- 1Department of Laboratory Medicine, State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University and Collaborative Innovation Center, Chengdu, China
- 2Department of Microbiology, West China School of Basic Medical Sciences and Forensic Medicine, Sichuan University, Chengdu, China
The prevalence of carbapenem-resistant Klebsiella pneumoniae (CRKP) is rapidly increasing worldwide in recent decades and poses a challenge for today’s clinical practice. Rapid detection of CRKP can avoid inappropriate antimicrobial therapy and save lives. Traditional detection methods for CRKP are extremely time-consuming; PCR and other sequencing methods are too expensive and technologically demanding, making it hard to meet the clinical demands. Nanopore assay has been used for screening biomarkers of diseases recently because of its high sensitivity, real-time detection, and low cost. In this study, we distinguished CRKP from carbapenem-sensitive K. pneumoniae (CSKP) by the detection of increasing amount of extracted 16S ribosomal RNA (16S rRNA) from bacterial culture with antibiotics imipenem, indicating the uninhibited growth of CRKP by the imipenem. Specific signals from single channel recording of 16S rRNA bound with probes by MspA nanopore allowed the ultra-sensitive and fast quantitative detection of 16S rRNA. We proved that only 4 h of CRKP culture time was needed for nanopore assay to distinguish the CRKP and CSKP. The time-cost of the assay is only about 5% of disk diffusion method while reaching the similar accuracy. This new method has the potential application in the fast screening of drug resistance in clinical microorganism samples.
Introduction
Klebsiella pneumoniae (KP) is one of the most important opportunistic pathogens in the clinical infection (Debby et al., 2012; Aracil-Garcia et al., 2017). KP usually presents in the intestines of humans and animals (Saidel-Odes et al., 2012). The infection of KP can cause serious clinical consequences, including central nervous system infection or abdominal infection, etc. (Khan et al., 2014; Chew et al., 2017). Antimicrobial agents are the major treatments for KP infection; early and proper using of antimicrobial agents is the key to KP infection cure (Nepka et al., 2016; Su et al., 2018). However, the widely use of broad-spectrum antimicrobial leads to the strong antimicrobial resistant characteristics of KP (Rojas et al., 2016; Lomonaco et al., 2018), which results in the prolonged course and failure in treatment. The carbapenem-resistance of K. pneumoniae HS11286 (PMID: 26169555) (Bi et al., 2015) may be caused by biofilm formation, active antimicrobial efflux, and β-lactamase generation (Chew et al., 2017).
Accurate and fast diagnosis of antimicrobial resistance of KP in the infected patients is very important for treatment, as it can help doctor choose proper kinds of antimicrobial agents, reduce the treatment cycle, and improve prognosis. Bacterial drug resistance phenotype detection, β-lactamase detection (Rojas et al., 2016), and drug resistance gene detection (Chiu et al., 2017) are the current major methods for drug resistance detection. The detection of bacterial drug resistance phenotype need sufficient time of KP culture, which is usually time-consuming; β-lactamase detection is fast, but it has a relative small detection range (Whichard et al., 2010; Rood and Li, 2017); gene detection for drug resistance has high precision, but it is costly and time-consuming as well (Frickmann et al., 2014).
Nanopore sensing is a novel biosensing and biodetection technology with single molecular sensitivity (Kasianowicz et al., 1996; Branton et al., 2008; Wanunu, 2012), which contributes to its widespread use in the third-generation DNA sequencing. The nanometer-sized protein pores are embedded in a phospholipid membrane, which separates the chambers into two parts (cis and trans). When a voltage is applied across the chamber containing a certain concentration of ion solution, the charged detection substance in the system is driven through the pores by the voltage to the other chamber. The patch clamp sensor detects the current change signals of the nanopore (Manara et al., 2015). Different molecules transported through the nanopore can cause corresponding blockage signals of the current flow (Howorka and Siwy, 2009; Ying et al., 2013). Using the specific translocation signal and translocation frequency, qualitative and quantitative analysis of the detected molecules can be achieved (Kim and Gale, 2008; Yang and Yamamoto, 2016). This nanopore sensing technology has the advantage of label-free, fast, real time, and high sensitivity with small volume of samples needed (Wang et al., 2013) in the detection process. These features are suitable for the rapid diagnosis of disease and detection of biomarker (Wang et al., 2011). The protein mycobacterium smegmatis porin A (MspA) nanopore is one of the outer membrane proteins of mycobacteria with a length of 9.6 nm and a diameter of 1.3 nm (Figure 1A; Faller, 2004). The nanopore is highly efficient to be incorporated into bilayer lipid membrane (Figure 1B) and allows the translocation of single-strand nucleic acids through the pore (Butler et al., 2008; Fleming et al., 2017). Due to its short and narrow channel constriction, MspA nanopore is ideally suitable for nanopore sequencing (Laszlo et al., 2016) such as identifying single nucleotides within random DNA (Manrao et al., 2011) or detecting and mapping 5-methylcytosine and 5-hydroxymethylcytosine within single strands of DNA (Laszlo et al., 2013).
