- 1Dipartimento di Scienze di Laboratorio e Infettivologiche, Fondazione Policlinico Universitario Agostino Gemelli IRCCS, Rome, Italy
- 2Dipartimento di Scienze Gastroenterologiche, Endocrino-Metaboliche e Nefro-Urologiche, Fondazione Policlinico Universitario Agostino Gemelli IRCCS, Rome, Italy
- 3Istituto di Patologia Medica e Semeiotica Medica, Università Cattolica del Sacro Cuore, Rome, Italy
- 4Istituto di Microbiologia, Università Cattolica del Sacro Cuore, Rome, Italy
Candida parapsilosis is the most frequent cause of catheter-related candidemia among non-Candida albicans species. This may be related to intrinsic capabilities as adhering and forming a biofilm on abiotic surfaces such as on medical devices. As previously demonstrated, patients infected with high biofilm-producing C. parapsilosis isolates had a greater mortality risk compared to patients infected with low biofilm-producing C. parapsilosis isolates. We developed the BIOF–HILO assay, a MALDI–TOF mass spectrometry (MS)-based assay, which compares mass spectra obtained from attached and suspended isolate cells during the early (i.e., 3-h) adhesion phase of in vitro biofilm formation. The composite correlation index (CCI) analysis was used to discriminate between mass spectra differences of the two cell types, classifying all 50 C. parapsilosis clinical isolates, included in the study, after only 3-h of testing, in high or low biofilm producers. All high (n = 25) or low (n = 25) biofilm producers had, according to CCI mass spectra comparison values, higher or lower than one CCI ratios, which were obtained by dividing the CCIsuspended cells by the CCIattached cells. In conclusion, the BIOF–HILO assay allows a rapid categorization of C. parapsilosis clinical isolates in high or low biofilm producers. This information, if timely provided to physicians, may improve treatment outcomes in patients with C. parapsilosis candidemia.
Introduction
Candida species are able to adhere and form a biofilm on abiotic surfaces, such as on indwelling medical devices (e.g., vascular or urinary catheters), prosthetic heart valves, and joint replacements. This ability may be responsible for the high mortality attributed to Candida infections in healthcare settings (Tumbarello et al., 2007; Rajendran et al., 2016). Key factors favoring colonization and/or infection by biofilm-forming Candida species include high biofilm cell concentration, altered expression of cell ergosterol biosynthesis genes, and extracellular polymeric substances in the biofilm matrix (Finkel and Mitchell, 2011; Taff et al., 2013; Zarnowski et al., 2016). Notably, sessile and planktonic cells within Candida biofilms are different in their antifungal drug susceptibility profiles (Ramage et al., 2012), because biofilm growth would prompt the sessile cells to acquire a reduced susceptibility against antifungal agents (Silva et al., 2017).
During the last decade, Candida parapsilosis or Candida glabrata are on first and second position of non-Candida albicans Candida (NCAC) species isolated from patients with symptomatic bloodstream infection causing increased candidemia incidence (Guinea, 2014). C. parapsilosis exhibits the highest capability of adhering to abiotic surfaces among NCAC species (including Candida tropicalis) (Cavalheiro and Teixeira, 2018) and, consequently, this species is considered the most causative agent for catheter-related infections (Bouza et al., 2014). High (and/or moderate) biofilm formation levels were a significant risk factor for in-hospital mortality in C. parapsilosis candidemia (Soldini et al., 2018). Therefore, rapid information on high or low biofilm-forming C. parapsilosis isolates would aid to improve treatment outcomes in infected patients (Sanguinetti and Posteraro, 2016).
High or low biofilm-producing C. parapsilosis isolates differ regarding attached or suspended cells in the early in vitro adhesion phase of biofilm formation (Pannanusorn et al., 2014). This difference is beneficial to discriminate between the two isolate types after their 3-h adhesion. With this in mind, we developed the BIOF–HILO assay, which uses MALDI–TOF mass spectrometry (MS) technology coupled with the composite correlation index (CCI) analysis of protein profiles, allowing a rapid (i.e., 3-h) identification of high- or low-biofilm-forming C. parapsilosis isolates. Specifically, we performed a quantitative comparison of the mass spectra obtained from attached and suspended C. parapsilosis isolates’ cells during the initial in vitro biofilm formation. The BIOF–HILO assay and MALDI–TOF MS-based identification assay are substantially similar when comparing isolates’ spectra. However, we did not employ any reference database of C. parapsilosis mass spectra for comparison purposes in this assay.
