
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 11 September 2019
Sec. Plant Pathogen Interactions
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.02005
The GacS/GacA two-component system is essential for virulence in many plant pathogenic bacteria, and thus represents a promising anti-virulence target. In the present study, we isolated and screened rhizobacteria that were capable of inhibiting the expression of the gacS gene in the phytopathogenic bacterium Pseudomonas syringae pv. tomato (Pto) DC3000. One candidate inhibitor bacterium, BR3 was obtained and identified as a Bacillus sp. strain based on 16s rRNA gene sequence analysis. Besides the gacS gene, the GacA-dependent small RNA genes rsmZ and rsmY were repressed transcriptionally when DC3000 was treated with an extract from strain BR3. Importantly, the extract also influenced bacterial motility, the expression of type three secretion system effector AvrPto, and the plant hypersensitive response triggered by strain DC3000. The results suggested that the extract from strain BR3 might offer an alternative method to control bacterial diseases in plants by targeting the GacS/GacA system.
To thrive and survive in complex environmental conditions, bacteria employ versatile signal transduction pathways to rapidly adapt to alterations in their surroundings. One of these pathways is the GacS/GacA two-component system, which detects and responds coordinately to external and internal stimuli, including different physiological state (Kay et al., 2005), metabolic levels (Chavez et al., 2010), and pH (Mondragón et al., 2006), and translates them into appropriate adaptive responses.
The GacS/GacA system is a conserved global regulatory system in many Gram-negative bacteria, which is comprised of the hybrid sensor kinase GacS and the cognate response regulator GacA. In the presence of an as-yet-unidentified environmental signal, the GacS sensor kinase autophosphorylates and then activates the cognate GacA response regulator via phosphotransfer (Haas and Défago, 2005). In Pseudomonas aeruginosa and Legionella pneumophila, the activated GacA exclusively regulates the expression of several small non-coding RNAs (sRNAs) RsmY and RsmZ (Brencic et al., 2009; Sahr et al., 2009). Mutation of all these sRNAs in many bacteria resulted in the same phenotypes as mutants of the GacS/GacA system (Kay et al., 2005; Brencic et al., 2009). GacA-activated sRNAs all have multiple GGA motifs for competitive binding to carbon storage regulator (CsrA)/regulator of secondary metabolites (RsmA) family proteins (Valverde et al., 2004). In addition, the CsrA/RsmA family proteins mediate either a negative or positive posttranscriptional effect by altering the rate of translation initiation or mRNA decay (Lapouge et al., 2008). A common consensus sequence (CANGGAYG) within the loop portion of a stem–loop structure in the 5′-untranslated region (UTR) is essential for RsmA/CsrA family proteins to bind target mRNAs (Kulkarni et al., 2014). Two histidine kinases, LadS and RetS, are involved in modulating the function of the GacS/GacA system. In P. aeruginosa PAK, RetS affects the phosphorylation state of GacS (Goodman et al., 2009). Crystallographic studies further indicated that RetS used the reversible unfolding of a helix, or helix cracking, to control interactions with GacS (Mancl et al., 2019). In contrast to RetS, LadS in P. aeruginosa activates the function of GacA under high calcium conditions (Broder et al., 2016).
Extensive studies have demonstrated that the GacS/GacA system and its homologs play an important role in coordinating the expression of virulence factors required for successful infection of many plant- and animal-pathogenic bacteria (Heeb and Haas, 2001). In P. syringae pv. tomato (Pto) DC3000 (hereafter termed Pto DC3000), GacA acts as master regulator to control carbon metabolism, motility, and production of virulence factors, syringomycin, and quorum-sensing (QS) signals (Chatterjee et al., 2003). Furthermore, GacA positively regulates the transcription of the pel, peh, and celV genes that are responsible for the production of pectate lyases, pectinases, and cellulases in Pectobacterium carotovorum subsp. carotovorum (Pcc), and mutation of gacA results in an avirulent phenotype (Cui et al., 2001). Production of these exoenzymes was under the control of ExpI-ExpR QS system (Pirhonen et al., 1993) and the QS system was positively regulated by the GacS/GacA system (Whitehead et al., 2002). The opportunistic pathogen P. aeruginosa caused extensive tissue damage on Arabidopsis and lettuce when infiltrated at high cell densities, while the gacA mutant sharply reduced the disease symptoms (Parkins et al., 2001). Moreover, the gacA or gacS mutants of P. aeruginosa are also much less virulent in several animal models compared with their wild-type (Pessi and Haas, 2001). In addition, the GacA homologs in human pathogens Salmonella enterica serovar Typhimurium and Vibrio cholerae act as key regulators of colonization, toxin production, and intracellular multiplication (Wong et al., 1998; Ahmer et al., 1999).
Hence, the GacS/GacA system represents a promising target for anti-infection drug development. Although the signaling circuit is well defined, little is known about the environmental signals that turn on the Gac/Rsm regulatory cascades. Short-chain fatty acids have been shown to induce the homologous systems in Escherichia coli and Salmonella typhimurium (Lawhon et al., 2002; Gonzalez Chavez et al., 2010). Bacterial culture supernatants and lysed kin cells could act as signals that are sensed by the GacS/GacA–CsrA/RsmA pathway in P. aeruginosa (Kay et al., 2005; LeRous et al., 2015). In addition, plant phenolic derivatives and the antibiotic azithromycin impaired the production of virulence factors in P. aeruginosa via the GacS/GacA system (Pérez-Martínez and Haas, 2011; Yamazaki et al., 2012).
In the present study, a Pto DC3000 (p970Gm-gacSDC3000p) transcriptional fusion reporter was developed to screen inhibitors of the GacS/GacA system from secondary metabolites produced by rhizobacteria. The extract of Bacillus sp. BR3 significantly repressed gacS expression and reduced the GacS protein level, and impaired GacA-dependent expression of small RNAs, motility, and the hypersensitive response (HR) triggered by Pto DC3000. These results contributed to our understanding of interspecies cell-to-cell communication in bacteria, and provided an additional method by which rhizobacteria might attenuate virulence factor production by plant and animal pathogenic bacteria.
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli was routinely grown in Luria-Bertani (LB) medium at 37°C. Pseudomonas syringae pv. tomato DC3000, P. carotovorum subsp. carotovorum (Pcc) Z3-3, Pseudomonas fluorescens 2P24, and Agrobacterium tumefaciens NTL4 (pZLR4) were cultured in LB medium, King’s Broth (KB) (King et al., 1954), or minimal medium ABM (Chilton et al., 1974) at 28°C. Type three secretion system (TTSS)-inducing minimal medium was used for immunoblotting analysis of AvrPto protein (Huynh et al., 1989). When necessary, growth media were supplemented with ampicillin (Ap) (50 μg ml–1), kanamycin (Km) (50 μg ml–1), gentamycin (Gm) (5 μg ml–1), tetracycline (Tet) (20 μg ml–1), or 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal) (40 μg ml–1).
Chromosomal DNA from strains Pto DC3000 and Bacillus sp. BR3, plasmid DNA extraction, and other molecular assays were performed according to standard procedures (Sambrook et al., 1989). Electroporation of Pto DC3000 and P. fluorescens cells with plasmid DNA was performed as described previously (Choi et al., 2006). The oligonucleotides used are listed in Table 1.
To ascertain its taxonomic position, the genomic DNA of strain BR3 was used as a templet for PCR amplification using primers 63F and 1387R (Marchesi et al., 1998). The PCR product was sequenced by the Sanger method at Sangon Biotech Co., Ltd., Shanghai, China. The obtained sequences were compared with sequences deposited in GenBank using a BLASTN algorithm (Altschul et al., 1997).
To construct the reporter strain Pto DC3000 (p970Gm-gacSDC3000p), the gacS promoter region from Pto DC3000 was PCR amplified using the primers gacS-promoterF/gacS-promoterR. The amplicon was then inserted into the BamHI site of plasmid pRG970Gm to create the gacS–lacZ transcriptional fusion plasmid p970Gm-gacSDC3000p, which was introduced into Pto DC3000 via electroporation (Smith and Iglewski, 1989). In the same way, the promoter regions of rsmY and rsmZ from Pto DC3000 were amplified by PCR using primer pairs RsmYBglII5p/RsmYBglII3p and rsmZp1DC3000/rsmZp2DC3000, respectively. The products were then cloned into plasmid pRG970Gm to obtain p970Gm-rsmYDC3000p and p970Gm-rsmZDC3000p. These plasmids were then introduced into appropriate Pto strains by electroporation.
