
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol., 04 October 2019
Sec. Virology
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.02297
This article is part of the Research TopicThe Interplay Between Innate Immunity And HerpesvirusesView all 16 articles
 Mariella Della Chiesa1,2
Mariella Della Chiesa1,2 Andrea De Maria2,3,4
Andrea De Maria2,3,4 Letizia Muccio1
Letizia Muccio1 Federica Bozzano5
Federica Bozzano5 Simona Sivori1,2
Simona Sivori1,2 Lorenzo Moretta5*
Lorenzo Moretta5*NK cells contribute to early defenses against viruses through their inborn abilities that include sensing of PAMPs and inflammatory signals such as cytokines or chemokines, recognition, and killing of infected cells through activating surface receptors engagement. Moreover, they support adaptive responses via Ab-dependent mechanisms, triggered by CD16, and DC editing. Their fundamental role in anti-viral responses has been unveiled in patients with NK cell deficiencies suffering from severe Herpesvirus infections. Notably, these infections, often occurring as primary infections early in life, can be efficiently cleared by NK, T, and B cells in healthy hosts. Herpesviruses however, generate a complicated balance with the host immune system through their latency cycle moving between immune control and viral reactivation. This lifelong challenge has contributed to the development of numerous evasion mechanisms by Herpesviruses, many of which devoted to elude NK cell surveillance from viral reactivations rather than primary infections. This delicate equilibrium can be altered in proportions of healthy individuals promoting virus reactivation and, more often, in immunocompromised subjects. However, the constant stimulus provided by virus-host interplay has also favored NK-cell adaptation to Herpesviruses. During anti-HCMV responses, NK cells can reshape their receptor repertoire and function, through epigenetic remodeling, and acquire adaptive traits such as longevity and clonal expansion abilities. The major mechanisms of recognition and effector responses employed by NK cells against Herpesviruses, related to their genomic organization will be addressed, including those allowing NK cells to generate memory-like responses. In addition, the mechanisms underlying virus reactivation or control will be discussed.
Human NK cells are innate lymphocytes that rapidly provide defenses against tumors and viral infections allowing pathogen elimination or limiting viral spread (Vivier et al., 2011; Della Chiesa et al., 2014b). Their fast responses mainly rely on the expression of multiple germ-line encoded activating receptors among which natural cytotoxicity receptors (NCRs) and NKG2D play the most relevant role in the recognition and killing of infected cells (Bottino et al., 2000; Moretta and Moretta, 2004; Lanier, 2015). The responses elicited by activating receptors are integrated and balanced by the engagement of inhibitory receptors mainly depending on those specific for HLA class I (HLA-I) molecules that include the Killer Ig-like Receptors (KIRs), able to distinguish among allotypic determinants of HLA-A, -B and -C (Bottino et al., 1996; Parham, 2005), the CD94/NKG2A heterodimer, specific for the non-classic HLA-I molecule HLA-E (Braud et al., 1998), and LILRB1 (or CD85j/ILT-2) broadly recognizing HLA-I alleles (Colonna et al., 1997).
Upon infection many viruses, including Herpesviruses, target T cell function via specific interactions with TCR and HLA-I molecules. Indeed, several viral products interfere with host TAP proteins and HLA-I expression, leading to reduced CTL-mediated recognition of infected cells, and decreased naïve T cell activation (Hill et al., 1995; Imai et al., 2013; Schuren et al., 2016). Conversely, downregulated HLA-I expression renders infected cells susceptible to NK-cell killing (Huard and Fruh, 2000; Tortorella et al., 2000). However, activating counterparts of HLA-I-specific receptors, namely activating KIRs (aKIRs), and CD94/NKG2C can also importantly contribute to defense against virus (Della Chiesa et al., 2015).
Human NK cells are usually divided in two major populations, the CD56bright subset expressing NKG2A, lacking KIRs and CD16 (i.e., a low affinity Fcγ Receptor) and the CD56dim subset expressing high CD16 and variable proportions of KIRs, NKG2A, LILRB1, CD57, and NKG2C (Cooper et al., 2001; Caligiuri, 2008; Freud et al., 2017). These two subsets differ in their proliferative potential, cytotoxic activity, cytokine production, and homing to peripheral tissues (Moretta, 2010; Castriconi et al., 2018) thus offering different anti-viral defenses. Notably, CD56dim NK cells, besides high cytotoxicity, can also rapidly produce IFN-γ and TNF-α upon receptor-induced cell triggering (De Maria et al., 2011).
The critical role of NK cells in viral defense has been disclosed by the higher susceptibility to viral infections, caused primarily by Herpesviruses, in individuals affected by congenital immunodeficiencies in which NK cells are absent or defective (Orange, 2002; Etzioni et al., 2005; Notarangelo and Mazzolari, 2006; Mace and Orange, 2019). Herpesviruses are a family of dsDNA viruses, divided in three subfamilies, i.e., α- (HSV-1, HSV-2, and VZV), β- (CMV, HHV6, and HHV7) and γ-Herpesvirus (EBV and KSHV), that differ for their genetic content, infection sites and pathogenesis, while sharing the ability to persist in the host in a latency status after resolution of a primary infection (De Pelsmaeker et al., 2018). The mechanisms by which Herpesviruses establish and maintain latency have not been completely elucidated.
In an evolutionary perspective, our immune system and Herpesviruses have co-evolved influencing reciprocally. During this process the generation of several viral immunoevasion mechanisms has been favored. Most of these mechanisms aim at limiting and suppressing NK-cell responses, which point again to the relevance of these lymphocytes in Herpesvirus control. Although viral immunoevasion strategies are crucial in NK-Herpesvirus interactions, they will not be specifically addressed here and have been exhaustively reviewed elsewhere (Corrales-Aguilar et al., 2014; De Pelsmaeker et al., 2018).
On the other hand, the host-Herpesvirus interaction has exerted a strong pressure on our immune system likely favoring the generation of unexpected memory responses by NK cells and their adaptation to Herpesviruses, in particular to CMV (Muntasell et al., 2013; Sun et al., 2014).
The main mechanisms by which NK cells can recognize and eliminate virus-infected cells involve the employ of (i) activating receptors for cellular ligands often overexpressed upon infection, (ii) activating receptors for virus-derived ligands, (iii) activating receptors, i.e., NKG2C and aKIRs, recognizing virus-modified HLA-I molecules, and (iv) CD16-mediated antibody-dependent cellular cytotoxicity (ADCC) (Hammer et al., 2018b). Almost all these mechanisms can be applied to NK cells in Herpesvirus control (Figures 1A–C).