Materials and Methods
Study Organisms
Fifty C. parapsilosis sensu stricto (hereafter referred to as C. parapsilosis) isolates collected from single patient candidemia episodes at the Fondazione Policlinico Universitario Agostino Gemelli IRCCS in Rome, Italy, were included in the study. The institutional Ethics Committee approved the study (no. 1401/16). No informed consent from patients was requested because testing was performed only on anonymized frozen isolates. These isolates had been characterized as high (n = 25) or low (n = 25) biofilm producers, i.e., as HP or LP, respectively, using the crystal-violet binding assay (Soldini et al., 2018). Isolates grew as biofilms in 96-well microtiter plates at 37°C for 24-h, starting from a standardized suspension (1 × 106 cells/ml) in RPMI 1640 broth medium (Sigma-Aldrich, St. Louis, MO, United States). As measured by the crystal violet retention, the optical density at 540 nm (OD540 nm) values were >1.17 for HP isolates and <0.44 for LP isolates, according to the OD540 nm cutoff values (<0.44, 0.44–1.17, and >1.17) used to classify Candida species isolates as low, moderate, and high biofilm level producers, respectively (Soldini et al., 2018).
MALDI–TOF MS Assay
Preliminarily, one HP and one LP C. parapsilosis isolate at different time points (1.5, 3, 4.5, 6, 8, and 24 h) were studied using biological (i.e., three from repeated cultivations on different days) and technical (i.e., two from each same cultivation) isolate replicates, obtained as described below. These experiments allowed us to find the optimal time at which the differences between the HP and LP isolates’ mass spectra could be correctly detected (data not shown).
Upon optimization of the experimental parameters, all the 50 C. parapsilosis isolates were blindly tested with the BIOF–HILO assay. First, frozen isolate stocks were sub-cultured on Sabouraud dextrose agar (SDA) plates. Then, the isolates were grown in yeast extract–peptone–dextrose broth, under agitation at 150 rpm, overnight at 37°C. For each isolate, cells were harvested, washed twice with 0.15 M phosphate-buffered saline (PBS; pH 7.2, Ca2+- and Mg2+-free), and were used to obtain a 2-McFarland suspension (equivalent to ∼0.5 × 107 cells/ml) in RPMI 1640 broth. After 3-h incubation in a 24-well cell culture plate, both the suspended (i.e., harvested by taking all the supernatant) and attached (i.e., harvested by scraping the well after addition of 200 μl of water) cells from each well were collected in two separate Eppendorf tubes, centrifuged, and twice washed in water. Randomly, a 0.1-ml aliquot of C. parapsilosis cells was plated in triplicate onto SDA and, then, the plates were read after 24-h incubation at 37°C for the CFU/ml determination. Expectedly, the number of cells attached to the microplate wells after 3-h was substantially higher than the number of cells suspended (i.e., non-attached) in HP isolates rather than in LP isolates (data not shown).
For MALDI–TOF MS analysis, the protein cell content was extracted from the 50 C. parapsilosis isolates, according to the fast formic-acid protocol previously developed (De Carolis et al., 2014b). The RNase B solution (1 mg/ml; Sigma-Aldrich, Milan, Italy) was used as an internal standard, which was added to each sample before MALDI–TOF mass spectra acquisition. Samples were spotted in duplicate on a MALDI polished target plate and measurements were performed with an MBT Smart mass spectrometer (Bruker Daltonics, Bremen, Germany) after calibration with a Bacterial Test Standard (Bruker Daltonics). The α-cyano-4-hydroxycinnamic acid (HCCA) matrix was used for sample crystallization. Spectra for each sample were generated from 500 laser shots acquired in automatic mode (100 laser shots at five different spot positions), and analyzed in a 3000–7000 Da range by dividing each spectrum into four intervals of the same size through the CCI tool of the MALDI BiotyperTM system (Bruker Daltonics). The mass spectra obtained from each isolate’s suspended or attached cells and from the RNase B alone were compared with each other using the MALDI Biotyper 3.1 software by the CCI analysis (De Carolis et al., 2012), and were automatically visualized in a CCI matrix view.