Plasmid pME-gacS expressing the gacS gene under the control of the isopropyl β-D-thiogalactoside (IPTG)-inducible tac promoter (Ptac) was constructed as follows. The gacS gene was amplified by PCR with the primers gacS-XhoI/gacS-EcoRI and cloned into pME6032, yielding pME-gacS. When required, 0.25 and 0.5 mM of IPTG were added to induce the Ptac promoter of the plasmid pME6032, respectively.
Soil samples were collected from various regions in Guangxi, China, and rhizobacteria were isolated using LB plates. The isolates were cultured in 5 ml of seed broth at 28°C for 7 days (Ling et al., 2015). One milliliter of the cultures was extracted using an equal volume of ethyl acetate. The ethyl acetate extracts were evaporated and dissolved in 200 μl of 100% dimethyl sulfoxide (DMSO). A portion (5 μl) of the extract was mixed with 1 ml of bacterial suspension of the reporter strain Pto DC3000 (p970Gm-gacSDC3000p). After 7 h of incubation at 28°C, the β-galactosidase activity was assayed using the Miller (1972) method.
Pto DC3000 and its gacA mutant were cultured overnight in KB liquid medium. Then, 5 μl of the stationary phase cultures was spotted onto SWM agar medium (0.4% agar) with BR3 extract (at final a concentration of 16 or 32 μg ml–1). The plates were incubated overnight at 28°C and then imaged (Chatterjee et al., 2003).
The procedures for HR testing in tobacco leaves were previously published (Cui et al., 1996). Young, fully expanded third and fourth leaves from approximately 8-week-old Nicotiana tabacum L. cv. Samsun were used for the HR test. Bacterial cells [1 × 107 colony forming units (CFU) ml–1] in the absence or presence of the BR3 extract were infiltrated into tobacco leaves as indicated in the figure captions. Images were acquired at 20 h after infiltration. DMSO was infiltrated as a control.
To construct a C-terminal vesicular stomatitis virus glycoprotein (VSV-G) epitope-GacS fusion, a PCR-generated fragment with the sequence 5′-TATACAGATATTGAAATGAATAGATTAG-3′ was inserted in-frame before the stop codon of gacS and cloned into p2P24Km (Zhang et al., 2018). The resulting plasmid was introduced into strain Pto DC3000 by electroporation, and the wild-type copy was replaced by the modified version after two recombination events under high-sucrose stress. Substitution was confirmed by sequencing.
To analyze the effect of the BR3 extract on the protein levels of GacS and the TTSS effector AvrPto, Pto DC3000 and its derivatives were cultured in TTSS-inducing minimal medium at 20°C for 16 h. Cell lysates were prepared by sonication and resuspended in SDS sample buffer. The samples were separated by SDS–PAGE and protein levels were analyzed using western blotting with anti-VSV antibodies (Sangon Biotech) or anti-AvrPto antibodies (Dijk et al., 1999).
Pseudomonas fluorescens 2P24 and its derivative strains were cultured in 20 ml of KBG (KB broth supplemented with 2% glucose) as described above. The antibiotic 2,4-Diacetylphloroglucinol (DAPG) was extracted from the culture supernatant and assayed using a previously described HPLC method (Shanahan et al., 1992).
Cultures of P. carotovorum subsp. carotovorum (Pcc) Z3-3 were grown overnight in LB medium with DMSO or the extract of strain BR3. One milliliter of the cultures was extracted with an equal volume of ethyl acetate. The ethyl acetate extracts were dried and resuspended in 50 μl of methanol. A portion (200 μl) of A. tumefaciens NTL4 (pZLR4) (OD600 = 0.8) with 5 μl of the extract solution was incubated at 28°C for 3 h, and then β-galactosidase activities were quantified (Miller, 1972).
Arabidopsis plants were grown in a growth room at 23°C and 70% relative humidity under a 14-h light cycle. Strain Pto DC3000 was grown on KB agar overnight at 28°C and resuspended in water with the extract from strain BR3 or DMSO at 105 CFU μl–1, 20 μl of the suspension was infiltrated into 4-week-old Arabidopsis leaves. Bacterial populations were determined at day 0 and day 3. Three 1-cm-diameter leaf disks were collected from three independent plants, and ground in 1 ml of 10 mM MgCl2. Bacterial colonies were then counted 2 days after plating 10 μl from serial dilutions on KB plates. For the pathogenicity test in Chinese cabbage, Pcc Z3-3 was grown in LB broth at 28°C for 12 h. Cells were harvested and resuspended in phosphate-buffered saline (PBS) to 108 CFU ml–1 with or without the BR3 extract. For each Chinese cabbage leaf, three wounds were punched and inoculated with 10 μl of bacterial suspension. Maceration symptoms were documented at 30 h post-inoculation, as described previously (Li et al., 2010).
All experiments were performed in triplicate. The data were analyzed and compared by performing two-sample independent t-tests using DPS v9.501.
Extracts from 5000 isolates obtained from soil were tested for their abilities to suppress the β-galactosidase activity of the gacS–lacZ transcriptional fusion in the strain Pto DC3000 that carrying reporter plasmid p970Gm-gacSDC3000p. The extract from a strain named BR3 showed significant inhibitory activity (Figure 1A). Analysis of the 16S rRNA gene sequence of BR3 indicated that this organism belongs to Bacillus sp. (data not shown). Introduction of pME-gacS into the wild-type DC3000 improved the promoter activities of the gacS gene (Figure 1A). However, the promoter activities of gacS were considerably induced in the extract-treated wild-type DC3000 with plasmid pME-gacS, indicating that the expression of gacS gene was repressed by the BR3 extract (Figure 1A). To further verify the effect of the BR3 extract on the expression of the gacS gene, the level of GacS protein was determined. Western blot analysis of the chromosomal gacS-vsv fusion strain showed that GacS protein production was significantly decreased in cells treated with the BR3 extract (Figure 1B). Our data also showed that the BR3 extract had no effect on the growth of Pto DC3000 (Figure 1C and Supplementary Figure S1). Taken together, these results suggested that the BR3 extract suppressed the expression of gacS gene.
