- 1Mycology Reference Laboratory, National Centre for Microbiology, Instituto de Salud Carlos III, Madrid, Spain
- 2Unitat de Microbiologia, Facultat de Medicina i Ciències de la Salut, Universitat Rovira i Virgili, Tarragona, Spain
- 3Spanish Network for the Research in Infectious Diseases (RD16CIII/0004/0003), Instituto de Salud Carlos III, Madrid, Spain
- 4Hospital Universitario Puerta de Hierro, Madrid, Spain
- 5Hospital Universitario 12 de Octubre, Madrid, Spain
The pathogenic yeast Candida glabrata has become a public health issue due to the increasing number of echinocandin resistant clinical strains reported. In this study, acquisition and development of resistance to this antifungal class were studied in serial C. glabrata isolates from five patients admitted in two Spanish hospitals with a resistant profile against echinocandins associated with different mutations in hot-spot 1 of FKS2 gene. For two of these patients susceptible FKS wild-type isolates obtained prior to resistant ones were also investigated. Isolates were genotyped using multilocus sequence typing and microsatellite length polymorphism techniques, which yielded comparable results. Susceptible and resistant isolates from the same patient had the same genotype, being sequence type (ST) 3 the most prevalent among them. Isolates with different FKS mutations but the same ST were present in the same patient. MSH2 gene alterations were also studied to investigate their correlation with antifungal resistance acquisition but no association was found with antifungal resistance nor with specific genotypes. In vitro exposure to increasing concentrations of micafungin to susceptible isolates developed colonies carrying FKS mutations in agar plates containing a minimum concentration of 0.06 mg/L of micafungin after less than 48 h of exposure. We investigated the correlation between development of resistance and genotype in a set of susceptible strains after being in vitro exposed to micafungin and anidulafungin but no correlation was found. Mutant prevention concentration values and spontaneous growth frequencies after selection with both echinocandins were statistically similar, although FKS mutant colonies were more abundant after micafungin exposure (p < 0.001). Mutation S663P and F659 deletion were the most common ones found after selection with both echinocandins.
Introduction
Infections caused by Candida species, extensively referred to as candidiasis, have been described as the most common fungal disease globally (Pappas et al., 2018). Although Candida albicans is the species causing the highest number of infections in clinical settings, an increasing prevalence of other Candida species has been reported in the last years, being Candida glabrata the second most common species isolated from invasive candidiasis in North America and central and northern countries in Europe (Pfaller et al., 2012; Asmundsdottir et al., 2013; Lortholary et al., 2014; Milazzo et al., 2014; Castanheira et al., 2016). In Spain, only C. albicans and Candida parapsilosis are more frequently isolated than C. glabrata from patients with fungemia (Guinea et al., 2014).
Echinocandins are the first line antifungal therapy against C. glabrata infections, as this species generally presents low susceptibility to azole drugs. Echinocandins non-competitively inhibit the 1–3-β-D-glucan synthase, which is responsible for the synthesis of β-glucan polymers that confer integrity to the fungal cell wall. Nevertheless, an ever-growing number of echinocandin resistant clinical isolates have been reported worldwide in the last years and population studies in the United States and Denmark have shown an increase in echinocandin resistance rate (Alexander et al., 2013; Farmakiotis et al., 2014; Vallabhaneni et al., 2015; Astvad et al., 2017), which is conferred by the presence of point mutations in specific regions (denominated as hot-spots) of FKS genes, which encode this enzyme’s catalytic subunits (Katiyar et al., 2012).
FKS mutations have been reported to correlate with elevated in vitro minimal inhibitory concentrations (MICs) and clinical failure (Alexander et al., 2013), yet an explanation for this increase in echinocandin resistant strains has not been proved. Several possibilities are being studied, such as strains proneness to acquire resistance as an answer to echinocandin exposure (Bordallo-Cardona et al., 2017, 2018a,c; Shields et al., 2019), the existence of hidden reservoirs in the human body of echinocandin resistant C. glabrata isolates (Shields et al., 2014; Grau et al., 2015; Jensen et al., 2015; Healey et al., 2017) or molecular mechanisms like MSH2 mutator phenotype (Delliere et al., 2016; Healey et al., 2016b; Byun et al., 2018; Hou et al., 2018; Singh et al., 2018; Bordallo-Cardona et al., 2019).
Multilocus sequence typing (MLST) and microsatellite length polymorphism (MLP) have been described as typing methodologies with high discrimination power (Dodgson et al., 2003; Foulet et al., 2005; Abbes et al., 2012) for assesing C. glabrata strain relatedness.
The objective of the present study was to investigate the antifungal susceptibility, molecular mechanisms of echinocandin resistance and strain relatedness of a series of C. glabrata sequentially isolated from patients admitted in two hospitals in Madrid; and also the potential development of echinocandin resistance of susceptible C. glabrata isolates collected from 2013 to 2017 after in vitro exposure to a range of micafungin and anidulafungin concentrations.
Materials and Methods
Yeast Isolates: Patients and Identification
Eighteen C. glabrata strains sequentially isolated from five patients admitted in two centers (Hospital Universitario Puerta de Hierro and Hospital Universitario 12 de Octubre, both located in Madrid, Spain) were selected for showing a resistance profile against echinocandins. For two of these patients previous susceptible isolates were also available and analyzed. 89% (16/18) of them were obtained from blood cultures, while the two remaining were isolated from a catheter (one isolate from Patient 1) and from ascitic liquid (one isolate from Patient 2) (Table 1). For in vitro exposure to micafungin and anidulafungin assays, 14 C. glabrata strains collected from 2012 to 2017 from Hospital Universitario Puerta de Hierro, all isolated from blood cultures except one from ascitic liquid, were chosen for being susceptible to echinocandin drugs. All strains were isolated during routine diagnostic procedures at the hospitals and received at the Mycology Reference Laboratory of the Spanish National Centre for Microbiology. Isolates were characterized by morphological features and confirmed as C. glabrata by amplification and sequencing of their ITS1-5.8S-ITS2 regions (White et al., 1990). According to the Law 14/2007 of 3rd July on Biomedical Research and the Recommendation CM/Rec(2016)6 of the Committee of Ministers to member States on research on biological materials of human origin, no informed consent was required as no work was performed neither with samples of human origin nor with clinical data. The Mycology Reference Laboratory directly received fungal strains, isolated from the patients as routine diagnostic procedures in the hospital and referred to the National Centre for Microbiology according to routine procedures.
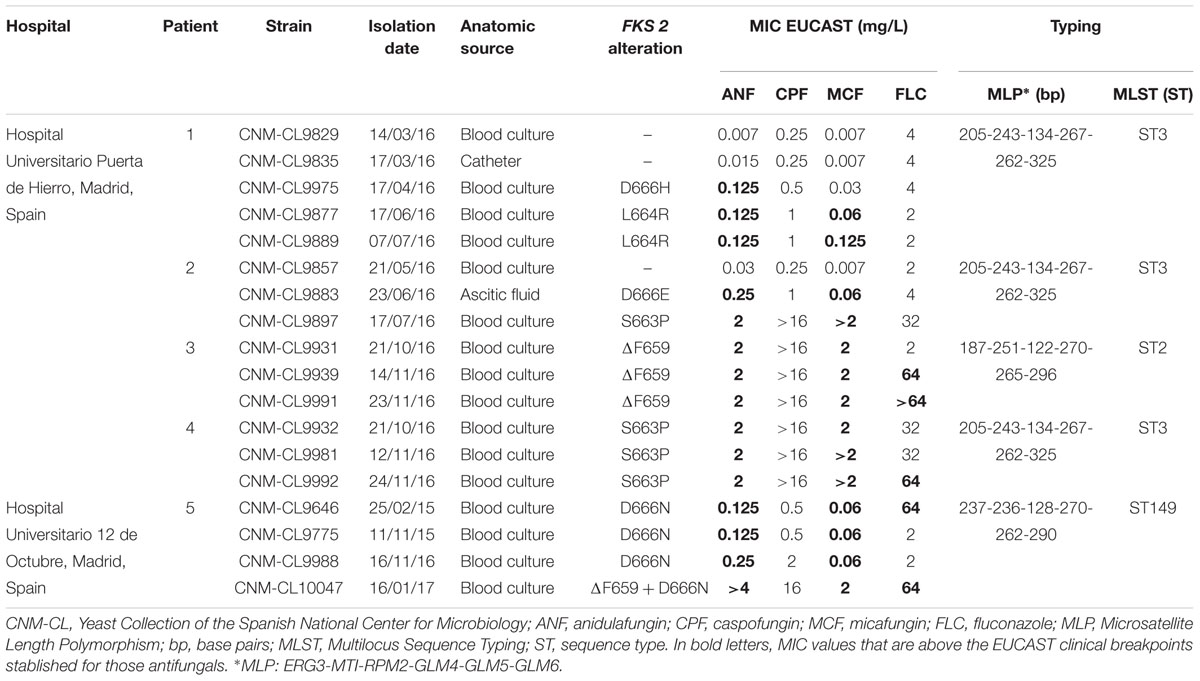
Table 1. Candida glabrata sequential isolates from five patients admitted in two hospitals in Madrid: isolation dates, anatomic sources, FKS2 alterations, in vitro susceptibility to echinocandins and fluconazole performed by EUCAST and genotyping results by MLST and MLP.
