- 1School of Food Science and Engineering, Wuhan Polytechnic University, Wuhan, China
- 2Molecular Characterization of Foodborne Pathogens Research Unit, United States Department of Agriculture, Agricultural Research Service, Eastern Regional Research Center (USDA-ARS-ERRC), USDA–MOST Joint Research Center for Food Safety, Wyndmoor, PA, United States
- 3State Key Laboratory of Microbial Metabolism, School of Agriculture and Biology, MOST-USDA Joint Research Center for Food Safety, Shanghai Jiao Tong University, Shanghai, China
Salmonella is a leading cause of foodborne disease and is often associated with the consumption of foods of animal origin. In this study, sixty-six Salmonella isolates were obtained from 631 raw meat samples purchased at small retail suppliers in Hubei Province, China. The most prevalent Salmonella serotypes were Thompson (18.2%) and Agona (13.6%). Frequent antimicrobial resistance was observed for the sulfonamides (43.9%), tetracycline (43.9%), and the β-lactams amoxicillin and ampicillin (36.4% for each). Interestingly, a high incidence of resistance to cephazolin was observed in strains of the most common serotype, S. Thompson. Class I integrons were found in 27.3% (18/66) of the isolates and five of these integrons contained different gene cassettes (aacA4C-arr-3-dfr2, dfrA12-aadA21, aadA2, dfrA12-aadA2, dfr17-aadA5). Additional antimicrobial resistance genes, including blaTEM–1, blaCTX–M–65, blaCTX–M–15, qnrB, and qnrS, were also identified among these Salmonella isolates. Results of replicon typing and conjugation experiments revealed that an integron with qnrB and blaCTX–M–15 genes was present on incH12 mobile plasmid in S. Thompson strain. Multilocus sequence typing (MLST) analysis revealed 32 sequence types, indicating that these isolates were phenotypically and genetically diverse, among which ST26 (18.2%) and ST541 (12.1%) were the predominant sequence types. The integrons, along with multiple antimicrobial resistance genes on mobile plasmids, are likely contributors to the dissemination of multidrug resistance in Salmonella.
Introduction
Salmonella is an important source of foodborne disease, resulting in morbidity and mortality worldwide (Park et al., 2015). In China, 70–80% of bacterial food poisoning cases are due to Salmonella that originates in poultry, eggs, beef and pork (Yang et al., 2013). Direct and indirect spread of Salmonella between animals and humans are threats to human health. Although more than 2610 serotypes of Salmonella have been identified, the majority of human Salmonella infections are caused by strains of only a few serotypes (Mancin et al., 2015). Therefore, serotype determination is an important aspect of epidemiological surveillance and disease assessment (Hung et al., 2017). Changes in the prevalence of specific serotypes can result from the movements of people, animals, and foods (Hendriksen et al., 2009). At present, serotypes Enteritidis and Typhimurium have been most frequently implicated in salmonellosis outbreaks from foods in Taiwan, Greece, Qatar and South Africa (Chang et al., 2016; Papadopoulos et al., 2016; Hung et al., 2017; Smith et al., 2017).
Though not as facile or rapid as serotyping, multilocus sequence typing (MLST) is a reliable and highly discriminatory molecular typing method. In this technique, the phylogenetic relationship between bacterial strains is determined based upon nucleotide sequence variations within several housekeeping and other conserved genes (Chang et al., 2016). MLST has many advantages compared with other genotyping techniques (Stepan et al., 2011). It is consistent and reproducible, and combines nucleotide sequencing and bioinformatics with a population genetics approach to produce accurate, reliable and highly portable results (Smith et al., 2017). Furthermore, it has been suggested that MLST is superior to serotyping for Salmonella surveillance and outbreak tracking (Achtman et al., 2012).
The widespread use of antibiotics poses problems for antimicrobial resistance, which leads to an increase in treatment costs and even to therapy failure (Hur et al., 2012). Multiple drug resistance (MDR) among Salmonella is prevalent. This applies not only to traditional antimicrobials such as sulfamethoxazole and chloramphenicol, but also against clinically important antimicrobials like the third generation β-lactams and fluoroquinolones (Agada et al., 2014). The selective pressure caused by the application of antimicrobials in poultry production and veterinary practice for growth promotion and prophylaxis has resulted in an increase in antibiotic resistance and an increase in the presence of genes conferring antimicrobial resistance to Salmonella (Zishiri et al., 2016). The spread of antimicrobial resistance in Salmonella is primarily due to integrons (Rajaei et al., 2014). Integrons are mobile DNA elements containing multiple antimicrobial resistant genes, which can move from one bacterium to another. This increases multi-drug resistance in bacterial populations (Asgharpour et al., 2014). Among various integrons, the class I integron is frequently observed among antimicrobial resistant Salmonella and plays a major role in the dissemination of resistance genes (Wright, 2010).
To the best of our knowledge, there is limited information regarding the incidence of Salmonella in retail raw meat products in Central China. Therefore, the objective of this study was to determine the prevalence, serotype distribution, genetic diversity, antimicrobial resistance, and the presence of class I integrons of Salmonella from retail raw meat products in Hubei Province, China. This data can be used for quantitative risk assessments of Salmonella isolates in retail meat and to help establish a series of prevention and control measures against foodborne pathogenic bacteria.
Materials and Methods
Sample Collection and Salmonella Isolation and Serotyping
We collected 631 retail raw meat samples, including 263 chicken meat, 102 duck meat, 80 pork and 186 fish products from 20 small markets, in Wuhan City of Hubei Province, China, from 2014 to 2017. All samples were transported to our laboratory on ice within 6 h of collection. Salmonella isolation and identification methods were carried out according to the standard procedures of National Food Safety Standard Food Microbiological Examination: Salmonella (GB 4789.4-2016). Biochemical identification was done using an HBI microbial biochemical identification kit (Hope Biotechnology, Qingdao, China).
Serotyping of Salmonella isolates was done according to the Kauffmann-White scheme using an O and H antigen slide agglutination kit (Tianrun Biopharmaceutical, Ningbo, China) (Zhao et al., 2017).
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing was performed using the Kirby-Bauer disk diffusion method as described by the Clinical and Laboratory Standards Institute (Clinical and Laboratory Standards Institute [CLSI], 2016). Antimicrobial disks (Oxoid Ltd., Basingstoke, United Kingdom) used for testing contained 10 μg amoxicillin, 10 μg ampicillin, 30 μg cefotaxime, 30 μg cefuroxime sodium, 30 μg cephazolin, 30 μg chloramphenicol, 5 μg ciprofloxacin, 30 μg nalidixic acid, 10 μg norfloxacin, 10 μg streptomycin, 10 μg gentamicin, 300 μg nitrofurantoin, 300 μg sulfonamide and 30 μg tetracycline. Escherichia coli strain ATCC 25922 was used as a reference strain. The standard break points were interpreted according to the guidelines of the Clinical and Laboratory Standards Institute (Clinical and Laboratory Standards Institute [CLSI], 2016).