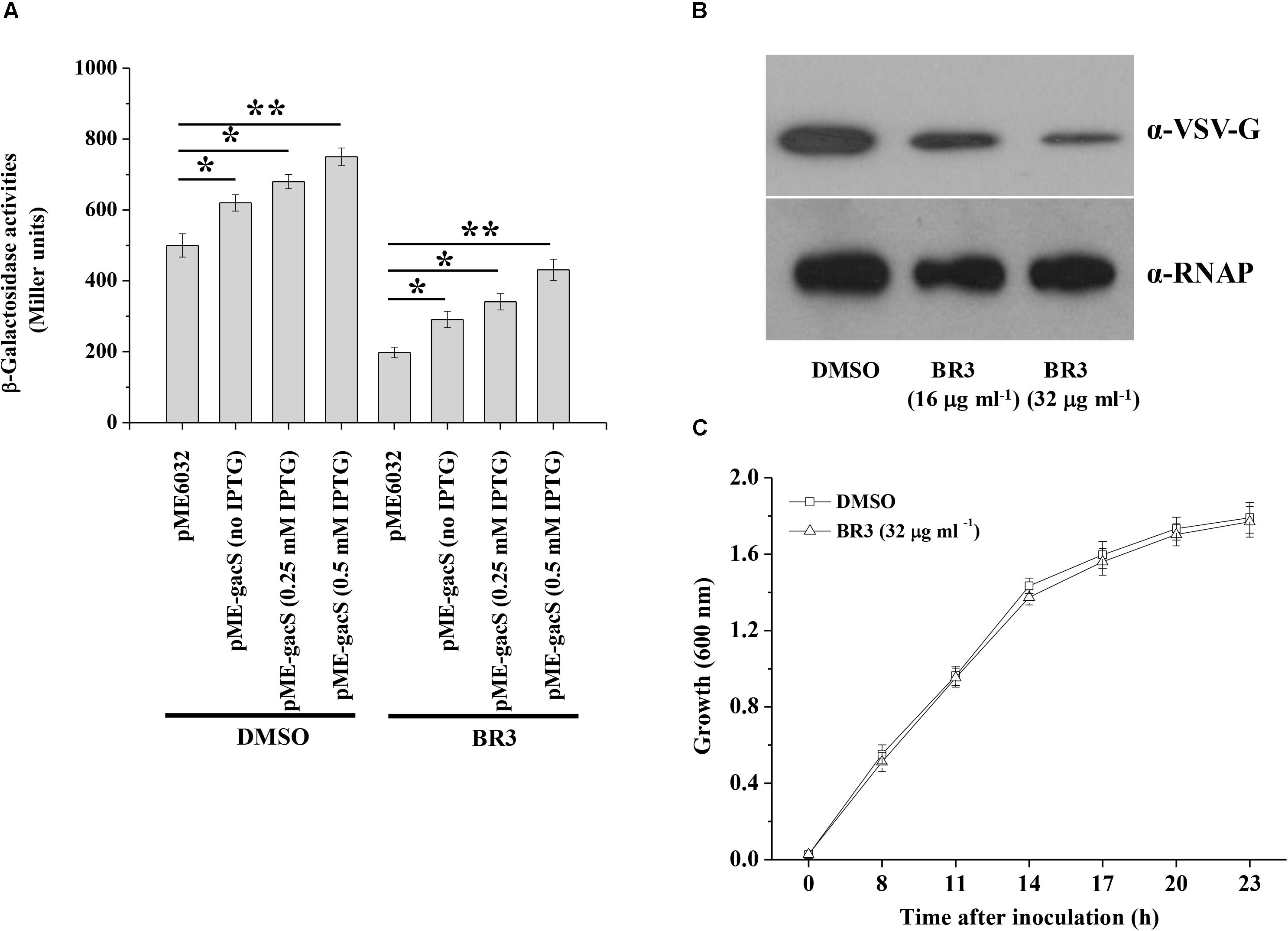
Figure 1. An extract of strain BR3 inhibits the expression of gacS and the GacS protein levels. (A) β-Galactosidase activities of the reporter fusion gacS–lacZ in Pto DC3000 (pME6032) and Pto DC3000 (pME-gacS) with DMSO or the BR3 extract (32 μg ml–1) were measured at 14 h after inoculation into KB medium. Different concentration of IPTG was added to induce the Ptac promoter of the pME6032 as indicated. The experiments were performed in triplicate; average values ± standard deviations are shown. ∗P < 0.05 and ∗∗P < 0.01. (B) Western blotting analysis of GacS-VSV in the absence or presence of the BR3 extract. An antibody directed against β subunit of RNA polymerase (RNAP) was used as a loading control in this and later blots. There independent experiments were performed; a representative blot is shown. (C) The extract of strain BR3 (32 μg ml–1) did not influence the growth of Pto DC3000. The experiments were performed in triplicate; average values ± standard deviations are shown.
RsmZ-like non-coding sRNAs are regulated by the GacS/GacA system (Kay et al., 2005; Moll et al., 2010); therefore, the expression of the rsmZ and rsmY genes was further analyzed in the presence or absence of the BR3 extract. The β-galactosidase activities of the rsmZ–lacZ fusion or the rsmY–lacZ fusion in wild-type Pto DC3000 were significantly higher than those in gacA mutants, suggesting that GacA positively regulated the rsmY and rsmZ genes expression (Figure 2). Addition of the BR3 extract (16 μg ml–1) reduced the expression of the rsmZ–lacZ and rsmY–lacZ fusions in the wild-type Pto DC3000 compared with that in the DMSO-treated control (Figure 2). Interestingly, the BR3 extract had no effect on the expression of rsmZ–lacZ and rsmY–lacZ fusions in the gacA mutant (Figure 2). These data indicated that the BR3 extract affected the expression of rsmZ and rsmY via the GacS/GacA system.
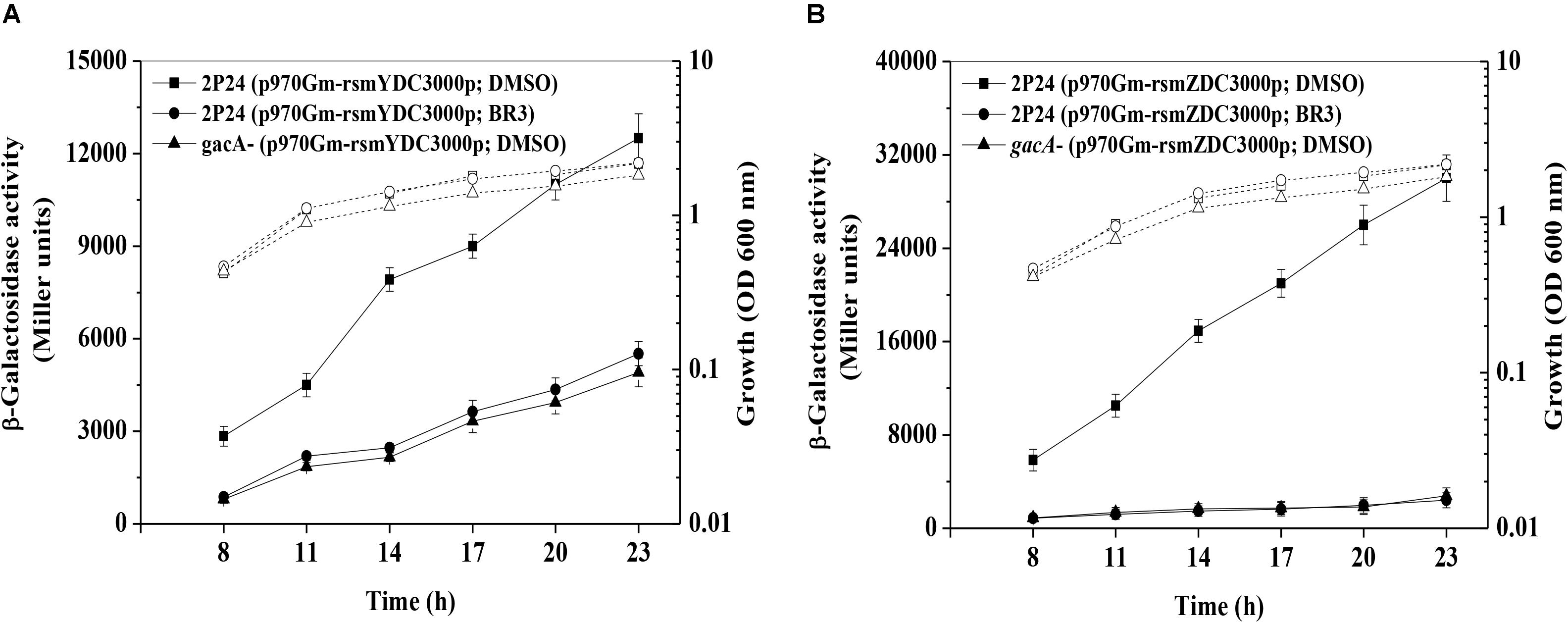
Figure 2. Regulation of rsmZ (A) and rsmY (B) expression in the presence or absence of the BR3 extract. β-Galactosidase activities of the reporter fusions rsmZ–lacZ (A) or rsmY–lacZ (B) in Pto strains with DMSO or the BR3 extract (16 μg ml–1) were measured at various time points tested after inoculation into KB medium. Growth is indicated by the dotted line. The experiments were performed in triplicate; average values ± standard deviations are indicated.
Previous studies indicated that the swarming motility of the gacA mutant was much reduced compared with that of wild-type Pto DC3000 (Chatterjee et al., 2003). Therefore, we checked whether the BR3 extract influenced the swarming motility of Pto DC3000. As expected, when treated with the extract, the wild-type displayed a swarming deficiency resembling that of its gacA mutant. While the swarming motility of the gacA mutant was not further effected upon the treatment of the BR3 extract (Figure 3A).
