
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CORRECTION article
Front. Microbiol. , 27 September 2019
Sec. Infectious Agents and Disease
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.02218
This article is a correction to:
Whole Genome Sequencing of Australian Candida glabrata Isolates Reveals Genetic Diversity and Novel Sequence Types
 Chayanika Biswas1,2
Chayanika Biswas1,2 Vanessa R. Marcelino2,3,4†
Vanessa R. Marcelino2,3,4† Sebastiaan Van Hal5†
Sebastiaan Van Hal5† Catriona Halliday6
Catriona Halliday6 Elena Martinez1,2
Elena Martinez1,2 Qinning Wang1,2
Qinning Wang1,2 Sarah Kidd7
Sarah Kidd7 Karina Kennedy8
Karina Kennedy8 Deborah Marriott9
Deborah Marriott9 C. Orla Morrissey10
C. Orla Morrissey10 Ian Arthur11
Ian Arthur11 Kerry Weeks12
Kerry Weeks12 Monica A. Slavin13
Monica A. Slavin13 Tania C. Sorrell2,3,4
Tania C. Sorrell2,3,4 Vitali Sintchenko1,2,3,4‡
Vitali Sintchenko1,2,3,4‡ Wieland Meyer1,2,3,4‡
Wieland Meyer1,2,3,4‡ Sharon C.-A. Chen1,2,3,4,6*‡
Sharon C.-A. Chen1,2,3,4,6*‡A Corrigendum on
Whole Genome Sequencing of Australian Candida glabrata Isolates Reveals Genetic Diversity and Novel Sequence Types
by Biswas, C., Marcelino, V. R., Van Hal, S., Halliday, C., Martinez, E., Wang, Q., et al. (2018). Front. Microbiol. 9:2946. doi: 10.3389/fmicb.2018.02946
In the original article, there were a small number of transcription errors in Figure 1 as published.
There are:
The label “Norway 6” (at position 6 o'clock) was incorrectly “positioned” with isolate WM_18.31 (ST45) and should have been positioned with “Norway 5.” This is now corrected.
Isolates Taiwan 1 and Taiwan 2 should be positioned together. In addition, the isolate WM_18.57 was inadvertently omitted from the ST16 “bubble.”
Isolate WM_18.51 should have been placed in the group under ST46 instead of ST59. The corrected Figure 1 appears below.
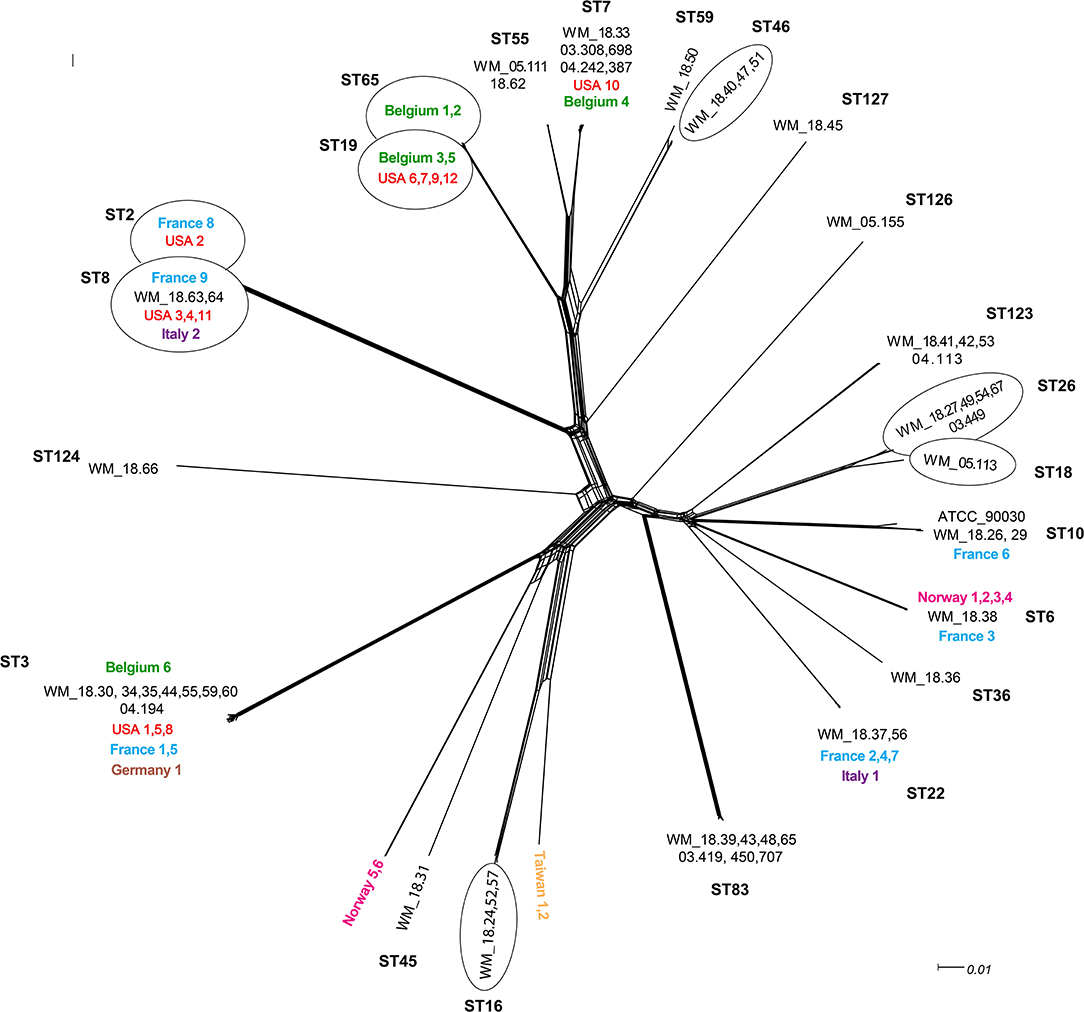
