
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol. , 21 August 2019
Sec. Microbial Physiology and Metabolism
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.01943
This article is part of the Research Topic Interdisciplinary Approaches to Face New Challenges on the N-Cycle View all 5 articles
Nitrous oxide (N2O) is the third most important greenhouse gas after carbon dioxide and methane, and a prominent ozone-depleting substance. Agricultural soils are the primary anthropogenic source of N2O because of the constant increase in the use of industrial nitrogen (N) fertilizers. The soybean crop is grown on 6% of the world’s arable land, and its production is expected to increase rapidly in the future. In this review, we summarize the current knowledge on N-cycle in the rhizosphere of soybean plants, particularly sources and sinks of N2O. Soybean root nodules are the host of dinitrogen (N2)-fixing bacteria from the genus Bradyrhizobium. Nodule decomposition is the main source of N2O in soybean rhizosphere, where soil organisms mediate the nitrogen transformations that produce N2O. This N2O is either emitted into the atmosphere or further reduced to N2 by the bradyrhizobial N2O reductase (N2OR), encoded by the nos gene cluster. The dominance of nos– indigenous populations of soybean bradyrhizobia results in the emission of N2O into the atmosphere. Hence, inoculation with nos+ or nos++ (mutants with enhanced N2OR activity) bradyrhizobia has proved to be promising strategies to reduce N2O emission in the field. We discussed these strategies, the molecular mechanisms underlying them, and the future perspectives to develop better options for global mitigation of N2O emission from soils.
The term, “rhizosphere” was first defined by Lorenz Hiltner as the soil compartment influenced by plant roots. Since then, the rhizosphere has received widespread attention from scientists in different disciplines as a hotspot for intra-microbial and plant-microbe interactions (Hartmann et al., 2008; Bakker et al., 2013). Nitrogen (N), an essential component in living organisms, is the most common limiting nutrient for plant growth in agricultural soils (LeBauer and Treseder, 2008). Fixation of dinitrogen (N2) into ammonia (NH3) by legume-associated endosymbiotic bacteria, generally known as rhizobia, is a major source of N in soils, and is an agriculturally and ecologically crucial process to reduce plant dependence on industrial N fertilizers. Rhizobia are hosted within the nodules formed in the roots as the result of the symbiosis; inside the nodules, rhizobial cells encounter oxygen-limiting conditions that are required for the synthesis and activity of nitrogenase that converts N2 into NH3 (Figure 1). Thus, rhizobia provide N to the host plant, which in return, supplies photosynthetically fixed carbon to the bacteria. Although rhizobia have been extensively studied as inhabitants of legume nodules, only few studies have focused on them as denitrifiers in the legume rhizosphere (Inaba et al., 2009, 2012; Shiina et al., 2014; Saeki et al., 2017).
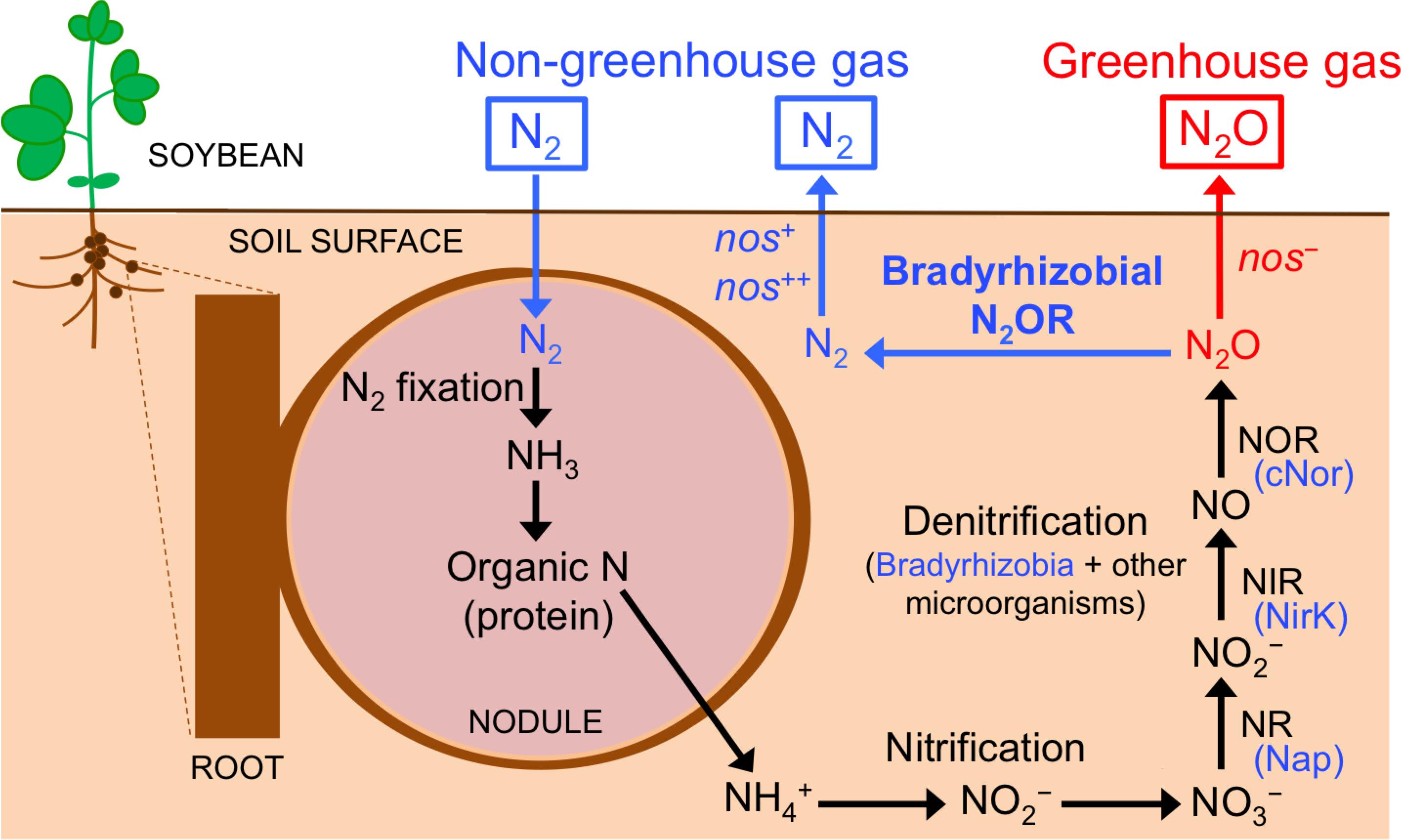
Figure 1. Nitrogen transformations in soybean rhizosphere. Organic nitrogen in the nodule is mineralized into NH4+ that will be transformed into N2O through nitrification and denitrification processes. Soybean bradyrhizobia (blue) and other microorganisms contribute to the denitrification process. The N2O formed is either emitted into the atmosphere (red) or further reduced to N2 exclusively by soybean bradyrhizobia that produce N2O reductase (N2OR; blue). Nase, nitrogenase; NR, dissimilatory nitrate reductase; Nap, periplasmic nitrate reductase; NIR, dissimilatory nitrite reductase; NirK, Cu-containing nitrite reductase; NOR, nitric oxide reductase; cNor, c-type nitric oxide reductase. See text for more details.