Antimicrobial Resistance Genes and Class I Integron Identification
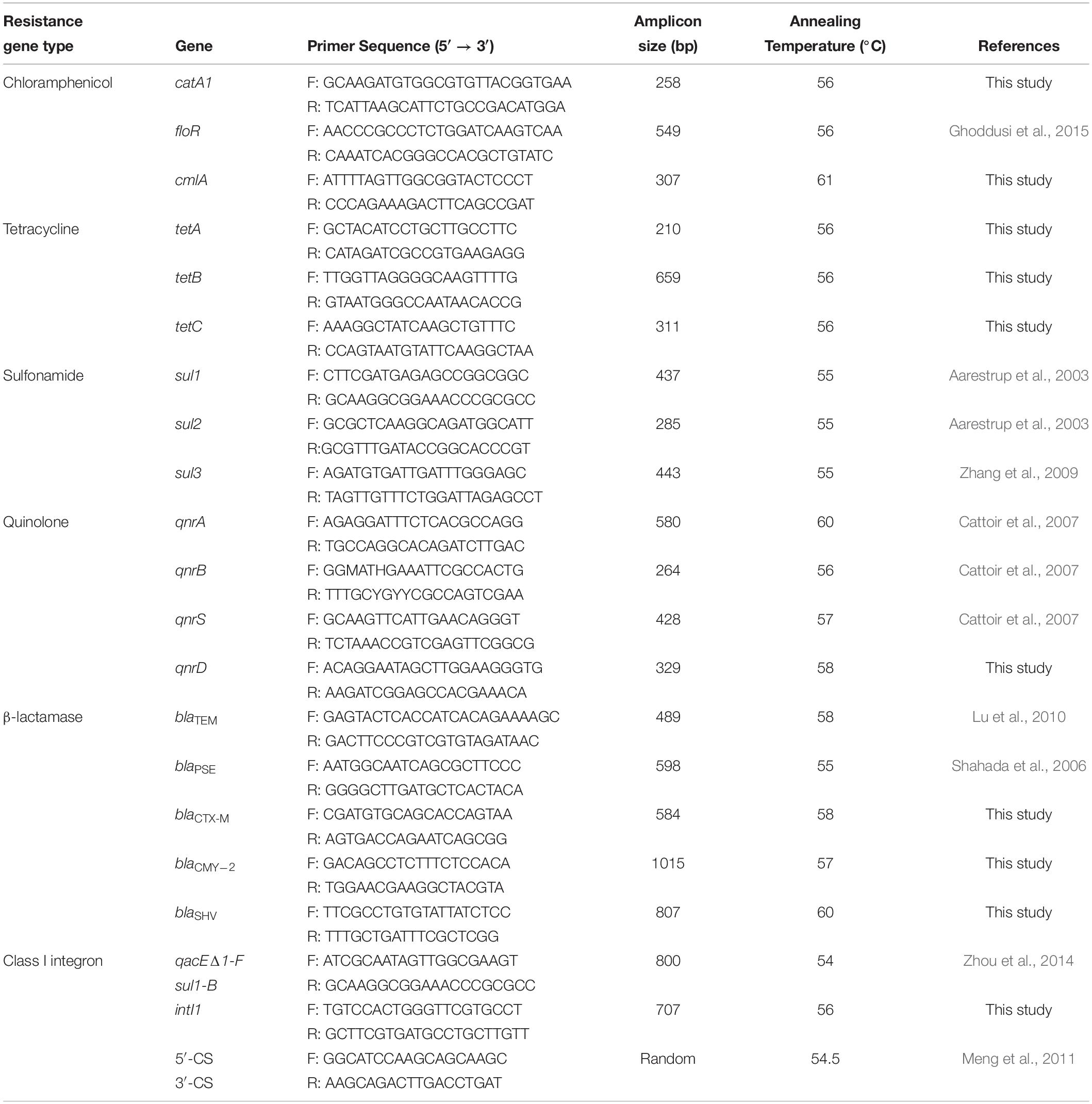
Bacterial DNA was extracted from Salmonella isolates cultured overnight in Luria–Bertani broth using the MiniBEST Bacteria Genomic DNA Extraction Kit Ver.3.0 (Takara, Beijing, China), according to the manufacturer’s instructions. Eighteen genes known to be associated with resistance to the antibiotics as well as Class I integrons were tested by PCR using the oligonucleotide primers listed in Table 1. The PCRs were carried out using a MyCyler Thermocycler (BioRad, Hercules, CA, United States) in a 25 μL mixture that contained 0.5 mM of each primer, 1 × PCR buffer (Takara), 250 μM of dNTP, 0.5 U of Taq DNA polymerase (Takara), 1.5 mM MgCl2, and 0.5 μL of template DNA. The cycling conditions were 94°C for 5 min, followed by 30 cycles of 94°C for 30 s, 54–61°C for 30 s and 72°C for 1 min, and a final extension of 72°C for 10 min. PCR amplification products were separated on agarose gels and the DNA was purified from agarose plugs using the Takara Agarose Gel DNA Purification Kit (Takara). BGI Tech Solutions (Wuhan, China) performed DNA sequencing, and this data was aligned and analyzed using the Basic Local Alignment Search Tool (BLAST).
Conjugation Experiments
To determine whether resistance determinants were present in transferable genetic elements in Salmonella isolates, conjugation experiments were carried out using the broth mating method as previously described (Olsen et al., 2004). The Salmonella isolates positive for blaCTX–M and/or integrons served as donors, and E. coli C600 was used as a recipient. Transconjugants were selected on Mueller–Hinton agar (Beijing Land Bridge Technology Co., Ltd.), containing 750 μg/ml rifampin and 100 μg/ml ampicillin. Susceptibility patterns of the transconjugants were determined by the methods described above. Co-transfer of resistance determinants was identified by amplifying the relevant genes from the transconjugants by PCR and determining the nucleotide sequencing of the PCR products. Plasmid DNA from transconjugants and donor strains was extracted utilizing a Tiangen Plasmid Purification Mini Kit (Tiangen Biotech, China) according to the manufacturer’s instructions. Replicon typing of plasmids was performed by a PCR-based method as previously described (Carattoli et al., 2005).
MLST
Seven housekeeping genes (aroC, dnaN, hemD, hisD, purE, sucA, and thrA) were used for MLST as described online1. All PCR products were purified from agarose gels and sequenced by BGI Tech Solutions as described above. Alleles and STs were assigned according to the MLST scheme.