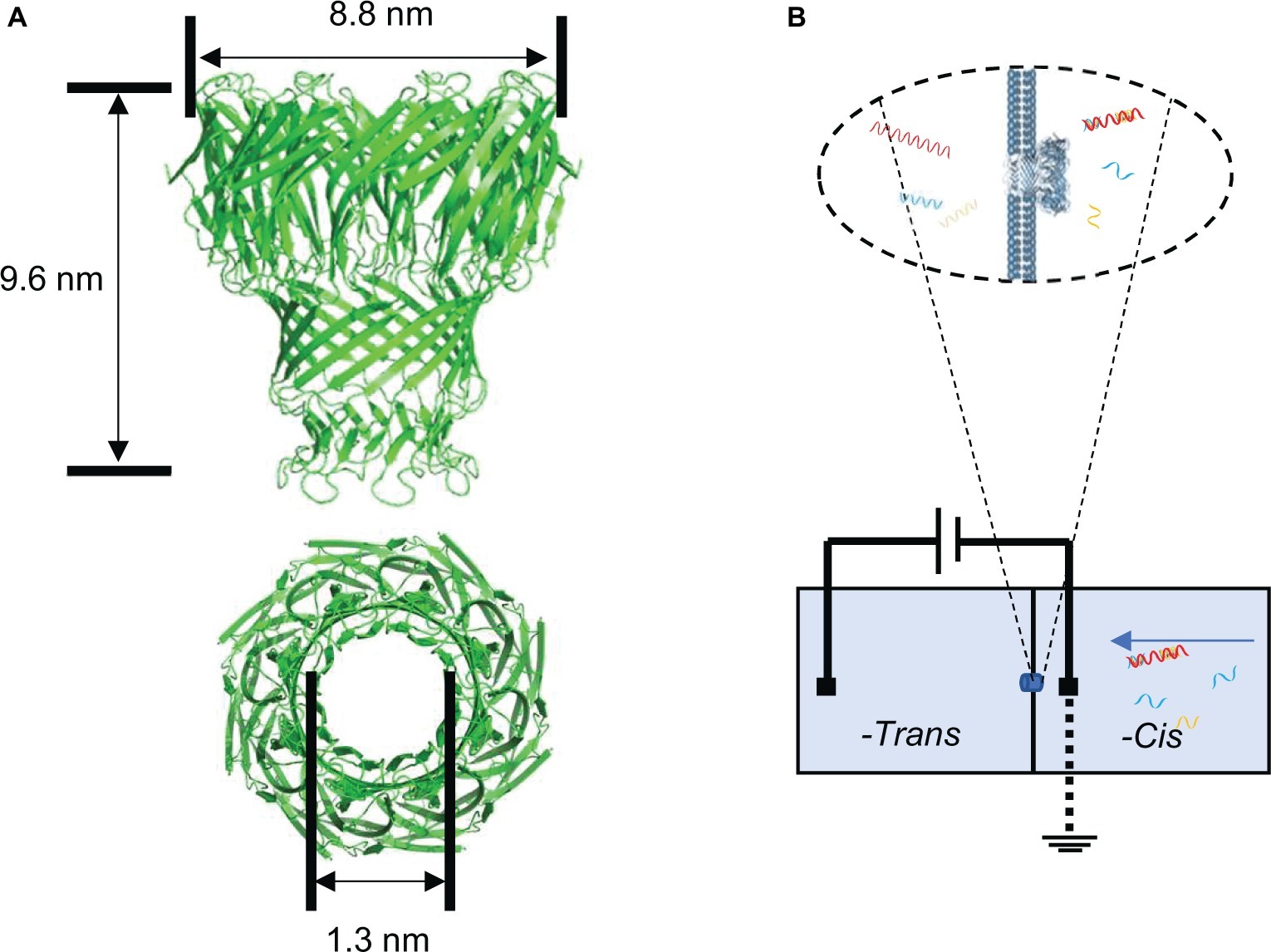
Figure 1. Structure of the nanopore used in the study and single-channel recording setup for nanopore assay. (A) Side view of MspA nanopore (top) and top view of MspA nanopore (bottom). (B) Schematic diagram of the single channel recording setup for 16S rRNA-probe complex detection.
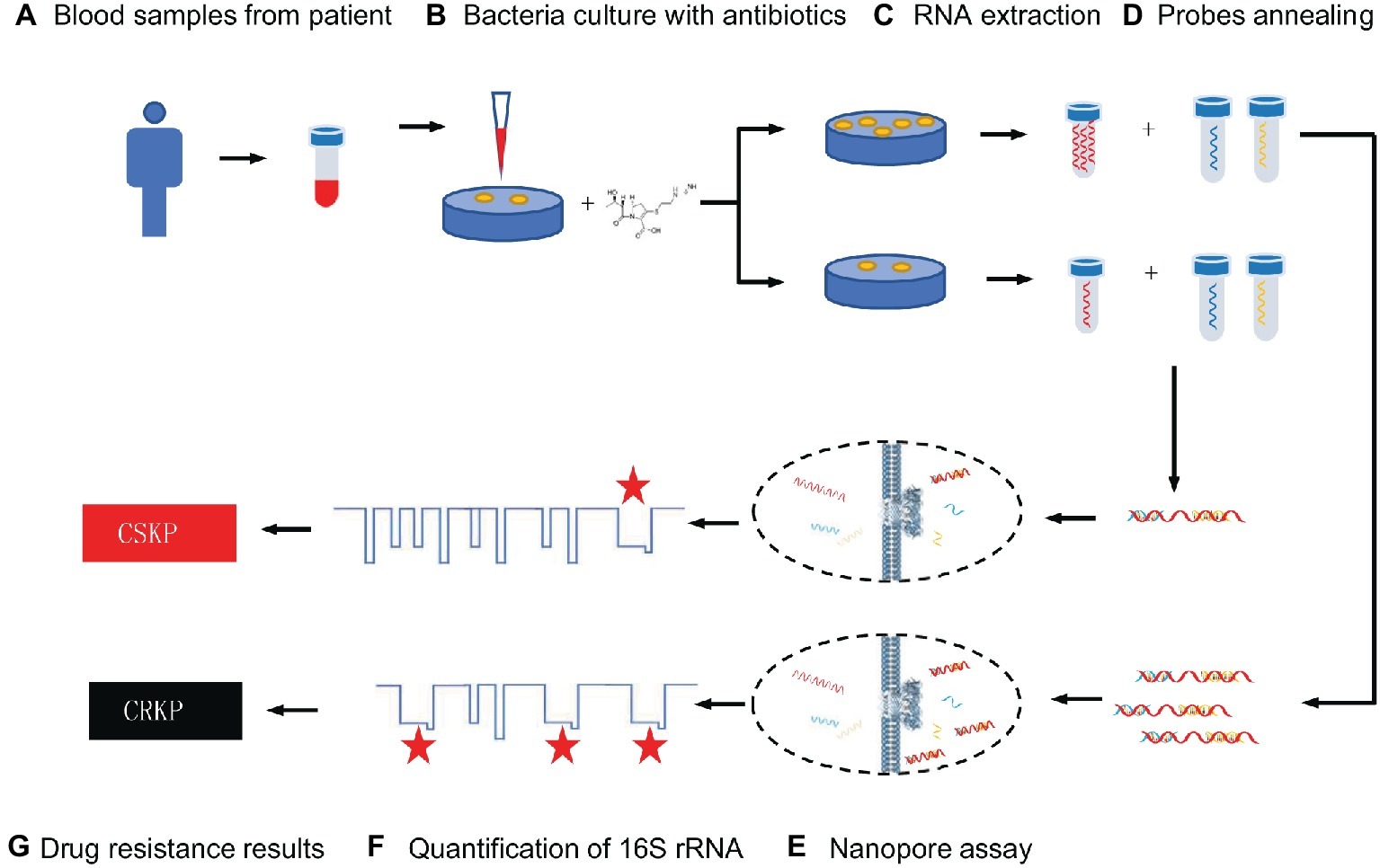
16S rRNA that presents in all bacteria is a component of the 30S subunit in the ribosome of prokaryotes, and its function does not change over time (Patel, 2001). 16S rRNA can be used to identify bacterial species because it contains a highly conserved region shared by all bacteria and a hypervariable region with differences in different bacteria (Kolbert and Persing, 1999; Pereira et al., 2010; Srinivasan et al., 2012). It has been proven to be a reliable genetic marker and is often used for bacterial classification (Liu et al., 2012; Deurenberg et al., 2017). There is literature demonstrating that it can be used for the identification of clinical pathogens (Schuurman et al., 2004; Srinivasan et al., 2012). In this study, we provide a novel, efficient, and rapid method based on the MspA nanopore to distinguish the CRKP and CSKP at the single molecular level (Scheme 1). These strains which identified by MALDI-TOF MS are separately cultured with imipenem for several hours. 16S rRNA is highly conserved and specific, which has become a powerful tool for pathogen detection and identification in the genetic testing technology (Kolbert and Persing, 1999; Liu et al., 2012; Deurenberg et al., 2017), so it was chosen as the parameter for the measurement of amount of live KP after culturing under the antibiotics. We use specific probes to combine with 16S rRNA in K. pneumoniae and record the nucleic acid reading processes by nanopore assay. The frequency of specific signals regarding the translocation of the target nucleic acid through nanopore reflects the quantities of live KP. Therefore, a quantitative analysis of remaining live KP against carbapenem can be established. We prove that only 4 h of bacterial culture was needed for the discrimination when using 4 MCF as the initial concentration of bacteria suspension. The accuracy of this method is 90%, and the time-cost is far less than cell culture method.
