- 1State Key Laboratory of Crop Stress Biology for Arid Areas, College of Life Sciences, Northwest A&F University, Yangling, China
- 2Shaanxi Key Laboratory of Agricultural and Environmental Microbiology, College of Life Sciences, Northwest A&F University, Yangling, China
- 3College of Life Sciences, Qufu Normal University, Qufu, China
Burkholderia thailandensis is a model organism for human pathogens Burkholderia mallei and Burkholderia pseudomallei. The study of B. thailandensis peroxiredoxin is helpful for understanding the survival, pathogenic infection, and antibiotic resistance of its homologous species. Alkyl hydroperoxide reductase subunit C (AhpC) is an important peroxiredoxin involved in oxidative damage defense. Here, we report that BthAhpC exhibits broad specificity for peroxide substrates, including inorganic and organic peroxides and peroxynitrite. AhpC catalyzes the reduction of oxidants using the N-terminal conserved Cys57 as a peroxidatic Cys and the C-terminal conserved Cys171 and Cys173 as resolving Cys. These three conserved Cys residues play critical roles in the catalytic mechanism. AhpD directly interacts with AhpC as an electron donor, and the conserved Cys residues in active site of AhpD are important for AhpC reduction. AhpC is directly repressed by OxyR as shown by identifying the OxyR binding site in the ahpC promoter with a DNA binding assay. This work sheds light on the function of AhpC in the peroxides and peroxynitrite damage response in B. thailandensis and homologous species.
Introduction
Obligate aerobes inescapably encounter endogenous sources or exogenous sources reactive oxygen species (ROS) like hydrogen peroxides (H2O2), alkyl hydroperoxide [ROOH, cumene hydroperoxide (CHP), and t-butyl hydroperoxide (t-BOOH)], the superoxide anion () and the hydroxyl radical (HO•) (Halliwell and Gutteridge, 1984; Chandra et al., 2000). Excess ROS can cause cell damage to lipids, nucleic acids, proteins, or metal cofactors (den Hengst and Buttner, 2008). Enzymatic and non-enzymatic systems are used to counteract ROS toxicity (Liochev, 2013). The peroxiredoxins are large peroxidase family found in most living organisms, and is responsible for antioxidant defense (Guimaraes et al., 2005). Alkyl hydroperoxide reductase subunit C (AhpC) is an important member of peroxiredoxin family that is widely conserved in prokaryotes.
AhpC plays a role in bacterial physiology involving ROS and reactive nitrogen species (RNI) resistance, biofilm formation, survival, persistence, and colonization (Loprasert et al., 2003; Cosgrove et al., 2007; Kimura et al., 2012; Oh and Jeon, 2014). AhpC belongs to the typical 2-Cys peroxiredoxins, important antioxidant enzymes for bacterial defense against oxidative damage caused by ROS. There are two or three conserved cysteine residues critical for enzymatic activity in the AhpC N-terminus and C-terminus (Bryk et al., 2002). AhpCs form a homodimer to catalyze the reduction of peroxides and peroxynitrite via the N-terminal conserved Cys (the peroxidatic Cys, CP) to form the intermediate cysteine sulfenic acid (Cys-SOH). A C-terminal conserved Cys (the resolving cysteine, CR) on the other AhpC in the dimer attacks the cysteine sulfenic acid to generate a stable disulfide bond and release water (Ellis and Poole, 1997; Guimaraes et al., 2005). In Mycobacterium tuberculosis, there is one conserved Cys residue as CP in the N-terminus, and two CR residues in the C-terminus in which another disulphide is to be attacked by external thiol (Wong et al., 2017). To complete the catalytic cycle, the oxidized AhpCs are reduced by alkyl hydroperoxide reductase AhpD, AhpF, or thioredoxin C (TrxC) (Bryk et al., 2002; Jaeger et al., 2004; Koshkin et al., 2004; Wong et al., 2017). In Mycobacteria, AhpC is reduced by AhpD instead of AhpF. AhpD recycles AhpC by linking dihydrolipoamide dehydrogenase (Lpd) and dihydrolipoamide succinyltransferase (SucB) to transfer electrons from NADH to AhpC. The study of AhpC is helpful for understanding of bacteria and useful for developing cancer prevention strategies in humans (Guo et al., 2017).
Transcription regulator OxyR plays important roles in redox sensing and protein expression regulation in response to oxidative stress. OxyR has a conserved Helix-Turn-Helix (HTH) DNA binding motif at the N-terminus and an oligomerization motif at the C-terminus, allowing it to bind to the promoter of target genes as a tetramer (Zheng et al., 1998). The reduced OxyR remains inactive and becomes active when oxidized to regulate target gene transcription, upon exposure to oxidative stress (Pomposiello and Demple, 2001). OxyR regulates more than 20 kinds of antioxidant genes like alkyl hydroperoxide reductase (ahpCF), hydroperoxidase I (katG), glutathione reductase (gorA), and glutaredoxin 1 (grxA) and others (den Hengst and Buttner, 2008). OxyR was reported to regulate ahpC expression in several bacteria species like E. coli, M. marinum, M. tuberculosis, and S. coelicolor (Pagan-Ramos et al., 1998; den Hengst and Buttner, 2008). The regulation of ahpC expression by OxyR is a field of active research.
Burkholderia thailandensis is a non-pathogenic gram-negative bacterium commonly used as a model organism for the related human pathogens Burkholderia mallei and Burkholderia pseudomallei (Haraga et al., 2008). B. mallei and B. pseudomallei are potential biological weapons because they can cause the highly contagious even fatal diseases, glanders, and melioidosis (Wiersinga et al., 2006; Saikh and Mott, 2017). B. thailandensis is studied as a model organism because of its non-pathogenic characteristics and relatively easy genetic manipulation tools (Garcia, 2017). AhpC is reported to be involved in ROS and RNI defense in B. pseudomallei and even in T cell immunity in pathogenic processes (Loprasert et al., 2003; Reynolds et al., 2015). Although many efforts have been made to the research of these bacteria, there are still many areas that must be explored.
We previously reported work on the manganese scavenging and oxidative stress response mediated by type VI secretion system in B. thailandensis (Si et al., 2017c). It was found that ahpC (BTH_I2092) played a role in protecting against oxidative stress in B. thailandensis. Here, we describe the following detailed work on the function of AhpC in vivo and in vitro. AhpC catalyzes the reduction of hydroperoxides and peroxynitrite in vitro and improves the defense activity of B. thailandensis against hydroperoxides and peroxynitrite. The conserved Cys57 is the peroxidatic cysteine residue (CP) that is critical for the AhpC activity like in other homologs. Cys171 and Cys173 work as resolving cysteine residues (CR) that play almost equal roles and could functionally substitute each other. The conserved Cys residues of the electron donor AhpD are important for the reducing AhpC. Thus, AhpC plays an important role in defending B. thailandensis from oxidative damages caused by ROS.
Materials and Methods
Bacterial Strains and Growth Conditions
All strains and plasmids used in this study are listed in Supplementary Table S1. The B. thailandensis E264 derivatives were mutations of the B. thailandensis E264 strain (Zhao and Shao, 2015). The B. thailandensis strains were cultured on LB plates or in LB medium in a rotary shaker (220 rpm) at 37°C.
Plasmid Construction
Genes encoding B. thailandensis E264 AhpC (BTH_I2092), dihydrolipoamide dehydrogenases Lpd (BTH_I2554), dihydrolipoamide acyltransferase SucB (BTH_I1865), TrxC (BTH_I2218), and thioredoxin reductase TrxR (BTH_I1560) were amplified using PCR with B. thailandensis E264 genomic DNA as the template. These DNA fragments were digested using corresponding restriction enzymes and cloned into pET-28a vectors to construct the plasmids pET-28a-ahpC, pET-28a-lpd, pET-28a-sucB, pET-28a-trxC, and pET-28a-trxR.
To prepare the ahpC (BTH_I2092) in-frame deletion mutant of B. thailandensis, pDM4-pheS-ΔahpC was constructed. The primer pairs ahpC-M1F SpeI/ahpC-M1R and ahpC-M2F/ahpC-M2R BglII were used to amplify the ahpC upstream and downstream fragments with B. thailandensis genomic DNA as the PCR template. The primer pair ahpC-M1F SpeI/ahpC-M2R BglII was used for overlap PCR to fuse the upstream and downstream fragments together. Finally, the PCR product was digested using SpeI and BglII and inserted into pDM4-phes to create the plasmid pDM4-pheS-ΔahpC. pME6032-ahpC was constructed to complement the ahpC mutant. The primers ahpC-F BamHI and ahpC-R XhoI were used to amplify the ahpC gene from B. thailandensis genomic DNA. The ahpC gene was digested by BamHI and XhoI and ligated into pME6032 and pGEX-6P-1, obtaining the plasmids pME6032-ahpC and pGEX-6P-1-ahpC, respectively. The point mutant ahpCC57S was created using overlap PCR (Zhang et al., 2013) to substitute a Ser residue for the Cys residue at position 57 of ahpC. The primer pairs ahpC-F BamHI/ahpCC57S R and ahpCC57S F/ahpC-R XhoI were used to amplify the upstream and downstream fragments of ahpC. The full-length ahpCC57S was obtained using overlap PCR. The other site-directed mutants such as ahpCC171S, ahpCC173S, ahpCC57SC171S, ahpCC57SC173S, and ahpCC171SC173S were created the same way. The ahpC and its mutant fragments were digested using BamHI and XhoI, and ligated into pET-28a to obtain the expression vectors like pET-28a-ahpC. The point mutants ahpDC131S and ahpDC134S were obtained using overlap PCR in the same way. The primer pairs are listed in Supplementary Table S2. ahpD (BTH_I2091) and its mutant fragments were then digested using BamHI and XhoI, and ligated into pET-21a to obtain the expression vectors pET-21a-ahpD, pET-21a-ahpDC131S, and pET-21a-ahpDC134S.
In-Frame Deletion and Complementation Mutant Construction in B. thailandensis
The in-frame deletion and complementation mutant construction in B. thailandensis was performed as described previously (Si et al., 2017c). The pDM4-pheS derivatives were transformed into B. thailandensis using E. coli SM10(λ pir)-mediated conjugational mating to carry out a single crossover. The transconjugants were plated on LB agar with chloramphenicol and streptomycin. The single clones were assayed using PCR. For complementation, the pME6032 derivatives were transformed into the ΔahpC mutant using electroporation. The expression was induced using 1 mM IPTG.