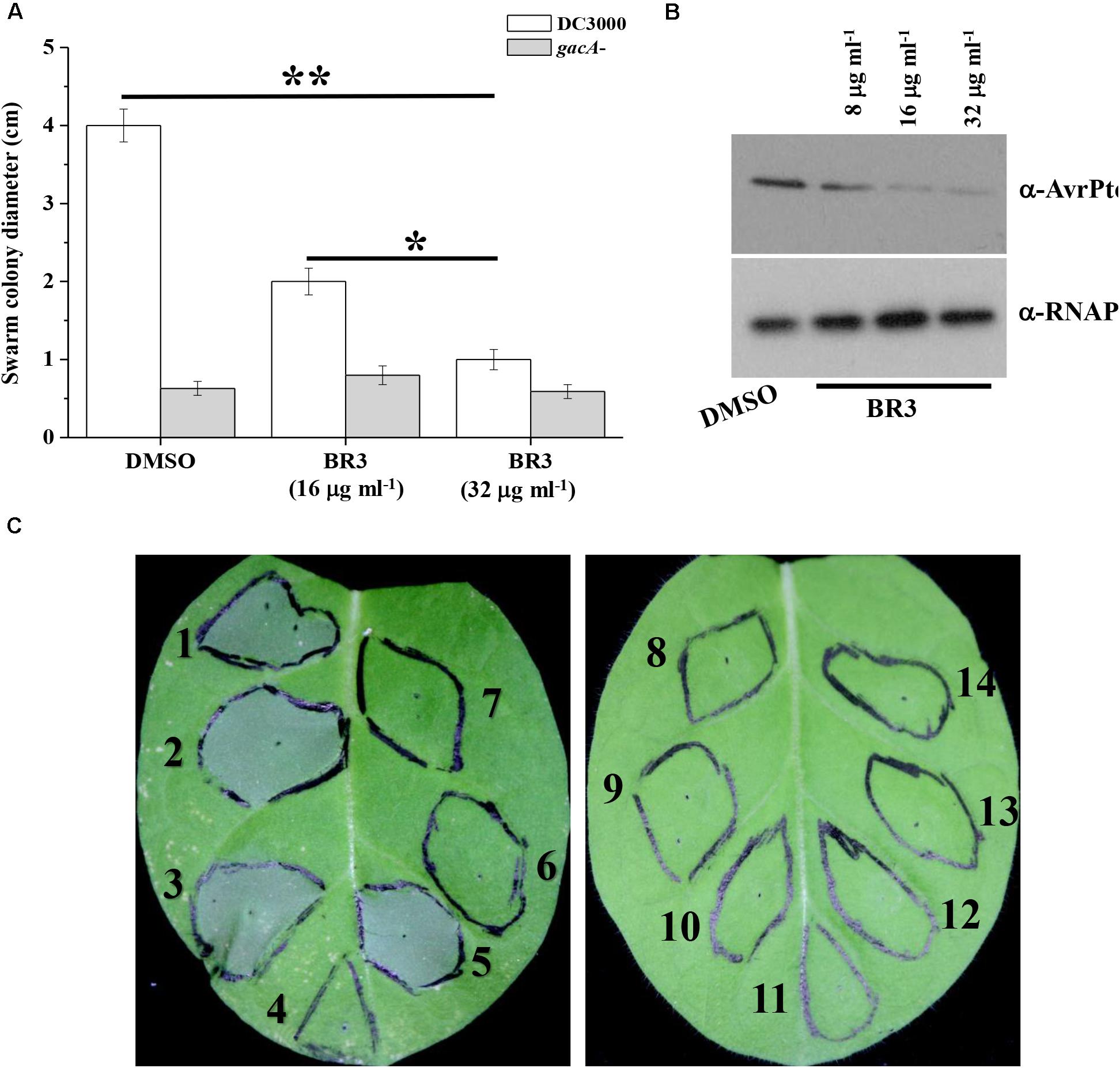
Figure 3. Influence of the BR3 extract on swarming motility, AvrPto production, and HR. (A) Swarming ability was tested on semisolid medium after 12 h of incubation at 28°C for Pto DC3000 in the absence or presence of the BR3 extract. Statistical significance was calculated using t-tests, ∗P < 0.05 and ∗∗P < 0.01. (B) Immunoblotting analysis with anti-AvrPto antibodies of strain Pto DC3000 in the absence or presence of the BR3 extract. (C) Effect of the BR3 extract on the elicitation of the hypersensitive response by Pto DC3000 in tobacco leaves. (1) DC3000 with 0 μl of DMSO; (2) DC3000 with 0 μl of the BR3 extract; (3) DC3000 with 5 μl of DMSO; (4) DC3000 with 5 μl of the BR3 extract (16 μg ml–1); (5) DC3000 with 10 μl of DMSO; (6) DC3000 with 10 μl of the BR3 extract (16 μg ml–1); (7) the hrcQ-U mutant with 0 μl of DMSO; (8) the hrcQ-U mutant with 0 μl of the BR3 extract; (9) the hrcQ-U mutant with 5 μl of DMSO; (10) the hrcQ-U mutant with 5 μl of the BR3 extract (16 μg ml–1); (11) the hrcQ-U mutant with 10 μl of DMSO; (12) the hrcQ-U mutant with 10 μl of the BR3 extract (16 μg ml–1); (13) 10 μl of DMSO; and (14) 10 μl of the BR3 extract (16 μg ml–1).
GacA regulates the expression of hrpL a master regulator of TTSS (Chatterjee et al., 2003); therefore, we further analyzed the protein level of the TTSS effector AvrPto using western blotting and observed a significant reduction in the level AvrPto in the extract-treated wild-type (Figure 3B). Pto DC3000 could elicit a typical HR in tobacco leaves when infiltrated at 1 × 107 CFU ml–1 of bacterial cells, whereas the BR3 extract-treated Pto DC3000 failed to trigger HR at the same cell concentration (Figure 3C). In addition, the hrcQ-U mutant was also incapable of showing HR with or without the BR3 extract (Figure 3C). Pto DC3000-triggered HR on tobacco leaves is TTSS-dependent, these data suggested that the BR3 extract possible impaired the function of the TTSS in Pto DC3000.
The GacS/Rsm system is conserved in many Gram-negative bacteria, and globally regulates the production of a large number of secondary metabolites, including antibiotic 2,4-DAPG in Pseudomonas spp. (Laville et al., 1992). We measured the influence of the BR3 extract on the expression of the sRNA gene rsmY2P24 and 2,4-DAPG biosynthetic gene phlA in P. fluorescens 2P24. The results showed that the BR3 extract reduced rsmY2P24 transcription and strongly inhibited the expression of the phlA–lacZ translational fusion (Figure 4). 2,4-DAPG production in the extract-treated cells reduced by approximately twofold (Figure 4C). In addition, the expression of rsmY2P24 and phlA was significantly decreased in the gacA2P24 mutant, but no further decrease was found when treated with the BR3 extract (Figures 4A,B). The expression of a housekeeping gene proC, which encodes the constitutive delta 1-pyrroline 5-carboxylate reductase (the third enzyme of proline biosynthesis), was not affected by the addition of the BR3 extract (Figure 4D; Savioz et al., 1993). In conclusion, our data suggested that the BR3 extract strongly inhibited the genes of the GacS/GacA regulon in P. fluorescens 2P24, and this inhibitory effect is GacA-dependent.
