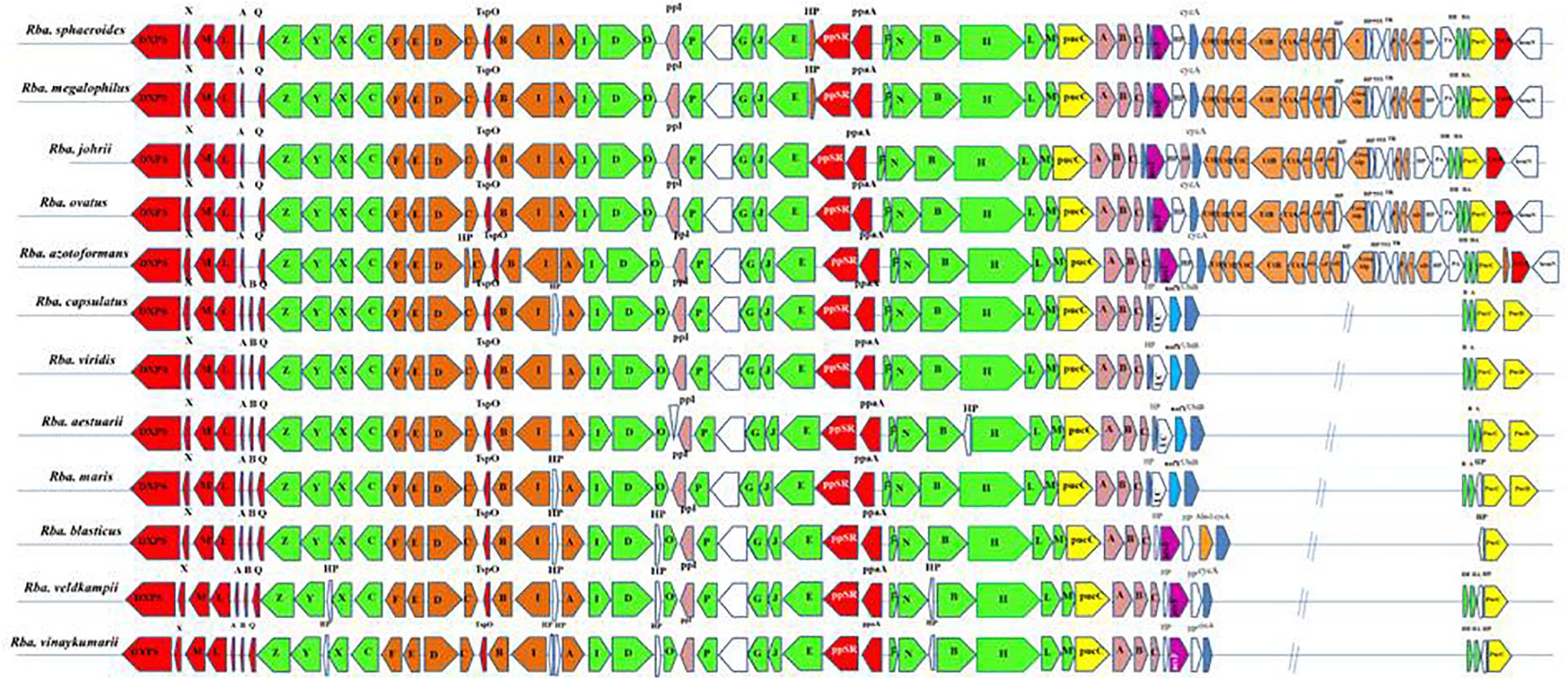
Figure 5. Arrangement of photosynthetic gene cluster and structure in Rhodobacter. Green, bacterial chlorophyll genes; red, puf and regulators genes; pink, puh genes; orange, carotenoid genes; blue, hem and cyc gene; yellow, lhaA gene; blank, uncertain or unrelated genes; gray, hypothetical protein.
The polar lipid composition of Rhodobacter spp. typically includes phosphatidylethanolamine, phosphatidylglycerol, uncharacterized aminolipid, phosphatidylcholine (except Rba. veldkampii) and other unidentified lipids. In addition, clade I members have DPG and an unidentified glycolipid (GL). Polar lipid profiles of the representative strains are shown in Supplementary Figure S6.
The use of genomics in taxonomic studies improves the credibility, reliability and reproducibility of the data, serves as an informative tool in establishing phylogenetic relationships among prokaryotes and helps in eliminating unreliable methods and subjective difficult-to-replicate data (Chun and Rainey, 2014; Chun et al., 2018; Pérez-Cataluña et al., 2018). To resolve the taxonomic conflict which have arisen due to heterogeneity among the Rhodobacter spp., genome based analysis was carried out in this study. The genome size of clade I members is relatively larger than that of the remaining clades, with the exception of Rba. ovatus. The clade I members have higher G+C content (68.2–69.1 mol%) when compared with clade II (61.0–66.6 mol%), clade III (66.4–66.5 mol%), clade IV (65.0 mol%) and clade V (68.2 mol%). According to Nouioui et al. (2018) genome size variation can be a genus specific taxonomic marker, indicating that the clade I members might be belong to a distinct genus from Rhodobacter sensu stricto (Clade II). This is also in agreement with the separation of Rba. capsulatus strains from Rba. sphaeroides in the phylogenomic analysis of Simon et al. (2017).
Taxonomy of bacteria based on core- and pan-genome analysis is a powerful extension of the polyphasic approach (Inglin et al., 2018). Pan-genome analysis of the genus Rhodobacter revealed that only 1239 core genes are present (Supplementary Table S4). An increased number of core genes was observed when the clade wise pan genome analysis performed, which indicated the inter cluster genomes were divergent.
Digital DDH values (Supplementary Table S5) among Rhodobacter members support the current species delineations, except between Rba. sphaeroides 2.4.1T and Rba. megalophilus JA194T which exceeds the cut off of 70% dDDH value for prokaryotic species delineation (Wayne et al., 1984). This suggests that Rba. megalophilus JA194T can be a sub-species of Rba. sphaeroides ATH 2.4.1T. With the advancement in next generation sequencing methods, ANI is increasingly being used to delineate closely related species (Kim et al., 2014). Bacterial strains having less than 95–96% of ANI are considered as distinct species (Goris et al., 2007; Richter and Rosselló-Móra, 2009; Rodriguez and Konstantinidis, 2014). In agreement with the dDDH data, OrthoANI values also confirm that all species of Rhodobacter are well described at species level, exception for Rba. megalophilus which gave an ANI score of 97.96% with Rba. sphaeroides 2.4.1T (Supplementary Table S5), confirming these two strains belong to the same species.
Qin et al. (2014) proposed that if the POCP values between two species were less than 50% then they belong to two different genera. However, this threshold does not appear to apply among members of Rhodobacter (Supplementary Table S6), suggesting that they all belong to a single genus. POCP cut off values are ineffective in delineating genera in the family ‘Rhodobacteraceae’ (Wirth and Whitman, 2018), Methylococcaceae (Orata et al., 2018) and Neisseriaceae (Li et al., 2017) suggesting a need to establish appropriate POCP cut-off values for different families.
Percentage of AAI below 60% between the compared genomes of species was also proposed to delineate genera (Rodriguez and Konstantinidis, 2014). However, the AAI values between members of related but different genera can vary between 60 and 80% (Luo et al., 2014; Orata et al., 2018). The similarity index of AAI between Rhodobacter spp. (Supplementary Table S6) strengthen the proposal for reclassifying them, while the phylogenetically related genera (chemotrophs) in the family ‘Rhodobacteraceae’ are well described and require no further reclassification. The phylogenomic tree topology using genomic DNA sequences (UBCG; Figure 3) or protein sequences (Figure 6 and Supplementary Figure S7) was in congruence with the 16S rRNA gene-based phylogeny (Figure 1). The phylogenetic tree based on the proteins (Supplementary Figures S3–S5) involved in photosynthesis also showed the polyphyletic assemblage of the Rhodobacter spp. which indicates the need of reclassification of some members of the genus Rhodobacter.