Antifungal Susceptibility Testing
Minimal inhibitory concentrations were determined and confirmed following EUCAST 7.3.1 reference method for yeasts1. Antifungals tested were anidulafungin (range 0.007–4 mg/L; Pfizer, Madrid, Spain), micafungin (range 0.004–2 mg/L; Astellas Pharma Inc., Tokyo, Japan), caspofungin (range 0.032–16 mg/L; Merck Sharp & Dohme, United Kingdom) and fluconazole (range 0.125–64 mg/L; Pfizer, Madrid, Spain).
Candida krusei ATCC 6258 and C. parapsilosis ATCC 22019 were used as quality control strains in all test performed. The optical density of the inoculated plates was determined after 24 and 48 h of incubation at 35°C in a humid atmosphere, and strains were classified as susceptible or resistant according to clinical breakpoints established by EUCAST for C. glabrata: MIC > 0.032 mg/L for micafungin, MIC > 0.064 mg/L for anidulafungin and MIC > 32 mg/L for fluconazole2.
DNA Extraction and FKS Sequencing
Genomic DNA of all isolates was extracted using the phenol-chloroform method (Tang et al., 1992). Molecular mechanisms of echinocandin resistance were studied by amplifying hot-spot regions 1 and 2 of FKS1 and FKS2 (Thompson et al., 2008; Zimbeck et al., 2010; Duran-Valle et al., 2012; Bizerra et al., 2013) as previously described with the following modifications: PCR reaction mixtures contained 25 ng of DNA, 0.2 μM of each primer, 0.2 μM of deoxynucleoside triphosphate (Roche, Spain), 5 μL of PCR 10× buffer (Applied Biosystems, Foster City, CA, United States), 2 mM of MgCl2 (Applied Biosystems, Foster City, CA, United States), 5.2% DMSO and 2.5 U of Taq DNA polymerase (Applied Biosystems, Foster City, CA, United States) in a final volume of 50 μL. PCRs conditions used were set as previously described (Duran-Valle et al., 2012), with an annealing temperature of 52°C for hot-spot regions 1 and 2 of FKS1, 53°C for hot-spot region 1 of FKS2 and 58°C for hot-spot region 2 of FKS2. PCR amplicons were purified using Illustra ExoProStar 1-step (GE Healthcare Life Science, United Kingdom), and were sequenced after by Sanger method with an ABI3730XLsequencer (Applied Biosystems, Foster City, CA, United States). DNA sequences were analyzed with DNAStar Lasergene 12 software (DNAStar Inc., United States), and queried against FKS1 (GenBank number CAGL0G01034g) and FKS2 (GenBank number CAGL0K04037g) sequences of the type strain CBS 1383.
Assessment of Pooled Reservoir of Mixed Resistant Isolates
For two patients with isolates harboring two different FKS mutations, the possible coexistence of diverse populations within the same sample was studied by randomly isolating ten colonies from the original samples sent from the hospital for DNA extraction and FKS amplification and sequencing.
Genotyping by MLST
Six housekeeping gene loci (FKS, LEU2, NMT1, TRP1, UGP1, and URA3) were studied for all isolates as previously described (Dodgson et al., 2003), with the following modifications: PCR reaction mixtures contained 25 ng of DNA, 1 μM of each primer, 0.05 μM of deoxynucleoside triphosphate, 5 μL of PCR 10× buffer, 2 mM of MgCl2, and 2.5 U of Taq DNA polymerase in a final volume of 50 μL. 5.2% of DMSO was added only to amplify NMT1. PCR conditions were set as described, but with an annealing temperature of 62°C for FKS and URA3. DNA sequences obtained were compared to C. glabrata MLST database4 to assign an allele number for each locus in order to define a sequence type (ST) or genotype according to the isolates’ allelic profile.
Genotyping by MLP
Six short tandem repeat markers described for C. glabrata (ERG3, MTI, RPM2, GLM4, GLM5, and GLM6) (Foulet et al., 2005; Abbes et al., 2012) were amplified by PCR for all isolates using forward labeled primers as previously described (Foulet et al., 2005; Abbes et al., 2012; Duran-Valle et al., 2012) with the following modifications: GLM5 was labeled with HEX and GLM6 with NED fluorochromes. PCR reaction mixtures contained 20 ng of DNA, 0.2 μM of deoxynucleoside triphosphate, 2 μL of PCR 10× buffer, 2.5 mM of MgCl2, 5 U of Taq DNA, 0.25 μM of RPM2 and ERG3 primers and 1 μM of MTI, GLM4, GLM5, and GLM6 primers in a final volume of 20 μL. PCR program used from amplifying all markers consisted on an initial step of 5 min at 95°C, followed by 40 cycles of 95°C for 30 s, 57°C for 1 min and 72°C for 1 min, and an additional step of 7 min at 72°C. Amplicons were sized by capillary electrophoresis using Hi-Di formamide (Applied Biosystems, Foster City, CA, United States) and ROX 500 (Applied Biosystems, Foster City, CA, United States) as internal size standard, as described (Duran-Valle et al., 2012). Reactions were analyzed in duplicate, and fragment sizes were calculated using Peak Scanner software 1.0 (Applied Biosystems, Foster City, CA, United States).
MSH2 Sequencing
MSH2 gene of sequentially isolated strains from patients was amplified and sequenced as previously described (Healey et al., 2016b), and DNA sequences were queried against MSH2 (Genbank number CAGL0I07733g) sequence of CBS 138 type strain.
In vitro Exposure to Growing Concentrations of Micafungin and Anidulafungin and Analysis of Generated Isolates
The potential development of micafungin and anidulafungin resistance of susceptible C. glabrata isolates was studied following a previously reported procedure (Bordallo-Cardona et al., 2017) with some modifications.
Adjusted inocula (2 × 109 to 4 × 109 CFU/mL) from overnight cultures in 7 mL of yeast extract-peptone-dextrose broth of these isolates were cultured on Sabouraud plates containing eight different echinocandin concentrations (from 0.015 to 2 mg/L). Two different sets of experiments were tested at first. Plates at all concentrations were stroked at once with 100 μL of inocula and checked for growth daily for up to 5 days at 35°C. For progressive exposure, the lowest concentration plate was inoculated and checked for growth after 24 h at 35°C. If isolates were observed, they were cultured on a plate containing the next twofold concentration. The procedure was repeated up to highest concentration available.
After exposure, mutant prevention concentration (MPC) and mutant selection window (MSW) were calculated for each isolate as previously reported (Zhao and Drlica, 2001; Drlica, 2003; Bordallo-Cardona et al., 2018c). Briefly, MPCs were defined as the lowest concentration that can totally inhibit fungal growth for each isolate after 5 days of incubation; for calculation purposes, MPC values that exceeded the highest concentration tested were transformed to the next dilution (i.e., if MPC >2 mg/L, it was changed to MPC = 4 mg/L). MSWs were defined as the range of concentrations between the MIC, obtained by EUCAST method, and the MPC for each isolate.
Spontaneous growth frequency was also calculated as the ratio of viable colonies growing on 2 mg/L echinocandin-containing plates and the initial inoculum stroked in them, as some plates containing lower concentrations did not allow the counting of individual colonies.
Micafungin and anidulafungin susceptibility of up to four isolates selected from each growing concentration was performed, and the hot-spot regions 1 and 2 of FKS1 and FKS2 genes were sequenced.
Statistical Analysis
All data obtained after in vitro exposure to echinocandins assays were compared using the Wilcoxon signed-rank test and the Fisher’s exact test (IBM SPSS Statistics for Windows, version 22.0; United States), considering as statistically significant a P-value of <0.05.
Results
In vitro Susceptibility and Determination of FKS Mutations of Sequentially Isolated Strains
Control strains were within the accepted ranges according to EUCAST QC ranges for all antifungals tested.
As shown in Table 1, a wide range of fluconazole MIC values was found among isolates tested. All strains isolated from Patients 3, 4, and 5 were echinocandin resistant, according to EUCAST breakpoints established for anidulafungin and micafungin. For Patients 1 and 2, echinocandin susceptible isolates were also available and analyzed. The first resistant isolate from Patient 1 was only resistant to anidulafungin.
All resistant isolates harbored an echinocandin resistance related mutation at hot-spot region 1 of FKS2 gene. Mutations found were S663P (n = 4), D666N (n = 4), ΔF659 (n = 4), L664R (n = 2), D666E (n = 1), and D666H (n = 1). No mutations were found at FKS1 nor at hot-spot region 2 of FKS2. Each mutation was related to a different echinocandin resistant profile, as S663P and ΔF659 showed higher MIC values than the rest of the isolates that harbored other mutations.
Isolates With Different FKS Mutations Can Be Present in the Same Patient
Resistant isolates with different FKS2 mutations were found in the same patient in two cases (Patient 1: D666H and L664R; and Patient 2: D666E and S663P). FKS2 gene sequencing of 10 randomly selected colonies from the original samples of those isolates sent from the hospital led to the same FKS mutation in all of them, so the absence of a mixed culture of resistant isolates was confirmed.
MLST and MLP Analysis
Three different STs were differentiated by MLST among the 18 isolates studied (Table 1). All isolates from the same patient had the same genotype. ST3 was found in three out of four patients of one hospital. The other two STs found were ST2 and a recently described ST149.
