
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 24 October 2019
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.02429
 Clara Cosgaya1
Clara Cosgaya1 Carlos Ratia1
Carlos Ratia1 Marta Marí-Almirall1
Marta Marí-Almirall1 Laia Rubio1
Laia Rubio1 Paul G. Higgins2,3
Paul G. Higgins2,3 Harald Seifert2,3
Harald Seifert2,3 Ignasi Roca1*
Ignasi Roca1* Jordi Vila1
Jordi Vila1The increased use of molecular identification methods and mass spectrometry has revealed that Acinetobacter spp. of the A. baumannii (Ab) group other than A. baumannii are increasingly being recovered from human samples and may pose a health challenge if neglected. In this study 76 isolates of 5 species within the Ab group (A. baumannii n = 16, A. lactucae n = 12, A. nosocomialis n = 16, A. pittii n = 20, and A. seifertii n = 12), were compared in terms of antimicrobial susceptibility, carriage of intrinsic resistance genes, biofilm formation, and the ability to kill Caenorhabditis elegans in an infection assay. In agreement with previous studies, antimicrobial resistance was common among A. baumannii while all other species were generally more susceptible. Carriage of genes encoding different efflux pumps was frequent in all species and the presence of intrinsic class D β-lactamases was reported in A. baumannii, A. lactucae (heterotypic synonym of A. dijkshoorniae) and A. pittii but not in A. nosocomialis and A. seifertii. A. baumannii and A. nosocomialis presented weaker pathogenicity in our in vitro and in vivo models than A. seifertii, A. pittii and, especially, A. lactucae. Isolates from the former species showed decreased biofilm formation and required a longer time to kill C. elegans nematodes. These results suggest relevant differences in terms of antibiotic susceptibility patterns among the members of the Ab group as well as highlight a higher pathogenicity potential for the emerging species of the group in this particular model. Nevertheless, the impact of such potential in the human host still remains to be determined.
Among the 62 validly named Acinetobacter species (List of Prokaryotic Names with Standing Nomenclature1, last accessed April 2019), Acinetobacter baumannii has, for many years, monopolized our attention. Responsible for bloodstream infections, ventilator-associated pneumonia, wound infections, and meningitis, among other infections, A. baumannii excels in its ability to persist and survive in the nosocomial setting (Peleg et al., 2008). In comparison, little is known regarding the closely related members of the so-called A. baumannii (Ab) group, such as Acinetobacter nosocomialis and Acinetobacter pittii, previously known members of the group, but also Acinetobacter lactucae (heterotypic synonym of A. dijkshoorniae) and Acinetobacter seifertii, whose species status has recently been acknowledged (Nemec et al., 2015; Cosgaya et al., 2016). Species other than A. baumannii among the Ab group are increasingly being isolated from human specimens, albeit not as frequently as A. baumannii, and they also possess an undeniable ability to cause disease (Espinal et al., 2011; Schleicher et al., 2013; Chen et al., 2018; Pailhories et al., 2018; Rocha et al., 2018). Unfortunately, they are often erroneously identified as A. baumannii by common phenotypic methods such as biochemical tests and semi-automated identification systems, thus underestimating their true prevalence and clinical relevance. Accurate species identification is achieved by molecular methods, such as ARDRA, rpoB and gyrB sequence analysis, Pasteur MLST scheme-based identification, whole genome sequencing (Vaneechoutte et al., 1995; Gundi et al., 2009; Diancourt et al., 2010; Higgins et al., 2010b; Fitzpatrick et al., 2016) and, more recently, mass spectrometry (Mari-Almirall et al., 2017).
Antimicrobial resistance studies containing information about A. lactucae, A. nosocomialis, A. pittii and A. seifertii are scarce, although existing data support their general susceptibility to most antibiotics (Chuang et al., 2011; Lai et al., 2012; Chen et al., 2018; Pailhories et al., 2018). In contrast, A. baumannii is considered a problem pathogen because of its frequently reported multi-drug resistant (MDR) phenotype. This pathogen has developed resistance to all antibiotics, including last resort antibiotics such as carbapenems, tigecycline and colistin (Roca et al., 2012; Nowak et al., 2017). Multi-drug resistance represents one of the best assets of A. baumannii and may account for its success in the hospital setting; nonetheless other pathogenic attributes may as well impact its ability to infect the host.
Likewise, there is plenty of information on the virulence traits of A. baumannii (Harding et al., 2018) but little is known regarding the other members of the Ab group. Studies regarding the virulence attributes of different Acinetobacter species are rare, and usually include only a few isolates (de Breij et al., 2010; Bitrian et al., 2012; Peleg et al., 2012; Kinsella et al., 2017) or are limited to bacteremic isolates from a unique region (Na et al., 2016). Noticeably, none of the abovementioned studies include information about A. lactucae and there is just one report that highlights its ability to cause disease and carry genetic determinants conferring carbapenem resistance (Espinal et al., 2015).
A few reports have suggested relevant differences in the clinical outcomes of infections caused by former members of the Ab group (Chuang et al., 2011; Wisplinghoff et al., 2012; Lee et al., 2013; Chusri et al., 2014), suggesting that, in order to understand the clinical relevance and the virulence potential of closely related Acinetobacter spp., further studies on susceptibility patterns and pathogenesis need to address them as distinct entities rather than as a homogeneous group. In this work we have gathered a collection of representative isolates from all five known Acinetobacter species of the Ab group and compared their antibiotic susceptibility profiles and carriage of resistance determinants as well as their pathogenic potential using both in vitro and in vivo assays.
Isolates used in this study belong to Acinetobacter baumannii (n = 16), A. lactucae (n = 12), A. nosocomialis (n = 16), A. pittii (n = 20) and A. seifertii (n = 12) (Supplementary Data Sheet S1). Species identification was performed by matrix-assisted laser desorption ionization-time of flight mass spectrometry (Mari-Almirall et al., 2017). The clonal relatedness was established either by repetitive element palindromic PCR (REP-PCR) or by pulsed-field gel electrophoresis (PFGE) of ApaI-digested DNA (Seifert et al., 2005). The sequence type (ST) was determined following the Pasteur multilocus sequence typing (MLST) scheme, as previously described (Diancourt et al., 2010). Bacterial cultures were routinely grown on Columbia sheep blood agar (Becton Dickinson, Heidelberg, Germany) at 37°C, unless stated otherwise.
The minimum inhibitory concentration (MIC) was determined by broth microdilution or gradient diffusion (AB bioMérieux, Solna, Sweeden) in Mueller Hinton II broth (CONDA, Madrid, Spain) or agar plates (Becton Dickinson, Heidelberg, Germany), respectively, and interpreted according to EUCAST guidelines when appropriate (EUCAST clinical breakpoints version 8.0, 2018). As no EUCAST susceptibility breakpoint for ceftazidime is available, the corresponding clinical breakpoint according to CLSI guidelines was used instead (susceptible ≤ 8 μg/mL, resistant ≥ 32 μg/mL; CLSI supplement M100-S28, 2018). The antibiotics tested were amikacin, ceftazidime, ciprofloxacin, colistin, gentamicin, imipenem, meropenem, tigecycline, and tobramycin. Presence of metallo-β-lactamases, blaKPC, blaOXA–23, blaOXA–24/40, and blaOXA–58 genes was determined by multiplex PCR (Bogaerts et al., 2013) or characterized in previous studies (Supplementary Data Sheet S1).
The presence of genes encoding efflux pumps from the resistance-nodulation-cell division (RND) family (adeABC, adeDE, adeFGH, and adeIJK), and intrinsic oxacillinases (OXA) from the class D β-lactamases was evaluated by PCR using the primers listed in Supplementary Table S1. In silico analysis of available genomic sequences at NCBI were performed in order to ensure amplification in all the Ab group species, otherwise, species-specific primers were designed. At the time of the analysis, only one A. seifertii genome and only two A. lactucae genomes were available at the NCBI databases. Intrinsic OXA β-lactamases were analyzed by DNA sequencing and cluster analysis of partial amino acid sequences (residues 9 to 267) using the MEGA version 6 software (Tamura et al., 2013). Briefly, sequences were aligned using MUSCLE (Edgar, 2004) and unrooted phylogenetic trees were constructed using the neighbor-joining method with bootstrap values based on 1000 replicates, with software default settings. When necessary, additional sequences were retrieved from public databases. The partial sequences of allelic variants used for cluster analysis are provided in the Supplementary Data Sheet S2.
Biofilm formation was assessed in non-treated 96-well polystyrene microtiter plates. Bacteria were grown overnight at 37°C with shaking in modified M63 medium consisting of KH2PO4 (12 g/l), K2HPO4 (7 g/l), (NH4)2SO4 (2 g/l), adjusted to pH 7 with KOH prior to autoclaving, and supplemented with glucose (0.2% w/v), MgSO4 (1 mM) and casaminoacids (0.5% w/v). Overnight cultures were adjusted to an OD600 of 0.1 and diluted 1:10 in modified M63 medium. For each isolate four wells were inoculated with 150 μL of the former suspension. Afterward, plates were incubated statically at 28 or 37°C. After 44 h, supernatants were removed with a 20 G needle, wells were washed once with PBS, and biofilm was fixed with methanol 99% for 10 min and evaporated at 65°C for 20 min. Biofilm was stained with crystal violet 2% for 20 min. Glacial acetic acid 33% was used to solubilize the dye and absorbance was recorded at 580 nm. The OD580/OD600 ratio was used to quantify biofilm formation and to normalize for bacterial growth differences. At least three biological replicas with a covariance for the OD580/OD600 ratio less than 45% were performed for each isolate. Isolates were considered biofilm producers when the OD580/OD600 ratio was greater than 1. Appropriate controls were added for all assays and all plates, and consisted of bacteria-free media and the inclusion of A. baumannii ATCC 19606 and A. baumannii ATCC 17978 bacterial strains as strong and weak biofilm producers, respectively.