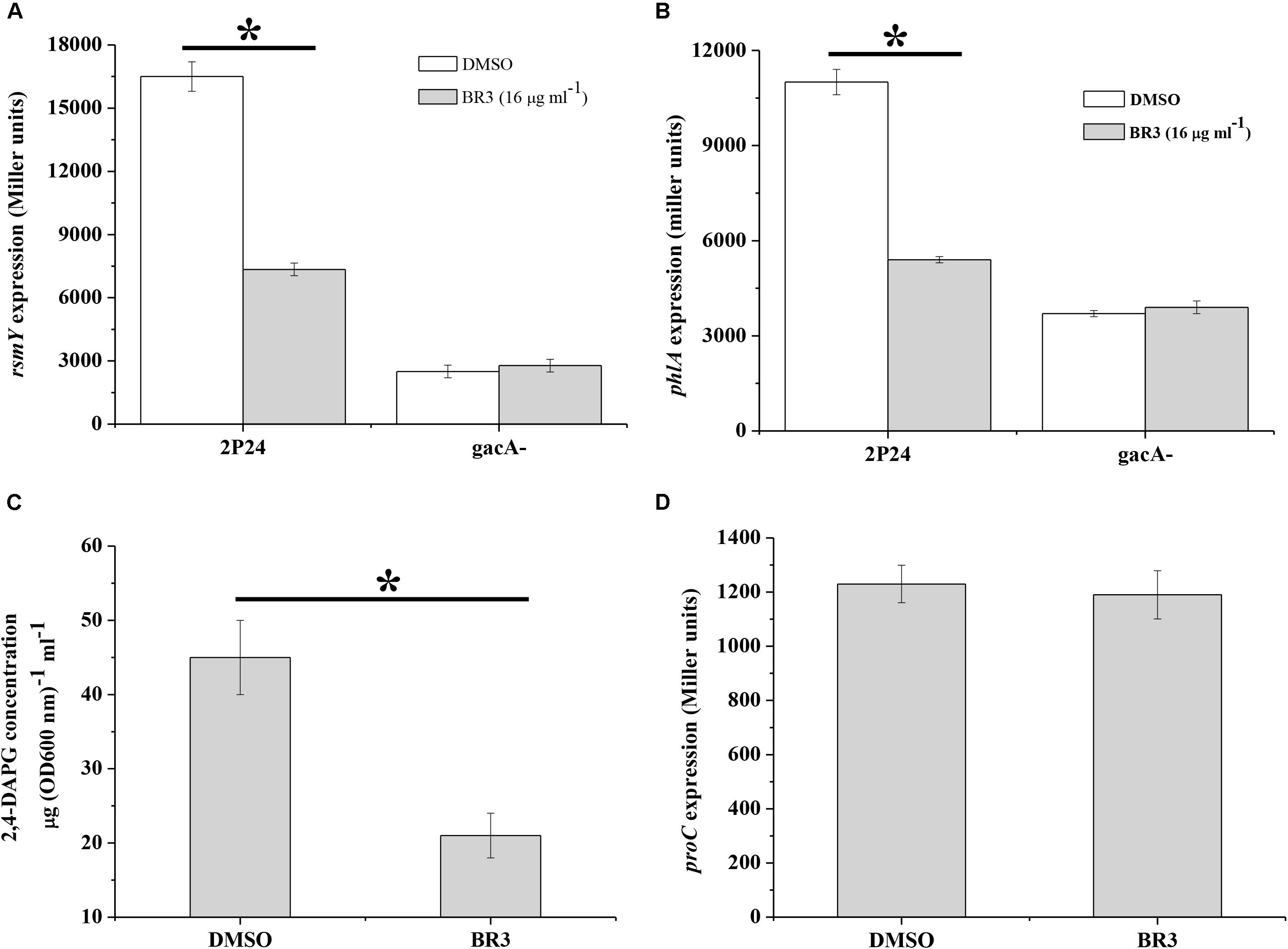
Figure 4. Regulation of rsmY, phlA, and proC of strain 2P24 in the presence or absence of the BR3 extract. The β-galactosidase activities of transcriptional rsmY–lacZ (A), translational phlA–lacZ (B), and proC–lacZ (D) fusions were determined in P. fluorescens 2P24 in the absence or presence of the BR3 extract. Biosynthesis of 2,4-DAPG of strain 2P24 in the absence or presence of the BR3 extract was assayed by HPLC (C). All experiments were performed in triplicate, and the mean values ± standard deviations are indicated, ∗P < 0.05.
In phytopathogenic bacteria, the Gac/Rsm system is associated with pathogenicity. The observation that the BR3 extract suppressed the activity of the Gac/Rsm system prompted us to determine whether it interfered with the GacS/GacA system-dependent virulence process. As predicted, the extract-treated Pto DC3000 displayed a significantly reduced in planta population compared with that of the Pto DC3000 strain treated with DMSO (Figure 5A).
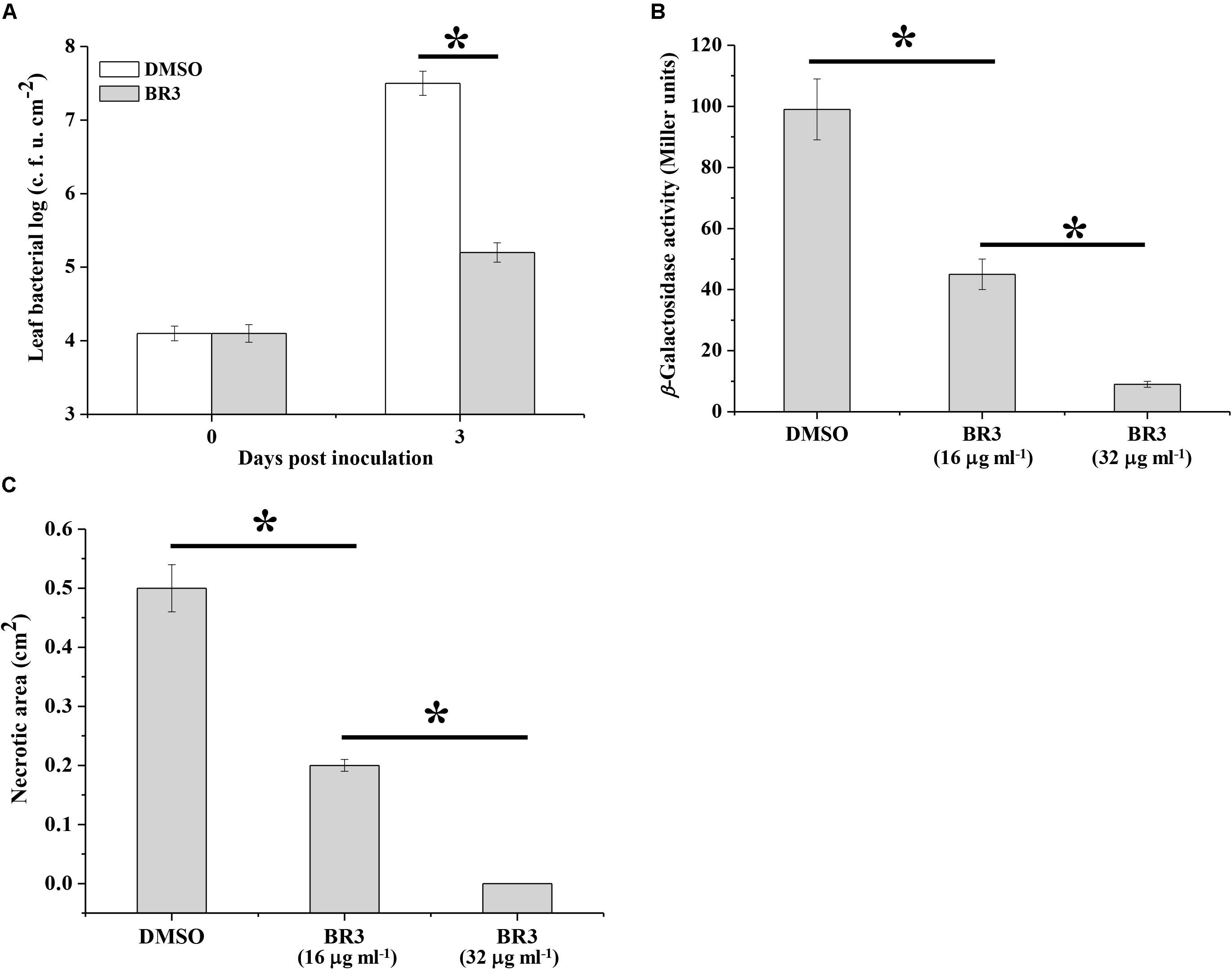
Figure 5. Effect of the extract of strain BR3 on the virulence function of Pto DC3000 and Pcc Z3-3. (A) Strain DC3000 in the absence or presence of the extract (16 μg ml–1) from strain BR3 was infiltrated into Arabidopsis leaves, and bacterial growth was determined. (B) Regulation by the BR3 extract on the production of AHL in Pcc Z3-3. The β-galactosidase activities of the traG–lacZ fusion in A. tumefaciens NTL4 (pZLR4) were measured via AHL signals that were extracted from strain Z3-3 in the absence or presence of the extract from strain BR3. (C) Disease lesions on Chinese cabbage caused by strain Z3-3 in the absence or presence of the BR3 extract. All experiments were performed in triplicate, and the mean values ± standard deviations are indicated, ∗P < 0.05.
