- 1Genetic Engineering Research Center, School of Life Sciences, Chongqing University, Chongqing, China
- 2Chongqing Engineering Research Center for Fungal Insecticide, Chongqing, China
- 3Key Laboratory of Gene Function and Regulation Technologies Under Chongqing Municipal Education Commission, Chongqing, China
- 4Chongqing Center for Disease Control and Prevention, Chongqing, China
As a C2H2 type zinc finger transcription factor, CreA is the key in Carbon Catabolism Repression (CCR) pathway, which negatively regulates the genes in carbon sources utilization. As conidiation in filamentous fungi is affected by nutritional conditions, CreA may contribute to fungal conidiation, which has been well studied in filamentous fungi, especially Aspergillus spp., but researches on entomopathogenic fungi are not enough. In this study, we found a homologous gene MaCreA in Metarhizium acridum, and the MaCreA deletion strain showed delayed conidiation, significant decrease in conidial yield, and 96.88% lower conidial production, when compared with the wild-type strain, and the normal conidiation and microcycle conidiation pattern shift was blocked. RT-qPCR showed that the transcription levels of the genes FlbD and LaeA (related to asexual development) were significantly altered, and those of most of the conidiation-related genes were higher in ΔMaCreA strain. The results of RNA-Seq revealed that MaCreA regulated the two conidiation patterns by mediating genes related to cell cycle, cell division, cell wall, and cell polarity. In conclusion, CreA, as a core regulatory gene in conidiation, provides new insight into the mechanism of conidiation in entomopathogenic fungi.
Introduction
Conidia, as the asexual propagules in many filamentous fungi, are start and end of fungal lifecycle (Adams et al., 1998; Papagianni, 2004). They are the infective form of entomopathogenic fungi, essential for their pathogenicity (Schrank and Vainstein, 2010). In filamentous fungi, normal conidiation and microcycle conidiation are the two patterns of asexual conidiation (Zhang et al., 2010; Jung et al., 2014; Wang Z. et al., 2016). In normal conidiation, asexual conidia are produced at the top or sides of the hyphae in a subsequent budding-like process for proper vegetative growth, whereas microcycle conidiation occurs when fungi are subjected to unfavorable environmental conditions. As a survival mechanism, microcycle conidiation does not comprise the hyphae extension stage, and the conidia are directly generated from the germ tubes or conidial cells (Hanlin, 1994).
Asexual conidiation in the filamentous fungus Aspergillus nidulans has been well researched. When mycelium grow to a certain extent, under the condition of external stimuli, the hyphae starts to transform into aerial mycelium, and thick-walled foot cells are formed at the tip or side of the aerial mycelium. Following successive emergence of the foot cells, multinuclear conidiophores develop. The outer layer of conidiophores comprise vesicle, metulae, phialides, and aerial spores or conidia, which are formed sequentially (Borkovich and Ebbole, 2010; Park and Yu, 2012). Conidiation is regulated by a central regulatory pathway containing three genes BrlA–AbaA–WetA (Mirabito et al., 1989), and other transcription factors, such as FluG, which is an upstream gene of the entire pathway and a major activating protein of conidiation in A. nidulans (Lee and Adams, 1994b). In contrast, SfgA, which is downstream of FluG but upstream of Flbs (such as FlbA, FlbB, FlbC, FlbD) and BrlA, plays a negative regulatory role in conidiation (Seo et al., 2006). FlbA encodes a protein with a RGS domain, regulates the activity of the GTPase of FadA, a Gα protein (Lee and Adams, 1994a; Yang et al., 2012), and participates in mycelial development and asexual conidiation (Yu et al., 1996). FlbB, FlbC, FlbD, and FlbE are the activators of brlA (Etxebeste et al., 2009; Garzia et al., 2009; Kwon et al., 2010). While orthologs of FlbA, FlbB, FlbC, FlbD, and FluG have been found in Metarhizium acridum, those of BrlA, AbaA, and WetA have not been identified.
Although molecular studies on conidiation in A. nidulans have been conducted, the core regulatory pathway of BrlA–AbaA–WetA has been investigated only in certain Aspergillus spp., Penicillium spp., and Talaromyces spp. (López et al., 2018). Most of the entomopathogenic fungi, such as Metarhizium spp. and Beauveria spp., do not possess core regulatory pathway, implying that the central regulatory pathway is not common in all fungi. Moreover, in M. acridum, the core three genes in the central regulatory pathway have not been detected.
The phenomenon of microcycle conidiation is not commonly observed among fungi, and only more than 100 fungal species are currently known to exhibit this process (Jung et al., 2014). For instance, Colletotrichum acutatum has been noted to present obvious microcycle conidiation when incubated at 21.3–32.7°C for 12–36 h (Leandro et al., 2003), and Penicillium variabile has been reported to display microcycle conidiation in liquid culture (Zhelifonova et al., 2006). Oidium longipes is the first powdery mildew pathogen in which microcycle conidiation was observed when incubated on petunia leaves (Kiss et al., 2008). Besides, deletion of WetA gene has been noted to result in microcycle conidiation (Son et al., 2014). In M. acridum, microcycle conidiation has been reported to produce 4–5-fold higher conidia yield, with the generated conidia exhibiting increased heat resistance and trehalose level, but similar UV-B resistance and virulence, when compared with those produced by normal conidiation (Zhang et al., 2010). A recent study revealed that regulation of conidiation pattern in M. acridum is a complex process involving cell cycle and metabolism (Wang Z. et al., 2016). Nevertheless, the mechanism of microcycle conidiation remains unclear.
In the present study, we identified a homologous gene CreA in M. acridum, MaCreA, which was found to be a C2H2-type zinc finger transcription factor playing an important role in carbon catabolism repression (CCR) pathway in microorganisms (Dowzer and Kelly, 1991; Mathieu and Felenbok, 1994; Adnan et al., 2017). Besides, deletion of MaCreA caused delayed conidiation, significant decrease in conidial yield, and 98.88% lower conidial production, when compared with the wild-type strain, suggesting that CreA plays a crucial role in conidiation.
Materials and Methods
Strains and Growth Conditions
Metarhizium acridum strain CQMa102 was obtained from Chongqing Engineering Research Center for Fungal Insecticide and stored at China General Microbiological Culture Collection Center (CGMCC, No. 0877), and cultured on 1/4 SDAY medium (1% dextrose, 0.25% mycological peptone, 2% agar, and 0.5% yeast extract) at 28°C. Escherichia coli DH5α cells, used for DNA manipulations and transformations, were purchased from Bioground (China), and grown on Luria-Bertani (LB) medium at 37°C. Agrobacterium tumefaciens AGL-1 was purchased from Bioground (China) and propagated on LB medium at 28°C and used for fungal transformations.
Construction of MaCreA Deletion Mutant and Complementation Mutant
The MaCreA disruption vector was constructed by homologous recombination based on the vector PK2-PB. About 1000-bp fragments upstream and downstream of the MaCreA open reading frame (ORF) were amplified from the WT genome by PCR, and primers used in this article is listed in Supplementary Table S1, then the two fragments were ligated to both the sides of the bar gene in PK2-PB to construct the knockout vector. The recombinant plasmid was transferred into the WT strain by Agrobacterium-mediated transformation. For the construction of complementation vector, the upstream 2000-bp fragment of the MaCreA ORF was amplified from the WT genome via PCR, ligated into the PK2-PB-SUR vector, and introduced into the knockout strain by Agrobacterium-mediated transformation. All the knockout and complementation strains were verified by Southern blot. The disruption and complementation of MaCreA were designed as shown in Supplementary Figures S1A,B.
Measurements of Conidial Yield and Germination
To measure the conidial yield of the WT, ΔMaCreA, and complemented transformant (CP), 800 μL of 1/4 SDAY medium were first added to each well of the 24-cell plates. 2 μL of 106 conidia/ml suspension of the WT, ΔMaCreA, and CP were inoculated into each well, and cultured at 28°C for 15 days. Three colonies of each strain were picked out every 2 days from the third day and suspended in sterile water for calculating conidial yield. The conidial germination rate of each strain was determined according to a previously described method (He and Xia, 2009). All experiments were performed in triplicate.