Figure 1. Unrooted network tree depicting the association between Australian Candida glabrata isolates and international isolates from seven countries based on their whole genome sequences. All clusters in the tree have been represented by different sequence types (STs) except Norway 5, Norway 6, Taiwan 1 and Taiwan 2 which have previously unassigned (new) STs. New sequence types (STs) from Australia are ST123, ST124, ST126 and ST127. Isolates representing a particular ST in branches, which contain multiple STs, are put in circles. The colors depict isolates from different countries: Black, Australia; Green, Belgium; Blue, France; Brown, Germany; Purple, Italy; Pink, Norway; Yellow, Taiwan; Red, United States. The Australian isolates have names starting with WM_ and the international isolates where named according to the country of origin, all followed by a numerical scheme. For isolates from same country in a cluster, the country name was followed by numerical identities of the isolates separated by commas. For example, in ST7 cluster, WM_18.33, 03.308,689, 04.242,387 (where 18, 03 and 04 are years of isolation followed by isolate number).
The authors apologize for these errors and state that they do not change the scientific findings of the article in any way. The original article has been updated.
Keywords: whole genome sequencing, Candida glabrata, MLST, sequence type, Australia
Citation: Biswas C, Marcelino VR, Van Hal S, Halliday C, Martinez E, Wang Q, Kidd S, Kennedy K, Marriott D, Morrissey CO, Arthur I, Weeks K, Slavin MA, Sorrell TC, Sintchenko V, Meyer W and Chen SC-A (2019) Corrigendum: Whole Genome Sequencing of Australian Candida glabrata Isolates Reveals Genetic Diversity and Novel Sequence Types. Front. Microbiol. 10:2218. doi: 10.3389/fmicb.2019.02218
Received: 09 August 2019; Accepted: 11 September 2019;
Published: 27 September 2019.
Edited by:
Sascha Brunke, Leibniz Institute for Natural Product Research and Infection Biology, GermanyReviewed by:
Volker U. Schwartze, Friedrich Schiller University Jena, GermanyCopyright © 2019 Biswas, Marcelino, Van Hal, Halliday, Martinez, Wang, Kidd, Kennedy, Marriott, Morrissey, Arthur, Weeks, Slavin, Sorrell, Sintchenko, Meyer and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sharon C.-A. Chen, c2hhcm9uLmNoZW5AaGVhbHRoLm5zdy5nb3YuYXU=
†These authors have contributed equally to this work
‡Equal senior authors
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.