Previous data indicated that the GacS/GacA system positively regulates the ExpI/ExpR QS system, which participates in the regulatory pathway for exoenzyme production in Pcc (Cui et al., 2001; Whitehead et al., 2002). Therefore, we further examined the influence of the BR3 extract on the pathogenicity of Pcc toward Chinese cabbage. The BR3 extract significantly inhibited QS signal molecules [N-acyl homoserine lactone (AHL)] production and the virulence of Pcc on Chinese cabbage (Figures 5B,C). Thus, the GacS/GacA system antagonist present in the BR3 extract is capable of disrupting GacS/GacA system-directed pathogenicity in phytopathogenic bacteria.
The interaction between pathogens and plants plays a critical role in plant development and fitness, including cellular mechanisms, growth, reproduction, hormonal signaling, and tolerance to environmental stresses are involved in this process (Saleem et al., 2017a). Furthermore, the interactions of bacterial species in the phyllosphere influence plant development and fitness (Saleem et al., 2017b). It is challenging to elucidate the detailed mechanisms of the effects of the microbiota on plant function (Haas and Défago, 2005).
Many plant growth-promoting rhizobacteria suppress plant disease by producing one or several antibiotic compounds to inhibit the growth or directly kill the pathogens (Haas and Défago, 2005). However, the resistance of pathogens developed quickly under the antibiotic stress and is becoming a potential challenge for the biological control of soil-borne pathogens (Spellberg and Shales, 2014; Deising et al., 2017). Other bacterial virulence-related factors, such as two-component systems, QS systems, disulfide bond forming enzymes, and TTSSs, have been considered as attractive targets of the chemical therapy (Cegelski et al., 2008; Landeta et al., 2015). The GacS/GacA system was one of these targets because of its global role in regulation of bacterial pathogenesis (Heeb and Haas, 2001).
In the present study, we developed a phytopathogenic reporter bacterium Pto DC3000 (p970Gm-gacSDC3000p) to screen the GacS/GacA system inhibitors from extracts of rhizobacteria. Our data indicated that the extract of Bacillus sp. BR3 inhibited the expression of gacS and its downstream genes rsmY and rsmZ, and the influence of rsmY and rsmZ expression is GacA-dependent, indicating the GacS–GacA system as the potential target of the BR3 extract (Figures 1, 2). In contrast, a previous genetic study of the human opportunistic pathogen P. aeruginosa suggested that the antibiotic azithromycin impaired the expression of rsmY and rsmZ regardless of the function of the GacS/GacA system, because azithromycin could reduce the expression of rsmY and rsmZ in the gacA mutant. Azithromycin might exert its function on genetic factors other than the GacS/GacA system, which then regulate the expression of rsmZ (Humair et al., 2010).
Although the GacS/GacA system plays a critical role in the production of extracellular factors, ecological fitness, and even primary metabolism (Heeb and Haas, 2001; Takeuchi et al., 2012), the chemical signals that stimulate the GacS/GacA system remain obscure. Dubuis and Haas (2007) found that extracts from culture supernatants of Pseudomonas sp. and Vibrio sp. induced the expression of sRNAs and the production of antibiotic compounds, indicating that the signals could be self-produced. Our study further demonstrated that the BR3 extract could interfere with multiple targets of the GacS/GacA regulon in different bacterial species. In addition, overexpression of the gacS gene could attenuate the inhibition effect of the extract BR3 on the promoter activities of gacS (Figure 1A). These data suggested that the BR3 extract inhibits the function of the GacS/GacA system by modulating the expression of gacS gene. To date, few regulatory elements have been identified to influence the expression of gacS (Hrabak and Willis, 1992). Our data showed that the BR3 extract could influence the GacS-dependent phenotypes in strains DC3000, 2P24, and Z3-3, whereas no conserved motif was found in the promoter regions of the gacS gene (data not shown), suggesting that the effect of the BR3 extract on gacS transcription may involve a potential regulator.
Although the method for isolation of the BR3 extract was similar to that of the AHL, the activity of the BR3 extract was not affected at pH 12 and 30°C for 1 h (data not shown). Under the same conditions, AHLs are be degraded (Swift et al., 2001). Moreover, the typical reporter system A. tumefaciens with the traG–lacZ fusion failed to show any reaction with the BR3 extract (Supplementary Figure S2A). These data suggested that the active ingredient from the BR3 extract does not belong to the AHL signals. Isolation of active ingredient from the BR3 extract showed that a pure compound named CX03 (MW = 413.2659) has the same inhibitory activity with the BR3 extract (Supplementary Figures S2B,C). Plant immunity is modulated by complex regulatory networks in response to abiotic and biotic factors (Jones and Dangl, 2006). Interestingly, our data showed that HR was restricted by the BR3 extract (Figure 3C). However, the BR3 extract had no effect on the transcript level of PR1 (At2g14610) in Arabidopsis (data not shown). Further studies are needed to solve the chemical formula of compound CX03 and to investigate the mechanism of CX03 on plant immunity.
Rhizobacteria deploy several strategies, such as the production of antibiotics, QS, and type six secretion systems, to disrupt or otherwise manipulate commensal bacteria to facilitate their colonization and survival (Quiñones et al., 2004; Vacheron et al., 2019). Our data explored the possibility that some bacteria produce specific secondary metabolites to influence the signal transduction of others. Bacillus species are ubiquitous in the soil environment, and some of them are beneficial to plant growth or suppression of plant diseases (Saleem et al., 2017b); therefore, our findings deepen our understanding of the biocontrol mechanisms used by root-associated Bacillus spp.
Collectively, the extract from strain BR3 caused dysfunction of the GacS/GacA system, leading to attenuated activity of virulence factors, thus it may form the basis of a new anti-virulence strategy.
The datasets generated for this study can be found in NCBI accession MK864268, https://www.ncbi.nlm.nih.gov/nuccore/MK864268.
BZ, YZ, FL, and XW conceived and designed the experiments. BZ, YZ, FL, and YM conduced most of the experiments. XW wrote the manuscript and all authors reviewed the manuscript critically.
This project was supported by grants from the National Key Research and Development Program of China (2017YFD02011083), the Science and Technology Major Project of Guangxi (AA17204041), the Natural Science Foundation of Guangxi (2016GXNSFCA380024 and 2017GXNSFAA198341), and the Chinese National Natural Science Foundation (31760533).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Hai-Lei Wei for the HR assays and Dr. Ching-Hong Yang for his thoughtful suggestions about the project.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02005/full#supplementary-material
FIGURE S1 | Bacillus sp. BR3 antagonism assays on agar plates. Strain DC3000 (A), Z3-3 (B), or 2P24 (C) were grown in LB at 28°C overnight and mixed with LB agar (ca. 50°C) to pour into the Petri dish, respectively. Five microliters of BR3 culture (1), the extract BR3 (32 μg ml–1) (2), and kanamycin (50 μg ml–1) (3) were spotted on the plates. The plates were inoculated at 28°C for 36 h. There independent experiments were performed; a representative result is shown.
FIGURE S2 | (A) The traG-lacZ fusion in A. tumefaciens was not induced by the BR3 extract. β-Galactosidase activities of a traG-lacZ fusion were determined in AHL reporter strain NLT4 with the BR3 extract (32 μg ml–1), and the 3-oxo-hexanoyl-homoserine lactone (3-oxo-C8-HSL) (3 μg ml–1). (B) Regulation of rsmY expression in the presence of the BR3 extract and CX03. β-Galactosidase activities of the reporter fusion rsmY-lacZ in strain DC3000 with DMSO, the BR3 extract (16 μg ml–1), or CX03 (2 μg ml–1) were measured. (C) Isolation of active ingredient from the BR3 extract using HPLC and LC–MS.