Motility plates consisted of tryptone (5 g/L), NaCl (2.5 g/L) and agarose (0.3% w/v) (CONDA, Madrid, Spain). Prior to pouring, media was cooled down to 50°C in a water bath. Twenty mL were dispensed in 90 mm diameter petri dishes and allowed to solidify for 45 min plus an additional 15 min for air-drying in a biosafety cabinet. Then, 1 μL of the same bacterial suspension used for biofilm assays was inoculated onto the center of the surface of the media, plates were sealed with parafilm and incubated at 37°C for 18 h. Isolates tended to grow concentrically from the inoculation point, thus surface-associated motility was recorded as the mean value (cm) of three diameter measurements per plate. Isolates were considered motile when the diameter exceeded 1 cm. In every assay, isolates were tested in duplicate, and at least four biological replicates were performed for all the isolates. In all the assays, A. baumannii ATCC 19606 and A. baumannii ATCC 17978 were used as non-motile and motile control strains, respectively.
The Fer-15 line of C. elegans, fertile at 15°C but not at 25°C and provided by the Caenorhabditis Genetics Center, was used for the infection model. Nematodes were routinely maintained at 15°C and fed with the non-virulent E. coli OP50 strain in Nematode Growth Medium agar. Assays were performed using L4 stage worms as previously described (Espinal et al., 2019). Live worms were scored every day for 15 days using a stereomicroscope. Worms were considered dead when unresponsive to touch and immotile for more than 20 s. The lethal time 50% (LT50), that is, the time (days) needed to kill 50% of the worms, of each isolate in each assay was obtained by extrapolation from the sigmoidal regression of the killing curves (R2 > 0.95) using GraphPad Prism version 5 for Windows (GrapPad Software, La Jolla, CA, United States). At least three biological replicates were performed for each isolate. In all the assays, E. coli OP50 was used as a non-virulent control strain.
All statistical analyses were performed using IBM SPSS Statistics for Windows, version 23 (IBM, Corp., Armonk, NY, United States). Kolmogorov–Smirnov test or Shapiro–Wilk test was used to assess normal distribution when n > 30 or n < 30, respectively. For the regression models, the Pearson correlation coefficient was determined when the variables followed a normal distribution. Whenever non-normally distributed variables or ordinal variables were analyzed, the Spearman rank correlation coefficient or the Kendall’s τ correlation coefficient were calculated, respectively. The Kruskal–Wallis test was performed to compare the distribution of variables among the different species; post hoc analyses were automatically performed by the software. Pairwise differences were compared using Mann–Whitney U-test. The Wilcoxon signed rank test was used to compare biofilm formation at 28 and 37°C using quantitative data, whereas the McNemar test was used to compare the proportions of biofilm-producing isolates at both temperatures. For all tests performed, P-values < 0.05 were considered to be statistically significant.
The experimental procedures described in this work do not involve the usage of biological samples from humans or animals. Clinical bacterial isolates studied here were obtained from clinical collections at several microbiology labs where they were initially recovered from clinical samples used for microbiological diagnosis. Informed consent was, therefore, not required. The protocol for this study was approved by the Ethics Committee on Clinical Research (CEIC) of the Hospital Clinic de Barcelona (HCB/2014/0499, HCB/2017/0923, and HCB/2017/0833).
We have characterized the susceptibility patterns of 76 isolates from 5 different Acinetobacter species within the Ab group, as shown in Table 1. Isolates other than A. baumannii were usually susceptible to almost all antibiotics tested, with only a few exceptions. The A. seifertii isolates from our collection were susceptible to all the agents tested, thus constituting the species with the lowest rates of resistance. Likewise, isolates of A. nosocomialis were susceptible to all agents except the aminoglycosides, for which rates of resistance were 25% for gentamicin and 5% for amikacin.
Carbapenem-resistance was identified in three A. pittii isolates and one A. lactucae isolate, and was associated with the presence of genes encoding New Delhi metallo-β-lactamase (NDM) or OXA-type enzymes (Supplementary Data Sheet S1). A. pittii and A. lactucae isolates, however, tested susceptible to almost all other antimicrobial agents but ceftazidime, with rates of resistance of 5 and 25%, respectively. One A. pittii isolate was resistant to ciprofloxacin (MIC > 32 μg/mL) and another isolate of the same species was also resistant to gentamicin (MIC of 6 μg/mL). In contrast, A. baumannii isolates presented higher rates of resistance (75% for ceftazidime, 68.8% for ciprofloxacin and gentamicin, 62.5% for imipenem, 56.3% for tobramycin, 43.8% for meropenem, and 31.5% for amikacin), and resistance to carbapenems was usually associated with the production of OXA-type carbapenemases (Supplementary Data Sheet S1). Non-susceptibility to at least one agent in all but two or fewer antimicrobial categories, i.e., XDR (Magiorakos et al., 2012), was determined in 68.75% of A. baumannii isolates, the NDM-1 producing A. pittii was resistant to three or more antimicrobial agents from different categories, i.e., MDR and all other isolates were overall considered as highly susceptible. Finally, all the species of the Ab group showed low MIC values of tigecycline (Table 1) and, among all the isolates of the Ab group, only one A. baumannii isolate was resistant to colistin with an MIC of 8 μg/mL.
The occurrence of the different RND efflux pumps in the species of the Ab group is shown in Figures 1A–D. All isolates encoded at least one of the RND efflux systems known to be associated with antimicrobial resistance, and it was common to detect three or even four different efflux pumps in isolates from all Acinetobacter species (Supplementary Data Sheet S1). adeG and adeJ genes were identified in all species and were commonly present in most isolates (80 to 100 and 50 to 100%, respectively). adeE was highly disseminated within A. nosocomialis (100%) and A. pittii (90%), and was also present in 75% of A. seifertii isolates. However, adeE was found in only one isolate of A. lactucae and it was missing in A. baumannii. The adeB gene, on the other hand, was absent in A. seifertii but was ubiquitous in A. baumannii (93.75%) and A. nosocomialis (100%). Interestingly, isolates from A. pittii and A. lactucae carried a distinct adeB-like gene with 75 and 86% similarity to the adeB gene from A. baumannii, respectively. This gene was present in 90 and 66.7% of A. pittii and A. lactucae isolates, respectively.
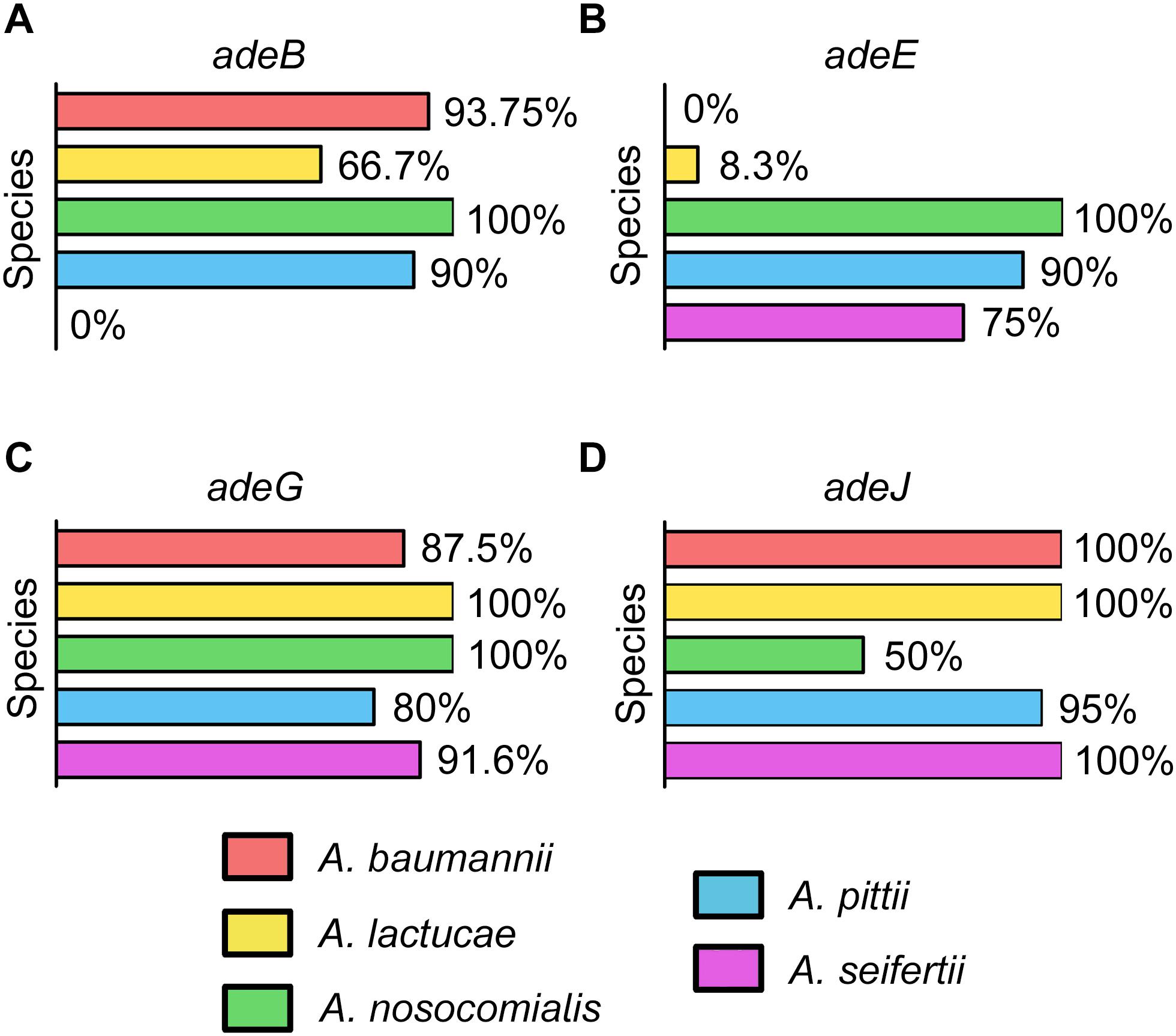
Figure 1. Occurrence of the genes encoding efflux pumps in the Ab group (A) adeABC, (B) adeDE, (C) adeFGH, and (D) adeIJK.