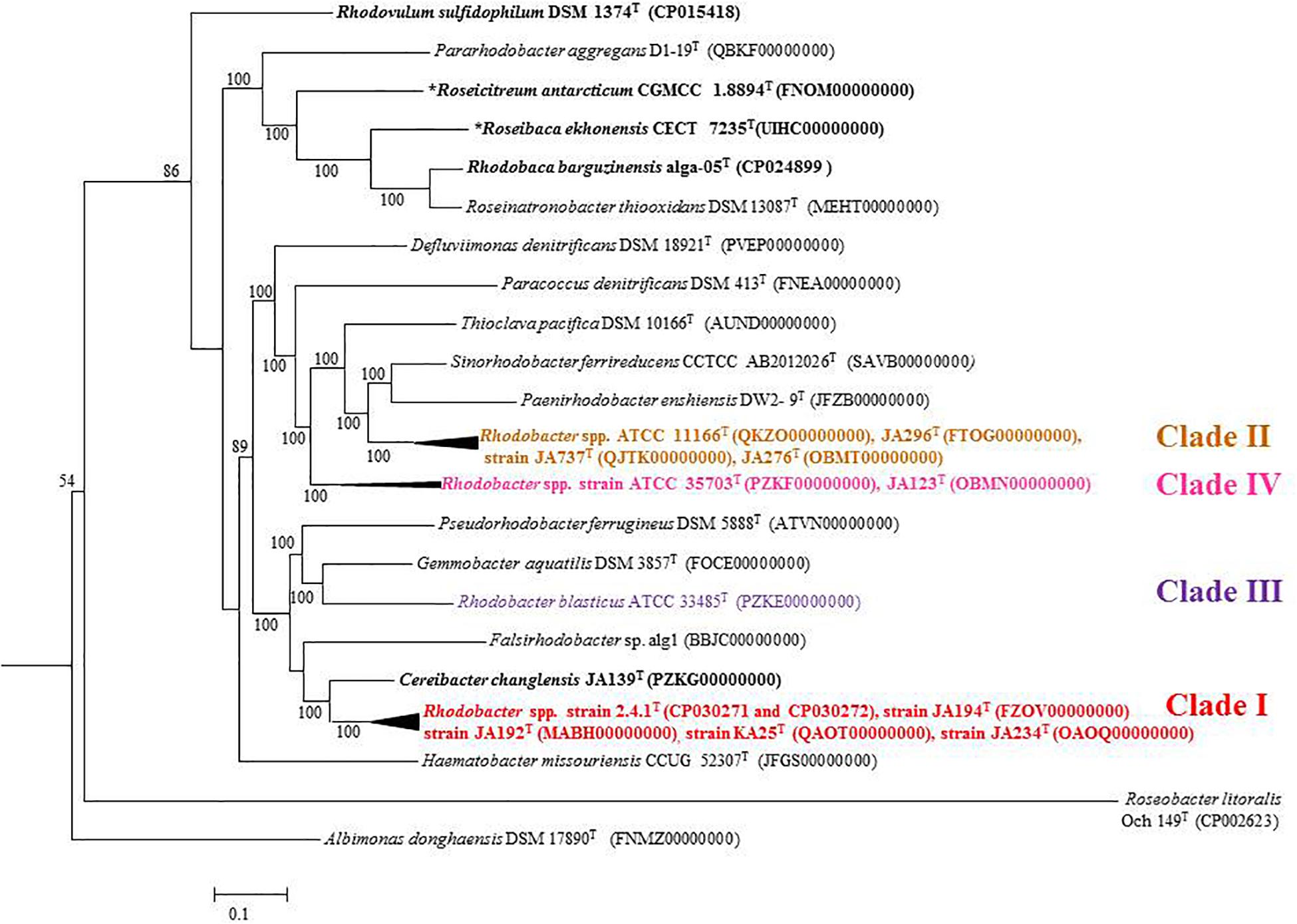
Figure 6. Phylogenomic tree of type strains of genus Rhodobacter and its nearest member with Escherichia coli and Pseudomonas as the out group. The tree was elucidated using the FastME from the whole proteome. The numbers above branches are pseudo-bootstrap support values from 100 replicates, only values above 50% are shown. Phototrophic bacteria are indicated by bold letters and aerobic anoxygenic phototrophic bacteria with bold letters and star.
The conflict observed in the current classification of the genus Rhodobacter is visible in single gene based trees, either in the 16S rRNA gene (Figure 1) or rpoB gene based tree (Gandham et al., 2018), which is strengthened by better resolved whole genome phylogenies. The results of phylogenomic analysis emphasize the need for reclassification of the taxonomically complex genus Rhodobacter. Housekeeping genes (such as rpoB) can be used to determine the phylogenetic relationships of species when the 16S rRNA gene sequence based phylogenetic tree is unable to resolve the phylogenetic position of taxa, based on the fact that the house keeping genes are less conversed and evolve faster than the 16S rRNA gene (Martens et al., 2008). Based on the 16S rRNA gene and rpoB gene phylogenetic trees we believe that genome sequences for the remaining four members (Rba. alkalitolerans, Rba. azollae, Rba. lacus and Rba. sediminis) are not essential to resolve the taxonomic positions of members of the genus Rhodobacter.
The members of Rba. sphaeroides clade (clade I) were isolated from freshwater environments only (Srinivas et al., 2008; Girija et al., 2010; Gandham et al., 2018), whereas Rba. capsulatus clade (clade II) members have been isolated from freshwater (Suresh et al., 2017), estuarine (Venkata Ramana et al., 2009) and marine habitats (Venkata Ramana et al., 2008). Rba. blasticus, which is assigned to clade III, and Rba. veldkampii (clade IV member), were isolated from freshwater habitats (Eckersley and Dow, 1980; Hansen and Imhoff, 1985), while Rba. vinaykumarii was isolated from a marine habitat (Srinivas et al., 2007). However, the isolation source is not the distinguishing feature between the clades. The members of clade II are motile (Suresh et al., 2017), whereas some of the members of clade I do not exhibit motility, and members of clade III, IV, and clade V are not motile (Eckersley and Dow, 1980; Hansen and Imhoff, 1985; Srinivas et al., 2007). Except for Rba. blasticus which multiplies by sessile budding and has a lamellar ICM, the remaining species of Rhodobacter multiply by binary fission and have a vesicular architecture of their ICMs (Table 1). Only the clade V member (Rba. vinaykumarii JA123T) has a requirement of NaCl for optimum growth and cannot assimilate sulfate (Srinivas et al., 2007). Only one member (Rhodobacter azotoformans) of clade I has denitrification ability (Hiraishi et al., 1996).