Ahmer, B. M., Reeuwijk, J. V., Watson, P. R., Wallis, T. S., and Heffron, F. (1999). Salmonella SirA is a global regulator of genes mediating enteropathogenesis. Mol. Microbiol. 31, 971–982. doi: 10.1046/j.1365-2958.1999.01244.x
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Brencic, A., McFarland, K. A., McManus, H. R., Castang, S., Mogno, I., Dove, S. L., et al. (2009). The GacS/GacA signal transduction system of Pseudomonas aeruginosa exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol. Microbiol. 73, 434–445. doi: 10.1111/j.1365-2958.2009.06782.x
Broder, U. N., Jaeger, T., and Jenal, U. (2016). LadS is a calcium-responsive kinase that induces acute-to-chronic virulence switch in Pseudomonas aeruginosa. Nat. Microbiol. 2:16184. doi: 10.1038/nmicrobiol.2016.184
Cegelski, L., Marshall, G. R., Eldridge, G. R., and Hultgren, S. J. (2008). The biology and future prospects of antivirulence therapies. Nat. Rev. Microbiol. 6, 17–27. doi: 10.1038/nrmicro1818
Chatterjee, A., Cui, Y., Yang, H., Collmer, A., Alfano, J. R., and Chatterjee, A. K. (2003). GacA, the response regulator of a two-component system, acts as a master regulator in Pseudomonas syringae pv. tomato DC3000 by controlling regulatory RNA, transcriptional activators, and alternate sigma factors. Mol. Plant Microbe Interact. 16, 1106–1117. doi: 10.1094/MPMI.2003.16.12.1106
Chavez, R. G., Alvarez, A. F., Romeo, T., and Georgellis, D. (2010). The physiological stimulus for the BarA sensor kinase. J. Bacteriol. 192, 2009–2012. doi: 10.1128/JB.01685-9
Chilton, M. D., Currier, T. C., Farrand, S. K., Bendich, A. J., Gordon, M. P., and Nester, E. W. (1974). Agrobacterium tumefaciens DNA and PS8 bacteriophage DNA not detected in crown gall tumors. Proc. Natl. Acad. Sci. U.S.A. 71, 3672–3676. doi: 10.1073/pnas.71.9.3672
Choi, K. H., Kumar, A., and Schweizer, H. P. (2006). A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods. 64, 391–397. doi: 10.1016/j.mimet.2005.06.001
Cui, Y., Chatterjee, A., and Chatterjee, A. K. (2001). Effects of the two-component system comprising GacA and GacS of Erwinia carotovora subsp. carotovora on the production of global regulatory rsmB RNA, extracellular enzymes, and HarpinEcc. Mol. Plant Microbe Interact. 14, 516–526. doi: 10.1094/MPMI.2001.14.4.516
Cui, Y., Madi, L., Mukherjee, A., Dumenyo, C. K., and Chatterjee, A. K. (1996). The RsmA- mutants of Erwinia carotovora subsp. carotovora strain Ecc71 overexpress hrpNEcc and elicit a hypersensitive reaction-like response in tobacco leaves. Mol. Plant Microbe Interact. 9, 565–573.
Deising, H. B., Gase, I., and Kubo, Y. (2017). The unpredictable risk imposed by microbial secondary metabolites: how safe is biological control of plant diseases? J. Plant Dis. Prot. 124, 413–419. doi: 10.1007/s41348-017-0109-5
Dijk, K. V., Fouts, D. E., Rehm, A. H., Hill, A. R., Collmer, A., and Alfano, J. R. (1999). The Avr (Effector) proteins HrmA (HopPsyA) and AvrPto are secreted in culture from Pseudomonas syringae pathovars via the Hrp (Type III) protein secretion system in a temperature- and pH-sensitive manner. J. Bacteriol. 181, 4790–4797.
Dubuis, C., and Haas, D. (2007). Cross-species GacA-controlled induction of antibiosis in Pseudomonads. Appl. Environ. Microbiol. 73, 650–654. doi: 10.1128/AEM.01681-06
Gonzalez Chavez, R., Alvarez, A. F., Romeo, T., and Georgellis, D. (2010). The physiological stimulus for the barA sensor kinase. J. Bacterial. 192, 2009–2012. doi: 10.1128/JB.01685-09
Goodman, A. L., Merighi, M., Hyodo, M., Ventre, I., Filloux, A., and Lory, S. (2009). Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev. 23, 249–259. doi: 10.1101/gad.1739009
Haas, D., and Défago, G. (2005). Biological control of soil-borne pathogens by fluorescent pseudomonads. Nature Rev. Microbiol. 3, 307–319. doi: 10.1038/nrmicro1129
Heeb, S., Blumer, C., and Haas, D. (2002). Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0. J Bacteriol. 184, 1046–1056. doi: 10.1128/jb.184.4.1046-1056.2002
Heeb, S., and Haas, D. (2001). Regulatory roles of the GacS/GacA two-component system in plant-associated and other Gram-Negative bacteria. Mol. Plant Microbe Interact. 14, 1351–1363. doi: 10.1094/MPMI.2001.14.12.1351
Hrabak, E. M., and Willis, D. K. (1992). The lemA gene required for pathogenicity of Pseudomonas syringae pv. syringae on bean is a member of a family of two-component regulators. J. Bacteriol. 174, 3011–3020. doi: 10.1128/jb.174.9.3011-3020.1992
Humair, B., Wackwitz, B., and Haas, D. (2010). GacA-controlled activation of promoters for small RNA genes in Pseudomonas fluorescens. Appl. Environ. Microbiol. 76, 1497–1506. doi: 10.1128/AEM.02014-09
Huynh, T. V., Dahlbeck, D., and Staskawicz, B. J. (1989). Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science 245, 1374–1377. doi: 10.1126/science.2781284
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kay, E., Dubuis, C., and Haas, D. (2005). Three small RNAs jointly ensure secondary metabolism and biocontrol in Pseudomonas fluorescens CHA0. Proc. Natl. Acad. Sci. U.S.A. 102, 17136–17141. doi: 10.1073/pnas.0505673102
King, E. O., Ward, M. K., and Raney, D. E. (1954). Two simple media for the demonstration of pyocyanin and fluorescein. J. Lab. Clin. Med. 44, 301–307.
Kulkarni, P. R., Jia, T., Kuehne, S. A., Kerkering, T. M., Morris, E. R., Searle, M. S., et al. (2014). A sequence-based approach for prediction of CsrA/RsmA targets in bacteria with experimental validation in Pseudomonas aeruginosa. Nucleic Acids Res. 42, 6811–6825. doi: 10.1093/nar/gku309
Kvitko, B. H., Ramos, A. R., Morello, J. E., Oh, H.-S., and Collmer, A. (2007). Identification of harpins in Pseudomonas syringae pv. tomato DC3000, which are functionally similar to HrpK1 in promoting translocation of type III secretion system effectors. J. Bacteriol. 189, 8059–8072. doi: 10.1128/JB.01146-07
Landeta, C., Blazyk, J. L., Hatahet, F., Meehan, B. M., Eser, M., Myrick, A., et al. (2015). Compounds targeting disulfide bond forming enzyme DsbB of Gram-negative bacteria. Nature Chem. Biol. 11, 292–298. doi: 10.1038/NCHEMBIO.1752
Lapouge, K., Schubert, M., Allain, F. H., and Haas, D. (2008). Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol. Microbiol. 67, 241–253. doi: 10.1111/j.1365-2958.2007.06042.x
Laville, J., Voisard, C., Keel, C., Maurhofer, M., Défago, G., and Haas, D. (1992). Global control in Pseudomonas fluorescens mediating antibiotic synthesis and suppression of black root rot of tobacco. Proc. Natl. Acad. Sci. U.S.A. 89, 1562–1566. doi: 10.1073/pnas.89.5.1562
Lawhon, S. D., Maurer, R., Suyemoto, M., and Altier, C. (2002). Intestinal short-chain fatty acids alter Salmonella typhimurium invasion gene expression and virulence through BarA/SirA. Mol. Microbiol. 46, 1451–1464. doi: 10.1046/j.1365-2958.2002.03268.x
LeRous, M., Kirkpatrick, R. L., Montauti, E. I., Tran, B. Q., Peterson, S. B., Harding, B. N., et al. (2015). Kin cell lysis is a danger signal that activates antibacterial pathways of Pseudomonas aeruginosa. eLife 4:e05701. doi: 10.7554/eLife.05701
Li, Y., Yamazaki, A., Zou, L., Biddle, E., Zeng, Q., Wang, Y., et al. (2010). ClpXP protease regulates the Type III secretion system of Dickeya dadantii 3937 and is essential for the bacterial virulence. Mol. Plant Microbe Interact. 23, 871–878. doi: 10.1094/MPMI-23-7-0871
Ling, L. L., Schneider, T., Peoples, A. J., Spoering, A. L., Engels, I., Conlon, B. P., et al. (2015). A new antibiotic kills pathogens without detectable resistance. Nature 517, 455–459. doi: 10.1038/nature14303
Liu, J., Zhang, W., Wu, X., and Zhang, L. (2013). Effect of retS gene on biosynthesis of 2,4-diacetylphloroglucinol in Pseudomonas fluorescens 2P24. Acta Microbiol. Sin. 53, 118–126.