MLP methodology yield comparable results to those of MLST (Table 1).
MSH2 Gene Sequencing
All isolates from the same patient harbored the same SNPs in MSH2 gene. Non-synonymous loss-of-function combined mutations V239L/A942T were found in all isolates coming from one patient, while the rest of isolates did not harbor any non-synonymous mutations in this gene.
Behavior of Echinocandin Susceptible C. glabrata Isolates From Patients 1 and 2 When in vitro Exposed to Growing Concentrations of Micafungin
In vitro exposure of echinocandin susceptible isolates CNM-CL9829, CNM-CL9835, and CNM-CL9857 from Patients 1 and 2 to micafungin generated FKS mutations that conferred echinocandin resistance after less than 48 h of incubation. Progressive exposure allowed the collection of isolates up to 2 mg/L, while colonies in direct exposure grew only up to 0.5 mg/L. All colonies obtained in plates containing 0.015 and 0.03 μg/mL were susceptible to micafungin and anidulafungin and had no mutations in hot-spot regions of FKS genes. All isolates growing from 0.06 μg/mL were resistant to both echinocandins and harbored the resistant related mutation S663P in FKS2 gene (Figure 1).
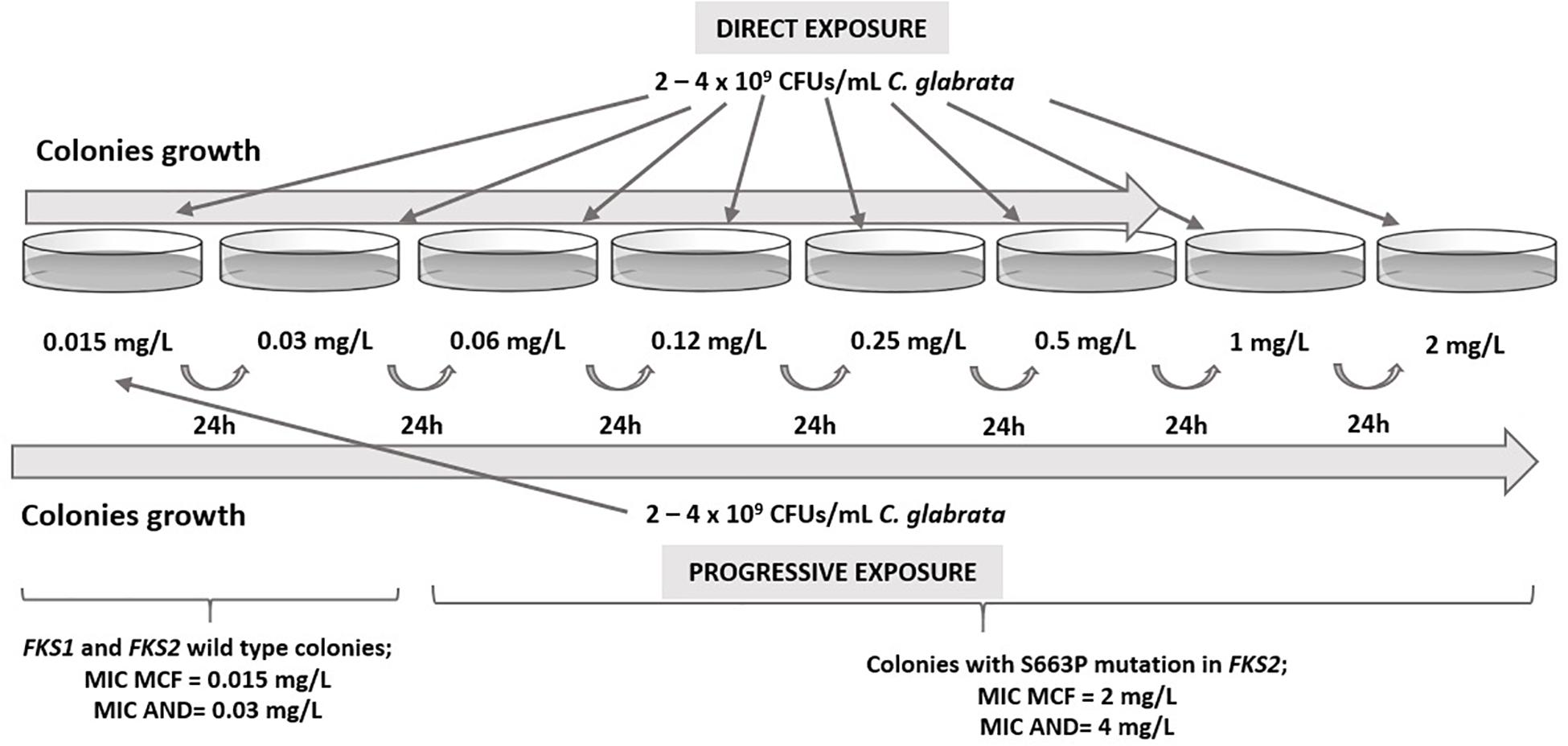
Figure 1. Results from in vitro direct and progressive exposure to micafungin of susceptible C. glabrata isolates from Patients 1 and 2.
Correlation Between Potential Development of Echinocandin Resistance After in vitro Exposure to Micafungin and Anidulafungin of Echinocandin Susceptible Isolates of C. glabrata and Their Genotype
Table 2 shows genotyping results by MLST and MLP, echinocandin susceptibility by EUCAST, MPC and MSW after in vitro direct exposure to micafungin and anidulafungin of fourteen echinocandin susceptible C. glabrata strains collected from 2012 to 2017 from Hospital Universitario Puerta de Hierro, total number of colonies analyzed per isolate (up to 4 colonies per concentration) after 5 days of incubation and FKS mutations found.
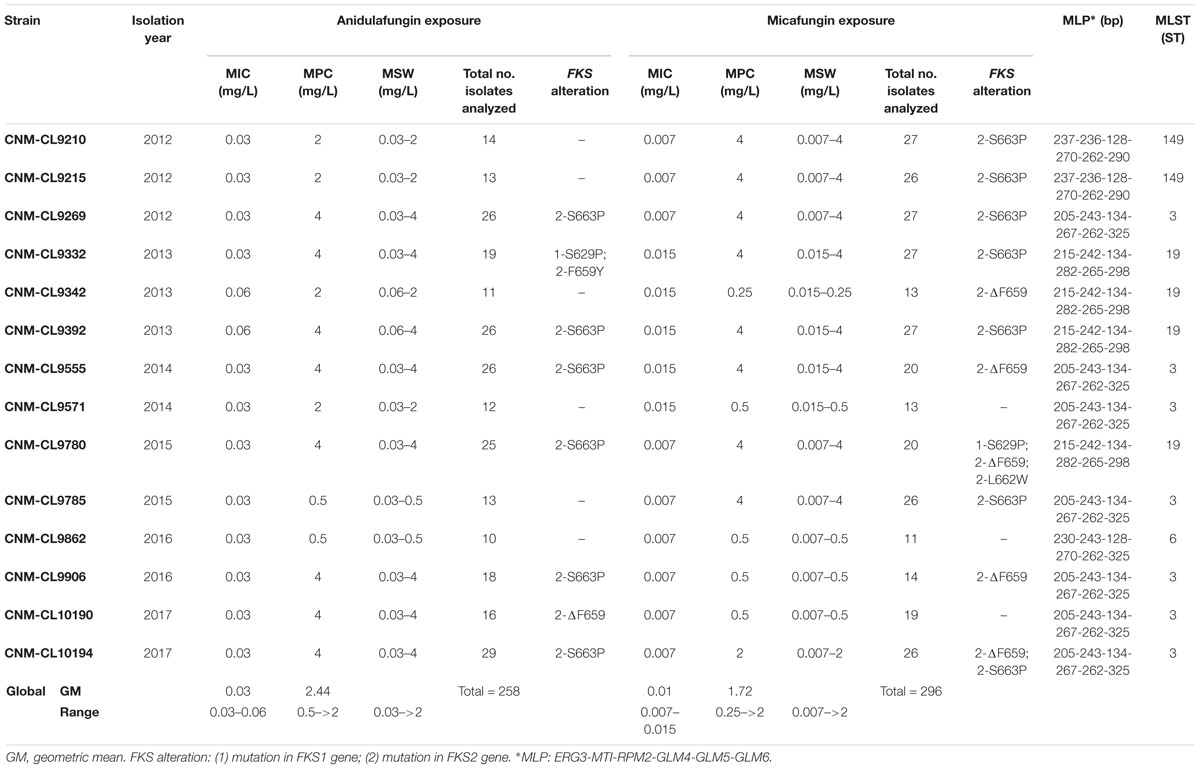
Table 2. Isolation year, micafungin and anidulafungin MIC values, mutant prevention concentration (MPC), mutant selection window (MSW), total number of colonies analyzed and FKS alterations found in them after 5 days of incubation for each isolate, and their genotype by MLP and MLST.
Four different STs were found among these strains. The most common one was ST3, found in half of the isolates (n = 7), followed by ST19 (29%, n = 4), ST149 (14%, n = 2), and ST6 (7%, n = 1).
Mutant prevention concentration values after anidulafungin and micafungin exposure differ widely between strains (Table 2), although no significant differences were found between the geometric mean of MPCs after anidulafungin exposure and after micafungin exposure after 5 days of incubation (2.44 mg/L versus 1.72 mg/L).