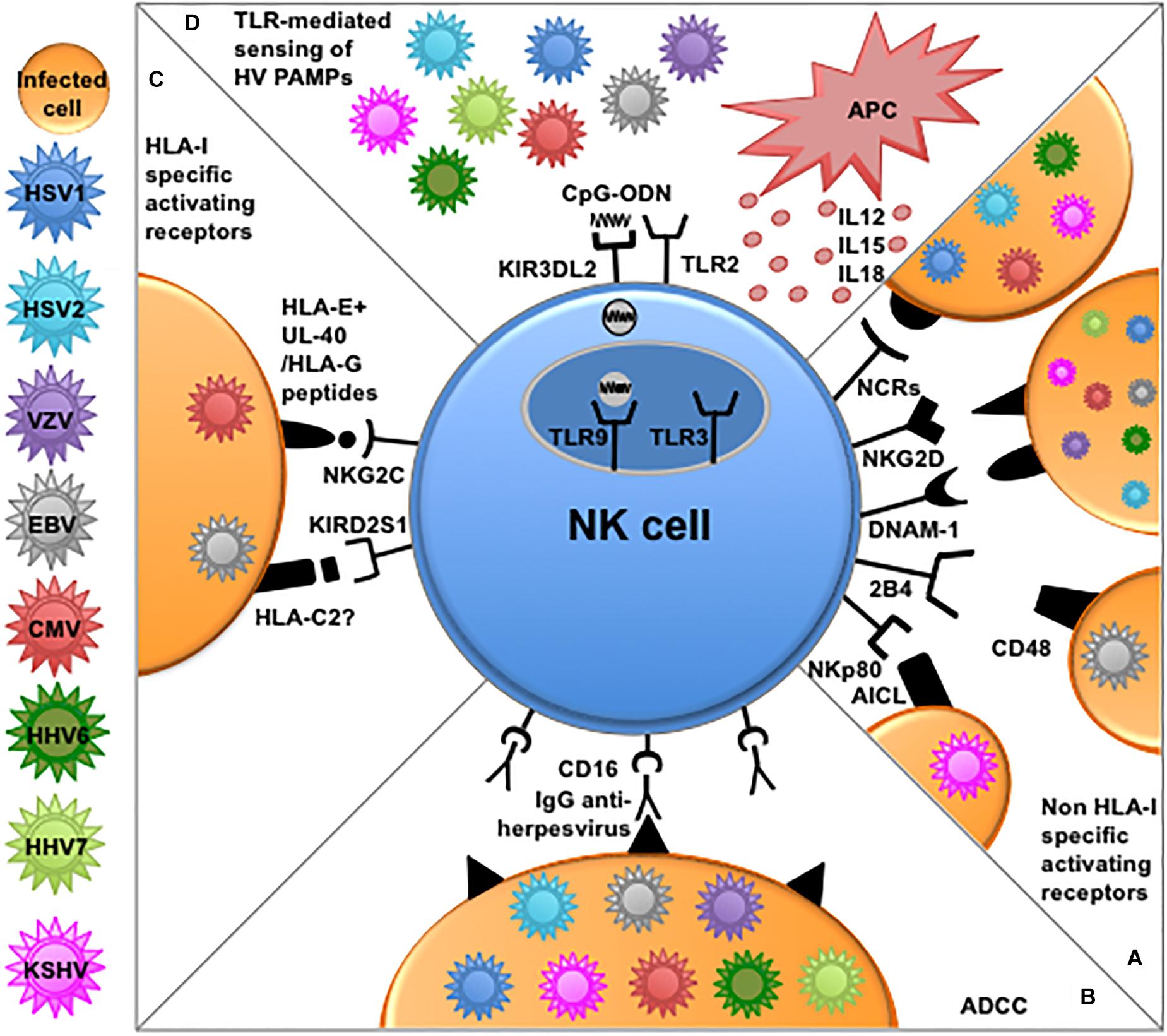
Figure 1. NK cell-mediated mechanisms of recognition and responses to Herpesviruses. (A) Several non-HLA-I-specific activating receptors and co-receptors, i.e., NCRs, NKG2D, DNAM-1, 2B4, and NKp80 play an important role in the elimination of cells infected by different Herpesviruses through the recognition of cellular ligands expressed on target cells. (B) NK cells can efficiently kill opsonized Herpesvirus-infected cells through antibody-dependent cellular cytotoxicity (ADCC) via CD16 engagement by the Fc fragment of anti-viral immunoglobulins. (C) NKG2C and aKIRs play a role mainly in the recognition of CMV-infected cells. The underlying recognition mechanisms are based on interactions with cognate HLA-I molecules. NKG2C shows enhanced interaction to HLA-E presenting peptides derived from viral UL-40 or HLA-G leader sequences, while, among aKIRs, KIR2DS1 seems to better recognize HLA-C2, modified upon CMV infection or presenting EBV-derived peptides. (D) NK cells express different functional TLRs involved in the recognition of PAMPs derived from Herpesviruses. In particular TLR2 alllows NK-mediated recognition of envelope glycoproteins from HSV and CMV, while TLR9 can recognize viral CpG sequences shuttled by KIR3DL2 from the surface of NK cells to endosomes. APC-derived cytokines and reciprocal interactions with these immune cells (e.g., dendritic cells and macrophages) can further enhance NK cells effector function against Herpesviruses.
The importance of certain activating receptors in Herpesvirus elimination has been indirectly revealed by the numerous proteins encoded by the different Herpesviruses aimed at limiting activating receptors function, in most cases by downregulating the respective cellular ligands on infected cells. In this context, the activating receptor NKG2D that recognizes stress-induced cellular ligands often overexpressed upon viral infection or tumor transformation (i.e., MIC-A, MIC-B, and ULBPs) (Lanier, 2015), is central in NK-mediated immune responses against virtually all Herpesviruses, namely HSV-1, VZV, CMV, HHV6, HHV7, KSHV, and EBV, all of which encode molecules downregulating NKG2D ligands (Wu et al., 2003; Thomas et al., 2008; Nachmani et al., 2009; Schneider and Hudson, 2011; Campbell et al., 2015; Schmiedel et al., 2016). Besides NKG2D, other non-HLA-I-specific activating receptors can play a role against several Herpesviruses suggesting a common strategy to eliminate these pathogens. In particular, the three NCRs (i.e., NKp46, NKp30, and NKp44) are involved in killing HSV-1-infected fibroblasts. The upregulation of cellular NCR ligands upon HSV-1 infection is resulted responsible for the increased susceptibility to NK-mediated cytotoxicity (Chisholm et al., 2007). Interestingly, NK-mediated killing was appreciable even before HLA-I downregulation had occurred, suggesting that, in NK-HSV-1 interactions, activating signals can overcome inhibitory receptors signaling (Chisholm et al., 2007). In this context, increased NCRs expression and function have been observed in NK cells differentiating in vitro from CD34+ precursors in the presence of HSV-infected myelomonocytes, further strengthening the relevance of the NCRs-NCR ligands axis against HSV (Costa et al., 2009).
The NCR NKp30 also participates in recognition and killing of CMV- and HHV6-infected cells. Its involvement is again testified by viral evasion mechanisms that downregulate B7-H6, a major NKp30 cellular ligand (Brandt et al., 2009), possibly expressed on infected cells (Schmiedel et al., 2016; Charpak-Amikam et al., 2017). In addition, NKp30 itself is the target of a CMV-encoded protein, pp65, that by binding to this NCR can induce its dissociation from the signaling molecule CD3ζ, thereby inhibiting NK-mediated killing of CMV-infected fibroblasts and dendritic cells (DCs) (Arnon et al., 2005). Along this line, a role for the NKp44-NKp44 ligand signaling pathway against KSHV is suggested by NKp44 ligand downregulation during lytic infection in KSHV-infected cells (Madrid and Ganem, 2012).
Similar to NKG2D and NCRs, the activating co-receptor DNAM1 recognizing PVR and Nectin-2 (CD112) (Bottino et al., 2003), plays a role against different Herpesviruses, i.e., CMV, EBV, and HSV-2 as demonstrated by different evasion strategies reducing DNAM-1 signaling (Tomasec et al., 2005; Prod’homme et al., 2010; Grauwet et al., 2014; Williams et al., 2015).
While NKG2D, DNAM-1, and NCRs serve against several Herpesviruses, other activating NK receptors are specifically involved in the recognition/elimination of cells infected only by a given Herpesvirus. An example is the co-receptor 2B4 (or CD244) which requires the adaptor protein SLAM-associated protein (SAP) to deliver activating signals upon engagement with its ligand CD48 (Nakajima et al., 1999; Bottino et al., 2000). 2B4 engagement is crucial to NK-mediated killing of EBV-infected B cells. Indeed, B cells that are CD48 high, represent a preferential target for this Herpesvirus (Chijioke et al., 2016). A role for 2B4 was actually revealed by the severe consequences of primary EBV infection in individuals suffering from X-linked lymphoproliferative disease (XLP-1), a congenital immunodeficiency in which SAP is absent or defective (Sayos et al., 1998), resulting in inhibitory signals from 2B4 impairing NK-mediated B-EBV elimination (Parolini et al., 2000). Interestingly, NK cells can respond efficiently to EBV-infected B cells in early lytic cycle and NK-mediated killing involves also NKG2D and DNAM-1 (Chijioke et al., 2013; Williams et al., 2015). However, EBV-infected B cells in latency or even in late lytic stages are resistant to NK attack, due to viral evasion mechanisms independent of NK cell function (Williams et al., 2015).
Finally, a role for the activating co-receptor NKp80 in the recognition of KSHV-infected cells was also proposed, based on the downregulation of its ligand AICL upon KSHV infection (Thomas et al., 2008).
Overall, in most instances, the activating receptors described above allow NK cells to eliminate infected cells by the recognition of cellular ligands expressed on target cells, while the engagement of activating receptors by virus-encoded ligands has not been demonstrated for Herpesviruses, at variance with influenza or vaccinia virus whose products hemagglutinin, and neuraminidase are directly recognized by NKp46 and NKp44 (Mandelboim et al., 2001; Ho et al., 2008). On the contrary, the HLA-I specific receptor NKG2C can recognize viral ligands although the mechanisms described so far are based on interactions with viral peptides bound to HLA-E molecules on CMV-infected cells. NKG2C is also involved in generating CMV-induced adaptive responses and will thus be discussed in more detail in the dedicated paragraph.