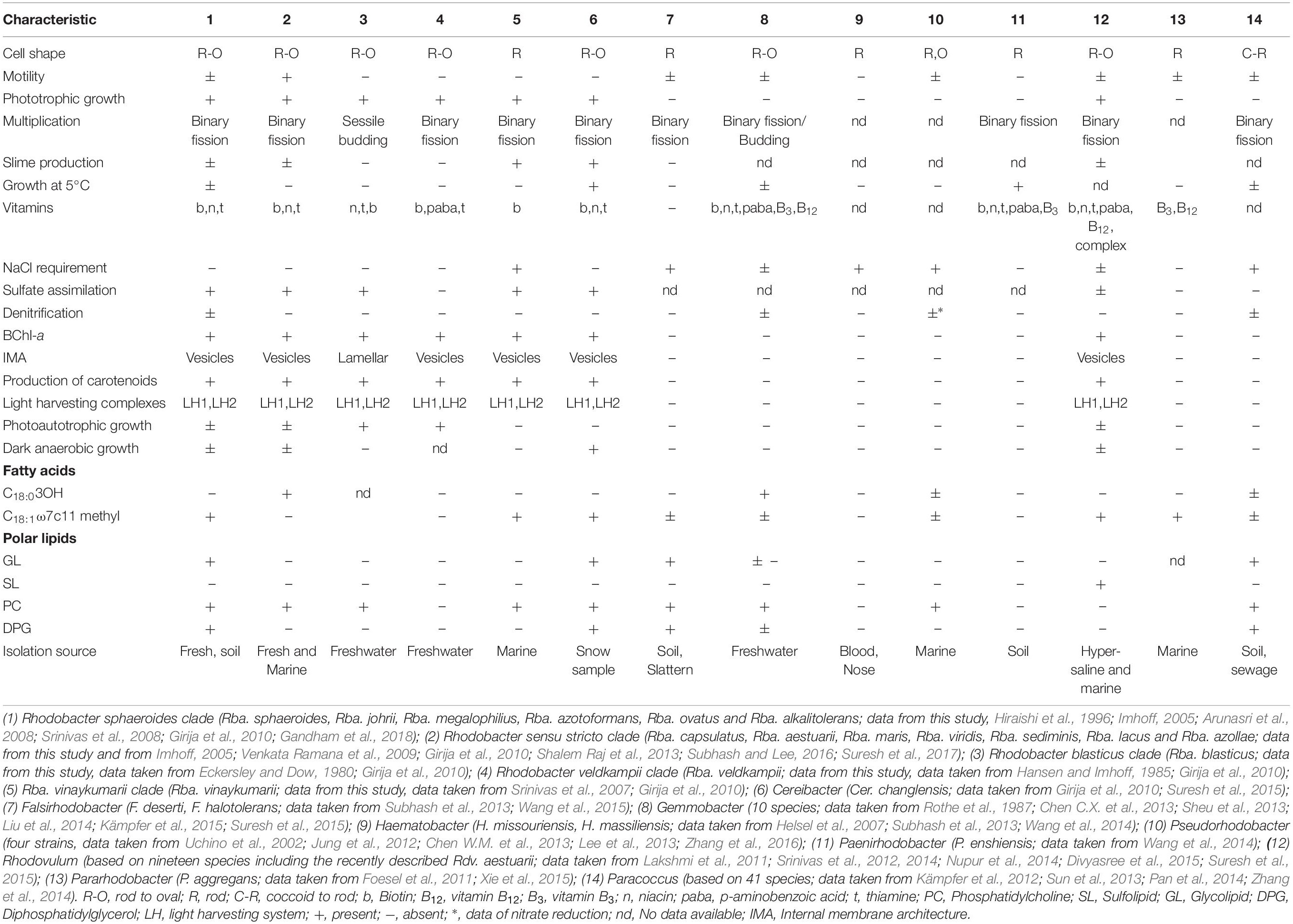
Table 1. Differentiating characters of taxa of the genus Rhodobacter and closely related genera of the family ‘Rhodobacteraceae.’
All the members of clade I (Rba. sphaeroides, Rba. johrii, Rba. megalophilus, Rba. azotoformans, Rba. ovatus and Rba. alkalitolerans) have glycolipid and DPG (Gandham et al., 2018; Supplementary Figure S6). Polar lipid analysis has been a central tool in chemotaxonomy and the presence/absence of glycolipid was considered as one of the genus differentiating characters in the reclassification of some species of the genus Rhodobium into a new genus, Afifella (Urdiain et al., 2008). Members of clade II, III, IV, and V do not have DPG or glycolipid (Suresh et al., 2017; Supplementary Figure S6). Except for some of members of clade II, all members of the new genera proposed below have spheroidene as their major carotenoid (Girija et al., 2010; Suresh et al., 2017; Gandham et al., 2018). The chemotaxonomic comparison between the genus Rhodobacter sensu stricto, the proposed novel genera and closely related genera is given in the Table 1.
Phenotypic and phylogenomic features support the separation of genus Rhodobacter into four different genera. Emended descriptions and reclassifications are given for these taxa. Apart from the tree topologies, with interspersing chemotrophic members, the branch lengths in the whole-genome and 16S rRNA trees indicated that the Rhodobacter species are too divergent to be placed into the same genus. Since its description, five out of 21 originally proposed Rhodobacter species have been reclassified into different genera and it has been emphasized that Rhodobacter is still heterogeneous and should be subdivided based on additional molecular taxonomic data (Hiraishi and Ueda, 1994; Lee et al., 2005; Helsel et al., 2007; Suresh et al., 2015).
16S rRNA gene and rpoB gene (Gandham et al., 2018) based phylogenetic trees shows that Rba. alkalitolerans JA916T together with Rba. johrii, Rba. megalophilus, Rba. sphaeroides, Rba. azotoformans and R. ovatus fall outside of the Rba. capsulatus clade and belong in what we define as the Rba. sphaeroides clade. We propose that the members of the Rba. sphaeroides clade be classified into a new genus for which we propose the name Luteovulum gen. nov. Moreover, as Rba. sphaeroides 2.4.1T and Rba. megalophilus JA194T have high genomic indices (OrthoANI and dDDH) we conclude that they are not two different species. Based on phenotypic differences (Arunasri et al., 2008), we propose Rba. megalophilus as a sub-species of Rba. sphaeroides.
Rba. viridis, Rba. azollae, Rba. sediminis, Rba. capsulatus, Rba. aestuarii, Rba. lacus and Rba. maris form a distinct clade in both the single gene trees (rpoB gene and 16S rRNA gene). This clade accommodates the type species of the genus Rhodobacter, Rba. capsulatus and members of this clade are proposed to be included in the genus Rhodobacter sensu stricto. Rba. veldkampii and Rba. vinaykumarii formed two different clusters with interspersing chemotrophs. However, in the genome-based trees (UBCG based, proteome based) they are clustered together. Genome based phylogenomic trees have better resolution than single gene-based trees, so for the time being we are not separating these two into two different genera, which can be done in future based on multiple species/strains. For the Rba. veldkampii clade accommodating Rba. vinaykumarii and Rba. veldkampii, we propose the new genus Phaeovulum gen. nov. To accommodate Rba. blasticus, we propose the new genus Fuscovulum gen. nov. The classification of Rba. blasticus itself controversial when it was transferred from the genus Rhodopseudomonas to Rba. blasticus, despite having lamellar ICM structures in contrast to all other members of genus Rhodobacter, which have vesicular ICM structures. It was proposed that ICM systems may be of questionable value as genus specific characters, because they are not coincident with the 16S rRNA gene phylogenetic relationships (Kawasaki et al., 1993). Later Hiraishi and Ueda (1994) suggested separating Rba. veldkampii and Rba. blasticus from the genus Rhodobacter into a novel genus based on the ICM architecture, mode of cell division of Rba. blasticus, and final oxidation product of sulfide in the case of Rba. veldkampii. Hiraishi and Ueda (1994) considered that mode of cell division and ICM types are genus specific characters. Here we propose the reclassification of each of these species into separate genera which, after two and half decades, helps to resolve the taxonomic conflict within the genus Rhodobacter, based on the genomic and chemotaxonomic information.