Soybean [Glycine max (L.) Merr.] is grown on 6% of the world’s arable land. Its production increased from 17 to 230 million metric tons in the past 50 years, and is expected to increase rapidly in the future due to an increased demand for soybean meal and oil (Uchida and Akiyama, 2013). Soybean generally hosts rhizobia from the genus Bradyrhizobium (Argaw, 2014). In addition to fixing N2, many soybean-associated Bradyrhizobium strains contain genes for some, or all of the four denitrification reductases. Denitrification is an alternate respiratory process in which the oxidized forms of N in the soil – nitrate (NO3–) and nitrite (NO2–) – are used as electron acceptors in oxygen limiting conditions. NO2– is reduced to nitric oxide (NO), nitrous oxide (N2O), and N2 gases, which are returned to the atmosphere (Figure 1). The complete denitrification pathway in soybean bradyrhizobia requires four enzymes, periplasmic NO3– reductase (Nap), copper (Cu)-containing NO2– reductase (NirK), c-type NO reductase (cNor), and N2O reductase (N2OR) (Figure 1). Bradyrhizobial denitrification is functional under both free-living (for example, in the soybean rhizosphere) and symbiotic (inside the root nodules) conditions (Sameshima-Saito et al., 2006a; Sánchez et al., 2011; Inaba et al., 2012).
N2O is the third most important greenhouse gas after carbon dioxide (CO2) and methane (CH4) and is currently the major ozone-depleting compound in the stratosphere (Hénault et al., 2012; Thomson et al., 2012). Terrestrial ecosystems are the main source of N2O, primarily due to the use of industrial N fertilizers in agriculture (Hénault et al., 2012; Thomson et al., 2012; Intergovernmental Panel on Climate Change [IPCC], 2014). The soybean rhizosphere is a hotspot for N transformations including production and removal of N2O. Nodule decomposition is a major source of N2O, particularly in soybean ecosystems, compared to other possible sources including aboveground plant residues. Inaba et al. (2009) showed that N2O is only emitted by decomposed nodules, but not by fresh nodules or roots. Studies showed that N2O emission not only occurs from decomposed nodules after soybean harvesting, but also starts before the harvest till the late growth period (Yang and Cai, 2005; Inaba et al., 2012; reviewed by Uchida and Akiyama, 2013). Biological N2 fixation is an important indirect source for N2O during nodule decomposition. Indeed, a 15N tracer experiment revealed that the N2O emitted from the soybean rhizosphere was almost entirely derived from N2 fixed symbiotically in the nodules (Inaba et al., 2012; Figure 1). During nodule decomposition, rhizospheric microbes are essential for N2O emission; organic N from the nodule is mineralized into ammonium (NH4+, Figure 1); N2O is then produced via nitrification and denitrification (Inaba et al., 2009, 2012; Figure 1). Although soybean bradyrhizobia are important players in denitrification (responsible for ∼41% of the total N2O produced), but other denitrifying microorganisms are also important contributors (∼59% of the total N2O produced; Inaba et al., 2012). Populations of nematodes, protozoans, and fungi were markedly enhanced in the soybean rhizosphere of decomposing nodules, suggesting that these organisms contributed to the complex N transformation (Inaba et al., 2009). N2O formed by denitrification is either emitted into the atmosphere or is further reduced to N2 by N2O reductase of soybean bradyrhizobia (Sameshima-Saito et al., 2004, 2006b; Inaba et al., 2012; Figure 1). In soybean fields, both N2- and N2O-producing soybean bradyrhizobia strains coexist; therefore soybean roots are infected with multiple bradyrhizobial strains that differ in denitrifying activity (Sameshima-Saito et al., 2004, 2006b; Shiina et al., 2014). Thus, the flux of N2O from soybean fields during nodule decomposition is partly determined by biotic factors like the balance between N2O emission by soil microbes whose denitrification produce N2O (including bradyrhizobia) and N2O uptake by soybean bradyrhizobia that produce N2OR (Inaba et al., 2012; Figure 1).
N2OR is a Cu-containing enzyme that catalyzes the two-electron reduction of N2O to N2, which is the only known pathway for the removal of N2O from ecosystems (Richardson et al., 2009). Therefore, the expression and activity of N2OR is a natural target to mitigate N2O emission from agricultural soils.