Another major mechanism employed by NK cells in controlling both primary viral infections, when adaptive immunity is already established, and secondary reactivations (either subclinical or clinical), relies on the activating receptor CD16 (FcγRIIIa), the low-affinity receptor for the immunoglobulin Fc fragment (Braud et al., 1998; Vivier et al., 2011). Upon CD16 engagement, NK cells can efficiently eliminate opsonized infected cells via ADCC. The relevance of this mechanism in providing defense against Herpesvirus is underlined by severe EBV and VZV infections associated to a dysfunctional mutated CD16 (de Vries et al., 1996; Grier et al., 2012). Furthermore, a polymorphism of the CD16 gene resulting in the surface expression of a high affinity CD16 receptor (i.e., the CD16A-158V/V polymorphism) is associated to enhanced NK-mediated ADCC and confers protection from clinical HSV-1 reactivation (Moraru et al., 2012, 2015). Not unexpectedly, this highly effective anti-viral mechanism is targeted by multiple evasion strategies, as both HSV and CMV encode Fcγ-binding proteins that act as decoy receptors interfering with IgG binding to CD16 and thus attenuating ADCC (Johnson et al., 1988; Atalay et al., 2002; Corrales-Aguilar et al., 2014; Costa-Garcia et al., 2015). However, it has been recently described that a viral Fcγ-binding protein, gE, which is expressed on the cell surface by HSV-infected cells, can react with non-specific IgG thus generating a “Fc-bridge” that instead favors NK-mediated ADCC responses (Dai et al., 2017; Dai and Caligiuri, 2018).
NK cells can importantly contribute to early viral defense not only by exerting cytolytic activity against infected cells but also through their ability to sense pathogens via toll-like receptors (TLRs) (Sivori et al., 2004). NK cells express different functional TLRs among which TLR2, TLR3, and TLR9 seem to be primarily involved in the recognition of pathogen-associated molecular patterns (PAMPs) derived from Herpesviruses, such as double stranded viral nucleic acids or structural proteins (Adib-Conquy et al., 2014; Della Chiesa et al., 2014b). In particular, NK cells can directly recognize envelope glycoproteins from both CMV and HSV virions through TLR2 (Kim et al., 2012; Muntasell et al., 2013). Upon TLR2 engagement, NK cells become activated, and produce IFN-γ, further promoting anti-viral immune responses. Indeed, NK cells have been detected in herpetic lesions in close contact with CD4 T cells, thus possibly contributing to directly shaping adaptive responses (Kim et al., 2012). Interestingly, TLR9 polymorphisms are associated with susceptibility to infection, with the T-1237C polymorphism that causes altered TLR9 expression, being predictive of susceptibility to CMV infection (Carvalho et al., 2009). NK cells could thus play a role in TLR9-mediated defense to CMV, as they can efficiently respond to TLR9 agonists such as CpG-ODNs. Remarkably, these TLR9 ligands can be bound at the cell surface by KIR3DL2, a member of the KIR family, and then shuttled by receptor internalization to endosomes where TLR9 is localized (Sivori et al., 2010).
Thus, in a scenario where NK cells are recruited to viral infection sites, their effector function (e.g., cytotoxicity, IFN-γ, and chemokine production) can be enhanced by combined exposure to microbial products and cytokines available in the inflammatory milieu, such as IL-12 or IL-18. In this context, TLRs- and/or cytokine-activated NK cells can reciprocally interact with other immune cells responding to the same PAMPs via TLRs, such as DCs or macrophages (Figure 1D). This cross-talk can occur in the early phases of anti-viral responses (Andrews et al., 2005; Vogel et al., 2014) and can also contribute to DC editing and/or promote DC maturation (Della Chiesa et al., 2005, 2014b; Ferlazzo and Morandi, 2014), thus possibly amplifying and regulating adaptive responses to Herpesviruses.
It should be noted however, that TLR-mediated sensing of viral PAMPs by NK cells has not been definitively settled yet, similar to the contribution of TLRs on DC and macrophages to the response to NK cells. A more extensive review work and additional original work will be needed to appropriately address this issue.
The conventional view of NK cells as short-lived innate lymphocytes, unable to retain any kind of memory has been considerably challenged in the last years, based on several studies demonstrating that NK cells are capable of adapting to viruses and keep memory of past infections (Sun and Lanier, 2009; Sun et al., 2011, 2014; Della Chiesa et al., 2015, 2016). Interestingly, the first evidence that NK cells can develop memory responses to pathogens was against the Herpesvirus CMV, initially in mice (Hadinoto et al., 2009) and later on in humans (Della Chiesa et al., 2012; Foley et al., 2012b; Muccio et al., 2016).
In CMV-seropositive individuals a striking expansion of NK cells expressing the HLA-E-specific activating receptor CD94/NKG2C was observed 15 years ago (Guma et al., 2004). Further studies on NK cells developing in hematopoietic stem cell transplation (HSCT) recipients showed that indeed CMV is a powerful driver of NK cell differentiation favoring the expansion of KIR+NKG2A–LILRB1+ mature NK cells expressing the marker of terminal differentiation CD57 (Della Chiesa et al., 2012; Foley et al., 2012a, b; Locatelli et al., 2018).
In the HSCT setting the CMV-induced reconfiguration also revealed features typical of adaptive immunity, i.e., virus-induced clonal expansions and long-term persistence that led to the concept of “adaptive” or “memory” NK cells (Sun et al., 2011; Della Chiesa et al., 2016; Rolle and Brodin, 2016). This peculiar CMV-driven NK cell subset is characterized by epigenetic modifications, altered expression of signaling molecules and transcription factors that modulate their phenotype and function (Luetke-Eversloh et al., 2014; Lee et al., 2015; Schlums et al., 2015). The generation of this population likely involves interactions between NKG2C and its ligand HLA-E that usually binds peptides derived from HLA-I leader sequences. However, in CMV-infected cells, HLA-I molecules are downregulated by viral evasion mechanisms, while HLA-E can be stabilized and upregulated by peptides derived from the viral-encoded protein UL40 leader sequence, thus stimulating NKG2C+ NK cells and favoring adaptive NK cells expansion (Guma et al., 2006; Rolle et al., 2014). Interestingly, recent studies demonstrated that NKG2C+ NK cells can distinguish subtle differences between peptides bound to HLA-E molecules, showing stronger responses to a particular peptide derived from rare variants of CMV-encoded UL40, precisely mimicking the peptide derived from HLA-G leader sequence (Hammer et al., 2018a; Rolle et al., 2018). This peptide-specificity and the avidity selection of NK cells during CMV infection recently reported in mice (Adams et al., 2019), further support the concept that CMV recognition by NK cells can elicit responses akin to T cell-adaptive responses.
In addition to NKG2C-HLA-E interactions, CD2-costimulation, and different cytokines such as IL-12, IL-18, and IL-15 are involved in adaptive NK cells generation and proliferation (Hammer et al., 2018a; Rolle et al., 2018).
Upon CMV-induced reconfiguration, NK cells display specialized effector function, showing in particular enhanced ADCC abilities. This increased response to Ab-coated targets has been associated to the downregulated expression of the signaling protein FcεRγ which represents a common feature in CMV-adapted NK cells (Lee et al., 2015; Schlums et al., 2015; Muntasell et al., 2016; Muccio et al., 2018). Although the generation of this subset seems to be promoted exclusively by CMV, its increased ability to eliminate Ab-coated infected cells through enhanced ADCC could keep under control infections and reactivations caused by other viruses, as suggested by studies reporting efficient ADCC-mediated killing of opsonized EBV- and HSV-infected targets by adaptive NKG2C+ NK cells (Costa-Garcia et al., 2015; Moraru et al., 2015).
Interestingly, adaptive NKG2C+ NK cells are also capable of presenting CMV antigens through HLA-DR to autologous memory CD4 T cells (Costa-Garcia et al., 2019), regulating T-cell mediated adaptive responses to CMV and possibly contributing to control viral reactivations.