Results and Discussion
Salmonella Serotype Prevalence Among the Animal Products
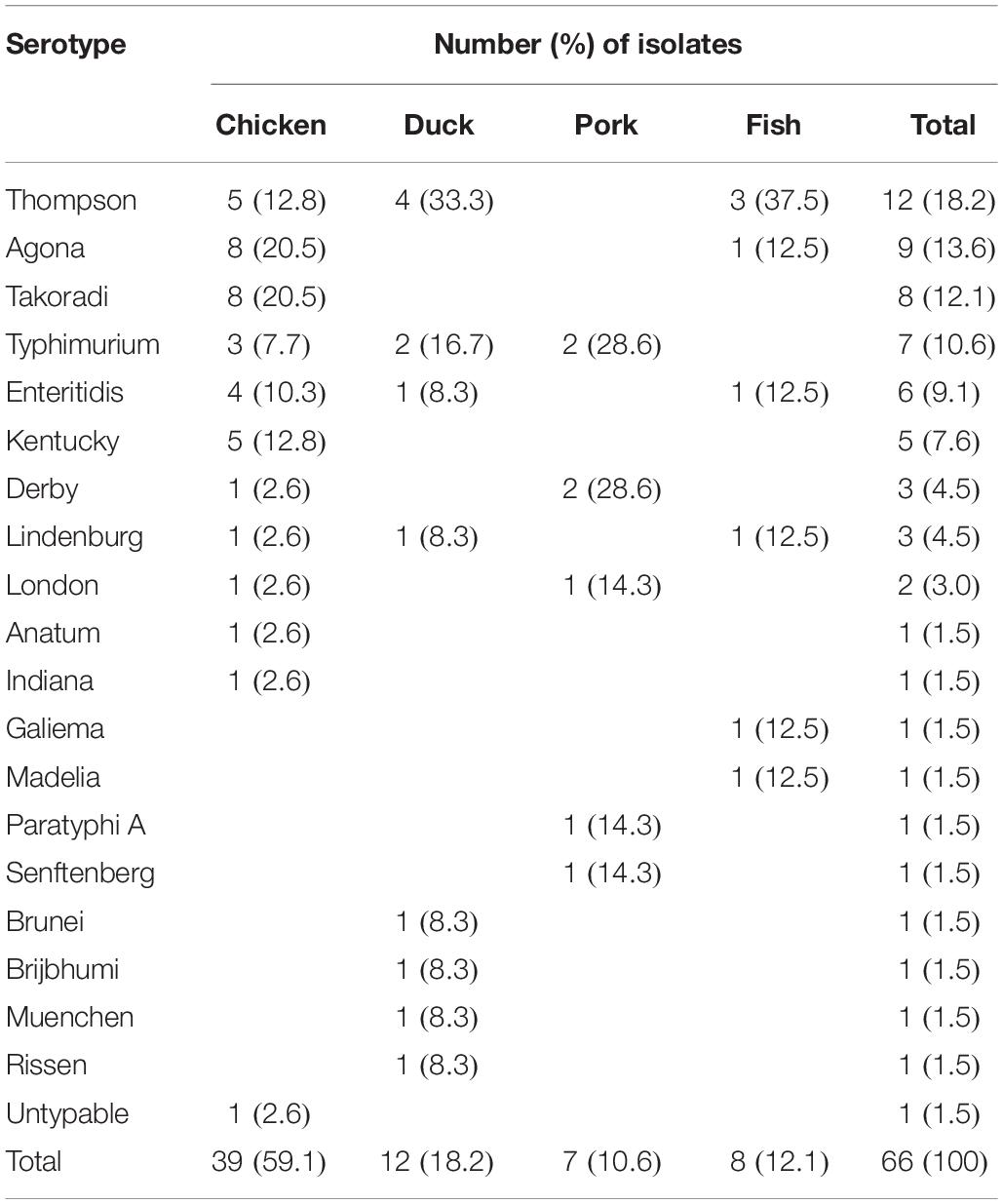
Out of the 631 meat samples, 66 (10.5%) tested positive for Salmonella, and chicken had the highest prevalence (14.8%; 39/263), followed by duck meat (11.8%; 12/102), pork (8.8%; 7/80), and fish (4.3%; 8/186).
A total of 19 serotypes were identified among 66 Salmonella isolates and one isolate from chicken was un-typeable (Table 2). The 19 serotypes included 11 from chicken, and 8, 5, and 6 from duck meat, pork and fish, respectively. S. Thompson was the serotype most frequently identified (18.2%; 12/66), followed by S. Agona (13.6%; 9/66), S. Takoradi (12.1%; 8/66), and S. Typhimurium (10.6%; 7/66). S. Thompson was not isolated from pork but it was the most common serotype isolated from both duck meat and fish with 33.3% (4/12) and 37.5% (3/8), respectively. Although S. Thompson was the most prevalent serotype overall and chicken was the most likely to be contaminated with Salmonella, S. Thompson was not the most common serotype isolated from chicken. Both S. Agona and S. Takoradi were isolated from chicken at a higher rate (20.5%; 8/39 for each of the two serotypes) than S. Thompson (12.8%; 5/39) (Table 2).
Salmonella is a major cause of foodborne illness worldwide. In this study, Salmonella were recovered from retail raw meat products in Hubei Provence, China. The overall prevalence of Salmonella in retail meat products sampled in this study (10.5%) agreed with that reported in Henan Province, China (9.7%) (Yu et al., 2014) but was higher than that reported from Xinjiang, China (6.8%) (Yin et al., 2016). However, a simple conclusion could not be made from direct comparisons among studies due to regional differences, sampling seasons and the types of products sampled. In this study, the higher contamination rate found may be related to fewer biosecurity and hygiene measures inside these small markets.
We also identified 19 serotypes among the 66 isolates. The most common serotype in retail raw meat products was S. Thompson. While this result was consistent to two previous studies conducted in Iran (Soltan-Dallal et al., 2010; Sodagari et al., 2015), it differed from other reports in which the dominant serotype was S. Hadar in Xinjiang, China (Yin et al., 2016), S. Enteritidis in Henan Province, China (Yang et al., 2013) and S. Typhimurium in Brazil (Ristori et al., 2017). Other serotypes recovered in this study included S. Agona, S. Typhimurium, S. Enteritidis, S. Kentucky, and S. Derby, which have all been previously found in food products (Ma̧ka et al., 2014; Shah et al., 2017; Wang et al., 2017). The differences in dominant serotypes may be due to geographical differences, sample types or seasons and serotype pathogenicity.
Antimicrobial Susceptibility
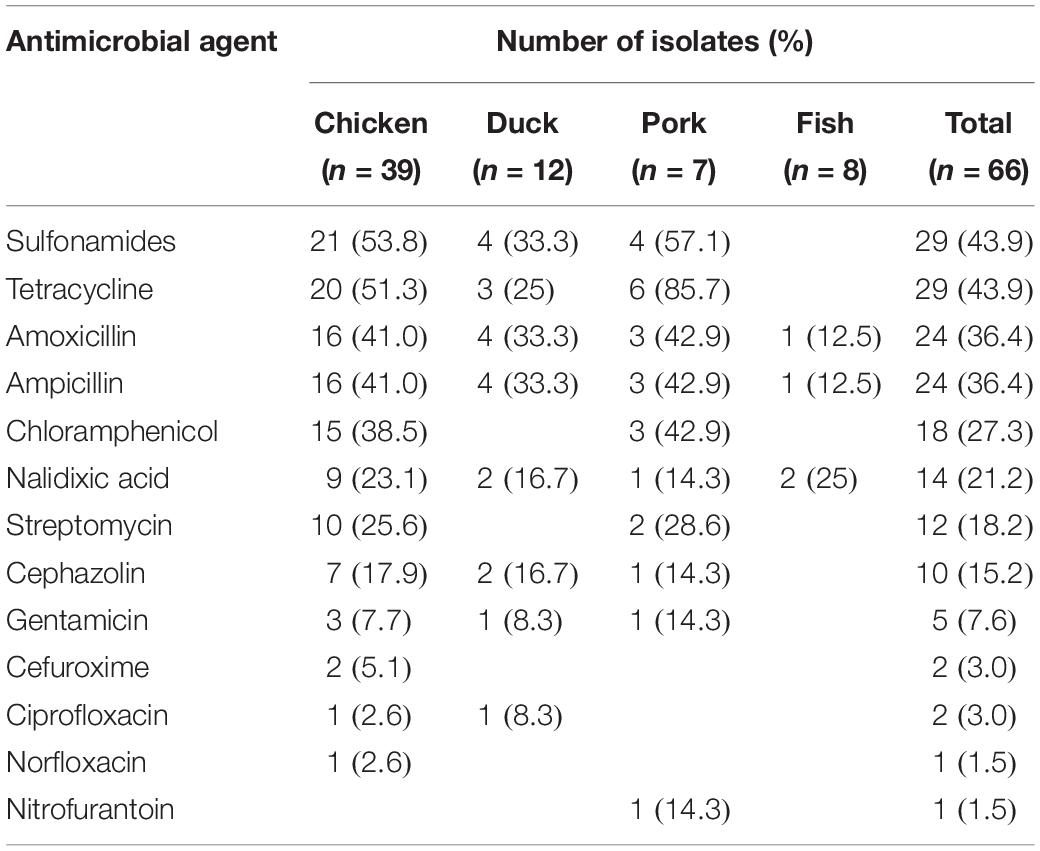
Among 66 Salmonella isolates, a high percentage of the isolates were resistant to sulfonamides (43.9%), tetracycline (43.9%), amoxicillin (36.4%), ampicillin (36.4%), chloramphenicol (27.3%), nalidixic acid (21.2%), streptomycin (18.2%), and cephazolin (15.2%) (Table 3). It was found that 17 (25.8%) of the 66 isolates were resistant to 1 antibiotic, 10 isolates (15.2%) were resistant to 2–3 antibiotics, 20 isolates (30.3%) were resistant to 4–6 antibiotics, 3 isolates (4.5%) were resistant to 7–9 antibiotics and a single isolate (1.5%) was resistant to 11 antibiotics (Table 4). Thus, 51 of the 66 Salmonella isolates (77.3%) were resistant to at least one antibiotic. Salmonella isolated from chicken were most likely to be resistant with 92.3% (36/39) of the isolates showing resistance to at least one antibiotic, and a single isolate exhibiting resistance to 11 antibiotics. Salmonella isolates from fish were the most susceptible to the antimicrobials tested with only 2 strains (25%) showing any resistance (Table 4).