Geometric mean of spontaneous growth frequency for micafungin-containing plates had no significant difference with that for anidulafungin-containing plates (8 × 10-8 versus 4.1 × 10-8; p = 0.78), and ranges were very similar for both of them (4.1 × 10-7 to 3.2 × 10-9 in the presence of micafungin and 3.7 × 10-7 to 5.3 × 10-9 in the presence of anidulafungin).
A total number of 296 and 258 isolates were analyzed after micafungin and anidulafungin exposure, respectively (Figure 2). The lowest concentrations of these antifungals in which resistant colonies harboring FKS mutations were found were 0.06 mg/L and 0.12 mg/L, respectively, while they appeared up to the highest concentration tested in both cases, 2 mg/L. 58% of the isolates yielded in micafungin plates harbored FKS mutations related to echinocandin resistance, which was a significantly higher number than the 42% of the isolates that did so after anidulafungin exposure (p < 0.0001). The most prevalent mutation found was S663P (no significant differences were found among the frequency of occurrence of this substitution after exposure to both echinocandins: n = 159 from MCF plates and n = 102 from AND plates; 92.4 and 93.5% of FKS mutant isolates generated, p = 0.8147), followed by ΔF659 (its appearance rate was close to be significantly different after exposure to both antifungals: n = 10 from MCF plates and n = 1 from AND plates, p = 0.0552), F659Y (n = 1 from AND plates) and L662W (n = 1 from MCF plates) in hot-spot 1 from FKS2 gene and S629P (n = 5 from AND plates and n = 2 from MCF plates) in hot-spot 1 from FKS1 gene. All of these isolates were echinocandin resistant by EUCAST, and their FKS mutations and resistance were stable and reproducible after subculturing on antifungal-free plates. The same isolate could develop different FKS mutations after exposure. Out of the total number of isolates analyzed after micafungin and anidulafungin exposure, 1 and 19%, respectively, were resistant to micafungin and/or anidulafungin but did not carry any FKS mutation.
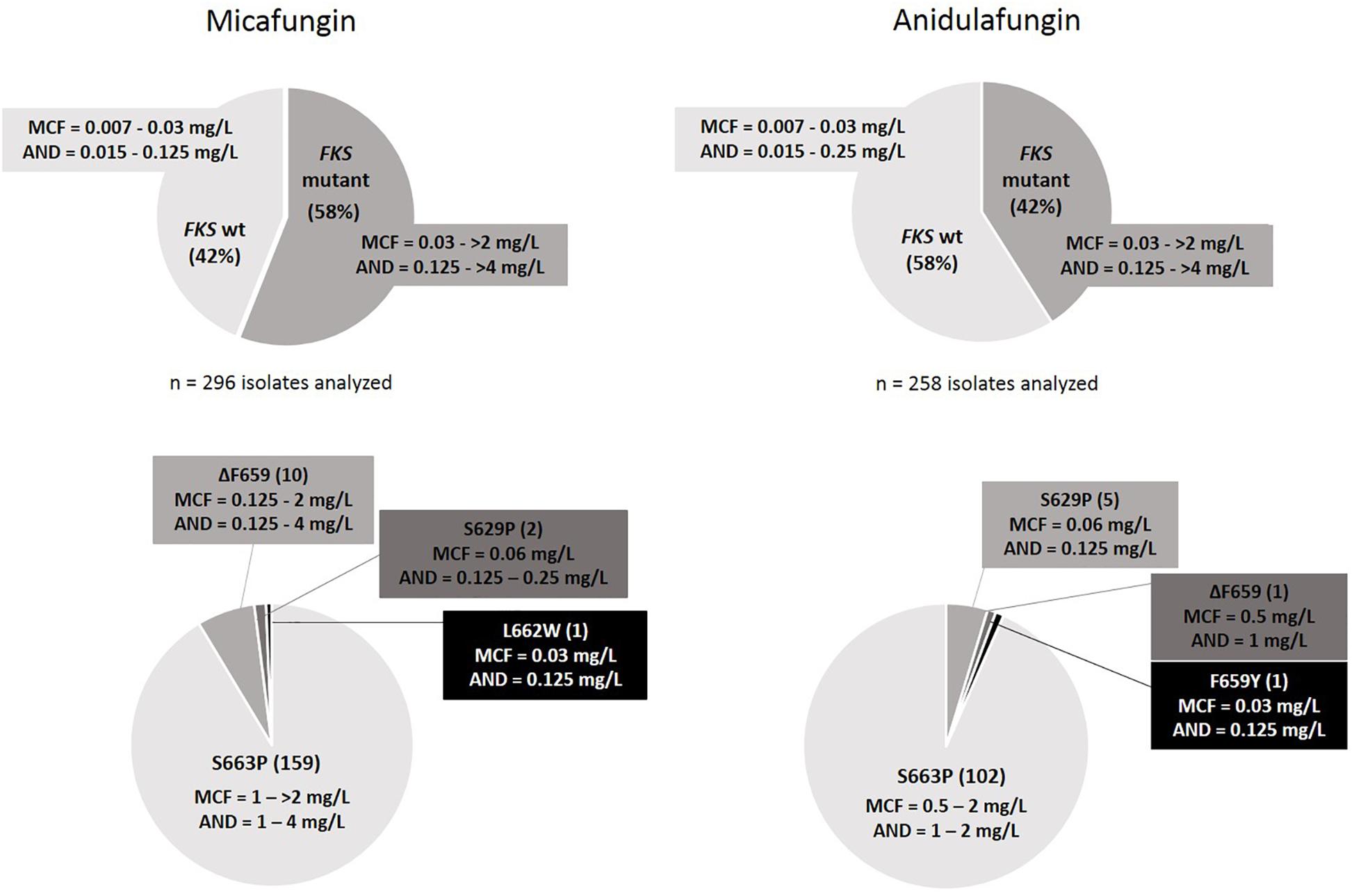
Figure 2. FKS mutant and wild-type colony rates with their MIC ranges (Top), and FKS mutations found with range of MIC values per mutation (Bottom) after in vitro exposure to micafungin and anidulafungin.
Discussion
The increasing number of C. glabrata clinical isolates reported showing decreased susceptibility for echinocandins is a growing concern. Recent studies indicate that echinocandin resistance rates among C. glabrata clinical isolates have risen worldwide (Kiraz et al., 2010; Bourgeois et al., 2014; Guinea et al., 2014; Orasch et al., 2014; Klotz et al., 2016; Chapman et al., 2017; Hou et al., 2017). Resistance has been reported to easily develop in vitro (Bordallo-Cardona et al., 2017, 2018a,c; Shields et al., 2019) and in patients after echinocandin exposure (Dannaoui et al., 2012; Shields et al., 2012; Alexander et al., 2013; Bizerra et al., 2014; Sasso et al., 2017), being conferred by the presence of point mutations in hot-spot regions of FKS1 and FKS2 genes (Castanheira et al., 2014; Pham et al., 2014) that have been associated with higher MICs and therapeutic failure (Shields et al., 2012). Our study provides a new insight into the development of echinocandin resistance of C. glabrata strains both sequentially isolated from several patients and after in vitro exposure to growing concentrations of micafungin and anidulafungin.
FKS Mutations
All isolates from five patients admitted in two Spanish hospitals that were resistant to at least one echinocandin carried mutations in FKS2 gene (Zimbeck et al., 2010; Castanheira et al., 2014; Locke et al., 2016). The most common ones found were S663P and ΔF659, as in previous studies (Zimbeck et al., 2010; Beyda et al., 2014; Castanheira et al., 2014). Some isolates carried less frequently found substitutions L664R, D666E, and D666N (Castanheira et al., 2014; Prigent et al., 2017; Bordallo-Cardona et al., 2018b) and, although it has been found after in vitro exposure to the novel antifungal rezafungin (Locke et al., 2016), we believe that this is the first time that D666H mutation is reported in a clinical isolate.
From two of the studied patients, echinocandin susceptible isolates without mutations in FKS genes were detected before the isolation of the resistant ones, indicating that echinocandin resistance could had been acquired due to therapy, as previously reported in other studies (Cleary et al., 2008; Thompson et al., 2008; Ishikawa et al., 2010; Costa-de-Oliveira et al., 2011; Dannaoui et al., 2012; Shields et al., 2012; Alexander et al., 2013; Lewis et al., 2013; Beyda et al., 2014). All isolates with FKS mutations had MIC values above the clinical breakpoints established by EUCAST both for anidulafungin and for micafungin, except one isolate that was resistant to anidulafungin but not to micafungin despite harboring an FKS mutation (D666H). This case has been described before (Arendrup et al., 2012; Shields et al., 2012; Jensen et al., 2015; Bordallo-Cardona et al., 2017) but should be taken into account when testing only one echinocandin to detect resistance.
Glucan synthase enzyme sensitivity has been described to be affected by FKS mutations on different degrees (Garcia-Effron et al., 2009). In this study, S663P and ΔF659 were associated with higher echinocandin MIC values, as previously reported (Arendrup and Perlin, 2014). Isolates harboring D666H, D666N, D666E, and L664R substitutions showed lower MIC values and conferred weaker echinocandin resistance. Patient 5 simultaneously carried ΔF659 and D666N substitutions in CNM-CL10047 isolate, which had high echinocandin MIC values. A double mutation in these two amino acid positions has been previously found (F659S and D666E) (Prigent et al., 2017), but we are reporting the association of ΔF659 and D666N for the first time.