Materials and Methods
Materials
4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES, purity > 99.5%, CAS#7365-45-9), potassium chloride (KCl, purity > 99.0%, CAS#7447-40-7), agarose (purity > 99.0%, CAS#: 9012-36-6), chloroform (purity > 99.0%, CAS: 67-66-3), isopropyl alcohol (purity > 99.0%, CAS#: 67-63-0), and alcohol (purity > 99.0%, CAS#: 64-17-5) was purchased from Sigma-Aldrich. RNase inhibitor (5 KU), pET-28b plasmid, and all the DNA were provided by Sangon Biotech. 1,2-Diphytanoyl-sn-glycero-3-phosphocholine (DPHPC) was purchased from Avanti. PrimeSTAR HS DNA polymerase was purchased from TaKaRa. Imipenem (CAS#: 64221-86-9) was purchased from MSD.
Preparation of Bacterial Extracts
Two groups of K. pneumoniae samples from clinical patients were provided by West China Hospital of Sichuan University. The KP samples were cultured to two different concentrations. The concentration of first group was 0.5 MCF, and the second group was 4 MCF. The final concentration of imipenem used in two groups was 16 mg/L at the beginning of culture. The total RNA of KP was extracted by TRIZOL method. First, 100 μl of bacteria solution is collected. The supernatant was removed after centrifugation subsequently (8,000g, 4°C, 2 min). The precipitation with lysozyme was incubated for 10 min at 37°C. The KP cells were lysed, and the total RNA was extracted and washed with ethanol. The centrifugal tube cap was removed, and the tube was dry at room temperature for 5–10 min, then the DEPC water was added or dissolved in RNase-free water. RNase inhibitor was added to the dissolved solution to a final concentration of 20 U/μl for storage.
Tested Samples Incubated With Probes
Probe A and probe B were synthesized by Sangon Biotech (Probe A: AGCAC AG AGA GCTTG CTCTC GGGTG ACGAG CGGCG GACGG GTGAG TAATG TCTGG GAAAC TGCCT GATGG AGGGG GATAA CTACT GGAAA CGGTA GCTAA TACCG CATAA CGTCG CAAGA CCAAG T, probe B: CCTTG AGGCG TGGCT TCCGG AGCTA ACGCG TTAAA TCGAC CGCCT GGGGA GTACG GCCGC AAGGT TAAAA CTCAA ATGAA TTGAC GGGGG CCCGC ACAAG CGGTG GAGGA TGTGG TTTAA TTCGA TGC). The probe A and probe B were annealed with tested samples using a PCR program (cooling of temperature from 95 to 4°C in 60 min) (Figure 2A). We used the agarose gel electrophoresis to verify the formation of probe A-probe B-target 16S rRNA complex.
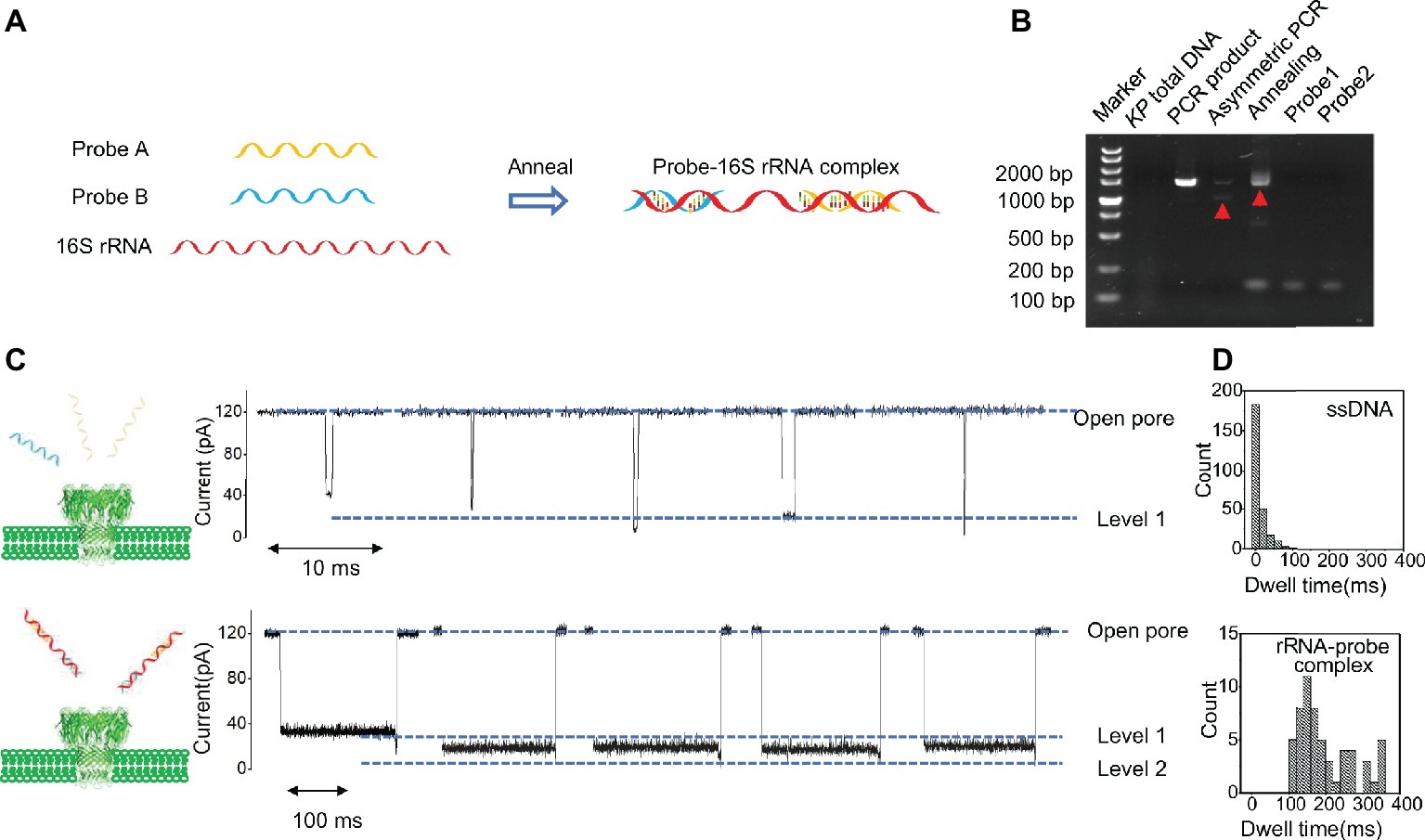
Figure 2. Probe-target RNA complex and its nanopore signal. (A) Probe A and probe B bound with 16S rRNA of carbapenem resistant KP. (B) Agarose gels result of probe A and probe B bound with CRKP 16S rRNA. (C) Signal of single-strand nucleic acid translocation and probe-16S rRNA complex translocation. (D) Dwell time distribution of the translocation events.
