
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 30 July 2019
Sec. Microbial Physiology and Metabolism
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.01716
Fusobacteria are common obligately anaerobic Gram-negative bacteria of the oral cavity that may act as a bridge between early and late colonizing bacteria in dental plaque and have a role in oral and extra-oral infections. Fusobacterium nucleatum has a crucial role in oral biofilm structure and ecology, as revealed in experimental and clinical biofilm models. The aim of this study was to investigate the impact of various Fusobacterium species on in vitro biofilm formation and structure in three different oral biofilm models namely a supragingival, a supragingival “feeding”, and a subgingival biofilm model. The standard six-species supragingival and “feeding” biofilm models employed contained Actinomyces oris, Candida albicans, Streptococcus mutans, Streptococcus oralis, Veillonella dispar, and Fusobacterium sp. The subgingival biofilm model contained 10 species (A. oris, Campylobacter rectus, F. nucleatum ssp. nucleatum, Porphyromonas gingivalis, Prevotella intermedia, Streptococcus anginosus, S. oralis, Tannerella forsythia, Treponema denticola, and V. dispar). Six different Fusobacterium species or subspecies, respectively, were tested namely F. nucleatum ssp. fusiforme, F. nucleatum ssp. nucleatum, F. nucleatum ssp. polymorphum, F. nucleatum ssp. vincentii, F. naviforme, and F. periodonticum). Biofilms were grown anaerobically on hydroxyapatite disks in 24-well culture dishes. After 64 h, biofilms were either harvested and quantified by culture analysis or proceeded to fluorescent in situ hybridization (FISH) and confocal laser scanning microscopy (CLSM). All Fusobacterium species tested established well in the biofilms, with CFUs ranging from 1.4E+04 (F. nucleatum ssp. fusiforme) to 5.6E+06 (F. nucleatum ssp. nucleatum). The presence of specific Fusobacterium sp./ssp. induced a significant decrease in C. albicans levels in the supragingival model and in V. dispar levels in the “feeding” supragingival model. In the subgingival model, the counts of A. oris, S. oralis, P. intermedia, P. gingivalis, and C. rectus significantly decreased in the presence of specific Fusobacterium sp./ssp. Collectively, this study showed variations in the growing capacities of different fusobacteria within biofilms, affecting the growth of surrounding species and potentially the biofilm architecture. Hence, clinical or experimental studies need to differentiate between Fusobacterium sp./ssp., as their biological properties may well vary.
The disposition of biofilms, which are composed of dynamic substratum-attached microbial networks, is an everyday phenomenon in nature (Hall-Stoodley et al., 2004; Marsh, 2005). In particular, the oral cavity is an excellent aquatic ecosystem allowing for the settlement of myriads of microorganisms on the salivary pellicle-coated tooth surfaces and epithelial layers of gingiva (Kolenbrander et al., 2002; Marsh, 2004). In fact, more than 700 different bacterial species embedded in extracellular polysaccharide-affluent matrix account for the formation of the multispecies oral communities (Foster and Kolenbrander, 2004; Keijser et al., 2008). The latter encounter 1,000-times more resistance to antimicrobial agents; host immunity; nutrient restriction; alternating oxygen-, nitrogen-, and carbon dioxide supply; and abrupt temperature fluctuation (Davies, 2003; Welin-Neilands and Svensater, 2007; Marsh and Devine, 2011).
Selective salivary-derived proteins allow for the initial adherence of the early bacterial colonizers, namely streptococci and actinomyces, upon direct adsorption onto the host surfaces (Takahashi and Nyvad, 2011; Nikitkova et al., 2013). The succession of intra-generic and intra-species microbial partnerships within the dental plaque biofilms is thereby promoted by co-adhesion of planktonic to immobilized microorganisms or co-aggregation among distinct bacterial species (Katharios-Lanwermeyer et al., 2014; Palmer, 2014). Thereafter, the maturation of oral biofilms is activated by the extracellular polymeric substances (EPS) formation and is mediated by Fusobacterium nucleatum, a Gram-negative microorganism, which is located in the interface of early and late colonizers such as obligate anaerobes and streptococci (He et al., 2012). Indeed, streptococci are found to prevail the first 6–48 h, whereas F. nucleatum-mediated coaggregation takes place after the first 48 h of oral biofilm formation (Dige et al., 2007).
F. nucleatum can be considered as a periodontal pathogen, due to its enhanced prevalence within the subgingival biofilm, which is retrieved from deep periodontal pockets (Gursoy et al., 2008). Specifically, F. nucleatum triggers the production of matrix metalloproteinases by the host contributing initially to periodontal inflammation and then to irreversible periodontal disease (Gursoy et al., 2010). Interestingly, F. nucleatum also consists of an adhesin-rich outer membrane enabling the adhesion to various salivary proteins, other microorganisms, and host substrata (Kaplan et al., 2009; Coppenhagen-Glazer et al., 2015). Additional virulence properties associated with F. nucleatum include its enhanced hemolytic activity, and the production of hydrogen sulfide (H2S) serves as key virulence traits employed by F. nucleatum (Gaetti-Jardim Júnior and Avila-Campos, 1999; Okamoto et al., 2000; Yoshida et al., 2009). F. nucleatum can also invade and reside within the human gastrointestinal tract (Strauss et al., 2011).
Due to the possession of numerous virulence properties, F. nucleatum may have an integral role in regulating oral biofilm formation and subsequent growth. Interestingly, an earlier report confirmed this assumption by revealing a significant decrease in the total colony forming units (CFU) of an in vitro supragingival biofilm model in the absence of F. nucleatum (Guggenheim et al., 2001a; Thurnheer et al., 2003). Reversely, the absence of streptococci or, streptococci and actinomyces altogether, from an in vitro subgingival biofilm model resulted in a less compact and more dispersed distribution of F. nucleatum within the biofilm mass (Ammann et al., 2013), confirming a structural functional interrelationship with the other species. Yet, given the importance of F. nucleatum as an essential “bridging” biofilm component, there is to date no evidence in literature how different Fusobacterium species can affect the formation and structure of multispecies oral biofilms in vitro. Moreover, to our knowledge, there is at present restricted evidence in literature of potential differences between F. nucleatum subspecies (closely related fusobacterial species) in the context of biofilm maturation. Therefore, six different Fusobacterium species or subspecies, namely F. nucleatum ssp. fusiforme, F. nucleatum ssp. nucleatum, F. nucleatum ssp. polymorphum, F. nucleatum ssp. vincentii, F. naviforme, and F. periodonticum were tested for their ability to integrate into and affect the growth of three different in vitro oral biofilm models (standard supragingival, supragingival “feeding” and subgingival), respectively. The null hypothesis of this study was that specific Fusobacterium species have no significant impact on the overall growth of in vitro oral biofilms and can all act as a bridge between their surrounding bacterial partners promoting biofilm maturation.
For this study, three different in vitro biofilm models were used namely the standard supragingival model, the supragingival “feeding” model, and the subgingival model. The procedures for biofilm production have been described in detail before (Shapiro et al., 2002; Ammann et al., 2012; Thurnheer et al., 2014b). In brief, the standard supragingival biofilm employed contained Actinomyces oris (OMZ 745), Candida albicans (OMZ 110), F. nucleatum ssp. nucleatum (OMZ 598), Streptococcus oralis SK 248 (OMZ 607), Streptococcus mutans (OMZ 918), and Veillonella dispar ATCC 17748T (OMZ 493). Six different inocula were used: as a control, the inoculum containing the six strains above was used, whereas in each of the further five inocula, one of the following Fusobacterium strains was used: (1) F. nucleatum ssp. fusiforme ATCC 51190 (OMZ 642), (2) F. nucleatum ssp. vincentii ATCC 49256T (OMZ 635), (3) F. nucleatum ssp. polymorphum ATCC 10953 (OMZ 595), (4) Fusobacterium naviforme (formerly F. nucleatum ssp. naviforme) NCTC 11464 (OMZ 594), and (5) Fusobacterium periodonticum ATCC 33693T (OMZ 636). The taxonomic standing of the used fusobacteria strains as well as the phylogenetic distances among them has previously been reported using monoclonal antibodies, ribotyping, and 16S rRNA sequencing, highlighting the individuality of the species (Thurnheer et al., 1999; Gmür et al., 2006).