Mancl, J. M., Ray, W. K., Helm, R. F., and Schubot, F. D. (2019). Helix cracking regulates the critical interaction between RetS and GacS in Pseudomonas aeruginosa. Structure 27, 785–793. doi: 10.1016/j.str.2019.02.006
Marchesi, J. R., Sato, T., Weightman, A. J., Martin, T. A., Fry, J. C., Hiom, S. J., et al. (1998). Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rDNA. Appl. Environ. Microbiol. 64, 795–799.
Miller, J. H. (1972). Experiments in Molecular Genetics. New York, NY: Cold Spring Harbor Laboratory.
Moll, S., Scheider, D. J., Stodghill, P., Myers, C. R., Cartinhour, S. W., and Filiatrault, M. J. (2010). Construction of an rsmX co-variance model and identification of five rsmX non-coding RNAs in Pseudomonas syrigae pv. tomato DC3000. RNA Biol. 7, 508–516. doi: 10.4161/rna.7.512687
Mondragón, V., Franco, B., Jonas, K., Suzuki, K., Romeo, T., and Melefors, Ö, et al. (2006). pH-Dependent activation of the BarA-UvrY two-component system in Escherichia coli. J. Bacteriol. 188, 8303–8306. doi: 10.1128/JB.01052-6
Parkins, M. D., Ceri, H., and Storey, D. G. (2001). Pseudomonas aeruginosa GacA, a factor in multihost virulence, is also essential for biofilm formation. Mol. Microbiol. 40, 1215–1226. doi: 10.1046/j.1365-2958.2001.02469.x
Pérez-Martínez, I., and Haas, D. (2011). Azithromycin inhibits expression of the GacA-dependent small RNAs RsmY and RsmZ in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 55, 3399–3405. doi: 10.1128/AAC.01801-10
Pessi, G., and Haas, D. (2001). Dual control of hydrogen cyanide biosynthesis by the global activator GacA in Pseudomonas aeruginosa PAO1. FEMS Microbiol. Lett. 200, 73–78. doi: 10.1111/j.1574-6968.2001.tb10695.x
Pirhonen, M., Flego, D., Heikinheimo, R., and Palva, E. T. (1993). A small diffusible signal molecule is responsible for the global control of virulence and exoenzyme production in the plant pathogen Erwinia carotovora. EMBO J. 12, 2467–2476. doi: 10.1002/j.1460-2075.1993.tb05901.x
Quiñones, B., Pujol, C. J., and Lindow, S. E. (2004). Regulation of AHL production and its contribution to epiphytic fitness in Pseudomonas syringae. Mol. Plant Microbe Interact. 17, 521–531. doi: 10.1094/MPMI.2004.17.5.521
Sahr, T., Bruggemann, H., Jules, M., Lomma, M., Albert-Weissenberger, C., Cazalet, C., et al. (2009). Two small ncRNAs jointly govern virulence and transmission in Legionella pneumophila. Mol. Microbiol. 72, 741–762. doi: 10.1111/j.1365-2958.2009.06677.x
Saleem, M., Ji, H., Amirullah, A. A., and Traw, M. B. (2017a). Pseudomonas syringae pv. tomato DC3000 growth in multiple gene knockouts predicts interactions among hormonal, biotic and abiotic stress responses. Eur. J. Plant Pathol. 149, 779–786. doi: 10.1007/s10658-017-1223-8
Saleem, M., Meckes, N., Pervaiz, Z. H., and Traw, M. B. (2017b). Microbial interactions in the phyllosphere increase plant performance under herbivore biotic stress. Front. Microbiol. 8:41. doi: 10.3389/fmicb.2017.00041
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd Edn. New York NY: Cold Spring Harbor Laboratory Press.
Savioz, A., Zimmermann, A., and Haas, D. (1993). Pseudomonas aeruginosa promoters which contain a conserved GG-N10-GC motif but appear to be RpoN-independent. Mol. Gen. Genet. 238, 74–80.
Shanahan, P., O’Sullivan, D. J., Simpson, P., Glennon, J. D., and O’Gara, F. (1992). Isolation of 2,4-diacetylphloroglucinol from a fluorescent pseudomonad and investigation of physiological parameters influencing its production. Appl. Environ. Microbiol. 58, 353–358.
Smith, A. W., and Iglewski, B. H. (1989). Transformation of Pseudomonas aeruginosa by electroporation. Nucleic Acids Res. 17:10509.
Spellberg, B., and Shales, D. (2014). Prioritized current unmet needs for antibacterial therapies. Clin. Pharmacol. Ther. 96, 151–153. doi: 10.1038/clpt.2014.106
Swift, S., Downie, J. A., Whitehead, N. A., Barnard, M. L., Salmond, G. P. C., and Williams, P. (2001). Quorum sensing as a population-density-dependent determinant of bacterial physiology. Adv. Microb. Physiol. 45, 199–270. doi: 10.1016/S0065-2911(01)45005-3
Takeuchi, K., Yamada, K., and Haas, D. (2012). ppGpp controlled by the Gac/Rsm regulatory pathway sustains biocontrol activity in Pseudomonas fluorescens CHA0. Mol. Plant Microbe Interact. 25, 1440–1449. doi: 10.1094/MPMI-02-12-0034-R
Vacheron, J., Péchy-Tarr, M., Brochet, S., Heiman, C. M., Stojiljkovic, M., Maurhofer, M., et al. (2019). T6SS contributes to gut microbiome invasion and killing of an herbivorous pest insect by plant-beneficial Pseudomonas protegens. ISME J. 13, 1318–1329. doi: 10.1038/s41396-019-0353-8
Valverde, C., Lindell, M., Wagner, E. G. H., and Haas, D. (2004). A repeated GGA motif is critical for the activity and stability of the riboregulatory RsmY of Pseudomonas fluorescens. J. Biol. Chem. 279, 25066–25074. doi: 10.1074/jbc.M401870200
Whitehead, N. A., Byers, J. T., Commander, P., Corbett, M. J., Coulthurst, S. J., Everson, L., et al. (2002). The regulation of virulence in phytophathogenic Erwinia species: quorum sensing, antibiotics and ecological considerations. Antonie Van Leeuwenhoek 81, 223–231.
Wong, S. M., Carroll, P. A., Rahme, L. G., Ausubel, F. M., and Calderwood, S. B. (1998). Modulation of expression of the ToxR regulon in Vibrio cholerae by a member of the two-component family of response regulators. Infect. Immun. 66, 5854–5861.
Wu, X., Liu, J., Zhang, W., and Zhang, L. (2012). Multiple-level regulation of 2,4-diacetylphloroglucinol production by the sigma regulator PsrA in Pseudomonas fluorescens 2P24. PLoS One 7:e50149. doi: 10.1371/journal.pone.0050149
Yamazaki, A., Li, J., Zeng, Q., Khokhani, D., Hutchins, W. C., Yost, A. C., et al. (2012). Derivatives of plant phenolic compound affect the type III secretion system of Pseudomonas aeruginosa via a GacS-GacA two-component signal transduction system. Antimicrob. Agents Chemother. 56, 36–43. doi: 10.1128/AAC.00732-11
Yan, X., Zhang, L., Yang, Z., and Tang, W. (2004). The role of regulatory gene gacA in the suppress ion of soil-borne diseases by Pseudomonas fluorescens 2P24. Acta Phyto. Pathol. Sin. 34, 272–279.
Keywords: the GacS/GacA two-component system, small non-coding RNA, Bacillus sp., Pseudomonas syringae pv. tomato, type three secretion system
Citation: Zhang B, Zhang Y, Liang F, Ma Y and Wu X (2019) An Extract Produced by Bacillus sp. BR3 Influences the Function of the GacS/GacA Two-Component System in Pseudomonas syringae pv. tomato DC3000. Front. Microbiol. 10:2005. doi: 10.3389/fmicb.2019.02005
Received: 15 April 2019; Accepted: 15 August 2019;
Published: 11 September 2019.
Edited by:
Dilantha Fernando, University of Manitoba, CanadaReviewed by:
Miguel Castañeda, Meritorious Autonomous University of Puebla, MexicoCopyright © 2019 Zhang, Zhang, Liang, Ma and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaogang Wu, NDE0NjMwODUwNUBvdXRsb29rLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.