In Bradyrhizobium diazoefficiens (reclassified from Bradyrhizobium japonicum by Delamuta et al., 2013), the N2OR (NosZ) and its accessory functions are encoded by nosRZDYFLX gene cluster (Velasco et al., 2004). The flavoproteins NosR and NosX form an electron transport pathway from the quinone pool to NosZ; NosR is also required for the transcription of nos genes (Velasco et al., 2004; Zumft and Kroneck, 2007). NosD, NosF, NosY, and NosL are involved in maturation of the CuZ site of NosZ (Zumft and Kroneck, 2007). Although the reduction of N2O to N2 by N2OR is integrated as the last step of the denitrification pathway, it can provide a benefit for N2O respiration as a separate module. When N2O is provided as the sole electron acceptor to B. diazoefficiens, anaerobic respiration and growth are sustained by reducing N2O to N2 (Zumft, 1997; Sánchez et al., 2013; Graf et al., 2014).
Bradyrhizobium diazoefficiens carries the nos gene cluster (nos+) and denitrifies NO3– to N2, whereas other soybean bradyrhizobia including B. japonicum lack the nos gene cluster (nos–) and cannot reduce N2O to N2 (Sameshima-Saito et al., 2004, 2006b; Inaba et al., 2012). Sameshima-Saito et al. (2006a) showed that soybean roots nodulated with B. diazoefficiens could scavenge very low concentrations of exogenous N2O, equivalent to the natural concentration of N2O in air (∼0.34 ppm; Badr and Probert, 1992). Later, pot studies demonstrated that soybean roots inoculated with nos+ strains have the potential to reduce N2O derived from decomposing nodules and other N sources from fertilizer and soil organic matter (Hénault and Revellin, 2011; Inaba et al., 2012; Uchida and Akiyama, 2013). Thus, Bradyrhizobium nos+ strain inoculation is a promising strategy for mitigating N2O emission at the field scale. This is likely effective in soybean soils that act as an N2O source, a condition that potentially arises from several situations like (i) indigenous bradyrhizobia community being dominated by nos– species (Sameshima-Saito et al., 2004, 2006b; Shiina et al., 2014), (ii) anoxic conditions such as waterlogging that induce N2O emissions from denitrification by Bradyrhizobium (Tortosa et al., 2016) and other microorganisms, and (iii) increased NO3– supply as a consequence of heavy N fertilization leading to increased N2O emission from intact soybean root systems via bradyrhizobial denitrification (Ciampitti et al., 2008; Hirayama et al., 2011; Inaba et al., 2012).
Among the N2O-mitigation options for agricultural soils, the first biological method for the field scale was described by Itakura et al. (2013). Mutants of B. diazoefficiens USDA110 with a high nos expression and N2OR activity (nos++ strains) were generated by a mutational strategy (Itakura et al., 2008). This strategy involved (1) introduction of a plasmid containing a mutated dnaQ gene (pKQ2) to enhance replication error on the B. diazoefficiens genome by disrupting the exonuclease proofreading activity of DNA polymerase, (2) enrichment culture under selection pressure favoring anaerobic N2O respiration, and (3) elimination of the pKQ2 plasmid by nodulation. Thus, the resulting mutants were not genetically modified organisms (GMOs). The nos++ mutants retained higher nos expression and N2OR activity in both free-living and symbiotic cells than the wild-type nos+ strains (Itakura et al., 2013; Sánchez et al., 2014). Comparative analysis of the nos++ mutant genomes revealed the mechanism underlying the nos++ phenotype, a point mutation in nasS gene encoding the NO3– sensor of the two-component NasST regulatory system (Sánchez et al., 2014, 2017), which will be discussed later.
The effectiveness of N2O emission mitigation by the nos++ mutant was first confirmed under simulated field conditions in a pot experiment with Andosol soil, which predominantly contains nos– bradyrhizobia (Itakura et al., 2013). N2O emission from the Andosol soil inoculated with the nos++ mutant strain was significantly reduced compared with that inoculated with wild-type nos+ strain. Itakura et al. (2013) demonstrated that inoculation of nos++ strains to growing soybean in Andosol soil reduced postharvest N2O emission by 43% in the lysimeter study and by 54% in the farm-scale study. However, reduction in postharvest N2O emission by inoculation with the nos++ strains was not significant in a Gleysol soil, which predominantly contains nos+ bradyrhizobia, although the nos++ strains clearly showed higher N2O-reducing potential than that of the nos+ strains under laboratory conditions (Itakura et al., 2013; Shiina et al., 2014). Thus, some factors present in the soybean rhizosphere of Gleysol soil limited the potential N2O mitigation ability of the nos++ strains.
A recent report showed that inoculation of soil with a mixed and enriched culture of indigenous nos+ strains of the B. diazoefficiens USDA110 group isolated from agricultural fields efficiently mitigated N2O emission (Akiyama et al., 2016). As in the nos++ approach above, inoculation with the mixed culture was successful in soils dominated by nos– bradyrhizobia. Additionally, this mixture is expected to be more competitive and adaptable to changing environmental factors than a single strain (Akiyama et al., 2016). This method is an alternative to GMOs and overcomes the problem of strong opposition to them.