Besides the central role played by NKG2C, aKIRs are also involved in CMV recognition and generation of adaptive responses (Beziat et al., 2013; Della Chiesa et al., 2015). Indeed, CMV infection can promote the expansion of mature NK cells expressing aKIRs in patients receiving Umbilical Cord Blood transplants from NKG2C–/– donors, thus lacking NKG2C expression (Della Chiesa et al., 2014a). The involvement of aKIRs is in line with observations in mice where NK cells expressing the activating receptor Ly49H, homolog of aKIR, expand in response to MCMV infection and confer long-term protection to secondary challenges through the recognition of the viral-encoded ligand m157 (Arase et al., 2002; Hadinoto et al., 2009). Moreover, in humans, a reduced risk of CMV reactivation was associated to the presence of aKIRs in both hematological and solid organ transplant patients supporting their role in anti-viral defense (Stern et al., 2008; Zaia et al., 2009; Mancusi et al., 2015). The exact mechanisms underlying the recognition of infected cells by aKIRs has not been precisely elucidated, however a role for KIR2DS1 in the recognition of its ligand HLA-C2, modified by CMV in infected fibroblasts, has been recently reported (van der Ploeg et al., 2017). Interestingly, KIR2DS1 tetramers were also described to efficiently interact with EBV-infected B cells expressing HLA-C2 (Stewart et al., 2005; Figure 1C).
Notably, in individuals lacking both NKG2C and aKIRs, CMV infection can still favor NK cell reconfiguration indicating that additional unknown mechanisms are responsible for CMV recognition and adaptive NK cell differentiation (Muntasell et al., 2016).
While in mice it has been reported that NK cells can maintain memory of prior encounters with HSV-2 and protect from reactivations (Abdul-Careem et al., 2012), in humans few reports suggest that Herpesviruses other than CMV can induce the generation of specific NK cell subsets with memory properties. Upon EBV infection an expansion of CD56brightNKG2A+CD62L– NK cells was observed in tonsils (Lunemann et al., 2013), whereas CD56dimNKG2A+KIR– NK cells accumulated in peripheral blood during infectious mononucleosis and were involved in lytic EBV-infected B cells elimination (Azzi et al., 2014). However, at variance with CMV-induced expansions, EBV-induced NK cells were not bearing a specific activating receptor and evidences for their epigenetic reprograming has not been provided (Chijioke et al., 2016).
Further studies are necessary to investigate the impact of NK-Herpesvirus interactions in inducing adaptive NK cell subsets outside the CMV context. The possibility to generate virus-specific NK cell populations could help in designing novel vaccine protocols against Herpesviruses, considering that only anti-VZV vaccines have been successfully developed (Arnold and Messaoudi, 2017). However, in the generation of novel vaccines, it should be considered that prolonged exposure to both VZV and HSV-1 can directly impair NK-cell effector function, through still unknown mechanisms, as recently described (Campbell et al., 2019).
As mentioned above, major defects in NK cell function in respect to human Herpesviruses have been described and become overwhelmingly manifest during primary infections that may be lethal upon first host-virus encounter [e.g., SAP defects, NK cell deficiencies (Sayos et al., 1998; Orange, 2002; Etzioni et al., 2005; Notarangelo and Mazzolari, 2006; Mace and Orange, 2019)]. These cases represent a very limited part of Herpesvirus-induced clinical syndromes, since most primary infections are controlled by the immune system often as asymptomatic infections and latency ensues in the vast majority of patients without further clinical reactivations in >70% of infected subjects in the absence of secondary immunodeficiencies (e.g., HIV infection, transplantation, immunosuppression) (Clark and Griffiths, 2003; Ljungman et al., 2011; Locatelli et al., 2016). For this reason, most NK cell evasion mechanisms are less relevant during this acute phase of primary infection. Herpesvirus latency (e.g., HSV/VZV in neuronal ganglia, EBV in B cells and epithelial cells or CMV in organ and BM macrophages) has been long considered a period of antigenic eclipse to the immune system, while reactivation with clinical symptoms (e.g., recurrent HSV, Zoster or shingles, transformation by EBV or KSHV) represent a possible failure of the immune system to control viral latency. Most virus-induced strategies to evade NK cell (and/or T cell) control may be active during these “clinical escape” or reactivation phases. This perspective, however, needs to be carefully reevaluated in view of the overwhelming evidence showing that exit from latency or virus reactivation routinely occurs for all Herpesviruses in infected hosts at subclinical levels (Ling et al., 2003; Hadinoto et al., 2009; Schiffer et al., 2009; Tronstein et al., 2011). Thus, clinically latent Herpesvirus infection actually has a continuous component of persistent immune stimulation due to virus replication in part of the infected cells pool. In this context, virus evasion mechanisms are likely to occur continuously, and are quantitatively more frequent and relevant than during primary infection. Indeed, the magnitude of the specific T cell response during CMV clinical latency is surprisingly high, with 10–20% of CD4 and CD8 CMV-specific circulating T cells, and 5–15% of NKG2C+ memory-like NK cells during clinical latency (Guma et al., 2004; Sylwester et al., 2005). For example, during latent EBV infection 5–10% of peripheral CD8 T cells are specific for latent or lytic epitopes (Tan et al., 1999; Hislop et al., 2002) and 20% of tonsil lymphocytes are EBV-specific (Hislop et al., 2005).
In view of these considerations, and of the participation of persistent Herpesvirus infection to the modulation of autoimmune, allergic, atopic and atherosclerotic events, Herpesviruses and the host may be regarded from an evolutionary-ecologic perspective as co-evolved symbionts with an evolutionary relationship (Virgin et al., 2009; Roossinck, 2011). It will be critical for future scientific focus to more precisely dissect which NK cell evasion mechanisms are functional to maintain this symbiontic equilibrium, from those that actually determine more severe, clinically relevant reactivations, particularly in immunosuppressed patients or in those with virus-induced tumor (e.g., NHL and KS).
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by the following grants: AIRC 5 × 1000, 2018 Project Code 21147 (LoM and SS); AIRC IG 2017, Project Code 19920 (LoM); and AIRC IG 2017 Project Code 20312 (SS). LeM was a recipient of a fellowship awarded by the Federazione Italiana Ricerca sul Cancro (FIRC).
