
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 02 July 2019
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.01504
This article is part of the Research Topic Evolution of Genetic Mechanisms of Antibiotic Resistance View all 16 articles
 Vincenzo Di Pilato1
Vincenzo Di Pilato1 Alberto Antonelli1
Alberto Antonelli1 Tommaso Giani1,2
Tommaso Giani1,2 Lucia Henrici De Angelis3
Lucia Henrici De Angelis3 Gian Maria Rossolini1,2
Gian Maria Rossolini1,2 Simona Pollini1,2*
Simona Pollini1,2*Acquisition of metallo-β-lactamases (MBLs) represents one of most relevant resistance mechanisms to all β-lactams, including carbapenems, ceftolozane and available β-lactamase inhibitors, in Pseudomonas spp. VIM-type enzymes are the most common acquired MBLs in Pseudomonas aeruginosa and, to a lesser extent, in other Pseudomonas species. Little is known about the acquisition dynamics of these determinants, that are usually carried on integrons embedded into chromosomal mobile genetic elements. To date, few MBL-encoding plasmids have been described in Pseudomonas spp., and their diversity and role in the dissemination of these MBLs remains largely unknown. Here we report on the genetic features of the VIM-1-encoding plasmid pMOS94 from P. mosselii AM/94, the earliest known VIM-1-producing strain, and of related elements involved in dissemination of MBL. Results of plasmid DNA sequencing showed that pMOS94 had a modular organization, consisting of backbone modules associated with replication, transfer and antibiotic resistance. Plasmid pMOS94, although not typable according to the PBRT scheme, was classifiable either in MOBF11 or MPFT plasmid families. The resistance region included the class I integron In70, carrying blaV IM-1, in turn embedded in a defective Tn402-like transposon. Comparison with pMOS94-like elements led to the identification of a defined plasmid lineage circulating in different Pseudomonas spp. of clinical and environmental origin and spreading different MBL-encoding genes, including blaIMP-63, blaBIM, and blaV IM-type determinants. Genetic analysis revealed that this plasmid lineage likely shared a common ancestor and had evolved through the acquisition and recombination of different mobile elements, including the MBL-encoding transposons. Our findings provide new insights about the genetic diversity of MBL-encoding plasmids circulating among Pseudomonas spp., potentially useful for molecular epidemiology purposes, and revealed the existence and persistence of a successful plasmid lineage over a wide spatio-temporal interval, spanning over five different countries among two continents and over 20-years.
Pseudomonas is a genetically and metabolically diverse genus of bacteria, which inhabit a wide variety of environments and can act as pathogens of humans, animals and plants. Among the wide variety of Pseudomonas species, only few have been recognized as human pathogens, with Pseudomonas aeruginosa being the most common cause of infections (Driscoll et al., 2007; Høiby et al., 2015; Peix et al., 2018).
The acquisition by pathogenic Pseudomonas species of β-lactamases able to degrade most anti-pseudomonas β-lactams and resistant to currently available β-lactamase inhibitors, such as the metallo-β-lactamases (MBLs), can provide a remarkable contribution to the emergence of strains with difficult-to-treat resistance (DTR) phenotypes (Hawkey et al., 2018). VIM-type enzymes are among the most widespread and prevalent acquired MBLs in P. aeruginosa (Hong et al., 2015), and have occasionally been reported also in other Pseudomonas spp. of clinical interest including Pseudomonas putida, Pseudomonas mosselii, Pseudomonas monteilii, Pseudomonas stutzeri, and Pseudomonas mendocina (Yan et al., 2001; Lombardi et al., 2002; Giani et al., 2012; Ocampo-Sosa et al., 2015; Almuzara et al., 2018).
Metallo-β-lactamase genes, and in particular blaV IM- and blaIMP-type genes, are typically carried on mobile gene cassettes inserted into integron platforms (Partridge et al., 2018). While among Enterobacterales MBL-encoding integrons are mostly associated with plasmid lineages of IncN-, IncI1- and IncHI2-type replicons (Carattoli, 2009; Villa et al., 2014), less is known about the genetic support of these elements among Pseudomonas spp. In these species, MBL-encoding integrons have been described both as chromosomal- (Lauretti et al., 1999; Perez et al., 2014; Di Pilato et al., 2015; Roy Chowdhury et al., 2016) and plasmid-borne, but only few plasmids have been completely sequenced (Li et al., 2008; Bonnin et al., 2013; Vilacoba et al., 2014; San Millan et al., 2015; Botelho et al., 2017, 2018; Partridge et al., 2018; Liapis et al., 2019), and information about prevalence and diversity of MBL-carrying plasmid lineages remains largely unknown.
In this study we have investigated the genetic support of the blaV IM-1 gene from a non-aeruginosa Pseudomonas strains of clinical origin, namely Pseudomonas mosselii AM/94, which represents the earliest known VIM-1-producing strain (Giani et al., 2012). In that strain, blaV IM-1 was previously shown to be carried on a gene cassette inserted into integron In70, but its genetic support was not further investigated. Here we report that in this strain the integron was plasmid-borne and describe the structure of the blaV IM-1-carrying plasmid from AM/94, named pMOS94. The results of a comparative genetic analysis with pMOS94, allowed the definition of a novel plasmid lineage involved in dissemination of MBL genes among Pseudomonas spp.
Pseudomonas mosselii AM/94 was isolated in 1994 from the lower respiratory tract of an inpatient in Genoa, Italy (Giani et al., 2012). P. aeruginosa C/53 and C/57 were isolated in 2014 from bloodstream infections from intensive care patients from Milan, Italy, and the complete genome has been previously determined (Giani et al., 2018). Identification to the species level and Sequence Type (ST) of C/57 strain were carried out in silico on whole genome sequence data. Antimicrobial susceptibility testing was carried out by reference broth-microdilution method and results were interpreted according to EUCAST clinical breakpoints (EUCAST breakpoint tables version 9, 20191).
Electrotransformation experiments were performed with electrocompetent E. coli DH10B and P. aeruginosa PAO-1 using plasmid DNA preparations, obtained as described previously (Docquier et al., 2003). The broad-host range vector plasmid pME6001 (Blumer et al., 1999) was used as positive control in electrotransformation experiments. Transformants were selected on LB agar containing ceftazidime (10 mg/L for E. coli and 20 mg/L for P. aeruginosa) or gentamicin (30 mg/L) for the pME6001 vector. Conjugation experiments were performed in solid medium as previously described (Docquier et al., 2003), using E. coli J53 AziR as the recipient strain. Ceftazidime (10 mg/L) was used for the selection of transconjugants and sodium azide (150 mg/L) for counterselection of the donor. Plasmid incompatibility groups were determined following the previously described PCR-based replicon typing (PBRT) schemes (Carattoli et al., 2005; García-Fernández et al., 2009; Villa et al., 2010; Johnson et al., 2012). In silico typing using plasmid relaxases (MOB) regions (Garcillán-Barcia et al., 2009) and the mating pair formation (MPF) apparatus (Smillie et al., 2010) was performed according to the previously proposed classification scheme (Guglielmini et al., 2014; Orlek et al., 2017).