Organism Categorization
For comparison analysis, the MALDI–TOF mass spectrum profiles of the suspended or attached cells from each isolate’s biological replicates were matched against the internal standard’s mass spectrum (unpaired t test; p < 0.05). Then, each isolate was categorized as HP or LP if the CCI obtained by matching the internal standard’s profile vs. the suspended cells’ profile (CCIsusp) was higher (i.e., in the case of HP) or lower (i.e., in the case of LP) than the CCI obtained by matching the internal standard’s profile vs. the attached cells’ profile (CCIattach), respectively. To take variation in the spectral profiles between HP and LP isolates into account CCI ratios were used, which were obtained by dividing the CCIsusp by the CCIattach (see Supplementary Table S1). Accordingly, an isolate was defined as HP if the CCIsusp/CCIattach ratio was higher than one or as LP if the CCIsusp/CCIattach ratio was lower than one. Results were visualized as scatter plots using GraphPad Prism version 8.0.0 (GraphPad Software, Inc., La Jolla, CA, United States).
Results and Discussion
Figures 1, 2 show the BIOF–HILO assay to discriminate between HP and LP C. parapsilosis isolates. Specifically, Figure 2 illustrates that the BIOF–HILO assay discriminated all clinical isolates (n = 50) within 3-h in line with the biofilm levels (high or low) as previously established by Soldini et al. (2018) (see also Supplementary Table S1). Figure 3 shows a comprehensive