Abdul-Careem, M. F., Lee, A. J., Pek, E. A., Gill, N., Gillgrass, A. E., Chew, M. V., et al. (2012). Genital HSV-2 infection induces short-term NK cell memory. PLoS One 7:e32821. doi: 10.1371/journal.pone.0032821
Adams, N. M., Geary, C. D., Santosa, E. K., Lumaquin, D., Le Luduec, J. B., Sottile, R., et al. (2019). Cytomegalovirus infection drives avidity selection of natural killer cells. Immunity 50, 1381–1390.e5. doi: 10.1016/j.immuni.2019.04.009
Adib-Conquy, M., Scott-Algara, D., Cavaillon, J. M., and Souza-Fonseca-Guimaraes, F. (2014). TLR-mediated activation of NK cells and their role in bacterial/viral immune responses in mammals. Immunol. Cell Biol. 92, 256–262. doi: 10.1038/icb.2013.99
Andrews, D. M., Andoniou, C. E., Scalzo, A. A., van Dommelen, S. L., Wallace, M. E., Smyth, M. J., et al. (2005). Cross-talk between dendritic cells and natural killer cells in viral infection. Mol. Immunol. 42, 547–555. doi: 10.1016/j.molimm.2004.07.040
Arase, H., Mocarski, E. S., Campbell, A. E., Hill, A. B., and Lanier, L. L. (2002). Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science 296, 1323–1326. doi: 10.1126/science.1070884
Arnold, N., and Messaoudi, I. (2017). Herpes zoster and the search for an effective vaccine. Clin. Exp. Immunol. 187, 82–92. doi: 10.1111/cei.12809
Arnon, T. I., Achdout, H., Levi, O., Markel, G., Saleh, N., Katz, G., et al. (2005). Inhibition of the NKp30 activating receptor by pp65 of human cytomegalovirus. Nat. Immunol. 6, 515–523. doi: 10.1038/ni1190
Atalay, R., Zimmermann, A., Wagner, M., Borst, E., Benz, C., Messerle, M., et al. (2002). Identification and expression of human cytomegalovirus transcription units coding for two distinct Fcgamma receptor homologs. J. Virol. 76, 8596–8608. doi: 10.1128/jvi.76.17.8596-8608.2002
Azzi, T., Lunemann, A., Murer, A., Ueda, S., Beziat, V., Malmberg, K. J., et al. (2014). Role for early-differentiated natural killer cells in infectious mononucleosis. Blood 124, 2533–2543. doi: 10.1182/blood-2014-01-553024
Beziat, V., Liu, L. L., Malmberg, J. A., Ivarsson, M. A., Sohlberg, E., Bjorklund, A. T., et al. (2013). NK cell responses to cytomegalovirus infection lead to stable imprints in the human KIR repertoire and involve activating KIRs. Blood 121, 2678–2688. doi: 10.1182/blood-2012-10-459545
Bottino, C., Augugliaro, R., Castriconi, R., Nanni, M., Biassoni, R., Moretta, L., et al. (2000). Analysis of the molecular mechanism involved in 2B4-mediated NK cell activation: evidence that human 2B4 is physically and functionally associated with the linker for activation of T cells. Eur. J. Immunol. 30, 3718–3722. doi: 10.1002/1521-4141(200012)30:12<3718::aid-immu3718>3.3.co;2-9
Bottino, C., Castriconi, R., Pende, D., Rivera, P., Nanni, M., Carnemolla, B., et al. (2003). Identification of PVR (CD155) and Nectin-2 (CD112) as cell surface ligands for the human DNAM-1 (CD226) activating molecule. J. Exp. Med. 198, 557–567. doi: 10.1084/jem.20030788
Bottino, C., Sivori, S., Vitale, M., Cantoni, C., Falco, M., Pende, D., et al. (1996). A novel surface molecule homologous to the p58/p50 family of receptors is selectively expressed on a subset of human natural killer cells and induces both triggering of cell functions and proliferation. Eur. J. Immunol. 26, 1816–1824. doi: 10.1002/eji.1830260823
Brandt, C. S., Baratin, M., Yi, E. C., Kennedy, J., Gao, Z., Fox, B., et al. (2009). The B7 family member B7-H6 is a tumor cell ligand for the activating natural killer cell receptor NKp30 in humans. J. Exp. Med. 206, 1495–1503. doi: 10.1084/jem.20090681
Braud, V. M., Allan, D. S., O’Callaghan, C. A., Soderstrom, K., D’Andrea, A., Ogg, G. S., et al. (1998). HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 391, 795–799. doi: 10.1038/35869
Caligiuri, M. A. (2008). Human natural killer cells. Blood 112, 461–469. doi: 10.1182/blood-2007-09-077438
Campbell, T. M., McSharry, B. P., Steain, M., Russell, T. A., Tscharke, D. C., Kennedy, J. J., et al. (2019). Functional paralysis of human natural killer cells by alphaherpesviruses. PLoS Pathog. 15:e1007784. doi: 10.1371/journal.ppat.1007784
Campbell, T. M., McSharry, B. P., Steain, M., Slobedman, B., and Abendroth, A. (2015). Varicella-zoster virus and herpes simplex virus 1 differentially modulate NKG2D Ligand expression during productive infection. J. Virol. 89, 7932–7943. doi: 10.1128/JVI.00292-15
Carvalho, A., Cunha, C., Carotti, A., Aloisi, T., Guarrera, O., Di Ianni, M., et al. (2009). Polymorphisms in Toll-like receptor genes and susceptibility to infections in allogeneic stem cell transplantation. Exp. Hematol. 37, 1022–1029. doi: 10.1016/j.exphem.2009.06.004
Castriconi, R., Carrega, P., Dondero, A., Bellora, F., Casu, B., Regis, S., et al. (2018). Molecular mechanisms directing migration and retention of natural killer cells in human tissues. Front. Immunol. 9:2324. doi: 10.3389/fimmu.2018.02324
Charpak-Amikam, Y., Kubsch, T., Seidel, E., Oiknine-Djian, E., Cavaletto, N., Yamin, R., et al. (2017). Human cytomegalovirus escapes immune recognition by NK cells through the downregulation of B7-H6 by the viral genes US18 and US20. Sci. Rep. 7:8661. doi: 10.1038/s41598-017-08866-2
Chijioke, O., Landtwing, V., and Munz, C. (2016). NK Cell Influence on the outcome of primary epstein-barr virus infection. Front. Immunol. 7:323. doi: 10.3389/fimmu.2016.00323
Chijioke, O., Muller, A., Feederle, R., Barros, M. H., Krieg, C., Emmel, V., et al. (2013). Human natural killer cells prevent infectious mononucleosis features by targeting lytic Epstein-Barr virus infection. Cell Rep. 5, 1489–1498. doi: 10.1016/j.celrep.2013.11.041
Chisholm, S. E., Howard, K., Gomez, M. V., and Reyburn, H. T. (2007). Expression of ICP0 is sufficient to trigger natural killer cell recognition of herpes simplex virus-infected cells by natural cytotoxicity receptors. J. Infect. Dis. 195, 1160–1168. doi: 10.1086/512862
Clark, D. A., and Griffiths, P. D. (2003). Human herpesvirus 6: relevance of infection in the immunocompromised host. Br. J. Haematol. 120, 384–395. doi: 10.1046/j.1365-2141.2003.04048.x
Colonna, M., Navarro, F., Bellon, T., Llano, M., Garcia, P., Samaridis, J., et al. (1997). A common inhibitory receptor for major histocompatibility complex class I molecules on human lymphoid and myelomonocytic cells. J. Exp. Med. 186, 1809–1818. doi: 10.1084/jem.186.11.1809
Cooper, M. A., Fehniger, T. A., and Caligiuri, M. A. (2001). The biology of human natural killer-cell subsets. Trends Immunol. 22, 633–640. doi: 10.1016/s1471-4906(01)02060-9
Corrales-Aguilar, E., Hoffmann, K., and Hengel, H. (2014). CMV-encoded Fcgamma receptors: modulators at the interface of innate and adaptive immunity. Semin. Immunopathol. 36, 627–640. doi: 10.1007/s00281-014-0448-2
Costa, P., Sivori, S., Bozzano, F., Martini, I., Moretta, A., Moretta, L., et al. (2009). IFN-alpha-mediated increase in cytolytic activity of maturing NK cell upon exposure to HSV-infected myelomonocytes. Eur. J. Immunol. 39, 147–158. doi: 10.1002/eji.200838532
Costa-Garcia, M., Ataya, M., Moraru, M., Vilches, C., Lopez-Botet, M., and Muntasell, A. (2019). Human cytomegalovirus antigen presentation by HLA-DR+ NKG2C+ adaptive NK cells specifically activates polyfunctional effector memory CD4+ T lymphocytes. Front. Immunol. 10:687. doi: 10.3389/fimmu.2019.00687
Costa-Garcia, M., Vera, A., Moraru, M., Vilches, C., Lopez-Botet, M., and Muntasell, A. (2015). Antibody-mediated response of NKG2Cbright NK cells against human cytomegalovirus. J. Immunol. 194, 2715–2724. doi: 10.4049/jimmunol.1402281
Dai, H. S., and Caligiuri, M. A. (2018). Molecular basis for the recognition of herpes simplex virus type 1 infection by human natural killer cells. Front. Immunol. 9:183. doi: 10.3389/fimmu.2018.00183
Dai, H. S., Griffin, N., Bolyard, C., Mao, H. C., Zhang, J., Cripe, T. P., et al. (2017). The FC domain of immunoglobulin is sufficient to bridge NK cells with virally infected cells. Immunity 47, 159–170.e10. doi: 10.1016/j.immuni.2017.06.019
De Maria, A., Bozzano, F., Cantoni, C., and Moretta, L. (2011). Revisiting human natural killer cell subset function revealed cytolytic CD56(dim)CD16+ NK cells as rapid producers of abundant IFN-gamma on activation. Proc. Natl. Acad. Sci. U.S.A. 108, 728–732. doi: 10.1073/pnas.1012356108
De Pelsmaeker, S., Romero, N., Vitale, M., and Favoreel, H. W. (2018). Herpesvirus Evasion of natural killer cells. J. Virol. 92, e2105–e2117. doi: 10.1128/JVI.02105-17
de Vries, E., Koene, H. R., Vossen, J. M., Gratama, J. W., von dem Borne, A. E., Waaijer, J. L., et al. (1996). Identification of an unusual FC gamma receptor IIIa (CD16) on natural killer cells in a patient with recurrent infections. Blood 88, 3022–3027.