Genomic DNA was extracted as previously described (Johnson, 1994). Plasmid DNA was extracted using Wizard® Plus SV Minipreps DNA purification system (Promega, Madison, United States) according to manufacturer’s instructions. Plasmid DNAs from strain AM/94 was subjected to complete sequencing with the Illumina MiSeq platform (Illumina Inc., San Diego, United States) with a 2 × 250 or 2 × 300 bp paired-end approach. A total of 171,952 high quality reads were generated and assembled using SPAdes 3.11 (Bankevich et al., 2012). Sequence annotation was performed using the RAST web-server (Aziz et al., 2008). Finishing of pMOS94 and pAER57 plasmid sequences was achieved through Sanger sequencing of PCR products spanning contigs’ gaps. Sequence comparisons were performed using BLAST2 and Mauve3 Core genome alignments were performed using Roary, using the non-paralog splitting method (Page et al., 2015; Rozwandowicz et al., 2017). Plasmid circular and linear maps were generated using the CGView Server and EasyFig tools, respectively (Grant and Stothard, 2008; Sullivan et al., 2011). Comparator plasmid sequences were selected from the NCBI-NIH database (non-redundant) to include all circular molecules with 100% sequence identity at nucleotide level of the repA-oriV region of plasmid pMOS94. A tentative plasmid classification based on replicase (rep) homology was inferred through nucleotide sequence alignments of rep genes from representative members of the IncN (García-Fernández et al., 2011), IncW (Suzuki et al., 2010; Aoki et al., 2018), and IncP9 replicons (Sevastsyanovich et al., 2008), collectively clustering into the MOBF11 relaxase family, to which pMOS94-like plasmids belong. Plasmids used in this analysis are reported in Supplementary Table S1. Sequences were aligned using the MAFFT v.7 server4 with the G-INS-1 progressive method, and the rep phylogeny was inferred using the Neighbor-joining method implemented by MAFFT. Phylogenetic trees were visualized and annotated through the Evolview software (He et al., 2016).
Pseudomonas mosselii AM/94 was previously shown to carry the blaV IM-1 gene cassette as part of an In70 integron platform (5′CS – blaV IM-1 – aacA4 – aphA15 – aadA15 – 3′CS) (Giani et al., 2012).
To investigate the genetic support of the blaV IM-1 cassette and of its cognate integron in P. mosselii AM/94, an S1 nuclease assay and hybridization with a blaV IM-specific probe were performed. Results mapped blaV IM to a 50 kb plasmid (data not shown), hereafter referred to as pMOS94.
Gene transfer experiments failed in yielding transconjugants using E. coli J53 as recipient in conjugation experiments, and transformants using E. coli DH10B and P. aeruginosa PAO-1 as recipients in electrotransformation experiments.
The PBRT assay (Carattoli et al., 2005; García-Fernández et al., 2009; Villa et al., 2010; Johnson et al., 2012), performed on total DNA purified from AM/94, could not identify any plasmid replicon.
In order to determine the genetic features of pMOS94, plasmid DNA was purified from AM/94 and completely sequenced. De novo sequence assembly yielded a single contig of 51,660 kb in length, which resulted in a 52,002 kb circular molecule, with a raw coverage of 380× and a mean G+C content of 59.6%, following sequence completion.
A total of 71 coding sequences (CDS) were identified by the RAST annotation pipeline, of which 37 had a predicted function consisting in: replication and stability (n = 7), transfer (n = 14), mobile genetic elements (n = 9) and antibiotic resistance (n = 7) (Figure 1).
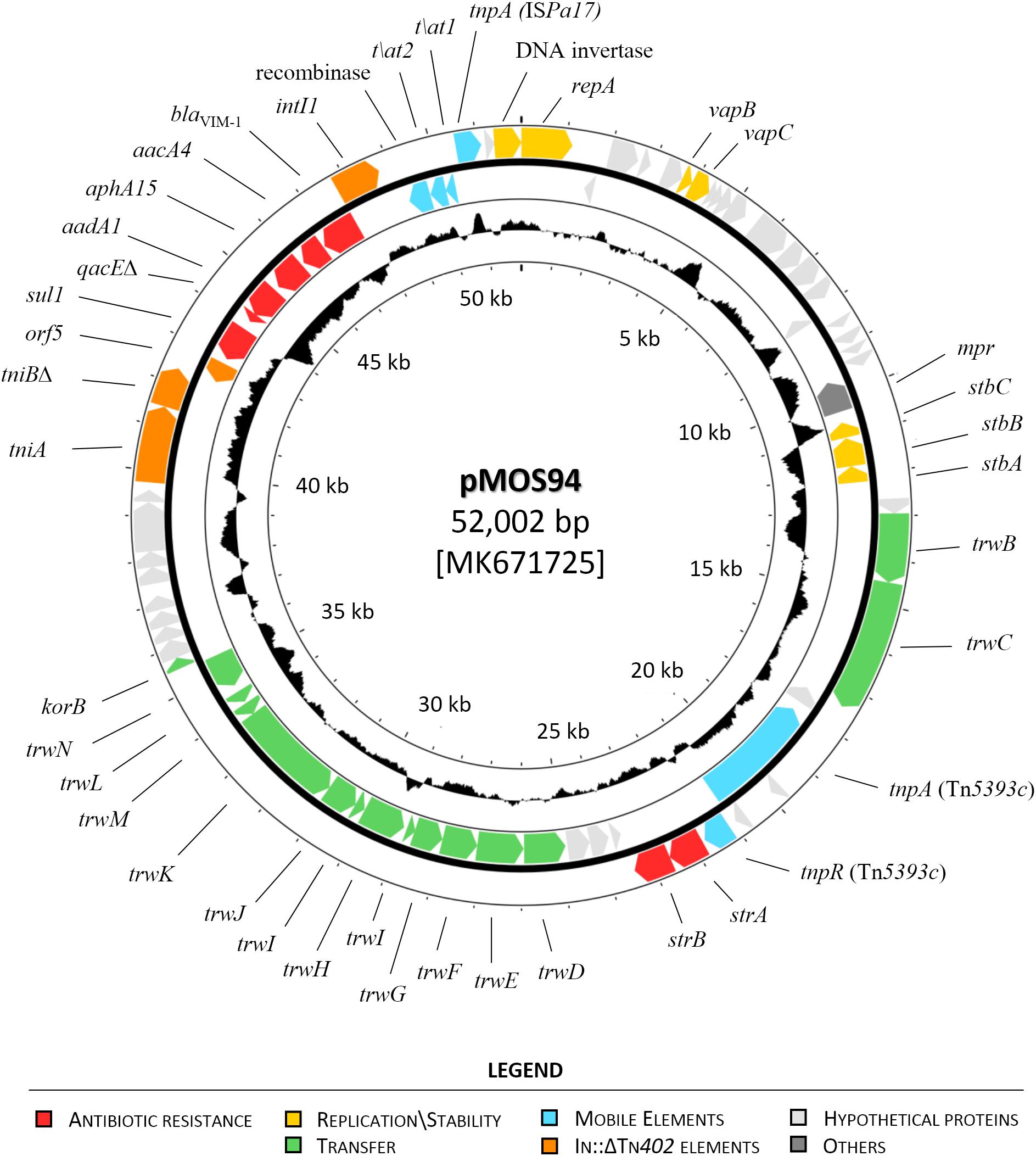
Figure 1. Circular map of plasmid pMOS94. The circles, from the outermost to the innermost, show: (i) coding DNA sequences (CDSs) encoded on the plus and minus DNA strands (annotation is reported for known genes only); and (ii) the G+C content, shown as deviation from the average G+C content of the entire molecule. Genes encoding proteins of known functions are in different colors, as detailed in the legend.
Plasmid pMOS94 showed a modular organization associated with the following functions: (i) plasmid replication; (ii) conjugal transfer; (iii) antibiotic resistance.
The replication module included genes essential for plasmid replication (repA, res) and stability (stbABC), as well as a type II toxin-antitoxin system (vapBC) for stable maintenance. A putative replication origin (oriV) was identified immediately adjacent to repA, characterized by four 19-bp (TTCGTCACTCCAGGGACCG) and seven 9-bp (TTCGTCACT) iteron repeats, suggesting a theta-type replication mechanism for pMOS94 (Chattoraj, 2000).