representation (from the sample to the result) of the BIOF–HILO assay. Based on our findings, BIOF–HILO assay, testing adhesions after 3-h, provides a substitute for the test after 24-h to measure the biofilm formed by clinical C. parapsilosis isolates. Consistent with our data, Pannanusorn et al. (2014) showed in an in vitro study that the initial adhesion to plastic surfaces (e.g., plate wells) after 3-h correlates with the different biofilm phenotypes displayed by clinical C. parapsilosis isolates, which in turn reflect their capacity to form high or low biofilm.
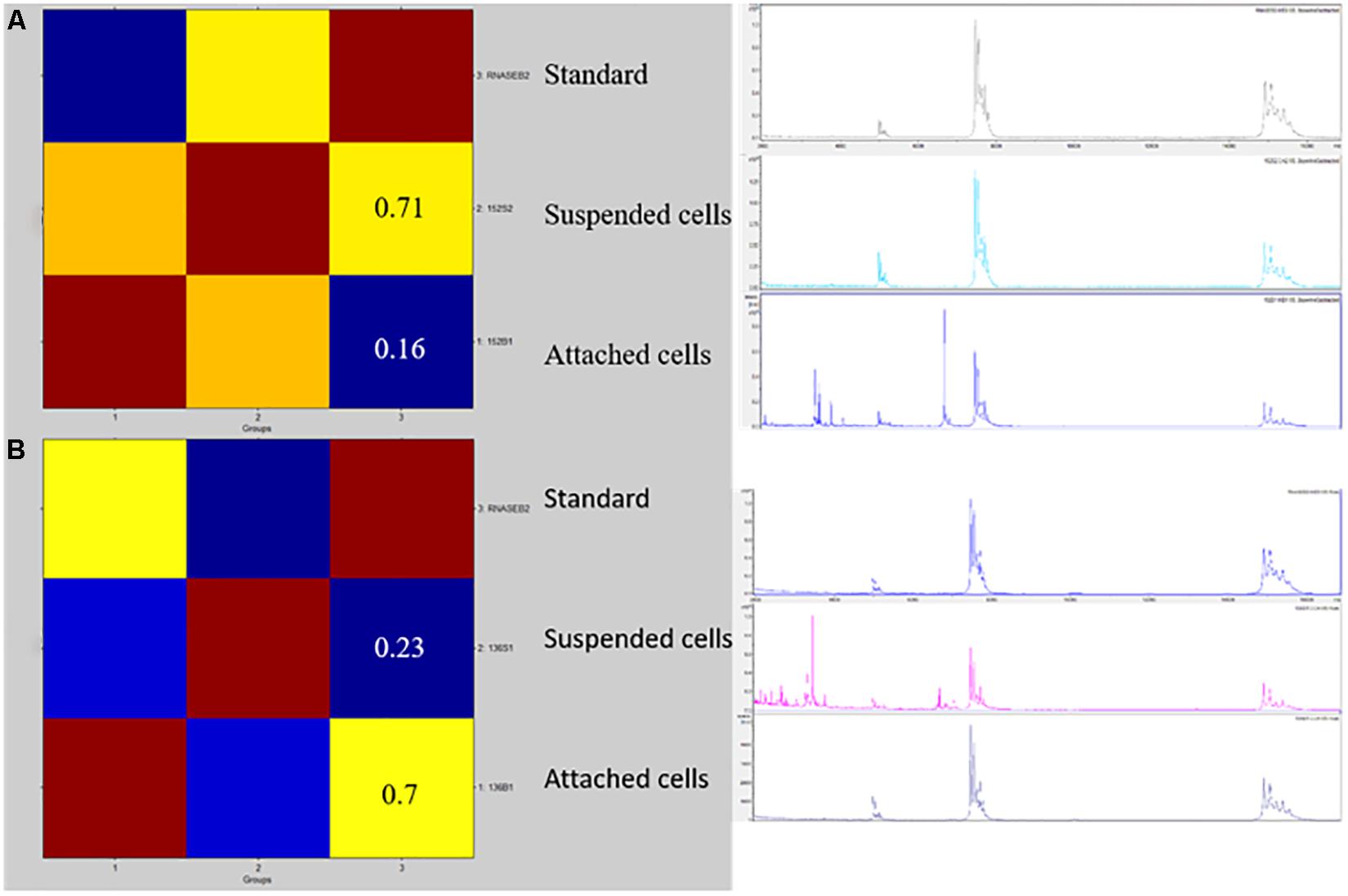
Figure 1. A CCI matrix based visualization for two representative isolates of HP (A) and LP (B) C. parapsilosis. Reddish colors indicate a close relationship between the isolates’ mass spectrum profiles. The CCI values refer to the suspended or attached cell profiles matched against the RNase-B internal standard’s profile, respectively. The right picture shows the row spectra profiles of the internal standard and the suspended or attached isolates’ cells used to create the CCI matrix.
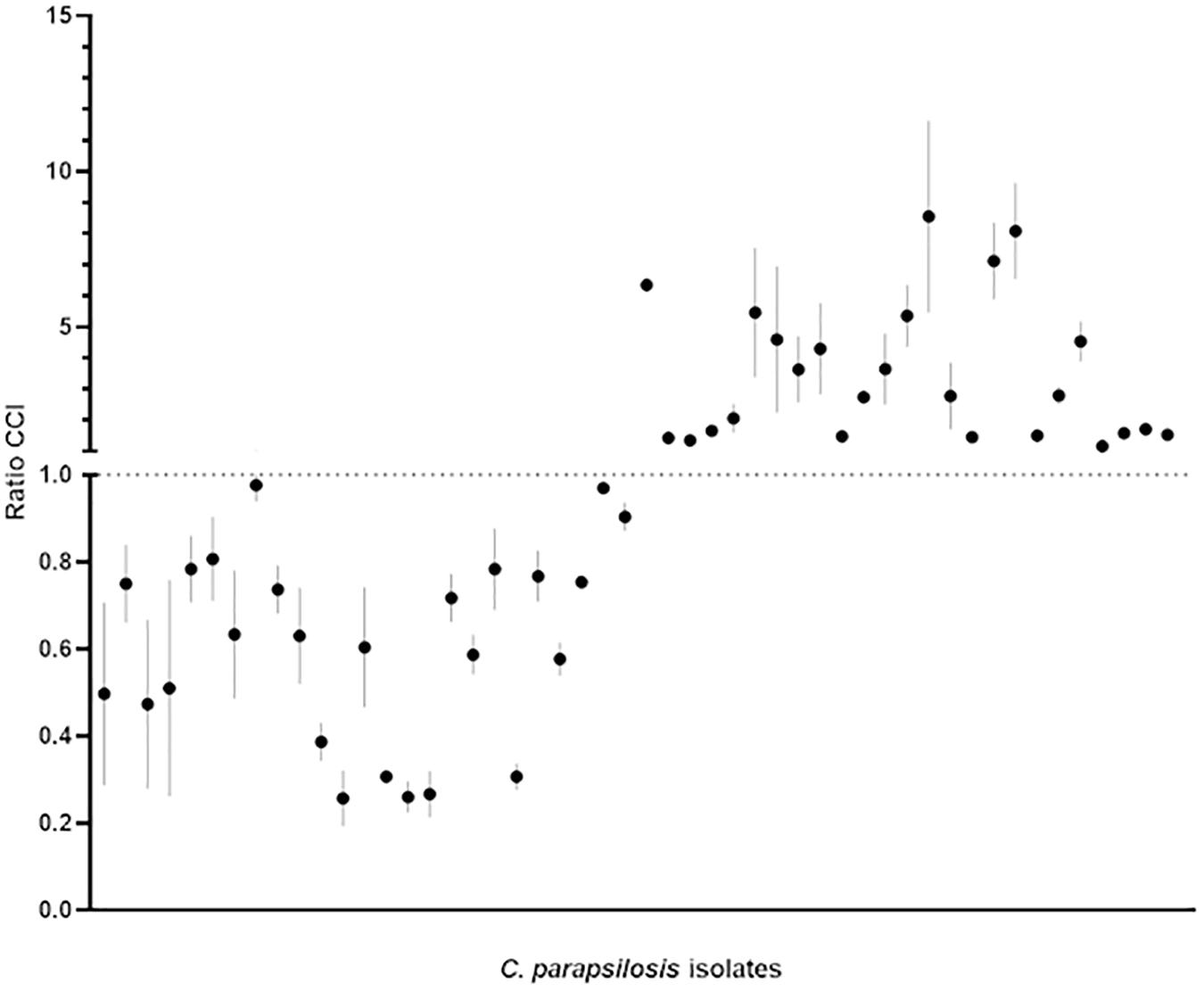
Figure 2. Categorization of the 50 C. parapsilosis isolates (25 HP and 25 LP) included in the study based on the CCIsusp/CCIattach ratio (for details, see Supplementary Table S1). Bars indicate the minimum and maximum CCI ratio obtained for each of the three biological replicates of each isolate, with the mean value marked as a dark dot. According to the discriminatory power of the BIOF–HILO assay, the HP and LP isolates settled in the upper or the lower part of the plot, respectively.