Table 4. Multidrug resistance strains identified among Salmonella isolates recovered from retail meat.
The increasing frequency of antimicrobial resistance among Salmonella has become an emerging challenge to public health. Our results indicated that 30.3% of isolates were resistant to 4–6 antimicrobial agents and 4.5% were resistant to 7–9 antimicrobials. This indicated that antimicrobial resistance among Salmonella isolates from retail meat in Hubei Province was lower than that in other areas of China (Zhu et al., 2014; Wang et al., 2017). In our study, the highest rates of antimicrobial resistance were to the sulfonamides, tetracycline, amoxicillin, ampicillin and chloramphenicol. These results were consistent to studies from other regions in China and other countries (Ma̧ka et al., 2014; Yu et al., 2014; Abdi et al., 2017). The reasons for this may lie in the continuous and extensive use of these antimicrobials in food animals for disease treatment and growth promotion.
Cephalosporins and fluoroquinolones have been effectively used for the treatment of invasive and systemic salmonellosis in humans and animals (Bhan et al., 2005). Our study revealed that 15.2 and 3.0% of Salmonella isolates were resistant to first-generation (cephazolin) and second-generation cephalosporins (cefuroxime), respectively, and two isolates showed intermediate resistance to third-generation cephalosporins (cefotaxime) (Table 5). Previous work has described a decreasing susceptibility of Salmonella from retail foods to these antimicrobials (Hindermann et al., 2017; Kim et al., 2017). Interestingly, our results indicated that a high prevalence of cephazolin resistance was observed in isolates of S. Thompson, the predominant serotype. In this study, Salmonella resistance to nalidixic acid (21.2%) was not very common, however, 31.8% of the isolates also showed intermediate resistance to nalidixic acid (Table 5). Only 3.0 and 1.5% of the isolates were resistant to ciprofloxacin and norfloxacin, respectively. These rates were lower than those reported in other studies from China (Pan et al., 2009; Yin et al., 2016).
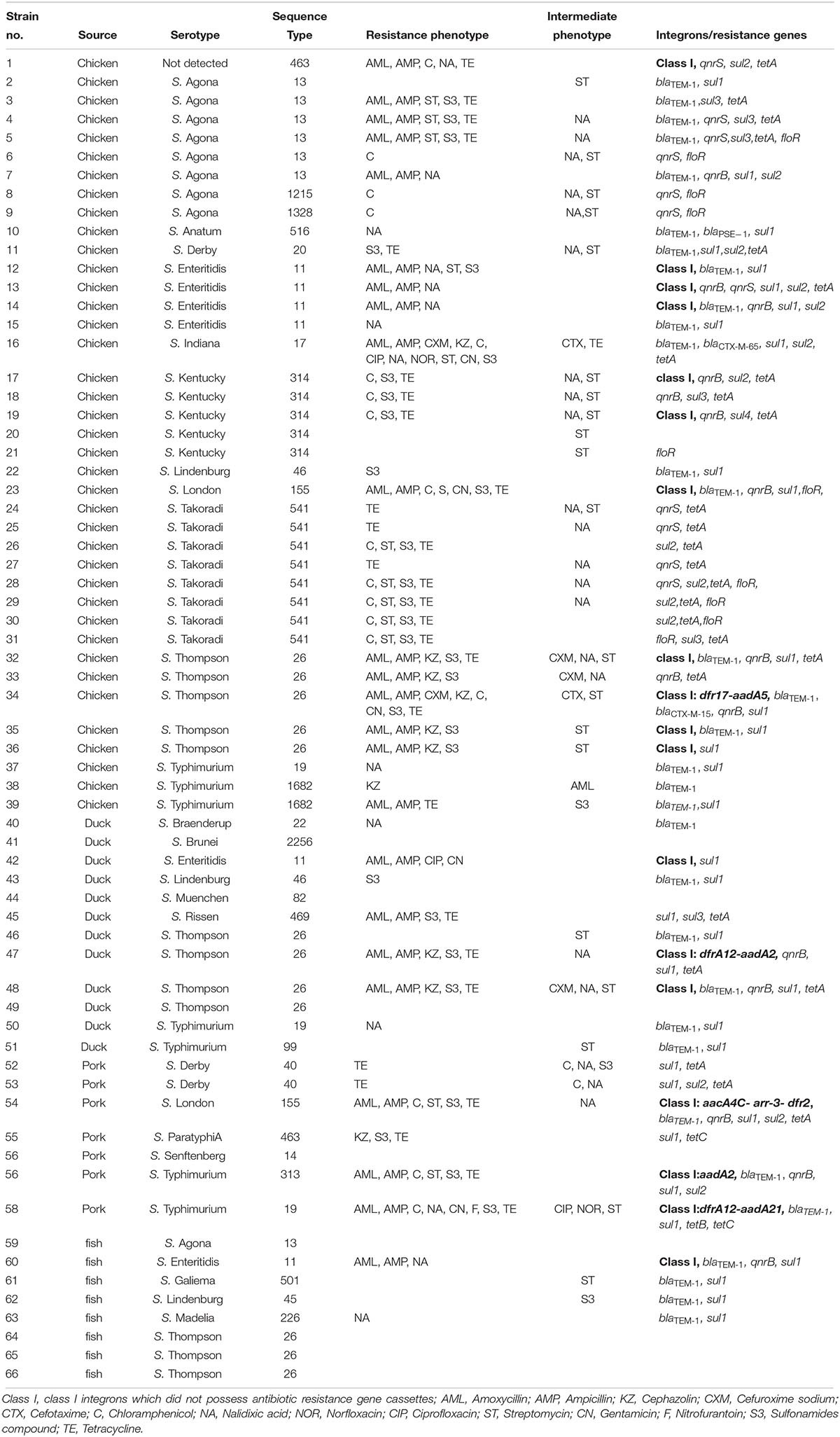
Table 5. Resistance phenotypes and antibiotic resistance genes from Salmonella isolates recovered from retail meat.