Biofilms were grown anaerobically in 24-well polystyrene cell culture plates on hydroxyapatite disks that had been preconditioned for pellicle formation in whole un-stimulated pooled saliva (in the following termed saliva) for 4 h. To initiate a biofilm experiment, disks were covered for the first 16 h with 1.6 ml of growth medium containing 70% saliva, 30% modified fluid universal medium (mFUM; Guggenheim et al., 2001a) supplemented with Sørensen’s buffer (final pH 7.2), and 200 μl of a cell suspension prepared from equal volumes and densities of each strain. The medium was changed after 16 and 40 h. For the first 16 h, the medium contained 0.3% glucose. After 16 h, the medium was replenished with one containing 0.15% glucose and 0.15% sucrose, instead of 0.3% glucose. In order to remove non-adherent microorganisms, biofilms were dipped three times in saline after 16, 20, and 24 h as well as after 40, 44, and 48 h. After 64 h of incubation, the biofilms were dip-washed again and either harvested for culture analyses by vigorous vortexing in 1 ml of 0.9% NaCl or proceeded to staining and confocal laser scanning microscopy (CLSM) (see below).
The supragingival “feeding” model in the following termed “feeding” model was established in order to mimic more accurately the fast and feast periods experienced by natural dental plaque (Thurnheer et al., 2006). Therefore, the standard experimental protocol described above was modified as follows: (1) the proportion of saliva and mFUM was reversed to 30% saliva and 70% mFUM and (2) exposure to this altered medium was time limited. That is, after inoculation the disks remained for only 45 min in the feeding solution containing 0.3% glucose. Thereafter, they were subjected to three consecutive 1-min washes in 2 ml 0.9% NaCl to remove growth medium and free-floating cells but not bacteria adhering firmly to HA disks. The biofilms were then further incubated in new wells containing 1.6 ml of saliva and no mFUM. Only after 16, 20, 24, 40, 44, and 48 h were biofilms pulse-fed by transferring the disks for 45 min into 30% saliva/70% mFUM with 0.15% glucose and 0.15% sucrose. Thereafter, they were washed as described above and re-incubated in saliva. Fresh saliva was provided after 16 and 40 h. After 64 h, biofilms were washed and processed for further analyses.
In order to grow subgingival in vitro biofilms, the protocol for standard supragingival biofilms described above was modified as follows: (1) 10 species were used instead of six, namely A. oris (OMZ 745), Campylobacter rectus (OMZ 388), F. nucleatum ssp. nucleatum (OMZ 598), Porphyromonas gingivalis ATCC 33277T (OMZ 925), Prevotella intermedia ATCC 25611T (OMZ 278), Streptococcus anginosus ATCC 9895 (OMZ 871), S. oralis SK 248 (OMZ 607), Tannerella forsythia (OMZ 1047), Treponema denticola ATCC 35405T (OMZ 661), and V. dispar ATCC 17748T (OMZ 493) and (2) the growth medium contained 60% saliva, 10% fetal bovine serum, and 30% FUM. To generate subgingival biofilms, the same procedure as described for the standard supragingival biofilm model was applied.
Total CFU, streptococci, and other taxa were assessed by culture using selective and non-selective media (Guggenheim et al., 2001a; Van Der Ploeg and Guggenheim, 2004; Thurnheer et al., 2014a). CFU data are not provided for T. denticola and T. forsythia, since these species do not grow on solid agar. Therefore, the CFU-related data provided are on the remaining eight species in the “subgingival” biofilm.
Biofilms were stained by fluorescence in situ hybridization following earlier described protocols (Thurnheer et al., 2004; Ammann et al., 2012). In brief, prehybridization (15 min, 46°C) was performed in 500 μl hybridization buffer with 40% formamide in the absence of any oligonucleotide probes. Thereafter, 500 μl of hybridization buffer (40% formamide) was used for each biofilm, supplemented with the genus-specific Cy3-labeled probe STR405 (5′-TAGCCGTCCCTTTCTGGT-3′) and Cy5-labeled probe FUS664 (5′-CTTGTAGTTCCGCYTACCTC-3′) to stain streptococci and fusobacteria, respectively, at a concentration of 20 ng/μl. The incubation time for the hybridization was at least 3 h at 46°C in the dark. After the incubation, biofilms were transferred into washing buffer preheated to 48°C and incubated for 20 min at this temperature. For counterstaining, biofilms were stained using a mixture of 3 μM YoPro 1 iodide (Invitrogen, Carlsbad, CA) and 15 μM Sytox green (Invitrogen; for 20 min, at room temperature in the dark), following the fluorescence in situ hybridization procedure. After staining, the samples were embedded upside down on chamber slides in 100 μl of Mowiol (Guggenheim et al., 2001b).
Stained biofilms were examined by confocal laser scanning microscopy using a Leica TCS SP5 microscope (Leica Microsystems, Wetzlar, Germany) with ×100/1.4 NA oil-immersion objective lens, in conjunction with an argon laser at 488 nm excitation, a DPSS diode laser at 561 nm, and a Helium-Neon laser at 633 nm excitation. Filters were set to 500–540 nm for YoPro/Sytox, to 570–600 nm for Cy3, and to 660–710 nm for Cy5, respectively. Biofilms were scanned in sequential mode, and z-series were generated by vertical optical sectioning using a step size of 1 μm. Image acquisition was done in ×8 line average mode, and scans were recombined and processed using IMARIS 7.6.5 software (Bitplane, Zurich, Switzerland), without any qualitative changes to the raw images.
Three individual experiments were performed and each group represented in triplicate biofilm cultures per experiment. A two-way analysis of variance in conjunction with Tukey’s multiple comparison text was used to evaluate the differences between the control and each experimental group. The significance level was set to p < 0.05. Values below the assay’s detection limit were ascribed the lowest detection limit value to allow for logarithmic transformation. The Prism v.7.0 statistical analysis software (GraphPad, La Jolla, CA, USA) was used to analyze the data.
A total of six Fusobacterium species/subspecies were tested in the three different experimental biofilm model variants. In the 70:30 model, the total bacterial numbers (CFU) yielded after the completion of the biofilm culture were similar irrespective of the Fusobacterium spp. used. Accordingly, no significant differences were observed between experimental groups among the other bacterial species in the biofilms (Figure 1). Nevertheless, the number of fusobacteria in the biofilm significantly differed, depending on the species or subspecies incorporated. In particular, levels of F. nucleatum ssp. vincenti, F. nucleatum ssp. polymorphum, F. naviforme, as well as that of F. periodonticum were significantly lower in the biofilm than the F. nucleatum ssp. nucleatum control, which is used in the standard biofilm model (Figure 1). The least incorporated in terms of numbers within the biofilm proved to be F. nucleatum ssp. vincentii. The numbers of C. albicans were also differentially affected according to the subspecies present in the biofilm, depending on the used fusobacteria, with a significant reduction in the presence of F. nucleatum ssp. polymorphum, F. naviforme, and F. periodonticum, compared to the F. nucleatum ssp. nucleatum control.
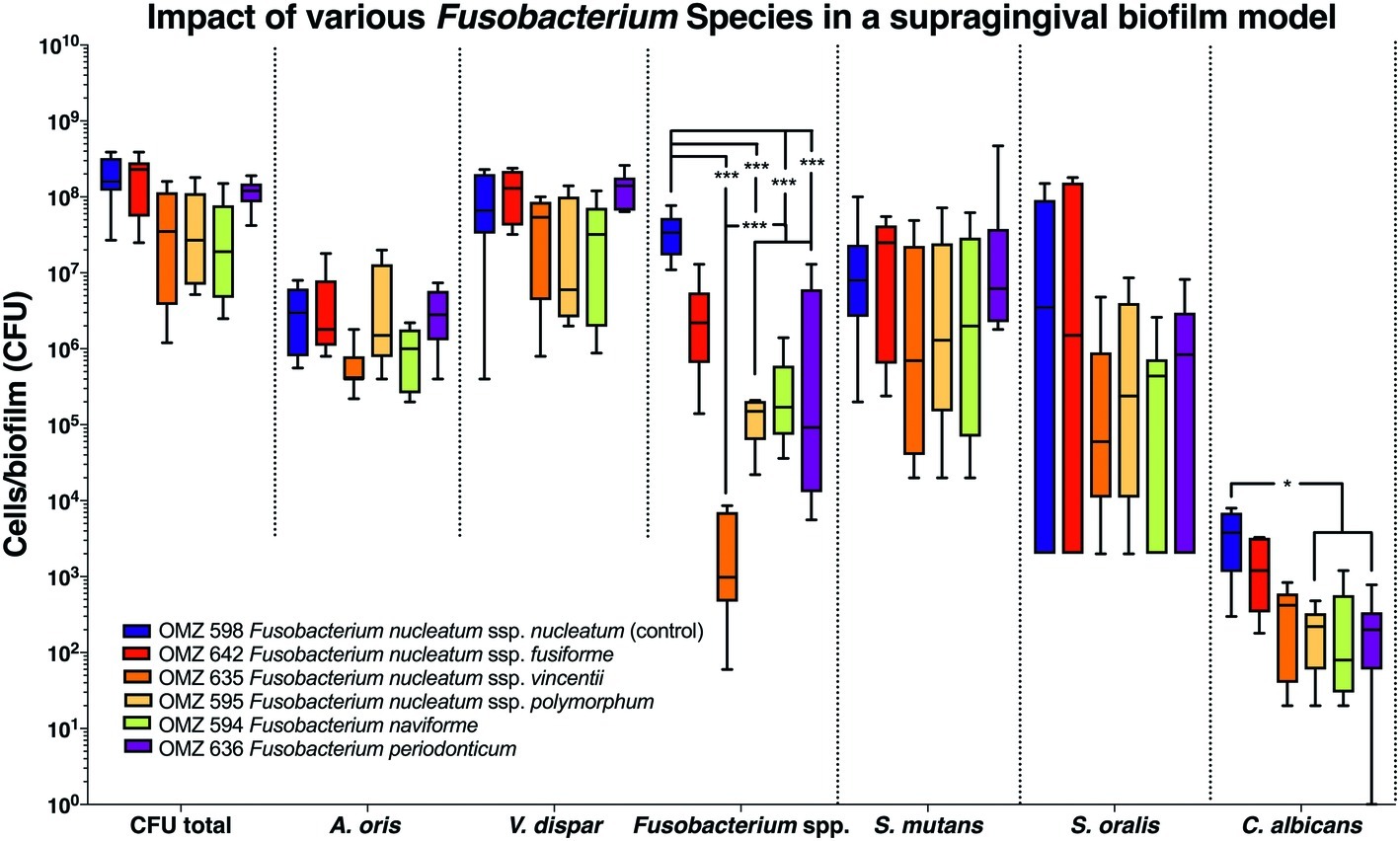
Figure 1. Fusobacterial growth in a supragingival biofilm model. Colony forming units (CFUs) of the supragingival biofilm model (A. oris, F. nucleatum, S. mutans, S. oralis, V. dispar, and C. albicans). The different colors correspond to experimental groups of different fusobacteria used, as indicated. Data derive from nine independent experiments in which every group was represented in triplicate biofilm cultures. Box plots represent the CFUs determined by selective agar plating, while horizontal lines indicate their median values. Undetectable values were ascribed the lowest detection limit value of the assay to allow for log transformation. Asterisks (*) represent significant difference compared with the control group (p < 0.05). Statistically significant differences compared with the control group are indicated with asterisks (*p < 0.05; ***p < 0.001).