Considering the importance of N2OR in N2O removal from ecosystems, significant progress has been made in understanding its genetic regulation in bacteria, especially in the denitrifying bacteria, Paracoccus denitrificans (reviewed by Gaimster et al., 2018), and B. diazoefficiens as a model for denitrification in legume-associated rhizobia. In the latter bacterium, the nosR gene is constitutively expressed at a low level from the promoter Pa in aerobiosis, but is strongly induced from the promoter Pd under denitrifying conditions (i.e., anoxia with NO3– as electron acceptor), which is dependent on the oxygen-responsive regulatory cascade FixLJ–FixK2 (Torres et al., 2016, 2017; Sánchez et al., 2017; Figure 2A). Decreasing oxygen level to 5% during a culture triggers ATP-dependent autophosphorylation of the heme-based sensor kinase, FixL to phosphorylate the response regulator FixJ, which activates FixK2, a transcriptional activator that directly interacts with the nosR promoter (Torres et al., 2016, 2017; Figure 2A). Although the FixLJ–FixK2 cascade has been considered as the main regulator for nos genes in bradyrhizobia for a long time, the NasST two-component system has been revealed as an important regulator of nos transcription in response to NO3– under both aerobic and anaerobic conditions (Sánchez et al., 2014, 2017; Figure 2A). The nasST operon encodes a NO3– and NO2– sensor/transcriptional antitermination regulatory system. This system was initially considered to be involved in the NO3–/NO2–-responsive regulation of the nas genes for the NO3– assimilation pathway in bacteria, including B. diazoefficiens (Romeo et al., 2012; Wang et al., 2012; Luque-Almagro et al., 2013; Cabrera et al., 2016). A recent transcriptomic study using RNA-seq has shown that most of the genes whose expression changed in the B. diazoefficiens ΔnasT mutant are related to N metabolism, especially amino acid transport (Sánchez et al., 2019).
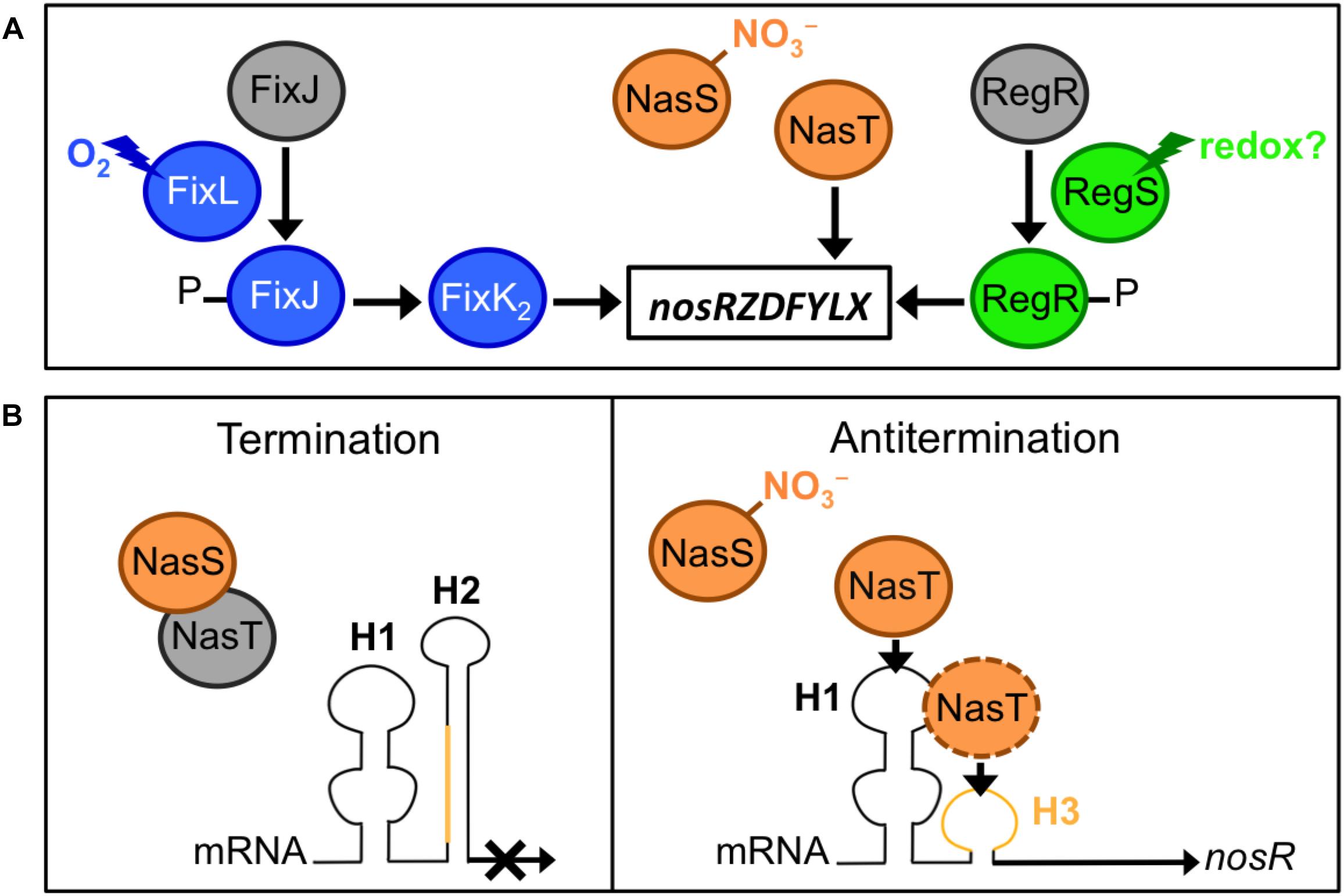
Figure 2. Regulation of N2O reductase genes in bradyrhizobia. (A) Environmental factors and regulatory proteins involved in the control of nos expression. (B) The mechanism for NasT-mediated transcriptional antitermination of the nos genes. In the absence of nitrate (NO3–), NasT is sequestered by NasS; thus in the absence of NasT, the native conformation of the nosR-leader mRNA, which contains the H1 and H2 hairpins, is responsible for the termination of the nos transcription. When a certain level of NO3– is sensed by NasS, NasT is dissociated from NasS–NasT complex; the binding of NasT to H1 (and likely to H3 region, in orange) results in a conformational change in the mRNA that allows the read-through transcription of nos genes.