The Occurrence of Antimicrobial Resistance Genes and Class I Integrons
All 66 Salmonella isolates were screened for the presence of antimicrobial resistance genes and class 1 integrons. Numerous oligonucleotide primer pairs (Table 1) were used to screen the 66 Salmonella isolates by PCR for the presence of genes encoding resistance to chloramphenicol, tetracycline, sulfonamides, quinolones, and β-lactam antibiotics. Overall, the results showed a strong correlation between antimicrobial resistance genotypes and phenotypes (Table 5).
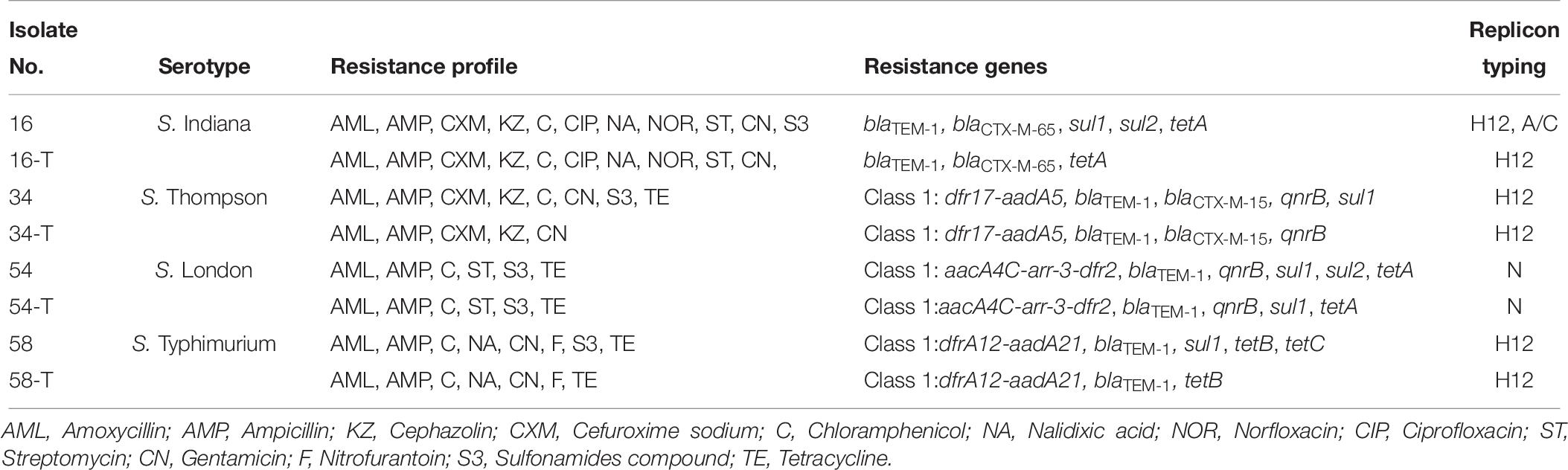
Among 26 β-lactam-resistant isolates in this study, four different β-lactamase genes were identified, including blaTEM–1, blaCTX–M–65, blaCTX–M–15, and blaPSE–1 (Table 5). The blaTEM–1 gene had the highest occurrence (69.2%, 18/26) among β-lactam-resistant isolates; a rate similar to that previously reported for Salmonella isolated from meat and dairy products in Egypt (Ahmed et al., 2014). The extended-spectrum β-lactamase (ESBL) genes, blaCTX–M–65 and blaCTX–M–15, were each observed in a single S. Indiana and S. Thompson isolate, respectively. The blaPSE–1 gene was also identified in only one isolate of S. Anatum. A cephalosporin resistant S. Thompson isolate carried the gene encoding CTX-M-15, which is the most prevalent ESBL worldwide (Chon et al., 2015). In this study, the rarely observed blaCTX–M–65 was present in a ciprofloxacin- and cefuroxime-resistant isolate of S. Indiana. Similarly, it was reported that the blaCTX–M–65 gene was harbored in ciprofloxacin- and cefotaxime-resistant S. Indiana isolates in Henan Province of China (Bai et al., 2016). In addition, the blaCTX–M–65 gene was identified previously in an outbreak of S. Infantis in Ecuador (Cartelle Gestal et al., 2016).
The qnr family of genes encodes a pentapeptide repeat protein that protects DNA gyrase and topoisomerase IV from quinolone and fluoroquinolone inactivation. Plasmids containing qnr can be transferred among bacteria via conjugation and this is a very important emerging public health concern (García-Fernández et al., 2009). Although qnr confers only low-level quinolone resistance, mutations in the quinolone resistance-determining region (QRDR) could result in higher-level resistance to fluoroquinolones, especially to ciprofloxacin. The existence of qnr genes has been reported in Salmonella from other regions in China and in other countries (Yang et al., 2013; Wasyl et al., 2014). In the current study, 62.9% (22/35) of the 14 nalidixic acid-resistant and 21 intermediate resistant Salmonella isolates harbored qnrB and/or qnrS genes (Table 5). The qnrB gene was present in 34.3% (12/35) of the nalidixic acid resistant or intermediate resistant Salmonella isolates while qnrS was present in 31.4% (11/35) of the isolates. Both qnrB and qnrS were found in only a single isolate of S. Enteritidis. The genes qnrA and qnrD were not found among any of the isolates. The blaCTX–M–15 gene was found in combination with qnrB, while the isolate with blaCTX–M–65 carried no quinolone resistance genes.
The tetracycline resistance genes (tetA, tetB, and tetC) and sulfonamide resistance genes (sul1, sul2, and sul3) were identified in 86.7 and 96.9% of the strains resistant or intermediate resistant to the cognate antibiotic. The floR gene was identified in 40% (8/20) of chloramphenicol-resistant isolates (n = 18) and intermediate resistant isolates (n = 2) (Table 5).
An integron is a mobile DNA element, which usually contains one or more antimicrobial resistance genes that can be transferred among bacteria. In this study, Class I integrons (intI1) were found in 18 isolates (27.3%) and 13 of them did not possess antimicrobial resistance gene cassettes. The remaining 5 intI1-positive isolates contained five groups of resistance gene cassettes including aacA4C-arr-3-dfr2, dfrA12-aadA21, aadA2, dfrA12-aadA2, and dfr17-aadA5 (Table 5). Our study also indicated that all of the class I integron-positive isolates were resistant to at least three classes of antimicrobials. This supports the hypothesis of the association between class I integrons and MDR in Salmonella isolates (Abraham et al., 2016; Meng et al., 2016).
Previous investigators identified class I integrons in foodborne Salmonella isolated in China at rates that ranged from 20 to 61% (Zhu et al., 2014; Meng et al., 2016; Li et al., 2017). In this study, the gene cassettes aacA4C-arr-3-dfr2 from S. London exhibited >98.0% nucleotide sequence identity to that of the variable region of Integron In1021 from a clinical E. coli isolated in China (accession no. KR338349), suggesting that some of its components may have originated from other species of Enterobacteriaceae such as Escherichia. The existence of class I integrons containing genes for antimicrobial resistance in Salmonella isolates poses a threat to public health and food safety.