Members can be isolated from freshwater ponds, estuarine and marine environments. Oval-to-rod shaped cells have vesicular ICM architecture. Cells are mostly motile with a single polar flagellum and multiply by binary fission. Catalase and oxidase positive. Primarily phototrophic and contain BChl-a and carotenoids of the spheroidene series. Aerobic growth occurs in most species. Mesophilic. Phototrophic growth occurs on a range of organic substrates. The growth factors biotin, niacin and thiamine, alone or in combination, are required for growth to occur. NaCl requirement is not obligatory, can tolerate up to 2–3%. C18:1ω7c/C18:1ω6C, C18:0, C16:0 and C16:1ω7C/C16:1ω6c are the major fatty acids. Most species have C10:03OH and C18:03OH as fatty acid alcohols. Phosphatidylethanolamine, phosphatidylglycerol and phosphatidylcholine are the major polar lipids. Glycolipid and diphosphatidylglycerol are absent. Hopanoids are not produced. Q-10 is the major quinone.
The type species is Rhodobacter capsulatus (Molisch, 1907) Imhoff et al. (1984).
Based on the OrthoANI and dDDH analysis, Rba. megalophilus is not a distinct species as the differences between Rba. sphaeroides and Rba. megalophilus represent intra-species divergence. It should be noted that Rba. sphaeroides and Rba. megalophilus differ with regard to growth at 5°C, absence of flagellar motility, cell suspension color, vitamins required for growth, ability to utilize sulfide or hydrogen as electron donors and ability to utilize citrate as carbon source (Arunasri et al., 2008). Based on chemotaxonomic data we propose that Rba. megalophilus be reclassified as a sub-species of Rba. sphaeroides.
Luteovulum [Lu.te.o’vu.lum. L. adj. luteus yellow; N.L. dim. neut. n. ovulum (from L. n. ovum, an egg) a small egg; N.L. neut. n. Luteovulum small yellow egg].
Members can be isolated from freshwater ponds, paddy soils, wastewater treatment plants, alkaline ponds and lake sediments. Gram-stain negative, oval-to-rod shaped cells and have vesicular ICM architecture. Cells are mostly motile with a single polar flagellum and multiply by binary fission. Catalase and oxidase positive. Primarily phototrophic and contain BChl-a and carotenoids of the spheroidene series. Facultative aerobes and mesophilic. Phototrophic growth occurs on a number of organic substrates. The growth factors biotin, niacin and thiamine, alone or in combination, are required for growth. NaCl requirement is not obligatory, can tolerate up to 2–3%. C18:1ω7c/C18:1ω6C, C18:0, C16:0, C10:0 3OH, C16:1ω7C/C16:1ω6c, C18;1ω7c11 methyl are the major fatty acids. Phosphatidylethanolamine, phosphatidylglycerol, diphosphatidylglycerol, phosphatidylcholine and an unidentified glycolipid are the major polar lipids. Hopanoids are not produced. Q10 is the major quinone. Delineation of the genus is based on 16S rRNA and rpoB gene-based phylogeny, phylogenomics, genome comparison and chemotaxonomic differences.
The type species is Luteovulum sphaeroides subsp. sphaeroides.
The description of Luteovulum sphaeroides is identical to that of Rba. sphaeroides (Imhoff et al., 1984; Imhoff, 2005) except for the following modifications. C17:0 is present in minor quantities. An unidentified aminolipid, an unidentified phospholipid and two unidentified lipids are additional polar lipids. Type strain is available from the DSMZ (DSM 158T) and LMG (LMG 2827T). The 16S rRNA gene sequence GenBank/EMBL/DDBJ accession number of the type strain is X53853 and CP030271, CP030272, CP030273, CP030274, CP030275, and CP030276 are the genome sequence accession numbers of the type strain.
Luteovulum megalophilum (me.ga.lo’phi.lum. Gr. adj. megas, wide; N.L. adj. philus -a -um (from Gr. adj. philos -ê -on) friend, loving; N.L. neut. adj. megalophilum, wide (temperature)-loving). The description of Luteovulum megalophilum is identical to that of Rba. megalophilus (Arunasri et al., 2008) except for the following modifications. 3-Hydroxy C10:0 and C12:0 fatty acids are present. The DNA G+C content of the type strain calculated from genome sequence is 68.8 mol%. The type strain is available from JCM (JCM 14598T) and KCTC (KCTC 5602T). The 16S rRNA gene sequence GenBank/EMBL/DDBJ accession number of the type strain AM421024 and that of the genome sequence is FZOV00000000.
Luteovulum johrii (joh’ri.i. N.L. masc. gen. n. johrii of B. N. Johri, an eminent and well-known Indian microbiologist).
Basonym: Rhodobacter johrii Girija et al. (2010).
The description of Luteovulum johrii is identical to that of Rba. johrii (Girija et al., 2010). The type strain is available from the JCM (JCM 14543T) and DSMZ (DSM 18678T). The 16S rRNA gene sequence GenBank/EMBL/DDBJ accession number of the type strain is and that of the genome sequence is MABH00000000.
Luteovulum ovatum (o.va’tum. L. neut. adj. ovatum, egg-shaped, ovate)
Basonym: Rhodobacter ovatus Srinivas et al. (2008).
The description of Luteovulum ovatum is identical to that of Rba. ovatus (Srinivas et al., 2008). Type strain is available at JCM (JCM 14779T) and CCUG (CCUG 55049T). AM690348 is the 16S rRNA gene sequence GenBank/EMBL/DDBJ accession number and OAOQ00000000 is the genome sequence accession number.
Luteovulum azotoformans (a.zt.to.for’mans. N.L. n. azotum [from French n. azote (from Gr. prep. a, not; Gr. n. zôê, life; N. Gr. n. azôê, not sustaining life)], nitrogen; N.L. pref. azo-, pertaining to nitrogen; L. part. adj. formans, forming; N.L. part. adj. azotoformans, nitrogen forming).
Basonym: Rhodobacter azotoformans Hiraishi et al. (1996).
The description of Luteovulum azotoformans is identical to that of Rba. azotofarmans (Hiraishi et al., 1996). The type strain is available from JCM (JCM 9340T) and NBRC (NBRC 16436T). The 16S rRNA gene sequence GenBank/EMBL/DDBJ accession number of the type strain is D70846 and that of the genome sequence is QAOT00000000.
Luteovulum alkalitolerans (al.ka.li.to’le.rans. N.L. n. alkali, alkali; L. part. adj. tolerans, tolerating; N.L. part. adj. alkalitolerans, alkali-tolerating).
Basonym: Rhodobacter alkalitolerans Gandham et al. (2018).
The description of Luteovulum alkalitolerans is identical to that of Rba. alkalitolerans (Gandham et al., 2018). The type strain is available from the KCTC (KCTC 15473T) and LMG (LMG 28749T). The 16S rRNA gene sequence GenBank/EMBL/DDBJ accession number of the type strain is LN810645.
Fuscovulum [Fusc.o’vu.lum. L. masc. adj. fuscus, tawny N.L. dim. neut. n. ovulum (from L. n. ovum, an egg), a small egg; N.L. neut. n. Fuscovulum small tawny egg].
Members can be isolated from freshwater habitats. Gram-stain negative, rod-to-oval shaped cells have lamellar ICM architecture. Cells are mostly non-motile and multiply by sessile budding. Catalase and oxidase positive. Primarily phototrophic and contain BChl-a and carotenoids of the spheroidene series. Growth occurs under aerobic, anaerobic and mesophilic conditions. Phototrophic growth occurs on a number of organic substrates. Nicotinic acid and thiamine are required for growth. NaCl is not required for growth. Q-10 is the major quinone. The G+C mol% of the type strain of the type species is 66.5%. Delineation of the genus is based on 16S rRNA and rpoB gene-based phylogeny, phylogenomics, genome comparison and chemotaxonomic differences.
The type species is Fuscovulum blasticum.
Fuscovulum blasticum (Gr. adj. blastikos -ê -on, budding, sprouting; N.L. masc. adj. blasticum, budding, apt to bud).
Basonym: Rhodobacter blasticus (Eckersley and Dow, 1980) Kawasaki et al. (1993).