The transfer module comprised a trw-type gene cluster encoding a T4SS machinery, composed by three structurally different components within the same segmented operon (Llosa and Alkorta, 2017) encoding: (i) the inner membrane complex (TrwMKIGB); (ii) the outer membrane core complex (TrwNHFE); (iii) the external pilus (TrwLJ). In silico analyses according to the MOB and MPF typing schemes, exploiting T4SS components for classification (Garcillán-Barcia et al., 2009; Smillie et al., 2010; Guglielmini et al., 2014; Orlek et al., 2017), revealed that pMOS94 was classifiable either in the MOBF11 or in the MPFT families.
The resistance region was organized in two non-contiguous segments, of which one was represented by the In70 integron platform carried on a defective Tn402-like transposon, whereas the second included Tn5393c, a Tn3-family transposon carrying the strA–strB genes, coding for phosphotransferases able to confer high level resistance to streptomycin (Chiou and Jones, 1993; Sunde and Norström, 2005). These segments were located downstream of and within the transfer module, respectively. The interruption of the trw operon by insertion of Tn5393c, apparently related with a transposition event generating 5-bp direct repeats (DRs) (5′-GAATA-3′) flanking the element, could explain the unability of pMOS94 for conjugational transfer. Conversely, the Tn402-like transposon carrying In70 was inserted downstream of the transfer module and upstream of the locus coding for a DNA invertase, a core component of the plasmid backbone associated with replication and stability functions.
In order to identify relatives of pMOS94 and to evaluate the possible host range and dissemination of a similar plasmid lineage, and its role in spreading clinically relevant antibiotic resistance determinants, an BLAST screening using the repA-oriV region was performed against publicly available databases in NCBI (non-redundant and wgs) (last accessed on April 2, 2019).
Altogether, seven records were identified showing 100% identity with the repA-oriV region, of which five were circular plasmids ranging in size from 24 to 80 kb, while two corresponded to putative plasmid sequences from a recently characterized collection of MBL-producing P. aeruginosa from Italy (Giani et al., 2018). Strains harboring the latter putative plasmids, namely P. aeruginosa C/53 and C/57, were available for further analysis, and gaps closure by PCR resulted in two circular elements showing a size of 102 kb. Since C/53 and C/57 were isolated from the same surveillance center in the same period (2014), and showed the same clonality (ST 532) and resistome (data not shown), C/57 was selected as representative for plasmid closure.
Interestingly, the analyzed plasmids were uniquely detected within members of the Pseudomonas genus, including strains of P. aeruginosa, P. putida, and P. mosselii. All of them carried an MBL-encoding gene (i.e., blaV IM, blaIMP, or blaBIM) in association with additional antibiotic resistance determinants (Table 1). In most of cases (6/7) the Pseudomonas strains were from clinical sources, with the exception of P. putida IEC33019, which was from an environmental source. Overall these strains were from five different countries, belonging to two continents, and had been isolated over a prolonged time period spanning over 20-years (Table 1).
Sequence analysis and comparison of pMOS94 with the other plasmids revealed an overall conserved backbone structure, containing genes involved in stability, maintenance and, in some cases, encoding the T4SS or parts thereof (Figure 2). Except for pAMBL2, where the transfer operon and the cognate relaxase were fully lacking, all plasmids were found to belong to the MOBF11/MPFT families and were untypeable by the replicon typing scheme, as pMOS94.
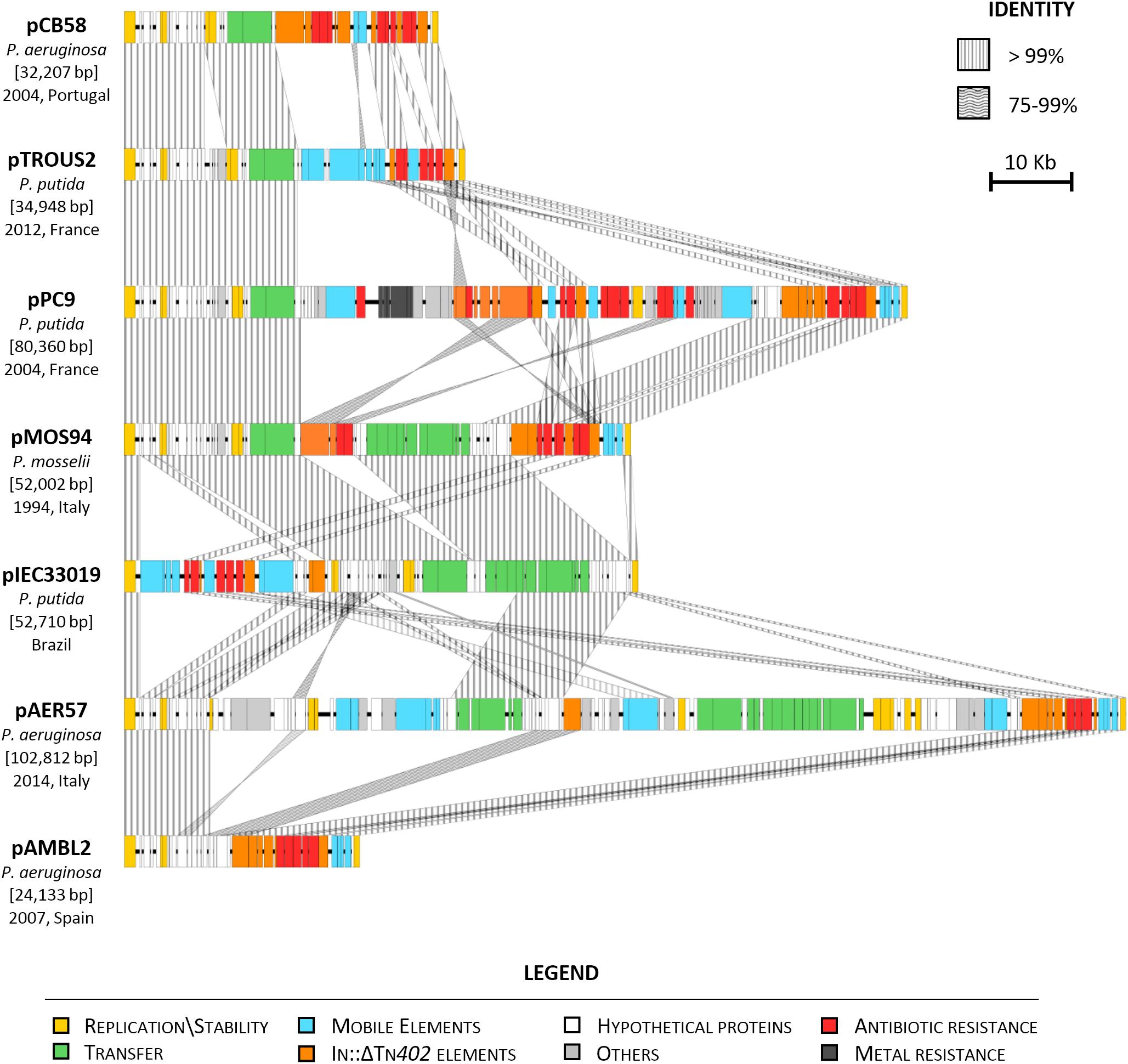
Figure 2. Linear map of VIM-1 encoding plasmids from different Pseudomonas spp. Year (when available), geographical region and species of isolation are indicated. Homologous segments are indicated by striped shading, representing 75–99% and ≥99% sequence identity. Genes encoding proteins of known functions are in different colors, as detailed in the legend.