Examination of Conidial Development
A total of 100 μL of 107conidia/mL suspension of WT and ΔMaCreA were respectively spread on 1/4 SDAY and SYA (3% sucrose, 0.5% yeast extract, 0.3% NaNO3, 0.05% MgSO4, 0.05% KCl, 0.1% KH2PO4, 0.001% FeSO4, 0.001% MnSO4, and 2% agar) plates and incubated at 28°C. Subsequently, the fungal cells were detected under a digital light microscope (MOTIC, China) every 2 h from 12 h after incubation.
Real-Time qPCR
For RNA extraction, the fungal cells were ground in liquid nitrogen, and the total RNA was isolated using Fungal RNA Kit (OMEGA) according to the manufacturer’s protocol. The quality of the extracted RNA was assessed by 2% agarose gel electrophoresis. To detect the expression levels of genes related to conidiation, reverse transcription of RNA was performed using PrimeScriptTM RT reagent Kit with gDNA Eraser. The genes VeA, MAPK, NsdD, MedA, FlbA, LreA, VosA, Mmc, StuA, NsdC, FadA, CatA, FlbD, FlbB, SfgA, GanB, PKA, FluG, LreB, and LaeA were chosen as target genes. qPCR was performed using SYBR® Premix Ex TaqTM (TaKaRa) according to the manufacturer’s protocol, and the relative expression levels of the target genes in the knockout strain and WT were computed by the 2−ΔΔCt method (Livak and Schmittgen, 2001). All the experimental procedures were performed in triplicate.
Digital Gene Expression Profiling and GO Analysis
RNA-Seq experiment based on Solexa sequencing was performed by Majorbio, Shanghai, China. The screening thresholds for differentially expressed genes (DEGs) are FDR ≤ 0.001 and Log2Ratio ≥ 1. All DEGs were mapped to the GO terms1 and KEGG pathways2. To verify the results of RNA-Seq, the transcript levels of 21 randomly selected DEGs were validated by RT-qPCR, and the results are shown in Supplementary Figure S2.
RNA-Seq Data Accession
RNA-Seq data are deposited in the NCBI Sequence Read Archive (SRA) database, and the accession numbers are SRR9089704, SRR9089705, SRR9089702, SRR9089703, SRR9089700, SRR9089701, SRR9089698, SRR9089699, SRR9089696, SRR9089697, SRR9089706, and SRR9089707.
Results
Sequence Analysis of MaCreA
Following BLAST search of the M. acridum genomic database, a CreA homologous gene was cloned and named as MaCreA (Accession number MK089826). The MaCreA comprised an ORF of 1131 bp and 376 amino acids, with predicted pI of 9.77 and molecular weight of 41 kDa3, and without signal peptide4. The gene exhibited two typical C2H2 zinc finger structures and other conserved domains (Figure 1A), similar to BbcreA in Beauveria bassiana. Phylogenetic analysis showed that MaCreA was closest to the near-origin Metarhizium anisopliae CRR1 gene (Figure 1B). Homology analysis revealed that the similarity of MaCreA to M. anisopliae (CAA71314.1), B. bassiana (PMB70895.1), and A. nidulans (XP_663799.1) was 96.83, 77.46, and 53.57%, respectively (Figure 1C), and investigation of C2H2 zinc finger domains revealed 99% similarity in the selected fungi, indicating that MaCreA is a CreA homologous gene in M. acridum.
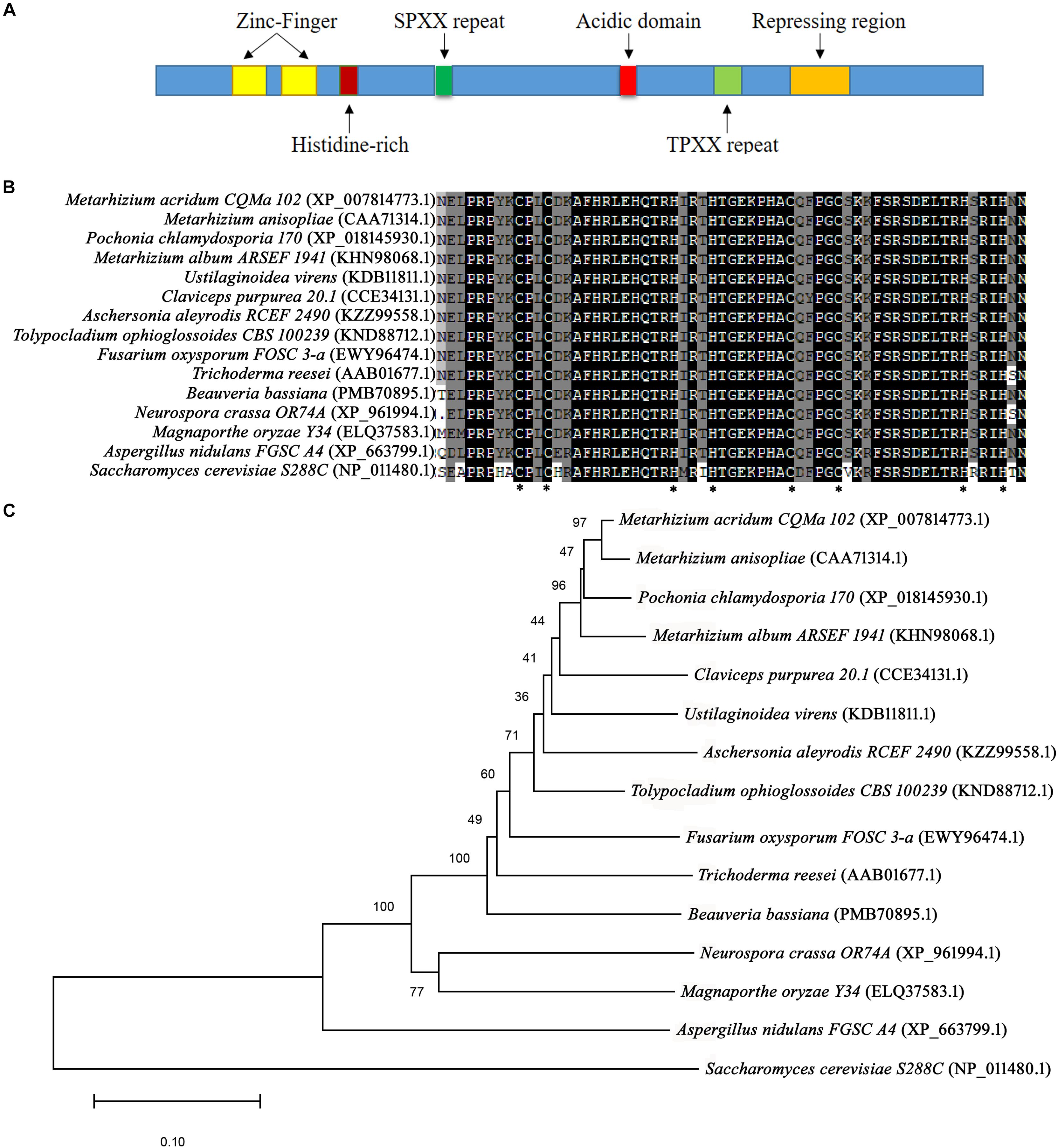
Figure 1. Sequences of CreA analysis. (A) Diagram of regions of Metarhizium acridum CreA. (B) Phylogenetic tree was reconstructed using the neighbor-joining method. (C) Comparison of C2H2 domain sequences of MaCreA with CreA homologs from other fungi.
MaCreA Is Required for Conidiation
To exclude random insertion of genes into the genome, all the transformed M. acridum strains with deletion and complementation of MaCreA were finally verified by Southern blot analysis (Supplementary Figure S1C). With regard to the phenotype of the deletion strains, after 7-day growth on 1/4 SDAY medium, the WT and CP colonies became obviously tawny with mass production of conidia, while the colonies of the knockout strain were fluffy and white with strong mycelia (Figure 2A). To explore the effects of MaCreA on the conidiation process of M. acridum, morphogenesis of conidia was observed on 1/4 SDAY (rich medium) and SYA (poor medium) using a digital camera. The results showed that the WT produced conidiophores at 20 h after incubation on 1/4 SDAY medium, and the mature conidia were shed at 36 h; in contrast, ΔMaCreA presented obvious delay in conidiation, and the conidia were not observed until 60 h (Figure 2B). Evaluation of the conidial yields of all the three strains every 2 days revealed that the conidial yield of the knockout strain ΔMaCreA was significantly decreased, with 96.88% lower conidial production than that of the WT (Figure 2C). Moreover, the germination rate of the knockout strain was significantly lower than that of the WT and CP (Figure 2D).