NasS contains a NO3–/NO2–-binding motif similar to that of NrtA, which is the periplasmic component of an ABC-type system for NO3– and NO2– uptake in cyanobacteria (Koropatkin et al., 2006). NasT is an ANTAR (AmiR and NasR transcription antitermination regulator)-family protein (Shu and Zhulin, 2002). NasS and NasT form a complex that dissociates when NasS senses NO3– in micromolar concentrations (Luque-Almagro et al., 2013; Sánchez et al., 2014; Hidaka et al., 2016). When NO3– is present, nos expression is markedly decreased (∼70%) in the ΔnasT background. In absence of NO3–, nos expression is induced in the ΔnasS background but such induction is abolished with the additional deletion of nasT. Thus, NO3– counteracts the NasS-mediated inhibition of nos by allowing the dissociation of the antiterminator NasT from the NasS-NasT complex (Sánchez et al., 2014, 2017; Figure 2). Then, the application of nos++ mutants (carrying a mutation in nasS) may be more effective than that of wild type nos+ alone if NO3– concentration in the rhizosphere is below the threshold for dissociation of the NasS-NasT complex. Although the concentration threshold in vitro is within the micromolar range (Hidaka et al., 2016), this concentration remains to be fixed under soil conditions.
When NasT is released from NasS, NasT interacts directly with a 5′-leader region of the nosR mRNA and interferes with the formation of a terminator structure, allowing a read-through transcription of nos genes (Sánchez et al., 2017; Figure 2B). The transcription terminator located upstream of nosR comprises two RNA-hairpin structures (H1 and H2); the binding of NasT to H1 induces a conformational change in the terminator and facilitates read-through transcription to induce nos expression (Sánchez et al., 2017; Figure 2B). Deletion of either H1 or H2 increases nos expression in the absence or presence of NO3– (Sánchez et al., 2014, 2017; Figure 2). Thus, theoretically, a B. diazoefficiens mutant defective in H1 (Figure 2B) would be an ideal nos++ inoculant, because (i) it is expected to specifically induce nos genes, whereas nos++ strains derived from nasS mutations affected other genes controlled by the NasST system, and (ii) nos induction is independent of soil NO3– concentration (Itakura et al., 2013; Sánchez et al., 2017, 2019). Mutation of H1 may be applicable to other agriculturally important nos+ bacteria such as Bradyrhizobium oligotrophicum S58, an endophyte of rice roots – where it potentially fixes N2 (Ohta and Hattori, 1983; Okubo et al., 2013; Sánchez et al., 2017; Sánchez and Minamisawa, 2018).
Furthermore, studies on P. denitrificans have shown that reduction of N2O to N2 is dependent on Cu, a key cofactor of the Nos enzyme. Thus, bacterial cultures lacking Cu accumulate significant amounts of N2O (Felgate et al., 2012). Cu deficiency results in a decreased expression of nosZ (Sullivan et al., 2013). Another key factor is the pH that significantly affects N2O emission from microbes. The expression of functional N2OR is difficult at low pH (Bakken et al., 2012). Sinorhizobium meliloti, the alfalfa endosymbiont, is unable to express N2OR at pH 6 (Bueno et al., 2015). In P. denitrificans, pH has little effect on the transcription of the nosZ, but may have a direct posttranslational effect on the assembly and/or activity of the N2OR holoenzyme (Bakken et al., 2012). The effect of Cu or pH on the reduction of N2O to N2 in B. diazoefficiens is currently unknown. Among the environmental factors that affect the bacterial N2OR activity, very little is known about the effect of availability and redox state of carbon sources. In this context, the response regulator RegR of the RegSR two-component regulatory system appears to induce nos expression in B. diazoefficiens, most likely in response to redox state (Torres et al., 2016; Figure 2A).
The understanding of N2O production in the soybean rhizosphere has been significantly advanced. A variety of techniques, such as functional omics, 15N isotope analysis, and zymography, will facilitate a better understanding of the players and processes for N transformation in the soybean rhizosphere of degrading nodules. In addition, further studies on soil factors that control the amount and distribution of soybean bradyrhizobia in the rhizosphere are required because they are key determinants for the flux of N2O during nodule decomposition (Inaba et al., 2012).
Shiina et al. (2014) reported that the soil type determines the occurrence of B. diazoefficiens (nos+) or B. japonicum (nos–) in Japanese soybean fields; the nos+ bradyrhizobia are predominant in Gleysol (wetland soils where water regime causes low-oxygen conditions), whereas the nos– bradyrhizobia are predominant in Andosols (volcanic soils containing porous sediments, resulting in more aerated conditions). Saeki et al. (2017) reported that the presence of nos in B. diazoefficiens confers a competitive advantage in flooded soils with low-oxygen conditions, similar to Gleysol soils. However, batch experiments suggested that B. japonicum may be less competitive compared to B. diazoefficiens due to energy depletion under anaerobic conditions, which is associated with a marked impairment of Nap activity in B. japonicum and not with the absence of nos (Siqueira et al., 2017). These findings emphasize the need for further research on how soil factors influence the relevance of the N2O reduction step in bradyrhizobial competition.
Significant advances have led to the use of bradyrhizobial N2OR as an N2O sink in soybean ecosystems. Following the work done by Itakura et al. (2013) and Akiyama et al. (2016), promising strategies for production of rhizobial inoculants for N2O mitigation would be the selection of superior native strains (in terms of adaptation to local environments and N2-fixing symbiotic efficiency) and the optimization of N2O reduction activity through appropriate genetic modification or management of soil chemical and physical properties. However, generating mutants requires more time, cost, and technical skill than isolating nos+ strains from local soybean fields (Itakura et al., 2013; Akiyama et al., 2016). Moreover, inoculating a mixture of native strains provides more adaptability than a single strain (Akiyama et al., 2016). Thus, isolating nos+ strains from local fields is more feasible for many soybean-producing countries and is potentially applicable to other ecosystems. Indeed, it has already been suggested the potential activity of Ensifer (formerly Sinorhizobium) meliloti, the alfalfa endosymbiont (Bueno et al., 2015).