In addition to the presence of different RND efflux pumps, we also evaluated the carriage of genes encoding intrinsic OXA enzymes. Intrinsic blaOXA genes were reported in all A. baumannii, A. lactucae and 16 out of 20 A. pittii isolates but we were not able to detect them in any of the A. nosocomialis and A. seifertii isolates.
Cluster analysis of all sequences revealed that the OXA variants harbored by members of each of the three Acinetobacter species clustered together with high bootstrap values (Figure 2), the only exception being the intrinsic OXA β-lactamase of the A. lactucae RUH-14531 isolate that clustered together with OXA sequences from A. pittii.
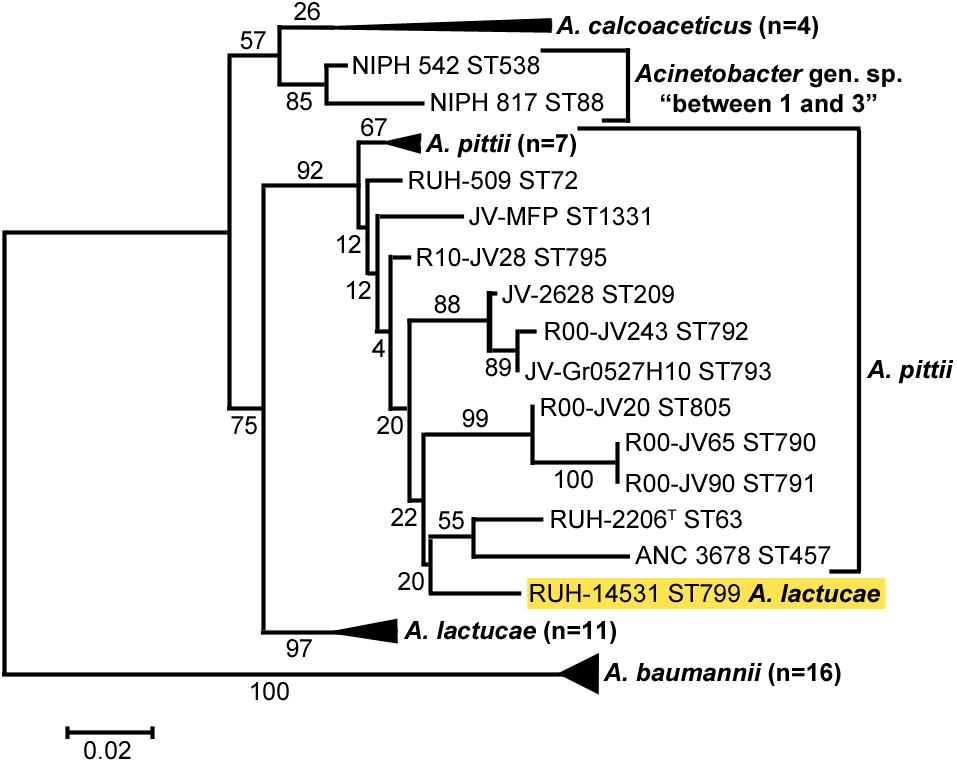
Figure 2. Cluster analysis of A. baumannii, A. lactucae, and A. pittii based on the partial amino acid sequence of their intrinsic OXA β-lactamases. Bootstrap values (%) are indicated in the branches. The scale bar indicates sequence divergence. The allele of A. lactucae clustering with those from A. pittii is highlighted in yellow. ST, sequence type; ND, not determined.
Biofilm formation values at both 28 and 37°C showed a high degree of variability among isolates of each of the five different Acinetobacter species included in the study (Figure 3A). No relevant differences in growth yields were observed among the different species or among members of the same species.
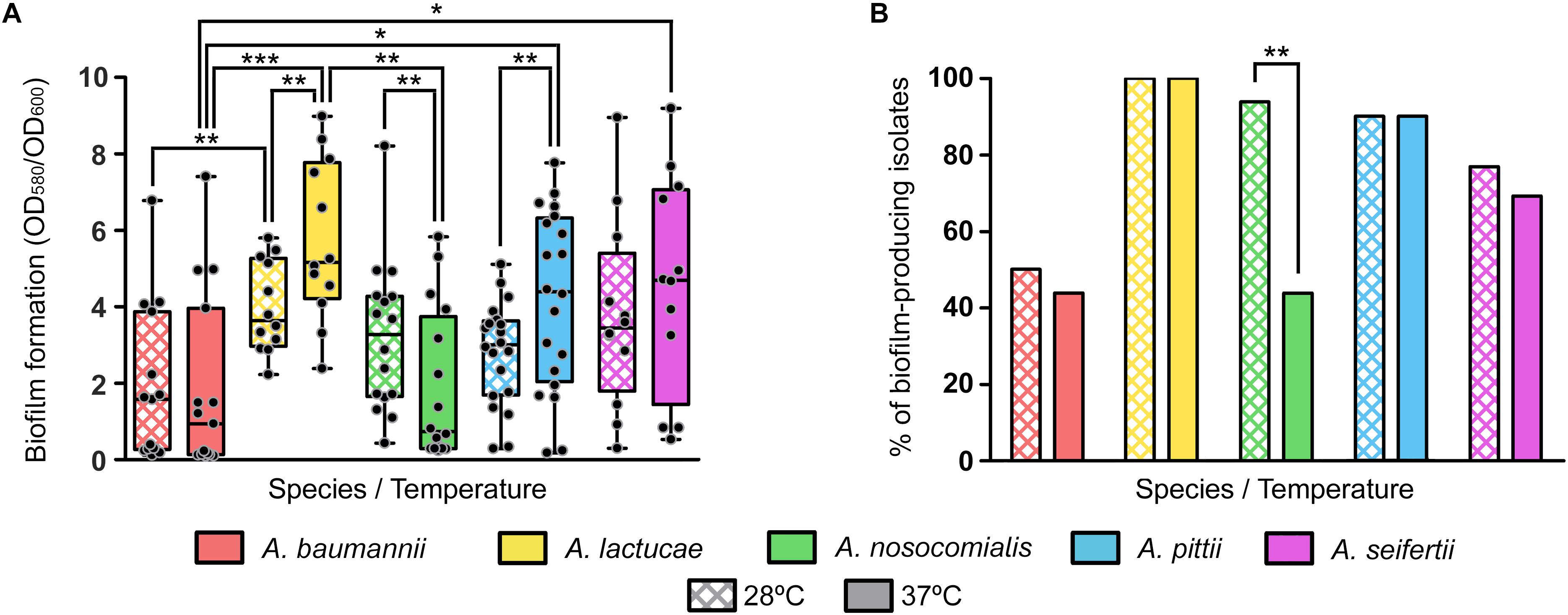
Figure 3. Biofilm formation at 28 and 37°C of the Ab group species. (A) Overlapping dot plot and box plot of the biofilm formation values of each species at 28 and 37°C. The boxes span from the first to the third quartile. The median is indicated as a segment inside the box. Whiskers indicate the minimum and maximum biofilm formation values. Each dot corresponds to the average biofilm formation value of an isolate after at least three biological replicates. Statistically significant differences upon temperature shift (Wilcoxon signed rank test) and between species (Kruskal–Wallis and post hoc tests) are highlighted with asterisks: (∗) if the P-value < 0.05, (∗∗) if the P-value ≤ 0.01, and (∗∗∗) if the P-value ≤ 0.001. (B) Percentage of biofilm-producing isolates of each species at 28 and 37°C. Isolates were considered as biofilm producers when the biofilm formation value was greater than 1. Statistically significant differences upon temperature shift (McNemar test) are highlighted with asterisks: (∗) if the P-value < 0.05 and (∗∗) if the P-value ≤ 0.01.