Our findings provide further insights into the clinical diagnostic possibilities of MALDI–TOF MS shown for Candida species (including C. parapsilosis). These possibilities encompass the in vitro assays for fungal identification, susceptibility/resistance detection, and typing, particularly for emerging Candida species (De Carolis et al., 2014a; Dhieb et al., 2015; Vella et al., 2017; Bao et al., 2018; Paul et al., 2018; Vatanshenassan et al., 2018, 2019). Very few studies, to date, tempted to apply the MALDI–TOF MS technology to the direct detection of biofilm-forming isolates of Candida species. Kubesová et al. (2012) used MALDI–TOF MS with intact cells and with sinapinic acid as a MALDI matrix to distinguish rapidly and reliably between two biofilm-negative and two biofilm-positive strains of C. parapsilosis (and Candida metapsilosis). Mlynáriková et al. (2016) used MALDI–TOF MS with ethanol/formic-acid extracted cells to differentiate between 12 biofilm-positive and 9 biofilm-negative C. parapsilosis strains using different MALDI matrices (HCCA, ferulic acid, etc.) and experimental conditions (cultivation times or media). However, the authors failed to group reliably the strains in two clusters within a MALDI–TOF mass spectra-based dendrogram, because mass spectra variations seemed to limit the reproducibility of the method.