Sensitivity Assay
A stationary phase strain was diluted 30-fold into M9 medium containing 0.25 mM CHP, 1 mM H2O2, 1 mM t-BOOH, or 20 mM 3-Morpholinosydnonimine (SIN-1) (Aladdin). After 30 min of treatment at 37°C, the culture was diluted to 10−4 and plated on LB agar. After 16 h of cultivation at 37°C, the colonies were counted and the survival percentage was calculated as described previously (Wang et al., 2015; Si et al., 2017b).
Measurement of Intracellular ROS
The fluorescent reporter dye 2′,7′-dichlorodihydrofluorescein diacetate (DCFHDA) (Invitrogen) was used to detect intracellular ROS as previously described (Si et al., 2014, 2017c). 1 mL of culture was collected, washed in PBS and resuspended in 1 mL PBS with 10 μM DCFHDA. The sample was incubated in the dark at 37°C for 40 min, pelleted and washed in PBS twice. 1 mM H2O2, 0.25 mM CHP, or 0.3 mM t-BOOH was added to the sample resuspended in LB and incubated for 30 min at 37°C. The cells were pelleted, washed in PBS and resuspended in 400 μL PBS. 200 μL of the resuspended sample was transferred into a dark 96-well plate. Fluorescence signals were measured using a SpectraMax M2 Plate Reader (Molecular Devices) with excitation/emission wavelengths of 495/520 nm.
Protein Expression and Purification
To obtain the His6-tagged and GST-tagged recombinant proteins, the pET-21a, pET-28a, and pGEX-6P-1 expression vectors were transformed into the E. coli BL21(DE3) host strain as described previously (Shen et al., 2005; Si et al., 2014, 2015a). The bacteria were cultured in 500 mL LB medium at 37°C to an OD of about 0.3, cooled to 26°C, and induced to express with 0.5 mM IPTG at 26°C for 8 h. The cell pellet was harvested, resuspended in 30 mL PBS and lysed using ultrasonication. After centrifugation, the supernatant was loaded onto 1 mL pre-equilibrated Ni-NTA (Qiagen) or the GST-Bind resin (Novagen). 5 column volume washing buffer was loaded onto Ni-NTA or GST-Bind resin to wash and elution buffer with 0.5 M imidazole or 10 mM glutathione (GSH) was used to elute the recombinant protein. The His6 tag was cleaved by adding 10 units of Enterokinase-Max (Invitrogen, Karlruhe, Germany) and incubating at 4°C overnight to conduct subsequent enzyme activity analysis. Ni-NTA agarose was used to remove the cleaved tag and uncleaved protein from the tag-free protein. All enzymes were purchased from Sigma-Aldrich (St. Louis, MO, United States). The pooled protein was determined using SDS-PAGE to be over 90% homogeneous, and its concentration was determined using Bradford assay according to the manufacturer’s instructions (Bio-Rad) with BSA as the standard.
Enzymatic Activity Assay of AhpC
As reported previously, Lpd, SucB, AhpD, and AhpC together sustained peroxide-dependent oxidation of NADH. The enzymatic activity of AhpC was determined by monitoring the consumption of NADH at 340 nm (Bryk et al., 2002; Si et al., 2015a,b, 2017a). The Trx reaction system was the same as described previously (Wong et al., 2017). The reaction mixture contained 100 mM KPi (pH 7.0), 1 mM EDTA, 150 μM NADH, 1 μM AhpC, 40 μM AhpD, 2 μM SucB, 2 μM Lpd and peroxides (1 mM H2O2 or 0.5 mM t-BOOH or CHP); or contained 50 mM Tris–HCl buffer (pH 8.0), 2 mM EDTA, 250 μM NADPH, 1 μM AhpC, 1 mM peroxides, and the reduced Trx-generating system [5 μM thioredoxin reductase (TrxR) and 40 μM TrxC]. After 5 min reaction, peroxides were added into the reaction mixture and then the NADH or NADPH consumption was monitored by the absorbance at 340 nm. The catalytic kinetic parameters for one substrate were acquired by changing its concentration in the case of saturating concentrations of the other substrate (between 0 and 500 μM for AhpD or TrxC, and between 0 and 1 mM for peroxide). The activity was determined after subtracting the spontaneous reduction rate detected without AhpC, and the micromoles amount of NADPH or NADH oxidized per second per micromolar of enzyme (i.e., turnover number, s−1) was calculated using the molar absorption coefficient of NADPH or NADH at 340 nm (ε340) of 6220 M−1 cm−1. Three independent experiments were performed at each substrate concentration. The kcat and Km values of AhpC were acquired from a non-linear fit to the Michaelis–Menten equation using the program GraphPad Prism 5.
Determination of the Peroxynitrite Reductase Activity of AhpC
The reaction rate of AhpC with peroxynitrite was examined using a competition approach by measuring the inhibitor effect of increasing AhpC concentrations on peroxynitrite-mediated HRP oxidation to compound I in a stopped-flow spectrophotometer (Applied Photophysics Ltd., United Kingdom) under our experimental conditions (100 mM phosphate buffer containing 0.1 mM DTPA, pH 7.4, and 25 ± 0.2°C) as previously described (Hayashi and Yamazaki, 1979; Floris et al., 1993; Ogusucu et al., 2007; Trujillo et al., 2008; Hugo et al., 2009; Manta et al., 2009; Demicheli et al., 2016; Reyes et al., 2016). In brief, 1 μM peroxynitrite-mediated HRP (5 μM) oxidation to Compound I in the absence or presence of increasing reduced AhpC concentrations (from 0 to 10 μM) was evaluated at 398 nm (Δε398 = 4.2 × 104 M−1 cm−1). The rate constant of peroxynitrite-mediated AhpC oxidation was calculated from HRP-Compound I yield obtained at different AhpC concentrations (0–10 μM) as previously described (Ogusucu et al., 2007; Hugo et al., 2009; Manta et al., 2009).
Protein Sulfenic Acid Analysis
To analyze the reaction intermediate Cys-SOH content, NBD chloride was used to label the site-directed mutant AhpCC57SC171S, AhpCC57SC173S, and AhpCC171SC173S as previously described (Baker and Poole, 2003; Si et al., 2015c). Each mutant protein (50 μM) was incubated with 5 mM DTT for 30 min. The redundant DTT was removed by ultrafiltration. The protein sample was then made anaerobic by flushing with argon gas for 30 min and separated into two parts. One was treated with 50 μM anaerobic CHP and the other kept as a control. The sample was incubated with 5 mM anaerobic NBD chloride for 30 min at 26°C in the dark. After removing excess NBD chloride by ultrafiltration, the protein sample was analyzed spectrophotometrically at 200–600 nm.
Protein Thiol Content Analysis
Free sulfhydryl groups in AhpC variants were determined using 5, 5′-dithio-bis (2-nitrobenzoic acid) (DTNB) as previously described (Ellman, 1959; Si et al., 2014, 2015c, 2017c). Proteins (25 μM) were treated with 5 mM CHP or 50 mM DTT at 26°C for 30 min. DTT or CHP was removed by ultrafiltration. The resulting proteins (10 μM) were added to DTNB (2 mM) in 50 mM Tris–HCl buffer (pH8.0) and the absorbance at 412 nm was measured against a 2 mM DTNB solution as the reference. The amounts of reactive sulfhydryl groups were determined using the molar absorption coefficient of TNB at 412 nm of 13,600 M−1s−1 (Eyer et al., 2003).
Electrophoretic Mobility Shift Assay (EMSA)
Electrophoretic mobility shift assay was performed using the method by Si et al. (2017c). To reduce non-specific binding in the EMSA assay, the ahpC promoter region (PahpC, 231 bp) containing the predicted OxyR binding site was amplified from the ahpC promoter region using primers PahpC-F/PahpC-R. Increasing concentrations of purified OxyR (0–16 μg) are incubated with 20 ng ahpC DNA promoter in EMSA buffer containing 20 mM Tris–HCl, pH7.4, 4 mM MgCl2, 100 mM NaCl, 1 mM DTT, and 10% glycerol. The binding reaction mixture is incubated at room temperature for 30 min, subjected to electrophoresis on a 6% native polyacrylamide gel with 5% glycerol in 0.5 × TBE electrophoresis buffer, and detected using SYBR Green. A 230 bp fragment from the ahpC coding region amplified with primers Control-F and Control-R instead of the 231 bp ahpC promoter in the binding assays was used as a negative control.
GST Pull-Down Assay
The GST pull-down assay was performed as previously described with minor modifications (Xu et al., 2014; Si et al., 2017b). Briefly, purified GST-AhpC was mixed with His6-AhpD in PBS on a rotator for 2 h at 4°C, and GST was used as a negative control. After adding 30 μL of prewashed glutathione beads slurry, binding proceeded for another 2 h at 4°C. The beads were then washed five times with TEN buffer (50 mM Tris–HCl, pH 8.0, 500 mM NaCl). Retained proteins were detected using immunoblot after SDS-PAGE with the anti-His antibody (Millipore).
Statistical Analysis
Student’s t-test was used to test for statistical significance. Statistical analyses were performed using GraphPad Prism Software (GraphPad Software, San Diego, CA, United States).
Results
The Antioxidant Function of AhpC in B. thailandensis
To examine the antioxidant functions of AhpC in B. thailandensis, the sensitivities of the wild type, the ΔahpC mutant, and the complementary strain ΔahpC(ahpC) to CHP (0.25 mM), H2O2 (1 mM), and t-BOOH (0.3 mM) were tested (Figure 1A). Concentrations of peroxides applied could reduce the survival rate of the wild type and increase the mortality rate from 50 to 70% (Supplementary Figure S1A). Although deletion of ahpC did not affect bacterial growth under normal conditions (Supplementary Figure S1B), the survival rates of the ΔahpC mutant decreased by about 31.4–52.6% compared to wild type. However, the oxidative stress sensitive phenotype of the ΔahpC mutant recovered to the wild type level after complementation with the ahpC gene. In addition, deletion of ahpC was shown to affect the survival rate of B. thailandensis in response to peroxynitrite (Supplementary Figure S1C).