Nanopore Electrophysiological Experiment
The nanopore detection experiment was carried out in the chamber provided by Warner Instrument. The nanopore electrophysiological assay experiments were carried out under 150 mV bias voltage. The conductive buffer solutions at both -cis side and -trans side are 400 mM KCl with 10 mM HEPES, pH 7.0. The bilayer lipid membrane (BLM) that painted on both sides of the 150 μm hole was formed by 1,2-diphytanoyl-sn-glycero-3-phosphocholine (DPHPC) (Montal and Muellertj, 1972). MspA was added to the solution in the -cis chamber, allowing MspA proteins to insert after BLM formation. A single MspA nanopore insertion will cause a current increase corresponding to a conductance of ca. 1.2 nS. Samples were added into the -cis side after the insertion of a single MspA nanopore current signals were recorded by Heka EPC-10 patch clamp (HEKA).
Data Analysis
We performed data analysis using software Clampfit 10.6 and Origin Pro 8.0. The current blockage was defined as ΔI/I 0, where I 0 was the current of a fully opened pore, and ΔI was the amplitude of current blockage caused by translocated molecules. The dwell time was collected by the single-channel search feature of the Clampfit 10.6. Those two parameters were applied for quantitative analysis of target 16S rRNA from the surviving live K. pneumoniae resistant to carbapenem. All the data were obtained from a 20-min electrophysiological recording, and the experimental group was repeated three times independently.
Clinical Specimens
Blood samples from 20 patients with KP infection were provided by the Department of Laboratory Medicine, West China Hospital of Sichuan University. This study was carried out in accordance with the recommendations of China National Measures for the Ethical Review of Biomedical Research Involving Humans and WMA Declaration of Helsinki. The protocol was approved by the Biomedical Ethics Committee of West China Hospital, Sichuan University.
The research used leftover specimens, that is, remnants of specimens collected for routine clinical care or analysis that would have been discarded, and complied with the criteria for a waiver of informed consent. The waiver for informed consent was granted by Biomedical Ethics Committee of West China Hospital, Sichuan University.
Results
Detection of 16S rRNA-Probe Complex
The result of agarose gel electrophoresis indicated that the rRNA-Probe complex was obtained successfully (Figure 2B). The dwell time of 16S rRNA-probe complex translocation signal in the carbapenem-resistant KP sample was in the range of 100–400 ms (Figures 2C,D) with a peak value of 196.98 ms, and the dwell time of single-stranded DNA translocation signal was in the range of 0–100 ms (Figures 2C,D) with a peak value of 12.03 ms. The dwell time of probe A and probe B was in the range of 0–70 ms (Supplementary Figure S1). Those results indicated that the signal with long dwell-time was attributed to the 16S rRNA-probe complex.
Optimization of Bacterial Concentration and Standard Sample Test
Two concentrations of KP were used to optimize the detection efficiency. In the samples of 0.5 MCF, the target RNA translocation signal frequency of the control group was 0.02 ± 0.02 per minute (n = 3), and the target RNA translocation signal frequency of CRKP group is 0.13 ± 0.05 per minute (n = 3) (Figure 3A). While in the samples of 4 MCF, target RNA translocation signal frequency of control group is 0 per minute (n = 3), and the translocation frequency of CRKP group is 0.33 ± 0.07 per minute (n = 3) (Figure 3A). Compared with 0.5 MCF sample, the 4 MCF sample showed improved detection performance in the nanopore assay.
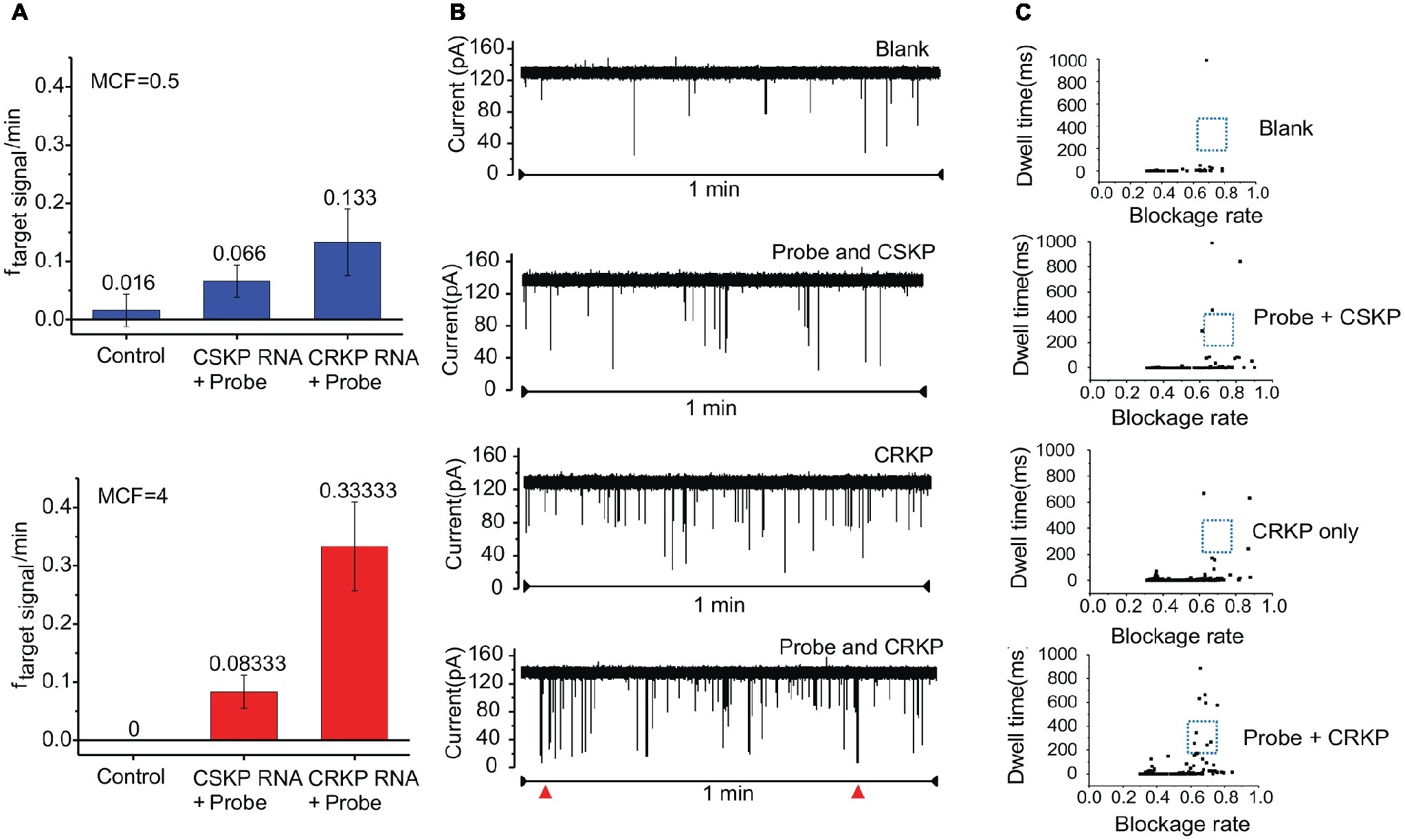
Figure 3. Distinguishing CRKP from CSKP from single channel recording signals. (A) Nanopore assay efficiency of 0.5 MCF sample and 4 MCF sample cultured for 4 h (n = 3 for each measurement). (B) Nanopore signal of control group, probe with carbapenem-sensitive KP, carbapenem-resistant KP, probe with carbapenem-resistant KP, respectively. (C) Scatter plot of translocation signal of different samples.