Next, the “feeding” model was investigated (Figure 2). The total bacterial numbers in the biofilm culture were similar, irrespective of the Fusobacterium ssp. used. Accordingly, no significant differences were detected between experimental groups among the other bacterial species or C. albicans in the biofilms (Figure 2). Apart from F. periodonticum, all other F. nucleatum ssp. were significantly reduced, compared to the ssp. nucleatum control. Interestingly, significant variations in numbers existed also among the different F. nucleatum ssp., indicating different incorporation capacities according to subspecies in this model. Among the other species in the biofilm, variations also existed in the numbers of V. dispar.
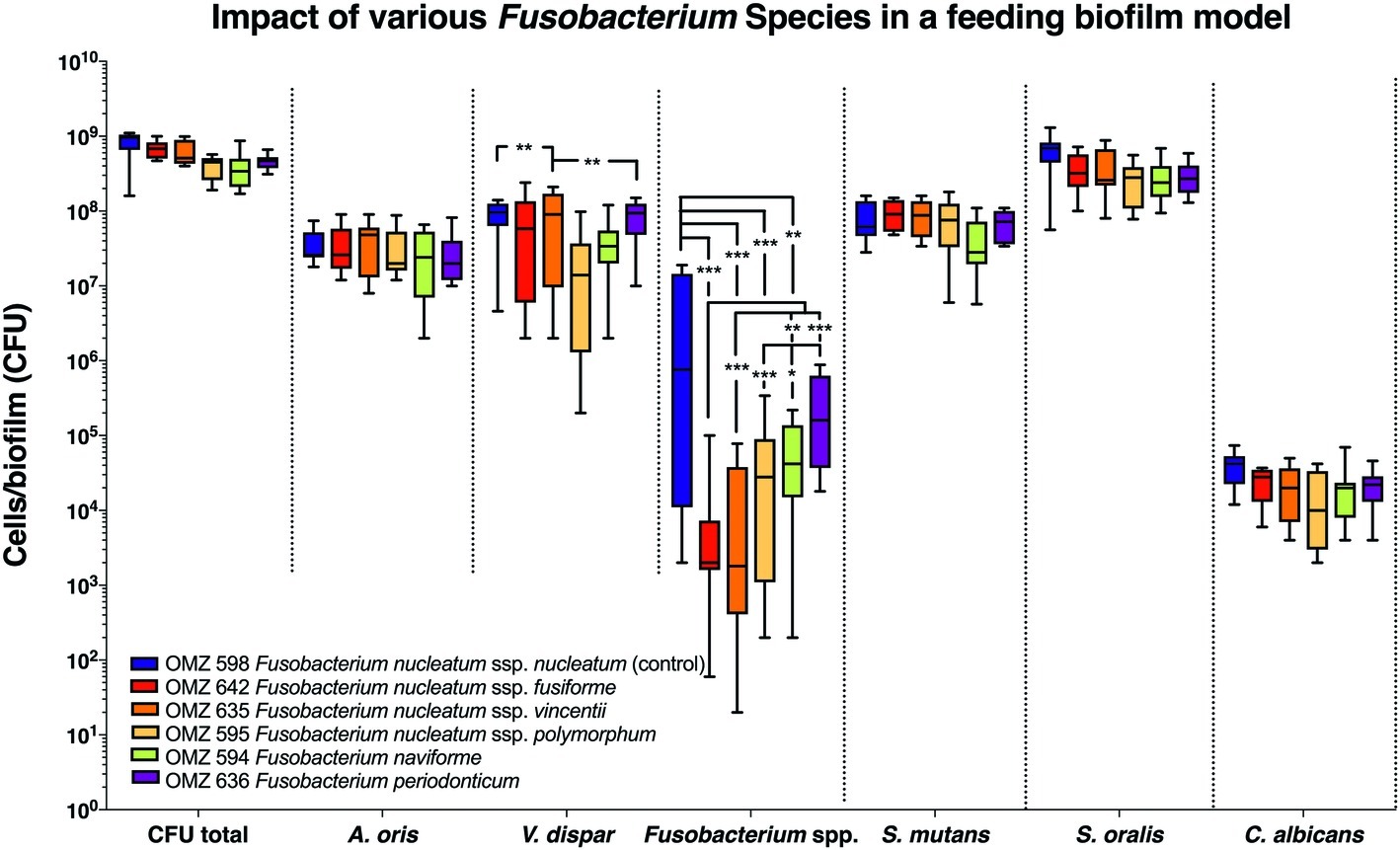
Figure 2. Fusobacterial growth in a supragingival “feeding” biofilm model. Colony forming units (CFUs) of the supragingival “feeding” biofilm model (A. oris, F. nucleatum, S. mutans, S. oralis, V. dispar, and C. albicans). The different colors correspond to experimental groups of different fusobacteria used, as indicated. Data derive from nine independent experiments in which every group was represented in triplicate biofilm cultures. Box plots represent the CFUs determined by selective agar plating, while horizontal lines indicate their median values. Undetectable values were ascribed the lowest detection limit value of the assay to allow for log transformation. Asterisks (*) represent significant difference compared with the control group (p < 0.05). Statistically significant differences compared with the control group are indicated with asterisks (*p < 0.05; **p < 0.01; ***p < 0.001).
In the “subgingival” model, the total bacterial numbers in the biofilm were significantly lower in the case of all Fusobacterium ssp. tested than the F. nucleatum control (Figure 3A). Accordingly, the numbers of all other Fusobacterium ssp. tested were also lower than the F. nucleatum control group, while significant variations also existed between the subspecies. A reduction in A. oris was also observed with all F. nucleatum ssp. tested but proved to be significant only in the case of F. periodonticum (Figure 3A). Significant reductions were also observed in the numbers of P. gingivalis, S. oralis, P. intermedia, and C. rectus with all F. nucleatum ssp. tested, compared to the F. nucleatum control (Figure 3B).