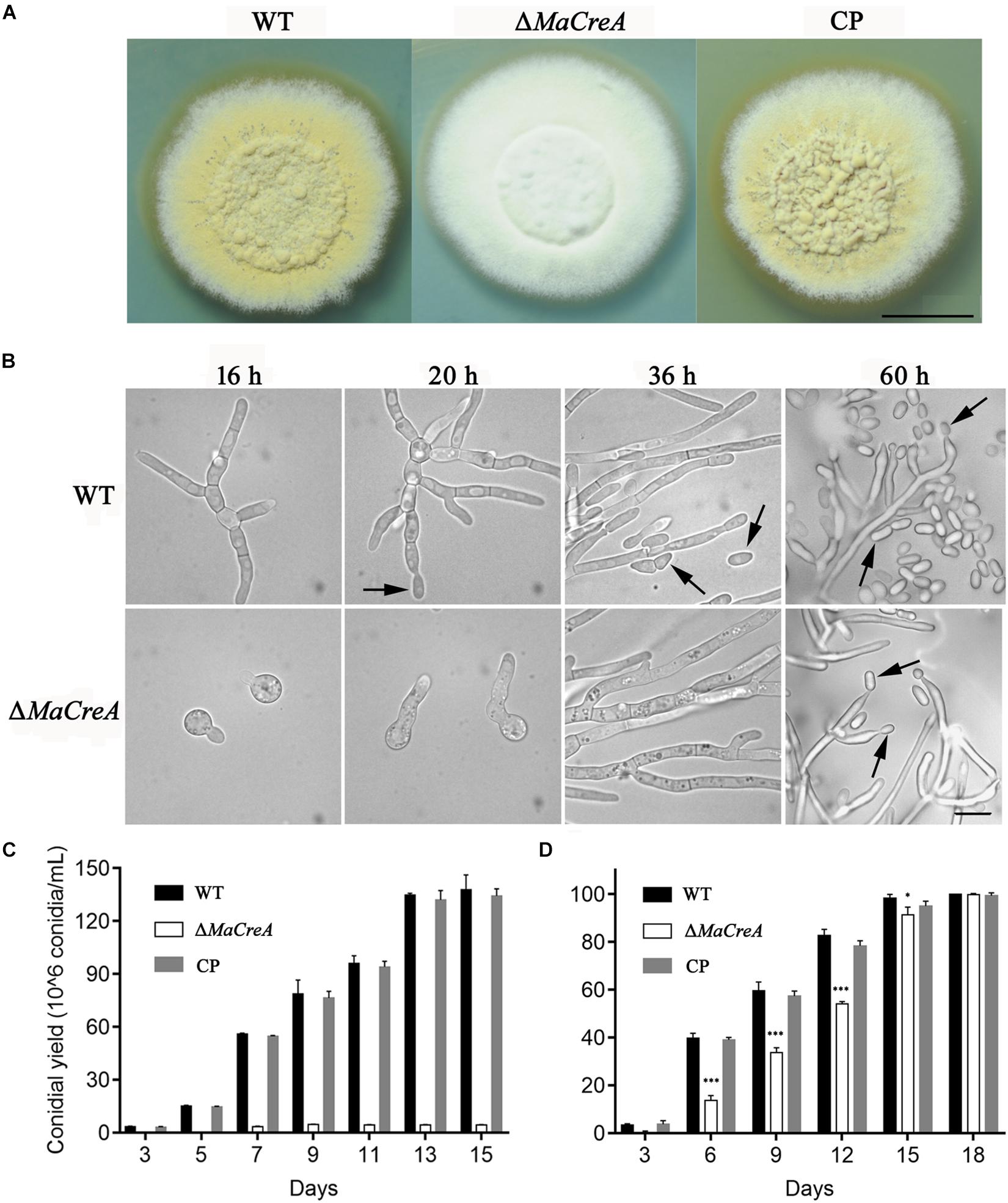
Figure 2. MaCreA deletion strain affects conidiation. (A) MaCreA deletion strain was grown for 7 days on 1/4 SDAY media, Scale bars indicate 1 cm. (B) Conidiation was postponed in MaCreA deletion strain on 1/4 SDAY. (C) Conidial yield of each stain on 1/4 SDAY media at 28°C, Scale bars indicate 10 μm, Black arrow: conidia. (D) Conidial germination of each stain on 1/4 SDAY media for 2, 4, 6, 8, 10, 12, and 14 h (t-text, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
MaCreA Is Required for Conidiation Pattern Shift
Two conidiation patterns are known in M. acridum, namely, normal conidiation and microcycle conidiation. In normal conidiation, conidia are produced at the top or sides of the hyphae, whereas in microcycle conidiation, conidia are directly generated from the germ tubes or conidial cells. In the present study, normal and microcycle conidiation patterns were observed in WT on 1/4 SDAY medium (rich medium) and SYA medium (poor medium), respectively (Figure 3A). However, after knocking out the MaCreA gene, the knockout strain exhibited a shift from microcycle conidiation to normal conidiation on SYA medium (Figure 3B), revealing that ΔMaCreA could not display microcycle conidiation under poor nutrient conditions (SYA medium), and that MaCreA gene played an important role in the shift of conidiation.
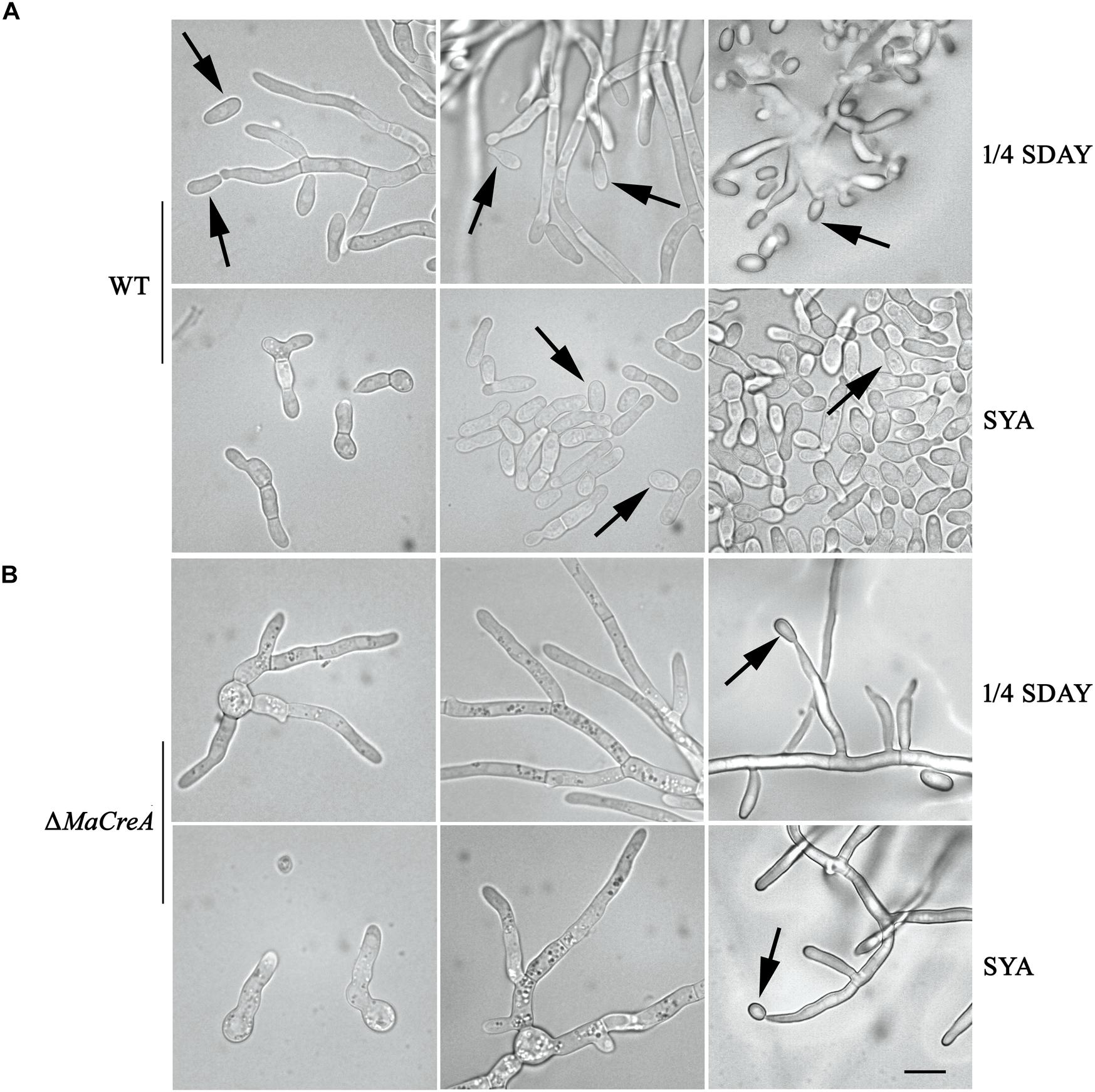
Figure 3. Conidiation pattern shift was lost in MaCreA deletion strain between 1/4 SDAY and SYA. Scale bars indicate 10 μm, Black arrow, conidia. (A) Conidiation of WT on 1/4 SDAY and SYA media. (B) Conidiation of ΔMaCreA on 1/4 SDAY and SYA media.