Della Chiesa, M., Falco, M., Bertaina, A., Muccio, L., Alicata, C., Frassoni, F., et al. (2014a). Human cytomegalovirus infection promotes rapid maturation of NK cells expressing activating killer IG-like receptor in patients transplanted with NKG2C-/- umbilical cord blood. J. Immunol. 192, 1471–1479. doi: 10.4049/jimmunol.1302053
Della Chiesa, M., Marcenaro, E., Sivori, S., Carlomagno, S., Pesce, S., and Moretta, A. (2014b). Human NK cell response to pathogens. Semin. Immunol. 26, 152–160. doi: 10.1016/j.smim.2014.02.001
Della Chiesa, M., Falco, M., Podesta, M., Locatelli, F., Moretta, L., Frassoni, F., et al. (2012). Phenotypic and functional heterogeneity of human NK cells developing after umbilical cord blood transplantation: a role for human cytomegalovirus? Blood 119, 399–410. doi: 10.1182/blood-2011-08-372003
Della Chiesa, M., Moretta, L., Muccio, L., Bertaina, A., Moretta, F., Locatelli, F., et al. (2016). Haploidentical haematopoietic stem cell transplantation: role of NK cells and effect of cytomegalovirus infections. Curr. Top. Microbiol. Immunol. 395, 209–224. doi: 10.1007/82_2015_450
Della Chiesa, M., Sivori, S., Carlomagno, S., Moretta, L., and Moretta, A. (2015). Activating KIRs and NKG2C in viral infections: toward NK cell memory? Front. Immunol. 6:573. doi: 10.3389/fimmu.2015.00573
Della Chiesa, M., Sivori, S., Castriconi, R., Marcenaro, E., and Moretta, A. (2005). Pathogen-induced private conversations between natural killer and dendritic cells. Trends Microbiol. 13, 128–136. doi: 10.1016/j.tim.2005.01.006
Etzioni, A., Eidenschenk, C., Katz, R., Beck, R., Casanova, J. L., and Pollack, S. (2005). Fatal varicella associated with selective natural killer cell deficiency. J. Pediatr. 146, 423–425. doi: 10.1016/j.jpeds.2004.11.022
Ferlazzo, G., and Morandi, B. (2014). Cross-talks between natural killer cells and distinct subsets of dendritic cells. Front. Immunol. 5:159. doi: 10.3389/fimmu.2014.00159
Foley, B., Cooley, S., Verneris, M. R., Curtsinger, J., Luo, X., Waller, E. K., et al. (2012a). Human cytomegalovirus (CMV)-induced memory-like NKG2C(+) NK cells are transplantable and expand in vivo in response to recipient CMV antigen. J. Immunol. 189, 5082–5088. doi: 10.4049/jimmunol.1201964
Foley, B., Cooley, S., Verneris, M. R., Pitt, M., Curtsinger, J., Luo, X., et al. (2012b). Cytomegalovirus reactivation after allogeneic transplantation promotes a lasting increase in educated NKG2C+ natural killer cells with potent function. Blood 119, 2665–2674. doi: 10.1182/blood-2011-10-386995
Freud, A. G., Mundy-Bosse, B. L., Yu, J., and Caligiuri, M. A. (2017). The broad spectrum of human natural killer cell diversity. Immunity 47, 820–833. doi: 10.1016/j.immuni.2017.10.008
Grauwet, K., Cantoni, C., Parodi, M., De Maria, A., Devriendt, B., Pende, D., et al. (2014). Modulation of CD112 by the alphaherpesvirus gD protein suppresses DNAM-1-dependent NK cell-mediated lysis of infected cells. Proc. Natl. Acad. Sci. U.S.A. 111, 16118–16123. doi: 10.1073/pnas.1409485111
Grier, J. T., Forbes, L. R., Monaco-Shawver, L., Oshinsky, J., Atkinson, T. P., Moody, C., et al. (2012). Human immunodeficiency-causing mutation defines CD16 in spontaneous NK cell cytotoxicity. J. Clin. Invest. 122, 3769–3780. doi: 10.1172/JCI64837
Guma, M., Angulo, A., Vilches, C., Gomez-Lozano, N., Malats, N., and Lopez-Botet, M. (2004). Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood 104, 3664–3671. doi: 10.1182/blood-2004-05-2058
Guma, M., Budt, M., Saez, A., Brckalo, T., Hengel, H., Angulo, A., et al. (2006). Expansion of CD94/NKG2C+ NK cells in response to human cytomegalovirus-infected fibroblasts. Blood 107, 3624–3631. doi: 10.1182/blood-2005-09-3682
Hadinoto, V., Shapiro, M., Sun, C. C., and Thorley-Lawson, D. A. (2009). The dynamics of EBV shedding implicate a central role for epithelial cells in amplifying viral output. PLoS Pathog. 5:e1000496. doi: 10.1371/journal.ppat.1000496
Hammer, Q., Ruckert, T., Borst, E. M., Dunst, J., Haubner, A., Durek, P., et al. (2018a). Peptide-specific recognition of human cytomegalovirus strains controls adaptive natural killer cells. Nat. Immunol. 19, 453–463. doi: 10.1038/s41590-018-0082-6
Hammer, Q., Ruckert, T., and Romagnani, C. (2018b). Natural killer cell specificity for viral infections. Nat. Immunol. 19, 800–808. doi: 10.1038/s41590-018-0163-6
Hill, A., Jugovic, P., York, I., Russ, G., Bennink, J., Yewdell, J., et al. (1995). Herpes simplex virus turns off the TAP to evade host immunity. Nature 375, 411–415. doi: 10.1038/375411a0
Hislop, A. D., Annels, N. E., Gudgeon, N. H., Leese, A. M., and Rickinson, A. B. (2002). Epitope-specific evolution of human CD8(+) T cell responses from primary to persistent phases of epstein-barr virus infection. J. Exp. Med. 195, 893–905. doi: 10.1084/jem.20011692
Hislop, A. D., Kuo, M., Drake-Lee, A. B., Akbar, A. N., Bergler, W., Hammerschmitt, N., et al. (2005). tonsillar homing of epstein-barr virus-specific CD8+ T cells and the virus-host balance. J. Clin. Invest. 115, 2546–2555. doi: 10.1172/JCI24810
Ho, J. W., Hershkovitz, O., Peiris, M., Zilka, A., Bar-Ilan, A., Nal, B., et al. (2008). H5-type influenza virus hemagglutinin is functionally recognized by the natural killer-activating receptor NKp44. J. Virol. 82, 2028–2032. doi: 10.1128/JVI.02065
Huard, B., and Fruh, K. (2000). A role for MHC class I down-regulation in NK cell lysis of herpes virus-infected cells. Eur. J. Immunol. 30, 509–515. doi: 10.1002/1521-4141(200002)30:2<509::aid-immu509>3.3.co;2-8
Imai, T., Koyanagi, N., Ogawa, R., Shindo, K., Suenaga, T., Sato, A., et al. (2013). Us3 kinase encoded by herpes simplex virus 1 mediates downregulation of cell surface major histocompatibility complex class I and evasion of CD8+ T cells. PLoS One 8:e72050. doi: 10.1371/journal.pone.0072050
Johnson, D. C., Frame, M. C., Ligas, M. W., Cross, A. M., and Stow, N. D. (1988). Herpes simplex virus immunoglobulin G FC receptor activity depends on a complex of two viral glycoproteins, GE and GI. J. Virol. 62, 1347–1354.