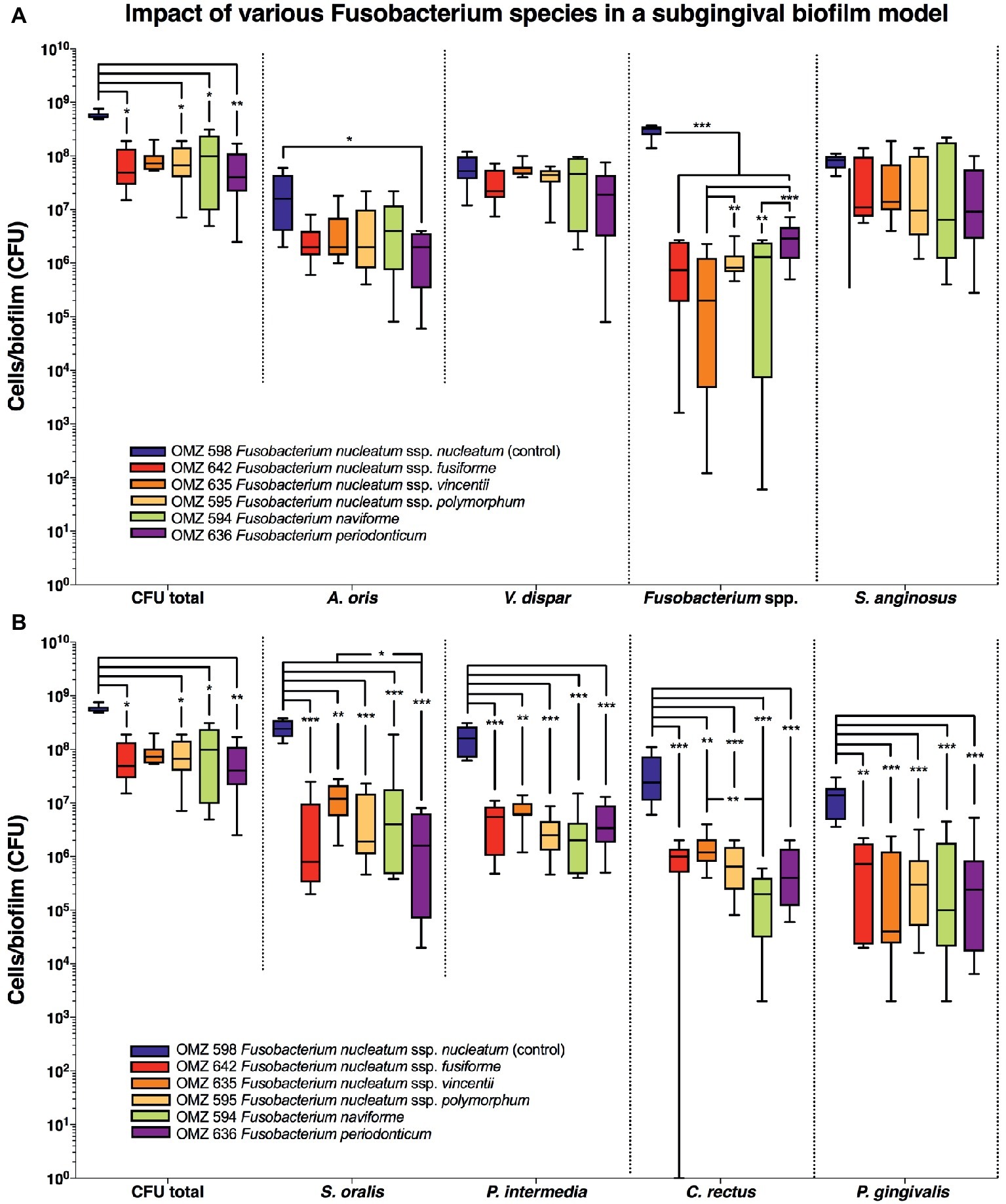
Figure 3. Fusobacterial growth in a subgingival biofilm model. Colony forming units (CFUs) of the subgingival biofilm model (A. oris, C. rectus, F. nucleatum, P. intermedia, P. gingivalis, S. anginosus, S. oralis, T. denticola, T. forsythia, and V. dispar). The total CFUs are provided in both (A) and (B), in order to facilitate their comparative assessment to all individual species. The different colors correspond to experimental groups of different fusobacteria used, as indicated. Data derive from nine independent experiments in which every group was represented in triplicate biofilm cultures. CFU data are not provided for T. denticola and T. forsythia, since these species do not grow on solid agar. Box plots represent the CFUs determined by selective agar plating, while horizontal lines indicate their median values. Undetectable values were ascribed the lowest detection limit value of the assay to allow for log transformation. Asterisks (*) represent significant difference compared with the control group (p < 0.05). Statistically significant differences compared with the control group are indicated with asterisks (*p < 0.05; **p < 0.01; ***p < 0.001).
Apart from the effect of the different subspecies on microbial numbers in the biofilms, their localization pattern within the biofilm was investigated by CLSM (Figure 4). In the supragingival biofilm model, a tighter distribution of F. nucleatum ssp. fusiforme was observed (Figures 4B,H), compared to F. nucleatum ssp. nucleatum control (Figures 4A,E), with a comparable distribution of streptococci. In case of F. nucleatum ssp. vincentii (Figure 4C) and F. nucleatum ssp. polymorphum (Figure 4D), the distribution of both fusobacteria and streptococci appeared to be sparser than the control. The distribution of F. periodonticum was characterized by more distinctive own cell clusters in the mass of the biofilm (Figure 4G).
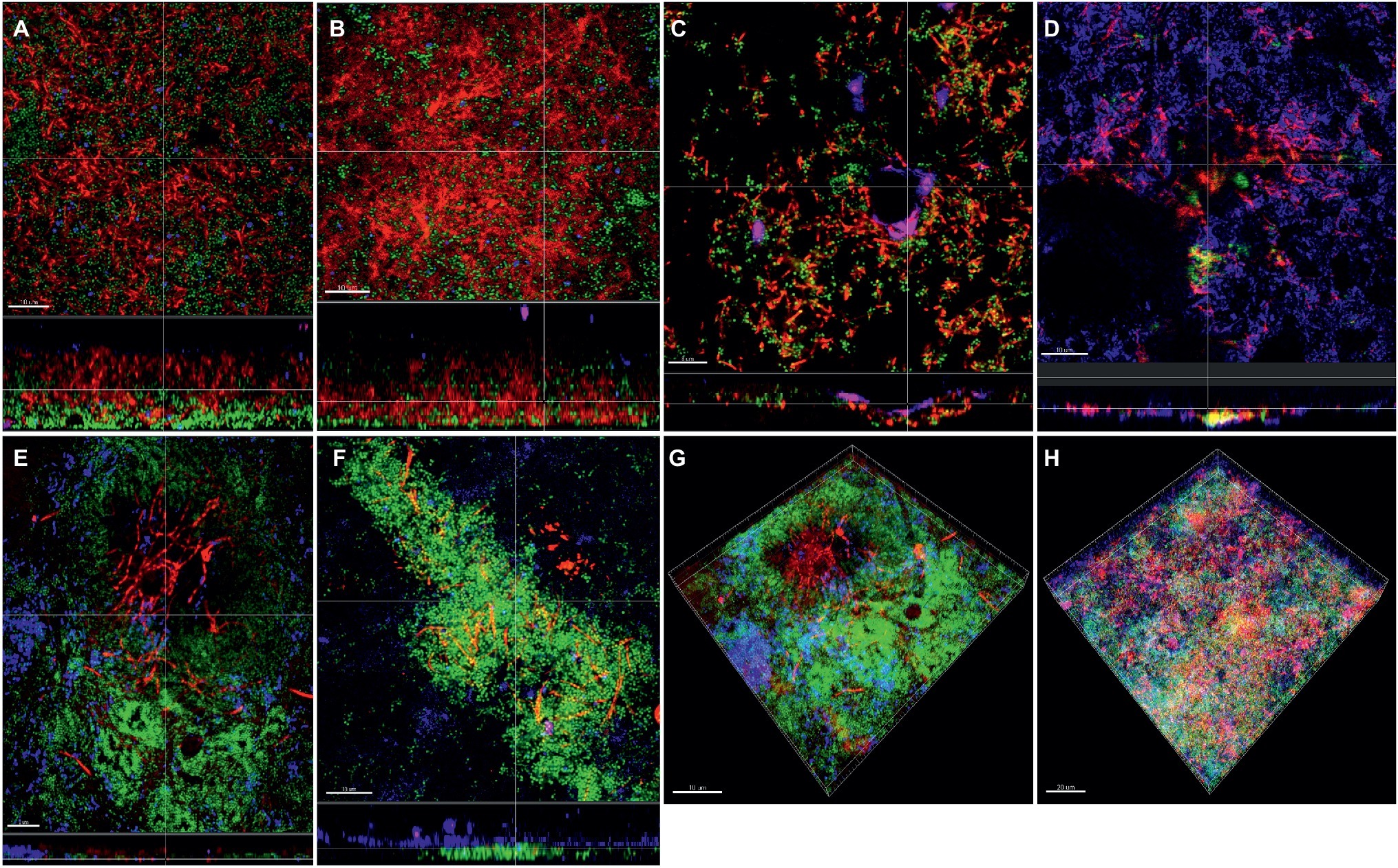
Figure 4. Incorporation of various Fusobacterium species in the supragingival biofilm model. Panels (A–F) depict confocal laser scanning microscopic (CLSM) image stack, while panels (G) and (H) represent 3D reconstructions of in vitro supragingival biofilms. Due to FISH staining, fusobacteria appear red and streptococci blue, and due to DNA staining with Sytox/YoPro 1, bacteria appear green. (A) F. nucleatum ssp. nucleatum (control), (B) F. nucleatum ssp. fusiforme, (C) F. nucleatum ssp. vincentii, (D) F. nucleatum ssp. polymorphum, (E) F. periodonticum, (F) F. naviforme, (G) F. periodonticum (3D), and (H) F. nucleatum ssp. fusiforme (3D); scale bars: 10 μm.