Compared to pMOS94, pIEC33019 exhibited the closest conserved architecture, with major differences due to the acquisition of mobile genetic elements targeting a different backbone region (Figure 2). Conversely, the most diverging elements were pAMBL2, pPC9 and pAER57. Plasmid pAMBL2 was a small (24.1 kb) deletion derivative of pMOS94, where the entire trw locus was lacking, carrying only genes essential for replication processes and a multiresistance region (Figure 2; San Millan et al., 2015). Plasmid pPC9 was characterized by a mosaic architecture, carrying mobile genetic elements and multiple genes coding for metal (e.g., merEA) and antibiotic resistance (e.g., In860) (Figure 2 and Table 1; Molina et al., 2014). Plasmid pAER57 was the largest identified element of this lineage (>100 kb), and was characterized by a more complex chimeric arrangement, including a 43 kb segment sharing 87% nucleotide identity with the archetypal IncP-9 pWW0 catabolic plasmid from P. putida (GenBank Acc. No. AJ344068) (Greated et al., 2002; Sevastsyanovich et al., 2008). The 43 kb segment, likely acquired through recombination with a pWW0-like plasmid, included genes coding for replication (e.g., res-oriV-rep), partitioning (e.g., parABC, korA), transfer (e.g., mpfABCDEFGHIJ) and unknown functions. Therefore, pAER57 was classifiable as a multi-replicon plasmid, being characterized by an additional replication locus (i.e., res-oriV-rep) 91% identical to that of pWW0.
Analysis of SNPs evaluated on core orthologous genes of pMOS94-like plasmids showed an overall limited sequence diversity. In detail, all plasmids differed only by 1–2 SNPs from each other, with the exception of pAER57 that was the more divergent, showing 13–14 SNPs differences.
Further sequence analysis revealed that members of this plasmid lineage were also able to integrate into the host chromosome as linear elements. Evidences of integration came from a recent study reporting on the structure of a chromosomal genomic island carrying an MBL determinant from a P. aeruginosa strain of clinical origin isolated in Prague in 2015 (GenBank acc. no. KY860572). In this case, the integrated element was apparently a deletion-derivative of pMOS94 (carrying only backbone essential regions coding for replication functions) inserted within a Tn2 family transposon (Tn6162), in turn embedded into a genomic island (PACS171b) previously described in high risk clones of P. aeruginosa (i.e., ST 235) as chromosomal support of MBL-encoding genes (Martinez et al., 2012; Di Pilato et al., 2015).
Noteworthy, a plasmid lineage closely related (84–87% identity of the repA-oriV locus and belonging to the MOBF11 and MPFT families) to the one identified in this study was detected in the NCBI non-redundant database and included elements preferentially circulating within the genus Pseudomonas. These elements included: (i) plasmid pCT14 encoding mercury resistance from Pseudomonas veronii (Bramucci et al., 2006); (ii) two unnamed plasmids from P. aeruginosa AR_0356 and 163940 producing a KPC-2 carbapenemase (GenBank acc. nos. CP027168.1 and CP029092.1); (iii) plasmid pTROUS1 from P. aeruginosa 163940 producing an IMP-63 carbapenemase (GenBank acc. no MK047610.1), and (iv) a non-circular molecule resembling a plasmid element (GenBank acc. no. LC350084.1) coding for a VIM-2 carbapenemase. Notably, most of these elements were associated with clinically relevant resistance determinants and, according to available metadata, were of clinical (pTROUS1 and LC350084.1) or environmental (pCT14) origin. Using the repA-oriV locus as probe, at least 96 records were found among Pseudomonas spp. draft genome sequences in NCBI wgs database (i.e., showing 100–90% identity), highlighting a potential wide dissemination of pMOS94-related elements. Nevertheless, the plasmidic nature of such records could not be verified due to their fragmentation and unfinished status. In order to infer possible phylogenetic relationships between pMOS94, pMOS94-related plasmids (i.e., those showing 84–87% identity of the repA-oriV locus) and known plasmid replicons belonging to the MOBF1relaxase family, to which pMOS94-like plasmids belong, a classification according to the rep locus was attempted. The analysis showed that pMOS94 and related plasmids clustered in a separate branch, that appeared more closely related to IncW than IncN and IncP9 replicons (Figure 3).
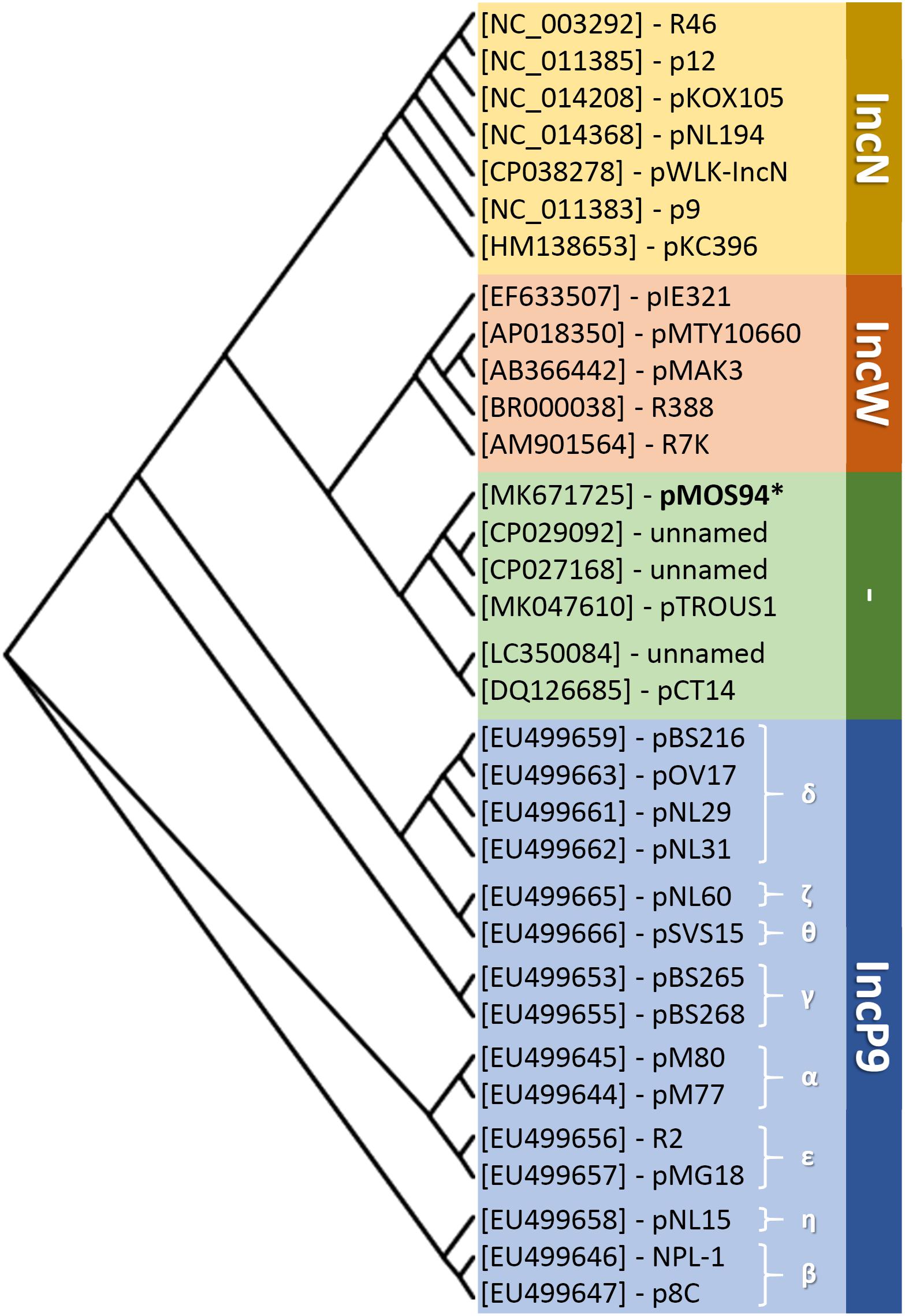
Figure 3. Neighbor-joining phylogenesis of replicase encoding genes (rep) from representative members of the IncN-, IncW-, and IncP9-type replicons, collectively clustering within the MOBF11 relaxase family. Selected plasmids and the corresponding host bacterial species are reported in Supplementary Table S1. IncP9 plasmid subgroups are indicated with Greek letters. ∗Plasmid pMOS94 was selected as representative of pMOS94-like plasmids, showing 100% identity with the repA-oriV region.