The total RNA from carbapenem-resistant KP and carbapenem-sensitive KP were incubated with probe A and probe B, and the incubation solution was detected by MspA nanopore, respectively (Figure 3B). The two parameters of the obtained signals, blockage rate, and dwell time, from nanopore assay of the samples were plotted into scattering plot (Figure 3C), and obvious difference in dwell time between different group can be observed, especially in the range of blockage rate 0.6–0.8 and dwell time 100–400 ms. Thus, signals in this range were chosen as specific signals for diagnosis. After comparing the number of 16S rRNA-probe signals in the given range from blank, control, CSKP and CRKP samples, f = 0.1 min−1 was used as the target events threshold to distinguish the carbapenem resistance of K. pneumoniae. In order to determine the minimum bacteria culture time needed to distinguish CSKP and CRKP, samples with different bacteria culture time including 2, 4, and 8 h were tested by MspA nanopore. Those results indicated that 4 h is the optimal bacteria culture time in considering both sensitivity and efficiency.
Double-Blind Test of Clinical Samples by MspA Nanopore
Based on the above results of standard CRKP and CSKP samples, further experiments were performed to verify the potential application of this method in clinical diagnosis. Bacteria in the blood samples from 20 patients with KP infection provided by the West China Hospital were cultured and the total RNA were extracted and used for double-blind experiments. Each sample was detected by MspA nanopore at least three times. After analysis, the number of 16S rRNA-probe signals within the blockage rate of 0.6–0.8 and dwell time of 100–400 ms were collected and compared with target events frequency threshold fthreshold.
Out of the 20 samples, nine of them were above the threshold (0.1·min−1) and were identified as carbapenem-resistant KP. The other 11 samples showed a lower target event frequency below 0.1·min−1, and these clinical samples were identified as carbapenem-sensitive KP (Figure 4A). Compared with the assay results obtained from standard clinical (disk diffusion method or PCR) method (Supplementary Tables S1, S2), results of 18 samples by nanopore assay were correct (Figure 4B), while two showed false negative results.
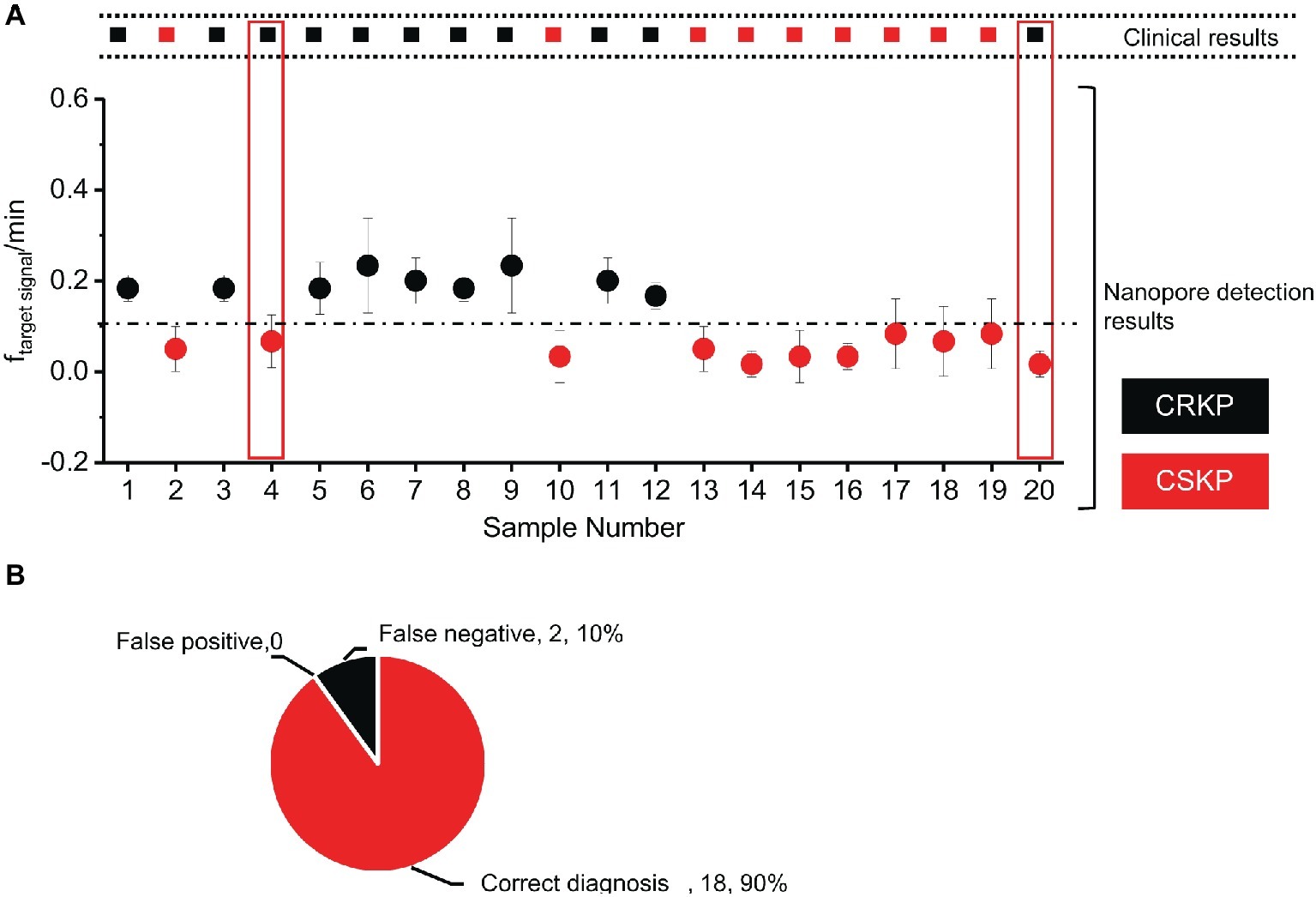
Figure 4. Double-blind test of clinical samples and evaluation of the assay accuracy. (A) Nanopore assay of 20 Klebsiella pneumoniae clinical samples. (B) Accuracy rate of the nanopore assay method. (n = 3 for each measurement).