We focused on the early development phase of the C. parapsilosis biofilm with MALDI–TOF MS analysis. Furthermore, we employed an automated CCI matrix-based algorithm facilitating the relatedness analysis between the mass spectra from HP or LP C. parapsilosis isolates. This algorithm allowed us to interpret objectively the MALDI–TOF MS analysis results and to achieve a reliable separation between the mass spectra profiles derived from experimental samples such as the attached or suspended cells of C. parapsilosis isolates, as a fingerprint matching technique previously described (De Carolis et al., 2012). We assessed the extent of this separation between the mass spectra of HP or LP C. parapsilosis isolates by quantifying their differences in mass peaks with respect to the RNase-B internal standard. In this context, we chose a molecular mass range analysis of 3000–7000 Da, which excluded the portion of the mass spectrum profile where only RNase-B mass peaks are detectable but allowed to assess the isolates’ mass peaks within the lower range.
MALDI–TOF MS assays are helpful to identify directly microbial organisms from clinical samples (Rodríguez-Sánchez et al., 2019). However, at least for Candida organisms, growing the isolates before the spectral profiles MALDI–TOF MS analysis is still a fundamental step when testing isolates for their capability of biofilm formation. Our assay seems to be particularly promising as it offers the possibility to combine MALDI–TOF MS-based identification with biofilm formation testing. In the latter case, only a short-incubation is required prior to testing; this, coupled with an unbiased capability of classifying the isolates, implies the assay to be advantageous compared with the conventional (Sanguinetti and Posteraro, 2016) or previously developed MALDI–TOF MS-based (Mlynáriková et al., 2016) biofilm detection assays. The rapid and inexpensive protein spectra acquisition using MALDI–TOF MS has led the technology to a standard tool in clinical settings. Currently, MALDI–TOF MS allows obtaining reliable identification and antimicrobial susceptibility testing results for the microbial pathogens within one working shift (Rodríguez-Sánchez et al., 2019). As these results are crucial for timely administration of effective antimicrobial treatment, the simultaneous additional information on the status of biofilm formation by pathogens in a quick manner may therefore enhance the likelihood that the administered antimicrobial treatment will be efficacious.
In conclusion, we ideated the BIOF–HILO assay with the purpose to provide a more rapid identification of C. parapsilosis isolates with high or low capacity of biofilm formation compared to the methods currently used in the clinical mycology laboratory (Sanguinetti and Posteraro, 2016). However, further studies using a larger number of C. parapsilosis isolates are needed, which can validate the present findings and additionally establish whether the BIOF–HILO assay may have in the near future a place in laboratory diagnostics.
Data Availability
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication. In particular, EDC conceived the study, participated in the experimental conduction of the study, and drafted the manuscript. SS, MLR, and FN performed the experiments and interpreted the data. BP and MS participated in the design and coordination of the study, and critically revised the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We wish to thank Franziska Lohmeyer for her English language assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02046/full#supplementary-material
TABLE S1 | Experimental data on MALDI–TOF MS analysis and biofilm formation for the HP or LP C. parapsilosis isolates included in the study.