Kim, M., Osborne, N. R., Zeng, W., Donaghy, H., McKinnon, K., Jackson, D. C., et al. (2012). Herpes simplex virus antigens directly activate NK cells via TLR2, thus facilitating their presentation to CD4 T lymphocytes. J. Immunol. 188, 4158–4170. doi: 10.4049/jimmunol.1103450
Lanier, L. L. (2015). NKG2D receptor and its ligands in host defense. Cancer Immunol. Res. 3, 575–582. doi: 10.1158/2326-6066.CIR-15-0098
Lee, J., Zhang, T., Hwang, I., Kim, A., Nitschke, L., Kim, M., et al. (2015). Epigenetic modification and antibody-dependent expansion of memory-like NK cells in human cytomegalovirus-infected individuals. Immunity 42, 431–442. doi: 10.1016/j.immuni.2015.02.013
Ling, P. D., Lednicky, J. A., Keitel, W. A., Poston, D. G., White, Z. S., Peng, R., et al. (2003). The dynamics of herpesvirus and polyomavirus reactivation and shedding in healthy adults: a 14-month longitudinal study. J. Infect. Dis. 187, 1571–1580. doi: 10.1086/374739
Ljungman, P., Hakki, M., and Boeckh, M. (2011). Cytomegalovirus in hematopoietic stem cell transplant recipients. Hematol. Oncol. Clin. North Am. 25, 151–169. doi: 10.1016/j.hoc.2010.11.011
Locatelli, F., Bertaina, A., Bertaina, V., and Merli, P. (2016). Cytomegalovirus in hematopoietic stem cell transplant recipients - management of infection. Expert Rev. Hematol. 9, 1093–1105. doi: 10.1080/17474086.2016.1242406
Locatelli, F., Pende, D., Falco, M., Della Chiesa, M., Moretta, A., and Moretta, L. (2018). NK cells mediate a crucial graft-versus-leukemia effect in haploidentical-HSCT to cure high-risk acute leukemia. Trends Immunol. 39, 577–590. doi: 10.1016/j.it.2018.04.009
Luetke-Eversloh, M., Hammer, Q., Durek, P., Nordstrom, K., Gasparoni, G., Pink, M., et al. (2014). Human cytomegalovirus drives epigenetic imprinting of the IFNG locus in NKG2Chi natural killer cells. PLoS Pathog. 10:e1004441. doi: 10.1371/journal.ppat.1004441
Lunemann, A., Vanoaica, L. D., Azzi, T., Nadal, D., and Munz, C. (2013). A distinct subpopulation of human NK cells restricts B cell transformation by EBV. J. Immunol. 191, 4989–4995. doi: 10.4049/jimmunol.1301046
Mace, E. M., and Orange, J. S. (2019). Emerging insights into human health and NK cell biology from the study of NK cell deficiencies. Immunol. Rev. 287, 202–225. doi: 10.1111/imr.12725
Madrid, A. S., and Ganem, D. (2012). Kaposi’s sarcoma-associated herpesvirus ORF54/dUTPase downregulates a ligand for the NK activating receptor NKp44. J. Virol. 86, 8693–8704. doi: 10.1128/JVI.00252-12
Mancusi, A., Ruggeri, L., Urbani, E., Pierini, A., Massei, M. S., Carotti, A., et al. (2015). Haploidentical hematopoietic transplantation from KIR ligand-mismatched donors with activating KIRs reduces nonrelapse mortality. Blood 125, 3173–3182. doi: 10.1182/blood-2014-09-599993
Mandelboim, O., Lieberman, N., Lev, M., Paul, L., Arnon, T. I., Bushkin, Y., et al. (2001). Recognition of haemagglutinins on virus-infected cells by NKp46 activates lysis by human NK cells. Nature 409, 1055–1060. doi: 10.1038/35059110
Moraru, M., Black, L. E., Muntasell, A., Portero, F., Lopez-Botet, M., Reyburn, H. T., et al. (2015). NK cell and IG interplay in defense against herpes simplex virus type 1: epistatic interaction of CD16A and IgG1 allotypes of variable affinities modulates antibody-dependent cellular cytotoxicity and susceptibility to clinical reactivation. J. Immunol. 195, 1676–1684. doi: 10.4049/jimmunol.1500872
Moraru, M., Cisneros, E., Gomez-Lozano, N., de Pablo, R., Portero, F., Canizares, M., et al. (2012). Host genetic factors in susceptibility to herpes simplex type 1 virus infection: contribution of polymorphic genes at the interface of innate and adaptive immunity. J. Immunol. 188, 4412–4420. doi: 10.4049/jimmunol.1103434
Moretta, L. (2010). Dissecting CD56dim human NK cells. Blood 116, 3689–3691. doi: 10.1182/blood-2010-09-303057
Moretta, L., and Moretta, A. (2004). Unravelling natural killer cell function: triggering and inhibitory human NK receptors. EMBO J. 23, 255–259. doi: 10.1038/sj.emboj.7600019
Muccio, L., Bertaina, A., Falco, M., Pende, D., Meazza, R., Lopez-Botet, M., et al. (2016). Analysis of memory-like natural killer cells in human cytomegalovirus-infected children undergoing alphabeta+T and B cell-depleted hematopoietic stem cell transplantation for hematological malignancies. Haematologica 101, 371–381. doi: 10.3324/haematol.2015.134155
Muccio, L., Falco, M., Bertaina, A., Locatelli, F., Frassoni, F., Sivori, S., et al. (2018). Late development of fcepsilonrgamma(neg) adaptive natural killer cells upon human cytomegalovirus reactivation in umbilical cord blood transplantation recipients. Front. Immunol. 9:1050. doi: 10.3389/fimmu.2018.01050
Muntasell, A., Costa-Garcia, M., Vera, A., Marina-Garcia, N., and Kirschning, C. J. (2013). Lopez-botet m. priming of NK cell anti-viral effector mechanisms by direct recognition of human cytomegalovirus. Front. Immunol. 4:40. doi: 10.3389/fimmu.2013.00040
Muntasell, A., Pupuleku, A., Cisneros, E., Vera, A., Moraru, M., Vilches, C., et al. (2016). Relationship of NKG2C copy number with the distribution of distinct cytomegalovirus-induced adaptive NK cell subsets. J. Immunol. 196, 3818–3827. doi: 10.4049/jimmunol.1502438
Nachmani, D., Stern-Ginossar, N., Sarid, R., and Mandelboim, O. (2009). Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe 5, 376–385. doi: 10.1016/j.chom.2009.03.003
Nakajima, H., Cella, M., Langen, H., Friedlein, A., and Colonna, M. (1999). Activating interactions in human NK cell recognition: the role of 2B4-CD48. Eur. J. Immunol. 29, 1676–1683. doi: 10.1002/(sici)1521-4141(199905)29:05<1676::aid-immu1676>3.0.co;2-y
Notarangelo, L. D., and Mazzolari, E. (2006). Natural killer cell deficiencies and severe varicella infection. J. Pediatr. 148, 563–564. doi: 10.1016/j.jpeds.2005.06.028
Orange, J. S. (2002). Human natural killer cell deficiencies and susceptibility to infection. Microbes Infect. 4, 1545–1558. doi: 10.1016/s1286-4579(02)00038-2
Parham, P. (2005). MHC class I molecules and KIRs in human history, health and survival. Nat. Rev. Immunol. 5, 201–214. doi: 10.1038/nri1570
Parolini, S., Bottino, C., Falco, M., Augugliaro, R., Giliani, S., Franceschini, R., et al. (2000). X-linked lymphoproliferative disease. 2B4 molecules displaying inhibitory rather than activating function are responsible for the inability of natural killer cells to kill Epstein-Barr virus-infected cells. J. Exp. Med. 192, 337–346. doi: 10.1084/jem.192.3.337
Prod’homme, V., Sugrue, D. M., Stanton, R. J., Nomoto, A., Davies, J., Rickards, C. R., et al. (2010). Human cytomegalovirus UL141 promotes efficient downregulation of the natural killer cell activating ligand CD112. J. Gen. Virol. 91(Pt 8), 2034–2039. doi: 10.1099/vir.0.021931-0
Rolle, A., and Brodin, P. (2016). Immune adaptation to environmental influence: the case of NK cells and HCMV. Trends Immunol. 37, 233–243. doi: 10.1016/j.it.2016.01.005
Rolle, A., Meyer, M., Calderazzo, S., Jager, D., and Momburg, F. (2018). Distinct HLA-E peptide complexes modify antibody-driven effector functions of adaptive NK cells. Cell Rep. 24, 1967–1976.e4. doi: 10.1016/j.celrep.2018.07.069
Rolle, A., Pollmann, J., Ewen, E. M., Le, V. T., Halenius, A., Hengel, H., et al. (2014). IL-12-producing monocytes and HLA-E control HCMV-driven NKG2C+ NK cell expansion. J. Clin. Invest. 124, 5305–5316. doi: 10.1172/JCI77440