Analysis of DEGs Involved in Conidiation in Fungal Cells Grown on Two Different Media
To determine the downstream genes and signaling pathways of MaCreA in conidiation, RNA-seq was used to identify significant up-regulated and down-regulated genes. The number of significant DEGs (twofold or greater, FDR ≤ 0.001) between ΔMaCreA and WT on 1/4 SDAY medium was 1099, among which 624 were up-regulated and 475 were down-regulated. However, 1743 DEGs (twofold or greater, FDR ≤ 0.001) were noted between ΔMaCreA and WT on SYA medium, of which 1039 were up-regulated and 704 were down-regulated. To investigate the conidiation pattern shift mechanisms involved in normal conidiation on 1/4 SDAY medium and microcycle conidiation on SYA medium, the differences and similarities between DEGs in fungal cells grown on the two media were examined. A total of 817 common DEGs in fungal cells grown on 1/4 SDAY and SYA media were noted, and 926 and 282 DEGs were detected in fungal cells grown on SYA medium and 1/4 SDAY medium, respectively. There are eight GO categories mainly enriched in biological process, eight in cellular component, and four in molecular function. Although the GO terms were similar in the two different media (Figure 4), 17 signaling pathways were only detected on SYA medium, suggesting that these pathways may respond to SYA medium, which are positive changes related to microcycle conidiation, such as lipid metabolism, carbohydrate metabolism, folding, sorting and degradation, signal transduction, and membrane transport (Supplementary Table S2).
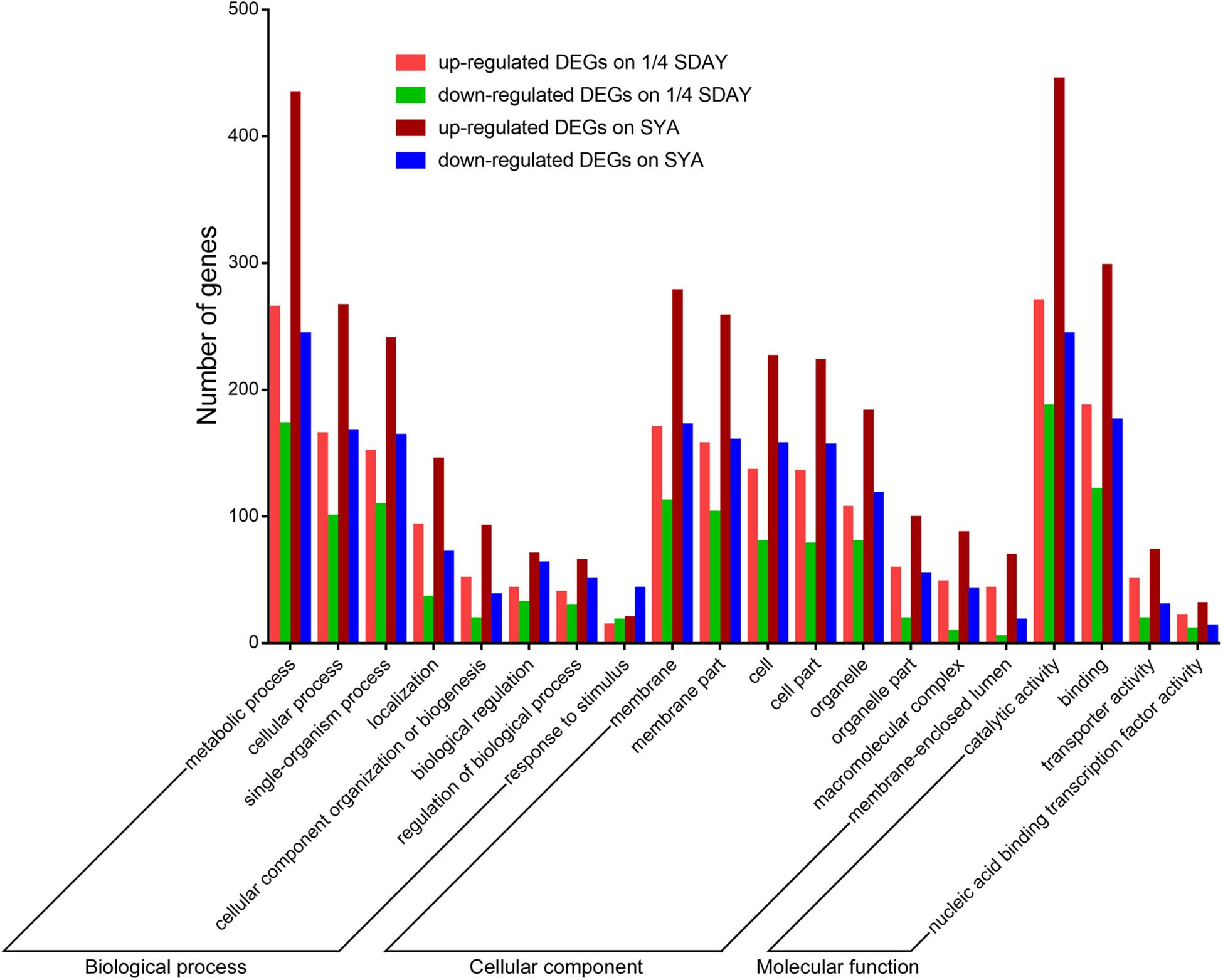
Figure 4. GO categories of DEGs on 1/4 SDAY and SYA. The lift Y-axis indicates the number of genes in a category; X-axis indicates the GO categories.
To determine the DEGs involved in conidiation, DEGs in fungal cells grown on the two types of media were analyzed based on their GO annotations. Some genes related to conidiation were found, which were noted to be involved in the cell cycle, cell division, cell wall, and cell polarity. For example, the developmental protein FluG (MAC_08691), sporulation protein RMD8 (MAC_07621), APSES transcription factor (MAC_03829), putative UDP-glucose 4-epimerase (MAC_02140), and putative methyltransferase LaeA (MAC_03279), which are known to play important roles in conidiation in other fungi. Conidial pigment polyketide synthase PksP/Alb1 (MAC_05385), laccase (MAC_05384), laccase Lcc2 (MAC_02006), and Lcc5 (MAC_04467) are involved in conidial pigment synthesis. Pescadillo (MAC_07265), tyrosine-protein phosphatase CDC14 (MAC_03011), cell division cycle protein 123 (MAC_04986), and cell division control protein CDC48 (MAC_02555) are involved in cell proliferation. Mucin (MAC_01612), WASP-like protein las17 (MAC_00912), and p21 activated kinase-like protein (MAC_08591) are associated with cell polarity. In addition, a large number of cell wall related genes were also found, such as putative WSC domain protein (MAC_03694), cell wall protein (MAC_06850), and glycine-rich cell wall structural protein 1 (MAC_06488). These genes all play a role in conidiation of M. acridium (Supplementary Table S3). Analysis of the expression levels of these genes (Figure 5) revealed that these genes were down-regulated in the knockout strain compared with those in the WT, suggesting that the CreA gene had a significant effect on the conidiation process.