In the “feeding” model, the distribution of fusobacteria was comparable between F. nucleatum ssp. nucleatum (Figures 5A,E), fusiforme, (Figure 5B), vincentii (Figure 5C), F. naviforme (Figure 5F) and F. periodonticum (Figure 5G), with scattered aggregate distribution through the biofilm and streptococcal clusters occasionally identified in close proximity. The distribution of F. nucleatum ssp. polymorphum (Figure 5D) displayed a more characteristic filamentous structure resulting from the deposition of multiple fusobacteria across the biofilm mass (Figures 5G,H).
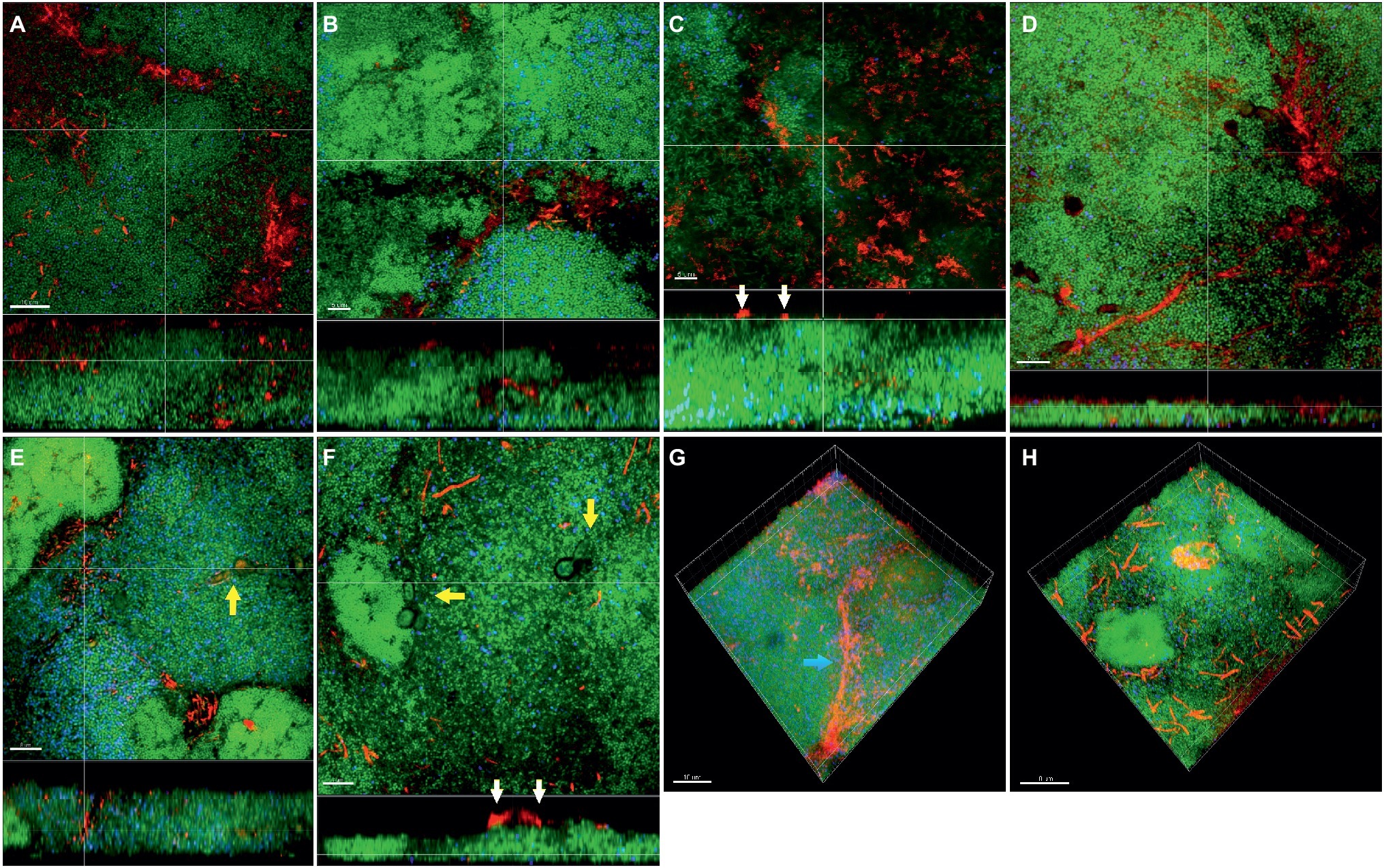
Figure 5. Incorporation of various Fusobacterium species in the feeding biofilm model. Panels (A–F) depict confocal laser scanning microscopic (CLSM) image stack, while panels (G) and (H) represent 3D reconstructions of in vitro “feeding” supragingival biofilms. Due to FISH staining, fusobacteria appear red and streptococci blue, and due to DNA staining with Sytox/YoPro 1, bacteria appear green. (A) F. nucleatum ssp. nucleatum (control), (B) F. nucleatum ssp. fusiforme, (C) F. nucleatum ssp. vincentii, (D) F. nucleatum ssp. polymorphum, (E) F. periodonticum, (F) F. naviforme, (G) F. nucleatum ssp. polymorphum (3D), and (H) F. periodonticum (3D); white arrows indicate the localization of fusobacteria [F. nucleatum ssp. vincentii and F. naviforme, panels (C) and (F), respectively] near the biofilm substratum interface, yellow arrows indicate the presence of C. albicans, and the blue arrow shows the filamentous structure resulting from the deposition of multiple F. nucleatum ssp. vincentii cells. Scale bars: 5–10 μm.
In the subgingival biofilm model, the distribution of F. nucleatum ssp. nucleatum (Figures 6A,E), polymorphum (Figures 6D,G), and F. periodonticum (Figures 6F,H) was scattered through the biofilm mass along with streptococci, whereas in the case of fusiforme (Figure 6B) and vincentii (Figure 6C), their distribution was also scattered, but the proximity with streptococci was less pronounced.