Nevertheless, statistically significant differences were observed between some species at each temperature; A. lactucae formed significantly more biofilm than A. baumannii at both temperatures and than A. nosocomialis at 37°C (Kruskal–Wallis test, post hoc P-value of 0.009, 0.001, and 0.004, respectively); and A. pittii and A. seifertii produced significantly more biofilm than A. baumannii at 37°C (Kruskal–Wallis test, post hoc P-values of 0.024 and 0.035, respectively). Despite the high dispersion of the data, and thus the lack of statistical significant differences for most pairwise comparisons, there were a few observations worth mentioning. A. baumannii isolates tended to form less biofilm than those of the other species, and biofilm formation was similar between A. lactucae, A. pittii and A. seifertii. Likewise, biofilm formation in A. nosocomialis resembled that of Ab group species other than A. baumannii at 28°C, but at 37°C it was comparable to that of A. baumannii.
Similar trends were observed when classifying isolates as biofilm producers and non-producers (Figure 3B). Our results showed that all A. lactucae isolates produced biofilm at both temperatures, and that the genetically related A. pittii species also presented a high proportion of biofilm-producing isolates (90%). In contrast, A. baumannii was the species with the least amount of biofilm-producing isolates, 50% or less, depending on the temperature. Also, upon shifting the temperature from 28 to 37°C, a reduction in the number of biofilm-producing isolates was observed in A. baumannii, A. nosocomialis and A. seifertii, while the number of biofilm-producing isolates remained unchanged in A. lactucae and A. pittii. Although this reduction was not statistically significant for A. baumannii and A. seifertii (from 50 to 43.8%, and from 83.3 to 75%, respectively), the effect of the temperature was greater in A. nosocomialis, in which shifting the temperature from 28 to 37°C decreased the percentage of biofilm-producing isolates from 93.8 to 43.8% (Figure 3B, McNemar test, P-value of 0.008). Thus, and under our experimental conditions, half of the isolates of this species were able to develop biofilm at 28°C but not at 37°C, and even in those isolates that did not lose the ability to produce biofilm at 37°C, a net reduction in the biofilm formation values was observed (Wilcoxon rank sum test, P-value of 0.01). On the other hand, the biofilm formation values of A. lactucae and A. pittii isolates were higher at 37°C than at 28°C (Wilcoxon rank sum test, P-value of 0.002 and 0.005, respectively; up to 2.3 fold changes, data not shown), although we did not detect biofilm non-producing isolates at 28°C turning into biofilm-producers at 37°C.
In our collection, surface-associated motility was observed in isolates of all the species studied, with greater intra-species variability for A. baumannii, A. nosocomialis and A. seifertii isolates (mean values ± SD (cm) of 2.315 ± 1.722, 2.078 ± 1.775, and 3.188 ± 1.755, respectively), than for A. pittii and A. lactucae (2.717 ± 0.829 and 1.653 ± 0.844, respectively) (Figure 4). Overall, differences in the distribution of surface-associated motility between species were not detected when taking into account the extent of motility. However, classification of the isolates into motile vs. non-motile according to the individual degree of motility showed higher rates of motility in A. pittii (95%) and A. lactucae (83.3%), followed by A. seifertii and A. baumannii (75 and 62.5%, respectively). A. nosocomialis was the least motile species of the group under the conditions tested with only half of the isolates being motile. Interestingly, rates of motility resembled those described above regarding biofilm formation at 37°C for A. nosocomialis. There was a positive correlation between motility and biofilm formation at 37°C in all species except for A. pittii (Spearman’s correlation coefficients between 0.622 and 0.7128, and P-values of 0.006 to 0.033), meaning that, in general, the more motile an isolate was, the more biofilm it tended to produce.
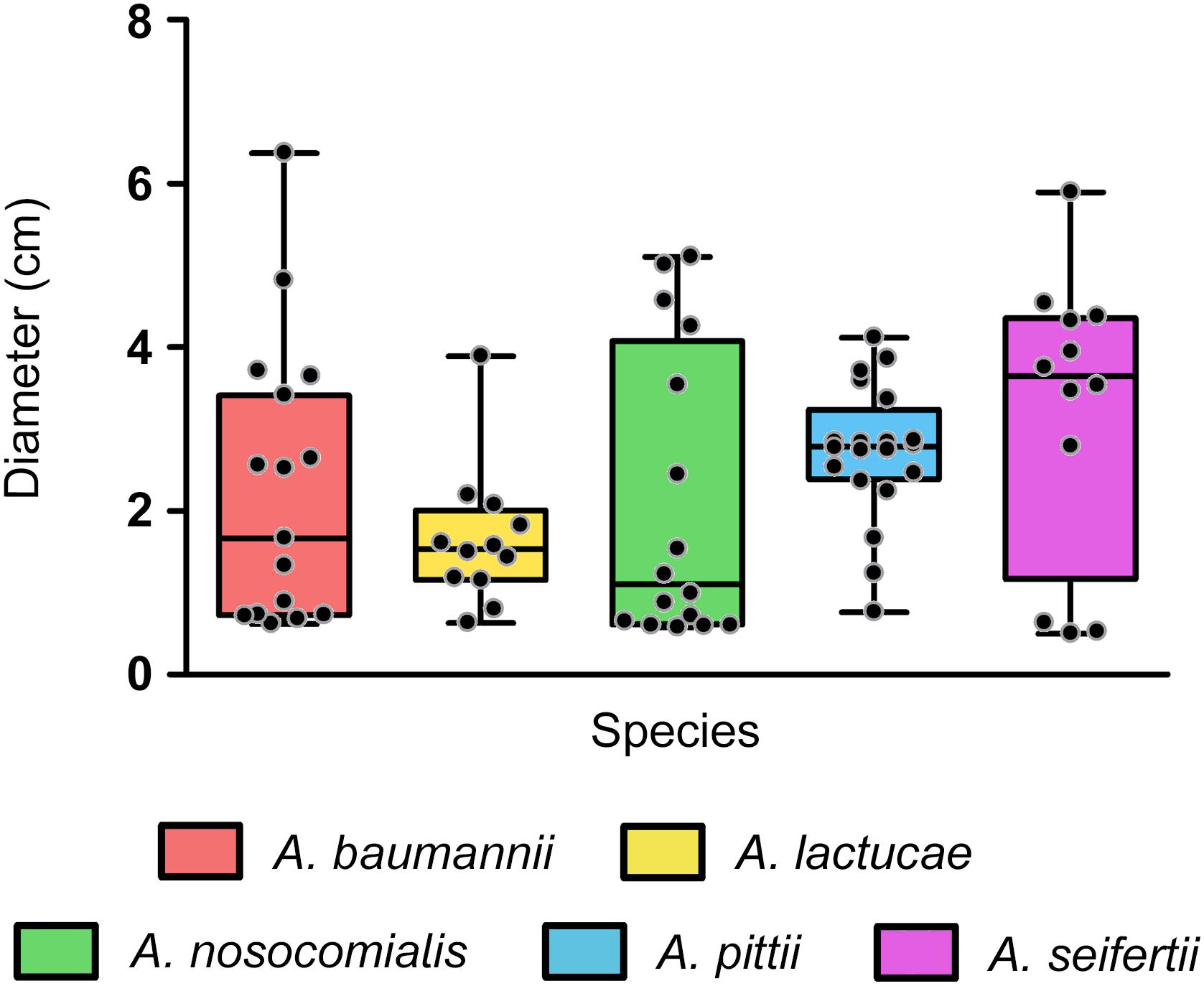
Figure 4. Surface associated motility in the Ab group species. Overlapping dot plot and box plot of the surface-associated motility of each species. Surface-associated motility was recorded as the diameter of growth after 18 h of incubation at 37°C using tryptone (5 g/L), NaCl (2.5 g/L), and agarose (0.3% w/v) motility media. The boxes span from the first to the third quartile. The median is indicated as a segment inside the box. Whiskers indicate the minimum and maximum diameters measured. Each dot corresponds to the average diameter measured for an isolate after at least four biological replicates. Statistically significant differences were not found (Kruskal–Wallis test, P-value > 0.05).
Interestingly, we also observed that all isolates displayed different surface-associated motility morphotypes, as previously described for A. baumannii (Skiebe et al., 2012), in a rather strain-specific than species-specific manner. A part from non-motile isolates (morphotype A), four additional different morphotypes (B, C, D and E, in order of abundance) were defined: morphotype B presented striated radiations from the inoculation point with well-defined edges; morphotype C radiated uniformly from the inoculation point and presented soft edges; morphotype D displayed densely-grown stria with thick edges; and morphotype E included the less abundant or unique morphotypes that could not be included in the any of the other groups (Supplementary Figure S1).
The daily survival of the nematodes was scored during 15 consecutive days in order to obtain LT50 values of each isolate. Determination of the LT50 values presented, once more, ample intra-species variability (Figure 5A). Nevertheless, all isolates could be easily grouped into two distinct clusters according to their LT50 values. Those considered as virulent presented LT50 values ranging from 0.48 to 1.90 days and those included in the non-virulent group showed LT50 values ranging from 4.10 to 8.19 days. In view of these results, an LT50 cut-off value of 3 days was selected to differentiate virulent (LT50 < 3 days) from non-virulent isolates (LT50 > 3 days). All A. baumannii isolates were included within the non-virulent cluster, presenting LT50 values ranging from 4.85 to 8.04 days. A. nosocomialis was also shown to be an overall non-virulent species under this model, with 87.5% of isolates included in the non-virulent cluster and a median LT50 value of 4.65 days. On the other hand, according to this model, A. lactucae turned out to be the most virulent species (median LT50 of 0.871 days), with only one isolate included in the non-virulent group (Figure 5A). Of note, this was the only isolate of A. lactucae that carried a carbapenemase gene. In terms of virulence, A. lactucae presented significant differences in virulence only with A. baumannii (Kruskal–Wallis test, post hoc P-value < 0.001).