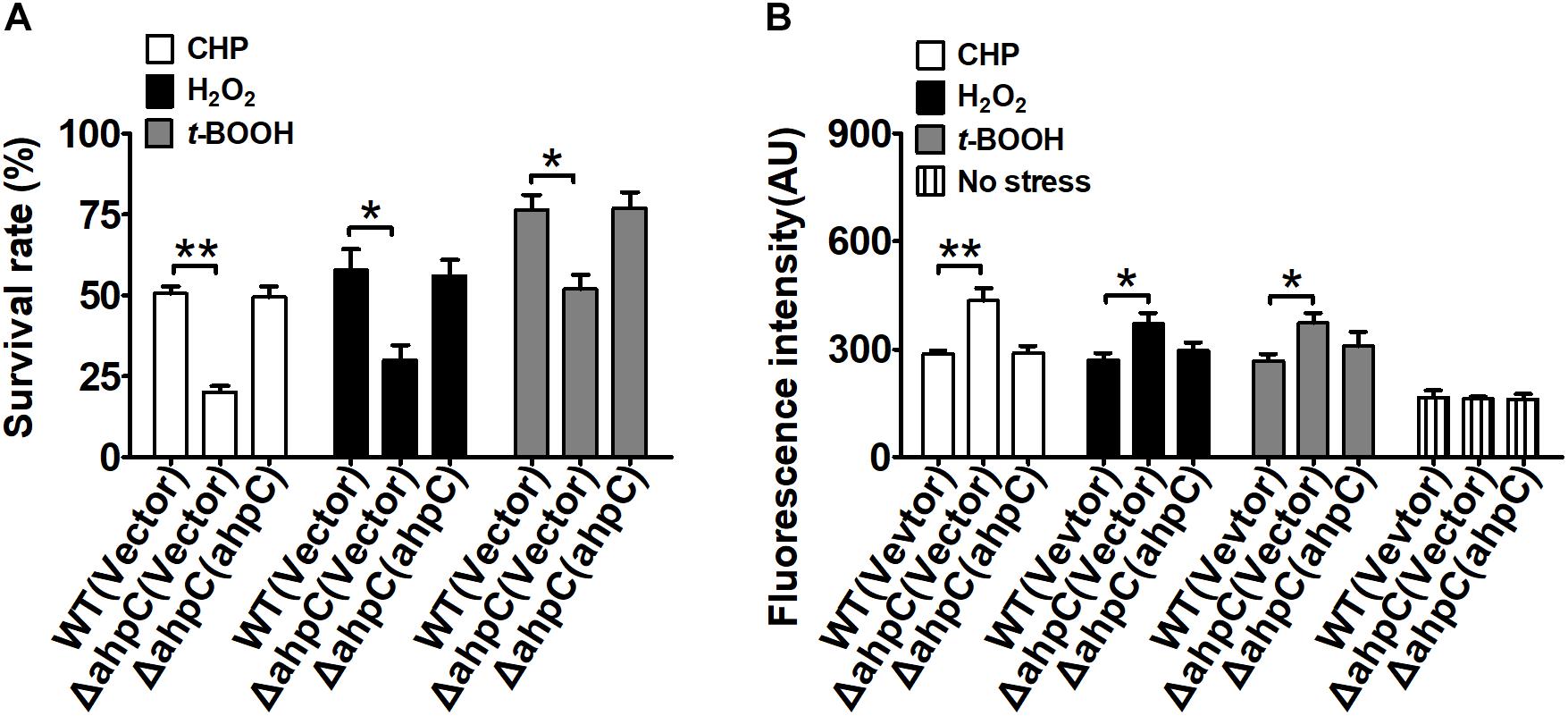
Figure 1. ahpC was required for cellular resistance to toxic agent-induced stresses. (A) The B. thailandensis WT(Vector), ΔahpC(Vector), and ΔahpC(ahpC) strains were exposed to H2O2 (1 mM), CHP (0.25 mM), and t-BOOH (0.3 mM) at 37°C for 30 min. The mutant lacking ahpC was sensitive to oxidative stress. Cell survival percentage was measured by viability assay. (B) The B. thailandensis WT(Vector), ΔahpC(Vector), and ΔahpC(ahpC) strains were exposed to H2O2 (1 mM), CHP (0.25 mM), and t-BOOH (0.3 mM). The ROS levels in B. thailandensis were measured using the DCF fluorescence determination assay after exposure to H2O2 (1 mM), CHP (0.25 mM), and t-BOOH (0.3 mM). The asterisk indicates a significant difference (∗p < 0.05; ∗∗p < 0.01, using the Student t-test).
Excess intracellular ROS is harmful in bacteria. To investigate the function of AhpC in reducing ROS under oxidative stress, the intracellular ROS levels were determined using the membrane-permeable fluorogenic probe 2′,7′-dichlorodihydrofluorescein diacetate (DCFHDA). As shown in Figure 1B, the ROS levels of all three strains under oxidative stress were higher than those of the unstressed strains, indicating ROS induction by oxidative stress in the strains. Moreover, the ΔahpC mutant had a significantly higher ROS level than the wild type after oxidative stress treatment. However, the ROS level of the ΔahpC mutant was completely restored to that of the wild type by complementation (Figure 1B). These results indicate that AhpC plays important roles in counteracting oxidative stress by reducing ROS in B. thailandensis.
The Peroxide Reductase Activity of AhpC in vitro
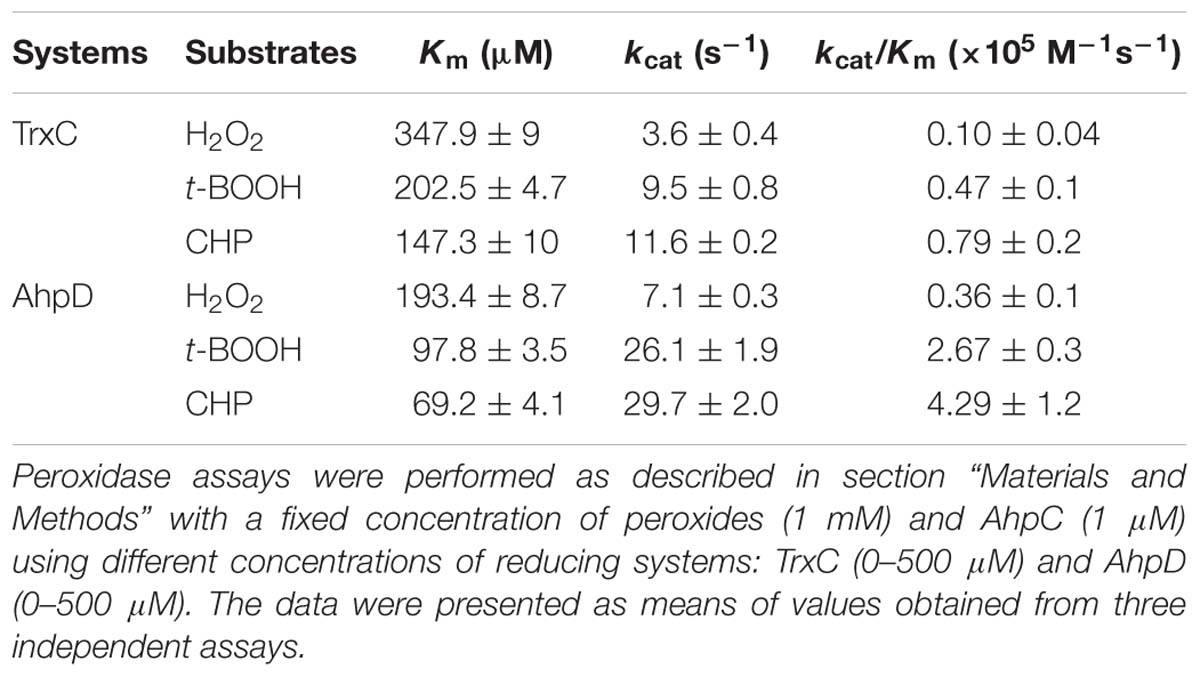
It was reported that AhpC metabolize peroxides and peroxynitrite via a conserved N terminal cysteine residue that is oxidized to form a disulfide bond. To complete the catalytic cycle, the Cys residue must be reduced. AhpC can be regenerated by the reducing agents Trxc and AhpD. Thus, to examine whether the in vitro peroxide reductase activity of B. thailandensis AhpC was supported by the AhpD system (Lpd, SucB, AhpD, and NADH) and Trx system (TrxC, TrxR, and NADPH), we measured the catalytic constants of B. thailandensis AhpC with TrxC or AhpD as recycling reductants under steady-state conditions at saturating concentrations of peroxides (1 mM) and varying concentrations of reductants (0–500 μM). As shown in Table 1, the apparent affinity of B. thailandensis AhpC toward AhpD obtained under steady-state conditions was, higher than the value determined with TrxC. The kcat values of AhpC-mediated CHP reduction with the TrxC and AhpD systems were 11.6 ± 0.2 s−1 and 29.7 ± 2 s−1, respectively; and the respective Km values were 147.3 ± 10 μM and 69.2 ± 4.1 μM, respectively. The catalytic efficiencies were 0.79 ± 0.2 × 105 M−1 s−1 and 4.29 ± 1.2 × 105M−1 s−1, respectively. So AhpD and TrxC were active with an apparent efficiency order AhpD > TrxC. Based on our biochemical studies of AhpC, we speculated that AhpC-dependent peroxide defense was supported primarily by NADH because of the preferred selectivity of AhpD for this cofactor. Moreover, AhpC showed a small preference for the larger hydroperoxides.