References
Bao, J. R., Master, R. N., Azad, K. N., Schwab, D. A., Clark, R. B., Jones, R. S., et al. (2018). Rapid, accurate identification of Candida auris by using a novel matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) database (Library). J. Clin. Microbiol. 56:e01700-17. doi: 10.1128/JCM.01700-1717
Bouza, E., Guinea, J., and Guembe, M. (2014). The role of antifungals against Candida biofilm in catheter-related candidemia. Antibiotics 4, 1–17. doi: 10.3390/antibiotics4010001
Cavalheiro, M., and Teixeira, M. C. (2018). Candida biofilms: threats, challenges, and promising strategies. Front. Med. 5:28. doi: 10.3389/fmed.2018.00028
De Carolis, E., Hensgens, L. A. M., Vella, A., Posteraro, B., Sanguinetti, M., Senesi, S., et al. (2014a). Identification and typing of the Candida parapsilosis complex: MALDI-TOF MS vs. AFLP. Med. Mycol. 52, 123–130. doi: 10.1093/mmy/myt009
De Carolis, E., Vella, A., Vaccaro, L., Torelli, R., Posteraro, P., Ricciardi, W., et al. (2014b). Development and validation of an in-house database for matrix-assisted laser desorption ionization-time of flight mass spectrometry-based yeast identification using a fast protein extraction procedure. J. Clin. Microbiol. 52, 1453–1458. doi: 10.1128/JCM.03355-3313
De Carolis, E., Vella, A., Florio, A. R., Posteraro, P., Perlin, D. S., Sanguinetti, M., et al. (2012). Use of matrix-assisted laser desorption ionization-time of flight mass spectrometry for caspofungin susceptibility testing of Candida and Aspergillus species. J. Clin. Microbiol 50, 2479–2483. doi: 10.1128/JCM.00224-12
Dhieb, C., Normand, A. C., Al-Yasiri, M., Chaker, E., El Euch, D., Vranckx, K., et al. (2015). MALDI-TOF typing highlights geographical and fluconazole resistance clusters in Candida glabrata. Med. Mycol. 53, 462–469. doi: 10.1093/mmy/myv013
Finkel, J. S., and Mitchell, A. P. (2011). Genetic control of Candida albicans biofilm development. Nat. Rev. Microbiol. 9, 109–118. doi: 10.1038/nrmicro2475
Guinea, J. (2014). Global trends in the distribution of Candida species causing candidemia. Clin. Microbiol. Infect. 20, 5–10. doi: 10.1111/1469-0691.12539
Kubesová, A., Šalplachta, J., Horká, M., Røužička, F., and Šlais, K. (2012). Candida “psilosis”–electromigration techniques and MALDI-TOF mass spectrometry for phenotypical discrimination. Analyst 137, 1937–1943. doi: 10.1039/c2an15931g
Mlynáriková, K., Šedo, O., Rùžièka, F., Zdráhal, Z., Holá, V., and Mahelová, M. (2016). Evaluation of capacity to detect ability to form biofilm in Candida parapsilosis sensu stricto strains by MALDI-TOF MS. Folia Microbiol. 61, 465–471. doi: 10.1007/s12223-016-0458-457
Pannanusorn, S., Ramírez-Zavala, B., Lünsdorf, H., Agerberth, B., Morschhäuser, J., and Römling, U. (2014). Characterization of biofilm formation and the role of BCR1 in clinical isolates of Candida parapsilosis. Eukaryot. Cell. 13, 438–451. doi: 10.1128/EC.00181-113
Paul, S., Singh, P., Shamanth, A. S., Rudramurthy, S. M., Chakrabarti, A., and Ghosh, A. K. (2018). Rapid detection of fluconazole resistance in Candida tropicalis by MALDI-TOF MS. Med. Mycol. 56, 234–241. doi: 10.1093/mmy/myx042
Rajendran, R., Sherry, L., Nile, C. J., Sherriff, A., Johnson, E. M., Hanson, M. F., et al. (2016). Biofilm formation is a risk factor for mortality in patients with Candida albicans bloodstream infection-Scotland, 2012-2013. Clin. Microbiol. Infect. 22, 87–93. doi: 10.1016/j.cmi.2015.09.018