Both authors have contributed equally to the discussion, writing, and approving the manuscript.
This work was supported by a Grant-in-Aid for Scientific Research (A) 26252065 and (B) 18H02112 from the Ministry of Education, Culture, Sports, Science and Technology, Japan, and by grants from the Project of the NARO Bio-oriented Technology Research Advancement Institution (Research Program on Development of Innovative Technology).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Akiyama, H., Hoshino, Y. T., Itakura, M., Shimomura, Y., Wang, Y., Yamamoto, A., et al. (2016). Mitigation of soil N2O emission by inoculation with a mixed culture of indigenous Bradyrhizobium diazoefficiens. Sci. Rep. 6:32869. doi: 10.1038/srep3286910.1038/srep32869
Argaw, A. (2014). Symbiotic effectiveness of inoculation with Bradyrhizobium isolates on soybean [Glycine max (L.) Merrill] genotypes with different maturities. Springer Plus. 3:753. doi: 10.1186/2193-1801-3-753
Badr, O., and Probert, S. D. (1992). Nitrous oxide in the Earth’s atmosphere. Appl. Energ. 41, 177–200. doi: 10.1016/0306-2619(92)90002-s
Bakken, L. R., Bergaust, L., Liu, B., and Frostegard, Å (2012). Regulation of denitrification at the cellular level: a clue to the understanding of N2O emissions from soils Phil. Trans. R. Soc. B. 367, 1224–1234. doi: 10.1098/rstb.2011.0321
Bakker, P. A., Berendsen, R. L., Doornbos, R. F., Wintermans, P. C., and Pieterse, C. M. (2013). The rhizosphere revisited: root microbiomics. Front. Plant Sci. 4:165. doi: 10.3389/fpls.2013.00165
Bueno, E., Mania, D., Frostegard, Å, Bedmar, E. J., Bakken, L. R., and Delgado, M. J. (2015). Anoxic growth of Ensifer meliloti 1021 by N2O-reduction, a potential mitigation strategy. Front. Microbiol. 6:537. doi: 10.3389/fmicb.2015.00537
Cabrera, J. J., Salas, A., Torres, M. J., Bedmar, E. J., Richardson, D. J., Gates, A. J., et al. (2016). An integrated biochemical system for nitrate assimilation and nitric oxide detoxification in Bradyrhizobium japonicum. Biochem. J. 473, 297–309. doi: 10.1042/BJ20150880
Ciampitti, I. A., Ciarlo, E. A., and Conti, M. E. (2008). Nitrous oxide emissions from soil during soybean [(Glycine max (L.) Merrill] crop phenological stages and stubbles decomposition period. Biol. Fertil. Soils 44, 581–588. doi: 10.1007/s00374-007-0241-7
Delamuta, J. R., Ribeiro, R. A., OrmeñÞo-Orrillo, E., Melo, I. S., Martínez-Romero, E., and Hungria, M. (2013). Polyphasic evidence supporting the reclassification of Bradyrhizobium japonicum group Ia strains as Bradyrhizobium diazoefficiens sp. nov. Int. J. Syst. Evol. Microbiol. 63, 3342–3351. doi: 10.1099/ijs.0.049130-0
Felgate, H., Giannopoulos, G., Sullivan, M. J., Gates, A. J., Clarke, T. A., Baggs, E., et al. (2012). The impact of copper, nitrate and carbon status on the emission of nitrous oxide by two species of bacteria with biochemically distinct denitrification pathways. Environ. Microbiol. 14, 1788–1800. doi: 10.1111/j.1462-2920.2012.02789.x
Gaimster, H., Alston, M., Richardson, D. J., Gates, A. J., and Rowley, G. (2018). Transcriptional and environmental control of bacterial denitrification and N2O emissions. FEMS Microbiol. Lett. 365:fnx277. doi: 10.1093/femsle/fnx277
Graf, D. R., Jones, C. M., and Hallin, S. (2014). Intergenomic comparisons highlight modularity of the denitrification pathway and underpin the importance of community structure for N2O emissions. PLoS One 9:e114118. doi: 10.1371/journal.pone.0114118
Hartmann, A., Rothballer, S., and Schmid, M. (2008). Lorenz Hiltner, a pioneer in rhizosphere microbial ecology and soil bacteriology research. Plant Soil 312, 7–14. doi: 10.1007/s11104-007-9514-z
Hénault, C., Grossel, A., Mary, B., Roussel, M., and Leonard, J. (2012). Nitrous oxide emission by agricultural soils: a review of spatial and temporal variability for mitigation. Pedosphere 22, 426–433. doi: 10.1016/s1002-0160(12)60029-0
Hénault, C., and Revellin, C. (2011). Inoculants of leguminous crops for mitigating soil emissions of the greenhouse gas nitrous oxide. Plant Soil 346, 289–296. doi: 10.1007/s11104-011-0820-0
Hidaka, M., Gotoh, A., Shimizu, T., Minamisawa, K., Imamura, H., and Uchida, T. (2016). Visualization of NO3-/NO2- dynamics in living cells by fluorescence resonance energy transfer (FRET) imaging employing a rhizobial two-component regulatory system. J. Biol. Chem. 291, 2260–2269. doi: 10.1074/jbc.M115.687632
Hirayama, J., Eda, S., Mitsui, H., and Minamisawa, K. (2011). Nitrate-dependent N2O emission from intact soybean nodules via denitrification by Bradyrhizobium japonicum bacteroids. Appl. Environ. Microbiol. 77, 8787–8790. doi: 10.1128/aem.06262-11
Inaba, S., Ikenishi, F., Itakura, M., Kikuchi, M., Eda, S., Chiba, N., et al. (2012). N2O emission from degraded soybean nodules depends on denitrification by Bradyrhizobium japonicum and other microbes in the rhizosphere. Microbes Environ. 27, 470–476. doi: 10.1264/jsme2.me12100
Inaba, S., Tanabe, K., Eda, S., Ikeda, S., Higashitani, A., Mitsui, H., et al. (2009). Nitrous oxide emission and microbial community in the rhizosphere of nodulated soybeans during the late growth period. Microbes Environ. 24, 64–67. doi: 10.1264/jsme2.me08544
Intergovernmental Panel on Climate Change [IPCC], (2014). Climate change 2014: Synthesis Report. Contribution of Working Groups I, II and III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Geneva: IPCC.