All seven plasmids of the pMOS94-like lineage described above carried an MBL-encoding gene, embedded in a class I integron platform (Figure 4), including blaV IM-1 in four plasmids, and the blaV IM-2, blaBIM, and blaIMP-63 in the remining ones. The blaV IM-1 gene was found embedded in the In70 integron (in pMOS94) or in its derivatives (e.g., In110, In860, In1167) carrying a modified version of the typical gene cassette array of this element (i.e., in pAMBL2, pAER57, and pPC9). On the other hand, the other MBL genes were carried in integrons of different structure (e.g., In58, In1326, and In1297; Figure 4). Despite their heterogeneity, the MBL-encoding integrons always carried aminoglycosides resistance determinants, with aacA4 variants being the most frequent (Figure 4). Most of the integrons carried typical 5′CS and 3′CS regions, except for In110 in pAER57, where the intI1 gene was partially deleted (Figure 4).
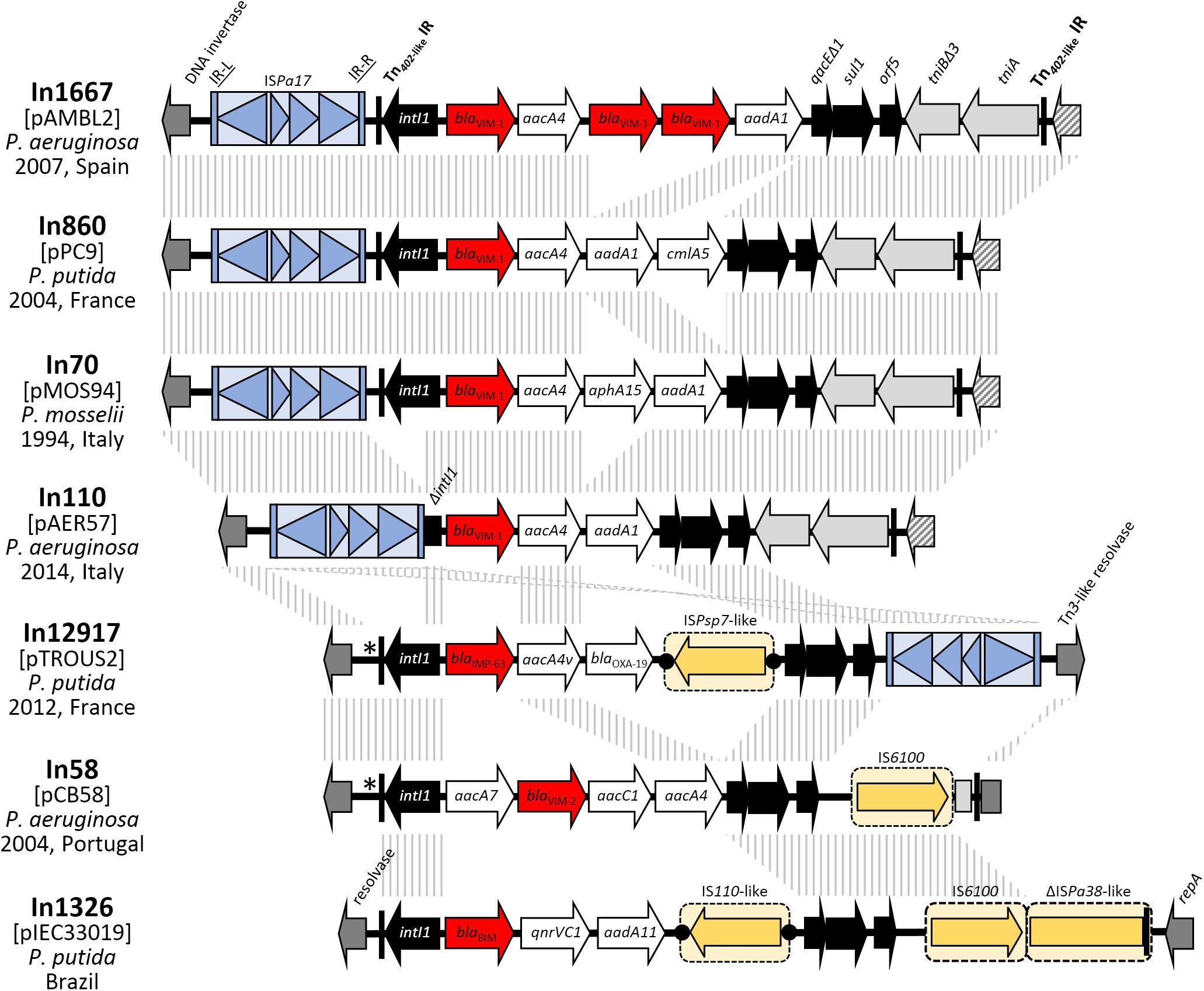
Figure 4. Structure of the cassette arrays and of the genetic context of MBL-harboring class 1 integrons carried on the analyzed plasmid. Integron name, year (when available), geographical region and species of isolation are indicated. MBL encoding gene cassettes are indicated by red arrows; gene cassettes encoding for other resistance enzymes (e.g., aminoglycosides and phenicols) are indicated by white arrows; intI1 integrase gene and 3′-conserved segments (3′CS) flanking the cassette array are indicated by black arrows; tni defective module is indicated by light gray arrows; genes flanking the Tn402-like insertion points (including repA gene and genes encoding for DNA invertase and resolvases) are indicated by dark gray arrows; hypothetical proteins are indicated by striped arrows. ISPa17 is shown as light blue boxed arrows bounded by light blue vertical bars. Other ISs and mobile elements are shown as yellow boxed arrows. Tn402-like transposon 25-bp IRs are indicated by a black vertical line. When present, DRs flanking the different mobile elements are shown as black dots. The diverse insertion point of Tn402-like transposons in pTROUS2 and pCB58 is indicated by an asterisk. Homologous segments (>99%) generated by a BLASTn comparison are shown as gray striped blocks that are connected across the different structures.
The MBL-encoding integrons were always associated with defective Tn402-like transposons. In most cases (namely pAMBL2, pPC9, pMOS94, pAER57, and pCB58), the Tn402 25 bp inverted repeats (IRi and IRt) were present at the boundaries of the defective transposons, while in the remaining plasmids (i.e., pTROUS2 and pIEC33019) IRi was present adjacent to the integrase gene, suggesting recombination events which deleted the IRt end. The Tn402 tni transposition module was present downstream the 3′CS region in a partially deleted form (in five plasmids), or completely absent (in the others) (Figure 4).