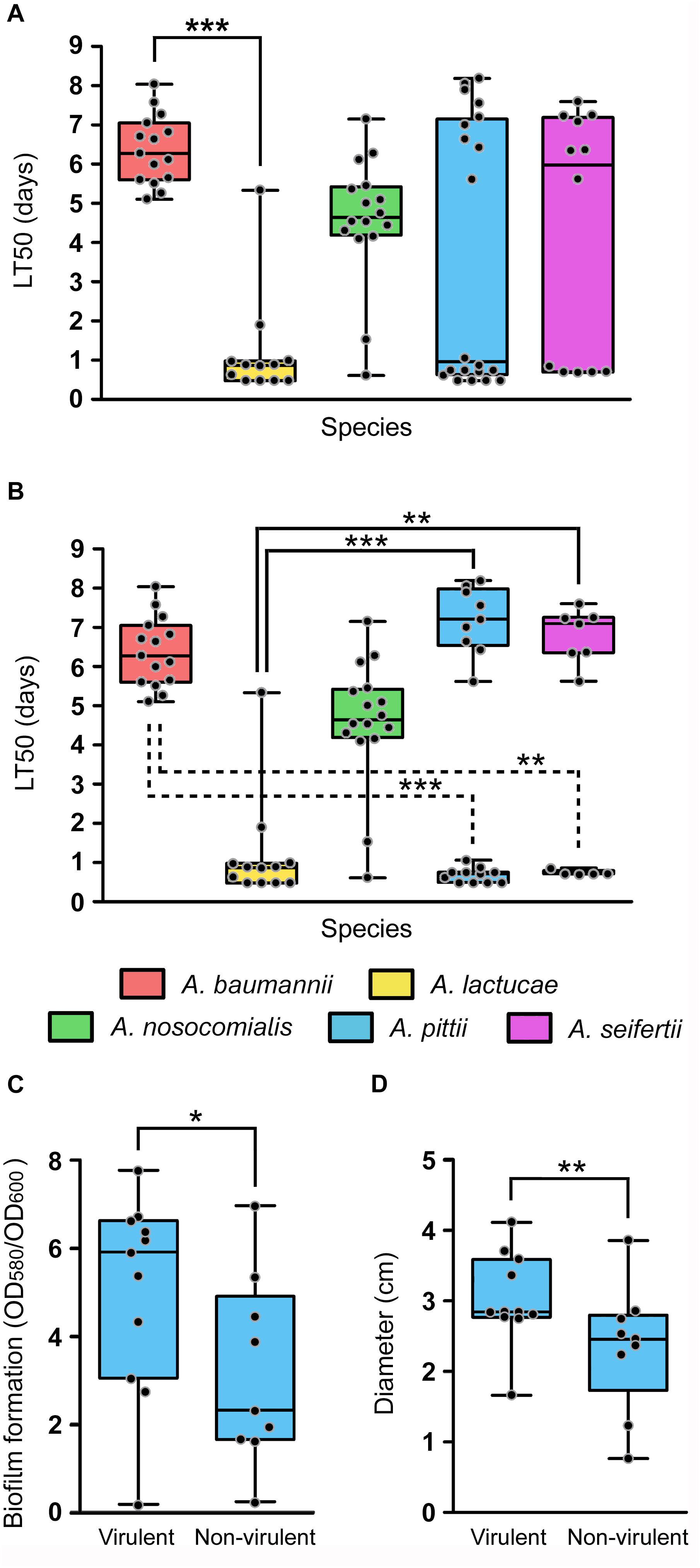
Figure 5. In vivo virulence of the Ab group species using the C. elegans infection model. Overlapping dot plot and box plot of (A) the overall LT50 values of each species; (B) the LT50 values of each species splitting A. pittii and A. seifertii in virulent (LT50 < 3 days) and non-virulent (LT50 > 3 days) subgroups; (C) the biofilm formation values at 28°C; and (D) the surface-associated motility values of A. pittii isolates divided into virulent and non-virulent subgroups. The boxes span from the first to the third quartile. The median is indicated as a segment inside the box. Whiskers indicate the minimum and maximum LT50 (A,B), biofilm formation values (C) and diameter (D). Each dot corresponds to the average phenotype value for an isolate after at least three biological replicates. Statistically significant differences between species (Kruskal–Wallis test) and subgroup species (Mann–Whitney U-test) are highlighted with asterisks: (∗) if the P-value < 0.05, (∗∗) if the P-value < 0.01, and (∗∗∗) if the P-value < 0.001.
Interestingly, A. pittii and A. seifertii, when considered as a whole, showed very large dispersion of the data and presented no significant differences compared to either A. baumannii or A. lactucae (Figure 5A). However, A. pittii and A. seifertii isolates could be clearly divided into virulent (with LT50 values comparable to those of A. lactucae) and non-virulent subgroups (with LT50 values similar to those of A. baumannii and A. nosocomialis). When considered as independent groups, the virulent isolates of both A. pittii and A. seifertii presented significant differences in terms of virulence when compared to A. baumannii (Kruskal–Wallis test, post hoc P-value < 0.001 and 0.011, respectively), and the non-virulent isolates also presented significant differences when compared to A. lactucae (Kruskal–Wallis test, post hoc P-value < 0.001 and 0.001, respectively) (Figure 5B). Pairwise comparisons of the growth rates at 25°C between the different species showed that there were no differences in the growth rates between A. baumannii and A. pittii/A. seifertii (Student’s t-test, P-value 0.521 and 0.122, respectively). In addition, evaluation of the fitness rates at 25°C in all the species showed that there was no significant correlation between the LT50 values and the growth rates at 25°C of the isolates (Spearman’s correlation coefficient −0.349, P-value 0.12), and no differences in the growth rates at 25°C were found when the non-virulent and virulent isolates were compared as two independent groups (Student’s t-test, P-value 0.217), thus suggesting that the outcome of the killing assays was not affected by the growth rate of the isolates at 25°C.
In view of these results, we wondered if there were intra-species differences between the virulent and non-virulent subgroups of A. pittii and A. seifertii in regard to the other virulence-related phenotypes studied above, i.e., biofilm formation and motility, instead of considering these species as a whole. In A. pittii, isolates included within the virulent group were also shown to produce more biofilm at 28°C as well as to be more motile than those isolates included in the non-virulent group (Figures 5C,D, Mann–Whitney U-test, P-value of 0.038 and 0.002, respectively). In A. seifertii, however, it was not possible to associate virulent and non-virulent isolates with any other phenotype as there were no significant differences between both groups.
Despite the phenotypic similarities that hinder an accurate identification at the species level of the members of the Ab group using conventional identification methods, here we have sought any relevant differences among them in terms of antibiotic susceptibility patterns and virulence potential. Our efforts are aimed at reinforcing the idea that reporting and studying them collectively may be misleading in the clinical setting, as infections caused by different members of the Ab group might have substantially different clinical implications, as it has been noted for A. pittii and A. nosocomialis but has not yet been thoroughly examined for A. lactucae and A. seifertii (Chuang et al., 2011; Chusri et al., 2014).
In terms of antimicrobial resistance, results from our study are in good agreement with those of previous investigations reporting overall less susceptibility to most antimicrobial agents in A. baumannii, while A. pittii and A. nosocomialis tended to be more susceptible (Chuang et al., 2011; Lai et al., 2012; Chen et al., 2018; Pailhories et al., 2018). Studies that report on the antimicrobial susceptibility of A. seifertii or A. lactucae, on the other hand, are almost non-existent, since both species have only recently been described. Nevertheless, a study by Karah et al. (2011) included 3 A. seifertii isolates that were susceptible to all antibiotics tested. Another report that identified 28/287 bloodstream A. seifertii isolates detected two carbapenem-resistant isolates with a MDR phenotype, including resistance to colistin (Park et al., 2012). Although some authors have claimed that A. seifertii might be intrinsically resistant to polymyxins (Cayo et al., 2016; Narciso et al., 2017), we did not find colistin-resistant isolates of this species among our collection. Indeed, our results suggest that the novel members of the Ab group might be susceptible to most antibiotics, resembling other non-baumannii Acinetobacter species.
Another characteristic of many Acinetobacter species is the carriage of naturally occurring oxacillinases, such as OXA-51 in A. baumannii, and it is widely accepted that members of the same Acinetobacter species seem to harbor OXA allelic variants that belong to the same OXA group (Perichon et al., 2014). Such intrinsic OXA groups may account for β-lactam resistance, especially when insertion sequences are found upstream from the blaOXA gene (Higgins et al., 2010a). Nevertheless, bacterial isolates belonging to A. nosocomialis and A. seifertii might be an exception since no intrinsic OXAs have yet been found in the genome sequences of A. nosocomialis and A. seifertii (Perichon et al., 2014), except for the presence of plasmid-borne intrinsic OXA from A. baumannii (Lee et al., 2012). In our study, also in good agreement with data from Perichon et al. (2014), we confirmed the presence of species-specific intrinsic OXA genes in A. baumannii, A. lactucae and A. pittii, and the lack of them in A. nosocomialis and A. seifertii (Figure 2).