As we found sequential isolates from the same patient carrying different FKS mutations, we studied the possible coexistence of a mixed population of resistant isolates within the same sample, confirming its absence. Nevertheless, this experiment had some limitations, as the original samples sent from the hospital could be an already isolated colony. Serial C. glabrata isolates from the same patient showing different antifungal resistance profiles due to the selective pressure induced by changes in antifungal treatment have been previously reported (Cho et al., 2015; Imbert et al., 2016).
Genotyping
Strain relatedness was determined by MLST and MLP, which led to similar results evidencing that both methodologies are equally useful for genotyping purposes, as previously reported (Brisse et al., 2009). Nevertheless, in other investigations a higher number of genotypes were obtained by MLP than by MLST (Hou et al., 2018; Bordallo-Cardona et al., 2019). Isolates from the same patient seemed to have a clonal origin by using these two typing techniques, although the use of next generation sequencing in order to compare their genomes would be necessary to prove if they are isogenic. The most frequent genotype among these patients was ST3, which has been reported as one of the most prevalent STs worldwide (Dodgson et al., 2003; Lott et al., 2012; Hou et al., 2017; Biswas et al., 2018; Byun et al., 2018; Mushi et al., 2018). No association between echinocandin resistance development and genetic type was found, which was in agreement with other reports (Dodgson et al., 2003; Abbes et al., 2012). A recent study has found a link between certain STs and reduced susceptibility to fluconazole (Mushi et al., 2018), something that we did not see when performing fluconazole susceptibility to all the isolates. Genotypes were also independent of the anatomic source of the isolates (Lin et al., 2007).
MSH2
MSH2 mismatch repair gene involved in DNA repair has been described as a promoter of the acquisition of resistance to antifungals of C. glabrata (Healey et al., 2016b), but in this study all isolates belonging to the same patient had the same MSH2 gene sequence, regardless their susceptibility pattern to echinocandins. Echinocandin susceptible isolates with and without MSH2 mutations yielded echinocandin resistant isolates with FKS mutations. Also, FKS mutant isolates for three patients had a wild-type MSH2 gene. Altogether, this supports that echinocandin resistance cannot be explained by MSH2 mutator phenotype, as previously reported (Delliere et al., 2016; Healey et al., 2016a; Biswas et al., 2018; Byun et al., 2018; Hou et al., 2018; Singh et al., 2018; Bordallo-Cardona et al., 2019). Likewise, no clear association between MSH2 sequence and increased fluconazole resistance or genotypes was detected on these isolates either (Biswas et al., 2018; Bordallo-Cardona et al., 2019), although a correlation with specific genetic types was previously described (Delliere et al., 2016; Byun et al., 2018; Hou et al., 2018). These results confirm that MSH2 substitutions may be constitutive variations from the gene rather than resistance-related or genotype-related mutations (Carrete et al., 2019). Still, it cannot be dismissed that MSH2 may just be one of a higher number of C. glabrata genes involved in mismatch repair mechanisms influencing on the development of antifungal resistance, as it happens for other yeasts (Legrand et al., 2007; Boyce et al., 2017).
In vitro Resistance Development
It is of interest to gain a deeper understanding of how C. glabrata isolates behave when in vitro exposed to echinocandins, and correlate these results with clinical findings or to anticipate to possible clinical cases. Susceptible isolates from Patients 1 and 2 were in vitro exposed to a range of growing concentrations of micafungin, obtaining echinocandin resistant and FKS mutant colonies after exposure to the lowest micafungin concentration considered resistant by EUCAST (0.06 mg/L) in less than 48 h of incubation. These results are in agreement with those previously reported (Bordallo-Cardona et al., 2017, 2018a). In our study, FKS mutations found after in vitro micafungin exposure were different from those isolated from the patients, something that evinces C. glabrata’s facility to develop resistance and to acquire different mutations under drug pressure.
It has been hypothesized that certain STs may have a better competence than others to acquire resistance through antifungal exposure at different frequencies (Lott et al., 2012; Hou et al., 2017). Therefore, we aimed to compare the potential development of in vitro echinocandin resistance of a set of susceptible C. glabrata isolates collected in 6 years from one hospital after exposure to a range of micafungin and anidulafungin concentrations, and to correlate this with their genotype. MLST and MLP revealed four different STs among these fourteen isolates. Not all strains isolated on the same year showed the same ST in all cases and no clear trend on the evolution of C. glabrata population in this center was found. No statistical differences were found among MPC values and spontaneous growth frequencies for both agents, something in concordance with previous results published (Bordallo-Cardona et al., 2018c). Results in our study varied between strains, being echinocandin resistant colonies harboring FKS mutations isolated from plates containing the first micafungin and anidulafungin concentrations considered as resistant by EUCAST. Nevertheless, a significantly higher FKS mutant rate was found after micafungin exposure than after anidulafungin selection.
In vitro micafungin and anidulafungin exposure allowed the selection of different FKS mutations grown at different concentrations and even at the same one for some strains. S663P was the most frequently found mutation following exposure to both echinocandins, although in previous in vitro selection studies ΔF659 was the most prevalent one (Bordallo-Cardona et al., 2018c; Shields et al., 2019), which was the second most common one in our study.
We found 53 colonies with MICs onefold or twofold above the established breakpoint for anidulafungin that did not carry any FKS mutations. This finding was especially detected after exposure to anidulafungin, as 50 colonies out of 258 isolated were anidulafungin resistant but FKS wild-type, while only three out of 296 colonies analyzed after micafungin exposure had this categorization. This similar case and also echinocandin susceptible isolates harboring FKS substitutions have been previously found, both in clinical isolates and after in vitro exposure (Castanheira et al., 2014; Pham et al., 2014; Shields et al., 2015, 2019; Locke et al., 2016). Precisely, anidulafungin resistance, according to EUCAST breakpoints, was vastly sensitive (100%; all colonies that were FKS mutant showed a MIC value above its clinical breakpoint for EUCAST) but showed lower specificity (80.6%; as 53 colonies out of a total of 273 that were FKS wild-type showed MICs above the EUCAST breakpoint) than in another study (Shields et al., 2019) for the identification of in vitro selected FKS mutant colonies. Micafungin showed higher sensitivity (99.3%; 279 out of 281 isolates that harbored FKS mutations were micafungin resistant according to EUCAST breakpoint) and similar specificity (99.8%; only one FKS wild-type colony was micafungin resistant) to those reported in that study. Taken together, this confirmed that both antifungals are suitable echinocandin resistance markers for C. glabrata, unlike caspofungin (Shields et al., 2013; Eschenauer et al., 2014).
We concluded that the development of echinocandin resistance in C. glabrata after in vitro exposure to micafungin and anidulafungin has no association with specific genotypes. Results obtained in all these in vitro studies on how echinocandin susceptible C. glabrata strains are able to develop resistance after exposure to low echinocandin concentrations supports the fact that C. glabrata is able to colonize and survive in certain reservoirs of the human body, such as the abdomen (Shields et al., 2014), the peritoneum (Grau et al., 2015), the gastrointestinal tract (Healey et al., 2017) or the mucosal surfaces (Jensen et al., 2015), due to long-term penetration of echinocandins in lower concentrations than those that prevent resistance acquisition. Since sometimes this required dose could lead to toxicity, the use of newly developed drugs that target the 1–3-β-D-glucan synthase, such as ibrexafungerp (Scynexis, Jersey City, NJ, United States), which has shown potential effectiveness against echinocandin resistant C. glabrata isolates (Wiederhold et al., 2018), or rezafungin (Cidara, San Diego, CA, United States), which has an extended-interval administration due to its improved pharmacodynamics (Sandison et al., 2017) could help to overcome echinocandin resistance. Nevertheless, C. glabrata strains have been in vitro exposed to both drugs, leading to similar results than those against other echinocandins (Locke et al., 2016; Jimenez-Ortigosa et al., 2017), so further research for new compounds that have a role on novel mechanisms of action is assured.
In summary, the present study analyzes the relevance of certain hypothesis raised on the increase of echinocandin resistance in C. glabrata, and sheds light on several important aspects related to its acquisition and development, both in genetically related serial isolates from the same patient and after in vitro exposure to micafungin and anidulafungin.
Data Availability
All datasets generated for this study are included in the manuscript and/or the supplementary files.
Author Contributions
AA-I and OR-M conceived and designed the experiments. OR-M, PN-R, LB-M, GM-C, and LL-P performed the experimental work. OR-M and AA-I analyzed the data. IS-R and AP-A provided clinical isolates. OZ provided materials. OR-M and AA-I wrote the manuscript. All authors read, edited, and approved the final version of the manuscript.
Funding
This work was supported by the Fondo de Investigación Sanitaria (Grant FI14CIII/00025 to OR-M and research projects PI13/02145 and PI16CIII/00035 to AA-I), and also supported by the Plan Nacional de I+D+i 2013–2016 and Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Ciencia, Innovación y Universidades, Spanish Network for Research in Infectious Diseases (REIPI RD16CIII/0004/0003) – co-financed by the European Development Regional Fund “A way to achieve Europe,” Operative Program Intelligent Growth 2014–2020.