Discussion
Accurate and fast diagnosis of KP’s carbapenem resistance is quite important in the clinical therapy process. Our objective is to develop a new KP carbapenem resistant detection method based on nanopore sensing technology to provide a potential solution to the challenges of the present methods that cannot fully meet the clinical demands.
Protein nanopore expression, purification and electrophysiological assay technology has been fast developed during the past 20 years, and various types of protein nanopores have been constructed for well-established nucleic acids assay (Haque et al., 2013). In this experiment, we designed two DNA probes to specifically bind to the 16S rRNA of carbapenem-resistant KP; the 16S rRNA-probe complex translocation through MspA nanopore will cause a much slower translocation dwell time between 100 and 400 ms. Based on the dwell time and blockage of translocation signals, we can figure out targeted 16S rRNA of carbapenem-resistant KP samples (Supplementary Figure S2). Both carbapenem resistant KP standard samples and carbapenem sensitive KP standard samples were detected, and the result showed that this method can be used to distinguish the carbapenem resistant KP and carbapenem sensitive KP with only 4 h of bacteria culture. Furthermore, 20 clinical samples provided by West China Hospital were assayed by MspA nanopore. Among the 11 carbapenem resistant KP clinical samples, nine samples were diagnosed correctly, while two samples were detected as false negative; among the nine carbapenem sensitive KP samples, nine samples were diagnosed correctly. The accuracy rate of nanopore diagnose method is 90%. RNA degradation during sample storage or transfer was the main reason leading to the 10% false negative diagnosis. Variation of the transportation of the clinical samples from hospital to research lab and time gap between the sample processing and nanopore assay increased the possibility of RNA degradation, leading to the decrease number of 16S rRNA and specific signals detected by nanopore.
The above results was a pilot and experimental study of new single-molecule diagnostic technology, and further validation studies aiming to increase the sensitivity could be carried out in several aspects: optimization of bacterial culture condition and RNA extraction; modification and reengineering of the protein nanopore for the 16S rRNA-probe complex detection; test with larger number of clinical samples; and improved statistics methods for data processing. Stable storage of the aliquoted MspA protein at −80°C enabled the batch production of the nanopore for future multiple tests.
In conclusion, our research provided a proof of concept nanopore sensing technology with potential application in the rapid clinical diagnosis of carbapenem resistant KP and carbapenem sensitive KP. Compared with the disk diffusion method or PCR method, two of the most widely used methods in the clinical diagnosis, nanopore sensing is a low-cost, efficient, and easy-operational method (Table 1). This method could be a complement to the existing diagnosis methods, thereby contributing to the clinical laboratory diagnosis. With the development of lab-on-chip nanopore devices (Quick et al., 2016), rapid detection of multiple clinical samples using nanopore array in an integrated POCT device in hospital could be realized.
Data Availability
All datasets generated for this study are included in the manuscript and the Supplementary Files.
Ethics Statement
Blood samples from 20 patients with KP infection were provided by the Department of Laboratory Medicine, West China Hospital of Sichuan University. This study was carried out in accordance with the recommendations of China National Measures for the Ethical Review of Biomedical Research Involving Humans and WMA Declaration of Helsinki. The protocol was approved by the Biomedical Ethics Committee of West China Hospital, Sichuan University. The research used leftover specimens, that is, remnants of specimens collected for routine clinical care or analysis that would have been discarded, and complied with the criteria for a waiver of informed consent. The waiver for informed consent was granted by Biomedical Ethics Committee of West China Hospital, Sichuan University.
Author Contributions
JG and MK designed the research. HN, CZ, and PZ took part in the experiment design. WZ, QL, YL, and SW performed total RNA extraction. HN, LW, ML, and HL finished the nanopore assay experiment. JG and MK wrote the manuscript. HN, CZ, LW, and QY assisted the manuscript preparation.
Funding
This work was supported by the National Key Research and Development Program of China (Grant No. 2016YFA0201400 and 2016YFC1200300), and the National Natural Science Foundation of China (Grant No. 81541133 and 30371267).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01672/full#supplementary-material
References
Aracil-Garcia, B., Oteo-Iglesias, J., Cuevas-Lobato, O., Lara-Fuella, N., Perez-Grajera, I., Fernandez-Romero, S., et al. (2017). Rapid increase in resistance to third generation cephalosporins, imipenem and co-resistance in Klebsiella pneumoniae from isolated from 7,140 blood-cultures (2010-2014) using EARS-Net data in Spain. Enferm. Infecc. Microbiol. Clin. 35, 480–486. doi: 10.1016/j.eirnc.2016.06.007
Bi, D., Jiang, X., Sheng, Z., Ngmenterebo, D., Tai, C., Wang, M., et al. (2015). Mapping the resistance-associated mobilome of a carbapenem-resistant Klebsiella pneumoniae strain reveals insights into factors shaping these regions and facilitates generation of a ‘resistance-disarmed’ model organism. J. Antimicrob. Chemother. 70, 2770–2774. doi: 10.1093/jac/dkv204