The description of Fuscovulum blasticum is identical to that of Rhodopseudomonas blastica (Eckersley and Dow, 1980) and Rhodobacter blasticus (Imhoff et al., 1984; Imhoff, 2005). The type species is available from the ATCC (ATCC 33485T) and NBRC (NBRC 16437T). The 16S rRNA gene sequence GenBank/EMBL/DDBJ accession number of the type strain is DQ342322 and that of the genome sequence is PZKE00000000.
Phaeovulum [Phae.o’vu.lum. N.L. neut. adj. phaeum (from Gr. neut. adj. phaion), brown; N.L. dim. neut. n. ovulum (from L. n. ovum, an egg) a small egg; N.L. neut. n. Phaeovulum small brown egg].
Members can be isolated from marine habitats including tidal waters, fresh water. Gram-stain negative, rod to oval shaped cells have vesicular ICM architecture. Cells are mostly non-motile and multiply by binary fission. Catalase and oxidase positive. Primarily phototrophic and contain BChl-a and carotenoids of the spheroidene series. Growth occurs under anaerobic and mesophilic conditions. Phototrophic growth occurs on a number of organic substrates. Biotin, para-aminobenzoate and thiamine are required for growth either singly or in combination. Some strains do not contain phosphatidylcholine. Some require NaCl for growth and can tolerate up to 4%. C18:1ω7c/C18:1ω6C, C18:0, C16:0, C10:0 3OH, C16:1ω7C/C16:1ω6c, C18;1ω7c11methyl are the major fatty acids. Q-10 is the major quinone. G+C mol% is 65–68.25. Delineation of the genus is based on 16S rRNA and rpoB gene based phylogeny, phylogenomics, genome comparison and chemotaxonomic differences.
The type species is Phaeovulum veldkampii.
Phaeovulum veldkampii (veld. kamp’i.i. N. L. gen. masc. n. veldkampii of Veldkamp; named for Hans Veldkamp, a Dutch microbiologist).
Basonym: Rhodobacter veldkampii Hansen and Imhoff (1985).
The description of Phaeovulum veldkampii is identical to that of Rhodobacter veldkampii (Hansen and Imhoff, 1985; Imhoff, 2005). The type strain is available from the ATCC (ATCC 35703T) and DSMZ (DSM 11550T). The 16S rRNA gene sequence GenBank/EMBL/DDBJ accession number is D16421 and that of the genome sequence is PZKF00000000.
Phaeovulum vinaykumarii (vi’nay.ku.ma’ri.i. N.L. masc. gen. n. vinaykumarii of Vinaykumar, named after the late Dr. M. Vinaykumar, an Indian microbiologist and research supervisor of CVR and CS, who initiated work on anoxygenic phototrophic bacteria in India).
Basonym: Rhodobacter vinaykumarii Srinivas et al. (2007).
The description of Phaeovulum vinaykumarii is identical to that of Rhodobacter vinaykumarii (Srinivas et al., 2007). The type strain is available from the JCM (JCM 14544T) and DSMZ (DSM 18714T). The 16S rRNA gene sequence GenBank/EMBL/DDBJ accession number of the type strain is AM408117 and that of the genome sequence is OBMN00000000.
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
CVR and CS designed the work. GS maintained the bacterial cultures. TL and GS genomic data retrieval from databases, pangenomic, phylogenomic, and phylogenetic data analysis. GS, TL, and BI calculated the genomic relatedness indices. All authors made the manuscript and accepted the work for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Professor Aharon Oren for his expert advice concerning the genus and species epithet and Latin etymology. We thank Prof. Meyer Terrance for sharing the genome sequences with us. GS thanks the UGC, BI thanks the CSIR, for fellowships. TL thanks Department of Biotechnology (DBT), Government of India for the project “Establishment of Centre of Excellence for NCMR.” CVR thanks the Department of Biotechnology, Government of India for the award of Tata Innovative fellowship and CSIR for financial support. Department of Science and Technology, University Grants Commission are acknowledged for providing infrastructural facilities as FIST and SAP (DRS). CS thanks UGC for award of mid-carrier award and TEQIP-III for financial support.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02480/full#supplementary-material
Aliyu, H., Lebre, P., Blom, J., Cowan, D., and De Maayer, P. (2016). Phylogenomic re-assessment of the thermophilic genus Geobacillus. Syst. Appl. Microbiol. 39, 527–533. doi: 10.1016/j.syapm.2016.09.004
Arunasri, K., Ramana, V. V., Spröer, C., Sasikala, Ch., and Ramana, Ch. V. (2008). Rhodobacter megalophilus sp. nov., a phototroph from the Indian Himalayas possessing a wide temperature range for growth. Int. J. Syst. Evol. Microbiol. 58, 1792–1796. doi: 10.1099/ijs.0.65642-0
Auch, A. F., Klenk, H. P., and Göker, M. (2012). Standard operating procedure for calculating genome-to-genome distances based on high-scoring segment pairs. Stand. Genomic. Sci. 2, 142–148. doi: 10.4056/sigs.541628
Breider, S., Scheuner, C., Schumann, P., Fiebig, A., Petersen, J., Pradella, S., et al. (2014). Genome-scale data suggest reclassifications in the Leisingera-Phaeobacter cluster including proposals for Sedimentitalea gen. nov. and Pseudophaeobacter gen. nov. Front. Microbiol. 5:416. doi: 10.3389/fmicb.2014.00416
Brinkmann, H., Göker, M., Koblížek, M., Wagner-Döbler, I., and Petersen, J. (2018). Horizontal operon transfer, plasmids, and the evolution of photosynthesis in Rhodobacteraceae. ISME J. 12, 1994–2010. doi: 10.1038/s41396-018-0150-9
Chaudhari, N. M., Gupta, V. K., and Dutta, C. (2015). BPGA- an ultra-fast pan-genome analysis pipeline. Sci. Rep. 6:24373. doi: 10.1038/srep24373
Chen, W. M., Cho, N. T., Huang, W. C., Young, C. C., and Sheu, S. Y. (2013). Description of Gemmobacter fontiphilus sp. nov., isolated from a freshwater spring, reclassification of Catellibacterium nectariphilum as Gemmobacter nectariphilus comb. nov., Catellibacterium changlense as Gemmobacter changlensis comb. nov., Catellibacterium aquatile as Gemmobacter aquaticus nom. nov., Catellibacterium caeni as Gemmobacter caeni comb. nov., Catellibacterium nanjingense as Gemmobacter nanjingensis comb. nov., and emended description of the genus Gemmobacter and of Gemmobacter aquatilis. Int. J. Syst. Evol. Microbiol. 63, 470–478. doi: 10.1099/ijs.0.042051-0