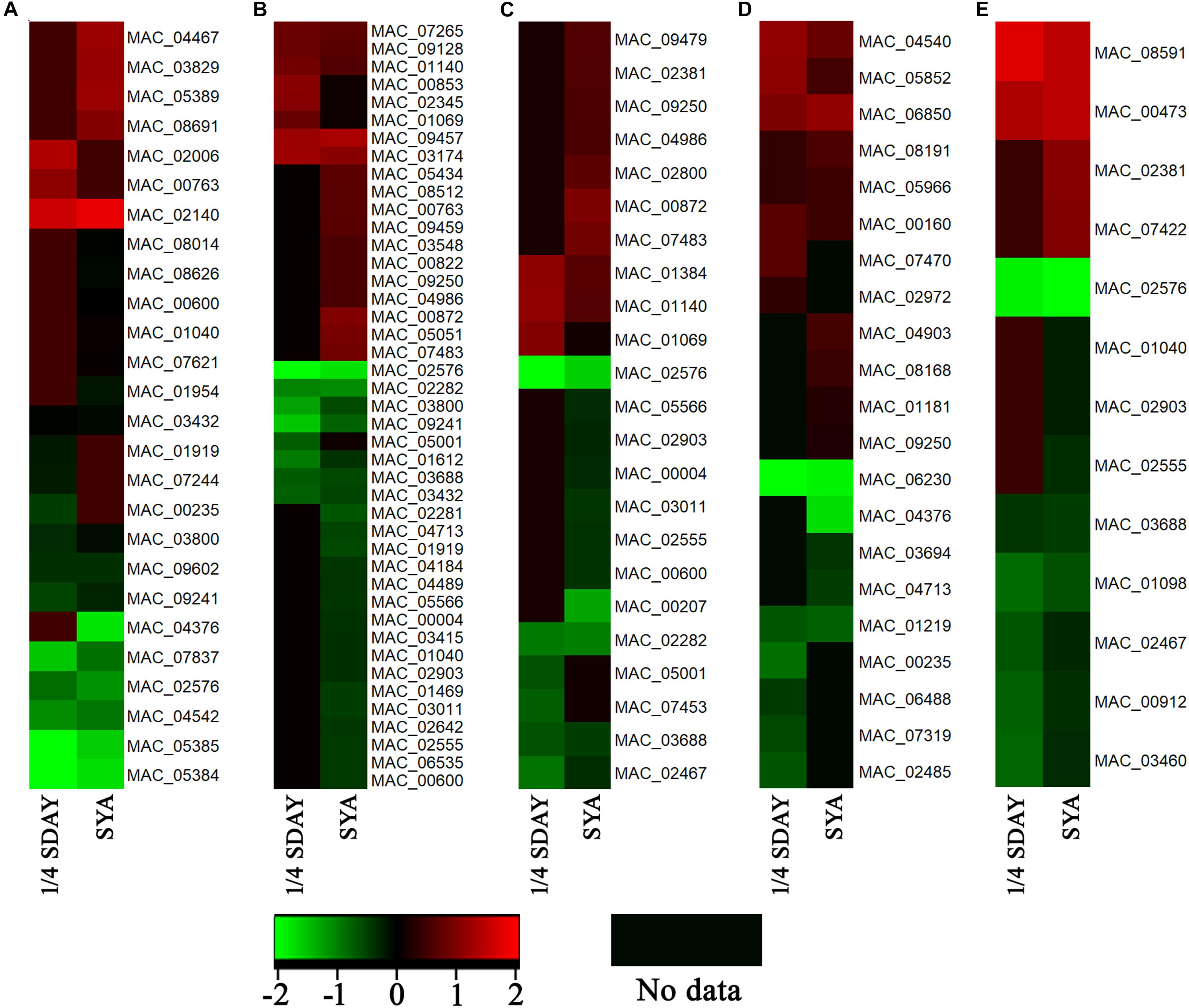
Figure 5. Expression profiling analysis of conidiation-relate genes in CreA deletion strain (ΔMaCreA) compared with wild-type strain (WT) in different culture states, namely, 1/4 SDAY and SYA. (A) genes involved in conidiation; (B) genes involved in cell cycle; (C) genes involved in cell division; (D) genes involved in cell wall; (E) genes involved in cell polarity.
MaCreA Affects Genes Related to Conidial Regulatory Pathway
To determine the relationship between MaCreA and conidiation pathway, the expression of conidiation-related homologous genes VeA, MAPK, NsdD, MedA, FlbA, LreA, VosA, Mmc, StuA, NsdC, FadA, CatA, FlbD, FlbB, SfgA, GanB, PKA, FluG, LreB, and LaeA in M. acridum was examined. To detect the expressions of conidiation-related genes in ΔMaCreA on 1/4 SDAY medium, RT-qPCR was performed, and the results showed that the expressions of MedA, FlbA, VosA, StuA, NsdC, FadA, FlbD, FlbB, and GanB in ΔMaCreA were higher than those in WT, with FlbD expression being significantly higher. In contrast, the expression of LaeA in ΔMaCreA was lower than those in WT, with significantly reduced. Furthermore, the expression of Mmc, which has been reported to be involved in microcycle conidiation (Liu et al., 2010), was also increased (Figure 6). It must be noted that three core genes, BrlA–AbaA–WetA, were not found in M. acridum as well as in other entomopathogenic fungi, but were only detected in Aspergillus and some Penicillium spp. (such as Penicillium brasilianum) (Supplementary Table S4). This indicated that the known asexual conidiation regulation pathway of A. nidulans was not common among fungi, and that different fungi may have unique conidiation regulation.