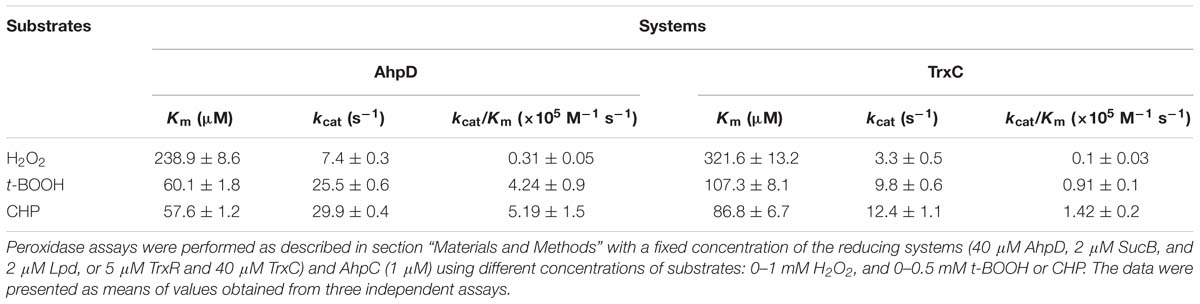
Next, enzymatic activities of B. thailandensis AhpC toward various oxidizing substrates were analyzed using the TrxC and AhpD system as the reducing power (Table 2). Results showed that B. thailandensis AhpC had strong reductase activity in treating peroxide H2O2, t-BOOH and CHP (the catalytic efficiencies kcat/Km in AhpD system were 0.31 ± 0.05 × 105 M−1s−1, 4.24 ± 0.9 × 105 M−1s−1, and 5.19 ± 1.5 × 105 M−1s−1, respectively; 0.1 ± 0.03 × 105 M−1 s−1, 0.91 ± 0.1 × 105 M−1 s−1, and 1.42 ± 0.2 × 105 M−1 s−1in TrxC system, respectively). The relatively high reactivity of AhpC with multiple substrates, at 105∼106 M−1 s−1, supports the view that it acts as a potent antioxidant defense across a range of peroxide substrates. Apart from these peroxides, peroxynitrite was also reported as the substrate of AhpC in M. tuberculosis, S. typhimurium, and H. pylori (Bryk et al., 2000). The capacity of AhpC to reduce peroxynitrite was analyzed using competition approaches as described (Ogusucu et al., 2007). 1 μM peroxynitrite in 10 mM NaOH was mixed with 5 μM HRP in the absence of or with varying concentrations of AhpC in 100 mM sodium phosphate buffer (pH 7.4) at 25°C. As shown in Figure 2A, mixing HRP (5 μM) with peroxynitrite (1 μM) without AhpC led to the formation of HRP-Compound I. However, addition of AhpC caused a decrease in HRP-Compound I formation yield. Kinetic analysis of the data indicated that the second order rate constant of peroxynitrite reduction by AhpC was (2.7 ± 0.53) × 106 M−1 s−1 at pH 7.4 and 25°C (Figure 2B), which was similar to those reported for other AhpCs (Bryk et al., 2000) Nature, or Prxs (Floris et al., 1993; Ogusucu et al., 2007; Manta et al., 2009; Reyes et al., 2016) in stopped-flow experiments.
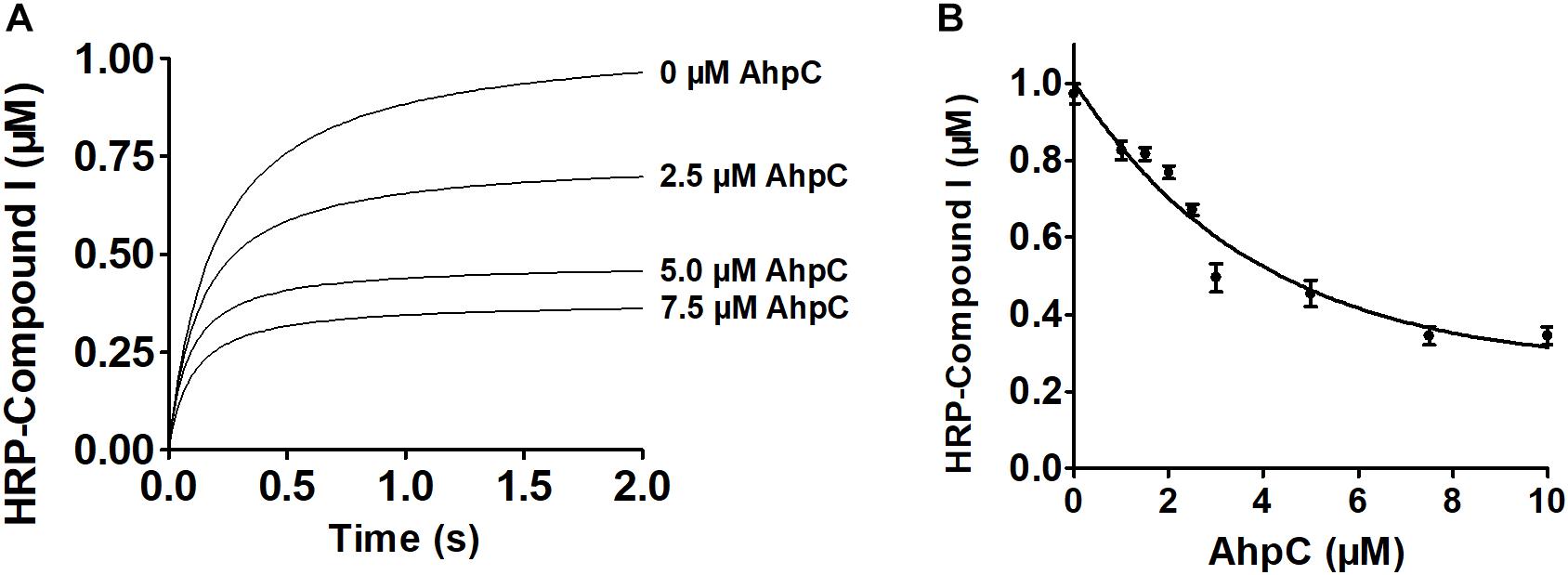
Figure 2. Kinetics of peroxynitrite reduction by AhpC. (A) Peroxynitrite (1 μM) in 10 mM NaOH was rapidly mixed with HRP (5 μM) with or without increasing concentrations of AhpC (0.0, 2.5, 5.0, 7.5 μM) in 100 mM sodium phosphate buffer (pH 7.4) at 25°C using a SX20 Stopped-Flow Spectrometer. (B) HRP-Compound I concentration formed was plotted compared to AhpC concentration. The continuous line represented HRP-Compound I yields simulated according to a simple competition system using the program GraphPad Prism 5, and the rate constant of AhpC oxidation was calculated.
The Function of Conserved Cysteine Residues in AhpC
Typical peroxiredoxin protein family members possess two conserved Cys residues, and mycobacterial AhpCs employ three Cys residues involved in catalysis (Guimaraes et al., 2005). The amino acid sequence alignment of AhpC homologs indicated that B. thailandensis AhpC possesses three conserved Cys residues: Cys57, Cys171, and Cys173. To investigate the function of Cys in AhpC, the site-directed mutations AhpCC57S, AhpCC171S, AhpCC173S, AhpCC171SC173S were produced, which substituted Ser residue for a Cys residue. The peroxidase activities of these proteins were measured in the presence of H2O2, t-BOOH and CHP. As shown in Figure 3A, the single mutant of Cys57 or double mutant of Cys171 and Cys173 resulted in a loss of AhpC catalytic activity. However, single mutant of Cys171 or Cys173 showed almost no obvious effect on the activity. Similar results were also observed when the Trx system was used as a reducing power. The result indicated that AhpC could be reduced by both AhpD and TrxC, and Cys171 and Cys173 could substitute each other in function. To examine the intracellular function of conserved Cys residues in AhpC, the sensitivity assay of B. thailandensis strains with ahpC in-frame deletion or serial mutant complementation were performed using H2O2 treatment. The result confirmed that ahpCC57S or ahpCC57SC171S mutant gene complementation almost could not increase the survival rate of the respective strains, but the ahpCC57S or ahpCC57S complementation could nearly recover the survival rate (Figure 3B).
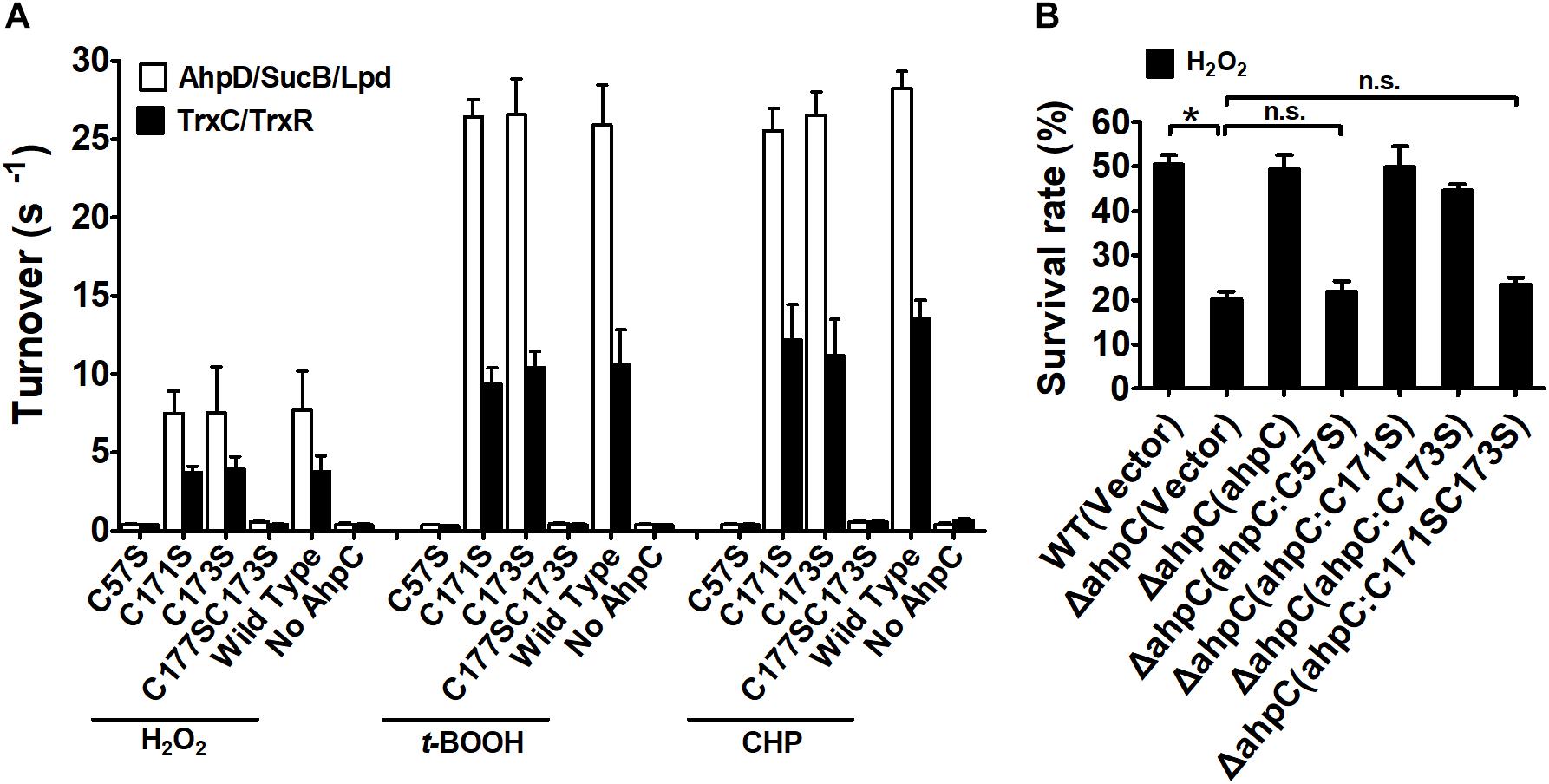
Figure 3. Analysis of the properties of AhpC point mutants. (A) Peroxidase activity of AhpC and the mutants in conserved Cys residues examined by AhpD/SucB/Lpd reaction system and TrxC reaction system. Peroxidase assays were performed as described in the section “Materials and Methods” using a fixed concentration of the reducing systems (0–500 μM AhpD, 2 μM SucB, and 2 μM Lpd, or 5 μM TrxR, and 0–500 μM TrxC), AhpC (WT and its variants 1 μM), and substrates (1 mM). (B) The B. thailandensis WT(Vector), ΔahpC(Vector), ΔahpC(ahpC), ΔahpC(ahpCC57S), ΔahpC(ahpCC171S), ΔahpC(ahpCC173S), and ΔahpC(ahpCC171SC173S) strains were exposed to H2O2 (1 mM) at 37°C for 30 min. The asterisk indicates a significant difference (∗p < 0.05; n.s., no significant difference using the Student t-test).