Chen, C. X., Zhang, X. Y., Liu, C., Yu, Y., Liu, A., Li, G. W., et al. (2013). Pseudorhodobacter antarcticus sp. nov., isolated from Antarctic intertidal sandy sediment, and emended description of the genus Pseudorhodobacter Uchino et al., 2002 emend. Jung et al., 2012. Int. J. Syst. Evolut. Microbiol. 63, 849–854. doi: 10.1099/ijs.0.042184-0
Chun, J., Oren, A., Ventosa, A., Christensen, H., Arahal, D. R., da Costa, M. S., et al. (2018). Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 68, 461–466. doi: 10.1099/ijsem.0.002516
Chun, J., and Rainey, F. A. (2014). Integrating genomics into the taxonomy and systematics of the Bacteria and Archaea. Int. J. Syst. Evol. Microbiol. 64, 316–324. doi: 10.1099/ijs.0.054171-0
Crouch, L., and Jones, M. (2012). Cross-species investigation of the functions of the Rhodobacter PufX polypeptide and the composition of the RC–LH1 core complex. Biochim. Biophys. Acta 2, 336–352. doi: 10.1016/j.bbabio.2011.10.009
Divyasree, B., Lakshmi, K. V. N. S., Bharti, D., Sasikala, Ch., and Ramana, Ch. V. (2015). Rhodovulum aestuarii sp. nov., isolated from a brackish waterbody. Int. J. Syst. Evol. Microbiol. 66, 165–171. doi: 10.1099/ijsem.0.000691
Eckersley, K., and Dow, C. S. (1980). Rhodopseudomonas blastica sp. nov.: a member of the Rhodospirillaceae. J. Gen. Microbiol. 119, 465–473. doi: 10.1099/00221287-119-2-465
Foesel, B., Drake, H., and Schramm, A. (2011). Defluviimonas denitrificans gen. nov., sp. nov., and Pararhodobacter aggregans gen. nov., sp. nov., non-phototrophic Rhodobacteraceae from the biofilter of a marine aquaculture. Syst. Appl. Microbiol. 34, 498–502. doi: 10.1016/j.syapm.2011.08.006
Gandham, S., Lodha, T., Sasikala, C., and Ramana, C. V. (2018). Rhodobacter alkalitolerans sp. nov., isolated from an alkaline brown pond. Arch. Microbiol. 200, 1487–1492. doi: 10.1007/s00203-018-1561-8
Girija, K. R., Sasikala, Ch., Ramana, Ch. V., Sproer, C., Takaichi, S., Thiel, V., et al. (2010). Rhodobacter johrii sp. nov., an endospore producing cryptic species isolated from semi-arid tropical soils. Int. J. Syst. Evol. Microbiol. 60, 2099–2107. doi: 10.1099/ijs.0.011718-0
Goris, J., Konstantinidis, K. T., Klappenbach, J. A., Coenye, T., Vandamme, P., and Tiedje, J. M. (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91. doi: 10.1099/ijs.0.64483-0
Gupta, R. S., Lo, B., and Son, J. (2018). Phylogenomics and comparative genomic studies robustly support division of the genus Mycobacterium into an emended genus Mycobacterium and four novel genera. Front. Microbiol. 9:67. doi: 10.3389/fmicb.2018.00067
Hansen, T. A., and Imhoff, J. F. (1985). Rhodobacter veldkampii, a new species of phototrophic purple nonsulfur bacteria. Int. J. Syst. Bacteriol. 35, 115–116. doi: 10.1099/00207713-35-1-115
Helsel, L. O., Hollis, D., Steigerwalt, A. G., Morey, R. E., Jordan, J., Aye, T., et al. (2007). Identification of “Haematobacter,”a new genus of aerobic Gram-negative rods isolated from clinical specimens, and reclassification of Rhodobacter massiliensis as “Haematobacter massiliensis comb. nov.”. J. Clin. Microbiol. 45, 1238–1243. doi: 10.1128/jcm.01188-06
Hiraishi, A., Muramatsu, K., and Ueda, Y. (1996). Molecular genetic analyses of Rhodobacter azotoformans sp. nov. and related species of phototrophic bacteria. Syst. Appl. Microbiol. 19, 168–177. doi: 10.1016/s0723-2020(96)80042-7
Hiraishi, A., and Ueda, Y. (1994). Intrageneric structure of the genus Rhodobacter: transfer of Rhodobacter sulfidophilus and related marine species to the genus Rhodovulum gen. nov. Int. J. Syst. Bacteriol. 44, 15–23. doi: 10.1099/00207713-44-1-15
Imhoff, J. F. (2005). “Genus Rhodobacter,” in Bergey’s Manual of Systematic Bacteriology, 2nd Edn, vol. 2, eds D. J. Brenner, N. R. Krieg, J. T. Staley, and G. M. Garrity, (New York, NY: Springer), 161–167.
Imhoff, J. F., Rahn, T., Künzel, S., and Neulinger, S. C. (2018). Photosynthesis is widely distributed among Proteobacteria as demonstrated by the phylogeny of PufLM reaction center proteins. Front. Microbiol. 8:2679. doi: 10.3389/fmicb.2017.02679
Imhoff, J. F., Trüper, H. G., and Pfennig, N. (1984). Rearrangement of the species and genera of the phototrophic ‘purple non sulfur bacteria’. Int. J. Syst. Bacteriol. 34, 340–343. doi: 10.1099/00207713-34-3-340
Inglin, R. C., Meile, L., and Stevens, M. J. A. (2018). Clustering of pan- and core-genome of Lactobacillus provides novel evolutionary insights for differentiation. BMC Genomics 19:284. doi: 10.1186/s12864-018-4601-5
Jung, Y. T., Ooh, K. H., Oh, T. K., and Yoon, J. H. (2012). Pseudorhodobacter aquimaris sp. nov. isolated from sea water, and emended description of the genus Pseudorhodobacter Uchino et al., 2002. Int. J. Syst. Evol. Microbiol. 62, 100–105. doi: 10.1099/ijs.0.029769-0
Kates, M. (1972). “Techniques in lipidology,” in Laboratory Techniques in Biochemistry and Molecular Biology, Vol. 3, eds T. S. Work, and E. Work, (New York, NY: American Elsevier Publishing Company), 355–356.
Kates, M. (1986). “Techniques of lipidology: isolation, analysis and identification of lipids,” in Laboratory Techniques in Biochemistry and Molecular Biology, Vol. 3, eds R. H. Burdon, and P. H. van Knippenberg, (Amsterdam: Elsevier), 100–112.
Kawasaki, H., Hoshino, Y., Hirata, A., and Yamasato, K. (1993). Is intracytoplasmic membrane structure a generic criterion? It does not coincide with phylogenetic interrelationships among phototrophic purple non-sulfur bacteria. Arch. Microbiol. 160, 358–362.