Branton, D., Deamer, D. W., Marziali, A., Bayley, H., Benner, S. A., Butler, T., et al. (2008). The potential and challenges of nanopore sequencing. Nat. Biotechnol. 26, 1146–1153. doi: 10.1038/nbt.1495
Butler, T. Z., Pavlenok, M., Derrington, I. M., Niederweis, M., and Gundlach, J. H. (2008). Single-molecule DNA detection with an engineered MspA protein nanopore. Proc. Natl. Acad. Sci. 105, 20647–20652. doi: 10.1073/pnas.0807514106
Chew, K. L., Lin, R. T. P., and Teo, J. W. P. (2017). Klebsiella pneumoniae in Singapore: hypervirulent infections and the carbapenemase threat. Front. Cell. Infect. Microbiol. 7, 1–9. doi: 10.3389/fcimb.2017.00515
Chiu, S.-K., Chan, M.-C., Huang, L.-Y., Lin, Y.-T., Lin, J.-C., Lu, P.-L., et al. (2017). Tigecycline resistance among carbapenem-resistant Klebsiella pneumoniae: clinical characteristics and expression levels of efflux pump genes. PLoS One 12:e0175140. doi: 10.1371/journal.pone.0175140
Debby, B. D., Ganor, O., Yasmin, M., David, L., Nathan, K., Ilana, T., et al. (2012). Epidemiology of carbapenem resistant Klebsiella pneumoniae colonization in an intensive care unit. Eur. J. Clin. Microbiol. Infect. Dis. 31, 1811–1817. doi: 10.1007/s10096-011-1506-5
Deurenberg, R. H., Bathoorn, E., Chlebowicz, M. A., Couto, N., Ferdous, M., García-Cobos, S., et al. (2017). Reprint of “Application of next generation sequencing in clinical microbiology and infection prevention”. J. Biotechnol. 250, 2–10. doi: 10.1016/j.jbiotec.2017.03.035
Faller, M. (2004). The structure of a mycobacterial outer-membrane channel. Science 303, 1189–1192. doi: 10.1126/science.1094114
Fleming, S. J., Lu, B., and Golovchenko, J. A. (2017). Charge, diffusion, and current fluctuations of single-stranded DNA trapped in an MspA nanopore. Biophys. J. 112, 368–375. doi: 10.1016/j.bpj.2016.12.007
Frickmann, H., Masanta, W. O., and Zautner, A. E. (2014). Emerging rapid resistance testing methods for clinical microbiology laboratories and their potential impact on patient management. Biomed. Res. Int. 2014, 1–19. doi: 10.1155/2014/375681
Haque, F., Geng, J., Montemagno, C., and Guo, P. (2013). Incorporation of a viral DNA-packaging motor channel in lipid bilayers for real-time, single-molecule sensing of chemicals and double-stranded DNA. Nat. Protoc. 8, 373–392. doi: 10.1038/nprot.2013.001
Hindiyeh, M., Smollan, G., Grossman, Z., Ram, D., Robinov, J., Belausov, N., et al. (2011). Rapid detection of blaKPC carbapenemase genes by internally controlled real-time PCR assay using bactec blood culture bottles. J. Clin. Microbiol. 49, 2480–2484. doi: 10.1128/JCM.00149-11
Howorka, S., and Siwy, Z. (2009). Nanopore analytics: sensing of single molecules. Chem. Soc. Rev. 38, 2360–2384. doi: 10.1039/b813796j
Kasianowicz, J. J., Brandin, E., Branton, D., and Deamer, D. W. (1996). Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl. Acad. Sci. 93, 13770–13773. doi: 10.1073/pnas.93.24.13770
Khan, F. Y., Abukhattab, M., Abu Kamar, M., and Anand, D. (2014). Adult Klebsiella pneumoniae meningitis in Qatar: clinical pattern of ten cases. Asian Pac. J. Trop. Biomed. 4, 669–672. doi: 10.12980/APJTB.4.201414B100
Kim, J., and Gale, B. K. (2008). Quantitative and qualitative analysis of a microfluidic DNA extraction system using a nanoporous AlOx membrane. Lab Chip 8, 1516–1523. doi: 10.1039/b804624g
Kim, J.-S., Kang, G.-E., Kim, H.-S., Kim, H. S., Song, W., and Lee, K. M. (2016). Evaluation of Verigene Blood Culture Test Systems for Rapid Identification of Positive Blood Cultures. Biomed Res. Int. 2016:1081536. doi: 10.1155/2016/1081536
Kolbert, C. P., and Persing, D. H. (1999). Ribosomal DNA sequencing as a tool for identification of bacterial pathogens. Curr. Opin. Microbiol. 2, 299–305. doi: 10.1016/S1369-5274(99)80052-6
Laszlo, A. H., Derrington, I. M., Brinkerhoff, H., Langford, K. W., Nova, I. C., Samson, J. M., et al. (2013). Detection and mapping of 5-methylcytosine and 5-hydroxymethylcytosine with nanopore MspA. Proc. Natl. Acad. Sci. 110, 18904–18909. doi: 10.1073/pnas.1310240110
Laszlo, A. H., Derrington, I. M., and Gundlach, J. H. (2016). MspA nanopore as a single-molecule tool: from sequencing to SPRNT. Methods 105, 75–89. doi: 10.1016/j.ymeth.2016.03.026
Liu, W., Li, L., Khan, M. A., and Zhu, F. (2012). Popular molecular markers in bacteria. Mol. Genet. Microbiol. Virol. 27, 103–107. doi: 10.3103/s0891416812030056
Lomonaco, S., Crawford, M. A., Lascols, C., Timme, R. E., Anderson, K., Hodge, D. R., et al. (2018). Resistome of carbapenem- and colistin-resistant Klebsiella pneumoniae clinical isolates. PLoS One 13:e0198526. doi: 10.1371/journal.pone.0198526
Manara, R. M. A., Jayne Wallace, E., and Khalid, S. (2015). DNA sequencing with MspA: molecular dynamics simulations reveal free-energy differences between sequencing and non-sequencing mutants. Sci. Rep. 5:12783. doi: 10.1038/srep12783
Manrao, E. A., Derrington, I. M., Pavlenok, M., Niederweis, M., and Gundlach, J. H. (2011). Nucleotide discrimination with DNA immobilized in the MSPA nanopore. PLoS One 6, 1–7. doi: 10.1371/journal.pone.0025723
Montal, M., and Muellertj, P. (1972). Formation of bimolecular membranes from lipid monolayers and a study of their electrical properties (membrane structure/membrane reconstitution/asymmetric membranes/lipid bilayers). Proc. Natl. Acad. Sci. USA 69, 3561–3566. Available at: http://www.pnas.org/content/pnas/69/12/3561.full.pdf
Nepka, M., Perivolioti, E., Kraniotaki, E., Politi, L., Tsakris, A., and Pournaras, S. (2016). In vitro bactericidal activity of trimethoprim-sulfamethoxazole alone and in combination with colistin against carbapenem-resistant Acinetobacter baumannii clinical isolates. Antimicrob. Agents Chemother. 60, 6903–6906. doi: 10.1128/AAC.01082-16