These results indicate that the conserved Cys57 in the N-terminus domain plays a critical role in the catalytic activity of AhpC in B. thailandensis like in other bacteria (Wong et al., 2017). The C-terminal Cys171 and Cys173 residues play important roles and they could functionally compensate for each other. For simplicity, we used the AhpD system only as the reducing power in the following experiments.
The Mechanism of Conserved Cys in AhpC Catalytic Activity
The peroxidate cysteine catalyzes the peroxidase reaction by temporarily producing sulfenic acids (Ellis and Poole, 1997). To investigate the functional difference between N-terminal cysteine and C-terminal cysteines in detail, AhpCC171SC173S, AhpCC57SC171S, and AhpCC57SC173S variants were generated and treated with NBD-Cl (4-chloro-7-nitrobenzo-2-oxa-1,3-diazole) with or without CHP. NBD-Cl can react with the thiol group and sulfenic acids. The reaction between NBD-Cl and the thiol group can generate absorption at 420 nm, and the reaction with sulfenic acids increases absorption at 347 nm (Baker and Poole, 2003). After the reaction with CHP, the AhpCC171SC173S variant exhibited Soret bands at 347 and 420 nm differing from the CHP untreated group. There was no peak change with or without CHP in the reaction of AhpCC57SC171S and AhpCC57SC173S variants (Figure 4A). The results suggest that Cys57 works as a peroxidatic cysteine in the active center of AhpC. The quantification of free thiol levels of AhpC variants was also performed with or without H2O2 treatment (Figure 4B). AhpCC57S showed two thiol groups per monomer with or without H2O2 treatment. But AhpCC171S and AhpCC173S lost two thiol groups, and AhpCC171SC173S lost one. These data indicate that the N-terminal Cys57 residue acts as a peroxidatic cysteine and the C-terminal Cys171 and Cys173 residues act as resolving cysteines like in other AhpC homologs (Guimaraes et al., 2005).
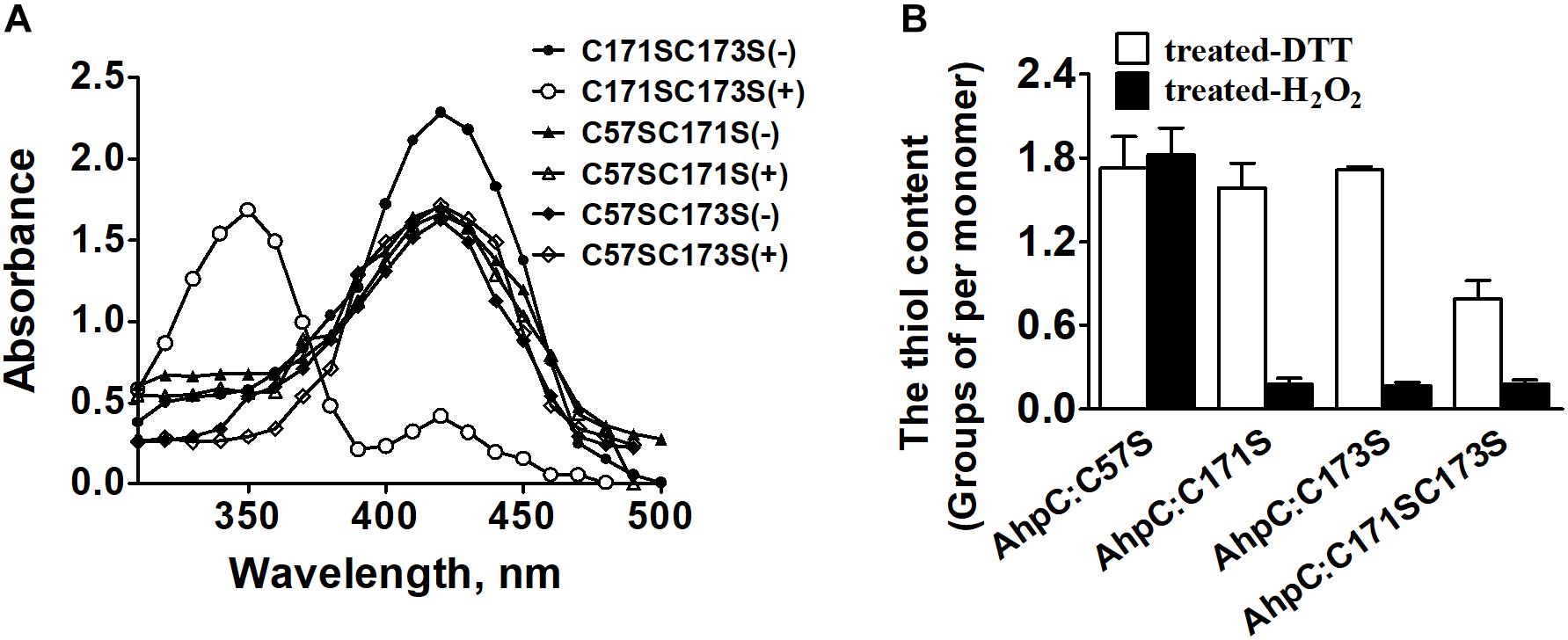
Figure 4. The thiol content and formation of H2O2-treated AhpC. (A) Spectrophotometric analysis of NBD-labeled AhpC variants. Proteins treated with (+) and without (−) H2O2 were modified with NBD chloride for 30 min. After removal of excess reagent by ultrafiltration, the labeled proteins were analyzed spectrophotometrically at 200–600 nm. (B) Free sulfhydryl groups in AhpC variants determined using 5, 5′-dithio-bio (2-nitrobenzoic acid).
C-Terminal Signature Motif Cys-X-X-Cys of AhpD Is Critical for the Catalytic Reaction With AhpC
In the AhpC/AhpD/SucB/Lpd peroxide reducing reaction system, AhpD is the electron donor for AhpC, which is important for the reaction. To test the direct interaction between AhpD and AhpC, the GST pulldown assay was performed as shown in Figure 5A. The results showed direct binding of AhpD to AhpC. AhpD possesses a signature motif at the C-terminus as Cys-X-X-Cys which is critical for the catalytic activity in M. tuberculosis (Bryk et al., 2002). To test the function of AhpD and the Cys-X-X-Cys motif (Figure 5B), Cys to Ser mutant proteins in the motif were designed and purified. AhpD and the variants were used to carry out the reductase assay with AhpC/SucB/Lpd against CHP, t-BOOH, H2O2 and peroxynitrite. The activities of AhpDCXXS decreased to about 10% of wild type, and the activities of AhpDSXXC nearly disappeared (Figure 5C). The results indicated that Cys131 is critical for AhpD the reductase activity, and Cys134 also had an important influence on the enzyme activity that differs from AhpD in M. tuberculosis (Koshkin et al., 2004). The Cys-X-X-Cys motif of AhpD plays a critical role in the peroxide and peroxynitrite reducing reaction with AhpC. As such, AhpD functions as a dithiol mixed disulfide reductase with the active site cysteines.