The prevalence of adeB in A. baumannii from our collection was high (93.75%) albeit within the range of previous reports (53–97%) (Chu et al., 2006; Nemec et al., 2007; Lin et al., 2009; Rajamohan et al., 2010; Modarresi et al., 2015; Nowak et al., 2015), but we acknowledge that isolates of A. baumannii in our study are the least polyclonal of all five species (Supplementary Data Sheet S1), although it is not clear what was the degree of clonality in previous studies. Interestingly, the adeB gene was present in all the A. nosocomialis isolates as well as in many A. lactucae and A. pittii isolates, confirming that this efflux system is not restricted to A. baumannii (Espinal et al., 2011). Likewise, the adeIJK and adeFGH operons were previously considered as being highly specific of A. baumannii, and adeDE as being specific of A. pittii. Here we have shown that adeJ and adeG genes are present in all species, and that adeE was present in all A. nosocomialis isolates and was detected among A. seifertii and A. lactucae, although it was missing in A. baumannii. Hou et al. (2012) reported a small number of A. baumannii isolates harboring adeE together with adeB, albeit species identification was performed with phenotypic methods which we now know may lead to misidentification.
The study of virulence-related phenotypes revealed that biofilm formation and surface-associated motility remained highly variable among isolates from the same species. Indeed, several authors have pointed out that the variability observed in biofilm formation might be due to this phenotype being clone-specific (Rodriguez-Baño et al., 2008; Wroblewska et al., 2008; de Breij et al., 2010; Giannouli et al., 2013; Kaliterna and Goic-Barisic, 2013; Gentile et al., 2014), unfortunately, there is still little knowledge regarding the population structure of Acinetobacter species other than A. baumannii and thus, it is not possible to compare major clonal groups for each of these species. In addition, and as previously described for A. baumannii (McQueary et al., 2012), we observed that motile isolates displayed different surface-associated morphotypes, which were not species-dependent but, again, strain-specific. Nonetheless, despite the intra-species variability observed, we were still able to find overall inter-species differences. In fact, the ability to produce biofilm not only differed between species but it was also temperature-dependent. Temperature modulation of biofilm formation, as well as motility and antibiotic resistance, have been previously observed in A. baumannii ATCC 17978, although the underlying regulatory mechanisms remain unclear (De Silva et al., 2018). Our results suggest different thermoregulation strategies of biofilm formation between isolates of A. baumannii, A. nosocomialis, and A. seifertii and those of A. lactucae and A. pittii.
Infection assays using the C. elegans animal model also revealed overall differences between isolates of A. baumannii and A. nosocomialis, being mostly non-virulent, and those of A. lactucae, being highly virulent, although statistical significance was only achieved between A. baumannii and A. lactucae. The infection assays highlight the existence of two subpopulations within our collection of A. pittii and A. seifertii isolates. It remains unclear whether this observation represents the intrinsic heterogeneity within each species, or it is an indication that the taxonomic delineation for these two species needs to be further investigated. It is worth mentioning that A. lactucae isolates were previously classified as “A. pittii-like” (Espinal et al., 2015), since both species are closely related, and the A. pittii taxon seems to be taxonomically difficult as it contains several related (“-like”) strains with no clear phenotypic/genotypic discontinuities (Perichon et al., 2014; Mari-Almirall et al., 2017). It is also plausible that our results reflect a process of adaptation to a particular ecological niche, such as the human host.
To summarize, we observed that species which are more related to the nosocomial environment, i.e., A. baumannii and A. nosocomialis, had non-virulent phenotypes assessed in the C. elegans infection model and tended to form less biofilm than the other species. In contrast, A. pittii, which is more ecologically diverse (Al Atrouni et al., 2016), showed greater virulence, together with the two recently described species A. lactucae and A. seifertii (Nemec et al., 2015; Mari-Almirall et al., 2017). Interestingly, the only study that so far attempted to compare the virulence traits of the different species of the Ab group (A. lactucae was not included) also reported that A. seifertii isolated from bloodstream infections showed enhanced in vitro virulence properties (Na et al., 2016).
These findings however, do not seem to correlate with clinical studies, where infections caused by A. baumannii in particular but also by A. nosocomialis seem to be associated with a less favorable outcome than that of patients with infections caused by other species (Lee et al., 2011; Wisplinghoff et al., 2012; Chusri et al., 2014; Lee et al., 2014; Fitzpatrick et al., 2015). There are a few studies that suggest that such observation might not be due to a higher pathogenicity of the former species and rather be the consequence of inappropriate early empirical treatment, since a higher proportion of A. baumannii infections are caused by MDR strains.
The underlying question we intended to address here is whether the novel Acinetobacter species present a higher pathogenicity potential than A. baumannii that may be currently masked by their overall antimicrobial susceptibility. If so, should we expect worse clinical outcomes upon the acquisition of resistance by these Acinetobacter species? Under this assumption, A. nosocomialis and A. baumannii might have suffered a reduction of intra-species diversity (population bottleneck) resulting from adaptation to the human host and/or antibiotic selection, while the variability observed among isolates of A. pittii, A. seifertii and A. lactucae might reflect their emergence and distinct degrees of adaptation as human nosocomial pathogens (Schleicher et al., 2013; Jain et al., 2016; Chen et al., 2018; Pailhories et al., 2018; Rocha et al., 2018). Nevertheless, it is important to acknowledge that the results provided in our study arise from the usage of in vitro and nematode infection models that do not entirely reflect more complex clinical scenarios and, thus, the extrapolation of these results to infer virulence in the human host must be taken with extreme caution. For instance, respiratory tract infections and in particular both community-acquired as well as nosocomial pneumonia are mainly attributed to A. baumannii and A. nosocomialis while there is little evidence that the other species are capable of causing such infections in humans (Wang et al., 2013; Larcher et al., 2017).
With the recent advances in bacterial identification methods it is likely that the number of reported infections caused by the novel species of Acinetobacter increases over time. This increase should provide more robust clinical data to evaluate the characteristics of infections caused by the different members of the Ab group and reveal their pathogenic potential in the human host.
The authors state that the dataset and partial protein sequences supporting the findings of this study are available within the article and its Supplementary Material.
CC, LR, PH, HS, IR, and JV collected the strains. CC, HS, IR, and JV conceived and designed the experiments. CC, CR, MM-A, LR, and PH performed the experiments. CC, CR, MM-A, PH, HS, IR, and JV analyzed and interpreted the data. CC, PH, HS, IR, and JV wrote the manuscript. All authors critically revised the manuscript for intellectual content and read and approved the final manuscript.
This study was supported by Planes Nacionales de I+D+i 2008–2011/2013–2016, Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Economía y Competitividad, Spanish Network for Research in Infectious Diseases (REIPI RD16/0016/0010); the 2017 call for Strategic Action on Health (PI17/01932 and PI17/01468), co-financed by European Development Regional Fund “A way to achieve Europe” and operative program Intelligent Growth 2014–2020; and grant 2017 SGR 0809 from the Departament d’Universitats, Recerca i Societat de la Informació of the Generalitat de Catalunya. CC and MM-A were supported by grants FPU 13/02564 and FPU 14/06357, respectively, from the Spanish Ministry of Education, Culture and Sports. IR was supported by the Department of Health, Generalitat de Catalunya, grant SLT002/16/00349. Part of the data has been presented as poster communications at the 10th International Symposium on the Biology of Acinetobacter, 3rd–5th June 2015, Athens (Greece), and at the XXVII European Congress of Clinical Microbiology and Infectious Diseases (ECCMID), 22nd–25th April 2017, Vienna (Austria).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank all the isolates donors listed in Supplementary Data Sheet S1 for kindly providing the isolates that made possible this work.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02429/full#supplementary-material