Nijhuis, R., Samuelsen, Ø., Savelkoul, P., and van Zwet, A. (2013). Evaluation of a new real-time PCR assay (Check-Direct CPE) for rapid detection of KPC, OXA-48, VIM, and NDM carbapenemases using spiked rectal swabs. Diagn. Microbiol. Infect. Dis. 77, 316–320. doi: 10.1016/j.diagmicrobio.2013.09.007
Patel, J. B. (2001). 16S rRNA gene sequencing for bacterial pathogen identification in the clinical laboratory. Mol. Diagn. 6, 313–321. doi: 10.2165/00066982-200106040-00012
Pereira, F., Carneiro, J., Matthiesen, R., van Asch, B., Pinto, N., Gusmão, L., et al. (2010). Identification of species by multiplex analysis of variable-length sequences. Nucleic Acids Res. 38:e203. doi: 10.1093/nar/gkq865
Quick, J., Loman, N. J., Duraffour, S., Simpson, J. T., Severi, E., Cowley, L., et al. (2016). Real-time, portable genome sequencing for Ebola surveillance. Nature 530, 228–232. doi: 10.1038/nature16996
Rojas, L. J., Salim, M., Cober, E., Richter, S. S., Perez, F., Salata, R. A., et al. (2016). Colistin resistance in carbapenem-resistant Klebsiella pneumoniae: laboratory detection and impact on mortality. Clin. Infect. Dis. 64, 711–718. doi: 10.1093/cid/ciw805
Rood, I. G. H., and Li, Q. (2017). Review: molecular detection of extended spectrum-β-lactamase- and carbapenemase-producing Enterobacteriaceae in a clinical setting. Diagn. Microbiol. Infect. Dis. 89, 245–250. doi: 10.1016/j.diagmicrobio.2017.07.013
Saidel-Odes, L., Polachek, H., Peled, N., Riesenberg, K., Schlaeffer, F., Trabelsi, Y., et al. (2012). A randomized, double-blind, placebo-controlled trial of selective digestive decontamination using oral gentamicin and oral Polymyxin E for eradication of carbapenem-resistant Klebsiella pneumoniae carriage. Infect. Control Hosp. Epidemiol. 33, 14–19. doi: 10.1086/663206
Schechner, V., Straus-Robinson, K., Schwartz, D., Pfeffer, I., Tarabeia, J., Moskovich, R., et al. (2009). Evaluation of PCR-based testing for surveillance of KPC-producing carbapenem-resistant members of the Enterobacteriaceae family. J. Clin. Microbiol. 47, 3261–3265. doi: 10.1128/JCM.02368-08
Schuurman, T., De Boer, R. F., Kooistra-Smid, A. M. D., and Van Zwet, A. A. (2004). Prospective study of use of PCR amplification and sequencing of 16S ribosomal DNA from cerebrospinal fluid for diagnosis of bacterial meningitis in a clinical setting. J. Clin. Microbiol. 42, 734–740. doi: 10.1128/JCM.42.2.734-740.2004
Srinivasan, L., Pisapia, J. M., Shah, S. S., Halpern, C. H., and Harris, M. C. (2012). Can broad-range 16S ribosomal ribonucleic acid gene polymerase chain reactions improve the diagnosis of bacterial meningitis? A systematic review and meta-analysis. Ann. Emerg. Med. 60, 609–620.e2. doi: 10.1016/j.annemergmed.2012.05.040
Su, C.-F., Chuang, C., Lin, Y.-T., Chan, Y.-J., Lin, J.-C., Lu, P.-L., et al. (2018). Treatment outcome of non-carbapenemase-producing carbapenem-resistant Klebsiella pneumoniae infections: a multicenter study in Taiwan. Eur. J. Clin. Microbiol. Infect. Dis. 37, 651–659. doi: 10.1007/s10096-017-3156-8
Swayne, R., Ellington, M. J., Curran, M. D., Woodford, N., and Aliyu, S. H. (2013). Utility of a novel multiplex TaqMan PCR assay for metallo-β-lactamase genes plus other TaqMan assays in detecting genes encoding serine carbapenemases and clinically significant extended-spectrum β-lactamases. Int. J. Antimicrob. Agents 42, 352–356. doi: 10.1016/j.ijantimicag.2013.06.018
van Dijk, K., Voets, G. M., Scharringa, J., Voskuil, S., Fluit, A. C., Rottier, W. C., et al. (2014). A disc diffusion assay for detection of class A, B and OXA-48 carbapenemases in Enterobacteriaceae using phenyl boronic acid, dipicolinic acid and temocillin. Clin. Microbiol. Infect. 20, 345–349. doi: 10.1111/1469-0691.12322
Wang, S., Haque, F., Rychahou, P. G., Evers, B. M., and Guo, P. (2013). Engineered nanopore of Phi29 DNA-packaging motor for real-time detection of single colon cancer specific antibody in serum. ACS Nano 7, 9814–9822. doi: 10.1021/nn404435v
Wang, Y., Zheng, D., Tan, Q., Wang, M. X., and Gu, L. Q. (2011). Nanopore-based detection of circulating microRNAs in lung cancer patients. Nat. Nanotechnol. 6, 668–674. doi: 10.1038/nnano.2011.147
Wanunu, M. (2012). Nanopores: a journey towards DNA sequencing. Phys Life Rev 9, 125–158. doi: 10.1016/j.plrev.2012.05.010
Whichard, J. M., Medalla, F., Hoekstra, R. M., McDermott, P. F., Joyce, K., Chiller, T., et al. (2010). Evaluation of antimicrobial resistance phenotypes for predicting multidrug-resistant salmonella recovered from retail meats and humans in the United States. J. Food Prot. 73, 445–451. doi: 10.4315/0362-028X-73.3.445
Yang, L., and Yamamoto, T. (2016). Quantification of virus particles using nanopore-based resistive-pulse sensing techniques. Front. Microbiol. 7:1500. doi: 10.3389/fmicb.2016.01500
Keywords: nanopore assay, carbapenem-resistant Klebsiella pneumoniae, rapid clinical detection, label-free, low-cost
Citation: Niu H, Zhang W, Wei L, Liu M, Liu H, Zhao C, Zhang P, Liao Q, Liu Y, Yuan Q, Wu S, Kang M and Geng J (2019) Rapid Nanopore Assay for Carbapenem-Resistant Klebsiella pneumoniae. Front. Microbiol. 10:1672. doi: 10.3389/fmicb.2019.01672
Edited by:
Bing Gu, Affiliated Hospital of Xuzhou Medical University, ChinaReviewed by:
Hong-Yu Ou, Shanghai Jiao Tong University, ChinaCarlos Henrique Camargo, Instituto Adolfo Lutz, Brazil
Copyright © 2019 Niu, Zhang, Wei, Liu, Liu, Zhao, Zhang, Liao, Liu, Yuan, Wu, Kang and Geng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mei Kang, a2FuZ21laUBzaW5hLmNvbQ==; Jia Geng, Z2VuZy5qaWFAc2N1LmVkdS5jbg==
†These authors have contributed equally to this work
 Haofu Niu
Haofu Niu Weili Zhang
Weili Zhang Liangwan Wei
Liangwan Wei Meng Liu
Meng Liu Hao Liu
Hao Liu Changjian Zhao
Changjian Zhao Peng Zhang
Peng Zhang Quanfeng Liao
Quanfeng Liao Ya Liu
Ya Liu Qingyue Yuan
Qingyue Yuan Siying Wu
Siying Wu Mei Kang
Mei Kang Jia Geng
Jia Geng