Itakura, M., Tabata, K., Eda, S., Mitsui, H., Murakami, K., Yasuda, J., et al. (2008). Generation of Bradyrhizobium japonicum mutants with increased N2O reductase activity by selection after introduction of a mutated dnaQ gene. Appl. Environ. Microbiol. 74, 7258–7264. doi: 10.1128/AEM.01850-08
Itakura, M., Uchida, Y., Akiyama, H., Takada-Hoshino, Y., Shimomura, Y., Morimoto, S., et al. (2013). Mitigation of nitrous oxide emissions from soils by Bradyrhizobium japonicum inoculation. Nat. Clim. Chan. 3, 208–212. doi: 10.1038/nclimate1734
Koropatkin, N. M., Pakrasi, H. B., and Smith, T. J. (2006). Atomic structure of a nitrate-binding protein crucial for photosynthetic productivity. Proc. Natl. Acad. Sci. U.S.A. 103, 9820–9825. doi: 10.1073/pnas.0602517103
LeBauer, D. S., and Treseder, K. (2008). Nitrogen limitation of net primary productivity in terrestrial ecosystems is globally distributed. Ecology 89, 371–379. doi: 10.1890/06-2057.1
Luque-Almagro, V. M., Lyall, V. J., Ferguson, S. J., Roldán, M. D., and Richardson, D. J. (2013). Nitrogen oxyanion-dependent dissociation of a two-component complex that regulates bacterial nitrate assimilation. J. Biol. Chem. 288, 29692–29702. doi: 10.1074/jbc.M113.459032
Ohta, H., and Hattori, T. (1983). Agromonas oligotrophica gen. nov., sp. nov., a nitrogen-fixing oligotrophic bacterium. Antonie Van Leeuwenhoek 49, 429–446.
Okubo, T., Fukushima, S., Itakura, M., Oshima, K., Longtonglang, A., Teaumroong, N., et al. (2013). Genome analysis suggests that the soil oligotrophic bacterium Agromonas oligotrophica (Bradyrhizobium oligotrophicum) is a nitrogen-fixing symbiont of Aeschynomene indica. Appl. Environ. Microbiol. 79, 2541–2551. doi: 10.1128/AEM.00009-13
Richardson, D. J., Felgate, H., Watmough, N., Thomson, A., and Baggs, E. (2009). Mitigating release of the potent greenhouse gas N2O from the nitrogen cycle-could enzymatic regulation hold the key? Trends Biotechnol. 27, 388–397. doi: 10.1016/j.tibtech.2009.03.009
Romeo, A., Sonnleitner, E., Sorger-Domenigg, T., Nakano, M., Eisenhaber, B., and Bläsi, U. (2012). Transcriptional regulation of nitrate assimilation in Pseudomonas aeruginosa occurs via transcriptional antitermination within the nirBD-PA1779-cobA operon. Microbiology 158, 1543–1552. doi: 10.1099/mic.0.053850-0
Saeki, Y., Nakamura, M., Mason, M. L. T., Yano, T., Shiro, S., Sameshima-Saito, R., et al. (2017). Effect of flooding and the nosZ gene in bradyrhizobia on bradyrhizobial community structure in the soil. Microbes Environ. 32, 154–163. doi: 10.1264/jsme2.ME16132
Sameshima-Saito, R., Chiba, K., Hirayama, J., Itakura, M., Mitsui, H., Eda, S., et al. (2006a). Symbiotic Bradyrhizobium japonicum reduces N2O surrounding the soybean root system via nitrous oxide reductase. Appl. Environ. Microbiol. 72, 2526–2532. doi: 10.1128/aem.72.4.2526-2532.2006
Sameshima-Saito, R., Chiba, K., and Minamisawa, K. (2006b). Correlation of denitrifying capability with the existence of nap, nir, nor and nos genes in diverse strains of soybean bradyrhizobia. Microbes Environ. 21, 174–184. doi: 10.1264/jsme2.21.174
Sameshima-Saito, R., Chiba, K., and Minamisawa, K. (2004). New method of denitrification analysis of Bradyrhizobium field isolates by gas chromatographic determination of 15N-labeled N2. Appl. Environ. Microbiol. 70, 2886–2891. doi: 10.1128/aem.70.5.2886-2891.2004
Sánchez, C., Bedmar, E. J., and Delgado, M. J. (2011). “Denitrification in legume-associated endosymbiotic bacteria,” in Nitrogen Cycling in Bacteria: Molecular Analysis, ed. J. W. B. Moir (Norfolk: Caister Academic Press), 197–210.