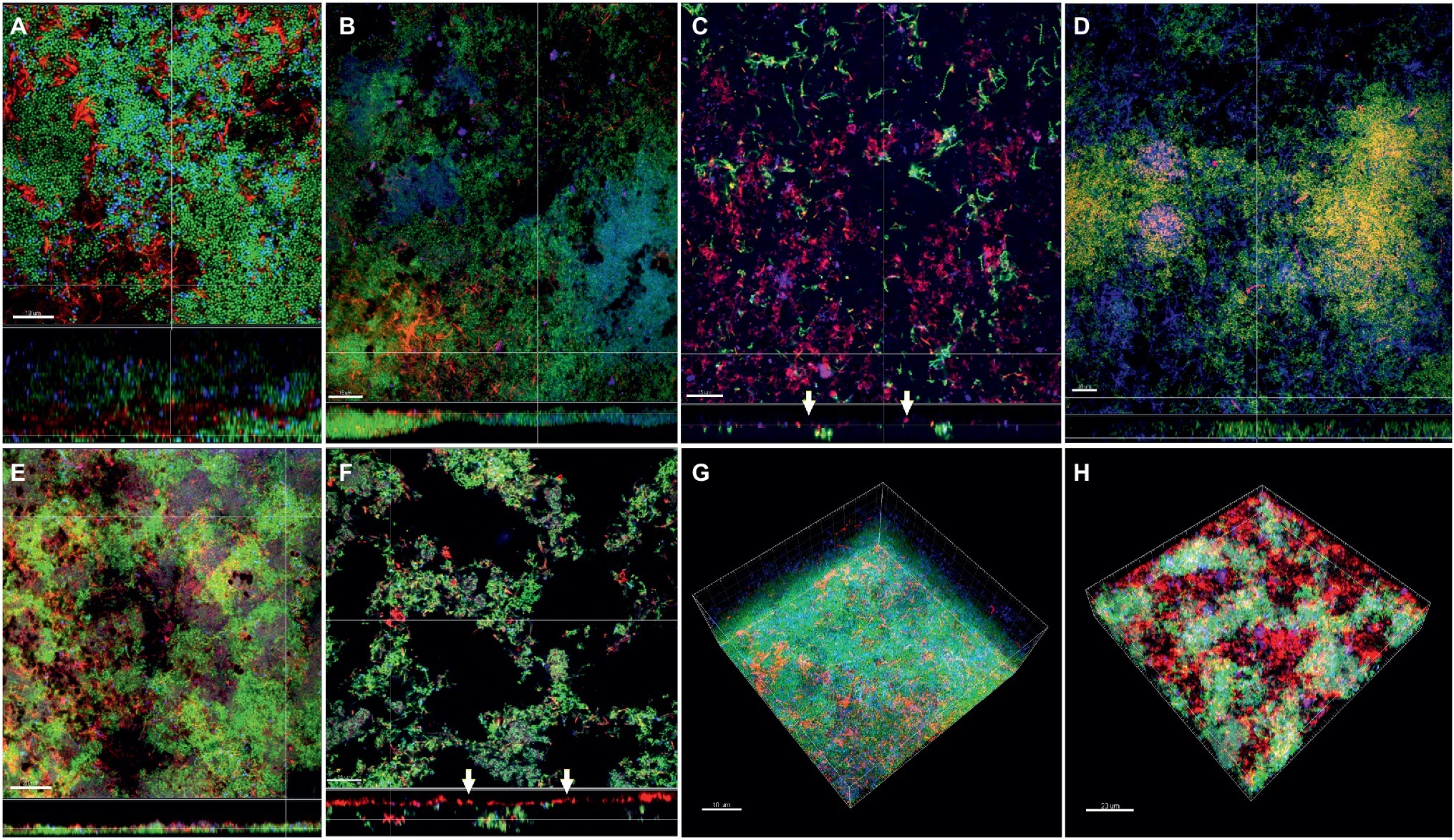
Figure 6. Incorporation of various Fusobacterium species in the subgingival biofilm model. Panels (A–F) depict confocal laser scanning microscopic (CLSM) image stack, while panels (G) and (H) represent 3D reconstructions of in vitro subgingival biofilms. Due to FISH staining, fusobacteria appear red and streptococci blue, and due to DNA staining with Sytox/YoPro 1, bacteria appear green. (A) F. nucleatum ssp. nucleatum (control), (B) F. nucleatum ssp. fusiforme, (C) F. nucleatum ssp. vincentii, (D) F. nucleatum ssp. polymorphum, (E) F. periodonticum, (F) F. naviforme, (G) F. nucleatum ssp. nucleatum (3D), and (H) F. naviforme (3D); white arrows indicate the localization of fusobacteria [F. nucleatum ssp. vincentii and F. naviforme, panels (C) and (F), respectively] near the biofilm substratum interface. Scale bars: 5–10 μm.
Fusobacteria are crucial microbial constituents of dental biofilms, with a crucial role as “bridging” microorganisms between early and late colonizing species (Bradshaw et al., 1998; Henne et al., 2018; Brennan and Garrett, 2019). F. nucleatum can communicate with other bacterial species in the biofilm via quorum-sensing signaling molecules (Frias et al., 2001), such as autoinducer-2 (Jang et al., 2013a), which has the capacity to regulate the interaction with other bacteria (Jang et al., 2013b). Apart for its potential involvement in periodontal disease, there is increasing evidence of extra-oral translocation of this species and association with extra-oral infections and systemic conditions (Han and Wang, 2013), to the extent that it is recently being considered as a potential oncobacterium (Brennan and Garrett, 2019). It can be classified in several subspecies (Jousimies-Somer, 1997), and it is not clear whether they display different metabolic versatility on a given microenvironmental niche, such as the supragingival or the subgingival biofilm environment (Rogers, 1998). Based on genomic evidence, it was recently proposed that F. nucleatum subsp. nucleatum, subsp. polymorphum, subsp. vincentii, and subsp. animalis should be classified as F. nucleatum, F. polymorphum, F. vincentii, and F. animalis, respectively (Kook et al., 2017). While two of the strains used in the present study, namely F. nucleatum ssp. fusiforme OMZ 642 (ATCC 51190) and F. nucleatum ssp. vincentii OMZ 635 (ATCC 49256), have been reported to have high genotypic similarity to a degree that they could be classified as a single subspecies (Kook et al., 2013), in our earlier work using ribotyping, antigenicity, and 16S rRNA sequencing, they are shown to be phenotypically and phylogenetically of adequate distance (Thurnheer et al., 1999; Gmür et al., 2006). An earlier study also developed a specialized qPCR assay in order to distinguish between these two subspecies species (annotated as F. nucleatum subsp. vincentii ATCC 49256 and subsp. fusiforme ATCC 51190(T), respectively; Shin et al., 2010, indicating that minute genomic differences may exist.
There is evidence that different F. nucleatum subspecies may differentially affect neutrophil function and the oxidative killing by neutrophils (Kurgan et al., 2017) denoting variations in their virulence properties. Yet, potential differences in their functional properties within biofilms have not yet been elucidated. The present study has addressed this issue by using an in vitro biofilm experimental approach and demonstrates differential biofilm behavior among the different subspecies. These may also be influenced by micro-ecological conditions, as portrayed by the three different biofilm models used here (i.e., “supragingival,” “feeding” and “subgingival”). Collectively, F. nucleatum ssp. nucleatum was the most well-adapted subspecies in the present in vitro biofilm models used, irrespective of their “supragingival” or “subgingival” profile. This is evident by the consistently higher numbers and evenly scattered distribution in the biofilm, compared to the other subspecies tested. The least favorably adapted subspecies in terms of numbers in the biofilm appeared to be vincentii, whereas the one behaving closest to F. nucleatum ssp. nucleatum appeared to be the species F. periodonticum. More detailed comparisons of their genomes and identification of genomic variations to ssp. nucleatum may help understand better their properties and capacities to adapt to different environmental pressures. It is worth further investigating the interkingdom interaction between C. albicans and F. nucleatum ssp. polymorphum, or F. naviforme and F. periodonticum species, as this yeast displayed reduced numbers in the “supragingival model.” Indeed, interaction between C. albicans and F. nucleatum may lead to a mutual attenuation of their virulence, resulting in a potential commensalism (Bor et al., 2016). C. albicans has been also shown to support the presence of strictly anaerobic bacteria under oxygen-rich conditions, including fusobacteria, in early in vitro biofilms (Janus et al., 2017), denoting its influence on the bacteriome in oral biofilms.
In the “feeding” oral biofilm model, variations were observed in the numbers of V. dispar depending on the fusobacterial species or subspecies used. As the exposure to nutritional constituents in this model deviates from that of the standard “supragingival” biofilm model, this difference may denote variations in the metabolic queues between V. dispar and fusobacterial subspecies, depending on the environmental exposures. It was recently shown that, via its catalase, Veillonella sp. can rescue the growth of F. nucleatum under microaerophilic conditions or the presence of S. gordonii (Zhou et al., 2017). S. gordonii has also displayed strong coaggregation properties to F. nucleatum (Mutha et al., 2018). While S. gordonii was not present in the model used here, S. mutans exposed to sugar may have altered the micro-environmental conditions in the biofilm, potentially affecting the interaction of F. nucleatum ssp. and V. dispar. It is worth investigating further the differential interactions of various F. nucleatum ssp. with V. dispar, which may account for the differences with the growth patterns observed in the present study. In regard to the biological significance of the intercommunication between fusobacteria and S. mutans, a recent study revealed that binding of F. nucleatum ssp. polymorphum to S. mutans can be attributed to recognition of the F. nucleatum ssp. polymorphum adhesin RadD by the S. mutans adhesin SpaP (Guo et al., 2017). Thus, the RaD/SpaP adhesin pair serves as a binding mechanism for fusobacteria and implies their high virulence potential, since S. mutans interacts with only a limited number of other bacterial species (Wang et al., 2011).
In the subgingival biofilm model, the periodontal pathogens P. gingivalis, P. intermedia, and C. rectus proved to grow better in the presence of F. nucleatum ssp. nucleatum than the other subspecies tested. This may imply that this subspecies is better adapted to co-exist synergistically with other periodontal pathogens in a biofilm environment. However, it could also reflect differences in the growth profile of the various F. nucleatum ssp. themselves, as they also proved to grow at lower numbers in the biofilm than the F. nucleatum ssp. nucleatum control. Since there are no studies describing oral fusobacterial interactions with other periodontal pathogens on a subspecies level in humans (Antiabong et al., 2013), we can only speculate that F. nucleatum ssp. nucleatum can bind to periopathogens such as P. gingivalis, P. intermedia, and C. rectus due to the presence of specific pairs of multifactorial proteins, which are not associated with other F. nucleatum subspecies.