Transferability of blaCTX–M Genes and Integrons
Six Salmonella strains harboring the ESBL blaCTX–M gene and integrons were selected as donor strains in conjugation experiments. These donor strains were mated with an E. coli C600 recipient strain. Four transconjugants were obtained and characterized. PCR based replicon typing revealed that plasmids belonging to incompatibility types H12 and N were transferred from Salmonella to the E. coli C600 (Table 6). However, an incA/C plasmid in a S. Indiana isolate was not transferred. Genes present in the transconjugants included class I integrons, blaTEM–1, blaCTX–M–65, blaCTX–M–15, qnrB, sul1, sul2, tetA, and tetB. The co-transfer of blaCTX–M–15 and qnrB was observed with an integron from an S. Thompson isolate to E. coli strain C600 (Table 6). When mated with the S. London donor, the E. coli C600 transconjugants showed an identical antimicrobial resistance phenotype to the donor. For the other Salmonella donors, the E. coli C600 recipients showed similar, but not identical antimicrobial resistances (Table 6).
Of concern, the findings of conjugation tests demonstrated the presence of genes including blaTEM–1, blaCTX–M–15, and qnrB on mobile plasmids. These resistance genes and integron were conjugally transferred from a Salmonella donor strain and conferred a MDR phenotype to a naïve E. coli recipient. Few studies have shown the co-existence of integron with plasmid-mediated quinolone resistance (PMQR) determinants and ESBL genes on transferable plasmids (Lai et al., 2013). It has been suggested that the transmissible elements containing these genes could be selected and transmitted during exposure to any of the antimicrobials (González and Araque, 2013; Jiang et al., 2014).
MLST Analysis
Thirty-two STs were identified among the 66 isolates, and ST26 (n = 12) was the most common followed by ST541 (n = 8), ST13 (n = 7), ST11 (n = 6), and ST314 (n = 5) (Table 5). The STs in this study were correlated with specific serotypes, such as ST11 with S. Enteritidis, ST13 with S. Agona, ST19 with S. Typhimurium, ST314 with S. Kentucky and ST26 with S. Thompson. Though a particular ST was always associated with the same serotype, some serotypes could be represented by multiple STs. For example, the isolates characterized as S. Derby belonged to ST20 and ST40.
The diversity of samples we collected was also reflected in a diversity of the 32 STs that we identified. ST26 was the most common especially among poultry samples and this ST corresponded to S. Thompson that has a low rate of occurrence in China (Ni et al., 2018). ST11 and ST19 were the most frequent sequence types recovered from meat samples and these STs are commonly associated with human salmonellosis outbreaks (Gunel et al., 2015; Ashton et al., 2016). Additionally, our results revealed that the serotypes and STs were highly correlated, which is consistent with previous research (Achtman et al., 2012). We did find sequence type differentiation within serotypes such as with S. Typhimurium and S. Derby. Therefore, MLST analysis provided a greater discriminatory power than serotype determination.
Conclusion
This study identified S. Thompson and S. Agona as the most common serotypes among Salmonella isolates recovered from retail raw meat products in Hubei Province, China. Antimicrobial resistance was prevalent and correlated well with the presence of cognate resistance genes. Salmonella isolates containing class I integrons showed broader antimicrobial resistance than those without them. In addition, conjugation experiments revealed a co-transfer of class I integron with PMQR and ESBL genes. The reasonable use of antibiotics in livestock breeding and management of the food supply chain is of importance to ensure food safety.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: http://enterobase.warwick.ac.uk/species/senterica/allele_st_search.
Author Contributions
XS and HW designed the study and revised the manuscript. MZ conducted the experiments and interpreted the results. XL and WH assisted in the completion of the experiments. GP rewrote the discussion and revised the manuscript.
Funding
This study was supported by the National Key R&D Program of China (Grant No. 2017YFC1600100), a grant from the Hubei Provincial Department of Education, China (Grant No. D20161702) and the National Natural Science Foundation of China (Grant No. 31471660).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Aarestrup, F. M., Lertworapreecha, M., Evans, M. C., Bangtrakulnonth, A., Chalermchaikit, T., Hendriksen, R. S., et al. (2003). Antimicrobial susceptibility and occurrence of resistance genes among Salmonella enterica serovar weltevreden from different countries. J. Antimicrob. Chemother. 52, 715–718. doi: 10.1093/jac/dkg426
Abdi, R. D., Mengstie, F., Beyi, A. F., Beyene, T., Waktole, H., Mammo, B., et al. (2017). Determination of the sources and antimicrobial resistance patterns of Salmonella, isolated from the poultry industry in southern ethiopia. BMC Infect. Dis. 17:352. doi: 10.1186/s12879-017-2437-2
Abraham, S., O’Dea, M., Trott, D. J., Abraham, R. J., Hughes, D., Pang, S., et al. (2016). Isolation and plasmid characterization of carbapenemase (imp-4) producing Salmonella enterica typhimurium from cats. Sci. Rep. 6:35527. doi: 10.1038/srep35527
Achtman, M., Wain, J., Weill, F. X., Nair, S., Zhou, Z., Sangal, V., et al. (2012). Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog. 8:e1002776. doi: 10.1371/journal.ppat.1002776
Agada, G. O. A., Abdullahi, I. O., Aminu, M., Odugbo, M., Chollom, S. C., Kumbish, P. R., et al. (2014). Prevalence and antibiotic resistance profile of Salmonella isolates from commercial poultry and poultry farm-handlers in Jos. Plateau State Nigeria. Br. Microbiol. Res. J. 4, 462–479. doi: 10.9734/BMRJ/2014/5872
Ahmed, A. M., Shimamoto, T., and Shimamoto, T. (2014). Characterization of integrons and resistance genes in multidrug-resistant Salmonella enterica isolated from meat and dairy products in Egypt. Int. J. Food Microbiol. 189, 39–44. doi: 10.1016/j.ijfoodmicro.2014.07.031
Asgharpour, F., Rajabnia, R., Ferdosi Shahandashti, E., Marashi, M. A., Khalilian, M., and Moulana, Z. (2014). Investigation of class I integron in Salmonella infantis and its association with drug resistance. Jundishapur J. Microbiol. 7:e10019. doi: 10.5812/jjm.10019
Ashton, P. M., Nair, S., Peters, T. M., Bale, J. A., Powell, D. G., Painset, A., et al. (2016). Identification of Salmonella for public health surveillance using whole genome sequencing. Peer J. 4, e1752. doi: 10.7717/peerj.1752
Bai, L., Zhao, J., Gan, X., Gan, X., Wang, J., Zhang, X., et al. (2016). Emergence and diversity of Salmonella enterica serovar Indiana isolates with concurrent resistance to ciprofloxacin and cefotaxime from patients and food-producing animals in China. Antimicrob. Agents Chemother 60, 3365–3371. doi: 10.1128/AAC.02849-15
Bhan, M. K., Bahl, R., and Bhatnagar, S. (2005). Typhoid and paratyphoid fever. Lancet 366, 749–762.
Carattoli, A., Bertini, A., Villa, L., Falbo, V., Hopkins, K. L., and Threlfall, E. J. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63, 219–228. doi: 10.1016/j.mimet.2005.03.018
Cartelle Gestal, M., Zurita, J., Paz, Y., Mino, A., Ortega-Paredes, D., and Alcocer, I. (2016). Characterization of a small outbreak of Salmonella enterica serovar Infantis that harbour CTX-M-65 in Ecuador. Braz. J. Infect. Dis. 20, 406–407. doi: 10.1016/j.bjid.2016.03.007
Cattoir, V., Poirel, L., Rotimi, V., Soussy, C. J., and Nordmann, P. (2007). Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J. Antimicrob. Chemother. 60, 394–397. doi: 10.1093/jac/dkm204
Chang, Y. C., Scaria, J., Ibraham, M., Doiphode, S., Chang, Y. F., Sultan, A., et al. (2016). Distribution and factors associated with Salmonella enterica genotypes in a diverse population of humans and animals in Qatar using multi-locus sequence typing (MLST). J.Infect. Public Health 9, 315–323. doi: 10.1016/j.jiph.2015.10.013
Chon, J. W., Jung, H. I., Kuk, M., Kim, Y. J., Seo, K. H., and Kim, S. K. (2015). High occurrence of extended-spectrum β-lactamase-producing Salmonella in broiler carcasses from poultry slaughterhouses in South Korea. Foodborne Pathog. Dis. 12, 190–196. doi: 10.1089/fpd.2014.1847
Clinical and Laboratory Standards Institute [CLSI] (2016). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-sixth Informational Supplement, M100-S26. Wayne, PA: Clinical and Laboratory Standards Institute.