In some cases, additional mobile elements were found within the Tn402 elements, targeting the cassette array or other regions (Figure 4). The ISPsp7-like and IS110-like insertion sequences, respectively targeting the cassette arrays of pTROUS2 and pIEC33019 were flanked by DRs, suggesting acquisition by transposition.
In all cases the Tn402-like elements carried by the pMOS94-like plasmids were not flanked by 5 bp DRs, that represent the molecular markers of the transposition of this kind of mobile elements, suggesting that acquisition had involved different recombination events, or that recombination events involving one transposon moiety had occurred after transposition.
In all plasmids but pIEC33019, the Tn402-like element was inserted into an A-T rich region located upstream of the locus coding for a DNA invertase. Within this region, all the Tn402-like elements carrying blaV IM-1 were inserted at the same site (i.e., 258 bp upstream of the DNA invertase gene), while the remaining ones were inserted slightly upstream (i.e., 281 bp upstream of the DNA invertase gene). On the other hand, in plasmid pIEC33019 the Tn402-like element was inserted in a different backbone region located downstream of repA (Figure 4).
Interestingly, all Tn402-like transposons carrying blaV IM-1 were flanked by a copy of the ISPa17 insertion sequence (Haines et al., 2005), located upstream of the IRi end (Figure 4). Conversely, a different ISPa17/Tn402-like transposon assembly was found in plasmid pTROUS2, where the IS element was inserted downstream of the class I integron 3′CS region and disrupted the tniA gene (Figure 4). This IS was already described as associated to MBL-encoding Tn402-like transposons aboard plasmids from GNNFs, where it likely contributed to the MBL gene platform acquisition through a novel transposition mechanism exploiting the similarity between the Tn402 IRs and their counterparts in ISPa17 (Di Pilato et al., 2014; Botelho et al., 2017; Liapis et al., 2019). Nevertheless, in pMOS94-like plasmids no evidence of an ISPa17-mediated mobilization was found, since no DRs bounding the ISPa17 IR-L and the Tn402 IRt end were detected (Figure 4).
In this work we characterized the blaV IM-1-carrying plasmid pMOS94 from P. mosselii AM/94 isolated in Italy in 1994, which represents the earliest known VIM-producing strain (Giani et al., 2012). Analysis of the complete sequence of pMOS94 led to the identification of a novel plasmid lineage characterized by an original replication origin (i.e., repA-oriV) and included in the MOBF11 and MPFT families, according to the typing schemes based on plasmid relaxases and on the MPF system, respectively (Guglielmini et al., 2014; Orlek et al., 2017). Interestingly, the MOBF11 family includes relaxases of plasmids belonging to IncW and IncN replicons, responsible for the spreading of antibiotic resistance determinants among enterobacteria, and to the IncP9 group, including metal-resistance and xenobiotic-biodegradation plasmids from Pseudomonas spp. (Garcillán-Barcia et al., 2009; Shintani et al., 2015). Based on replicase sequence homology, pMOS94-like plasmids showed a closer relatedness with the IncW replicon family. Further transfer experiments will be needed to evaluate the incompatibility behavior of these plasmids.
Sequence analyses revealed that members of this novel plasmid lineage were involved in spread of MBL-encoding genes, and accounted for a broad dissemination of such resistance determinants among pseudomonads of clinical and environmental origin, spanning over a time period of at least 20 years, among at least five different countries from two continents. However, the association of pMOS94 and pMOS94-like plasmids with resistance genes could be somewhat biased by the fact that the main purpose of bacterial sequencing focuses on antibiotic resistance.
The comparison of pMOS94-like plasmids highlighted a high phylogenetic relatedness within the lineage, which had likely evolved from a common ancestor. This point was also supported by the low sequence divergence observed among core backbone genes with the exception of pAER57, which was slightly more divergent. Nonetheless, the different insertion points of Tn402-like transposons and cognate integrons within the accessory region might represent a molecular marker of different evolutionary pathways for pMOS94 and pMOS94-like plasmids. In this scenario, different hypotheses could be made on the evolution of MGEs associated with this plasmid lineage: (i) the Tn402-like transposons could have been acquired following different insertion events that, in some cases (e.g., plasmids harboring blaVIM-1), occurred at the same site; (ii) at least three insertion events (occurring at different sites) led to the acquisition of the Tn402-like transposons, that subsequently evolved independently by genetic rearrangements and gene cassettes loss/acquisition. The number of the analyzed plasmids and sequence data publicly available for comparison, however, are limited and the chronological order of these events (i.e., the evolution of the backbone core genes and the acquisition of the accessory resistance regions) remains to be clarified.
Recently, a number of studies have described novel plasmid supports for different MBL genes among pseudomonads (Li et al., 2008; Bonnin et al., 2013; Vilacoba et al., 2014; San Millan et al., 2015; Botelho et al., 2017, 2018; Partridge et al., 2018; Liapis et al., 2019), reporting on the structure of single plasmids or on structures circulating in a restricted geographical area or within a relatively short time period.
The present study contributed to extend current knowledge about the diversity and phylogeny of plasmids circulating in Pseudomonas spp., reporting on a successful plasmid lineage carrying MBL genes. Although mobilization experiments were not successful under the experimental conditions used in this study, the heterogeneous range of species carrying pMOS94-like plasmids suggests that these elements are able to be transferred. Therefore, further experiments are needed to evaluate their mobilization potential among GNNFs and eventually Enterobacterales, in order to better understand the possible contribution in disseminating antibiotic resistance traits by this novel plasmid lineage.
The complete nucleotide sequence of pMOS94 from P. mosselii AM/94 and pAER57 from P. aeruginosa C/57 have been registered in GenBank under accession numbers MK671725 and MK671726, respectively.
The nucleotide sequences described for the first time in this study, namely pMOS94 and pAER57 sequences, have been deposited in GenBank database and will be made available to other researchers following publication. The remaining plasmid nucleotide sequences analyzed in this study are publicly available in the GenBank database under the accession numbers cited in the manuscript.
VDP and AA did plasmid sequencing, analyzed the data, and drafted the manuscript. TG and LHDA produced the phenotypic data and handled the samples. GR and SP coordinated the experiments, drafted and edited the manuscript.