Interestingly, a reduction in numbers of S. oralis, a commensal microorganism, was also observed. We have shown earlier that F. nucleatum growth was significantly enhanced following the late addition of the streptococci when compared to their absence (Ammann et al., 2013). Others have also identified interaction networks between F. nucleatum and Streptococcus mitis (Zhang et al., 2019), or P. gingivalis (Mohammed et al., 2017). Nevertheless, variations depending on the subspecies of F. nucleatum have been rarely taken under consideration.
In conclusion, differences in the growth and localization patters of F. nucleatum in biofilms may be observed based on the subspecies. Subspecies other than nucleatum, or species F. periodonticum, appear to be less efficiently growing in the biofilm and exhibit a more condensed localization pattern. Differences in the genotypic, phenotypic, and metabolic properties among fusobacteria may account for their different behavioral patterns. It is therefore important to consider further and define more accurately fusobacteria at the subspecies level, in order to understand their role not only in biofilm-associated oral diseases but also in light of their involvement in extra-oral infections and malignancies.
All datasets generated for this study are included in the manuscript.
TT conceived the idea for this manuscript. MF conducted the experiments. LK was involved in the data analysis. GB was involved in study design, data interpretation, and manuscript drafting. All authors critically reviewed the manuscript and approved its final version.
The study was supported by Institutional funds of the University of Zurich.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are grateful to Helga Lüthi-Schaller and Elly Plattner for excellent technical assistance. We thank the Center of Microscopy and Image Analysis (ZMB) of the University of Zurich for their support with confocal microscopy.
Ammann, T. W., Belibasakis, G. N., and Thurnheer, T. (2013). Impact of early colonizers on in vitro subgingival biofilm formation. PLoS One 8:e83090. doi: 10.1371/journal.pone.0083090
Ammann, T., Gmur, R., and Thurnheer, T. (2012). Advancement of the 10-species subgingival Zurich biofilm model by examining different nutritional conditions and defining the structure of the in vitro biofilms. BMC Microbiol. 12:227. doi: 10.1186/1471-2180-12-227
Antiabong, J. F., Boardman, W., Smith, I., Brown, M. H., Ball, A. S., and Goodman, A. E. (2013). A molecular survey of a captive wallaby population for periodontopathogens and the co-incidence of Fusobacterium necrophorum subspecies necrophorum with periodontal diseases. Vet. Microbiol. 163, 335–343. doi: 10.1016/j.vetmic.2013.01.012
Bor, B., Cen, L., Agnello, M., Shi, W., and He, X. (2016). Morphological and physiological changes induced by contact-dependent interaction between Candida albicans and Fusobacterium nucleatum. Sci. Rep. 6:27956. doi: 10.1038/srep27956
Bradshaw, D. J., Marsh, P. D., Watson, G. K., and Allison, C. (1998). Role of Fusobacterium nucleatum and coaggregation in anaerobe survival in planktonic and biofilm oral microbial communities during aeration. Infect. Immun. 66, 4729–4732.
Brennan, C. A., and Garrett, W. S. (2019). Fusobacterium nucleatum – symbiont, opportunist and oncobacterium. Nat. Rev. Microbiol. 17, 156–166. doi: 10.1038/s41579-018-0129-6
Coppenhagen-Glazer, S., Sol, A., Abed, J., Naor, R., Zhang, X., Han, Y., et al. (2015). Fap2 of Fusobacterium nucleatum is a galactose-inhibitable adhesin involved in coaggregation, cell adhesion, and preterm birth. Infect. Immun. 83, 1104–1113. doi: 10.1128/IAI.02838-14
Davies, D. (2003). Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discov. 2, 114–122. doi: 10.1038/nrd1008
Dige, I., Nilsson, H., Kilian, M., and Nyvad, B. (2007). In situ identification of streptococci and other bacteria in initial dental biofilm by confocal laser scanning microscopy and fluorescence in situ hybridization. Eur. J. Oral Sci. 115, 459–467. doi: 10.1111/j.1600-0722.2007.00494.x
Foster, J. S., and Kolenbrander, P. E. (2004). Development of a multispecies oral bacterial community in a saliva-conditioned flow cell. Appl. Environ. Microbiol. 70, 4340–4348. doi: 10.1128/AEM.70.7.4340-4348.2004
Frias, J., Olle, E., and Alsina, M. (2001). Periodontal pathogens produce quorum sensing signal molecules. Infect. Immun. 69, 3431–3434. doi: 10.1128/IAI.69.5.3431-3434.2001
Gaetti-Jardim Júnior, E., and Avila-Campos, M. (1999). Haemagglutination and haemolysis by oral Fusobacterium nucleatum. New Microbiol. 22, 63–67.
Gmür, R., Munson, M., and Wade, W. (2006). Genotypic and phenotypic characterization of fusobacteria from Chinese and European patients with inflammatory periodontal diseases. Syst. Appl. Microbiol. 29, 120–130. doi: 10.1016/j.syapm.2005.07.011
Guggenheim, B., Giertsen, E., Schüpbach, P., and Shapiro, S. (2001a). Validation of an in vitro biofilm model of supragingival plaque. J. Dent. Res. 80, 363–370. doi: 10.1177/00220345010800011201
Guggenheim, M., Shapiro, S., Gmür, R., and Guggenheim, B. (2001b). Spatial arrangements and associative behavior of species in an in vitro oral biofilm model. Appl. Environ. Microbiol. 67, 1343–1350. doi: 10.1128/AEM.67.3.1343-1350.2001
Guo, L., Shokeen, B., He, X., Shi, W., and Lux, R. (2017). Streptococcus mutans SpaP binds to RadD of Fusobacterium nucleatum ssp. polymorphum. Mol. Oral Microbiol. 32, 355–364. doi: 10.1111/omi.12177
Gursoy, U., Könönen, E., and Uitto, V. (2008). Intracellular replication of fusobacteria requires new actin filament formation of epithelial cells. APMIS 116, 1063–1070. doi: 10.1111/j.1600-0463.2008.00868.x
Gursoy, U., Pollanen, M., Kononen, E., and Uitto, V. (2010). Biofilm formation enhances the oxygen tolerance and invasiveness of Fusobacterium nucleatum in an oral mucosa culture model. J. Periodontol. 81, 1084–1091. doi: 10.1902/jop.2010.090664
Hall-Stoodley, L., Costerton, J., and Stoodley, P. (2004). Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2, 95–108. doi: 10.1038/nrmicro821
Han, Y. W., and Wang, X. (2013). Mobile microbiome: oral bacteria in extra-oral infections and inflammation. J. Dent. Res. 92, 485–491. doi: 10.1177/0022034513487559
He, X., Hu, W., Kaplan, C., Guo, L., Shi, W., and Lux, R. (2012). Adherence to streptococci facilitates Fusobacterium nucleatum integration into an oral microbial community. Microb. Ecol. 63, 532–542. doi: 10.1007/s00248-011-9989-2
Henne, K., Schilling, H., Stoneking, M., Conrads, G., and Horz, H. P. (2018). Sex-specific differences in the occurrence of Fusobacterium nucleatum subspecies and Fusobacterium periodonticum in the oral cavity. Oncotarget 9, 20631–20639. doi: 10.18632/oncotarget.25042
Jang, Y., Choi, Y., Lee, S., Jun, H., and Choi, B. (2013a). Autoinducer 2 of Fusobacterium nucleatum as a target molecule to inhibit biofilm formation of periodontopathogens. Arch. Oral Biol. 58, 17–27. doi: 10.1016/j.archoralbio.2012.04.016
Jang, Y. J., Sim, J., Jun, H. K., and Choi, B. K. (2013b). Differential effect of autoinducer 2 of Fusobacterium nucleatum on oral streptococci. Arch. Oral Biol. 58, 1594–1602. doi: 10.1016/j.archoralbio.2013.08.006
Janus, M. M., Crielaard, W., Volgenant, C. M., Van Der Veen, M. H., Brandt, B. W., and Krom, B. P. (2017). Candida albicans alters the bacterial microbiome of early in vitro oral biofilms. J. Oral Microbiol. 9:1270613. doi: 10.1080/20002297.2016.1270613
Jousimies-Somer, H. (1997). Recently described clinically important anaerobic bacteria: taxonomic aspects and update. Clin. Infect. Dis. 25(Suppl. 2), S78–S87. doi: 10.1086/516227