García-Fernández, A., Fortini, D., Veldman, K., Mevius, D., and Carattoli, A. (2009). Characterization of plasmids harbouring qnrS1, qnrB2 and qnrB19 genes in Salmonella. J. Antimicrob. Chemother. 63, 274–281. doi: 10.1093/jac/dkn470
Ghoddusi, A., Nayeri Fasaei, B., Karimi, V., Ashrafi Tamai, I., Moulana, Z., and Zahraei Salehi, T. (2015). Molecular identification of Salmonella Infantis isolated from backyard chickens and detection of their resistance genes by PCR. Iran. J. Vet. Res. 16, 293–297.
González, F., and Araque, M. (2013). Association of transferable quinolone resistance determinant qnrB19 with extended-spectrumβ-lactamases in Salmonella Give and Salmonella Heidelberg in Venezuela. Int. J. Microbiol. 2013:628185. doi: 10.1155/2013/628185
Gunel, E., Polat Kilic, G., Bulut, E., Durul, B., Acar, S., Alpas, H., et al. (2015). Salmonella surveillance on fresh produce in retail in Turkey. Int.J. Food Microbiol. 199, 72–77. doi: 10.1016/j.ijfoodmicro.2015.01.010
Hendriksen, R. S., Mikoleit, M., Carlson, V. P., Karlamose, S., Vieira, A. R., Jensen, A. B., et al. (2009). WHO Global Salm-Surv external quality assurance system for serotyping of Salmonella isolates from 2000 to 2007. J. Clin. Microbiol. 47, 2729–2736. doi: 10.1128/JCM.02437-08
Hindermann, D., Gopinath, G., Chase, H., Negrete, F., Althaus, D., Zurfluh, K., et al. (2017). Salmonella enterica serovar Infantis from food and human infections, Switzerland, 2010-2015: poultry-related multidrug resistant clones and an emerging ESBL producing clonal lineage. Front. Microbiol. 8:1322. doi: 10.3389/fmicb.2017.01322
Hung, Y. T., Lay, C. J., Wang, C. L., and Koo, M. (2017). Characteristics of nontyphoidal Salmonella gastroenteritis in Taiwanese children: a 9-year period retrospective medical record review. J. Infect. Public Health 10, 518–521. doi: 10.1016/j.jiph.2016.09.018
Hur, J., Jawale, C., and Lee, J. H. (2012). Antimicrobial resistance of Salmonella, isolated from food animals: a review. Food Res. Int. 45, 819–830. doi: 10.1016/j.foodres.2011.05.014
Jiang, H., Song, L., Liu, J., Zhang, X., Ren, Y., Zhang, W., et al. (2014). Multiple transmissible genes encoding fluoroquinolone and third-generation cephalosporin resistance co-located in non-typhoidal Salmonella isolated from food-producing animals in China. Int. J. Antimicrob. Agents 43, 242–247. doi: 10.1016/j.ijantimicag.2013.12.005
Kim, J. S., Kim, S., Park, J., Shin, E., Yun, Y. S., Lee, D. Y., et al. (2017). Plasmid-mediated transfer of CTX-M-55 extended-spectrum beta-lactamase among different strains of Salmonella and Shigella spp. in the republic of korea. Diagn. Microbiol. Infect. Dis. 89, 86–88. doi: 10.1016/j.diagmicrobio.2017.03.014
Lai, J., Wang, Y., Shen, J., Li, R., Han, J., Foley, S. L., et al. (2013). Unique class 1 integron and multiple resistance genes co-located on IncHI2 plasmid is associated with the emerging multidrug resistance of Salmonella Indiana isolated from Chicken in China. Foodborne Pathog. Dis. 10, 581–588. doi: 10.1089/fpd.2012.1455
Li, S., Zhou, Y., and Miao, Z. (2017). Prevalence and antibiotic resistance of non-typhoidal Salmonella isolated from raw chicken carcasses of commercial broilers and spent hens in Tai’an. China. Front. Microbiol. 8:2106. doi: 10.3389/fmicb.2017.02106
Lu, H., Wang, X., Lang, X., Wang, Y., Dang, Y., Zhang, F., et al. (2010). Preparation and application of microarrays for the detection of antibiotic resistance genes in samples isolated from Changchun. China. Mol. Biol. Rep. 37, 1857–1865. doi: 10.1007/s11033-009-9621-4
Ma̧ka, Ł., Mackiw, E., Sciezynska, H., Pawłowska, K., and Popowska, M. (2014). Antimicrobial susceptibility of Salmonella strains isolated from retail meat products in Poland between 2008 and 2012. Food Control 36, 199–204. doi: 10.1016/j.foodcont.2013.08.025
Mancin, M., Barco, L., Saccardin, C., and Ricci, A. (2015). Proposed statistical analysis to evaluate qualitative profificiency testing of Salmonella serotyping. Accred. Qual. Assur. 20, 305–310. doi: 10.1007/s00769-015-1129-0
Meng, H., Zhang, Z., Chen, M., Su, Y., Li, L., Miyoshi, S., et al. (2011). Characterization and horizontal transfer of class 1 integrons in Salmonella strains isolated from food products of animal origin. Int. J. Food Microbiol. 149, 274–277. doi: 10.1016/j.ijfoodmicro.2011.07.006
Meng, X., Zhang, Z., Li, K., Wang, Y., Xia, X., Wang, X., et al. (2016). Antibiotic susceptibility and molecular screening of class I integron in Salmonella isolates recovered from retail raw chicken carcasses in China. Microb. Drug Resist. 23, 230–235. doi: 10.1089/mdr.2015.0359
Ni, P., Xu, Q., Yin, Y., Liu, D., Zhang, J., Wu, Q., et al. (2018). Prevalence and characterization of Salmonella serovars isolated from farm products in Shanghai. Food Control 85, 269–275. doi: 10.1016/j.foodcont.2017.10.009
Olsen, J. E., Brown, D. J., Thomsen, L. E., Platt, D. J., and Chadfield, M. S. (2004). Differences in the carriage and the ability to utilize the serotype associated virulence plasmid in strains of Salmonella enterica serotype typhimurium investigated by use of a self-transferable virulence plasmid, pOG669. Microb. Pathog. 36, 337–347. doi: 10.1016/j.micpath.2004.02.005
Pan, Z., Wang, X., Zhang, X., Geng, S., Chen, X., Pan, W., et al. (2009). Changes in antimicrobial resistance among Salmonella enterica subspecies enterica serovar Pullorum isolates in China from 1962 to 2007. Vet. Microbiol. 136, 387–392. doi: 10.1016/j.vetmic.2008.11.015
Papadopoulos, T., Petridou, E., Zdragas, A., Mandilara, G., Nair, S., Peters, T., et al. (2016). Comparative study of all Salmonella enterica, serovar Enteritidis strains isolated from food and food animals in Greece from 2008 to 2010 with clinical isolates. Eur. J. Clin. Microbiol. Infect. Dis. 35, 741–746. doi: 10.1007/s10096-016-2591-2