Al Atrouni, A., Joly-Guillou, M. L., Hamze, M., and Kempf, M. (2016). Reservoirs of non-baumannii Acinetobacter species. Front. Microbiol. 7:49. doi: 10.3389/fmicb.2016.00049
Bitrian, M., Solari, C. M., Gonzalez, R. H., and Nudel, C. B. (2012). Identification of virulence markers in clinically relevant strains of Acinetobacter genospecies. Int. Microbiol. 15, 79–88. doi: 10.2436/20.1501.01.161
Bogaerts, P., Rezende de Castro, R., de Mendonca, R., Huang, T. D., Denis, O., and Glupczynski, Y. (2013). Validation of carbapenemase and extended-spectrum beta-lactamase multiplex endpoint PCR assays according to ISO 15189. J. Antimicrob. Chemother. 68, 1576–1582. doi: 10.1093/jac/dkt065
Cayo, R., Rodrigues-Costa, F., Pereira Matos, A., Godoy Carvalhaes, C., Dijkshoorn, L., and Gales, A. C. (2016). Old clinical isolates of Acinetobacter seifertii in Brazil producing OXA-58. Antimicrob. Agents Chemother. 60, 2589–2591. doi: 10.1128/aac.01957-15
Chen, L., Yuan, J., Xu, Y., Zhang, F., and Chen, Z. (2018). Comparison of clinical manifestations and antibiotic resistances among three genospecies of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex. PLoS One 13:e0191748. doi: 10.1371/journal.pone.0191748
Chu, Y. W., Chau, S. L., and Houang, E. T. (2006). Presence of active efflux systems AdeABC, AdeDE and AdeXYZ in different Acinetobacter genomic DNA groups. J. Med. Microbiol. 55, 477–478. doi: 10.1099/jmm.0.46433-0
Chuang, Y. C., Sheng, W. H., Li, S. Y., Lin, Y. C., Wang, J. T., Chen, Y. C., et al. (2011). Influence of genospecies of Acinetobacter baumannii complex on clinical outcomes of patients with acinetobacter bacteremia. Clin. Infect. Dis. 52, 352–360. doi: 10.1093/cid/ciq154
Chusri, S., Chongsuvivatwong, V., Rivera, J. I., Silpapojakul, K., Singkhamanan, K., McNeil, E., et al. (2014). Clinical outcomes of hospital-acquired infection with Acinetobacter nosocomialis and Acinetobacter pittii. Antimicrob. Agents Chemother. 58, 4172–4179. doi: 10.1128/aac.02992-14
Cosgaya, C., Mari-Almirall, M., Van Assche, A., Fernandez-Orth, D., Mosqueda, N., Telli, M., et al. (2016). Acinetobacter dijkshoorniae sp. nov., a member of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex mainly recovered from clinical samples in different countries. Int. J. Syst. Evol. Microbiol. 66, 4105–4111. doi: 10.1099/ijsem.0.001318
de Breij, A., Dijkshoorn, L., Lagendijk, E., van der Meer, J., Koster, A., Bloemberg, G., et al. (2010). Do biofilm formation and interactions with human cells explain the clinical success of Acinetobacter baumannii? PLoS One 5:e10732. doi: 10.1371/journal.pone.0010732
De Silva, P. M., Chong, P., Fernando, D. M., Westmacott, G., and Kumar, A. (2018). Effect of incubation temperature on antibiotic resistance and virulence factors of Acinetobacter baumannii ATCC 17978. Antimicrob. Agents Chemother. 62, e1514–e1517. doi: 10.1128/aac.01514-17
Diancourt, L., Passet, V., Nemec, A., Dijkshoorn, L., and Brisse, S. (2010). The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 5:e10034. doi: 10.1371/journal.pone.0010034
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Espinal, P., Mosqueda, N., Telli, M., van der Reijden, T., Rolo, D., Fernandez-Orth, D., et al. (2015). Identification of NDM-1 in a putatively novel Acinetobacter species (“NB14”) closely related to Acinetobacter pittii. Antimicrob. Agents Chemother. 59, 6657–6660. doi: 10.1128/aac.01455-15
Espinal, P., Pantel, A., Rolo, D., Marti, S., Lopez-Rojas, R., Smani, Y., et al. (2019). Relationship between different resistance mechanisms and virulence in Acinetobacter baumannii. Microb. Drug Resist. 25, 752–760. doi: 10.1089/mdr.2018.0182
Espinal, P., Roca, I., and Vila, J. (2011). Clinical impact and molecular basis of antimicrobial resistance in non-baumannii Acinetobacter. Future Microbiol. 6, 495–511. doi: 10.2217/fmb.11.30
Fitzpatrick, M. A., Ozer, E., Bolon, M. K., and Hauser, A. R. (2015). Influence of ACB complex genospecies on clinical outcomes in a U.S. hospital with high rates of multidrug resistance. J. Infect. 70, 144–152. doi: 10.1016/j.jinf.2014.09.004
Fitzpatrick, M. A., Ozer, E. A., and Hauser, A. R. (2016). Utility of whole-genome sequencing in characterizing Acinetobacter epidemiology and analyzing hospital outbreaks. J. Clin. Microbiol. 54, 593–612. doi: 10.1128/jcm.01818-15
Gentile, V., Frangipani, E., Bonchi, C., Minandri, F., Runci, F., and Visca, P. (2014). Iron and Acinetobacter baumannii biofilm formation. Pathogens 3, 704–719. doi: 10.3390/pathogens3030704
Giannouli, M., Antunes, L. C., Marchetti, V., Triassi, M., Visca, P., and Zarrilli, R. (2013). Virulence-related traits of epidemic Acinetobacter baumannii strains belonging to the international clonal lineages I-III and to the emerging genotypes ST25 and ST78. BMC Infect. Dis. 13:282. doi: 10.1186/1471-2334-13-282
Gundi, V. A., Dijkshoorn, L., Burignat, S., Raoult, D., and La Scola, B. (2009). Validation of partial rpoB gene sequence analysis for the identification of clinically important and emerging Acinetobacter species. Microbiology 155, 2333–2341. doi: 10.1099/mic.0.026054-0
Harding, C. M., Hennon, S. W., and Feldman, M. F. (2018). Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat. Rev. Microbiol. 16, 91–102. doi: 10.1038/nrmicro.2017.148
Higgins, P. G., Dammhayn, C., Hackel, M., and Seifert, H. (2010a). Global spread of carbapenem-resistant Acinetobacter baumannii. J. Antimicrob. Chemother. 65, 233–238. doi: 10.1093/jac/dkp428
Higgins, P. G., Lehmann, M., Wisplinghoff, H., and Seifert, H. (2010b). gyrB multiplex PCR to differentiate between Acinetobacter calcoaceticus and Acinetobacter genomic species 3. J. Clin. Microbiol. 48, 4592–4594. doi: 10.1128/jcm.01765-10
Hou, P. F., Chen, X. Y., Yan, G. F., Wang, Y. P., and Ying, C. M. (2012). Study of the correlation of imipenem resistance with efflux pumps AdeABC, AdeIJK, AdeDE and AbeM in clinical isolates of Acinetobacter baumannii. Chemotherapy 58, 152–158. doi: 10.1159/000335599
Jain, A. L., Harding, C. M., Assani, K., Shrestha, C. L., Haga, M., Leber, A., et al. (2016). Characteristics of invasive Acinetobacter species isolates recovered in a pediatric academic center. BMC Infect. Dis. 16:346. doi: 10.1186/s12879-016-1678-9
Kaliterna, V., and Goic-Barisic, I. (2013). The ability of biofilm formation in clinical isolates of Acinetobacter baumannii belonging to two different European clones causing outbreaks in the Split University Hospital, Croatia. J. Chemother. 25, 60–62. doi: 10.1179/1973947812y.0000000052
Karah, N., Haldorsen, B., Hegstad, K., Simonsen, G. S., Sundsfjord, A., and Samuelsen, O. (2011). Species identification and molecular characterization of Acinetobacter spp. blood culture isolates from Norway. J. Antimicrob. Chemother. 66, 738–744. doi: 10.1093/jac/dkq521
Kinsella, R. L., Lopez, J., Palmer, L. D., Salinas, N. D., Skaar, E. P., Tolia, N. H., et al. (2017). Defining the interaction of the protease CpaA with its type II secretion chaperone CpaB and its contribution to virulence in Acinetobacter species. J. Biol. Chem. 292, 19628–19638. doi: 10.1074/jbc.M117.808394
Lai, C. C., Hsu, H. L., Tan, C. K., Tsai, H. Y., Cheng, A., Liu, C. Y., et al. (2012). Recurrent bacteremia caused by the Acinetobacter calcoaceticus-Acinetobacter baumannii complex. J. Clin. Microbiol. 50, 2982–2986. doi: 10.1128/jcm.01194-12
Larcher, R., Pantel, A., Arnaud, E., Sotto, A., and Lavigne, J. P. (2017). First report of cavitary pneumonia due to community-acquired Acinetobacter pittii, study of virulence and overview of pathogenesis and treatment. BMC Infect. Dis. 17:477. doi: 10.1186/s12879-017-2589-0
Lee, H. Y., Chen, C. L., Wu, S. R., Huang, C. W., and Chiu, C. H. (2014). Risk factors and outcome analysis of Acinetobacter baumannii complex bacteremia in critical patients. Crit. Care Med. 42, 1081–1088. doi: 10.1097/ccm.0000000000000125
Lee, Y. C., Huang, Y. T., Tan, C. K., Kuo, Y. W., Liao, C. H., Lee, P. I., et al. (2011). Acinetobacter baumannii and Acinetobacter genospecies 13TU and 3 bacteraemia: comparison of clinical features, prognostic factors and outcomes. J. Antimicrob. Chemother. 66, 1839–1846. doi: 10.1093/jac/dkr200