Kim, M., Oh, H. S., Park, S. C., and Chun, J. (2014). Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 64, 346–351. doi: 10.1099/ijs.0.059774-0
Kimura, M. (1980). A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/bf01731581
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kämpfer, P., Jerzak, L., Wilharm, G., Golke, J., Büsse, H., and Glaeser, S. (2015). Gemmobacter intermedius sp. nov., isolated from a white stork (Ciconia ciconia). Int. J. Syst. Evol. Microbiol. 65, 778–783. doi: 10.1099/ijs.0.000012
Kämpfer, P., Lai, W. A., Arun, A. B., Young, C. C., Rekha, P. D., Martin, K., et al. (2012). Paracoccus rhizosphaerae sp. nov., isolated from the rhizosphere of the plant Crossostephium chinense (L.) Makino (Seremban). Int. J. Syst. Evol. Microbiol. 62, 2750–2756. doi: 10.1099/ijs.0.039057-0
Lakshmi, K. V. N. S., Sasikala, Ch., Takaichi, S., and Ramana, Ch. V. (2011). Phaeospirillum oryzae sp. nov., a spheroplast forming phototrophic alphaproteobacterium from a paddy soil. Int. J. Syst. Evol. Microbiol. 61, 1656–1661. doi: 10.1099/ijs.0.025544-0
Lee, K. B., Liu, C. T., Anzai, Y., Kim, H., Aono, T., and Oyaizu, H. (2005). The hierarchical system of the “Alphaproteobacteria”: description of Hyphomonadaceae fam. nov., Xanthobacteraceae fam. nov. and Erythrobacteraceae fam. nov. Int. J. Syst. Evol. Microbiol. 55, 1907–1919. doi: 10.1099/ijs.0.63663-0
Lee, M., Lee, S., Jung, Y., Park, S., and Yoon, J. J. (2013). Pseudorhodobacter wandonensis sp. nov., isolated from wood falls, and emended description of the genus Pseudorhodobacter. Int. J. Syst. Evol. Microbiol. 63, 1479–1484. doi: 10.1099/ijs.0.042879-0
Li, Y., Xue, H., Sang, S. Q., Lin, C. L., and Wang, X. Z. (2017). Phylogenetic analysis of family Neisseriaceae based on genome sequences and description of Populibacter corticis gen. nov., sp. nov., a member of the family Neisseriaceae, isolated from symptomatic bark of Populus euramericana canker. PLoS One 12:e0174506. doi: 10.1371/journal.pone.0174506
Liu, J. J., Zhang, X. Q., Chi, F. T., Pan, J., Sun, C., and Wu, M. (2014). Gemmobacter megaterium sp. nov., isolated from coastal planktonic seaweeds. Int. J. Syst. Evol. Microbiol. 64, 66–71. doi: 10.1099/ijs.0.050955-0
Lopes-Santos, L., Castro, D. B. A., Ferreira-Tonin, M., Corrêa, D. B. A., Weir, B. S., Park, D., et al. (2017). Reassessment of the taxonomic position of Burkholderia andropogonis and description of Robbsia andropogonis gen. nov., comb. nov. Antonie van Leeuwenhoek 110, 727–736. doi: 10.1007/s10482-017-0842-6
Luo, C., Rodriguez-R, L. M., and Konstantinidis, K. T. (2014). MyTaxa: an advanced taxonomic classifier for genomic and metagenomic sequences. Nucleic Acids Res. 42:e73. doi: 10.1093/nar/gku169
Martens, M., Dawyndt, P., Coopman, R., Monique, G., Vos, P. D., and Willems, A. (2008). Advantages of multilocus sequence analysis for taxonomic studies: a case study using 10 housekeeping genes in the genus Ensifer (including former Sinorhizobium). Int. J. Syst. Evol. Microbiol. 58, 200–214. doi: 10.1099/ijs.0.65392-0
Meier-Kolthoff, J. P., and Göker, M. (2019). TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 10:2182. doi: 10.1038/s41467-019-10210-3
Na, S. I., Kim, Y. O., Yoon, S. H., Ha, S. M., Baek, I., and Chun, J. (2018). UBCG: up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J. Microbiol. 56, 281–285. doi: 10.1007/s12275-018-8014-6
Nouioui, I., Carro, L., Garcia-Lopez, M., Meier-Kolthoff, J. P., Woyke, T., Kyrpides, N. C., et al. (2018). Genome-based taxonomic classification of the phylum Actinobacteria. Front. Microbiol. 9:2007. doi: 10.3389/fmicb.2018.02007
Nupur, P., Srinivas, T. N. R., Takaichi, S., and Anil, K. P. (2014). Rhodovulum mangrove sp. nov. a phototrophic alphaproteobacterium isolated from a mangrove forest sediment sample. Int. J. Syst. Evol. Microbiol. 64, 3168–3173. doi: 10.1099/ijs.0.059857-0
Orata, F. D., Meier-Kolthoff, J. P., Sauvageau, D., and Stein, L. Y. (2018). Phylogenomic analysis of the gammaproteobacterial methanotrophs (Order Methylococcales) calls for the reclassification of members at the genus and species levels. Front. Microbiol. 9:3162. doi: 10.3389/fmicb.2018.03162
Oren, A., Duker, S., and Ritter, S. (1996). The polar lipid composition of Walsby’s square bacterium. FEMS. Microbiol. Lett. 138, 135–140. doi: 10.1016/0378-1097(96)00085-7
Pan, J., Sun, C., Zhang, X., Huo, Y., Zhu, X., and Wu, M. (2014). Paracoccus sediminis sp. nov., isolated from Pacific Ocean marine sediment. Int. J. Syst. Evol. Microbiol. 64, 2512–2516. doi: 10.1099/ijs.0.051318-0
Pérez-Cataluña, A., Collado, L., Salgado, O., Lefiñanco, V., and Figueras, M. (2018). A polyphasic and taxogenomic evaluation uncovers Arcobacter cryaerophilus as a species complex that embraces four genomovars. Front. Microbiol. 9:805. doi: 10.3389/fmicb.2018.00805
Pujalte, M. J., Lucena, T., Ruvira, M. A., Arahal, D. R., and Macián, M. C. (2014). “The family Rhodobacteraceae,” in The Prokaryotes, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson, (Berlin: Springer), 439–512. doi: 10.1007/978-3-642-30197-1_377
Qian, P., Martin, E. C., Ng, I. W., and Hunter, C. N. (2017). The C-terminus of PufX plays a key role in dimerisation and assembly of the reaction center light-harvesting 1 complex from Rhodobacter sphaeroides. Biochim. Biophys. Acta Bioenerg. 1858, 795–803. doi: 10.1016/j.bbabio.2017.06.001
Qin, Q., Xie, B., Zhang, X., Chen, X., Zhou, B., Zhou, J., et al. (2014). A proposed genus boundary for the prokaryotes based on genomic insights. J. Bacteriol. 196, 2210–2215. doi: 10.1128/jb.01688-14
Richter, M., and Rosselló-Móra, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 106, 19126–19131. doi: 10.1073/pnas.0906412106
Rodriguez, L. M., and Konstantinidis, K. T. (2014). Bypassing cultivation to identify bacterial species. Microbe. 9, 111–117.
Rosselló-Móra, R., and Amann, R. (2015). Past and future species definitions for Bacteria and Archaea. Syst. Appl. Microbiol. 38, 209–216. doi: 10.1016/j.syapm.2015.02.001
Rothe, B., Fischer, A., Hirsch, P., Sittig, M., and Stackebrandt, E. (1987). The phylogenetic position of the budding bacteria Blastobacter aggregates and Gemmobacter aquatilis gen. nov., sp. nov. Arch. Microbiol. 147, 92–99. doi: 10.1007/bf00492911
Shalem Raj, P., Ramaprasad, E. V. V., Vaseef, S., Sasikala, Ch., and Ramana, Ch. V (2013). Rhodobacter viridis sp. nov., a phototrophic bacterium isolated from mud of a stream. Int. J. Syst. Evol. Microbiol. 63, 181–186. doi: 10.1099/ijs.0.038471-0
Sheu, S. Y., Sheu, D. S., Sheu, F. S., and Chen, W. M. (2013). Gemmobacter tilapiae sp. nov., a poly-β-hydroxybutyrate accumulating bacterium isolated from a freshwater culture pond. Int. J. Syst. Evol. Microbiol. 63, 1550–1556. doi: 10.1099/ijs.0.044735-0
Simon, M., Scheuner, C., Meier-Kolthoff, J. P., Brinkhoff, T., Wagner-Döbler, I., Ulbrich, M., et al. (2017). Phylogenomics of Rhodobacteraceae reveals evolutionary adaptation to marine and non-marine habitats. ISME J. 11, 1483–1499. doi: 10.1038/ismej.2016.198