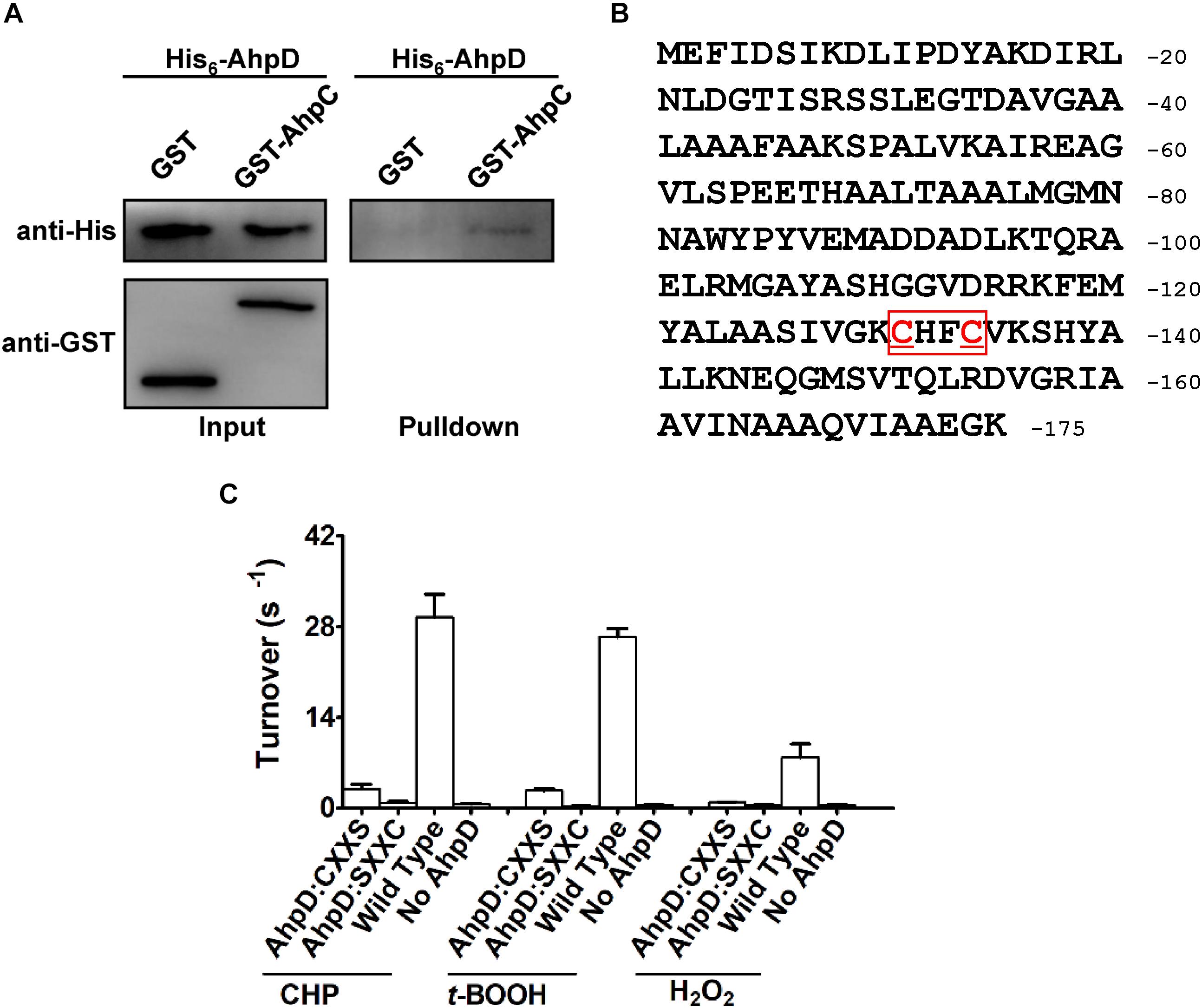
Figure 5. Interaction mechanism between AhpC and AhpD. (A) Direct binding of AphC to AphD. His6-AphD was incubated with GST-AphC or GST, and the protein complexes captured with glutathione beads were detected using western blotting. (B) Cys-X-X-Cys motif sequence in AhpD. (C) Cys in AhpD is required for AhpC’s peroxidase activity. Peroxidase assays were performed as described in section “Materials and Methods” with peroxides (1 mM), AhpC (1 μM), 2 μM SucB, 2 μM Lpd, and AhpD (WT or its variants for 0–500 μM). The data are presented as means of values obtained from three independent assays. Mean values with standard deviations (error bars) from at least three repeats are shown.
The Regulation of AhpC by OxyR in B. thailandensis
The OxyR protein is a well-known regulator for oxidative stress in bacteria. In our previous study, the negative regulation of AhpC by OxyR was discovered using RNA sequencing (RNA-seq)-based transcriptomic analysis and quantitative reverse transcriptase PCR (qRT-PCR) (Si et al., 2017c). To explore the regulation between OxyR and the expression of ahpC, promoter sequence analysis was performed and an OxyR binding site in the promoter region of ahpC was identified using the online software Virtual Footprint1 (Figure 6A). To examine the direct interaction between OxyR and ahpC promoter DNA, a promoter region DNA fragment containing the putative OxyR binding site was amplified using PCR with PahpC-F/PahpC-R primer pair was incubated with varying concentrations of purified OxyR protein in vitro. The reaction mixture was analyzed using an EMSA. As shown in Figure 6B, OxyR directly bound to the ahpC promoter region DNA fragment, but the negative control unrelated DNA without the OxyR binding site showed no interaction with OxyR. These data provided direct evidence that OxyR binds to the promoter region of ahpC to regulate its expression.
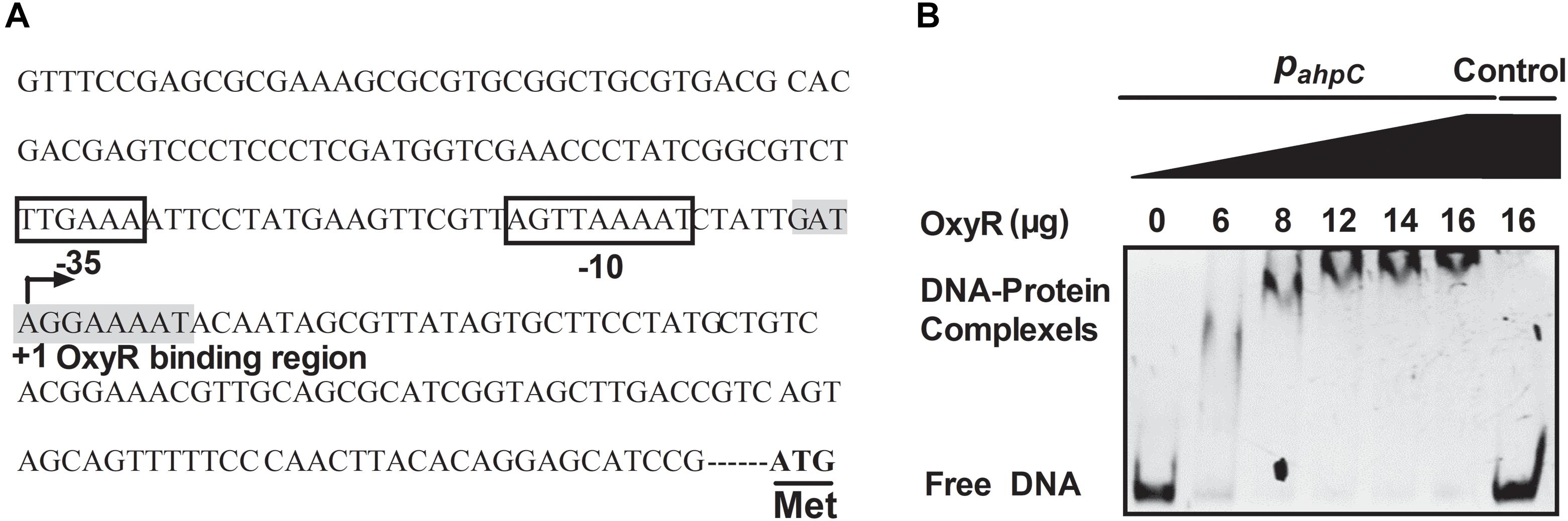
Figure 6. OxyR directly binds to the AhpC promoter. (A) ahpC promoter structure analysis. ATG was the starting codon. The shadow part was the binding region of OxyR. The binding regions of OxyR were labeled by square box. +1: The transcriptional starting point. (B) Gel migration experiment verified the binding of OxyR to ahpC promoter. Gradient concentration (0, 6, 8, 12, 14, 16 μg) of OxyR were mixed with PahpC (ahpC primer DNA containing putative OxyR binding region) and separated by EMSA. DNA without the OxyR binding region was mixed with OxyR as control.
Discussion
Reactive oxygen species are involved in aerobic metabolism in living organisms. In addition to causing harmful stresses in plant and animal, ROS also play a role as signaling during stress defense and aging (Apel and Hirt, 2004; Liochev, 2013; Moloney and Cotter, 2018). In bacteria, ROS play important roles in host-pathogen interactions, infection and antibiotic resistance (Marrakchi et al., 2014; Paiva and Bozza, 2014). Abnormal ROS levels activate a regulator, like OxyR and OhrR, and increase the expression and activity of peroxidase for antioxidant defense AhpC or KatG (Pagan-Ramos et al., 1998). The study of the function and catalytic mechanism of peroxidase AhpC is helpful for understanding the mechanism of bacterial survival, infection and antibiotic resistance.
AhpC was confirmed to play a critical role in B. thailandensis oxidant defense. The deletion of the ahpC gene in B. thailandensis caused an ROS level increase and a survival rate decrease in vivo after the exposure of the cells to peroxides. ahpC gene complementation could recover the ROS level and survival rate (Figure 1). The AhpC activity in vitro also showed its function in oxidant defense like in other bacteria including E. coli, M. tuberculosis, and M. marinum (Pagan-Ramos et al., 1998; Ritz et al., 2001; Seaver and Imlay, 2001). AhpC in B. thailandensis can also catalyze the conversion of peroxynitrite to nitrite as reported in M. tuberculosis, S. typhimurium, M. tuberculosis, and H. pylori (Bryk et al., 2000). The difference between the activities against peroxides and peroxynitrite of AhpC may reflect the partial functional complementation of KatG, SOD, and other antioxidant enzymes indicating various antioxidant mechanisms in living organisms.
AhpC of B. thailandensis is a 2-Cys peroxiredoxin and the conserved Cys residues are important for its function. Cys57 was proved to be the peroxidatic Cys residue that was critical for the function of AhpC (Figures 3, 4). Cys171 and Cys173 were both recognized as resolving Cys residues and together play important roles like the resolving Cys residues of MtbAhpC and MbAhpC (Guimaraes et al., 2005; Wong et al., 2017). Cys171 and Cys173 worked nearly equally as resolving residues. However, the survival rate of ahpCC171S complementation was higher than ahpCC173S indicating that Cys173 plays a more important role than Cys171. The apparent functions of these two Cys residues differ from the resolving Cys residues in MtbAhpC, wherein Cys176 could only substitute for Cys174 to some extent (Koshkin et al., 2004). To explore the reason for the difference, detailed structural biology data are needed for BthAhpC.