Conflict of Interest Statement
In the past five years, AA-I has received grants from GILEAD, F2G, and Scynexis and personal fees for educational conferences from Pfizer Astellas, MSD, and GILEAD outside the submitted work.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
- ^ http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/AFST/Files/EUCAST_E_Def_7_3_1_Yeast_testing__definitive.pdf (accessed April 8, 2019).
- ^ http://www.eucast.org/astoffungi/clinicalbreakpointsforantifungals/
- ^ http://www.ncbi.nlm.nih.gov/
- ^ https://pubmlst.org/cglabrata/
References
Abbes, S., Sellami, H., Sellami, A., Hadrich, I., Amouri, I., Mahfoudh, N., et al. (2012). Candida glabrata strain relatedness by new microsatellite markers. Eur. J. Clin. Microbiol. Infect. Dis. 31, 83–91. doi: 10.1007/s10096-011-1280-4
Alexander, B. D., Johnson, M. D., Pfeiffer, C. D., Jimenez-Ortigosa, C., Catania, J., Booker, R., et al. (2013). Increasing echinocandin resistance in Candida glabrata: clinical failure correlates with presence of FKS mutations and elevated minimum inhibitory concentrations. Clin. Infect. Dis. 56, 1724–1732. doi: 10.1093/cid/cit136
Arendrup, M. C., and Perlin, D. S. (2014). Echinocandin resistance: an emerging clinical problem? Curr. Opin. Infect. Dis. 27, 484–492. doi: 10.1097/QCO.0000000000000111
Arendrup, M. C., Perlin, D. S., Jensen, R. H., Howard, S. J., Goodwin, J., and Hope, W. (2012). Differential in vivo activities of anidulafungin, caspofungin, and micafungin against Candida glabrata isolates with and without FKS resistance mutations. Antimicrob. Agents Chemother. 56, 2435–2442. doi: 10.1128/AAC.06369-11
Asmundsdottir, L. R., Erlendsdottir, H., and Gottfredsson, M. (2013). Nationwide study of candidemia, antifungal use, and antifungal drug resistance in Iceland, 2000 to 2011. J. Clin. Microbiol. 51, 841–848. doi: 10.1128/JCM.02566-12
Astvad, K. M. T., Johansen, H. K., Roder, B. L., Rosenvinge, F. S., Knudsen, J. D., Lemming, L., et al. (2017). Update from 12-year nationwide fungaemia surveillance: increasing intrinsic and acquired resistance causes concern. J. Clin. Microbiol. 56:e1564-17. doi: 10.1128/JCM.01564-17
Beyda, N. D., John, J., Kilic, A., Alam, M. J., Lasco, T. M., and Garey, K. W. (2014). FKS mutant Candida glabrata: risk factors and outcomes in patients with candidemia. Clin. Infect. Dis. 59, 819–825. doi: 10.1093/cid/ciu407
Biswas, C., Marcelino, V. R., Van Hal, S., Halliday, C., Martinez, E., Wang, Q., et al. (2018). Whole genome sequencing of Australian Candida glabrata isolates reveals genetic diversity and novel sequence types. Front. Microbiol. 9:2946. doi: 10.3389/fmicb.2018.02946
Bizerra, F. C., Jimenez-Ortigosa, C., Queiroz-Telles, F., Perlin, D. S., and Lopes Colombo, A. (2013). “A Point Mutation in the FKS2 gene of Candida glabrata confers reduced sensitivity to echinocandin drugs,” in Poster P991 at the European Congress of Clinical Microbiology and Infectious Diseases (ECCMID), Berlin.
Bizerra, F. C., Jimenez-Ortigosa, C., Souza, A. C., Breda, G. L., Queiroz-Telles, F., Perlin, D. S., et al. (2014). Breakthrough candidemia due to multidrug-resistant Candida glabrata during prophylaxis with a low dose of micafungin. Antimicrob. Agents Chemother. 58, 2438–2440. doi: 10.1128/AAC.02189-13
Bordallo-Cardona, M. A., Agnelli, C., Gomez-Nunez, A., Sanchez-Carrillo, C., Bouza, E., Munoz, P., et al. (2019). MSH2 gene point mutations are not antifungal resistance markers in Candida glabrata. Antimicrob. Agents Chemother. 63:e1876-18. doi: 10.1128/AAC.01876-18
Bordallo-Cardona, M. A., Escribano, P., de la Pedrosa, E. G., Marcos-Zambrano, L. J., Canton, R., Bouza, E., et al. (2017). In vitro exposure to increasing micafungin concentrations easily promotes echinocandin resistance in Candida glabrata isolates. Antimicrob. Agents Chemother. 61:e1542-16. doi: 10.1128/AAC.01542-16
Bordallo-Cardona, M. A., Escribano, P., Marcos-Zambrano, L. J., Diaz-Garcia, J., de la Pedrosa, E. G., Canton, R., et al. (2018a). Low and constant micafungin concentrations may be sufficient to lead to resistance mutations in FKS2 gene of Candida glabrata. Med. Mycol. 56, 903–906. doi: 10.1093/mmy/myx124
Bordallo-Cardona, M. A., Marcos-Zambrano, L. J., Sanchez-Carrillo, C., Bouza, E., Munoz, P., Escribano, P., et al. (2018b). Resistance to echinocandins in candida can be detected by performing the etest directly on blood culture samples. Antimicrob. Agents Chemother. 62:e162-18. doi: 10.1128/AAC.00162-18
Bordallo-Cardona, M. A., Marcos-Zambrano, L. J., Sanchez-Carrillo, C., de la Pedrosa, E. G. G., Canton, R., Bouza, E., et al. (2018c). Mutant prevention concentration and mutant selection window of micafungin and anidulafungin in clinical Candida glabrata isolates. Antimicrob. Agents Chemother. 62:e1982-17. doi: 10.1128/AAC.01982-17
Bourgeois, N., Laurens, C., Bertout, S., Balard, Y., Krasteva, D., Rispail, P., et al. (2014). Assessment of caspofungin susceptibility of Candida glabrata by the Etest(R), CLSI, and EUCAST methods, and detection of FKS1 and FKS2 mutations. Eur. J. Clin. Microbiol. Infect. Dis. 33, 1247–1252. doi: 10.1007/s10096-014-2069-z
Boyce, K. J., Wang, Y., Verma, S., Shakya, V. P. S., Xue, C., and Idnurm, A. (2017). Mismatch repair of DNA replication errors contributes to microevolution in the pathogenic fungus Cryptococcus neoformans. mBio 8:e595-17. doi: 10.1128/mBio.00595-17
Brisse, S., Pannier, C., Angoulvant, A., de Meeus, T., Diancourt, L., Faure, O., et al. (2009). Uneven distribution of mating types among genotypes of Candida glabrata isolates from clinical samples. Eukaryot. Cell 8, 287–295. doi: 10.1128/EC.00215-08
Byun, S. A., Won, E. J., Kim, M. N., Lee, W. G., Lee, K., Lee, H. S., et al. (2018). Multilocus Sequence Typing (MLST) genotypes of Candida glabrata bloodstream isolates in Korea: association with antifungal resistance, mutations in mismatch repair gene (MSH2), and clinical outcomes. Front. Microbiol. 9:1523. doi: 10.3389/fmicb.2018.01523
Carrete, L., Ksiezopolska, E., Gomez-Molero, E., Angoulvant, A., Bader, O., Fairhead, C., et al. (2019). Genome comparisons of Candida glabrata serial clinical isolates reveal patterns of genetic variation in infecting clonal populations. Front. Microbiol. 10:112. doi: 10.3389/fmicb.2019.00112
Castanheira, M., Messer, S. A., Rhomberg, P. R., and Pfaller, M. A. (2016). Antifungal susceptibility patterns of a global collection of fungal isolates: results of the SENTRY antifungal surveillance program (2013). Diagn. Microbiol. Infect. Dis. 85, 200–204. doi: 10.1016/j.diagmicrobio.2016.02.009
Castanheira, M., Woosley, L. N., Messer, S. A., Diekema, D. J., Jones, R. N., and Pfaller, M. A. (2014). Frequency of FKS mutations among Candida glabrata isolates from a 10-year global collection of bloodstream infection isolates. Antimicrob. Agents Chemother. 58, 577–580. doi: 10.1128/AAC.01674-13
Chapman, B., Slavin, M., Marriott, D., Halliday, C., Kidd, S., Arthur, I., et al. (2017). Changing epidemiology of candidaemia in Australia. J. Antimicrob. Chemother. 72, 1103–1108. doi: 10.1093/jac/dkw422
Cho, E. J., Shin, J. H., Kim, S. H., Kim, H. K., Park, J. S., Sung, H., et al. (2015). Emergence of multiple resistance profiles involving azoles, echinocandins and amphotericin B in Candida glabrata isolates from a neutropenia patient with prolonged fungaemia. J. Antimicrob. Chemother. 70, 1268–1270. doi: 10.1093/jac/dku518
Cleary, J. D., Garcia-Effron, G., Chapman, S. W., and Perlin, D. S. (2008). Reduced Candida glabrata susceptibility secondary to an FKS1 mutation developed during candidemia treatment. Antimicrob. Agents Chemother. 52, 2263–2265. doi: 10.1128/AAC.01568-07
Costa-de-Oliveira, S., Marcos Miranda, I., Silva, R. M., Pinto, E. S. A., Rocha, R., Amorim, A., et al. (2011). FKS2 mutations associated with decreased echinocandin susceptibility of Candida glabrata following anidulafungin therapy. Antimicrob. Agents Chemother. 55, 1312–1314. doi: 10.1128/AAC.00589-10
Dannaoui, E., Desnos-Ollivier, M., Garcia-Hermoso, D., Grenouillet, F., Cassaing, S., Baixench, M. T., et al. (2012). Candida spp. with acquired echinocandin resistance, France, 2004-2010. Emerg. Infect. Dis. 18, 86–90. doi: 10.3201/eid1801.110556