This work was supported in part by EvoTAR (No. HEALTH-F3-2011-2011-123282004) to GR.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01504/full#supplementary-material
Almuzara, M., Montaña, S., Carulla, M., Sly, G., Fernandez, J., Hernandez, M., et al. (2018). Clinical cases of VIM-producing Pseudomonas mendocina from two burned patients. J. Glob. Antimicrob. Resist. 14, 273–274. doi: 10.1016/j.jgar.2018.08.002
Aoki, K., Harada, S., Yahara, K., Ishii, Y., Motooka, D., Nakamura, S., et al. (2018). Molecular Characterization of IMP-1-Producing Enterobacter cloacae Complex Isolates in Tokyo. Antimicrob. Agents Chemother. 23, e2091–17. doi: 10.1128/AAC.02091-2017
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Blumer, C., Heeb, S., Pessi, G., and Haas, D. (1999). Global GacA-steered control of cyanide and exoprotease production in Pseudomonas fluorescens involves specific ribosome binding sites. Proc. Natl. Acad. Sci. U.S.A. 96, 14073–14078. doi: 10.1073/pnas.96.24.14073
Bonnin, R. A., Poirel, L., Nordmann, P., Eikmeyer, F. G., Wibberg, D., Pühler, A., et al. (2013). Complete sequence of broad-host-range plasmid pNOR-2000 harbouring the metallo-β-lactamase gene blaV IM-2 from Pseudomonas aeruginosa. J. Antimicrob. Chemother. 68, 1060–1065. doi: 10.1093/jac/dks526
Botelho, J., Grosso, F., and Peixe, L. (2017). Characterization of the pJB12 plasmid from Pseudomonas aeruginosa reveals Tn6352, a novel putative transposon associated with mobilization of the blaV IM-2-harboring In58 integron. Antimicrob. Agents Chemother. 61, e2532–16. doi: 10.1128/AAC.02532-16
Botelho, J., Grosso, F., Quinteira, S., Brilhante, M., Ramos, H., and Peixe, L. (2018). Two decades of blaV IM-2-producing Pseudomonas aeruginosa dissemination: an interplay between mobile genetic elements and successful clones. J. Antimicrob. Chemother. 73, 873–882. doi: 10.1093/jac/dkx517
Bramucci, M., Chen, M., and Nagarajan, V. (2006). Genetic organization of a plasmid from an industrial wastewater bioreactor. Appl. Microbiol. Biotechnol. 71, 67–74. doi: 10.1007/s00253-005-0119-112
Carattoli, A. (2009). Resistance Plasmid Families in Enterobacteriaceae. Antimicrob. Agents Chemother. 53, 2227–2238. doi: 10.1128/AAC.01707-1708
Carattoli, A., Bertini, A., Villa, L., Falbo, V., Hopkins, K. L., and Threlfall, E. J. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63, 219–228. doi: 10.1016/j.mimet.2005.03.018
Chattoraj, D. K. (2000). Control of plasmid DNA replication by iterons: no longer paradoxical. Mol. Microbiol. 37, 467–476. doi: 10.1046/j.1365-2958.2000.01986.x
Chiou, C. S., and Jones, A. L. (1993). Nucleotide sequence analysis of a transposon (Tn5393) carrying streptomycin resistance genes in Erwinia amylovora and other gram-negative bacteria. J. Bacteriol. 175, 732–740. doi: 10.1128/jb.175.3.732-740.1993
Di Pilato, V., Pollini, S., and Rossolini, G. M. (2014). Characterization of plasmid pAX22, encoding VIM-1 metallo-β-lactamase, reveals a new putative mechanism of In70 integron mobilization. J. Antimicrob. Chemother. 69, 67–71. doi: 10.1093/jac/dkt311
Di Pilato, V., Pollini, S., and Rossolini, G. M. (2015). Tn6249, a new Tn6162 transposon derivative carrying a double-integron platform and involved with acquisition of the blaV IM-1 metallo-β-lactamase gene in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 59, 1583–1587. doi: 10.1128/AAC.04047-4014
Docquier, J.-D., Riccio, M. L., Mugnaioli, C., Luzzaro, F., Endimiani, A., Toniolo, A., et al. (2003). IMP-12, a new plasmid-encoded metallo-β-lactamase from a Pseudomonas putida clinical isolate. Antimicrob. Agents Chemother. 47, 1522–1528. doi: 10.1128/aac.47.5.1522-1528.2003
Driscoll, J. A., Brody, S. L., and Kollef, M. H. (2007). The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa Infections. Drugs 67, 351–368. doi: 10.2165/00003495-200767030-200767033
García-Fernández, A., Fortini, D., Veldman, K., Mevius, D., and Carattoli, A. (2009). Characterization of plasmids harbouring qnrS1, qnrB2 and qnrB19 genes in Salmonella. J. Antimicrob. Chemother. 63, 274–281. doi: 10.1093/jac/dkn470
García-Fernández, A., Villa, L., Moodley, A., Hasman, H., Miriagou, V., Guardabassi, L., et al. (2011). Multilocus sequence typing of IncN plasmids. J. Antimicrob. Chemother. 66, 1987–1991. doi: 10.1093/jac/dkr225
Garcillán-Barcia, M. P., Francia, M. V., and de La Cruz, F. (2009). The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol. Rev. 33, 657–687. doi: 10.1111/j.1574-6976.2009.00168.x
Giani, T., Arena, F., Pollini, S., Di Pilato, V., D’Andrea, M. M., Henrici De Angelis, L., et al. (2018). Italian nationwide survey on Pseudomonas aeruginosa from invasive infections: activity of ceftolozane/tazobactam and comparators, and molecular epidemiology of carbapenemase producers. J. Antimicrob. Chemother. 73, 664–671. doi: 10.1093/jac/dkx453
Giani, T., Marchese, A., Coppo, E., Kroumova, V., and Rossolini, G. M. (2012). VIM-1-producing Pseudomonas mosselii isolates in Italy, predating known VIM-producing index strains. Antimicrob. Agents Chemother. 56, 2216–2217. doi: 10.1128/AAC.06005-11
Grant, J. R., and Stothard, P. (2008). The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 36, W181–W184. doi: 10.1093/nar/gkn179
Greated, A., Lambertsen, L., Williams, P. A., and Thomas, C. M. (2002). Complete sequence of the IncP-9 TOL plasmid pWW0 from Pseudomonas putida. Environ. Microbiol. 4, 856–871. doi: 10.1046/j.1462-2920.2002.00305.x
Guglielmini, J., Néron, B., Abby, S. S., Garcillán-Barcia, M. P., de la Cruz, F., and Rocha, E. P. C. (2014). Key components of the eight classes of type IV secretion systems involved in bacterial conjugation or protein secretion. Nucleic Acids Res. 42, 5715–5727. doi: 10.1093/nar/gku194
Haines, A. S., Jones, K., Cheung, M., and Thomas, C. M. (2005). The IncP-6 Plasmid Rms149 consists of a small mobilizable backbone with multiple large insertions. J. Bacteriol. 187, 4728–4738. doi: 10.1128/JB.187.14.4728-4738.2005
Hawkey, P. M., Warren, R. E., Livermore, D. M., McNulty, C. A. M., Enoch, D. A., Otter, J. A., et al. (2018). Treatment of infections caused by multidrug-resistant Gram-negative bacteria: report of the British society for antimicrobial chemotherapy/healthcare infection society/british infection association joint working party. J. Antimicrob. Chemother. 73, iii2–iii78. doi: 10.1093/jac/dky027
He, Z., Zhang, H., Gao, S., Lercher, M. J., Chen, W. H., and Hu, S. (2016). Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 44, W236–W241. doi: 10.1093/nar/gkw370
Høiby, N., Ciofu, O., and Bjarnsholt, T. (2015). “Pseudomonas,” in Manual of Clinical Microbiology, Eleventh Edn, eds J. Jorgensen, M. A. Pfaller, K. C. Carroll, G. Funke, M. L. Landry, S. Richter, et al. (Washington, DC: ASM Press).
Hong, D. J., Bae, I. K., Jang, I.-H., Jeong, S. H., Kang, H.-K., and Lee, K. (2015). Epidemiology and characteristics of metallo-β-lactamase-producing Pseudomonas aeruginosa. Infect. Chemother. 47, 81–97. doi: 10.3947/ic.2015.47.2.81
Johnson, J. L. (1994). “Similarity analysis of DNAs,” in Methods for General and Molecular Bacteriology, eds P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (Washington, DC: American Society for Microbiology), 655–682.