The oxidized AhpC was reported to be reduced by AhpD, AhpF, TrxB, or TrxC in bacteria. AhpC is reduced by AhpD and TrxC instead of AhpF and TrxB in mycobacteria, respectively (den Hengst and Buttner, 2008). Here we showed that AhpC can be reduced by AhpD and TrxC in B. thailandensis like in M. tuberculosis (Figure 3A). As an electron donor, AhpD reduces AhpC via the Cys-X-X-Cys motif in the active site (Bryk et al., 2002). In the MtbAhpD active site, Cys133 but not Cys130 directly interacts with Cys61 of AhpC to form a disulfide cross-link, because Cys133 lies at the base of active site cleft and easily interacts with other molecules while Cys130 is partially buried in the fold (Bryk et al., 2002; Koshkin et al., 2004). In B. thailandensis, the Cys-X-X-Cys motif of AhpD plays a critical role in reducing AhpC, and Cys131 had a much more important role than Cys134 (Figure 5). The BthAhpD active site Cys-X-X-Cys motif has a similar function mechanism to MtbAhpD but it may differ in structure. More effort in the structural biology of BthAhpD is needed to illuminate the mechanism in future.
The ahpC gene is negatively regulated by OxyR in B. thailandensis based on detection of the transcriptional level of ahpC in an oxyR-null mutant strain (Si et al., 2017c). The OxyR binding site in the ahpC promoter was identified and shown using EMSA to be especially bound by OxyR (Figure 6). The OxyR binding site in the ahpC promoter of B. thailandensis has a sequence signature like the sequences in other bacteria such as N. meningitidis, M. tuberculosis, M. marinum, M. leprae, and some other mycobacterium (Pagan-Ramos et al., 1998; Ieva et al., 2008). OxyR negatively regulates AhpC in B. thailandensis differently than in E. coli, S. coelicolor and M. marinum, where OxyR activates ahpC gene expression (den Hengst and Buttner, 2008), and similarly to its negative regulator activity for catalase in N. gonorrhoeae and C. diphtheriae (Tseng et al., 2003; Kim and Holmes, 2012). The inducing regulation of AhpC by OxyR is important for the bacteria’s oxidation stress response.
In conclusion, this study reveals that AhpC plays an important role in peroxide and peroxynitrite defense in B. thailandensis. AhpC catalyzes the reducing activity via the three conserved Cys residues and AhpC reduction is affected by the conserved Cys residues in the active site of AhpD.
Data Availability
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
BZ and HG contributed equally to the design of the study and the experiments. HB and CZ performed the statistical analysis. YY wrote the first draft of the manuscript. HG and MS wrote the sections of the manuscript. XS and TS designed the study and are responsible for this work. All authors contributed to the revision, read and approved the final version of the manuscript for submission.
Funding
This work was supported by the National Natural Science Foundation of China (Nos. 31725003 and 31670053).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01483/full#supplementary-material
FIGURE S1 | The growth conditions of B. thailandensis and variants in cultivation with or without oxidants. (A) The survival rate of B. thailandensis against different concentrations of peroxides. (B) The growth curve of B. thailandensis and variant without oxidant. (C) The survival rate of B. thailandensis and variants against 20 mM SIN-1. The asterisk indicates a significant difference (∗p < 0.05, using the Student t-test).
TABLE S1 | Bacterial strains and plasmids used in this study.
TABLE S2 | Primers used in this study.
Footnotes
References
Apel, K., and Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. doi: 10.1146/annurev.arplant.55.031903.141701
Baker, L. M. S., and Poole, L. B. (2003). Catalytic mechanism of thiol peroxidase from Escherichia coli sulfenic acid formation and overoxidation of essential CYS61. J. Biol. Chem. 278, 9203–9211. doi: 10.1074/jbc.m209888200
Bryk, R., Griffin, P., and Nathan, C. (2000). Peroxynitrite reductase activity of bacterial peroxiredoxins. Nature 407, 211–215. doi: 10.1038/35025109
Bryk, R., Lima, C. D., Erdjument-Bromage, H., Tempst, P., and Nathan, C. (2002). Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein. Science 295, 1073–1077. doi: 10.1126/science.1067798
Chandra, J., Samali, A., and Orrenius, S. (2000). Triggering and modulation of apoptosis by oxidative stress. Free Radic. Biol. Med. 29, 323–333. doi: 10.1016/s0891-5849(00)00302-6
Cosgrove, K., Coutts, G., Jonsson, I. M., Tarkowski, A., Kokai-Kun, J. F., Mond, J. J., et al. (2007). Catalase (KatA) and alkyl hydroperoxide reductase (AhpC) have compensatory roles in peroxide stress resistance and are required for survival, persistence, and nasal colonization in Staphylococcus aureus. J. Bacteriol. 189, 1025–1035. doi: 10.1128/jb.01524-06
Demicheli, V., Moreno, D. M., Jara, G. E., Lima, A., Carballal, S., Rios, N., et al. (2016). Mechanism of the reaction of human manganese superoxide dismutase with peroxynitrite: nitrationnitration of critical tyrosine. Biochemistry 55, 3403–3417. doi: 10.1021/acs.biochem.6b00045
den Hengst, C. D., and Buttner, M. J. (2008). Redox control in actinobacteria. Biochim. Biophys. Acta 1780, 1201–1216. doi: 10.1016/j.bbagen.2008.01.008
Ellis, H. R., and Poole, L. B. (1997). Roles for the two cysteine residues of AhpC in catalysis of peroxide reduction by alkyl hydroperoxide reductase from Salmonella typhimurium. Biochemistry 28, 13349–13356. doi: 10.1021/bi9713658
Ellman, G. L. (1959). Tissue sulfhydryl groups. Arch. Biochem. Biophys. 82, 70–77. doi: 10.1016/0003-9861(59)90090-6
Eyer, P., Worek, F., Kiderlen, D., Sinko, G., Stuglin, A., Simeon-Rudolf, V., et al. (2003). Molar absorption coefficients for the reduced ellman reagent: reassessment. Anal. Biochem. 312, 224–227. doi: 10.1016/s0003-2697(02)00506-7
Floris, R., Piersma, S. R., Yang, G., Jones, P., and Wever, R. (1993). Interaction of myeloperoxidase with peroxynitrite. a comparison with lactoperoxidase, horseradish peroxidase and catalase. Eur. J. Biochem. 215, 767–775. doi: 10.1111/j.1432-1033.1993.tb18091.x
Garcia, E. C. (2017). Burkholderia thailandensis: genetic manipulation. Curr. Protoc. Microbiol. 45, 4C.2.1–4C.2.15. doi: 10.1002/cpmc.27
Guimaraes, B. G., Souchon, H., Honore, N., Saint-Joanis, B., Brosch, R., Shepard, W., et al. (2005). Structure and mechanism of the alkyl hydroperoxidase AhpC, a key element of the Mycobacterium tuberculosis defense system against oxidative stress. J. Biol. Chem. 280, 25735–25742. doi: 10.1074/jbc.M503076200
Guo, S. H., Wang, H. F., Nian, Z. G., Wang, Y. D., Zeng, Q. Y., and Zhang, G. (2017). Immunization with alkyl hydroperoxide reductase subunit C reduces Fusobacterium nucleatum load in the intestinal tract. Sci. Rep. 7:10566. doi: 10.1038/s41598-017-11127-x
Halliwell, B., and Gutteridge, J. (1984). Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem. J. 219, 1–14. doi: 10.1042/bj2190001
Haraga, A., West, T. E., Brittnacher, M. J., Skerrett, S. J., and Miller, S. I. (2008). Burkholderia thailandensis as a model system for the study of the virulence-associated type III secretion system of Burkholderia pseudomallei. Infect Immun. 76, 5402–5411. doi: 10.1128/IAI.00626-08
Hayashi, Y., and Yamazaki, I. (1979). The oxidation-reduction potentials of compound I/compound II and compound II/ferric couples of horseradish peroxidases A2 and C. J. Biol. Chem. 254, 9101–9106.
Hugo, M., Turell, L., Manta, B., Botti, H., Monteiro, G., Netto, L. E., et al. (2009). Thiol and sulfenic acid oxidation of AhpE, the one-cysteine peroxiredoxin from Mycobacterium tuberculosis: kinetics, acidity constants, and conformational dynamics. Biochemistry 48, 9416–9426. doi: 10.1021/bi901221s
Ieva, R., Roncarati, D., Metruccio, M. M. E., Seib, K. L., Scarlato, C., and Delany, I. (2008). OxyR tightly regulates catalase expression in Neisseria meningitidis through both repression and activation mechanisms. Mol. Microbiol. 70, 1152–1165. doi: 10.1111/j.1365-2958.2008.06468.x
Jaeger, T., Budde, H., Flohe, L., Menge, U., Singh, M., Trujillo, M., et al. (2004). Multiple thioredoxin mediated routes to detoxify hydroperoxides in Mycobacterium tuberculosis. Arch. Biochem. Biophys. 423, 182–191. doi: 10.1016/j.abb.2003.11.021
Kim, J. S., and Holmes, R. K. (2012). Characterization of OxyR as a negative transcriptional regulator that represses catalase production in Corynebacterium diphtheriae. PLoS One 7:e31709. doi: 10.1371/journal.pone.0031709
Kimura, A., Yuhara, S., Ohtsubo, Y., Nagata, Y., and Tsuda, M. (2012). Suppression of pleiotropic phenotypes of a Burkholderia multivorans fur mutant by oxyR mutation. Microbiology 158(Pt 5), 1284–1293. doi: 10.1099/mic.0.057372-0
Koshkin, A., Knudsen, G. M., and Ortiz De Montellano, P. R. (2004). Intermolecular interactions in the AhpC/AhpD antioxidant defense system of Mycobacterium tuberculosis. Arch. Biochem. Biophys. 427, 41–47. doi: 10.1016/j.abb.2004.04.017
Liochev, S. I. (2013). Reactive oxygen species and the free radical theory of aging. Free Radic. Biol. Med. 60, 1–4. doi: 10.1016/j.freeradbiomed.2013.02.011
Loprasert, S., Sallabhan, R., Whangsuk, W., and Mongkolsuk, S. (2003). Compensatory increase in ahpC gene expression and its role in protecting Burkholderia pseudomallei against reactive nitrogen intermediates. Arch. Microbiol. 180, 498–502. doi: 10.1007/s00203-003-0621-9
Manta, B., Hugo, M., Ortiz, C., Ferrer-Sueta, G., Trujillo, M., and Denicola, A. (2009). The peroxidase and peroxynitrite reductase activity of human erythrocyte peroxiredoxin 2. Arch. Biochem. Biophys. 484, 146–154. doi: 10.1016/j.abb.2008.11.017
Marrakchi, M., Liu, X., and Andreescu, S. (2014). “Oxidative Stress and Antibiotic Resistance in Bacterial Pathogens: State of the Art, Methodologies, and Future Trends,” in Advancements of Mass Spectrometry in Biomedical Research. Advances in Experimental Medicine and Biology, Vol. 806, eds A. Woods and C. Darie (Cham: Springer).