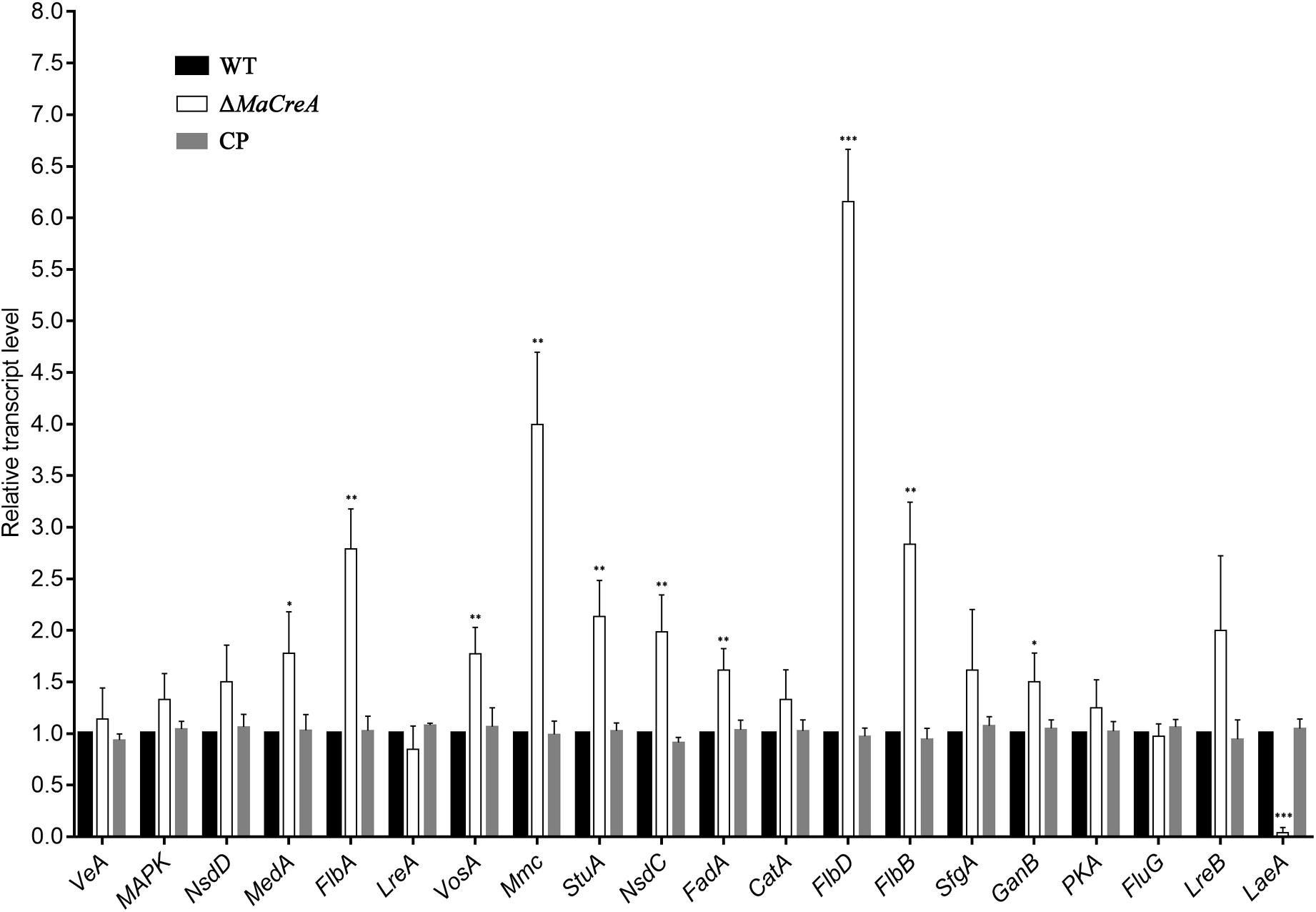
Figure 6. Relative expression of conidiation-related genes in ΔMaCreA (t-text, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
Discussion
CreA, a gene involved in carbon sources utilization, plays a pivotal role in the CCR pathway with a wide range of regulation. This gene is a C2H2 transcription factor and its zinc finger structure is highly conserved from yeasts to human pathogens, implying that CreA plays an important role among different species. In Aspergillus niger, the feruloyl esterase gene faeA, alpha-glucuronidase gene aguA, endoxylanase gene xlnB, and beta-xylosidase gene xlnD in xylose metabolism have been noted to be repressed by CreA (de Vries et al., 1999). In A. nidulans, CreA has been observed to mediate indirect repression of xlnR, which controls the production of xylanolytic enzymes (Tamayo et al., 2008). In B. bassiana, deletion of BbcreA has been demonstrated to result in nutrient toxicity, conidial yield reduction, and defects in virulence (Luo et al., 2014).
While most of the previous studies on CreA had focused on carbon sources metabolism, only a few works had examined the effects of this gene on conidiation in fungi. In the present study, the colonies of ΔMaCreA were noted to be fluffy and white, with strong mycelium, obviously delayed conidiation, and severe decrease in conidial yield. In addition, CreA deletion strain of the Magnaporthe oryzae and Penicillium oxalicum also demonstrated a decrease in conidial yield (Cao et al., 2016; Zhang et al., 2016). The quantitative PCR (qPCR) experiments showed that the expressions of LaeA decreased significantly in ΔMaCreA while the Flbs (FlbA, FlbB, and FlbD) increased significantly, suggesting that CreA positively regulates LaeA and negatively regulates Flbs. The Flb genes have homologous genes in entomopathogenic fungus B. bassiana, and all of them are involved in the conidiation process (Chu et al., 2017). MoRgs1, as a homologous gene of FlbA, has a positive role in conidiation in M. oryzae (Zhang et al., 2011). Chlae1, a homologous gene of LaeA, is present in plant pathogenic fungi Cochliobolus heterostrophus, which clearly affects its asexual development (Wu et al., 2012). Moreover, StuA has been recently identified to regulate conidiation in M. robertsii (Yang et al., 2018). The expressions of these conidiation related genes had been altered in CreA deletion strain, Therefore, it is reasonable to speculate that CreA is involved in the conidiation regulatory pathway of M. acridium.
While asexual conidiation of filamentous fungi has been extensively studied, systematic investigations have been conducted only on Aspergillus spp., especially A. nidulans, in which the functions of genes for asexual conidiation have been elucidated. In the present study, analysis of the genomes of entomopathogenic fungi for homologs of asexual conidiation genes showed that most of the fungi did not possess related core homologous genes, consistent with that reported in previous study (López et al., 2018). Similarly, a recent research on Zymoseptoria tritici showed that asexual conidiation observed in A. nidulans is only partially relevant in Z. tritici, suggesting the presence of uncharacterized genes that control asexual conidiation in this pathogen (Tiley et al., 2018). and indicating that entomopathogenic fungi have their own unique asexual conidiation system. In nature, entomopathogenic fungi have evolved highly diversified survival strategies with co-evolutionary relationships between pathogens and insect hosts (Gao et al., 2011; Wang J.B. et al., 2016; Wang and Wang, 2017), implying rapid changes in their genome sequences to accommodate their own survival. Therefore, asexual conidiation in entomopathogenic fungi also vary.
In the present study, on SYA medium, conidiation was significantly delayed and microcycle conidiation was absent in ΔMaCreA, indicating that MaCreA regulates the shift of the two conidiation patterns. The researches on the mechanism of microcycle conidiation is still scarce. Recently, studies about microcycle conidiation mostly focus on the phenotypic observation in different fungi (Souza-Paccola et al., 2015; van Heerden et al., 2016; Rosli et al., 2018). In the preliminary work in our lab, we found that normal conidiation and microcycle conidiation are involved in lipid metabolism, sugar chain biosynthesis, and metabolism and translation (Wang Z. et al., 2016). Nevertheless, the mechanism of the shifts of the two conidiation patterns remains unclear. Some DEGs are involved in the conidiation process in filamentous fungi, such as fluG, sporulation protein RMD8, APSES transcription factor, and putative UDP-glucose 4-epimerase (Seo et al., 2006; El-Ganiny et al., 2010; Gao et al., 2011; Yang et al., 2018). There are four genes involved in pigmentation synthesis, such as PksP/Alb1, laccase, and laccase Lcc2 and Lcc5 (Langfelder et al., 1998; Rivera-Hoyos et al., 2013). After conidia germination, changes in cell polarity occur (Momany, 2002; Harris, 2006). DEGs of Mucin, WASP-like protein las17, and p21 activated kinase-like protein are involved in cell polarity (Goehring et al., 2003; Pitoniak et al., 2009; Robertson et al., 2009). Therefore, we have reason to conclude that conidiation process also related to polarity changes, when the conidiation process begins, the hyphal polarity maintenance state will converted into isotropic expansion of the daughter conidial cells. Together, these genes are downstream of MaCreA and affect the conidiaion process of M. acridium.
Conclusion
In general, the deletion strain ΔMaCreA exhibited a significant decrease in conidiation yield, indicating that MaCreA played an important role in the conidiation process in the entomopathogenic fungus M. acridum and was a core conidiation regulatory gene. RT-qPCR revealed that MaCreA had certain effections on other known conidiation regulatory genes. High-throughput sequencing results confirmed that deletion of MaCreA altered the expressions of a large number of genes involved in cell cycle, cell division, conidiation, and cell polarity, demonstrating that this gene had a significant function in the process of conidiation. Thus, these results indicated that MaCreA was a core conidiation regulatory gene in M. acridum, providing a new insight into the process of conidiation in entomopathogenic fungi.
Data Availability
The datasets generated for this study can be accessed from the RNA-Seq data, are deposited in the NCBI Sequence Read Archive (SRA) database, and the accession numbers are SRR9089704, SRR9089705, SRR9089702, SRR9089703, SRR9089700, SRR9089701, SRR9089698, SRR9089699, SRR9089696, SRR9089697, SRR9089706, and SRR9089707.
Author Contributions
DS and YS contributed equally to this work. HJ analyzed the RNA-Seq data. All authors read and approved the final manuscript.