Delliere, S., Healey, K., Gits-Muselli, M., Carrara, B., Barbaro, A., Guigue, N., et al. (2016). Fluconazole and echinocandin resistance of Candida glabrata correlates better with antifungal drug exposure rather than with MSH2 mutator genotype in a French cohort of patients harboring low rates of resistance. Front. Microbiol. 7:2038. doi: 10.3389/fmicb.2016.02038
Dodgson, A. R., Pujol, C., Denning, D. W., Soll, D. R., and Fox, A. J. (2003). Multilocus sequence typing of Candida glabrata reveals geographically enriched clades. J. Clin. Microbiol. 41, 5709–5717. doi: 10.1128/jcm.41.12.5709-5717.2003
Drlica, K. (2003). The mutant selection window and antimicrobial resistance. J. Antimicrob. Chemother. 52, 11–17. doi: 10.1093/jac/dkg269
Duran-Valle, M. T., Gago, S., Gomez-Lopez, A., Cuenca-Estrella, M., Jimenez Diez-Canseco, L., Gomez-Garces, J. L., et al. (2012). Recurrent episodes of candidemia due to Candida glabrata with a mutation in hot spot 1 of the FKS2 gene developed after prolonged therapy with caspofungin. Antimicrob. Agents Chemother. 56, 3417–3419. doi: 10.1128/AAC.06100-11
Eschenauer, G. A., Nguyen, M. H., Shoham, S., Vazquez, J. A., Morris, A. J., Pasculle, W. A., et al. (2014). Real-world experience with echinocandin MICs against candida species in a multicenter study of hospitals that routinely perform susceptibility testing of bloodstream isolates. Antimicrob. Agents Chemother. 58, 1897–1906. doi: 10.1128/AAC.02163-13
Farmakiotis, D., Tarrand, J. J., and Kontoyiannis, D. P. (2014). Drug-resistant Candida glabrata infection in cancer patients. Emerg. Infect. Dis. 20, 1833–1840. doi: 10.3201/eid2011.140685
Foulet, F., Nicolas, N., Eloy, O., Botterel, F., Gantier, J. C., Costa, J. M., et al. (2005). Microsatellite marker analysis as a typing system for Candida glabrata. J. Clin. Microbiol. 43, 4574–4579. doi: 10.1128/JCM.43.9.4574-4579.2005
Garcia-Effron, G., Lee, S., Park, S., Cleary, J. D., and Perlin, D. S. (2009). Effect of Candida glabrata FKS1 and FKS2 mutations on echinocandin sensitivity and kinetics of 1,3-beta-D-glucan synthase: implication for the existing susceptibility breakpoint. Antimicrob. Agents Chemother. 53, 3690–3699. doi: 10.1128/AAC.00443-09
Grau, S., Luque, S., Campillo, N., Samso, E., Rodriguez, U., Garcia-Bernedo, C. A., et al. (2015). Plasma and peritoneal fluid population pharmacokinetics of micafungin in post-surgical patients with severe peritonitis. J. Antimicrob. Chemother. 70, 2854–2861. doi: 10.1093/jac/dkv173
Guinea, J., Zaragoza, O., Escribano, P., Martin-Mazuelos, E., Peman, J., Sanchez-Reus, F., et al. (2014). Molecular identification and antifungal susceptibility of yeast isolates causing fungemia collected in a population-based study in Spain in 2010 and 2011. Antimicrob. Agents Chemother. 58, 1529–1537. doi: 10.1128/AAC.02155-13
Healey, K. R., Jimenez Ortigosa, C., Shor, E., and Perlin, D. S. (2016a). Genetic drivers of multidrug resistance in Candida glabrata. Front. Microbiol. 7:1995. doi: 10.3389/fmicb.2016.01995
Healey, K. R., Zhao, Y., Perez, W. B., Lockhart, S. R., Sobel, J. D., Farmakiotis, D., et al. (2016b). Prevalent mutator genotype identified in fungal pathogen Candida glabrata promotes multi-drug resistance. Nat. Commun. 7:11128. doi: 10.1038/ncomms11128
Healey, K. R., Nagasaki, Y., Zimmerman, M., Kordalewska, M., Park, S., Zhao, Y., et al. (2017). The gastrointestinal tract is a major source of echinocandin drug resistance in a murine model of Candida glabrata colonization and systemic dissemination. Antimicrob. Agents Chemother. 61, e1412–e1417. doi: 10.1128/AAC.01412-17
Hou, X., Xiao, M., Chen, S. C., Kong, F., Wang, H., Chu, Y. Z., et al. (2017). Molecular epidemiology and antifungal susceptibility of Candida glabrata in China (August 2009 to July 2014): a multi-center study. Front. Microbiol. 8:880. doi: 10.3389/fmicb.2017.00880
Hou, X., Xiao, M., Wang, H., Yu, S. Y., Zhang, G., Zhao, Y., et al. (2018). Profiling of PDR1 and MSH2 in Candida glabrata bloodstream isolates from a multicenter study in China. Antimicrob. Agents Chemother. 62:e153-18. doi: 10.1128/AAC.00153-18
Imbert, S., Castain, L., Pons, A., Jacob, S., Meyer, I., Palous, M., et al. (2016). Discontinuation of echinocandin and azole treatments led to the disappearance of an FKS alteration but not azole resistance during clonal Candida glabrata persistent candidaemia. Clin. Microbiol. Infect. 22, 891.e5–891.e8. doi: 10.1016/j.cmi.2016.07.025
Ishikawa, T., Takata, T., Akamatsu, S., Hatakenaka, K., Matsumoto, S., Maki, K., et al. (2010). Analysis of Candida glabrata strains with reduced sensitivity to micafungin in vitro isolated from a patient with persistent candidemia. Jpn. J. Infect. Dis. 63, 332–337.
Jensen, R. H., Johansen, H. K., Soes, L. M., Lemming, L. E., Rosenvinge, F. S., Nielsen, L., et al. (2015). Posttreatment antifungal resistance among colonizing candida isolates in candidemia patients: results from a systematic multicenter study. Antimicrob. Agents Chemother. 60, 1500–1508. doi: 10.1128/AAC.01763-15
Jimenez-Ortigosa, C., Perez, W. B., Angulo, D., Borroto-Esoda, K., and Perlin, D. S. (2017). De novo acquisition of resistance to SCY-078 in Candida glabrata involves FKS mutations that both overlap and are distinct from those conferring echinocandin resistance. Antimicrob. Agents Chemother. 61:e833-17. doi: 10.1128/AAC.00833-17
Katiyar, S. K., Alastruey-Izquierdo, A., Healey, K. R., Johnson, M. E., Perlin, D. S., and Edlind, T. D. (2012). Fks1 and Fks2 are functionally redundant but differentially regulated in Candida glabrata: implications for echinocandin resistance. Antimicrob. Agents Chemother. 56, 6304–6309. doi: 10.1128/AAC.00813-12
Kiraz, N., Dag, I., Oz, Y., Yamac, M., Kiremitci, A., and Kasifoglu, N. (2010). Correlation between broth microdilution and disk diffusion methods for antifungal susceptibility testing of caspofungin, voriconazole, amphotericin B, itraconazole and fluconazole against Candida glabrata. J. Microbiol. Methods 82, 136–140. doi: 10.1016/j.mimet.2010.05.002
Klotz, U., Schmidt, D., Willinger, B., Steinmann, E., Buer, J., Rath, P. M., et al. (2016). Echinocandin resistance and population structure of invasive Candida glabrata isolates from two university hospitals in Germany and Austria. Mycoses 59, 312–318. doi: 10.1111/myc.12472
Legrand, M., Chan, C. L., Jauert, P. A., and Kirkpatrick, D. T. (2007). Role of DNA mismatch repair and double-strand break repair in genome stability and antifungal drug resistance in Candida albicans. Eukaryot. Cell 6, 2194–2205. doi: 10.1128/EC.00299-07
Lewis, J. S. II, Wiederhold, N. P., Wickes, B. L., Patterson, T. F., and Jorgensen, J. H. (2013). Rapid emergence of echinocandin resistance in Candida glabrata resulting in clinical and microbiologic failure. Antimicrob. Agents Chemother. 57, 4559–4561. doi: 10.1128/AAC.01144-13
Lin, C. Y., Chen, Y. C., Lo, H. J., Chen, K. W., and Li, S. Y. (2007). Assessment of Candida glabrata strain relatedness by pulsed-field gel electrophoresis and multilocus sequence typing. J. Clin. Microbiol. 45, 2452–2459. doi: 10.1128/JCM.00699-07
Locke, J. B., Almaguer, A. L., Zuill, D. E., and Bartizal, K. (2016). Characterization of in vitro resistance development to the novel echinocandin CD101 in Candida species. Antimicrob. Agents Chemother. 60, 6100–6107. doi: 10.1128/AAC.00620-16
Lortholary, O., Renaudat, C., Sitbon, K., Madec, Y., Denoeud-Ndam, L., Wolff, M., et al. (2014). Worrisome trends in incidence and mortality of candidemia in intensive care units (Paris area, 2002-2010). Intensive Care Med. 40, 1303–1312. doi: 10.1007/s00134-014-3408-3
Lott, T. J., Frade, J. P., Lyon, G. M., Iqbal, N., and Lockhart, S. R. (2012). Bloodstream and non-invasive isolates of Candida glabrata have similar population structures and fluconazole susceptibilities. Med. Mycol. 50, 136–142. doi: 10.3109/13693786.2011.592153
Milazzo, L., Peri, A. M., Mazzali, C., Grande, R., Cazzani, C., Ricaboni, D., et al. (2014). Candidaemia observed at a university hospital in milan (northern Italy) and review of published studies from 2010 to 2014. Mycopathologia 178, 227–241. doi: 10.1007/s11046-014-9786-9
Mushi, M. F., Gross, U., Mshana, S. E., and Bader, O. (2018). High diversity of Candida glabrata in a tertiary hospital-Mwanza, Tanzania. Med. Mycol. doi: 10.1093/mmy/myy151 [Epub ahead of print].