Moloney, J. N., and Cotter, T. G. (2018). ROS signalling in the biology of cancer. Semin. Cell Dev. Biol. 80, 50–64. doi: 10.1016/j.semcdb.2017.05.023
Ogusucu, R., Rettori, D., Munhoz, D. C., Netto, L. E., and Augusto, O. (2007). Reactions of yeast thioredoxin peroxidases I and II with hydrogen peroxide and peroxynitrite: rate constants by competitive kinetics. Free Radic. Biol. Med. 42, 326–334. doi: 10.1016/j.freeradbiomed.2006.10.042
Oh, E., and Jeon, B. (2014). Role of alkyl hydroperoxide reductase (AhpC) in the biofilm formation of Campylobacter jejuni. PLoS One 9:e87312. doi: 10.1371/journal.pone.0087312
Pagan-Ramos, E., Song, J., McFalone, M., Mudd, M. H., and Deretic, V. (1998). Oxidative stress response and characterization of the oxyR-ahpC and furA-katG loci in Mycobacterium marinum. J. Bacteriol. 180, 4856–4864.
Paiva, C. N., and Bozza, M. T. (2014). Are reactive oxygen species always detrimental to pathogens? Antioxid. Redox Signal. 20, 1000–1037. doi: 10.1089/ars
Pomposiello, P. J., and Demple, B. (2001). Redox-operated genetic switches: the SoxR and OxyR transcription factors. Trends Biotechnol. 19, 109–114. doi: 10.1016/s0167-7799(00)01542-0
Reyes, A. M., Vazquez, D. S., Ari Zeida, A., Hugo, M., Piñeyro, M. D., Armas, M. I. D., et al. (2016). PrxQ B from Mycobacterium tuberculosis is a monomeric, thioredoxin dependent and highly efficient fatty acid hydroperoxide reductase. Free Radic. Biol. Med. 101, 249–260. doi: 10.1016/j.freeradbiomed.2016.10.005
Reynolds, C., Goudet, A., Jenjaroen, K., Sumonwiriya, M., Rinchai, D., Musson, J., et al. (2015). T cell immunity to the alkyl hydroperoxide reductase of Burkholderia pseudomallei: a correlate of disease outcome in acute melioidosis. J. Immunol. 194, 4814–4824. doi: 10.4049/jimmunol.1402862
Ritz, D., Lim, J., Reynolds, C. M., Poole, L. B., and Beckwith, J. (2001). Conversion of a peroxiredoxin into a disulfide reductase by a triplet repeat expansion. Science 294, 158–160. doi: 10.1126/science.1063143
Saikh, K. U., and Mott, T. M. (2017). Innate immune response to Burkholderia mallei. Curr. Opin. Infect Dis. 30, 297–302. doi: 10.1097/QCO.0000000000000362
Seaver, L. C., and Imlay, J. A. (2001). Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J. Bacteriol. 183, 7173–7181. doi: 10.1128/JB.183.24.7173-7181
Shen, X. H., Jiang, C. Y., Huang, Y., Liu, Z. P., and Liu, S. J. (2005). Functional identification of novel genes involved in the glutathione-independent gentisate pathway in Corynebacterium glutamicum. Appl. Environ. Microbiol. 71, 3442–3452. doi: 10.1128/AEM.71.7.3442-3452.2005
Si, M., Wang, J., Xiao, X., Guan, J., Zhang, Y., Ding, W., et al. (2015a). Ohr Protects Corynebacterium glutamicum against organic hydroperoxide induced oxidative stress. PLoS One 10:e0131634. doi: 10.1371/journal.pone.0131634
Si, M., Xu, Y., Wang, T., Long, M., Ding, W., Chen, C., et al. (2015b). Functional characterization of a mycothiol peroxidase in Corynebacterium glutamicum that uses both mycoredoxin and thioredoxin reducing systems in the response to oxidative stress. Biochem. J. 469, 45–57. doi: 10.1042/BJ20141080
Si, M., Zhang, L., Chaudhry, M. T., Ding, W., Xu, Y., Chen, C., et al. (2015c). Corynebacterium glutamicum methionine sulfoxide reductase A uses both mycoredoxin and thioredoxin for regeneration and oxidative stress resistance. Appl. Environ. Microbiol. 81, 2781–2796. doi: 10.1128/AEM.04221-14
Si, M., Wang, T., Pan, J., Lin, J., Chen, C., Wei, Y., et al. (2017a). Graded response of the multifunctional 2-Cysteine peroxiredoxin, CgPrx, to increasing levels of hydrogen peroxide in Corynebacterium glutamicum. Antioxid. Redox Signal. 26, 1–14. doi: 10.1089/ars.2016.6650
Si, M., Wang, Y., Zhang, B., Zhao, C., Kang, Y., Bai, H., et al. (2017b). The Type VI secretion system engages a redox-regulated dual-functional heme transporter for zinc acquisition. Cell Rep. 20, 949–959. doi: 10.1016/j.celrep.2017.06.081
Si, M., Zhao, C., Burkinshaw, B., Zhang, B., Wei, D., Wang, Y., et al. (2017c). Manganese scavenging and oxidative stress response mediated by type VI secretion system in Burkholderia thailandensis. Proc. Natl. Acad. Sci. U.S.A. 114, E2233–E2242. doi: 10.1073/pnas.1614902114
Si, M. R., Zhang, L., Yang, Z. F., Xu, Y. X., Liu, Y. B., Jiang, C. Y., et al. (2014). NrdH redoxin enhances resistance to multiple oxidative stresses by acting as a peroxidase cofactor in Corynebacterium glutamicum. Appl. Environ. Microbiol. 80, 1750–1762. doi: 10.1128/AEM.03654-13
Trujillo, M., Ferrer-Sueta, G., and Radi, R. (2008). Kinetic studies on peroxynitrite reduction by peroxiredoxins. Method Enzymol. 441, 173–196. doi: 10.1016/S0076-6879(08)01210-X
Tseng, H.-J., McEwan, A. G., Apicella, M. A., and Jennings, M. P. (2003). OxyR acts as a repressor of catalase expression in Neisseria gonorrhoeae. Infect. Immun. 71, 550–556. doi: 10.1128/iai.71.1.550-556.2003
Wang, T., Si, M., Song, Y., Zhu, W., Gao, F., Wang, Y., et al. (2015). Type VI secretion system transports Zn2+ to combat multiple stresses and host immunity. PLoS Pathog. 11:e1005020. doi: 10.1371/journal.ppat.1005020
Wiersinga, W. J., van der Poll, T., White, N. J., Day, N. P., and Peacock, S. J. (2006). Melioidosis: insights into the pathogenicity of Burkholderia pseudomallei. Nat. Rev. Microbiol. 4, 272–282. doi: 10.1038/nrmicro1385
Wong, C. F., Shin, J., Subramanian Manimekalai, M. S., Saw, W. G., Yin, Z., Bhushan, S., et al. (2017). AhpC of the mycobacterial antioxidant defense system and its interaction with its reducing partner Thioredoxin-C. Sci. Rep. 7:5159. doi: 10.1038/s41598-017-05354-5
Xu, S., Peng, Z., Cui, B., Wang, T., Song, Y., Zhang, L., et al. (2014). FliS modulates FlgM activity by acting as a non-canonical chaperone to control late flagellar gene expression, motility and biofilm formation in Yersinia pseudotuberculosis. Environ. Microbiol. 16, 1090–1104. doi: 10.1111/1462-2920.12222
Zhang, W., Wang, Y., Song, Y., Wang, T., Xu, S., Peng, Z., et al. (2013). A type VI secretion system regulated by OmpR in Yersinia pseudotuberculosis functions to maintain intracellular pH homeostasis. Environ. Microbiol. 15, 557–569. doi: 10.1111/1462-2920.12005
Zhao, Y., and Shao, F. (2015). The NAIP-NLRC4 inflammasome in innate immune detection of bacterial flagellin and type III secretion apparatus. Immunol. Rev. 265, 85–102. doi: 10.1111/imr.12293
Keywords: alkyl hydroperoxide reductase subunit C, Burkholderia thailandensis, oxidative stress, reactive nitrogen species, molecular mechanism
Citation: Zhang B, Gu H, Yang Y, Bai H, Zhao C, Si M, Su T and Shen X (2019) Molecular Mechanisms of AhpC in Resistance to Oxidative Stress in Burkholderia thailandensis. Front. Microbiol. 10:1483. doi: 10.3389/fmicb.2019.01483
Received: 16 February 2019; Accepted: 14 June 2019;
Published: 02 July 2019.
Edited by:
Haike Antelmann, Freie Universität Berlin, GermanyReviewed by:
Derek Parsonage, Wake Forest School of Medicine, United StatesMadia Trujillo, Universidad de la República, Uruguay
Copyright © 2019 Zhang, Gu, Yang, Bai, Zhao, Si, Su and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tao Su, dmluY2VudHRhbzJAMTYzLmNvbQ==; Xihui Shen, eGlodWlzaGVuQG53c3VhZi5lZHUuY24=
†These authors have contributed equally to this work
 Bing Zhang1,2†
Bing Zhang1,2† Huawei Gu
Huawei Gu Yantao Yang
Yantao Yang Meiru Si
Meiru Si Xihui Shen
Xihui Shen