Srinivas, A., Rahul, K., Ramprasad, E. V. V., Sasikala, Ch., and Ramana, Ch. V. (2012). Rhodovulum bhavnagarense sp. nov., a phototrophic alphaproteobacterium isolated from a pink pond. Int. J. Syst. Evol. Microbiol. 62, 2528–2532. doi: 10.1099/ijs.0.036152-0
Srinivas, A., Vinay Kumar, B., Divya Sree, B., Tushar, L., Sasikala, Ch., and Ramana, Ch. V. (2014). Rhodovulum salis sp. nov. and Rhodovulum viride sp. nov., phototrophic Alphaproteobacteria isolated from marine habitats. Int. J. Syst. Evol. Microbiol. 64, 957–962. doi: 10.1099/ijs.0.058974-0
Srinivas, T. N. R., Anil Kumar, P., Sasikala, Ch., Ramana, Ch. V., and Imhoff, J. F. (2007). Rhodobacter vinaykumarii sp. nov., a marine phototrophic alphaproteobacterium from tidal waters, and emended description of the genus Rhodobacter. Int. J. Syst. Evol. Microbiol. 57, 1984–1987. doi: 10.1099/ijs.0.65077-0
Srinivas, T. N. R., Anil Kumar, P., Sasikala, Ch., Spröer, C., and Ramana, Ch. V. (2008). Rhodobacter ovatus sp. nov., an alphaproteobacterium isolated from industrially polluted freshwater pond. Int. J. Syst. Evol. Microbiol. 58, 1379–1383. doi: 10.1099/ijs.0.65619-0
Subhash, Y., and Lee, S. S. (2016). Rhodobacter sediminis sp. nov. isolated from lagoon sediments. Int. J. Syst. Evol. Microbiol. 66, 2965–2970. doi: 10.1099/ijsem.0.001130
Subhash, Y., Tushar, L., Sasikala, Ch., and Ramana, Ch. V. (2013). Falsirhodobacter halotolerans gen. nov., sp. nov., isolated from dry soils of a solar saltern. Int. J. Syst. Evol. Microbiol. 63, 2132–2137. doi: 10.1099/ijs.0.044107-0
Sun, L. N., Zhang, J., Kwon, S. W., He, J., Zhou, S. G., and Li, S. P. (2013). Paracoccus huijuniae sp. nov., an amide pesticide-degrading bacterium isolated from activated sludge of a wastewater biotreatment system. Int. J. Syst. Evol. Microbiol. 63, 1132–1137. doi: 10.1099/ijs.0.044180-0
Suresh, G., Sailaja, B., Ashif, A., Dave, B. P., Sasikala, Ch, and Ramana, Ch. V. (2017). Description of Rhodobacter azollae sp. nov. and Rhodobacter lacus sp. nov. Int. J. Syst. Evol. Microbiol. 67, 3289–3295. doi: 10.1099/ijsem.0.002107
Suresh, G., Sasikala, Ch., and Ramana, Ch. V. (2015). Reclassification of Gemmobacter changlensis to a new genus as Cereibacter changlensis gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 65, 794–798. doi: 10.1099/ijs.0.000016
Tindall, B. J. (1990b). Lipid composition of Halobacterium lacusprofundi. FEMS. Microbiol. Lett. 66, 199–202. doi: 10.1016/j.bbamem.2016.08.010
Tindall, B. J. (1990a). A comparative study of the lipid composition of Halobacterium saccharovorum from various sources. Syst. Appl. Microbiol. 13, 128–130. doi: 10.1016/s0723-2020(11)80158-x
Uchino, Y., Hamada, T., and Yokota, A. (2002). Proposal of Pseudorhodobacter ferrugineus gen. nov., comb. nov., for a non-photosynthetic marine bacterium, Agrobacterium ferrugineum, related to the genus Rhodobacter. J. Gen. Appl. Microbiol. 48, 309–320.
Urdiain, M., López-López, A., Gonzalo, C., Busse, H. J., Langer, S., Kämpfer, P., et al. (2008). Reclassification of Rhodobium marinum and Rhodobium pfennigii as Afifella marina gen. nov. comb. nov. and Afifella pfennigii comb. nov., a new genus of photoheterotrophic Alphaproteobacteria and emended descriptions of Rhodobium, Rhodobium orientis and Rhodobium gokarnense. Syst. Appl. Microbiol. 31, 339–351. doi: 10.1016/j.syapm.2008.07.002
Venkata Ramana, V., Kumar, A. P., Srinivas, T. N. R., Sasikala, Ch., and Ramana, Ch. V. (2009). Rhodobacter aestuarii sp. nov., a phototrophic alphaproteobacterium isolated from an estuarine environment. Int. J. Syst. Evol. Microbiol. 59, 1133–1136. doi: 10.1099/ijs.0.004507-0
Venkata Ramana, V., Sasikala, Ch., and Ramana, Ch. V. (2008). Rhodobacter maris sp. nov., a phototrophic alphaproteobacterium isolated from a marine habitat of India. Int. J. Syst. Evol. Microbiol. 58, 1719–1722. doi: 10.1099/ijs.0.65638-0
Wang, D., Liu, H., Zheng, S., and Wang, G. (2014). Paenirhodobacter enshiensis gen. nov., sp. nov., a non-photosynthetic bacterium isolated from soil, and emended descriptions of the genera Rhodobacter and Haematobacter. Int. J. Syst. Evol. Microbiol. 64, 551–558. doi: 10.1099/ijs.0.050351-0
Wang, L., Zhou, Z., Wu, G., Chen, M., Lin, M., Zhang, W., et al. (2015). Falsirhodobacter deserti sp. nov., isolated from sandy soil. Int. J. Syst. Evol. Microbiol. 65, 650–655. doi: 10.1099/ijs.0.068262-0
Wayne, L. G., Brenner, D. J., Colwell, R. R., Grimont, P. A. D., Kandler, O., Krichevsk, M. I., et al. (1984). International committee on systematic bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J. Syst. Bacteriol. 37, 463–464.
Wirth, J. S., and Whitman, W. B. (2018). Phylogenomic analyses of a clade within the roseobacter group suggest taxonomic reassignments of species of the genera Aestuariivita, Citreicella, Loktanella, Nautella, Pelagibaca, Ruegeria, Thalassobius, Thiobacimonas and Tropicibacter, and the proposal of six novel genera. Int. J. Syst. Evol. Microbiol. 68, 2393–2411. doi: 10.1099/ijsem.0.002833
Xie, B. S., Lv, X. L., Cai, M., Tang, Y. Q., Wang, Y. N., Cui, H. L., et al. (2015). Plastorhodobacter daqingensis gen. nov., sp. nov.: a non-phototrophic Bacterium isolated from Daqing Oilfield. Curr. Microbiol. 70, 657–664. doi: 10.1007/s00284-014-0769-3
Yoon, S. H., Ha, S. M., Lim, J. M., Kwon, S. J., and Chun, J. (2017). A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie van Leeuwenhoek 110, 1281–1286. doi: 10.1007/s10482-017-0844-4
Zhang, G., Yang, Y., Yin, X., and Wang, S. (2014). Paracoccus pacificus sp. nov., isolated from the Western Pacific Ocean. Antonie van Leeuwenhoek. 106, 725–731. doi: 10.1007/s10482-014-0242-0
Zhang, Y., Jiang, F., Chang, X., Qiu, X., Ren, L., Qu, Z., et al. (2016). Pseudorhodobacter collinsensis sp. nov., isolated from a till sample of an icecap front. Int. J. Syst. Evol. Microbiol. 66, 178–183. doi: 10.1099/ijsem.0.000693
Keywords: Rhodobacter, taxogenomics, Rhodobacter sensu stricto, gen. nov., Rhodobacter reclassification, phylogenomics, proposal of 3 new phototrophic genera, photosynthetic gene cluster
Citation: Suresh G, Lodha TD, Indu B, Sasikala C and Ramana CV (2019) Taxogenomics Resolves Conflict in the Genus Rhodobacter: A Two and Half Decades Pending Thought to Reclassify the Genus Rhodobacter. Front. Microbiol. 10:2480. doi: 10.3389/fmicb.2019.02480
Received: 28 May 2019; Accepted: 15 October 2019;
Published: 31 October 2019.
Edited by:
Iain Sutcliffe, Northumbria University, United KingdomReviewed by:
Barny Whitman, University of Georgia, United StatesCopyright © 2019 Suresh, Lodha, Indu, Sasikala and Ramana. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ch. Sasikala, c2FzaTQ0OUB5YWhvby5pZQ==; Ch. V. Ramana, Y3ZyNDQ5QGdtYWlsLmNvbQ==
†These authors have contributed equally to this work
‡Present address: Tushar D. Lodha, National Centre for Microbial Resource, National Centre for Cell Science, Pune, India
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.