Johnson, T. J., Bielak, E. M., Fortini, D., Hansen, L. H., Hasman, H., Debroy, C., et al. (2012). Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid 68, 43–50. doi: 10.1016/j.plasmid.2012.03.001
Lauretti, L., Riccio, M. L., Mazzariol, A., Cornaglia, G., Amicosante, G., Fontana, R., et al. (1999). Cloning and characterization of blaV IM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 43, 1584–1590. doi: 10.1128/aac.43.7.1584
Li, H., Toleman, M. A., Bennett, P. M., Jones, R. N., and Walsh, T. R. (2008). Complete sequence of p07-406, a 24,179-base-pair plasmid harboring the blaV IM-7 metallo-β-lactamase gene in a Pseudomonas aeruginosa isolate from the United States. Antimicrob. Agents Chemother. 52, 3099–3105. doi: 10.1128/AAC.01093-1097
Liapis, E., Bour, M., Triponney, P., Jové, T., Zahar, J.-R., Valot, B., et al. (2019). Identification of diverse integron and plasmid structures carrying a novel carbapenemase among Pseudomonas Species. Front. Microbiol. 10:404. doi: 10.3389/fmicb.2019.00404
Llosa, M., and Alkorta, I. (2017). Coupling proteins in type IV secretion. Curr. Top. Microbiol. Immuno. 413, 143–168. doi: 10.1007/978-3-319-75241-9_6
Lombardi, G., Luzzaro, F., Docquier, J.-D., Riccio, M. L., Perilli, M., Coli, A., et al. (2002). Nosocomial infections caused by multidrug-resistant isolates of Pseudomonas putida producing VIM-1 metallo-β-Lactamase. J. Clin. Microbiol. 40, 4051–4055. doi: 10.1128/JCM.40.11.4051-4055.2002
Martinez, E., Marquez, C., Ingold, A., Merlino, J., Djordjevic, S. P., Stokes, H. W., et al. (2012). Diverse mobilized class 1 integrons are common in the chromosomes of pathogenic Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 56, 2169–2172. doi: 10.1128/AAC.06048-6011
Molina, L., Udaondo, Z., Duque, E., Fernández, M., Molina-Santiago, C., Roca, A., et al. (2014). Antibiotic resistance determinants in a Pseudomonas putida strain isolated from a Hospital. PLoS One 9:e81604. doi: 10.1371/journal.pone.0081604
Ocampo-Sosa, A. A., Guzmán-Gómez, L. P., Fernández-Martínez, M., Román, E., Rodríguez, C., Marco, F., et al. (2015). Isolation of VIM-2-producing Pseudomonas monteilii clinical strains disseminated in a tertiary Hospital in northern Spain. Antimicrob. Agents Chemother. 59, 1334–1336. doi: 10.1128/AAC.04639-14
Orlek, A., Phan, H., Sheppard, A. E., Doumith, M., Ellington, M., Peto, T., et al. (2017). A curated dataset of complete Enterobacteriaceae plasmids compiled from the NCBI nucleotide database. Data Brief 12, 423–426. doi: 10.1016/j.dib.2017.04.024
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T. G., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. doi: 10.1093/bioinformatics/btv421
Partridge, S. R., Kwong, S. M., Firth, N., and Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31, e88–17. doi: 10.1128/CMR.00088-17
Peix, A., Ramírez-Bahena, M.-H., and Velázquez, E. (2018). The current status on the taxonomy of Pseudomonas revisited: an update. Infect. Genet. Evol. 57, 106–116. doi: 10.1016/j.meegid.2017.10.026
Perez, F., Hujer, A. M., Marshall, S. H., Ray, A. J., Rather, P. N., Suwantarat, N., et al. (2014). Extensively drug-resistant Pseudomonas aeruginosa isolates containing blaV IM-2 and elements of Salmonella genomic island 2: a new genetic resistance determinant in Northeast Ohio. Antimicrob. Agents Chemother. 58, 5929–5935. doi: 10.1128/AAC.02372-2314
Roy Chowdhury, P., Scott, M., Worden, P., Huntington, P., Hudson, B., Karagiannis, T., et al. (2016). Genomic islands 1 and 2 play key roles in the evolution of extensively drug-resistant ST235 isolates of Pseudomonas aeruginosa. Open Biol. 6:150175. doi: 10.1098/rsob.150175
Rozwandowicz, M., Brouwer, M. S., Zomer, A. L., Bossers, A., Harders, F., Mevius, D. J., et al. (2017). Plasmids of Distinct IncK Lineages Show Compatible Phenotypes. Antimicrob. Agents Chemother. 61, e1954–16. doi: 10.1128/AAC.01954-1916
San Millan, A., Toll-Riera, M., Escudero, J. A., Cantón, R., Coque, T. M., and Craig MacLean, R. (2015). Sequencing of plasmids pAMBL1 and pAMBL2 from Pseudomonas aeruginosa reveals a blaV IM-1 amplification causing high-level carbapenem resistance. J. Antimicrob. Chemother. 70, 3000–3003. doi: 10.1093/jac/dkv222
Sevastsyanovich, Y. R., Krasowiak, R., Bingle, L. E. H., Haines, A. S., Sokolov, S. L., Kosheleva, I. A., et al. (2008). Diversity of IncP-9 plasmids of Pseudomonas. Microbiology 154, 2929–2941. doi: 10.1099/mic.0.2008/017939-17930
Shintani, M., Sanchez, Z. K., and Kimbara, K. (2015). Genomics of microbial plasmids: classification and identification based on replication and transfer systems and host taxonomy. Front. Microbiol. 6:242. doi: 10.3389/fmicb.2015.00242
Smillie, C., Garcillan-Barcia, M. P., Francia, M. V., Rocha, E. P. C., and de la Cruz, F. (2010). Mobility of Plasmids. Microbiol. Mol. Biol. Rev. 74, 434–452. doi: 10.1128/MMBR.00020-10
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Sunde, M., and Norström, M. (2005). The genetic background for streptomycin resistance in Escherichia coli influences the distribution of MICs. J. Antimicrob. Chemother. 56, 87–90. doi: 10.1093/jac/dki150
Suzuki, H., Yano, H., Brown, C. J., and Top, E. M. (2010). Predicting plasmid promiscuity based on genomic signature. J. Bacteriol. 192, 6045–6055. doi: 10.1128/JB.00277-210
Vilacoba, E., Quiroga, C., Pistorio, M., Famiglietti, A., Rodríguez, H., Kovensky, J., et al. (2014). A blaV IM-2 plasmid disseminating in extensively drug-Resistant clinical Pseudomonas aeruginosa and Serratia marcescens isolates. Antimicrob. Agents Chemother. 58, 7017–7018. doi: 10.1128/AAC.02934-14
Villa, J., Viedma, E., Brañas, P., Orellana, M. A., Otero, J. R., and Chaves, F. (2014). Multiclonal spread of VIM-1-producing Enterobacter cloacae isolates associated with In624 and In488 integrons located in an IncHI2 plasmid. Int. J. Antimicrob. Agents 43, 451–455. doi: 10.1016/j.ijantimicag.2014.02.006
Villa, L., García-Fernández, A., Fortini, D., and Carattoli, A. (2010). Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J. Antimicrob. Chemother. 65, 2518–2529. doi: 10.1093/jac/dkq347
Keywords: metallo-β-lactamase, mobile genetic elements, integron, antibiotic resistance, plasmid
Citation: Di Pilato V, Antonelli A, Giani T, Henrici De Angelis L, Rossolini GM and Pollini S (2019) Identification of a Novel Plasmid Lineage Associated With the Dissemination of Metallo-β-Lactamase Genes Among Pseudomonads. Front. Microbiol. 10:1504. doi: 10.3389/fmicb.2019.01504
Received: 08 April 2019; Accepted: 14 June 2019;
Published: 02 July 2019.
Edited by:
Raffaele Zarrilli, University of Naples Federico II, ItalyReviewed by:
Remy A. Bonnin, Université Paris-Saclay, FranceCopyright © 2019 Di Pilato, Antonelli, Giani, Henrici De Angelis, Rossolini and Pollini. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Simona Pollini, c2ltb25hLnBvbGxpbmlAdW5pZmkuaXQ=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.