Funding
This work was supported by the National Key R&D Program of China (No. 2017YFD0201200), the National Natural Science Foundation of China (Nos. 31572057 and 31071730) and the Technological Innovation and Application Demonstration Project of CQ (cstc2018jscx-msybX0222).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01946/full#supplementary-material
Footnotes
- ^ http://www.geneontology.org/
- ^ https://www.kegg.jp/
- ^ https://web.expasy.org/protparam/
- ^ http://www.cbs.dtu.dk/services/SignalP/
References
Adams, T. H., Wieser, J. K., and Yu, J. H. (1998). Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 62, 545–545.
Adnan, M., Zheng, W., Islam, W., Arif, M., Abubakar, Y. S., Wang, Z., et al. (2017). Carbon catabolite repression in filamentous fungi. Int. J. Mol. Sci. 19:E48. doi: 10.3390/ijms19010048
Borkovich, K. A., and Ebbole, D. J. (2010). Cellular and Molecular Biology of Filamentous Fungi. Washington, DC: American Society for Microbiology Press.
Cao, H. J., Huang, P. Y., Zhang, L. L., Shi, Y. K., Sun, D. D., Yan, Y. X., et al. (2016). Characterization of 47 Cys(2)-His(2) zinc finger proteins required for the development and pathogenicity of the rice blast fungus Magnaporthe oryzae. New Phytol. 211, 1035–1051. doi: 10.1111/nph.13948
Chu, Z. J., Sun, H. H., Ying, S. H., and Feng, M. G. (2017). Vital role for cyclophilin B (CypB) in asexual development, dimorphic transition and virulence of Beauveria bassiana. Fungal Genet. Biol. 105, 8–15. doi: 10.1016/j.fgb.2017.05.004
de Vries, R. P., Visser, J., and de Graaff, L. H. (1999). CreA modulates the XlnR-induced expression on xylose of Aspergillus niger genes involved in xylan degradation. Res. Microbiol. 150, 281–285. doi: 10.1016/S0923-2508(99)80053-9
Dowzer, C. E. A., and Kelly, J. M. (1991). Analysis of the CreA gene, a regulator of carbon catabolite repression in Aspergillus nidulans. Mol. Cell. Biol. 11, 5701–5709. doi: 10.1128/Mcb.11.11.5701
El-Ganiny, A. M., Sheoran, I., Sanders, D. A. R., and Kaminskyj, S. G. W. (2010). Aspergillus nidulans UDP-glucose-4-epimerase UgeA has multiple roles in wall architecture, hyphal morphogenesis, and asexual development. Fungal Genet. Biol. 47, 629–635. doi: 10.1016/j.fgb.2010.03.002
Etxebeste, O., Herrero-Garcia, E., Araujo-Bazan, L., Rodriguez-Urra, A. B., Garzia, A., Ugalde, U., et al. (2009). The bZIP-type transcription factor FlbB regulates distinct morphogenetic stages of colony formation in Aspergillus nidulans. Mol. Microbiol. 73, 775–789. doi: 10.1111/j.1365-2958.2009.06804.x
Gao, Q., Jin, K., Ying, S. H., Zhang, Y., Xiao, G., Shang, Y., et al. (2011). Genome sequencing and comparative transcriptomics of the model entomopathogenic fungi Metarhizium anisopliae and M. acridum. PLoS Genet. 7:e1001264. doi: 10.1371/journal.pgen.1001264
Garzia, A., Etxebeste, O., Herrero-Garcia, E., Fischer, R., Espeso, E. A., and Ugalde, U. (2009). Aspergillus nidulans FlbE is an upstream developmental activator of conidiation functionally associated with the putative transcription factor FlbB. Mol. Microbiol. 71, 172–184. doi: 10.1111/j.1365-2958.2008.06520.x
Goehring, A. S., Mitchell, D. A., Tong, A. H. Y., Keniry, M. E., Boone, C., and Sprague, G. F. (2003). Synthetic lethal analysis implicates Ste20p, a p21-activated protein kinase, in polarisome activation. Mol. Biol. Cell 14, 1501–1516. doi: 10.1091/mbc.E02-06-0348
Hanlin, R. T. (1994). Microcycle conidiation – A review. Mycoscience 35, 113–123. doi: 10.1007/BF02268539
Harris, S. D. (2006). Cell polarity in filamentous fungi: shaping the mold. Int. Rev. Cytol. 251, 41–77. doi: 10.1016/S0074-7696(06)51002-2
He, M., and Xia, Y. X. (2009). Construction and analysis of a normalized cDNA library from metarhizium anisopliae var. acridum germinating and differentiating on locusta migratoria wings. FEMS Microbiol. Lett. 291, 127–135. doi: 10.1111/j.1574-6968.2008.01447.x
Jung, B., Kim, S., and Lee, J. (2014). Microcyle conidiation in filamentous fungi. Mycobiology 42, 1–5. doi: 10.5941/Myco.2014.42.1.1
Kiss, L., Jankovics, T., Kovacs, G. M., and Daughtrey, M. L. (2008). Oidium longipes, a new powdery mildew fungus on petunia in the USA: a potential threat to ornamental and vegetable solanaceous crops. Plant Dis. 92, 818–825. doi: 10.1094/Pdis-92-5-0818
Kwon, N. J., Garzia, A., Espeso, E. A., Ugalde, U., and Yu, J. H. (2010). FlbC is a putative nuclear C2H2 transcription factor regulating development in Aspergillus nidulans. Mol. Microbiol. 77, 1203–1219. doi: 10.1111/j.1365-2958.2010.07282.x
Langfelder, K., Jahn, B., Gehringer, H., Schmidt, A., Wanner, G., and Brakhage, A. A. (1998). Identification of a polyketide synthase gene (pksP) of Aspergillus fumigatus involved in conidial pigment biosynthesis and virulence. Med. Microbiol. Immunol. 187, 79–89. doi: 10.1007/s004300050077
Leandro, L. F. S., Gleason, M. L., Nutter, F. W., Wegulo, S. N., and Dixon, P. A. (2003). Influence of temperature and wetness duration on conidia and appressoria of Colletotrichum acutatum on symptomless strawberry leaves. Phytopathology 93, 513–520. doi: 10.1094/Phyto.2003.93.4.513
Lee, B. N., and Adams, T. H. (1994a). Overexpression of FlbA, an early regulator of Aspergillus asexual sporulation, leads to activation of BrlA and premature initiation of development. Mol. Microbiol. 14, 323–334. doi: 10.1111/j.1365-2958.1994.tb01293.x
Lee, B. N., and Adams, T. H. (1994b). The Aspergillus nidulans FluG Gene Is Required for production of an extracellular developmental signal and is related to prokaryotic glutamine-synthetase I. Genes Dev. 8, 641–651. doi: 10.1101/gad.8.6.641
Liu, J., Cao, Y. Q., and Xia, Y. X. (2010). Mmc, a gene involved in microcycle conidiation of the entomopathogenic fungus Metarhizium anisopliae. J. Invertebr. Pathol. 105, 132–138. doi: 10.1016/j.jip.2010.05.012
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-CT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
López, M. O., Chen, W., Eagle, C. E., Gutiérrez, G., and Dyer, P. S. (2018). Evolution of asexual and sexual reproduction in the Aspergilli. Stud. Mycol. 91, 37–59. doi: 10.1016/j.simyco.2018.10.002
Luo, Z. B., Qin, Y. Q., Pei, Y., and Keyhani, N. O. (2014). Ablation of the creA regulator results in amino acid toxicity, temperature sensitivity, pleiotropic effects on cellular development and loss of virulence in the filamentous fungus Beauveria bassiana. Environ. Microbiol. 16, 1122–1136. doi: 10.1111/1462-2920.12352
Mathieu, M., and Felenbok, B. (1994). The Aspergillus nidulans CreA protein mediates glucose repression of the ethanol regulon at various levels through competition with the alcr-specific transactivator. EMBO J. 13, 4022–4027. doi: 10.1002/j.1460-2075.1994.tb06718.x