Sánchez, C., Itakura, M., Mitsui, H., and Minamisawa, K. (2013). Linked expressions of nap and nos genes in a Bradyrhizobium japonicum mutant with increased N2O reductase activity. Appl. Environ. Microbiol. 79, 4178–4180. doi: 10.1128/AEM.00703-13
Sánchez, C., Itakura, M., Okubo, T., Matsumoto, T., Yoshikawa, H., Gotoh, A., et al. (2014). The nitrate-sensing NasST system regulates nitrous oxide reductase and periplasmic nitrate reductase in Bradyrhizobium japonicum. Environ. Microbiol. 16, 3263–3274. doi: 10.1111/1462-2920.12546
Sánchez, C., and Minamisawa, K. (2018). Redundant roles of Bradyrhizobium oligotrophicum Cu-type (NirK) and cd1-type (NirS) nitrite reductase genes under denitrifying conditions. FEMS Microbiol. Lett. 365:fny015. doi: 10.1093/femsle/fny015
Sánchez, C., Mitsui, H., and Minamisawa, K. (2017). Regulation of nitrous oxide reductase genes by NasT-mediated transcription antitermination in Bradyrhizobium diazoefficiens. Environ. Microbiol. Rep. 9, 389–396. doi: 10.1111/1758-2229.12543
Sánchez, C., Siqueira, A. F., Mitsui, H., and Minamisawa, K. (2019). Identification of genes regulated by the antitermination factor NasT during denitrification in Bradyrhizobium diazoefficiens. Microbes Environ. doi: 10.1111/1758-2229.12543 [Epub ahead of print].
Shiina, Y., Itakura, M., Choi, H., Seaki, Y., Hayatsu, M., and Minamisawa, K. (2014). Relationship between soil type and N2O reductase genotype (nosZ) of indigenous soybean bradyrhizobia: nosZ-minus populations are dominant in Andosols. Microbes Environ. 29, 420–426. doi: 10.1264/jsme2.ME14130
Shu, C. Y. J., and Zhulin, I. B. (2002). ANTAR: an RNA-binding domain in transcription antitermination regulatory proteins. Trends Biochem. Sci. 27, 3–5. doi: 10.1016/s0968-0004(01)02036-9
Siqueira, A. F., Minamisawa, K., and Sánchez, C. (2017). Anaerobic reduction of nitrate to nitrous oxide is lower in Bradyrhizobium japonicum than in Bradyrhizobium diazoefficiens. Microbes Environ. 32, 398–401. doi: 10.1264/jsme2.me17081
Sullivan, M. J., Gates, A. J., Appia-Ayme, C., Rowley, G., and Richardson, D. J. (2013). Copper control of bacterial nitrous oxide emission and its impact on vitamin B12-dependent metabolism. Proc. Natl. Acad. Sci. U.S.A. 110, 19926–19931. doi: 10.1073/pnas.1314529110
Thomson, A. J., Giannopoulos, G., Pretty, J., Baggs, E. M., and Richardson, D. J. (2012). Biological sources and sinks of nitrous oxide and strategies to mitigate emissions. Phil. Trans. R. Soc. B. 367, 1157–1168. doi: 10.1098/rstb.2011.0415
Torres, M. J., Bueno, E., Jiménez-Leiva, A., Cabrera, J. J., Bedmar, E. J., Mesa, S., et al. (2017). FixK2 is the main transcriptional activator of Bradyrhizobium diazoefficiens nosRZDYFLX genes in response to low oxygen. Front. Microbiol. 8:1621. doi: 10.3389/fmicb.2017.01621
Torres, M. J., Simon, J., Rowley, G., Bedmar, E. J., Richardson, D. J., Gates, A. J., et al. (2016). Nitrous oxide metabolism in nitrate-reducing bacteria: physiology and regulatory mechanisms. Adv. Microb. Physiol. 68, 353–432. doi: 10.1016/bs.ampbs.2016.02.007
Tortosa, G., Hidalgo, A., Salas, A., Bedmar, E. J., Mesa, S., and Delgado, M. J. (2016). Nitrate and flooding induce N2O emissions from soybean nodules. Symbiosis 67, 125–133. doi: 10.1007/s13199-015-0341-3
Uchida, Y., and Akiyama, H. (2013). Mitigation of postharvest nitrous oxide emissions from soybean ecosystems: a review. Soil Sci. Plant Nutr. 59, 477–487. doi: 10.1080/00380768.2013.805433
Velasco, L., Mesa, S., Xu, C. A., Delgado, M. J., and Bedmar, E. J. (2004). Molecular characterization of nosRZDFYLX genes coding for denitrifying nitrous oxide reductase of Bradyrhizobium japonicum. Antonie Van Leeuwenhoek 85, 229–235. doi: 10.1023/b:anto.0000020156.42470.db
Wang, B., Pierson, L. S. III, Rensing, C., Gunatilaka, M. K., and Kennedy, C. (2012). NasT-mediated antitermination plays an essential role in the regulation of the assimilatory nitrate reductase operon in Azotobacter vinelandii. Appl. Environ. Microbiol. 78, 6558–6567. doi: 10.1128/AEM.01720-12
Yang, L., and Cai, Z. (2005). The effect of growing soybean (Glycine max. L.) on N2O emission from soil. Soil Biol. Biochem. 37, 1205–1209. doi: 10.1016/j.soilbio.2004.08.027
Zumft, W. G. (1997). Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. R. 61, 533–616.
Keywords: Bradyrhizobium, soybean, rhizosphere, denitrification, N2O reductase, nos regulation, greenhouse gas, mitigation strategies
Citation: Sánchez C and Minamisawa K (2019) Nitrogen Cycling in Soybean Rhizosphere: Sources and Sinks of Nitrous Oxide (N2O). Front. Microbiol. 10:1943. doi: 10.3389/fmicb.2019.01943
Received: 26 June 2019; Accepted: 07 August 2019;
Published: 21 August 2019.
Edited by:
Rosa María Martínez-Espinosa, University of Alicante, SpainReviewed by:
Hiroko Akiyama, National Agriculture and Food Research Organization (NARO), JapanCopyright © 2019 Sánchez and Minamisawa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cristina Sánchez, Y3Jpc3RpbmEuc2Fnb0BnbWFpbC5jb20=; Kiwamu Minamisawa, a2l3YW11QGlnZS50b2hva3UuYWMuanA=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.