Ramage, G., Rajendran, R., Sherry, L., and Williams, C. (2012). Fungal biofilm resistance. Int. J. Microbiol. 2012:528521. doi: 10.1155/2012/528521
Rodríguez-Sánchez, B., Cercenado, E., Coste, A. T., and Greub, G. (2019). Review of the impact of MALDI-TOF MS in public health and hospital hygiene, 2018. Euro Surveill. 24:1800193. doi: 10.2807/1560-7917.ES.2019.24.4.1800193
Sanguinetti, M., and Posteraro, B. (2016). Diagnostic of fungal infections related to biofilms. Adv. Exp. Med. Biol. 931, 63–82. doi: 10.1007/5584_2016_9
Silva, S., Rodrigues, C. F., Araújo, D., Rodrigues, M. E., and Henriques, M. (2017). Candida species biofilms’ antifungal resistance. J. Fungi. 3:E8. doi: 10.3390/jof3010008
Soldini, S., Posteraro, B., Vella, A., De Carolis, E., Borghi, E., Falleni, M., et al. (2018). Microbiologic and clinical characteristics of biofilm-forming Candida parapsilosis isolates associated with fungaemia and their impact on mortality. Clin. Microbiol. Infect. 24, 771–777. doi: 10.1016/j.cmi.2017.11.005
Taff, H. T., Mitchell, K. F., Edward, J. A., and Andes, D. R. (2013). Mechanisms of Candida biofilm drug resistance. Future Microbiol. 8, 1325–1337. doi: 10.2217/fmb.13.101
Tumbarello, M., Posteraro, B., Trecarichi, E. M., Fiori, B., Rossi, M., Porta, R., et al. (2007). Biofilm production by Candida species and inadequate antifungal therapy as predictors of mortality for patients with candidemia. J. Clin. Microbiol. 45, 1843–1850. doi: 10.1128/JCM.00131-137
Vatanshenassan, M., Boekhout, T., Lass-Flörl, C., Lackner, M., Schubert, S., Kostrzewa, M., et al. (2018). Proof of concept for MBT ASTRA, a rapid matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS)-based method to detect caspofungin resistance in Candida albicans and Candida glabrata. J. Clin. Microbiol. 56:e00420-18. doi: 10.1128/JCM.00420-418
Vatanshenassan, M., Boekhout, T., Meis, J. F., Berman, J., Chowdhary, A., Ben-Ami, R., et al. (2019). Candida auris identification and rapid antifungal susceptibility testing against echinocandins by MALDI-TOF MS. Front. Cell. Infect. Microbiol. 9:20. doi: 10.3389/fcimb.2019.00020
Vella, A., De Carolis, E., Mello, E., Perlin, D. S., Sanglard, D., Sanguinetti, M., et al. (2017). Potential use of MALDI-ToF mass spectrometry for rapid detection of antifungal resistance in the human pathogen Candida glabrata. Sci. Rep. 7:9099. doi: 10.1038/s41598-017-09329-9324
Keywords: MALDI-based assay, Candida parapsilosis, biofilm formation, candidemia, composite correlation index
Citation: De Carolis E, Soldini S, La Rosa M, Nucci F, Posteraro B and Sanguinetti M (2019) BIOF–HILO Assay: A New MALDI–TOF Mass Spectrometry Based Method for Discriminating Between High- and Low-Biofilm-Producing Candida parapsilosis Isolates. Front. Microbiol. 10:2046. doi: 10.3389/fmicb.2019.02046
Received: 06 June 2019; Accepted: 20 August 2019;
Published: 30 August 2019.
Edited by:
Armand Paauw, Netherlands Organisation for Applied Scientific Research (TNO), NetherlandsReviewed by:
Macit Ilkit, Çukurova University, TurkeyAleksandra Barac, University of Belgrade, Serbia
Copyright © 2019 De Carolis, Soldini, La Rosa, Nucci, Posteraro and Sanguinetti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maurizio Sanguinetti, bWF1cml6aW8uc2FuZ3VpbmV0dGlAdW5pY2F0dC5pdA==; bWF1cml6aW8uc2FuZ3VpbmV0dGlAcG9saWNsaW5pY29nZW1lbGxpLml0
 Elena De Carolis
Elena De Carolis Silvia Soldini1
Silvia Soldini1 Marilisa La Rosa
Marilisa La Rosa Maurizio Sanguinetti
Maurizio Sanguinetti