Orasch, C., Marchetti, O., Garbino, J., Schrenzel, J., Zimmerli, S., Muhlethaler, K., et al. (2014). Candida species distribution and antifungal susceptibility testing according to european committee on antimicrobial susceptibility testing and new vs. old clinical and laboratory standards institute clinical breakpoints: a 6-year prospective candidaemia survey from the fungal infection network of switzerland. Clin. Microbiol. Infect. 20, 698–705. doi: 10.1111/1469-0691.12440
Pappas, P. G., Lionakis, M. S., Arendrup, M. C., Ostrosky-Zeichner, L., and Kullberg, B. J. (2018). Invasive candidiasis. Nat. Rev. Dis. Primers 4:18026. doi: 10.1038/nrdp.2018.26
Pfaller, M. A., Castanheira, M., Lockhart, S. R., Ahlquist, A. M., Messer, S. A., and Jones, R. N. (2012). Frequency of decreased susceptibility and resistance to echinocandins among fluconazole-resistant bloodstream isolates of Candida glabrata. J. Clin. Microbiol. 50, 1199–1203. doi: 10.1128/JCM.06112-11
Pham, C. D., Iqbal, N., Bolden, C. B., Kuykendall, R. J., Harrison, L. H., Farley, M. M., et al. (2014). Role of FKS mutations in Candida glabrata: MIC values, echinocandin resistance, and multidrug resistance. Antimicrob. Agents Chemother. 58, 4690–4696. doi: 10.1128/AAC.03255-14
Prigent, G., Ait-Ammar, N., Levesque, E., Fekkar, A., Costa, J. M., El Anbassi, S., et al. (2017). Echinocandin resistance in Candida species isolates from liver transplant recipients. Antimicrob. Agents Chemother. 61:e1229-16. doi: 10.1128/AAC.01229-16
Sandison, T., Ong, V., Lee, J., and Thye, D. (2017). Safety and pharmacokinetics of CD101 IV, a novel echinocandin, in healthy adults. Antimicrob. Agents Chemother. 61:e1627-16. doi: 10.1128/AAC.01627-16
Sasso, M., Roger, C., and Lachaud, L. (2017). Rapid emergence of FKS mutations in Candida glabrata isolates in a peritoneal candidiasis. Med. Mycol. Case Rep. 16, 28–30. doi: 10.1016/j.mmcr.2017.04.004
Shields, R. K., Kline, E. G., Healey, K. R., Kordalewska, M., Perlin, D. S., Nguyen, M. H., et al. (2019). Spontaneous mutational frequency and FKS mutation rates vary by echinocandin agent against Candida glabrata. Antimicrob. Agents Chemother. 63:e1692-18. doi: 10.1128/AAC.01692-18
Shields, R. K., Nguyen, M. H., Press, E. G., and Clancy, C. J. (2014). Abdominal candidiasis is a hidden reservoir of echinocandin resistance. Antimicrob. Agents Chemother. 58, 7601–7605. doi: 10.1128/AAC.04134-14
Shields, R. K., Nguyen, M. H., Press, E. G., Cumbie, R., Driscoll, E., Pasculle, A. W., et al. (2015). Rate of FKS mutations among consecutive Candida isolates causing bloodstream infection. Antimicrob. Agents Chemother. 59, 7465–7470. doi: 10.1128/AAC.01973-15
Shields, R. K., Nguyen, M. H., Press, E. G., Kwa, A. L., Cheng, S., Du, C., et al. (2012). The presence of an FKS mutation rather than MIC is an independent risk factor for failure of echinocandin therapy among patients with invasive candidiasis due to Candida glabrata. Antimicrob. Agents Chemother. 56, 4862–4869. doi: 10.1128/AAC.00027-12
Shields, R. K., Nguyen, M. H., Press, E. G., Updike, C. L., and Clancy, C. J. (2013). Anidulafungin and micafungin MIC breakpoints are superior to that of caspofungin for identifying FKS mutant Candida glabrata strains and echinocandin resistance. Antimicrob. Agents Chemother. 57, 6361–6365. doi: 10.1128/AAC.01451-13
Singh, A., Healey, K. R., Yadav, P., Upadhyaya, G., Sachdeva, N., Sarma, S., et al. (2018). Absence of azole or echinocandin resistance in Candida glabrata isolates in India despite background prevalence of strains with defects in the DNA mismatch repair pathway. Antimicrob. Agents Chemother. 62:e195-18. doi: 10.1128/AAC.00195-18
Tang, C. M., Cohen, J., and Holden, D. W. (1992). An Aspergillus fumigatus alkaline protease mutant constructed by gene disruption is deficient in extracellular elastase activity. Mol. Microbiol. 6, 1663–1671. doi: 10.1111/j.1365-2958.1992.tb00891.x
Thompson, G. R. III, Wiederhold, N. P., Vallor, A. C., Villareal, N. C., Lewis, J. S. II, and Patterson, T. F. (2008). Development of caspofungin resistance following prolonged therapy for invasive candidiasis secondary to Candida glabrata infection. Antimicrob. Agents Chemother. 52, 3783–3785. doi: 10.1128/AAC.00473-08
Vallabhaneni, S., Cleveland, A. A., Farley, M. M., Harrison, L. H., Schaffner, W., Beldavs, Z. G., et al. (2015). Epidemiology and risk factors for echinocandin nonsusceptible Candida glabrata bloodstream infections: data from a large multisite population-based candidemia surveillance program, 2008-2014. Open Forum Infect. Dis. 2:ofv163. doi: 10.1093/ofid/ofv163
White, T. J., Bruns, T., and Lee, S. B. (1990). Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. San Diego, CA: Academic Press.
Wiederhold, N. P., Najvar, L. K., Jaramillo, R., Olivo, M., Pizzini, J., Catano, G., et al. (2018). Oral glucan synthase inhibitor SCY-078 is effective in an experimental murine model of invasive candidiasis caused by WT and echinocandin-resistant Candida glabrata. J. Antimicrob. Chemother. 73, 448–451. doi: 10.1093/jac/dkx422
Zhao, X., and Drlica, K. (2001). Restricting the selection of antibiotic-resistant mutants: a general strategy derived from fluoroquinolone studies. Clin. Infect. Dis. 33(Suppl. 3), S147–S156. doi: 10.1086/321841
Keywords: Candida glabrata, echinocandins, antifungal resistance, FKS, MSH2, genotyping, anidulafungin, micafungin
Citation: Rivero-Menendez O, Navarro-Rodriguez P, Bernal-Martinez L, Martin-Cano G, Lopez-Perez L, Sanchez-Romero I, Perez-Ayala A, Capilla J, Zaragoza O and Alastruey-Izquierdo A (2019) Clinical and Laboratory Development of Echinocandin Resistance in Candida glabrata: Molecular Characterization. Front. Microbiol. 10:1585. doi: 10.3389/fmicb.2019.01585
Received: 18 April 2019; Accepted: 25 June 2019;
Published: 11 July 2019.
Edited by:
Miguel Cacho Teixeira, University of Lisbon, PortugalReviewed by:
Marie Desnos-Ollivier, Institut Pasteur, FranceCheshta Sharma, The University of Tennessee Health Science Center (UTHSC), United States
Dominique Sanglard, Université de Lausanne, Switzerland
Copyright © 2019 Rivero-Menendez, Navarro-Rodriguez, Bernal-Martinez, Martin-Cano, Lopez-Perez, Sanchez-Romero, Perez-Ayala, Capilla, Zaragoza and Alastruey-Izquierdo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ana Alastruey-Izquierdo, YW5hYWxhc3RydWV5QGlzY2lpaS5lcw==
 Olga Rivero-Menendez
Olga Rivero-Menendez Patricia Navarro-Rodriguez
Patricia Navarro-Rodriguez Leticia Bernal-Martinez
Leticia Bernal-Martinez Gema Martin-Cano1
Gema Martin-Cano1 Javier Capilla
Javier Capilla Oscar Zaragoza
Oscar Zaragoza Ana Alastruey-Izquierdo
Ana Alastruey-Izquierdo