Mirabito, P. M., Adams, T. H., and Timberlake, W. E. (1989). Interactions of three sequentially expressed genes control temporal and spatial specificity in Aspergillus development. Cell 57, 859–868. doi: 10.1016/0092-8674(89)90800-3
Momany, M. (2002). Polarity in filamentous fungi: establishment, maintenance and new axes. Curr. Opin. Microbiol. 5, 580–585. doi: 10.1016/S1369-5274(02)00368-5
Papagianni, M. (2004). Fungal morphology and metabolite production in submerged mycelial processes. Biotechnol. Adv. 22, 189–259. doi: 10.1016/j.biotechadv.2003.09.005
Park, H. S., and Yu, J. H. (2012). Genetic control of asexual sporulation in filamentous fungi. Curr. Opin. Microbiol. 15, 669–677. doi: 10.1016/j.mib.2012.09.006
Pitoniak, A., Birkaya, B., Dionne, H. M., Vadaie, N., and Cullen, P. J. (2009). The signaling mucins Msb2 and Hkr1 differentially regulate the filamentation mitogen-activated protein kinase pathway and contribute to a multimodal response. Mol. Biol. Cell 20, 3101–3114. doi: 10.1091/mbc.E08-07-0760
Rivera-Hoyos, C. M., Morales-Alvarez, E. D., Poutou-Pinales, R. A., Pedroza-Rodriguez, A. M., Rodriguez-Vazquez, R., and Delgado-Boada, J. M. (2013). Fungal laccases. Fungal Biol. Rev. 27, 67–82. doi: 10.1016/j.fbr.2013.07.001
Robertson, A. S., Allwood, E. G., Smith, A. P. C., Gardiner, F. C., Costa, R., Winder, S. J., et al. (2009). The WASP homologue Las17 activates the novel actin-regulatory activity of Ysc84 to promote endocytosis in yeast. Mol. Biol. Cell 20, 1618–1628. doi: 10.1091/mbc.E08-09-0982
Rosli, H., Batzer, J. C., Harrington, T. C., and Gleason, M. L. (2018). Peltaster gemmifer: a new species in the sooty blotch and flyspeck species complex from the United States. Mycologia 110, 822–834. doi: 10.1080/00275514.2018.1486679
Schrank, A., and Vainstein, M. H. (2010). Metarhizium anisopliae enzymes and toxins. Toxicon 56, 1267–1274. doi: 10.1016/j.toxicon.2010.03.008
Seo, J. A., Guan, Y. J., and Yu, J. H. (2006). FluG-dependent asexual development in Aspergillus nidulans occurs via derepression. Genetics 172, 1535–1544. doi: 10.1534/genetics.105.052258
Son, H., Kim, M. G., Min, K., Lim, J. Y., Choi, G. J., Kim, J. C., et al. (2014). WetA is required for conidiogenesis and conidium maturation in the ascomycete fungus Fusarium graminearum. Eukaryot. Cell 13, 87–98. doi: 10.1128/Ec.00220-13
Souza-Paccola, E. A. D., Bomfeti, C. A., Tanaka, F. A. O., Massola Junior, N. S., Colauto, N. B., Figueiredo, J. E. F., et al. (2015). Novel insights into the early stages of infection by oval conidia of Colletotrichum sublineolum. Sci. Agr. 72, 351–355. doi: 10.1590/0103-9016-2014-0409
Tamayo, E. N., Villanueva, A., Hasper, A. A., de Graaff, L. H., Ramon, D., and Orejas, M. (2008). CreA mediates repression of the regulatory gene xlnR which controls the production of xylanolytic enzymes in Aspergillus nidulans. Fungal Genet. Biol. 45, 984–993. doi: 10.1016/j.fgb.2008.03.002
Tiley, A. M. M., Foster, G. D., and Bailey, A. M. (2018). Exploring the genetic regulation of asexual sporulation in Zymoseptoria tritici. Front. Microbiol. 9:1859. doi: 10.3389/fmicb.2018.01859
van Heerden, A., Mouton, M., Postma, F., van Wyk, P. W., Lerm, B., Van Zyl, W. H., et al. (2016). The microcyclic conidial stage of Coniochaeta pulveracea and its effect on selected biological interactions. Folia Microbiol. 61, 319–328. doi: 10.1007/s12223-015-0441-8
Wang, C., and Wang, S. (2017). Insect pathogenic fungi: genomics, molecular interactions, and genetic improvements. Annu. Rev. Entomol. 62, 73–90. doi: 10.1146/annurev-ento-031616-035509
Wang, J. B., St Leger, R. J., and Wang, C. (2016). Advances in genomics of entomopathogenic fungi. Adv. Genet. 94, 67–105. doi: 10.1016/bs.adgen.2016.01.002
Wang, Z., Jin, K., and Xia, Y. (2016). Transcriptional analysis of the conidiation pattern shift of the entomopathogenic fungus Metarhizium acridum in response to different nutrients. BMC Genomics 17:586. doi: 10.1186/s12864-016-2971-0
Wu, D., Oide, S., Zhang, N., Choi, M. Y., and Turgeon, B. G. (2012). ChLae1 and ChVel1 regulate T-toxin production, virulence, oxidative stress response, and development of the maize pathogen Cochliobolus heterostrophus. PLoS Pathog. 8:e1002542. doi: 10.1371/journal.ppat.1002542
Yang, W., Wu, H., Wang, Z., Sun, Q., Qiao, L., and Huang, B. (2018). The APSES Gene MrStuA regulates sporulation in Metarhizium robertsii. Front. Microbiol. 9:1208. doi: 10.3389/fmicb.2018.01208
Yang, Y. S., Li, L., Li, X., Shao, Y. C., and Chen, F. S. (2012). mrflba, encoding a putative FlbA, is involved in aerial hyphal development and secondary metabolite production in monascus ruber M-7. Fungal Biol. 116, 225–233. doi: 10.1016/j.funbio.2011.11.005
Yu, J. H., Wieser, J., and Adams, T. H. (1996). The Aspergillus flbA RGS domain protein antagonizes G protein signaling to block proliferation and allow development. EMBO J. 15, 5184–5190. doi: 10.1002/j.1460-2075.1996.tb00903.x
Zhang, H., Tang, W., Liu, K., Huang, Q., Zhang, X., Yan, X., et al. (2011). Eight RGS and RGS-like proteins orchestrate growth, differentiation, and pathogenicity of magnaporthe oryzae. PLoS Pathog. 7:e1002450. doi: 10.1371/journal.ppat.1002450
Zhang, S. Z., Peng, G. X., and Xia, Y. X. (2010). Microcycle conidiation and the conidial properties in the entomopathogenic fungus Metarhizium acridum on agar medium. Biocontrol Sci. Technol. 20, 809–819. doi: 10.1080/09583157.2010.482201
Zhang, X. J., Zhu, Y. Y., Bao, L. F., Gao, L. W., Yao, G. S., Li, Y. N., et al. (2016). Putative methyltransferase LaeA and transcription factor CreA are necessary for proper asexual development and controlling secondary metabolic gene cluster expression. Fungal Genet. Biol. 94, 32–46. doi: 10.1016/j.fgb.2016.07.004
Keywords: creA, normal conidiation, microcycle conidiation, conidiation pattern shift, Metarhizium acridum
Citation: Song D, Shi Y, Ji HQ, Xia Y and Peng G (2019) The MaCreA Gene Regulates Normal Conidiation and Microcycle Conidiation in Metarhizium acridum. Front. Microbiol. 10:1946. doi: 10.3389/fmicb.2019.01946
Received: 22 June 2019; Accepted: 07 August 2019;
Published: 21 August 2019.
Edited by:
Chengshu Wang, Institute of Plant Physiology and Ecology (SIBS-CAS), ChinaCopyright © 2019 Song, Shi, Ji, Xia and Peng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guoxiong Peng, gxpeng@cqu.edu.cn; Yuxian Xia, yuxianxia@cqu.edu.cn
†These authors have contributed equally to this work
‡ORCID: Guoxiong Peng, orcid.org/0000-0001-5551-703X; Yuxian Xia, orcid.org/0000-0003-2443-8691
 Dongxu Song
Dongxu Song Youhui Shi1,2,3†
Youhui Shi1,2,3† Yuxian Xia
Yuxian Xia Guoxiong Peng
Guoxiong Peng