Kaplan, C., Lux, R., Haake, S., and Shi, W. (2009). The Fusobacterium nucleatum outer membrane protein RadD is an arginine-inhibitable adhesin required for inter-species adherence and the structured architecture of multispecies biofilm. Mol. Microbiol. 71, 35–47. doi: 10.1111/j.1365-2958.2008.06503.x
Katharios-Lanwermeyer, S., Xi, C., Jakubovics, N., and Rickard, A. (2014). Mini-review: microbial coaggregation: ubiquity and implications for biofilm development. Biofouling 30, 1235–1251. doi: 10.1080/08927014.2014.976206
Keijser, B., Zaura, E., Huse, S., Van Der Vossen, J., Schuren, F., Montijn, R., et al. (2008). Pyrosequencing analysis of the oral microflora of healthy adults. J. Dent. Res. 87, 1016–1020. doi: 10.1177/154405910808701104
Kolenbrander, P. E., Andersen, R. N., Blehert, D. S., Egland, P. G., Foster, J. S., and Palmer, R. J. Jr. (2002). Communication among oral bacteria. Microbiol. Mol. Biol. Rev. 66, 486–505. doi: 10.1128/MMBR.66.3.486-505.2002
Kook, J. K., Park, S. N., Lim, Y. K., Cho, E., Jo, E., Roh, H., et al. (2017). Genome-based reclassification of Fusobacterium nucleatum subspecies at the species level. Curr. Microbiol. 74, 1137–1147. doi: 10.1007/s00284-017-1296-9
Kook, J. K., Park, S. N., Lim, Y. K., Choi, M. H., Cho, E., Kong, S. W., et al. (2013). Fusobacterium nucleatum subsp. fusiforme Gharbia and Shah 1992 is a later synonym of Fusobacterium nucleatum subsp. vincentii Dzink et al. 1990. Curr. Microbiol. 66, 414–417. doi: 10.1007/s00284-012-0289-y
Kurgan, S., Kansal, S., Nguyen, D., Stephens, D., Koroneos, Y., Hasturk, H., et al. (2017). Strain-specific impact of Fusobacterium nucleatum on neutrophil function. J. Periodontol. 88, 380–389. doi: 10.1902/jop.2016.160212
Marsh, P. D. (2004). Dental plaque as a microbial biofilm. Caries Res. 38, 204–211. doi: 10.1159/000077756
Marsh, P. (2005). Dental plaque: biological significance of a biofilm and community life-style. J. Clin. Periodontol. 32(Suppl. 6), 7–15. doi: 10.1111/j.1600-051X.2005.00790.x
Marsh, P., and Devine, D. (2011). How is the development of dental biofilms influenced by the host? J. Clin. Periodontol. 38(Suppl. 11), 28–35. doi: 10.1111/j.1600-051X.2010.01673.x
Mohammed, M. M. A., Pettersen, V. K., Nerland, A. H., Wiker, H. G., and Bakken, V. (2017). Quantitative proteomic analysis of extracellular matrix extracted from mono- and dual-species biofilms of Fusobacterium nucleatum and Porphyromonas gingivalis. Anaerobe 44, 133–142. doi: 10.1016/j.anaerobe.2017.03.002
Mutha, N. V. R., Mohammed, W. K., Krasnogor, N., Tan, G. Y. A., Choo, S. W., and Jakubovics, N. S. (2018). Transcriptional responses of Streptococcus gordonii and Fusobacterium nucleatum to coaggregation. Mol. Oral Microbiol. 33, 450–464. doi: 10.1111/omi.12248
Nikitkova, A., Haase, E., and Scannapieco, F. (2013). Taking the starch out of oral biofilm formation: molecular basis and functional significance of salivary α-amylase binding to oral streptococci. Appl. Environ. Microbiol. 79, 416–423. doi: 10.1128/AEM.02581-12
Okamoto, A., Gaetti-Jardim, E., Cai, S., and Avila-Campos, M. (2000). Influence of antimicrobial subinhibitory concentrations on hemolytic activity and bacteriocin-like substances in oral Fusobacterium nucleatum. New Microbiol. 23, 137–142.
Palmer, R. (2014). Composition and development of oral bacterial communities. Periodontol. 64, 20–39. doi: 10.1111/j.1600-0757.2012.00453.x
Rogers, A. H. (1998). Studies on fusobacteria associated with periodontal diseases. Aust. Dent. J. 43, 105–109. doi: 10.1111/j.1834-7819.1998.tb06098.x
Shapiro, S., Giertsen, E., and Guggenheim, B. (2002). An in vitro oral biofilm model for comparing the efficacy of antimicrobial mouthrinses. Caries Res. 36, 93–100. doi: 10.1159/000057866
Shin, H. S., Kim, M. J., Kim, H. S., Park, S. N., Kim, D. K., Baek, D. H., et al. (2010). Development of strain-specific PCR primers for the identification of Fusobacterium nucleatum subsp. fusiforme ATCC 51190(T) and subsp. vincentii ATCC 49256(T). Anaerobe 16, 43–46. doi: 10.1016/j.anaerobe.2009.04.003
Strauss, J., Kaplan, G., Beck, P., Rioux, K., Panaccione, R., Devinney, R., et al. (2011). Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm. Bowel Dis. 17, 1971–1978. doi: 10.1002/ibd.21606
Takahashi, N., and Nyvad, B. (2011). The role of bacteria in the caries process: ecological perspectives. J. Dent. Res. 90, 294–303. doi: 10.1177/0022034510379602
Thurnheer, T., Belibasakis, G., and Bostanci, N. (2014a). Colonisation of gingival epithelia by subgingival biofilms in vitro: role of “red complex” bacteria. Arch. Oral Biol. 59, 977–986. doi: 10.1016/j.archoralbio.2014.05.023
Thurnheer, T., Gmür, R., and Guggenheim, B. (2004). Multiplex FISH analysis of a six-species bacterial biofilm. J. Microbiol. Methods 56, 37–47. doi: 10.1016/j.mimet.2003.09.003
Thurnheer, T., Gmür, R., Shapiro, S., and Guggenheim, B. (2003). Mass transport of macromolecules within an in vitro model of supragingival plaque. Appl. Environ. Microbiol. 69, 1702–1709. doi: 10.1128/AEM.69.3.1702-1709.2003
Thurnheer, T., Guggenheim, B., Gruica, B., and Gmür, R. (1999). Infinite serovar and ribotype heterogeneity among oral Fusobacterium nucleatum strains? Anaerobe 5, 79–92. doi: 10.1006/anae.1999.0188
Thurnheer, T., Rohrer, E., Belibasakis, G., Attin, T., and Schmidlin, P. (2014b). Static biofilm removal around ultrasonic tips in vitro. Clin. Oral Investig. 18, 1779–1784. doi: 10.1007/s00784-013-1157-2
Thurnheer, T., Van Der Ploeg, J., Giertsen, E., and Guggenheim, B. (2006). Effects of Streptococcus mutans gtfC deficiency on mixed oral biofilms in vitro. Caries Res. 40, 163–171. doi: 10.1159/000091065
Van Der Ploeg, J. R., and Guggenheim, B. (2004). Deletion of gtfC of Streptococcus mutans has no influence on the composition of a mixed-species in vitro biofilm model of supragingival plaque. Eur. J. Oral Sci. 112, 433–438. doi: 10.1111/j.1600-0722.2004.00158.x
Wang, R. K., He, X. S., Hu, W., Lux, R., Li, J. Y., Zhou, X. D., et al. (2011). Analysis of interspecies adherence of oral bacteria using a membrane binding assay coupled with polymerase chain reaction-denaturing gradient gel electrophoresis profiling. Int. J. Oral Sci. 3, 90–97. doi: 10.4248/IJOS11033
Welin-Neilands, J., and Svensater, G. (2007). Acid tolerance of biofilm cells of Streptococcus mutans. Appl. Environ. Microbiol. 73, 5633–5638. doi: 10.1128/AEM.01049-07
Yoshida, A., Yoshimura, M., Ohara, N., Yoshimura, S., Nagashima, S., Takehara, T., et al. (2009). Hydrogen sulfide production from cysteine and homocysteine by periodontal and oral bacteria. J. Periodontol. 80, 1845–1851. doi: 10.1902/jop.2009.090012
Zhang, Y., Shi, W., Song, Y., and Wang, J. (2019). Metatranscriptomic analysis of an in vitro biofilm model reveals strain-specific interactions among multiple bacterial species. J. Oral Microbiol. 11:1599670. doi: 10.1080/20002297.2019.1599670
Keywords: Fusobacterium nucleatum, subspecies, supra- and subgingival biofilm models, bacterial growth, fluorescent in situ hybridization, confocal laser scanning microscopy
Citation: Thurnheer T, Karygianni L, Flury M and Belibasakis GN (2019) Fusobacterium Species and Subspecies Differentially Affect the Composition and Architecture of Supra- and Subgingival Biofilms Models. Front. Microbiol. 10:1716. doi: 10.3389/fmicb.2019.01716
Received: 28 May 2019; Accepted: 11 July 2019;
Published: 30 July 2019.
Edited by:
Daniela De Biase, Sapienza University of Rome, ItalyReviewed by:
Kazuyuki Ishihara, Tokyo Dental College, JapanCopyright © 2019 Thurnheer, Karygianni, Flury and Belibasakis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thomas Thurnheer, dGhvbWFzLnRodXJuaGVlckB6em0udXpoLmNo
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.