Lee, Y. T., Kuo, S. C., Chiang, M. C., Yang, S. P., Chen, C. P., Chen, T. L., et al. (2012). Emergence of carbapenem-resistant non-baumannii species of Acinetobacter harboring a blaOXA–51-like gene that is intrinsic to A. baumannii. Antimicrob. Agents Chemother. 56, 1124–1127. doi: 10.1128/aac.00622-11
Lee, Y. T., Kuo, S. C., Yang, S. P., Lin, Y. T., Chiang, D. H., Tseng, F. C., et al. (2013). Bacteremic nosocomial pneumonia caused by Acinetobacter baumannii and Acinetobacter nosocomialis: a single or two distinct clinical entities? Clin. Microbiol. Infect. 19, 640–645. doi: 10.1111/j.1469-0691.2012.03988.x
Lin, L., Ling, B. D., and Li, X. Z. (2009). Distribution of the multidrug efflux pump genes, adeABC, adeDE and adeIJK, and class 1 integron genes in multiple-antimicrobial-resistant clinical isolates of Acinetobacter baumannii-Acinetobacter calcoaceticus complex. Int. J. Antimicrob. Agents 33, 27–32. doi: 10.1016/j.ijantimicag.2008.06.027
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Mari-Almirall, M., Cosgaya, C., Higgins, P. G., Van Assche, A., Telli, M., Huys, G., et al. (2017). MALDI-TOF/MS identification of species from the Acinetobacter baumannii (Ab) group revisited: inclusion of the novel A. seifertii and A. dijkshoorniae species. Clin. Microbiol. Infect. 23, 210.e1–210.e9. doi: 10.1016/j.cmi.2016.11.020
McQueary, C. N., Kirkup, B. C., Si, Y., Barlow, M., Actis, L. A., Craft, D. W., et al. (2012). Extracellular stress and lipopolysaccharide modulate Acinetobacter baumannii surface-associated motility. J. Microbiol. 50, 434–443. doi: 10.1007/s12275-012-1555-1
Modarresi, F., Azizi, O., Shakibaie, M. R., Motamedifar, M., Valibeigi, B., and Mansouri, S. (2015). Effect of iron on expression of efflux pump (adeABC) and quorum sensing (luxI, luxR) genes in clinical isolates of Acinetobacter baumannii. APMIS 123, 959–968. doi: 10.1111/apm.12455
Na, I. Y., Chung, E. S., Jung, C. Y., Kim, D. H., Shin, J., Kang, K., et al. (2016). Comparison of the virulence-associated phenotypes of five species of Acinetobacter baumannii complex. J. Microbiol. Biotechnol. 26, 171–179. doi: 10.4014/jmb.1507.07076
Narciso, A. C., Martins, W., Cayo, R., Pereira de Matos, A., Santos, S. V., Ramos, P. L., et al. (2017). Detection of OXA-58-producing Acinetobacter seifertii recovered from a black-necked Swan at a Zoo Lake. Antimicrob. Agents Chemother. 61:e01360-17. doi: 10.1128/aac.01360-17
Nemec, A., Krizova, L., Maixnerova, M., Sedo, O., Brisse, S., and Higgins, P. G. (2015). Acinetobacter seifertii sp. nov., a member of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex isolated from human clinical specimens. Int. J. Syst. Evol. Microbiol. 65, 934–942. doi: 10.1099/ijs.0.000043
Nemec, A., Maixnerova, M., van der Reijden, T. J., van den Broek, P. J., and Dijkshoorn, L. (2007). Relationship between the AdeABC efflux system gene content, netilmicin susceptibility and multidrug resistance in a genotypically diverse collection of Acinetobacter baumannii strains. J. Antimicrob. Chemother. 60, 483–489. doi: 10.1093/jac/dkm231
Nowak, J., Seifert, H., and Higgins, P. G. (2015). Prevalence of eight resistance-nodulation-division efflux pump genes in epidemiologically characterized Acinetobacter baumannii of worldwide origin. J. Med. Microbiol. 64, 630–635. doi: 10.1099/jmm.0.000069
Nowak, J., Zander, E., Stefanik, D., Higgins, P. G., Roca, I., Vila, J., et al. (2017). High incidence of pandrug-resistant Acinetobacter baumannii isolates collected from patients with ventilator-associated pneumonia in Greece, Italy and Spain as part of the MagicBullet clinical trial. J. Antimicrob. Chemother. 72, 3277–3282. doi: 10.1093/jac/dkx322
Pailhories, H., Tiry, C., Eveillard, M., and Kempf, M. (2018). Acinetobacter pittii isolated more frequently than Acinetobacter baumannii in blood cultures: the experience of a French hospital. J. Hosp. Infect. 99, 360–363. doi: 10.1016/j.jhin.2018.03.019
Park, Y. K., Jung, S. I., Park, K. H., Kim, S. H., and Ko, K. S. (2012). Characteristics of carbapenem-resistant Acinetobacter spp. other than Acinetobacter baumannii in South Korea. Int. J. Antimicrob. Agents 39, 81–85. doi: 10.1016/j.ijantimicag.2011.08.006
Peleg, A. Y., de Breij, A., Adams, M. D., Cerqueira, G. M., Mocali, S., Galardini, M., et al. (2012). The success of Acinetobacter species; genetic, metabolic and virulence attributes. PLoS One 7:e46984. doi: 10.1371/journal.pone.0046984
Peleg, A. Y., Seifert, H., and Paterson, D. L. (2008). Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21, 538–582. doi: 10.1128/cmr.00058-07
Perichon, B., Goussard, S., Walewski, V., Krizova, L., Cerqueira, G., Murphy, C., et al. (2014). Identification of 50 class D beta-lactamases and 65 Acinetobacter-derived cephalosporinases in Acinetobacter spp. Antimicrob. Agents Chemother. 58, 936–949. doi: 10.1128/aac.01261-13
Rajamohan, G., Srinivasan, V. B., and Gebreyes, W. A. (2010). Novel role of Acinetobacter baumannii RND efflux transporters in mediating decreased susceptibility to biocides. J. Antimicrob. Chemother. 65, 228–232. doi: 10.1093/jac/dkp427
Roca, I., Espinal, P., Vila-Farres, X., and Vila, J. (2012). The Acinetobacter baumannii oxymoron: commensal hospital dweller turned pan-drug-resistant menace. Front. Microbiol. 3:148. doi: 10.3389/fmicb.2012.00148
Rocha, G. A., Lima, D. F., Rodrigues, E. R., Leao, R. S., Folescu, T. W., Firmida, M. C., et al. (2018). Species distribution, sequence types and antimicrobial resistance of Acinetobacter spp. from cystic fibrosis patients. Epidemiol. Infect. 146, 524–530. doi: 10.1017/s0950268817002849
Rodriguez-Baño, J., Marti, S., Soto, S., Fernandez-Cuenca, F., Cisneros, J. M., Pachon, J., et al. (2008). Biofilm formation in Acinetobacter baumannii: associated features and clinical implications. Clin. Microbiol. Infect. 14, 276–278. doi: 10.1111/j.1469-0691.2007.01916.x
Schleicher, X., Higgins, P. G., Wisplinghoff, H., Korber-Irrgang, B., Kresken, M., and Seifert, H. (2013). Molecular epidemiology of Acinetobacter baumannii and Acinetobacter nosocomialis in Germany over a 5-year period (2005-2009). Clin. Microbiol. Infect. 19, 737–742. doi: 10.1111/1469-0691.12026
Seifert, H., Dolzani, L., Bressan, R., van der Reijden, T., van Strijen, B., Stefanik, D., et al. (2005). Standardization and interlaboratory reproducibility assessment of pulsed-field gel electrophoresis-generated fingerprints of Acinetobacter baumannii. J. Clin. Microbiol. 43, 4328–4335. doi: 10.1128/jcm.43.9.4328-4335.2005
Skiebe, E., de Berardinis, V., Morczinek, P., Kerrinnes, T., Faber, F., Lepka, D., et al. (2012). Surface-associated motility, a common trait of clinical isolates of Acinetobacter baumannii, depends on 1,3-diaminopropane. Int. J. Med. Microbiol. 302, 117–128. doi: 10.1016/j.ijmm.2012.03.003
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Vaneechoutte, M., Dijkshoorn, L., Tjernberg, I., Elaichouni, A., de Vos, P., Claeys, G., et al. (1995). Identification of Acinetobacter genomic species by amplified ribosomal DNA restriction analysis. J. Clin. Microbiol. 33, 11–15.
Wang, X., Chen, T., Yu, R., Lu, X., and Zong, Z. (2013). Acinetobacter pittii and Acinetobacter nosocomialis among clinical isolates of the Acinetobacter calcoaceticus-baumannii complex in Sichuan, China. Diagn. Microbiol. Infect. Dis. 76, 392–395. doi: 10.1016/j.diagmicrobio.2013.03.020
Wisplinghoff, H., Paulus, T., Lugenheim, M., Stefanik, D., Higgins, P. G., Edmond, M. B., et al. (2012). Nosocomial bloodstream infections due to Acinetobacter baumannii, Acinetobacter pittii and Acinetobacter nosocomialis in the United States. J. Infect. 64, 282–290. doi: 10.1016/j.jinf.2011.12.008
Wroblewska, M. M., Sawicka-Grzelak, A., Marchel, H., Luczak, M., and Sivan, A. (2008). Biofilm production by clinical strains of Acinetobacter baumannii isolated from patients hospitalized in two tertiary care hospitals. FEMS Immunol. Med. Microbiol. 53, 140–144. doi: 10.1111/j.1574-695X.2008.00403.x
Keywords: Acinetobacter, multi-drug resistance, virulence, biofilm formation, C. elegans, efflux pumps, oxacillinases
Citation: Cosgaya C, Ratia C, Marí-Almirall M, Rubio L, Higgins PG, Seifert H, Roca I and Vila J (2019) In vitro and in vivo Virulence Potential of the Emergent Species of the Acinetobacter baumannii (Ab) Group. Front. Microbiol. 10:2429. doi: 10.3389/fmicb.2019.02429
Received: 18 June 2019; Accepted: 08 October 2019;
Published: 24 October 2019.
Edited by:
Paolo Visca, Roma Tre University, ItalyReviewed by:
Raffaele Zarrilli, University of Naples Federico II, ItalyCopyright © 2019 Cosgaya, Ratia, Marí-Almirall, Rubio, Higgins, Seifert, Roca and Vila. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ignasi Roca, SWduYXNpLnJvY2FAaXNnbG9iYWwub3Jn
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.