Park, S., Choi, S., Kim, H., Kim, Y., Kim, B. S., Beuchat, L. R., et al. (2015). Fate of mesophilic aerobic bacteria and Salmonella enterica on the surface of eggs as affected by chicken feces, storage temperature, and relative humidity. Food Microbiol. 48, 200–205. doi: 10.1016/j.fm.2015.01.003
Rajaei, B., Siadat, S. D., Rad, N. S., Badmasti, F., Razavi, M. R., Aghasadeghi, M. R., et al. (2014). Molecular detection of antimicrobial resistance gene cassettes associated with class 2 integron in Salmonella serovars isolated in Iran. Br. Microbiol. Res. J. 4, 132–141. doi: 10.9734/BMRJ/2014/4639
Ristori, C. A., Rowlands, R. E. G., Martins, C. G., Barbosa, M. L., Dos Santos, L. F., Jakabi, M., et al. (2017). Assessment of consumer exposure to Salmonella spp., Campylobacter spp., and Shiga Toxin-Producing Escherichia coli in meat products at retail in the city of Sao Paulo, Brazil. Foodborne Pathog. Dis. 14, 447–453. doi: 10.1089/fpd.2016.2270
Shah, D. H., Paul, N. C., Sischo, W. C., Crespo, R., and Guard, J. (2017). Population dynamics and antimicrobial resistance of the most prevalent poultry-associated Salmonella serotypes. Poult. Sci. 96, 687–702. doi: 10.3382/ps/pew342
Shahada, F., Chuma, T., Tobata, T., Okamoto, K., Sueyoshi, M., and Takase, K. (2006). Molecular epidemiology of antimicrobial resistance among Salmonella enterica serovar Infantis from poultry in Kagoshima. Japan. Int. J. Antimicrob. Agents 28, 302–307. doi: 10.1016/j.ijantimicag.2006.07.003
Smith, A. M., Smouse, S. L., Tau, N. P., Bamford, C., Moodley, V. M., Jacobs, C., et al. (2017). Laboratory-acquired infections of Salmonella enterica serotype typhi in South Africa: phenotypic and genotypic analysis of isolates. Bmc Infect. Dis. 17:656. doi: 10.1186/s12879-017-2757-2
Sodagari, H. R., Mashak, Z., and Ghadimianazar, A. (2015). Prevalence and antimicrobial resistance of Salmonella serotypes isolated from retail chicken meat and giblets in Iran. J. Infect. Dev. Ctries. 9, 463–469. doi: 10.3855/jidc.5945
Soltan-Dallal, M., Doyle, M., Rezadehbashi, M., Dabiri, H., Sanael, M., Modarresi, S., et al. (2010). Prevalence and antimicrobial resistance profiles of Salmonella serotypes, Campylobacter and Yersinia spp. isolated from retail chicken and beef, Tehran, Iran. Food Control 21, 388–392. doi: 10.1016/j.foodcont.2009.06.001
Stepan, R. M., Sherwood, J. S., Petermann, S. R., and Logue, C. M. (2011). Molecular and comparative analysis of Salmonella enterica Senftenberg from humans and animals using PFGE. MLST and NARMS. BMC Microbiol. 11:153. doi: 10.1186/1471-2180-11-153
Wang, Y., Cao, C., Alali, W. Q., Cui, S., Li, F., Zhu, J., et al. (2017). Distribution and antimicrobial susceptibility of foodborne Salmonella serovars in eight provinces in China from 2007 to 2012 (Except 2009). Foodborne Pathog. Dis. 14, 393–399. doi: 10.1089/fpd.2016.2237
Wasyl, D., Hoszowski, A., and Zaja̧c, M. (2014). Prevalence and characterisation of quinolone resistance mechanisms in Salmonella spp. Vet. Microbiol. 171, 307–314. doi: 10.1016/j.vetmic.2014.01.040
Wright, G. D. (2010). Antibiotic resistance in the environment: a link to the clinic? Curr. Opin. Microbiol. 13, 589–594. doi: 10.1016/j.mib.2010.08.005
Yang, B., Qiao, L., Zhang, X., Cui, Y., Xia, X., and Cui, S. (2013). Serotyping, antimicrobial susceptibility, pulse field gel electrophoresis analysis of Salmonella isolates from retail foods in Henan Province. China. Food Control 32, 228–235. doi: 10.1016/j.foodcont.2012.11.022
Yin, M., Yang, B., Wu, Y., Wang, L., Wu, H., Zhang, T., et al. (2016). Prevalence and characterization of Salmonella enterica serovar in retail meats in market place in Uighur, Xinjiang, China. Food Control 64, 165–172. doi: 10.1016/j.foodcont.2015.12.029
Yu, T., Jiang, X., Zhou, Q., Wu, J., and Wu, Z. (2014). Antimicrobial resistance, class I integrons, and horizontal transfer in Salmonella isolated from retail food in Henan. China. J. Infect. Dev. Ctries. 8, 705–711. doi: 10.3855/jidc.4190
Zhang, A., Wang, H., Tian, G., Zhang, Y., Yang, X., Xia, Q., et al. (2009). Phenotypic and genotypic characterisation of antimicrobial resistance in faecal bacteria from 30 Giant pandas. Int. J. Antimicrob. Agents 33, 456–460. doi: 10.1016/j.ijantimicag.2008.10.030
Zhao, X., Ye, C., Chang, W., and Sun, S. (2017). Serotype distribution, antimicrobial resistance, and class 1 integrons profiles of Salmonella from animals in slaughterhouses in Shandong province, China. Front. Microbiol 8:1049. doi: 10.3389/fmicb.2017.01049
Zhou, R., Li, L., Su, J., Li, B., and Xu, Z. (2014). Detection of class I integron from foodborne Salmonella and analysis on the drug resistance gene cassettes. Sci. Technol. Food Ind. 35, 279–278.
Zhu, Y., Yi, Y., Liu, F., Lv, N., Yang, X., Li, J., et al. (2014). Distribution and molecular profiling of class 1 integrons in MDR Acinetobacter baumannii isolates and whole genome-based analysis of antibiotic resistance mechanisms in a representative strain. Microbiol. Res. 169, 811–816. doi: 10.1016/j.micres.2014.04.002
Keywords: Salmonella, MLST, class I integrons, blaCTX–M–65, blaCTX–M–15, qnrB
Citation: Zhou M, Li X, Hou W, Wang H, Paoli GC and Shi X (2019) Incidence and Characterization of Salmonella Isolates From Raw Meat Products Sold at Small Markets in Hubei Province, China. Front. Microbiol. 10:2265. doi: 10.3389/fmicb.2019.02265
Received: 24 May 2019; Accepted: 17 September 2019;
Published: 04 October 2019.
Edited by:
Arun K. Bhunia, Purdue University, United StatesReviewed by:
Ashraf Khan, National Center for Toxicological Research (FDA), United StatesHaiyan Zeng, Guangdong Academy of Science, China
Copyright © 2019 Zhou, Li, Hou, Wang, Paoli and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongxun Wang, d2FuZ2hvbmd4dW5odXN0QDE2My5jb20=; Xianming Shi, eG1zaGlAc2p0dS5lZHUuY24=
 Min Zhou1
Min Zhou1 Xianming Shi
Xianming Shi