Roossinck, M. J. (2011). The good viruses: viral mutualistic symbioses. Nat. Rev. Microbiol. 9, 99–108. doi: 10.1038/nrmicro2491
Sayos, J., Wu, C., Morra, M., Wang, N., Zhang, X., Allen, D., et al. (1998). The X-linked lymphoproliferative-disease gene product SAP regulates signals induced through the co-receptor SLAM. Nature 395, 462–469. doi: 10.1038/26683
Schiffer, J. T., Abu-Raddad, L., Mark, K. E., Zhu, J., Selke, S., Magaret, A., et al. (2009). Frequent release of low amounts of herpes simplex virus from neurons: results of a mathematical model. Sci. Transl. Med. 1:7ra16. doi: 10.1126/scitranslmed.3000193
Schlums, H., Cichocki, F., Tesi, B., Theorell, J., Beziat, V., Holmes, T. D., et al. (2015). Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function. Immunity 42, 443–456. doi: 10.1016/j.immuni.2015.02.008
Schmiedel, D., Tai, J., Levi-Schaffer, F., Dovrat, S., and Mandelboim, O. (2016). Human Herpesvirus 6b downregulates expression of activating ligands during lytic infection to escape elimination by natural killer cells. J. Virol. 90, 9608–9617. doi: 10.1128/JVI.01164-16
Schneider, C. L., and Hudson, A. W. (2011). The human herpesvirus-7 (HHV-7) U21 immunoevasin subverts NK-mediated cytoxicity through modulation of MICA and MICB. PLoS Pathog. 7:e1002362. doi: 10.1371/journal.ppat.1002362
Schuren, A. B., Costa, A. I., and Wiertz, E. J. (2016). Recent advances in viral evasion of the MHC class I processing pathway. Curr. Opin. Immunol. 40, 43–50. doi: 10.1016/j.coi.2016.02.007
Sivori, S., Falco, M., Carlomagno, S., Romeo, E., Soldani, C., Bensussan, A., et al. (2010). A novel KIR-associated function: evidence that CpG DNA uptake and shuttling to early endosomes is mediated by KIR3DL2. Blood 116, 1637–1647. doi: 10.1182/blood-2009-12-256586
Sivori, S., Falco, M., Della Chiesa, M., Carlomagno, S., Vitale, M., Moretta, L., et al. (2004). CpG and double-stranded RNA trigger human NK cells by Toll-like receptors: induction of cytokine release and cytotoxicity against tumors and dendritic cells. Proc. Natl. Acad. Sci. U.S.A. 101, 10116–10121. doi: 10.1073/pnas.0403744101
Stern, M., Elsasser, H., Honger, G., Steiger, J., Schaub, S., and Hess, C. (2008). The number of activating KIR genes inversely correlates with the rate of CMV infection/reactivation in kidney transplant recipients. Am. J. Transplant. 8, 1312–1317. doi: 10.1111/j.1600-6143.2008.02242.x
Stewart, C. A., Laugier-Anfossi, F., Vely, F., Saulquin, X., Riedmuller, J., Tisserant, A., et al. (2005). Recognition of peptide-MHC class I complexes by activating killer immunoglobulin-like receptors. Proc. Natl. Acad. Sci. U.S.A. 102, 13224–13229. doi: 10.1073/pnas.0503594102
Sun, J. C., and Lanier, L. L. (2009). Natural killer cells remember: an evolutionary bridge between innate and adaptive immunity? Eur. J. Immunol. 39, 2059–2064. doi: 10.1002/eji.200939435
Sun, J. C., Lopez-Verges, S., Kim, C. C., DeRisi, J. L., and Lanier, L. L. (2011). NK cells and immune memory. J. Immunol. 186, 1891–1897. doi: 10.4049/jimmunol.1003035
Sun, J. C., Ugolini, S., and Vivier, E. (2014). Immunological memory within the innate immune system. EMBO J. 33, 1295–1303. doi: 10.1002/embj.201387651
Sylwester, A. W., Mitchell, B. L., Edgar, J. B., Taormina, C., Pelte, C., Ruchti, F., et al. (2005). Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J. Exp. Med. 202, 673–685. doi: 10.1084/jem.20050882
Tan, L. C., Gudgeon, N., Annels, N. E., Hansasuta, P., O’Callaghan, C. A., Rowland-Jones, S., et al. (1999). A re-evaluation of the frequency of CD8+ T cells specific for EBV in healthy virus carriers. J. Immunol. 162, 1827–1835.
Thomas, M., Boname, J. M., Field, S., Nejentsev, S., Salio, M., Cerundolo, V., et al. (2008). Down-regulation of NKG2D and NKp80 ligands by Kaposi’s sarcoma-associated herpesvirus K5 protects against NK cell cytotoxicity. Proc. Natl. Acad. Sci. U.S.A. 105, 1656–1661. doi: 10.1073/pnas.0707883105
Tomasec, P., Wang, E. C., Davison, A. J., Vojtesek, B., Armstrong, M., Griffin, C., et al. (2005). Downregulation of natural killer cell-activating ligand CD155 by human cytomegalovirus UL141. Nat. Immunol. 6, 181–188. doi: 10.1038/ni1156
Tortorella, D., Gewurz, B. E., Furman, M. H., Schust, D. J., and Ploegh, H. L. (2000). Viral subversion of the immune system. Ann. Rev. Immunol. 18, 861–926. doi: 10.1146/annurev.immunol.18.1.861
Tronstein, E., Johnston, C., Huang, M. L., Selke, S., Magaret, A., Warren, T., et al. (2011). Genital shedding of herpes simplex virus among symptomatic and asymptomatic persons with HSV-2 infection. JAMA 305, 1441–1449. doi: 10.1001/jama.2011.420
van der Ploeg, K., Chang, C., Ivarsson, M. A., Moffett, A., Wills, M. R., and Trowsdale, J. (2017). Modulation of human leukocyte antigen-C by human cytomegalovirus stimulates KIR2DS1 recognition by natural killer cells. Front. Immunol. 8:298. doi: 10.3389/fimmu.2017.00298
Virgin, H. W., Wherry, E. J., and Ahmed, R. (2009). Redefining chronic viral infection. Cell 138, 30–50. doi: 10.1016/j.cell.2009.06.036
Vivier, E., Raulet, D. H., Moretta, A., Caligiuri, M. A., Zitvogel, L., Lanier, L. L., et al. (2011). Innate or adaptive immunity? The example of natural killer cells. Science 331, 44–49. doi: 10.1126/science.1198687
Vogel, K., Thomann, S., Vogel, B., Schuster, P., and Schmidt, B. (2014). Both plasmacytoid dendritic cells and monocytes stimulate natural killer cells early during human herpes simplex virus type 1 infections. Immunology 143, 588–600. doi: 10.1111/imm.12337
Williams, L. R., Quinn, L. L., Rowe, M., and Zuo, J. (2015). Induction of the lytic cycle sensitizes epstein-barr virus-infected B cells to nk cell killing that is counteracted by virus-mediated NK cell evasion mechanisms in the late lytic cycle. J. Virol. 90, 947–958. doi: 10.1128/JVI.01932-15
Wu, J., Chalupny, N. J., Manley, T. J., Riddell, S. R., Cosman, D., and Spies, T. (2003). Intracellular retention of the MHC class I-related chain B ligand of NKG2D by the human cytomegalovirus UL16 glycoprotein. J. Immunol. 170, 4196–4200. doi: 10.4049/jimmunol.170.8.4196
Zaia, J. A., Sun, J. Y., Gallez-Hawkins, G. M., Thao, L., Oki, A., Lacey, S. F., et al. (2009). The effect of single and combined activating killer immunoglobulin-like receptor genotypes on cytomegalovirus infection and immunity after hematopoietic cell transplantation. Biol. Blood Marrow Transplant. 15, 315–325. doi: 10.1016/j.bbmt.2008.11.030
Keywords: NK cells, Herpesvirus, activating receptors, TLRs, memory responses, viral reactivation
Citation: Della Chiesa M, De Maria A, Muccio L, Bozzano F, Sivori S and Moretta L (2019) Human NK Cells and Herpesviruses: Mechanisms of Recognition, Response and Adaptation. Front. Microbiol. 10:2297. doi: 10.3389/fmicb.2019.02297
Received: 06 August 2019; Accepted: 20 September 2019;
Published: 04 October 2019.
Edited by:
Santo Landolfo, University of Turin, ItalyReviewed by:
Sebastian Voigt, Charité – Berlin University of Medicine, GermanyCopyright © 2019 Della Chiesa, De Maria, Muccio, Bozzano, Sivori and Moretta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lorenzo Moretta, bG9yZW56by5tb3JldHRhQG9wYmcubmV0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.